Abstract
Liver injuries induced by various stimuli share in common an acute inflammatory response, in which circulating macrophages home to the liver parenchyma to participate in the regulation of repair, regeneration, and fibrosis. In the present study we investigated the role of hepatocyte-derived C–C motif ligand 7 (CCL7) in macrophage migration during liver injury focusing on its transcriptional regulation. We report that CCL7 expression was up-regulated in the liver by lipopolysaccharide (LPS) injection (acute liver injury) or methionine-and-choline-deficient (MCD) diet feeding (chronic liver injury) paralleling increased macrophage infiltration. CCL7 expression was also inducible in hepatocytes, but not in hepatic stellate cells or in Kupffer cells, by LPS treatment or exposure to palmitate in vitro. Hepatocyte-specific deletion of Brahma-related gene 1 (BRG1), a chromatin remodeling protein, resulted in a concomitant loss of CCL7 induction and macrophage infiltration in the murine livers. Of interest, BRG1-induced CCL7 transcription and macrophage migration was completely blocked by the antioxidant N-acetylcystine. Further analyses revealed that BRG1 interacted with activator protein 1 (AP-1) to regulate CCL7 transcription in hepatocytes in a redox-sensitive manner mediated in part by casein kinase 2 (CK2)-catalyzed phosphorylation of BRG1. Importantly, a positive correlation between BRG1/CCL7 expression and macrophage infiltration was identified in human liver biopsy specimens. In conclusion, our data unveil a novel role for BRG1 as a redox-sensitive activator of CCL7 transcription.
Keywords: Macrophage infiltration, Liver injury, Transcriptional regulation, Chemokine, Epigenetics
1. Introduction
Inflammation is a key process and a driving force of liver injuries [[1], [2], [3], [4], [5]]. Macrophages represent a heterogenous population of cells that play versatile roles modulating liver injuries [6,7]. During liver injury, there is an expansion of hepatic macrophage population typically paralleling the initiation and perpetuation of inflammatory response and contributing to the loss of hepatic functions [[8], [9], [10]]. Congruently, macrophage depletion has been observed to confer hepatoprotection in animal models of both acute and chronic liver injuries. Zeng et al. have reported that macrophage depletion via clodronate injection attenuates LPS induced liver injury by suppressing hepatic inflammation and hepatocyte necrosis in mice, which is presumably mediated by the signaling lymphocyte activating molecule (SLAM) family of receptors [11]. Similarly, clodronate-mediated macrophage depletion attenuates liver injury induced by bile duct ligation (BDL), ischemia-reperfusion challenge, high-fat diet feeding, and excessive alcohol consumption [[12], [13], [14], [15]].
Navigation of circulating monocytes/macrophages to the liver to promote inflammation is guided by chemokines [16,17]. C–C motif ligand (CCL) family of polypeptides, currently comprising 28 members, plays predominant roles steering macrophages to their destinations [18]. CCL proteins exert the immunomodulatory roles by binding to their cognate receptors (CCR) structurally related to the G-protein coupled receptor (GPCR) superfamily [19]. Genetic manipulation or pharmacological blockade of CCRs has been reported to ameliorate liver injury in model animals. For instance, deletion of CCR1 [20], CCR2 [21], CCR5 [22], CCR8 [23], or CCR9 [24], in mice is associated with reduced liver injury and improved liver function. Concordantly, administration of the dual CCR2/CCR5, to which CCL2 and CCL5 bind, inhibitor cenicriviroc alleviates alcoholic liver injury in mice [25]. Monocyte/macrophage chemoattractant proteins, which include MCP-1 (CCL2), MCP-2 (CCL8), and MCP-3 (CCL7), can be categorized into the same sub-family of CCL chemokines owing to structural similarities [26]. CCL2 expression is up-regulated during liver injury and has a well-documented role in promoting macrophage infiltration to sustain the pro-inflammatory response [[27], [28], [29]]. The role of CCL7 in this process is not well understood.
Brahma related gene 1 (BRG1) and the closely related BRM are core components of the mammalian SWI/SNF chromatin remodeling complex [30]. Encoded by Smarca4, BRG1 plays a wide range of roles regulating cellular proliferation, differentiation, migration, and death by orchestrating lineage- and cue-specific transcriptional programs [31]. Mice with hepatocyte-conditional ablation of BRG1 are phenotypically indistinguishable from their wild type littermates [32,33] suggesting that BRG1 is dispensable for normal liver functions probably due to compensation by BRM. Under stress conditions, however, the hepatocyte BRG1-null mice are more resistant to liver injury than the WT littermates [[34], [35], [36], [37]]. In the present study, we investigated the regulation of CCL7 transcription by BRG1 and the implication in macrophage infiltration during liver injury. Our data indicate that activation of CCL7 by BRG1 in hepatocytes may contribute to macrophage infiltration during liver injury.
2. Materials and methods
2.1. Animals
All animal experiments were reviewed and approved by the intramural Nanjing Medical University Ethics Committee on Humane Treatment of Experimental Animals. Hepatocyte conditional BRG1 knockout (LKO) mice were obtained by crossing the Smarca4f/f mice [38] with the Alb-Cre mice [39]. Hepatocyte conditional BRG1 over-expression mice (LKI) were obtained by crossing the RosaBrg1/+ mice [40] with the Alb-Cre mice. To induce liver injury, 8-week male mice were fasted overnight before receiving a single injection of LPS (Sigma) at 15 mg/kg [41]. Alternatively, 8-week male mice were fed a methionine- and choline-deficient (MCD) diet (A02082002B, Research Diets) for 4 consecutive weeks as previously described [42,43].
2.2. Cell culture, plasmids, and transient transfection
Human hepatoma cells (HepG2) and mouse macrophage-like cells (RAW264.7) were maintained in DMEM supplemented with 10 % fetal bovine serum (FBS, Hyclone). Primary hepatocytes were isolated and cultured as previously described [44]. Human CCL7 promoter-luciferase constructs [45] and BRG1 expression constructs [46] have been previously described. Small interfering RNAs were purchased from Dharmacon. Transient transfections were performed with Lipofectamine 2000. Luciferase activities were assayed 24–48 h after transfection using a luciferase reporter assay system (Promega) as previously described [[47], [48], [49], [50]]. For conditioned media (CM) collection, the cells were switched to and incubated with serum-free media overnight. The next day, the media were collected, centrifuged at 4000×g for 30min at 4 °C using 3-kDa MW cut-off filter units (Millipore) and sterilized through a 0.4-μm filter.
2.3. Protein extraction and Western blot
Whole cell lysates were obtained by re-suspending cell pellets in RIPA buffer (50 mM Tris pH7.4, 150 mM NaCl, 1 % Triton X-100) with freshly added protease inhibitor (Roche) as previously described [[51], [52], [53], [54]]. Western blot analyses were performed with anti-BRG1 (Santa Cruz, sc-10768), anti-c-Jun (Santa Cruz, sc-1694), anti-Fos (Santa Cruz, sc-166940), anti-Phospho-CK2 substrate (Cell Signaling Tech, 8738), anti-CK2α1 (Proteinch, 10992–1), anti-CK2α2 (Proteinch, 10606–1), anti-CK2β (Proteintech, 20234–1), and anti-β-actin (Sigma, A2228) antibodies.
2.4. RNA isolation and real-time PCR
RNA was extracted with the RNeasy RNA isolation kit (Qiagen). Reverse transcriptase reactions were performed using a SuperScript First-strand Synthesis System (Invitrogen) as previously described [[55], [56], [57], [58]]. Real-time PCR reactions were performed on an ABI Prism 7500 system with the following primers: human CCL7, 5′-GACAAGAAAACCCAAACTCCAAAG-3′ and 5′-TCAAAACCCACCAAAATCCA-3’; human BRG1, 5′-TCATGTTGGCGAGCTATTTCC-3′ and 5′-GGTTCCGAAGTCTCAACGATG-3’; mouse Ccl7, 5′-GCAGTCTGAAGGCACAGCAA-3′ and 5′-GGTTGGCACAGACCTGGAAC-3’. Ct values of target genes were normalized to the Ct values of house keekping control gene (18s, 5′-CGCGGTTCTATTTTGTTGGT-3′ and 5′-TCGTCTTCGAAACTCCGACT-3′ for both human and mouse genes) using the ΔΔCt method and expressed as relative mRNA expression levels compared to the control group which is arbitrarily set as 1.
2.5. Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described before [50,51,[59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75]]. In brief, chromatin in control and treated cells were cross-linked with 1 % formaldehyde. Cells were incubated in lysis buffer (150 mM NaCl, 25 mM Tris pH 7.5, 1 % Triton X-100, 0.1 % SDS, 0.5 % deoxycholate) supplemented with protease inhibitor tablet and PMSF. DNA was fragmented into ~200 bp pieces using a Branson 250 sonicator. Aliquots of lysates containing 200 μg of protein were used for each immunoprecipitation reaction with anti-BRG1 (Santa Cruz, sc-10768), anti-c-Jun (Santa Cruz, sc-1694), anti-Fos (Santa Cruz, sc-166940), or pre-immune IgG. For re-ChIP, immune complexes were eluted with the elution buffer (1 % SDS, 100 mM NaCO3), diluted with the re-ChIP buffer (1 % Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris pH 8.1), and subject to immunoprecipitation with a second antibody of interest. Precipitated DNA was amplified by the following primers: human CCL7 promoter (−266/-65), 5′-ACTTTGGTATCCCTGATTCCTTCC-3′ and 5′-ATGGAAGCAGAGAGGATGAATC-3’; human GAPDH promoter (−197/+39), 5′-GGGTTCCTATAAATACGGACTGC-3′ and 5′-CTGGCACTGCACAAGAAGA-3’.
2.6. Macrophage migration assay
Macrophage migration was measured using the Boyden chamber inserts (5 μm, Corning) as previously described [58,64]. Briefly, RAW264.7 cells were added to the upper chamber whereas the conditioned media collected from endothelial cells were added to the lower chamber. The number of migrated macrophages in the lower chamber was counted in five randomly chosen fields using an inverted microscope. In certain experiments, recombinant human CCL7 (20 ng/ml, R&D) was directly added to the conditioned media. Migrated macrophages were counted in at least 5 different fields for each well. All experiments were performed in triplicates and repeated three times.
2.7. Enzyme-linked immunosorbent assay
Secreted CCL7 levels were examined by ELISA using commercially available kits according to vendor's recommendations (for human CCL7, ab193769, Abcam, and for murine CCL7, ab205571, Abcam).
2.8. Luminescence ROS assay
Quantitative measurements of intracellular ROS were performed with a ROS-Glo system (Promega). Briefly, a luminescence substrate solution was added to and incubated with cultured cells for 6 h followed by the addition of the diction solution. Luminescence was measured using a microplate reader. Data were expressed as relative ROS levels compared to the control group.
2.9. Human specimen collection
Liver biopsies were collected from patients with NASH referring to Nanjing Drum Tower Hospital. Written informed consent was obtained from subjects or families of liver donors. All procedures that involved human samples were approved by the Ethics Committee of Nanjing Drum Tower Hospital and adhered to the principles outlined in the Declaration of Helsinki.
2.10. Histology
Histological analyses were performed essentially as described before. Briefly, the paraffin embedded sections were blocked with 10 % normal goat serum for 1 h at room temperature and then incubated with an anti-CD68 (Proteintech, 66231–2) antibody or an anti-Ly6C antibody (Novus, NBP2-11784). Staining was visualized by incubation with anti-rabbit secondary antibody and developed with a streptavidin-horseradish peroxidase kit (Pierce) for 20min. Pictures were taken using an Olympus IX-70 microscope.
2.11. Statistical analysis
One-way ANOVA with post-hoc Scheff'e analyses were performed by SPSS software (IBM SPSS v18.0, Chicago, IL, USA). Unless otherwise specified, values of p<0.05 were considered statistically significant.
3. Results
3.1. Up-regulation of CCL7 accompanies macrophage infiltration in the murine livers
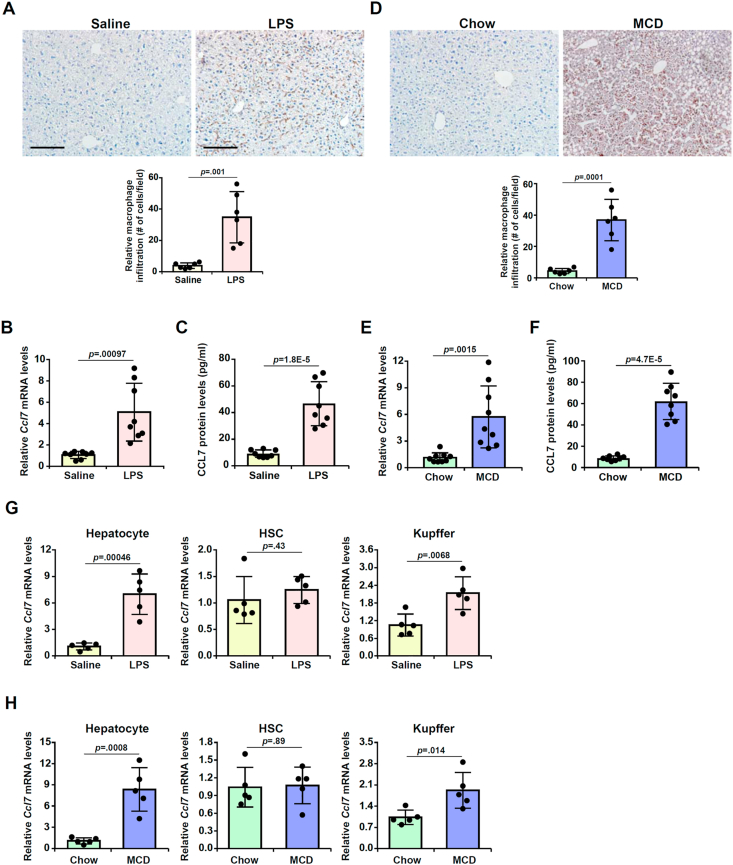
We first evaluated the relationship between CCL7 expression and macrophage infiltration in two different animal models of liver injury. In the first model of acute liver injury in which the mice were injected peritoneally with LPS, it was observed that there was massive infiltration of CD68+ macrophages in the murine livers 12 h following the injection of LPS compared to the livers injected with saline as examined by immunohistochemical staining (Fig. 1A). QPCR (Fig. 1B) and ELISA (Fig. 1C) experiments showed that CCL7 mRNA and protein levels were robustly up-regulated in the murine livers by LPS injection compared to saline injection. In the second model of (chronic) steatotic injury in which the mice were fed an MCD diet for 4 weeks, immunohistochemical staining detected marked increase in the number of CD68+ macrophages in the murine livers fed with the MCD diet compared to those fed the control diet (Fig. 1D). Consistently, qPCR (Fig. 1E) and Western (Fig. 1F) confirmed that CCL7 expression was dramatically up-regulated in the MCD-fed murine livers compared to the chow diet-fed livers. When hepatic expression levels of several other C–C motif ligand chemokines were examined, we noted that CCL2, CCl5, and CCL8 were significantly up-regulated to various extents in either the LPS model or the MCD model whereas CCL3 and CCL4 were not altered (Fig. S1).
Fig. 1.
Up-regulation of CCL7 in hepatocytes accompanies macrophage infiltration in the murine livers. (A-C) C57/BL6 mice were injected with LPS or saline as described in the Methods. Macrophages were stained with anti-CD68. CCL7 expression was examined by qPCR and ELISA. (D-F) C57/BL6 mice were fed an MCD diet or a control diet as described in the Methods. Macrophages were stained with anti-CD68. CCL7 expression was examined by qPCR and ELISA. (G) C57/BL6 mice were injected with LPS or saline as described in Methods. Primary hepatocytes, hepatic stellate cells, and Kupffer cells were isolated and CCL7 expression was examined by qPCR. N = 5 mice for each group. (H) C57/BL6 mice were fed an MCD diet or a control diet as described in Methods. Primary hepatocytes, hepatic stellate cells, and Kupffer cells were isolated and CCL7 expression was examined by qPCR. N = 5 mice for each group. Scale bar, 100 μm.
3.2. CCL7 expression is up-regulated by injurious stimuli in hepatocytes
Next, we determined the cellular origin(s) in which CCL7 levels were most significantly up-regulated by injurious stimuli. To this end, primary hepatocytes, hepatic stellate cells (HSCs), and F4/80+CD11b+ liver resident macrophages (Kupffer cells) were isolated from the murine livers by either gradient centrifugation or FACS sorting. As shown in Fig. 1G, CCL7 expression was up-regulated in both hepatocytes and Kupffer cells but not in HSCs following LPS injection. However, CCL7 expression was up-regulated by more than 6-fold in hepatocytes whereas it was only up-regulated by just over 2-fold in Kupffer cells. Similarly, in the model of steatotic liver injury, more than 7-fold increase in CCL7 expression was detected in hepatocytes isolated from the MCD diet-fed mice compared to those from the chow diet-fed mice. On the contrary, less than 2-fod increase in CCL7 expression was recorded in Kupffer cells whereas no significant changes in CCL7 expression were found in HSCs (Fig. 1H). Therefore, it appears that hepatocyte might be the major source from which CCL7 is derived in the injured liver.
3.3. Hepatocyte-specific BRG1 deficiency represses CCL7 expression and dampens macrophage infiltration in the murine livers
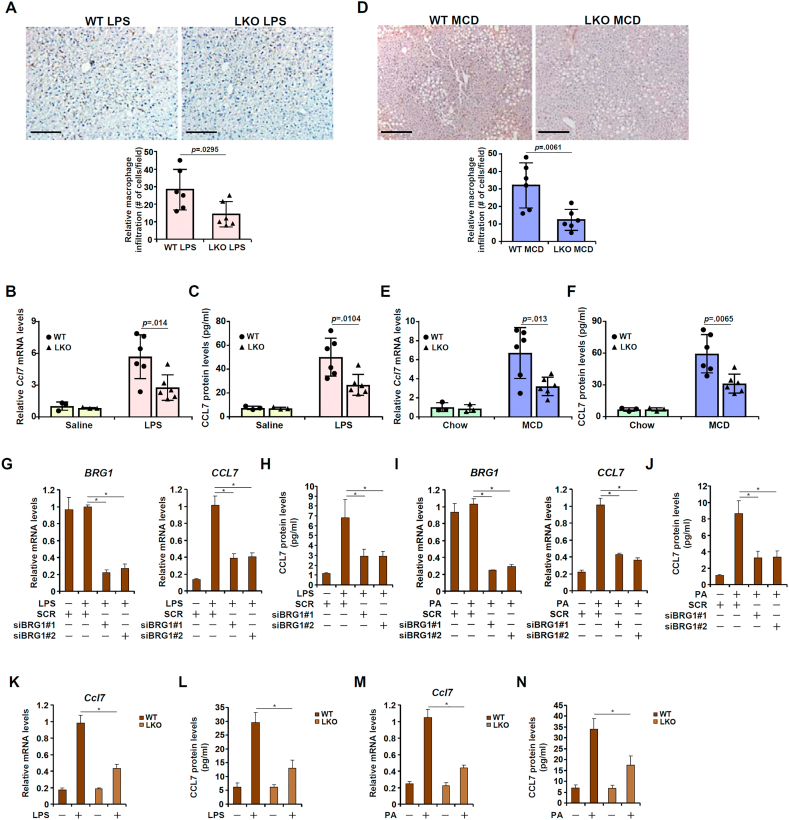
We have previously shown that BRG1, a chromatin remodeling protein, is a key mediator of liver injury [[34], [35], [36], [37],76]. We asked whether BRG1 might be involved in CCL7 induction and macrophage infiltration during liver injury. Hepatocyte-specific BRG1 knockout (LKO) mice and wild type (WT) littermates were injected with LPS or saline and sacrificed 12 h after the injection. Compared to the LPS-injected WT livers, there was a significant reduction in the number of infiltrated macrophages in the LPS-injected LKO livers (Fig. 2A). In addition, BRG1 deficiency down-regulated CC7 induction by 51 % as measured by qPCR (Fig. 2B) and by 44 % as measured by ELISA (Fig. 2C). In the second model wherein liver injury was induced by MCD diet feeding, BRG1 deficiency likewise dampened macrophage infiltration (Fig. 2D) and repressed CCL7 induction (Fig. 2E and F). Because CCL7 has also been noted for its role in monocyte chemotaxis, immunohistochemical staining was performed with an antibody that recognizes the monocyte surface marker Ly6C. The results indicated that BRG1 deficiency attenuated monocyte infiltration in the liver in both the LPS model and the MCD model (Fig. S2).
Fig. 2.
Hepatocyte-specific BRG1 deficiency represses CCL7 expression and dampens macrophage infiltration. (A-C) BRG1 conditional knockout (LKO) mice and wild type littermates were injected with LPS for 12 h. CCL7 expression was examined by qPCR and ELISA. Macrophages were stained with anti-CD68. (D-F) BRG1 conditional knockout (LKO) mice and wild type littermates were fed an MCD diet for 4 weeks. CCL7 expression was examined by qPCR and ELISA. Macrophages were stained with anti-CD68. (G, H) HepG2 cells were transfected with siRNA targeting BRG1 or scrambled siRNA (SCR) followed by treatment with LPS for 6 h. CCL7 expression was examined by qPCR and ELISA. (I, J) HepG2 cells were transfected with siRNA targeting BRG1 or scrambled siRNA (SCR) followed by treatment with palmitate for 12 h. CCL7 expression was examined by qPCR and ELISA. (K, L) Primary hepatocytes were isolated from WT or LKO mice and treated with or without LPS for 6 h. CCL7 expression was examined by qPCR and ELISA. (M, N) Primary hepatocytes were isolated from WT or LKO mice and treated with palmitate for 12 h. CCL7 expression was examined by qPCR and ELISA.
Next we examined the effect of BRG1 deficiency on CCL7 induction in hepatocytes in vitro. LPS treatment markedly stimulated CCL7 expression in HepG2 cells; BRG1 depletion by siRNA attenuated CCL7 induction by LPS as assessed by both qPCR (Fig. 2G) and ELISA (Fig. 2H). Similarly, exposure to palmitate (PA) also led to robust induction of CCL7 in HepG2 cells whereas BRG1 knockdown suppressed PA-induced CCL7 expression (Fig. 2I and J). Primary hepatocytes isolated from the LKO mice exhibit reduced sensitivity to LPS treatment by producing less CCL7 at the mRNA level (Fig. 2K) and the protein (Fig. 2L) level than those isolated from the WT mice. Finally, PA treatment up-regulated CCL7 expression less potently in primary hepatocytes isolated from the LKO mice than from the WT mice (Fig. 2M, N).
3.4. BRG1-dependent hepatocyte-derived CCL7 promotes macrophage migration
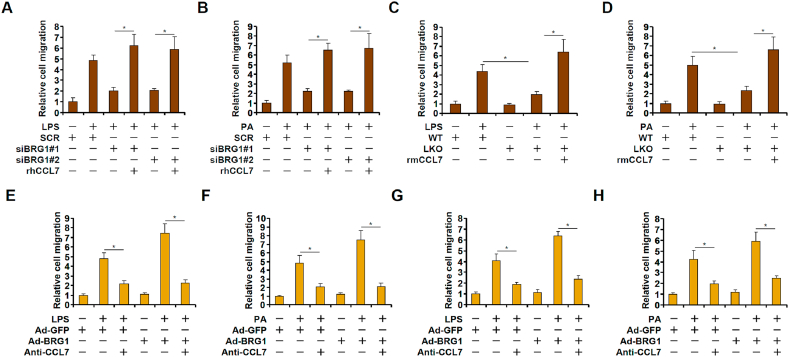
To determine whether induction of CCL7 by BRG1 might contribute to macrophage migration, we performed Boyden chamber transwell assay. Conditioned media (CM) collected from HepG2 cells treated with LPS (Fig. 3A) or PA (Fig. 3B) exhibited strong chemotacic activity to promote macrophage migration. BRG1 knockdown in HepG2 cells dulled the response of macrophages to the conditioned media following either LPS (Fig. 3A) or PA (Fig. 3B) treatment, which could be fully restored by the addition of recombinant CCL7. In a similar set of experiments, conditioned media were collected from primary hepatocytes isolated from the BRG1 LKO mice and the WT mice. The WT CM promoted the migration of macrophages better than the LKO CM; recombinant CCL7 revitalized the ability of the LKO CM bringing it up to the level of the WT CM (Fig. 3C and D). On the contrary, BRG1 over-expression enhanced the ability of LPS- or PA-treated CM to promote macrophage migration whereas an anti-CCL7 neutralizing antibody completely blocked the effect of BRG1 over-expression (Fig. 3E–H).
Fig. 3.
BRG1-dependent hepatocyte-derived CCL7 promotes macrophage migration. (A) HepG2 cells were transfected with siRNA targeting BRG1 or scrambled siRNA (SCR) followed by treatment with LPS for 6 h. Conditioned media were collected and chemotaxis assay was performed as described in Methods. (B) HepG2 cells were transfected with siRNA targeting BRG1 or scrambled siRNA (SCR) followed by treatment with palmitate for 12 h. Conditioned media were collected and chemotaxis assay was performed as described in Methods. (C) Primary hepatocytes were isolated from WT or LKO mice and treated with or without LPS for 6 h. Conditioned media were collected and chemotaxis assay was performed as described in Methods. (D) Primary hepatocytes were isolated from WT or LKO mice and treated with palmitate for 12 h. Conditioned media were collected and chemotaxis assay was performed as described in Methods. (E) HepG2 cells were infected with BRG1 adenovirus (Ad-BRG1) or the control adenovirus (Ad-GFP) followed by treatment with LPS for 6 h. Conditioned media were collected and chemotaxis assay was performed in the presence or absence of a CCL7-neutralizing antibody. (F) HepG2 cells were infected with BRG1 adenovirus (Ad-BRG1) or the control adenovirus (Ad-GFP) followed by treatment with palmitate for 12 h. Conditioned media were collected and chemotaxis assay was performed in the presence or absence of a CCL7-neutralizing antibody. (G) Primary hepatocytes were infected with BRG1 adenovirus (Ad-BRG1) or the control adenovirus (Ad-GFP) followed by treatment with LPS for 6 h. Conditioned media were collected and chemotaxis assay was performed in the presence or absence of a CCL7-neutralizing antibody. (H) Primary hepatocytes were infected with BRG1 adenovirus (Ad-BRG1) or the control adenovirus (Ad-GFP) followed by treatment with palmitate for 12 h. Conditioned media were collected and chemotaxis assay was performed in the presence or absence of a CCL7-neutralizing antibody.
3.5. BRG1-mediated CCL7 induction and macrophage infiltration is redox-sensitive
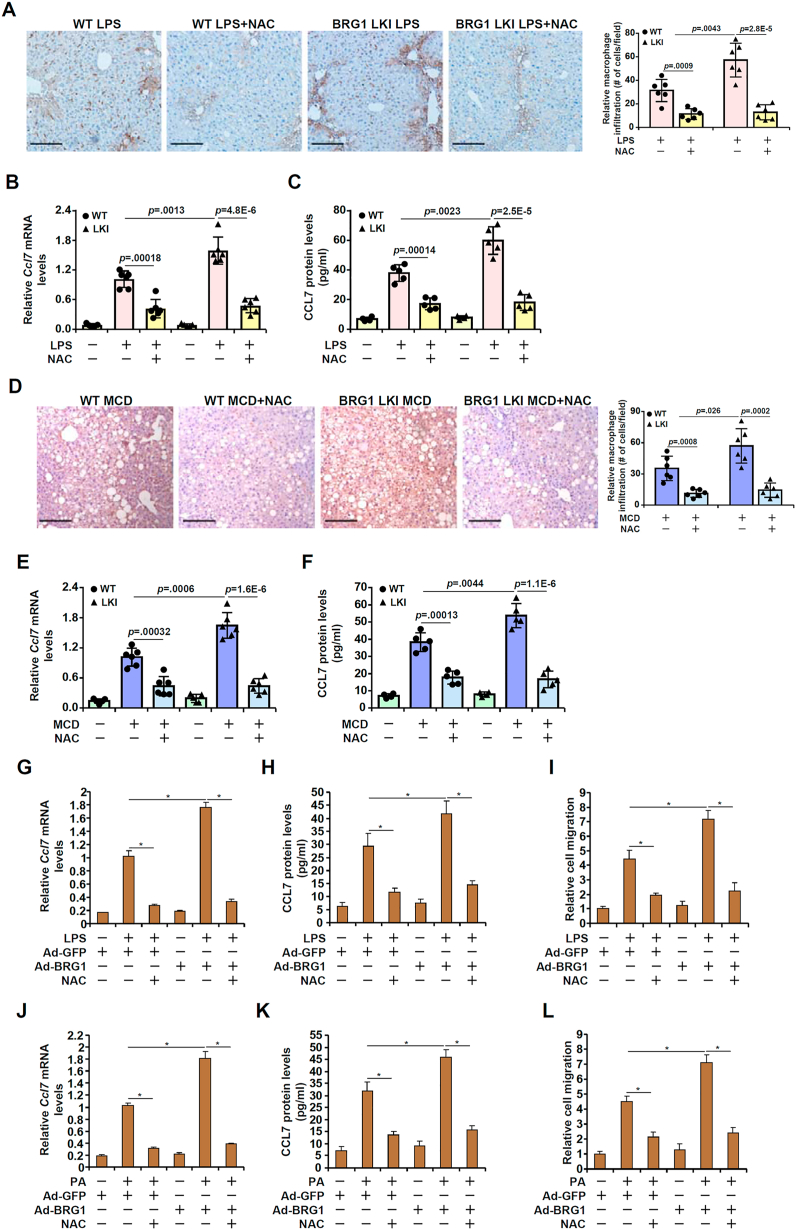
Mounting evidence suggests that BRG1 is involved in redox-sensitive regulation of cellular pathophysiological events. To verify whether BRG1-mediated CCL7 induction and macrophage infiltration is a function of cellular redox status, we generated a hepatocyte-specific BRG1 over-expression mouse strain (LKI) by crossing the RosaBrg1/+ strain [40] with the Alb-Cre strain. Compared to the control mice injected with LPS, more infiltrated macrophages were detected in the BRG1 LKI mice injected with LPS; injection with an antioxidant NAC neutralized the augmentative effect of BRG1 over-expression (Fig. 4A). Congruently, CCL7 expression was enhanced by BRG1 over-expression but dampened by NAC injection (Fig. 4B and C). Similar observations were made in the MCD model (Fig. 4D–F). In addition, over-expression of BRG1 in cultured hepatocytes potentiated the induction of CCL7 by LPS treatment (Fig. 4G–I) or PA treatment (Fig. 4J–L), which was attenuated by the addition of NAC.
Fig. 4.
BRG1-mediated CCL7 induction and macrophage infiltration is redox-sensitive. (A-C) BRG1 conditional knock-in (LKI) mice and wild type littermates were injected with LPS for 12 h with or without NAC. CCL7 expression was examined by qPCR and ELISA. Macrophages were stained with anti-CD68. (D-F) BRG1 conditional knock-in (LKI) mice and wild type littermates were fed an MCD diet for 4 weeks with or without NAC. CCL7 expression was examined by qPCR and ELISA. Macrophages were stained with anti-CD68. (G-I) Primary hepatocytes were infected with BRG1 adenovirus (Ad-BRG1) or the control adenovirus (Ad-GFP) followed by treatment with LPS and/or NAC. CCL7 expression was examined by qPCR and ELISA. Conditioned media were collected and chemotaxis assay was performed as described in Methods. (J-L) Primary hepatocytes were infected with BRG1 adenovirus (Ad-BRG1) or the control adenovirus (Ad-GFP) followed by treatment with palmitate and/or NAC. CCL7 expression was examined by qPCR and ELISA. Conditioned media were collected and chemotaxis assay was performed as described in Methods. Scale bar, 100 μm.
3.6. BRG1 interacts with AP-1 to activate CCL7 transcription
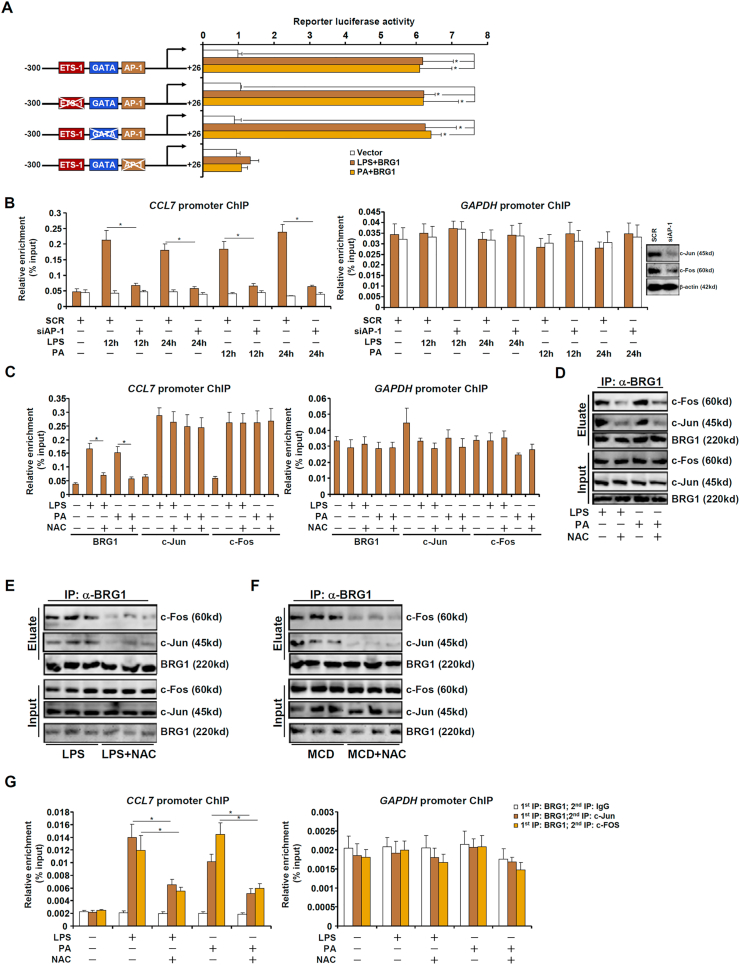
To further explore the mechanism whereby BRG1 contributes to the redox-sensitive induction of CCL7 transcription, a CCL7 promoter-luciferase construct (−300/+26) was transfected into HepG2 cells. BRG1 over-expression in the presence of LPS treatment or PA treatment greatly potentiated the CCL7 promoter activity (Fig. 5A). ChIP assay confirmed that LPS treatment or PA treatment promoted the association of BRG1 with the proximal CCL7 promoter but not the GAPDH promoter (Fig. 5B). Several binding motifs were identified within the CCL7 promoter and mutagenesis studies found that disruption of the most proximal AP-1 site completely abrogated the response of the CCL7 promoter to LPS or PA treatment whereas neither the ETS1 site nor the GATA site contributed to the induction of the CCL7 promoter activity (Fig. 5A). Indeed, depletion of endogenous AP-1 by siRNA blocked BRG1 recruitment to the CCL7 promoter (Fig. 5B). Of note, NAC treatment largely blocked the binding of BRG1 to the CCL7 promoter without altering AP-1 binding (Fig. 5C), suggesting that the interaction between BRG1 and AP-1 might be dependent on cellular redox status. Co-immunoprecipitation showed that NAC treatment dampened the association of BRG1 with AP-1 in both cultured hepatocytes (Fig. 5D) and in the murine livers (Fig. 5E and F). Importantly, treatment with LPS or PA promoted the formation of an AP-1-BRG1 complex on the CCL7 promoter, as judged by Re-ChIP assay, which became disassembled by the addition of NAC (Fig. 5G).
Fig. 5.
BRG1 interacts with AP-1 to activate CCL7 transcription in a redox-sensitive manner. (A) CCL7 promoter-luciferase constructs were transfected into HepG2 cells with or without BRG1 followed by treatment with LPS or PA. Luciferase activities were normalized by protein concentration and GFP fluorescence. (B) HepG2 cells were transfected with siRNA targeting AP-1 or scrambled siRNA (SCR) followed by treatment with LPS or PA. ChIP assays were performed with anti-BRG1 or IgG. (C) HepG2 cells were treated with LPS or PA in the presence or absence of NAC. ChIP assays were performed with anti-c-Jun, anti-c-Fos, or anti-BRG1. (D) HepG2 cells were treated with LPS or PA in the presence or absence of NAC. Immunoprecipitation was performed with anti-BRG1 or IgG. (E) C57/BL6 mice were injected with LPS in the presence or absence of NAC for 12 h. Immunoprecipitation was performed with anti-BRG1. (F) C57/BL6 mice were fed an MCD diet in the presence or absence of NAC for 4wk. Immunoprecipitation was performed with anti-BRG1. (G) HepG2 cells were treated with LPS or PA for 12 h. Re-ChIP assay was performed with indicated antibodies.
3.7. CK2-mediated BRG1 phosphorylation promotes its interaction with AP-1
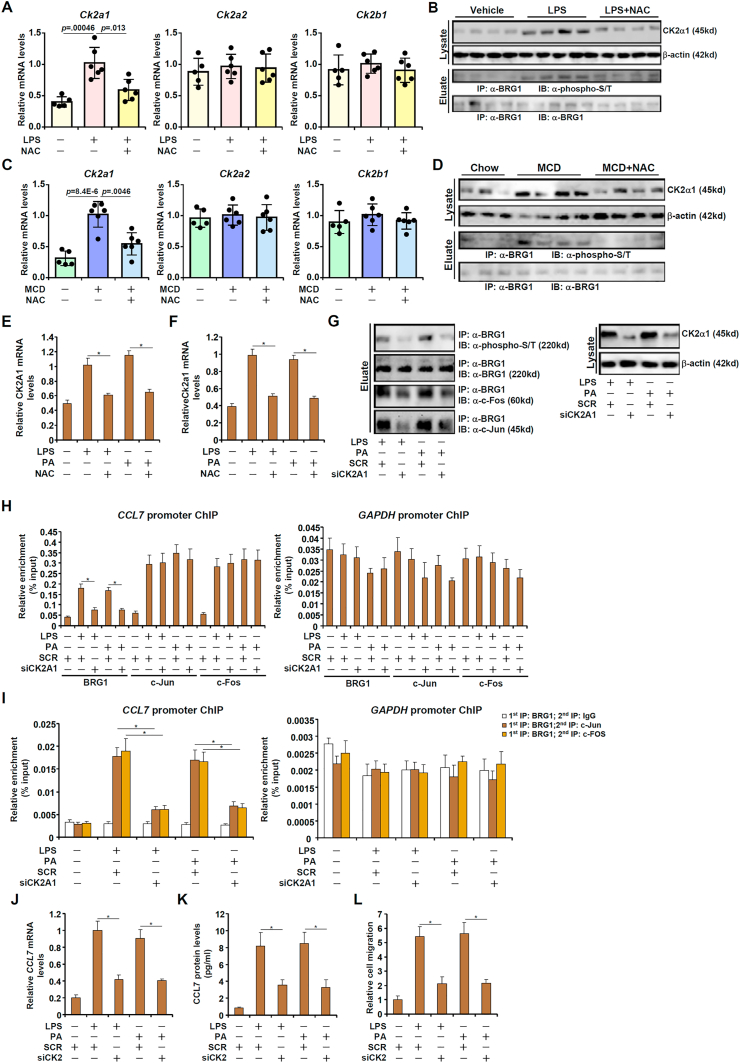
Several reports suggest that BRG1 can be phosphorylated by casein kinase 2. Of interest, expression of CK2A1, encoding one of the CK2 catalytic subunits, was up-regulated in the injured livers compared to the control livers whereas NAC administration suppressed CK2A1 induction (Fig. 6A–D). Inducible CK2A1 expression by injurious stimuli was also attenuated by NAC treatment in cultured hepatocytes (Fig. 6E and F). Of note, NAC treatment dampened ROS accumulation in the cells consistent with the changes in CK2A1 expression (Fig. S3). In keep with these observations, BRG1 phosphorylation levels appeared to be oscillating with CK2 expression in the liver (Fig. 6B and D). We therefore hypothesized that redox-sensitive CK2 induction may regulate BRG1 phosphorylation and its interaction with AP-1. A string of evidence supported this hypothesis: 1) CK2 knockdown inhibited BRG1 phosphorylation and disrupted the interaction between BRG1 and AP-1 (Fig.6G); 2) CK2 knockdown blocked the recruitment of BRG1 to the CCL7 promoter (Fig.6H); 3) CK2 knockdown interfered with the BRG1-AP-1 complex formation on the CCL7 promoter (Fig. 6I); and 4) CK2 knockdown repressed CCL7 expression and antagonized macrophage migration (Fig. 6J–L).
Fig. 6.
CK2-mediated BRG1 phosphorylation promotes its interaction with AP-1. (A, B) C57/BL6 mice were injected with LPS in the presence or absence of NAC for 12 h. CK2 expression levels in the liver were examined by qPCR and Western. Immunoprecipitation was performed with anti-BRG1. (C, D) C57/BL6 mice were fed an MCD diet in the presence or absence of NAC for 4wk. CK2 expression levels in the liver were examined by qPCR and Western. Immunoprecipitation was performed with anti-BRG1. (E) HepG2 cells were treated with LPS or PA in the presence or absence of NAC. CK2 expression levels were examined by qPCR. (F) Primary murine hepatocytes were treated with LPS or PA in the presence or absence of NAC. CK2 expression levels were examined by qPCR. (G) HepG2 cells were transfected with siRNA targeting CK2 or scrambled siRNA (SCR) followed by treatment with LPS or PA. Immunoprecipitation was performed with anti-BRG1. (H) HepG2 cells were transfected with siRNA targeting CK2 or scrambled siRNA (SCR) followed by treatment with LPS or PA. ChIP assay was performed with anti-BRG1. (I) HepG2 cells were transfected with siRNA targeting CK2 or scrambled siRNA (SCR) followed by treatment with LPS or PA. Re-ChIP assay was performed with indicated antibodies. (J-L) HepG2 cells were transfected with siRNA targeting CK2 or scrambled siRNA (SCR) followed by treatment with LPS or PA. CCL7 expression was examined by qPCR and ELISA. Conditioned media were collected and chemotaxis assay was performed as described in Methods.
3.8. Silmitasertib attenuates CCL7 expression and macrophage infiltration
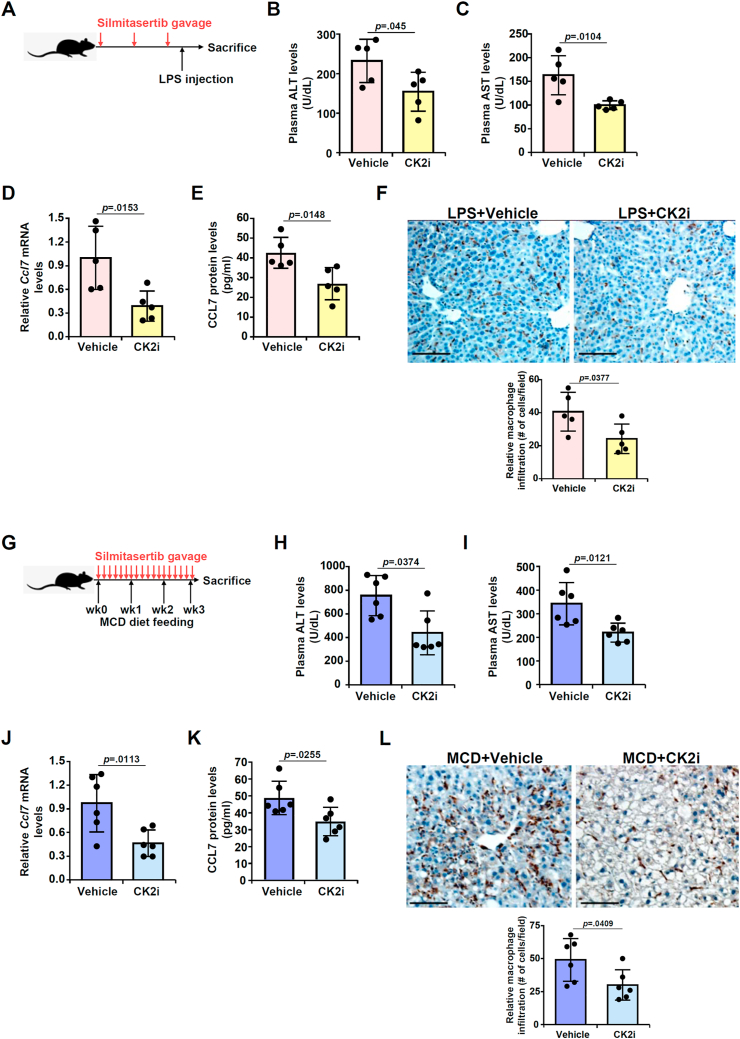
Next, we sought to determine whether inhibition of CK2 activity by administration of a specific CK2 inhibitor silmitasertib could influence CCL7 expression and macrophage infiltration in mice. To this end, silmitasertib was administered in C57/BL6 mice via oral gavage (Fig. 7A). Measurements of plasma ALT (Fig. 7B) and AST (Fig. 7C) levels indicate that CK2 inhibition slightly but significantly mitigated liver injury. More importantly, macrophage infiltration as evidenced by CD68 staining (Fig. 7D) as well as monocyte infiltration as evidenced by Ly6C staining (Fig. S4A) was attenuated by silmitasertib administration. This could be explained, at least in part, by a reduction in CCL7 expression (Fig. 7E and F). The effect of CK2 inhibition was also evaluated in a second model of liver injury induced by MCD feeding (Fig. 7G). Similar observations were made to suggest that CK2 inhibition by silmitasertib administration ameliorated liver injury (Fig. 7H and I), and blockaded macrophage infiltration (Fig. 7J) and monocyte infiltration (Fig. S4B), which were possibly attributable to repression of CCL7 expression (Fig. 7K, L).
Fig. 7.
CK2 inhibition by silmitasertib attenuates CCL7 expression and macrophage infiltration. (A-F) C57/BL6 mice were injected with LPS in the presence or absence of silmitasertib. Scheme of protocol (A). Plasma ALT (B) and AST (C) levels were examined by colorimetric kits. Macrophages were stained with anti-CD68 (D). CCL7 expression was examined by qPCR (E) and ELISA (F). (G-L) C57/BL6 mice were fed the MCD diet in the presence or absence of silmitasertib. Scheme of protocol (G). Plasma ALT (H) and AST (I) levels were examined by colorimetric kits. Macrophages were stained with anti-CD68 (J). CCL7 expression was examined by qPCR (K) and ELISA (L). Scale bar, 50 μm.
3.9. Correlation between BRG1 expression, CCL7 expression and hepatic macrophage accumulation in humans
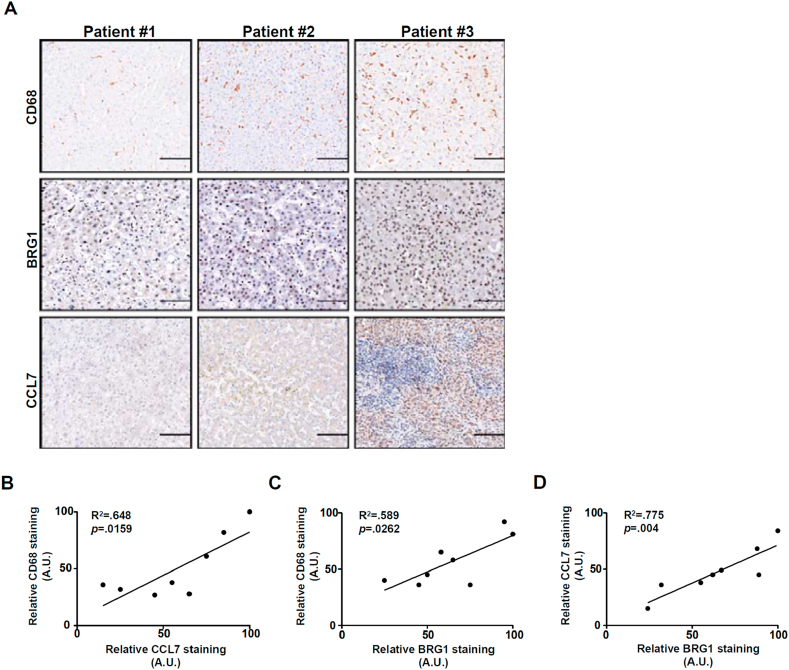
We finally verified the pathophysiological relevance of our model in which BRG1 induced CCL7 expression promotes macrophage recruitment in liver biopsy specimens of non-alcoholic steatohepatitis (NASH) patients. As shown in Fig. 8A, immunohistochemical staining showed that there generally appeared to be a trend where more macrophage infiltration was detected in the liver that exhibited stronger CCL7 expression. Linear regression analysis revealed a significant positive correlation between CCL7 expression, BRG1 expression, and CD68+ macrophage accumulation in the NASH livers (Fig. 8B–D).
Fig. 8.
Correlation between BRG1, CCL7 and macrophage infiltration in humans. (A) Representative images of CD68 staining, BRG1 staining, and CCL7 staining in steatotic human liver biopsy specimens. (B-D) Linear regression was performed with Graphpad Prism. Scale bar, 200 μm.
4. Discussion
Macrophage infiltration is a hallmark event in a wide range of liver pathologies that encompass viral hepatitis [77], steatohepatitis [78], cholestasis [79], and ischemia-reperfusion injury [80]. Chemokines play a crucial role in navigating macrophages to the liver to participate in the modulation of liver injuries. Here we report that the chromatin remodeling protein BRG1 interacts with AP-1 to activate the transcription of CCL7, which in turn promotes macrophage accumulation in the liver. The present study leaves open several key questions with regard to the regulation of macrophage behavior by CCL7. First, it remains to be determined whether CCL7 is mandatory for macrophage infiltration in the process of liver injury. CCL7-null mice are viable and can be distinguished from their WT littermates by the dramatic decrease in peripheral 7/4briLy-6G– leukocytes [81]. Further analyses have revealed an essential role for CCL7 in macrophage recruitment in mouse models of obstructive nephropathy [82], cutaneous leishmaniasis [83], and colitis [84]. Future studies should employ a CCL7 knockout strain, ideally spatiotemporally controllable, to investigate its role in hepatic macrophage trafficking. Second, it is unclear whether immune cell lineages other than macrophages can be modulated by CCL7 induction in liver injury. Mercer et al. have demonstrated that intranasal administration of recombinant CCL7 promotes pulmonary neutrophil infiltration in a mouse model of acute lung injury [85]. Zhang et al. have observed that CCL7 may play a role in eosinophil recruitment to the nasal mucosa in a model of allergic rhinitis [86]. Intratumoral CCL7 over-expression is also implicated in the recruitment of T lymphocytes, dendritic cells, and natural killer (NK) cells [87]. A comprehensive profiling of immune cell sub-populations in the CCL7-null livers in the setting of liver injury would help clarify its role in hepatic immunity.
Mounting evidence suggests that BRG1 exerts immunomodulatory effects in the pathogenesis of human diseases. Whereas it is abundantly clear that BRG1 can directly regulate the autonomous behaviors of immune cells, BRG1 can also influence the phenotype of immune cells non-autonomously via cell-cell crosstalk by producing and releasing immunomodulatory substances from non-immune cells [[88], [89], [90]]. We have previously demonstrated that BRG1 activates two different endothelium-derived chemokines, colony stimulating factor 1 (CSF1) and migration inhibitory factor-related protein 8 (MRP8), to regulate macrophage migration in the context of aortic aneurysm [91] and cardiac hypertrophy [58], respectively. More recently, Liu et al. have discovered that intestinal epithelial cell (IEC) conditional BRG1 knockout mice develop spontaneous colitis owing to augmented expression of chemokines in IECs and increased intestinal infiltration of macrophages/lymphocytes/neutrophils [40]. While our finding here that BRG1-induced CCL7 in hepatocyte is associated with macrophage recruitment in liver injury certainly adds another layer of regulation by which BRG1 contributes to immune cell trafficking, it begs for far more questions than it intended to answer. For instance, whether these BRG1-induced chemoattractive substances, CCL7 included, act in tandem or in parallel is not clear. Whether these proteins act redundantly and can substitute for one another is equally ambiguous and needs further clarification.
Our data suggest that the ability of BRG1 to control CCL7 production and, by extension, hepatic macrophage trafficking relies on CK2-mediated phosphorylation. This observation, however, does not foreclose the possibility that other post-translational modifications may regulate this process. Adenuga et al. have reported that CK2 can directly phosphorylate HDAC2 to target HDAC2 for proteasomal degradation, which unleashes pro-inflammatory response in the lungs in response to cigarette smoking [92]. Alternatively, Cho et al. have shown that CK2-mediated repression of Ikaros leads to up-regulation of iNOS, a prototypical inflammatory mediator [93]. There is also evidence to suggest that phosphorylation of liver X receptor (LXR) by CK2 contributes to altered expression of CCL24, a remote sibling of CCL7, in macrophages [94]. A phosphoproteomics-based approach applied in beige adipocytes has uncovered key CK2 substrates that may contribute to thermogenesis [95]. Similar strategies should be considered to systemically examine the dynamic spectrum of CK2 substrates involved in the regulation of hepatic inflammation.
Here we show that CK2 inhibition is associated with diminished macrophage infiltration in the murine livers likely owing to reduced CCL7 expression (Fig. 7). This observation echoes previous reports that point to a key role for CK2 in the regulation of liver homeostasis. Choi et al. have reported that obesity induced CK2 promotes SIRT1 phosphorylation and its nuclear exclusion thereby leading to steatosis [96]. CK2-mediated phosphorylation of farnesoid X receptor (FXR), a master regulator of hepatic metabolism, couples FXR SUMOylation to its ubiquitination and degradation thus skewing bile acid metabolism and liver regeneration [97]. It is of intrigue to note that there appears to be a striking similarity in terms of the pathobiological processes that BRG1 and CK2 seem to co-regulate although no prior study has ever linked one to the other. BRG1 and CK2 can both regulate SREBP-dependent lipogenesis [35,98], liver regeneration [32,99], and hepatocellular carcinoma [100,101]. Further studies are warranted to address whether there is a universal co-dependency between BRG1 and CK2 in liver diseases.
There are several major limitations regarding the present study. First, hepatic macrophages could be derived from circulating monocytes as well as from in situ proliferation of resident macrophages (Kupffer cell). Because the method used in the present study (CD68 immunostaining) does not differentiate between myelomonocytic cell-derived and Kupffer cell-derived macrophages, the contribution of these two sub-populations of cells to hepatic inflammation cannot be ascertained. Equally, the effect of CCL7 on the phenotype and function of these cells remains ambiguous. Second, the conclusion that newly synthesized CCL7 in the course of liver injury mainly originates from hepatocytes is based on the freshly isolated primary murine hepatocytes. Many primary cells undergo rapid transcriptomic and phenotypic alterations once detached from the in vivo microenvironment. Therefore, more accurate techniques (e.g., lineage tracing or in situ hybridization) should be employed to confirm this finding. Third, no effort was made in the present study to place this BRG1-CCL7 axis in a broader context. Recent investigations harnessing next-gen sequencing techniques have generated a plethora of datasets encompassing a wide range of liver pathologies [102,103]. Mining these publicly available datasets may bring novel perspective to the model as proposed here. Fourth, several other C–C motif ligand chemokines, in addition to CCL7, were up-regulated in the animal models examined. Whether these chemokines are similarly regulated by BRG1 in a redox-sensitive and CK2-dependent manner remains to be investigated. Until and unless this issue is clarified, the assertion that CCL7 trans-activation alone is sufficient to account for hepatic inflammation cannot be ascertained. Finally, it was also observed that infiltration of monocytes in the liver was similarly influenced by BRG1 deficiency (Fig. S2) and CK2 inhibition (Fig. S4) as macrophages, pointing to the possibility that multiple different immune cell populations may be influenced by BRG1 and/or CK2 collectively contributing to the dynamic composition of hepatic inflammatory milieu. A comprehensive profiling of the immune cell makeup in the liver under these experimental conditions would be of great help to determine the precise role of BRG1 and/or CK2 in the modulation of hepatic inflammation.
In summary, we identify CCL7 as a novel transcriptional target for BRG1 in hepatocytes. Dual targeting of the CCL7 siblings CCL2 and CCL5 has been proved effective in pre-clinical models of liver injury [25]. Our data likely fuel renewed incentive for screening small-molecule CCL7 inhibitors/antagonists in the attempt to rein in hepatic inflammation and ameliorate liver injury.
Declaration of competing interest
None.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81725001, 81971504, and 81700554), Post-doctoral Special Foundation of China (2020M670065ZX), Post-Doctoral Foundation of Jiangsu Province (2020Z021), and Changzhou Society Development Funding (CE20205038).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102079.
Contributor Information
Yunjie Lu, Email: yjluforresearch@126.com.
Zilong Li, Email: Miracle_lee1114@sina.com.
Yong Xu, Email: yjxu@njmu.edu.cn.
Data availability statement
The data that support the findings of this study are available upon reasonable request.
Author contributions
ZL Li and Y Xu conceived the project; all authors designed experiments; M Kong, WH Dong, YW Zhu, ZW Fan, XL Miao, Y Guo, CP Li, ZL Li, YF Duan, and YJ Lu performed experiments, collected data, and analyzed data; Y Xu and ZL Li wrote the manuscript; YJ Lu secured funding; Y Xu provided supervision.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Robinson M.W., Harmon C., O'Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell. Mol. Immunol. 2016;13(3):267–276. doi: 10.1038/cmi.2016.3. Epub 2016/04/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernsmeier C., van der Merwe S., Perianin A. Innate immune cells in cirrhosis. J. Hepatol. 2020;73(1):186–201. doi: 10.1016/j.jhep.2020.03.027. Epub 2020/04/03. [DOI] [PubMed] [Google Scholar]
- 3.Gao B., Jeong W.I., Tian Z. Liver: an organ with predominant innate immunity. Hepatology. 2008;47(2):729–736. doi: 10.1002/hep.22034. Epub 2008/01/02. [DOI] [PubMed] [Google Scholar]
- 4.Seki E., Schwabe R.F. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61(3):1066–1079. doi: 10.1002/hep.27332. Epub 2014/07/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S., Tan H.Y., Wang N., Feng Y., Wang X. Recent insights into the role of immune cells in alcoholic liver disease. Front. Immunol. 2019;10:1328. doi: 10.3389/fimmu.2019.01328. Epub 2019/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wajant H., Siegmund D. TNFR1 and TNFR2 in the control of the life and death balance of macrophages. Front. cell Develop. Biol. 2019;7:91. doi: 10.3389/fcell.2019.00091. Epub 2019/06/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shan Z., Ju C. Hepatic macrophages in liver injury. Front. Immunol. 2020;11:322. doi: 10.3389/fimmu.2020.00322. Epub 2020/05/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao B., Ahmad M.F., Nagy L.E., Tsukamoto H. Inflammatory pathways in alcoholic steatohepatitis. J. Hepatol. 2019;70(2):249–259. doi: 10.1016/j.jhep.2018.10.023. Epub 2019/01/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oates J.R., McKell M.C., Moreno-Fernandez M.E., Damen M., Deepe G.S., Jr., Qualls J.E. Macrophage function in the pathogenesis of non-alcoholic fatty liver disease: the Mac Attack. Front. Immunol. 2019;10:2893. doi: 10.3389/fimmu.2019.02893. Epub 2020/01/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmermann H.W., Trautwein C., Tacke F. Functional role of monocytes and macrophages for the inflammatory response in acute liver injury. Front. Physiol. 2012;3:56. doi: 10.3389/fphys.2012.00056. Epub 2012/10/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng X., Liu G., Peng W., He J., Cai C., Xiong W. Combined deficiency of SLAMF8 and SLAMF9 prevents endotoxin-induced liver inflammation by downregulating TLR4 expression on macrophages. Cell. Mol. Immunol. 2020;17(2):153–162. doi: 10.1038/s41423-018-0191-z. Epub 2018/12/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canbay A., Feldstein A.E., Higuchi H., Werneburg N., Grambihler A., Bronk S.F. Kupffer cell engulfment of apoptotic bodies stimulates death ligand and cytokine expression. Hepatology. 2003;38(5):1188–1198. doi: 10.1053/jhep.2003.50472. Epub 2003/10/28. [DOI] [PubMed] [Google Scholar]
- 13.Reiling J., Bridle K.R., Schaap F.G., Jaskowski L., Santrampurwala N., Britton L.J. The role of macrophages in the development of biliary injury in a lipopolysaccharide-aggravated hepatic ischaemia-reperfusion model. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2018;1864(4 Pt B):1284–1292. doi: 10.1016/j.bbadis.2017.06.028. Epub 2017/07/16. [DOI] [PubMed] [Google Scholar]
- 14.Rivera C.A., Adegboyega P., van Rooijen N., Tagalicud A., Allman M., Wallace M. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J. Hepatol. 2007;47(4):571–579. doi: 10.1016/j.jhep.2007.04.019. Epub 2007/07/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng B., Jiao P., Nie Y., Kim T., Jun D., van Rooijen N. Clodronate liposomes improve metabolic profile and reduce visceral adipose macrophage content in diet-induced obese mice. PloS One. 2011;6(9) doi: 10.1371/journal.pone.0024358. Epub 2011/09/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saiman Y., Friedman S.L. The role of chemokines in acute liver injury. Front. Physiol. 2012;3:213. doi: 10.3389/fphys.2012.00213. Epub 2012/06/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marra F., Tacke F. Roles for chemokines in liver disease. Gastroenterology. 2014;147(3):577–594 e1. doi: 10.1053/j.gastro.2014.06.043. Epub 2014/07/30. [DOI] [PubMed] [Google Scholar]
- 18.Bachelerie F., Ben-Baruch A., Burkhardt A.M., Combadiere C., Farber J.M., Graham G.J. International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol. Rev. 2014;66(1):1–79. doi: 10.1124/pr.113.007724. Epub 2013/11/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nibbs R.J., Graham G.J. Immune regulation by atypical chemokine receptors. Nat. Rev. Immunol. 2013;13(11):815–829. doi: 10.1038/nri3544. Epub 2013/12/10. [DOI] [PubMed] [Google Scholar]
- 20.Seki E., De Minicis S., Gwak G.Y., Kluwe J., Inokuchi S., Bursill C.A. CCR1 and CCR5 promote hepatic fibrosis in mice. J. Clin. Invest. 2009;119(7):1858–1870. doi: 10.1172/JCI37444. Epub 2009/07/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seki E., de Minicis S., Inokuchi S., Taura K., Miyai K., van Rooijen N. CCR2 promotes hepatic fibrosis in mice. Hepatology. 2009;50(1):185–197. doi: 10.1002/hep.22952. Epub 2009/05/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi D.Y., Ban J.O., Kim S.C., Hong J.T. CCR5 knockout mice with C57BL6 background are resistant to acetaminophen-mediated hepatotoxicity due to decreased macrophages migration into the liver. Arch. Toxicol. 2015;89(2):211–220. doi: 10.1007/s00204-014-1253-3. Epub 2014/04/29. [DOI] [PubMed] [Google Scholar]
- 23.Heymann F., Hammerich L., Storch D., Bartneck M., Huss S., Russeler V. Hepatic macrophage migration and differentiation critical for liver fibrosis is mediated by the chemokine receptor C-C motif chemokine receptor 8 in mice. Hepatology. 2012;55(3):898–909. doi: 10.1002/hep.24764. Epub 2011/10/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamoto N., Ebinuma H., Kanai T., Chu P.S., Ono Y., Mikami Y. CCR9+ macrophages are required for acute liver inflammation in mouse models of hepatitis. Gastroenterology. 2012;142(2):366–376. doi: 10.1053/j.gastro.2011.10.039. Epub 2011/11/15. [DOI] [PubMed] [Google Scholar]
- 25.Ambade A., Lowe P., Kodys K., Catalano D., Gyongyosi B., Cho Y. Pharmacological inhibition of CCR2/5 signaling prevents and reverses alcohol-induced liver damage, steatosis, and inflammation in mice. Hepatology. 2019;69(3):1105–1121. doi: 10.1002/hep.30249. Epub 2018/09/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proost P., Wuyts A., Van Damme J. Human monocyte chemotactic proteins-2 and -3: structural and functional comparison with MCP-1. J. Leukoc. Biol. 1996;59(1):67–74. doi: 10.1002/jlb.59.1.67. Epub 1996/01/01. [DOI] [PubMed] [Google Scholar]
- 27.Cai S.Y., Ouyang X., Chen Y., Soroka C.J., Wang J., Mennone A. Bile acids initiate cholestatic liver injury by triggering a hepatocyte-specific inflammatory response. JCI insight. 2017;2(5) doi: 10.1172/jci.insight.90780. Epub 2017/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandrekar P., Ambade A., Lim A., Szabo G., Catalano D. An essential role for monocyte chemoattractant protein-1 in alcoholic liver injury: regulation of proinflammatory cytokines and hepatic steatosis in mice. Hepatology. 2011;54(6):2185–2197. doi: 10.1002/hep.24599. Epub 2011/08/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galastri S., Zamara E., Milani S., Novo E., Provenzano A., Delogu W. Lack of CC chemokine ligand 2 differentially affects inflammation and fibrosis according to the genetic background in a murine model of steatohepatitis. Clin. Sci. (Lond.) 2012;123(7):459–471. doi: 10.1042/CS20110515. Epub 2012/05/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khavari P.A., Peterson C.L., Tamkun J.W., Mendel D.B., Crabtree G.R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366(6451):170–174. doi: 10.1038/366170a0. Epub 1993/11/11. [DOI] [PubMed] [Google Scholar]
- 31.Trotter K.W., Archer T.K. The BRG1 transcriptional coregulator. Nucl. Recept. Signal. 2008;6:e004. doi: 10.1621/nrs.06004. Epub 2008/02/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li N., Kong M., Zeng S., Hao C., Li M., Li L. Brahma related gene 1 (Brg1) contributes to liver regeneration by epigenetically activating the Wnt/beta-catenin pathway in mice. Faseb. J. : official publication of the Federation of American Societies for Experimental Biology. 2019;33(1):327–338. doi: 10.1096/fj.201800197R. Epub 2018/07/13. [DOI] [PubMed] [Google Scholar]
- 33.Wang B., Kaufmann B., Engleitner T., Lu M., Mogler C., Olsavszky V. Brg1 promotes liver regeneration after partial hepatectomy via regulation of cell cycle. Sci. Rep. 2019;9(1):2320. doi: 10.1038/s41598-019-38568-w. Epub 2019/02/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L., Hong W., Li M., Ren H., Wang J., Xu H. A cross talk between BRG1 and males absent on the first contributes to reactive oxygen species production in a mouse model of nonalcoholic steatohepatitis. Antioxidants Redox Signal. 2019;30(12):1539–1552. doi: 10.1089/ars.2016.6822. Epub 2018/07/03. [DOI] [PubMed] [Google Scholar]
- 35.Li N., Li M., Hong W., Shao J., Xu H., Shimano H. Brg1 regulates pro-lipogenic transcription by modulating SREBP activity in hepatocytes. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2018;1864(9 Pt B):2881–2889. doi: 10.1016/j.bbadis.2018.05.022. Epub 2018/06/02. [DOI] [PubMed] [Google Scholar]
- 36.Li N., Kong M., Zeng S., Xu Z., Li M., Hong W. The chromatin remodeling protein BRG1 regulates APAP-induced liver injury by modulating CYP3A11 transcription in hepatocyte. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2018;1864(10):3487–3495. doi: 10.1016/j.bbadis.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Kong M., Chen X., Xu H., Wenping, Fang M., Xu Y. Hepatocyte-specific deletion of Brg1 alleviates methionine-and-choline-deficient diet (MCD) induced non-alcoholic steatohepatitis in mice. Biochem. Biophys. Res. Commun. 2018;503(1):344–351. doi: 10.1016/j.bbrc.2018.06.027. Epub 2018/06/12. [DOI] [PubMed] [Google Scholar]
- 38.Dong W., Kong M., Zhu Y., Shao Y., Wu D., Lu J. Activation of TWIST transcription by chromatin remodeling protein BRG1 contributes to liver fibrosis in mice. Front. cell Develop. Biol. 2020;8:340. doi: 10.3389/fcell.2020.00340. Epub 2020/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan Z., Kong M., Li M., Hong W., Fan X., Xu Y. Brahma related gene 1 (Brg1) regulates cellular cholesterol Synthesis by acting as a Co-factor for SREBP2. Front. cell Develop. Biol. 2020;8:259. doi: 10.3389/fcell.2020.00259. Epub 2020/06/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu M., Sun T., Li N., Peng J., Fu D., Li W. BRG1 attenuates colonic inflammation and tumorigenesis through autophagy-dependent oxidative stress sequestration. Nat. Commun. 2019;10(1):4614. doi: 10.1038/s41467-019-12573-z. Epub 2019/10/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamesch K., Borkham-Kamphorst E., Strnad P., Weiskirchen R. Lipopolysaccharide-induced inflammatory liver injury in mice. Lab. Anim. 2015;49(1 Suppl):37–46. doi: 10.1177/0023677215570087. Epub 2015/04/04. [DOI] [PubMed] [Google Scholar]
- 42.Kong M., Hong W., Shao Y., Lv F., Fan Z., Li P. Ablation of serum response factor in hepatic stellate cells attenuates liver fibrosis. J. Mol. Med. (Berl.) 2019;97(11):1521–1533. doi: 10.1007/s00109-019-01831-8. Epub 2019/08/23. [DOI] [PubMed] [Google Scholar]
- 43.Kong M., Chen X., Lv F., Ren H., Fan Z., Qin H. Serum response factor (SRF) promotes ROS generation and hepatic stellate cell activation by epigenetically stimulating NCF1/2 transcription. Redox biology. 2019;26:101302. doi: 10.1016/j.redox.2019.101302. Epub 2019/08/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan Z., Li N., Xu Z., Wu J., Fan X., Xu Y. An interaction between MKL1, BRG1, and C/EBPbeta mediates palmitate induced CRP transcription in hepatocytes. Biochimica et biophysica acta Gene regulatory mechanisms. 2019;1862(9):194412. doi: 10.1016/j.bbagrm.2019.194412. Epub 2019/07/30. [DOI] [PubMed] [Google Scholar]
- 45.Murakami K., Nomiyama H., Miura R., Follens A., Fiten P., Van Coillie E. Structural and functional analysis of the promoter region of the human MCP-3 gene: transactivation of expression by novel recognition sequences adjacent to the transcription initiation site. DNA Cell Biol. 1997;16(2):173–183. doi: 10.1089/dna.1997.16.173. Epub 1997/02/01. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y., Liu L., Li M., Cheng X., Fang M., Zeng Q. The chromatin remodeling protein BRG1 links ELOVL3 trans-activation to prostate cancer metastasis. Biochimica et biophysica acta Gene regulatory mechanisms. 2019;1862(8):834–845. doi: 10.1016/j.bbagrm.2019.05.005. Epub 2019/06/04. [DOI] [PubMed] [Google Scholar]
- 47.Yang Y., Liu L., Fang M., Bai H., Xu Y. The chromatin remodeling protein BRM regulates the transcription of tight junction proteins: implication in breast cancer metastasis. Biochimica et biophysica acta Gene regulatory mechanisms. 2019;1862(5):547–556. doi: 10.1016/j.bbagrm.2019.03.002. Epub 2019/04/05. [DOI] [PubMed] [Google Scholar]
- 48.Weng X., Zhang Y., Li Z., Yu L., Xu F., Fang M. Class II transactivator (CIITA) mediates IFN-gamma induced eNOS repression by enlisting SUV39H1. Biochimica et biophysica acta Gene regulatory mechanisms. 2019;1862(2):163–172. doi: 10.1016/j.bbagrm.2019.01.005. Epub 2019/02/05. [DOI] [PubMed] [Google Scholar]
- 49.Shao J., Weng X., Zhuo L., Yu L., Li Z., Shen K. Angiotensin II induced CSF1 transcription is mediated by a crosstalk between different epigenetic factors in vascular endothelial cells. Biochimica et biophysica acta Gene regulatory mechanisms. 2019;1862(1):1–11. doi: 10.1016/j.bbagrm.2018.10.001. Epub 2018/10/15. [DOI] [PubMed] [Google Scholar]
- 50.Chen B., Fan Z., Sun L., Chen J., Feng Y., Fan X. Epigenetic activation of the small GTPase TCL contributes to colorectal cancer cell migration and invasion. Oncogenesis. 2020;9(9):86. doi: 10.1038/s41389-020-00269-9. Epub 2020/10/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mao L., Liu L., Zhang T., Qin H., Wu X., Xu Y. Histone deacetylase 11 contributes to renal fibrosis by repressing KLF15 transcription. Front. cell Develop. Biol. 2020;8:235. doi: 10.3389/fcell.2020.00235. Epub 2020/05/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Y., Lv F., Kong M., Chen X., Duan Y., Sun D. A cAbl-MRTF-A feedback loop contributes to hepatic stellate cell activation. Front. cell Develop. Biol. 2019;7:243. doi: 10.3389/fcell.2019.00243. Epub 2019/11/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z., Kong X., Zhang Y., Yu L., Guo J., Xu Y. Dual roles of chromatin remodeling protein BRG1 in angiotensin II-induced endothelial-mesenchymal transition. Cell Death Dis. 2020;11(7):549. doi: 10.1038/s41419-020-02744-y. Epub 2020/07/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y., Yang G., Yu L., Lin L., Liu L., Fang M. An interplay between MRTF-A and the histone acetyltransferase TIP60 mediates hypoxia-reoxygenation induced iNOS transcription in macrophages. Front. cell Develop. Biol. 2020;8:484. doi: 10.3389/fcell.2020.00484. Epub 2020/07/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Q., Yang J., Chen H., Li J., Que L., Zhu G. Peli1 induction impairs cardiac microvascular endothelium through Hsp90 dissociation from IRE1alpha. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2019;1865(10):2606–2617. doi: 10.1016/j.bbadis.2019.06.017. Epub 2019/07/02. [DOI] [PubMed] [Google Scholar]
- 56.Liu L., Mao L., Wu X., Wu T., Liu W., Yang Y. BRG1 regulates endothelial-derived IL-33 to promote ischemia-reperfusion induced renal injury and fibrosis in mice. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2019;1865(9):2551–2561. doi: 10.1016/j.bbadis.2019.06.015. Epub 2019/06/23. [DOI] [PubMed] [Google Scholar]
- 57.Lv F., Li N., Kong M., Wu J., Fan Z., Miao D. CDKN2a/p16 antagonizes hepatic stellate cell activation and liver fibrosis by modulating ROS levels. Front. cell Develop. Biol. 2020;8:176. doi: 10.3389/fcell.2020.00176. Epub 2020/04/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z., Zhang Y., Yu L., Xiao B., Li T., Kong X. BRG1 stimulates endothelial derived alarmin MRP8 to promote macrophage infiltration in an animal model of cardiac hypertrophy. Front. cell Develop. Biol. 2020;8:569. doi: 10.3389/fcell.2020.00569. Epub 2020/08/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Z., Xia J., Fang M., Xu Y. Epigenetic regulation of lung cancer cell proliferation and migration by the chromatin remodeling protein BRG1. Oncogenesis. 2019;8(11):66. doi: 10.1038/s41389-019-0174-7. Epub 2019/11/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z., Chen B., Dong W., Kong M., Shao Y., Fan Z. The chromatin remodeler Brg1 integrates ROS production and endothelial-mesenchymal transition to promote liver fibrosis in mice. Front Dev Cell Biol. 2019;7:245. doi: 10.3389/fcell.2019.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z., Chen B., Dong W., Kong M., Fan Z., Yu L. MKL1 promotes endothelial-to-mesenchymal transition and liver fibrosis by activating TWIST1 transcription. Cell Death Dis. 2019;10(12):899. doi: 10.1038/s41419-019-2101-4. Epub 2019/11/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun L., Chen B., Wu J., Jiang C., Fan Z., Feng Y. Epigenetic regulation of a disintegrin and metalloproteinase (ADAM) promotes colorectal cancer cell migration and invasion. Front. cell Develop. Biol. 2020;(8):581692. doi: 10.3389/fcell.2020.581692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen B., Yuan Y., Sun L., Chen J., Yang M., Yin Y. MKL1 mediates TGF-β induced RhoJ transcription to promote breast cancer cell migration and invasion. Front. cell Develop. Biol. 2020;8:832. doi: 10.3389/fcell.2020.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li N., Liu S., Zhang Y., Yu L., Hu Y., Wu T. Transcriptional activation of matricellular protein Spondin2 (SPON2) by BRG1 in vascular endothelial cells promotes macrophage chemotaxis. Front. cell Develop. Biol. 2020;8:794. doi: 10.3389/fcell.2020.00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen B., Zhao Q., Xu T., Yu L., Zhuo L., Yang Y. BRG1 activates PR65A transcription to regulate NO bioavailability in vascular endothelial cell. Front. cell Develop. Biol. 2020;8:774. doi: 10.3389/fcell.2020.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Y., Li Z., Guo J., Xu Y. Deacetylation of MRTF-A by SIRT1 defies senescence induced down-regulation of collagen type I in fibroblast cells. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866(5):165723. doi: 10.1016/j.bbadis.2020.165723. Epub 2020/02/18. [DOI] [PubMed] [Google Scholar]
- 67.Kong M., Zhu Y., Shao J., Fan Z., Xu Y. The chromatin remodeling protein BRG1 regulates SREBP maturation by activating SCAP transcription in hepatocytes. Front. cell Develop. Biol. 2021;9:622866. doi: 10.3389/fcell.2021.622866. Epub 2021/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong W., Kong M., Qi M., Bai H., Fan Z., Zhang Z. BRG1 mediates nephronectin activation in hepatocytes to promote T lymphocyte infiltration in ConA-induced hepatitis. Front. cell Develop. Biol. 2021;8:587502. doi: 10.3389/fcell.2020.587502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu X., Dong W., Zhang T., Ren H., Wang J., Shang L. Epiregulin (EREG) and myocardin related transcription factor A (MRTF-A) form a feedforward loop to drive hepatic stellate cell activation. Front. cell Develop. Biol. 2020;8:591246. doi: 10.3389/fcell.2020.591246. Epub 2021/02/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu T., Wang H., Xin X., Yang J., Hou Y., Fang M. An MRTF-A-sp1-PDE5 Axis mediates angiotensin-II-induced cardiomyocyte hypertrophy. Front. cell Develop. Biol. 2020;8:839. doi: 10.3389/fcell.2020.00839. Epub 2020/10/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dong W., Zhu Y., Zhang Y., Fan Z., Z Z., Fan X. BRG1 links TLR4 trans-activation to LPS-induced SREBP1a expression and liver injury. Front. cell Develop. Biol. 2021;9:617073. doi: 10.3389/fcell.2021.617073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Z., Chen B., Zhu Y., Zhang T., Zhang X., Yuan Y. The Jumonji domain-containing histone demethylase homolog 1D/lysine demethylase 7A (JHDM1D/KDM7A) is an epigenetic activator of RHOJ transcription in breast cancer cells. Front. cell Develop. Biol. 2021;9:664375. doi: 10.3389/fcell.2021.664375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu L., Zhao Q., Lin L., Yang G., Yu L., Zhuo L. Myeloid MKL1 disseminates cues to promote cardiac hypertrophy in mice. Front. cell Develop. Biol. 2021;9:583492. doi: 10.3389/fcell.2021.583492. Epub 2021/04/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu L., Zhao Q., Kong M., Mao L., Yang Y., Xu Y. Myocardin-related transcription factor A (MRTF-A) regulates integrin beta 2 transcription to promote macrophage infiltration and cardiac hypertrophy in mice. Cardiovasc. Res. 2021 doi: 10.1093/cvr/cvab110. Epub 2021/03/23. [DOI] [PubMed] [Google Scholar]
- 75.Chen B., Zhu Y., Chen J., Feng Y., Xu Y. Activation of TCL transcription by lysine demethylase KDM4B in colorectal cancer cells. Front. cell Develop. Biol. 2021;9:617549. doi: 10.3389/fcell.2021.617549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Z., Lv F., Dai C., Wang Q., JIang C., Fang M. Activation of galectin-3 (LGALS3) transcription by injurious stimuli in the liver is commonly mediated by BRG1. Front. cell Develop. Biol. 2019;7:310. doi: 10.3389/fcell.2019.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heydtmann M. Macrophages in hepatitis B and hepatitis C virus infections. J. Virol. 2009;83(7):2796–2802. doi: 10.1128/JVI.00996-08. Epub 2008/10/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miura K., Yang L., van Rooijen N., Ohnishi H., Seki E. Hepatic recruitment of macrophages promotes nonalcoholic steatohepatitis through CCR2. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302(11):G1310–G1321. doi: 10.1152/ajpgi.00365.2011. Epub 2012/03/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li M., Cai S.Y., Boyer J.L. Mechanisms of bile acid mediated inflammation in the liver. Mol. Aspect. Med. 2017;56:45–53. doi: 10.1016/j.mam.2017.06.001. Epub 2017/06/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu T.F., Yang T.H., Zhong C.P., Shen C., Lin W.W., Gu G.X. Dual effect of hepatic macrophages on liver ischemia and reperfusion injury during liver transplantation. Immune network. 2018;18(3):e24. doi: 10.4110/in.2018.18.e24. Epub 2018/07/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsou C.L., Peters W., Si Y., Slaymaker S., Aslanian A.M., Weisberg S.P. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J. Clin. Invest. 2007;117(4):902–909. doi: 10.1172/JCI29919. Epub 2007/03/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gonzalez J., Mouttalib S., Delage C., Calise D., Maoret J.J., Pradere J.P. Dual effect of chemokine CCL7/MCP-3 in the development of renal tubulointerstitial fibrosis. Biochem. Biophys. Res. Commun. 2013;438(2):257–263. doi: 10.1016/j.bbrc.2013.07.025. Epub 2013/07/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ford J., Hughson A., Lim K., Bardina S.V., Lu W., Charo I.F. CCL7 is a negative regulator of cutaneous inflammation following leishmania major infection. Front. Immunol. 2018;9:3063. doi: 10.3389/fimmu.2018.03063. Epub 2019/01/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He J., Song Y., Li G., Xiao P., Liu Y., Xue Y. Fbxw7 increases CCL2/7 in CX3CR1hi macrophages to promote intestinal inflammation. J. Clin. Invest. 2019;129(9):3877–3893. doi: 10.1172/JCI123374. Epub 2019/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mercer P.F., Williams A.E., Scotton C.J., Jose R.J., Sulikowski M., Moffatt J.D. Proteinase-activated receptor-1, CCL2, and CCL7 regulate acute neutrophilic lung inflammation. Am. J. Respir. Cell Mol. Biol. 2014;50(1):144–157. doi: 10.1165/rcmb.2013-0142OC. Epub 2013/08/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y.L., Han D.H., Kim D.Y., Lee C.H., Rhee C.S. Role of interleukin-17a on the chemotactic responses to CCL7 in a murine allergic rhinitis model. PloS One. 2017;12(1) doi: 10.1371/journal.pone.0169353. Epub 2017/01/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y., Cai Y., Liu L., Wu Y., Xiong X. Crucial biological functions of CCL7 in cancer. PeerJ. 2018;6:e4928. doi: 10.7717/peerj.4928. Epub 2018/06/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Q., Cao X. Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 2019;19(7):417–432. doi: 10.1038/s41577-019-0151-6. Epub 2019/03/29. [DOI] [PubMed] [Google Scholar]
- 89.Henning A.N., Roychoudhuri R., Restifo N.P. Epigenetic control of CD8(+) T cell differentiation. Nat. Rev. Immunol. 2018;18(5):340–356. doi: 10.1038/nri.2017.146. Epub 2018/01/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ansel K.M., Lee D.U., Rao A. An epigenetic view of helper T cell differentiation. Nat. Immunol. 2003;4(7):616–623. doi: 10.1038/ni0703-616. Epub 2003/06/28. [DOI] [PubMed] [Google Scholar]
- 91.Zhang X., Liu S., Weng X., Wu T., Yu L., Xu Y. Brg1 trans-activates endothelium-derived colony stimulating factor to promote calcium chloride induced abdominal aortic aneurysm in mice. J. Mol. Cell. Cardiol. 2018;125:6–17. doi: 10.1016/j.yjmcc.2018.10.012. Epub 2018/10/20. [DOI] [PubMed] [Google Scholar]
- 92.Adenuga D., Yao H., March T.H., Seagrave J., Rahman I. Histone deacetylase 2 is phosphorylated, ubiquitinated, and degraded by cigarette smoke. Am. J. Respir. Cell Mol. Biol. 2009;40(4):464–473. doi: 10.1165/rcmb.2008-0255OC. Epub 2008/10/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cho S.J., Huh J.E., Song J., Rhee D.K., Pyo S. Ikaros negatively regulates inducible nitric oxide synthase expression in macrophages: involvement of Ikaros phosphorylation by casein kinase 2. Cell. Mol. Life Sci. : CMLS. 2008;65(20):3290–3303. doi: 10.1007/s00018-008-8332-7. Epub 2008/09/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Torra I.P., Ismaili N., Feig J.E., Xu C.F., Cavasotto C., Pancratov R. Phosphorylation of liver X receptor alpha selectively regulates target gene expression in macrophages. Mol. Cell Biol. 2008;28(8):2626–2636. doi: 10.1128/MCB.01575-07. Epub 2008/02/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shinoda K., Ohyama K., Hasegawa Y., Chang H.Y., Ogura M., Sato A. Phosphoproteomics identifies CK2 as a negative regulator of beige adipocyte thermogenesis and energy expenditure. Cell Metabol. 2015;22(6):997–1008. doi: 10.1016/j.cmet.2015.09.029. Epub 2015/11/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Choi S.E., Kwon S., Seok S., Xiao Z., Lee K.W., Kang Y. Obesity-linked phosphorylation of SIRT1 by casein kinase 2 inhibits its nuclear localization and promotes fatty liver. Mol. Cell Biol. 2017;37(15) doi: 10.1128/MCB.00006-17. Epub 2017/05/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bilodeau S., Caron V., Gagnon J., Kuftedjian A., Tremblay A. A CK2-RNF4 interplay coordinates non-canonical SUMOylation and degradation of nuclear receptor FXR. J. Mol. Cell Biol. 2017;9(3):195–208. doi: 10.1093/jmcb/mjx009. Epub 2017/02/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Viscarra J.A., Wang Y., Hong I.H., Sul H.S. Transcriptional activation of lipogenesis by insulin requires phosphorylation of MED17 by CK2. Sci. Signal. 2017;10(467) doi: 10.1126/scisignal.aai8596. Epub 2017/02/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cavin L.G., Romieu-Mourez R., Panta G.R., Sun J., Factor V.M., Thorgeirsson S.S. Inhibition of CK2 activity by TGF-beta1 promotes IkappaB-alpha protein stabilization and apoptosis of immortalized hepatocytes. Hepatology. 2003;38(6):1540–1551. doi: 10.1016/j.hep.2003.09.019. Epub 2003/12/03. [DOI] [PubMed] [Google Scholar]
- 100.Chen M.C., Chen C.H., Chuang H.C., Kulp S.K., Teng C.M., Chen C.S. Novel mechanism by which histone deacetylase inhibitors facilitate topoisomerase IIalpha degradation in hepatocellular carcinoma cells. Hepatology. 2011;53(1):148–159. doi: 10.1002/hep.23964. Epub 2011/01/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang P., Song X., Cao D., Cui K., Wang J., Utpatel K. Oncogene-dependent function of BRG1 in hepatocarcinogenesis. Cell Death Dis. 2020;11(2):91. doi: 10.1038/s41419-020-2289-3. Epub 2020/02/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Argemi J., Latasa M.U., Atkinson S.R., Blokhin I.O., Massey V., Gue J.P. Defective HNF4alpha-dependent gene expression as a driver of hepatocellular failure in alcoholic hepatitis. Nat. Commun. 2019;10(1):3126. doi: 10.1038/s41467-019-11004-3. Epub 2019/07/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barbier-Torres L., Fortner K.A., Iruzubieta P., Delgado T.C., Giddings E., Chen Y. Silencing hepatic MCJ attenuates non-alcoholic fatty liver disease (NAFLD) by increasing mitochondrial fatty acid oxidation. Nat. Commun. 2020;11(1):3360. doi: 10.1038/s41467-020-16991-2. Epub 2020/07/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available upon reasonable request.