Abstract
Mitochondrial biogenesis and energy metabolism are essential for regulating the inflammatory state of monocytes. This state is partially controlled by peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), a coactivator that regulates mitochondrial biogenesis and energy metabolism. Disruption of these processes can also contribute to the initiation of chronic inflammatory diseases, such as pulmonary fibrosis, atherosclerosis, and rheumatoid arthritis. Methyltransferase-like 3 (METTL3)-dependent N6-methyladenosine (m6A) methylation has recently been shown to regulate a variety of inflammatory processes. However, the role of m6A mRNA methylation in affecting mitochondrial metabolism in monocytes under inflammation is unclear, nor is there an established relationship between m6A methylation and PGC-1α. In this study, we identified a novel mechanism by which METTL3 acts during oxidized low-density lipoprotein (oxLDL)-induced monocyte inflammation, where METTL3 and YTH N6-methyladenosine RNA binding protein 2 (YTHDF2) cooperatively modify PGC-1α mRNA, mediating its degradation, decreasing PGC-1α protein levels, and thereby enhancing the inflammatory response. METTL3 coordinated with YTHDF2 to suppress the expression of PGC-1α, as well as that of cytochrome c (CYCS) and NADH:ubiquinone oxidoreductase subunit C2 (NDUFC2) and reduced ATP production and oxygen consumption rate (OCR). This subsequently increased the accumulation of cellular and mitochondrial reactive oxygen species (ROS) and the levels of proinflammatory cytokines in inflammatory monocytes. These data may provide new insights into the role of METTL3-dependent m6A modification of PGC-1α mRNA in the monocyte inflammation response. These data also contribute to a more comprehensive understanding of the pathogenesis of monocyte-macrophage inflammation-associated diseases, such as pulmonary fibrosis, atherosclerosis, and rheumatoid arthritis.
Keywords: METTL3, PGC-1α, m6A modification, RNA methylation, mitochondria, monocytes, inflammation, post-transcriptional regulation
Abbreviations: CYCS, cytochrome c; ICAM1, intercellular cell adhesion molecule 1; LPS, lipopolysaccharide; m6A, N6-methyladenosine; METTL3, methyltransferase like 3; NDUFA6, NADH: ubiquinone oxidoreductase subunit A6; NDUFA12, NADH: ubiquinone oxidoreductase subunit A12; NDUFAF1, NADH: ubiquinone oxidoreductase complex assembly factor 1; NDUFB6, NADH: ubiquinone oxidoreductase subunit B6; NDUFC2, NADH: ubiquinone oxidoreductase subunit C2; NDUFV3, NADH: ubiquinone oxidoreductase subunit V3; oxLDL, oxidized low-density lipoprotein; PGC-1α, PPARγ coactivator 1 alpha; PGC-1β, PPARγ coactivator 1 beta; PPARγ, peroxisome proliferator-activated receptor gamma; ROS, reactive oxygen species; TNF-α, tumor necrosis factor-α; UTR, untranslated region; YTHDF1/2/3, YTH N6-methyladenosine RNA binding protein1/2/3
Monocytes play an important role in the innate immune system and exhibit phagocytic activity to resist viral, bacterial, and fungal infections (1, 2). Circulating monocytes leave the bloodstream and migrate to inflamed tissues, where they can differentiate into macrophages or dendritic cells following stimulation by interaction with cytokines and/or microbial molecules (e.g., complement proteins, bacterial flagellin, lipopolysaccharide) (2). Recruitment of monocytes is essential for effective control and clearance of pathogen infections, but persistent monocyte infiltration also contributes to the pathogenesis of chronic inflammatory and degenerative diseases, such as glomerulonephritis, pulmonary fibrosis, atherosclerosis, rheumatoid arthritis, and Alzheimer's disease (3, 4, 5, 6, 7). OxLDL is immunogenic and activates endothelial cells, monocytes/macrophages and T cells (8, 9, 10), but is toxic and can cause cell death at higher concentrations (8, 9, 10). Mitochondrial function and bioenergetics have gained recent attention for their contributions to chronic inflammatory diseases, as these processes—beyond their conventional role in generating energy—have been demonstrated to play critical roles in immunity (11, 12). For example, mitochondrial constituents (e.g., mtDNA, mtROS, ATP, cardiolipin, N-formyl peptides) can activate innate immune receptors (e.g., FPR1, NLRP3, NLRC4 and TLR9) to promote inflammatory responses. In addition, a change in mitochondrial metabolism can induce monocyte differentiation into different types of macrophages: M1 (proinflammatory) and M2 (anti-inflammatory) macrophages. M1 macrophages rely on glycolysis for energy production and, as such, have a lower ratio of oxidative phosphorylation to glycolysis. In contrast, M2 macrophages preferentially utilize oxidative phosphorylation and have a higher ratio of oxidative phosphorylation to glycolysis. PGC-1α (also known as PPARGC1A), a member of the peroxisome proliferator-activated receptor γ (PPARγ) coactivator family (13), transcriptionally regulates ~70% of respiratory chain complexes, as well as ATP synthases, tricarboxylic acid cycle intermediary enzymes, and fatty acid oxidation (14). Dysregulation of PGC-1α may induce aging, metabolic and degenerative diseases, as well as cancer in humans by suppressing mitochondrial biogenesis (15, 16). Interestingly, some studies have shown that PGC-1α may regulate inflammatory responses, such as its role in reprogramming energy metabolism of monocyte-macrophages during sepsis, beginning in the early stages of acute inflammation to later stages (17).
m6A is one of the most prevalent posttranscriptional modifications in eukaryotic mRNA transcripts and impacts a variety of physiological events (18). The level of m6A methylation is dynamically mediated through the activity of methyltransferases (METTL3 and METTL14) and demethylases (FTO and ALKBH5) (19, 20). The m6A modification functions by recruiting YT521-B homology (YTH) domain family proteins (e.g., YTHDF1/2/3, YTHDC1/2) and IGF2BP family (IGF2BP1/2/3) proteins, as well as other factors (e.g., eIF3) (20, 21). METTL3 is the core catalytic subunit of the m6A methyltransferase complex, which plays a major catalytic role in m6A modification. These m6A-specific binding proteins either directly bind to m6A or through indirect mechanisms to mediate RNA function or metabolism, including mRNA export, mRNA stability, mRNA splicing, mRNA translation, and mRNA decay (20, 21). Previous studies have demonstrated that m6A modifications affect a diverse range of physiological and pathological processes, including spermatogenesis, neurogenesis, tumorigenesis, and immunoregulation (21, 22, 23, 24, 25). More recently, m6A has been shown to play important roles in innate immunity. For example, METTL3 deficiency sustains long-chain fatty acid absorption by blocking the TRAF6-dependent inflammation response in lipopolysaccharide (LPS)-stimulated intestinal porcine epithelial cells (26). Another study has implicated METTL3-mediated alternative splicing of MyD88 in the regulation of LPS-induced inflammatory response in human dental pulp cells. A shorter MyD88 mRNA (MyD88S), lacking the second exon, exerts a dominant-negative effect on LPS-induced, TLR4-mediated signaling pathways and inhibits inflammatory cytokine production (27). Moreover, METTL3-mediated mRNA m6A methylation promotes dendritic cell activation during LPS stimulation (28). These and other observations have suggested that the expression and functional role of METTL3 are complex and depend upon cell context, tissue type, individual developmental stage, and inflammation-stimulating factors. Although the role of METTL3 in inflammatory response in a variety of cells has been reported, the role of METTL3-mediated mRNA m6A modification in mitochondrial function and metabolism during monocyte inflammation is unclear, especially, the functional linking between METTL3 and PGC-1α is not yet known.
In the present study, the association between METTL3 and mitochondrial metabolism in the context of monocyte inflammation was investigated. METTL3 associated with PGC-1α mRNA, thereby suppressing translation and protein expression. Moreover, knockdown of METTL3 or YTHDF2 increased both PGC-1α mRNA lifetime and expression, which reduced mitochondrial dysfunction and oxLDL-induced monocyte inflammation. These results suggest that the physical and functional interaction between METTL3 and PGC-1α is involved in monocyte inflammatory response. These data may contribute to a comprehensive understanding of the pathogenesis of monocyte-macrophage inflammation-associated diseases.
Result
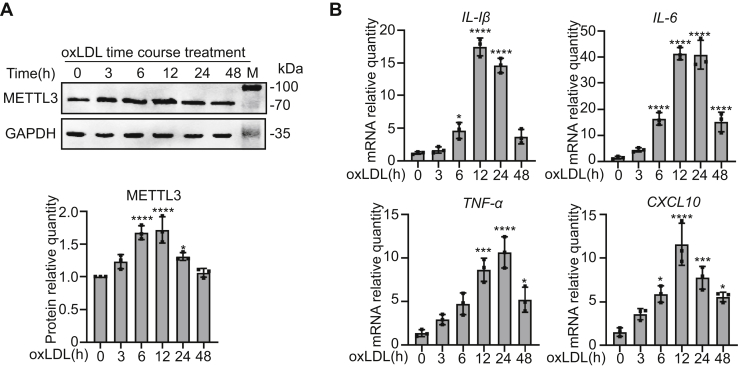
METTL3 is upregulated in oxLDL-induced THP1 monocyte inflammation
To investigate the role of METTL3 in monocyte inflammation, THP-1 cells were treated with 20 μg/ml oxLDL and METTL3 was detected by immunoblot. Significantly higher levels of METTL3 were observed after oxLDL treatment for both 6 and 12 h (Fig. 1A). To confirm that oxLDL promotes inflammation, we measured mRNA levels of proinflammatory genes—including IL-1β, IL-6, TNF-α, and CXCL10—and found all four transcripts were significantly increased and reached a peak after being exposed to oxLDL for 12 and 24 h (Fig. 1B). METTL3 levels increased prior to oxLDL-induced inflammatory markers, suggesting a potential regulatory role in inflammation.
Figure 1.
METTL3 is upregulated in oxLDL-induced THP1 monocyte inflammation.A and B, THP-1 cells were treated with 20 μg/ml oxLDL for 0, 3, 6, 12, 24, and 48 h. A, immunoblot detection of METTL3 protein levels of untreated and oxLDL-treated cells, with GAPDH as a loading control (n = 3). B, qRT-PCR determination of mRNA levels of IL-1β, IL-6, TNF-α, and CXCL10 in response to oxLDL (n = 3). Data are represented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 in all figures.
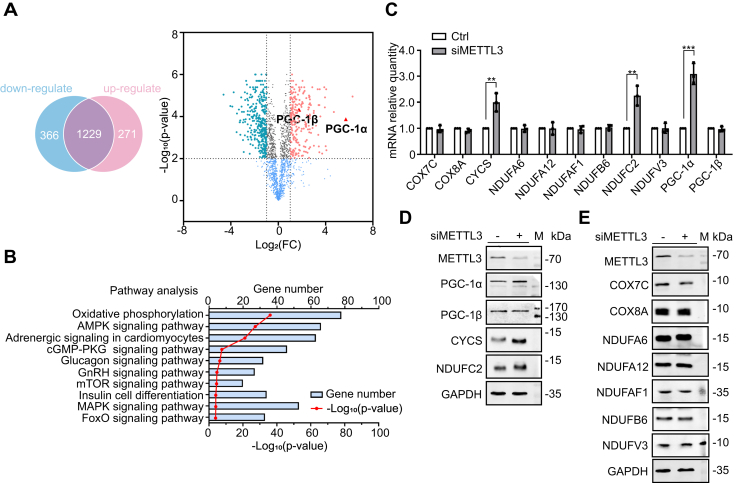
METTL3 inhibits the expression of energy metabolism genes
To probe downstream target genes of METTL3, the RNA-seq data set GES98623 (gene expression database GEO) was used to identify differentially expressed genes (DEGs) between control and silenced METTL3 MOLM13 monocytes. From this analysis, 637 DEGs were identified, with 366 downregulated and 271 upregulated genes. Interestingly, the expression of both PGC-1α and PGC-1β was upregulated (Fig. 2A). The CLIP-seq database POSTAR was used to identify 2121 genes that could be directly recognized and bound by METTL3, followed by pathway enrichment analysis using the online analysis tool DAVID that indicated correlation with the oxidative phosphorylation signaling pathway (Fig. 2B). Comparing the genes pulled from the POSTAR database to those correlated in pathway enrichment analysis and incorporating the DEGs from METTL3-silenced MOLM13 cells, we identified 11 candidate genes for interaction with METTL3 that may be regulated by METTL3 (Table S1).
Figure 2.
METTL3 inhibits the expression of energy metabolism genes.A, Venn diagram of upregulated and downregulated DEGs of siMETTL3 cells versus control cells. B, POSTAR online database pathway enrichment analysis. C, qRT-PCR determination of mRNA levels of various proinflammatory genes in METTL3 knockdown cells (n = 3). D and E, immunoblot detection of various pro-inflammatory protein levels in control cells and METTL3 knockdown cells (n = 3). Data are represented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗p < 0.0001 in all figures.
To explore the role of the 11 candidate genes, METTL3 expression was silenced in THP-1 monocytes by METTL3 siRNA. After silencing METTL3 (Fig. S1A), the mRNA and protein levels of PGC-1α, CYCS, and NDUFC2 were upregulated, whereas COX7C, COX8A, NDUFA6, NDUFA12, NDUFAF1, NDUFB6, NDUFV3, and PGC-1β were unchanged (Fig. 1, C–E). These data suggest that METTL3 may suppress the expression of PGC-1α and nuclear-encoded mitochondrial respiratory chain complex proteins CYCS and NDUFC2, thereby regulating energy metabolism.
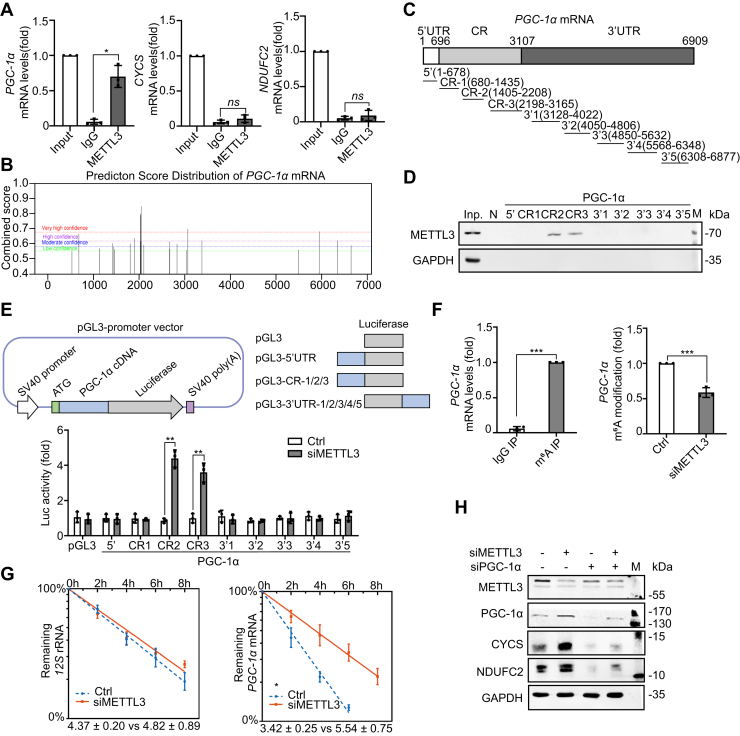
METTL3 binds to the coding region of PGC-1α mRNA and decreases its stability
METTL3-dependent m6A modification can influence mRNA stability, translation, and translocation (20, 21). As METTL3 ablation increased both mRNA and protein levels of PGC-1α, CYCS, and NDUFC2, we investigated the role of METTL3 in regulating PGC-1α, CYCS, and NDUFC2 expression at the posttranscriptional level. An anti-METTL3 antibody and primers specific to PGC-1α, CYCS, and NDUFC2 were used in an RNA immunoprecipitation-qRT-PCR assay, revealing that PGC-1α mRNA, but not CYCS and NDUFC2 mRNA, coprecipitated with METTL3 (Fig. 3A). Putative METTL3-binding sites on PGC-1α mRNA were identified using a sequence-based N6-methyladenosine (m6A) modification site predictor (29). The predicted sites were mainly enriched in the coding sequence and 3′UTR of PGC-1α mRNA (Fig. 3B). These sequences were divided into nine segments—the 5′UTR, three segments of the coding region (CR), and five segments of the 3′UTR (Fig. 3C). RNA pull-down assays were performed using in vitro-transcribed, biotinylated fragments of these segments, revealing that METTL3 associated with the CR-2 and CR-3 fragments of PGC-1α mRNA (Fig. 3D). To test the functional association of METTL3 with the PGC-1α mRNA segments, pGL3-derived reporters bearing fragments of PGC-1α mRNAs were constructed to conduct an siRNA-repressible luciferase assay (Fig. 3E). 293T cells were transfected with the pGL3-PGC-1α constructs and a luciferase reporter construct. Luciferase activity increased after silencing METTL3 in pGL3-derived reporters bearing PGC-1α CR-2 and CR-3 segments, but not with reporters containing segments that did not interact with METTL3 (Fig. 3E). This suggested that METTL3 expression was negatively associated with PGC-1α expression and functioned through direct physical interaction in the coding region of PGC-1α mRNA. The role of METTL3-mediated m6A modification in the regulation of PGC-1α mRNA was investigated using an m6A-methylated RNA immunoprecipitation assay in order to analyze changes in m6A modification in PGC-1α mRNA. In an METTL3 knockdown model, there was a ~41% decrease in m6A methylation for PGC-1α mRNA compared with the control group (Fig. 3F), indicating that METTL3 knockdown reduced levels of PGC-1α mRNA m6A modification.
Figure 3.
METTL3 binds to the coding region of PGC-1α mRNA and decreases its stability. A, RNA immunoprecipitation of various mRNAs with an anti-METTL3 antibody (n = 3). B, prediction sites of PGC-1α mRNA binding to METTL3 as produced from SRAMP. C, schematic of PGC-1α mRNA segments. D, RNA pull-down assays using THP-1 cell lysates and in vitro-transcribed RNA segments. Antisense sequence of METTL3 served as the negative control [N], as well as a 5-μg aliquot input [Inp.] and GAPDH (n = 2). E, relative luciferase activities with in vitro-transcribed RNA segments in control and METTL3 knockdown cells (n = 3). F, PGC-1α mRNA collected from a m6A RNA immunoprecipitation assay with control and METTL3 knockdown THP-1 cells using an IgG antibody or an anti-m6A antibody (n = 3). G, degradation of PGC-1α mRNA and 12S rRNA in control and METTL3 knockdown cells, which was used to determine half-life (n = 3). H, immunoblot detection of various proteins in control, METTL3 knockdown, PGC-1α knockdown, and METTL3/PGC-1α knockdown cells, with GAPDH as a loading control (n = 3). Data are represented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
As m6A methylation regulates mRNA stability (21, 30), the enhanced PGC-1α mRNA expression in METTL3-silenced cells may have been due to increased stability of the unmethylated mRNA transcript. Silencing METTL3 in THP-1 cells prolonged the half-life of PGC-1α mRNA from 3.4 h to 5.5 h, indicating that expression of METTL3 may reduce the stability of PGC-1α mRNA (Fig. 3G), which is consistent with upregulated PGC-1α mRNA and protein levels in METTL3-silenced cells. Although PGC-1α mRNA stability was linked to interaction with METTL3, the mechanism by which CYCS and NDUFC2 were upregulated after silencing METTL3 is unknown. PGC-1α siRNA was used to silence PGC-1α expression in THP-1 cells (Fig. 3H), which decreased CYCS and NDUFC2 protein levels, indicating that the expression of CYCS and NDUFC2 was dependent on the presence of PGC-1α and is indirectly affected by the presence of METTL3.
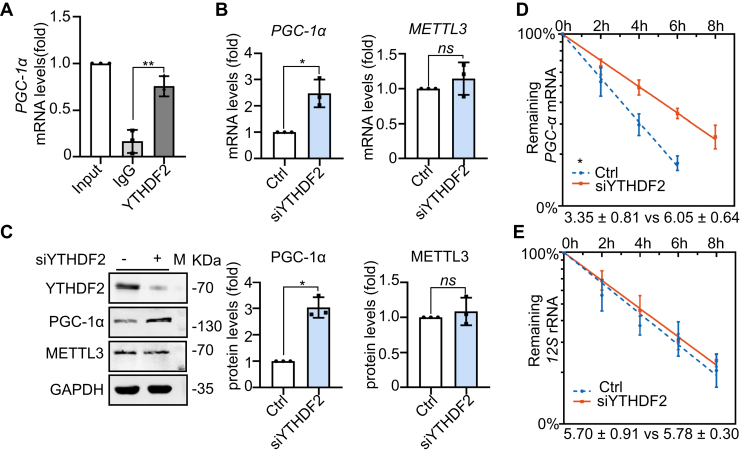
METTL3's regulation of PGC-1α is dependent on YTHDF2-based recognition of m6A
YTHDF2 is an m6A-binding protein that recognizes and destabilizes m6A-modified mRNA (30). YTHDF2 recognition of PGC-1α mRNA was investigated by an RNA immunoprecipitation qRT-PCR assay, revealing that YTHDF2 can directly bind to PGC-1α mRNA (Fig. 4A). Similar to METTL3 silencing, YTHDF2 knockdown increased PGC-1α mRNA and protein levels (Fig. 4, B and C). RNA stability assays further demonstrated that YTHDF2 knockdown increased the half-life of PGC-1α mRNA from 3.4 h to 6.1 h (Fig. 4, D and E). These results indicated that YTHDF2 may recognize the PGC-1α mRNA m6A modification and induce degradation, a mechanism by which YTHDF2 and METTL3 may coordinately regulate PGC-1α mRNA stability and translation.
Figure 4.
METTL3's regulation of PGC-1α is dependent on YTHDF2-based recognition of m6A. A, PGC1-α mRNA collected from an RNA immunoprecipitation assay with THP-1 cells using an IgG antibody or an anti-YTHDF2 antibody (n = 3). B, qRT-PCR determination of mRNA levels of PGC-1α and METTL3 in control and YTHDF2 knockdown cells was detected by qRT-PCR (n = 3). C, immunoblot detection of various protein levels in control and YTHDF2 knockdown cells lines, with GAPDH as a loading control (n = 3). D and E, half-lives of PGC-1α mRNA and 12S rRNA in control and YTHDF2 knockdown cells, which were used to determine half-life (n = 3). Data are represented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
METTL3 and YTHDF2 promote oxLDL-induced mitochondrial dysfunction in monocytes
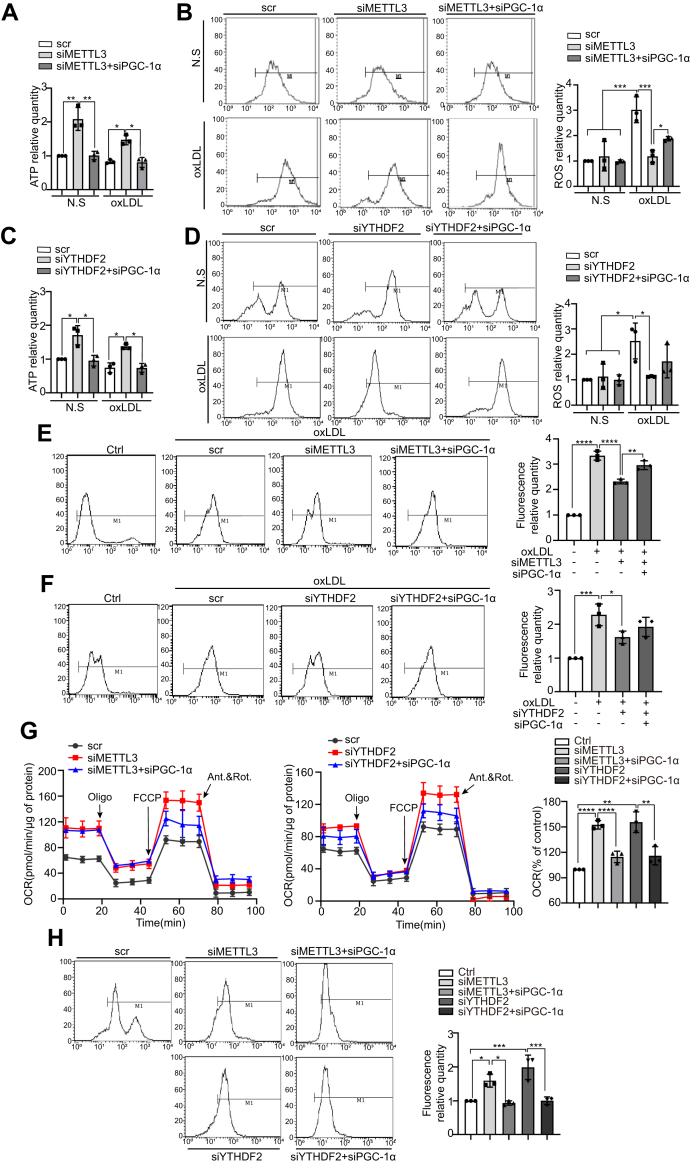
As PGC-1α has a role in mitochondrial biogenesis and energy metabolism, METTL3 and YTHDF2 might regulate ATP synthesis and ROS production of monocytes by targeting and destabilizing PGC-1α mRNA. THP-1 cell lines were generated by siRNA to knockdown METTL3 alone, METTL3/PGC-1α, YTHDF2 alone, and YTHDF2/PGC-1α and measured for ATP levels under treatment with a negative control or oxLDL. METTL3 knockdown increased cellular ATP levels by 2-fold under both treatments, while knocking down both PGC-1α and METTL3 attenuated this increase in ATP levels (Fig. 5A). Similar changes in ATP levels were observed in both the YTHDF2 knockdown and knocking down both YTHDF2 and PGC-1α (Fig. 5C). As increased levels of reactive oxygen species (ROS) are a marker of mitochondrial injury, flow cytometry was used to detect cellular and mitochondrial ROS levels. Interestingly, compared with untreated cells, cellular ROS levels in oxLDL-treated cells were increased by more than 2-fold, yet this increase was attenuated in METTL3 or YTHDF2 knockdowns when compared with oxLDL-treated cells transfected with scrambled (control) RNA. Meanwhile, knocking down PGC-1α at least partially abrogated the reduced cellular ROS levels in the METTL3 or YTHDF2 knockdowns (Fig. 5, B and D). Similar changes in mitochondrial levels were observed in both knockdown METTL3 alone, METTL3/PGC-1α, YTHDF2 alone, and YTHDF2/PGC-1α (Fig. 5, E and F). To assess the role of METTL3 and YTHDF2 in mitochondrial function, we investigated oxygen consumption rate (OCR) in THP-1 cell knockdown of METTL3 alone, METTL3/PGC-1α, YTHDF2 alone, and YTHDF2/PGC-1α, which were detected by Seahorse extracellular flux analysis. As shown in Figure 5G, knockdown of METTL3 or YTHDF2 increased OCR and knockdown of METTL3/PGC-1α or YTHDF2/PGC-1α could reverse this effect. MitoTracker was also used to analyze mitochondria function in control cells, METTL3 knockdown cells, and YTHDF2 knockdown cells. As shown in Figure 5H, knocking down METTL3 or YTHDF2 caused increased fluorescence, indicating enhanced mitochondrial function, while the additional knockdown of PGC-1α impaired mitochondrial function. These results suggested that knocking down METTL3 and YTHDF2 promoted mitochondrial function by allowing PGC-1α translation.
Figure 5.
METTL3 and YTHDF2 promote oxLDL-induced mitochondrial dysfunction in monocytes. A, intracellular ATP levels in control, METTL3 knockdown, and METTL3/PGC-1α knockdown cells both untreated and treated with oxLDL (n = 3). B, cellular ROS levels in control, METTL3 knockdown, and METTL3/PGC-1α knockdown cells both untreated and treated with oxLDL (n = 3). C, intracellular ATP levels in control, YTHDF2 knockdown, and YTHDF2/PGC-1α knockdown cells both untreated and treated with oxLDL (n = 3). D, cellular ROS levels in control, YTHDF2 knockdown, and YTHDF2/PGC-1α cells both untreated and treated with oxLDL (n = 3). E, mitochondrial ROS levels in control untreated with oxLDL, control, METTL3 knockdown, and METTL3/PGC-1α knockdown cells treated with oxLDL (n = 3). F, mitochondrial ROS levels in control untreated with oxLDL, control, YTHDF2 knockdown, and YTHDF2/PGC-1α cells treated with oxLDL (n = 3). G, OCR in control, METTL3 knockdown, and METTL3/PGC-1α cells (n = 3) and OCR in control, YTHDF2 knockdown, and YTHDF2/PGC-1α knockdown cells (n = 3). H, THP-1 cells were transfected with scrambled RNA, METTL3 siRNA, YTHDF2 siRNA, and PGC-1α siRNA for 48 h and then were labeled with MitoTracker fluorescence intensity in control, METTL3 knockdown, METTL3/PGC-1α, YTHDF2 knockdown, and YTHDF2/PGC-1α knockdown cell (n = 3). Data are represented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
METTL3 promotes oxLDL-induced inflammation of monocytes
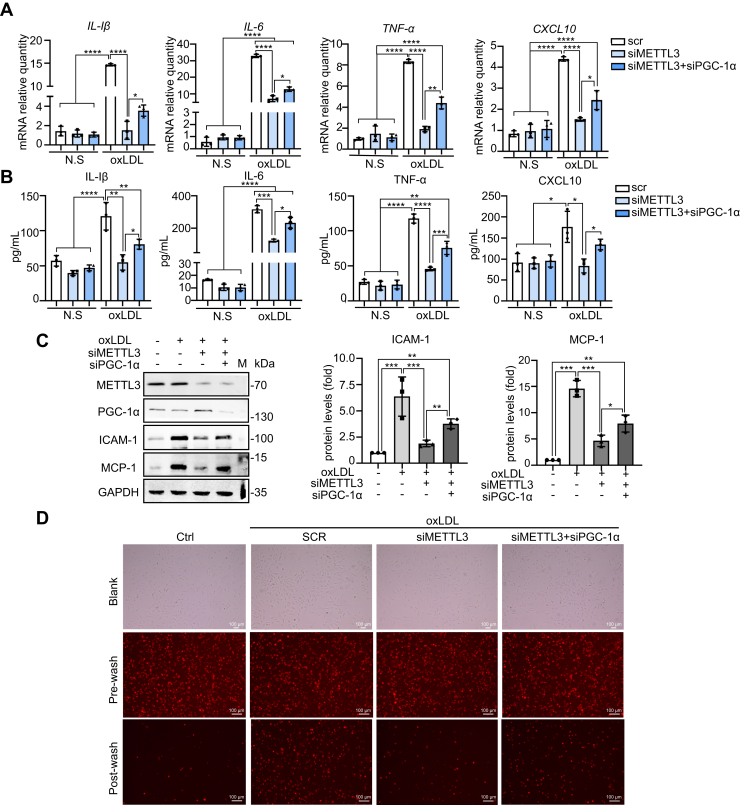
METTL3 expression increased during oxLDL-induced monocyte inflammation (Fig. 1A). To investigate the role of METTL3 in monocyte inflammation, the mRNA and protein expression levels of proinflammatory cytokines IL-1β, IL-6, TNF-α, and CXCL10 were measured in THP-1 cell knockdowns of METTL3 and METTL3/PGC-1α. qRT-PCR assays revealed that IL-1β, IL-6, TNF-α, and CXCL10 mRNA levels increased by 3.5–30-fold in control cells after 24 h of oxLDL treatment, while the METTL3 knockdown decreased mRNA levels after oxLDL treatment, which was partially reversed in the PGC-1α double knockdown (Fig. 6A). Similar results were obtained via ELISA assay (Fig. 6B). These results indicated that METTL3 promoted, while PGC-1α suppressed, the inflammatory response induced by oxLDL treatment. In addition to proinflammatory cytokines, inflammatory monocytes express certain adhesion molecules under inflammation conditions (2, 5); therefore, we measured the expression of adhesion molecules ICAM-1 and MCP-1 by immunoblot. ICAM-1 and MCP-1 were markedly increased after oxLDL treatment, while knocking down METTL3 significantly reduced the oxLDL-induced ICAM-1 and MCP-1 protein levels. This reduction was partially reversed in the PGC-1α double knockdown (Fig. 6C). In addition, monocyte-endothelial adhesion assays revealed decreased interaction between METTL3 knockdown THP-1 cells and HUVEC cells, and this effect was reversed in the PGC-1α double knockdown (Fig. 6D). Taken together, these results indicated that METTL3 promoted oxLDL-induced mitochondrial dysfunction and inflammation of monocytes.
Figure 6.
METTL3 promotes oxLDL-induced inflammation of monocytes.A, qRT-PCR determination of mRNA levels of IL-1β, IL-6, TNF-α, and CXCL10 in METTL3 knockdown and METTL3/PGC-1α knockdown cells both untreated and treated with oxLDL (n = 3). B, ELISA detection of protein levels of IL-1β, IL-6, TNF-α, and CXCL10 in control, METTL3 knockdown, and METTL3/PGC-1α knockdown cells both untreated and treated with oxLDL (n = 3). C, immunoblot detection of various protein levels in control, METTL3 knockdown, and METTL3/PGC-1α knockdown cells both untreated and treated with oxLDL (n = 3). D, monocyte-endothelial adhesion assay and fluorescence of untreated control cells and control METTL3 knockdown, and METTL3/PGC-1α knockdown cells treated with oxLDL (n = 3, scale bar: 100 μm). Data are represented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Discussion
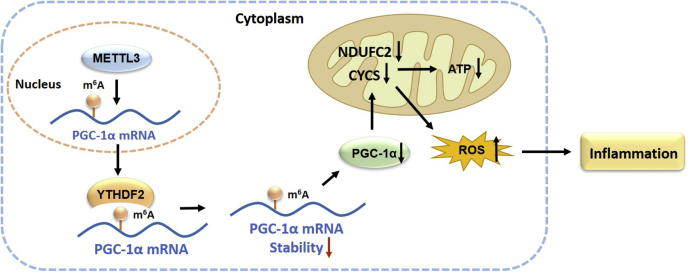
In addition to mediating host antimicrobial defense, monocytes also contribute to many inflammatory diseases (3, 4, 5, 6, 7, 31). Recently, METTL3 was shown to be involved in inflammatory responses in preosteoblasts (32), dental pulp cells (27), and epithelial cells (26), although little is known about the role of METTL3 in monocyte inflammation. In this study, we demonstrate that inflammatory stimulator-induced METTL3 promotes mitochondrial dysfunction and inflammatory response in monocytes via suppressing PGC-1α. We have identified a new mechanism underlying METTL3 acting in monocyte inflammation, in which METTL3 interacts with PGC-1α mRNA to mediate its m6A modification. YTHDF2 selectively recognizes m6A modification to mediate PGC-1α mRNA degradation. Subsequently, suppression of PGC-1α by METTL3 leads to inhibiting the expression of nuclear-encoded mitochondrial respiratory chain protein-related genes CYCS and NDUFC2, thereby reducing ATP production and increasing ROS accumulation in inflammatory monocytes (Fig. 7). These data may provide new insight into the role of METTL3-dependent m6A modification of PGC-1α mRNA in monocyte inflammatory response and aid in better understanding of pathogenesis of monocyte-macrophage inflammation-related diseases.
Figure 7.
A working model depicting the role and the mechanism of METTL3 in promoting mitochondrial dysfunction and monocyte inflammation by PGC-1α suppression. METTL3 acts in monocyte inflammation, in which METTL3 interacts with PGC-1α mRNA to mediate m6A modification; YTHDF2 selectively recognizes m6A modification to mediate PGC-1α mRNA degradation, by which YTHDF2 and METTL3 coordinately reduce the lifetime of PGC-1α mRNA. Subsequently, suppression of PGC-1α by METTL3 leads to inhibition of the expression of nuclear-encoded mitochondrial respiratory chain protein-related genes CYCS and NDUFC2, thereby reducing ATP production and increasing ROS accumulation in inflammatory monocytes.
The functional outcomes of m6A modification are extensive and determined by the dynamic balance of m6A RNA-specific binding proteins (m6A readers) and the type of modified RNAs (20, 30). The most common m6A readers, YTHDF1, YTHDF2, and YTHDF3, are mainly distributed in the cytoplasm and have different functions. YTHDF1 enhances translation by interacting with translation initiating factors and ribosomes. YTHDF3 interacts with both YTHDF1 and YTHDF2 to assist with regulation of mRNA fate. YTHDF2 specifically recognizes mRNA m6A sites in the stop codon region, the 3′UTR, and the coding region to promote target m6A RNA degradation (30). The C-terminal domain of YTHDF2 selectively binds to m6A RNAs, while the N-terminus brings the YTHDF2-mRNA complex to RNA decay sites in the cytosol and recruits the CCR4-NOT deadenylase complex to the RNA (33). As observed with other mRNA transcripts, YTHDF2 targeted m6A methylation and mediated PGC-1α mRNA degradation, thereby decreasing PGC-1α mRNA lifetime and protein expression. In contrast, knocking down either METTL3 or YTHDF2 increased mRNA lifetime and protein levels. Study data indicated that the METTL3 regulatory effects on PGC-1α expression depended upon the m6A reading function of YTHDF2. YTHDF3 may also play a role in this process. Knocking down the demethylase FTO increases the m6A methylation level of PGC-1α mRNA and decreases the expression of PGC-1α (34). As such, other factors may participate in METTL3-mediated PGC-1α mRNA degradation.
PGC-1α regulates gene networks involved in mitochondrial biogenesis and energy metabolism by interacting with a number of coactivator complexes containing chromatin remodeling activities (e.g., CBP/p300 and GCN5), transcription factor families (NRF-1, PPARα, YY1, and MEF2C), and nuclear hormone receptor families (ERRs) (35). Dysregulation of PGC-1α may lead to mitochondrial dysfunction and disturb mitochondrial oxidative phosphorylation, TCA cycle reactions, and fatty acid oxidation (14, 35). In this study, METTL3 methylation was determined to be necessary for YTHDF2 suppression of the expression of PGC-1α, CYCS, and NDUFC2, which was related to mitochondrial dysfunction in oxLDL-stimulated monocytes. The mitochondrial dysfunction may be due to PGC-1α-induced downregulation of CYCS and NDUFC2 genes in monocytes during oxLDL-induced acute inflammation. CYCS is a central component of the electron transport chain, i.e., the ubiquinol cytochrome c reductase (mitochondrial complex III), transferring electrons from cytochrome b (one subunit of complex III) to the cytochrome oxidase complex (complex IV). The nuclear respiratory factor 1 (NRF-1) bound to the upstream region of the CYCS promoter is required for CYCS gene activation (36). PGC-1α can directly interact with NRF-1 to activate the CYCS gene (35). NDUFC2 is one of the subunits of mitochondrial complex I, and the CCAAT/enhancer-binding protein alpha (C/EBPα) can trigger aberrant expression of NADH dehydrogenase-encoding genes, including NDUFC2 and NDUFA1 (37). The prediction of the transcription factor binding sites by QIAGEN in the NDUFC2 gene promoter shows potential binding motifs for PPARγ, c-Myc, HFH-1, HOXA3, Max, NF-1, and Pax-5. PGC-1α has been identified as a coactivator of PPARγ (38) and increases PPARα transcriptional activity, as well as promotes interactions with a variety of transcription factors and proteins (39). Therefore, it is reasonable to infer that NDUFC2 could be a potential target of PGC-1α, although the relationship between the two molecules is not clear. Taken together, these findings suggest that CYCS and NDUFC2 were downregulated in oxLDL-induced inflammatory monocytes when PGC-1α was suppressed by METTL3 with YTHDF2. Reduction of PGC-1α, CYCS, and NDUFC2 impairs electron transport and oxidative phosphorylation (a major source for ATP production), thereby reducing mitochondrial ATP synthesis and increasing ROS production, ultimately causing inflammation (40).
In summary, we demonstrated that METTL3 promoted mitochondrial dysfunction and inflammatory response during oxLDL-induced inflammation in monocytes. METTL3 caused m6A methylation of PGC-1α mRNA, as knockdown of METTL3 suppressed oxLDL-induced monocyte inflammation and protected mitochondrial function against oxLDL-induced inflammation. These data may provide new insights into the role of METTL3-dependent m6A modification of PGC-1α mRNA in monocyte inflammation and provide additional information into the pathogenesis of monocyte-macrophage inflammation-related diseases
Experimental procedures
Cell culture and treatment
THP-1 cells were cultured in RPMI-1640 (Roswell Park Memorial Institute) media (Invitrogen) with 1.5 g NaHCO3/L and supplemented with 10% fetal bovine serum (FBS) at 37 °C in 5% CO2, with 100 U/ml penicillin and 100 μg/ml streptomycin. 293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) with 1.5 g NaHCO3/L and supplemented with 10% FBS at 37 °C in 5% CO2, with 100 U/ml penicillin and 100 μg/ml streptomycin. Human umbilical vein endothelial cells (HUVECs) were isolated from segments of human umbilical cord vein by collagenase digestion and cultured in medium 199 (Gibco) supplemented with 10% FBS. Cells between passages 2 and 4 were used for the experiments.
For plasmid transfections, Lipofectamine 2000 (Invitrogen) was used following the manufacturer's instructions. Cells were harvested 48 h after transfection for further analysis. METTL3 (1# 5′-AGGAGCCAGCCAAGAAAUCAATT-3′, 2# 5′-GCACTTGGATCTACGGAAT-3′) siRNA, PGC-1α (1# 5′-CCAAGACUCUAGACAACUATT-3′, 2# 5′-UUUCUGGGUGGAUUGAAGUGGUGUA-3′) siRNA, YTHDF2 (1# 5′-CAAGGAAACAAAGTGCAAA-3′, 2# 5′- AAGGACGTTCCCAATAGCCAA-3′) siRNA or a control siRNA (AAGAGGCAAUUACCAGUUUCA) was transfected using RNAimax (Invitrogen) following the manufacturer's instructions. Cells were harvested 72 h after transfection for further analysis.
For preparing monocyte inflammation model, THP-1 cells were treated with 20 μg/ml oxLDL (Meilunbio) for given hour(s) and harvested for the next experiments.
Data resources and bioinformatics analysis
RNA-seq of human myeloid leukemia cells was obtained from data set GSE98623 in the GEO Database (https://www.ncbi.nlm.nih.gov/geo/). CLIP-seq of METTL3 was from the database POSTAR (http://lulab.life.tsinghua.edu.cn/postar/), and pathway enrichment analysis was conducted using the online analysis tool DAVID (https://david.ncifcrf.gov/).
Western blot analysis
Western blot analysis was performed using standard procedures (41). Briefly, the collected cells were washed three times with cold PBS on ice and placed in 3-fold cold lysis buffer. The suspension was vortexed. The lysate was centrifugated at 4 °C, 12,000 rpm for 20 min in an Eppendorf microcentrifuge, and the supernatant was collected for measurement of protein concentration by BCA Protein Assay Kit (Thermo Fisher Scientific-CN). Then after, 6× protein loading buffer was added and incubated in 98 °C water for 10 min. Thirty micrograms of proteins were subjected to 10% SDS-PAGE and transferred onto nitrocellulose membranes, blocked with 5% nonfat milk in TBS-T (20 mM Tris, 500 mM NaCl, and 0.1% Tween 20) at room temperature for 2 h with rocking. The membranes were probed with specific antibodies overnight at 4 °C. After washing with 5% nonfat milk/TBS-T three times for 15 min each, membranes were incubated with fluorescently labeled secondary antibodies (LICOR, USA) in 5% nonfat milk/TBS-T at room temperature for 1 h. After washing with TBS-T, the protein–antibody complex was detected by Odyssey two-color infrared laser imaging system (LICOR, USA).
Monoclonal anti-METTL3, monoclonal anti-GAPDH, and monoclonal anti-YTHDF2 were bought from Abcam. Monoclonal anti-PGC-1β, monoclonal anti-CYCS, monoclonal anti-MCP-1, monoclonal anti-NDUFC2, and monoclonal anti-NDUFB6 were bought from Santa Cruz. Monoclonal anti-COX8A, monoclonal anti-NDUFA6, monoclonal anti-NDUFA12, and monoclonal anti-ICAM-1 antibodies were bought from Abclonal. Monoclonal anti-PGC-1α antibodies were from Merck Millipore.
RNA isolation, reverse transcription, and quantitative real-time qPCR analysis
Total cellular RNA was extracted using TRIzol (Invitrogen) and treated with reverse transcriptase and a mixture of oligo-dT and random hexamers (Invitrogen). To quantify the levels of different mRNAs, qRT-PCR analysis was performed using the primer pairs (Table S2) listed in the supporting information.
Preparation of transcripts
cDNA was used as a template for PCR amplification of RNA fragments. All primers contained the T7 promoter sequence (5′-CCAAGCTTCTAATACGACTCACTATAGGGAGA-3′). The in vitro transcription reaction (1 μl 10 mmol/L ATP, 1 μl 10 mmol/L GTP, 1 μl 10 mmol/L CTP, 0.5 μl 10 mmol/L UTP, 0.5 μl 10 mmol/L Biotin-11-UTP, 1 μg DNA templet, 4 μl 5× buffer, 1 μl T7 RNA polymerase, 2 μ1 100 mmol/L DTT, 1 μl RNA inhibitor, added DEPC H2O to 50 μl) was performed in a 37 °C water bath for 6–8 h. and then, biotin-labeled RNA probes were purified and the concentration of probes were determined.
Constructs and luciferase reporter assay
To construct pGL3-derived reporter vectors, the fragments of PGC-1α mRNA 5′UTR were inserted between the HindIII and NcoI sites of a pGL3-promoter vector (Promega). The CR and 3′UTR fragments were inserted into the FseI and XbaI sites of a pGL3-promoter vector (Promega). 293T cells were seeded in 24-well plates (4 × 104 cells per well). Cells were transfected with siRNA targeting METTL3. After 24 h, cells were transfected with each of the pGL3-derived reporters and cultured for an additional 48 h. Firefly and renilla luciferase activities were measured with a double luciferase assay system (Promega) following the manufacturer's instructions. All firefly luciferase measurements were normalized to renilla luciferase measurements from the same sample.
RNA pull-down assays
PCR-amplified DNA fragments were used as templates to transcribe biotinylated RNA by using T7 RNA polymerase in the presence of biotin-UTP. One microgram of purified biotinylated transcripts was incubated with 30 mg of cell lysates for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal), and the pull-down material was analyzed by western blot.
RNA immunoprecipitation
A total of 5 × 107 THP-1 cells were collected and lysed in a buffer containing 150 mmol/L KCl, 10 mmol/L HEPES (pH 7.6), 2 mmol/L EDTA, 0.5% CA-630, 0.5 mmol/L DTT, 1:100 protease inhibitor cocktail, and 400 U/ml RNase inhibitor. The cell lysate was incubated with METTL3 beads at 4 °C for 4 h, followed by washing the beads with NT2 buffer, and incubation in elution solution. After washing, bound RNAs were extracted using TRIzol reagent and analyzed by qRT-PCR.
m6A-IP
Total RNA was extracted from THP-1 cells with TRIzol (Invitrogen), and additional DNase I was applied to all samples to avoid DNA contamination. Enrichment of m6A-containing transcript segments was analyzed through qRT-PCR.
RNA half-life analysis
To determine the half-life of PGC-1α mRNAs, actinomycin D (final concentration 2 μg/ml, Amresco) was added to THP-1 cell cultures. Following 48 h of siRNA transfection, the total RNAs were extracted at 0, 2, 4, 6, and 8 h after actinomycin D treatment. The mRNA levels at different times were analyzed by qRT-PCR.
Cytokine ELISA assays
Cells were incubated in RPMI1640 with 10% FBS and were treated with 20 μg/ml oxLDL for 24 h. Postculture media was then collected and subjected to ELISA assays specific for IL-1β, IL-6, TNF-α, and CXCL10 (Abclonal).
ATP level determination
For measuring ATP levels, an ATP Bioluminescent Assay Kit (Sigma) was used following the manufacturer's instructions.
Measurement of cellular ROS
For live cell analysis, THP-1 cells were cultured in a 6-well plate at a density of 2 × 105 cells/well and incubated in RPMI1640/10% FBS in the presence of 10 μmol/L carboxy-DCFDA (Beyotime) for 20 min at 37 °C to assess total cellular ROS. At the end of incubation, cells were collected and suspended in 200 μl buffer and immediately subjected to flow cytometry analysis. Carboxy-DCFDA fluorescence was detected with the FITC channel.
Measurement of mitochondrial ROS
For live cell analysis, THP-1 cells were cultured in a 6-well plate at a density of 2 × 105 cells/well and were transfected with siRNA for 48 h. Before the experiment, the cells were stained with 5 μM MitoSox Red (yeasen) for mitochondria. They were then treated per the manufacturer's protocol in the cellular retention assay. Data were collected using flow cytometry analysis. The fluorescence of MitoTracker Red (red) was excited at 510 nm and measured at 580 nm.
MitoTracker staining
For live cell analysis, THP-1 cells were cultured in a 6-well plate at a density of 2 × 105 cells/well and were transfected with siRNA for 48 h. Before the experiment, the cells were stained with 50 nM MitoTracker Red (Solarbio) for mitochondria and then were treated per the manufacturer's protocol in the cellular retention assay. Data were collected by using flow cytometry analysis. The fluorescence of MitoTracker Red (red) was excited at 579 nm and collected at 599 nm.
Monocytes energetics
THP-1 cells were seeded into the specialized XFe24 cell culture microplate (Seahorse Bioscience) at about 50,000–80,000 cells/well. THP-1 cells were transfected with METTL3 and PGC-1α siRNA for 24 h and treated with 20 μg/ml oxLDL for 24 h. The OCR was measured using a Seahorse Bioscience Extracellular Flux Analyzer (Agilent) and Seahorse XF Cell Mito Stress Test Kit (Agilent). Cells were then lysed in RIPA buffer and subjected to Bradford protein assay (Bio-Rad). OCR values were normalized by protein values in each well.
Adhesion assay
In total, 3 × 104 HUVECs were plated per well in a 24-well plate (Corning) in M199 and grown to complete confluence. THP-1 cells were transfected with METTL3 and PGC-1α siRNA for 24 h and were treated with 20 μg/ml oxLDL for 24 h. THP-1 monocytes were labeled with 5 mmol/L of CellTracker CM-DiI (Invitrogen) in parallel according to the manufacturer's instructions. A total of 0.5 × 106 labeled THP-1 monocytes were added per well in a 24-well plate and incubated for 30 min in 5% CO2 at 37 °C. After incubation, each well was washed three times with 500 μl M199, and ten high-power field digital images were taken using an inverted fluorescence microscope (Lecia).
Statistics
All data are presented as mean ± SD and were analyzed using GraphPad Prism version 8.0 software. The paired t test and one-way ANOVA analysis of variance were used for statistical analysis. p < 0.05 was considered statistically significant.
Data availability
All data are contained within the article and the supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We would like to thank Guohui Dang for providing THP-1 cells. This work was supported by the National Natural Science Foundation of China (grant 81571368).
Author contributions
X. Z. and X. L. investigation; X. Z. and X. L. methodology; G. A. project administration; H. J. and J. N. supervision; X. Z. visualization; X. Z. and X. L. writing-original draft; H. J. and J. N. writing-review and editing.
Edited by Dennis Voelker
Supporting information
References
- 1.Shi C., Pamer E.G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auffray C., Sieweke M.H., Geissmann F. Blood monocytes: Development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 3.Duffield J.S. Macrophages and immunologic inflammation of the kidney. Semin. Nephrol. 2010;30:234–254. doi: 10.1016/j.semnephrol.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misharin A.V., Morales-Nebreda L., Reyfman P.A., Cuda C.M., Walter J.M., McQuattie-Pimentel A.C., Chen C.-I., Anekalla K.R., Joshi N., Williams K.J.N., Abdala-Valencia H., Yacoub T.J., Chi M., Chiu S., Gonzalez-Gonzalez F.J. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J. Exp. Med. 2017;214:2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woollard K.J., Geissmann F. Monocytes in atherosclerosis: Subsets and functions. Nat. Rev. Cardiol. 2010;7:77–86. doi: 10.1038/nrcardio.2009.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katschke K.J., Rottman J.B., Ruth J.H., Qin S., Wu L., LaRosa G., Ponath P., Park C.C., Pope R.M., Koch A.E. Differential expression of chemokine receptors on peripheral blood, synovial fluid, and synovial tissue monocytes/macrophages in rheumatoid arthritis. Arthritis Rheum. 2001;44:1022–1032. doi: 10.1002/1529-0131(200105)44:5<1022::AID-ANR181>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 7.Zuroff L., Daley D., Black K.L., Koronyo-Hamaoui M. Clearance of cerebral Aβ in Alzheimer's disease: Reassessing the role of microglia and monocytes. Cell Mol. Life Sci. 2017;74:2167–2201. doi: 10.1007/s00018-017-2463-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frostegård J., Nilsson J., Haegerstrand A., Hamsten A., Wigzell H., Gidlund M. Oxidized low density lipoprotein induces differentiation and adhesion of human monocytes and the monocytic cell line U937. Proc. Natl. Acad. Sci. U. S. A. 1990;87:904–908. doi: 10.1073/pnas.87.3.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frostegård J., Wu R., Giscombe R., Holm G., Lefvert A.K., Nilsson J. Induction of T-cell activation by oxidized low density lipoprotein. Arterioscler. Thromb. 1992;12:461–467. doi: 10.1161/01.atv.12.4.461. [DOI] [PubMed] [Google Scholar]
- 10.Berliner J.A., Territo M.C., Sevanian A., Ramin S., Kim J.A., Bamshad B., Esterson M., Fogelman A.M. Minimally modified low density lipoprotein stimulates monocyte endothelial interactions. J. Clin. Invest. 1990;85:1260–1266. doi: 10.1172/JCI114562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Yang M., Huang W., Chen W., Zhao Y., Schulte M.L., Volberding P., Gerbec Z., Zimmermann M.T., Zeighami A., Demos W., Zhang J., Knaack D.A., Smith B.C., Cui W. Mitochondrial metabolic reprogramming by CD36 signaling drives macrophage inflammatory responses. Circ. Res. 2019;125:1087–1102. doi: 10.1161/CIRCRESAHA.119.315833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuo H., Wan Y. Metabolic reprogramming in mitochondria of myeloid cells. Cells. 2019;9:5. doi: 10.3390/cells9010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villena J.A. New insights into PGC-1 coactivators: Redefining their role in the regulation of mitochondrial function and beyond. FEBS J. 2015;282:647–672. doi: 10.1111/febs.13175. [DOI] [PubMed] [Google Scholar]
- 14.Dorn G.W., Kitsis R.N. The mitochondrial dynamism-mitophagy-cell death interactome: Multiple roles performed by members of a mitochondrial molecular ensemble. Circ. Res. 2015;116:167–182. doi: 10.1161/CIRCRESAHA.116.303554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finck B.N., Kelly D.P. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin J., Handschin C., Spiegelman B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Liu T.F., Vachharajani V.T., Yoza B.K., McCall C.E. NAD+-dependent sirtuin 1 and 6 proteins coordinate a switch from glucose to fatty acid oxidation during the acute inflammatory response. J. Biol. Chem. 2012;287:25758–25769. doi: 10.1074/jbc.M112.362343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., Sorek R., Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 19.Frye M., Harada B.T., Behm M., He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vu L.P., Cheng Y., Kharas M.G. The biology of mA RNA methylation in normal and malignant hematopoiesis. Cancer Discov. 2019;9:25–33. doi: 10.1158/2159-8290.CD-18-0959. [DOI] [PubMed] [Google Scholar]
- 21.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu K., Yang Y., Feng G.-H., Sun B.-F., Chen J.-Q., Li Y.-F., Chen Y.-S., Zhang X.-X., Wang C.-X., Jiang L.-Y., Liu C., Zhang Z.-Y., Wang X.-J., Zhou Q., Yang Y.-G. Mettl3-mediated mA regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27:1100–1114. doi: 10.1038/cr.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., Zhang Y.-C., Huang C., Shen H., Sun B., Cheng X., Zhang Y.-J., Yang Y.-G., Shu Q., Yang Y., Li X. mA regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genomics Proteomics Bioinformatics. 2019;17:154–168. doi: 10.1016/j.gpb.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L., Wang J., Sun G., Wu Q., Ma J., Zhang X., Huang N., Bian Z., Gu S., Xu M., Yin M., Sun F., Pan Q. mA mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol. Cancer. 2019;18:188. doi: 10.1186/s12943-019-1119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C., Fu J., Zhou Y. A review in research progress concerning m6A methylation and immunoregulation. Front. Immunol. 2019;10:922. doi: 10.3389/fimmu.2019.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zong X., Zhao J., Wang H., Lu Z., Wang F., Du H., Wang Y. Mettl3 deficiency sustains long-chain fatty acid absorption through suppressing Traf6-dependent inflammation response. J. Immunol. 2019;202:567–578. doi: 10.4049/jimmunol.1801151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Z., Li Q., Meng R., Yi B., Xu Q. METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J. Cell. Mol. Med. 2018;22:2558–2568. doi: 10.1111/jcmm.13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., Hu X., Huang M., Liu J., Gu Y., Ma L., Zhou Q., Cao X. Mettl3-mediated mRNA mA methylation promotes dendritic cell activation. Nat. Commun. 2019;10:1898. doi: 10.1038/s41467-019-09903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y., Zeng P., Li Y.-H., Zhang Z., Cui Q. SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., Ren B., Pan T., He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chimen M., Evryviadou A., Box C.L., Harrison M.J., Hazeldine J., Dib L.H., Kuravi S.J., Payne H., Price J.M.J., Kavanagh D., Iqbal A.J., Lax S., Kalia N., Brill A., Thomas S.G. Appropriation of GPIbα from platelet-derived extracellular vesicles supports monocyte recruitment in systemic inflammation. Haematologica. 2020;105:1248–1261. doi: 10.3324/haematol.2018.215145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Gu X., Li D., Cai L., Xu Q. METTL3 regulates osteoblast differentiation and inflammatory response via Smad signaling and MAPK signaling. Int. J. Mol. Sci. 2019;21:199. doi: 10.3390/ijms21010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Huang N., Yang M., Wei D., Tai H., Han X., Gong H., Zhou J., Qin J., Wei X., Chen H., Fang T., Xiao H. FTO is required for myogenesis by positively regulating mTOR-PGC-1α pathway-mediated mitochondria biogenesis. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans M.J., Scarpulla R.C. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF, and intron Sp1 recognition sequences. J. Biol. Chem. 1989;264:14361–14368. [PubMed] [Google Scholar]
- 37.Li X., Yang H., Sun H., Lu R., Zhang C., Gao N., Meng Q., Wu S., Wang S., Aschner M., Wu J., Tang B., Gu A., Kay S.A., Chen R. Taurine ameliorates particulate matter-induced emphysema by switching on mitochondrial NADH dehydrogenase genes. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E9655–E9664. doi: 10.1073/pnas.1712465114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 39.Soyal S., Krempler F., Oberkofler H., Patsch W. PGC-1alpha: A potent transcriptional cofactor involved in the pathogenesis of type 2 diabetes. Diabetologia. 2006;49:1477–1488. doi: 10.1007/s00125-006-0268-6. [DOI] [PubMed] [Google Scholar]
- 40.Arena G., Cissé M.Y., Pyrdziak S., Chatre L., Riscal R., Fuentes M., Arnold J.J., Kastner M., Gayte L., Bertrand-Gaday C., Nay K., Angebault-Prouteau C., Murray K., Chabi B., Koechlin-Ramonatxo C. Mitochondrial MDM2 regulates respiratory complex I activity independently of p53. Mol. Cell. 2018;69:594–609.e8. doi: 10.1016/j.molcel.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hnasko T.S., Hnasko R.M. The Western blot. Methods Mol. Biol. 2015;1318:87–96. doi: 10.1007/978-1-4939-2742-5_9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and the supporting information.