Summary
We recently established an in vitro culture system in which mammary gland organoid undergoes directional migration in response to an FGF10 concentration gradient. Here, we describe a step-by-step protocol for preparing organoids, the setup of the 3D culture system, and the image acquisition approach. The technical difficulties in conducting the 3D migration assay are choosing epithelial organoids of appropriate sizes and manually paring organoids and beads pre-soaked in FGF10 within a desirable distance (∼100 μm).
For complete details on the use and execution of this protocol, please refer to Lu et al. (2020).
Subject areas: Cell Biology, Cell culture, Developmental biology, Microscopy, Organoids
Graphical abstract

Highlights
-
•
Organoid epithelium directionally migrates up the FGF10-concentration gradient
-
•
A 3D in vitro culture system to study epithelial collective migration
-
•
A reliable setup for reproducible results on the epithelium directional migration
We recently established an in vitro culture system in which mammary gland organoid undergoes directional migration in response to an FGF10 concentration gradient. Here, we describe a step-by-step protocol for preparing organoids, the setup of the 3D culture system, and the image acquisition approach. The technical difficulties in conducting the 3D migration assay are choosing epithelial organoids of appropriate sizes and manually paring organoids and beads pre-soaked in FGF10 within a desirable distance (∼ 100 μm).
Before you begin
Timing: 2 h
Increasing evidence shows that vertebrate epithelia are more motile than historically believed. Understanding the cellular and molecular mechanisms by which vertebrate epithelia migrate is essential for understanding organ development and etiologies that cause disease conditions. We recently established a culture system in which mammary gland organoid undergoes directional migration (Zhang et al., 2014a) (Zhang et al., 2014b) (Lu et al., 2020). Here, we present a detailed protocol that tackles several challenging aspects of the technique so that the assay can be reliably and successfully conducted by scientists not familiar with the system.
Because of Matrigel’s unique physical properties, where it is liquid at ∼ 4°C–10˚C but solidifies once it is warmed up to the room temperature, one must work relatively fast and be able to complete pairing epithelial organoids and beads before Matrigel solidifies. This requires that the experimenters be familiar with the protocol and have all the reagents prepared in advance before the day of the experiment.
-
1.Soak heparin beads with FGF10
-
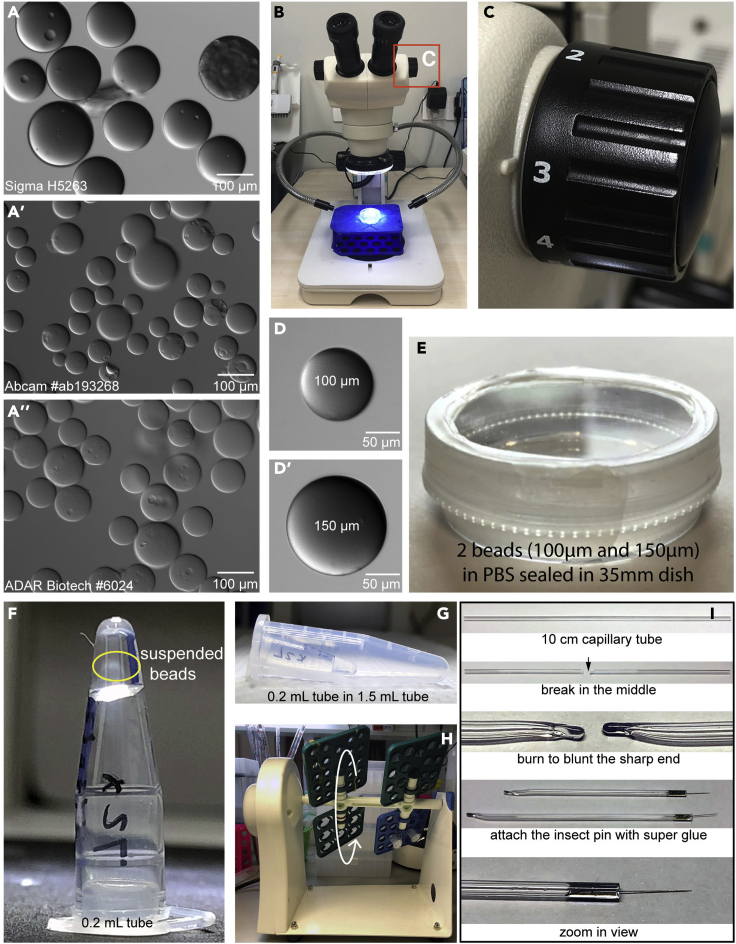
a.Prepare reference beadsReference beads serve two purposes: first, they assist the experimenters in choosing beads of relatively uniform sizes, which, at least in theory, can pre-soak a similar amount of FGF10 proteins; second, they help the experimenters readily determine under the stereoscope the distance between the FGF10-beads and organoids, and the FGF10 concentration gradient hereby created, and help keep the distances relatively uniform between each bead-organoid pair.
-
i.Transfer 20 μL beads into a 35 mm tissue culture dish containing 2 mL of PBS.
-
ii.Pick several beads (Figure 1B) of different sizes and put them in a 96-well culture plate containing 100 μL of PBS per well, and limit one bead in each well.
-
iii.Take pictures and check the size of each bead under a microscope.
-
iv.Transfer two beads, one 100 μm and the other 150 μm in diameter, into a 35 mm dish containing 2 mL of PBS (Figures 1D and 1D′).
-
v.Seal the dish with parafilm (Figure 1E). These two beads will be used as references for checking the sizes and distances of beads and organoids.Note: The beads are stored in a 50% slurry. Thus, 20 μL of slurry contains ∼ 10 μL of beads and ∼ 10 μL of PBS. Mix the tube first before transferring the mixture. Three brands of heparin beads have been compared (Figures 1A–1A’’). In our hands, there are no noticeable differences when they are used to stimulate organoid migration. We prefer using the Sigma beads (Sigma, #H5263, Figure 1A) because they are opaque and more noticeable than the other two brands, both of which are transparent. However, the sigma beads have been discontinued. The zoom knob was set at 3 for all steps (Figure 1C).
-
i.
-
b.Pick the beads between 100 - 150 μm in diameter, based on their comparison with the reference beads, under a stereo microscope. Use a tally counter to count the number of beads that you will have.
-
c.Soak the picked beads with FGF10
-
i.Transfer ∼ 30 beads (<40 beads so that each has been saturated by FGF10 proteins after soaking) into a 0.2 mL tube.
-
ii.Add 200 μL of 0.1% BSA in PBS (see recipe in materials and equipment), mix gently, and then spin briefly.
-
iii.Remove supernatant, add 10 μL of FGF10 stock solution (see recipe in materials and equipment), rotate at 18 rpm at 4°C overnight on a rocker (Kylin-Bell, BE-1100) (Figure 1H). The beads should be used within one week.Note: The unused beads can be saved in 200 μL of 0.1% BSA in PBS in a 35 mm dish or a 0.2 mL tube, and stored at 4°C up to a week without a noticeable decrease in performance.
-
i.
-
a.
Figure 1.
Prepare FGF10 soaked bead and needles for migration assay
(A–A'') Heparin beads of different brands. (A) from sigma company, Cat#H5263, (A′) from Abcam company, Cat#ab193268, and (A'') from ADAR Biotech company, Cat#6024.
(B) A stereo microscope used to pick up suitable beads, organoids and to set up the migration assay.
(C) The zoom knob was set at 3 for all steps.
(D and D′) Picture of Heparin bead 100 μm (D) and 150 μm (D′).
(E) A 100 μm and a 150 μm beads were sealed in PBS in a 35 mm dish. These two beads will serve as a reference for picking beads and setting up steps.
(F) Picked beads in a 0.2 mL tube and suspended in 20 μL of FGF10 solution. When upside down, the solution should not move along the tube wall, yet the bead will move freely in the solution.
(G) 0.2 mL tube containing the picked beads was secured in a capless 1.5 mL tube with tape.
(H) the 1.5 mL tube was rotated on a mixer.
(I) Steps of preparing needles for migration assay.
Make sure that there is no liquid on the tube wall to ensure that the beads are suspended in the liquid while on the rocker (Figures 1F–1H).
-
2.
Rather than weigh collagenase and take a small amount (0.02 g) each time, we find it is more convenient to weigh it once and store multiple aliquots (0.02 g/tube) at −20°C until future use.
-
3.
The day before or on the day of the experiment, warm 9.3 mL of DMEM/F12 medium in a tissue culture incubator or a 37°C water bath to make the collagenase digestion solution.
-
4.
Place Matrigel in the refrigerator one night ahead so that it thaws overnight.
Alternatively, thaw the Matrigel on the day of the experiment in a 4°C–10°C water bath or running tap water. However, once melted, Matrigel should be put on the ice so that it does not solidify as it would when over-heated to the RT.
-
5.
Prepare two fine needles for pairing organoids and beads: first, break a 10 cm capillary tube in the middle. Use a Bunsen burner to seal the sharp end. Then apply one drop of superglue to the open end of the broken capillary tube, to which insert an insect pin. Leave the needle on the benchtop one day for the glue to dry (Figure 1I). Needles can be stored in 75% EtOH and washed in cold PBS before use.
-
6.
Make sure that there are enough reagents listed in the key resource table for the experiments.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD49f (Integrin alpha 6) APC | Invitrogen | Cat#17-0495-82 RRID: AB_2016694 |
| SMA-FITC | Sigma | Cat#F3777 |
| Chemicals, peptides, and recombinant proteins | ||
| FGF10 | GenScript | Cat#Z03155-10 or Cat#Z03155-50 |
| Collagenase | Sigma | Cat#C5138 |
| Trypsin | Thermo Fisher Scientific | Cat#27250-018 |
| Insulin | Yeasen | Cat#40107ES60 |
| Gentamicin | Meilunbio | Cat#MA0322 |
| FBS | Gemini | Cat#900-108 |
| DMEM/F12 | Thermo/Life/Invitrogen | Cat#C11330500CP |
| DNase | Sigma | Cat#D4263 |
| ITS | Sigma | Cat#I3146 |
| Penicillin-streptomycin | Life/Invitrogen | Cat#15070063 |
| L-Glutamine, 200 mM | HyClone | Cat#SH30034.01 |
| PBS without Ca2+, Mg2+ | Meilunbio | Cat#MA0015 |
| BSA | Sigma | Cat#B2064 |
| DAPI | Sigma | Cat#D9542-10MG |
| Paraformaldehyde | Sigma | Cat#158127 |
| Glycine | MdBio | Cat#G007-500g |
| Triton X-100 | BBI Life Sciences | Cat#A600198-0500 |
| Tween-20 | BBI Life Sciences | Cat#A600560-0500 |
| Goat serum | Meilunbio | Cat#MB4508 |
| 10× PBS | Life/Invitrogen | Cat#70011-044 |
| Heparin-Acrylic beads | Sigma | Cat#H5263 |
| Heparin Sepharose | Abcam | Cat#ab193268 |
| Heparin beads | Adar Biotech | Cat#6024 |
| Matrigel (Growth Factor Reduced) | Corning | Cat#354230 |
| Experimental models:Organisms/strains | ||
| FVB mouse strains | Vital River Laboratory Animal Technology Co., Ltd. (Beijing) | Vital River: 215 |
| Software and algorithms | ||
| Fiji | https://imagej.net/ | RRID: SCR_003070 |
| Other supplies | ||
| 24-Well glass bottom plate | Cellvis | Cat#P24-1.5H-N |
| 8-Well chambered cover glass | Cellvis | Cat#C8-1.5H-N |
| Multi-well plate cover with #1 (0.13–0.16 mm) cover glass for DIC (Differential interference contrast) imaging | Cellvis | Cat#L001 |
| Minutiens (Insect Pin) | Austerlitz Insect Pin | 0.10mm |
| 2.5 mm Capillary tube | Wilmad | Cat#WG-1365-2.5A-1EA |
Materials and equipment
Trypsin solution
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| Trypsin | 100 mg/mL | 100 mg |
| DMEM/F12 | n/a | 1 mL |
| Total | n/a | 1 mL |
Make 200 μL aliquots. Store at −20°C.
Collagenase digestion solution
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| Collagenase | 2 mg/mL | 0.02g |
| Trypsin 100 mg/mL | 2 mg/mL | 200 μL |
| FBS | 5% (vol/vol) | 0.5 mL |
| Gentamicin 50 mg/mL | 50 μg/mL | 10 μL |
| Insulin 2 mg/mL | 5 μg/mL | 25 μL |
| DMEM/F12 | n/a | 9.3 mL |
| Total | n/a | 10 mL |
Prepare freshly every time. Filter sterilize.
DNase solution
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| DNase | 2 U/μL | 2,000 U |
| NaCl | 0.15M | 1 mL |
| Total | n/a | 1 mL |
Use sterilized NaCl solution. Make 40 μL aliquots. Store at −20°C.
Basal organoid culture medium
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| ITS 100× | 1× | 100 μL |
| Penicillin-Streptomycin 100× | 1× | 100 μL |
| L-Glutamine 200 mM | 2 mM | 100 μL |
| DMEM/F12 | n/a | 9.7 mL |
| Total | n/a | 10 mL |
Store at 4°C, use within one week.
2.5% BSA
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| BSA | 2.5% (w/vol) | 1.25 g |
| PBS without Ca2+, Mg2+ | n/a | 50 mL |
| Total | n/a | 50 mL |
Mix the tube on end over end rotator for 30 min. Filter sterilize. Make 10 mL aliquots. The stock can be stored for up to six months at 4°C.
0.1% BSA
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| BSA 2.5% (w/vol) | 0.1% (w/vol) | 0.4 mL |
| PBS without Ca2+, Mg2+ | n/a | 9.6 mL |
| Total | n/a | 10 mL |
Make 1 mL aliquots. Store at −20°C.
Insulin solution
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| Insulin | 5 mg/mL | 100 mg |
| HCL 5 mM | n/a | 20 mL |
| Total | n/a | 20 mL |
Filter the solution through the 0.22-μm filter. Aliquoted and stored at −20°C. Multiple freeze-thaw cycles should be avoided. Dilute the 5 mg/mL aliquot to 2 mg/mL with PBS or medium. The 2 mg/mL stock can be stored for up to six months at 2°C–8°C.
80% Matrigel
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| Matrigel (Growth Factor Reduced) | 80% (vol/vol) | 0.8 mL |
| Basal organoid culture medium | n/a | 0.2 mL |
| Total | n/a | 1 mL |
After freezing thaw cycles, Matrigel tends to have undissolvable precipitates. Spin the tube briefly at 6000 rpm for 30 s. Transfer the supernatant into a precooled tube and leave the visible pellet behind. The amount for setting up one well is 35 μL. Only prepare the amount for one experiment.
FGF10
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| FGF10 | 100 μg/mL | 50 μg |
| ddH2O | n/a | 0.5 mL |
| Total | n/a | 0.5 mL |
Briefly spin the FGF10 tube, then add ddH2O. Vortex the tube for 30 s, mix up and down to dissolve any FGF10 powder on the cap. Briefly spin the tube. Aliquot 10 μL per tube. Store at −80°C. Upon reconstitution, the aliquot should be used within six months.
4% Paraformaldehyde (PFA)
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| Paraformaldehyde | 4% (w/vol) | 2 g |
| PBS 10× | n/a | 5 mL |
| H2O | n/a | 40 mL |
| Total | n/a | 50 mL |
Add 200 μL of 1N NaOH, heated in a 60°C water bath. Let the solution cool down to room temperature. Adjust pH to 7.4 with HCL. Bring the final volume to 50 mL. Filter through a 0.45 μm filter or fluted filter paper to remove any particle matter. A 0.22 μm filter can be used to sterilize the solution, but the filter will be easily blocked, and it will be challenging to push the liquid through. Aliquot and freeze at −20°C. The solution should be used within three months.
Note: Paraformaldehyde is toxic. Use only in a fume hood. The buffer containing paraformaldehyde must be disposed of safely per applicable local regulations.
Quenching solution
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| Glycine | 100 mM | 0.375 g |
| PBS | n/a | 50 mL |
| Total | n/a | 50 mL |
Aliquot 5 mL per tube, and store in −20°C.
Permeabilizing buffer
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| Triton X-100 10% | 0.5% (vol/vol) | 2.5 mL |
| PBS 10× | n/a | 5 mL |
| H2O | n/a | 42.5 mL |
| Total | n/a | 50 mL |
Aliquot 5 mL per tube, and store in −20°C.
Blocking buffer
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| Tween-20 10% | 0.2% (vol/vol) | 0.02 mL |
| PBS 10× | n/a | 0.1 mL |
| H2O | n/a | 0.78 mL |
| Goat serum | 10% (vol/vol) | 0.1 mL |
| Total | n/a | 1 mL |
Prepare freshly each time.
DAPI
| Reagent | Final concentration (mM or μM) | Amount |
|---|---|---|
| DAPI | 10 mg/mL | 10 mg |
| H2O | n/a | 1 mL |
| Total | n/a | 1 mL |
Add autoclaved H2O 1 mL, cover tube with aluminum foil, and mix on end over end rotator 30 min. Aliquot in a black tube, 100 μL per tube, and store at −20°C.
Note: DAPI is a known mutagen and should be handled with care. The dye must be disposed of safely and in accordance with applicable local regulations.
Step-by-step method details
Mammary gland harvest and organoid preparation
Timing: 2.5 h
Preparation of properly-sized organoids.
Based on our experience, this migration assay works the best when the size of the organoids is between 100 and 200 μm. Mincing of mammary glands (MGs) at around 300–400 times per mouse is optimal for getting enough properly-sized organoids. Moreover, minimizing the number of dead cells and maximizing the vitality of the organoids is essential for the success of the migration assay. Thus, the entire organoid preparation procedures, which are shortened when compared to the most commonly used organoid preparation protocols, need to complete as fast as one possibly can.
-
1.
To harvest mammary glands, first put two 10 cm Petri dishes and surgery tools on the surgery table and turn on the dry glass bead sterilizer.
-
2.
Turn on 37°C heat block and 37°C shaker in tissue culture room.
-
3.
Warm-up DMEM/F12 bottle to room temperature in tissue culture hood.
-
4.
Prepare collagenase digestion solution:
Add 0.02 g collagenase to a 15 mL tube containing warm 9.3 mL of DMEM/F12, and put it on a 37°C shaker.
Note: Before mincing the mammary glands: add 0.5 mL of FBS, 200 μL of 100 mg/mL trypsin, 10 μL of 50 mg/mL gentamicin, 25 μL of 2 mg/mL insulin. Sterilize the solution by running it through a 0.22 μm vacuum filter.
CRITICAL: Collagenase does not dissolve well. So add it to the medium early to allow more time for it to dissolve. Add the trypsin stock solution last, i.e., right before the filtration step, to minimize collagenase digestion by trypsin. Collagenase digestion solution should be freshly prepared each time.
-
5.
Prepare to harvest the MGs from a mature virgin female mouse of 6–10 weeks of age.
Sterile techniques should be used on an open bench during mammary gland harvest.-
a.Wipe down the dissection station with 70% ethanol.
-
b.Disinfect the dissecting tools by heat in a glass bead sterilizer at 280°C for 1 min. Cool the surgical tools on an open Petri dish.
-
c.Sterilize all the surgical supplies, a surgical board (a Styrofoam board covered in an aluminum foil ), and pins with 70% alcohol. Use sterilized cotton swabs during mammary gland harvest.
-
a.
-
6.
Euthanize the mouse in a CO2-saturated chamber for 3–5 min, followed by cervical dislocation.
-
7.
With the carcass on its back, pin it to the surgical board. Then spray with 70% ethanol until the fur is soaking wet.
-
8.
To harvest the #3, 4, and 5 MGs from the mouse, identify the midline between the two hind legs. Pinch a small piece of the skin with a tweezer (JZ, Cat# JD1080), and make a small incision on the abdominal skin with sharp scissors (SCRC, Cat# 92360147), then extend the cut along the ventral midline up to the neck (Figures 2A and 2B). Be careful to cut only the skin and not the peritoneum underneath.
-
9.
Extend the cuts laterally towards the legs and arms. Use the tweezer and a cotton swab to release the skin from the peritoneum. Pull the skin away and stretch it tight before pinning it down (Figure 2B'). Remove the lymph nodes (Figure 2C) located at the intersection of three blood vessels in gland #4 (Figure 2C'). Then harvest the remaining #4 MGs similarly as the #5 MGs using forceps (JZ, Cat# JD1050) and scissors to grasp and cut them from the skin (Figure 2E). For harvesting the #3 MGs (Figure 2D), pull up a thin layer of muscle located on top of gland #3 to expose the mammary gland (Figure 2D’), and use a cotton swab to push the #3 MG together (Figure 2D''). This will reduce the contamination by muscle tissue.
-
10.
Collect and pool the MG tissues on the lid of a Petri dish (Figure 4A). The edge of the lid is shallow, thus making the tissues more accessible to the blades and easy to mince.
-
11.
Bring the dissected MG tissue to a tissue culture hood.
-
12.
Prepare collagenase solution as mentioned in step 4.
-
13.
Mince the glands in the Petri dish lid using two surgical blades (Jinhuan, surgical blade #22, Cat# V500620, scalpel knife handle, Cat# K6-20). Rotate the lid after mincing every ten times so that the tissues will be chopped again in a different orientation. The MGs will have been finely minced after chopping with two scalpels for ∼300–400 times per mouse (Figure 4A′).
Note: The mincing conditions have been optimized for this assay. For harvesting organoids, fibroblast cells for FGF2 induced branching assay, chopping should be less than 120 times. If over-chopped or under-chopped, the organoids might be too small or too big, either of which will negatively affect the performance of the assay.
-
14.
Transfer the minced tissue into the collagenase solution, mix well (Figure 4B). Place it horizontally in a 37°C shaker to ensure even mixing. Shake for 22 min.
Note: While waiting for digestion, warm 4 mL of the medium for the DNase solution (see recipe in materials and equipment). Coat the inner surfaces of a 15 mL tube (Figure 3A) and two 35 mm dishes (Figure 3B) with 2.5% BSA (see recipe in materials and equipment). Without the coating, organoids tend to stick on the plastic surfaces. So all serological pipettes (Figure 3D) and pipette tips (Figure 3C) must be coated with the BSA solution. To coat the tubes, mix BSA solution thoroughly to cover the inner walls, then it can be removed. A short coating with BSA works well enough, though more prolonged exposure does not seem to pose adverse effects either.
Figure 2.
Collection of the mammary glands from a mouse
(A) Schematic diagram of isolation and three-dimensional culture of mouse mammary organoids for migration assay.
(B) Use surgical scissors to cut the mouse skin, and the incision sequence is marked with numbers.
(B′) Expose glands #3, 4, and 5 by pushing the abdomen with a cotton swab toward the mouse’s center. The solid black line indicates the general area of the mammary gland corresponding to the label.
(C and C′) Remove lymph node from the #4 mammary gland with scissors and forceps.
(D–D'') A thin layer of muscle partially covers gland #3 (D). Pick up the muscle with forceps, and then use the cotton swab to push gland #3(D′) below the muscle to isolate gland #3 (D''). The solid black line represents the approximate area of gland #3.
(E) Use scissors and tweezers to remove gland #5 below the thigh. The solid yellow line represents the approximate area of gland #5.
Figure 4.
Isolation of mammary organoids
(A and A ′) Collected mammary tissue was placed in the lid of a Petri dish (A). The MG tissue was minced (A ′) with double surgical blades about 300 times.
(B and C) (B) Transfer mammary gland to collagenase solution, (C) incubated at 37°C for 22 min with rotation, thereby breaking down the fat pad into relatively dispersed pieces.
(C′) After digestion, DNase was added, and the suspension was pipette up and down ten times with a 10 mL serological pipette.
(C'') Centrifugation separates the suspension into three layers, the top layer is an opaque fatty layer, the middle is a transparent aqueous phase, and the bottom precipitation is the organoid pellet.
(D) The organoid pellet was resuspended in 4 mL DMEM/F12 containing DNase and transferred to new tubes.
(D′) DIC images of the resuspended pellet with many single cells.
(E) Pellet of grayish-white organoids after four differential centrifugations.
(E′) DIC images of the resuspended pellet. Single cells were removed.
(F) DIC image of picked organoids. The proper size is about 50 μm diameter × 100–200 μm length.
(G and G′) Pictures of organoid epithelium under different focus levels. It is hard to distinguish endothelium from organoid epithelium under the stereo microscope.
Figure 3.
Precoating the tube, dish, and pipette tip with BSA
Fresh organoids tend to stick on the uncoated plastic surface, so it is necessary to precoat all the plastic surfaces with 2.5% BSA.
(A) Precoat a 15 mL tube by adding BSA solution, mixing up and down to coat all plastic surfaces, and removing the BSA solution.
(B) Precoat a 35 mm dish by adding 2 mL of BSA solution and removing it.
(C) Precoat a 1 mL pipette tip by taking up BSA solution and eject out.
(D) Use the same way to coat a 10 mL pipette.
-
15.
After digestion (Figure 4C), add 40 μL of DNase solution to the mixture. Pipette it up and down ten times with a 10 mL serological pipette (Figure 4C').
Note: DNase treatment will digest the viscous genomic DNA leaked out of the dead cells during digestion and thus will make the solution clearer and less sticky.
-
16.
Spin the tube at 560 g for 10 min at 25°C, accel/brake set as 8 (Eppendorf centrifuge 5810 R).
Note: After centrifugation, the solution in the tube will form three layers: a fatty layer on the top, an aqueous layer in the middle, a pellet layer of organoids at the bottom (Figure 4C'').
-
17.
Remove the fatty layer and the aqueous layer.
-
18.
Tap on the tube wall to loosen up the pellet, transfer it into the BSA-coated 15 mL tube containing 4 mL of warm DMEM/F12 medium, add 40 μL of DNase solution (Figures 4D and 4D′). Mix well by pipetting up and down, and let the tube sit in the hood for 5 min. The cotton-like genomic DNA will slowly disappear.
-
19.
Add 6 mL DMEM/F12 and mix gently.
-
20.Differential centrifugation
-
a.Pulse at 450 g (accel/brake set 8), then stop as quick as possible.
-
b.Pour out the supernatant.
-
c.Resuspend pellet in 10 mL of DMEM/F12 with 250 μL of 2.5% BSA solution.
-
d.Repeat the above steps for three more times.
-
a.
Note: The purpose of differential centrifugation is to wash out enzymes and remove unwanted single cells from organoids. It is crucial to add extra BSA after each centrifugation. If BSA is not added, organoids will still stick to the tube, and the yield will be compromised.
-
21.
The remaining pellet (Figure 4E) should be mostly organoids. Resuspend it in 2 mL of basal medium (see recipe in materials and equipment) (Figure 4E’), then place the mixture on ice.
-
22.
Transfer the organoids into a 35 mm dish coated with 2.5% BSA and place it on ice (Figure 5B).
Figure 5.
Prepare the equipment and materials for the experiment
(A) Full view of the workbench. Materials that necessary for the migration experiment are on the table. The table is set in a sterilized room.
(B) Experimental materials on ice. Coated heparin beads and fresh organoids are stored in a 35 mm dish. PBS, BSA, Matrigel, and needles are store in tubes.
(C–D''') Setting up of the stereo microscope during the operation of the migration experiment. (C) Align the center of the working well with one cold block hole and the stereo microscope views. (D) A blue rack was used for picking up the beads and organoids. Use the top light on the stereo microscope. (D′–D''') The black disk was removed when arranging the beads and organoids in a 24-well plate. Use the cold light source.
(E and E′) Water condensation on the cold block surface (E), wipe the water off the surface (E′).
Set up 3D culture for migration assays
Timing: 2–6 h
Pairing organoids with FGF10-coated beads.
Manually juxtapose the organoids with the FGF10-coated beads under a stereo microscope. The goal is to arrange five pairs in each well.
-
23.
Wash FGF10-coated beads three times with 0.1% BSA in PBS, 200 μL per wash, spin 15 s on a mini centrifuge. Transfer washed beads into a 35 mm dish containing 2 mL of 0.1% BSA in PBS. Let the samples sit on ice.
-
24.Juxtapose a bead with an organoid in 80% Matrigel (see recipe in materials and equipment).Note: Before starting the experiment, prepare the working station (Figures 5A and 5B).
Before handling the Matrigel, pipette tips should be cooled by soaking in ice-cold PBS for 20 s. Before handling organoids, pipette tips should be coated with 2.5% BSA in PBS.
Both 60% and 80% Matrigel works for this assay. However, in 60% Matrigel, there is a higher chance that the organoid may settle and touch the glass bottom, at which point it will spread on the bottom surface rather than directionally migrate toward FGF10 beads. This assay is sensitive to Matrigel concentration. When the concentration of Matrigel is too high, organoid migration may be hindered.
When opening a new vial of Matrigel, record the concentration of the gel. The Matrigel protein concentration between 5.7 mg/mL to 7.6 mg/mL works well for this assay. The calculation is based on the protein concentration of different lot numbers provided by the manufacturer.-
a.Choose five organoids of appropriate sizes under the stereo microscope (Figures 5D and 6A).Note: The proper size is about 50 μm diameter × 100–200 μm length (Figures 4F, 4G, and 4G′). When an organoid length is less than 50 μm, its morphology tends to look defective. Organoids first undergo a sealing process to form a cyst (Figure 7B’). If the organoid length is longer than 200 μm, the area center of the cyst will be more than 150 μm away from the bead. Such organoids often respond poorly to FGF10 stimulation and do not migrate well toward the bead.
-
b.Place a glass-bottom plate on a cold block for 1 min (Figure 6B).
-
c.Use a cold tip (10 μL tip), transfer 7.5 μL of 80% Matrigel, and coat a small circle (1 cm diameter) on a 24-well glass-bottom plate (Figures 6B and 6B′). Make sure the tip does not touch the wall. Add an extra 7.5 μL of 80% Matrigel in the coating area. So the coating is finished with a total of 15 μL gel (Figure 6B’’). It is easier to spread the gel when using a small volume.Note: When leaving the chamber on the ice block for too long, water condensation might form in the well. If this occurs, water condensation can be removed with a vacuum before laying on the coating gel.If the sample will be used for subsequent immunofluorescence (IF) staining, an 8-well glass-bottom chamber can be used. In this case, 2 μL of coating gel should be used. The coating area is a 0.6 cm diameter cycle.
-
d.Put the plate on a 37°C heat block for 1–2 min to solidify the Matrigel (Figure 6B).
-
e.Transfer the 24-well glass-bottom plate on the cold block for 2 min (Figure 6B).
-
f.Use a cold tip to transfer 20 μL of 80% Matrigel onto the coating gel (Figure 6C).
-
g.Add 5 organoids to the right side of the top gel (Figure 6D).
- h.
-
i.Line up the beads with a fine needle prepared earlier, and allow >300 μm of distance between beads. Juxtapose the organoids with the beads and allow a 50–100 μm of distance between them. Repeat the above steps until five organoid-bead pairs have been set up for each well (Figures 6D and 6E).
-
j.Let them sit still for 1 min.
-
k.Transfer to 37°C heat block for 8 min (Figure 6F).Note: Double-check and make sure all of the wells have the organoids and beads in the right places before transferring them to the 37°C heat block. While waiting for the gel to solidify, find the right-sized organoids under the stereo microscope to get ready for the next round of set-up. Four wells of samples can be set up together for each experiment.
-
l.Setting up one well usually takes about 1 h. And setting up four wells together takes approximately 1.5–2 h.
-
m.Add 1 mL of basal organoid culture medium to each well (Figure 6F).
-
n.Repeat the above steps for the next well (Figures 6A–6F).
-
a.
-
25.
When finished, add autoclaved water with P/S in the empty wells to maintain humidity.
-
26.
Samples can be cultured in the incubator and imaged at different time points. The usual time points include 0, 24, 36, and 72 h (Figures 6G, 7A–7A’’’, and 7B–7B’’’).
Figure 6.
Schematic diagram of the migration experiment steps
(A) Organoids of appropriate size were selected under a stereo microscope and placed in a 35 mm dish for later use.
(B) Side view of the process of laying Matrigel on a 24-well plate. First, place the plate on the cold block for 1 min, draw a 1 cm circle on the bottom using a 10 μL tip containing 7.5 μL Matrigel, fill the circle, then add an extra 7.5 μL Matrigel.
(B ′ and B'') A top view diagram of the above process. Once Matrigel is laid, the plate is heated on the metal block at 37°C for 2 min and then cooled down on the cold block for 2 min. All subsequent operations are performed on the cold block.
(C) Add 20 μL top Matrigel to the solidified coating gel.
(D) Aspirate five organoids at a time with a P2 pipette setting at 1.2 μL and add them to the right side of the top Matrigel. In the same way, aspirate five beads and add them to the left side of the top Matrigel. Under the dissection microscope, line up the beads with the fine needle, >300 μm apart between beads. Then, the organoid was juxtaposed to the right side of the bead in a one-to-one manner, 50 μm–100 μm apart between the bead and organoid.
(E) After completion, the overall view of the position and proportion.
(F) The plate was heated on the metal block at 37°C for 8 min to solidify the Matrigel, and then 1 mL basal medium was added.
(G) For time-course images or IF staining, the sample can be cultured in an incubator at 37°C. The sample can also be maintained in time-lapse microscopy.
(H) Flip the image horizontally when present the data.
Figure 7.
Different responses of epithelial organoids to beads pre-soaked in BSA (A–A'''), or FGF10 (B–B''')
(A–A''') Organoids did not migrate toward the beads pre-soaked in BSA.
(B–B''') Time course of directional migration of epithelial organoid toward the FGF10 bead. The beads of ~100 μm in diameter were juxtaposed with mammary organoids at a distance of ~100 μm. Scale bars, 50 μm.
(C–D′) (C and D) Immunofluorescence examination of cell markers in organoids at the migrating cyst stages. IF of CD49f (Integrin alpha 6, staining with 1:100 dilution) on organoid juxtaposed with BSA (C) or FGF10 (C′) soaked bead. IF of SMA (staining with 1:200 dilution) on organoid juxtaposed with BSA (D) or FGF10 (D′) soaked bead.
Alternatively, the samples can be imaged via differential interference contrast (DIC) microscopy for the whole course of 72 h and by taking images every 15 min.
Live cell imaging
Timing: 3 days
Organoid collective migration is monitored by time course or time-lapse microscopy.
The migration status of the organoid can also be evaluated by time-lapse imaging.
-
27.
For time-lapse imaging, DIC microscopy can be performed on a Zeiss Cell Observer SD spinning disk confocal microscope using an EC Plan-Neofluar 10×/0.3 Ph1 M27 objective lens (Item no.: 420341-9911-000), an Analy DIC Transmission light reflector, an sCMOS camera at 37°C with 5% CO2. Images were taken every 15 min using ZEN (blue edition) software. Refocus was performed every 12 h. Images can be assembled using Fiji software (Methods video S1).
DIC movie of directional migration of epithelial organoid toward an FGF10-bead. Images were recorded using time-lapse microscopy over 86 hours and 45 minutes. Images were taken every 15 minutes, and the playing speed is 20 frames per second.
Immunofluorescence staining of 3D cultured samples
Timing: 3 days
-
28.
For IF staining, the samples are set up in an 8-well glass-bottom chamber. Samples are harvested 36 h later. Remove medium from the chamber.
-
29.
Fix Matrigel and organoid by incubating the samples with 300 μL of 4% PFA solution (see recipe in materials and equipment) for 15 min.
Note: In the PFA solution, Matrigel shrinks and becomes fragile. The shrinkage will reduce the thickness of the coating gel and make it easier for imaging. The lowest percentage of the Matrigel is 80%. If the concentration gets lower even more, the gel will fall apart during fixation. Different lot numbers of Matrigel might have different concentrations of protein. This will also influence the gel stability in the PFA solution. If the pH of the 4% PFA solution is not correct, the Matrigel can also be dissolved. A pilot experiment should be done to check if the Matrigel is stable upon fixation.
For if your protein of interest shows weak IF signals, putting the sample on a rocking shaker at 30 rpm for all the incubation steps can improve the signal/noise ratio.
-
30.
Remove PFA solution, and wash 3-times, 15 min each, with quenching solution (see recipe in materials and equipment).
-
31.
Permeabilize the gel with permeabilizing buffer (see recipe in materials and equipment) for 3 h at room temperature.
-
32.
After permeabilization, wash 3-times 15 min each with PBS.
-
33.
Immediately block samples with blocking buffer (see recipe in materials and equipment) for 2 h at room temperature or overnight at 4°C. Remove the blocking solution, add primary antibody diluted in blocking buffer at the desired ratio, and incubate for 1–2 days at 4°C.
Note: To avoid spilling of the primary antibody over to the next well, remove the chamber’s lid. And put the chamber in a wet box. Seal the box with a zip bag.
-
34.
Remove the primary antibody solution, and wash 3-times 15 min each with PBS at room temperature.
-
35.
Add secondary antibody in blocking buffer at the desired ratio, and incubate for 1–2 days at 4°C.
-
36.
Stain with DAPI 1:2000 in PBS for 15 min.
-
37.
Wash 3-times 15 min each with PBS at room temperature.
-
38.
Store samples in PBS at 4°C.
-
39.
Confocal images should be taken within three days. Still confocal images can be acquired using a Leica SP8 STED 3× Leica microscope using an HC PL APO CS2 40×/1.30 OIL objective. The magnification is 40×. Image resolution is 1,024 × 1,024 square pixels, with a calibration factor 0.098 μm/pixel (Figures 7C and 7D), 0.117 μm/pixel (Figure 7C’), and 0.125 μm/pixel (Figure 7D’). Red-green-blue (RGB) images can be assembled using Fiji software (Figure 6H, 7C, 7C′, 7D, and 7D′). For imaging CD49f, a laser line with a 631 nm of wavelength, 32% of intensity, 132% of HyD detector gain is used, and the image processing threshold range is 200–2000. For imaging SMA, a laser line with a 498 nm of wavelength, 2% of intensity, 146% of HyD detector gain is used, and image processing threshold range is 100–3000. For imaging DAPI, a laser line with a 405 nm of wavelength, 4% of intensity, 100% of HyD detector gain, and image processing threshold range is 100–3000.
Expected outcomes
Heparan beads pre-soaked in FGF10 but not in bovine serum albumin attracted organoid migration (Figures 7A’’’ and 7B’’’). The organoid first underwent a sealing process to form a cyst (Figure 7B'). By the time the organoid was about to migrate at ∼36 h, the side of the organoid closer to the FGF10 bead, which we refer to as the “organoid front,” was noticeably thicker than the opposite, or rear, side (Figures 7B'', 7C', and 7D′). Directional migration from FGF10 stimulation is a multi-stage process by which a simple epithelium forms a polarized cyst, which then undergoes epithelial sealing, differential cell proliferation and stratification, and subsequent migration (Lu et al., 2020).
Limitations
One limitation of the current protocol is that it has only been optimized for mammary gland epithelium, and not for other vertebrate epithelia. Conditions, e.g., growth factors used, will need to be changed/optimized depending on the epithelia of interest.
Likewise, another limitation is that the heparin beads, which are useful for soaking and then delivering FGF10, might not be suitable for other growth factors. Users will need to find the best method to deliver their proteins/growth factors of interest.
Troubleshooting
Problem 1
Organoid floating on the Matrigel, at step 24: i.
Potential solution
When pairing organoids with beads, if organoids are found floating on Matrigel’s surface, use a fine needle to gently push them into the gel. Once they enter the Matrigel, they will slowly sink to the bottom.
Problem 2
It is challenging to manually arrange the distance of 100 μm between beads and organoids, at step 24: i.
Potential solution
First, to determine the distance under a stereo microscope relatively accurately, a reference bead of 100 μm in diameter is required (Figure 1D, D' and E). A distance less than 100 μm but greater than 50 μm between the bead and the organoid is acceptable. Furthermore, try to keep the beads and organoids in as the middle of the Matrigel drop and away from the edges as possible. The length from the first pair to the fifth pair should be half of the diameter of the entire coating gel (Figure 6E). Finally, each organoid-bead pair should be at least 300 μm apart to minimize the impact between each pair.
When using the needle to adjust the bead and organoids in the Matrigel, the needle does not need to touch them directly. Let the needle flick the Matrigel that is directly above or by the side of the bead or organoid. If the needle touches them directly, it tends to scratch the solidifying coating gel or the organoids and cause unnecessary damages to them. Practice will until you are skilled at these manual operations. Start from working one well each time. Once you are familiar with the procedure, then you can start working on four wells each time.
If the room temperature is higher than 25 Celsius or the operation time is too long, Matrigel will solidify. If the bead or organoid cannot be pushed around in Matrigel, it is because the gel has solidified. If so, discard this well and start over.
Problem 3
Beads and organoids are initially well arranged, but become scattered or misarranged once the Matrigel has solidified, at step 24: m.
Potential solution
This annoyance could happen when working with multiple wells of samples at once. Any vibrations to the wells could cause the samples to move and become misarranged in the unsolidified Matrigel. When moving the icebox and the 24-well plate together, the black disk on the stereo microscope might interfere with smooth movement (Figure 5D'). Thus, we find it helpful to remove the black disk when working with the icebox (Figures 5D'' and 5D’’’). Moreover, water condensation on the cold block could also cause noticeable vibrations when moving the 24-well plate. Wipe clean any water condensation before putting the 24-well plate on the block (Figures 5E and 5E′).
Problem 4
Organoids touch the glass bottom of the culture plate, at step 26.
Potential solution
If the coating gel is not well solidified, the organoid will tend to touch the culture plate bottom, which causes the organoid to spread, rather than to directionally migration. If the ambient temperature is lower than 20oC, the step of placing the plate on a cold block for 1 min can be omitted (step 24: b, Figure 6B).
Alternatively, leave the coated plate on the 37°C heat block for up to 3 min. Be careful, however, that leaving it on the heat block for too long may cause excessive drying of the gel, thus changing gel concentration and compromise organoid migration.
Problem 5
Organoids do not migrate towards FGF10-coated beads, at steps 26 and 27.
Potential solution
First, the organoids should not be too big or too small. Their lengths, when derived from mammary gland ductal epithelium, should be between 100 to 200 μm. Second, the organoid front, which refers to the side closer to the FGF10 bead, should be 50–100 μm apart from the bead. This means that the area center of the organoid should be 100–150 μm apart from the bead. Third, multiple (three to five) pairs of juxtaposed organoid-bead should be used to determine the migration efficiency. If the above conditions have all been met, but the conclusion still is that there is no migration, then the possible solution is to look at the critical reagents one by one, especially the Matrigel and FGF10.
Regarding Matrigel, make sure you know the lot# of the Matrigel and that it has been stored at -20°C. Moreover, the freeze-thaw cycle of leftover Matrigel should be less than five times. According to the experimental requirements, usually the aliquot volume of Matrigel is 250 or 500 μL per tube. It is best to use the newly aliquoted Matrigel when performing the 3D migration assay for the first time, which will reduce the difficulty of troubleshooting. We only used Corning's Matrigel (Corning, Cat#354230) in this experiment and haven't tried any other company's basement membrane extracts. When you use other company's products, it is a good practice to use Corning's Matrigel as a positive control to ensure that other brands can provide the same migration results.
Regarding FGF10, buying a large quantity of FGF10 is not recommended. Store lyophilized FGF10 at −80°C until use. Follow the manufacturer's instructions to reconstitute it. Make small aliquots, 5 or 10 μL per tube, so that one aliquot is used up each time if possible. Store at −80°C. Upon reconstitution, the aliquots should be used within six months. We have compared FGF10 provided by different manufacturers and have found that rmFGF10 (GenScript, Cat#Z03155) worked well.
Organoids should be used within 7 h. For a skilled experimenter, it usually takes 1.5 h to arrange a well containing five organoid-bead pairs. Most of the steps of this protocol have been optimized, although we recognize there is still room for improvement.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Pengfei Lu (lvpf@shanghaitech.edu.cn).
Materials availability
This protocol did not generate new unique reagents.
Acknowledgments
We thank the Mouse Core Facility at the National Institute for Protein Science Center at Shanghai and Jiabo Biotechnology. We also thank the Molecular Imaging Core Facility (MICF) at ShanghaiTech University. This work was supported by the National Natural Science Foundation of China (NSF) grant 31671494 (to P.L.), the Ministry of Science and Technology of China grant 2017YFA0103502 (to P.L.), and a start-up grant from Shanghai-Tech University (to P.L.).
Author contributions
Conceptualization, P.L. and Y.L.; investigation, Y.L.; writing – original draft, Y.L.; writing – figure legend, R.D. and H.Y.; writing – review & editing, P.L.; funding acquisition, P.L.; supervision, P.L.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100778.
Contributor Information
Yunzhe Lu, Email: lvyz@shanghaitech.edu.cn.
Pengfei Lu, Email: lvpf@shanghaitech.edu.cn.
Data and code availability
This protocol did not generate datasets or code.
References
- Lu Y., Deng R., You H., Xu Y., Antos C., Sun J., Klein O.D., Lu P. Asymmetric stratification-induced polarity loss and coordinated individual cell movements drive directional migration of vertebrate epithelium. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Martinez D., Koledova Z., Qiao G., Streuli C.H., Lu P. FGF ligands of the postnatal mammary stroma regulate distinct aspects of epithelial morphogenesis. Development. 2014;141:3352–3362. doi: 10.1242/dev.106732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Qiao G., Lu P. Modulation of fibroblast growth factor signaling is essential for mammary epithelial morphogenesis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DIC movie of directional migration of epithelial organoid toward an FGF10-bead. Images were recorded using time-lapse microscopy over 86 hours and 45 minutes. Images were taken every 15 minutes, and the playing speed is 20 frames per second.
Data Availability Statement
This protocol did not generate datasets or code.