Abstract
Evolution of novel enzymes has fueled the diversification of life on earth for billions of years. Insights into events that set the stage for evolution of a new enzyme can be obtained from ancestral reconstruction and laboratory evolution. Ancestral reconstruction can reveal the emergence of a promiscuous activity in a pre-existing protein and the impact of subsequent mutations that enhance a new activity. Laboratory evolution provides a more holistic view by revealing mutations elsewhere in the genome that indirectly enhance the level of a newly important enzymatic activity. This review will highlight recent studies that probe the early stages of evolution of a new enzyme from these complementary points of view.
Enzymes catalyze chemical reactions by up to 26 orders of magnitude [1]. The evolutionary origins of these magnificent catalysts have fascinated biochemists for decades. This review will highlight two experimental approaches that probe the early stages of enzyme evolution. Ancestral reconstruction [2,3] allows characterization of the level of physiologically irrelevant promiscuous activities in ancestral proteins and the impact of mutations on structure and function as a new enzymatic activity evolves. This approach was proposed by Pauling and Zuckerkandl in 1963 [4], but due to technological limitations, was not brought to fruition by the Benner group for nearly thirty years [5]. Since then, scores of papers have examined the evolution of steroid hormone receptors [6–8], opsins [9–12], transcriptional regulators [13], elongation factors [14], fluorescent proteins [15], globins [16], enzymes [17–20] and metabolic pathways [21].
Since Pauling’s and Zuckerkandl’s seminal article, ever more sophisticated computational methods for ancestral reconstruction have been developed. Recent reviews describe the theory and application of various methods, as well as their strengths and limitations [2,22–24]. In brief, ancestral sequence reconstruction requires a multiple sequence alignment of a set of homologous protein sequences that adequately represents the diversity of extant proteins. Maximum likelihood methods infer ancestral sequences based upon a phylogenetic tree and an evolutionary model that describes the probability of substitution at each aligned position in the protein. Bayesian methods estimate the phylogenetic tree and the parameters of the evolutionary model simultaneously. In either case, the output is a list of the probabilities of each amino acid at each position for each ancestral node. The most likely ancestral sequence has the highest probability amino acid at each position.
A limitation of ancestral reconstruction is that, due to ambiguities in ancestral reconstruction, even the most likely sequence has a low probability of being the real ancestral sequence. For example, a 100-amino acid protein for which each position has been reconstructed with a probability of 95% has a probability of only 0.006 [(0.95)100]. In practice, many positions are predicted with a probability of <50%, particularly if the extant proteins are highly diverged. Thus, the maximum likelihood ancestor is usually highly unlikely. This problem is typically addressed by reconstructing multiple possible ancestral proteins to ensure that their properties are similar and therefore likely to be representative of the true ancestor [22,23]. Using this approach, Bar-Rogosvky et al. [25] found that the most probable ancestor of mammalian PONs exhibited enzymatic activities with several substrates that were representative of an ensemble of probable sequences that varied at the ambiguous positions most likely to affect function. In contrast, the most probable ancestor of vertebrate PONs, which had more ambiguously reconstructed positions, exhibited only one of many enzymatic phenotypes represented in the ensemble. This work is an important cautionary tale for efforts to reconstruct and characterize very ancient proteins.
A further limitation of ancestral reconstruction is its focus upon a single protein. It cannot address the cellular context in which a new enzyme evolved, or other mechanisms that helped cells respond to a new selective pressure. Laboratory evolution of microbes in which a promiscuous enzyme has been recruited to serve a new function offers the opportunity to observe the early processes in evolution of a new enzyme starting from a known progenitor. Laboratory evolution is carried out by growing an organism in a defined medium for hundreds or thousands of generations. Long-term cultivation is enabled by either serial dilution or use of continuous cultivation devices such as chemostats or turbidostats. Addition of mutagens, which would sprinkle mutations throughout the genome and obscure the identification of adaptive mutations, is not necessary. Mutants with adaptive mutations typically begin to accumulate within tens to hundreds of generations [26–32]. However, evolution of a highly efficient and properly regulated new enzyme may require an impractically long time frame. Thus, these two approaches provide complementary perspectives on the emergence of new enzymes in nature.
Evolution of catalytic activity in a non-catalytic protein
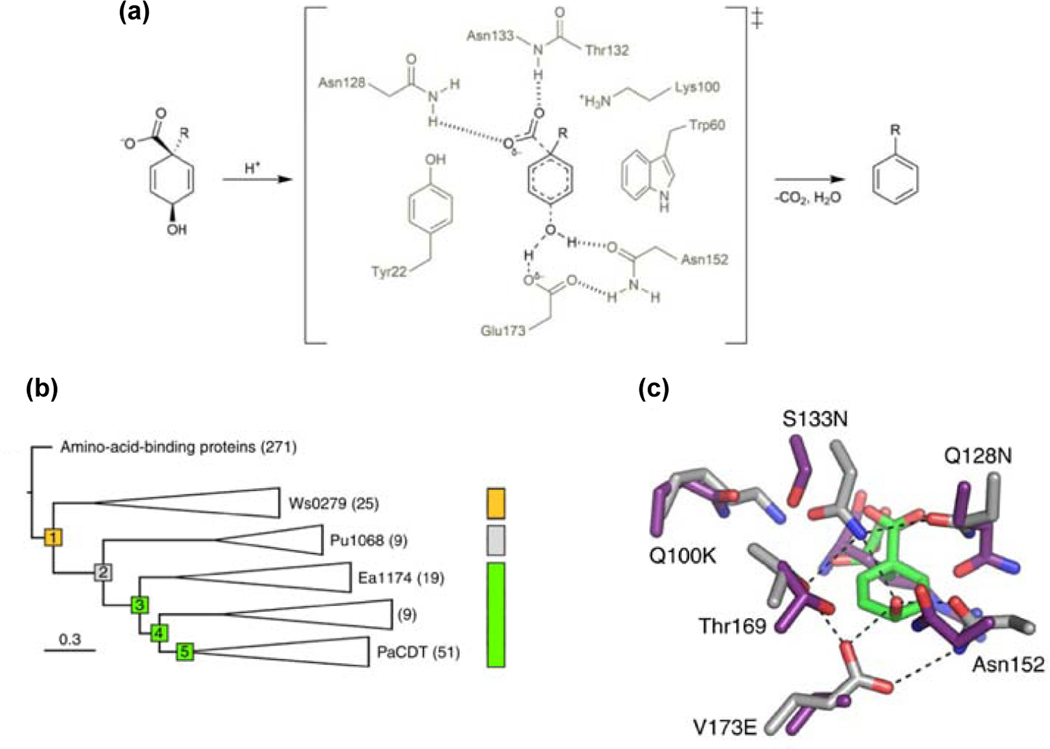
The existence of large superfamilies of enzymes [33–38] suggests that most enzymes have evolved from previously existing enzymes by gene duplication and divergence. But how does catalytic activity evolve in the first place? A recent study tracked mutations that conferred enzymatic activity on a non-catalytic protein. Cyclohexadienyl dehydratase (CDT), which converts prephenate to phenylpyruvate (Figure 1a) and L-arogenate to L-phenylalanine, is distantly related to non-catalytic periplasmic amino acid-binding proteins that deliver solutes to inner-membrane ABC transporters [39]. Clifton et al. [40] resurrected ancestral proteins at nodes of a phylogenetic tree connecting CDTs and related solute-binding proteins (Figure 1b). The earliest reconstructed protein, AncCDT1, had high affinity for cationic amino acids. This protein had three residues, Asp29, Asn152 and Thr169, that would later be important in the CDT active site, but no catalytic activity. AncCDT2 had lost affinity for cationic amino acids, but, like its descendant, the extant solute-binding protein Pu1068, had weak affinity for negatively charged ligands. AncCDT2 had also acquired several changes that would later be important for enzymatic activity. Substitution of Glu for Val173 provided the general acid that protonates the departing hydroxyl group of prephenate in CDT (Figure 1c). Two changes (D19T and A20G) altered the position of Trp60 and reshaped the ligand-binding site. Three additional changes introduced residues (Lys100, Asn128 and Asn133) that would later form hydrogen bonds with the departing carboxylate of prephenate. Despite these changes, AncCDT2 lacked catalytic activity. Weak CDT activity (kcat/KM = 6 M−1 s−1) appeared in AncCDT3. The only change near the active site between AncCDT2 and AncCDT3 (L198K) does not confer catalytic activity, so unidentified changes at remote sites must also have been important.
Figure 1.
Evolution of CDT activity in a non-catalytic protein. (a) The reaction catalyzed by CDT. (b) A phylogenetic tree connecting extant CDTs and related amino-acid binding proteins. Yellow, affinity for cationic amino acids; grey, affinity for negatively charged ligands; green, CDT activity. (c) Comparison of the ligand binding site of AncCDT1 (purple) and the active site of extant P. aeruginosa CDT (grey). Reprinted by permission from Springer Nature Nature Chemical Biology 14:542–547, Evolution of cyclohexadienyl dehydratase from an ancestral solute-binding protein, Clifton BE, Kaczmarski JA, Carr PD, Gerth ML, Tokuriki N, Jackson CJ.
A crystal structure of P188L AncCDT3, which has a modestly higher kcat/KM (155 M−1 s−1), showed that the positions of active site residues are nearly identical to those in Pseudomonas aeruginosa CDT (kcat/KM = 9.8 × 105 M−1 s−1). (The beneficial P188L change was identified by shuffling AncCDT3 genes predicted by two different reconstruction methods.) Because simply assembling the catalytic machinery did not suffice to generate high activity, Clifton et al. hypothesized that a change in the dynamics of a hinge that opens and closes the active site might have been required. Indeed, MD simulations showed that AncCDT1 prefers the open conformation, which favors ligand binding and delivery to the ABC transporter, but P. aeruginosa CDT prefers the catalytically competent closed conformation.
Three intriguing findings emerged from this study. First, several residues required for catalysis appeared before the protein acquired any enzymatic activity. Second, an unsuspected novel noncatalytic function evolved between the earliest ancestor, AncCDT1, and CDT. Finally, efficient catalysis required adjustment of protein dynamics.
Exploitation of pre-existing buried aromatic residues enabled more efficient degradation of lignin
Evolution of new catalytic abilities usually exploits a previously existing promiscuous activity that utilizes features of the ancestral active site. Recent studies of the evolution of ligninases [41] revealed that aromatic residues in the hydrophobic core of an ancestral enzyme, rather than its active site, set the stage for a novel electron transfer pathway that changed the efficiency of lignin degradation.
Lignin, a complex polymer formed by radical condensation reactions of phenylpropanoid precursors, provides structural integrity to plant cell walls. Wood-rotting fungi secrete three types of heme-dependent peroxidases that degrade lignin. In each case, the resting enzyme is oxidized by H2O2, yielding an Fe(IV)-oxo/porphyrin cation radical complex [42]. Manganese peroxidases (MnPs) oxidize Mn2+ at a site near the heme [43,44], releasing Mn3+ to diffuse to the surface of lignin and initiate radical depolymerization reactions by oxidizing phenolic residues. In lignin peroxidases (LiPs), a surface tryptophanyl radical formed by transfer of an electron to the porphyrin cation radical can access lignin directly and is powerful enough to oxidize the nonphenolic component of lignin [45]. “Versatile” peroxidases use both strategies [46].
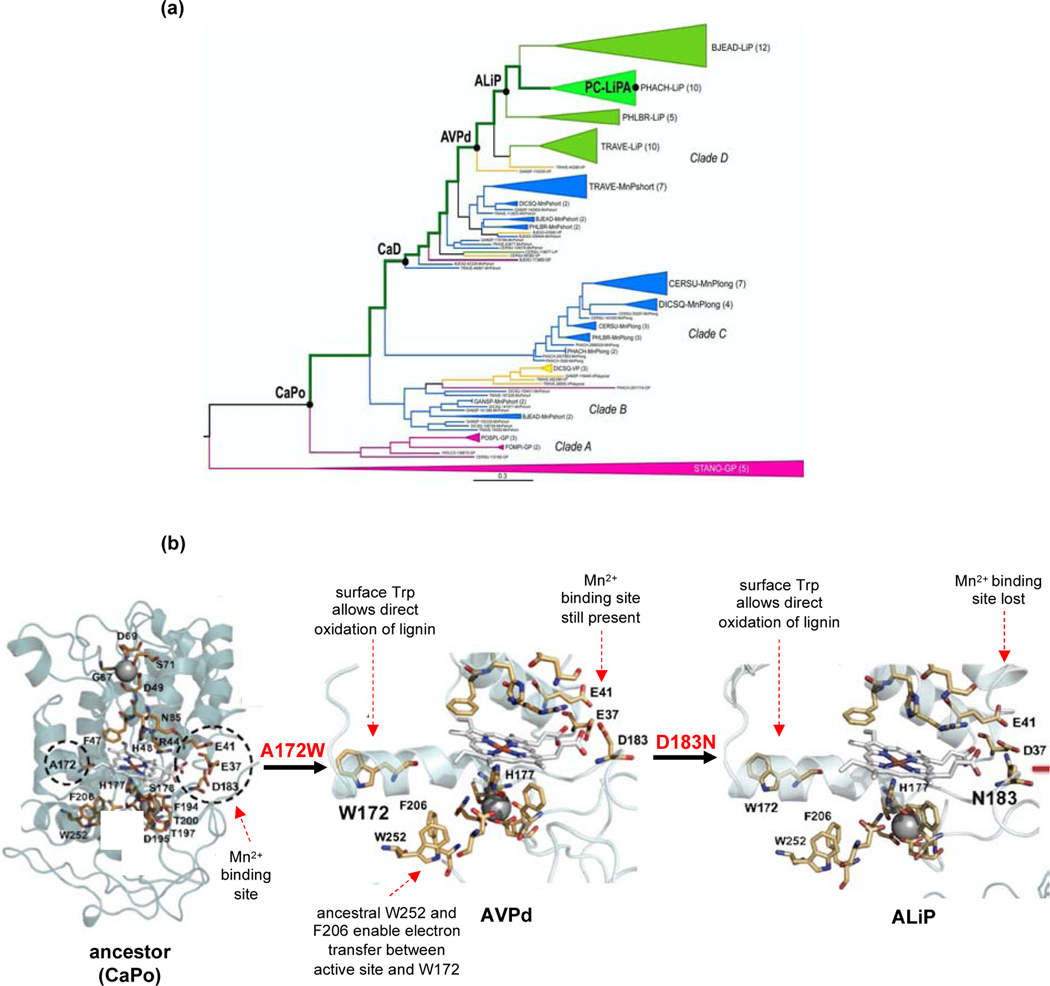
The Martinez group used ancestral reconstruction to investigate the evolution of these three classes of peroxidases in Polyporales fungi [41]. The earliest reconstructed enzyme was a MnP. A surface tryptophan was acquired independently in Clades B and D (Figure 2). The ability to oxidize lignosulfonate (a soluble form of lignin produced by treating wood pulp with sulfite) increased at each stage of the Clade D lineage with the exception of the ancestor of lignin peroxidases (ALiP) [47]. This anomaly might truly reflect a decrease in activity, but might also be due to a peculiarity of the sequence reconstructed for either the ancestral VP in clade D (AVPd) or the ALiP. Only one sequence was resurrected at each node, so one or both might not be representative of the true ancestral enzyme. Notably, the enzymes became progressively stronger oxidants along the evolutionary trajectory [48]. 1H-NMR data suggest that the bond between His177 and the Fe atom became progressively weaker, destabilizing the Fe(IV)-oxo species and making it a stronger oxidant.
Figure 2.
Evolution of lignin peroxidases. (a) Phylogenetic tree of fungal lignin peroxidases. CaPo, common ancestor of Polyporales peroxidases; CaD, common ancestor of Clade D peroxidases; AVPd, ancestral VP in Clade D; ALiP, ancestral lignin peroxidase; PCLiPA, P. chrysosporium lignin peroxidase A. Reprinted from ref. [47] under Creative Commons license 4.0 CC BY-NC-ND. (b) Homology models of ancestral peroxidases. Dashed circles on the CaPo structure show the positions that will be mutated as evolution toward the extant lignin peroxidases proceeds. Reprinted with modifications from ref. [41] under Creative Commons license 4.0 CC BY-NC-ND.
An intriguing aspect of this study was the observation that an incipient path for electron transfer from the heme to the enzyme surface was already present in the ancestor. Trp252 and Phe205 formed part of the hydrophobic core of the ancestral protein, but played no role in catalysis. However, their fortuitous location between the heme and the surface of the enzyme set the stage for emergence of a new catalytic strategy—transfer of electrons between the active site and the surface—as a result of a change of Ala172 to Trp in Clade D, and of Ala172 to Asp and then to Trp in Clade B.
Evolution of enzymes that degrade anthropogenic pollutants
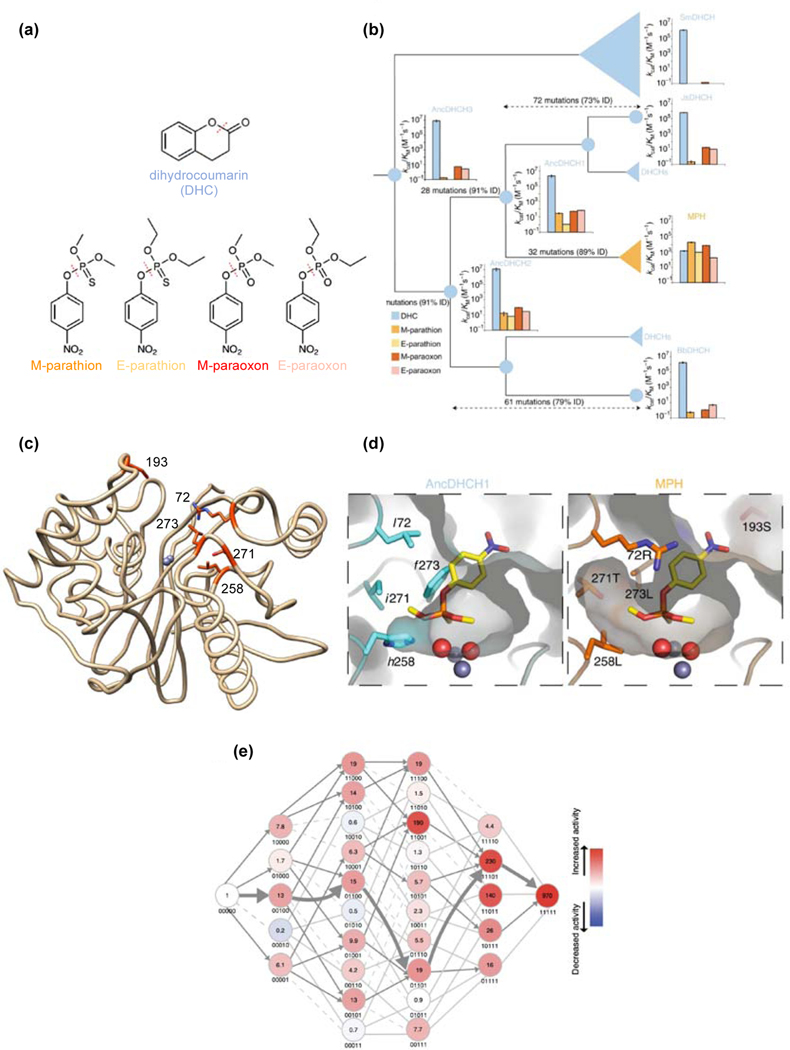
In response to novel selective pressures presented by large-scale use of anthropogenic pesticides since the 1940s, enzymes that detoxify or degrade compounds such as DDT [49], atrazine [50], pentachlorophenol [51] and organophosphate insecticides [52] have evolved in an evolutionary blink of an eye. Yang et al. [53] examined the evolutionary origin of methyl parathion hydrolase (MPH). MPH was first identified in Pseudomonas sp. WBC-3, which was isolated from contaminated soil near a plant that manufactured methyl parathion [54]. MPH is a member of the metallo-β-lactamase superfamily, and is most closely related to bacterial dihydrocoumarin hydrolases (DHCHs). Yang et al. reconstructed enzymes at ancestral nodes separating extant MPHs and DHCHs. The ancestral DHCHs have high efficiency for hydrolysis of DHC (Figure 3). Each also has an inefficient ability to hydrolyze methyl parathion and the related organophosphates ethyl parathion, methyl paraoxon and ethyl paraoxon, substrates that appeared in the environment only in the 20th century.
Figure 3.
Evolution of MPH from a DHCH. (a) Substrates for ancestral and extant enzymes. (b) Phylogenetic tree connecting MPHs and DHCHs. Catalytic efficiencies are shown for reconstructed ancestral proteins and extant DHCHs from Serratia marcescens (SmDHCH), Janthinobacterium sp. HH01 (JsDHCH), Pseudomonas sp. WBC-3 MPH and Burkholderiales bacterium JOSHI_001 (BbDHCH). Bar colors correspond to substrates shown in (a). (c) Crystal structure of Pseudomonas sp. WBC-3 MPH (PDB 1P9E). Five residues that improved MPH activity by 970-fold when introduced into AncDHCH1 are highlighted in orange. (d) Comparison of the active sites of AncCDT1 and MPH, showing that remodeling of the active site allowed binding of methyl parathion. (e) The fitness landscape for evolution of MPH from AncCDT1. Each node is designated by the absence (0) or presence (1) of sequence changes at five positions in the following order: 72, 193, 258, 271 and 273. Fold-changes in MPH activity are indicated in each node. Parts a, b, d and e reprinted with permission from Springer Nature Nature Chemical Biology 15:1120–1128, Higher-order epistasis shapes the fitness landscape of a xenobiotic-degrading enzyme, Yang G, Anderson DW, Baier F, Dohmen E, Hong N, Carr PD, Kamerlin SCL, Jackson CJ, Bornberg-Bauer E, Tokuriki N.
Pseudomonas sp. WBC-3 MPH is 89% identical to the most recent common ancestor of DHCHs and MPHs, differing at 32 positions (Figure 3b). Introduction of just 5 changes near the active site (Figure 3c) into the ancestral enzyme improved MPH activity by 970-fold and yielded an enzyme with kinetic properties nearly identical to those of extant MPH. Four of these changes enlarge the substrate binding site, allowing methyl parathion to bind (Figure 3d). Deletion of Ser193 in a nearby loop alters the conformation of the loop.
The recent emergence of MPH made it possible to identify five mutations that enhanced MPH activity in an ancestral DHCH, but the order in which the mutations occurred cannot be discerned based upon the ancestral reconstruction. To explore this question, the authors characterized 32 variants with all possible combinations of the five critical sequence changes. Only 19 of the 120 trajectories for accumulation of the five mutations were feasible (Figure 3e). The remainder involved at least one step where an additional mutation decreased the evolving MPH activity. Productive trajectories toward improved MPH activity could have begun with four of the five changes. Whereas one trajectory (broad arrow in Figure 3e) was most propitious, the availability of 19 others increases the probability of a successful outcome in a relatively short period of time.
The early stages of recruitment of a promiscuous enzyme to serve a new function
Ancestral reconstruction provides the opportunity to trace the evolution of improved function as an enzyme evolves. However, it cannot answer many questions about the process of evolving a new enzyme. For example, what promiscuous activities were available in the organism in which a new enzyme evolved? If there was more than one suitable starting point, why was one “chosen”? What changes in the environment or the genome enabled recruitment of a promiscuous enzyme to serve a new function? Was gene duplication/amplification involved? Laboratory evolution experiments allow many of these questions to be addressed. The environment and the genome of the parental strain are known, and genomes of intermediate strains can be sequenced to identify mutations acquired during adaptive evolution. Transcriptomic, proteomic and metabolomic analyses can be employed to identify physiological changes in the system as a whole, rather than in the protein alone, as evolution of a new enzyme proceeds.
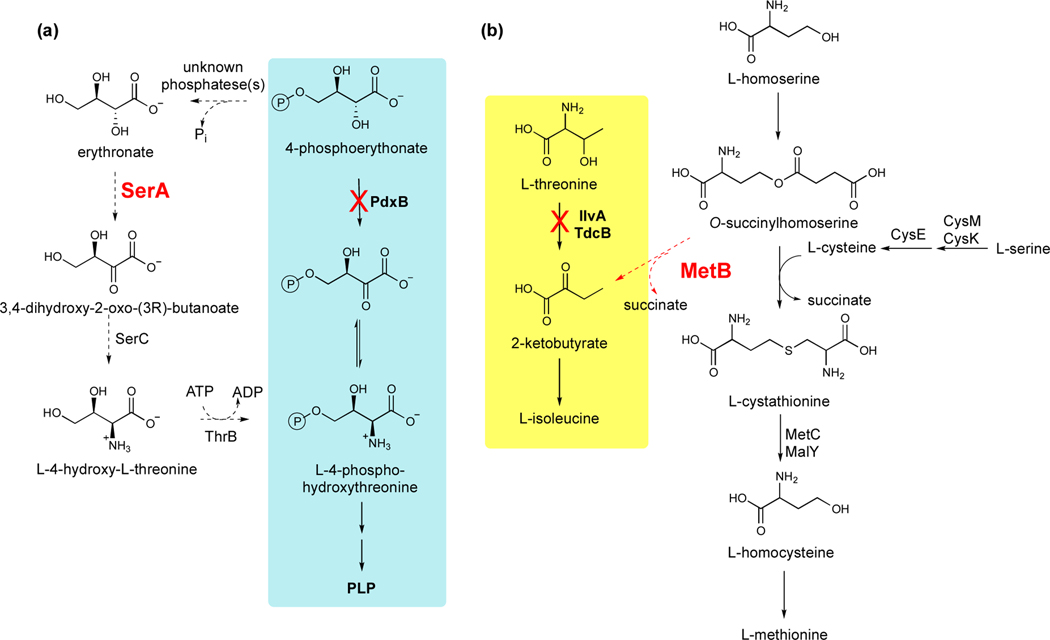
Three recent studies have shown that evolution of a new enzyme begins with recruitment of a promiscuous enzyme to serve a new function, as expected, but that the earliest mutations increase the cellular activity of the weak-link enzyme indirectly rather than modifying the enzyme itself. Kim et al. [28] identified a four-step bypass pathway (Figure 4a) that emerges within 150 generations when pyridoxal 5’-phosphate (PLP) synthesis in E. coli is blocked by deletion of pdxB. This pathway depends critically on a promiscuous erythronate dehydrogenase activity of SerA, which normally oxidizes 3-phosphoglycerate in the first step of serine biosynthesis. The erythronate dehydrogenase activity of SerA is compromised by competition from the native substrate, 3-phosphoglycerate, and also by feedback inhibition of the enzyme by serine. Mutations abolished feedback inhibition by serine in three of 10 evolved strains, allowing PLP synthesis to continue even when adequate serine was present. However, most of the evolved strains had no mutations in serA. In one of these strains, mutations in genes encoding enzymes in glycolysis and the pentose phosphate pathway led to diversion of flux away from glycolysis, resulting in low levels of 3-phosphoglycerate, a glycolytic intermediate, and its downstream product, serine. Elevating the productivity of the promiscuous reaction by minimizing competition from the native substrate and feedback inhibition clearly provided the most accessible way to immediately improve PLP synthesis.
Figure 4.
Promiscuous enzyme activities recruited to serve new functions in laboratory evolution experiments in E. coli. (a) A four-step bypass pathway restores synthesis of PLP after deletion of pdxB [28]. (b) Recruitment of MetB to synthesize 2-ketobutyrate restores isoleucine synthesis after deletion of ilvA and tdcB [55]. Dashed lines indicate promiscuous reactions.
Cotton et al. [55] carried out a similar experiment in E. coli by deleting genes encoding two threonine deaminases that produce 2-ketobutyrate in the isoleucine synthesis pathway. Evolution of this strain on glucose resulted in recruitment of MetB (cystathionine γ-synthase) to convert O-succinylhomoserine to 2-ketobutyrate (Figure 4b). MetB catalyzes this reaction quite efficiently (kcat/KM = 1.6 × 104 M−1s−1) in the absence of the usual co-substrate, cysteine. However, in the presence of cysteine, O-succinylhomoserine is converted to L-cystathionine. Although one might expect that a mutation in metB could increase the efficiency of O-succinylhomoserine conversion to 2-ketobutyrate, none of the evolved strains acquired a mutation in metB. Rather, the evolved strains appear to have favored the cleavage reaction by altering the ratio of O-succinylhomoserine to cysteine.
Six of nine evolved strains increased the level of O-succinylhomoserine by deleting metC. MetC is one of two enzymes in E. coli that convert L-cystathionine to L-homocysteine in the methionine synthesis pathway. Deletion of metC in a “Δ5” strain lacking the two threonine deaminases and also three serine deaminases that might have substituted for the missing threonine deaminases (but didn’t) caused a partial block in methionine synthesis and consequently a 3-fold increase in the level of O-succinylhomoserine upstream of the block.
One evolved strain decreased the level of cysteine. A mutation in cysE, which encodes serine acetyl transferase, decreased kcat/KM,acetyl CoA by 17-fold. Introduction of this mutation into the Δ5 strain resulted in a 2-fold decrease in the level of cysteine. Further, the decreased availability of cysteine impaired methionine synthesis, leading to 5-fold upregulation of metB, providing a second mechanism for increasing 2-ketobutyrate production.
An increase in fitness due to mutations elsewhere in the genome was also observed after evolution of a strain of E. coli in which a promiscuous activity of E383A ProA (γ-glutamyl phosphate reductase)(ProA*) had been recruited to substitute for ArgC (N-acetylglutamyl phosphate reductase) in arginine synthesis [27]. proA* amplified within 200 generations of growth of the ΔargC proA* strain on glucose + proline to select for mutations that improved arginine synthesis. Mutations that indirectly enhanced the neo-ArgC activity of ProA*occurred in all eight replicate populations. Some increased expression of the enzyme upstream of the weak-link ProA*, likely pushing material through the compromised pathway. Others abolished feedback inhibition of an enzyme that produces a co-substrate for a later enzyme, likely pulling material through the pathway. In just one population, a mutation that changed Phe372 to Ala boosted the inefficient neo-ArgC activity of ProA*.
A common feature of these cases is that the effectiveness of a newly important activity was increased indirectly by mutations elsewhere in the genome, rather than mutation of the gene encoding the enzyme itself. The rarity of mutations that enhance the catalytic efficiency of newly recruited enzymes in the short term is likely due to differences in mutational target size. Most of the beneficial mutations in these three experiments altered the metabolic network due to total or partial loss of function of an enzyme. Loss of function is easily achieved by insertions, deletions, or point mutations at many places in a gene. In contrast, increasing the efficiency of a new activity may require surgically precise modification of specific active site residues.
Summary
Ancestral reconstruction experiments have provided a fascinating glimpse into how mutations set the stage for emergence of new activities as well as the structural basis for improvements in a new activity. Experimental evolution studies place the process of evolving a new enzyme in a cellular context, and have revealed unexpected complexities surrounding the initial recruitment of a promiscuous enzyme to serve a new function. Both approaches contribute to our expanding understanding of enzyme evolution and could potentially be used in conjunction by carrying out experimental evolution under selective pressure for improvement of a predicted ancestral enzyme. Major gaps and challenges still remain, however. Evolution of a new enzyme requires not only improvements in catalytic activity, but also evolution of proper regulation. In particular, we have little insight into the evolution of allostery, a major mechanism for post-translational regulation of enzyme activity. One recent study used ancestral reconstruction to investigate the evolution of allostery [56], but the findings are controversial [57,58]. The prevalence of loss-of-function mutations in laboratory evolution experiments is well-recognized [59–62], but how mutations that indirectly improve the efficiency of newly recruited enzymes at a cost to previously well-evolved functions are repaired after evolution of a new enzyme has not yet been addressed.
Acknowledgments
Funding: This work was supported by the National Institutes of Health R01GM134044 and R01 GM135364.
Footnotes
The author declares no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edwards DR, Lohman DC, Wolfenden R: Catalytic proficiency: the extreme case of S-O cleaving sulfatases. J Am Chem Soc 2012, 134:525–531. [DOI] [PubMed] [Google Scholar]
- 2.Garcia AK, Kacar B: How to resurrect ancestral proteins as proxies for ancient biogeochemistry. Free Radic Biol Med 2019, 140:260–269. [DOI] [PubMed] [Google Scholar]
- 3.Hochberg GKA, Thornton JW: Reconstructing ancient proteins to understand the causes of structure and function. Annu Rev Biophys 2017, 46:247–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pauling L, Zuckerkandl E: Chemical paleogenetics: Molecular “restoration studies” of extinct forms of life. Acta Chem Scandinavica 1963, 17:S9–S16. [Google Scholar]
- 5.Stackhouse J, Presnell SR, McGeehan GM, Nambiar KP, Benner SA: The ribonuclease from an extinct bovid ruminant. FEBS Lett 1990, 262:104–106. [DOI] [PubMed] [Google Scholar]
- 6.Thornton JW, Need E, Crews D: Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science 2003, 301:1714–1717. [DOI] [PubMed] [Google Scholar]
- 7.Eick GN, Colucci JK, Harms MJ, Ortlund EA, Thornton JW: Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLoS Genet. 2012, 8:e1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harms MJ, Eick GN, Goswami D, Colucci JK, Griffin PR, Ortlund EA, Thornton JW: Biophysical mechanisms for large-effect mutations in the evolution of steroid hormone receptors. Proc Natl Acad Sci U S A 2013, 110:11475–11480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang BS, Jonsson K, Kazmi MA, Donoghue MJ, Sakmar TP: Recreating a functional ancestral archosaur visual pigment. Mol Biol Evol 2002, 19:1483–1489. [DOI] [PubMed] [Google Scholar]
- 10.Chinen A, Matsumoto Y, Kawamura S: Reconstitution of ancestral green visual pigments of zebrafish and molecular mechanism of their spectral differentiation. Mol Biol Evol 2005, 22:1001–1010. [DOI] [PubMed] [Google Scholar]
- 11.Chinen A, Matsumoto Y, Kawamura S: Spectral differentiation of blue opsins between phylogenetically close but ecologically distant goldfish and zebrafish. J Biol Chem 2005, 280:9460–9466. [DOI] [PubMed] [Google Scholar]
- 12.Simoes BF, Foley NM, Hughes GM, Zhao H, Zhang S, Rossiter SJ, Teeling EC: As blind as a bat? Opsin phylogenetics illuminates the evolution of color vision in bats. Mol Biol Evol 2019, 36:54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perica T, Kondo Y, Tiwari SP, McLaughlin SH, Kemplen KR, Zhang X, Steward A, Reuter N, Clarke J, Teichmann SA: Evolution of oligomeric state through allosteric pathways that mimic ligand binding. Science 2014, 346:1254346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaucher EA, Thomson JM, Burgan MF, Benner SA: Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 2003, 425:285–288. [DOI] [PubMed] [Google Scholar]
- 15.Ugalde JA, Chang BS, Matz MV: Evolution of coral pigments recreated. Science 2004, 305:1433. [DOI] [PubMed] [Google Scholar]
- 16.Pillai AS, Chandler SA, Liu Y, Signore AV, Cortez-Romero CR, Benesch JLP, Laganowsky A, Storz JF, Hochberg GKA, Thornton JW: Origin of complexity in haemoglobin evolution. Nature 2020, 581:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaltenbach M, Burke JR, Dindo M, Pabis A, Munsberg FS, Rabin A, Kamerlin SCL, Noel JP, Tawfik DS: Evolution of chalcone isomerase from a noncatalytic ancestor. Nature Chemical Biology 2018, 14:548-+. [DOI] [PubMed] [Google Scholar]
- 18.Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM: Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian beta-lactamases. J Am Chem Soc 2013, 135:2899–2902. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Damry AM, Petrie JR, Vanhercke T, Singh SP, Jackson CJ: Consensus mutagenesis and ancestral reconstruction provide insight into the substrate specificity and evolution of the front-end delta6-desaturase family. Biochemistry 2020, 59:1398–1409. [DOI] [PubMed] [Google Scholar]
- 20. Chaloupkova R, Liskova V, Toul M, Markova K, Sebestova E, Hernychova L, Marek M, Pinto GP, Pluskal D, Waterman J, et al. : Light-emitting dehalogenases: Reconstruction of multifunctional biocatalysts. ACS Catal. 2019, 9:4810–4823. *Identification of catalytic promiscuity in an ancestor of haloalkane dehalogenases and luciferases, related enzymes that catalyze very different reactions.
- 21.Huang R, O’Donnell AJ, Barboline JJ, Barkman TJ: Convergent evolution of caffeine in plants by co-option of exapted ancestral enzymes. Proc Natl Acad Sci U S A 2016, 113:10613–10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merkl R, Sterner R: Ancestral protein reconstruction: techniques and applications. Biol Chem 2016, 397:1–21. [DOI] [PubMed] [Google Scholar]
- 23.Aadland K, Kolaczkowski B: Alignment-integrated reconstruction of ancestral sequences improves accuracy. Genome Biol Evol 2020, 12:1549–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selberg AGA, Gaucher EA, Liberles DA: Ancestral sequence reconstruction: From chemical paleogenetics to maximum likelihood algorithms and beyond. J Mol Evol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bar-Rogovsky H, Stern A, Penn O, Kobl I, Pupko T, Tawfik DS: Assessing the prediction fidelity of ancestral reconstruction by a library approach. Protein Eng Des Sel 2015, 28:507–518. [DOI] [PubMed] [Google Scholar]
- 26.Kristofich J, Morgenthaler AB, Kinney WR, Ebmeier CC, Snyder DJ, Old WM, Cooper VS, Copley SD: Synonymous mutations make dramatic contributions to fitness when growth is limited by a weak-link enzyme. PLoS Genet 2018, 14:e1007615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morgenthaler AB, Kinney WR, Ebmeier CC, Walsh CM, Snyder DJ, Cooper VS, Old WM, Copley SD: Mutations that improve efficiency of a weak-link enzyme are rare compared to adaptive mutations elsewhere in the genome. Elife 2019, 8. *Fitness of a cell in which a weak-link enzyme in arginine synthesis limits growth rate is most easily improved by gene amplification and mutations that either push or pull material through the compromised pathway rather than mutations in the gene encoding the weak-link enzyme itself.
- 28. Kim J, Flood JJ, Kristofich MR, Gidfar C, Morgenthaler AB, Fuhrer T, Sauer U, Snyder D, Cooper VS, Ebmeier CC, et al. : Hidden resources in the Escherichia coli genome restore PLP synthesis and robust growth after deletion of the essential gene pdxB. Proc Natl Acad Sci U S A 2019, 116:24164–24173. **The efficiency of a novel 4-step pathway assembled from promiscuous enzymes can be improved without mutations in any of the genes encoding the promiscuous enzymes.
- 29.Knoppel A, Knopp M, Albrecht LM, Lundin E, Lustig U, Nasvall J, Andersson DI: Genetic adaptation to growth under laboratory conditions in Escherichia coli and Salmonella enterica. Front Microbiol 2018, 9:756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, et al. : Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nature Genetics 2006, 38:1406–1412. [DOI] [PubMed] [Google Scholar]
- 31.Lee DH, Palsson BO: Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a nonnative carbon source, L-1,2-propanediol. Appl Environ Microbiol 2010, 76:4158–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charusanti P, Conrad TM, Knight EM, Venkataraman K, Fong NL, Xie B, Gao Y, Palsson BO: Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene. PLoS Genet 2010, 6:e1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akiva E, Copp JN, Tokuriki N, Babbitt PC: Evolutionary and molecular foundations of multiple contemporary functions of the nitroreductase superfamily. Proc Natl Acad Sci U S A 2017, 114:E9549-E9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Loo B, Bayer CD, Fischer G, Jonas S, Valkov E, Mohamed MF, Vorobieva A, Dutruel C, Hyvonen M, Hollfelder F: Balancing specificity and promiscuity in enzyme evolution: Multidimensional activity transitions in the alkaline phosphatase superfamily. J Am Chem Soc 2019, 141:370–387. [DOI] [PubMed] [Google Scholar]
- 35.Holliday GL, Akiva E, Meng EC, Brown SD, Calhoun S, Pieper U, Sali A, Booker SJ, Babbitt PC: Atlas of the radical SAM superfamily: Divergent evolution of function Using a “plug and play” domain. Methods Enzymol 2018, 606:1–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerlt JA, Babbitt PC, Jacobson MP, Almo SC: Divergent evolution in enolase superfamily: strategies for assigning functions. J Biol Chem 2012, 287:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baier F, Tokuriki N: Connectivity between catalytic landscapes of the metallo-beta-lactamase superfamily. J Mol Biol 2014, 426:2442–2456. [DOI] [PubMed] [Google Scholar]
- 38.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L: Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol 2006, 361:1003–1034. [DOI] [PubMed] [Google Scholar]
- 39.Tam R, Saier MH Jr., : A bacterial periplasmic receptor homologue with catalytic activity: cyclohexadienyl dehydratase of Pseudomonas aeruginosa is homologous to receptors specific for polar amino acids. Res Microbiol 1993, 144:165–169. [DOI] [PubMed] [Google Scholar]
- 40. Clifton BE, Kaczmarski JA, Carr PD, Gerth ML, Tokuriki N, Jackson CJ: Evolution of cyclohexadienyl dehydratase from an ancestral solute-binding protein. Nat Chem Biol 2018, 14:542–547. **Tracks changes in sequence, structure and function as an enzyme evolves from a noncatalytic ancestral protein.
- 41.Ayuso-Fernandez I, Ruiz-Duenas FJ, Martinez AT: Evolutionary convergence in lignin-degrading enzymes. Proc Natl Acad Sci U S A 2018, 115:6428–6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poulos TL: Heme enzyme structure and function. Chem Rev 2014, 114:3919–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pozdnyakova NN, Leontievsky AA, Golovleva LA: Extracellular oxidases from solid-state culture of the ligninolytic fungus panus tigrinus 8/18. Biochemistry (Mosc) 1999, 64:442–447. [PubMed] [Google Scholar]
- 44.Sundaramoorthy M, Kishi K, Gold MH, Poulos TL: The crystal structure of manganese peroxidase from Phanerochaete chrysosporium at 2.06-A resolution. J Biol Chem 1994, 269:32759–32767. [PubMed] [Google Scholar]
- 45.Mester T, Ambert-Balay K, Ciofi-Baffoni S, Banci L, Jones AD, Tien M: Oxidation of a tetrameric nonphenolic lignin model compound by lignin peroxidase. J Biol Chem 2001, 276:22985–22990. [DOI] [PubMed] [Google Scholar]
- 46.Camarero S, Sarkar S, Ruiz-Duenas FJ, Martinez MJ, Martinez AT: Description of a versatile peroxidase involved in the natural degradation of lignin that has both manganese peroxidase and lignin peroxidase substrate interaction sites. J Biol Chem 1999, 274:10324–10330. [DOI] [PubMed] [Google Scholar]
- 47. Ayuso-Fernandez I, Rencoret J, Gutierrez A, Ruiz-Duenas FJ, Martinez AT: Peroxidase evolution in white-rot fungi follows wood lignin evolution in plants. Proc Natl Acad Sci U S A 2019, 116:17900–17905. *Traces the emergence of a new catalytic strategy in peroxidases and the improvement in the ability to degrade lignosulfonate during evolution of lignin peroxidases from Polyporales fungi.
- 48. Ayuso-Fernandez I, De Lacey AL, Canada FJ, Ruiz-Duenas FJ, Martinez AT: Increase of redox potential during the evolution of enzymes degrading recalcitrant lignin. Chemistry 2019, 25:2708–2712. *Biophysical studies of ancestral and extant peroxidases that demonstrate an increase in the heme redox potential correlated with a change in binding between the proximal histidine and the Fe atom.
- 49.Sudharshan S, Naidu R, Mallavarapu M, Bolan N: DDT remediation in contaminated soils: a review of recent studies. Biodegradation 2012, 23:851–863. [DOI] [PubMed] [Google Scholar]
- 50.de Souza ML, Seffernick J, Martinez B, Sadowsky MJ, Wackett LP: The atrazine catabolism genes atzABC are widespread and highly conserved. J. Bacteriol 1998, 180:1951–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Copley SD, Rokicki J, Turner P, Daligault H, Nolan M, Land M: The whole genome sequence of Sphingobium chlorophenolicum L-1: insights into the evolution of the pentachlorophenol degradation pathway. Genome Biol Evol 2012, 4:184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schenk G, Mateen I, Ng T-K, Pedroso MM, Mitić N, Jafelicci J,M, Marques RFC Gahan LR, Ollis DL: Organophosphate-degrading metallohydrolases: Structure and function of potent catalysts for bioremediation. Coordination Chem Rev 2016, 317:122–131. [Google Scholar]
- 53. Yang G, Anderson DW, Baier F, Dohmen E, Hong N, Carr PD, Kamerlin SCL, Jackson CJ, Bornberg-Bauer E, Tokuriki N: Higher-order epistasis shapes the fitness landscape of a xenobiotic-degrading enzyme. Nat Chem Biol 2019, 15:1120–1128. **Identifies 19 possible trajectories for evolution of a xenobiotic-degrading enzyme, methyl parathion hydrolase, from an ancestral dihydrocoumarin hydrolase via 5 sequence changes.
- 54.Chen Y, Zhang X, Liu H, Wang Y, Xia X: [Study on Pseudomonas sp. WBC-3 capable of complete degradation of methylparathion]. Wei Sheng Wu Xue Bao 2002, 42:490–497. [PubMed] [Google Scholar]
- 55. Cotton CA, Bernhardsgrutter I, He H, Burgener S, Schulz L, Paczia N, Dronsella B, Erban A,Toman S, Dempfle M, et al. : Underground isoleucine biosynthesis pathways in E. coli. Elife 2020, 9. **Recruitment of MetB to produce 2-ketobutyrate from O-succinylhomoserine for isoleucine synthesis requires mutations elsewhere in the genome.
- 56.Hadzipasic A, Wilson C, Nguyen V, Kern N, Kim C, Pitsawong W, Villali J, Zheng Y, Kern D: Ancient origins of allosteric activation in a Ser-Thr kinase. Science 2020, 367:912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park Y, Patton JEJ, Hochberg GKA, Thornton JW: Comment on “Ancient origins of allosteric activation in a Ser-Thr kinase”. Science 2020, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson C, Kern D: Response to Comment on “Ancient origins of allosteric activation in a Ser-Thr kinase”. Science 2020, 370. [DOI] [PubMed] [Google Scholar]
- 59.Hottes AK, Freddolino PL, Khare A, Donnell ZN, Liu JC, Tavazoie S: Bacterial adaptation through loss of function. PLoS Genet 2013, 9:e1003617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Behe MJ: Experimental evolution, loss-of-function mutations, and “the first rule of adaptive evolution”. Q Rev Biol 2010, 85:419–445. [DOI] [PubMed] [Google Scholar]
- 61.Lang GI, Desai MM: The spectrum of adaptive mutations in experimental evolution. Genomics 2014, 104:412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venkataram S, Dunn B, Li Y, Agarwala A, Chang J, Ebel ER, Geiler-Samerotte K, Herissant L, Blundell JR, Levy SF, et al. : Development of a comprehensive genotype-to-fitness map of adaptation-driving mutations in yeast. Cell 2016, 166:1585–1596 e1522. [DOI] [PMC free article] [PubMed] [Google Scholar]