Abstract
The natural history and treatment landscape of primary brain tumours are complicated by the varied tumour behaviour of primary or secondary gliomas (high-grade transformation of low-grade lesions), as well as the dilemmas with identification of radiation necrosis, tumour progression, and pseudoprogression on MRI. Radiomics and radiogenomics promise to offer precise diagnosis, predict prognosis, and assess tumour response to modern chemotherapy/immunotherapy and radiation therapy. This is achieved by a triumvirate of morphological, textural, and functional signatures, derived from a high-throughput extraction of quantitative voxel-level MR image metrics. However, the lack of standardisation of acquisition parameters and inconsistent methodology between working groups have made validations unreliable, hence multi-centre studies involving heterogenous study populations are warranted. We elucidate novel radiomic and radiogenomic workflow concepts and state-of-the-art descriptors in sub-visual MR image processing, with relevant literature on applications of such machine learning techniques in glioma management.
Subject terms: Medical imaging, Cancer
Background
Primary brain tumours account for about 2% of all cancers in the US with an incidence of about 23 per 100,000. Gliomas account for 80.6% of all malignant brain tumours.1 The incidence is highest for glioblastoma (3.21 per 100,000 population), followed by diffuse astrocytoma (0.46 per 100,000 population). The age-adjusted mortality rate is 4.4 per 100,000 and the 5-year survival rate is 35%.1 However, the rate varies significantly by age at diagnosis and the histology of the tumour.
Advances in our understanding of the molecular pathogenesis of gliomas has prompted significant changes to the World Health Organization (WHO) classification of central nervous system (CNS) tumours in 2016.2 Previously, the classification criteria was based solely on microscopic features.3 The new criteria reclassifies entities with the incorporation of genetic information in certain tumours. These changes were incorporated because of the impact genetic factors have on tumorigenesis and subsequent therapy.
In today’s era of modern imaging, accurate non-invasive prediction of glioma grade/type, survival, and treatment response remains challenging. Stereotactic brain biopsy, despite being invasive and costly, remains the reference standard for histological and genetic classification; however, pathological diagnosis may still remain inconclusive in 7–15% of patients.4 This necessitates imaging surrogates to characterise tumour heterogeneity. Recently, multiple studies have shown strong association between morphological features from multiparametric magnetic resonance imaging (MRI) and survival.5–10 Similarly, functional imaging techniques such as perfusion weighted MRI and magnetic resonance spectroscopy (MRS) have been shown to be beneficial when used along with morphological features, but with limited success and reproducibility.11–16 The limitations in current imaging techniques provide an opportunity for more sophisticated sub-visual feature analysis to augment the morphological features and current functional imaging capabilities.
Radiomics refers to the computerised extraction of quantifiable data from radiological images in the form of radiographical cues that are usually sub-visual.17,18 The extraction of these data creates mineable databases from radiological images which can be used for diagnosis, prognosis characterisation, and to assess or predict response to certain therapies.19–21 Genetic mutations often determine the aggressiveness of the tumour and have been shown to be associated with a lesion’s growth pattern and response to therapy. Radiomic features have been shown to identify genomic alterations within tumour DNA and RNA. The integrated study of data from radiographical and the genomic scales is termed radiogenomics.
In this review, we describe the applications of radiomics and radiogenomics from the perspective of neuroradiologists, neurosurgeons, and neuro-oncologists. Specifically, we review work that highlights the importance of the evolving field for diagnosing and predicting prognosis of individuals with different brain tumour types. Additionally, we discuss the potential and importance of integrating these applications into radiological workflows to improve patient care and outcome.
Overview of radiomics and radiogenomics workflow
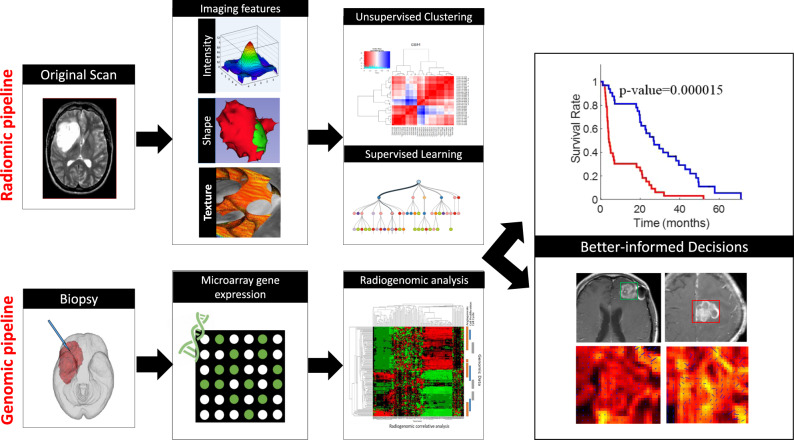
Radiomics is an emergent field that involves converting radiological images into high-dimensional mineable data in a high-throughput fashion. This multi-step process involves (a) image acquisition and reconstruction, (b) image pre-processing, (c) identification of regions of interest, (d) feature extraction and quantification, (e) feature selection, and (f) building predictive and prognostic models using machine learning (Fig. 1).22 To account for MRI intensity non-uniformity, inter- and intra-site scanner variability, image processing routines, such as intensity normalisation, voxel intensity calibration and bias field correction, are used as a precursor to radiomic feature extraction.23–25 The segmentation of the regions of interest (ROI) can be achieved by manual, semi-automated or fully-automated methods.19,26–29 Radiomic features are then extracted from the identified ROIs. Common features can be divided into the following groups: morphological radiomics, textural radiomics, and functional radiomics.
Fig. 1. Radiomics and Radiogenomics workflow.
Use of Radiomics and Radiogenomics pipelines in personalized medicine.
Following feature extraction, different statistical methods are used to select a subset of top features that correlate with the expected outcome.30 Commonly used feature selection algorithms include minimum redundancy maximum relevance (mRMR) algorithm31,32 and sequential feature selection methods.33 Feature selection is performed in order to reduce potential model overfitting associated with the high dimensionality of the radiomic feature set. Once top features are identified, machine learning classifiers and other statistical methods such as the Cox-proportional Hazards modelling techniques34 are used to build predictive and prognostic models. Sanduleanu et al. proposed a “radiomics quality score” tool to assess the quality of the radiomics research study linking tumour biology; however, interpretability of the outcomes of those scores is still questionable.35
The recent advent of radiogenomics has also accelerated the integration of multi-omic data for accurate diagnosis and improved personalised cancer treatments. The first step of the radiogenomic pipeline in neuro-oncology (Fig. 1) is to acquire genomic material via a fresh frozen paraffin embedded (FFPE) sample or a tissue microarray (TMA) sample obtained from a stereotactic brain biopsy from within the brain tumour. Next, bioinformatics techniques, such as sequencing, can detect single-gene mutations. For instance, epidermal growth factor receptor (EGFR) amplification, O6-methylguanine-methyltransferase (MGMT) methylation can be detected by analysing the proteins through immunohistochemical (IHC) analysis and next-generation sequencing (NGS) techniques such as mRNA sequencing. mRNA sequencing, whole-exome sequencing, and whole-genome sequencing can help detect multi-gene expression anomalies. The decisive goal of radiogenomic analysis involves associating gene mutations and pathways directly with distinct imaging phenotypes.
Radiomic feature groups
Morphological radiomics
Morphological radiomic features are used to quantify lesion topology induced by the proliferating boundaries. These can be further divided into global and local morphological features. Global features characterise the contour of the lesion by extracting measurements such as roundness, perimeter, diameters of major and minor axes, and elongation factor. Local morphological features characterise the surface curvature attributes derived from isosurfaces.26,36 These comprise quantitative measurements such as degree of curvature (curvedness) and degree of sharpness.
Textural radiomics
Structural texture analysis
Structural methods describe texture by identifying structural primitives and their placement rules. Multi-scale, multi-resolution steerable bandpass filters like Gabor filter banks37,38 are among the most widely used orientation-based structural descriptors. Gabor descriptors are modelled to mimic the way the human visual system deciphers object appearances, by decomposing the original image into filter responses of a sinusoidal wave of multiple frequencies and orientations. Gabor filters have been shown to distinguish pathologies on histology samples as demonstrated by Doyle et al.39
Statistical texture analysis
Statistical methods analyse the spatial distribution of grey values by computing local features at each image point and deriving a set of statistics from the distribution of local features. One commonly used statistical technique for identifying shape-based object classes is histogram of oriented gradients (HOG).40 Traditionally, the applicability of HOG has been demonstrated for detection of human forms in cluttered images. Multi-coordinate HOG can distinguish different categories of lung tissues in high-resolution tomography images. It characterises local object appearance and shape by computing distribution of local intensity gradients. Grey level co-occurrence matrix (GLCM) features, popularly known as Haralick features41 and originally designed for aerial photography, utilise the values of distance and angle for a combination of grey levels.
Texture analysis using a combination of statistical and structural techniques
Local binary patterns (LBP) is a textural operator that combines statistical and structural methods in appearance classification. LBP is robust with regards to illumination changes and has been shown to be useful in medical datasets which are corrupted by patient motion artefacts. This feature presents texture information as a joint distribution of the intensity of a central pixel and that of its neighbors.42 Li et al.43 demonstrated the use of LBP along with neural networks to classify endoscopic images. Another feature that combines statistical and structural techniques is the co-occurrence of local anisotropic gradient orientations (CoLlAGe) descriptor which seeks to capture and exploit local anisotropic differences in voxel-level gradient orientations to distinguish similar appearing phenotypes.44
Functional radiomics
A critical obstacle to the clinical adoption of traditional radiomic features is its low biological interpretability. To qualify as a biomarker, an attribute should not only be measurable and reproducible, but also be reflective of the underlying anatomy or physiology. It is imperative to discover radiomic signatures that are biologically relevant. Functional radiomic markers are a new class of markers which specifically target the issue of ‘interpretability’ by modelling features that directly capture underlying physiological properties such as angiogenesis. Properties of vessels feeding the lesions (such as convolutedness, density) play an important role in the drugs’ ultimate response. Recently, tortuosity-based features capturing local and global disorder in vessel network arrangement have been shown to be effective in diagnosis and treatment response assessment.45 Deformation descriptors are another class of functional radiomics markers which seek to measure tissue deformation in the brain parenchyma due to mass effect.46 These features provide an insight into the microenvironment outside the visible surgical margins.
Vessel architecture imaging (VAI) MRI is a technique that non-invasively measures parameters to describe structural heterogeneity of brain microvasculature.47–49 The different gradient echo (GE) and spin echo (SE) images produce an apparent different variation in the MRI readout based on the structural and physiological properties of the vessels. Stadlbauer et al.47 examined gliomas (n = 60) using vascular architectural mapping (VAM). They introduced three new VAM biomarkers (i) microvessel type indicator (MTI), (ii) vascular-induced bolus peak time shift (VIPS), and (iii) the curvature (Curv) and adapted known parameters, microvessel radius (RU) and density (NU). MTI and VIPS parameters were helpful in detecting neovascularisation, especially in the tumour core of the HGGs, whereas curvature showed peritumoral vasogenic oedema which correlated with neovascularisation in the tumour core of HGGs. These biomarkers gave insight into complexity and heterogeneity of vascular changes in gliomas to differentiate HGGs versus LGGs.50 Furthermore, combining multiparametric quantitative blood oxygenation level-dependent approach (qBOLD) with VAM parameters helped distinguish LGGs versus HGGs and identify isocitrate dehydrogenase (IDH) mutation status with higher sensitivity.50 Stadlbauer et al.51 also performed analysis of vascular hysteresis loop (VHLs) in combination with the VAM biomarkers to assess response of glioblastoma to anti-angiogenic therapy. MTI was found to be useful to predict responding versus non-responding regions, whereas, Curv was better to assess severity of vasogenic oedema. Price et al.52 used diffuse tensor imaging (DTI) with MR perfusion and MRS imaging to determine changes in the invasive versus non-invasive margins of glioblastomas to better predict treatment efficacy and overall survival.53,54
Semantic features
Semantic features, such as tumour location, shape, and geometric properties on structural MRI,19,55 are qualitative features used by neuroradiologists to describe the tumour environment. Previous studies have found that semantic features are related to the genetic phenotype of brain tumours.56 The Visually AcceSAble Rembrandt Images (VASARI) project by TCIA established a feature set to enable consistent description of gliomas using a set of defined visual features and controlled vocabulary.57 Studies have shown that these features are highly reproducible and provide meaningful guidance in glioblastomas.5 Semantic features are also robust to changes in image acquisition parameters and noise and can be used along with more sophisticated radiomic features in machine learning settings.22
Diagnostic applications
Differentiating tumours based on texture analysis
Many studies have shown the application of textural analysis for differentiating HGGs from LGGs. Skogen et al.58 applied a filtration-histogram technique for characterising tumour heterogeneity. In a cohort of 95 patients (27-grade II, 34-grade III, and 34-grade IV), by using standard deviation (SD) at a fine texture scale, they were able to distinguish LGGs from HGGs with sensitivity and specificity of 93% and 81% (AUC 0.91, P < 0.0001). Tian59 et al. applied textural analysis on multiparametric MRI of 153 patients and reported an accuracy of 96.8% for classifying LGGs from HGGs and 98.1% for classifying grade III from grade IV using an SVM classifier. Xie et al.60 evaluated five GLCM features from (DCE)-MRI of 42 patients with gliomas. They reported that entropy (AUC = 0.885) and IDM (AUC = 0.901) were able to differentiate grade III from grade IV and grade II from grade III gliomas, respectively.
Suh et al.61 used MRI radiomics-based machine-learning algorithms in differentiating central nervous system lymphoma (PCNSL; n = 54) from non-necrotic atypical glioblastoma (n = 23). Over 6000 multi-sequence and multi-regional radiomic features including shape, volume, and texture were first obtained. AUCs achieved statistical significance for each of the three machines used, demonstrating a higher diagnostic performance than three radiologists.
Characterising tumour molecular subtypes of low-grade gliomas (LGG)
Adult LGGs with IDH1 mutation are associated with a better prognosis and longer survival than are IDH1-wild-type adult LGGs (i.e., negative for IDH1 mutation), which act much more aggressively.2 1p/19q codeletion has been associated with good response to chemotherapy and a very favourable prognosis. Non-invasive radiogenomic molecular phenotyping may allow for personalised therapeutic decisions in LGG.
Multiple groups have evaluated radiomic features to determine molecular phenotype of gliomas.62 Zhang et al.63 extracted 15 optimal radiomic features (n = 152) using SVM-recursive feature elimination (SVM-RFE) that could detect IDH mutation with accuracy of 82.2%. Han et al.64 extracted radiomic features from 42 patients with histopathologically confirmed gliomas. They showed joint variables derived from T1-weighted image (T1WI), T2 weighted image (T2WI), and contrast-enhanced T1WI imaging histograms and GLCM features could be used to detect IDH1-mutated gliomas. The AUC of joint variableT1WI+C for predicting IDH1 mutation was 0.984, and the AUC of joint variableT1WI for predicting the IDH1 mutation was 0.927. Jakola et al.65 reported that textural homogeneity could discriminate between LGG patients with IDH mutation and IDH wild-type (P = 0.005). The AUC for combined parameters (tumour homogeneity and tumour volume) was 0.940 for predicting IDH mutation. However, this method could not differentiate LGG with IDH mutation with or without 1p19q codeletion. Bahrami et al.66 measured tissue heterogeneity and edge contrast (EC) on FLAIR images of 61 patients and reported that patients with IDH wild-type tumours showed higher signal heterogeneity (P = 0.001) and lower EC (P = 0.008) compared with IDH mutant type. Among patients with IDH mutant tumours, 1p/19q codeleted tumours had greater signal heterogeneity (P = 0.002) and lower EC (P = 0.005), and MGMT-methylated tumours showed lower EC (P = 0.03).
Metabolic alteration of D-2-hydroxyglutarate (D-2HG) production is a hallmark for IDH mutation in gliomas.67 Recently, a number of groups have demonstrated reliable detection of D-2HG using in vivo 1H MRS.68–70 Andronesi et al.68 reported apparent in vivo detection of D-2HG using 2D correlation spectroscopy (COSY) and J-difference spectroscopy in IDH1-mutated gliomas. Rohle et al.71 identified a selective R132H-IDH1 inhibitor via a high-throughput screening, which in a dose-dependent manner can inhibit the production of R-2HG, providing further avenues for targeted therapies.
Differentiating treatment effects (radiation necrosis, pseudoprogression) and tumour recurrence
A major challenge in the management of glioblastoma is the difficulty of accurately assessing the response to treatment with several entities that can mimic tumour recurrence or progression on structural MRI, namely pseudoprogression and radiation necrosis (RN).26,72–74 Visual diagnosis is often ambiguous and remains extremely challenging, clinically.75 Functional MRI, such as MR perfusion and MRS improves the diagnostic accuracy; however, this may not be universally available and are often difficult to reproduce.72,76–79 Radiomics provides a non-invasive approach to reliably distinguish tumour recurrence from treatment effects and can potentially help prevent unnecessary biopsies.26,36,80
By combining 3D shape and surface radiomic features extracted from both T1WI-enhancing lesions and T2WI/FLAIR hyperintense perilesional area, Ismail et al.36 were able to differentiate between true progression and pseudoprogression with 90.2% accuracy (n = 105). The two most discriminative features were found to be local features capturing total curvature of the enhancing lesion and curvedness of the T2WI/FLAIR hyperintense peritumoral region. The differential expression patterns may be attributed to the alteration of white matter structure via infiltration, resulting in surface shape irregularities.
Similar to pseudoprogression, distinguishing brain tumour recurrence (RT) from RN can be challenging on routine MRI due to the lack of objective methods of assessment. A texture analysis in conjunction with support vector machine approach presented by Larroza et al.74 could differentiate brain metastasis from RN (n = 115) with an AUC of >0.9. CoLlAGe features were shown to express differentially across different grades of RN and tumours.44,81 A novel aspect of this study was the inclusion of pure cerebral RN from nasopharyngeal carcinoma. CoLlAGe entropy values were found to be skewed toward higher values for the predominant tumour cases compared with the pure cerebral RN or predominant cerebral RN.44,81,82 Prasanna et al.81 further demonstrated that incorporating CoLlAGe features from pure RN in the training set resulted in improved classification performance of the predominant RN/RT, compared with using features from the predominant RN/RT alone. This potentially demonstrates, to some extent, the similarity in structural and morphological properties between pure RN and its mixed presentations.
Hu et al.80 presented an automated technique to identify RN at high spatial resolution using multi-parametric MR features. The classification feature vector comprised eight parameters derived from the multiple sequences, including contrast-enhanced T1, T2, FLAIR, PD, ADC, rCBF, rCBV and MTT. The mean AUC obtained on n = 31 (RT = 15, RN = 16) cases was 0.94. Interestingly, the performance using ADC features was significantly better than those using conventional MRI measures. Combining textural measures of heterogeneity with tracer uptake kinetics was shown to be more effective in distinguishing brain metastasis recurrence from radiation injury.83 The diagnostic accuracy when using tumour-to-brain ratios (TBRs) of 18F-FET uptake was 83% which increased to 85% upon combining with textural parameters such as coarseness, short-zone emphasis, or correlation.83 This shows the potential complementary diagnostic information that the texture attributes may provide along with other modalities.
Prognostic applications
Survival stratification in glioblastomas
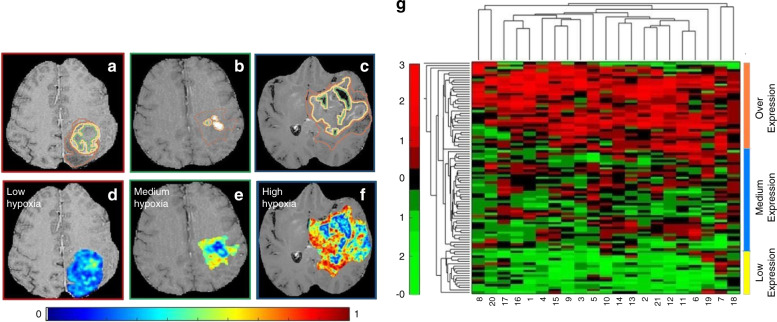
Over 40% of glioblastoma patients do not respond to conventional chemo-radiation therapy and show progression within 6–9 months.84 Hypoxia in glioblastoma multiforme is a key pathway known to promote tumour neovascularisation and invasion of healthy tissue as well as driving treatment resistance leading to poor prognosis.85 Multiple pathways such as cellular proliferation, apoptosis and increased angiogenesis are also known to contribute towards poor progression-free survival (PFS) outcome. Currently, there is a lack of well-validated biomarkers to monitor levels of hypoxia and predict treatment response to anti-angiogenic agents.86 Beig et al.87 showed that surrogate radiomic descriptors can capture the extent of hypoxia of glioblastoma on pre-treatment MRI and predict survival (Fig. 2). In this radiogenomic study, the authors used microarray expression data from 85 glioblastoma patients to construct a hypoxia enrichment score (HES). Next, a radiomic model was trained that correlated with HES, and then used to stratify glioblastoma based on their overall survival (OS). On a validation set of n = 30 patients, the radiomic features which were strongly associated with HES, could also distinguish short-term survivors (OS < 7 months) from long-term survivors (OS > 16 months) (P = 0.003). Another study by Kickingereder et al.88 (n = 119) extracted over 12,190 radiomic features of glioblastoma and concluded that an 11-feature radiomic signature allowed for the prediction of PFS and OS.
Fig. 2. Construction of Hypoxia Enrichment Score.
a–c show a 2D Gd-T1w MRI slice with expert-annotated necrosis (outlined in green), enhancing tumour (yellow) and oedematous regions (brown) in three different GBM patients that exhibited low, medium, and high hypoxia enrichment score (HES) respectively. The corresponding inverse difference moment (Haralick) feature map has been overlaid on the manually annotated tumour regions, for HESlow (d), HESmedium (e), and HEShigh (f). g Unsupervised clustering of the RNAseq data from the 21 hypoxia associated genes clustered as high hypoxia (HEShigh—shown in navy blue, medium hypoxia (HESmedium—shown in teal) and low hypoxia (HESlow—shown in yellow). The x axis in the clustergram represents the 21 genes and y axis represents the patient population of 97 GBM cases. Figure from Beig et al.87; licensed under a Creative Commons Attribution (CC BY) license.
Jain et al.89 investigated imaging, genomic, and haemodynamic parameters obtained from the non-enhancing region (NER) of glioblastoma on multi-sequence enhanced MRI. Increased relative cerebral blood volume of the NER (rCBVNER) and NER crossing the midline were found to be associated with poor survival. Wild-type EGFR mutation was the only genomic alteration that was associated with significantly poor survival in patients with high rCBVNER.
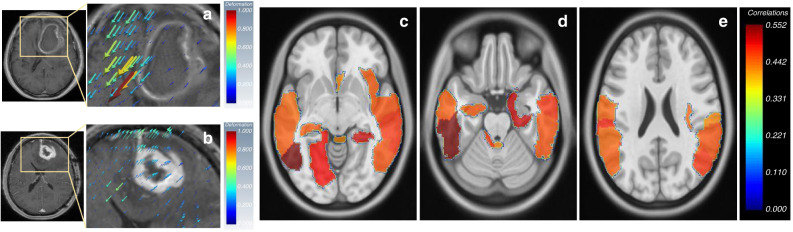
Prasanna et al.46 discussed how mass effect-induced deformation heterogeneity (MEDH) from glioblastoma on multi-sequence MRI affects survival. High expression of MEDH in the areas of language comprehension, social cognition, visual perception, emotion, somatosensory, cognitive and motor-control functions were found to be associated with worse survival (Fig. 3). McGarry et al.90 were able to use multi-sequence MRI radiomic profiles (RPs) of newly diagnosed glioblastoma in order to further stratify patient prognosis. Each voxel examined was assigned an RP. Five such RPs were predictive of overall survival prior to therapy initiation.
Fig. 3. Deformation Radiomics.
a, b Deformation vectors representing tissue displacement are shown as volume rendered 3D quivers overlaid on an image slice of right-hemispheric GBMs. The deformation magnitude is proportional to the size of quivers. Higher value of deformation magnitude is represented by ‘red’ and lower value by “blue” colour respectively. The quivers also show the direction of tissue displacement. c The AAL regions in which, the MEDH negatively correlated with survival with P < 0.05 for right-hemispheric tumour group (neurological view). The colormaps show the negative correlation values (shown as positive for easier representation). Figure from Prasanna et al.46; licensed under a Creative Commons Attribution (CC BY) license.
Survival stratification in lower-grade gliomas
Liu et al.91 developed a radiomics signature to predict PFS in LGGs. The radiomic risk score (RRS) was calculated and the genetic characteristics of the group with high-risk scores were identified by radiogenomic analysis. Biological processes of cell adhesion, cell proliferation, differentiation and angiogenesis were identified to be positively associated with the RRS. A nomogram combining the RRS with other clinical features improved patient stratification and resulted in better assessment of PFS.
Zhou at al.92 built radiomic models using automated texture analysis and VASARI features to predict IDH1 mutation (AUC = 0.86), 1p/19q codeletion status (AUC = 0.96), histological grade (AUC = 0.86) and tumour progression (AUC = 0.80) in patients (n = 165) with diffuse low- and intermediate-grade gliomas. They found that on MRI images no enhancement and a smooth non-enhancing margin were predictive of longer PFS; smooth non-enhancing margins were also a significant predictor of longer OS in LGGs.
Li et al.93 selected nine radiological features that could predict Ki-67 expression level and achieved accuracies of 83.6% and 88.6% in the training (n = 78) and validation (n = 39) sets, respectively. Only spherical disproportion (SDp) feature was found to be a prognostic factor with patients in the high SDp group. Ki-67 expression level and SDp were independent prognostic factors in the multivariate Cox regression analysis.
Applications in selecting optimal therapy
Stratifying anti-angiogenic treatment response for recurrent glioblastomas
Anti-angiogenic treatment is the treatment of choice for recurrent glioblastoma. Kickingereder et al.94 investigated imaging biomarkers that may be able to predict treatment outcome. By utilising multi-sequence MRI with recurrent glioblastoma prior to anti-angiogenic treatment with bevacizumab, radiomic MRI features were extracted and analysed from 172 patients. Using these radiomic features, a model was created to predict median PFS and OS in low-risk and high-risk groups. The authors concluded that radiomics might be able to identify patients who would most benefit from bevacizumab therapy.
Bahrami et al.95 evaluated 33 HGG patients before and after initiation of bevacizumab treatment. They segmented volume of interest (VOI) within FLAIR hyperintense region and extracted edge contrast (EC) magnitude for each VOI using gradients of the 3D-FLAIR images. They reported that lower EC of the FLAIR hyperintense region was associated with poor PFS (P = 0.009) and OS (P = 0.022). Other relevant literature has been summarised in Tables 1–3.
Table 2.
Prognostic applications.
| Author | No. of patients | Magnet strength/MRI sequence | Segmentation manual vs automatic | Texture software | Type of radiomic analysis | Best discriminating features | Machine learning/statistical approach | Results |
|---|---|---|---|---|---|---|---|---|
| Beig et al.87 | 115 (GBM) | T1WI, T2WI, FLAIR | Manual 2D | Matlab | 30 features: Laws, Gabor, GLCM | Laws energy (R5R5, E5E5, S5S5) from enhancing tumour and oedema | RF model | Features from the enhancing and oedematous regions were predictive of the extent of hypoxia. Features on the validation set were also found to be prognostic of overall survival |
| Kickingereder et al.88 | 119 (GBM) | 3 T/T1-CE, FLAIR | Manual 3D | Medical Imaging Toolkita |
Histogram volume and shape features; texture features; wavelet analysis |
11 features of 12,190 features | SPC analysis | SPC analysis performed better than clinical (age and Karnofsky Performance Score) and radiologic (rCBV and ADC) parameters |
| Molina et al.128 | 79 (GBM) | 1.5–3 T/T1-CE | Manual 3D | Matlab | Five GLCM features, 11 GLRLM features |
GLCM (entropy, homogeneity, contrast, dissimilarity) GLRLM (LRE, HGLRE, LRHGE, RPC) |
Kaplan–Meier curves and Cox-proportional hazards analysis | Patients had better prognosis when high LRHGE, low RPC, low entropy, high homogeneity, and low dissimilarity were present (P < 0.05) |
| Chaddad et al.129 | 73 (GBM)/ TCIA | T1-CE, FLAIR | Semi-automatic 2D | – | GLCM JIM |
JIM (entropy, inverse-variance) in necrosis region and (entropy, contrast) in oedema region |
RF model | JIM of T1-CE and FLAIR images significantly predicted survival outcomes with moderate correlation; nine features were found to be associated with glioblastoma survival, P < 0.05, accuracy of 68%-70%; AUC of 77.56% with P = 0.003 was achieved when combining JIM, GLCM, and gene expression features into a single radiogenomic signature. |
| Liu et al.130 | 119 (GBM)/TCGA |
T1WI, T2WI, FLAIR, T1-CE |
Manual 2D | – | Histogram, GLCM, GLRLM | 13 textural features | SVM-RFE | T1-CE sequence performed best, with AUC of 0.7915 and accuracy of 80.67% |
| Yang et al.131 | 82 (GBM)/TCGA | T1-CE, T2WI, FLAIR | Manual 3D | Matlab | SFTA, RLM, LBP, HOG, Haralick texture features | – | RF model | Molecular subtypes and 12-month survival was predicted by several features; SFTA features on T1-CE were most predictive of survival and proneural subtype (AUC = 0.82); RLM features on T2 FLAIR axial for neural (AUC = 0.75); Haralick features on T1-CE for classic (AUC = 0.72); HOG features on T2 FLAIR axial for mesenchymal (AUC = 0.70) |
| Bahrami et al.95 | 33 (HGG) | 3 T/FLAIR | Semi-automatic 3D | – | EC | – | Kaplan–Meier curves | Reported that lower EC of the FLAIR hyperintense region was associated with poor PFS (P = 0.009) and OS (P = 0.022) status post-bevacizumab therapy |
| Jain et al.89 | 45 (GBM) | 1.5/3 T, T1, T2w/perfusion, FLAIR | Manual 2D | Rb | VASARI features, rCBV statistics | rCBVNER | Random Survival Forest | Increased maximum rCBVCER found to be associated with increased risk of death |
| McGarry et al.90 | 81 (GBM) | 1.5/3 T, T1, CE-T1, T2, FLAIR | Semi-automatic 3D | – | 81 radiomic profiles (5 RPs): 4-digit code assigned to each voxel representing the intensity-based segmentation | Five RPs correlated with survival when thresholded by volume | Cox Regression | Presented a method for creating radiographic profiles by combining intensity information from multiple MRI scans. Pathologically validated that voxels indicated by one of the RPs contained hypercellular tumour and necrosis. |
| Lao et al.117 | 112 (GBM) | 1.5/3 T, T1, T1-CE, T2, FLAIR | Manual 2D | Python CAFFE | 1403 handcrafted (HC) features: geometry, intensity, texture and 98304 deep features using transfer learning | 150 HC and deep features | LASSO Cox Regression | The radiomics signature achieved a C-Index of 0.731 for the discovery dataset, and 0.710 for the independent validation set. |
| Li et al.132 | 92 (GBM)/TCIA, local data | T1, T1-CE, T2, FLAIR | Automated using Matlab | Rb | Texture at different voxel size, quantization and grey levels | - | Cox regression model | The multiparametric signature achieved better performance for OS prediction (C-Index = 0.705) |
| Sun et al.133 | 542 (training n = 285; validation =66; test=191; LGG,HGG)/BRATS 2018 | T1, T1-CE, T2, FLAIR | 3D CNN architectures | Pyradiomics toolbox | Shape, texture, first-order statistics | Age, 14 selected features | RF regression | Achieved 61% accuracy in predicting survival outcome. |
| Sanghani et al.134 | 163 (GBM)/BRATS 2017 | T1, T1-CE, T2, FLAIR | Segmentation masks from BraTS 2017 dataset | – | Texture, shape, volumetric, age | Top 150 selected features out of 2200 | SVM-RFE/fivefold cross-validation | The 2-class and 3-class OS group prediction accuracy obtained were 98.7% and 88.95% respectively. |
| Beig et al.135 | 460 (GBM), Male = 290, female = 170 | T1, T1-CE, T2, FLAIR | Semi-automatically with Slicer 3Dd | Matlab | 2850 features: Laws, Gabor, shape based | Five Laws energy, two Gabor wavelets; one shape based | Cox regression model | Sexually dimorphic radiomic risk score (RRS) models that are prognostic of overall survival (OS) in primary GBM |
| Han et al.136 | 178 (HGG)/TCIA, local dataset | T1-CE | Manual segmentation | Matlab, Kerasc, R | 348 handcrafted - volume, size, texture, intensity, first-order statistical + 8192 deep CNN features | – | Elastic net-Cox modelling | The combined feature analysis framework classified the patients into long- and short-term survivor groups with a log-rank test P < 0.001 |
EC Edge contrast, JIM joint intensity matrices, RF random forest, TCGA The Cancer Genome Atlas, TCIA The Cancer Imaging Archive, LRE long-run emphasis, HGLRE high grey-level run emphasis, LRHGE long-run high grey-level emphasis, RPC run percentage, SFTA segmentation-based fractal texture analysis, LBP local binary pattern, HOG histogram-oriented gradient, SPC supervised principal component.
ahttp://www.mitk.org/wiki/The_Medical_Imaging_Interaction_Toolkit_(MITK).
bR statistical and computing software (http://www.r-project.org).
cKeras (www.keras.io).
Table 1.
Diagnostic applications.
| Author | No. of patients | Magnet strength/MRI Sequences | Segmentation manual vs automatic |
Software | Type of radiomic analysis | Best discriminating features | Machine learning/statistical approach | Results |
|---|---|---|---|---|---|---|---|---|
| Ismail et al.36 | 105 (Training n = 59, test n = 46; GBM) | 1.5 T/T1-CE, T2WI/FLAIR | Manual 2D | Matlaba | 30 shape features; 14 “global” contour characteristics and 16 “local” curvature |
Top two most discriminative features including the SD of S and the mean of KT |
SVM | 3D shape attributes from the lesion habitat can be differentially expressed across pseudoprogression and tumour progression and could be used to distinguish these radiographically similar pathologies |
| Larroza et al.74 | 115 (RN = 32, radiation treated mets = 23, untreated mets = 60) | T1-CE | Manual 2D | Mazda | 179 features; histogram, gradient, GLCM, GRLM, wavelets | Intensity 90th percentile | SVM | High classification accuracy (AUC > 0.9) was obtained using texture features and a support vector machine classifier to differentiate between brain metastasis and RN |
| Prasanna et al.81 | 75 (different grades of RN) | T1-CE | Manual 2D | Matlab | Four CoLlAGe entropy | Collage entropy skewness | RF | COLLAGE features exhibited decreased skewness for patients with pure and predominant RN and were statistically significantly different from those in patients with predominant recurrent tumours. |
| Hu et al.80 | 31 (RT = 15, RN = 16) | T1-CE, T2, FLAIR, PD, ADC, rCBF, rCBV and MTT | Manual 2D | – | Eight parameters derived from the multiple MR sequences: CE-T1, T2, FLAIR, PD, ADC, rCBF, rCBV and MTT. | rCBV | OC-SVMs | Greater value of advanced MRI DWI and PWI derived measures as compared to conventional imaging for discrimination of RN from viable tumour |
| Tiwari et al.26 | 58 (training n = 43, test n = 15; GBM) | T1-CE, T2, FLAIR | Manual 2D | Matlab | 119 texture maps: Haralick, Laws, Laplacian pyramid, histogram of gradient orientations | GLCM and Laws features in the lower Laplacian scale | SVM | Laplacian pyramid features were identified to be most discriminative, possibly because these emphasize edge-related differences between RT and RN at lower resolutions. |
| Skogen et al.58 | 95 (HGG = 68, LGG = 27) | 3 T/T1-CE | Manual 2D | TexRADb | Histogram metric | Fine textures scale | – | LGGs and HGGs were best discriminated using SD at fine-texture scale, with a sensitivity and specificity of 93% and 81% (AUC = 0.910, P < 0.0001) |
| Xie et al.60 | 42 (HGG = 27, LGG = 15) | 3 T/dynamic contrast-enhanced | Manual 2D | OmniKineticsc | Five GLCM features - energy, entropy, inertia, correlation, and inverse difference moment (IDM) | Entropy and IDM | – | Evaluated five GLCM features from (DCE)-MRI of 42 patients with gliomas. They reported that entropy (AUC = 0.885) and IDM (AUC = 0.901) were able to differentiate grade III from grade IV and grade II from grade III gliomas, respectively.; no feature was able to distinguish subtypes of grade II and grade III gliomas. |
| Qi et al[.126 | 39 (HGG = 26, LGG = 13) | 3 T/DWI/DKI | Manual 2D | ImageJd | Histogram metric | Mean kurtosis (MK) | – | Histogram parameters on DKI were significant in differentiating high- (grade III and IV) from low-grade (II) gliomas (P < .05); mean kurtosis was the best independent predictor of differentiating glioma grades with AUC = 0.925 |
| Tian et al.59 | 153 (Grade II = 42, III = 33, IV = 78) |
3 T/Multiparametric (T1WI, T1-CE, T2WI, DWI, ASL) |
Manual 2D-VOI | Matlab |
GLCM, GLGCM histogram mean |
30 and 28 Optimal features of 420 texture and 90 histogram features | SVM-RFE | Texture features were statistically significant over histogram parameters for glioma grading; AUC for classifying LGGs versus HGGs was 0.987, while it was 0.992 for grade III versus IV gliomas |
| Zacharaki et al.96 | 102 (mets = 24, meningiomas = 4, grade II gliomas = 22, grade III gliomas = 18, GBMs = 34) | 3 T (T1, T2, FLAIR, DTI, perfusion) | Manual 2D | – | 161: tumour shape, image intensity, Gabor texture | Different features for each pair-wise classification task, mainly comprising intensity from T1, T2, rCBV statistics and Gabor texture from FLAIR | LDA, kNN, SVM | The binary SVM classification achieved via a leave-one-out cross-validation reported accuracy, sensitivity, and specificity of 85%, 87%, and 97% for discrimination of metastases from gliomas and 88%, 85%, and 96% for discrimination of high-grade (grade III and IV) from low-grade (grade II) neoplasms. |
| Suh et al.61 | 77 (GBM = 23, PCNSLs = 54) | 3 T/T1-CE, T2, FLAIR | Manual 2D | Python package, PyRadiomics 1.2.0e | Shape, volume, 1st order, GLCM, GLRLM, mGLSZM, and wavelet transform | A total of 6366 radiomics features subjected to recursive feature elimination and random forest analysis with nested cross-validation | SVM | In comparing diagnostic performances, the AUC (0.921) and accuracy (90%) of the radiomics classifier was significantly higher than those of the 3 radiologists (P < 0.001) |
| Beig et al.127 | medulloblastomas (n = 22), ependymomas (n = 12), and gliomas (n = 25) | T1, T2, FLAIR | Manual 2D | Matlab | 52 CoLlAGe features | sum variance and entropy of CoLlAGe on T2 | RF | Medulloblastomas exhibited higher CoLlAGe entropy values than ependymomas and Gliomas for the paediatric brain tumour cases. |
OC one class, SVM support vector machine, RFE recursive feature elimination, RF Random Forest, rCBV relative cerebral blood volume, RLM run-length matrix, ADC apparent diffusion coefficient, EC edge contrast, – not available, GLSZM grey-level size-zone matrix, GLCM grey level co-occurrence matrix, GLRLM grey level run-length matrix features, LDA linear discriminant analysis, S sharpness, KT measure of the total curvature, VOI volume of interest, kNN k-nearest neighbours.
aMathWorks, Natick, Massachusetts.
bhttps://imagingendpoints.com/texrad-software/.
chttp://www.omnikinetics.com/.
dNational Institutes of Health, Bethesda, Maryland.
ehttps://pyradiomics.readthedocs.io/en/latest/modules/radiomics/ngtdm.html.
Table 3.
Applications in selecting optimal therapy.
| Author | No. of patients | Magnet strength/MRI sequence | Segmentation manual vs automatic | Texture software | Type of radiomic analysis | Best discriminating features | Machine learning/statistical approach | Results |
|---|---|---|---|---|---|---|---|---|
| Beig et al.137 | 83 LGGs | T2w/FLAIR | Manual 2D | Matlab | GLCM, Gabor | 3D Gabor | SVM | Initial results indicate that radiomic features from non-enhancing regions on T2 and infiltrative edges on FLAIR can segregate the 3 subgroups. |
| Zhang et al.63 | 152 (IDH mutant = 92, wild-type = 60) | 1.5T–3 T/T1-CE, T2WI, FLAIR | Manual 3D | Matlab | GLCM GLGCM | 15 Optimal features from 168 Haralick features | SVM-RFE | AUC 0.841 and accuracy of 82.2% for non-invasively discriminating IDH mutation of patient with glioma |
| Hsieh et al.138 | 39 (IDH mutant = 7, wild-type = 32; TCGA) | 1.5T–3 T/T1-CE | Manual 2D | CAD system | Morphological intensity-GLCM | 14 GLCM | Binary logistic regression classifier | Textural features describing local patterns yielded an accuracy of 85% in detecting the IDH status |
| Han et al.64 | 42 (IDH mutant = 21, wild-type = 21) | 3 T/T1WI, T2WI, 3D-T1-CE | Manual 3D | OmniKinetics | 29 Texture features from first-order and GLCM | Inertia, cluster prominence, GLCM entropy | – | Showed joint variables derived from T1WI, T2WI, and contrast-enhanced T1WI imaging histograms and GLCM features could be used to detect IDH1-mutated gliomas. The AUC of Joint VariableT1WI+C for predicting IDH1 mutation was 0.984, and the AUC of Joint VariableT1WI for predicting the IDH1 mutation was 0.927 |
| Jakola et al.65 | 25 (IDH mutant = 20, wild-type = 5) | 3 T/3D-FLAIR | Semiautomatic 3D | ImageJ | Homogeneity, energy, entropy, correlation, inertia | Homogeneity | – | Homogeneity discriminated patients with LGG in IDH mutant and IDH wild-type (P = 0.005), AUC for combined parameters was 0.940 for predicting IDH mutation; authors could not separate IDH-mutant tumours on basis of 1p/19q-codeletion status |
| Bahrami et al.66 | 61 (IDH mutant = 43, wild-type = 11); 7 unknown) | 3 T/Pre- and post-T1-CE, FLAIR | Semiautomatic 3D | 3D-co-occurrence matrix | Histogram, GLCM | Homogeneity, pixel correlation, EC | Logistic regression with LASSO regularization | Greater signal heterogeneity and lower EC noted in IDH wild-type tumours; IDH mutant tumours with 1p/19q-codeleted status; lower EC in MGMT-methylated tumours |
| Shofty et al.139 | 47 LGGs | 1.5T–3 T/FLAIR, T2, TI-CE | Automatic using FSLa, 3D | Matlab | Histogram, contrast, correlation, energy, entropy, homogeneity | 39 of 152 textural features | 17 classifiers | Ensemble of bagged trees classifier achieved the best performance (Accuracy = 87%; AUC = 0.87) for the detection of 1p/19q codeletion; majority of differences detected for T2 and T1-CE |
| Kickingereder et al.94 | 172 (GBM) | 3 T/Pre- and post-T1-CE, FLAIR | Semiautomatic 3D | Medical imaging Toolkitb | 188 imaging features, 17 first-order features (FO), 9 volume and shape features (VSF) and 162 texture features (GLCM,) GLRLM). | – | Supervised principal component (superpc) analysis | The superpc predictor stratified patients in the validated set into a low or high-risk group for PFS (HR = 1.85, P = 0.030) and OS (HR = 2.60, P = 0.001). |
| Grossmann et al.100 | 126 (GBM) |
Baseline and follow-up MRI (1 and 6 wks), T1WI, T2WI, FLAIR, T1-CE |
Semi-automatically with Slicer 3Dd |
R version 3.1.0c |
First-order statistics of the voxel intensity histogram; tumour shape; tumour texture |
Information correlation | PCA | Radiomics provides prognostic value for survival and progression in patients with recurrent glioblastoma receiving bevacizumab treatment; features derived from postcontrast T1WI yielded higher prognostic power compared with T2WI |
| Wu et al.140 | 126 (grade II-III = 43 and grade IV = 83)/TCIA | T1, T1-CE, T2, FLAIR | Semiautomatic approach | R version 3.3.1c | GLCM texture, Volume, intensity, histogram, diffusion | 20 of 704 radiomic features | SVM, kNN, RF, NB, NN, FDA, Adaboost/tenfold cross-validation | Random Forest (RF) showed high predictive performance for identifying IDH genotype (AUC = 0.931, accuracy=88.5%) |
| Zhou et al.141 | 744 (LGG, HGG)/ TCIA, local datasets | T1-CE, T2-FLAIR | Semi-automatically with Slicer 3Dd | Matlab | Histogram, texture, age, shape | Top 15 features out of 127 | RF/Train-test model | The overall accuracy for 3 group prediction (IDH-wild type, IDH-mutant and 1p19q co-deletion, IDH-mutant and 1p19q non-codeletion) was 78.2% |
| Lee et al.142 | 123 (GBM) | T1, T2, T1-CE, FLAIR, PWI, DWI | Manual segmentation | Nordi-cICE | Volume, ADC map, CBV | Four of 31 radiological features | kNN, SVM, RF, Adaboost, decision tree, NB, LDA, gradient boosting | Prediction rate of IDH1 mutation status with 66.3–83.4% accuracy |
| Sudre et al.143 | 333 (IDH mutant = 151, wild-type = 182); | 1.5 T/T1, T2, FLAIR, DSC MRI | Olea Sphere, Version 3 | – | Shape, intensity, texture | Nine histogram features, 11 texture features | RF/cross-validation | Gliomas were correctly stratified 53% for grade classification and 71% for IDH classification |
GLCM grey level co-occurrence matrix, GLRLM grey level run-length matrix features, LDA linear discriminant analysis, PCA principal component analysis, Adaboost adaptive boosting, NB Naive Bayes, FDA flexible discriminant analysis, NN neural network.
aFSL (http://www.fmrib.ox.ac.uk/fsl).
bhttp://www.mitk.org/wiki/The_Medical_Imaging_Interaction_Toolkit_(MITK).
cR statistical and computing software (http://www.r-project.org).
Discussion
Conventional structural MRI, although popular as a universally available imaging modality, has often failed in distinguishing tumour recurrence, pseudoprogression and radionecrosis with classic criteria such as enhancement, mass effect and perilesional oedema.11 The enhancement patterns like ‘soap bubble’, ‘Swiss cheese, and moving wave front are considered therapy-induced, but they lack reproducibility and mandate further texture analysis, spectroscopy or blood flow studies for validation.96 The dilemmas and delays in detection of progression versus pseudoprogression versus mixed lesions can delay treatment or cause discontinuation of treatment.26,36
Presently, stereotactic brain biopsy remains the gold standard for histological and genetic classification. However, the high tumour heterogeneity in gliomas may decrease the accuracy of biopsies and render pathological diagnosis inconclusive in about 7–15% of patients.97 The ability to assess local imaging presentations of tumours, based on the underlying genotype, could potentially mitigate bias associated with tissue sampling during biopsy procedures. The potential for radiomic analysis to distinguish glioma molecular subtypes non-invasively would not only provide additional prognostic information, but would also assist in the selection of targeted chemotherapy in patients with multiple genetic mutations and potentially high-grade tumour types.84
The median survival of glioblastoma with surgical debulking is 15 months, with the clinical outcome depending on the extent of initial resection and response to chemo-radiation therapy.98 Radiomic risk models can therefore be utilised to better predict treatment response, PFS and OS. By obtaining the radiogenomic profile of a tumour non-invasively,99 the effect of anti-angiogenic therapies, such as bevacizumab, can be assessed without harm to the patient.100 To date, the efficacy of anti-angiogenic therapies is primarily monitored with MRI and MRS. Tumour pseudo response to anti-angiogenic therapies becomes problematic in that standard enhancement characteristics of a tumour may appear falsely reduced after the administration of anti-angiogenic medications.101 The rapid decrease in contrast enhancement and vasogenic oedema suggests anti-angiogenic response when the tumour may be stable or has progressed. The alterations in MR characterisation of tumours after anti-angiogenic therapy is primarily related to changes in blood–brain barrier permeability. By utilising radiomics in such cases, the actual tumour response may be monitored, and treatment strategies can be further tailored prior to tumour progression.
Gliomas are genetically highly heterogenous. The broad genetic alterations, coupled with microenvironment biochemistry, manifest in characteristic appearances both on gross histology and on the radiological scale. In addition to the work reviewed in the boarder context of IDH, MGMT and EGFR differences, other associations have also been supported. Contrast enhancement has been shown to be associated with genes implicated in hypoxia–angiogenesis pathway,102 such as vascular endothelial growth factor (VEGF). Radiomic attributes capturing abnormal intensity patterns in the internal capsule have been shown to be correlated with MYC oncogene expression.103 Interestingly, multiple radiogenomic correlation experiments have revealed strong associations of imaging phenotypes with pathways that are implicated in extracellular matrix destruction, cell invasion and metabolism.
Furthermore, radiomics offers an opportunity to perform an analysis on complete tumour that could mitigate the limitation of sampling errors and inability of complete molecular and histopathological assessment by neuropathologists given the lack of tumour sample.104–106 With quantitative mutation values rather than binary designations, radiomics can help neuro-oncologists and neurosurgeons make personalised therapy decisions and reliably predict response to therapies.
Limitations
A major feature limiting radiomic quantification is poor reproducibility secondary to variability and lack of consistency attributed to the absence of standardised acquisition parameters and radiomic approaches.107 The accuracy of radiomic signatures typically varies when tested on different datasets. Multiple studies have addressed impact of different acquisition parameters on textural analysis. Magnet strength, flip angles, different spatial/matrix size, TR/TE variations in T1WI and T2WI, and different scanner platforms can affect texture features.108–112 Molina et al.112 found that no textural measures were robust under dynamic range changes, entropy was the only robust feature under spatial resolution changes. Buch et al.109 concluded that some of the features were more robust and some of the features were more susceptible to different acquisition parameters, necessitating the need for standardised MRI techniques for textural analysis. Furthermore, variation in usability of textural analysis software add complexity to standardisation and reproducibility. Multiple studies used indigenous software with varying algorithms making reproducibility and repeatability of these studies almost impossible. Future studies are needed to assess accuracy of these results from different type of software to help with standardisation.
Scarcity of publicly available databases with annotated radiological studies for specific clinical domains limit capability of researchers to conduct large sample size studies. Small sample size and a high number of prediction variables often leads to overfitting, a major limitation in machine learning models. To prevent overfitting, it is recommended to have sample size 6–10 times larger than the analysed variables or conducting analysis with a few preselected robust variables only. Collaboration among research universities is required to create professionally annotated standardised datasets for larger cohort studies which can be split into training, testing, and validation datasets to avoid overfitting. This would also allow the researchers to test their algorithms on external cohorts and validating robustness of their solutions. A recent development towards achieving this is the use of federated learning which facilitates multi-institutional validation of machine learning models without explicit sharing of data using a distributed framework.113
Variability in the selection of appropriate regions of interest for feature extraction can affect certain radiomic attributes, such as shape-based measures. There are no existing guidelines for radiologists to report quantitative imaging features, making huge existing image repositories inaccessible for curation. For generating high-quality data with segmented and annotated appropriate regions of interest, radiologists need to be integral part of data quantification and curation.114
The lack of routinely acquired gene expression profiles and tissue sampling errors impose limitations to the application of radiogenomics in current clinical workflow.115 It is difficult for a single institute to create a large imaging database with auxiliary data such as genomic profile, demographics, treatment information and their outcomes. The Cancer Genome Atlas (TCGA) has made cancer datasets publicly available with a comprehensive catalogue of genomic profiles to address this issue. The clinical translation of radiogenomics is also hindered by spatial and temporal heterogeneity within a given brain tumour. However, ability of radiomics to perform an analysis on complete tumour might address this limitation.
Deep learning algorithms that facilitate automated feature learning have recently shown great promise in tasks ranging from tumour segmentation116 to survival prediction.117 Methods combining radiology and pathology datasets have been proposed to distinguish gliomas into oligodendroglioma and astrocytoma classes.118 Such methods, however, require large datasets for training purposes besides often lacking transparency and interpretability. Uncertainty and interpretability of deep learning networks in the field of medical imaging is an active area of research. A significant challenge for the translation of radiomics and deep learning algorithms into clinical workflow as clinical decision support systems stems from a regulatory perspective. US Food and Drug Administration (FDA) has closely regulated CAD (computer-aided detection) systems that rely on machine learning and pattern-recognition techniques; machine learning models present new regulatory challenges and require specialised guidance for submissions seeking approval (https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device). Furthermore, new AI models keep evolving even after going to market as they are exposed to more data. It is vital to adopt periodic testing requirements over specific time intervals to make sure the adaptive changes of these models follow forecasted projections.
Future directions
Recent initiatives such as the image biomarker standardisation initiative have proposed certain guidelines based on results obtained on radiomics phantoms.119 Test–retest experimental settings have also been widely proposed to facilitate selection of stable and robust radiomic measures. In one of the first studies of this kind, the repeatability of CT radiomics was ascertained in a “coffee-break” test–retest setting with scans obtained from the same scanner within an interval of 15 min.120 Similar settings are warranted for brain imaging to identify suitable radiomic features for clinical applications. The Cancer Imaging Archive hosts imaging datasets of brain tumour collections (HGGs and LGGs), among other cancers, obtained from several institutions. Such datasets have been widely used by the research community to develop and validate radiomics and radiogenomics tools.
One obvious deficiency in virtually all the retrospective radiogenomic studies is lack of information regarding the location of the biopsy sample vis-à-vis pixels in the patient’s images.121 Image-localised biopsies and subsequent imaging-pathology co-registration are essential steps in mitigating biases associated with locating biopsy region on MRI. Hu et al.122 have previously co-registered MRI scans and corresponding texture maps with biopsy locations to study regional genetic variation with spatially matched imaging descriptors. In a follow-up study, Hu et al.123 have proposed a Gaussian process and transductive learning based probabilistic model to quantify spatial uncertainty in radiogenomic pipelines. The sparse availability of ground truth labels in radiogenomic models can be modelled as a ‘weak supervision’ or ‘incomplete supervision’ task. Multi-instance learning techniques may be implemented to address this limitation.124
The field of radiomics promises to elevate the role of medical imaging by enabling objective tumour characterisation. In oncology, radiomics can provide prognostic information non-invasively via biomarker utilisation.19 In neurosurgery, it can be used for improved pre- and post-operative treatment planning. Emerging companies are now providing software that delivers web-based radiological analysis with the PACS viewing system.125 Support from academic institutions, such as the American College of Radiology, is growing. Such efforts can readily facilitate the transition of radiomic research to clinical practice.
Conclusions
Radiomics is not intended to replace radiologists in the future, but rather improve disease diagnosis and characterisation with greater precision. It is imperative for the future of neuroradiology, neurosurgery and neuro-oncology to utilise advances in radiomics and radiogenomics in order to provide less invasive and tumour-specific precision treatment strategies and to ultimately optimise patient care. In order for this field to continue evolving and make its way into clinical practice, it is vital to develop more standardised and reproducible methods of data interpretation, maintain publicly available databases of radiological studies, and conduct prospective large-scale multi-institutional clinical trials. In the future, the fields of radiomics and radiogenomics promise to improve the utility of already available imaging modalities and channel them towards personalised medicine.
Acknowledgements
The authors wish to thank Maria Figueroa, for her assistance with logistics and proofreading.
Author contributions
Conceptualisation, G.D.S., P.P. and V.S.; methodology, G.D.S., P.P. and V.S.; validation, G.D.S., S.M., P.P. and V.S.; formal analysis, G.D.S., S.M., P.P. and V.S.; investigation, G.D.S., S.M., P.P. and V.S.; resources, G.D.S., P.P. and V.S.; data curation, G.D.S., N.S., A.T., A.H.W. and P.P.; writing—original draft preparation, G.D.S., A.H.W., P.P. and V.S.; writing—review and editing, S.M., N.S., A.T., A.V., J.M. and P.P.; visualisation, G.D.S., B.S., A.T. and P.P.; supervision, S.M., A.V., J.M., P.P. and V.S.; project administration, G.D.S., S.M., P.P. and V.S. All authors have read and agreed to the published version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Data availability
Not applicable.
Competing interests
The authors declare no competing interests.
Funding information
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Prateek Prasanna, Vadim Spektor
References
- 1.Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20:iv1–iv86. doi: 10.1093/neuonc/noy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DR, Guerin JB, Giannini C, Morris JM, Eckel LJ, Kaufmann TJ. 2016 updates to the WHO brain tumor classification system: what the radiologist needs to know. Radiographics. 2017;37:2164–2180. doi: 10.1148/rg.2017170037. [DOI] [PubMed] [Google Scholar]
- 4.Gulsen S. Achieving higher diagnostic results in stereotactic brain biopsy by simple and novel technique. Open Access Maced. J. Med. Sci. 2015;3:99–104. doi: 10.3889/oamjms.2015.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutman DA, Cooper LA, Hwang SN, Holder CA, Gao J, Aurora TD, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267:560–569. doi: 10.1148/radiol.13120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez-Beteta J, Molina-García D, Martínez-González A, Henares-Molina A, Amo-Salas M, Luque B, et al. Morphological MRI-based features provide pretreatment survival prediction in glioblastoma. Eur. Radiol. 2019;29:1968–1977. doi: 10.1007/s00330-018-5758-7. [DOI] [PubMed] [Google Scholar]
- 7.Pérez-Beteta, J., Molina-García, D., Villena, M., Rodríguez, M. J., Velásquez, C., Martino, J. et al. Morphologic features on MR imaging classify multifocal glioblastomas in different prognostic groups. Am. J. Neuroradiol.10.3174/ajnr.A6019 (2019). [DOI] [PMC free article] [PubMed]
- 8.Henker C, Kriesen T, Glass Ä, Schneider B, Piek J. Volumetric quantification of glioblastoma: experiences with different measurement techniques and impact on survival. J. Neurooncol. 2017;135:391–402. doi: 10.1007/s11060-017-2587-5. [DOI] [PubMed] [Google Scholar]
- 9.Pérez-Beteta J, Molina-García D, Ortiz-Alhambra JA, Fernández-Romero A, Luque B, Arregui E, et al. Tumor surface regularity at MR imaging predicts survival and response to surgery in patients with glioblastoma. Radiology. 2018;288:218–225. doi: 10.1148/radiol.2018171051. [DOI] [PubMed] [Google Scholar]
- 10.Nicolasjilwan M, Hu Y, Yan C, Meerzaman D, Holder CA, Gutman D, et al. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. J. Neuroradiol. 2015;42:212–221. doi: 10.1016/j.neurad.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law M, Yang S, Wang H, Babb JS, Johnson G, Cha S, et al. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am. J. Neuroradiol. 2003;24:1989–1998. [PMC free article] [PubMed] [Google Scholar]
- 12.Conte GM, Castellano A, Altabella L, Iadanza A, Cadioli M, Falini A, et al. Reproducibility of dynamic contrast-enhanced MRI and dynamic susceptibility contrast MRI in the study of brain gliomas: a comparison of data obtained using different commercial software. Radiol. Med. 2017;122:294–302. doi: 10.1007/s11547-016-0720-8. [DOI] [PubMed] [Google Scholar]
- 13.Usinskiene J, Ulyte A, Bjørnerud A, Venius J, Katsaros VK, Rynkeviciene R, et al. Optimal differentiation of high- and low-grade glioma and metastasis: a meta-analysis of perfusion, diffusion, and spectroscopy metrics. Neuroradiology. 2016;58:339–350. doi: 10.1007/s00234-016-1642-9. [DOI] [PubMed] [Google Scholar]
- 14.Goh V, Schaeffter T, Leach M. Reproducibility of dynamic contrast-enhanced MR imaging: why we should care. Radiology. 2013;266:698–700. doi: 10.1148/radiol.12122447. [DOI] [PubMed] [Google Scholar]
- 15.Viallon M, Cuvinciuc V, Delattre B, Merlini L, Barnaure-Nachbar I, Toso-Patel S, et al. State-of-the-art MRI techniques in neuroradiology: principles, pitfalls, and clinical applications. Neuroradiology. 2015;57:441–467. doi: 10.1007/s00234-015-1500-1. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Liu H, Tong H, Wang S, Yang Y, Liu G, et al. Clinical applications of contrast-enhanced perfusion MRI techniques in gliomas: recent advances and current challenges. Contrast Media Mol. Imaging. 2017;2017:7064120. doi: 10.1155/2017/7064120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magn. Reson. Imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou M, Scott J, Chaudhury B, Hall L, Goldgof D, Yeom KW, et al. Radiomics in brain tumor: image assessment, quantitative feature descriptors, and machine-learning approaches. Am. J. Neuroradiol. 2018;39:208–216. doi: 10.3174/ajnr.A5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotrotsou A, Zinn PO, Colen RR. Radiomics in brain tumors: an emerging technique for characterization of tumor environment. Magn. Reson. Imaging Clin. N. Am. 2016;24:719–729. doi: 10.1016/j.mric.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Thawani R, McLane M, Beig N, Ghose S, Prasanna P, Velcheti V, et al. Radiomics and radiogenomics in lung cancer: a review for the clinician. Lung Cancer. 2018;115:34–41. doi: 10.1016/j.lungcan.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Um H, Tixier F, Bermudez D, Deasy JO, Young RJ, Veeraraghavan H. Impact of image preprocessing on the scanner dependence of multi-parametric MRI radiomic features and covariate shift in multi-institutional glioblastoma datasets. Phys. Med. Biol. 2019;64:165011. doi: 10.1088/1361-6560/ab2f44. [DOI] [PubMed] [Google Scholar]
- 24.Stringfield O, Arrington JA, Johnston SK, Rognin NG, Peeri NC, Balagurunathan Y, et al. Multiparameter MRI predictors of long-term survival in glioblastoma multiforme. Tomography. 2019;5:135–144. doi: 10.18383/j.tom.2018.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Velden BHM, van Rijssel MJ, Lena B, Philippens MEP, Loo CE, Ragusi MAA, et al. Harmonization of quantitative parenchymal enhancement in T(1)-weighted breast MRI. J. Magn. Reson. Imaging. 2020;52:1374–1382. doi: 10.1002/jmri.27244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiwari P, Prasanna P, Wolansky L, Pinho M, Cohen M, Nayate AP, et al. Computer-extracted texture features to distinguish cerebral radionecrosis from recurrent brain tumors on multiparametric MRI: a feasibility study. Am. J. Neuroradiol. 2016;37:2231–2236. doi: 10.3174/ajnr.A4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juan-Albarracín J, Fuster-Garcia E, Manjón JV, Robles M, Aparici F, Martí-Bonmatí L, et al. Automated glioblastoma segmentation based on a multiparametric structured unsupervised classification. PLoS ONE. 2015;10:e0125143. doi: 10.1371/journal.pone.0125143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prasanna P, Karnawat A, Ismail M, Madabhushi A, Tiwari P. Radiomics-based convolutional neural network for brain tumor segmentation on multiparametric magnetic resonance imaging. J. Med. Imaging. 2019;6:024005. doi: 10.1117/1.JMI.6.2.024005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereira S, et al. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging. 2016;35.5:1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 30.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts H. Machine learning methods for quantitative radiomic biomarkers. Sci. Rep. 2015;5:13087. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radovic M, Ghalwash M, Filipovic N, Obradovic Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinform. 2017;18:9. doi: 10.1186/s12859-016-1423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng H, Long Fuhui, Ding Chris. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Machine Intell. 2005;8:1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 33.Marcano-Cedeño, A., Quintanilla-Domínguez, J., Cortina-Januchs, M. G. & Andina, D. Feature selection using sequential forward selection and classification applying artificial metaplasticity neural network. in IECON 2010–36th Annual Conference on IEEE Industrial Electronics Society. 2845–2850 (IEEE, 2010).
- 34.Fox, J. & Weisberg, S. Cox Proportional-Hazards Regression for Survival Data. An R and S-PLUS Companion to Applied Regression. https://socialsciences.mcmaster.ca/jfox/Books/Companion/appendices/Appendix-Cox-Regression.pdf (2002).
- 35.Sanduleanu S, Woodruff HC, de Jong EEC, van Timmeren JE, Jochems A, Dubois L, et al. Tracking tumor biology with radiomics: a systematic review utilizing a radiomics quality score. Radiother. Oncol. 2018;127:349–360. doi: 10.1016/j.radonc.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 36.Ismail M, Hill V, Statsevych V, Huang R, Prasanna P, Correa R, et al. Shape features of the lesion habitat to differentiate brain tumor progression from pseudoprogression on routine multiparametric MRI: a multisite study. AJNR Am. J. Neuroradiol. 2018;39:2187–2193. doi: 10.3174/ajnr.A5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, W., Mao, K., Zhang, H. & Chai, T. Designing compact Gabor filter banks for efficient texture feature extraction. in 11th International Conference on Control Automation Robotics & Vision. 1193–1197. 10.1109/ICARCV.2010.5707806 (2010).
- 38.Jain AK, Farrokhnia F. Unsupervised texture segmentation using Gabor filters. Pattern Recogn. 1991;24:1167–1186. doi: 10.1016/0031-3203(91)90143-S. [DOI] [Google Scholar]
- 39.Doyle, S., Agner, S., Madabhushi, A., Feldman, M. & Tomaszewski, J. Automated grading of breast cancer histopathology using spectral clustering with textural and architectural image features. in 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 496–499 (IEEE, 2008).
- 40.Dalal, N. & Triggs, B. Histograms of oriented gradients for human detection. In 2005IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'05). Vol. 881, 886–893 (IEEE, 2005).
- 41.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans. Syst. Man, Cybernet. 1973;SMC-3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 42.Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Machine Intell. 2002;24:971–987. doi: 10.1109/TPAMI.2002.1017623. [DOI] [Google Scholar]
- 43.Li B, Meng MQH. Automatic polyp detection for wireless capsule endoscopy images. Expert Syst. Appl. 2012;39:10952–10958. doi: 10.1016/j.eswa.2012.03.029. [DOI] [Google Scholar]
- 44.Prasanna P, Tiwari P, Madabhushi A. Co-occurrence of local anisotropic gradient orientations (CoLlAGe): a new radiomics descriptor. Sci. Rep. 2016;6:37241. doi: 10.1038/srep37241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braman, N., Prasanna, P., Alilou, M., Beig, N. & Madabhushi, A. Vascular network organization via hough transform (VaNgOGH): a novel radiomic biomarker for diagnosis and treatment response. in Medical Image Computing and Computer Assisted Intervention—MICCAI 2018. (eds Alejandro, F. F. et al.) 803–811 (Springer International Publishing, 2018).
- 46.Prasanna P, Mitra J, Beig N, Nayate A, Patel J, Ghose S, et al. Mass effect deformation heterogeneity (MEDH) on gadolinium-contrast T1-weighted MRI is associated with decreased survival in patients with right cerebral hemisphere glioblastoma: a feasibility study. Sci. Rep. 2019;9:1145. doi: 10.1038/s41598-018-37615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stadlbauer A, Zimmermann M, Heinz G, Oberndorfer S, Doerfler A, Buchfelder M, et al. Magnetic resonance imaging biomarkers for clinical routine assessment of microvascular architecture in glioma. J. Cereb. Blood Flow Metab. 2017;37:632–643. doi: 10.1177/0271678X16655549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emblem KE, Mouridsen K, Bjornerud A, Farrar CT, Jennings D, Borra RJ, et al. Vessel architectural imaging identifies cancer patient responders to anti-angiogenic therapy. Nat. Med. 2013;19:1178–1183. doi: 10.1038/nm.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K, Yun SD, Triphan SMF, Sturm VJ, Buschle LR, Hahn A, et al. Vessel architecture imaging using multiband gradient-echo/spin-echo EPI. PLoS ONE. 2019;14:e0220939. doi: 10.1371/journal.pone.0220939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stadlbauer A, Zimmermann M, Kitzwögerer M, Oberndorfer S, Rössler K, Dörfler A, et al. MR imaging-derived oxygen metabolism and neovascularization characterization for grading and IDH gene mutation detection of gliomas. Radiology. 2017;283:799–809. doi: 10.1148/radiol.2016161422. [DOI] [PubMed] [Google Scholar]
- 51.Stadlbauer A, Zimmermann M, Oberndorfer S, Doerfler A, Buchfelder M, Heinz G, et al. Vascular hysteresis loops and vascular architecture mapping in patients with glioblastoma treated with antiangiogenic therapy. Sci. Rep. 2017;7:8508. doi: 10.1038/s41598-017-09048-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price SJ, Young AM, Scotton WJ, Ching J, Mohsen LA, Boonzaier NR, et al. Multimodal MRI can identify perfusion and metabolic changes in the invasive margin of glioblastomas. J. Magn. Reson. Imaging. 2016;43:487–494. doi: 10.1002/jmri.24996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boonzaier NR, Larkin TJ, Matys T, van der Hoorn A, Yan JL, Price SJ, Multiparametric MR. Imaging of diffusion and perfusion in contrast-enhancing and nonenhancing components in patients with glioblastoma. Radiology. 2017;284:180–190. doi: 10.1148/radiol.2017160150. [DOI] [PubMed] [Google Scholar]
- 54.Li, C., Wang, S., Yan, J. L., Torheim, T., Boonzaier, N. R., Sinha, R. et al. Characterizing tumor invasiveness of glioblastoma using multiparametric magnetic resonance imaging. J. Neurosurg. 10.3171/2018.12.Jns182926, 1–8 (2019). [DOI] [PubMed]
- 55.Mazurowski MA, Desjardins A, Malof JM. Imaging descriptors improve the predictive power of survival models for glioblastoma patients. Neuro Oncol. 2013;15:1389–1394. doi: 10.1093/neuonc/nos335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc. Natl Acad. Sci. USA. 2008;105:5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiki for the VASARI feature set The National Cancer Institute Web site. Available at https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project (2020).
- 58.Skogen K, Schulz A, Dormagen JB, Ganeshan B, Helseth E, Server A. Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur. J. Radiol. 2016;85:824–829. doi: 10.1016/j.ejrad.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 59.Tian Q, Yan LF, Zhang X, Zhang X, Hu YC, Han Y, et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J. Magn. Reson. Imaging. 2018;48:1518–1528. doi: 10.1002/jmri.26010. [DOI] [PubMed] [Google Scholar]
- 60.Xie T, Chen X, Fang J, Kang H, Xue W, Tong H, et al. Textural features of dynamic contrast-enhanced MRI derived model-free and model-based parameter maps in glioma grading. J. Magn. Reson. Imaging. 2018;47:1099–1111. doi: 10.1002/jmri.25835. [DOI] [PubMed] [Google Scholar]
- 61.Suh HB, Choi YS, Bae S, Ahn SS, Chang JH, Kang S-G, et al. Primary central nervous system lymphoma and atypical glioblastoma: differentiation using radiomics approach. Eur. Radiol. 2018;28:3832–3839. doi: 10.1007/s00330-018-5368-4. [DOI] [PubMed] [Google Scholar]
- 62.Soni N, Priya S, Bathla G. Texture analysis in cerebral gliomas: a review of the literature. AJNR Am. J. Neuroradiol. 2019;40:928–934. doi: 10.3174/ajnr.A6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, X., Tian, Q., Wu, Y.-X., Xu, X.-P., Li, B.-J., Liu, Y.-X. et al. IDH Mutation Assessment of Glioma Using Texture Features of Multimodal MR Images. Vol. 10134 MI (SPIE, 2017).
- 64.Han L, Wang S, Miao Y, Shen H, Guo Y, Xie L, et al. MRI texture analysis based on 3D tumor measurement reflects the IDH1 mutations in gliomas—a preliminary study. Eur. J. Radiol. 2019;112:169–179. doi: 10.1016/j.ejrad.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 65.Jakola AS, Zhang YH, Skjulsvik AJ, Solheim O, Bo HK, Berntsen EM, et al. Quantitative texture analysis in the prediction of IDH status in low-grade gliomas. Clin. Neurol. Neurosurg. 2018;164:114–120. doi: 10.1016/j.clineuro.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 66.Bahrami N, Hartman SJ, Chang YH, Delfanti R, White NS, Karunamuni R, et al. Molecular classification of patients with grade II/III glioma using quantitative MRI characteristics. J. Neurooncol. 2018;139:633–642. doi: 10.1007/s11060-018-2908-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leather, T., Jenkinson, M. D., Das, K. & Poptani, H. Magnetic resonance spectroscopy for detection of 2-hydroxyglutarate as a biomarker for IDH mutation in gliomas. Metabolites7, 29 (2017). [DOI] [PMC free article] [PubMed]
- 68.Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci. Transl. Med. 2012;4:116ra114. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat. Med. 2012;18:624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verma G, Mohan S, Nasrallah MP, Brem S, Lee JY, Chawla S, et al. Non-invasive detection of 2-hydroxyglutarate in IDH-mutated gliomas using two-dimensional localized correlation spectroscopy (2D L-COSY) at 7 Tesla. J. Transl. Med. 2016;14:274. doi: 10.1186/s12967-016-1035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu LS, Eschbacher JM, Heiserman JE, Dueck AC, Shapiro WR, Liu S, et al. Reevaluating the imaging definition of tumor progression: perfusion MRI quantifies recurrent glioblastoma tumor fraction, pseudoprogression, and radiation necrosis to predict survival. Neuro Oncol. 2012;14:919–930. doi: 10.1093/neuonc/nos112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abbasi AW, Westerlaan HE, Holtman GA, Aden KM, van Laar PJ, van der Hoorn A. Incidence of tumour progression and pseudoprogression in high-grade gliomas: a systematic review and meta-analysis. Clin. Neuroradiol. 2018;28:401–411. doi: 10.1007/s00062-017-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larroza A, Moratal D, Paredes-Sanchez A, Soria-Olivas E, Chust ML, Arribas LA, et al. Support vector machine classification of brain metastasis and radiation necrosis based on texture analysis in MRI. J. Magn. Reson. Imaging. 2015;42:1362–1368. doi: 10.1002/jmri.24913. [DOI] [PubMed] [Google Scholar]
- 75.Raimbault A, Cazals X, Lauvin MA, Destrieux C, Chapet S, Cottier JP. Radionecrosis of malignant glioma and cerebral metastasis: a diagnostic challenge in MRI. Diagn. Interv. Imaging. 2014;95:985–1000. doi: 10.1016/j.diii.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 76.Thust SC, van den Bent MJ, Smits M. Pseudoprogression of brain tumors. J. Magn. Resonance Imaging. 2018;48:571–589. doi: 10.1002/jmri.26171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Catalaa I, Henry R, Dillon WP, Graves EE, McKnight TR, Lu Y, et al. Perfusion, diffusion and spectroscopy values in newly diagnosed cerebral gliomas. NMR Biomed. 2006;19:463–475. doi: 10.1002/nbm.1059. [DOI] [PubMed] [Google Scholar]
- 78.Pope WB, Kim HJ, Huo J, Alger J, Brown MS, Gjertson D, et al. Recurrent glioblastoma multiforme: ADC histogram analysis predicts response to bevacizumab treatment. Radiology. 2009;252:182–189. doi: 10.1148/radiol.2521081534. [DOI] [PubMed] [Google Scholar]
- 79.van Dijken BRJ, van Laar PJ, Smits M, Dankbaar JW, Enting RH, van der Hoorn A, Perfusion MRI. in treatment evaluation of glioblastomas: clinical relevance of current and future techniques. J. Magn. Reson. Imaging. 2019;49:11–22. doi: 10.1002/jmri.26306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu X, Wong KK, Young GS, Guo L, Wong ST. Support vector machine multiparametric MRI identification of pseudoprogression from tumor recurrence in patients with resected glioblastoma. J. Magn. Reson. Imaging. 2011;33:296–305. doi: 10.1002/jmri.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prasanna P, Rogers L, Lam TC, Cohen M, Siddalingappa A, Wolansky L, et al. Disorder in pixel-level edge directions on T1WI is associated with the degree of radiation necrosis in primary and metastatic brain tumors: preliminary findings. AJNR. Am. J. Neuroradiol. 2019;40:412–417. doi: 10.3174/ajnr.A5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prasanna P, Tiwari P, Madabhushi A. Co-occurrence of local anisotropic gradient orientations (CoLIAGe): distinguishing tumor confounders and molecular subtypes on MRI. Med. Image Comput. Comput. Assist. Interv. 2014;17:73–80. doi: 10.1007/978-3-319-10443-0_10. [DOI] [PubMed] [Google Scholar]
- 83.Lohmann P, Kocher M, Ceccon G, Bauer EK, Stoffels G, Viswanathan S, et al. Combined FET PET/MRI radiomics differentiates radiation injury from recurrent brain metastasis. NeuroImage. Clin. 2018;20:537–542. doi: 10.1016/j.nicl.2018.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mallick S, Benson R, Hakim A, Rath GK. Management of glioblastoma after recurrence: a changing paradigm. J. Egypt Natl. Canc. Inst. 2016;28:199–210. doi: 10.1016/j.jnci.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 85.Monteiro AR, Hill R, Pilkington GJ, Madureira PA. The role of hypoxia in glioblastoma invasion. Cells. 2017;6:45. doi: 10.3390/cells6040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rapisarda A, Melillo G. Overcoming disappointing results with antiangiogenic therapy by targeting hypoxia. Nat. Rev. Clin. Oncol. 2012;9:378. doi: 10.1038/nrclinonc.2012.64. [DOI] [PubMed] [Google Scholar]
- 87.Beig N, Patel J, Prasanna P, Hill V, Gupta A, Correa R, et al. Radiogenomic analysis of hypoxia pathway is predictive of overall survival in Glioblastoma. Sci. Rep. 2018;8:7. doi: 10.1038/s41598-017-18310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kickingereder P, Burth S, Wick A, Gotz M, Eidel O, Schlemmer HP, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280:880–889. doi: 10.1148/radiol.2016160845. [DOI] [PubMed] [Google Scholar]
- 89.Jain R, Poisson LM, Gutman D, Scarpace L, Hwang SN, Holder CA, et al. Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology. 2014;272:484–493. doi: 10.1148/radiol.14131691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McGarry SD, Hurrell SL, Kaczmarowski AL, Cochran EJ, Connelly J, Rand SD, et al. Magnetic resonance imaging-based radiomic profiles predict patient prognosis in newly diagnosed glioblastoma before therapy. Tomography. 2016;2:223–228. doi: 10.18383/j.tom.2016.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu X, Li Y, Qian Z, Sun Z, Xu K, Wang K, et al. A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. NeuroImage: Clin. 2018;20:1070–1077. doi: 10.1016/j.nicl.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou H, Vallieres M, Bai HX, Su C, Tang H, Oldridge D, et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol. 2017;19:862–870. doi: 10.1093/neuonc/now256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Y, Qian Z, Xu K, Wang K, Fan X, Li S, et al. Radiomic features predict Ki-67 expression level and survival in lower grade gliomas. J. Neurooncol. 2017;135:317–324. doi: 10.1007/s11060-017-2576-8. [DOI] [PubMed] [Google Scholar]
- 94.Kickingereder, P., Götz, M., Muschelli, J., Wick, A., Neuberger, U., Shinohara, R. T. et al. Large-scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for stratifying anti-angiogenic treatment response. Clin. Cancer Res. 10.1158/1078-0432.ccr-16-0702 (2016). [DOI] [PMC free article] [PubMed]
- 95.Bahrami N, Piccioni D, Karunamuni R, Chang YH, White N, Delfanti R, et al. Edge contrast of the FLAIR hyperintense region predicts survival in patients with high-grade gliomas following treatment with bevacizumab. AJNR Am. J. Neuroradiol. 2018;39:1017–1024. doi: 10.3174/ajnr.A5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zacharaki EI, Wang S, Chawla S, Soo Yoo D, Wolf R, Melhem ER, et al. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. 2009;62:1609–1618. doi: 10.1002/mrm.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Akay A, Rüksen M, Islekel S. Magnetic resonance imaging-guided stereotactic biopsy: a review of 83 cases with outcomes. Asian. J. Neurosurg. 2019;14:90–95. doi: 10.4103/ajns.AJNS_81_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thakkar JP, Dolecek TA, Horbinski C, Ostrom QT, Lightner DD, Barnholtz-Sloan JS, et al. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol. Biomarkers Prev. 2014;23:1985–1996. doi: 10.1158/1055-9965.EPI-14-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fathi Kazerooni, A., Bakas, S., Saligheh Rad, H. & Davatzikos, C. Imaging signatures of glioblastoma molecular characteristics: a radiogenomics review. J. Magn. Reson. Imaging10.1002/jmri.26907 (2019). [DOI] [PMC free article] [PubMed]
- 100.Grossmann P, Narayan V, Chang K, Rahman R, Abrey L, Reardon DA, et al. Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro Oncol. 2017;19:1688–1697. doi: 10.1093/neuonc/nox092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arevalo OD, Soto C, Rabiei P, Kamali A, Ballester LY, Esquenazi Y, et al. Assessment of glioblastoma response in the era of bevacizumab: longstanding and emergent challenges in the imaging evaluation of pseudoresponse. Front. Neurol. 2019;10:460–460. doi: 10.3389/fneur.2019.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fischer I, Gagner JP, Law M, Newcomb EW, Zagzag D. Angiogenesis in gliomas: biology and molecular pathophysiology. Brain Pathol. 2005;15:297–310. doi: 10.1111/j.1750-3639.2005.tb00115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Colen RR, Vangel M, Wang J, Gutman DA, Hwang SN, Wintermark M, et al. Imaging genomic mapping of an invasive MRI phenotype predicts patient outcome and metabolic dysfunction: a TCGA glioma phenotype research group project. BMC Med. Genomics. 2014;7:30. doi: 10.1186/1755-8794-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chaddad A, Kucharczyk MJ, Daniel P, Sabri S, Jean-Claude BJ, Niazi T, et al. Radiomics in glioblastoma: current status and challenges facing clinical implementation. Front. Oncol. 2019;9:374–374. doi: 10.3389/fonc.2019.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kros JM, van Run PR, Alers JC, Avezaat CJ, Luider TM, van Dekken H. Spatial variability of genomic aberrations in a large glioblastoma resection specimen. Acta Neuropathol. 2001;102:103–109. doi: 10.1007/s004010000327. [DOI] [PubMed] [Google Scholar]
- 106.Woodworth GF, McGirt MJ, Samdani A, Garonzik I, Olivi A, Weingart JD. Frameless image-guided stereotactic brain biopsy procedure: diagnostic yield, surgical morbidity, and comparison with the frame-based technique. J. Neurosurg. 2006;104:233–237. doi: 10.3171/jns.2006.104.2.233. [DOI] [PubMed] [Google Scholar]
- 107.Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. Radiographics. 2017;37:505–515. doi: 10.1148/rg.2017160130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ford J, Dogan N, Young L, Yang F. Quantitative radiomics: impact of pulse sequence parameter selection on MRI-based textural features of the brain. Contrast Media Mol. Imaging. 2018;2018:1729071. doi: 10.1155/2018/1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buch K, Kuno H, Qureshi MM, Li B, Sakai O. Quantitative variations in texture analysis features dependent on MRI scanning parameters: a phantom model. J. Appl. Clin. Med. Phys. 2018;19:253–264. doi: 10.1002/acm2.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Waugh SA, Lerski RA, Bidaut L, Thompson AM. The influence of field strength and different clinical breast MRI protocols on the outcome of texture analysis using foam phantoms. Med. Phys. 2011;38:5058–5066. doi: 10.1118/1.3622605. [DOI] [PubMed] [Google Scholar]
- 111.Mayerhoefer ME, Szomolanyi P, Jirak D, Materka A, Trattnig S. Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: an application-oriented study. Med. Phys. 2009;36:1236–1243. doi: 10.1118/1.3081408. [DOI] [PubMed] [Google Scholar]
- 112.Molina D, Pérez-Beteta J, Martínez-González A, Martino J, Velasquez C, Arana E, et al. Lack of robustness of textural measures obtained from 3D brain tumor MRIs impose a need for standardization. PLoS ONE. 2017;12:e0178843. doi: 10.1371/journal.pone.0178843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sheller MJ, Reina GA, Edwards B, Martin J, Bakas S. Multi-institutional deep learning modeling without sharing patient data: a feasibility study on brain tumor segmentation. Brainlesion. 2019;11383:92–104. doi: 10.1007/978-3-030-11723-8_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jang K, Russo C, Di Ieva A. Radiomics in gliomas: clinical implications of computational modeling and fractal-based analysis. Neuroradiology. 2020;62:771–790. doi: 10.1007/s00234-020-02403-1. [DOI] [PubMed] [Google Scholar]
- 115.Gore, S., Chougule, T., Jagtap, J., Saini, J. & Ingalhalikar, M. A review of radiomics and deep predictive modeling in glioma characterization. Acad. Radiol. 10.1016/j.acra.2020.06.016 (2020). [DOI] [PubMed]
- 116.Bakas, S., Reyes, M., Jakab, A., Bauer, S., Rempfler, M., Crimi, A. et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. Preprint at arXivhttps://ui.adsabs.harvard.edu/abs/2018arXiv181102629B (2018).
- 117.Lao J, Chen Y, Li ZC, Li Q, Zhang J, Liu J, et al. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci. Rep. 2017;7:10353. doi: 10.1038/s41598-017-10649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kurc T, Bakas S, Ren X, Bagari A, Momeni A, Huang Y, et al. Segmentation and classification in digital pathology for glioma research: challenges and deep learning approaches. Front. Neurosci. 2020;14:27. doi: 10.3389/fnins.2020.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zwanenburg A, Vallieres M, Abdalah MA, Aerts H, Andrearczyk V, Apte A, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Balagurunathan Y, Kumar V, Gu Y, Kim J, Wang H, Liu Y, et al. Test-retest reproducibility analysis of lung CT image features. J. Digit. Imaging. 2014;27:805–823. doi: 10.1007/s10278-014-9716-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lo Gullo R, Daimiel I, Morris EA, Pinker K. Combining molecular and imaging metrics in cancer: radiogenomics. Insights Imaging. 2020;11:1. doi: 10.1186/s13244-019-0795-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hu LS, Ning S, Eschbacher JM, Baxter LC, Gaw N, Ranjbar S, et al. Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro Oncol. 2017;19:128–137. doi: 10.1093/neuonc/now135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hu, L., Wang, L., Hawkins-Daarud, A., Eschbacher, J. M., Singleton, K. W., Jackson, P. R. et al. Uncertainty quantification in the radiogenomics modeling of EGFR amplification in glioblastoma. Sci Rep 11, 3932. 10.1038/s41598-021-83141-z (2021). [DOI] [PMC free article] [PubMed]
- 124.Zhou, Z.-H. & Zhang, M.-L. Neural networks for multi-instance learning. in Proceedings of the International Conference on Intelligent Information Technology, Beijing, China, 455–459 (Association for Computing Machinery (ACM) New York, NY, United States, 2002). http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.87.1254&rep=rep1&type=pdf.
- 125.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat. Rev. Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qi XX, Shi DF, Ren SX, Zhang SY, Li L, Li QC, et al. Histogram analysis of diffusion kurtosis imaging derived maps may distinguish between low and high grade gliomas before surgery. Eur. Radiol. 2018;28:1748–1755. doi: 10.1007/s00330-017-5108-1. [DOI] [PubMed] [Google Scholar]
- 127.Beig N, Correa R, Thawani R, Prasanna P, Badve C, Gold D, et al. MEDU-48. MRI textural features can differentiate pediatric posterior possa tumors. Neuro Oncol. 2017;19:iv47–iv47. doi: 10.1093/neuonc/nox083.197. [DOI] [Google Scholar]
- 128.Molina D, Pérez-Beteta J, Luque B, Arregui E, Calvo M, Borrás JM, et al. Tumour heterogeneity in glioblastoma assessed by MRI texture analysis: a potential marker of survival. Br. J. Radiol. 2016;89:20160242. doi: 10.1259/bjr.20160242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chaddad A, Daniel P, Desrosiers C, Toews M, Abdulkarim B. Novel radiomic features based on joint intensity matrices for predicting glioblastoma patient survival time. IEEE J. Biomed. Health Inform. 2019;23:795–804. doi: 10.1109/JBHI.2018.2825027. [DOI] [PubMed] [Google Scholar]
- 130.Liu Y, Zhang X, Feng N, Yin L, He Y, Xu X, et al. The effect of glioblastoma heterogeneity on survival stratification: a multimodal MR imaging texture analysis. Acta Radiol. 2018;59:1239–1246. doi: 10.1177/0284185118756951. [DOI] [PubMed] [Google Scholar]
- 131.Yang D, Rao G, Martinez J, Veeraraghavan A, Rao A. Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma. Med. Phys. 2015;42:6725–6735. doi: 10.1118/1.4934373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li Q, Bai H, Chen Y, Sun Q, Liu L, Zhou S, et al. A fully-automatic multiparametric radiomics model: towards reproducible and prognostic imaging signature for prediction of overall survival in glioblastoma multiforme. Sci. Rep. 2017;7:14331. doi: 10.1038/s41598-017-14753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sun L, Zhang S, Chen H, Luo L. Brain tumor segmentation and survival prediction using multimodal MRI scans with deep learning. Front. Neurosci. 2019;13:810. doi: 10.3389/fnins.2019.00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sanghani P, Ang BT, King NKK, Ren H. Overall survival prediction in glioblastoma multiforme patients from volumetric, shape and texture features using machine learning. Surg. Oncol. 2018;27:709–714. doi: 10.1016/j.suronc.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 135.Beig, N., Singh, S., Bera, K., Prasanna, P., Singh, G., Chen, J. et al. Sexually dimorphic radiogenomic models identify distinct imaging and biological pathways that are prognostic of overall survival in Glioblastoma. Neuro Oncol. 10.1093/neuonc/noaa231 (2020). [DOI] [PMC free article] [PubMed]
- 136.Han W, Qin L, Bay C, Chen X, Yu KH, Miskin N, et al. Deep transfer learning and radiomics feature prediction of survival of patients with high-grade gliomas. AJNR Am. J. Neuroradiol. 2020;41:40–48. doi: 10.3174/ajnr.A6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Beig, N., Correa, R., Prasanna, P., Mitra, J., Nayate, A., Madabhushi, A. & Tiwari, P. Radiogenomic analysis of distinct tumor sub-compartments on T2 and FLAIR predict distinct molecular subtypes in lower grade gliomas. Int. Soc. Magn. Reson. Med. ISMRM 25th Annual Meeting Session, 4232–4255 (2017).
- 138.Hsieh KL, Chen CY, Lo CM. Radiomic model for predicting mutations in the isocitrate dehydrogenase gene in glioblastomas. Oncotarget. 2017;8:45888–45897. doi: 10.18632/oncotarget.17585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shofty B, Artzi M, Ben Bashat D, Liberman G, Haim O, Kashanian A, et al. MRI radiomics analysis of molecular alterations in low-grade gliomas. Int. J. Comput. Assist. Radiol. Surg. 2018;13:563–571. doi: 10.1007/s11548-017-1691-5. [DOI] [PubMed] [Google Scholar]
- 140.Wu S, Meng J, Yu Q, Li P, Fu S. Radiomics-based machine learning methods for isocitrate dehydrogenase genotype prediction of diffuse gliomas. J. Cancer Res. Clin. Oncol. 2019;145:543–550. doi: 10.1007/s00432-018-2787-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhou H, Chang K, Bai HX, Xiao B, Su C, Bi WL, et al. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J. Neurooncol. 2019;142:299–307. doi: 10.1007/s11060-019-03096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee MH, Kim J, Kim ST, Shin HM, You HJ, Choi JW, et al. Prediction of IDH1 mutation status in glioblastoma using machine learning technique based on quantitative radiomic data. World Neurosurg. 2019;125:e688–e696. doi: 10.1016/j.wneu.2019.01.157. [DOI] [PubMed] [Google Scholar]
- 143.Sudre CH, Panovska-Griffiths J, Sanverdi E, Brandner S, Katsaros VK, Stranjalis G, et al. Machine learning assisted DSC-MRI radiomics as a tool for glioma classification by grade and mutation status. BMC Med. Inform. Decis. Mak. 2020;20:149. doi: 10.1186/s12911-020-01163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.