Abstract
Progressive fibrosis leads to loss of organ function and affects many organs as a result of excessive extracellular matrix production. The ubiquitous matrix polysaccharide hyaluronan (HA) is central to this through association with its primary receptor, CD44, which exists as standard CD44 (CD44s) or multiple splice variants. Mediators such as profibrotic transforming growth factor (TGF)-β1 and proinflammatory interleukin (IL)-1β are widely associated with fibrotic progression. TGF-β1 induces myofibroblast differentiation, while IL-1β induces a proinflammatory fibroblast phenotype that promotes fibroblast binding to monocyte/macrophages. CD44 expression is essential for both responses. Potential CD44 splice variants involved, however, are unidentified. The TGF-β1-activated CD44/epidermal growth factor receptor complex induces differentiation of metastatic cells through interactions with the matrix metalloproteinase inducer, CD147. This study aimed to determine the CD44 variants involved in TGF-β1- and IL-1β-mediated responses and to investigate the potential profibrotic role of CD147. Using immunocytochemistry and quantitative PCR, standard CD44s were shown to be essential for both TGF-β1-induced fibroblast/myofibroblast differentiation and IL-1β-induced monocyte binding. Co-immunoprecipitation identified that CD147 associated with CD44s. Using CD147-siRNA and confocal microscopy, we also determined that incorporation of the myofibroblast marker, αSMA, into F-actin stress fibers was prevented in the absence of CD147 and myofibroblast-dependent collagen gel contraction was inhibited. CD147 did not associate with HA, but removal of HA prevented the association of CD44s with CD147 at points of cell–cell contact. Taken together, our data suggest that CD44s/CD147 colocalization is essential in regulating the mechanical tension required for the αSMA incorporation into F-actin stress fibers that regulates myofibroblast phenotype.
Keywords: transforming growth factor-β1, differentiation, hyaluronan, CD44, EMMPRIN, CD147
Abbreviations: αSMA, α-smooth muscle actin; CFM, confocal microscopy; CKD, chronic kidney disease; Co-IP, coimmunoprecipitation; ECL, enhanced chemiluminescence; ECM, extracellular matrix; EGFR, epidermal growth factor receptor; HA, hyaluronan; ICAM, intercellular adhesion molecule; ICC, immunocytochemistry; IL, interleukin; PIC, protease inhibitor cocktail; RT-qPCR, real time–quantitative polymerase chain reaction; TGF, transforming growth factor
Fibrosis underlies several organ-specific diseases and contributes to the burden of chronic diseases, such as chronic kidney disease (CKD), liver cirrhosis, pulmonary fibrosis, cardiac failure, and degenerative joint disease (1, 2, 3, 4, 5). Myofibroblasts are the principal effector cells driving fibrosis, and their accumulation in tissues is a fundamental marker of fibrosis. These cells are derived from the differentiation of fibroblasts, fibrocytes, or epithelial cells under the influence of circulating profibrotic growth factors such as transforming growth factor–β1 (TGF-β1) (6, 7, 8, 9). We have identified the increased synthesis and accumulation of the linear polysaccharide hyaluronan (HA) as central to this event through its interactions with its principal receptor, CD44 (10).
HA is a glycosaminoglycan consisting of repeating D-glucuronic acid and N-acetyl-glucosamine disaccharide units. It influences distinct cellular functions depending on its manner of synthesis, assembly, protein-binding interactions, cell localization and organization. HA is synthesized by three HA-synthase-(HAS)-isoenzymes (HAS1, HAS2, HAS3) (11), and broken down by hyaluronidase-(HYAL) enzymes (HYAL1, HYAL2) (12). In vitro experiments have shown that TGF-β1 drives HAS2-specific synthesis of HA-pericellular matrices, tethered to the principal HA receptor (CD44) at the cell surface (13, 14). This HA-CD44 binding induces the migration of CD44 in the plasma membrane and promotes interactions with the epidermal growth factor receptor (EGFR) in lipid rafts leading to MAPK/ERK signaling (15). This, in parallel with the canonical Smad-signaling pathway, drives differentiation of fibroblasts to α-smooth muscle actin (αSMA)-positive, contractile myofibroblasts that generate the fibrous collagen matrix, characteristic of fibrosis and scarring (16). IL-1β is a proinflammatory mediator that induces fibroblasts and other cells to adopt an inflammatory phenotype, which includes the upregulation of intercellular adhesion molecules. This phenotypic change is also dependent on an HA/CD44 interaction. This change, however, involves the formation of HA protrusions that bind monocytes/macrophages to HA/CD44/intercellular adhesion molecule (ICAM)-1 complexes in non-lipid-raft regions of the plasma membrane. The complex induces the activation of a downstream p38 pathway (10). Similarly, to the TGF-β1 response observed in myofibroblasts, IL-1β-mediated HA protrusions are HAS2-dependent.
CD44 is a transmembrane glycoprotein, which can exist as multiple variant isoforms due to alternative splicing (17). It is broadly distributed in tissues and is involved in diverse physiological processes. Its aberrant function is implicated widely in pathology, with key roles in cancer, inflammation, immune dysregulation, vascular disease, fibrosis, and wound healing (18, 19, 20, 21, 22, 23). In humans, 19 exons encode for CD44 pre-mRNA. The first five exons (1, 2, 3, 4, 5) and the last five exons (15, 16, 17, 18, 19) are common to all the CD44 variants. The variability of CD44 is the result of alternative splicing of nine exons (6, 7, 8, 9, 10, 11, 12, 13, 14) that are situated between these common regions. The simplest CD44 variant has only the common exons translated into protein and is known as CD44 standard or CD44s. This is the most abundant of all the CD44 variants and is present in most cell types. In contrast, the largest CD44 variant, which has all the variable exons translated into protein and is known as CD44v2-10, is expressed much less widely. We have recently described the expression of a variant of CD44 (CD44v7/8), which is strongly induced by bone morphogenetic protein (BMP)-7. This variant becomes highly expressed at the cell surface and has antifibrotic effects in fibroblasts, preventing their TGF-β1-dependent differentiation to myofibroblasts (22). However, we have not yet identified which and how many of the other potential CD44 variants may be involved in the profibrotic myofibroblastic differentiation or the proinflammatory responses of fibroblasts.
CD44 undergoes multiple interactions at the cell surface and an important regulator of the interaction of CD44 with EGFR is CD147 (24). CD147, also known as extracellular matrix metalloproteinase inducer, EMMPRIN or Basigin, is a type-1 transmembrane glycoprotein and a member of the immunoglobulin superfamily (25). The CD147 gene encodes for a protein, which has a 185 amino acid extracellular region, a 24 amino acid highly conserved transmembrane region, and a 39 amino acid cytoplasmic domain (26, 27). The extracellular region is composed of two Immunoglobulin domains with similarity to other members of the immunoglobulin superfamily. The translated CD147 protein is 28 kDa, whereas the observed molecular weight ranges between 32 and 66 kDa, due to posttranslational glycosylation.
In vivo experiments have identified that CD147 may be a major contributor to fibrosis. CD147 expression has increased correlation and colocalization with hepatic stellate cells in the space of Disse as they undergo EMT to myofibroblasts, in carbon tetrachloride-induced mouse liver fibrosis (28). The role of CD147 in fibrosis and its potential as a target for prevention and reversal has recently been described in corneal opacity. The dysregulated and increased expression of myofibroblasts and increased αSMA expression contribute to the formation of corneal opacity, which contributes to blindness, following an initial insult. Using SP-8356, an inhibitor of CD147, topically on rat corneas that had undergone alkali burns, inhibited αSMA expressing myofibroblasts and other fibrotic-associated ECM components such as collagen and MMP9 (29). Basigin (bsg)+/− mice had less cardiac interstitial fibrosis following transverse aortic constriction compared with control mice. The response of cardiac fibroblasts from bsg ± and bsg+/+ was also assessed following angiotensin II mechanical stretch stimuli and CD147 expression increased significantly in bsg+/+ cells compared with cells isolated from bsg ± mice (30). In chronic kidney models, Bsg +/+ mice that had undergone unilateral ureteral obstruction had increased kidney interstitial fibrosis compared with bsg−/− mice with increased MMP activation, macrophage infiltration, and upregulation of αSMA and hyaluronan by fibroblasts (31). Regardless of the primary underlying kidney disease or its pathogenesis in CKD patients, there is also a significant relationship between the degree of proteinuria and urinary CD147 (32). In addition, it has been suggested that in CKD there may be a “molecular circuit” involving CD147, MMPs, and transforming growth factor-β and that this may be involved in the pathogenesis of progressive fibrosis through hyaluronan production (33).
CD147 induces breast cancer invasiveness through association with CD44 and EGFR in lipid rafts (34) and is a well-established inflammatory mediator (35), in addition to its involvement in pathogenesis. Since myofibroblast differentiation also involves CD44/EGFR association in lipid rafts (15) and IL-1β induces CD44/I-CAM-1 association outside of lipid rafts (14), this study examined a potential mechanistic role for CD147 in our in vitro models and further aimed to investigate the hypothesis that there were also specific variants of CD44 involved.
Results
The effect of TGF-β1 and IL-1β on CD44 variant expression
CD44 variants with a single exon insertion
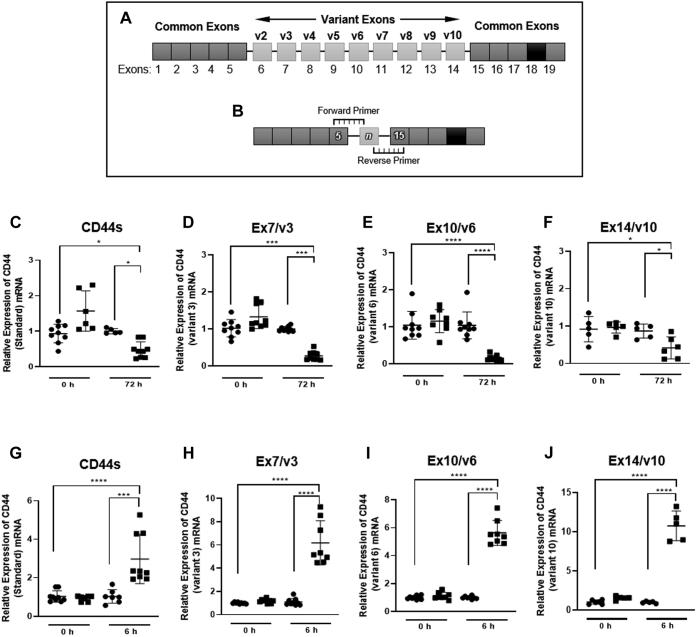
CD44 variants that contained just the common regions and a single spliced exon were amplified using custom-designed primers (Fig. 1, A and B). Using RT-qPCR, the highest expressed CD44 variant was CD44s, followed in descending order by CD44v3, CD44v6, CD44v10 (Fig. 1, C–J). To a lesser extent CD44 variants, CD44v8, CD44v4, CD44v2, CD44v9, and CD44v7 were also expressed (see Figs. S1 and S2). The cytokines TGF-β1 and IL-1β are associated with profibrotic and proinflammatory responses, respectively (15, 16, 36). Both responses have been shown to be mediated by the association of HA with CD44 (14, 15). Using the highest expressed variant CD44s to determine a timepoint for analysis, it was determined that the expression of CD44s decreased following 72 h of stimulation with TGF-β1 (Fig. 1C). In a separate experiment stimulation of fibroblasts with IL-1β increased the expression of CD44s following only 6 h of activation (Fig. 1G). Subsequently all CD44 variants containing a single exon between the common regions were examined at 72 h following TGF-β1 stimulation (Fig. 1, C–F) or 6 h following IL-1β treatment (Fig. 1, G–J). All the variants identified had a response similar to that observed for CD44s when treated with either TGF-β1 or IL-1β. These data suggest that all CD44 variant expression transcription is supressed by TGF-β1 and promoted by IL-1β.
Figure 1.
The expression of CD44 variants containing a single exon insert by untreated TGF-β1and IL-1β-treated fibroblasts. Custom primers were designed to target variants that express one of the variable exons (6–14) between the common regions. Schematic (A) demonstrates the exon arrangement of CD44. Exons 1–5 and 15–19 are common to all CD44 variants. The variable region of CD44 is coded by exons 6–14 (labeled below exons). Corresponding variant nomenclature is highlighted in bold (labeled above exons 6–14). Primers were designed to span the exon boundary of the common region and the variant of interest. Forward primers overlapped the 3′ end of exon 5 in the common region with the 5′ end of the exon of interest. Reverse primers overlapped the 5′ end of exon 15 with the 3′ end of the exon of interest (B). Assessment of CD44 variants containing a single exon insert was carried out using qPCR. The expression of each CD44 variant in fibroblast (black circles) was compared with myofibroblasts (black squares) at 0 h and 72 h timepoints. (C–F) represent; CD44 (standard) (C), CD44v3 (D), CD44v6 (E), CD44v10 (F). The expression of each CD44 variant in fibroblasts (black circles) was compared with IL-1β (1.0 ng/ml) stimulated fibroblasts (black squares) at 0 h and 6 h timepoints. G–J represent; CD44 (standard) (G), CD44v3 (H), CD44v6 (I) and CD44v10 (J). For the mRNA expression of other CD44 variants in basal fibroblasts TGF-β1 and IL1-β stimulation including CD44v2, CD44v4, CD44v7, CD44v8, and CD44v9, see Figures S1 and S2. CD44v5 was not expressed in fibroblasts or myofibroblasts. Data are shown as mean ± SD from three experimental repeats with statistical significance represented by; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001.
In contrast, western blot analysis of the highest expressed variants CD44s, CD44v3, CD44v6, and CD44v10 over a 24–72 h time course showed no effect of TGF-β1 or IL-1β on the protein expression of these variants (see Figs. S3 and S4).
CD44 variants with multiple exon insertions
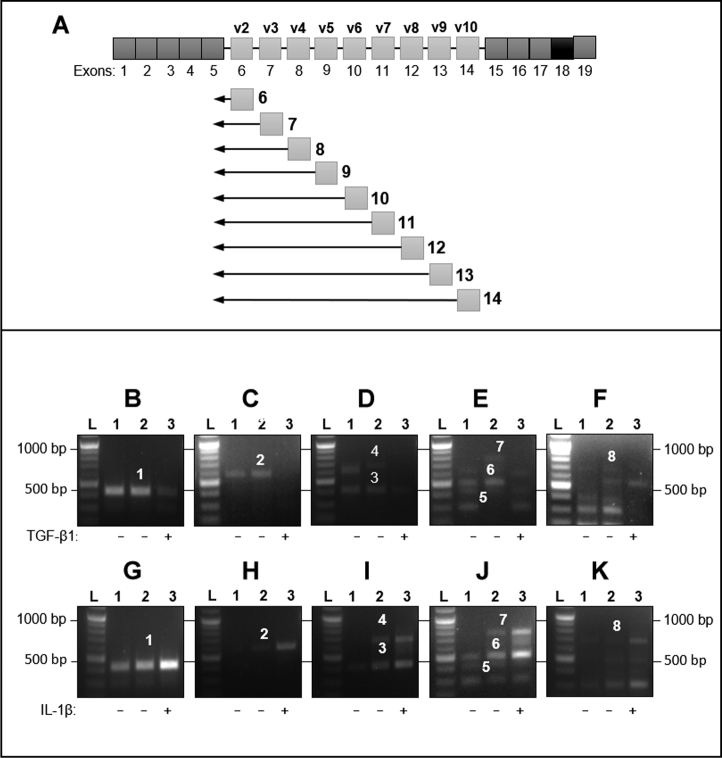
A panel of reverse primers and a common forward primer were used (Fig. 2A) to identify CD44 variants containing multiple exons between the CD44 common regions. All amplifications were carried out using a common forward primer situated in exon 5. When the common forward primer was used with a reverse primer situated in the common downstream region (exon 17), a single product was amplified and identified as CD44s. A single product was also amplified when each of the reverse primers situated in exons 6, 7, and 8 were amplified with the common forward primer. The products were sequenced as CD44v2, CD44v3, and CD44v4, respectively (Fig. S5). Amplification of the common forward primer with separate reverse primers situated within exons 10, 11, 12 and 13 amplified sequentially larger amplicons with each successive primer position (Fig. 2). DNA sequencing of bands 1–7 (Fig. 2, B–E and G–J) was used to identify each amplicon that resulted from each primer combination (Table 1). The final large amplicon (band 7) was identified as a sequential amplicon of CD44 exons 6–10 of approximately 720 bp (Fig. 2, F and K (band 8)).
Figure 2.
Large CD44 spliced variant expression by fibroblasts stimulated with TGF-β1 or IL-1β. Schematic (A) shows the primers designed for PCR. A common forward primer located in exon 5 was used for all targets. A panel of reverse primers that were located within spliced variant exons (6–14) was used to amplify CD44 spliced variants (v2-v10). A separate reverse primer, located in exon 17, was used to identify CD44 standard. Fibroblasts were grown to 80% confluence, then growth-arrested in serum-free medium for 48 h. Cells were treated with TGF-β1 (10 ng/ml, 72 h), IL-1β (1 ng/ml, 6 h), or serum-free media (control samples). RNA extraction, touch-down PCR, gel extraction, and sequencing were all as described in the methods section. Lane 1: control extracted 48 h after growth arrest; Lane 2 control treated with serum-free medium; Lane 3 TGF-β1 stimulated or IL-1β stimulated fibroblasts. Gels shown are representative of those from three independent experiments. Table 1 shows the band number, the position of reverse primers used in TD-PCR, the product size, and the identity, determined by DNA sequencing, of bands 1–8.
Table 1.
Forward and reverse primer exon position for amplification of larger CD44 variants
| Identified product | Band number | Exon position of forward primer | Exon position of reverse primer | Target size |
|---|---|---|---|---|
| CD44v6 | 1 | Exon 5 | Exon 10 | 424 bp |
| CD44 v6-7 | 2 | Exon 5 | Exon 11 | 573 bp |
| CD44v8 | 3 | Exon 5 | Exon 12 | 408 bp |
| CD44v6-8 | 4 | Exon 5 | Exon 12 | 674 bp |
| CD44v9 | 5 | Exon 5 | Exon 13 | 402 bp |
| CD44v8-9 | 6 | Exon 5 | Exon 13 | 496 bp |
| CD44v6-9 | 7 | Exon 5 | Exon 13 | 757 bp |
| CD44v6-10 | 8 | Exon5/Exon10 Boundary | Exon15/Exon14 Boundary | 720 bp |
CD44 standard (CD44s) is the principle CD44 variant involved in TGF-β1-induced differentiation and IL-1β-stimulated monocyte binding
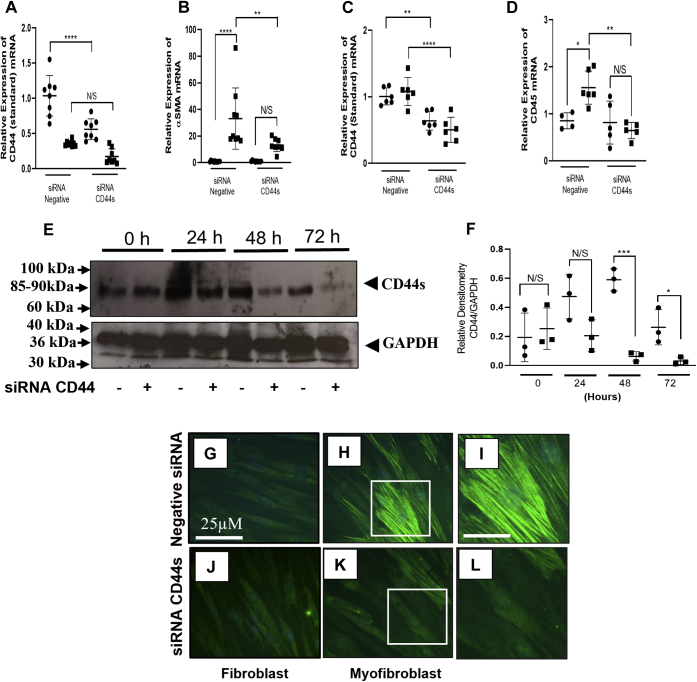
Our laboratory has previously shown that CD44 via its association with HA was required for both TGF-β1-induced fibroblast to myofibroblast differentiation and IL-1β-induced fibroblast monocyte binding (15, 36). However, it is not understood which of the CD44 variants are involved in these pathways. Investigation of the importance of each variant identified was carried out using a process of elimination. Firstly, CD44 variants that were highly expressed (CD44s, CD44v3, CD44v6, and CD44v10) were silenced using custom-designed siRNA oligonucleotides. Additionally, an siRNA to CD44v8 was used to confirm sufficient knockdown of the larger variant CD44v6-10. Silencing mRNA of CD44v3, CD44v6, CD44v8, and CD44v10 had no effect on the expression of αSMA following TGF-β1 stimulation or CD45, (a marker for bound monocytes) following IL-1β treatment (see Figs. S6 and S7). In contrast, silencing CD44s (Fig. 3, A and C) decreased αSMA mRNA expression in myofibroblasts compared with the negative siRNA control (Fig. 3B) and IL-1β-induced monocyte binding (Fig. 3D). However, there was no effect on the expression of CD44 variants CD44v3, CD44v6, CD44v8 or CD44v10 when CD44s was silenced (Figs. S8 and S9). CD44s knockdown at the protein level was confirmed using western blot (Fig. 3E) and quantified using densitometry (Fig. 3F). Using ICC to observe αSMA protein expression demonstrated that cells transfected with siRNA to CD44s had a marked reduction in αSMA stress fiber formation following TGF-β1 induction (Fig. 3, K and L) compared with those cells transfected with the negative siRNA control (Fig. 3, H and I). There was no effect of silencing CD44s by fibroblasts (Fig. 3J) compared with those transfected with the negative siRNA control (Fig. 3G).
Figure 3.
CD44s regulates fibroblast to myofibroblast differentiation and IL-1β-induced monocyte binding. Fibroblasts were cultured in 6-well plates until approx. 50–60% confluent. Cells were growth-arrested for 48 h before transfection with an siRNA to CD44s or a negative siRNA control. Cells were treated with either TGF-β1 (10 ng/ml) or IL-1β (1 ng/ml) for 72 h. Control fibroblasts were treated with serum-free media alone. RNA was extracted and RTqPCR performed firstly to show the knockdown of CD44s (A and C). The effect of CD44s knockdown on αSMA expression (a marker of myofibroblasts) and CD45 (a marker of monocytes) can be observed in (B and D), respectively. Data compare cells transfected with CD44s siRNA to cells transfected with a negative siRNA control and compare control cells (black circles) and cells treated with TGF-β1 (10 ng/ml) (B) or IL-1β (1 ng/ml) (C) for 72 h (black squares). CD44s protein expression was knocked down over a time course of 0–72 h. Cells were treated with siRNA CD44s (+) or with an siRNA negative control (−). Western blot was used to identify CD44s in cell lysates and GAPDH as a loading control (E). Western blots were quantified using Image J software to perform densitometry (using CD44/GAPDH ratios). Proteins from fibroblasts transfected with negative siRNA controls (black circles) were compared with protein from fibroblasts transfected with an siRNA to CD44 (black squares) over a 0–72 h time course (F). Data are shown as mean ± SD from three experimental repeats with statistical significance represented by; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001. ICC shows Fibroblasts (G and J) and myofibroblasts (H, I, K, and L) following CD44s siRNA transfection (J–L) compared with cells transfected with a negative siRNA (G–I). Scale bar for figures (I and L) is 100 μm. Images are representative of each cell population from three independent experiments, magnification ×400.
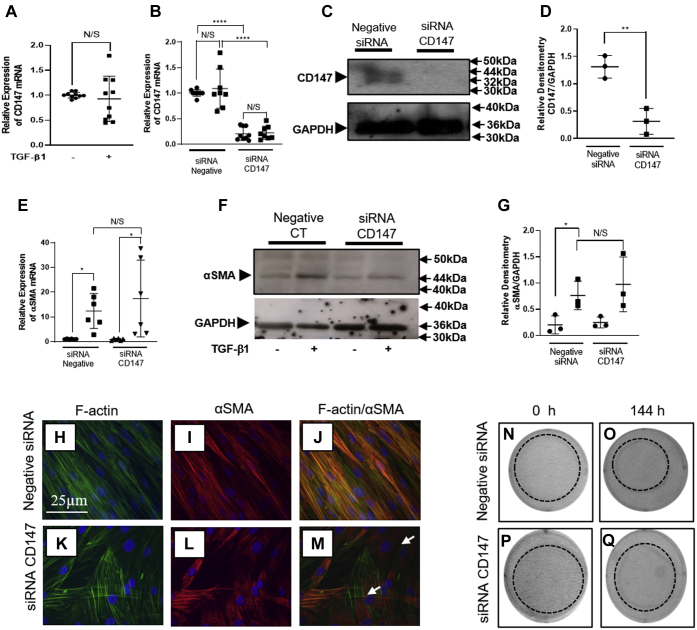
Inhibiting CD147 protein expression prevents the formation of TGF-β1-induced αSMA stress fibers and contractility associated with the myofibroblast phenotype
CD147 is a cell-surface protein that has been previously identified to interact with HA/CD44/EGFR complexes and modulate cellular function (34). In addition, it has long been established that TGF-β1 mediates fibroblast to myofibroblast differentiation in an HA/CD44/EGFR-dependent manner (15). Therefore, this study investigated whether CD147 was involved with the CD44s-dependent induction of the myofibroblast phenotype. Future work will investigate the role of CD147 and CD44s in monocyte binding following IL-1β stimulation. The mRNA for CD147 was highly expressed in our fibroblast model. TGF-β1 induction of the myofibroblast phenotype, however, did not alter the expression of CD147 mRNA (Fig. 4A). To investigate whether CD147 was involved in TGF-β1-dependent myofibroblast differentiation, we used siRNA to CD147. It was determined that the knockdown of the CD147 mRNA was greater than 80% following 72 h of transient transfection (Fig. 4B). However, 144 h of transfection with CD147 siRNA was required for complete knockdown of protein expression (Fig. 4C) and was confirmed using densitometry (Fig. 4D). Therefore, fibroblasts were transfected with an siRNA to CD147 for 72 h and then treated with TGF-β1 for a further 72 h. Control fibroblasts were transfected with a negative siRNA. Transfection for 144 h with siRNA to CD147 did not prevent the induction of αSMA mRNA in TGF-β1-treated myofibroblasts (Fig. 4E). In addition, the expression of αSMA protein was not inhibited following CD147 knockdown (Fig. 4, F–G). Previously, it has been shown that TGF-β1 induction of the myofibroblast phenotype induces the incorporation of αSMA into F-actin stress fibers. In myofibroblasts these stress fibers are arranged cortically throughout the cell and are characteristic of the myofibroblast (37). Transient transfection using a negative siRNA showed that F-actin (green stain; Fig. 4H) and αSMA (red stain; Fig. 4I) colocalized forming yellow cortical stress fibers (Fig. 4J). Myofibroblasts that were transfected with an siRNA to CD147 also had cortical F-actin stress fibers (green stain; Fig. 4K). These data suggest that CD147 does not have a role in regulating the rearrangement of F-actin in fibroblast to myofibroblast differentiation. Interestingly, although αSMA (red stain Fig. 4L) was still expressed, it was not incorporated into cortical stress fibers of the myofibroblasts. Instead, αSMA was situated within the cytoplasmic region of the cell in an irregular pattern. There was limited colocalization when F-actin (Fig. 4K) and αSMA (Fig. 4L) images were merged (Fig. 4M). The expression of αSMA-positive stress fibers is often associated with the contractile phenotype of myofibroblasts (38). The lack of uniform αSMA-positive stress fiber formation in myofibroblasts transfected with siRNA targeting CD147 suggested that the contractile ability of the myofibroblasts would be impaired. Collagen gels were used to assess the effect of silencing CD147 on the contractility of myofibroblasts. Cells were seeded on to the collagen gel and growth arrested for 24 h. Transient transfection was then carried out overnight with an siRNA to CD147 or a negative control siRNA. TGF-β1 (10 ng/ml) was then added with fresh serum-free media and replenished after 72 h. Control cells were treated with media alone and are labeled 0 h. Collagen gels that contained cells transfected with the siRNA to CD147 consistently had impaired contraction when compared with the negative siRNA control cells (Fig. 4, N–Q).
Figure 4.
CD147 regulates αSMA incorporation into F-actin stress fibers in myofibroblasts. Fibroblasts were grown to ~80% and growth arrested for 48 h. Cells were then treated with fresh serum-free media containing TGF-β1 (10 ng/ml) or serum-free media alone (control). CD147 mRNA expression was assessed by RTqPCR in fibroblasts (black circles) and TGF-β1 induced myofibroblasts (black squares) (A). Using siRNA to CD147, there was >80% knockdown in CD147 at the mRNA level (B) in both fibroblasts (black circles) and TGF-β1-induced myofibroblasts (black squares). Western blot and densitometry analysis were used to assess CD147 protein expression following 144 h siRNA treatment compared with control siRNA. C and D, following specific siCD147 knockdown, fibroblasts were treated with TGF-β1 (10 ng/ml) (black squares) or serum-free media alone (control) (black circles). The RNA from cells was then extracted and RTqPCR carried out for αSMA mRNA expression (E). Cells were also extracted for protein and western blot for αSMA was performed (F) and quantified using densitometry (G). Data are displayed as CD147/GAPDH. Protein from fibroblasts transfected with siRNA negative control (black circles) was compared with protein from fibroblasts. For all qPCR and western blot analysis, data are shown as mean ± SD from three experimental repeats with statistical significance represented by; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001. Confocal microscopy was used to analyze αSMA association with the F-actin cytoskeleton following CD147 knockdown. Images show F-actin ([green stain] [H and K]) and αSMA ((red stain) (I and L)) following transfection with either a negative control (H and I) or an siRNA to CD147 (K and L). F-actin and αSMA images were overlaid to determine colocalization. Colocalization was observed in merged images (yellow stain) (J and M). Cells that did not exhibit colocalization are indicated by (white arrows) (M). A representative picture was taken of each cell population under each condition. Original magnification x 400. The role of CD147 in myofibroblast contraction was assessed using collagen gels. Briefly, fibroblasts were seeded onto premade collagen gels and grown to 50–60% confluence. Following growth arrest fibroblasts were transfected with an siRNA targeting CD147 or a negative control siRNA. Fibroblasts were further growth arrested before being treated with serum-free media (control cells) or serum-free media containing TGF-β1 (10 ng/ml). Collagen gels were photographed at 0 h and 144 h, following TGF-β1 stimulation. N and O show contraction gel of fibroblasts transfected with a scrambled control siRNA. P and Q show fibroblasts transfected with an siRNA targeting CD147.
CD147 does not colocalize with EGFR in myofibroblasts
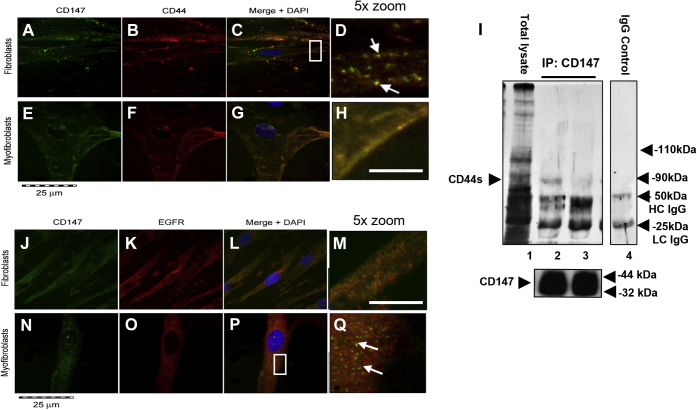
To investigate whether the inhibition seen above was due to a change in CD147 association with CD44 and EGFR, we examined its cell surface localization by immunocytochemistry. CD147 (green stain) was highly expressed on the cell surface and intracellularly in fibroblasts and myofibroblasts (Fig. 5, A and E). CD44 (red stain) was also highly associated with the cell membrane in fibroblasts and myofibroblasts (Fig. 5, B and F). CD147/CD44 colocalization in fibroblasts and myofibroblasts was assessed by merging the CD147 and CD44 images (Fig. 5, C, D, G and H). CD147/CD44 association in fibroblasts was limited and CD147 formed independent clusters from CD44 throughout the cell (Fig. 5D (white arrows)). Conversely, myofibroblasts showed an almost complete association of CD147 with CD44 (Fig. 5H). To confirm this association in myofibroblasts, CD147 was immunoprecipitated and its association with CD44 observed (Fig. 5I). Using a Pan CD44 antibody as a detection antibody, all CD44 variants were detectable as shown by total lysate (lane 1). Interestingly, when coimmunoprecipitated with CD147, only one band was detected with the Pan-CD44 antibody at an MW of ∼85–90 kDa, indicative of the standard form of CD44. The association of CD44s with CD147 was increased in myofibroblasts (lane 2) compared with fibroblasts (lane 3). These confirmed results were observed using ICC. An IgG negative control is shown (lane 4). To investigate the CD147 interactions further, ICC was used to analyze CD147 (green stain) and its potential association with EGFR (red stain) in fibroblasts (Fig. 5, J–M) and myofibroblasts (Fig. 5, N–Q). CD147 had some colocalization with EGFR in fibroblasts (Fig. 5, L and M). In contrast, myofibroblasts had separate populations of CD147 and EGFR (Fig. 5, P and Q). Furthermore, CD147 formed clusters throughout the cell, which were separated from EGFR-rich areas (Fig. 5Q) (white arrows).
Figure 5.
CD147 associates with CD44s but not EGFR. Immunocytochemistry analysis was used to assess CD147/CD44 and CD147/EGFR colocalization on the membrane of fibroblasts (A–D and J–M) and myofibroblasts (E–H and N–Q). Fibroblasts were grown to approximately 50% confluence in DMEM/F12 medium containing 10% v/v FCS. Following growth arrest cells were treated with serum-free medium containing TGF-β1 (10 ng/ml) or fresh serum-free medium alone (control fibroblasts) for 72 h prior to cells being fixed in 4% paraformaldehyde. Images show ICC florescence staining of CD147 (green stain) (A, E, J, and N), CD44 (red stain) (B, F, K, and O). Merged staining for CD147/CD44 association in fibroblasts is shown in (C and D). Images (G and H) show CD147/CD44 colocalization in myofibroblasts. Original magnification ×400. Confirmation of CD147/CD44 was determined using coimmunoprecipitation (I). Image shows total lysate of CD44 (lane 1). Localization of CD44s and CD147 in fibroblasts (Lane 1) and myofibroblasts (Lane 2). An IgG control is shown in Lane 4. Images demonstrating EGFR/CD147 colocalization by fibroblasts and myofibroblasts show CD147 (J and N) (green stain), EGFR (K and O) (red stain) and merged images (L and M) fibroblasts and (P and Q) myofibroblasts. CD147 clusters in CD44/CD147 fibroblasts and CD147/EGFR myofibroblasts are depicted by the white arrows in enlarged images (D and Q). The scale bar for enlarged images (D, H, M and Q) (5 × zoom) is equal to 125 μm.
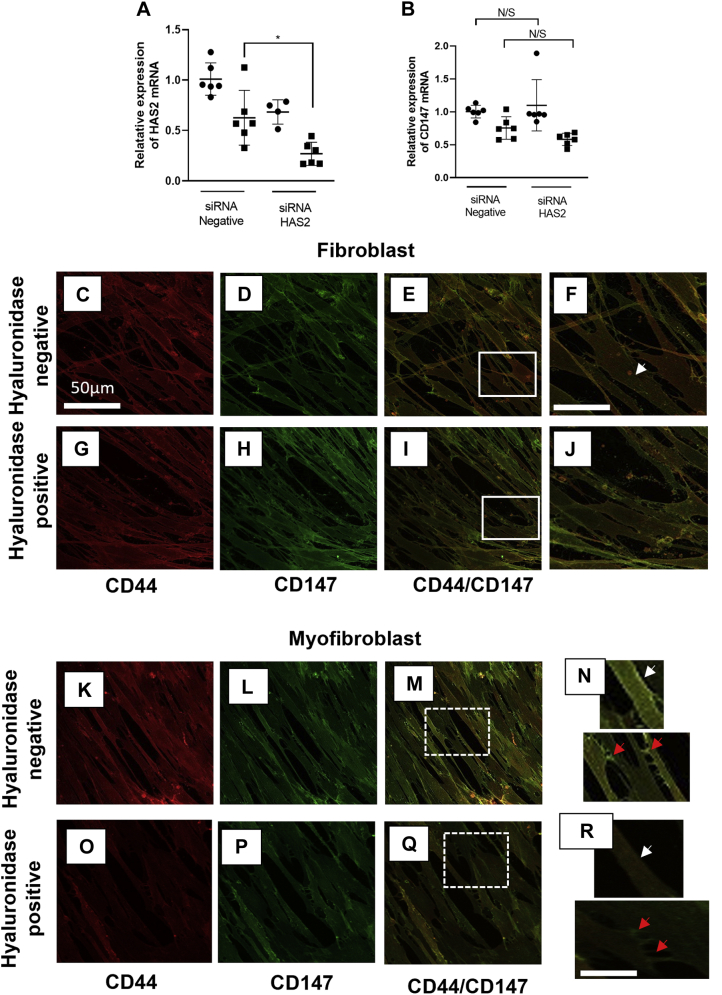
HAS2 and CD147 do not synergistically regulate the mRNA expression of the other and HA mediates the CD147/CD44 association via movement though the membrane and cell–cell contacts
HAS2 is the principal HA synthase involved in fibroblast to myofibroblast TGF-β1-induced differentiation (39). The two other HA synthases HAS1 and HAS3 have previously been shown to have no important role in the differentiation process (13). As HAS 2 has such an essential role in regulating and maintaining the myofibroblast phenotype and data from this study identifies CD147 as an important regulator of the myofibroblast phenotype, it was deemed important to assess if the two had a synergistic role in their regulation of mRNA transcription. Using an siRNA to HAS2 (Fig. 6A), there was significant knockdown of HAS2 in myofibroblasts; however, the decreased HAS2 expression did not affect the overall expression of CD147 mRNA (Fig. 6B). The CD147/CD44 colocalization identified in this paper regulates the incorporation of αSMA into stress fibers. HA associates with its principal receptor CD44, but HA also has a regulatory role with CD147 and other receptors (including CD44) in forming multireceptor complexes in lipid raft regions of the plasma membrane (24) To investigate the role of HA and CD44 in this study, hyaluronidase was used to remove extracellular and membrane-bound HA and the coassociation of CD44 and CD147 in the membrane was assessed. CD44 (Fig. 6, C and G (red stain)) and CD147 (Fig. 6, D and H (green stain)) were highly expressed in fibroblasts. Furthermore, CD44 and CD147 did not colocalize in cells untreated with hyaluronidase (Fig. 6, E and F). This is in line with previous data shown in (Fig. 5, D and C). Removing HA using hyaluronidase had no effect on the lack of CD44/CD147 colocalization in fibroblasts (Fig. 6, I and J). Control myofibroblasts that had not been treated with hyaluronidase showed that CD44 (Fig. 6K) (red stain) and CD147 (Fig. 6L) green stain had increased colocalization (Fig. 6M) (yellow merge). Interestingly, the colocalization of CD44/CD147 was present throughout the cell membrane (Fig. 6N (white arrows)) and on closer inspection, there was an increased colocalization observed at the points of cell–cell contact (Fig. 6N (red arrows)). Using hyaluronidase to remove HA reduced CD44 (Fig. 6O (red stain)) and CD147 (Fig. 6P (green stain)) colocalization CD44/CD147 (Fig. 6Q). This reduction of colocalization was observed in both the membrane (Fig. 6R (white arrows)) and at cell–cell contact regions (Fig. 6R (red arrows)).
Figure 6.
HA mediates CD44/CD147 colocalization in the cell membrane and at cell–cell contact regions. qPCR was used to show the knockdown of HAS2 expression and CD147 mRNA in myofibroblasts (A and B) respectively. Cells were grown to approximately 50–60% confluence and growth arrested for 48 h. They were then transfected with either an siRNA to HAS2 or CD147 or a negative siRNA. Following transfection cells were treated with TGF-β1 (10 ng/ml) for 72 h (black squares) or with serum-free media alone for control fibroblasts (black circles). Data are shown as mean ± SD from three experimental repeats with statistical significance represented by; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001. Immunocytochemistry analysis was used to assess CD147/CD44 colocalization in fibroblasts (C–J) and myofibroblasts (K–R). Control cells (C–F) and (K–N) were compared with cells that had HA removed using hyaluronidase (G–J) and (O–R). Fibroblasts were grown to approximately 50% confluence in DMEM/F12 medium containing 10% v/v FCS. Following a 48 h growth arrest period cells were treated with Hyaluronidase (100 μg/ml) or fresh serum-free medium and incubated at 37 °C, 5% CO2 for 2 h. The media was then removed and replaced with either serum-free media containing TGF-β1 (10 ng/ml) or fresh serum-free medium alone (control fibroblasts) for 72 h. Cells were then fixed in 4% paraformaldehyde. Images show ICC florescence staining of CD44 (red stain) (C, G, K, and O) and CD147 (green stain) (D, H, L, and P). Merged staining (E, I, M, and Q) represents CD44/CD147 association in fibroblasts. Images (M, Q, N, and R) show CD147/CD44 colocalization in myofibroblasts. Enlarged images for fibroblasts (N) and myofibroblasts (R) show cell membrane regions (red arrows) and cell–cell contact regions (white arrows). Scale bar for enlarged images F&J is equal to 100 μm. Scale bar for enlarged images N&R is equal to 125 μm Original magnification ×400.
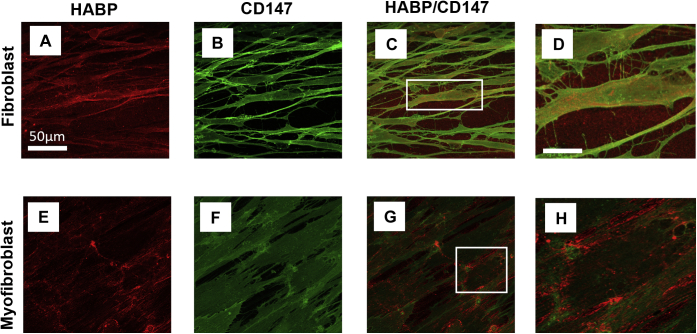
Extracellular HA does not bind CD147
Removing pericellular HA from myofibroblasts prevents the movement of CD44 in the membrane and inhibits its essential associations with other membrane-bound receptors (15). Although we have a clear understanding of the HA/CD44 association in myofibroblasts, we do not know if CD147 binds HA in a similar manner. To determine if HA binds CD147 in fibroblasts and myofibroblasts, ICC was carried out and observed using confocal microscopy. Extracellular HA was observed using HABP in fibroblasts (Fig. 7A) and in myofibroblasts (Fig. 7E cultures (red stain)). As expected, extracellular HA increased in myofibroblast cultures. Consistent with previous data in this research, CD147 was expressed by fibroblasts and myofibroblasts (Fig. 7, B and F (green stain)). Merging HABP and CD147 images did not show any association of HABP with CD147 in fibroblasts (Fig. 7, C and D) or myofibroblasts (Fig. 7, G and H). These data suggest that any CD44/CD147 association is modulated by HA association with CD44.
Figure 7.
HA does not bind CD147. Immunocytochemistry analysis was used to assess HA binding to CD147 in fibroblasts (A–D) and myofibroblasts (E–H). Fibroblasts were grown to approximately 50% confluence in DMEM/F12 medium containing 10% v/v FCS. Following a 48 h growth arrest period cells were treated with TGF-β1(10 ng/ml) or fresh serum-free medium and incubated at 37 °C, 5% CO2 for 72 h. Cells were then fixed in 4% paraformaldehyde and stained with HABP (A and E) (red stain) or CD147 (B and F) (Green stain). HABP/CD147 association can be observed in fibroblasts (C and D) and myofibroblasts (G and H). Scale bar for enlarged images (D and H) is equal to150 μm. Original magnification ×400.
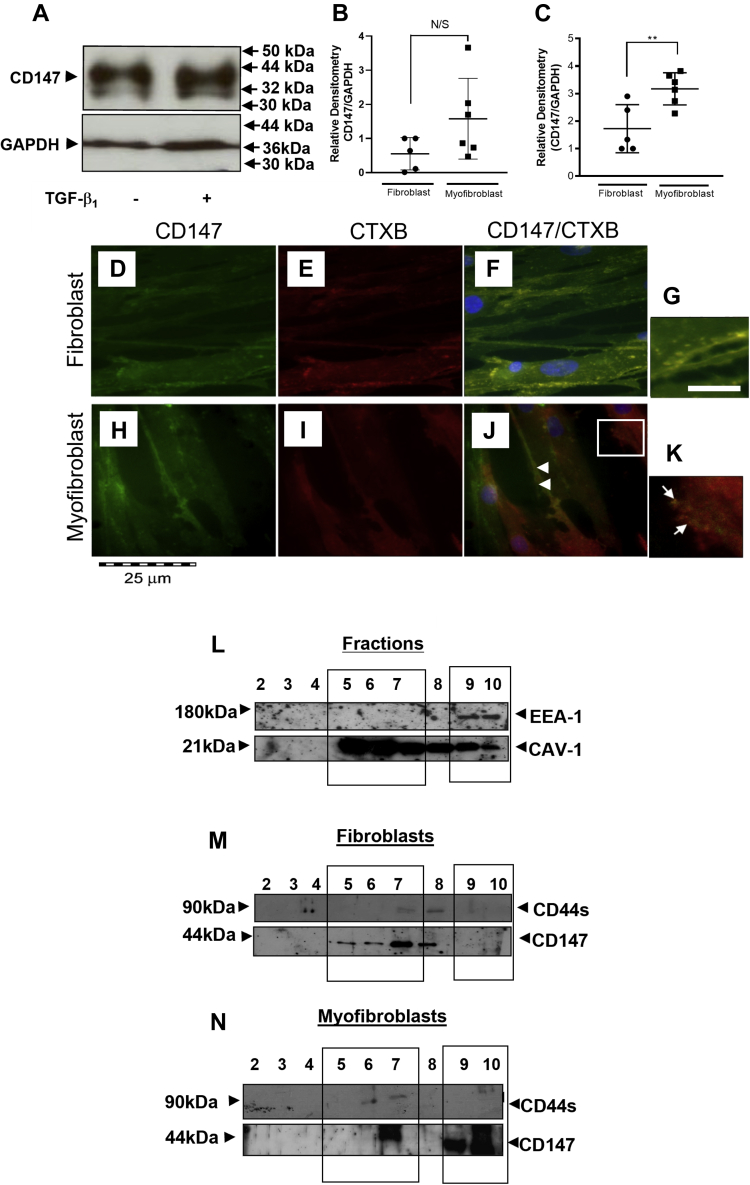
CD147 exists in and out of lipid raft regions of the plasma membrane in myofibroblasts
CD147 exists in multiple glycosylated forms that mediate different functions and have varying expression between cell types (40). Western blot analysis of fibroblasts and myofibroblasts identified two bands of 32 and 44 kDa molecular mass (Fig. 8A), suggesting that the less glycosylated form (LG-CD147) (32 kDa) and a highly glycosylated form (HG-CD147) (44 kDa) of CD147 were expressed in fibroblasts and myofibroblasts. Densitometry analysis compared the expression of each glycosylated CD147 form between fibroblasts and myofibroblasts (Fig. 8, B and C). There was no significant difference in LG-CD147 expression between fibroblasts and myofibroblasts (Fig. 8B). However, HG-CD147 had an increased expression in myofibroblasts compared with fibroblasts (Fig. 8C). CD147 (green stain) and CTXB (binds to lipid rafts; red stain) indicated CD147 and its location in the plasma membrane in fibroblasts and myofibroblasts (Fig. 8, D–K). CD147 and CTXB staining was ubiquitous throughout fibroblasts (Fig. 8, D and E) and myofibroblasts (Fig. 8, H and I). Complete colocalization between CD147 and CTXB was observed in fibroblasts (yellow merge) (Fig. 8, F and G). Conversely, myofibroblasts had two distinct populations of CD147. The first showed some areas of colocalization of CD147 with CTXB (white block arrows) (Fig. 8J). Additionally, CD147 was observed to form independent clusters that were separate from CTXB staining (lipid raft regions) (Fig. 8K (white arrows)). To confirm this, a density gradient fractionation analysis of the membrane was performed. Our previous studies have identified cholesterol-rich lipid rafts to be important for CD44/EGFR colocalization (15). These are abundant regions within the membrane, containing various lipids and caveolin (41, 42). CD147 association with lipid raft and non-lipid-raft regions was investigated using CAV-1 (band MW ∼21 kDa) and early endosomal antigen 1 (EEA-1) (band MW ∼170 kDa), respectively. Ten fractions were extracted from the gradient sample and analyzed using western blot. The first sample was discarded as waste and fractions 2–10 were used to determine membrane regions. Fig. 8L shows that CAV-1 expression was the highest in fractions 5–7 and EEA-1 expression was only present in fractions 9–10 indicative of nonraft regions. Fibroblasts expressed CD147 mainly in CAV-1rich regions in fractions 5–8 and CD44s was expressed in fractions 7–8. CD147 was not localized to nonraft regions (fractions 9–10) (Fig. 8M). Myofibroblasts had two distinct populations of CD147: one that was situated in CAV-1-positive lipid raft regions (fraction 7), together with CD44; and another seen in EEA-1-positive nonraft regions (fractions 9–10) (Fig. 8N).
Figure 8.
CD147 and CD44s are both localized in raft and nonraft regions of the plasma membrane in myofibroblasts. CD147 glycosylation forms that were expressed by fibroblasts and myofibroblasts were assessed using SDS PAGE followed by western blot analysis (A). Total lysate was extracted from cell culture using RIPA cell lysis buffer before separation by 7.5% SDS-PAGE. Proteins were transferred onto a nitrocellulose membrane before being probed with an antibody to CD147. GAPDH was used as a loading control. Densitometry was used to compare LMW bands (B) and HMW bands (C) between fibroblasts (black circles) and myofibroblasts (black squares). Data are shown as mean ± SD from three experimental repeats with statistical significance represented by; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001. D–K show the position of CD147 in fibroblasts (D–G) and myofibroblasts (H–K). Images show CD147 (green stain) location in fibroblasts and myofibroblasts (D and H), respectively. Cholera toxin B (red stain) was used to show lipid raft regions in fibroblasts (E) and myofibroblasts (I). Merging the images highlighted areas of colocalization (yellow stain) of CD147 and CTX in fibroblasts (F and G) and myofibroblasts (J and K). Areas of CD147 cluster formation are depicted by white arrows in the enlarged image (K). Scale bar for enlarged images (G and K) is equal to 100 μm. Where in the plasma membrane CD44s and CD147 were positioned in fibroblasts and myofibroblasts was determined using density gradient ultracentrifugation. Briefly, the density gradient had five layers containing various percentages of OptiPrep Medium. These were (starting from the lowest layer) 35%, 30%, 25%, 20%, and 0% made up to 1 ml with lysis buffer. The cell lysate was added to the final 0% layer. Samples were the centrifuged at 200,000g for 14 h at 4 °C. Ten 500 ml aliquots were extracted from the final gradient tube. The aliquots were labeled fractions 1–10 (with fraction 1 being the first aliquot taken from the top of the ultracentrifugation tube). Following collection the fractions were separated using SDS-PAGE. To determine which of the fractions were lipid-rich rafts, an antibody to Caveolin-1 was used (L). Non-lipid-raft regions were identified using an antibody to EEA-1 (L). Fractions 5–7 were identified as Cav-1-rich lipid raft fractions. Fractions 9–10 were identified as nonraft regions. Fraction analysis of CD44 and CD147 was carried out in fibroblasts (M) and myofibroblasts (N).
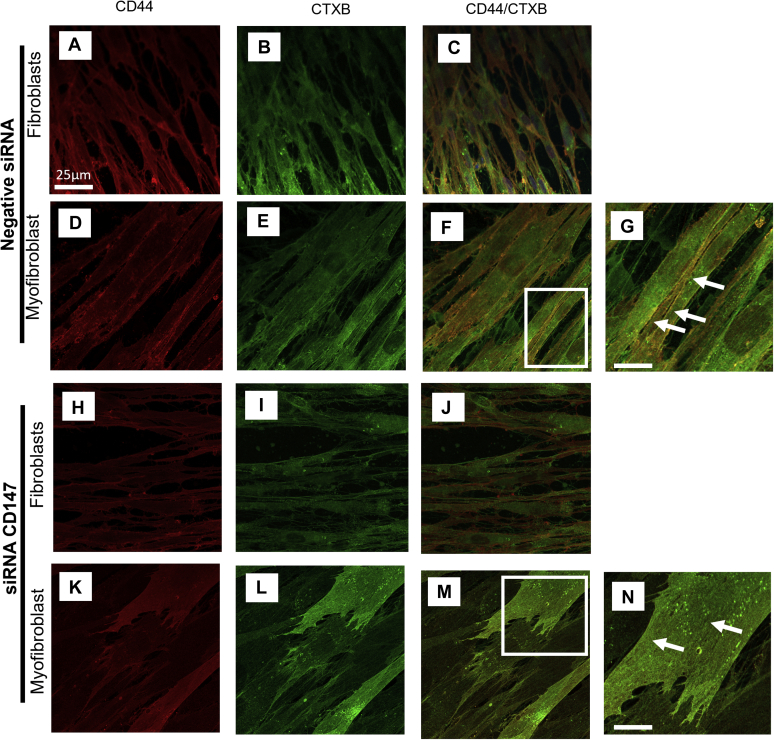
CD147 regulates CD44 distribution throughout the membrane
This study identified that CD44 had increased colocalization with CD147 in myofibroblasts (Fig. 5, G and H). It was also identified that CD147 existed in and out of lipid raft regions in the plasma membrane in areas where CD44 was also situated. Here, we have investigated how silencing CD147 affected the CD44/CTXB association, which has previously been shown to be important in myofibroblast differentiation (15) Fibroblasts transfected with the negative siRNA showed CD44 (red stain) to have a high expression in fibroblasts (Fig. 9A). Myofibroblasts had increased CD44 expression localized to the membrane (Fig. 9D). Similarly, CTXB (lipid rafts; green stain) had an increased membrane association in myofibroblasts (Fig. 9E) compared with fibroblasts (Fig. 9B). In line with our previous work, the membrane association of CD44 with lipid raft regions had an increased association in myofibroblasts (Fig. 9, F and G (white arrows)) (yellow; merged image), compared with fibroblasts (Fig. 9C). Silencing CD147 protein did not affect the level of CD44 (red stain) or CTXB (green stain) expression by fibroblasts (Fig. 9, H and I). Merger of CD44 with CTXB did not show colocalization in fibroblasts (Fig. 9J) transfected with siRNA to CD147. CD44 (red stain) was situated diffusely throughout the membrane in myofibroblasts that had CD147 expression silenced (Fig. 9K). Interestingly CTXB was also diffusely situated in the membrane (Fig. 9L), and the colocalization observed was increased across the total cell (Fig. 9, M and N) not just select areas in the plasma membrane as observed in siRNA control myofibroblasts (Fig. 9, F and G).
Figure 9.
Increased colocalization is seen in myofibroblasts with CD147 protein expression silenced. Cells were transfected with a negative siRNA control (A–G) or with an siRNA that targeted CD147 (H–N). Fibroblasts were then treated with TGF-β1 (10 ng/ml) (D–G) and (K–N) or with serum-free media (A–C) and (H–J) and incubated for 72 h. Using ICC cells were stained for CD44 (red stain) (A, D, H, and K) or Cholera toxin B (green stain) (to identify lipid raft regions) (B, E, I, and L). Merged images show colocalization (yellow stain) between CD44 and CTX in fibroblasts (C and J) and myofibroblasts (F and M). Enlarged images show areas of CD44/CTXB colocalization in myofibroblasts transfected with negative siRNA CT (G) or an siRNA to CD147 (N) (white arrows). Scale bar for enlarged images equals 100 μm.
Discussion
Hyaluronan and its principal receptor CD44 mediate profibrotic and proinflammatory responses following TGF-β1 and IL-1β activation in fibroblasts. The association of HA/CD44 complexes with membrane receptors EGFR and ICAM1, respectively, mediates these responses by modulating multifunctional pericellular HA coats (14, 15). CD44 is expressed as multiple variants, resulting from alternative splicing of its stem region (43). Moreover, the expression of CD44 variants is well documented to be dependent on cell type (reviewed by (44)), cytokine activation, and environmental changes associated with pathology (45, 46, 47). This study investigated which CD44 variants were expressed by fibroblasts and demonstrates the effects of TGF- β1 and IL-1β on their expression.
CD44s is the simplest CD44 variant, it is comprised of only the extracellular region and the transmembrane/cytoplasmic domains; known as the common regions (regions that are common to all CD44 variants). It is well established that CD44s is the most abundant and widely distributed isoform of CD44 and is most commonly associated with HA-binding functions (48). This study identified CD44s as also being the most highly expressed CD44 variant in normal resting fibroblasts. CD44 variants containing a single exon insertion into the stem region situated between the two common regions were identified in descending order of expression as: CD44v3, CD44v6, CD44v10, CD44v8, CD44v4, CD44v2, CD44v9, and CD44v7. A larger, multiple exon variant CD44v6-10 was also identified. Although all CD44 variants express the same HA-binding domain, situated within the amino domain they display different HA-binding affinity, often associated with CD44 cluster formation within the plasma membrane (49, 50). This study identified CD44s as the variant that was essential for myofibroblast differentiation following TGF-β1 activation and for IL-1β induction of monocyte binding. Silencing CD44s prevented the increased expression of αSMA stress fiber formation, giving cell morphology that was more representative of a fibroblast than a myofibroblast. CD44s is well documented to be associated with pathology (51, 52), and this study identified CD44s as the important CD44 variant in the HA/CD44/EGFR complex, known to be induced following TGF-β1 induction (15). Interestingly, all identified CD44 variants had an attenuated expression following induction with TGF-β1. An explanation for this may be that HA/CD44 interaction alters in myofibroblasts compared with fibroblasts. In fibroblasts, CD44 is diffused throughout the plasma membrane. When TGF-β1 induces differentiation, however, CD44 is moved into clusters by HA, resulting in the formation of an HA/CD44-dependent pericellular coat (16, 53). This different orientation of CD44 alters the affinity of the HA/CD44 complex, a factor that is known to alter the cellular response (54).
Conversely, the proinflammatory cytokine IL-1β increased the expression of all identified CD44 variants. It is well established that a continued inflammatory response plays a role in the underlying pathology of fibrotic progression by mediating the influx of healing mediators to the site of injury including monocytes/macrophages and fibroblasts (Reviewed by (55)). The association of fibroblasts with monocytes/macrophages has been demonstrated previously to play a role in fibrogenesis (56). Our studies have implicated IL-1β in the induction of fibroblast–monocyte binding in an HA/CD44-dependent manner (14). After IL-1β stimulation, fibroblasts form cell membrane protrusions that associate with linear spikes of HA. CD44 is central to this formation and accumulates within the protrusions, closely associated with ICAM-1 and mediating monocyte binding (14). This study demonstrated that CD44s was also the variant involved in this interaction.
CD147 is well established as an MMP inducer (57, 58, 59) and has been shown to have multifunctional roles in many progressive diseases and in animal models of disease (28, 29, 30, 31), including, cardiovascular disease, Alzheimer's, and more recently as an entry route into cells for SARs-CoV-2 (COVID 19) (60, 61, 62). Its involvement in CKD has been well described (32, 33) and in the Figure S10, we show its changing expression in a mouse model of CKD. CD147 is strongly expressed by the tubular epithelium of both proximal and distal tubules but is absent from the interstitium in healthy kidneys. In chronic fibrosis, there is loss of CD147 expression associated with injured tubules, while its expression is increased in interstitial regions associated with myofibroblast expansion and collagen deposition. Despite this wealth of in vivo evidence, there is little known about its potential involvement and associated mechanisms in the control of cell phenotype. Interestingly, CD147 has also been shown to associate with an HA/CD44/EGFR complex in breast cancer cells, increasing their invasiveness (34). Our study identified that CD147 was highly expressed in fibroblasts, but its transcription was not altered by stimulation with TGF-β1. This is in contrast to previous reports that TGF-β1 induced CD147 expression in corneal fibroblasts (63). We did, however, see a difference at the protein level. CD147 exists in multiple glycosylated forms dependent on cell type and function (40). High (HG) and low (LG) glycosylated forms of CD147 have been reported to associate with lipid rafts (42). Interestingly, the HG-CD147 form has previously been identified to have increased expression under fibrotic conditions (64). Similarly, this study identified a high and low glycosylated form of CD147 to be expressed by fibroblasts and myofibroblasts. However, only HG-CD147 expression was increased in myofibroblasts, suggesting that the high glycosylated form of CD147 may alter its position in myofibroblasts in a manner that contributes to fibrotic progression.
Myofibroblasts can be characterized by the increased expression of αSMA stress fibers. Silencing CD147 did not affect αSMA transcription or αSMA protein expression. However, silencing CD147 prevented αSMA incorporation into F-actin to form stress fibers. The incorporation of αSMA into the F-actin cytoskeleton by the NH2 terminal motif AC-EED requires the correct mechanical tension mediated by the combination of correct mature focal adhesion formation, cell–cell contact, cell–ECM contact, and intracellular cytoskeletal rearrangement (38, 65, 66). In line with this, inhibiting CD147 limited myofibroblasts contractility. That the contractility was not totally impaired can be explained by a study showing that γ-actin and β-actin partly compensate for the functional role in the absence of αSMA in myofibroblasts (67).
This study established that CD147 had an increased association with CD44s in myofibroblasts but did not associate with EGFR. Therefore, CD147 did not have a role in the previously described TGF-β1-activated HA/CD44/EGFR pathway. CD44 and CD147 are both known to regulate MMP transcription and activation (59, 68), and CD44 has been documented to act as a platform for MMPs (69), the association of CD44 with CD147 (a known MMP inducer) may allow for a synergistic relationship where CD44 positions MMPs and mediates CD147 activation. Elevated mechanical tension has been shown to exacerbate pulmonary lung fibrosis following TGF-β1 activation (70). CD44 and CD147 have both been documented to be involved in mechanotransduction (71). An important factor in obtaining the correct mechanical tension is ECM rearrangement and as both CD44 and CD147 modulate MMPs, it is likely that both contribute to mechanism.
CD147 is able to regulate HA function by forming multireceptor complexes. Indeed, the stability of cell membrane lipid rafts is dependent on the formation of such receptor complexes and CD147 has been identified as essential for raft integrity, reviewed in (72). This study shows that CD147 binds CD44s, and these receptors together form clusters within cell–cell contact areas of adjacent myofibroblasts. Both CD147 and CD44 have previously been identified to mediate cell–cell contact, which is an important contributor to mechanical tension (73). Removing HA before differentiation inhibited TGF-β1-induced association of CD147 with CD44 at points of cellular contact. Furthermore, HA did not associate with CD147 in fibroblasts or myofibroblasts. As HA is well established to mediate the movement and function of CD44, this study suggests that CD44 is moved through the membrane where it forms a complex with CD147, which in turn stabilizes the complex through association with the plasma membrane.
In fibroblasts CD147 was situated in CAV-1-positive lipid raft regions, while it was also situated in nonraft regions in myofibroblasts with CD44. These findings suggest that the increased CD147/CD44 colocalization, observed in myofibroblasts, could occur both in and out of raft regions. Out of raft regions CD147 forms clusters and its MMP role is reestablished due to a lack of CAV-1 association. CD147 cluster formation is commonly associated with the N-glycosylations associated with HG-CD147 (42, 74). This study showed an increased expression of the HG-CD147 and silencing CD147 increased the association of CD44 with lipid-rich regions in myofibroblasts. These data together suggest that clusters of CD147 may be essential for a CD147/CD44 complex formation outside of raft regions and that removal of HA also prevents this complex formation.
We have further investigated the presence of CD147 in our rat AA model compared with a control sham model.
In conclusion, this current study has revealed the central role of CD44s in two important cellular transformations: proinflammatory and profibrotic. For the purposes of this research study, only the profibrotic pathway of myofibroblastic differentiation was followed further, to investigate the potential mechanisms involved. In subsequent work we are, however, following up our results with a similar investigation of the IL-1β pathway. Having described for the first time that CD147, outside lipid raft areas, may have a role to play in myofibroblastic differentiation, we have also begun preliminary studies to examine the potential interactions involved. Interestingly, while two more important biological roles for CD44s have been confirmed, it will also be intriguing to investigate what, if any roles, the other variants identified in this study may have.
Experimental Procedures
Materials and reagents
All reagents were purchased from Sigma-Aldrich (Poole), GIBCO/Thermo-Fisher scientific, or Life Technologies/Thermo-fisher scientific, unless otherwise stated.
Cell culture
Primary Human Fibroblasts (no. AG02262) were purchased from Coriell Institute for Medical Research. The cells were cultured in Dulbecco's Modified Eagle Medium nutrient mixture F-12 Ham's Medium ((DMEM/F12) 1:1 ratio), which was supplemented with 2 mM L-Glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 10% fetal bovine serum (FBS) (Biological Industries Ltd). Cells were incubated at 37 °C in 5% CO2. Prior to experimentation cells were serum starved for 48 h. U397 cells (human histiocytic lymphoma cell line) were purchased from ATCC and cultured in RPMI-1640 medium supplemented with 2 mM L-Glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS. Cells were incubated at 37 °C in 5% CO2 until they reached a high cell density in 75 cm2 flasks.
Cell treatments
Cells were incubated in serum-free DMEM/F12 containing either TGF-β1 (10 ng/ml) or IL-1β (1 ng/ml) (R&D Systems). Unstimulated control fibroblasts were incubated in fresh serum-free medium at the time of stimulation unless otherwise stated.
Real time–quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was used to determine CD44 variant (CD44v) expression, αSMA (ACTA2), CD45 (PTPRC), and CD147 (EMMPRIN) mRNA expression. Primers were purchased from Thermo Fisher Scientific and were either custom-designed or commercially available (see Tables S1–S3). Experiments were carried out in 35 mm dishes. Briefly, cells were washed with PBS and total RNA was extracted using Trizol (used according to manufacturer's protocol). Reverse transcription was carried out using high-capacity cDNA reverse transcription kits, according to the manufacturer's protocols (Thermo Fisher Scientific). As a negative control dH2O was used in the place of RNA during the reverse transcription reaction. RT-qPCR using either TaqMan Fast Universal PCR master mix (x2 No AmpErase UNG) or Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) was performed according to the manufacture's protocol. One microliter of RNA converted to cDNA was used for all qPCR reactions. A negative control containing RNase-free deionized H2O in the place of the cDNA was included in each experiment. Endogenous controls, rRNA (TaqMan) or GAPDH (SYBR green) that were not affected by the treatments were amplified simultaneously with the gene target to be used as a reference gene. Expression analysis was carried out using the ViiA-7 real-time PCR system from Thermo Fisher Scientific. The amplification program used a cycle of 95 °C for 15 s and 60 °C for 1 min for 40 cycles, followed by a melt-curve stage at 95 °C for 15 s, 60 °C for 1 min, and a final step dissociation step of 95 °C for 15 s. Relative quantification was calculated using the comparative CT method. The CT value (the threshold cycle where the amplification is in the linear range of the amplification curve) of the standard endogenous control reference gene was subtracted from the CT value of the target gene to obtain a (ΔCT) value. The mean ΔCT was then calculated for control experiments. The relative quantification (RQ) for the experimental target genes was then calculated using the mean of the control experiments with the following equation .
Transient transfection
Transient transfection was carried out using either custom-designed siRNA that targeted specific CD44 variants or predesigned siRNA to CD147 (siRNA I.D. s2098) and HAS2 (siRNA I.D. s6457) purchased from Life technologies. Fibroblasts were grown to 50–60% confluence in 35 mm dishes before a growth arrest period of 48 h. The siRNA was then diluted to 2% v/v in Opti-mem (Invitrogen) and Lipofectamine 2000 was also diluted to 2% v/v in Opti-mem. The two solutions were incubated at room temperature for 15 min, combined and mixed thoroughly before addition to the cells (final siRNA concentration was 33 nM). The cells were incubated in 5 % CO2 at 37.5 °C for 6 h. A total of 1 ml of fresh DMEM/F12 containing 20 % v/v FBS was then added to each well and samples were incubated for a further 24 h. Following transfection, the medium was removed and fresh serum-free DMEM/F12 was added (1 ml/well) to growth arrest cells for 48 h prior to experimentation. A negative control, siRNA I.D. AM4613 (Ambion) (A nonsense sequence, bearing no resemblance to any known human mRNA sequence) was included in all transfection experiments.
Touch-down conventional PCR (TD-PCR)
Total RNA was extracted using Trizol (according to manufacturer's protocol). Reverse transcription was carried out using high-capacity cDNA reverse transcription kits, according to the manufacturer's protocols (ThermoFisher Scientific). The cDNA was amplified using custom-designed primers to amplify CD44 variants (Table S3), according to the Phusion DNA polymerase kit protocol, Bio-RAD. Briefly, 2 μl of cDNA was added to a solution containing: 1 μl dNTPs (10 mM), 2.5 μl of each 10 mM primer (forward and reverse), 10 μl of Phusion buffer, and 0.5 μl of DNA polymerase. RNase-free dH2O was added to each sample to give a final volume of 50 μl. Samples were amplified using an ATC-225, Peltier Thermal Cycler. The calculated melting temperature (Tm) +10 °C was used as the initial annealing temperature, the temperature was then decreased by 1 °C for each additional cycle for the first ten cycles, then remained consistent for the remaining 22 cycles. The samples were mixed with a gel loading dye (Qiagen) and loaded on a 1% agarose gel containing ethidium bromide. Gels were submerged in 1X Tris-acetate buffer containing ethidium bromide and separated by electrophoresis. Bands were visualized and extracted from the gel and DNA was isolated using a QIAquick Gel Extraction Kit (Qiagen) and quantified using a Nanodrop 3300 (Thermo Scientific). Product identity was confirmed by DNA sequencing (BioCore Sequencing, Cardiff University).
Immunocytochemistry and confocal microscopy
Immunocytochemistry (ICC) and confocal microscopy (CFM) were used to analyze αSMA stress fiber formation (CFM) and the colocalization of CD147 with CD44, EGFR (ICC), respectively. All experiments were carried out in 8-well Permanox chamber slides (Nunc; Thermo Fisher Scientific). Cells were fixed using 4 % (wt/vol) paraformaldehyde solution for 10 min. For intracellular analysis cells were treated with 0.1 % (v/v) TRITONX-100 for 5 min at room temperature. The cells were washed with 0.1% (w/v) BSA/PBS. Samples were blocked using 200 μl of 1 % (w/v) BSA/PBS per well for 1 h to prevent nonspecific binding. Samples were then washed and treated with 200 μl of the appropriate primary antibody overnight at 4 °C. Following a further wash step, a secondary antibody conjugated to a fluorescent tag was added to the well for 1 h, at RT. Wells were washed again and left to air dry. The slides were then mounted using Vector shield mounting media for fluorescence containing a DAPI nuclear stain (Vector laboratories Inc). Samples were visualized and examined under UV-light using a Leica Dialux 20 fluorescent microscope or Zeiss LSM880 upright confocal microscope with Airyscan. Primary antibodies used and dilution factors were mouse monoclonal anti-α-SMA antibody 1A4, (1:100) (ThermoFisher), mouse monoclonal anti-CD147 ((BD Pharminogen; Dorcan) (1:50)), monoclonal rat anti-CD44 (A020) (Merck Millipore; Milton) (1:100) (monoclonal mouse anti-EGFR (528)) (Merck Millipore) (1:25). Biotinylated HABP (Merck Lifesciences) (1:200) Secondary antibodies used were polyclonal goat anti-mouse (IgG) AlexaFlour 488 (FITC) (1:500) and goat polyclonal anti-rat (IgG) AlexaFlour 555 (TRITC) (1:500) (Sigma-Aldrich). For F-Actin and lipid raft visualization, a phalloidin conjugate Alexa Fluor 555 (1:100) (Sigma-Aldrich), Cholera Toxin B subunit conjugated with Alexa Fluor 548 (1:100) (Sigma-Aldrich), and a Streptavidin Alexa fluor 596 conjugate (1:500) (Sigma-Aldrich) were used, respectively.
Density gradient lipid raft fraction analysis
Caveolae raft analysis was performed using a Raft Isolation Kit (Sigma-Aldrich; product code CS0750). Cells were lysed in 1 ml of ice-cold lysis buffer containing 1 % v/v TRITON X- 100 and 1 % v/v of protease inhibitor cocktail (PIC). Cells were harvested and incubated on ice for 30 min before transfer into a precooled 2 ml Eppendorf. Lysed samples were centrifuged at 450 rpm for 5 min at 4 °C and the supernatant was discarded. The cell pellet was carefully washed twice in ice-cold PBS and resuspended in 1 ml of lysis buffer containing 1 % TRITON–X100 and 1% PIC. The density gradient consisted of 5 x 1 ml layers of 35%, 30%, 25%, 20%, and 0%. OptiPrep Medium (Sigma-Aldrich). Samples were centrifuged at 200,000g for 14 h at 4 °C using an Optima-Max ultracentrifuge (Beckman Coulter). Fractions were then carefully collected into 500 μl fractions. To precipitate the protein from each fraction, samples were treated with 10% v/vTrichloroacetic acid (TCA) for 30 min, on ice. Samples were then centrifuged at 13,400g for 10 min. The pellets were washed in 50:50 v/v ethanol/ether and analysed by western blot.
SDS PAGE/western blot
Cells were dissociated from the plate and lysed using Radio Immunoprecipitation Assay (RIPA) Kit (Santa Cruz, Biotechnology) according to manufacturer's protocol. Protein was quantified before being separated using SDS-PAGE, by BioRad Mini Protein II (Bio-Rad Laboratories). Protein was transferred onto a nitrocellulose membrane (GE Healthcare). The membrane was blocked with 5 % (w/v) skimmed milk in 0.5% (v/v) Tween/PBS for 1 h. The membrane was then washed in 0.1 % (v/v) tween/PBS three times for 5 min. A primary antibody prediluted in 0.1% (v/v) Tween/PBS containing 1% (w/v) BSA was added to the membrane and left at 4 °C, overnight. The membrane was washed before the addition of the secondary antibody conjugated to Horse Radish Peroxidase (HRP) diluted in 0.1 % (v/v) Tween/PBS containing 1 % (w/v) BSA. The Enhanced Chemiluminescence (ECL) method using ECL reagent (GE Healthcare) and HyperFilm X-ray film (GE Healthcare) was used to detect transferred protein. The film was developed using Curix-60 developer (AGFA Healthcare). Primary antibodies used were mouse monoclonal anti-αSMA antibody 1A4 (1:500) (Thermo Fisher Scientific), mouse monoclonal anti-CD147 (BD Pharminogen), rabbit polyclonal anti-Caveolin-1 (Sigma-Aldrich), mouse monoclonal anti-CD44 (v3) (1:1000) (Invitrogen), mouse monoclonal anti CD44 (v6) (1:1000) (Invitrogen), mouse monoclonal anti-CD44 (v10) (Invitrogen (1:1000), mouse monoclonal anti-EEA-1 (1:1000) (BD Bioscience), mouse monoclonal anti-GAPDH (1:5000) (Santa Cruz), and rat monoclonal anti-CD44 (A020) (1:500) (Merck Millipore). Secondary antibodies used were goat polyclonal anti-mouse (IgG) HRP (1:5000) (Abcam), goat polyclonal anti-rabbit (IgG) HRP (1:5000) (Abcam), goat polyclonal anti-rat (IgG) HRP (Abcam). Images were quantified using Image J software. Densitometry used target protein/control protein ratio.
Coimmunoprecipitation (Co-IP)
Following protein extraction, Co-IP was carried out using MagnaBind Goat anti-Mouse (IgG) magnetic beads (Thermoscientific). Briefly, beads were washed in PBS and blocked using 0.1% w/v BSA/PBS for 1 h. A total of 10 μg of mouse monoclonal anti-CD147 (BD Pharminogen) was added to 200 μl of beads and incubated at 4 °C for 2 h. The beads/antibody complex was washed in PBS and 5 μg of sample was then added to the beads/antibody complex and incubated at 4 °C overnight. The beads were washed with PBS followed by 1 % (v/v) Nonidet P40 detergent solution. The beads were dissociated from the antibody-sample complex by boiling for 5 min at 95 °C under reducing conditions. The beads were then removed using a magnetic holder and coprecipitated proteins were identified using SDS-PAGE/western blot analysis.
Collagen gel
Rat tail Collagen Type 1 (5 mg/ml) (Gibco) was diluted to 4 mg/ml. Briefly, 8 ml of collagen was added to 1 ml 10× PBS, 0.20 ml 1 M NaOH, and 0.8 ml water. The subsequent solution was then slowly mixed to achieve the optimal pH 7.0. Once neutralized, fibroblasts (5 × 10-5) were added to the solution, and the suspension was added to 22 mm dishes. The gels were incubated at 37 °C for 40 min until the gel was firm and fresh growth media added. Following a 24 h growth arrest period, cells were transfected with siRNA targeting CD147 or a scrambled negative control for 72 h before being stimulated with TGF-β1 for a further 72 h. Nonstimulated fibroblast cultures were used as experimental controls. Images were captured at 0 h and 144 h timepoints.
Hyaluronidase treatment
Fibroblasts were grown in 35 mm dishes until 65–70% confluent and growth arrested in serum-free media for 48 h. Fresh serum-free media containing hyaluronidase (ICN Pharmaceuticals Ltd) (100 μg/ml) or fresh serum-free media alone (control samples) were added to the culture dish. Cells were then incubated at 37⁰C, 5% CO2 for 2 h. The media was removed and fresh serum-free media containing TGF-β1 (10 ng/ml) or serum-free media alone was added. Cells were incubated for a further 72 h.
Statistical analysis
To assess two experimental groups, a two tailed unpaired student t test was performed. For multiple experimental groups with multiple test conditions a two-way analysis of variance was performed followed by the Tukey Multiple comparison analysis. All data are expressed as mean ± SD unless otherwise stated. Data were analyzed using Graph Pad v8 (GraphPad Software). ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001 were considered statistically significant.
Animal experiments
Animal experiments provided in the Figure S10 were carried out using 8–9-week-old C57BL/6 male mice bred by Charles River Laboratories. Experiments were performed in line with institutional and UK Home Office guidelines under the authority of an appropriate project licence.
Data availability
All the data are included within the manuscript or supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that there is no conflict of interest.
Acknowledgments
Author contributions
E. L. W. investigation; C. V. M. B. and Y.-a. L. resources; A. O. P., T. B., S. M. and R. S. supervision; E. L. W. writing-original draft; E. L. W., I. G. V. G., A. C. M.; Y.-a. L., and R. S. writing-review and editing.
Funding and additional information
E. L.W. was funded by the School of Medicine, Cardiff University.
Edited by Gerald Hart
Footnotes
Present address for Adam C. Midgley: Key Laboratory of Bioactive Materials (Ministry of Education), Rongxiang Xu Center for Regenerative Life Science, College of Life Sciences, Nankai University, Tianjin, China.
Supporting information
References
- 1.Eddy A.A. Progression in chronic kidney disease. Adv. Chronic Kidney Dis. 2005;12:353–365. doi: 10.1053/j.ackd.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 2.Schuppan D., Afdhal N.H. Liver cirrhosis. Lancet. 2008;371:838–851. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gross T.J., Hunninghake G.W. Idiopathic pulmonary fibrosis. N Engl J Med. 2001;345:517–525. doi: 10.1056/NEJMra003200. [DOI] [PubMed] [Google Scholar]
- 4.Segura A.M., Frazier O.H., Buja L.M. Fibrosis and heart failure. Heart Fail. Rev. 2014;19:173–185. doi: 10.1007/s10741-012-9365-4. [DOI] [PubMed] [Google Scholar]
- 5.Marenzana M., Vande Velde G. Refine, reduce, replace: Imaging of fibrosis and arthritis in animal models. Best Pract. Res. Clin. Rheumatol. 2015;29:715–740. doi: 10.1016/j.berh.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Mori L., Bellini A., Stacey M.A., Schmidt M., Mattoli S. Fibrocytes contribute to the myofibroblast population in wounded skin and originate from the bone marrow. Exp. Cell Res. 2005;304:81–90. doi: 10.1016/j.yexcr.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Micallef L., Vedrenne N., Billet F., Coulomb B., Darby I.A., Desmoulière A. The myofibroblast, multiple origins for major roles in normal and pathological tissue repair. Fibrogenesis Tissue Repair. 2012;5:S5. doi: 10.1186/1755-1536-5-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibb A.A., Lazaropoulos M.P., Elrod J.W. Myofibroblasts and fibrosis: Mitochondrial and metabolic control of cellular differentiation. Circ. Res. 2020;127:427–447. doi: 10.1161/CIRCRESAHA.120.316958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willis B.C., duBois R.M., Borok Z. Epithelial origin of myofibroblasts during fibrosis in the lung. Proc. Am. Thorac. Soc. 2006;3:377–382. doi: 10.1513/pats.200601-004TK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meran S., Steadman R. Fibroblasts and myofibroblasts in renal fibrosis. Int. J. Exp. Pathol. 2011;92:158–167. doi: 10.1111/j.1365-2613.2011.00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itano N., Sawai T., Yoshida M., Lenas P., Yamada Y., Imagawa M., Shinomura T., Hamaguchi M., Yoshida Y., Ohnuki Y., Miyauchi S., Spicer A.P., McDonald J.A., Kimata K. Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties. J. Biol. Chem. 1999;274:25085–25092. doi: 10.1074/jbc.274.35.25085. [DOI] [PubMed] [Google Scholar]
- 12.Sampson P.M., Rochester C.L., Freundlich B., Elias J.A. Cytokine regulation of human lung fibroblast hyaluronan (hyaluronic acid) production. Evidence for cytokine-regulated hyaluronan (hyaluronic acid) degradation and human lung fibroblast-derived hyaluronidase. J. Clin. Invest. 1992;90:1492–1503. doi: 10.1172/JCI116017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meran S., Thomas D., Stephens P., Martin J., Bowen T., Phillips A., Shinomura T., Hamaguchi M., Yoshida Y., Ohnuki Y., Miyauchi S., Spicer A.P., McDonald J.A., Kimata K. Involvement of hyaluronan in regulation of fibroblast phenotype. J. Biol. Chem. 2007;282:25687–25697. doi: 10.1074/jbc.M700773200. [DOI] [PubMed] [Google Scholar]
- 14.Meran S., Martin J., Luo D.D., Steadman R., Phillips A. Interleukin-1β induces hyaluronan and CD44-dependent cell protrusions that facilitate fibroblast-monocyte binding. Am. J. Pathol. 2013;182:2223–2240. doi: 10.1016/j.ajpath.2013.02.038. [DOI] [PubMed] [Google Scholar]
- 15.Midgley A.C., Rogers M., Hallett M.B., Clayton A., Bowen T., Phillips A.O., Steadman R. Transforming growth factor-beta 1 (TGF-beta 1)-stimulated fibroblast to myofibroblast differentiation is mediated by hyaluronan (HA)-facilitated epidermal growth factor receptor (EGFR) and CD44 co-localization in lipid rafts. J. Biol. Chem. 2013;288:14824–14838. doi: 10.1074/jbc.M113.451336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webber J., Meran S., Steadman R., Phillips A. Hyaluronan orchestrates transforming growth factor-β1-dependent maintenance of myofibroblast phenotype. J. Biol. Chem. 2009;284:9083–9092. doi: 10.1074/jbc.M806989200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tölg C., Hofmann M., Herrlich P., Ponta H. Splicing choice from ten variant exons establishes CD44 variability. Nucleic Acids Res. 1993;21:1225–1229. doi: 10.1093/nar/21.5.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C., Zhao S., Karnad A., Freeman J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018;11:64. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aghajanian H., Kimura T., Rurik J.G., Hancock A.S., Leibowitz M.S., Li L., Scholler J., Monslow J., Lo A., Han W., Wang T., Bedi K., Morley M.P., Linares Saldana R.A., Bolar N.A. Targeting cardiac fibrosis with engineered T cells. Nature. 2019;573:430–433. doi: 10.1038/s41586-019-1546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothenberg M.E. CD44—a sticky target for asthma. J. Clin. Invest. 2003;111:1460–1462. doi: 10.1172/JCI18392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slevin M., Krupinski J., Gaffney J., Matou S., West D., Delisser H., Savani R.C., Kumar S. Hyaluronan-mediated angiogenesis in vascular disease: Uncovering RHAMM and CD44 receptor signaling pathways. Matrix Biol. 2007;26:58–68. doi: 10.1016/j.matbio.2006.08.261. [DOI] [PubMed] [Google Scholar]
- 22.Midgley A.C., Oltean S., Hascall V., Woods E.L., Steadman R., Phillips A.O., Meran S. Nuclear hyaluronidase 2 drives alternative splicing of CD44 pre-mRNA to determine profibrotic or antifibrotic cell phenotype. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aao1822. [DOI] [PubMed] [Google Scholar]
- 23.Govindaraju P., Todd L., Shetye S., Monslow J., Puré E. CD44-dependent inflammation, fibrogenesis, and collagenolysis regulates extracellular matrix remodeling and tensile strength during cutaneous wound healing. Matrix Biol. 2019;75:314–330. doi: 10.1016/j.matbio.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grass G.D., Dai L., Qin Z., Parsons C., Toole B.P. CD147: Regulator of hyaluronan signaling in invasiveness and chemoresistance. Adv. Cancer Res. 2014;123:351–373. doi: 10.1016/B978-0-12-800092-2.00013-7. [DOI] [PubMed] [Google Scholar]
- 25.Liang L., Major T., Bocan T. Characterization of the promoter of human extracellular matrix metalloproteinase inducer (EMMPRIN) Gene. 2002;282:75–86. doi: 10.1016/s0378-1119(01)00847-2. [DOI] [PubMed] [Google Scholar]
- 26.Fossum S., Mallett S., Barclay A.N. The MRC-OX-47 antigen is a member OF the immunoglobulin superfamily with an unusual transmembrane sequence. Eur. J. Immunol. 1991;21:671–679. doi: 10.1002/eji.1830210320. [DOI] [PubMed] [Google Scholar]
- 27.Biswas C., Zhang Y., DeCastro R., Guo H., Nakamura T., Kataoka H., Nabeshima K. The human tumor cell-derived collagenase stimulatory factor (renamed EMMPRIN) is a member of the immunoglobulin superfamily. Cancer Res. 1995;55:434–439. [PubMed] [Google Scholar]
- 28.Zhang D.-W., Zhao Y.-X., Wei D., Li Y.-L., Zhang Y., Wu J., Xu J., Chen C., Tang H., Zhang W., Gong L., Han Y., Chen Z.N., Bian H. HAb18G/CD147 promotes activation of hepatic stellate cells and is a target for antibody therapy of liver fibrosis. J. Hepatol. 2012;57:1283–1291. doi: 10.1016/j.jhep.2012.07.042. [DOI] [PubMed] [Google Scholar]
- 29.Joung C., Noh H., Jung J., Song H.Y., Bae H., Pahk K., Kim W.K. A novel CD147 inhibitor, SP-8356, attenuates pathological fibrosis in alkali-burned rat cornea. Int. J. Mol. Sci. 2020;21:2990. doi: 10.3390/ijms21082990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki K., Satoh K., Ikeda S., Sunamura S., Otsuki T., Satoh T., Kikuchi N., Omura J., Kurosawa R., Nogi M., Numano K., Sugimura K., Aoki T., Tatebe S., Miyata S. Basigin promotes cardiac fibrosis and failure in response to chronic pressure overload in mice. Arterioscler. Thromb. Vasc. Biol. 2016;36:636–646. doi: 10.1161/ATVBAHA.115.306686. [DOI] [PubMed] [Google Scholar]
- 31.Kato N., Kosugi T., Sato W., Ishimoto T., Kojima H., Sato Y., Sakamoto K., Maruyama S., Yuzawa Y., Matsuo S., Kadomatsu K. Basigin/CD147 promotes renal fibrosis after unilateral ureteral obstruction. Am. J. Pathol. 2011;178:572–579. doi: 10.1016/j.ajpath.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagaya H., Kosugi T., Maeda-Hori M., Maeda K., Sato Y., Kojima H., Hayashi H., Kato N., Ishimoto T., Sato W., Yuzawa Y., Matsuo S., Kadomatsu K., Maruyama S. CD147/basigin reflects renal dysfunction in patients with acute kidney injury. Clin. Exp. Nephrol. 2014;18:746–754. doi: 10.1007/s10157-013-0916-3. [DOI] [PubMed] [Google Scholar]
- 33.Kosugi T., Maeda K., Sato W., Maruyama S., Kadomatsu K. CD147 (EMMPRIN/basigin) in kidney diseases: From an inflammation and immune system viewpoint. Nephrol. Dial. Transpl. 2015;30:1097–1103. doi: 10.1093/ndt/gfu302. [DOI] [PubMed] [Google Scholar]
- 34.Grass G.D., Tolliver L.B., Bratoeva M., Toole B.P. CD147, CD44 and the EGFR signaling pathway cooperate to regulate breast epithelial cell invasiveness. J. Biol. Chem. 2013;288:26089–26104. doi: 10.1074/jbc.M113.497685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X., Song Z., Zhang S., Nanda A., Li G. CD147: A novel modulator of inflammatory and immune disorders. Curr. Med. Chem. 2014;21:2138–2145. doi: 10.2174/0929867321666131227163352. [DOI] [PubMed] [Google Scholar]
- 36.Webber J., Meran S., Jenkins R., Phillips A., Steadman R. Maintenance of the myofibroblastic phenotype: A role for hyaluronan. Int. J. Exp. Pathol. 2009;90:A95–A. [Google Scholar]
- 37.Hinz B., Dugina V., Ballestrem C., Wehrle-Haller B., Chaponnier C. Alpha-smooth muscle actin is crucial for focal adhesion maturation in myofibroblasts. Mol. Biol. Cell. 2003;14:2508–2519. doi: 10.1091/mbc.E02-11-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinz B., Mastrangelo D., Iselin C.E., Chaponnier C., Gabbiani G. Mechanical tension controls granulation tissue contractile activity and myofibroblast differentiation. Am. J. Pathol. 2001;159:1009–1020. doi: 10.1016/S0002-9440(10)61776-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson R.M., Meran S., Thomas D., Stephens P., Bowen T., Steadman R., Phillips A. Age-related changes in pericellular hyaluronan organization leads to impaired dermal fibroblast to myofibroblast differentiation. Am. J. Pathol. 2009;175:1915–1928. doi: 10.2353/ajpath.2009.090045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai Y., Huang W., Ma L.-T., Jiang J.-L., Chen Z.-N. Importance of N-glycosylation on CD147 for its biological functions. Int. J. Mol. Sci. 2014;15:6356–6377. doi: 10.3390/ijms15046356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quest A.F.G., Leyton L., Parraga M. Caveolins, caveolae, and lipid rafts in cellular transport, signaling, and disease. Biochem. Cell Biology-Biochimie Biologie Cellulaire. 2004;82:129–144. doi: 10.1139/o03-071. [DOI] [PubMed] [Google Scholar]
- 42.Tang W., Chang S.B., Hemler M.E. Links between CD147 function, glycosylation, and caveolin-1. Mol. Biol. Cell. 2004;15:4043–4050. doi: 10.1091/mbc.E04-05-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackay C.R., Terpe H.-J., Stauder R., Marston W.L., Stark H., Günthert U. Expression and modulation of CD44 variant isoforms in humans. J. Cell Biol. 1994;124:71–82. doi: 10.1083/jcb.124.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thapa R., Wilson G.D. The importance of CD44 as a stem cell biomarker and therapeutic target in cancer. Stem Cells Int. 2016;2016:2087204. doi: 10.1155/2016/2087204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bourguignon L.Y.W., Singleton P.A., Zhu H., Diedrich F. Hyaluronan-mediated CD44 interaction with RhoGEF and Rho kinase promotes Grb2-associated binder-1 phosphorylation and phosphatidylinositol 3-kinase signaling leading to cytokine (Macrophage-Colony stimulating factor) production and breast tumor progression. J. Biol. Chem. 2003;278:29420–29434. doi: 10.1074/jbc.M301885200. [DOI] [PubMed] [Google Scholar]
- 46.Barshishat M., Ariel A., Cahalon L., Chowers Y., Lider O., Schwartz B. TNFα and IL-8 regulate the expression and function of CD44 variant proteins in human colon carcinoma cells. Clin. Exp. Metastasis. 2002;19:327–337. doi: 10.1023/a:1015528314970. [DOI] [PubMed] [Google Scholar]
- 47.Pinner E., Gruper Y., Ben Zimra M., Kristt D., Laudon M., Naor D., Zisapel N. CD44 splice variants as potential players in Alzheimer's disease pathology. J. Alzheimers Dis. 2017;58:1137–1149. doi: 10.3233/JAD-161245. [DOI] [PubMed] [Google Scholar]
- 48.Bourguignon L.Y.W., Zhu D., Zhu H. CD-44 isoform-cytoskeleton interaction in oncogenic signaling and tumor progression. Front. Biosci. 1998;3:D637–D649. doi: 10.2741/a308. [DOI] [PubMed] [Google Scholar]
- 49.Yang C., Cao M., Liu H., He Y., Xu J., Du Y., Liu Y., Wang W., Cui L., Hu J., Gao F. The high and low molecular weight forms of hyaluronan have distinct effects on CD44 clustering. J. Biol. Chem. 2012;287:43094–43107. doi: 10.1074/jbc.M112.349209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lesley J., Hascall V.C., Tammi M., Hyman R. Hyaluronan binding by cell surface CD44. J. Biol. Chem. 2000;275:26967–26975. doi: 10.1074/jbc.M002527200. [DOI] [PubMed] [Google Scholar]
- 51.Vis A., Van Rhijn B., Noordzij M., Schröder F., van der Kwast T.H. Value of tissue markers p27kip1, MIB-1, and CD44s for the pre-operative prediction of tumour features in screen-detected prostate cancer. J. Pathol. 2002;197:148–154. doi: 10.1002/path.1084. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y.-J., Yan P.-S., Li J., Jia J.-F. Expression and significance of CD44s, CD44v6, and nm23 mRNA in human cancer. World J. Gastroenterol. 2005;11:6601. doi: 10.3748/wjg.v11.i42.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Midgley A.C., Duggal L., Jenkins R., Hascall V., Steadman R., Phillips A.O., Meran S. Hyaluronan regulates bone morphogenetic protein-7-dependent prevention and reversal of myofibroblast phenotype. J. Biol. Chem. 2015;290:11218–11234. doi: 10.1074/jbc.M114.625939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iida N., Bourguignon L.Y.W. Coexpression of CD44 variant (v10/ex14) and CD44S in human mammary epithelial cells promotes tumorigenesis. J. Cell Physiol. 1997;171:152–160. doi: 10.1002/(SICI)1097-4652(199705)171:2<152::AID-JCP5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 55.Glaros T., Larsen M., Li L. Macrophages and fibroblasts during inflammation, tissue damage and organ injury. Front. Biosci. 2009;14:3988–3993. doi: 10.2741/3506. [DOI] [PubMed] [Google Scholar]
- 56.Song E., Ouyang N., Hörbelt M., Antus B., Wang M., Exton M.S. Influence of alternatively and classically activated macrophages on fibrogenic activities of human fibroblasts. Cell Immunol. 2000;204:19–28. doi: 10.1006/cimm.2000.1687. [DOI] [PubMed] [Google Scholar]
- 57.Sier C.F., Zuidwijk K., Zijlmans H.J., Hanemaaijer R., Mulder-Stapel A.A., Prins F.A., Dreef E.J., Kenter G.G., Fleuren G.J., Gorter A. EMMPRIN-induced MMP-2 activation cascade in human cervical squamous cell carcinoma. Int. J. Cancer. 2006;118:2991–2998. doi: 10.1002/ijc.21778. [DOI] [PubMed] [Google Scholar]
- 58.Kim J.-Y., Kim W.-J., Kim H., Suk K., Lee W.-H. The stimulation of CD147 induces MMP-9 expression through ERK and NF-kappaB in macrophages: Implication for atherosclerosis. Immune Netw. 2009;9:90–97. doi: 10.4110/in.2009.9.3.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun J., Hemler M.E. Regulation of MMP-1 and MMP-2 production through CD147/extracellular matrix metalloproteinase inducer interactions. Cancer Res. 2001;61:2276. [PubMed] [Google Scholar]
- 60.Sturhan H., Ungern-Sternberg SNIv, Langer H., Gawaz M., Geisler T., May A.E., Seizer P. Regulation of EMMPRIN (CD147) on monocyte subsets in patients with symptomatic coronary artery disease. Thromb. Res. 2015;135:1160–1164. doi: 10.1016/j.thromres.2015.03.022. [DOI] [PubMed] [Google Scholar]
- 61.Zhou S., Zhou H., Walian P.J., Jap B.K. CD147 is a regulatory subunit of the γ-secretase complex in Alzheimer's disease amyloid β-peptide production. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7499–7504. doi: 10.1073/pnas.0502768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., Gong L., Zhang Y., Cui H.-Y., Geng J.-J., Wang B., Sun X.-X., Wang C.-F., Yang X., Lin P. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. BioRxiv. 2020 doi: 10.1101/2020.03.14.988345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huet E., Vallée B., Szul D., Verrecchia F., Mourah S., Jester J.V., Hoang-Xuan T., Menashi S., Gabison E.E. Extracellular matrix metalloproteinase inducer/CD147 promotes myofibroblast differentiation by inducing α-smooth muscle actin expression and collagen gel contraction: Implications in tissue remodeling. FASEB J. 2008;22:1144–1154. doi: 10.1096/fj.07-8748com. [DOI] [PubMed] [Google Scholar]
- 64.Barth K., Blaesche R., Kasper M. Lack of evidence for caveolin-1 and CD147 interaction before and after bleomycin-induced lung injury. Histochem. Cell Biol. 2006;126:563–573. doi: 10.1007/s00418-006-0192-3. [DOI] [PubMed] [Google Scholar]
- 65.Hinz B., Gabbiani G., Chaponnier C. The NH2-terminal peptide of α–smooth muscle actin inhibits force generation by the myofibroblast in vitro and in vivo. J. Cell Biol. 2002;157:657–663. doi: 10.1083/jcb.200201049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clement S., Hinz B., Dugina V., Gabbiani G., Chaponnier C. The N-terminal Ac-EEED sequence plays a role in alpha-smooth-muscle actin incorporation into stress fibers. J. Cell Sci. 2005;118:1395–1404. doi: 10.1242/jcs.01732. [DOI] [PubMed] [Google Scholar]
- 67.Tomasek J.J., Haaksma C.J., Schwartz R.J., Howard E.W. Whole animal knockout of smooth muscle alpha-actin does not alter excisional wound healing or the fibroblast-to-myofibroblast transition. Wound Repair Regen. 2013;21:166–176. doi: 10.1111/wrr.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murray D., Morrin M., McDonnell S. Increased invasion and expression of MMP-9 in human colorectal cell lines by a CD44-dependent mechanism. Anticancer Res. 2004;24:489–494. [PubMed] [Google Scholar]
- 69.Seiki M. The cell surface: The stage for matrix metalloproteinase regulation of migration. Curr. Opin. Cell Biol. 2002;14:624–632. doi: 10.1016/s0955-0674(02)00363-0. [DOI] [PubMed] [Google Scholar]
- 70.Wu H., Yu Y., Huang H., Hu Y., Fu S., Wang Z., Shi M., Zhao X., Yuan J., Li J., Yang X., Bin E., Wei D., Zhang H., Zhang J. Progressive pulmonary fibrosis is caused by elevated mechanical tension on alveolar stem cells. Cell. 2020;180:107–121.e17. doi: 10.1016/j.cell.2019.11.027. [DOI] [PubMed] [Google Scholar]
- 71.Qazi H., Palomino R., Shi Z.-D., Munn L.L., Tarbell J.M. Cancer cell glycocalyx mediates mechanotransduction and flow-regulated invasion. Integr. Biol. 2013;5:1334–1343. doi: 10.1039/c3ib40057c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grass G.D., Toole Bryan P. How, with whom and when: An overview of CD147-mediated regulatory networks influencing matrix metalloproteinase activity. Biosci. Rep. 2016;36 doi: 10.1042/BSR20150256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mauris J., Woodward A.M., Cao Z., Panjwani N., Argüeso P. Molecular basis for MMP9 induction and disruption of epithelial cell-cell contacts by galectin-3. J. Cell Sci. 2014;127(Pt 14):3141–3148. doi: 10.1242/jcs.148510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang W., Hemler M.E. Caveolin-1 regulates matrix metalloproteinases-1 induction and CD147/EMMPRIN cell surface clustering∗. J. Biol. Chem. 2004;279:11112–11118. doi: 10.1074/jbc.M312947200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are included within the manuscript or supporting information.