ABSTRACT
Facioscapulohumeral muscular dystrophy (FSHD) is caused by misexpression of DUX4 in skeletal myocytes. As DUX4 is the key therapeutic target in FSHD, surrogate biomarkers of DUX4 expression in skeletal muscle are critically needed for clinical trials. Although no natural animal models of FSHD exist, transgenic mice with inducible DUX4 expression in skeletal muscles rapidly develop myopathic phenotypes consistent with FSHD. Here, we established a new, more-accurate FSHD-like mouse model based on chronic DUX4 expression in a small fraction of skeletal myonuclei that develops pathology mimicking key aspects of FSHD across its lifespan. Utilizing this new aged mouse model and DUX4-inducible mouse models, we characterized the DUX4-related microRNA signatures in skeletal muscles, which represent potential biomarkers for FSHD. We found increased expression of miR-31-5p and miR-206 in muscles expressing different levels of DUX4 and displaying varying degrees of pathology. Importantly, miR-206 expression is significantly increased in serum samples from FSHD patients compared with healthy controls. Our data support miR-31-5p and miR-206 as new potential regulators of muscle pathology and miR-206 as a potential circulating biomarker for FSHD.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: DUX4, FSHD, Biomarker, miR-206, miRNA
Summary: Candidate miRNA biomarkers for facioscapulohumeral muscular dystrophy (FSHD) were identified using FSHD-like mouse models that present cumulative pathology from chronic expression of DUX4 in skeletal muscles and confirmed in FSHD patient serum.
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD) is a progressive myopathy that affects ∼1 in 8300-15,000 people worldwide (Deenen et al., 2014; Orphanet, 2021; Padberg et al., 1995). FSHD affects women and men of all ages; however, it is characterized by high variability in clinical presentation with disease onset, progression and severity differing greatly between individuals and particularly within families (Jones et al., 2012; Mul et al., 2018b; Statland et al., 2015a; Tawil et al., 1996; Tonini et al., 2004; Wang and Tawil, 2016). FSHD-specific clinical trials have recently begun, but there is still no treatment or cure. Importantly, these clinical trials have brought to the forefront that one of the greatest unmet needs in the search for an FSHD therapeutic is a validated circulating surrogate biomarker for pathogenic gene expression or disease progression that can be used to readily monitor clinical efficacy of a treatment.
The root cause of FSHD is misexpression of the double homeobox 4 (DUX4) retrogene in skeletal muscle (Gabriëls et al., 1999; Kowaljow et al., 2007; Lemmers et al., 2010; Snider et al., 2010; Tassin et al., 2013). Residing within the chromosome 4q35 D4Z4 macrosatellite repeat array, DUX4 encodes a critical transcription factor that drives the early stages of normal embryonic development (De Iaco et al., 2017; Gabriëls et al., 1999; Hendrickson et al., 2017; Kowaljow et al., 2007; Rickard et al., 2015; Snider et al., 2010). In contrast to its embryonic role, DUX4 is under strong epigenetic silencing in most adult somatic cells and paradoxically pathogenic when expressed in these cells (Gabriëls et al., 1999; Snider et al., 2010). In all forms of FSHD, expression of the pathogenic DUX4 mRNA full-length isoform (DUX4-fl) is induced upon epigenetic de-repression of the 4q35 D4Z4 locus brought about by genetic mutations (de Greef et al., 2009; Jones et al., 2015; Snider et al., 2010; van Overveld et al., 2003). This epigenetic dysregulation can be induced either by contraction of the D4Z4 array (FSHD1) (van Deutekom et al., 1993; van Overveld et al., 2003; Wijmenga et al., 1992) or by mutations in genes encoding epigenetic repressors of the D4Z4 array (FSHD2) (Hamanaka et al., 2020; Lemmers et al., 2012; van den Boogaard et al., 2016). Thus, the aberrant expression of DUX4 is the prime target for FSHD therapeutic development.
A major obstacle facing FSHD clinical trials targeting DUX4 expression is the lack of any independently validated circulating molecules that can reproducibly be used as predictive, prognostic and/or pharmacodynamic biomarkers (Banerji, 2020; Harafuji et al., 2013; Heier et al., 2020; Matsuzaka et al., 2014; Petek et al., 2016; Signoelli et al., 2020; Statland et al., 2014). Circulating biomarkers are ideal candidates as these are non-invasive, typically inexpensive to assess and allow multiple points of analysis over time, compared with biomarkers that must be detected using muscle biopsies or magnetic resonance imaging (MRI). Three previous studies identified three distinctly different candidate protein biomarkers in serum: creatine kinase MB isoform (CKMB), epidermal growth factor and the inflammatory regulatory factor S100A8 (Heier et al., 2020; Petek et al., 2016; Statland et al., 2014). As an alternative to circulating protein biomarkers, a global gene expression analysis was performed using blood from two well-controlled patient cohorts, but failed to find gene expression profiles differentially expressed in FSHD patients (Signoelli et al., 2020). Additional non-invasive approaches such as MRI and ultrasound techniques have been developed as imaging biomarkers of disease progression, and display the sensitivity to define disease severity and motor function (Mul et al., 2018a, 2017; Wang et al., 2019, 2021b). Importantly MRI measurements have shown a correlation with both pathological changes and expression of DUX4 target genes (Wang et al., 2019), but not with immunosuppressive treatments for FSHD (Wang et al., 2021a). However, using MRI to track drug efficacy in a large clinical trial could be severely limiting due to the cost, limited accessibility and inconsistency of equipment across trial sites. Thus, a DUX4-responsive circulating biomarker is still needed in FSHD clinical trials for drugs targeting DUX4 expression.
Recently, microRNAs (miRNAs) have gained considerable attention as potential circulating disease biomarkers (Dai et al., 2019; Huang, 2017; Leidinger et al., 2013; Zhao et al., 2019). miRNAs are a class of highly conserved small non-coding RNAs that function in the post-transcriptional regulation of gene expression and are important for the healthy development of many organs, including skeletal muscle (Dai et al., 2019; Huang, 2017; Leidinger et al., 2013; Zhao et al., 2019). They function to tune gene expression by binding to target mRNAs and either inducing their degradation or inhibiting their translation (Galagali and Kim, 2020). Importantly, miRNAs can be transported extracellularly and are found in serum, where their levels may indicate specific disease states (Dai et al., 2019; Huang, 2017; Leidinger et al., 2013; Zhao et al., 2019). For example, microRNA-206 (miR-206), one of the original myomiRs, or muscle-specific miRNAs, is a key regulator of myogenic differentiation during muscle repair (Cacchiarelli et al., 2011b; Chen et al., 2010; Greco et al., 2009; Horak et al., 2016; Kim et al., 2006; Liu et al., 2012; Ma et al., 2015; Yuasa et al., 2008). Interestingly, miR-206 levels are elevated in muscle biopsies from the mdx mouse model of Duchenne muscular dystrophy (DMD) and in serum and muscle biopsies isolated from DMD patients (Cacchiarelli et al., 2011b; Mizuno et al., 2011; Roberts et al., 2012, 2013). Other miRNAs have been shown to play major roles in skeletal myogenesis and regeneration, further supporting the use of miRNAs as biomarkers for muscle diseases (Buckingham and Rigby, 2014; Liu and Olson, 2010; McCarthy, 2008; Sempere et al., 2004). For example, a large miRNA profile performed using patient muscle biopsies demonstrated that miR-146b, miR-221, miR-155, miR-214 and miR-222 are consistently dysregulated in many muscular dystrophies (Eisenberg et al., 2007). With respect to miRNAs in FSHD, characterization of quadriceps muscle biopsies taken from fetal tissue revealed that miR-34a, miR-330, miR-331-5p, miR-380-3p, miR-516b, miR-517, miR-582-5p and miR-625 are exclusively expressed in FSHD1 compared with control fetal tissue (Portilho et al., 2015). In addition, a recent miRNA screen of FSHD patient serum using a panel of 384 miRNAs identified eight as being increased in FSHD patients compared with healthy controls, thus warranting further investigation (Heier et al., 2020). Overall, despite recent advances, no circulating disease biomarkers, including miRNAs, have been validated as being DUX4 responsive for use in FSHD clinical trials.
To address this need, we used our recently developed tamoxifen (TMX)-inducible DUX4 bi-transgenic mouse line, ACTA1-MCM;FLExD, that recapitulates many aspects of FSHD pathophysiology of varying severity in skeletal muscles (Jones et al., 2020). Interestingly, in the absence of induction, these mice show chronic mosaic DUX4 expression in all skeletal muscles over their lifetime, developing FSHD-like pathophysiology that accumulates with age, thereby representing a new FSHD-like model of chronic DUX4 expression. Here, we used this model as a discovery tool for identifying the DUX4-reponsive miRNA signatures associated with short-term and chronic exposure to DUX4 in skeletal muscles and demonstrated that miR-31-5p and miR-206 levels are consistently dysregulated. Importantly, levels of miR-206 are also increased in serum isolated from FSHD patients, thus validating miR-206 as a potential circulating DUX4-responsive disease biomarker for FSHD.
RESULTS
Immune/regeneration-related muscle miRNA profile in DUX4-induced FSHD-like mouse model
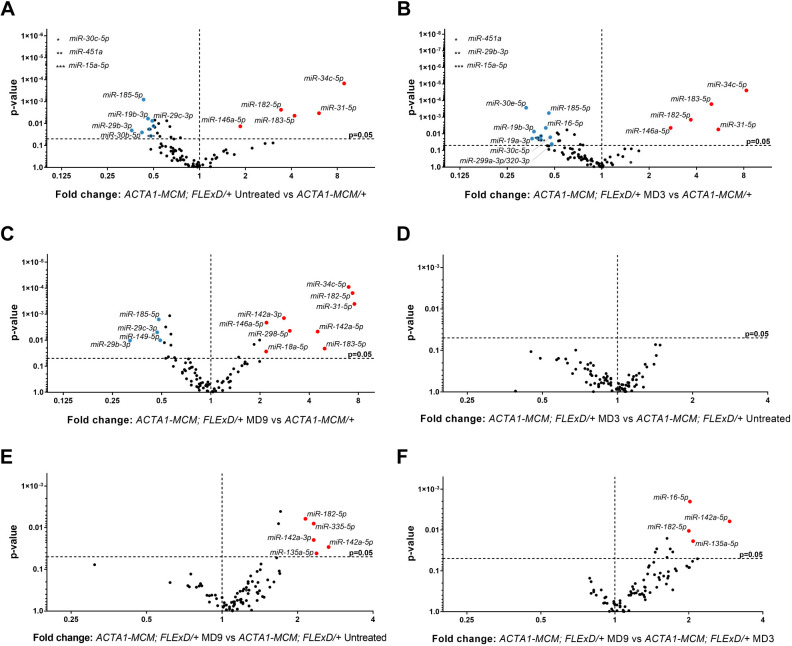
We used FSHD-like mouse models as a primary discovery tool to identify miRNAs altered by variable DUX4 expression in skeletal muscles for evaluation as FSHD biomarkers in serum analysis. We first used our laboratory's moderate FSHD-like mouse model (Jones et al., 2020) to evaluate the effect of DUX4 levels on the muscle miRNA signature. As previously published, DUX4 expression is very low in muscles of untreated 3-month-old ACTA1-MCM;FLExD/+ mice [no TMX in Jones et al. (2020)]. Following TMX induction (single dose, 5 mg/kg), increased DUX4 protein is first detectable by day 3 [moderate day (MD)3], with levels increasing until day 14 (MD14). The increase in DUX4 is accompanied by increased immune cell infiltration, damaged/split myofibers showing variable diameter, centrally localized nuclei and embryonic myosin expression in murine skeletal muscles, all consistent with an FSHD-like phenotype (Jones et al., 2020). Thus, based on this shared histopathology between our FSHD-like mouse and FSHD patients, we chose an immune-related miRNA panel and screened the gastrocnemius muscles from these mice over a time course of DUX4 induction (Fig. 1; Fig. S1, Table S1). Comparing the miRNA profiles from the gastrocnemius muscles, we found that miR-31-5p, miR-34c-5p, miR-146a-5p, miR-182-5p and miR-183-5p were significantly upregulated by more than 2-fold in the muscles from untreated low-DUX4 mice compared with those of age-matched DUX4-negative control ACTA1-MCM/+ mice (Fig. 1A). When moderate expression of DUX4 was induced by TMX injection, these miRNAs remained upregulated in MD3 and MD9 muscles compared with control muscles (Fig. 1B,C). In addition, the levels of miR-18a-5p, miR-142a-3p, miR-142-5p and miR-298-5p were also upregulated in MD9 muscles (Fig. 1C). To see whether the miRNA profile is sensitive to the progressive increase in DUX4 levels after DUX4 induction, we compared the miRNA profile of ACTA1-MCM;FLExD/+ MD3 and MD9 muscles with the profile of low DUX4-expressing untreated ACTA1-MCM;FLExD/+ muscles (Fig. 1D-F). We found no significant miRNA dysregulation at MD3 compared with untreated ACTA1-MCM;FLExD/+ mice, which suggests that subtle changes in DUX4 expression do not immediately affect certain miRNAs (Fig. 1D). However, as expected, muscles from the ACTA1-MCM;FLExD/+ MD9 mice expressed higher levels of immune-related miR-142a-5p and miR-182-5p, as well as miR-135a-5, which are associated with muscle regeneration (Fig. 1E,F), consistent with the DUX4-related histopathology (Jones et al., 2020). This indicates that their increases in expression similarly coincide with the increased levels of DUX4 expression, inflammation, regeneration and overall muscle pathology (Fig. 1E,F) (Jones et al., 2020).
Fig. 1.
The miRNA profile in gastrocnemius muscle from DUX4-induced FSHD-like mouse models. (A-C) Volcano plots depict the miRNA profile in gastrocnemius muscle from FSHD-like models compared with ACTA1-MCM/+ controls for 3-month-old untreated (no TMX) ACTA1-MCM;FLExD/+ mice (A), moderate FSHD-like model mice, day 3 (MD3) (B) and moderate FSHD-like model mice, day 9 (MD9) (C). (D,E) Volcano plots for the miRNA profiles for MD3 (D) and MD9 (E) compared with ACTA1-MCM FLExD/+ untreated mice. (F) The miRNA profile for MD9 compared with MD3 gastrocnemius muscle. Red and blue circles depict miRNAs for which levels are significantly upregulated or downregulated by ≥2-fold, respectively. All studies were performed with n=8 animals for each experimental group. Volcano plots represent two-tailed unpaired Student's t-test analysis.
The miRNA profile analysis of the moderate FSHD-like mouse model revealed significantly increased expression of multiple miRNAs after DUX4 induction (Fig. 1 and Fig. 2A). Although several of the dysregulated miRNAs did not appear to be sensitive to DUX4 levels, there was a general trend toward increasing levels for seven of the 12 dysregulated miRNAs (e.g. miR-142a-3p and miR-182-5p), with increasing levels of DUX4 in the muscles (Fig. 2A). The most striking upregulation was found with the immune- and regeneration-related miR-31-5p, the levels of which were increased 6-fold in untreated muscles and 8-fold in MD9 muscles (Fig. 2A). By using more-sensitive TaqMan probe quantification, we confirmed the significant upregulation of miR-31-5p levels by 15-fold in untreated and MD9 muscles compared with ACTA1-MCM/+ (DUX4-negative) controls, validating miR-31-5p as a potential early biomarker for DUX4 expression levels (Fig. 2B). However, miR-31-5p levels continued to increase by 35-fold in the MD28 ACTA1-MCM;FLExD/+ muscles, which have very low DUX4 expression (Jones et al., 2020). This suggests that although increased levels of miR-31-5p are DUX4 dependent, expression of this miRNA is not DUX4 responsive (i.e. does not decrease with decreasing DUX4 levels) and thus may reflect the activation of DUX4-induced pathology.
Fig. 2.
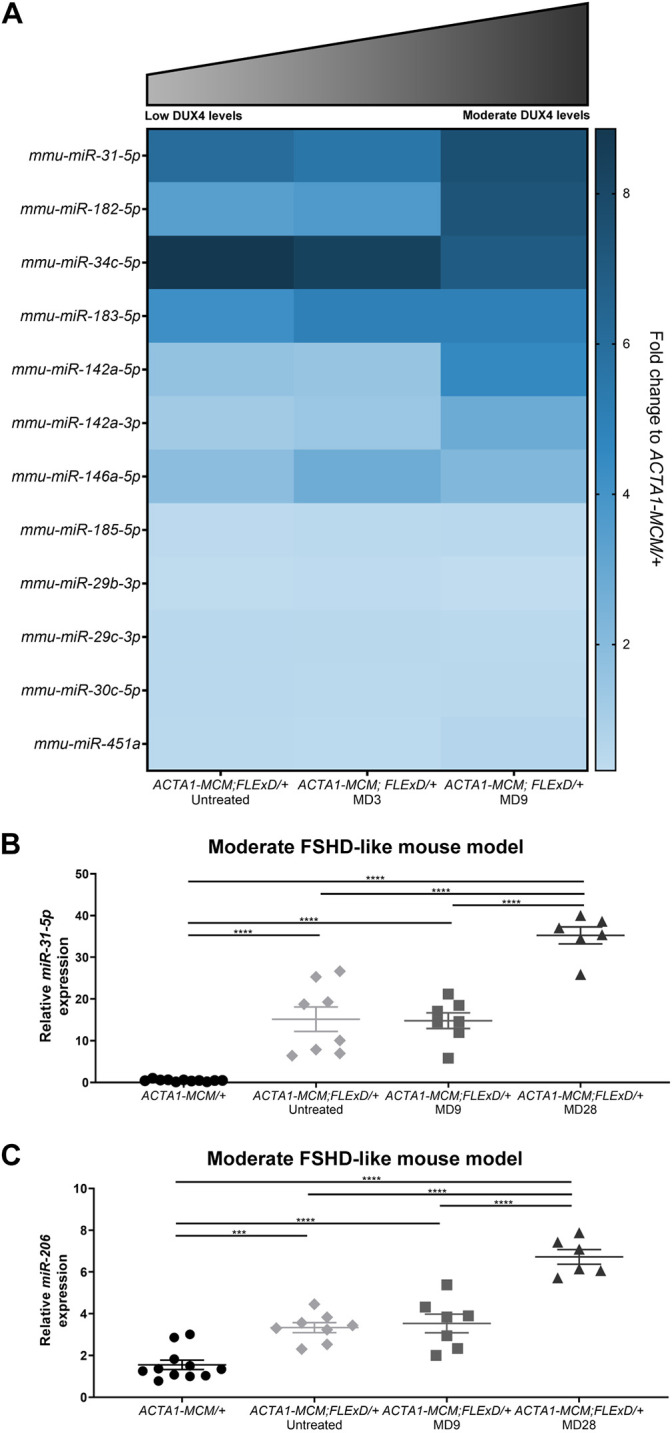
The miRNA profile dynamics in DUX4-induced FSHD-like mouse models. (A) Heat map for significantly dysregulated miRNAs in untreated and DUX4-induced ACTA1-MCM;FLExD/+ models (n=8 animals for each experimental group). Schematic depicts DUX4 levels in untreated and DUX4-induced ACTA1-MCM;FLExD/+ models. (B,C) TaqMan qRT-PCR quantification of miR-31-5p (B) and miR-206 (C) in gastrocnemius muscle from 3-month-old ACTA1-MCM/+ control mice, untreated (no TMX) ACTA1-MCM;FLExD/+ mice, moderate FSHD-like model mice, day 9 (MD9) and moderate FSHD-like model mice, day 28 (MD28) (n=6-11 for each group). Statistical analysis was performed with two-tailed one-way ANOVA (uncorrected Fisher's LSD) with n=11 animals for ACTA1-MCM/+, n=8 animals for ACTA1-MCM;FLExD/+ untreated, n=7 animals for ACTA1-MCM;FLExD/+ MD9 and n=6 animals for ACTA1-MCM;FLExD/+ MD28 experimental groups. Data are presented as mean±s.e.m.; ***P<0.001, ****P<0.0001.
In addition to the immune/regeneration-related miRNA array, we also analyzed another regeneration-related miRNA, miR-206, which has been proposed as a biomarker for DMD and is a key regulator of myogenesis, muscle differentiation and muscle repair (Cacchiarelli et al., 2011b; Mizuno et al., 2011; Roberts et al., 2012, 2013). Interestingly, the TaqMan probe assay revealed that miR-206 was increased by 3- and 4-fold in untreated and MD9 ACTA1-MCM;FLExD/+ muscles, respectively, compared with ACTA1-MCM/+ control, indicating that even low DUX4 expression is sufficient to induce a significant dysregulation of miR-206 expression (Fig. 2C). As with miR-31-5p, levels of miR-206 were further increased in MD28 ACTA1-MCM;FLExD/+ muscles (Fig. 2C), similarly suggesting that its expression may correlate with DUX4-induced pathology.
The miRNA profile analysis also identified several miRNAs for which levels were significantly decreased in DUX4-induced muscle (Fig. 1). Expression levels for miR-15a-5p, miR-19b-3p, miR-29b-3p, miR-185-5p and miR-451a were decreased in both untreated and MD3 ACTA1-MCM;FLExD/+ muscles (Fig. 1A,B), and miR-29c-3p and miR-149-5p levels were uniquely downregulated in MD9 ACTA1-MCM;FLExD/+ muscles (Fig. 1C). However, only miR-29b-3p and miR-185-5p levels were consistently decreased in all DUX4-expressing muscles (Fig. 1A-C), and therefore they are also potential candidates for FSHD muscle biomarkers. Overall, our results reveal that DUX4 expression results in a dynamic miRNA profile in murine skeletal muscle.
Chronic DUX4-expressing FSHD-like mouse model
The short-term increase in mosaic bursting of DUX4 expression in skeletal muscles caused by TMX induction in the moderate ACTA1-MCM;FLExD FSHD-like mouse model was useful for pushing the system and enabling candidate DUX4-reponsive biomarker discovery; however, it is not very representative of the situation found in FSHD patients, who experience chronic, low levels of mosaic DUX4 expression in skeletal muscles, leading to accumulated muscle damage over their lifetimes. Fortunately, the ACTA1-MCM;FLExD/+ mouse similarly expresses very low levels of mosaic DUX4 expression in skeletal muscles throughout its lifetime in the absence of TMX induction (Jones and Jones, 2018; Jones et al., 2020). This chronic, skeletal muscle-specific expression of DUX4 is a result of the ACTA1 [or human skeletal actin promoter (HSA)]-regulated mutated estrogen receptor dimer fused with cre (MCM) protein leaking into a small fraction of myonuclei of ACTA1-MCM;FLExD mice. This leads to sporadic transgene recombination in the absence of TMX induction exclusively in skeletal muscles, resulting in a mosaic expression pattern in rare myonuclei, and suggests a potentially better model for recapitulating FSHD (Jones et al., 2020).
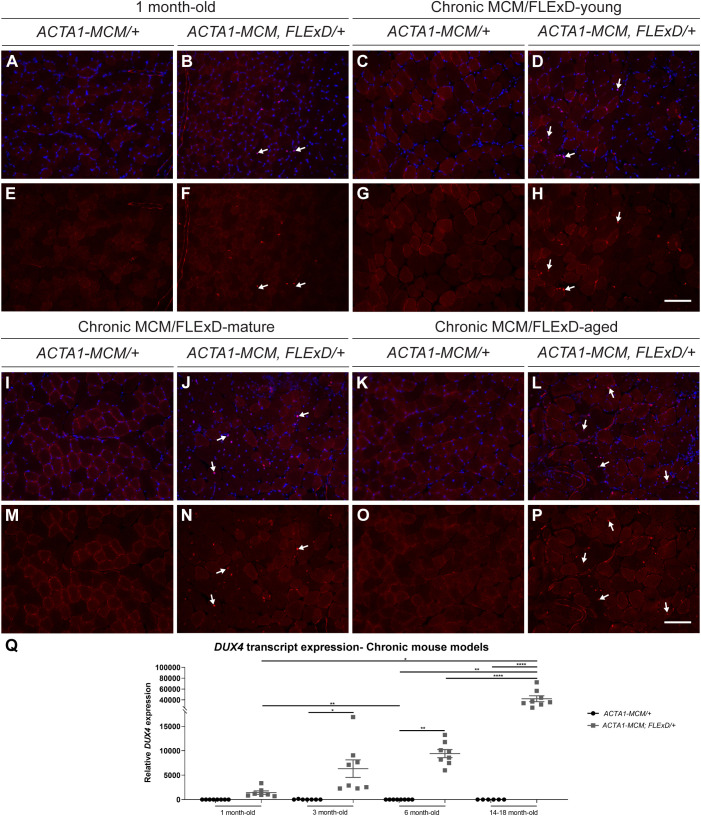
We quantified DUX4 expression in the gastrocnemius muscle of 1-, 3-, 6- and 14- to 18-month-old ACTA1-MCM;FLExD/+ mice and their age-matched ACTA1-MCM/+ controls using immunohistochemistry and quantitative reverse transcription PCR (qRT-PCR) (Fig. 3). Hereafter, we will refer to 3-month-old ACTA1-MCM;FLExD/+ mice as chronic MCM;FLExD-young (equivalent to human age 20 years), whereas 6-month-old mice will be referred to as chronic MCM;FLExD-mature (equivalent to human age 30 years) and 14- to 18-month-old mice as chronic MCM;FLExD-aged (equivalent to human age 45-55 years) (Flurkey et al., 2007). Analysis of body weight (Fig. S2A) and the weight of different skeletal muscles [tibialis anterior, gastrocnemius, quadriceps, extensor digitorum longus (EDL) and soleus muscles; Fig. S2B-F] showed a significant body and muscle weight decrease in the chronic MCM/FLExD-mature and -aged animals compared with age-matched ACTA1-MCM/+ controls. Histological analysis of DUX4 immunostaining revealed the presence of scattered DUX4-positive fibers in 1-month-old muscle and a gradual qualitative increase in the number of DUX4-positive nuclei in chronic MCM;FLExD-young, -mature and -aged muscles (Fig. 3A-P). Although quantitative analysis of DUX4 protein expression by immunostaining was hindered by frequent high background staining in the myofibers and additional unspecific staining in interstitial cells, potentially immune cells, in MCM;FLExD-mature and -aged animals, overall, we observed that the number of recombined DUX4-expressing myonuclei increased over time, consistent with recent data that endogenous DUX4 expression is not immediately cytotoxic in FSHD myogenic cells (Chau et al., 2021). Similarly, DUX4-fl levels increased significantly as the ACTA1-MCM;FLExD/+ mice aged. Young, mature and aged mice displayed significantly higher DUX4-fl expression compared with age-matched ACTA1-MCM/+ controls (Fig. 3Q); however, it should be noted that DUX4-fl levels were highly variable in chronic MCM;FLExD-mature and -aged muscles, which needs to be considered in downstream analyses (Fig. 3). Overall, DUX4-fl levels progressively increased with age in the bi-transgenic ACTA1-MCM;FLExD/+ mice, indicating that chronic expression of DUX4 leads to a cumulative increase in DUX4-positive myonuclei and DUX4 mRNA levels, suggesting that many aged DUX4-positive myofibers were accumulating, rather than dying (Fig. 3).
Fig. 3.
DUX4 expression in chronic FSHD-like mouse models. (A-P) Immunofluorescence staining for DUX4 (red) and DAPI (blue) in gastrocnemius muscle isolated from 1-month-old MCM/FLExD mice (A,B,E,F), 3-month-old or chronic MCM/FLExD-young mice (C,D,G,H), 6-month-old or chronic MCM/FLExD-mature mice (I,J,M,N) and 14- to 18-month-old or chronic MCM/FLExD-aged mice (K,L,O,P), compared with control ACTA1-MCM/+ mice. Representative images from n=3 animals for each experimental group. Scale bars: 100 µm. (Q) DUX4 expression in the gastrocnemius muscle isolated from 1 month-old, chronic MCM/FLExD-young, chronic MCM/FLExD-mature and chronic MCM/FLExD-aged mice, compared with age-matched ACTA1-MCM/+ controls. Statistical analysis was performed with two-tailed one-way ANOVA (uncorrected Fisher's LSD) with n=8 animals for ACTA1-MCM/+ 1-month-old, n=7 animals for ACTA1-MCM;FLExD/+ 1-month-old, n=7 animals for ACTA1-MCM/+ 3-month-old, n=8 animals for chronic MCM/FLExD-young, n=8 animals for ACTA1-MCM/+ 6-month-old, n=8 animals for chronic MCM/FLExD-mature, n=8 animals for ACTA1-MCM/+ 14- to 18-month-old and n=8 animals for chronic MCM/FLExD-aged experimental groups. Data are presented as mean±s.e.m.; *P<0.05, **P<0.01, ****P<0.0001.
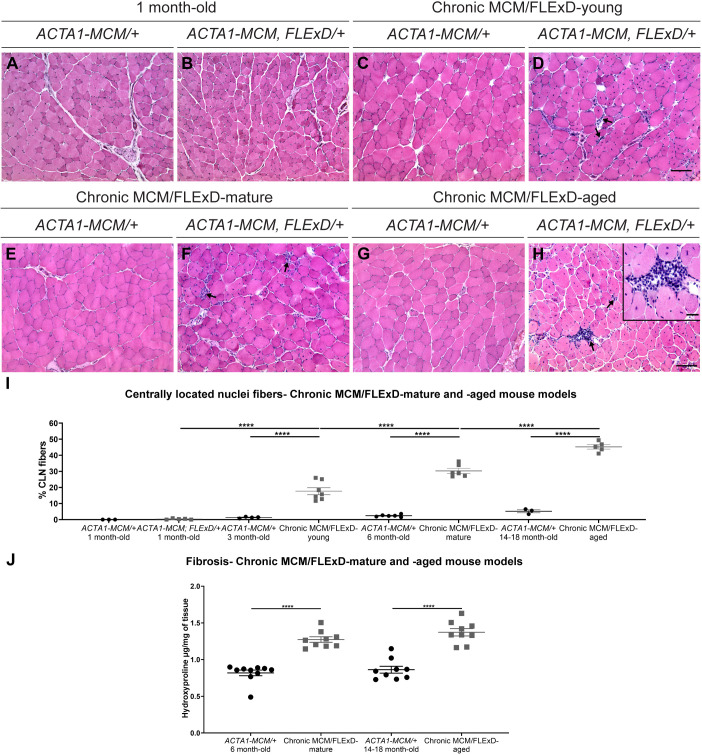
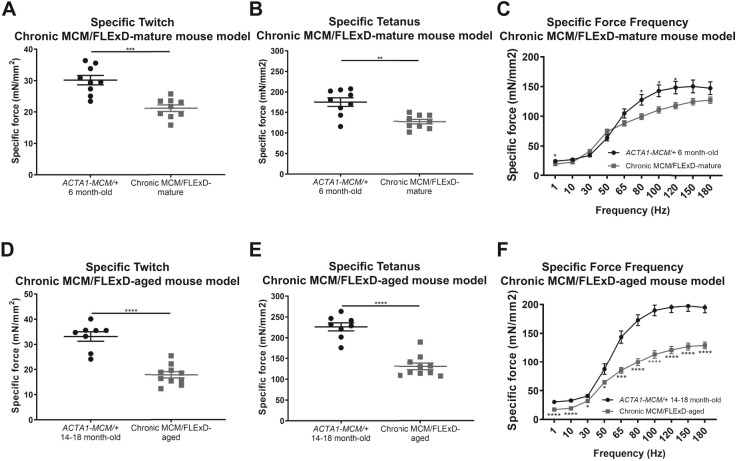
To assess the effects of chronic, low, mosaic DUX4 expression on skeletal muscle pathology, we analyzed muscles by immunofluorescence and Hematoxylin and Eosin (H&E) staining (Figs 4 and 5). Histological analysis of the gastrocnemius muscle revealed that chronic exposure to DUX4 leads to a steady increase in the percentage of damaged/regenerated fibers marked by centrally localized nuclei from 0% to 45%, and also an increase in immune cell infiltration over a period of 1.5 years. Control ACTA1-MCM/+ muscles did not show any significant increase in regenerated myofibers or immune cell infiltration over the same period (Fig. 4A-H,I). An equivalent significant increase in centrally localized nuclei was detected in the tibialis anterior muscle (Fig. S2G-K). Furthermore, fibrosis levels, measured by the muscle hydroxyproline content, were significantly increased in MCM;FLExD-mature and -aged muscles compared with age-matched ACTA1-MCM/+ controls (Fig. 4J). This significant increase in the percentage of fibers with centralized nuclei and fibrosis in chronic MCM;FLExD-aged muscles compared with chronic MCM;FLExD-mature muscles correlates with an increase in DUX4 expression (Fig. 3 and Fig. 4I,J). However, analysis of embryonic myosin, a marker for muscle regeneration, found that the percentage of embryonic myosin-positive fibers in both older populations of ACTA1-MCM;FLExD/+ DUX4-expressing muscles corresponded, on average, to <2% of the total number of fibers (Fig. 5A-H,I). This suggests that chronically increased levels of DUX4 expression lead to sporadic muscle regeneration events, as opposed to massive muscle regeneration, and, ultimately, to cumulative muscle pathology. We then analyzed muscle physiology of the EDL muscle from the chronic MCM/FLExD-mature and -aged mice (Fig. 6). As expected, chronic MCM;FLExD-mature and -aged muscles displayed significantly lower strength in specific twitch (Fig. 6A,D), tetanus (Fig. 6B,E), and force-frequency (Fig. 6C,F) measurements compared with age-matched ACTA1-MCM/+ controls. Taken together, these results indicate that aged ACTA1-MCM;FLExD/+ muscles exposed to chronic, low levels of DUX4 develop a complex pathology that more accurately recapitulates FSHD than previous models.
Fig. 4.
Histological analysis of the chronic DUX4 mouse models reveals FSHD-like histopathological changes in skeletal muscle. (A-H) Hematoxylin and Eosin staining of gastrocnemius muscles from 1-month-old MCM/FLExD mice (A,B), chronic MCM/FLExD-young mice (C,D), chronic MCM/FLExD-mature mice (E,F) and chronic MCM/FLExD-aged mice (G,H), compared with age-matched control ACTA1-MCM/+ mice. Arrows show immune cell infiltration marked by interstitial aggregation of mononuclear cells, which is often observed with myofiber necrosis. High-magnification image inset in H scale bar: 10 µm. Representative images are shown for each experimental group (n=3 animals). Scale bars: 100 µm. (I) Quantification of centrally localized nuclei-positive myofibers in gastrocnemius muscles isolated from 1-month-old, chronic MCM/FLExD-young, chronic MCM/FLExD-mature and chronic MCM/FLExD-aged mice compared with age-matched control ACTA1-MCM/+ mice. Statistical analysis was performed with two-tailed one-way ANOVA (uncorrected Fisher's LSD) with n=3 animals for ACTA1-MCM/+ 1-month-old, n=5 animals for ACTA1-MCM;FLExD/+ 1-month-old, n=4 animals for ACTA1-MCM/+ 3-month-old, n=7 animals for chronic MCM/FLExD-young, n=6 animals for ACTA1-MCM/+ 6-month-old, n=6 animals for chronic MCM/FLExD-mature, n=3 animals for ACTA1-MCM/+ 14- to 18-month-old and n=6 animals for chronic MCM/FLExD-aged experimental groups. (J) Fibrosis was analyzed by hydroxyproline quantification in gastrocnemius muscles isolated from 1-month-old, chronic MCM/FLExD-young, chronic MCM/FLExD-mature and chronic MCM/FLExD-aged mice compared with age-matched control ACTA1-MCM/+ mice. Statistical analysis was performed with two-tailed one-way ANOVA (uncorrected Fisher's LSD) with n=10 animals for ACTA1-MCM/+ 6-month-old, n=9 animals for chronic MCM/FLExD-mature, n=9 animals for ACTA1-MCM/+ 14- to 18-month-old and n=9 animals for chronic MCM/FLExD-aged experimental groups. Data are presented as mean±s.e.m.; ****P<0.0001.
Fig. 5.
Histological analysis of muscle regeneration in the chronic DUX4 mouse models. (A-H) Immunofluorescence staining for dystrophin (green), embryonic myosin (red) and DAPI (blue) in gastrocnemius muscles from 1-month-old MCM/FLExD mice (A,B), chronic MCM/FLExD-young mice (C,D), chronic MCM/FLExD-mature mice (E,F) and chronic MCM/FLExD-aged mice (G,H), compared with age-matched control ACTA1-MCM/+ mice. Representative images are shown for each experimental group (n=3 animals). Scale bars: 100 µm. (I) Quantification of embryonic myosin-positive myofibers in gastrocnemius muscles isolated from chronic MCM/FLExD-mature mice and chronic MCM/FLExD-aged mice compared with age-matched control ACTA1-MCM/+ mice. Statistical analysis was performed with two-tailed Kruskal–Wallis test (no Dunn's test) with n=6 animals for ACTA1-MCM/+ 6-month-old, n=6 animals for chronic MCM/FLExD-mature, n=3 animals for ACTA1-MCM/+ 14- to 18-month-old and n=5 animals for chronic MCM/FLExD-aged experimental groups. Data are presented as mean±s.e.m.; ***P<0.001.
Fig. 6.
Ex vivo physiology analysis of extensor digitorum longus in chronic DUX4-expressing FSHD-like mouse models. (A-F) Physiology measurements for specific twitch (A,D), specific tetanus (B,E) and force frequency (C,F) in the chronic MCM/FLExD-mature and -aged mouse model. Statistical analysis was performed with two-tailed Welch's t-test and two-way ANOVA with n=9 animals for ACTA1-MCM/+ 6-month-old, n=9 animals for chronic MCM/FLExD-mature, n=8 animals for ACTA1-MCM/+ 14- to 18-month-old and n=10 animals for chronic MCM/FLExD-aged experimental groups. Data are presented as mean±s.e.m.; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
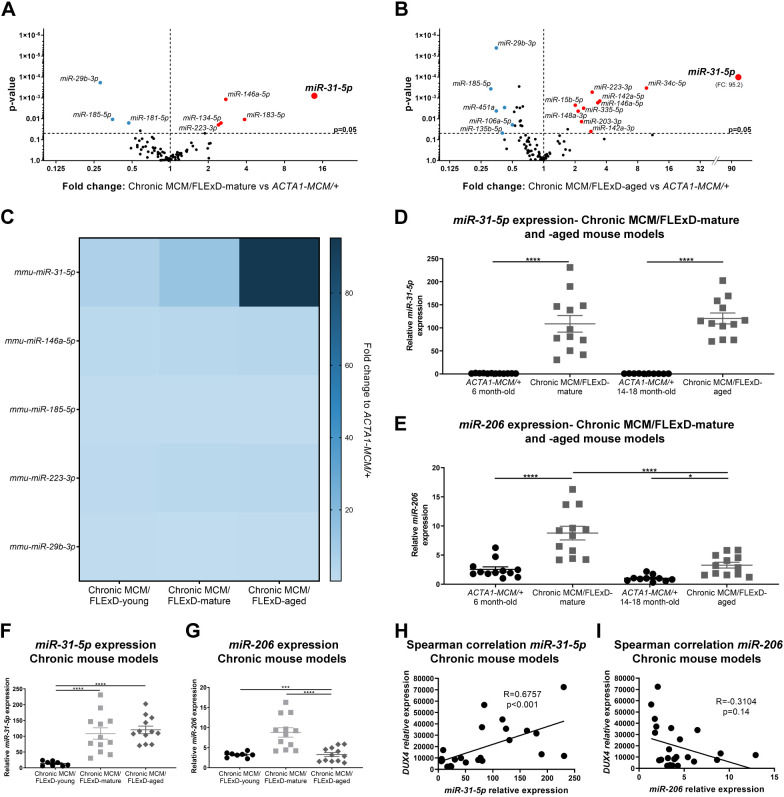
Immune/regeneration-related muscle miRNA profile in chronic DUX4-expressing FSHD-like mouse models
Late-onset and progressive muscle pathology in FSHD patients is thought to be initiated by chronic, low, sporadic expression of DUX4 throughout one's lifetime (Haynes et al., 2018; Jones et al., 2012; Kowaljow et al., 2007; Rickard et al., 2015; Snider et al., 2010; Tassin et al., 2013). Because we found that our chronic DUX4-expressing mouse model also develops late-onset, accumulative FSHD-like pathology including immune infiltration, we further investigated the immune/regeneration-related miRNA profile in the later phases of pathogenesis after 1.5 years of exposure to chronic DUX4 expression in skeletal muscle. We analyzed gastrocnemius muscle from the chronic MCM;FLExD-mature and -aged mouse models (Fig. 7; Fig. S1B, Table S1) and found six notable categories of dysregulated miRNAs: category 1, miRNAs for which upregulation correlates with pathogenesis progression (miR-31-5p and miR-146a-5p); category 2: miRNAs for which upregulation throughout pathogenesis does not correlate with pathology severity (miR-206); category 3, miRNAs upregulated during the early to mid phase of pathogenesis (miR-183-5p); category 4, miRNAs upregulated during the mid to late phase of pathogenesis (miR-223-3p); category 5, miRNAs upregulated in the late phase of pathogenesis (miR-15b-5p, miR-148a-3p, miR-335-5p, miR-203-3p); and category 6, miRNAs downregulated throughout pathogenesis (miR-29b-3p and miR-185-5p). Overall, the levels of immune-related miRNAs were consistently increased in chronic MCM;FLExD muscles compared with age-matched control, whereas the levels of miR-29b-3p, a hallmark of atrophic muscles (Hu et al., 2014), were consistently downregulated (Fig. 7A,B).
Fig. 7.
Targeted miRNA profile in the chronic DUX4 FSHD-like mouse models shows dysregulation of miRNAs in skeletal muscle. (A,B) Volcano plots depict the miRNA profile in gastrocnemius muscle from the chronic MCM/FLExD-mature model (A) and the chronic MCM/FLExD-aged model (B) compared with age-matched control ACTA1-MCM/+ mice (n=6-9 animals for each experimental group). Red and blue circles depict miRNAs for which levels are significantly upregulated and downregulated by ≥2-fold, respectively. Volcano plots represent two-tailed unpaired Student's t-test analysis with n=7 animals for ACTA1-MCM/+ 6-month-old, n=9 animals for chronic MCM/FLExD-mature, n=6 animals for ACTA1-MCM/+ 14- to 18-month-old and n=8 animals for chronic MCM/FLExD-aged experimental groups. (C) Heat map for significantly dysregulated miRNAs in the gastrocnemius muscle isolated from the chronic MCM/FLExD-young, -mature and -aged mouse models (n=6-9 for each group). (D,E) qRT-PCR quantification of miR-31-5p (D) and miR-206 (E) in gastrocnemius muscle isolated from chronic MCM/FLExD-mature and chronic MCM/FLExD-aged mouse models. (F,G) Combined expression profiles for miR-31-5p (F) and miR-206 (G) in the chronic MCM/FLExD-young, -mature and -aged mouse models. Statistical analysis was performed with two-tailed one-way ANOVA test (uncorrected Fisher's LSD) with n=8 animals for chronic MCM/FLExD-young, n=12 animals for ACTA1-MCM/+ 6-month-old, n=12 animals for chronic MCM/FLExD-mature, n=10 animals for ACTA1-MCM/+ 14- to 18-month-old and n=12 animals for chronic MCM/FLExD-aged experimental groups. Data are presented as mean±s.e.m.; *P<0.05, ***P<0.001, ****P<0.0001. (H,I) Spearman correlation for miR-31-5p (H) or miR-206 (I) and DUX4 expression in gastrocnemius muscle from the chronic MCM/FLExD-mature and -aged mouse models.
Perhaps the most interesting finding in this miRNA analysis of chronic DUX4-expressing FSHD mouse models was that miR-31-5p, the levels of which were altered in the short-term DUX4-induced moderate model, was in category 1. Strikingly, miR-31-5p expression levels were steeply upregulated by 6-fold, 14-fold and 95-fold in chronic MCM;FLExD-young, -mature and -aged muscles, respectively (Fig. 1A and Fig. 7A-C). TaqMan probe quantification of miR-31-5p confirmed that its levels were significantly increased by 97-fold, on average, in chronic MCM;FLExD-mature muscles and by 139-fold in chronic MCM;FLExD-aged muscles compared with ACTA1-MCM/+ control muscles (Fig. 7D,F), and significantly higher than in muscles from the chronic MCM;FLExD-young mice (Fig. 2B and Fig. 7F). Interestingly, miR-31-5p levels were significantly different in females and males in the chronic MCM/FLExD-mature animals, but this difference was not detected in the older chronic MCM/FLExD-aged animals (Fig. S3A). To determine whether miR-31-5p levels correlate with DUX4-fl levels in chronic DUX4-expressing muscles, we tested for potential associations between DUX4-fl and miR-31-5p expression using Spearman's correlation. We found a significant correlation between miR-31-5p and DUX4-fl expression, suggesting that dysregulation of miR-31-5p is DUX4 dependent and may be DUX4 responsive (Fig. 7H). Regardless, the strongly increased upregulation of miR-31-5p over the progressive FSHD-like pathogenesis in this chronic DUX4 model suggests that miR-31-5p is a potential FSHD disease biomarker for muscle biopsy. We also assessed miR-206 in the chronic DUX4-expressing FSHD-like mouse models, as this was a promising candidate biomarker from our analysis using the DUX4-induced ACTA1-MCM;FLExD/+ models. We found that the levels of miR-206, as determined by TaqMan qRT-PCR, were also significantly increased in gastrocnemius muscle from chronic MCM/FLExD-mature and -aged muscles by 7- and 3-fold, respectively, compared with chronic MCM/FLExD-young muscles (Fig. 7E,G). miR-206 expression was similar in females and males from chronic MCM/FLExD-mature and -aged models (Fig. S3B). We then tested for potential associations between miR-206 and DUX4-fl expression levels using Spearman's correlation, but did not find a significant correlation (Fig. 7I). Nevertheless, because miR-31-5p and miR-206 levels are increased in skeletal muscle throughout the course of DUX4-induced pathogenesis, they are very attractive potential FSHD biomarkers for further evaluation.
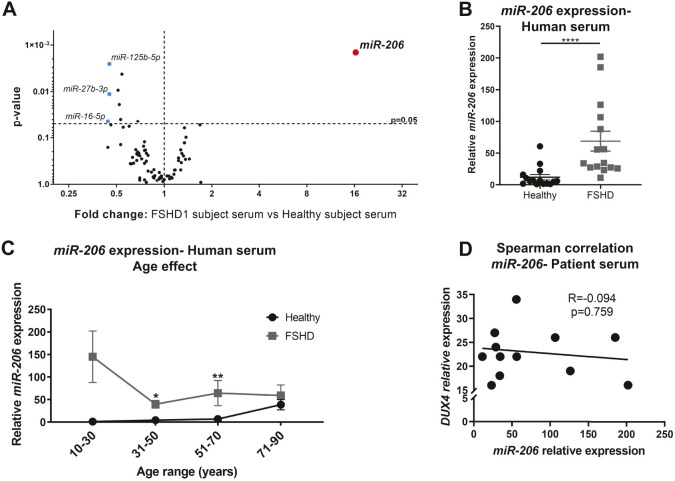
miR-206 is a candidate FSHD serum biomarker
Discovery work using the above mouse models identified a number of DUX4-dependent miRNAs that are attractive candidates for FSHD biomarkers. However, although these were identified from muscle tissue, the goal of this study was to identify circulating biomarkers for FSHD for non-invasive assessment in clinical trials. Therefore, the candidate miRNA profiles were validated using serum samples isolated from FSHD patients and healthy controls. We collected serum samples from 15 healthy subjects and 15 clinically affected and genetically confirmed FSHD patients and processed these samples for the equivalent human immunopathology miRNA profile used for the murine muscle samples (Fig. 8; Table S2). Surprisingly, we found that miR-206 was the only miRNA significantly upregulated in FSHD patient serum compared with serum from healthy subjects (Fig. 8A). Further analysis by TaqMan probe assay confirmed that although miR-206 levels were highly variable in the FSHD study population, they were nonetheless significantly elevated in FSHD patient serum (Fig. 8B). Because miR-206 expression is known to increase with age in healthy mice (Hamrick et al., 2010), and inclusion of older subjects could therefore bias the analysis of biomarkers, we stratified the study population by age and verified that the significant increase in miR-206 levels in serum is only significant in the 31-50 and 51-70 age-range groups (Fig. 8C). Furthermore, miR-206 expression did not correlate with age or D4Z4 deletion size (Fig. 8D), just FSHD clinical status. An equivalent analysis of mouse serum revealed that the miRNA levels in serum are too low for a reliable quantification (>30 Cq considered unreliable for analysis of pre-amplified products by the manufacturer) (Fig. S4). Overall, these data support that miR-206 is a strong candidate DUX4-dependent circulating biomarker for the presence of muscle damage such at that found in clinical FSHD.
Fig. 8.
Expression of miRNA-206 is significantly upregulated in FSHD patient serum. (A) Volcano plot for the miRNA profile in FSHD patient serum (n=7 FSHD patients and n=10 healthy subjects). Red and blue circles depict miRNAs for which levels are significantly upregulated and downregulated by ≥2-fold, respectively. Volcano plots depict two-tailed unpaired Student's t-test analysis. (B) miR-206 expression in human serum from all patients in the study (n=14 FSHD patients and n=15 healthy subjects). Statistical analysis was performed using the two-tailed Mann–Whitney U-test with n=15 FSHD patients and n=15 healthy subjects. (C) miR-206 expression in human serum depicted according to patient age. Statistical analysis was performed using the two-tailed Mann–Whitney U-test with n=15 FSHD patients and n=15 healthy subjects. Data are presented as mean±s.e.m.; *P<0.05, **P<0.01, ****P<0.0001. (D) Spearman correlation of miR-206 expression and DUX4 D4Z4 reduced allele size (EcoRI/Blnl-kb) with n=14 FSHD1 patients.
DISCUSSION
Identifying reliable and independently validated circulating DUX4-dependent biomarkers is one of the great unmet needs in the FSHD field, as none have been described to date. The discovery of circulating serum biomarkers could greatly impact the prediction of disease progression and the determination of therapeutic efficacy in clinical trials, and thus is of utmost need. With the recent development of FSHD-like mouse models (Bosnakovski et al., 2017; Giesige et al., 2018; Jones and Jones, 2018; Jones et al., 2018 preprint), research on FSHD disease biomarkers has become technically more feasible. Here, we describe new aged FSHD-like mouse models of chronic DUX4 expression that recapitulate many important aspects of FSHD pathology and characterize the muscle miRNA signature in these models. We demonstrate that miR-31-5p and miR-206 levels are consistently increased in the muscle of FSHD-like mouse models and that miR-206 expression is significantly increased in serum samples from FSHD patients. We propose miR-206 as a new candidate circulating disease biomarker for FSHD.
Chronic DUX4-expressing FSHD-like mouse model
Although multiple strains of inducible FSHD-like mouse models have now been generated (Bosnakovski et al., 2017, 2020; Giesige et al., 2018; Jones and Jones, 2018; Jones et al., 2020), these either rely on acute induction of DUX4 to produce massive pathology or on long-term small-molecule treatments resulting in chronic expression of DUX4 in both muscle and non-muscle tissues. None of these models mimics the typical situation in FSHD in which pathology is caused by sporadic, but chronic, expression of DUX4 in skeletal muscles throughout one's lifetime (Haynes et al., 2018; Jones et al., 2012; Rickard et al., 2015; Snider et al., 2010; Tassin et al., 2013). Here, we characterized two aged models recapitulating chronic skeletal muscle-specific DUX4 expression: one at 6 months old (chronic MCM;FLExD-mature model) and one at 14-18 months old (chronic MCM;FLExD-aged model) using the ACTA1-MCM;FLExD/+ mice. Importantly, generation of these chronic FSHD-like models does not require any treatments to activate the chronic expression of DUX4 in either male or female animals and thus provides an important advantage for consistency and reproducibility in long-term studies. We demonstrated that DUX4 expression increases progressively in chronic MCM;FLExD-young, -mature and -aged muscles compared with 1-month-old asymptomatic muscles and age-matched control ACTA1-MCM/+ muscles. The data also revealed that chronic DUX4 expression induced hallmarks of FSHD pathology such as increased centrally nucleated myofibers (damaged, regenerated myofibers), immune infiltration and reduced muscle regeneration (Arahata et al., 1995; Banerji et al., 2020; Frisullo et al., 2011; Honda et al., 1987; Padberg, 1982; Statland et al., 2015b). Accordingly, immunostaining for DUX4 protein in the muscles of chronic FSHD-like mice revealed the presence of unspecific staining in interstitial cells, possibly immune cells, and prevented an accurate quantification. In addition, this suggests that the muscle from these mice is more immunoreactive than that from the age-matched controls.
In contrast to most other muscular dystrophies such as DMD, in which the underlying primary defects cause structural myofiber disruption, leading to high levels of degeneration and regeneration, FSHD is a slowly progressive disease caused by expression of a toxic protein, DUX4, exclusively in a very few myonuclei. The ultimate result is the development of a myopathy with many general hallmarks of muscle degeneration that coincide with the other muscular dystrophies, but also with several features that are distinct to FSHD as well. That is evidenced by differences in muscle regeneration levels as assessed by embryonic myosin staining. Whereas DMD muscle biopsies display 24-33% regenerating fibers (Janghra et al., 2016; Schiaffino et al., 1986), FSHD muscle biopsies exhibit only 0.48% and 1.72% regenerating fibers in the quadriceps and tibialis anterior, respectively (Banerji et al., 2020). This strikingly low level of regeneration is also common to myotonic muscular dystrophy type 2, another slowly progressing muscle disease (Banerji et al., 2020). Thus, our chronic FSHD-like mouse model better recapitulates both the skeletal muscle-specific mosaic expression of DUX4 and the hallmark pathophysiology of FSHD. It is important when using mouse models for biomarker discovery and investigations into disease mechanisms to minimize the interference of tissues not affected in the human condition as well as to avoid degeneration/regeneration-related artifacts. For comparison, an alternative transgenic mouse model for chronic FSHD has recently been proposed; however, unlike in the human disease situation, the model uses constitutive tetracycline treatment to induce DUX4 at low levels in both muscle and non-muscle tissues of female mice (Bosnakovski et al., 2020). By contrast, our aged chronic FSHD-like mouse models based on the ACTA1-MCM;FLExD/+ bi-transgenic animals more accurately recapitulate FSHD pathogenic mechanisms and pathophysiology, and are thus ideally suited to identify disease biomarkers, study potential disease mechanisms and determine the effects of potential therapeutics.
DUX4-induced miRNA dysregulation in FSHD-like mouse models
miRNA profiles can be informative in understanding both healthy and pathogenic mechanisms as well as revealing biomarkers for disease. For example, immune-related miRNAs such as miR-146b, miR-155, miR-214, miR-221 and miR-222 are consistently found to be dysregulated in the muscular dystrophies, including FSHD (Eisenberg et al., 2007). Portilho et al. (2015) identified miR-330, miR-331-5p, miR-34a, miR-380-3p, miR-516b, miR-582-5p, miR-517 and miR-625 as being exclusively expressed in human FSHD1 quadriceps biopsies obtained from fetuses compared with healthy muscles. However, Heier et al. (2020) recently identified a very different set of eight miRNAs in FSHD patient serum, with only miR-34a and miR-146b overlapping with either the Portilho et al. (2015) or Eisenberg et al. (2007) study, respectively. Thus, there has been great variability between studies on miRNAs using different types of FSHD clinical samples, and no studies have shown any miRNA as being DUX4 dependent.
Here, we used DUX4-expressing, FSHD-like model mice to identify DUX4-dependent dysregulated miRNAs in skeletal muscle. The data revealed that multiple immune-related miRNAs were dysregulated, supporting a role for the immune response in contributing to DUX4-dependent FSHD muscle pathology and degeneration. Interestingly, none of these miRNAs were significantly dysregulated in the serum from FSHD patients, which suggests that these immune-related miRNAs are part of a local response to muscle damage. This is consistent with a recent study that reported dysregulation of miR-142-3p and miR-146b levels in mildly affected FSHD patients, but failed to show significance upon larger validation (Heier et al., 2020).
The most strikingly dysregulated miRNA in both of our models is miR-31-5p, which is expressed in immune cells as well as in regenerating skeletal muscle (Cacchiarelli et al., 2011a; Greco et al., 2009; Alrafas et al., 2020; Xue et al., 2013; Zhu et al., 2019). Whether the DUX4-dependent increase in miR-31-5p levels takes place in DUX4-expressing myofibers or in infiltrating non-muscle cells has yet to be determined. If the dysregulation takes place in myofibers, miR-31-5p may serve as a useful interstitial muscle fluid biomarker of disease. Considering that muscle microdialysis was already successfully implemented in the study of muscle fluid from FSHD patients (Tasca et al., 2017), this alternative methodology holds a promising new avenue for FSHD human muscle analysis. Although miR-31-5p levels are sensitive to DUX4 expression in muscle, this miRNA may not be a direct target of DUX4. The potential involvement of miR-31-5p in DUX4 downstream cascades is supported by the dysregulation of miR-31-5p in several inflammatory-related disorders such as cancer (Cho et al., 2015; Necela et al., 2011; Yi et al., 2019), psoriasis (Abdallah and Pichon, 2019; Xu et al., 2013), experimental autoimmune encephalomyelitis (Zhang et al., 2015), inflammatory bowel disease (Olaru et al., 2011), lupus (Dai et al., 2010) and asthma (Shi et al., 2019). In particular, miR-31-5p is an important regulator of cytokine/chemokine production (Dai et al., 2019; Xu et al., 2013) and neutrophil function and T-cell activation during inflammation (Alrafas et al., 2020; Rovira-Llopis et al., 2018; Suárez et al., 2010; Xue et al., 2013), and is dysregulated in the mdx mouse model and muscle biopsies from DMD patients (Cacchiarelli et al., 2011a; Fiorillo et al., 2015), likely as a consequence of its high expression in regenerating fibers (Cacchiarelli et al., 2011a; Greco et al., 2009). However, the low levels of muscle regeneration in our FSHD-like mouse models suggest that the role of miR-31-5p in response to DUX4 expression may be different from that in mdx/DMD. As with the other immune-related miRNAs, miR-31-5p levels were not dysregulated in FSHD patient serum, similar to previous findings (Heier et al., 2020), again suggesting that this dysregulation may be a local response. Further studies will be important to clarify whether miR-31-5p levels are elevated in FSHD patient biopsies and in which cells miR-31-5p levels are upregulated.
miR-206 is a potential new disease biomarker for FSHD
Several miRNAs have been described as circulating biomarkers for neuromuscular diseases such as amyotrophic lateral sclerosis (Si et al., 2018; Toivonen et al., 2014) and DMD (Cacchiarelli et al., 2011b; Mizuno et al., 2011; Roberts et al., 2012, 2013), and unrelated diseases such as cancer (Wang et al., 2014). Yet, although miRNA profiling in the current study revealed the dysregulation of multiple immune-related miRNAs in DUX4-expressing muscle, none of these were validated as circulating FSHD biomarkers. Interestingly, the levels of miR-206, one of the original myomiRs and a major regulator of muscle myogenesis and regeneration (Chen et al., 2010; Greco et al., 2009; Horak et al., 2016; Kim et al., 2006; Ma et al., 2015; Yuasa et al., 2008), were consistently increased in the muscle from all FSHD-like mouse models and in serum from FSHD patients. Although miR-206 levels were found to be similar in muscle biopsies from healthy and FSHD fetuses (Portilho et al., 2015), this is not necessarily at odds with our findings in adult FSHD-like mice. Because miR-206 is a major regulator of myogenesis, the mechanisms controlling its expression as well as its DUX4 dependence are likely to be very different in developing versus adult muscle. In fact, miR-206 levels progressively increase in later stages of human fetal skeletal muscle development, suggesting that miR-206 has a different impact during different phases of skeletal muscle development in utero, and its dynamic profile has to be taken into consideration when comparing developing and adult muscle (Koutsoulidou et al., 2011). Additionally, although a previous study reported that miR-206 levels were unchanged in FSHD muscle biopsies (Eisenberg et al., 2007), incongruities regarding miRNA profiles have been reported previously in DMD and, as discussed previously, technical differences might account for differences in the reported miRNA signatures (Roberts et al., 2012). Our current study relies on a miRNA array profile further validated by TaqMan qRT-PCR quantification and thus provides consistent support for miR-206 dysregulation in mouse muscle biopsies and human serum samples. Unfortunately, the most recent report on FSHD circulating biomarkers did not include miR-206 in the analysis (Heier et al., 2020). Thus, further independent studies still need to be performed and will be important to validate miR-206 as a circulating FSHD biomarker. Nonetheless, miR-206 has been proposed as a serum biomarker for DMD and DM1 (Myotonic dystrophy type 1) (Cacchiarelli et al., 2011b; Koutsoulidou et al., 2015, 2017; Mizuno et al., 2011; Roberts et al., 2012, 2013), and we propose it as a strong candidate for FSHD as well.
Taken together, we describe two new FSHD-like mouse models of chronic DUX4 expression that recapitulate important hallmarks of FSHD gene expression and cumulative pathology. We provide a thorough characterization of the miRNA profile in muscles of these models, as well as in serum samples from FSHD patients. We identified miR-31-5p and miR-206 as prospective regulators of muscle disease in response to DUX4 expression and validated miR-206 as a candidate DUX4-dependent serum biomarker for FSHD. Future studies on the mechanisms underlying the involvement of miR-206 in FSHD pathology, as well as independent analyses of larger cohorts of FSHD patients and healthy controls, will be important for further validation of this miRNA as a useful clinical biomarker for FSHD.
MATERIALS AND METHODS
Human patients
This project was approved by the University of Nevada, Reno Institutional Review Board (#1316095). Demographic and clinical information are provided in Table S2. This clinical investigation was conducted according to the principles expressed in the Declaration of Helsinki. All patient sera were de-identified and processed in a randomized and blinded manner.
Animals
All procedures involving animals were approved by University of Nevada, Reno Institutional Animal Care and Use Committee (protocol #20-09-1078). The care and use of experimental animals complied with relevant institutional and national animal welfare laws, guidelines and policies. The ACTA1-MCM line (McCarthy et al., 2012) was purchased from The Jackson Laboratory (Bar Harbor, ME, USA). ACTA1-MCM;FLExD mice were generated and genotyped as previously described (Jones et al., 2020). ACTA1-MCM;FLExD mice were injected with one 5 mg/kg dose of TMX, as described (Jones et al., 2020), to create the moderate FSHD-like model, and were analyzed at day 3, 9 and 28 after TMX administration. Mice designated as 1 and 3 months old correspond to 4- and 13- to 14-week-old mice, respectively. Mice were sacrificed under deep anesthesia (3% isoflurane). Analysis of mouse tissue was performed in a blinded manner.
Total RNA extraction
Gastrocnemius muscles were freshly dissected and immediately snap-frozen in liquid nitrogen. A portion of gastrocnemius mid belly was collected for total RNA (including small RNAs) extraction with an RNeasy Mini Kit as indicated by the manufacturer (Qiagen, 74104). Briefly, muscle was homogenized in ten times the volume of the sample using Trizol reagent in a tissue lyser for 10 min at 50 Hz. The lysates were then incubated at room temperature for 7 min and RNA was separated with 0.2× total volume of chloroform (room temperature incubation for 3 min). After phase separation (12,000 g for 15 min at 4°C), the aqueous phase was mixed with 1.5× total volume of 100% ethanol. The sample was then transferred to an RNeasy MinElute column and centrifuged for 30 s at 10,000 g. The column was then washed with Buffer RWT and treated with DNase. After a second Buffer RWT wash, the column was washed twice with Buffer RPE and twice with 80% ethanol. The RNeasy MinElute column was dried for 5 min, and RNA was eluted with RNase-free water. Mouse and human serum samples were collected after blood centrifugation at 1000 g for 10 min and 15 min, respectively, at 15°C. Total RNA (including small RNAs) was extracted from 500 µl human serum and 200 µl mouse serum with the miRNeasy Serum/Plasma Advanced Kit, as indicated by the manufacturer (Qiagen, 217204).
Reverse transcription and quantitative PCR (qPCR) reactions for miScript profile
Reverse transcription reaction was performed according to the manufacturer's instructions with 250 ng RNA (Qiagen, 218161). The reverse transcription reaction mixture was then diluted in 200 µl RNase-free water and 100 µl of the resulting solution was subsequently used for the qPCR reaction. miScript miRNA profiles for the immunopathology panel (MIMM-104ZD for mouse, and MIHS-104ZD for human) were prepared with SYBR Green (Qiagen, 218076) according to the manufacturer's instructions in a 96-well plate (Qiagen, 218076). The cycling conditions were set up as indicated by the manufacturer. All samples were compared after baseline cycle and threshold value standardization. RNA levels were normalized to Snord68/SNORD68 (Small Nucleolar RNA, C/D Box 68) (Zanotti et al., 2018) and global Ct mean for mouse gastrocnemius muscle and human serum, respectively.
TaqMan qRT-PCR analysis
The reverse transcription reaction was performed with 10 ng total RNA from gastrocnemius muscle and 3 µl serum according to the manufacturer's instructions (Thermo Fisher Scientific, 4366596). Pre-amplification was performed for serum-extracted RNA (Thermo Fisher Scientific, 4391128). A maximum of 350 ng total RNA was incubated at 16°C for 30 min and then at 42°C for 30 min for the reverse transcription reaction. The reaction was concluded with a 5 min incubation at 85°C. The reverse transcription reaction product was pre-amplified with the Taqman PreAmp Master Mix and 5× primer pool (including miR-31-5p, miR-206 and miR-191-5p). The pre-amplification reaction was performed with the following protocol: 95°C for 10 min, 55°C for 2 min, 72°C for 2 min; 12× cycle of a two-step cycling protocol with a 95°C for 15 s denaturation, 60°C for 4 min annealing/extension and enzyme inactivation at 99.9°C for 10 min. The pre-amplified product was then diluted by a 12.5× factor and used for the qPCR reaction. The qPCR reaction was performed with the TaqMan Fast Advanced Master Mix according to the manufacturer's instructions for CFX96 real-time detection system (Thermo Fisher Scientific, 4444557). Briefly, each reaction was run with the following run method: UNG activation (50°C for 2 min); polymerase activation (95°C for 20 s) and 40× cycles of a denaturation (95°C for 3 s) and annealing/extension (60°C for 30 s) two-step cycling. Normalization was performed to the U6 non-coding small nuclear RNA (snRNA) (Fiorillo et al., 2015) and miR-191-5p for RNA extracted from gastrocnemius muscle and serum RNA-extracted samples, respectively. The following miRNA assays were used: mmu-miR-31-5p Assay ID 000185; U6 snRNA Assay ID 001973; has-miR-206 Assay ID 000510; hsa-miR-31-5p Assay ID 002279; and hsa-miR-191-5p Assay ID 002299.
Reverse transcription and qPCR reactions for DUX4-fl mRNA
Quantification of DUX4-fl transcription was performed with a nested qRT-PCR as previously described (Jones et al., 2017). Primers used for ten-cycle pre-amplification were DUX4-fl For, 5′-GCTCTGCTGGAGGAGCTTTAGGA-3′ and DUX4-fl Rev1, 5′-CGCACTGCTCGCAGGTCTGCWGGT-3′. Primers used for qPCR were DUX4-fl For and DUX4-fl Rev2, 5′-GCAGGTCTGCWGGTACCTGG-3′. Expression of DUX4-fl was normalized to 18S rRNA using 10 ng cDNA.
H&E staining
Gastrocnemius muscle was dissected and then freshly frozen in Tissue-Tek OCT compound (Sakura Finetek, 4583) in cold isopentane. Briefly, 12 μm cryosections were fixed in cold acetone at −20°C for 5 min and stained with Hematoxylin for 3 min. The cryosections were subsequently incubated with differentiation solution for 20 s and Scott's Bluing reagent for 1 min. The stained cryosections were rinsed three times in a 1:2 Eosin to 70% ethanol solution, and dehydrated with a series of 30 s ethanol washes (70%, 95%) and 1 min 100% ethanol wash. The stained cryosections were cleared with xylene for 5 min and mounted. A series of three non-consecutive sections were imaged on a Leica DM2000 and analyzed with LAS 4.12 software (Leica Microsystems).
Hydroxyproline quantification
The hydroxyproline assay was performed on gastrocnemius muscle, as previously described (Heydemann et al., 2005). Briefly, the tissue was minced overnight in 2 ml of 6 M hydrochloric acid at 110°C and the resulting hydrolysate (10 µl) was mixed with 150 µl isopropanol. Hydroxyproline oxidation was then performed for 10 min at room temperature with the addition of 72 µl of 1.4% cholramin-T in citrate buffer. Ehrlich's reagent (1 ml) was then added for 30 min at 55°C. The extinction measurement was performed at 558 nm.
Immunofluorescence
Immunofluorescence was performed on 10 μm cryosections from OCT-embedded frozen gastrocnemius muscle as described (Jones et al., 2020). Cryosections were fixed with 4% paraformaldehyde/PBS on ice for 10 min and permeabilized with 0.25% Triton X-100/PBS for 10 min. When using mouse primary antibodies, the sections were incubated with Mouse on Mouse Detection Kit (Vector Laboratories; BMK-2202) IgG blocking solution for 1 h. Sections were subsequently incubated with a 5% normal goat serum, 2% bovine serum albumin, 0.01% TritonX-100/PBS blocking solution for 30 min and with primary antibodies at 4°C overnight. The cryosections were then incubated with secondary antibody at room temperature for 45 min on the following day and then mounted in ProLong Gold with 4′,6-diamidino-2-phenylindole (DAPI) nuclear staining. A series of three non-consecutive sections were acquired using a Leica DMi8, DFC365 FX camera and LAS X Expert software (Leica Microsystems) and used for quantification in plantaris, and medial and lateral gastrocnemius muscles. Quantifications were performed in the entire plantaris muscle, whereas quantifications in gastrocnemius muscle were performed in 1183.11 µm square crops (one crop for each gastrocnemius muscle). Images were analyzed with ImageJ 1.52i software. Primary antibodies used were as follows: anti-DUX4 (clone E5-5, Abcam, ab124699), diluted 1:200; anti-dystrophin rabbit polyclonal antibody (Abcam, ab15277), diluted 1:200; and anti-embryonic fast myosin heavy chain mouse monoclonal antibody (Developmental Studies Hybridoma Bank, F1.652), diluted 1:20. The F1.652 monoclonal antibody, developed by the Baxter Laboratory for Stem Cell Biology, Stanford University, was obtained from the Developmental Studies Hybridoma Bank, created by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health and maintained at The University of Iowa (Iowa City, IA, USA).
Ex vivo muscle physiology
Specific muscle twitch, tetanus and force-frequency ex vivo physiology measurements were performed and analyzed in the EDL as previously described (Jones et al., 2020).
Statistical analysis
All miScript two-group statistical analyses for miRNA profiles were performed with unpaired Student's t-test using the Qiagen data analysis Excel spreadsheet and according to Qiagen handbook instructions. The remaining statistical analysis was performed using GraphPad Prism 8 and described for each set of data. n=7-8 animals with an even number of males and females per group was used for analysis. P<0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank the FSHD families that donated their time and themselves to further this work. We thank Danielle Eaton, Jennifer Sanchez and Valerie Smith of the University of Nevada, Reno School of Medicine (UNR Med) Clinical Research Center for help obtaining the clinical samples, and Dr Charis L. Himeda for critical discussions and editing the manuscript. We thank the William R. Lewis Family and Mick Hitchcock, PhD, for their support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.M.N., T.I.J., P.L.J.; Methodology: A.M.N., M.R., T.I.J.; Formal analysis: A.M.N., M.R., T.I.J.; Data curation: A.M.N., M.R., T.I.J., P.L.J.; Writing - original draft: A.M.N., T.I.J., P.L.J.; Writing - review & editing: A.M.N, M.R., T.I.J., P.L.J., M.R.; Supervision: T.I.J., P.L.J.; Project administration: P.L.J.; Funding acquisition: T.I.J., P.L.J.
Funding
This work was funded by a grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01 AR070432) to P.L.J., and by the Mick Hitchcock, Ph.D. Endowed Chair in Medical Biochemistry at University of Nevada, Reno School of Medicine.
References
- Abdallah, F. and Pichon, C. (2019). Evidence on the direct correlation between miR-31 and IL-22 axis in IMQ-induced psoriasis. Exp. Dermatol. 28, 1336-1340. 10.1111/exd.14001 [DOI] [PubMed] [Google Scholar]
- Alrafas, H. R., Busbee, P. B., Nagarkatti, M. and Nagarkatti, P. S. (2020). Resveratrol downregulates miR-31 to promote T regulatory cells during prevention of TNBS-induced colitis. Mol. Nutr. Food Res. 64, 1900633. 10.1002/mnfr.201900633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arahata, K., Ishihara, T., Fukunaga, H., Orimo, S., Lee, J. H., Goto, K. and Nonaka, I. (1995). Inflammatory response in facioscapulohumeral muscular dystrophy (FSHD): immunocytochemical and genetic analyses. Muscle Nerve 18, S56-S66. 10.1002/mus.880181312 [DOI] [PubMed] [Google Scholar]
- Banerji, C. R. S. (2020). PAX7 target gene repression associates with FSHD progression and pathology over 1 year. Hum. Mol. Genet. 29, 2124-2133. 10.1093/hmg/ddaa079 [DOI] [PubMed] [Google Scholar]
- Banerji, C. R. S., Henderson, D., Tawil, R. N. and Zammit, P. S. (2020). Skeletal muscle regeneration in facioscapulohumeral muscular dystrophy is correlated with pathological severity. Hum. Mol. Genet. 29, 2746-2760. 10.1093/hmg/ddaa164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski, D., Chan, S. S. K., Recht, O. O., Hartweck, L. M., Gustafson, C. J., Athman, L. L., Lowe, D. A. and Kyba, M. (2017). Muscle pathology from stochastic low level DUX4 expression in an FSHD mouse model. Nat. Commun. 8, 550. 10.1038/s41467-017-00730-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosnakovski, D., Shams, A. S., Yuan, C., da Silva, M. T., Ener, E. T., Baumann, C. W., Lindsay, A. J., Verma, M., Asakura, A., Lowe, D. A.et al. (2020). Transcriptional and cytopathological hallmarks of FSHD in chronic DUX4-expressing mice. J. Clin. Invest. 130, 2465-2477. 10.1172/JCI133303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham, M. and Rigby, P. W. (2014). Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell 28, 225-238. 10.1016/j.devcel.2013.12.020 [DOI] [PubMed] [Google Scholar]
- Cacchiarelli, D., Incitti, T., Martone, J., Cesana, M., Cazzella, V., Santini, T., Sthandier, O. and Bozzoni, I. (2011a). miR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. EMBO Rep. 12, 136-141. 10.1038/embor.2010.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacchiarelli, D., Legnini, I., Martone, J., Cazzella, V., D'Amico, A., Bertini, E. and Bozzoni, I. (2011b). miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol. Med. 3, 258-265. 10.1002/emmm.201100133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau, J., Kong, X., Nguyen, N., Williams, K., Tawil, R., Kiyono, T., Mortazavi, A. and Yokomori, K. (2021). Relationship of DUX4 and target gene expression in FSHD myocytes. Hum. Mutat. 42, 421-433. 10.1002/humu.24171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.-F., Tao, Y., Li, J., Deng, Z., Yan, Z., Xiao, X. and Wang, D.-Z. (2010). microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 190, 867-879. 10.1083/jcb.200911036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, J.-H., Dimri, M. and Dimri, G. P. (2015). MicroRNA-31 is a transcriptional target of histone deacetylase inhibitors and a regulator of cellular senescence. J. Biol. Chem. 290, 10555-10567. 10.1074/jbc.M114.624361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, R., Zhang, Y., Khan, D., Heid, B., Caudell, D., Crasta, O. and Ahmed, S. A. (2010). Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PLoS ONE 5, e14302. 10.1371/journal.pone.0014302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Y., Liu, S., Xie, X., Ding, M., Zhou, Q. and Zhou, X. (2019). MicroRNA-31 promotes chondrocyte proliferation by targeting C-X-C motif chemokine ligand 12. Mol. Med. Rep. 19, 2231-2237. 10.3892/mmr.2019.9859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greef, J. C., Lemmers, R. J. L. F., van Engelen, B. G. M., Sacconi, S., Venance, S. L., Frants, R. R., Tawil, R. and van der Maarel, S. M. (2009). Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum. Mutat. 30, 1449-1459. 10.1002/humu.21091 [DOI] [PubMed] [Google Scholar]
- De Iaco, A., Planet, E., Coluccio, A., Verp, S., Duc, J. and Trono, D. (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 49, 941-945. 10.1038/ng.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deenen, J. C. W., Arnts, H., van der Maarel, S. M., Padberg, G. W., Verschuuren, J. J. G. M., Bakker, E., Weinreich, S. S., Verbeek, A. L. and van Engelen, B. G. M. (2014). Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology 83, 1056-1059. 10.1212/WNL.0000000000000797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, I., Eran, A., Nishino, I., Moggio, M., Lamperti, C., Amato, A. A., Lidov, H. G., Kang, P. B., North, K. N., Mitrani-Rosenbaum, S.et al. (2007). Distinctive patterns of microRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. USA 104, 17016-17021. 10.1073/pnas.0708115104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorillo, A. A., Heier, C. R., Novak, J. S., Tully, C. B., Brown, K. J., Uaesoontrachoon, K., Vila, M. C., Ngheim, P. P., Bello, L., Kornegay, J. N.et al. (2015). TNF-α-induced microRNAs control dystrophin expression in becker muscular dystrophy. Cell Rep. 12, 1678-1690. 10.1016/j.celrep.2015.07.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey, K., Currer, J. M. and Harrison, D. E. (2007). The Mouse in Aging Research. Burlington, MA: Elsevier. [Google Scholar]
- Frisullo, G., Frusciante, R., Nociti, V., Tasca, G., Renna, R., Iorio, R., Patanella, A. K., Iannaccone, E., Marti, A., Rossi, M.et al. (2011). CD8+ T cells in facioscapulohumeral muscular dystrophy patients with inflammatory features at muscle MRI. J. Clin. Immunol. 31, 155-166. 10.1007/s10875-010-9474-6 [DOI] [PubMed] [Google Scholar]
- Gabriëls, J., Beckers, M.-C., Ding, H., De Vriese, A., Plaisance, S., van der Maarel, S. M., Padberg, G. W., Frants, R. R., Hewitt, J. E., Collen, D.et al. (1999). Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236, 25-32. 10.1016/S0378-1119(99)00267-X [DOI] [PubMed] [Google Scholar]
- Galagali, H. and Kim, J. K. (2020). The multifaceted roles of microRNAs in differentiation. Curr. Opin. Cell Biol. 67, 118-140. 10.1016/j.ceb.2020.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesige, C. R., Wallace, L. M., Heller, K. N., Eidahl, J. O., Saad, N. Y., Fowler, A. M., Pyne, N. K., Al-Kharsan, M., Rashnonejad, A., Chermahini, G. A.et al. (2018). AAV-mediated follistatin gene therapy improves functional outcomes in the TIC-DUX4 mouse model of FSHD. JCI Insight 3, e123538. 10.1172/jci.insight.123538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco, S., De Simone, M., Colussi, C., Zaccagnini, G., Fasanaro, P., Pescatori, M., Cardani, R., Perbellini, R., Isaia, E., Sale, P.et al. (2009). Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J. 23, 3335-3346. 10.1096/fj.08-128579 [DOI] [PubMed] [Google Scholar]
- Hamanaka, K., Šikrova, D., Mitsuhashi, S., Masuda, H., Sekiguchi, Y., Sugiyama, A., Shibuya, K., Lemmers, R. J. L. F., Goossens, R., Ogawa, M.et al. (2020). Homozygous nonsense variant in LRIF1 associated with facioscapulohumeral muscular dystrophy. Neurology 94, e2441-e2447. 10.1212/WNL.0000000000009617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamrick, M. W., Herberg, S., Arounleut, P., He, H.-Z., Shiver, A., Qi, R.-Q., Zhou, L., Isales, C. M. and Mi, Q.-S. (2010). The adipokine leptin increases skeletal muscle mass and significantly alters skeletal muscle miRNA expression profile in aged mice. Biochem. Biophys. Res. Commun. 400, 379-383. 10.1016/j.bbrc.2010.08.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harafuji, N., Schneiderat, P., Walter, M. C. and Chen, Y.-W. (2013). miR-411 is up-regulated in FSHD myoblasts and suppresses myogenic factors. Orphanet J. Rare Dis. 8, 55. 10.1186/1750-1172-8-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, P., Bomsztyk, K. and Miller, D. G. (2018). Sporadic DUX4 expression in FSHD myocytes is associated with incomplete repression by the PRC2 complex and gain of H3K9 acetylation on the contracted D4Z4 allele. Epigenetics Chromatin 11, 47. 10.1186/s13072-018-0215-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heier, C. R., Zhang, A., Nguyen, N. Y., Tully, C. B., Panigrahi, A., Gordish-Dressman, H., Pandey, S. N., Guglieri, M., Ryan, M. M., Clemens, P. R.et al. (2020). Multi-omics identifies circulating miRNA and protein biomarkers for facioscapulohumeral dystrophy. J. Pers Med. 10, 236. 10.3390/jpm10040236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, P. G., Doráis, J. A., Grow, E. J., Whiddon, J. L., Lim, J.-W., Wike, C. L., Weaver, B. D., Pflueger, C., Emery, B. R., Wilcox, A. L.et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49, 925-934. 10.1038/ng.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydemann, A., Huber, J. M., Demonbreun, A., Hadhazy, M. and McNally, E. M. (2005). Genetic background influences muscular dystrophy. Neuromuscul. Disord. 15, 601-609. 10.1016/j.nmd.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Honda, H., Mano, Y. and Takahashi, A. (1987). Inflammatory changes in affected muscles of facioscapulohumeral dystrophy. J. Neurol. 234, 408-411. 10.1007/BF00314086 [DOI] [PubMed] [Google Scholar]
- Horak, M., Novak, J. and Bienertova-Vasku, J. (2016). Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 410, 1-13. 10.1016/j.ydbio.2015.12.013 [DOI] [PubMed] [Google Scholar]
- Hu, Z., Klein, J. D., Mitch, W. E., Zhang, L., Martinez, I. and Wang, X. H. (2014). MicroRNA-29 induces cellular senescence in aging muscle through multiple signaling pathways. Aging 6, 160-175. 10.18632/aging.100643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W. (2017). MicroRNAs: biomarkers, diagnostics, and therapeutics. Methods Mol. Biol. 1617, 57-67. 10.1007/978-1-4939-7046-9_4 [DOI] [PubMed] [Google Scholar]
- Janghra, N., Morgan, J. E., Sewry, C. A., Wilson, F. X., Davies, K. E., Muntoni, F. and Tinsley, J. (2016). Correlation of utrophin levels with the dystrophin protein complex and muscle fibre regeneration in Duchenne and Becker muscular dystrophy muscle biopsies. PLoS ONE 11, e0150818. 10.1371/journal.pone.0150818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T. and Jones, P. L. (2018). A cre-inducible DUX4 transgenic mouse model for investigating facioscapulohumeral muscular dystrophy. PLoS ONE 13, e0192657. 10.1371/journal.pone.0192657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T. I., Chen, J. C. J., Rahimov, F., Homma, S., Arashiro, P., Beermann, M. L., King, O. D., Miller, J. B., Kunkel, L. M., Emerson, C. P.Jr.. et al. (2012). Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet. 21, 4419-4430. 10.1093/hmg/dds284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T. I., King, O. D., Himeda, C. L., Homma, S., Chen, J. C. J., Beermann, M. L., Yan, C., Emerson, C. P., Jr., Miller, J. B., Wagner, K. R.et al. (2015). Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clin. Epigenetics 7, 37. 10.1186/s13148-015-0072-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T. I., Himeda, C. L., Perez, D. P. and Jones, P. L. (2017). Large family cohorts of lymphoblastoid cells provide a new cellular model for investigating facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 27, 221-238. 10.1016/j.nmd.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T. I., Chew, G.-L., Barraza-Flores, P., Schreier, S., Ramirez, M., Wuebbles, R. D., Burkin, D. J., Bradley, R. K. and Jones, P. L. (2020). Transgenic mice expressing tunable levels of DUX4 develop characteristic facioscapulohumeral muscular dystrophy-like pathophysiology ranging in severity. Skelet. Muscle 10, 8. 10.1186/s13395-020-00227-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. K., Lee, Y. S., Sivaprasad, U., Malhotra, A. and Dutta, A. (2006). Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 174, 677-687. 10.1083/jcb.200603008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsoulidou, A., Mastroyiannopoulos, N. P., Furling, D., Uney, J. B. and Phylactou, L. A. (2011). Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev. Biol. 11, 34. 10.1186/1471-213X-11-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsoulidou, A., Kyriakides, T. C., Papadimas, G. K., Christou, Y., Kararizou, E., Papanicolaou, E. Z. and Phylactou, L. A. (2015). Elevated muscle-specific miRNAs in serum of myotonic dystrophy patients relate to muscle disease progress. PLoS ONE 10, e0125341. 10.1371/journal.pone.0125341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsoulidou, A., Photiades, M., Kyriakides, T. C., Georgiou, K., Prokopi, M., Kapnisis, K., Łusakowska, A., Nearchou, M., Christou, Y., Papadimas, G. K.et al. (2017). Identification of exosomal muscle-specific miRNAs in serum of myotonic dystrophy patients relating to muscle disease progress. Hum. Mol. Genet. 26, 3285-3302. 10.1093/hmg/ddx212 [DOI] [PubMed] [Google Scholar]
- Kowaljow, V., Marcowycz, A., Ansseau, E., Conde, C. B., Sauvage, S., Matteotti, C., Arias, C., Corona, E. D., Nuñez, N. G., Leo, O.et al. (2007). The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 17, 611-623. 10.1016/j.nmd.2007.04.002 [DOI] [PubMed] [Google Scholar]
- Leidinger, P., Backes, C., Deutscher, S., Schmitt, K., Mueller, S. C., Frese, K., Haas, J., Ruprecht, K., Paul, F., Stähler, C.et al. (2013). A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 14, R78. 10.1186/gb-2013-14-7-r78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers, R. J. L. F., van der Vliet, P. J., Klooster, R., Sacconi, S., Camano, P., Dauwerse, J. G., Snider, L., Straasheijm, K. R., van Ommen, G. J., Padberg, G. W.et al. (2010). A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329, 1650-1653. 10.1126/science.1189044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers, R. J., Tawil, R., Petek, L. M., Balog, J., Block, G. J., Santen, G. W. E., Amell, A. M., van der Vliet, P. J., Almomani, R., Straasheijm, K. R.et al. (2012). Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 44, 1370-1374. 10.1038/ng.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N. and Olson, E. N. (2010). MicroRNA regulatory networks in cardiovascular development. Dev. Cell 18, 510-525. 10.1016/j.devcel.2010.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N., Williams, A. H., Maxeiner, J. M., Bezprozvannaya, S., Shelton, J. M., Richardson, J. A., Bassel-Duby, R. and Olson, E. N. (2012). microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J. Clin. Invest. 122, 2054-2065. 10.1172/JCI62656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, G., Wang, Y., Li, Y., Cui, L., Zhao, Y., Zhao, B. and Li, K. (2015). MiR-206, a key modulator of skeletal muscle development and disease. Int. J. Biol. Sci. 11, 345-352. 10.7150/ijbs.10921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaka, Y., Kishi, S., Aoki, Y., Komaki, H., Oya, Y., Takeda, S.-i. and Hashido, K. (2014). Three novel serum biomarkers, miR-1, miR-133a, and miR-206 for limb-girdle muscular dystrophy, facioscapulohumeral muscular dystrophy, and Becker muscular dystrophy. Environ. Health Prev. Med. 19, 452-458. 10.1007/s12199-014-0405-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, J. J. (2008). MicroRNA-206: the skeletal muscle-specific myomiR. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 1779, 682-691. 10.1016/j.bbagrm.2008.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, J. J., Srikuea, R., Kirby, T. J., Peterson, C. A. and Esser, K. A. (2012). Inducible Cre transgenic mouse strain for skeletal muscle-specific gene targeting. Skelet. Muscle 2, 8. 10.1186/2044-5040-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, H., Nakamura, A., Aoki, Y., Ito, N., Kishi, S., Yamamoto, K., Sekiguchi, M., Takeda, S. and Hashido, K. (2011). Identification of muscle-specific microRNAs in serum of muscular dystrophy animal models: promising novel blood-based markers for muscular dystrophy. PLoS ONE 6, e18388. 10.1371/journal.pone.0018388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mul, K., Vincenten, S. C. C., Voermans, N. C., Lemmers, R. J. L. F., van der Vliet, P. J., van der Maarel, S. M., Padberg, G. W., Horlings, C. G. C. and van Engelen, B. G. M. (2017). Adding quantitative muscle MRI to the FSHD clinical trial toolbox. Neurology 89, 2057-2065. 10.1212/WNL.0000000000004647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mul, K., Heatwole, C., Eichinger, K., Dilek, N., Martens, W. B., Van Engelen, B. G. M., Tawil, R. and Statland, J. M. (2018a). Electrical impedance myography in facioscapulohumeral muscular dystrophy: a 1-year follow-up study. Muscle Nerve 58, 213-218. 10.1002/mus.26127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mul, K., Voermans, N. C., Lemmers, R. J. L. F., Jonker, M. A., van der Vliet, P. J., Padberg, G. W., van Engelen, B. G. M., van der Maarel, S. M. and Horlings, C. G. C. (2018b). Phenotype-genotype relations in facioscapulohumeral muscular dystrophy type 1. Clin. Genet. 94, 521-527. 10.1111/cge.13446 [DOI] [PubMed] [Google Scholar]
- Necela, B. M., Carr, J. M., Asmann, Y. W. and Thompson, E. A. (2011). Differential expression of microRNAs in tumors from chronically inflamed or genetic (APCMin/+) models of colon cancer. PLoS ONE 6, e18501. 10.1371/journal.pone.0018501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaru, A. V., Selaru, F. M., Mori, Y., Vazquez, C., David, S., Paun, B., Cheng, Y., Jin, Z., Yang, J., Agarwal, R.et al. (2011). Dynamic changes in the expression of MicroRNA-31 during inflammatory bowel disease-associated neoplastic transformation. Inflamm. Bowel Dis. 17, 221-231. 10.1002/ibd.21359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanet (2021). Prevalence and incidence of rare diseases: Bibliographic data. In Orphanet Report Series: Rare Diseases Collection, p. 35. https://www.orpha.net/orphacom/cahiers/docs/GB/Prevalence_of_rare_diseases_by_alphabetical_list.pdf, Inserm. [Google Scholar]
- Padberg, G. W. (1982). Facioscapulohumeral Disease, PhD thesis, pp. 178-187. Leiden University, Leiden, The Netherlands. [Google Scholar]
- Padberg, G. W., Frants, R. R., Brouwer, O. F., Wijmenga, C., Bakker, E. and Sandkuijl, L. A. (1995). Facioscapulohumeral muscular dystrophy in the Dutch population. Muscle Nerve 18, S81-S84. 10.1002/mus.880181315 [DOI] [PubMed] [Google Scholar]
- Petek, L. M., Rickard, A. M., Budech, C., Poliachik, S. L., Shaw, D., Ferguson, M. R., Tawil, R., Friedman, S. D. and Miller, D. G. (2016). A cross sectional study of two independent cohorts identifies serum biomarkers for facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul. Disord. 26, 405-413. 10.1016/j.nmd.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portilho, D. M., Alves, M. R., Kratassiouk, G., Roche, S., Magdinier, F., de Santana, E. C., Polesskaya, A., Harel-Bellan, A., Mouly, V., Savino, W.et al. (2015). miRNA expression in control and FSHD fetal human muscle biopsies. PLoS ONE 10, e0116853. 10.1371/journal.pone.0116853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard, A. M., Petek, L. M. and Miller, D. G. (2015). Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet. 24, 5901-5914. 10.1093/hmg/ddv315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, T. C., Blomberg, K. E. M., McClorey, G., El Andaloussi, S., Godfrey, C., Betts, C., Coursindel, T., Gait, M. J., Smith, C. I. and Wood, M. J. A. (2012). Expression analysis in multiple muscle groups and serum reveals complexity in the microRNA transcriptome of the mdx mouse with implications for therapy. Mol. Ther. Nucleic Acids 1, e39. 10.1038/mtna.2012.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, T. C., Godfrey, C., McClorey, G., Vader, P., Briggs, D., Gardiner, C., Aoki, Y., Sargent, I., Morgan, J. E. and Wood, M. J. Y. (2013). Extracellular microRNAs are dynamic non-vesicular biomarkers of muscle turnover. Nucleic Acids Res. 41, 9500-9513. 10.1093/nar/gkt724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira-Llopis, S., Escribano-Lopez, I., Diaz-Morales, N., Iannantuoni, F., Lopez-Domenech, S., Andújar, I., Jover, A., Pantoja, J., Pallardo, L. M., Bañuls, C.et al. (2018). Downregulation of miR-31 in diabetic nephropathy and its relationship with inflammation. Cell Physiol. Biochem. 50, 1005-1014. 10.1159/000494485 [DOI] [PubMed] [Google Scholar]
- Schiaffino, S., Gorza, L., Dones, I., Cornelio, F. and Sartore, S. (1986). Fetal myosin immunoreactivity in human dystrophic muscle. Muscle Nerve 9, 51-58. 10.1002/mus.880090108 [DOI] [PubMed] [Google Scholar]
- Sempere, L. F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E. and Ambros, V. (2004). Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5, R13. 10.1186/gb-2004-5-3-r13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Z. G., Sun, Y., Wang, K. S., Jia, J. D., Yang, J. and Li, Y. N. (2019). Effects of miR-26a/miR-146a/miR-31 on airway inflammation of asthma mice and asthma children. Eur. Rev. Med. Pharmacol. Sci. 23, 5432-5440. 10.26355/eurrev_201906_18212 [DOI] [PubMed] [Google Scholar]
- Si, Y., Cui, X., Crossman, D. K., Hao, J., Kazamel, M., Kwon, Y. and King, P. H. (2018). Muscle microRNA signatures as biomarkers of disease progression in amyotrophic lateral sclerosis. Neurobiol. Dis. 114, 85-94. 10.1016/j.nbd.2018.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoelli, M., Mason, A. G., Mul, K., Evangelista, T., Mei, H., Voermans, N., Tapscott, S. J., Tsonaka, R., van Engelen, B. G. M., van der Maarel, S. M.et al. (2020). Evaluation of blood gene expression levels in facioscapulohumeral muscular dystrophy patients. Sci. Rep. 10, 17547. 10.1038/s41598-020-74687-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider, L., Geng, L. N., Lemmers, R. J. L. F., Kyba, M., Ware, C. B., Nelson, A. M., Tawil, R., Filippova, G. N., van der Maarel, S. M., Tapscott, S. J.et al. (2010). Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 6, e1001181. 10.1371/journal.pgen.1001181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statland, J., Donlin-Smith, C. M., Tapscott, S. J., van der Maarel, S. and Tawil, R. (2014). Multiplex screen of serum biomarkers in facioscapulohumeral muscular dystrophy. J. Neuromuscul. Dis. 1, 181-190. 10.3233/JND-140034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statland, J. M., Donlin-Smith, C. M., Tapscott, S. J., Lemmers, R. J. L. F., van der Maarel, S. M. and Tawil, R. (2015a). Milder phenotype in facioscapulohumeral dystrophy with 7-10 residual D4Z4 repeats. Neurology 85, 2147-2150. 10.1212/WNL.0000000000002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statland, J. M., Odrzywolski, K. J., Shah, B., Henderson, D., Fricke, A. F., van der Maarel, S. M., Tapscott, S. J. and Tawil, R. (2015b). Immunohistochemical characterization of facioscapulohumeral muscular dystrophy muscle biopsies. J. Neuromuscul. Dis. 2, 291-299. 10.3233/JND-150077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez, Y., Wang, C., Manes, T. D. and Pober, J. S. (2010). Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J. Immunol. 184, 21-25. 10.4049/jimmunol.0902369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasca, G., Monforte, M., Corbi, M., Granata, G., Lucchetti, D., Sgambato, A. and Ricci, E. (2017). Muscle microdialysis to investigate inflammatory biomarkers in facioscapulohumeral muscular dystrophy. Mol. Neurobiol. 55, 2959-2966. 10.1007/s12035-017-0563-x [DOI] [PubMed] [Google Scholar]
- Tassin, A., Laoudj-Chenivesse, D., Vanderplanck, C., Barro, M., Charron, S., Ansseau, E., Chen, Y.-W., Mercier, J., Coppée, F. and Belayew, A. (2013). DUX4 expression in FSHD muscle cells: how could such a rare protein cause a myopathy? J. Cell. Mol. Med. 17, 76-89. 10.1111/j.1582-4934.2012.01647.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawil, R., Forrester, J., Griggs, R. C., Mendell, J., Kissel, J., McDermott, M., King, W., Weiffenbach, B., Figlewicz, D. and The FSH-DY Group. (1996). Evidence for anticipation and association of deletion size with severity in facioscapulohumerd muscular dystrophy. Ann. Neurol. 39, 744-748. 10.1002/ana.410390610 [DOI] [PubMed] [Google Scholar]
- Toivonen, J. M., Manzano, R., Oliván, S., Zaragoza, P., García-Redondo, A. and Osta, R. (2014). MicroRNA-206: a potential circulating biomarker candidate for amyotrophic lateral sclerosis. PLoS ONE 9, e89065. 10.1371/journal.pone.0089065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonini, M. M. O., Passos-Bueno, M. R., Cerqueira, A., Matioli, S. R., Pavanello, R. and Zatz, M. (2004). Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul. Disord. 14, 33-38. 10.1016/j.nmd.2003.07.001 [DOI] [PubMed] [Google Scholar]
- van den Boogaard, M. L., Lemmers, R. J. L. F., Balog, J., Wohlgemuth, M., Auranen, M., Mitsuhashi, S., van der Vliet, P. J., Straasheijm, K. R., van den Akker, R. F. P., Kriek, M.et al. (2016). Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am. J. Hum. Genet. 98, 1020-1029. 10.1016/j.ajhg.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom, J. C. T., Wijmenga, C., van Tienhoven, E. A. E., Gruter, A.-M., Hewitt, J. E., Padberg, G. W., van Ommen, G.-J. B., Hofker, M. H. and Frants, R. R. (1993). FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 2, 2037-2042. 10.1093/hmg/2.12.2037 [DOI] [PubMed] [Google Scholar]
- van Overveld, P. G. M., Lemmers, R. J. F. L., Sandkuijl, L. A., Enthoven, L., Winokur, S. T., Bakels, F., Padberg, G. W., van Ommen, G.-J. B., Frants, R. R. and van der Maarel, S. M. (2003). Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 35, 315-317. 10.1038/ng1262 [DOI] [PubMed] [Google Scholar]
- Wang, L. H. and Tawil, R. (2016). Facioscapulohumeral dystrophy. Curr. Neurol. Neurosci. Rep. 16, 66. 10.1007/s11910-016-0667-0 [DOI] [PubMed] [Google Scholar]
- Wang, J., Zhang, K.-Y., Liu, S.-M. and Sen, S. (2014). Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules 19, 1912-1938. 10.3390/molecules19021912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. H., Friedman, S. D., Shaw, D., Snider, L., Wong, C.-J., Budech, C. B., Poliachik, S. L., Gove, N. E., Lewis, L. M., Campbell, A. E.et al. (2019). MRI-informed muscle biopsies correlate MRI with pathology and DUX4 target gene expression in FSHD. Hum. Mol. Genet. 28, 476-486. 10.1093/hmg/ddy364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. H., Johnstone, L. M., Bindschadler, M., Tapscott, S. J. and Friedman, S. D. (2021a). Adapting MRI as a clinical outcome measure for a facioscapulohumeral muscular dystrophy trial of prednisone and tacrolimus: case report. BMC Musculoskelet. Disord. 22, 56. 10.1186/s12891-020-03910-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. H., Shaw, D. W. W., Faino, A., Budech, C. B., Lewis, L. M., Statland, J., Eichinger, K., Tapscott, S. J., Tawil, R. N. and Friedman, S. D. (2021b). Longitudinal study of MRI and functional outcome measures in facioscapulohumeral muscular dystrophy. BMC Musculoskelet. Disord. 22, 262. 10.1186/s12891-021-04134-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijmenga, C., Hewitt, J. E., Sandkuijl, L. A., Clark, L. N., Wright, T. J., Dauwerse, H. G., Gruter, A.-M., Hofker, M. H., Moerer, P., Williamson, R.et al. (1992). Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 2, 26-30. 10.1038/ng0992-26 [DOI] [PubMed] [Google Scholar]
- Xu, N., Meisgen, F., Butler, L. M., Han, G., Wang, X.-J., Söderberg-Nauclér, C., Ståhle, M., Pivarcsi, A. and Sonkoly, E. (2013). MicroRNA-31 is overexpressed in psoriasis and modulates inflammatory cytokine and chemokine production in keratinocytes via targeting serine/threonine kinase 40. J. Immunol. 190, 678-688. 10.4049/jimmunol.1202695 [DOI] [PubMed] [Google Scholar]
- Xue, F., Li, H., Zhang, J., Lu, J., Xia, Y. and Xia, Q. (2013). miR-31 regulates interleukin 2 and kinase suppressor of ras 2 during T cell activation. Genes Immunol. 14, 127-131. 10.1038/gene.2012.58 [DOI] [PubMed] [Google Scholar]
- Yi, S. J., Liu, P., Chen, B. L., Ou-Yang, L., Xiong, W. M. and Su, J. P. (2019). Circulating miR-31-5p may be a potential diagnostic biomarker in nasopharyngeal carcinoma. Neoplasma 66, 825-829. 10.4149/neo_2018_181109N847 [DOI] [PubMed] [Google Scholar]
- Yuasa, K., Hagiwara, Y., Ando, M., Nakamura, A., Takeda, S. and Hijikata, T. (2008). MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct. Funct. 33, 163-169. 10.1247/csf.08022 [DOI] [PubMed] [Google Scholar]
- Zanotti, S., Gibertini, S., Blasevich, F., Bragato, C., Ruggieri, A., Saredi, S., Fabbri, M., Bernasconi, P., Maggi, L., Mantegazza, R.et al. (2018). Exosomes and exosomal miRNAs from muscle-derived fibroblasts promote skeletal muscle fibrosis. Matrix Biol. 74, 77-100. 10.1016/j.matbio.2018.07.003 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Ke, F., Liu, Z., Bai, J., Liu, J., Yan, S., Xu, Z., Lou, F., Wang, H., Zhu, H.et al. (2015). MicroRNA-31 negatively regulates peripherally derived regulatory T-cell generation by repressing retinoic acid-inducible protein 3. Nat. Commun. 6, 7639. 10.1038/ncomms8639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Chen, M., Lian, D., Li, Y., Wang, J., Deng, S., Yu, K. and Lian, Z. (2019). Non-coding RNA regulates the myogenesis of skeletal muscle satellite cells, injury repair and diseases. Cells 8, 988. 10.3390/cells8090988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Zhang, S., Li, Z., Wang, H., Li, Z., Hu, Y., Chen, H., Zhang, X., Cui, L., Zhang, J.et al. (2019). miR-125b-5p and miR-99a-5p downregulate human γδ T-cell activation and cytotoxicity. Cell Mol. Immunol. 16, 112-125. 10.1038/cmi.2017.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.