Abstract
Rpb4 and Rpb7 are two yeast RNA polymerase II (Pol II) subunits whose mechanistic roles have recently started to be deciphered. Although previous data suggest that Rpb7 can stably interact with Pol II only as a heterodimer with Rpb4, RPB7 is essential for viability, whereas RPB4 is essential only during some stress conditions. To resolve this discrepancy and to gain a better understanding of the mode of action of Rpb4, we took advantage of the inability of cells lacking RPB4 (rpb4Δ, containing Pol IIΔ4) to grow above 30°C and screened for genes whose overexpression could suppress this defect. We thus discovered that overexpression of RPB7 could suppress the inability of rpb4Δ cells to grow at 34°C (a relatively mild temperature stress) but not at higher temperatures. Overexpression of RPB7 could also partially suppress the cold sensitivity of rpb4Δ strains and fully suppress their inability to survive a long starvation period (stationary phase). Notably, however, overexpression of RPB4 could not override the requirement for RPB7. Consistent with the growth phenotype, overexpression of RPB7 could suppress the transcriptional defect characteristic of rpb4Δ cells during the mild, but not during a more severe, heat shock. We also demonstrated, through two reciprocal coimmunoprecipitation experiments, a stable interaction of the overproduced Rpb7 with Pol IIΔ4. Nevertheless, fewer Rpb7 molecules interacted with Pol IIΔ4 than with wild-type Pol II. Thus, a major role of Rpb4 is to augment the interaction of Rpb7 with Pol II. We suggest that Pol IIΔ4 contains a small amount of Rpb7 that is sufficient to support transcription only under nonstress conditions. When RPB7 is overexpressed, more Rpb7 assembles with Pol IIΔ4, enough to permit appropriate transcription also under some stress conditions.
Transcriptional response to ever-changing environmental conditions is commonly found in nature. RNA polymerase II core enzyme (Pol II), which is composed of 12 subunits, is known to play an active role in regulating transcription (28). Rpb4, the fourth largest subunit, exhibits some unique features that distinguish it from the other subunits. As for Rpb7 (23a), but unlike other subunits, the stoichiometry of Rpb4 is dependent on growth conditions. In optimally growing cells, the fraction of Pol II molecules containing Rpb4 is about 20% (7, 17), and it gradually increases after the shift to starvation-induced postlogarithmic phases. Thus, in stationary phase, virtually all Pol II molecules contain Rpb4 (7). RPB4 is not essential for cell viability (27). Under optimal growth conditions, in liquid rich medium at moderate temperatures (18 to 22°C), cells lacking RPB4 (designated herein rpb4Δ cells) grow similarly to their wild-type counterparts and show almost normal transcriptional activity (7). However, as they experience higher or lower temperatures, rpb4Δ cells rapidly lose their capacity for efficient growth and global transcription (7). In addition to its requirement during temperature extremes, Rpb4 is also required for efficient transcription and for maintaining viability during starvation in the stationary phase (7). Using a promoter-independent transcription reaction assay, we recently demonstrated that Rpb4, and probably also Rpb7, is required for Pol II enzymatic activity at temperature extremes but not at moderate temperatures (21a). This observation is in accord with a direct and stress-specific role that Rpb4 plays in the overall transcriptional activity of Pol II.
The pattern of RPB4 expression differs from the pattern of expression of the other Pol II subunit genes. Whereas mRNA and protein levels of other subunits are reduced after the shift from log to postlog phases, RPB4 mRNA and protein levels remain constitutively high (5, 6). Furthermore, during starvation, but not during optimal growth conditions, Rpb4 protein level is regulated posttranscriptionally. Thus, under optimal growth conditions, when Rpb4 is dispensable, the Rpb4 protein level is directly proportional to the RPB4 mRNA level. However, during starvation, when Rpb4 is essential for maintaining viability, Rpb4 protein level is little affected by artificial changes in its mRNA level (6). Taken together, the unusual phenotype of rpb4Δ cells and the pattern of RPB4 expression indicate that Rpb4 plays a vital role specifically during certain stress conditions.
Rpb4 is known to interact with an essential Pol II subunit, Rpb7. Together, they readily dissociate from Pol II in vitro as a heterodimer (9), and their physical interaction in vivo was demonstrated by a two-hybrid assay (16). Furthermore, Rpb7 was not detected in Pol II which was immunoprecipitated (17) or chemically purified (9) from cells lacking RPB4; moreover, Pol II, purified to homogeneity from the rpb4Δ strain, contained no detectable Rpb7 and could form high-quality crystals (8). All these results suggested that Rpb7 is associated with Pol II only as a heterodimer with Rpb4. Yet RPB7 is an essential gene (20), whereas RPB4 is not (27). One possible explanation for this discrepancy was to hypothesize an additional function for Rpb7, one unrelated to its association with Pol II (20). This hypothetical function is the essential one. It was not clear, therefore, whether the association of Rpb7 with Pol II was essential for viability.
Previous attempts to crystallize Pol II, purified from logarithmically grown cells, were unsuccessful due to the substoichiometric amounts of Rpb4 and Rpb7 which resulted in heterogeneity that interfered with the crystallization. The demonstration that Pol II purified from stationary-phase cells contains the full complement of Rpb4 and Rpb7 (7) enabled the two-dimensional crystallization of the wild-type Pol II (2, 14). Comparison of the crystal structure of the wild-type Pol II with that of Pol II lacking both Rpb4 and Rpb7 (pol IIΔ4/7) revealed that the Rpb4-Rpb7 heterodimer is located at the floor of the DNA binding clef. Association of the heterodimer imposes a slight movement of the protein domain surrounding the clef. Jensen et al. (14) suggested that this structural change is associated with a closure of the Pol II clef after entry of the DNA into the active center. They also proposed that the Rpb4-Rpb7 heterodimer stabilizes the paused Pol II, which had been demonstrated previously, in nonstressed Drosophila melanogaster, to be located downstream of heat shock (HS) promoters (21). This proposed function might be essential for the stress response, explaining the requirement for Rpb4 during stress (14).
Here we show that Rpb7 can functionally interact with Pol II complex independently of Rpb4. This interaction is detectable only when RPB7 is overexpressed. Overproduction of Rpb7 not only resulted in its detectable association with Pol IIΔ4 but also partially suppressed the various stress phenotypes of rpb4Δ cells, including their inability to transcribe non-HS genes under mild HS (a shift from 22 to 34°C). Surprisingly, during the mild HS of cells lacking RPB4, when the transcription of non-HS genes was strongly reduced, the transcription of HS genes was largely unaffected.
MATERIALS AND METHODS
Yeast strains and growth conditions.
Yeast strains are described in Table 1. Cells were grown either in synthetic and selective media containing the full complement of the amino acids uracil and adenine (SC) but lacking the component to select for the presence of a plasmid (24) or they were grown in YPD (2% Bacto Peptone, 1% yeast extract [Difco Laboratories], 2% dextrose) or YPG (2% Bacto Peptone, 1% yeast extract [Difco Laboratories], 2% galactose) medium at the indicated temperature.
TABLE 1.
List of strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| SUB62 | MATa lys2-801 leu2-3,2-112 ura3-52 his3-Δ200 trp1-1(am) | D. Botstein |
| MC11-1 | MATa lys2-801 leu2-3,2-112 ura3-52 his3-Δ200 trp1-1(am)rpb4Δ1::HIS3 | This laboratory (7) |
| MC12-1 | MATα ura3-52 lys2-801(am)ade2-101(am)trp1-Δ1 his3Δ200 leu2-Δ1 rpb4Δ1::HIS3 | This laboratory |
| WY-73 | MATa ura3-52 his3Δ200 leu2-3,112 lys2Δ201 ade2 rpb7Δ1::LEU2 (pRP729 [RPB7 URA3 CEN]) | N. Woychik (20) |
| AS1 | MATa lys2-801 leu2-3,2-112 ura3-52 his3-Δ200 trp1-1(am)rpb4Δ1::HIS3 (pGAL1p-RPB7 URA3) | This work |
| AS24 | MATa ura3-52 his3Δ200 leu2-3,112 lys2Δ201 ade2 rpb7Δ1::LEU2 (pRP729 [RPB7 URA3 CEN]) (pMC120 [RPB4 2μm HIS3]) | This work |
| AS25 | MATa ura3-52 his3Δ200 leu2-3,112 lys2Δ201 ade2 rpb7Δ1::LEU2 (pRP729 [RPB7 URA3 CEN]) (pMC121 [tetp-RPB7 2μm HIS3]) | This work |
| AS30 | MATa lys2-801 leu2-3,2-112 ura3-52 his3-Δ200 trp1-1(am) (pMC117 [tetp-RPB7-myc2 2μm URA3]) | This work |
| AS33 | MATa lys2-801 leu2-3,2-112 ura3-52 his3-Δ200 trp1-1(am)rpb4Δ1::HIS3 (pMC116 [tetp-RPB7 2μm URA3]) | This work |
| AS35 | MATa lys2-801 leu2-3,2-112 ura3-52 his3-Δ200 trp1-1(am)rpb4Δ1::HIS3 (pMC117 [tetp-RPB7-myc2 2μm URA3]) | This work |
Yeast transformation.
Transformation of the yeast cDNA library (19) was carried out by electroporation according to a previously published protocol (3). After the transformation, plates were incubated overnight at room temperature and then incubated at 34°C for several days. Other transformation experiments were done by the lithium acetate procedure (22).
Plasmid constructions. (i) RPB7-myc2 plasmid (pMC117).
A RPB7-myc2 fragment was synthesized by PCR with the following oligonucleotides. The forword primer is OMC67 (5′-CGG GATCCCTCTAACATTGGGTCGATCAGG-3′; the BamHI region is underlined). It is a forward oligonucleotide containing the sequence located 170 bp upstream of the RPB7 open reading frame (ORF) (located at the 5′ end of the untranslated region of RPB7 mRNA). The reverse primer is OMC70 (5′-ATGAATTCGCGGCCGCTTAGAGATCTTCCTCACT GATAAGCTTTTGCTCCGGGAGATCTTCCTCACTGATAAGCTTTTGCT CCGGAGCGCGTGCCGCAATAGCACCCAAATAATCTTC-3′; the NotI region is underlined). This oligonucleotide contains two myc epitope repeats separated by a proline codon, a Pro(Ala)4 linker, and a sequence capable of hybridizing to the end of the ORF. The epitope tags were thus introduced in frame and downstream to the last codon of RPB7 ORF by the reverse oligonucleotide. A TAA stop codon, encoded by the reverse oligonucleotide, was introduced just downstream of the second tag. The fragment, bordered by BamHI and NotI sites, was introduced into the BamHI and NotI site of pCM190. This 2μm-derived plasmid contains a strong hybrid tetO-CYC1 promoter (designated herein tetp) and tetR-VP16 fusion gene encoding the tetracycline-repressible activator (11). In the absence of tetracycline, this promoter provides an overexpression level which is comparable to that observed with the GAL1-driven promoters (11). The RPB7-myc2 fragment was placed downstream of the hybrid promoter. Thus, expression of the tagged RPB7 is repressible by tetracycline.
(ii) RPB7 plasmid (pMC116).
Control plasmid pMC116 is identical to pMC117 except that the epitope tag is missing. This plasmid was constructed by using OMC67 as the forward oligonucleotide and OMC68, instead of OMC70, as the reverse oligonucleotide. The OMC68 sequence is 5′-ATAAGAATGCGGCCGCTTAAATAGCACCCAAATAATCTTC-3′ (the NotI region is underlined).
(iii) pMC120 (RPB4 2μm HIS3).
pMC120 is an HIS3 derivative of the previously described pMC42μ plasmid (6).
(iv) pMC121 (tetp-RPB7 HIS3).
pMC121 is an HIS3 derivative of the pMC116 plasmid described above.
Antibodies and immunological procedures.
Anti-Rpb7 antibodies and the affinity-purified anti-Rpb4 and anti-Rpb2 antibodies were a generous gift of A. Sentenac (13). Western analysis was done as described previously (7). For the immunoprecipitations (IP) two antibodies were used to immunoprecipitate Pol II. A total of 5 to 10 μg of anti-Rpb1 C-terminal-domain monoclonal antibody (8WG16 MAb) (a generous gift of Nancy Thompson and Richard Burgess [25]) was used to immunoprecipitate Pol II from 200 μg of whole-cell extract as described previously (7). To immunoprecipitate the epitope-tagged Rpb7 from 500 μg of whole-cell protein extract, 10 μg of 9E10 antibodies were used as described previously (7). The immunoprecipitates were electrophoresed through a 5 to 15% gradient sodium dodecyl sulfate-polyacrylamide gel; this was followed by electrotransfer of the proteins to a nitrocellulose filter, and the relevant proteins were then detected by Western analysis (7).
RNA extraction and analyses.
RNA extraction, dot blot analysis, and Northern blot hybridization analyses were done as described previously (7).
RESULTS
Sensitivity of rpb4Δ cells to nonpermissive temperatures and to starvation is suppressed by overexpression of RPB7.
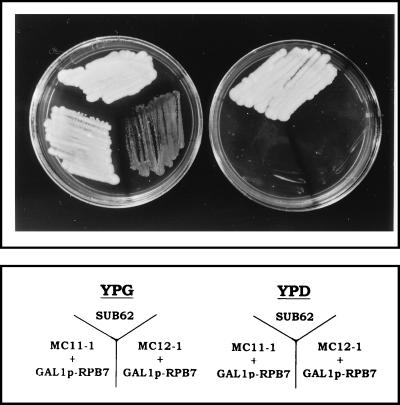
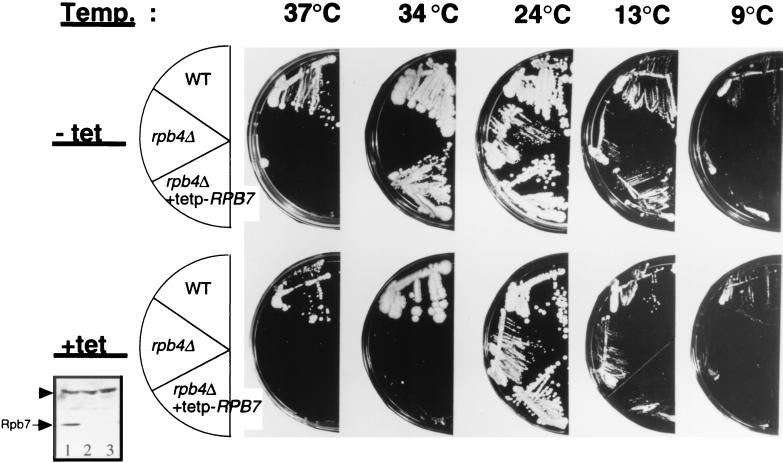
Cells lacking RPB4 can grow similarly to wild-type cells under optimal conditions at moderate temperatures. However, unlike wild-type cells, they do not grow at temperatures above 30°C or below 12°C. In order to study the role of Rpb4 during temperature stress, we selected for high-copy-number suppressors of the temperature-sensitive phenotype. To this end, we introduced into rpb4Δ cells a yeast cDNA library expressed under the strong GAL1 promoter, which is induced by galactose and repressed by dextrose (19). Transformants were tested for their ability to grow at 34°C on galactose-containing media but not on dextrose-containing media. One of the suppressors, RPB7, was selected several times during our screens and was further studied. Figure 1 shows that GAL1p-RPB7 plasmid, recovered from one of the original transformants and reintroduced into two different rpb4Δ cells, rescued the growth defect at 34°C independently of the genetic background. Growth could be rescued only under inducing conditions (when galactose was the major carbon source) and not under repressing conditions (when dextrose was the major carbon source). To ascertain the high-copy-number suppression by other means, RPB7 was placed in the pCM190 plasmid downstream of a strong tetracycline-repressible promoter (designated tetp). Expression from the promoter is comparable to that from the GAL1 promoter. Yet, this expression is not dependent on the carbon source but is instead repressed when tetracycline is present in the medium (11). The resulting tetp-RPB7 plasmid is called pMC116. Figure 2 (34°C) shows that rpb4Δ cells carrying pMC116 could grow at 34°C. The presence of tetracycline abolished the growth. However, the high-copy-number suppression was not complete, since rpb4Δ cells carrying pMC116 could not grow at 37°C (Fig. 2, 37°C). The inset in Fig. 2 shows that the steady-state level of Rpb7 was much higher in cells carrying pMC116 (lane 1) than in the wild-type cells (lane 3) and that overexpression was prevented when tetracycline was present in the medium (lane 2). We also noticed that the growth temperature had no significant effect on the steady-state level of the overexpressed Rpb7 (results not shown).
FIG. 1.
Overexpression of RPB7 suppresses the growth defect of rpb4Δ cells at 34°C independently of the genetic background. A high-copy-number suppression approach was taken, with a yeast cDNA library under GAL1 promoter in a URA3 plasmid, to select for genes which could rescue rpb4Δ growth defect at 34°C (see Results). RPB7 was thus selected. pGAL1-RPB7 from one of the positive transformants was recovered, propagated in Escherichia coli, and introduced into MC11-1 or MC12-1 (see genotypes in Table 1) by the lithium acetate transformation method (see Materials and Methods). URA3 colonies were selected at 25°C. Transformants were then allowed to grow at 34°C on either YPG (inducing condition, left plate) or YPD (repressing condition, right plate).
FIG. 2.
Overexpression of RPB7 suppresses the growth defect of rpb4Δ cells at both high and low temperatures. SUB62 (WT), MC11-1 (rpb4Δ), and AS33 (rpb4Δ strain carrying pMC116 [tetp-RPB7 URA3 plasmid]) were streaked onto YPD plates lacking tetracycline (designated “−tet” at the left of the figure), or containing 20 μg of tetracycline per ml (designated “+tet” at the left). Plates were incubated at the various temperatures indicated at the top. Pictures were obtained either after 3 days of incubation (at 24, 34, and 37°C) or after 7 days (at 9 and 13°C). After the 9°C picture was taken, the wild-type colonies continued to grow, whereas those of the other strains did not. The inset (lower left) shows a Western analysis (see Materials and Methods) of 50-μg extracts probed with anti-Rpb7 antibodies. Lanes: 1, extract from AS33 which had been grown on YPD lacking tetracycline; 2, extract from AS33 which had been grown on YPD containing 20 μg of tetracycline per ml; 3, extract from SUB62 cells which had been grown on YPD lacking tetracycline. The band seen above Rpb7 and marked by an arrowhead is nonspecific. It is shown to demonstrate equal loading.
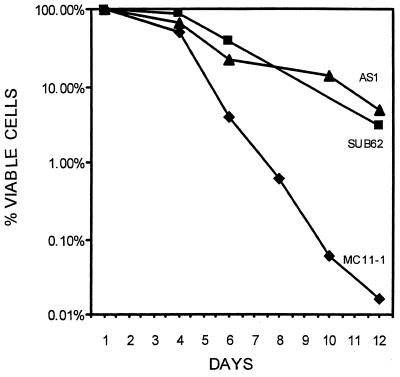
Next, we tested whether overexpression of RPB7 can rescue other defects characteristic of rpb4Δ cells. Cells lacking RPB4 are unable to grow at ≤13°C (7, 26). We have found that rpb4Δ cells overexpressing RPB7 through the GAL1 promoter (AS1 cells) could grow better than the parental rpb4Δ cells (MC11-1 cells) on galactose-containing plates, but not on glucose-containing plates, at 13°C (results not shown). Likewise, rpb4Δ cells carrying pMC116 (tetp-RPB7 plasmid) could grow better than MC11-1 at 13°C on a plate lacking tetracycline (Fig. 2, 13°C). Thus, overexpression of RPB7 can suppress growth defects not only at high temperatures but also at low temperatures. However, at a lower temperature (9°C), overexpression of RPB7 could not suppress the growth defect of rpb4Δ cells (Fig. 2, 9°C). Another characteristic of rpb4Δ cells is their inability to maintain viability in the stationary phase (7). To determine whether overexpression of RPB7 can rescue this defect, wild-type cells (SUB62), rpb4Δ cells (MC11-1), and rpb4Δ cells overexpressing RPB7 (AS1) were allowed to grow until stationary phase. Aliquots were taken at various time points after entry into stationary phase to determine the proportion of viable cells. The results, summarized in Fig. 3, demonstrate that whereas MC11-1 cells died during the stationary phase more rapidly than SUB62 cells, AS1 cells survived at a rate comparable to that of SUB62. Thus, overexpression of RPB7 rescued starved rpb4Δ cells.
FIG. 3.
Overexpression of RPB7 prevents the accelerated death of rpb4Δ cells during starvation in stationary phase. AS1 (rpb4Δ strain overexpressing RPB7) cells were grown at 28°C on SC medium lacking uracyl and containing 2% galactose. SUB62 (wild-type) and MC11-1 (rpb4Δ) cells were grown in the same medium supplemented with uracyl. The viability was determined, at various time points after entry into stationary phase, by evaluating the plating efficiency on YPG at 25°C. Symbols: ■, SUB62; ⧫, MC11-1; ▴, AS1. The graph represents an average of three different experiments. The deviation was less than 25%.
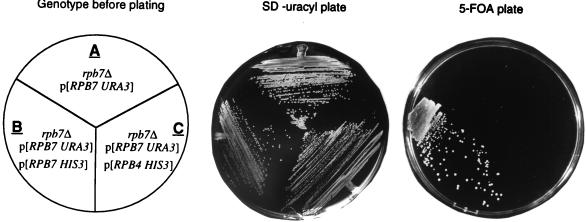
Rpb4 and Rpb7 are known to interact with Pol II as a heterodimer. Yet, RPB4 is essential only during some stress conditions, whereas RPB7 is essential under all of the growth conditions tested thus far. Our finding, i.e., that overexpression of RPB7 can override the requirement for RPB4 during various stresses, raised the inverse possibility that overexpression of RPB4 can override the requirement for RPB7. To test this possibility, we overexpressed RPB4 by using two overexpression approaches. The first uses a high-copy-number, 2μm-derived RPB4 plasmid. The level of Rpb4 expressed from this plasmid is 24-fold higher than that in the wild type (6). The RPB4 plasmid was introduced into a strain lacking the chromosomal copy of RPB7 but containing an RPB7 gene on a URA3 plasmid (WY-73 strain). As expected, WY-73 could not grow on a plate containing 5-fluoro-orotic acid (5-FOA) (4) because it could not lose the URA3 plasmid containing the only copy of the essential RPB7 (Fig. 4A, 5-FOA plate), unless the plasmid was replaced by a non-URA3 plasmid carrying RPB7 (Fig. 4B, 5-FOA plate). Figure 4C (5-FOA plate) shows that RPB7 was still essential in cells overexpressing RPB4, since this strain could not lose the RPB7 plasmid and could not grow on a 5-FOA plate. The other overexpression strategy was to use the GAL1 promoter instead of using a high-copy-number plasmid. Western analysis showed that this system yielded even higher levels of Rpb4 than that obtained by the high-copy-number system (results not shown). Results of this second overexpression experiment (not shown) led to the same conclusion. Therefore, although Rpb4 and Rpb7 function as a heterodimer, and although Rpb7 can function independently of Rpb4, Rpb4 does not seem to be able to replace Rpb7, at least as determined by using the high-copy-number and the GAL1 approaches.
FIG. 4.
Overexpressed RPB4 cannot replace RPB7. WY-73 (A), AS25 (B), and AS24 (C), whose genotypes are depicted in the left panel, were grown on appropriate selective media until mid-log phase. Similar numbers of cells were then streaked on either a selective plate lacking uracyl or on an SC plate containing 50 μg of 5-FOA as indicated at the top of the figure. Plates were incubated at 30°C for 3 days (SD-uracil plate) or for 5 days (5-FOA plate). Other experiments, similar to the one described here except that the plates were incubated at various temperatures, ranging from 25 to 34°C, gave similar results.
Rpb7 can bind to Pol II independently of Rpb4.
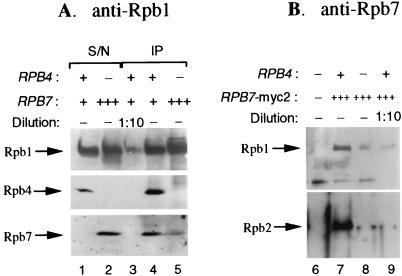
The interaction of Rpb7 with the other Pol II subunits was assumed to be dependent on Rpb4 for the following reasons. (i) Both Rpb4 and Rpb7 were removed from the Pol II complex by treatment with 2 M urea, and these subunits were fractionated as a single peak in high-pressure liquid chromatography fractionation, probably as a heterodimer (9). (ii) Pol II, which was either immunoprecipitated (reference 17 and our unpublished observations) or purified (9) from extract of rpb4Δ cells, contained no detectable amount of Rpb7. (iii) Pol II purified to homogeneity from rpb4Δ strain contained no detectable Rpb7 and could form high-quality crystals (8). Our observation that overexpression of Rpb7 could suppress phenotypes characteristic of rpb4Δ cells suggested that Rpb7 can associate with Pol II independently of Rpb4. To examine this possibility, we immunoprecipitated Pol II extracted from rpb4Δ cells overexpressing RPB7 (AS1 or AS35). Two IP approaches were taken. In one, we used antibodies against the C-terminal domain of Rpb1 to immunoprecipitate Pol IIΔ4 and to examine whether Rpb7 could be coimmunoprecipitated with it. These antibodies had been used previously to immunoprecipitate or to purify Pol II containing stoichiometric amounts of Rpb4 and Rpb7 (7, 9). The reciprocal experiment was to immunoprecipitate the Rpb7 and to determine whether Rpb1 and Rpb2 could be coimmunoprecipitated with it. Figure 5A shows the results of the experiment with the anti-Rpb1. Whereas Pol II immunoprecipitated from an extract of rpb4Δ cells carrying the chromosomal copy of RPB7 contained no detectable Rpb7 (data not shown; see also references 9 and 17), Pol II immunoprecipitated from an extract of AS1 cells contained a detectable level of Rpb7 (Fig. 5A, lane 5). As expected, the level of Rpb7 left in the AS1 supernatant of the IP was much higher than that left in the wild-type IP supernatant (Fig. 5A, compare lanes 1 and 2). The level of Pol II-associated Rpb7 in the extract of AS1 was lower than that found in extract of the wild-type cells (carrying one chromosomal copy each of RPB4 and RPB7) (Fig. 5A, compare lanes 4 and 5). To estimate the amount of Rpb7 in the immunoprecipitated Pol IIΔ4, we compared it to the amount present in a 10-fold dilution of the Rpb4-containing Pol II (Fig. 5A, lane 3). This comparison revealed that there was more Rpb7 in the Pol IIΔ4 than in a 1:10 dilution of that of the Rpb4-containing Pol II (Fig. 5A, compare lanes 3 and 5). Thus, a significant portion of Pol II molecules (>10% of that present in the wild-type Pol II) could stably interact with Rpb7 in the absence of Rpb4. To ascertain the Rpb4-independent interaction of Rpb7 with Pol II, RPB7 was tagged with the c-myc epitope at its C-terminal end (see Materials and Methods). The epitope-tagged RPB7 (designated RPB7-myc2) was cloned in the high-copy-number pCM190 plasmid downstream of the strong tetracycline-repressible promoter (see description of the plasmid above). The plasmid carrying RPB7-myc2 is termed pMC117. As a control, we used a similar plasmid, pMC116, carrying the wild-type untagged RPB7. These plasmids were introduced into rpb4Δ cells, and the resulting transformants are named AS33 (carrying pMC116) and AS35 (carrying pMC117). Both transformants, which overproduce the plasmid-borne Rpb7, could grow at 34°C on plates or in liquid media lacking tetracycline but not on tetracycline-containing media (see Fig. 2; results not shown). These results indicate that the epitope-tagged Rpb7 can function like the wild-type Rpb7 in its ability to suppress the temperature-sensitive phenotype of rpb4Δ cells. Furthermore, a HIS3 derivative of pMC117 could replace the URA3-RPB7 plasmid in WY-73, as determined by their abilities to grow on 5-FOA plates (results not shown). Thus, the Rpb7-myc2 can replace the wild-type protein. Figure 5B demonstrates that Rpb1 and Rpb2 were immunoprecipitated with the anti-myc antibodies both from extracts of RPB4+ (lane 7) or rpb4Δ (lane 8) cells. The control experiment shows that these Pol II subunits could not be immunoprecipitated with the anti-myc antibodies if the overproduced Rpb7 was not epitope tagged (lane 6). Note, however, that the IP efficiency was strongly affected by Rpb4. This is consistent with the results shown in Fig. 5A demonstrating that, in the absence of Rpb4, the interaction of the overproduced Rpb7 was less efficient than the interaction of the naturally produced Rpb7 with wild-type Pol II. Nevertheless, the interaction of Rpb7 with Pol IIΔ4 was relatively efficient, as there was more Rpb1 and Rpb2 in the Pol IIΔ4 than in a 1:10 dilution of that of the Rpb4-containing Pol II (Fig. 5B, compare lanes 8 and 9). Consistent with the results of the other IP experiment (Fig. 5A), the results in Fig. 5B demonstrate that >10% of Pol IIΔ4 molecules interact stably with Rpb7. Taken together, the results of these IP experiments reinforce the model in which Rpb4 enhances the interaction of Rpb7 with Pol II. However, they demonstrate that the association of Rpb7 with Pol II is not strictly dependent on Rpb4 and, upon RPB7 overexpression, a significant portion of Pol IIΔ4 can stably interact with Rpb7.
FIG. 5.
Coimmunoprecipitation of the overproduced Rpb7 with Pol IIΔ4. (A) IP of Pol II with anti-Rpb1. Equal amounts of protein (200 μg), extracted from logarithmically growing SUB62 (wild-type) or AS1 (rpb4Δ GAL1p-RPB7) cells, were used for IP with the anti-C-terminal-domain antibody of Rpb1 (8WG16 MAb). These antibodies were chosen because they had already been demonstrated to be suitable for IP of the full complement of Pol II, including stoichiometric amounts of Rpb4 and Rpb7 (7, 9). IP followed by Western analysis was carried out as described in Materials and Methods. Subunits detected by specific antibodies (see Materials and Methods) are marked by arrows. Inclusion of nonspecific antibodies, instead of 8WG16 MAb, produced no detectable signals of Rpb1, Rpb4, or Rpb7 (results not shown). Lanes 1 (SUB62) and 2 (AS1) show the supernatant (S/N) left after the IP (due to technical limitations only 20 of 200 μg was loaded onto the gel). Lane 4 shows the IP of SUB62; lane 5 shows the IP of AS1. Lane 3 shows a 1:10 dilution of the material used in lane 4 in order to estimate the ratio between the Rpb7 band intensity in lane 5 to that in lane 4. (B) IP of Pol II with anti-myc antibodies that recognize the Rpb7-myc2. Equal amounts of protein (500 μg) extracted from logarithmically growing AS33 (lane 6), AS30 (lanes 7 and 9), and AS35 (lane 8) were taken for IP with 10 μg of the 9E10 MAb as described for panel A. Subunits, detected by the respective specific antibodies (see Materials and Methods), are marked by arrows. Lane 9 shows a 1:10 dilution of the material used in lane 7. This is shown to estimate the ratio between the Rpb1 and Rpb2 band intensities in lane 8 to those in lane 7. The band underneath Rpb1 is nonspecific. The absence of a gene or a plasmid (−), the presence of one copy of a gene (+), or the overexpression of RPB7 or RPB7-myc2 (+++) are indicated.
Overexpression of RPB7 partially recovers the transcriptional defect of rpb4Δ cells.
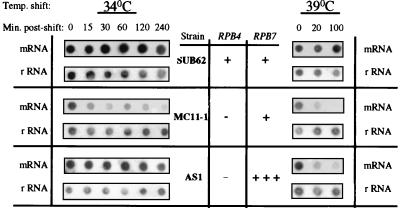
We have demonstrated previously that RPB4 is required for Pol II transcription upon HS (7). The relatively efficient interaction of the overproduced Rpb7 with Pol IIΔ4 and the RPB7 high-copy-number suppression of rpb4Δ phenotypes suggested that overproduction of Rpb7 might improve the transcriptional capacity of Pol IIΔ4 during HS. The global transcriptional effect of RPB7 overexpression on Pol II transcription during HS was monitored by hybridizing 32P-labeled poly(dT) to equal amounts of RNA dot blotted onto a nitrocellulose filter (5). Since the majority of mRNAs are of non-HS genes, this assay monitors the transcription of the non-HS genes. It is important to note that during HS the kinetics of the decrease in the global mRNA levels, as detected by this assay, paralleled the kinetics of Pol IIΔ4 inactivation (7). Two HS temperatures were used. First, a shift from 23 to 34°C was used. At 34°C both the wild-type strain and the strain lacking RPB4 and overexpressing RPB7 (AS1) can grow efficiently. Strains lacking RPB4 and carrying the only single chromosomal copy of RPB7 cannot grow at this temperature. Second, a shift from 23 to 39°C was used. At this temperature, only the wild type and neither of the other two strains can grow (see Fig. 2). Results of the dot blots are shown in Fig. 6. Consistent with results obtained previously (see reference 7 and references therein), the exposure of wild-type cells to HS had little or no effect on the global mRNA level (Fig. 6, SUB62 strain). In contrast, the global mRNA level in the rpb4Δ mutant decreased gradually after the shift from 22 to 34°C and more dramatically after the shift to 39°C (Fig. 6, MC11-1 strain), a finding consistent with previous results (7). The gradual decline in mRNA levels of MC11-1 cells demonstrates the transcriptional defect characteristic of RNA Pol II lacking Rpb4. Overexpression of RPB7 partially suppressed this defect. Thus, after the shift from 22 to 34°C, the mRNA level in AS1 showed a behavior similar, although not identical, to that found in the wild type (Fig. 6). After the shift from 22 to 39°C, the mRNA level in AS1 was higher than that detected in MC11-1. However, the effect of Rpb7 overproduction under this severe HS was small (Fig. 6) and apparently was not sufficient to support cell growth (see Fig. 2, 37°C).
FIG. 6.
Steady-state level of the global poly(A)+ mRNA during mild and severe HS: effect of RPB4 deletion and overexpression of RPB7. SUB62 (wild-type), MC11-1 (rpb4Δ), and AS1 (rpb4Δ GAL1p-RPB7) strains were grown at 22°C on galactose-containing medium as described in Materials and Methods. In mid-log phase (at 107 cells/ml), cells were shifted to either 34 or 39°C and harvested at the indicated time points after the shift. RNA was extracted, and 1 μg of the RNA samples was dot blotted onto nitrocellulose filter in duplicate. One set of dots was hybridized with 32P-labeled poly(dT) to detect the poly(A)-containing mRNA (designated mRNA at the left or right of the figure) and the other set was hybridized with 32P-labeled rDNA to detect the rRNA (designated rRNA at the left or right of the figure). This procedure was described in detail previously (5). The absence of a gene (−), the presence of one copy of a gene (+), and the overexpression of a gene (+++) are indicated.
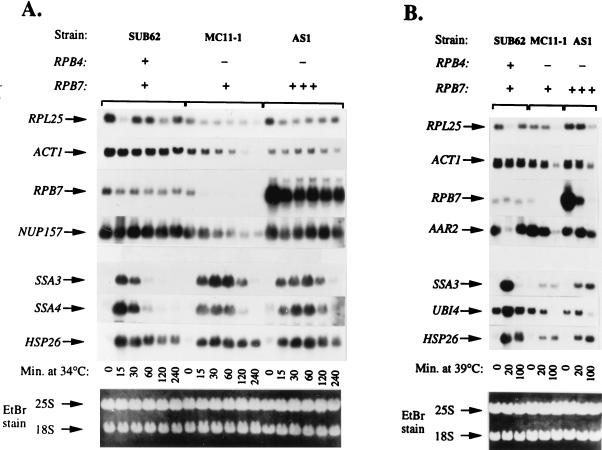
To gain additional insights into the transcriptional defects that occur in rpb4Δ cells after exposure to HS and into how the overexpression of RPB7 can affect this, we investigated the effect of HS on the accumulation of specific mRNA species by using Northern blot hybridization analysis. As in the experiments shown in Fig. 6, two HS conditions were tested: a shift from 23 to 34°C (Fig. 7A) and a shift from 23 to 39°C (Fig. 7B). The levels of the non-HS mRNAs ACT1, RPB7, RPL25, NUP157, and AAR2 and of the HS mRNAs SSA3, SSA4, HSP26, and UBI4 were monitored in wild type (SUB62), in rpb4Δ mutant (MC11-1), and in rpb4Δ1 mutant overexpressing RPB7 (AS1). In the wild-type strain shifted from 23 to 34°C, the mRNA levels of most non-HS genes tested were transiently decreased at 15 to 20 min post-temperature shiftup, followed by a recovery to the pre-HS level (Fig. 7A and B, SUB62 strain). In contrast, the levels of these mRNAs in strain MC11-1 decreased gradually after the temperature shiftup (Fig. 7A, MC11-1). Interestingly, the level of RPB7 mRNA decreased very rapidly after the temperature shiftup of the MC11-1 strain. This result suggests that the RPB7 mRNA half-life is very short. Thus, the level of RPB7 mRNA seems to be a good probe for monitoring changes in Pol II activity. The results obtained with SUB62 and MC11-1 strains are consistent with previous observations. The gradual decline in mRNA levels of MC11-1 cells demonstrates the transcriptional defect characteristic of RNA Pol II lacking Rpb4 (7). Monitoring the mRNA levels of the non-HS genes after the shift of AS1 strain from 23 to 34°C shows kinetics similar to this in the wild type. A transient decline in the mRNA levels shortly after the temperature shiftup was followed by a recovery to almost pre-HS levels at later stages (Fig. 7A, AS1 strain). In contrast to their transcription competence during mild HS (34°C), the ability of AS1 cells to transcribe non-HS genes after the shift from 23 to 39°C was impaired. Although the levels of these mRNA in the AS1 strain were higher than those found in MC11-1, they could not be recovered to the nearly pre-HS levels (Fig. 7B, AS1 strain). These results are consistent with those shown in Fig. 6 and with the growth defects characteristic of AS1 at 39°C. Cumulatively, these results demonstrate the inability of the overproduced Rpb7 to provide a full recovery of Pol II activity in severe HS conditions.
FIG. 7.
Levels of specific mRNAs during mild or severe HS: effect of RPB4 deletion and overexpression of RPB7. Strains, cell growth, HS at 34°C (A) or at 39°C (B), and RNA extraction procedures were as described for Fig. 6. RNA samples (8 μg) were analyzed by Northern blot hybridization as described previously (5, 7). The application and transfer of equal amounts of RNA was verified by ethidium bromide staining (shown at the bottom). The same filter was used for sequential hybridization with the indicated DNA probes. The absence of a gene (−), the presence of one copy of a gene (+), and the overexpression of a gene (+++) are indicated.
The transient and dramatic transcriptional induction of HS genes in response to HS is a well-documented phenomenon (see, for example, reference 7 and references therein). We have demonstrated previously that cells lacking RPB4 are defective in the transcription of HS genes upon exposure to 39°C (7). Figure 7B shows similar results. Whereas levels of SSA3, UBI4, and HSP26 mRNAs in the wild-type cells transiently and dramatically increased after the shift from 23 to 39°C, the increase in these mRNAs was severely impaired in MC11-1 cells. Overexpression of RPB7 partially enhanced the transcription of these genes (Fig. 7B, compare lanes for AS1 with those for MC11-1). Monitoring the transcriptional induction of the HS genes SSA3, SSA4, and HSP26 in wild-type cells after the shift from 23 to 34°C revealed an induction similar to that observed during the severe HS. The mRNA levels increased dramatically at 15 min post-temperature shiftup and gradually decreased at later time points (see Fig. 7A, lanes for SUB62). Unexpectedly, cells lacking RPB4 could also transcribe HS genes during this mild HS. Figure 7A (lanes for MC11-1) shows that the levels of SSA3, SSA4, and HSP26 mRNAs in MC11-1 cells increased substantially after the shift to 34°C. However, the mutant cells differed from their wild-type counterparts in the kinetics of their transcription. Whereas in the wild-type cells the levels of these mRNAs peaked at 15 min post-temperature shiftup, in the rpb4Δ strain they exhibited a broader peak, which centered at ca. 30 min after the temperature shiftup. Surprisingly, overexpression of RPB7 in rpb4Δ strains had only little effect or no effect at all on both the kinetics and the extent of transcriptional induction of HS genes during the mild HS (Fig. 7A, compare lanes for AS1 with those for MC11-1).
In summary, during the mild HS, the main defect of Pol IIΔ4 is demonstrated by the inefficient transcription of non-HS genes. This defect is efficiently suppressed by the overexpression of RPB7. During more severe HS, Pol IIΔ4 is incapable of transcribing both HS and non-HS genes, and a high level of Rpb7 can only little suppress this defect. A strong correlation exists between the ability of AS1 cells to efficiently transcribe genes and their ability to grow.
DISCUSSION
In an attempt to understand the essential role of Rpb4 during some stresses, a high-copy-number suppression approach was taken. In this way we found that overproduction of Rpb7 enabled rpb4Δ cells to grow at otherwise lethal high (34°C) or low (13°C) temperatures. Moreover, the rapid death of rpb4Δ cells in stationary phase was prevented by the overexpression of RPB7. Thus, it seems that high levels of Rpb7 can recuperate several stress phenotypes of rpb4Δ cells. However, the suppression was not complete: AS1 and AS33 (rpb4Δ cells which overexpress RPB7 by two different expression systems [see Results]) could not grow at temperatures above 34°C or below 13°C, whereas their isogenic RPB4+ strain can grow even above 39°C or below 9°C (results not shown).
Previous attempts to demonstrate an association of Rpb7 with Pol IIΔ4, when Rpb7 was expressed from the single chromosomal copy, were unsuccessful (9, 17). This result raised the question of whether Rpb7 interacts with Pol II in the absence of Rpb4 (20). Our results demonstrate that Rpb7 can interact with Pol IIΔ4. Detection of this interaction was made possible by overexpressing RPB7. This interaction is stable enough to withstand the long process of the IP procedure, and the duration of the washing procedure of the IP process had no significant effect on the results (results not shown). It seems that once Rpb7 assembles with Pol IIΔ4, the complex is relatively stable and can endure, at least to some degree, in vitro manipulations. The interaction of Rpb7 with Pol IIΔ4 indicates that, in addition to its interaction with Rpb4, Rpb7 can interact with other Pol II subunit(s). Interaction of the human homolog of Rpb7, hsRpb7, with three Pol II subunits (hsRpb1, hsRpb3, and hsRpb5) has been previously observed. This interaction can occur in the absence of hsRpb4 (1). In view of the many functional and structural similarities between the human and yeast Pol II molecules (28), it is likely that similar interactions also occur in the yeast Pol II. Interestingly, the S. pombe homolog of Rpb7, spRpb7, interacts with Pol II independently of spRpb4 (23). We suggest that, in S. cerevisiae, the interaction of Rpb7 with Pol II subunits is mediated by Rpb4. In the absence of the Rpb4, only a small fraction of Rpb7 stably associates with Pol II complex. This Pol II fraction in cells expressing RPB7 from the single chromosomal copy could not be detected by the techniques used thus far.
Overexpression of RPB7 suppresses the transcriptional defects characteristic of rpb4Δ cells at 34°C. Under this condition, a high level of Rpb7 recuperates the overall transcriptional activity in rpb4Δ cells, as indicated by analyzing the poly(A)+ mRNA levels and by analyzing the levels of specific mRNAs (Fig. 6 and 7). This global effect of Rpb7 overproduction on transcription at 34°C is consistent with a direct effect of Rpb7 on Pol IIΔ4 activity. Moreover, not only does the high-copy-number suppression of the stress-sensitive phenotype correlate with an increased assembly of Rpb7 with Pol IIΔ4 but it also correlates with the increased transcriptional capacity of this polymerase at 34°C. Taken together, the global effect and these correlations strongly suggest that assembly of Rpb7p with Pol IIΔ4 improves its transcriptional capacity.
In light of the previous inability to detect an association between Rpb7 and Pol IIΔ4, it was not clear whether the essential role of Rpb7 was related to its interaction with Pol II (see introduction). Our observations that Rpb7 can interact with Pol IIΔ4 and that the extent of the interaction correlates with the extent of the transcriptional capabilities of Pol IIΔ4 lend support to the notion that the essential function of Rpb7p is related to its interaction with Pol II. Alternatively, it is possible that Rpb7 carries some other essential role that is not related to its association with Pol II. In order to explain the transcriptional recovery of rpb4Δ strain at 34°C in accord with the latter possibility, we need to hypothesize that overproduction of Rpb7 indirectly leads to transcriptional recovery of Pol IIΔ4. According to this possibility the increased assembly of Rpb7 with Pol IIΔ4 has no direct effect on Pol IIΔ4 transcriptional activity. We regard this possibility as unlikely because of the strong correlation observed between the binding of Rpb7 to Pol IIΔ4 and the transcription activity. However, it is quite possible that Rpb7 has other roles in addition to its essential role in transcription.
We have noticed unexpected differences between the involvement of Rpb4 and Rpb7 in the transcription of non-HS genes and their involvement in the transcription of HS genes. Our results clearly demonstrate that, during HS, efficient transcription of non-HS genes is dependent upon RPB4. In the absence of RPB4 the transcription of non-HS genes was inefficient during the mild HS (a shift from 22 to 34°C) and even more so during the more severe HS (from 22 to 39°C). Overexpression of RPB7 could suppress this defect in both cases. Nevertheless, during the mild HS the suppression was efficient, whereas during the severe HS it was less efficient. The involvement of Rpb4 and Rpb7 in the transcription of HS genes seems to be more complex. During the severe HS, cells lacking RPB4 could hardly transcribe the HS genes SSA3, UBI4, and HSP26. This result is in agreement with previously published results (7). However, during the mild HS the transcription of SSA3, UBI4, and HSP26 by Pol IIΔ4 was surprisingly efficient. Interestingly, however, the kinetics of the transcriptional induction of these HS genes was slower in rpb4Δ cells compared to the fast induction observed in the wild type. Equally surprising was the observation that RPB7 overexpression had little or no effect either on the transcription efficiency or on the slow kinetics. The reason for the slow induction kinetics of HS genes and why this is not affected by the overexpression of RPB7 remains to be elucidated. Thus, whereas the current data strongly support the model that Rpb4 and Rpb7 are involved in the transcription of non-HS genes, we still do not know whether there is a single unifying theme that governs the function of Rpb4 and Rpb7 in the transcription of non-HS and HS genes. It is possible that the inability of cells lacking RPB4 to transcribe HS genes during severe HS is an effect secondary to the inefficient expression of a specific non-HS gene(s). The finding that the overexpression of RPB7 could only inefficiently suppress the transcription defect of Pol IIΔ4 during the severe HS provides a good explanation for the growth defect of AS1 and AS33 at 39°C.
Interestingly, although deletion of RPB4 has a severe and global effect on transcription under some stress conditions (e.g., cold stress, heat stress, and starvation), the absence of RPB4 is tolerable under some other stress conditions (e.g., osmotic stress and growth on glycerol). Are there other factors that are responsible for sustaining Pol II activity under these other stresses? Rox3, an essential component of Pol II mediator complex (12), might have been considered as one candidate because its truncation renders cells sensitive to osmotic stress and incapable of growing on glycerol as the main carbon source (10). However, unlike deletion of RPB4, truncation of ROX3 has only a selective effect on transcription. Thus, whereas, during osmotic stress, the transcription of CYC7 is impaired in rox3 mutants, that of ACT1 and RTS1 is unaffected by this mutation (10). It therefore seems that the stress phenotype of rox3 mutants is due to a defect in the transcription of a specific group of genes rather than to a global transcriptional defect. Therefore, Rox3 does not seem to be an analog or a substitute of Rpb4 and Rpb7 during some stress conditions.
Recently, a model which suggests that a major defect of Pol IIΔ4/7 during stress is its inability to support the transcription induction of HS genes was proposed. According to this model, the interaction of Rpb4 and Rpb7 with Pol II enhances cell resistance to stresses by facilitating the accumulation of the paused Pol II at promoter proximal locations of HS genes (14). We show here that, in response to the mild HS, cells lacking RPB4 can efficiently transcribe at least three HS genes. Our results suggest that under some stress conditions the major defect of Pol IIΔ4 is the failure to appropriately transcribe non-HS genes rather than HS genes.
Whereas overexpression of RPB7 can suppress the transcriptional defects of Pol IIΔ4 during mild HS, overexpression of RPB4 cannot replace RPB7. Thus, the two subunits are not interchangeable. This study focused our attention on Rpb7 as a key element in the Rpb4-Rpb7 heterodimer, which can interact with the rest of the Pol II subunits and function in transcription during nonstress conditions and during moderate temperature stresses independently of Rpb4. Rpb4 is likely to function as a mediator that facilitates the recruitment of Rpb7. Whereas Rpb7 is essential for viability during all growth conditions, cells can survive well in the absence of Rpb4. As discussed above, in the absence of Rpb4, the number of Rpb7-containing Pol II is smaller than in the presence of Rpb4. This inefficient interaction of Rpb7 with Pol II is tolerable under optimal growth conditions, so that rpb4Δ cells can grow similarly to their wild-type counterparts in rich medium at 18 to 22°C (7). However, during various stress conditions an efficient interaction is crucial for viability. Is Rpb4 required only to recruit Rpb7 or does it have other functions? The answer to this question awaits future experiments. The growing number of organisms that have been demonstrated to carry two distinct RPB4 and RPB7 homologs (15, 18), such as S. cerevisiae, suggests that there was a selective advantage in maintaining the two functions separately. Interestingly, Pol II purified from S. pombe does not contain a detectable level of an Rpb4 homolog (23). It is possible that, unlike the Rpb7 in S. cerevisiae, which interacts inefficiently with Pol IIΔ4 (Fig. 5), the S. pombe Rpb7 can interact efficiently with Pol II without a requirement for an Rpb4 homolog (at least under the standard laboratory growth conditions). S. pombe may serve as an extreme example for a separation of the two functions. Because overexpression of RPB7 in S. cerevisiae lacking RPB4 could not rescue phenotypes associated with severe temperature stresses, it is possible that during severe stresses Rpb4 is required for functions other than the recruitment of Rpb7.
ACKNOWLEDGMENTS
We thank J. Lis for advice, A. Sentenac and N. Thompson for antibodies, and N. Woychik for yeast strains and the RPB7 disruption plasmid.
This work was supported by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities to M.C.
REFERENCES
- 1.Acker J, de Graaff M, Cheynel I, Khazak V, Kedinger C, Vigneron M. Interactions between the human RNA polymerase II subunits. J Biol Chem. 1997;272:16815–16821. doi: 10.1074/jbc.272.27.16815. [DOI] [PubMed] [Google Scholar]
- 2.Asturias F J, Meredith G D, Poglitsch C L, Kornberg R D. Two conformations of RNA polymerase II revealed by electron crystallography. J Mol Biol. 1997;272:536–540. doi: 10.1006/jmbi.1997.1273. [DOI] [PubMed] [Google Scholar]
- 3.Becker D M, Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 5.Choder M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991;5:2315–2326. doi: 10.1101/gad.5.12a.2315. [DOI] [PubMed] [Google Scholar]
- 6.Choder M. A growth rate-limiting process in the last growth phase of the yeast life cycle involves RPB4, a subunit of RNA polymerase II. J Bacteriol. 1993;175:6358–6363. doi: 10.1128/jb.175.19.6358-6363.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choder M, Young R A. A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Mol Cell Biol. 1993;13:6984–6991. doi: 10.1128/mcb.13.11.6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darst S A, Kubalek E W, Edwards A M, Kornberg R D. Two-dimensional and epitaxial crystallization of a mutant form of yeast RNA polymerase II. J Mol Biol. 1991;221:347–357. doi: 10.1016/0022-2836(91)80223-h. [DOI] [PubMed] [Google Scholar]
- 9.Edwards A M, Kane C M, Young R A, Kornberg R D. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem. 1991;266:71–75. [PubMed] [Google Scholar]
- 10.Evangelista C C, Jr, Rodriguez-Torres A M, Limbach M P, Zitomer R S. Rox3 and Rts1 function in the global stress response pathway in baker’s yeast. Genetics. 1996;142:1083–1093. doi: 10.1093/genetics/142.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 12.Gustafsson C M, Myers L C, Li Y, Redd M J, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. Identification of Rox3 as a component of mediator and RNA polymerase II holoenzyme. J Biol Chem. 1997;272:48–50. doi: 10.1074/jbc.272.1.48. [DOI] [PubMed] [Google Scholar]
- 13.Huet J, Sentenac A, Fromageot P. Spot-immunodetection of conserved determinants in eukaryotic RNA polymerases. J Biol Chem. 1982;257:2613–2618. [PubMed] [Google Scholar]
- 14.Jensen G J, Meredith G, Bushnell D A, Kornberg R D. Structure of wild-type yeast RNA polymerase II and location of Rpb4 and Rpb7. EMBO J. 1998;17:2353–2358. doi: 10.1093/emboj/17.8.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khazak V, Estojak J, Cho H, Majors J, Sonoda G, Testa J R, Golemis E A. Analysis of the interaction of the novel RNA polymerase II (pol II) subunit hsRPB4 with its partner hsRPB7 and with pol II. Mol Cell Biol. 1998;18:1935–1945. doi: 10.1128/mcb.18.4.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khazak V, Sadhale P P, Woychik N A, Brent R, Golemis E A. Human RNA polymerase II subunit hsRPB7 functions in yeast and influences stress survival and cell morphology. Mol Biol Cell. 1995;6:759–775. doi: 10.1091/mbc.6.7.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolodziej P A, Woychik N, Liao S M, Young R A. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol Cell Biol. 1990;10:1915–1920. doi: 10.1128/mcb.10.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin R M, Guilfoyle T J. Two small subunits in arabidopsis RNA polymerase II are related to yeast RPB4 and RPB7 and interact with one another. J Biol Chem. 1998;273:5631–5637. doi: 10.1074/jbc.273.10.5631. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKune K, Richards K L, Edwards A M, Young R A, Woychik N A. RPB7, one of two dissociable subunits of yeast RNA polymerase II, is essential for cell viability. Yeast. 1993;9:295–299. doi: 10.1002/yea.320090309. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen E B, Lis J T. Short transcripts of the ternary complex provide insight into RNA polymerase II elongational pausing. J Mol Biol. 1995;252:522–535. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]
- 21a.Rosenheck S, Choder M. Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. J Bacteriol. 1998;180:6187–6192. doi: 10.1128/jb.180.23.6187-6192.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 23.Sakurai H, Ishihama A. Gene organization and protein sequence of the small subunits of Schizosaccharomyces pombe RNA polymerase II. Gene. 1997;196:165–174. doi: 10.1016/s0378-1119(97)00222-9. [DOI] [PubMed] [Google Scholar]
- 23a.Sheffer, A., and M. Choder. Unpublished observations.
- 24.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 25.Thompson N E, Steinberg T H, Aronson D B, Burgess R R. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- 26.Woychik N A, Lane W S, Young R A. Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. J Biol Chem. 1991;266:19053–19055. [PubMed] [Google Scholar]
- 27.Woychik N A, Young R A. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol Cell Biol. 1989;9:2854–2859. doi: 10.1128/mcb.9.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young R A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]