Summary
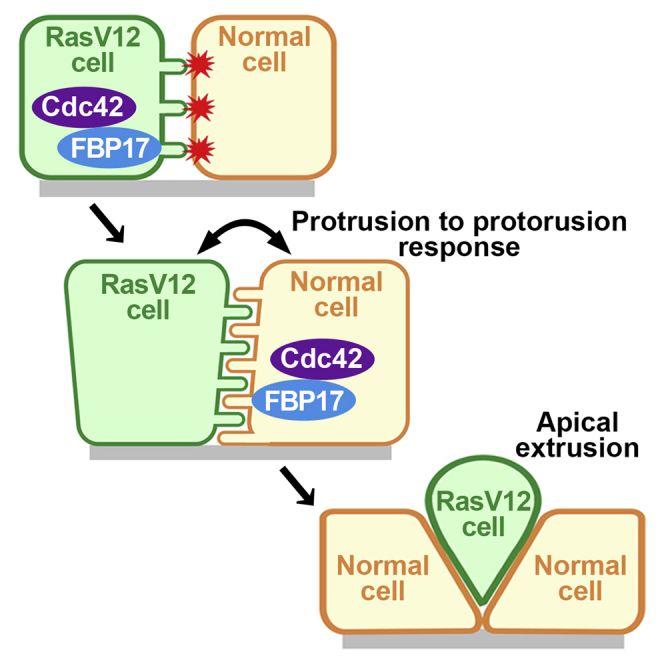
At the initial stage of carcinogenesis, cell competition often occurs between newly emerging transformed cells and the neighboring normal cells, leading to the elimination of transformed cells from the epithelial layer. For instance, when RasV12-transformed cells are surrounded by normal cells, RasV12 cells are apically extruded from the epithelium. However, the underlying mechanisms of this tumor-suppressive process still remain enigmatic. We first show by electron microscopic analysis that characteristic finger-like membrane protrusions are projected from both normal and RasV12 cells at their interface. In addition, FBP17, a member of the F-BAR proteins, accumulates in RasV12 cells, as well as surrounding normal cells, which plays a positive role in the formation of finger-like protrusions and apical elimination of RasV12 cells. Furthermore, cdc42 acts upstream of these processes. These results suggest that the cdc42/FBP17 pathway is a crucial trigger of cell competition, inducing “protrusion to protrusion response” between normal and RasV12-transformed cells.
Subject areas: Optics, Cell biology, Organizational aspects of cell biology
Graphical abstract
Highlights
-
•
EM analysis shows finger-like membrane protrusions between normal and RasV12 cells
-
•
Cdc42/FBP17 regulate the formation of the finger-like membrane protrusions
-
•
Cdc42/FBP17-mediated finger-like protrusions promote elimination of RasV12 cells
-
•
‘Protrusion to protrusion response’ triggers cell competition
Optics; Cell biology; Organizational aspects of cell biology
Introduction
Throughout the lifespan of multicellular organisms, oncogenic mutations sporadically occur in single cells within their tissues. Newly emerging transformed cells and the surrounding normal cells often compete with each other for survival and space; loser cells in the competition are eliminated from the cellular society, whereas winner cells proliferate and fill the vacant spaces (Amoyel and Bach, 2014; Baker, 2011; Bowling et al., 2019; Claveria and Torres, 2016; Johnston, 2009; Levayer, 2020; Merino et al., 2016; Morata and Calleja, 2020; Ohsawa et al., 2018; Vincent et al., 2013; Wagstaff et al., 2013). This process is termed cell competition, which was originally identified in Drosophila (Morata and Ripoll, 1975), but recent studies have shown that cell competition can also occur in mammals (Maruyama and Fujita, 2017). For example, when oncogenic Ras mutation (e.g. RasV12) occurs within the monolayer of epithelial tissues, RasV12-transformed cells are often apically extruded in a cell death-independent manner via competitive interaction with the surrounding normal cells (Hogan et al., 2009; Kon et al., 2017; Sasaki et al., 2018; Wu et al., 2014). During this apical extrusion process, various non-cell-autonomous changes are observed in both normal and transformed cells at their boundaries. In transformed cells surrounded by normal cells, the activity of cdc42 and myosin-II is elevated, which positively regulate the apical elimination of transformed cells (Hogan et al., 2009). In addition, in normal cells contacting transformed cells, cytoskeletal protein Filamin is accumulated at the interface with transformed cells, which generates physical forces that are required for apical extrusion (Kajita et al., 2014). These results imply a new concept that normal epithelia have intrinsic anti-tumor activity which does not involve immune systems. This homeostatic, tumor-suppressive machinery is named epithelial defense against cancer (EDAC) (Kajita et al., 2014). The underlying molecular mechanisms of EDAC are not fully understood yet, although several lines of evidence suggest that direct interaction between normal and transformed cells is required (Hogan et al., 2009; Kajita et al., 2010, 2014; Wu et al., 2014). However, the intercellular recognition machinery between normal and transformed cells and the trigger of their competitive interactions remain obscure. Electron microscopic (EM) analyses have contributed to gain insights into biological mechanism of interactions between heterogeneous cell populations such as osteoblast and osteoclast (Everts et al., 2002; Matsuo and Irie, 2008), neuromuscular junction (Heuser and Reese, 1973), an effector cytotoxic T cell and a target cell (Zagury et al., 1975), and macrophage and pathogen (Niedergang et al., 2003). To our knowledge, cell-cell adhesion sites between winner and loser cells in cell competition have not been studied in depth at the ultrastructural level.
The surface of plasma membranes encompasses various deformed structures including protrusions and invaginations. F-BAR and I-BAR, subfamilies of the BAR domain proteins, are crucial regulators for convex or concave membrane deformations such as lamellipodia, filopodia, clathrin-coated pit and caveolae (Frost et al., 2009; Saarikangas et al., 2009; Suetsugu et al., 2014). At the inner surface of plasma membranes, the BAR domain proteins bind to lipids or membrane proteins, and the crescent shape of BAR proteins leads to the deformation of the membrane by assembling actin cytoskeleton (Suetsugu et al., 2014; Takenawa and Suetsugu, 2007). FBP17 is a prototype F-BAR protein, which is involved in the formation of membrane invagination and also known as one of the downstream targets of cdc42, a member of the Rho family small GTPase (Frost et al., 2009; Itoh et al., 2005). During the induction of membrane deformation, FBP17 molecules first bind to membrane lipids and bend the cell membranes to negative curvature (Frost et al., 2009; Shimada et al., 2007). Then, FBP17 at the plasma membrane enhances cdc42-, WASP/N-WASP-, and Arp2/3-dependent actin nucleation and polymerization (Padrick et al., 2008; Takano et al., 2008), which generate forces and further extend the membrane deformation (Takenawa and Suetsugu, 2007).
In this study, we have performed the EM analyses on the intercellular regions between normal and RasV12-transformed epithelial cells. The results reveal that the cdc42/FBP17 pathway induces characteristic protrusive structures at the interface between normal and RasV12 cells, which would play a positive role in the apical extrusion of RasV12 cells from epithelia.
Results
The formation of characteristic membrane protrusions is promoted between normal and RasV12-transformed epithelial cells, as well as between RasV12 cells
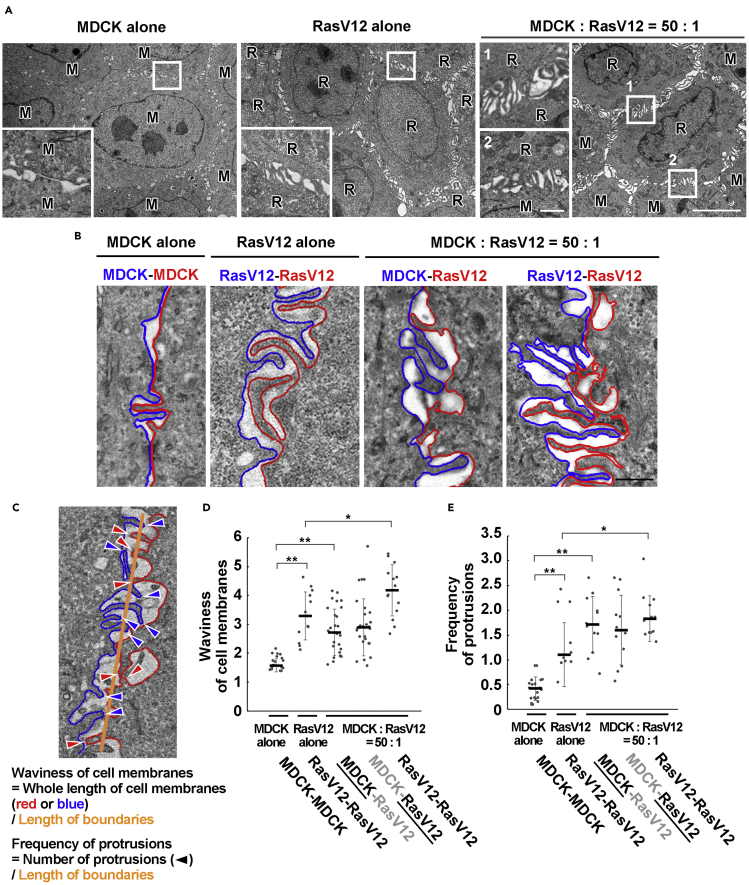
To gain structural insights into cell competition, we examined the ultrastructure of intercellular adhesion sites between normal and RasV12-transformed Madin-Darby canine kidney (MDCK) epithelial cells by transmission electron microscopy. For RasV12-transformed cells, we used MDCK-pTR GFP-RasV12 cells in which expression of GFP-RasV12 can be induced by tetracycline treatment. First, we cultured cells under three different conditions: (1) normal cells alone, (2) RasV12 cells alone, (3) mix culture of normal and RasV12 cells. Cells were cultured on a collagen matrix in the absence of tetracycline until a monolayer was formed. Subsequently, GFP-RasV12 expression was induced with tetracycline. In previous studies, we have reported that under the mix culture condition, around 16 hr of tetracycline addition, various non-cell-autonomous changes occur in both normal and RasV12 cells, and at 16-24 hr RasV12 cells are apically extruded from the monolayer of normal cells (Hogan et al., 2009; Kajita et al., 2014). To identify GFP-RasV12-expressing cells in the mix culture during the EM analyses, we used correlative light and electron microscopy in which differential interference contrast (DIC), fluorescence, and EM images were overlaid (Kon et al., 2017). In normal cells alone culture, cell-cell adhesions were rather flat and smooth with intermittent short protrusions (Figures 1A and 1B). In contrast, in RasV12 cells alone culture, numerous longer protrusions were interdigitated along cell-cell contacts (Figures 1A and 1B). To clarify the differences in the intercellular membrane structures, two parameters were quantified: waviness of cell membranes and frequency of protrusions (Figure 1C). We showed that both membrane waviness and protrusion formation were significantly elevated by RasV12 expression (Figures 1D and 1E). In addition, under the mix culture condition, at intercellular junctions between RasV12 cells surrounded by normal cells the formation of interdigitated protrusions was further promoted (Figures 1A, 1B, 1D, and 1E). Moreover, at the interface between normal and RasV12 cells, the membrane waviness and protrusion formation were elevated in normal cells compared with those in alone culture (Figures 1A, 1B, 1D, and 1E). X-Z image analyses showed that protrusions were formed at the whole cell-cell adhesions except apicalmost tight junction regions (Figure S1A). Collectively, these data indicate that the formation of finger-like protrusions is regulated between normal and RasV12 cells, as well as between RasV12 cells.
Figure 1.
The formation of characteristic membrane protrusions is promoted in RasV12-transformed cells and the neighboring normal cells
(A) EM images showing the alone cultures or mix culture of normal and RasV12-transformed MDCK cells. Cells were fixed at 16 hr after the induction of RasV12 and sectioned along the X-Y axis. The areas in the white boxes are shown as insets at higher magnification, demonstrating intercellular regions. Scale bars, 5 μm; 1 μm (insets).
(B) Tracings of cell membranes at cell-cell contacts in alone or mix culture. Scale bar, 0.5 μm.
(C) An example for analyses of waviness of cell membranes and frequency of protrusions. A protrusive plasma membrane of which length is longer than the width is defined as “protrusion”.
(D and E) Waviness of cell membranes (D) or frequency of protrusions (E) at each cell-cell contact in alone or mix culture. As for MDCK-RasV12 boundary, the data on the underlined side is shown. Data are median ±SD. ∗p < 0.05; ∗∗p < 0.01 (unpaired two-tailed Student's t-tests).
Accumulated FBP17 in RasV12 cells positively regulates the formation of finger-like protrusions
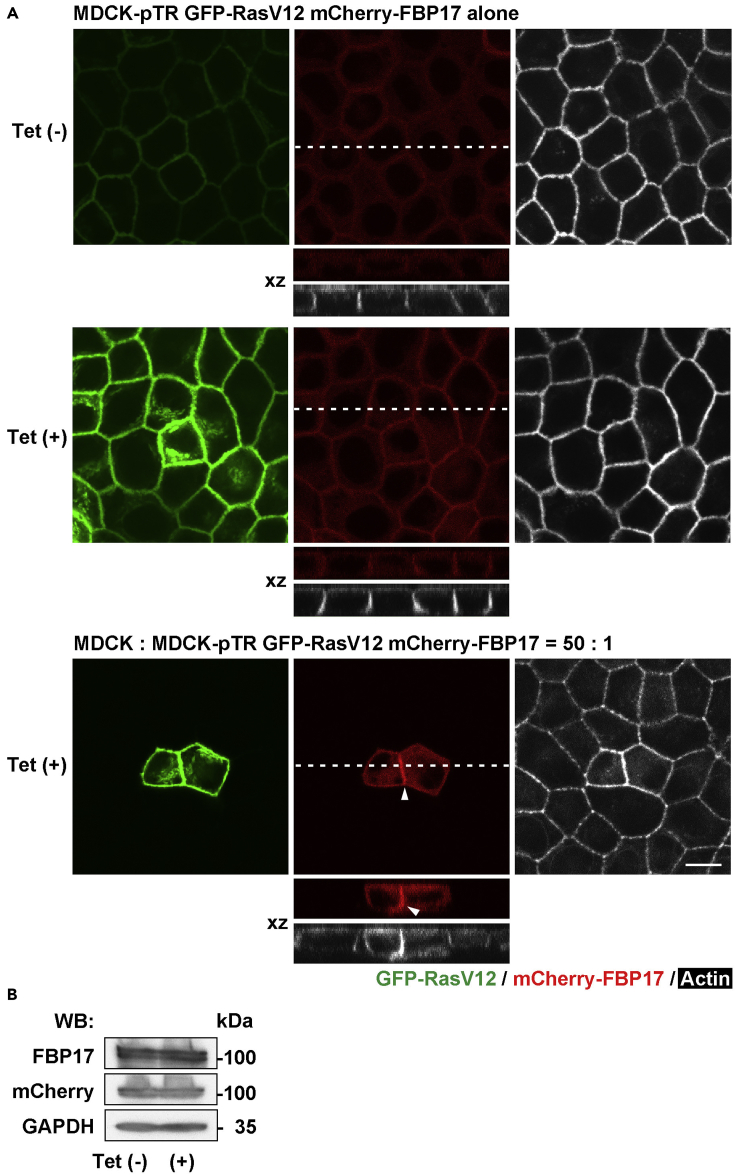
The formation of finger-like membrane protrusions can be regulated by F-BAR proteins such as FBP17 (Itoh et al., 2005; Tsujita et al., 2015). We thus examined whether the localization of FBP17 was controlled by RasV12 expression. We established MDCK-pTR GFP-RasV12 cells stably expressing mCherry-FBP17 (Figure S1B). In the absence of tetracycline where RasV12 expression was not induced, mCherry-FBP17 was diffusely localized within the cells (Figure 2A). Upon tetracycline treatment, FBP17 was substantially accumulated at the cell-cell contacts between RasV12-expressing cells (Figures 2A and S1C). The immuno-EM analysis showed that FBP17 was localized along and at the base of the intercellular protrusions (Figure S1D). The expression level of FBP17 proteins was not affected by tetracycline (Figure 2B), implying that RasV12 expression influences the localization, but not expression of FBP17. In addition, the stronger accumulation of FBP17 was observed at cell-cell adhesions between RasV12 cells under the mix culture condition (Figures 2A, arrowheads, and S1C).
Figure 2.
FBP17 accumulates at cell-cell contacts between RasV12-transformed cells
(A) MDCK-pTR GFP-RasV12 cells stably expressing mCherry-FBP17 were cultured alone (upper and middle panels) or mixed with normal cells (lower panels) in the absence (upper panels) or presence (middle and lower panels) of tetracycline for 16 hr. Arrowheads indicate accumulated FBP17 between RasV12 cells in the mix culture (lower panels). Scale bar, 10 μm.
(B) Western blotting analysis of MDCK-pTR GFP-RasV12 mCherry-FBP17 cells cultured without or with tetracycline for 16 hr
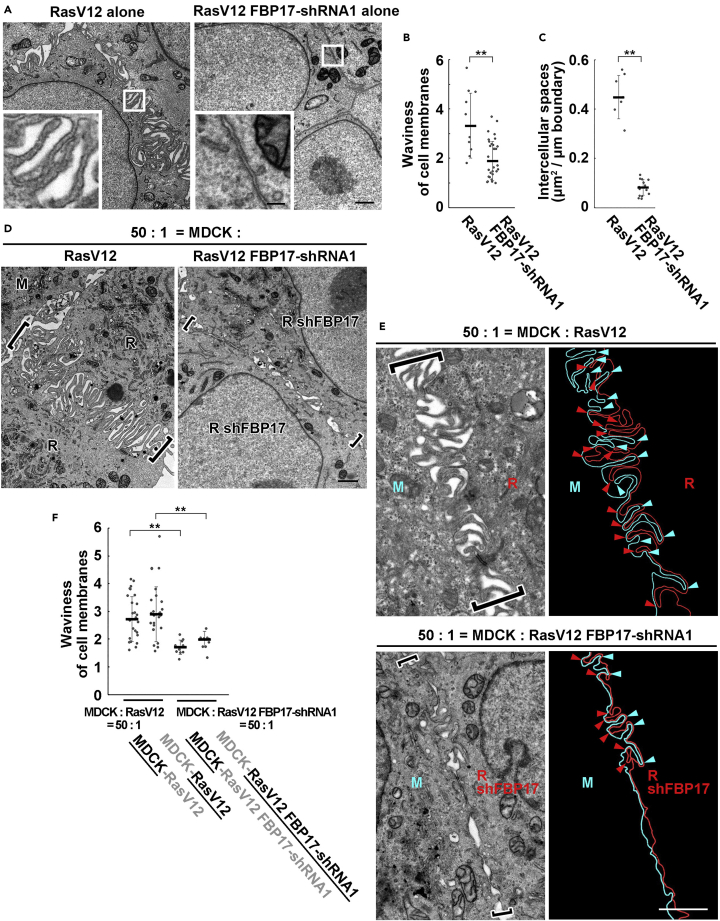
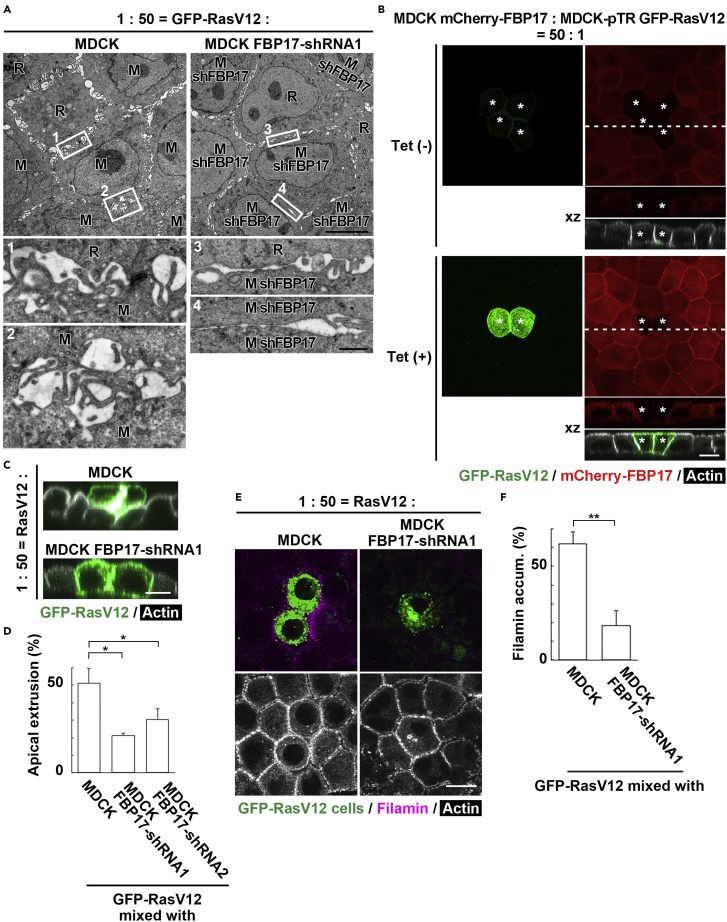
To elucidate the functional role of FBP17, we established two FBP17-knockdown RasV12 cell lines (Figure S2A). We have obtained comparable results with these two cell lines, thus the data using shRNA1-expressing cells are mainly shown below. Knockdown of FBP17 did not influence the localization of adherens junction protein E-cadherin or tight junction protein ZO-1 (Figure S2B). The EM analysis revealed that in FBP17-knockdown RasV12 cells, the formation of protrusions was strongly suppressed at cell-cell contact sites, which were utterly flattened with much lesser membrane waviness (Figures 3A, 3B, S2C, and S2D). Accordingly, the intercellular spaces were diminished (Figures 3A, 3C, S2C, and S2E). Similarly, the interdigitated protrusions disappeared between RasV12 cells surrounded by normal cells (Figure 3D). Knockdown of FBP17 also decreased waviness of cell membranes or intercellular spaces in normal cells, but the effect was milder than that in FBP17-knockdown RasV12 cells (Figures S3A–S3D). In addition, at the interface between normal and FBP17-knockdown RasV12 cells, protrusion formation was diminished at the normal side as well as the FBP17-knockdown RasV12 side (Figures 3E and 3F). Collectively, these data indicate that accumulated FBP17 in RasV12 cells regulates the formation of finger-like protrusions cell-autonomously (in RasV12 cells) and non-cell-autonomously (in the neighboring normal cells).
Figure 3.
FBP17 in RasV12-transformed cells positively regulates the waviness of cell membranes at cell-cell contacts
(A) EM images of RasV12-transformed cells or RasV12 cells stably expressing FBP17-shRNA1. Cells were fixed at 24 hr after the induction of RasV12. The areas in the white boxes are shown as insets at higher magnification, demonstrating intercellular regions.
(B and C) Quantification of waviness of cell membranes (B) and intercellular spaces (C) in RasV12-transformed cells or RasV12 FBP17-shRNA1 cells.
(D) Interdigitation of cell membranes between RasV12 cells in mix culture is diminished by FBP17 knockdown. Cells were fixed at 24 hr after the induction of RasV12. Brackets represent the position of cell-cell contacts.
(E) EM images and tracings of cell membranes showing the interface between normal cells and RasV12 cells (upper panels) or FBP17-knockdown RasV12 cells (lower panels). Cell membranes of normal and RasV12 or RasV12 FBP17-shRNA1 cells are colored in blue and red, respectively. The brackets and arrowheads indicate the position of cell-cell contacts and finger-like protrusions, respectively. (A, D, E) Scale bars, 1 μm; 0.2 μm (inset).
(F) Waviness of cell membranes at each cell-cell contact. The data on the underlined side is shown. (B, C, F) Data are median ±SD. ∗∗p < 0.01 (unpaired two-tailed Student's t-tests).
FBP17 in RasV12-transformed cells plays a positive role in apical extrusion
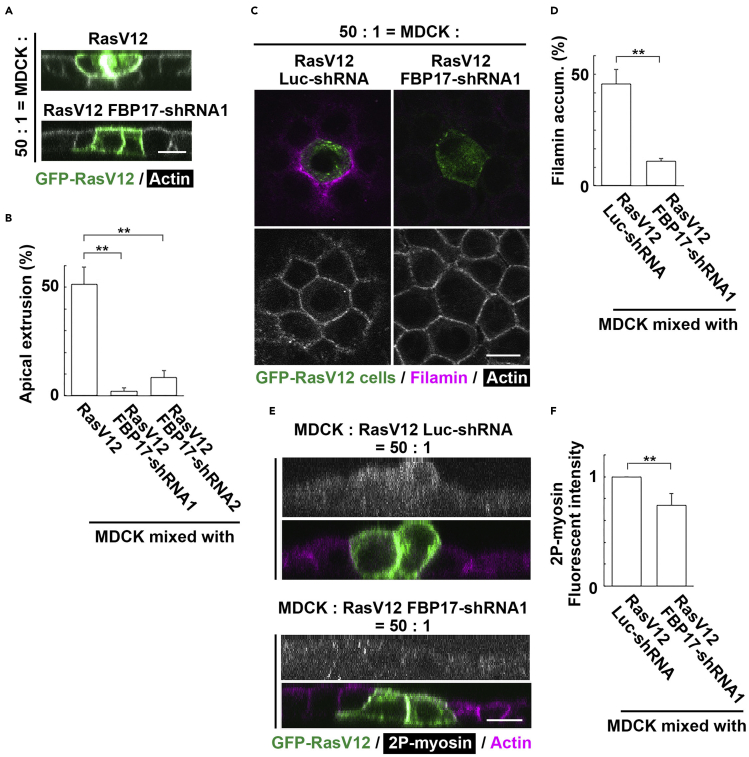
Next, we examined the effect of FBP17 knockdown on the behavior and fate of the mix-cultured normal and RasV12 cells. We found that when FBP17-knockdown RasV12 cells were surrounded by normal cells, apical extrusion was profoundly suppressed (Figures 4A and 4B). We then analyzed how FBP17 knockdown influences the intercellular communication between normal and RasV12 cells. Previous studies have shown that when RasV12-transformed cells are surrounded by normal cells, various non-cell-autonomous changes occur in both normal and RasV12 cells. For instance, cytoskeletal protein Filamin accumulates in normal cells at the boundary with RasV12 cells, generating physical forces for apical extrusion of RasV12 cells (Kajita et al., 2014). In addition, myosin-II is activated in RasV12 cells, which also plays a positive role in their extrusion (Hogan et al., 2009). Around FBP17-knockdown RasV12 cells, accumulation of Filamin, a crucial regulator of EDAC, was substantially suppressed (Figures 4C and 4D). Furthermore, in FBP17-knockdown RasV12 cells surrounded by normal cells, non-cell-autonomous elevation of myosin-II activity was significantly diminished (Figures 4E and 4F). Collectively, these data suggest that the increased membrane localization of FBP17 in RasV12-transformed cells induces non-cell-autonomous changes in both normal and RasV12 cells, thereby promoting apical extrusion.
Figure 4.
FBP17 in RasV12-transformed cells plays a positive role in apical extrusion
(A) X-Z fluorescence images of RasV12 or FBP17-knockdown RasV12 cells surrounded by normal cells.
(B) Effect of FBP17 knockdown in RasV12-transformed cells on apical extrusion in mix cultures.
(C) Immunofluorescence images of Filamin in mix cultures of RasV12 or RasV12 FBP17-shRNA1 cells surrounded by normal cells.
(D) Effect of FBP17 knockdown in RasV12 cells on Filamin accumulation in the neighboring normal cells.
(E) X-Z immunofluorescence images of di-phosphorylated myosin (2P-myosin) in RasV12 or RasV12 FBP17-shRNA1 cells surrounded by normal cells. (A, C, E) Scale bars, 10 μm.
(F) Effect of FBP17 knockdown in RasV12 cells surrounded by normal cells on 2P-myosin accumulation. (B, D, F) Data are mean ± SD. ∗∗p < 0.01 (unpaired two-tailed Student's t-tests).
Accumulated FBP17 in the surrounding normal cells also plays a positive role in apical extrusion
As shown in Figures 1A and 1B, at the interface between normal and RasV12 cells, finger-like protrusion formation was promoted from normal cells as well (also shown in Figure 5A: white box 1). We realized that the protrusion formation was also enhanced between the first and second rows of normal cells around RasV12 cells (Figure 5A: white box 2). The membrane protrusions were observed between the second and third rows of normal cells to a lesser extent and diminished in further distant normal cells. To examine the functional involvement of FBP17 in normal cells, we analyzed MDCK cells stably expressing mCherry-FBP17 or FBP17-shRNA (Figures S3A and S3E). First, mCherry-FBP17-expressing MDCK cells were mix-cultured with MDCK-pTR GFP-RasV12 cells, and the effect of RasV12 expression on the localization of FBP17 in the surrounding cells was examined. In the absence of tetracycline where RasV12 expression was not induced, mCherry-FBP17 was diffusely localized within the surrounding normal cells (Figures 5B and S3F). Upon tetracycline treatment, FBP17 was accumulated at the intercellular regions between the surrounding cells (Figures 5B and S3F), indicating that the expression of RasV12 induces membrane accumulation of FBP17 in the surrounding normal cells in a non-cell-autonomous fashion. In addition, FBP17-knockdown in normal cells surrounding RasV12 cells profoundly suppressed the formation of finger-like protrusions (Figure 5A: white boxes 3 and 4). Furthermore, when RasV12 cell were surrounded by FBP17-knockdown cells, apical extrusion was significantly suppressed (Figures 5C and 5D), accompanied by diminished Filamin accumulation at the interface with RasV12 cells (Figures 5E and 5F). Thus, membrane accumulation of FBP17 in the surrounding normal cells also facilitates the formation of membrane protrusions and apical extrusion.
Figure 5.
FBP17 accumulating in the surrounding normal cells plays a positive role in apical extrusion
(A) EM images of MDCK-pTR GFP-RasV12 cells surrounded by normal MDCK or MDCK FBP17-shRNA1 cells. The intercellular regions in the white boxes 1-4 are shown below at higher magnification: (1) between normal and RasV12 cells, (2) between the first and second row of normal cells, (3) between a RasV12 cell and the first row of an FBP17-knockdown cell, (4) between the first and second row of FBP17-knockdown cells. Scale bars, 5 μm; 0.5 μm (lower panels).
(B) Fluorescence images of mix cultures of MDCK-pTR GFP-RasV12 and MDCK mCherry-FBP17 cells in the absence or presence of tetracycline. X-Y images show z stack.
(C) X-Z fluorescence images of RasV12-transformed cells surrounded by normal or FBP17-knockdown cells.
(D) Effect of FBP17 knockdown in normal cells on apical extrusion.
(E) Immunofluorescence images of Filamin in mix cultures of RasV12 cells and normal or FBP17-knockdown cells. (B, C, E) Scale bars, 10 μm.
(F) Effect of FBP17 knockdown in the surrounding normal cells on Filamin accumulation. (D, F) Data are mean ± SD. ∗p < 0.05, ∗∗p < 0.01 (unpaired two-tailed Student's t-tests).
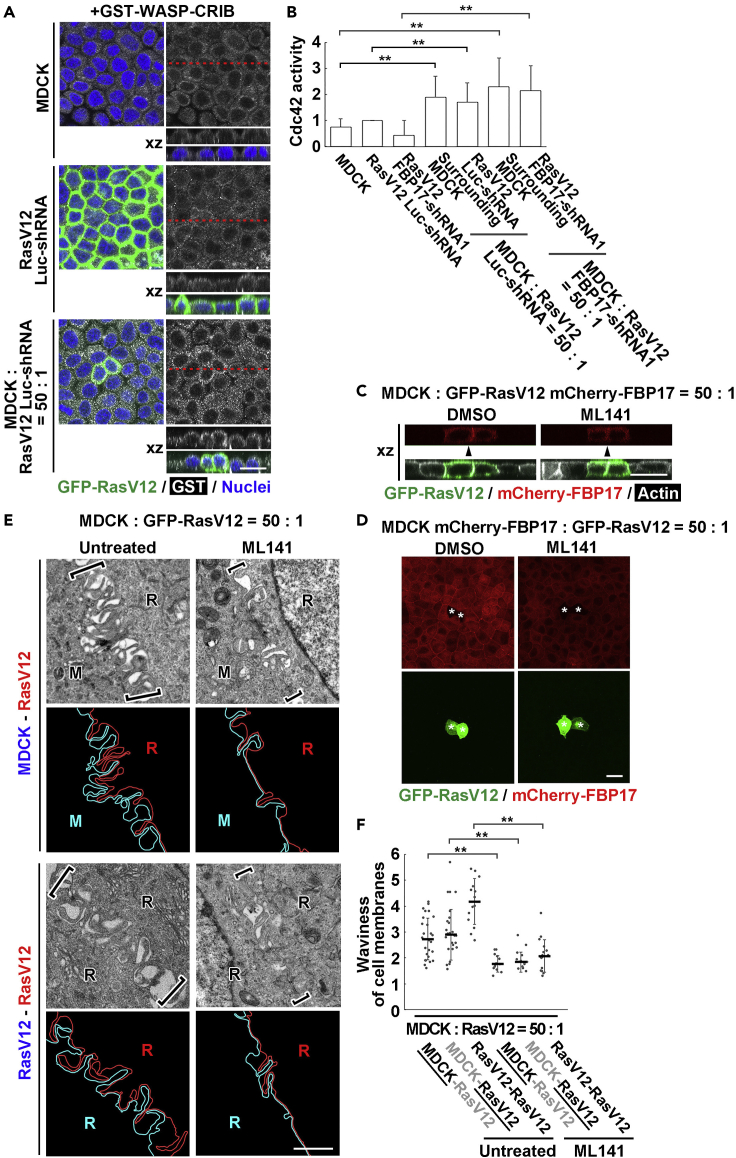
Cdc42 functions upstream of FBP17-mediated protrusion formation
Previous studies have demonstrated that the elevated activity of cdc42 promotes membrane localization and membrane-deforming activity of FBP17 (Chan Wah Hak et al., 2018; Itoh et al., 2005; Tsujita et al., 2015; Watson et al., 2016). To examine the cdc42 activity, we performed indirect-immunofluorescence analysis using a recombinant protein for the Cdc42-binding domain (CRIB) of WASP (Hogan et al., 2009). When normal cells or RasV12 cells were cultured alone, the cdc42 activity remained at a low level (Figures 6A and 6B). In contrast, when RasV12 cells were surrounded by normal cells, the cdc42 activity was significantly elevated in both RasV12 cells and the surrounding normal cells (Figures 6A, 6B, and S4A). Knockdown of FBP17 in RasV12 cells did not affect the activity of cdc42 (Figure 6B). We then examined the effect of cdc42 inhibitor ML141, which suppressed the elevated cdc42 activity in RasV12 cells and the surrounding normal cells under the mix culture condition (Figure S4B). The treatment with ML141 diminished the accumulation of FBP17 between RasV12 cells or between surrounding normal cells under the mix culture condition (Figures 6C, 6D, S4C, and S4D), resulting in the decreased membrane waviness and intercellular spaces (Figures 6E and 6F). These data indicate that cdc42 acts as a crucial upstream regulator for intercellular structures upon cell competition between normal and transformed cells.
Figure 6.
Cdc42 functions upstream of FBP17 not only in RasV12-transformed cells but also in the neighboring normal cells
(A) Cdc42 activity assay in single or mix cultures of MDCK and MDCK-pTR GFP-RasV12 cells. At 16 hr after the induction of RasV12, cells were treated with GST-WASP-CRIB proteins and then stained with anti-GST antibody. RasV12 cells stably expressing Luciferase (Luc)-shRNA were used as control for RasV12 FBP17-shRNA1 cells.
(B) Quantification of the cdc42 activity. Values of pixel intensity in the cytoplasm were measured and expressed as a ratio relative to RasV12 Luc-shRNA. Differences of pixel intensity between GST-WASP-CRIB and GST were calculated in each condition. Data are mean ± SD. ∗∗p < 0.01 (unpaired two-tailed Student's t-tests).
(C and D) Fluorescence images of RasV12-transformed (C) or normal (D) cells expressing mCherry-FBP17 in the mix culture without or with the cdc42 inhibitor ML141. (D) z stack images are shown. The asterisks indicate RasV12 cells. (A, C, D) Scale bars, 20 μm.
(E) EM images of the interface between normal and RasV12 cells (upper panels) or between RasV12 cells (lower panels) in the mix culture without or with ML141. Tracings of cell membranes were shown in blue or red. The brackets indicate the position of cell-cell contacts. Scale bar, 1 μm.
(F) Waviness of cell membranes at each cell-cell contact in mix culture without or with ML141. As for MDCK-RasV12 boundary, the data on the underlined side is shown. Data are median ±SD. ∗∗p < 0.01 (unpaired two-tailed Student's t-tests).
Discussion
This is the first report, to our knowledge, that demonstrates the ultrastructures on the boundary between winner and loser cells in cell competition. Our EM analyses have revealed various alterations at plasma membranes such as enhanced membrane waviness, characteristic finger-like protrusion formation and increased intercellular spaces, which have not been identified by conventional light microscopy analyses. We first demonstrate that RasV12 expression enhances the formation of finger-like protrusions. In addition, at the interface between normal and RasV12-transformed cells, the formation of finger-like protrusions is also substantially promoted from normal cells in a non-cell-autonomous manner. Knockdown of FBP17 in RasV12 cells profoundly diminished the number of finger-like protrusions in both RasV12 cells and the surrounding normal cells. These data imply the presence of “protrusion to protrusion response” between normal and transformed cells; the protrusions from transformed cells are sensed by the neighboring normal cells, which in turn stick back protrusions toward transformed cells (Figure S5A). Thus, the visualization of ultrastructures by a series of EM analyses has revealed that the FBP17-mediated protrusion formation is implicated in the intercellular recognition and the following response between loser and winner cells in cell competition.
The finger-like membrane protrusions have specific morphological and structural features. In various cell types including fibroblasts, macrophages, nerve growth cones, and epithelial cells, filopodia are straight, 0.1–0.3 μm in a diameter and ∼10 μm in length (Mattila and Lappalainen, 2008; Vasioukhin et al., 2000; Welch and Mullins, 2002) that are filled with tight parallel bundles of filamentous actin. Very short filopodia (0.1–0.3 μm in a diameter and 0.5–1 μm in length) that are almost completely embedded in the cell cortex or leading edge are thought to be precursors of filopodia, often termed microspikes (Mattila and Lappalainen, 2008; Svitkina et al., 2003). Unlike filopodia or microspikes, finger-like protrusions observed in this study represent irregular shapes, often bent or curved, and are thinner (40-50 nm in diameter), and often extended just ∼0.4 μm. In addition, no obvious actin bundles are observed inside the finger-like protrusions (e.g. Figure 3A, inset). The treatment with cdc42 inhibitor suppresses the formation of finger-like protrusions in both RasV12 and the neighboring normal cells under the mix culture condition (Figures 6E and 6F), indicating that cdc42 is a crucial upstream regulator of the membrane protrusions. The cdc42 activity is elevated in both RasV12 and the neighboring normal cells under the mix culture condition but not in RasV12 cells cultured alone, suggesting that cdc42-independent mechanism might be also involved in finger-like protrusions under the latter condition. Previous studies have shown that active cdc42 is involved in the stabilization of E-cadherin-based cell-cell adhesions (Braga, 2000; Fukata et al., 2002), but our data imply that cdc42 regulates intercellular adhesions between normal and RasV12 cells in a distinct manner. Around RasV12 cells, not only in directly contacting normal cells but also in normal cells at the second or third row, membrane localization of FBP17 and cdc42 activity as well as protrusion formation are promoted. These results suggest that certain types of signaling pathways are propagated across the surrounding normal cells. Recently, we have found that the production of reactive oxygen species (ROS) is increased in a wide range of normal cells surrounding transformed cells (Unpublished data). Indeed, addition of the antioxidant Trolox significantly suppresses the cdc42 activity in both RasV12 and the surrounding normal cells (Figure S4E). Hence, the link between ROS and cdc42 as well as the functional involvement of other upstream regulators of cdc42/FBP17 should be further elucidated. Under the mix culture condition, knockdown of FBP17 in either RasV12 cells or the surrounding normal cells diminishes the accumulation of Filamin and frequency of apical extrusion, indicating that FBP17-mediated protrusions from both cells play a key role in these processes. However, it remains unknown whether and how the membrane protrusions act as signaling hub (Figure S5B). Signaling molecules might be enriched on the finger-like protrusions, or physical stimuli (poking or pushing) by protrusions could be recognized by mechanosensors on the opposite cells. In addition, the diffuse activation of cdc42 in the large areas of surrounding normal cells might suggest the involvement of certain soluble factors (Figure S5B). Moreover, we need to consider a possibility that cdc42/FBP17 might affect apical extrusion of RasV12 cells by regulating additional cellular processes, independently of finger-like protrusions. The functional modes of cdc42/FBP17 and the finger-like protrusions need to be clarified in future studies.
Limitations of the study
In this study, we present several lines of evidence suggesting the intercellular recognition between normal and RasV12-transformed cells via cdc42/FBP17-mediated finger-like membrane protrusions. We demonstrate using cdc42 activity assay that antioxidant Trolox treatment suppresses the cdc42 activity in both RasV12 and the surrounding normal cells. In future studies, we should further elucidate the link between ROS and cdc42 as well as the functional involvement of other upstream regulators of cdc42/FBP17.
Cdc42 can affect a variety of cellular processes including endocytosis and cell-cell adhesions in addition to FBP17-mediated membrane protrusions. Thus, a possibility cannot be ruled out that cdc42 also regulates cell competition by affecting the other cellular processes than the formation of finger-like protrusion. We should clarify functional modes of cdc42/FBP17 to further validate the proposed “protrusion to protrusion response” model.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-FBP17 antibody | Itoh et al. (2005) | |
| rabbit anti-GST antibody | Santa Cruz Biotechnology | Cat#sc-459; RRID: AB_631586 |
| rabbit anti-DsRed antibody | Clontech | Cat#632496; RRID: AB_10013483 |
| rabbit anti-RFP antibody | MBL | Cat#PM005; RRID: AB_591279 |
| mouse anti-GAPDH antibody | Millipore | Cat#MAB374; RRID: AB_2107445 |
| rat anti-E-cadherin antibody | Life Technologies | Cat#13-1900; RRID: AB_86571 |
| rabbit anti-ZO-1 antibody | Thermo Fisher Scientific | Cat#61-7300; RRID: AB_2533938 |
| mouse anti-Filamin antibody | Sigma | Cat#F6682; RRID: 2106432 |
| rabbit anti-phospho-Myosin light chain 2 (Thr18/Ser19) antibody | Cell Signaling Technology | Cat#95777 |
| Alexa Fluor-568-conjugated phalloidin | Invitrogen | Cat#A12380 |
| Alexa Fluor-647-conjugated phalloidin | Invitrogen | Cat#A22287 |
| Alexa Fluor-568 donkey anti-mouse IgG (H+L) | Life Technologies | Cat#A10037; RRID:AB_2534013 |
| Alexa Fluor-568 donkey anti-rat IgG (H+L) | Life Technologies | Cat#A11077; RRID:AB_2534121 |
| Alexa Fluor-647 donkey anti-rabbit IgG (H+L) | Life Technologies | Cat#A31573; RRID:AB_2536183 |
| 20-nm gold goat anti-rabbit IgG (H+L) | British BioCell International | Cat#EMGAR20 |
| Chemicals, peptides, and recombinant proteins | ||
| ML141 | Tocris Bioscience | Cat#4266 |
| Trolox | Cayman Chemical | Cat#10011659 |
| Blasticidin | Invivogen | Cat#ant-bl-1 |
| Zeocin | Invivogen | Cat#ant-zn-1 |
| G418 (Geneticin) | GIBCO | Cat#10131027 |
| Tetracycline | Sigma-Aldrich | Cat#T7660 |
| Paraformaldehyde | Sigma-Aldrich | Cat#6148 |
| Glutaraldehyde | Electron Microscopy Sciences | Cat#16020 |
| osmium tetroxide | TAAB | Cat#O003 |
| Critical commercial assays | ||
| MycoAlert | Lonza | Cat#LT07-118 |
| Hoechst 33342 | Life Technologies | Cat#H3570 |
| Type I collagen | Nitta Gelatin | N/A |
| Nucleofector 2b Kit L | Lonza | Cat#VACA-1005 |
| Araldite502/Embed812 kit | Electron Microscopy Sciences | Cat#13940 |
| LR-white resin (medium) | Nisshin EM | Cat#3962 |
| Experimental models: Cell lines | ||
| MDCK | Dr. W. Birchmeier | N/A |
| Oligonucleotides | ||
| canine FBP17-shRNA1 forward: 5′-GATCCCCGGATCGGAGAAT CAATGAATTCAAGAGATTCATTGATTCTCCGATCCTTTTTC-3′ |
This paper | N/A |
| canine FBP17-shRNA1 reverse: 5′-TCGAGAAAAAGGATCGGAG AATCAATGAATCTCTTGAATTCATTGATTCTCCGATCCGGG-3′ |
This paper | N/A |
| canine FBP17-shRNA2 forward: 5′-GATCCCCGGGAAATGCTTG GATGAAATTCAAGAGATTTCATCCAAGCATTTCCCTTTTTC-3′ |
This paper | N/A |
| canine FBP17-shRNA2 reverse: 5′-TCGAGAAAAAGGGAAATGC TTGGATGAAATCTCTTGAATTTCATCCAAGCATTTCCCGGG-3′ |
This paper | N/A |
| Software and algorithms | ||
| ImageJ | NIH Image | https://imagej.nih.gov/ij/download.html |
| 3DMOD | Kremer et al. (1996) | https://bio3d.colorado.edu/imod/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yasuyuki Fujita (fujita@monc.med.kyoto-u.ac.jp).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
MDCK cells were used in this study. The parental MDCK cells were a gift from Dr. Walter Birchmeier. Mycoplasma contamination is regularly tested for all cell lines using a commercially available kit (MycoAlert, Lonza).
Method details
Antibodies and materials
Rabbit anti-FBP17 antibody was generated in a previous study (Itoh et al., 2005). Rabbit anti-GST antibody (sc-459) was purchased from Santa Cruz Biotechnology. Rabbit anti-DsRed antibody (632496) from Clontech and Rabbit anti-RFP antibody (PM005) from MBL were used for western blotting and immuno-EM, respectively. Mouse anti-GAPDH antibody (6C5, MAB374) was from Millipore. Rat anti-E-cadherin antibody (ECCD-2, 13-1900) and rabbit anti-ZO-1 antibody (61-7300) were from Thermo Fisher Scientific. Mouse anti-Filamin antibody (F6682) was from Sigma. Rabbit anti-phospho-Myosin light chain 2 (Thr18/Ser19) antibody (E2J8F, 95777) was from Cell Signaling Technology. Alexa Fluor-568- or -647-conjugated phalloidin (Invitrogen) was used at 1.0 U/ml. Alexa Fluor-568- and -647-conjugated secondary antibodies were from Life Technologies. Hoechst 33342 (Life Technologies) was used at a dilution of 1:5,000. For immunofluorescence, primary antibodies were used at 1:100, except for GST antibody that was used at 1:50, and all secondary antibodies were used at 1:200. For western blotting, peroxidase-conjugated anti-rabbit and anti-mouse secondary antibodies were from GE Healthcare and Jackson ImmunoReseach, respectively. The selective Cdc42 inhibitor ML141 (20 μM) and the antioxidant Trolox (1 mM) were from Tocris Bioscience and Cayman Chemical, respectively.
Cell culture
MDCK cells were cultured as previously described (Hogan et al., 2009). MDCK-pTR GFP-RasV12 cells were cultured in the medium containing 10% FBS, 1% GlutaMAX (Gibco), 5 μg/ml of blasticidin (Invivogen) and 400 μg/ml of zeocin (Invivogen) (Hogan et al., 2009). To establish MDCK or MDCK-pTR GFP-RasV12 cells stably expressing mCherry-FBP17, MDCK or MDCK-pTR GFP-RasV12 cells were transfected with pPB-EF1-MCS-IRES-Neo-mCherry-canine FBP17 by nucleofection (Nucleofector 2b Kit L, Lonza), followed by selection in the medium containing 800 μg/ml of G418 (Geneticin, Gibco). MDCK or MDCK-pTR GFP-RasV12 cells stably expressing canine FBP17-shRNA were established as follows; canine FBP17-shRNA oligonucleotides (canine FBP17-shRNA1: 5′-GATCCCCGGATCGGAGAATCAATGAATTCAAGAGATTCATTGATTCTCCGATCCTTTTTC-3′ and 5′-TCGAGAAAAAGGATCGGAGAATCAATGAATCTCTTGAATTCATTGATTCTCCGATCCGGG-3′ or canine FBP17-shRNA2: 5′-GATCCCCGGGAAATGCTTGGATGAAATTCAAGAGATTTCATCCAAGCATTTCCCTTTTTC-3′ and 5′-TCGAGAAAAAGGGAAATGCTTGGATGAAATCTCTTGAATTTCATCCAAGCATTTCCCGGG-3′) were cloned into the BglII and XhoI sites of pSUPER.neo + gfp (Oligoengine). MDCK or MDCK-pTR GFP-RasV12 cells were transfected with pSUPER.neo + gfp canine FBP17-shRNA by nucleofection, followed by selection in the medium containing 800 μg/ml of G418. ML141 or Trolox was added together with tetracycline.
Immunofluorescence and western blotting
For immunofluorescence, RasV12-transformed cells were mixed with non-transformed cells at a ratio of 1:50. The mixture of cells were cultured on coverslips (Matsunami) coated with the collagen matrix as previously described (Hogan et al., 2009). Cells were incubated for 8-12 h until a monolayer was formed. The expression of GFP-RasV12 was then induced by 2 μg/ml of tetracycline (Sigma-Aldrich) for 16 h for analyses of non-cell-autonomous changes occurring prior to apical extrusion or for 24 h for analyses of apical extrusions. Cells were fixed with 4% paraformaldehyde (PFA) in PBS and permeabilized as previously described (Hogan et al., 2009), except for Filamin staining, for which cells were fixed in methanol for 2.5 min at -20°C, followed by blocking in 10% BSA/PBS for 1 h. Immunofluorescence images were analyzed at 0.5-μm X-Z-intervals by the FV1000 or FV1200 system (Olympus) equipped with a 60× oil-immersion objective lens (1.35 NA). Image acquisition was performed by the Olympus FV10-ASW software. Fluorescent images were quantified by ImageJ. For Figures S1C, S3F, S4C, and S4D, to quantify the ratio of mCherry-FBP17 fluorescence (cell-cell contact site/cytosol), the pixel intensity was quantified within three regions (0.76 μm-square each) at cell-cell boundary or cytosol. The average values were used for calculations of the fluorescence ratio. For quantification of apical extrusion of RasV12-transformed cells, 1-5 transformed cells that were surrounded by normal cells were analyzed. More than 50 colonies of transformed cells were analyzed for each condition, and then the ratio of apically extrusion of transformed cells was quantified. To determine the level of active GTP-bound Cdc42 in cells, Cdc42 activity assay using a GST fusion protein containing the Cdc42-interacting domain of WASP (WASP-CRIB) was performed as previously described (Hogan et al., 2009). Briefly, after fixation by PFA, permeabilization and blocking, cells were incubated with 25 μg/ml of GST or GST-WASP-CRIB protein in blocking buffer at 4°C for overnight. After washings with blocking buffer, cells were stained with anti-GST antibody, Alexa-647-conjugated secondary antibody, and Hoechst. Western blotting was performed as previously described (Hogan et al., 2004). Primary antibodies were used at 1:500 except anti-GAPDH antibody at 1:2,000. Western blotting data were analyzed using ImageQuant LAS4010 (GE Healthcare) or LuminoGraph WSE-6100 (ATTO).
Electron microscopy
EM analyses were carried out as previously described (Kon et al., 2017). Briefly, cells were cultured in collagen-gel-coated, grid-imprinted plastic dishes (μ-Dish 35 mm Grid-500, Ibidi). Cells were fixed by 2% glutaraldehyde (GA) (Electron Microscopy Sciences) in HEPES buffer (pH 7.4; 30 mM HEPES, 0.1 M NaCl and 2 mM CaCl2). Fluorescence and DIC images of GFP-RasV12 cells and the surrounding cells were captured together with the underlying, numbered grid information. Cells were then post-fixed in 2% osmium tetroxide in 0.1 M imidazole (pH 7.4), and en bloc staining was performed by 1% uranyl acetate (UA). Samples were embedded in Araldite502/Embed812 resin (Electron Microscopy Sciences). During these procedures, GFP fluorescence was lost, but the grid was imprinted onto the resin. Ultrathin sections (∼70 nm) were made by EM-UC7 (Leica) and mounted on formvar-coated copper grids (SP3 slit mesh, Nisshin EM). Sections were stained with 4% UA and 0.4% lead citrate, and cells were imaged with a transmission electron microscope (TEM, JEM-1400; JEOL) operating at 80 kV. TEM images were analyzed with the 3DMOD program (Kremer et al., 1996).
For immunoelectron microscopic analyses, MDCK-pTR GFP-RasV12 mCherry-FBP17 cells were cultured on sapphire glasses (Naruse Koueki) and fixed by 4% PFA containing 0.2% GA in PBS. Samples were embedded in LR-white resin (Nisshin EM). Sections (∼80 nm) were mounted on formvar-coated nickel grids (SP2 slit mesh, Nisshin EM). After blocking with 4% BSA/PBS, the sections were treated with anti-RFP antibody (1:300, MBL) and 20-nm gold-labeled secondary antibody (1:50, British BioCell International), followed by fixation in 1% GA and staining with 4% UA.
Quantification and statistical analysis
For statistics analyses, unpaired two-tailed Student’s t-tests were used to determine p values. p values less than 0.05 were considered to be significant. No statistical method was used to predetermine sample size.
The numbers of the analyzed samples in each figure are as follows. For Figures 1D and 1E, 20, 13, 26, 26, 14 boundaries for waviness of cell membranes (D; left to right in the graph) and 20, 10, 12, 12, 12 boundaries for frequency of protrusions (E; left to right in the graph) were analyzed from two (MDCK-MDCK) or three (the others) independent experiments. For Figures 3B, 3C, and 3F, 10 (RasV12) or 30 (RasV12 FBP17-shRNA1) boundaries (B) or 7 (RasV12) or 15 (RasV12 FBP17-shRNA1) boundaries (C) or 10 (MDCK in the mix culture of MDCK and RasV12 FBP17-shRNA1 cells) or 10 (RasV12 FBP17-shRNA1 in the mix culture of MDCK and RasV12 FBP17-shRNA1 cells) boundaries (F) from three independent experiments. For Figure 4B, 155, 137, 140 cells for RasV12, 177, 160, 145 cells for RasV12 FBP17-shRNA1, and 116, 131, 147 cells for RasV12 FBP17-shRNA2 were analyzed from three independent experiments. For Figure 4D, 132, 140, 158 cells for RasV12, and 134, 127 cells for RasV12 FBP17-shRNA1 were analyzed from three or two independent experiments, respectively. For Figure 4F, 34, 32, 36 cells for RasV12 Luc-shRNA, and 30, 36, 38 cells for RasV12 FBP17-shRNA1 were analyzed from three independent experiments. For Figure 5D, 155, 137, 140 cells for MDCK, 145, 164, 151 cells for MDCK FBP17-shRNA1, and 133, 122, 168, 179 cells for MDCK FBP17-shRNA2 were analyzed from three (MDCK and MDCK FBP17-shRNA1) or four (MDCK FBP17-shRNA2) independent experiments. For Figure 5F, 136, 134, 138 cells for MDCK and 149, 152, 148 cells for MDCK FBP17-shRNA1 were analyzed from three independent experiments. For Figure 6B, 45, 45, 45, 57, 48, 57, 59 cells for GST (left to right in the graph) and 63, 63, 63, 68, 66, 66, 63 cells for GST-WASP-CRIB (left to right in the graph) were analyzed from three independent experiments. For Figure 6F, 13, 13, 16 boundaries for waviness of cell membranes (ML141; left to right in the graph) were analyzed from two independent experiments. For Figure S1C, 47, 47, 29 boundaries (left to right in the graph) from three independent experiments. For Figures S2D and S2E, 10 (RasV12) or 24 (RasV12 FBP17-shRNA2) boundaries (D) or 7 (RasV12) or 12 (RasV12 FBP17-shRNA2) boundaries (E) from three independent experiments. For Figures S3C and S3D, 20 (MDCK) or 28 (MDCK FBP17-shRNA1) boundaries (C) or 10 (MDCK) or 14 (MDCK FBP17-shRNA1) boundaries (D) from three independent experiments. For Figure S3F, 48 (Tet (-)) or 48 (Tet (+)) boundaries from three independent experiments. For Figure S4B, 60, 51, 51, 41 cells for GST (left to right in the graph) and 60, 50, 65, 48 cells for GST-WASP-CRIB (left to right in the graph) from three independent experiments. For Figures S4C and S4D, 37 (DMSO) or 25 (ML141) boundaries (C) or 31 (DMSO) or 23 (ML141) boundaries (D) from three independent experiments. For Figure S4E, 57, 47, 66, 47 cells for GST (left to right in the graph) and 54, 49, 60, 53 cells for GST-WASP-CRIB (left to right in the graph) from three independent experiments.
Acknowledgments
We thank K. Tanaka and T. Kishimoto for the support for EM analyses. This work was supported by Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (S) 21H05039, JSPS Bilateral Joint Research Projects (The Royal Society) JPJSBP1 20215703, JSPS Grant-in-Aid for Challenging Research (Pioneering) 20K21411, Japan Science and Technology Agency (JST) (Moonshot R&D: Grant Number JPMJPS2022), the Takeda Science Foundation, and SAN-ESU GIKEN CO. LTD (to Y.F.) and by Grant-in-Aid for Young Scientists (B) 17K15589, Grant-in-Aid for JSPS Fellows 19J40132, Grant-in-Aid for Scientific Research (C) 20K07559, Kazato Research Foundation, Shiseido Female Researcher Science Grant, Hokkaido University Promotion Office of Research Environment for Diversity, and the Akiyama Life Science Foundation (to T.K.).
Author contributions
T.K. designed the experiments and generated most of the data. Y.Mi., S.I., K.H., K.K., N.T., Y.Mo., and R.U. performed and assisted experiments. M.T. and T.N. assisted light microscopic analyses. S.S. and T.I. assisted experiments concerning FBP17. Y.F. conceived and designed the study. The manuscript was written by T.K. and Y.F. with assistance from the other authors.
Declaration of interests
The authors declare no competing interests.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102994.
Contributor Information
Tomoko Kamasaki, Email: tkamasaki@igm.hokudai.ac.jp.
Yasuyuki Fujita, Email: fujita@monc.med.kyoto-u.ac.jp.
Supplemental information
Data and code availability
-
•
This paper does not report original code
-
•
All data associated with the paper is available upon request.
References
- Amoyel M., Bach E.A. Cell competition: how to eliminate your neighbours. Development. 2014;141:988–1000. doi: 10.1242/dev.079129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.E. Cell competition. Curr. Biol. 2011;21:R11–R15. doi: 10.1016/j.cub.2010.11.030. [DOI] [PubMed] [Google Scholar]
- Bowling S., Lawlor K., Rodriguez T.A. Cell competition: the winners and losers of fitness selection. Development. 2019;146:dev167486. doi: 10.1242/dev.167486. [DOI] [PubMed] [Google Scholar]
- Braga V. Epithelial cell shape: cadherins and small GTPases. Exp. Cell Res. 2000;261:83–90. doi: 10.1006/excr.2000.5050. [DOI] [PubMed] [Google Scholar]
- Chan Wah Hak L., Khan S., Di Meglio I., Law A.L., Lucken-Ardjomande Hasler S., Quintaneiro L.M., Ferreira A.P.A., Krause M., McMahon H.T., Boucrot E. FBP17 and CIP4 recruit SHIP2 and lamellipodin to prime the plasma membrane for fast endophilin-mediated endocytosis. Nat. Cell Biol. 2018;20:1023–1031. doi: 10.1038/s41556-018-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claveria C., Torres M. Cell competition: mechanisms and physiological roles. Annu. Rev. Cell Dev. Biol. 2016;32:411–439. doi: 10.1146/annurev-cellbio-111315-125142. [DOI] [PubMed] [Google Scholar]
- Everts V., Delaisse J.M., Korper W., Jansen D.C., Tigchelaar-Gutter W., Saftig P., Beertsen W. The bone lining cell: its role in cleaning Howship's lacunae and initiating bone formation. J. Bone Miner. Res. 2002;17:77–90. doi: 10.1359/jbmr.2002.17.1.77. [DOI] [PubMed] [Google Scholar]
- Frost A., Unger V.M., De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M., Nakagawa M., Kuroda S., Kaibuchi K. Effects of Rho family GTPases on cell-cell adhesion. Methods Mol. Biol. 2002;189:121–128. doi: 10.1385/1-59259-281-3:121. [DOI] [PubMed] [Google Scholar]
- Heuser J.E., Reese T.S. Evidence for recycling of synaptic vesicle membrane during transmitter release at the frog neuromuscular junction. J. Cell Biol. 1973;57:315–344. doi: 10.1083/jcb.57.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C., Dupre-Crochet S., Norman M., Kajita M., Zimmermann C., Pelling A.E., Piddini E., Baena-Lopez L.A., Vincent J.P., Itoh Y. Characterization of the interface between normal and transformed epithelial cells. Nat. Cell Biol. 2009;11:460–467. doi: 10.1038/ncb1853. [DOI] [PubMed] [Google Scholar]
- Hogan C., Serpente N., Cogram P., Hosking C.R., Bialucha C.U., Feller S.M., Braga V.M., Birchmeier W., Fujita Y. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol. Cell. Biol. 2004;24:6690–6700. doi: 10.1128/MCB.24.15.6690-6700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Erdmann K.S., Roux A., Habermann B., Werner H., De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Johnston L.A. Competitive interactions between cells: death, growth, and geography. Science. 2009;324:1679–1682. doi: 10.1126/science.1163862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M., Hogan C., Harris A.R., Dupre-Crochet S., Itasaki N., Kawakami K., Charras G., Tada M., Fujita Y. Interaction with surrounding normal epithelial cells influences signalling pathways and behaviour of Src-transformed cells. J. Cell Sci. 2010;123:171–180. doi: 10.1242/jcs.057976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M., Sugimura K., Ohoka A., Burden J., Suganuma H., Ikegawa M., Shimada T., Kitamura T., Shindoh M., Ishikawa S. Filamin acts as a key regulator in epithelial defence against transformed cells. Nat. Commun. 2014;5:4428. doi: 10.1038/ncomms5428. [DOI] [PubMed] [Google Scholar]
- Kon S., Ishibashi K., Katoh H., Kitamoto S., Shirai T., Tanaka S., Kajita M., Ishikawa S., Yamauchi H., Yako Y. Cell competition with normal epithelial cells promotes apical extrusion of transformed cells through metabolic changes. Nat. Cell Biol. 2017;19:530–541. doi: 10.1038/ncb3509. [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Levayer R. Solid stress, competition for space and cancer: the opposing roles of mechanical cell competition in tumour initiation and growth. Semin. Cancer Biol. 2020;63:69–80. doi: 10.1016/j.semcancer.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Fujita Y. Cell competition in mammals - novel homeostatic machinery for embryonic development and cancer prevention. Curr. Opin. Cell Biol. 2017;48:106–112. doi: 10.1016/j.ceb.2017.06.007. [DOI] [PubMed] [Google Scholar]
- Matsuo K., Irie N. Osteoclast-osteoblast communication. Arch. Biochem. Biophys. 2008;473:201–209. doi: 10.1016/j.abb.2008.03.027. [DOI] [PubMed] [Google Scholar]
- Mattila P.K., Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- Merino M.M., Levayer R., Moreno E. Survival of the fittest: essential roles of cell competition in development, aging, and cancer. Trends Cell Biol. 2016;26:776–788. doi: 10.1016/j.tcb.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Morata G., Calleja M. Cell competition and tumorigenesis in the imaginal discs of Drosophila. Semin. Cancer Biol. 2020;63:19–26. doi: 10.1016/j.semcancer.2019.06.010. [DOI] [PubMed] [Google Scholar]
- Morata G., Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev. Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- Niedergang F., Colucci-Guyon E., Dubois T., Raposo G., Chavrier P. ADP ribosylation factor 6 is activated and controls membrane delivery during phagocytosis in macrophages. J. Cell Biol. 2003;161:1143–1150. doi: 10.1083/jcb.200210069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa S., Vaughen J., Igaki T. Cell extrusion: a stress-responsive force for good or evil in epithelial homeostasis. Dev. Cell. 2018;44:532. doi: 10.1016/j.devcel.2018.02.007. [DOI] [PubMed] [Google Scholar]
- Padrick S.B., Cheng H.C., Ismail A.M., Panchal S.C., Doolittle L.K., Kim S., Skehan B.M., Umetani J., Brautigam C.A., Leong J.M. Hierarchical regulation of WASP/WAVE proteins. Mol. Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J., Zhao H., Pykalainen A., Laurinmaki P., Mattila P.K., Kinnunen P.K., Butcher S.J., Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Sasaki A., Nagatake T., Egami R., Gu G., Takigawa I., Ikeda W., Nakatani T., Kunisawa J., Fujita Y. Obesity suppresses cell-competition-mediated apical elimination of RasV12-transformed cells from epithelial tissues. Cell Rep. 2018;23:974–982. doi: 10.1016/j.celrep.2018.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A., Niwa H., Tsujita K., Suetsugu S., Nitta K., Hanawa-Suetsugu K., Akasaka R., Nishino Y., Toyama M., Chen L. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Suetsugu S., Kurisu S., Takenawa T. Dynamic shaping of cellular membranes by phospholipids and membrane-deforming proteins. Physiol. Rev. 2014;94:1219–1248. doi: 10.1152/physrev.00040.2013. [DOI] [PubMed] [Google Scholar]
- Svitkina T.M., Bulanova E.A., Chaga O.Y., Vignjevic D.M., Kojima S., Vasiliev J.M., Borisy G.G. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano K., Toyooka K., Suetsugu S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 2008;27:2817–2828. doi: 10.1038/emboj.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T., Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Tsujita K., Takenawa T., Itoh T. Feedback regulation between plasma membrane tension and membrane-bending proteins organizes cell polarity during leading edge formation. Nat. Cell Biol. 2015;17:749–758. doi: 10.1038/ncb3162. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Yin M., Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- Vincent J.P., Fletcher A.G., Baena-Lopez L.A. Mechanisms and mechanics of cell competition in epithelia. Nat. Rev. Mol. Cell Biol. 2013;14:581–591. doi: 10.1038/nrm3639. [DOI] [PubMed] [Google Scholar]
- Wagstaff L., Kolahgar G., Piddini E. Competitive cell interactions in cancer: a cellular tug of war. Trends Cell Biol. 2013;23:160–167. doi: 10.1016/j.tcb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Watson J.R., Fox H.M., Nietlispach D., Gallop J.L., Owen D., Mott H.R. Investigation of the interaction between Cdc42 and its effector TOCA1: HANDOVER OF Cdc42 TO THE ACTIN REGULATOR N-WASP IS FACILITATED BY DIFFERENTIAL BINDING AFFINITIES. J. Biol. Chem. 2016;291:13875–13890. doi: 10.1074/jbc.M116.724294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M.D., Mullins R.D. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 2002;18:247–288. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
- Wu S.K., Gomez G.A., Michael M., Verma S., Cox H.L., Lefevre J.G., Parton R.G., Hamilton N.A., Neufeld Z., Yap A.S. Cortical F-actin stabilization generates apical-lateral patterns of junctional contractility that integrate cells into epithelia. Nat. Cell Biol. 2014;16:167–178. doi: 10.1038/ncb2900. [DOI] [PubMed] [Google Scholar]
- Zagury D., Bernard J., Thierness N., Feldman M., Berke G. Isolation and characterization of individual functionally reactive cytotoxic T lymphocytes: conjugation, killing and recycling at the single cell level. Eur. J. Immunol. 1975;5:818–822. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper does not report original code
-
•
All data associated with the paper is available upon request.