Abstract
Objectives
The Human Telomerase enzyme has become a drug target in the treatment of cancers and age-related disorders. This study aims to identify potential natural inhibitors of the Human Telomerase from compounds derived from edible African plants.
Materials and methods
A library of 1,126 natural compounds was molecularly docked against the Telomerase Reverse Transcriptase (PDB ID: 5ugw), the catalytic subunit of the target protein. Curcumin, a known Telomerase inhibitor was used as the standard. The front-runner compounds were screened for bioavailability, pharmacokinetic properties, and bioactivity using the SWISSADME, PKCSM, and Molinspiration webservers respectively. The molecular dynamic simulation and analyses of the apo and holo proteins were performed by the Galaxy supercomputing webserver.
Results
The results of the molecular docking and virtual screening reveal Augustamine and Camptothecin as lead compounds. Augustamine has better drug-likeness and pharmacokinetic properties while Camptothecin showed better bioactivity and stronger binding affinity (-8.2 kcal/mol) with the target. The holo structure formed by Camptothecin showed greater inhibitory activity against the target with a total RMSF of 169.853, B-Factor of 20.164, and 108 anti-correlating residues.
Conclusion
Though they both act at the same binding site, Camptothecin induces greater Telomerase inhibition and better molecular stability than the standard, Curcumin. Further tests are required to investigate the inhibitory activities of the lead compounds.
Keywords: Telomerase, Molecular dynamic simulation, Pharmacokinetic, Augustamine, Camptothecin
Telomerase, Molecular dynamic simulation, Pharmacokinetic, Augustamine, Camptothecin.
1. Introduction
Immortality is a common distinguishing feature of cancers, and the overexpression of Telomerase has been associated with the hyperproliferation of tumor cells [1]. The replicative capabilities of cells are essentially associated with the lengthening of telomeres which are found at the end of chromosomes. Telomerase is a ribonucleoprotein polymerase that maintains the telomere ends by adding the repetitive DNA sequences, TTAGGG [2]. The Telomerase complex mainly consists of a catalytic subunit, human Telomerase Reverse Transcriptase (hTERT) and a Telomerase RNA component (TERC) [3].
Due to the inability of DNA to replicate fully its end during replication, the telomere length decreases gradually after every cell division [5]. The correlation between ageing and Telomerase is also confirmed by the evidence that fibroblast telomeres shortened with increasing age until it reaches a point where cells experience a non-proliferative status otherwise known as replicative senescence. Replicative senescence has now been associated with the ageing process [4]. The abnormal lengthening of the telomeres can result in aberrant cell replication mechanisms resulting in conditions such as premature ageing and cancer [5]. Cancer cells have an increased Telomerase activity more than normal somatic cells as seen in prostate carcinoma [6], kidney carcinoma [7], colorectal carcinoma [8], thyroid carcinoma [9], bladder carcinoma [10, 11]. This is due to the upregulation of the hTERT [12].
Consequently, Telomerase has become a potential therapeutic target. Studies have shown that the inhibition of Telomerase, as a form of therapeutic intervention, is considerably sufficient in tackling malignancies and many age-related diseases [4]. Specifically, targeting the hTERT catalytic activity has shown good promise and some reported inhibitors with relative clinical effectiveness includes Nucleoside reverse transcriptase inhibitors (NRTI), Non-nucleoside reverse transcriptase inhibitors (NNRTI), BIBR1532, GRN163L (imtelstat), and some selected Tyrosine Kinase Inhibitors (TKI) [13].
There are also a variety of plant products and plants secondary metabolites that have been used as Telomerase inhibitors. These include sesquiterpene, polyphenols, and alkaloids [14]. Curcumin, a polyphenolic compound, can be found in the Turmeric spice and has been revealed to have telomerase inhibitory activity. In a dose-dependent manner, Curcumin directly inhibited the proliferation of human cancer cell lines (Bel7402, SGC7901 and HL60) by suppressing telomerase activity [15].
As a potential source of cancer therapy, plants consumed as part of human diet contain rich bioactive phytoconstituents which have fewer or no side-effects, easier availability, and cheaper cost than conventional anticancer drugs [16]. However, most African vegetables, spices, and fruits have not been explored and profiled for their anti-telomerase activity; they could be the key to the discovery of potential drug candidates that would serve as telomerase inhibitory agents. Therefore, more research work is required to uncover more natural products with anti-telomerase activity. Consequently, this study aims to identify leads from a library of natural compounds derived from edible African plants that can directly inhibit the hTERT catalytic subunit of the holoenzyme, using in silico/structural-based methodologies. Curcumin was used as a reference compound.
2. Materials and methods
2.1. Analysis and validation of the Human Telomerase structure
The 3D structure of the target protein, the Human Telomerase thumb domain (PDBID: 5ugw), was downloaded from the Protein Data Bank [17]. The water molecules and co-crystallized ligand were removed from the target protein using the visualization software, PyMol [18]. The freed protein was minimized using Chiron [19]. The architecture was analysed using the web server VADAR 1.8 [20], and the Ramachandran plot determined from the MolProbity web server [21].
2.2. Preparation of ligands
From the PubChem database [22], a group of 1,126 compounds derived mainly from plants such as vegetables, spices, and fruits were downloaded. The compounds had been previously screened for Lipinski rule (log P value ≤5, hydrogen bond acceptor (HBA) value ≤10, molecular weight value ≤500, and hydrogen bond donor (HBD) value ≤ 5), and Veber rule (polar surface area (PSA) ≤ 140, and rotatable bonds value ≤10 [23]. The 3D configurations of all the compounds, and that of the standard, Curcumin (PubChem CID: 969516) were obtained from PubChem in the structure-data file (sdf) format.
2.3. Molecular docking of ligands against Human Telomerase and virtual screening
For docking, the Open Babel plug-in tool of the PyRx (version 0.8) software was used to upload ligands and changed from sdf to Partial Charge, Protein Data Bank, and Atom Type (pdbqt) format [24]. For conformational stability, the energy minimisation parameter was represented as Universal Force Field (UFF) and optimisation algorithm represented as conjugate gradient descent. The standard and the ligands were docked against the targeted protein by the PyRx's AutoDock Vina plug-in tool using the following grid parameters: Centre X = -50.955, Y = 42.898, Z = 162.221 and Dimensions (Angstrom): X = 49.829, Y = 27.918, Z = 20.524. Using the Microsoft Excel software, the docked results were exported in comma-separated values (csv) format and screened using the docking score of Curcumin (-7.1 kcal/mol) as cut-off. The SWISSADME server was used to predict the bioavailability of the compounds. The pkCSM and Molinspiration webservers were used to calculate the pharmacokinetic properties and bioactivity of the ligands respectively [25, 26]. The SMILES for the ligand and standard, Curcumin were obtained from PubChem.
2.4. Binding site analyses
The PyMol software was utilised to superimpose the docked poses of all the front-runner compounds with the target protein. The resultant protein-ligand complexes were evaluated for hydrogen bonds, salt bridges, and other interactions by using the Protein-Ligand Interaction Profiler (PLIP) webserver [27]. The bond distances, exhaustiveness, bond angles, names, and numbers of the residues were also included in the analyses. The Fpocket web server was employed in analysing the location and parameters of the binding pockets of the target protein [28].
2.5. Molecular dynamic simulations (MDS) and analyses
Versions 2019.1 and 2019.1.4 of the GROMACS [29] software on the Galaxy server [30] were used to perform a 2-nanoseconds MDS of the holo and apo structures of the Telomerase. GROMACS-compatible topology files for small molecules were generated by the LigParGen server [31] for ligand parameterisation. The force field parameter used was OPLS-AA/1.14∗CM1A. A 1,000,000-step MDS was carried out following the initial minimisation and equilibration of energy (NVT and NPT), solvation, and conversion of topology files. The Galaxy supercomputing BIO 3D tool was used to analyse the trajectories which include the Dynamical Cross-Correlation Matrix (DCCM), Root Mean Square Deviation (RMSD) of atomic positions, the per residue Root Mean Square Fluctuation (RMSF) of the protein backbone, and the Principal Component Analysis (PCA) [30]. Analysis of the B factor was carried out by the MDWeb webserver [32].
3. Results
3.1. Analysis and validation of the Human Telomerase structure
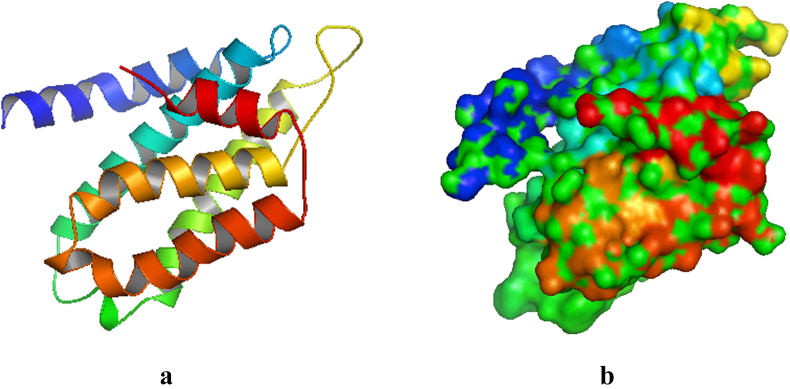
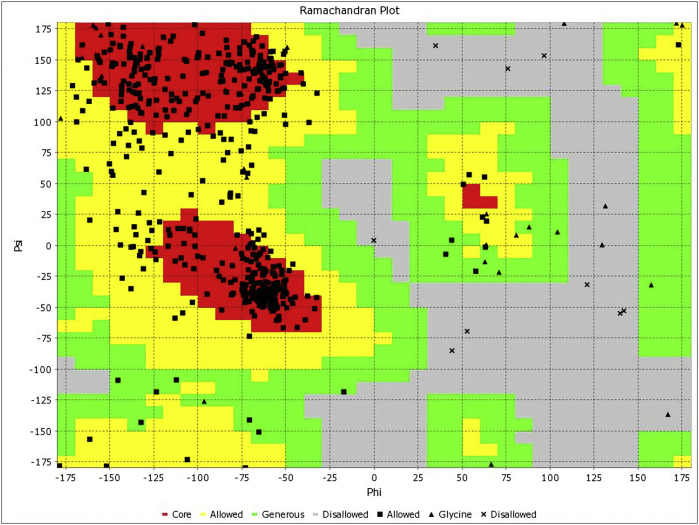
The architectural confirmation of the targeted Human Telomerase protein prior to docking exercise is hereby presented. The crystal structure of Human Telomerase (PDB ID 5ugw) has 158 amino acids with the following X ray diffraction data: Resolution: 2.31 Å, R-Value Free: 0.209, and R-Value Work: 0.197. The length of the unit cell is a = 92.425Å, b = 92.425Å, and c = 50.178Å with the following angles: α = 90°, β = 90°, and γ = 120°. The secondary structure has the following constituents: α helix 74%, beta sheets 0%, Coil 25%, and Turns 27% (Figure 1). The Total Accessible Solvent Area (ASA) is 9551.3 (Å) [2]. The geometry of Human Telomerase (PDB ID 5ugw) reveals 2.26% poor rotamers, 89.47% favoured rotamers, 0.00% Ramachandran outliers, 96.79% Ramachandran favoured, 1.33% Carbon Beta deviations (>0.25Å), 0.00% bad bonds, and 0.06% bad angles (Figure 2). The Peptide omegas of Human Telomerase (PDB ID 5ugw) include 0.00% Cis Prolines, and the low-resolution criteria include 0.6% CaBLAM outliers and 0.65% CA Geometry outliers.
Figure 1.
Cartoon model of the crystal structure of Human Telomerase (PDB ID: 5ugw). b: Surface representation.
Figure 2.
Ramachandran plot for Human Telomerase (PDB ID: 5ugw).
With respect to drug-likeness, the values of the molecular descriptors of the standard and lead compounds were within acceptable thresholds. They include molecular weight ≤500 g/mol, hydrogen bond donors ≤5, hydrogen bond acceptors ≤10, log P ≤ 5, molar refractivity between 40 and 130, the number of rotatable bonds ≤10, and the topological polar surface area (TPSA) ≤ 140. The standard has higher values XLogP3 than the lead compounds. Augustamine has the lowest TPSA value. All the compounds are predicted to have no PAIN alerts. However, Curcumin has a saturation value less than 0.25. For enzyme inhibition, the bioactivity scores of all the compounds are greater than 0.00 while Camptothecin has the highest value.
3.2. Pharmacokinetic properties of ligands
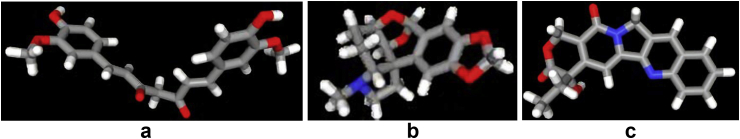
The properties of the ligands targeting the Human Telomerase protein are enumerated in Figure 3. The standard and lead compounds have skin permeability (log Kp) values less than -2.5; and human intestinal absorption (%) values greater than 30%. However, the predicted Caco2 permeability (log Papp in 10–6 cm/s) value of the standard is less than 0.9; and its water solubility (log mol/L) value is less than -4.0. The lead compounds are non-inhibitors of P-glycoprotein II while the standard is predicted to be an inhibitor of this protein. The standard and Camptothecin are P-glycoprotein substrates and non-inhibitors of P-glycoprotein I but Augustamine is not.
Figure 3.
The 3D chemical structures (stick model) of standard and lead compounds a: Curcumin b: Augustamine c: Camptothecin.
For drug distribution properties, the VDss (log L/kg) value for Augustamine is higher than 0.45, Camptothecin is slightly lower than 0.45, and the standard is predicted to be lower than -0.15. The predicted value of Fraction unbound for the standard is less than 0.1 while those of Augustamine and Camptothecin are higher than 0.1. The predicted values of CNS permeability (log PS) in the lead compounds and the standard are greater than -3.0 but less than -2.0. The predicted values of BBB permeability (log BB) for the lead compounds, and the standard are less than 0.3 and greater than -1.0.
For the parameters of drug metabolism, the two lead compounds, and the standard are predicted to be substrates of CYP3A4, non-substrates of CYP2D6, and non-inhibitors of CYP2D6. Unlike the standard, the two lead compounds are non-inhibitors of CYP3A4, and CYP2C19. Unlike Augustamine, the standard and Camptothecin are inhibitors of the CYP2C9, and CYP1A2 enzymes.
For the drug excretion profile, the value for total clearance (log ml/min/kg) is predicted to be highest in Augustamine and the lowest in the standard. Only Camptothecin is predicted to be a Renal OCT2 substrate.
The standard, and the two lead compounds are predicted to have no AMES toxicity, no hERG I & II inhibition, and no skin sensitization. Similarly, the value for the maximum tolerated dose (log mg/kg/day) is predicted to be lower than 0.477, and the T. Pyriformis toxicity values (log μg/L) greater than -0.5 in all the compounds. Only Camptothecin is predicted to be hepatotoxic with a minnow toxicity (log mM) value less than -0.3. Augustamine has the highest value of Oral Rat Acute Toxicity (LD50) (mol/kg).
The evaluation of the pharmacokinetics properties suggests that the lead compounds are good drug candidates.
3.3. Molecular docking of ligands against Human Telomerase
With a binding affinity score of -8.2 kcal/mol, Camptothecin showed the least free energy when bound with the target protein (Table 1). The docking results suggest that Camptothecin is potentially a good inhibitor of the Human Telomerase.
Table 1.
Molecular docking scores of ligands against Human Telomerase.
| Ligand | Binding (Kcal/mol) affinity |
|---|---|
| Curcumin (standard) | -7.1 |
| Augustamine | -7.5 |
| Camptothecin | -8.2 |
3.4. Binding site analyses
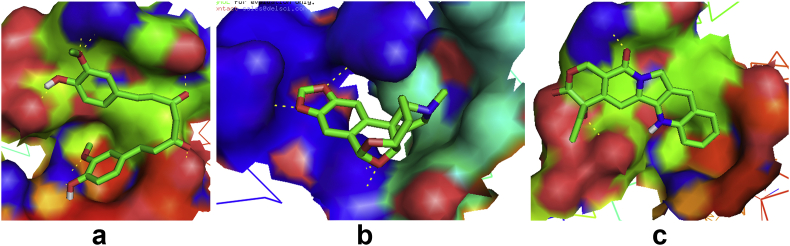
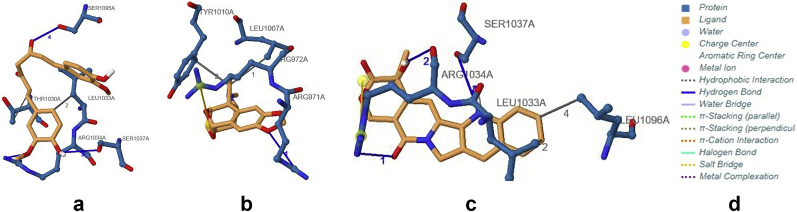
The analysis of the target –ligand interactions at the binding pockets are presented. From Figures 4 and 5, and Table 2, results reveal that Curcumin and Camptothecin interact with the protein through the same binding pocket. Both compounds each have four intermolecular hydrogen bonds in this binding pocket. However, only Camptothecin forms a bond angle greater than 130° at SER1037A. Augustamine binds in a different pocket and has only one intermolecular hydrogen bond at ARG971A with an angle less than 130°. Regarding the donor to acceptor distance, all intramolecular hydrogen bonds of the compounds are fall within a range of 3.2–4.0 Å except the interaction of Curcumin at SER1095A which is within the range of 2.5–3.2 Å.
Figure 4.
Binding site of Human Telomerase interacting with standard and lead compounds a: hTERT-Curcumin complex b: hTERT-Augustamine c: hTERT-Camptothecin complex.
Figure 5.
Protein-Ligand interactions of Human Telomerase with standard and lead compounds a: hTERT-Curcumin complex b: hTERT-Augustamine c: hTERT-Camptothecin complex.
Table 2.
Hydrogen bond analysis of Human Telomerase with standard and lead compounds.
| Complex | Number of bonds | Residues | Distance (H-A) | Distance (D-A) | Bond angle |
|---|---|---|---|---|---|
| hTERT-Curcumin | 4 | ARG1034A | 2.71 | 3.29 | 117.72 |
| SER1037A | 2.87 | 3.52 | 124.74 | ||
| SER1037A | 2.98 | 3.52 | 116.18 | ||
| SER1095A | 2.23 | 2.86 | 121.31 | ||
| hTERT-Augustamine | 1 | ARG971A | 2.97 | 3.65 | 126.6 |
| hTERT-Camptothecin | 4 | ARG1034A | 2.69 | 3.3 | 120.01 |
| ARG1034A | 3.15 | 3.86 | 129.8 | ||
| SER1037A | 3.69 | 4.06 | 105.3 | ||
| SER1037A | 3.08 | 4.06 | 161.49 |
From Table 3, hTERT-Camptothecin complex has four hydrophobic interactions and one salt bridge; hTERT-Curcumin complex has three hydrophobic interactions and no salt bridge; and the hTERT-Augustamine complex has two hydrophobic interactions and one salt bridge.
Table 3.
Other Protein-ligand interactions.
| Complex | Hydrophobic Int. |
Salt bridge |
p-Stacking |
|||
|---|---|---|---|---|---|---|
| Residue | Distance | Residue | Distance | Residue | Distance | |
| hTERT -Curcumin | THR1030A | 3.64 | ||||
| LEU1033A | 3.89 | |||||
| LEU1033A | 3.92 | |||||
| hTERT-Augustamine | LEU1007A | 3.95 | ARG972A | 4.54 | ||
| TYR1010A | 3.62 | |||||
| hTERT -Camptothecin | LEU1033A | 3.7 | ||||
| LEU1033A | 3.58 | |||||
| ARG1034A | 3.74 | ARG1034A | 4.75 | |||
| LEU1096A | 3.87 | |||||
3.5. Molecular dynamic simulations (MDS) and analyses
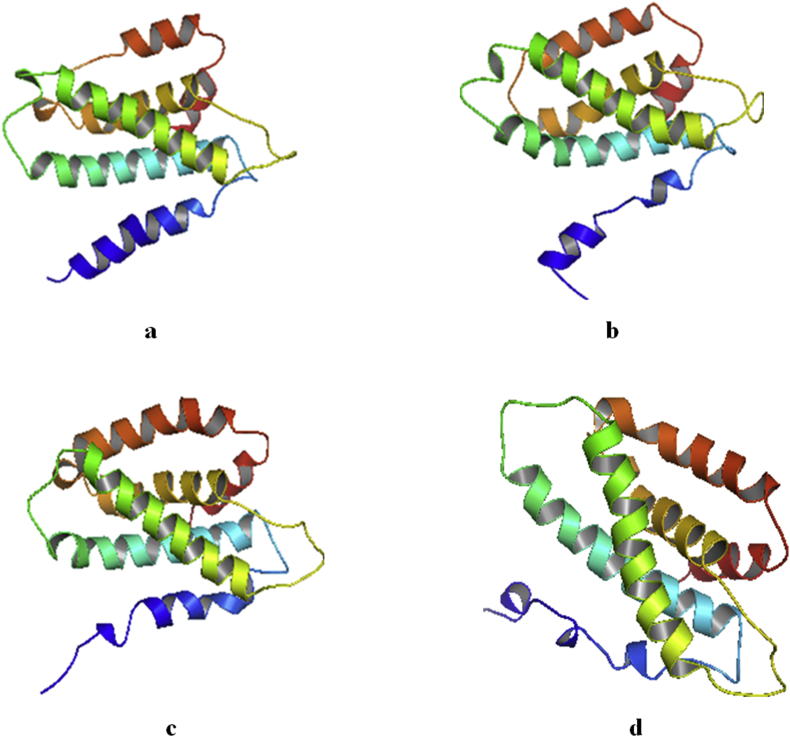
The MDS of the holo and apo structures of the Telomerase are presented. Figure 6 showed a distortion of the alpha helix of the holo structure of the hTERT-Camptothecin complex. A less severe distortion is also seen in the hTERT-Curcumin complex.
Figure 6.
Cartoon model of the crystal structure of Human Telomerase Apo and Holo structures (without water and ions) after molecular dynamics simulation. Beta sheets (yellow), Alpha helix (red) and Loops (green) a: hTERT b: hTERT-Curcumin complex c: hTERT-Augustamine complex d: hTERT-Camptothecin complex.
3.5.1. Root Mean Square Deviation of atomic positions (RMSD)
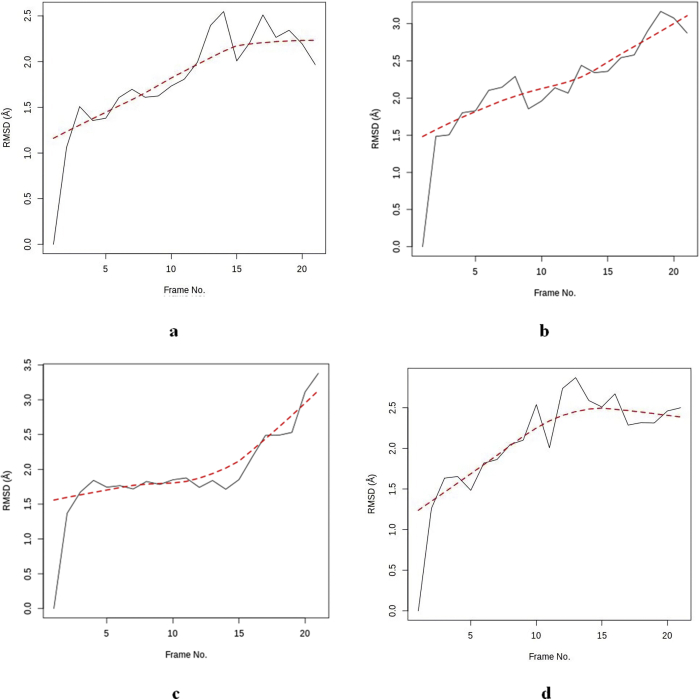
From the RMSD of all the holo structures, the hTERT-Augustamine complex has the least total and average RMSD values suggesting the least structural distortion during the simulation period (Table 4). However, from Figure 7, the RMSD slopes of hTERT-Augustamine and hTERT- Curcumin complexes are steep suggesting that the RMSD values will increase with more simulation time. This is also revealed in the time frame of the highest RMSD which is 21 and 19 respectively. The slope of the hTERT-Camptothecin complex suggests a downward trend in values with more simulation time as the time frame of highest RMSD is 13.
Table 4.
Summary of data from Molecular Dynamics Simulations of apo and holo structures of Human Telomerase.
| RMSD | hTERT-Apo | hTERT-Curcumin | hTERT-Augustamine | hTERT-Camptothecin |
|---|---|---|---|---|
| Total RMSD | 37.815 | 45.452 | 40.772 | 43.652 |
| Average RMSD | 1.801 | 2.164 | 1.942 | 2.079 |
| Lowest RMSD | 0 | 0 | 0 | 0 |
| Highest RMSD | 2.548 | 3.165 | 3.38 | 2.87 |
| Time Frame of Highest RMSD | 14 | 19 | 21 | 13 |
| Time Frame of Lowest RMSD | 1 | 1 | 1 | 1 |
| RMSD Peak Distribution | ||||
| 0.00–0.49A | 1 | 1 | 1 | 1 |
| 0.50–0.99A | 0 | 0 | 0 | 0 |
| 1.00–1.49A | 3 | 1 | 1 | 2 |
| 1.50–1.99A | 9 | 5 | 13 | 4 |
| 2.00–2.49A | 6 | 8 | 3 | 8 |
| 2.50–2.99A | 2 | 4 | 1 | 6 |
| 3.00–3.49A | 0 | 2 | 2 | 0 |
| 3.50–3.99 | 0 | 0 | 0 | 0 |
| 4.00–4.49 | 0 | 0 | 0 | 0 |
| 4.50–4.99 | 0 | 0 | 0 | 0 |
| 5.00–5.49 | 0 | 0 | 0 | 0 |
| RMSF | ||||
| Total Global RMSF | 167.828 | 174.503 | 188.648 | 169.853 |
| Average Global RMSF | 1.062 | 1.104 | 1.194 | 1.075 |
| Least Fluctuation | 0.413 | 0.375 | 0.404 | 0.387 |
| Highest Fluctuation | 5.191 | 8.189 | 6.749 | 5.012 |
| Range of RMSF | 4.778 | 7.814 | 6.345 | 4.625 |
| B Factor | ||||
| Global Average B Factor |
256.14 |
270.7 |
325.433 |
207.164 |
|
PCA |
Total (Mean) |
Total (Mean) |
Total (Mean) |
Total (Mean) |
| Total global motions (PC1-3) | 28.63 (9.543) | 26.786 (8.929) | 29.511 (9.837) | 27.583 (9.194) |
| Average global motions (PC1-3) | 0.181 (0.06) | 0.17 (0.057) | 0.188 (0.063) | 0.171 (0.057) |
| PC1 Eigenvalue (%) | 47.91 | 49.64 | 53.39 | 31.7 |
| PC2 Eigenvalue (%) | 12.28 | 12.25 | 11.28 | 28.79 |
| PC3 Eigenvalue (%) | 7.77 | 7.34 | 8.24 | 10.21 |
| Total (%) | 67.96 | 69.23 | 72.91 | 70.7 |
| DCC Analysis | ||||
| Total No of anti-correlating residues | 110 | 96 | 97 | 108 |
| Number of H Bonds per cycle | ||||
| Average | 124.68 | 131.23 | 124.41 | 127.09 |
| Maximum no of H bonds | 145 | 148 | 145 | 142 |
| Minimum no of bonds | 102 | 99 | 109 | 109 |
| Range | 43 | 49 | 36 | 33 |
| H Bond Occupancy | ||||
| No of bonds Found | 2309 | 2364 | 2235 | 2287 |
| Donor | SOL488-Side | SOL537-Side | SOL955-Side | SOL490-Side |
| Acceptor | ASP988-Side | ASP988-Side | ALA1077-Main | ASP988-Side |
| % Occupancy | 81.82% | 86.36% | 59.09% | 86.36% |
Figure 7.
RMSD for Apo and Holo structures a: hTERT b: hTERT-Curcumin complex c: hTERT-Augustamine complex d: hTERT-Camptothecin complex.
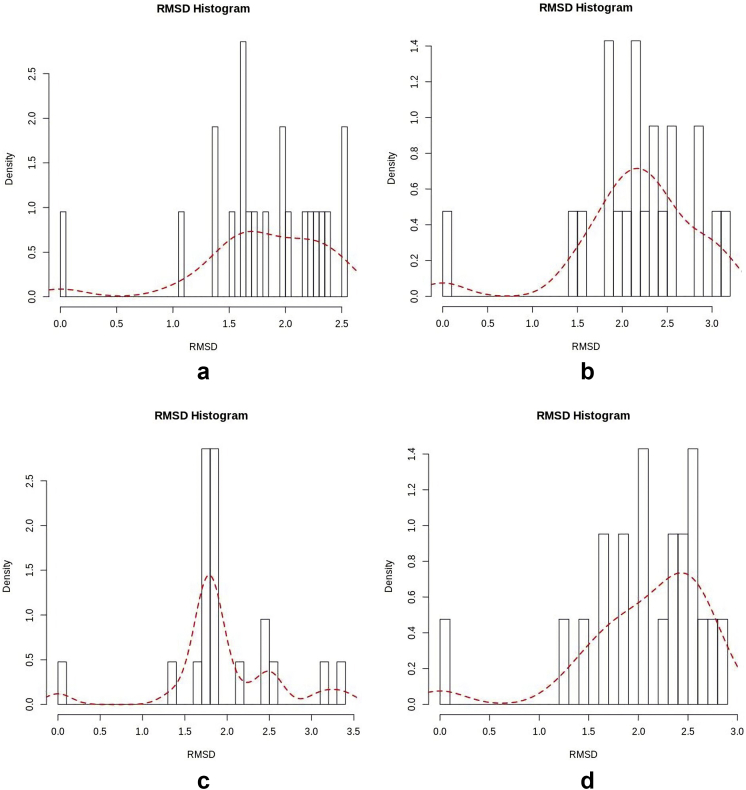
From Figure 8 and Table 4, the distribution of RMSD values of the Apo and holo structures reveals that all the 21 peaks of the hTERT-Camptothecin complex are skewed within 0.00–3.00Å. Within this range, the hTERT-Curcumin and hTERT-Augustamine complexes each have 19 peaks. The remaining 2 peaks are found within 3.00–3.49Å. This distribution pattern suggests that the hTERT-Camptothecin complex has the least deviation to the right from its reference structure. In this regard, the hTERT-Augustamine complex has more peaks than hTERT-Curcumin below 2.5Å.
Figure 8.
RMSD histogram for Apo and Holo structures a: hTERT b: hTERT-Curcumin complex c: hTERT-Augustamine complex d: hTERT-Camptothecin complex.
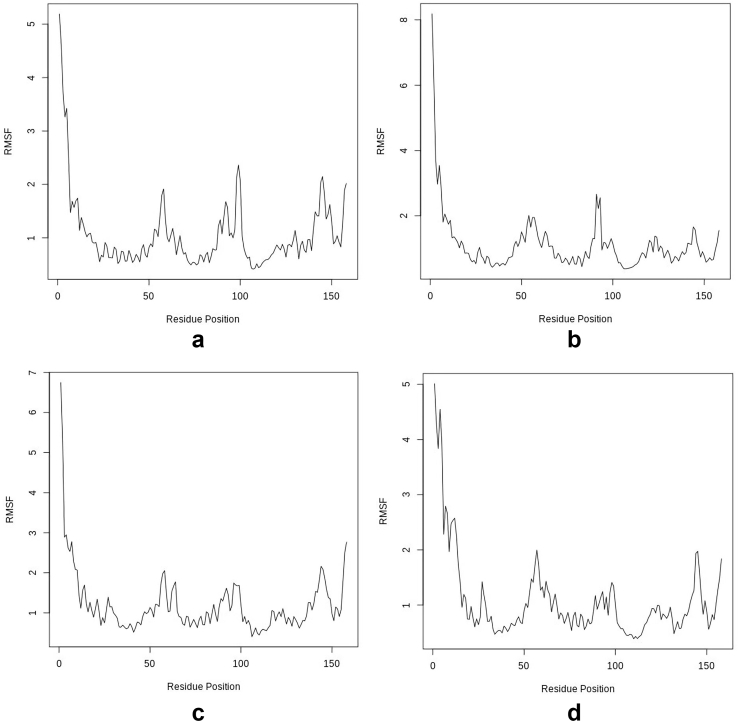
3.5.2. Root Mean Square Fluctuation (RMSF)
From Figure 9 and Table 4, the total and average global RMSF are the least in the hTERT-Camptothecin complex, followed by the hTERT-Curcumin complex. In a similar vein, the least fluctuation and least range of RMSF were found in the hTERT-Camptothecin complex and this is followed by the hTERT-Augustamine complex. The hTERT-Curcumin complex (Standard) had the highest values. The greatest fluctuations are seen at the N-termini of the apo and holo structures. The fluctuation pattern of the hTERT-Camptothecin complex most resembles the apo structure.
Figure 9.
Per-residue RMSF for Apo and Holo structures a: hTERT b: hTERT-Curcumin complex c: hTERT-Augustamine complex d: hTERT-Camptothecin complex.
3.5.3. B-factor
From Table 4, the hTERT-Camptothecin complex had the least average B factor value of all the holo structures followed closely by the hTERT-Curcumin complex.
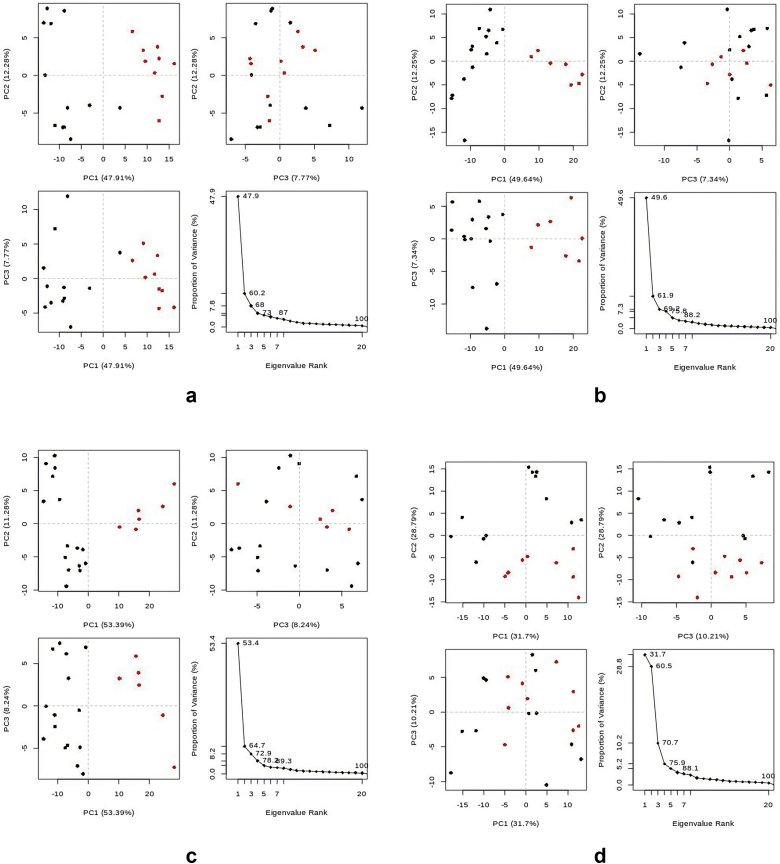
3.5.4. Principal components (PC) analysis
From Table 4 and Figure 10, the hTERT-Curcumin complex has the least total global motions of all the holo structures, and this is followed closely by the hTERT-Camptothecin complex. However, the values of the average global motions for both complexes were the same and less than that of the hTERT-Augustamine complex. Specifically, based on the least motions, the best global conformations are PC2 of the Apo protein, and PC1 for all the holo structures (Supplementary data).
Figure 10.
Principal component analysis cluster plot of Apo and Holo structures. The projection of trajectory onto 1st few eigenvectors for: a: hTERT b: hTERT-Curcumin complex c: hTERT-Augustamine complex d: hTERT-Camptothecin complex.
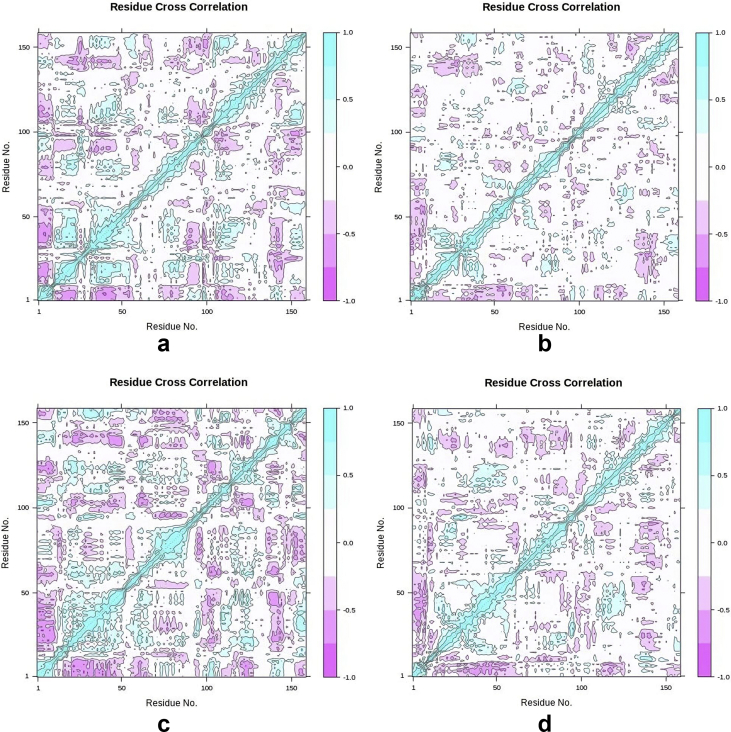
3.5.5. The dynamic cross-correlation (DCC) analysis
From the DCC data (Figure 11, Table 4, & supplementary data) of the apo and holo structures, there are three major sites of anti-correlation which include residues 973 to 1064; residues 1068 to 1099; and residues 1104 to 1121 covering about 88.5% of the target protein. Of all the holo structures, the hTERT-Camptothecin complex had the highest number of anti-correlating residues.
Figure 11.
Dynamic cross correlation map apo and holo structures of SARS-CoV-2 2′ OMT. Purple represents anti-correlated, dark cyan represents fully correlated while white and cyan represent moderately and uncorrelated respectively. 1.0 = correlated; 0 is non-correlated; and -1 is anti-correlated. a: hTERT b: hTERT-Curcumin complex c: hTERT-Augustamine complex d: hTERT-Camptothecin complex.
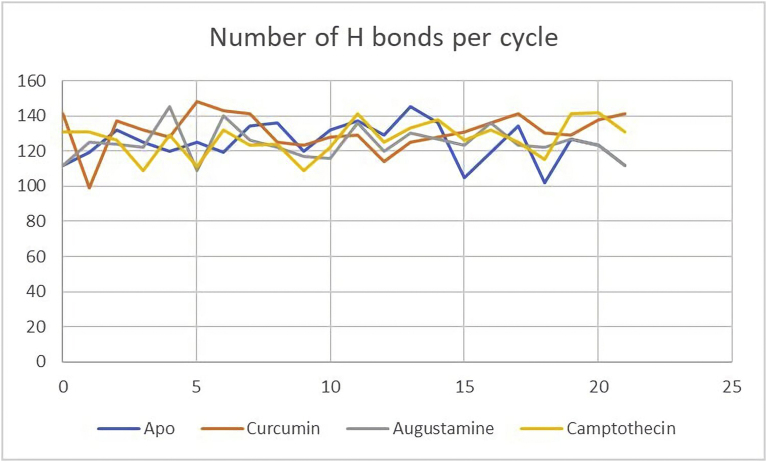
3.5.6. Hydrogen bond analysis
Residue ASP988 provided the acceptor atom for the hydrogen bond with the highest percentage occupancy in the apo protein (81.82%), hTERT-Curcumin (86.36%), and the hTERT-Camptothecin (86.36%) complexes. In the hTERT-Augustamine complex, the residue ASP988 provided the acceptor atom for the hydrogen bond with the third highest percentage occupancy (45.45%). The holo structures each had two residues that formed hydrogen bonds with percentage occupancy greater than 50% (Supplementary data). From Table 4, the highest occupancy occurred at the hTERT-Curcumin and the hTERT-Camptothecin complexes. As seen in Table 4 and Figure 12 which show the number of Hydrogen bonds per frame, the hTERT-Camptothecin complex has the least range of hydrogen bond fluctuations.
Figure 12.
Hydrogen Bond Stability during MDS. hTERT Apo (Blue), hTERT-Curcumin complex (Red), hTERT-Augustamine complex (Grey), hTERT-Camptothecin complex (Yellow).
4. Discussion
4.1. Chemoinformatic profile of ligands (Figure 3, Table 5)
Table 5.
Chemo-informatic properties of standard and lead compounds.
| Curcumin (standard) | Augustamine | Camptothecin | |
|---|---|---|---|
| Molecular Weight (g/mol) | 368.4 | 301.34 | 348.4 |
| XLogP3 | 3.2 | 1.8 | 1 |
| Hydrogen Bond Donors | 2 | 0 | 1 |
| Hydrogen bond acceptors | 6 | 5 | 5 |
| # heavy atoms | 27 | 22 | 26 |
| # rotatable bonds | 8 | 0 | 1 |
| TPSA (angstrom) | 93.1 | 40.2 | 79.7 |
| Molar Refractivity | 102.80 | 81.30 | 95.31 |
| Saturation (fraction csp3) | 0.14 | 0.65 | 0.25 |
| PAIN Alert | 0 | 0 | 0 |
| GCPR ligand | -0.06 | 0.36 | 0.46 |
| Ion channel modulator | -0.20 | 0.35 | -0.15 |
| Kinase Inhibitor | -0.26 | 0.02 | 0.27 |
| Nuclear Receptor Ligand | 0.12 | -0.07 | 0.07 |
| Protease Inhibitor | -0.14 | -0.01 | -0.10 |
| Enzyme Inhibitor | 0.08 | 0.18 | 1.11 |
The oral bioavailability of a drug determines its penetration into the biological target and consequently its efficacy [33]. In the field of medicinal chemistry, certain guidelines are applied in determining good oral bioavailability and they include the Ghose, Lipinski, and Veber rules. The molecular descriptors of these drug-likeness rules put together include a molar refractivity between 40 to 130, log P ≤ 5, hydrogen bond donors ≤5, hydrogen bond acceptors ≤10, molecular weight ≤500 g/mol, polar surface area (PSA) ≤ 140, and the number of rotatable bonds ≤10 [34].
Table 5 reveals that the standard and all lead compounds did not violate the Ghose, Lipinski, and Veber rules suggesting that they all have good oral bioavailability and are therefore good drug candidates [35]. The standard has the highest Log P value signifying that it is the most lipophilic of all the compounds. All the compounds are predicted to be non-promiscuous as they showed no PAIN alert. This suggests that these compounds do not have problematic structural moieties and are less likely to aggregate in biological assays giving false positive results. This minimizes the risk of unintended interactions with other targets leading to undesirable side effects [36]. The standard, Curcumin, has a saturation value less than the 0.25 threshold [37] It is considered to have the least molecular stability of all the compounds as revealed in the ratio of sp hybridized carbons to the total carbon count (Fraction Csp3) [37]. Augustamine has the lowest TPSA value suggesting that it would have the highest blood-brain barrier permeation and intestinal absorption of all the compounds [38].
Based on bioactivity, drugs are classified into six major classes which include kinase inhibitors, protease inhibitors, enzyme inhibitors, GPCR ligands, ion channel modulators, and nuclear receptor ligands [26]. A bioactivity score less than -5.0 implies the drug or compound is inactive; between −5.0 and 0.0 is moderately active; and greater than 0.00 is considered active [26]. All the compounds are active enzyme inhibitors with Camptothecin showing the highest activity [26].
4.2. Pharmacokinetic properties of ligands
Good in silico ADMET predictions can save cost and time in the drug development process. To avoid drug failures, compounds with poor ADMET predictions are eliminated because of safety and efficacy concerns. Absorption affects the bioavailability of drugs at their targets and consequently their efficacy [39].
From Table 6, all compounds show high intestinal absorption (>30%), and high skin permeability (<-2.5). At values of water solubility (log mol/L) less than -4.0, compounds are considered poorly soluble. The results suggest that Curcumin has poor solubility, and this would make its formulation more difficult [40]. Augustamine and Camptothecin have high caco2 permeability (>0.9) while the standard would be poorly absorbed by human colorectal adenocarcinoma cells [25].
Table 6.
Pharmacokinetic properties of standard and lead compounds.
| Curcumin (standard) | Augustamine | Camptothecin | |
|---|---|---|---|
| Water solubility (log mol/L) | -4.01 | -2.426 | -4.688 |
| Caco2 permeability (log Papp in 10–6 cm/s) | -0.093 | 1.429 | 0.125 |
| Human Intestinal absorption (% Absorbed) | 82.19 | 97.237 | 92.007 |
| Skin Permeability (log Kp) | -2.764 | -3.508 | -3.275 |
| P-glycoprotein substrate (Yes/No) | Yes | No | Yes |
| P-glycoprotein I inhibitor (Yes/No) | Yes | No | No |
| P-glycoprotein II inhibitor (Yes/No) | Yes | No | No |
| VDss (human) (log L/kg) | -0.215 | 0.8 | 0.028 |
| Fraction unbound (human) (Fu) | 0 | 0.332 | 0.502 |
| BBB permeability (log BB) | -0.562 | 0.288 | -0.451 |
| CNS permeability (log PS) | -2.99 | -2.579 | -3.017 |
| CYP2D6 substrate (Yes/No) | No | No | No |
| CYP3A4 substrate (Yes/No) | Yes | Yes | Yes |
| CYP1A2 inhibitor (Yes/No) | Yes | No | No |
| CYP2C19 inhibitor (Yes/No) | Yes | No | No |
| CYP2C9 inhibitor (Yes/No) | Yes | No | No |
| CYP2D6 inhibitor (Yes/No) | No | No | No |
| CYP3A4 inhibitor (Yes/No) | Yes | No | No |
| Total Clearance (log ml/min/kg) | -0.002 | 1.21 | 1.237 |
| Renal OCT2 substrate (Yes/No) | No | No | No |
| AMES toxicity (Yes/No) | No | No | No |
| Max. Tolerated dose (human) (log mg/kg/day) | 0.081 | -0.221 | -0.581 |
| hERG I inhibitor (Yes/No) | No | No | No |
| hERG II inhibitor (Yes/No) | No | No | No |
| Oral Rat Acute Toxicity (LD50) (mol/kg) | 1.833 | 2.986 | 2.865 |
| Oral Rat Chronic Toxicity (log mg/kg_bw/day) | 2.228 | 1.532 | 1.047 |
| Hepatotoxicity (Yes/No) | No | No | Yes |
| Skin Sensitization (Yes/No) | No | No | No |
| T. Pyriformis toxicity (log ug/L) | 0.494 | 0.355 | 0.482 |
| Minnow toxicity (log mM) | -0.081 | 1.323 | 2.272 |
The P-glycoprotein I and II are cell surface proteins that pump out foreign substances out of cells. This ATP-dependent transmembrane efflux pump pumps its substrate from inside to outside the cell [41]. From Table 6, Curcumin and Camptothecin are predicted to be P-glycoprotein substrates suggesting that they must be co-administered with a P-glycoprotein inhibitor to allow for drugs to be retained at the site of action [42]. Curcumin is a PgP I and II inhibitor and this would affect the way foreign substances are pumped out of the cell. Camptothecin also inhibits PgP [43, 44]. Augustamine is neither a substrate nor an inhibitor of PgP making it a good drug candidate.
From Table 6, the standard and the lead compounds are partially permeable to the CNS (permeable Log PS > -2; poor Log PS < -3) [37]. Also, all the compounds are partially permeable to the blood brain barrier with Augustamine having the best value (permeable: Log BBB >0.3; poor: Log BBB < -1) [25]. The Volume of distribution steady state (VDSS values) for Curcumin and Augustamine are low and high respectively. The VDss value for Camptothecin is within pharmacological range (Low: Log VDss < - 0.15; High: Log VDss> 0.45) [25]. This can be adjusted by dosage. The fraction of Curcumin bound to plasma is very high and this would impact on the volume of distribution and clearance. However, this can be altered by the concentration of drug, other plasma binding drugs, and the amount of plasma protein [45].
The biotransformation of drugs and bioactive compounds take place in the liver through the activity of the CYP450 enzymes. The inhibition of these enzymes results in the toxic accumulation of drugs and substrates in the cellular spaces. Drugs that are CYP450 substrates should not be administered together with inhibitors to facilitate their metabolism [46]. From Table 6, the metabolic profile of the standard and lead compounds reveals that they are non-substrates and non-inhibitors of CYP2D6. All CYP450 enzyme inhibitors should not be administered with their respective substrates as it would lead to the toxic accumulation of the substrate due to lack of metabolism [47].
Renal Organic Cation Transporter 2 is a transporter protein involved in the renal clearance of drugs and endogenous compounds. It facilitates their elimination from the blood into the proximal tubular cell [23]. Camptothecin is a renal OCT2 substrate and should be co-administered with an OCT2 inhibitor [23]. The predicted values for Total Clearance show that the standard would be the most slowly excreted of all the compounds [23].
From Table 6, the standard, and the lead compounds are predicted to be non-genotoxic as they are unlikely to have a mutagenic effect on the DNA of bacteria, Salmonella Typhimurium. Similarly, they are also all predicted to be non-dermatotoxic and non-cardiotoxic because they would not inhibit the potassium ion channel protein, hERG that conducts the electrical activity of the heart [25]. Camptothecin is predicted to be hepatotoxic, but this is could be dose related [47].
The maximum recommended tolerated dose determines the dosage to be used in the first phase of clinical trials. The values for the standard and lead compounds are low (Low: <0.477 log mg/kg/day; High: > 0.477 log mg/kg/day) [25]. Also, the predicted Oral Rat Acute Toxicity values suggest that Augustamine is the safest of all the compounds [48]. The predicted values for Oral Rat Acute Toxicity and Oral Rat Chronic Toxicity should be considered alongside factors such as dose, concentration of drug, and the duration of administration [48].
All the compounds are considered as toxic against the protozoan bacterium, T. pyriformis (Toxic: greater than -0.5 log Ug/L) but this does not indicate that they are toxic to eucaryoticcells [48].
Only Camptothecin is considered toxic to the cells of flathead Minnows (High acute toxicity: less than -0.3 log mM) [48].
4.3. Molecular docking analyses of ligands against Human Telomerase
Through molecular docking, the binding affinity score of a ligand in the binding pocket of a protein target is obtained. The ligands with the poses with the least binding energies are considered the best drug candidates [49]. With a binding affinity score of -8.2 kcal/mol (Table 1), Camptothecin showed the greatest potency.
4.4. Binding site analyses
Hydrogen bonds play a significant role in drug discovery as it determines the specificity and direction in which the ligand would bind at the active site of the protein. This would affect the molecular recognition and affinity of the drug. Hydrogen bonds replace water molecules to enhance ligand binding [50].
Table 2 reveals that Curcumin and Camptothecin interact with the protein through the same binding pocket. Augustamine binds at a pocket likely to be at the N-terminal end (TEN domain) of the hTERT [51]. Curcumin and Camptothecin each have four intermolecular hydrogen bonds in this binding pocket. However, only Camptothecin forms a strong (greater than 130°) bond at SER1037A [52]. Augustamine binds in a different pocket and has only one weak intermolecular hydrogen bond [52]. Regarding the donor-to-acceptor distance, all intramolecular hydrogen bonds of the compounds are weak (3.2–4.0 Å) except the interaction of Curcumin at SER1095A which is moderate (2.5–3.2 Å) [52]. Hydrophobic interactions and salt bridges add strength and stability to the protein-ligand complexes. This is because the binding affinity is favored by the interaction between the hydrophobic areas of the drug target and the lipophilic surface of the drug [53].
4.5. Analysis of MDS
As seen in Figure 6, there is a mechanical unfolding of part of the alpha-helices of the hTERT-Curcumin and hTERT-Camptothecin holo structures. This gradual conformational change into random coils is caused by the external force of simulation and ligand-induced binding which destabilizes the intramolecular hydrogen bond of the alpha helices [54].
4.5.1. Root Mean Square Deviation of atomic positions (RMSD)
The protein backbone holds the protein's tertiary structure together and maintains its shape. The RMSD is essentially the measurement of the difference (distance between the alpha carbon atoms) in the core backbone of the reference structure of the protein relative to its final position after simulation [55]. The deviation is a function of the stability of the protein. The reference has an RMSD of zero, and the higher the deviation, the lesser the stability of the protein. When dealing with human proteins, a lower RMSD is generally preferable to a higher one [56].
From Table 4, the results suggest that the hTERT-Augustamine complex has the least structural distortion during the simulation period. However, an ascending gradient for the RMSD slopes of hTERT-Augustamine and hTERT-curcumin complexes (Figure 7 and Table 4) suggest that the RMSD values will increase with more simulation time. The hTERT-Camptothecin complex shows the greatest stability as revealed by the least slope gradient. From Figure 8 and Table 4, the distribution pattern of the RMSD values of the apo and holo structures suggests that the hTERT-Camptothecin complex has the least deviation to the right from its reference structure. In this regard, the hTERT-Augustamine complex is more stable than the hTERT-Curcumin complex.
4.5.2. RMSF
The dynamics of a protein is a major determinant of its function. The RMSF evaluates the residual fluctuations of the protein using the alpha-carbon atoms as a reference for measurement. The greatest fluctuations are at the N-termini, C-termini, and the loops [57]. From Figure 9 and Table 4, Camptothecin induced the least fluctuation on the target protein and hence its holo structure is the most stable. This is shown in the data and the fluctuation patterns which most closely resembles that of the apo structure. In this regard the next most stable is the hTERT-Curcumin complex.
4.5.3. B-factor
The B-Factor or Temperature factor evaluates the thermostability of the protein [58]. Internal motions in the protein structure could be caused by X-ray or ligand-induced binding. This flexibility or rigidity can affect the stereochemical quality of the protein [59]. Results from Table 4 suggest that the hTERT-Camptothecin complex is the most thermally stable of all the holo structures causing the least dynamic disorder of the hTERT stereochemistry. This is followed closely by the hTERT-Curcumin complex.
4.5.4. Principal components analysis (PCA)
During MDS, new conformations of apo and holo structures are generated. PCA is used to determine the statistical significance of these conformations in a bid to pick structures that are good representatives of the clusters [60]. From Table 4 and Figure 10, PCA results suggest that Curcumin and Camptothecin induced the least motions and hence had the greatest stability in their respective holo structures.
4.5.5. The dynamic cross-correlation (DCC) analysis
The DCC matrix calculates and analyses the cross correlation of the atomic fluctuations obtained from MDS. This DCC map captures the data as positive and negative correlations depicting the multimodal characteristics of atoms of the residues of the target protein [61]. From DCCM data from the Apo and holo structures, the large coverage of anti-correlating residues further goes to prove that the hTERT is the catalytic domain of the telomerase enzyme (Figure 11, Table 4) [51]. The anti-correlated motion of the residues within the hTERT is suggestive of inhibition of the enzyme [62]. Consequently, the hTERT-Camptothecin shows the greatest inhibitory activity of all the holo structures having the highest number of residues with anti-correlation motions. This significant external perturbation of hTERT is due to the ligand binding of Camptothecin.
4.5.6. Hydrogen bond analysis
During molecular dynamics simulation, conformational changes in the residues leads to the formation of the hydrogen bonds with the solvent. These bonds affect protein-ligand specificity and consequently the binding of the inhibitors in the active site of the enzyme [63]. Hydrogen bond analysis calculates the number and occupancy of the hydrogen bonds during the simulation period. These singular hydrogen bonds are formed between donor and acceptor within a cut-off distance of 3 Å and an angle of 120°. From Table 4, the highest occupancies were seen in the hTERT-Curcumin and the hTERT-Camptothecin complexes showing that during the simulation period these two holo structures have higher number of interactions, higher structural stability, and consequently provide a better scaffold for inhibiting the target enzyme than the hTERT-Augustamine complex. As seen in Table 4 and Figure 12, the hTERT-Camptothecin complex has the least range of hydrogen bond fluctuations suggesting it is the most stable holo structure during the simulation.
Put together, Augustamine is a better drug candidate than Curcumin (standard) and Camptothecin regarding drug-likeness and pharmacokinetic properties. Specifically, Camptothecin proved to be better than the standard and Augustamine regarding bioactivity, binding affinity, and molecular stability during time resolved MDS. Augustamine binds to the hTERT at a pocket likely to be in the N-terminal end of the protein and this is different for that of Camptothecin which shares the same binding site as the standard [51].
Augustamine can be obtained from the aqueous extract of the leaves of the water leaf plant, Talinumtriangulare [64]. The anti-telomerase activity of Augustamine is unknown. Curcumin is a natural compound obtained from the Turmeric plant, Curcuma Longa, and its antiproliferative activity on cancer cells has been well established [65]. Specifically, it has been shown to downregulate hTERT expression in breast cancer cells [12]. Camptothecin is obtained from the bark and seeds of a Chinese tree, Camptotheca acuminata [66]. As a natural anti-cancer drug, Camptothecin inhibits DNA Topoisomerase I enzyme which also shows catalytic activity at the G-rich Telomeric DNA strand [67]. Camptothecin has also been shown to directly inhibit telomerase activity and induce apoptosis in human leukemia HL-60 cells [68]. The implication of this study is that plants that contain the lead compounds can be used in the production of nutraceuticals for the management of cancer patients.
4.6. Study limitations
Natural compounds derived from tropical plants were investigated for their inhibitory properties against the Human Telomerase. Augustamine and Camptothecin showed promising results as potential inhibitors of the target Telomerase enzyme. Compounds from tropical plants are to be further investigated for enzyme inhibitory potentials to validate the in-silico predictions.
5. Conclusion
Beyond the Topoisomerase-inhibiting activity of Camptothecin, this study reveals another possible anticancer activity which is Telomerase inhibition. Like Curcumin, Camptothecin also binds at the TEN functional domains of hTERT suggesting a similar mechanism of inhibitory activity. At this site, Camptothecin induces greater Telomerase inhibition with better molecular stability than Curcumin. Further evaluation of the efficacy and toxicity of the lead compounds through in vivo and in vitro tests are required.
Declarations
Author contribution statement
Adekunle Babajide Rowaiye: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Analyzed and interpreted the data; Wrote the paper.
Yoroshi Joana Teca Mendes: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Samson Ayodeji Olofinsae: Performed the experiments; Wrote the paper.
John Breakthrough Oche; Oluwakemi Hannah Oladipo; Okiemute Ajiroghene Okpalefe; Joyce Oloaigbe Ogidigo: Performed the experiments.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors wish to thank the management of the National Biotechnology Development Agency, Abuja, Nigeria for providing an enabling environment for this research.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Duesberg P., McCormack A. Immortality of cancers. A consequence of inherent karyotypic variations and selections for autonomy. Cell Cycle. 2013;12:783–802. doi: 10.4161/cc.23720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jafri M.A., Ansari S.A., Alqahtani M.H., Shay J.W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016;8(1):69. doi: 10.1186/s13073-016-0324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubtsova M., Dontsova O. Human Telomerase RNA: telomerase component or more? Biomolecules. 2020;10(6):873. doi: 10.3390/biom10060873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ait-Aissa K., Ebben J.D., Kadlec A.O., Beyer A.M. Friend or foe? Telomerase as a pharmacological target in cancer and cardiovascular disease. Pharmacol. Res. 2016;111:422–433. doi: 10.1016/j.phrs.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Jesus B.B., Blasco M.A. Telomerase at the intersection of cancer and aging. Trends Genet. 2013;29(9):513–520. doi: 10.1016/j.tig.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glybochko P.V., Zezerov E.G., Glukhov A.I., Alyaev Y.G., Severin S.E., Polyakovsky K.A., Varshavsky V.A., Severin E.S., Vinarov A.Z. Telomerase as a tumor marker in diagnosis of prostatic intraepithelial neoplasia and prostate cancer. Prostate. 2014;74:1043–1051. doi: 10.1002/pros.22823. [DOI] [PubMed] [Google Scholar]
- 7.Wills L.P., Schnellmann R.G. Telomeres and telomerase in renal health. J. Am. Soc. Nephrol.: JASN (J. Am. Soc. Nephrol.) 2011;22(1):39–41. doi: 10.1681/ASN.2010060662. [DOI] [PubMed] [Google Scholar]
- 8.Chen C.H., Chen R.J. Prevalence of telomerase activity in human cancer. J. Formos. Med. Assoc. 2011;110:275–289. doi: 10.1016/S0929-6646(11)60043-0. [DOI] [PubMed] [Google Scholar]
- 9.McKelvey B.A., Umbricht C.B., Zeiger M.A. Telomerase reverse transcriptase (TERT) regulation in thyroid cancer: a review. Front. Endocrinol. (Lausanne) 2020 Jul 31;11:485. doi: 10.3389/fendo.2020.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koonrungsesomboon N., Wadagni A.C., Mbanefo E.C. Molecular markers and Schistosoma-associated bladder carcinoma: a systematic review and meta-analysis. Cancer Epidemiol. 2015;39:487–496. doi: 10.1016/j.canep.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Lou X., Zhuang Y., Zuo X., Jia Y., Hong Y., Min X., Zhang Z., Xu X., Liu N., Xia F. Real-time, quantitative lighting-up detection of telomerase in urines of bladder cancer patients by AIEgens. Anal. Chem. 2015;87:6822–6827. doi: 10.1021/acs.analchem.5b01099. [DOI] [PubMed] [Google Scholar]
- 12.Nasiri M., Zarghami N., Koshki K.N., Mollazadeh M., Moghaddam M.P., Yamchi M.R., Esfahlan R.J., Barkhordari A., Alibakhshi A. Curcumin and silibinin inhibit telomerase expression in T47D human breast cancer cells. Asian Pac. J. Cancer Prev. APJCP. 2013;14(6):3449–3453. doi: 10.7314/apjcp.2013.14.6.3449. [DOI] [PubMed] [Google Scholar]
- 13.Ait-Aissa K., Ebben J.D., Kadlec A.O., Beyer A.M. Friend or foe? Telomerase as a pharmacological target in cancer and cardiovascular disease. Pharmacol. Res. 2016 Sep;111:422–433. doi: 10.1016/j.phrs.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J.L.Y., Sperry J., Ip N.Y., Brimble M.A. Natural products targeting telomere maintenance. Med. Chem. Comm. 2011;2(4):229–245. [Google Scholar]
- 15.Ganesan K., Xu B. Telomerase inhibitors from natural products and their anticancer potential. Int. J. Mol. Sci. 2018 Jan;19(1):13. doi: 10.3390/ijms19010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenwell M., Rahman P.K.S.M. Medicinal plants: their use in anticancer treatment. Int. J. Pharmaceut. Sci. Res. 2015 Oct 1;6(10):4103–4112. doi: 10.13040/IJPSR.0975-8232.6(10).4103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021;49:D437–D445. doi: 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
- 19.Ramachandran S., Kota P., Ding F., Dokholyan N.V. Automated minimization of steric clashes in protein structures. Proteins. 2011;79(1):261–270. doi: 10.1002/prot.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willard L., Ranjan A., Zhang H., Monzavi H., Boyko R.F., Sykes B.D., Wishart D.S. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003 July 1;31(13):3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams C.J., Hintze B.J., Headd J.J., Moriarty N.W., Chen V.B., Jain S., Prisant M.G., Lewis S.M., Videau L.L., Keedy D.A., Deis L.N., Arendall W.B., III, Verma V., Snoeyink J.S., Adams P.D., Lovell S.C., Richardson J.S., Richardson D.C. More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A., Wang J., Yu B., Zhang J., Bryant S.H. PubChem substance and compound databases. Nucleic Acids Res. 2016 Jan 4;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atatreh N.A., Rawashdah S.A., Neyadi S.S., Abuhamdah S.M., Ghattas M.A. Discovery of new butyrylcholinesterase inhibitors via structure-based virtual screening. J. Enzym. Inhib. Med. Chem. 2019;34:1373–1379. doi: 10.1080/14756366.2019.1644329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dallakyan S., Olson A.J. Small-Molecule library screening by docking with PyRx. Methods Mol. Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 25.Pires D.E.V., Tom L., Blundell D.B., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic properties using graph-based signatures. J. Med. Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molinspiration . 2015. Calculation of Molecular Properties and Bioactivity Score.http://www.molinspiration.com/cgi-bin/properties [Google Scholar]
- 27.Salentin S., Schreiber S., Haupt V.J., Adasme M.F., Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015 Jul 1;43(W1):W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinf. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1(1):19–25. [Google Scholar]
- 30.Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Grüning B.A., Guerler A., Hillman-Jackson J., Hiltemann S., Jalili V., Rasche H., Soranzo N., Goecks J., Taylor J., Nekrutenko A., Blankenberg D. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018 Jul 2;46(W1):W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dodda L.S., Cabeza de Vaca I., Tirado-Rives J., Jorgensen W.L. LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res. 2017 Jul 3;45(W1):W331–W336. doi: 10.1093/nar/gkx312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hospital A., Andrio P., Fenollosa C., Cicin-Sain D., Orozco M., Gelpí J.L. MDWeb and MDMoby: an integrated web-based platform for molecular dynamics simulations. Bioinformatics. 2012 May 1;28(9):1278–1279. doi: 10.1093/bioinformatics/bts139. [DOI] [PubMed] [Google Scholar]
- 33.Yi T., Tang D., Wang F., Zhang J., Zhang J., Wang J., Xu X., Zhang J. Enhancing both oral bioavailability and brain penetration of puerarin using borneol in combination with preparation technologies. Drug Deliv. 2017 Nov;24(1):422–429. doi: 10.1080/10717544.2016.1259372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bickerton G.R., Paolini G.V., Besnard J., Muresan S., Hopkins A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012;4(2):90–98. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al Wasidi A.S., Hassan A.S., Naglah A.M. In vitro cytotoxicity and drug-likeness of pyrazolines and pyridines bearing benzofuran moiety. J. Appl. Pharmaceut. Sci. 2020;10(4):142–148. [Google Scholar]
- 36.Feldmann C., Miljković F., Yonchev D., Bajorath J. Identifying promiscuous compounds with activity against different target classes. Molecules. 2019;24(22):4185. doi: 10.3390/molecules24224185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness, and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark D.E. What has polar surface area ever done for drug discovery? Future Med. Chem. 2011;(4):469–484. doi: 10.4155/fmc.11.1. [DOI] [PubMed] [Google Scholar]
- 39.Price G., Patel D.A. StatPearls Publishing; 2020. Drug Bioavailability. Treasure Island (FL) [PubMed] [Google Scholar]
- 40.Savjani K.T., Gajjar A.K., Savjani J.K. Drug solubility: importance and enhancement techniques. ISRN Pharmaceut. 2012;2012 doi: 10.5402/2012/195727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nanayakkara A.K., Follit C.A., Chen G., Williams N.S., Vogel P.D., Wise J.G. Targeted inhibitors of P-glycoprotein increase chemotherapeutic-induced mortality of multidrug resistant tumor cells. Sci. Rep. 2018;8(1):967. doi: 10.1038/s41598-018-19325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amin M.L. P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights. 2013;7:27–34. doi: 10.4137/DTI.S12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srivalli K.M., Lakshmi P.K. Overview of P-glycoprotein inhibitors: a rational outlook. Brazilian J. Pharmaceut. Sci. 2012 Sep;48(3):353–367. [Google Scholar]
- 44.Ondieki G., Nyagblordzro M., Kikete S., Liang R., Wang L., He X. Cytochrome P450 and P-Glycoprotein-Mediated interactions involving African herbs indicated for common noncommunicable diseases. Evid.-Based Compl. Altern. Med.: eCAM. 2017:2582463. doi: 10.1155/2017/2582463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Min Ye M.S., Nagar S., Korzekwa K.A. Physiologically based pharmacokinetic model to predict the pharmacokinetics of highly protein-bound drugs and impact of errors in plasma protein binding. Biopharm Drug Dispos. 2016;37(3):123–141. doi: 10.1002/bdd.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonnell A.M., Dang C.H. Basic review of the cytochrome p450 system. J. Adv. Practit. Oncol. 2013;4(4):263–268. doi: 10.6004/jadpro.2013.4.4.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ćurčić M., Tanasković S., Stanković S., Janković S., Antunović M., Djordjević S., Kilibarda V., Vučnić S., Antonijević B. Relationship of hepatotoxicity and the target tissue dose of decabrominated diphenyl ether in subacutely exposed Wistar rats. Vojnosanit. Pregl. 2015 May;72(5):405–413. [PubMed] [Google Scholar]
- 48.Sliwoski G., Kothiwale S., Meiler J., Lowe E.W. Computational methods in drug discovery. Pharmacol. Rev. 2013;66:334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tatu P., Antti P. Binding affinity via docking: fact and fiction. Molecules. 2018;23(8):1899. doi: 10.3390/molecules23081899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen D., Oezguen N., Urvil P., Ferguson C., Dann S.M., Savidge T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016;2(3) doi: 10.1126/sciadv.1501240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robart A.R., Collins K. Human Telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol. Cell. 2011;42(3):308–318. doi: 10.1016/j.molcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desiraju G., Steiner T. Structural Chemistry and Biology. Oxford University Press; Oxford: 2001. The weak hydrogen bond. [Google Scholar]
- 53.Khurana L., Fu B.Q., Duddupudi A.L., Liao Y.H., Immadi S.S., Kendall D.A., Lu D. Pyrimidinylbiphenylureas: identification of new lead compounds as allosteric modulators of the cannabinoid receptor CB1. J. Med. Chem. 2017;60(3):1089–1104. doi: 10.1021/acs.jmedchem.6b01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergues-Pupo A.E., Lipowsky R., Vila Verde A. Unfolding mechanism and free energy landscape of single, stable, alpha helices at low pull speeds. Soft Matter. 2020 Nov 11;16(43):9917–9928. doi: 10.1039/d0sm01166e. [DOI] [PubMed] [Google Scholar]
- 55.Aier I., Varadwaj P.K., Raj U. Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci. Rep. 2016;6(1):1–10. doi: 10.1038/srep34984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Irina K., Ruben A. Methods of protein structure comparison. Methods Mol. Biol. 2012;857:231–257. doi: 10.1007/978-1-61779-588-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fuglebakk E., Echave J., Reuter N. Measuring and comparing structural fluctuation patterns in large protein datasets. Bioinformatics. 2012;28(19):2431–2440. doi: 10.1093/bioinformatics/bts445. [DOI] [PubMed] [Google Scholar]
- 58.Sun Z., Liu Q., Qu G., Feng Y., Reetz M.T. Utility of B-factors in protein science: interpreting rigidity, flexibility, and internal motion and engineering thermostability. Chem. Rev. 2019;119(3):1626–1665. doi: 10.1021/acs.chemrev.8b00290. [DOI] [PubMed] [Google Scholar]
- 59.Zheng H., Hou J., Zimmerman M.D., Wlodawer A., Minor W. The future of crystallography in drug discovery. Expet Opin. Drug Discov. 2014;9(2):125–137. doi: 10.1517/17460441.2014.872623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.David C.C., Donald J., Jacobs D.J. Principal component analysis: a method for determining the essential dynamics of proteins. Methods Mol. Biol. 2014;1084:193–226. doi: 10.1007/978-1-62703-658-0_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kasahara K., Fukuda I., Nakamura H.A. Novel approach of dynamic cross correlation analysis on molecular dynamics simulations and its application to Ets1 dimer–DNA complex. PloS One. 2014;9(11):e112419. doi: 10.1371/journal.pone.0112419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agoni C., Ramharack P., Soliman M.E.S. Allosteric inhibition induces an open WPD-loop: a new avenue towards glioblastoma therapy. RSC Adv. 2018;8:40187–40197. doi: 10.1039/c8ra08427k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nittinger E., Inhester T., Bietz S., Meyder A., Schomburg K.T., Lange G., Klein R., Rarey M. Large-scale Analysis of hydrogen bond interaction patterns in protein–ligand interfaces. J. Med. Chem. 2017;60(10):4245–4257. doi: 10.1021/acs.jmedchem.7b00101. [DOI] [PubMed] [Google Scholar]
- 64.Ikewuchi C.C., Ikewuchi J.C., Ifeanacho M.O. Bioactive phytochemicals in an aqueous extract of the leaves of Talinum triangulare. Food Sci. Nutr. 2017;5(3):696–701. doi: 10.1002/fsn3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rahmani A.H., Alsahli M.A., Aly S.M., Khan M.A., Aldebasi Y.H. Role of Curcumin in disease prevention and treatment Advanced biomedical research. 2018;7:38. doi: 10.4103/abr.abr_147_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qu X., Pu X., Chen F., Yang Y., Yang L., Zhang G., Luo Y. Molecular cloning, heterologous expression, and functional characterization of an NADPH-cytochrome P450 reductase gene from Camptotheca acuminata, a camptothecin-producing plant. PloS One. 2015;10(8) doi: 10.1371/journal.pone.0135397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gokduman K. Strategies targeting DNA topoisomerase I in cancer chemotherapy: camptothecins, nanocarriers for camptothecins, organic non-camptothecin compounds and metal complexes. Curr. Drug Targets. 2016;17(16):1928–1939. doi: 10.2174/1389450117666160502151707. [DOI] [PubMed] [Google Scholar]
- 68.Zhou Y., Zhao H.Y., Jiang D., Wang L.Y., Xiang C., Wen S.P., Fan Z.C., Zhang Y.M., Guo N., Teng Y.O., Yu P. Low toxic and high soluble camptothecin derivative 2–47 effectively induces apoptosis of tumor cells in vitro. Biochem. Biophys. Res. Commun. 2016;472(3):477–481. doi: 10.1016/j.bbrc.2016.02.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.