Abstract
Purpose:
The registration of multiple imaging studies to radiation therapy computed tomography simulation, including magnetic resonance imaging, positron emission tomography-computed tomography, etc. is a widely used strategy in radiation oncology treatment planning, and these registrations have valuable roles in image guidance, dose composition/accumulation, and treatment delivery adaptation. The NRG Oncology Medical Physics subcommittee formed a working group to investigate feasible workflows for a self-study credentialing process of image registration commissioning.
Methods and Materials:
The American Association of Physicists in Medicine (AAPM) Task Group 132 (TG132) report on the use of image registration and fusion algorithms in radiation therapy provides basic guidelines for quality assurance and quality control of the image registration algorithms and the overall clinical process. The report recommends a series of tests and the corresponding metrics that should be evaluated and reported during commissioning and routine quality assurance, as well as a set of recommendations for vendors. The NRG Oncology medical physics subcommittee working group found incompatibility of some digital phantoms with commercial systems. Thus, there is still a need to provide further recommendations in terms of compatible digital phantoms, clinical feasible workflow, and achievable thresholds, especially for future clinical trials involving deformable image registration algorithms. Nine institutions participated and evaluated 4 commonly used commercial imaging registration software and various versions in the field of radiation oncology.
Results and Conclusions:
The NRG Oncology Working Group on image registration commissioning herein provides recommendations on the use of digital phantom/data sets and analytical software access for institutions and clinics to perform their own self-study evaluation of commercial imaging systems that might be employed for coregistration in radiation therapy treatment planning and image guidance procedures. Evaluation metrics and their corresponding values were given as guidelines to establish practical tolerances. Vendor compliance for image registration commissioning was evaluated, and recommendations were given for future development.
Introduction
Image registration has a variety of applications in radiation oncology, from simulation to treatment delivery, and plays a central role in image guidance and treatment adaptation. Multiple imaging studies may be brought in congruence to help better define the location and the extent of the tumor, and image registration can be employed to propagate contours between different studies. The treatment planning process and the plan quality may benefit from using image registration as a tool for dose accumulation by projecting previous treatment plans (encompassing various time spans) onto a reference patient anatomy. Rigid image registration (RIR) is a standard tool for patient alignment, and daily imaging studies may be used to estimate daily doses. Bringing image registration into any of the scenarios listed previously will have direct implications on the achievable and achieved accuracy of the dose received during radiation therapy, which reinforces the need for proper evaluation of the image registration performance, more so in the context of clinical trials. Image registration is the process by which homologous points, most often identified with image voxels, from multitemporal, mono- or multimodal, anatomic, or functional image sets are mapped onto each other. The process is described by a mathematical transformation, the complexity of which depends on how different the 2 image sets are. The image registration validation process must include 2 components: first, the performance of an image registration platform needs to be validated against well-defined standards, to ensure accurate results in a controlled environment (using phantom image data or patient data with known geometric transformations); second, the accuracy of the image registration has to be acceptable and suitable for a given anatomic site of a given patient, depending on the intended use of the image registration. For clinical trials, the former should be the subject of a “credentialing” or self-study evaluation methodology, whereas the latter should be specifically defined per protocol and part of a pretreatment review process (much like one is required, for example, to submit a treatment plan for review, despite having received credentialing for that treatment planning modality).
The American Association of Physicists in Medicine (AAPM) Task Group 132 (TG132) on the use of image registration and fusion algorithms in radiation therapy was published in May 2017,1 providing basic guidelines for quality assurance (QA) and quality control of image registration operation for the overall clinical process. The TG132 recommends a series of tests and corresponding metrics that should be evaluated and reported during commissioning and routine QA, as well as a set of recommendations for vendor software improvements. However, members in the present committee found incompatibility of some digital phantoms provided by the TG132 report with commercial software, thus practical guidelines for clinical implementation are still needed, especially for deformable image registration (DIR) tests.
NRG Oncology medical physics subcommittee formed a work group consisting of 9 institutions to evaluate 4 commonly used systems in radiation oncology. The goals of this report are 2-fold: (1) to evaluate the image registration (rigid and deformable) performance and the compliance with TG132 guidelines (addressed in the section Commercial Systems and User Testing); and (2) to present a workflow for self-credentialing a clinical system for rigid/deformable image registration and obtain group consensus in recommendations for future NRG Oncology trials that involve image registration, dose accumulation, and adaptive radiation therapy (addressed in the section Self-Evaluation and Patient Specific QA Recommendations).
This report serves as a guidance for self-study evaluation of institutional image registration for NRG Oncology and IROC (Imaging and Radiation Oncology Core) Clinical Trial participants. Case examples are used in the present manuscript, including TG132 provided basic geometric and anatomy phantoms, as well as a thorax data set for DIR validation. Individual clinical sites are encouraged to evaluate their imaging registration systems with the methodology/digital phantoms before enrolling in any protocol that might involve image registration, rigid or deformable. In general, the NRG Oncology and IROC rigorously review all initial cases submitted for trial participation, with the acknowledgment that successful self-study in the TG132 and related RIR/DIR exercises presented here will be instructive for enrolling in clinical trials. It is important to note that because there are a wide array of commercial systems currently available and/or in the development for imaging registration, it is more practical and expeditious for each individual clinic to undertake a self-study evaluation of their software with the phantoms and analytical tools provided in this report than it would be to have the entire credentialing process routed through IROC in the traditional way.
Image Registration Methodology and QA Considerations
Image registration in radiation therapy
Computed tomography (CT), cone beam computed tomography (CBCT), magnetic resonance imaging (MRI), and positron emission tomography (PET) are the anatomic and functional imaging modalities of most interest in radiation oncology. The images to be registered can be mono-modal (eg, CTs acquired at 2 points in time) or multimodal (CT-MR, CT-PET, CT-CBCT).
Rigid registration is a global match between image sets that preserves the relative distance between every pair of points from the patient’s anatomy. The deformable registration is a computational process in which an image similarity measure function and a transformation model are defined for the images of interest, then an optimization algorithm is used to adjust the transformation model in a way that maximizes the similarity function. The transformation models currently available include spline and demons, elastic, fluid, finite element model, and free form deformations.
Whether the images should be aligned rigidly or nonrigidly depends on the nature of the differences that exist between 2 imaging studies and the purpose the registration transformation will serve. Rigid registration is limited to 6° of freedom (3 rotations and 3 translations), and the user input typically consists of setting up a registration volume around the region of interest (ROI) to be transformed rigidly. Nonrigid transformations require many more degrees of freedom and rely more on the user input (and the prerequisite step of rigid registration) to drive the registration accuracy locally. Image registration is most often a succession of automatic and interactive iterations, with visual inspection being used as the first-line evaluator of the overall matching integrity. Multimodality registration poses additional challenges, because, to be able to correlate the morphologic and/or functional features between image sets, the registration algorithm needs to first establish the image intensity correspondences – not trivial, given the different appearance of the same anatomic entity on different imaging studies. The registration volume may be the entire imaged volume or only a subset of the available image set (boxed volume around the ROI), to facilitate a more accurate match locally, where needed. As a bonus, the likely gain in the registration quality with smaller volumes may also be complemented by a decrease in the computational time.
Quantitative morphologic image registration QA
The undertaking of image registration is to establish a bijective morphologic correspondence between the image elements of the data sets under consideration, usually referred to as “target image set” (aka, primary, fixed) and “source image set” (aka, secondary, movable). QA for image registration entails the computation and the evaluation of metrics that describe the realization of the morphologic correspondence; any mismatch between the target and the source image content will degrade the metrics used to evaluate the similarity of the images. There are multiple causes for the mismatch. For example, the cause can be the registration algorithm itself, depending on how sensitive that is to data sampling, interpolation, histogram binning, and so on. Other reasons of mismatching include largely deformed anatomic structures, image artifacts, lack of accurate multimodal correlations (ie, CT-MRI), image truncation (ie, CBCT), and so forth. In the case of rigid registration, a matrix of translations and rotations fully describes the transformation that will bring the source and the target in congruence, whereas deformable transformation will be characterized by a deformation vector field (DVF) that establishes the displacement of every voxel pair of the 2 image sets considered. Of note, in the case of deformable image registration, the similarity is quantified between the target image and the “deformed (warped) source image” created by applying the DVF to the source image. Several metrics are commonly used to evaluate the registration performance, that is, the degree of similarity between images, and they are briefly summarized next.
Morphologic correspondence
Landmarks.
The most intuitive quantitative way of validating the registration is to identify homologous landmarks in the target and the source images and to establish how accurately the registration transformation describes the target landmark coordinates given the source landmark coordinates. The method is time consuming and usually less effective in low-contrast regions due to the difficulty in identifying exact locations on both images. The performance of the registration for the landmark points, no matter how good (or bad), is not necessarily met with a similar performance in other regions of the volume, whether remote or proximal to the landmarks. This is due to the landmarks being visible in the image, which lends itself to driving the registration accuracy. The quantity used to describe the landmark registration error, typically referred to as “target registration error” (TRE),2,3 represents the distance between the fiducial location on an image set and the transformed location of the corresponding fiducial from the other image set (along any axis or the total displacement). A value of 0 indicates a perfect match between the homologous points considered. TRE does not include the uncertainties in identifying the landmarks, whether this is accomplished manually or automatically.
Contours.
Contours of structures identified on the target image can be compared with contours of the homologous structures on the source image by evaluating the overlap between the warped structures from the source image and the structures from the target image. The overlap is usually characterized by metrics such as Hausdorff distance (HD),4,5 HD quantiles (ie, 90% and 95%), the Dice similarity coefficient (DSC),6 the mean distance to agreement (MDA),7,8 and the Jaccard (Jac) index.9,10 HD is defined as the largest distance of a set A to the nearest point in another set B and is very sensitive to outliers. Percentage HD metrics can be used, for example, HD95 or HD90, to provide a similar measure but excluding 5% or 10% of the outliers when identifying the 2 surface distances. The mean surface distance, or MDA, is the mean distance of the closest approach used in defining the HD and was introduced to alleviate its outlier issue. DSC, a spatial overlap index, equals twice the number of elements common to both sets divided by the sum of the elements in both sets, with 0 indicating no overlap and 1 indicating complete overlap. The Jac similarity index also measures the overlap of 2 sets, defined as the elements common in both sets divided by elements in either set; it has a value of 0 if the 2 sets are disjoint and 1 if they are identical.9,10 Therefore, Jac and DSC are mathematically correlated through the equation DSC = 2Jac / (Jac + 1). Note that any of the contour-based metrics do not address the quality of the image registration inside the contours.
Deformation vector field and inverse consistency error
The DVF describes the displacements of every voxel from 1 image to the other image. In mathematical terms, the DVFs are physically possible when the Jacobian J of the deformation field is positive, describing compressions (0 < J < 1) or expansions (J > 1); folding of the structures, for example, is not physically possible and is quantified as a negative Jacobian. As an exception, local negative values of the Jacobian may exist where interface tissue sliding occurs.
The inverse consistency erro validation method establishes the extent to which the forward registration of the “source” to the “target” is described by the same transformation as the inverse registration from the “target” to the “source.” The consistency requirement is necessary for a perfect registration; however, the fulfilment of this condition is not sufficient to ensure accuracy.
Qualitative morphologic image registration QA
Visual inspection and human deliberation are non-standardized QA methods for image registration validation, which provide the user with the opportunity to evaluate the registration and to ponder the potential consequences of poor registration on a patient-specific basis and with a clear clinical endpoint in mind. Visual registration is, for all practical purposes, a landmark congruence assessment and strictly qualitative. The process relies on the clinicians’ experience and lacks consistency. Various image fusion verification options are available, to facilitate the visual verification of the alignment: overlays, checker boards, image differences, and spy glasses.
Quantitative dosimetric image registration QA
The level of required accuracy (assuming that it can be rigorously and consistently quantified) will vary depending on the endpoint; for example, although a poor registration of a region irradiated with a homogeneous dose distribution will likely have no clinical consequences, small uncertainties may be far-reaching if they happen when one plans to boost a high-risk portion of the target based on molecular imaging. Various methods have been investigated to evaluate the dosimetric relevance of the registration errors, including, but not limited to, the use of virtual phantoms, the evaluation of mean doses within an ROI using mass conserving deformations, and the analysis of a distance-to-dose difference tool to investigate the DVF accuracy.11–16 To date, none of these approaches have emerged or been established as a method sufficiently robust and efficient for routine clinical use.
Commercial Systems and User Testing
Overview of commercial systems
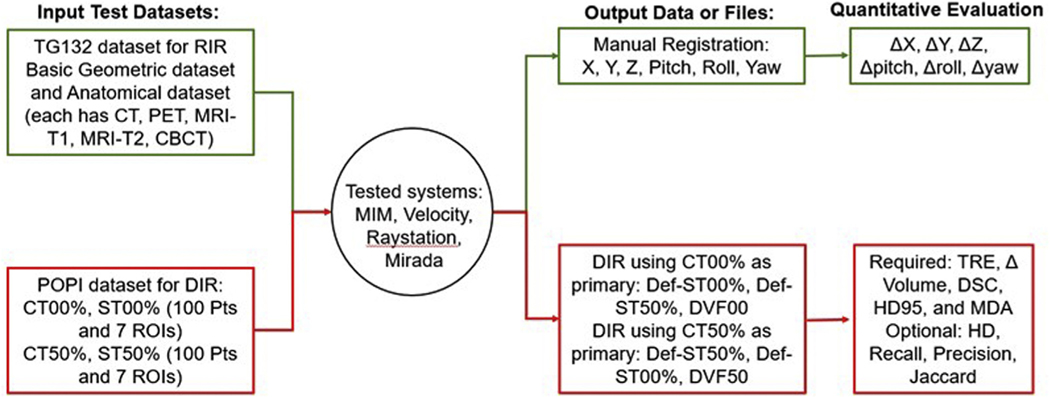
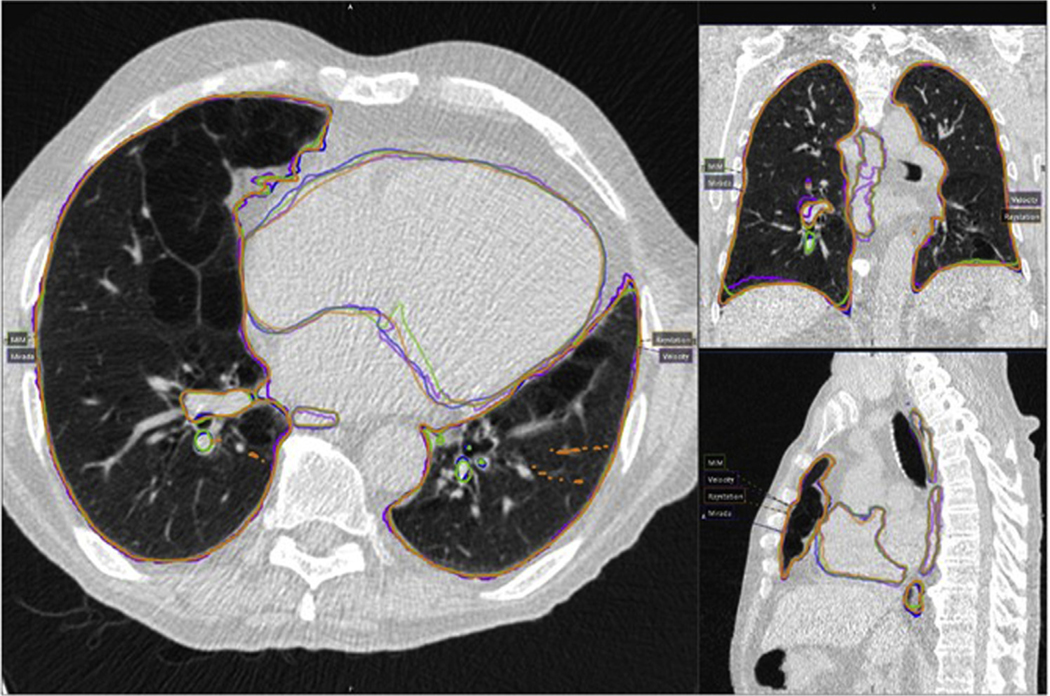
There exists a variety of software or systems that provide algorithms for rigid and deformable image registration and fusion, including open-source codes, in-house executables, and commercialized systems that are made specifically for the radiation oncology field. Commonly used commercial systems in radiation oncology are either stand-alone image processing systems or treatment planning systems with image registration applications. The systems we evaluated in this working group include 3 commercial stand-alone image processing systems, MIM Maestro (MIM Software Inc, Cleveland, OH) (8 user evaluations), Velocity (Varian Medical Systems, Palo Alto, CA) (3 user evaluations), and Mirada (Mirada Medical, Oxford, UK) (1 user evaluation), as well as 1 treatment planning system with imaging registration modules, Raystation (RaySearch Laboratories, Stockholm, Sweden) (2 user evaluations). These 4 systems are commonly used commercial systems in the field of radiation oncology, and were evaluated by members of this working group as indicated previously (note that some institutions have multiple systems). The detailed system functionality comparison is elaborated in the section Current System Limitations, Recommendations to Vendors, and Future Work for Phantoms. The workflow of the system testing is shown in Figure 1. For the rigid registration accuracy, TG132 proposed 2 data sets created using ImSimQA (basic phantom data set and basic anatomic data set) for various modalities (CT, CBCT, PET, MRI-T1, and MRI-T2). Translation and rotation accuracy were evaluated. For DIR accuracy assessment, due to the difficulties (ie, not compatible for the tested commercial systems) in using the deformation phantom and the corresponding DVF file provided by TG132, this working group adopted the POPI model for deformation testing17 (downloaded at https://www.creatis.insa-lyon.fr/rio/popi-model?action=show&redirect=popi). The POPI data set17,18 contains a set of end-of-inhalation and end-of-exhalation CTs, structure contours on both CTs, and 100 anatomic landmarks in the lung identified by expert radiologists for both CTs. Figure 2 shows an example of propagated structures from CT 50% phase to CT 00% phase extracted from all 4 commercial systems and overlaid on the same CT for comparison. The purpose of showing this example is to provide a visual demonstration of the variation in DIR-related operations, that is, structure propagation, using different systems/algorithms. This variation should be taken into consideration when designing acceptance tolerance for future adaptive radiation therapy trials.
Figure 1.
Workflow of rigid and deformable image registration with input data sets and output files and equalization metrics.
Figure 2.
An example of the propagated structures from CT 50% phase to CT 00% phase comparing all 4 commercial systems. Magenta: Velocity; blue: Mirada; green: MIM; orange: Raystation. (A color version of this figure is available at https://doi.org/10.1016/j.prro.2021.02.007.)
Rigid image registration results
Table E1 shows the list of basic geometric and anatomic phantom data sets provided and updated by TG132. Detailed image scanning parameters and sequences are listed, as well as the manual translations and rotations applied to those testing phantoms (served as the ground truth). RIR tests were performed using basic geometric and anatomic data sets with and without taking into account rotations. Rotation definitions are shown in Figure E1. Overall average and standard deviation of image registration errors are calculated across 4 algorithms in 9 institutions, as listed in Table E2 for translation/rotation test and Table E3 for translation only. Table E3 results exclude the velocity system because it does not provide the translations-only option when performing automated registration. We noticed higher deviation along the anterior-posterior direction for PET, MRI-T1, and MRI-T2 images, with PET having the highest deviation. For all tests involving CBCT data set, the 4 tested commercial systems reported 2 different shift values along the inferior-superior direction (Raystation/Mirada reported 1.5 cm while MIM/Velocity reported 3.0 cm), but this result was consistent among users of the same system. We believe this is due to a glitch in data creation in the older version of ImSimQA software, thus the CBCT RIR results were discarded from further analysis.
The TG132 proposed tolerance is half of the voxel dimension of the images for registration. For the tested data set, voxel dimensions are 0.91 × 0.91 × 3.0 mm for CT/PET/CBCT and 1.83 × 1.83 × 3.0 mm for MRI. Therefore, TG132 proposed tolerance would be 0.46 × 0.46 × 1.5 mm for CT/PET/CBCT and 0.92 × 0.92 × 1.5 mmfor MRI. As shown with the bolded values from the testing results across 9 institutions and 4 commercial systems (Tables E2 and E3), averaged translational discrepancies exceeded the proposed tolerance when involving PET and MRI images. Results are consistent with very small standard deviation across all institutions and tested systems, suggesting that TG132 recommended tolerance is achievable with the given PET and MRI data sets. We recommend using 1 voxel dimension as a more practical tolerance when participating in clinical trials.
TG132 did not specify tolerance recommendations for rotations, even though the group 2 tests involve translation and rotation. We performed forward and inverse registration for basic phantom group 2 and reported our results in Table E2. All pitch values for the group 2 tests exceeded 1° in average rotation discrepancy, whereas they were all within 1° in the corresponding inverse direction tests. In reality, the reported results in matching 2 images when allowing both translational and rotational operations may vary if the center of rotation is not fixed. Therefore, we recommend complementing any quantitative analysis with qualitative visual inspection when matching 2 images with rotation involved.
Deformable image registration results
The POPI data set selected for the DIR tests includes 2 sets of thorax CTs (CT00% and CT50%), 2 sets of manually segmented structures (esophagus, left lung, right lung, left ventricle, spinal cord, and trachea) on the corresponding CT (ST00% and ST50%), as well as 100 paired homologous points identified on both CTs based on tissue landmarks (included in the POPI model). However, DVF comparison tests (Jacobian and Inverse consistency) were not performed by this working group, due to the inconsistency of DVF format exported from each individual commercial system. Only the landmark-based and contour-based evaluation metrics, including TRE of 100 points, HD, HD95, DSC, MDA, and Jac were performed. Due to the limitations of commercial systems in implementing TG132 recommended metrics, an in-house software was developed to calculate metrics from Digital Imaging and Communications in Medicine (DICOM) exported files.
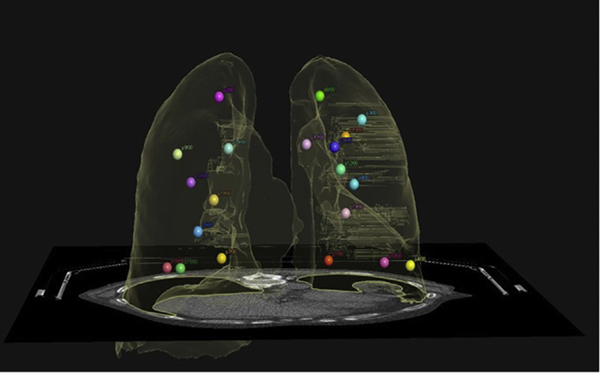
For TRE analysis, the current version of Mirada (v. RTX1.8) does not support automatic point propagation; as such, the TRE analysis can only be performed manually point by point in the system. Table 1a shows the min, max, mean, and standard deviation of the TRE of 20 points calculated in the same direction of deformation on all 4 systems. The locations of these 20 points were reviewed and confirmed to include upper, middle, and lower lobes of both lungs, as shown in Figure 3. The ranges of motion of these 20 pairs of points were confirmed to be close to the overall average range of motion of 100 points. Table 1b shows the TRE analysis performed on 100-point pairs with the in-house software for all 3 systems that allow DICOM export of the 100 pairs of matching points. Additional TRE analysis is shown in Table E4 for 100-point pairs with forward and inversed registration directions on both MIM and Velocity systems. The discrepancy between the corresponding metrics for the forward and inverse registrations is indicative of the DVF inverse inconsistency.
Table 1.
TRE analysis of minimum, maximum, mean, and standard deviation (SD) values (mm). (a) TRE analysis for all 4 systems from the first 20 pairs of matching points. (b) TRE analysis for 3 systems from all 100 point pairs (unit mm)
| (a) | MIM | Velocity | RayStation | Mirada | |||
|---|---|---|---|---|---|---|---|
| Min | 0.4 | 0.0 | 0.5 | 0.4 | |||
| Max | 17.6 | 21.5 | 23.3 | 16.8 | |||
| Mean | 3.0 | 6.0 | 4.3 | 3.3 | |||
| SD | 4.0 | 6.0 | 5.5 | 4.2 | |||
| (b) | MIM | Velocity | RayStation | ||||
|
| |||||||
| Min | 0.3 | 0.0 | 0.4 | ||||
| Max | 29.1 | 28.8 | 23.9 | ||||
| Mean | 2.8 | 6.8 | 4.0 | ||||
| SD | 4.4 | 6.7 | 5.3 | ||||
Abbreviations: SD = standard deviation; TRE = target registration error.
Figure 3.
The 3-dimensional (3D) illustration of the 20-point pairs selected for comparison evenly distributed in both lungs.
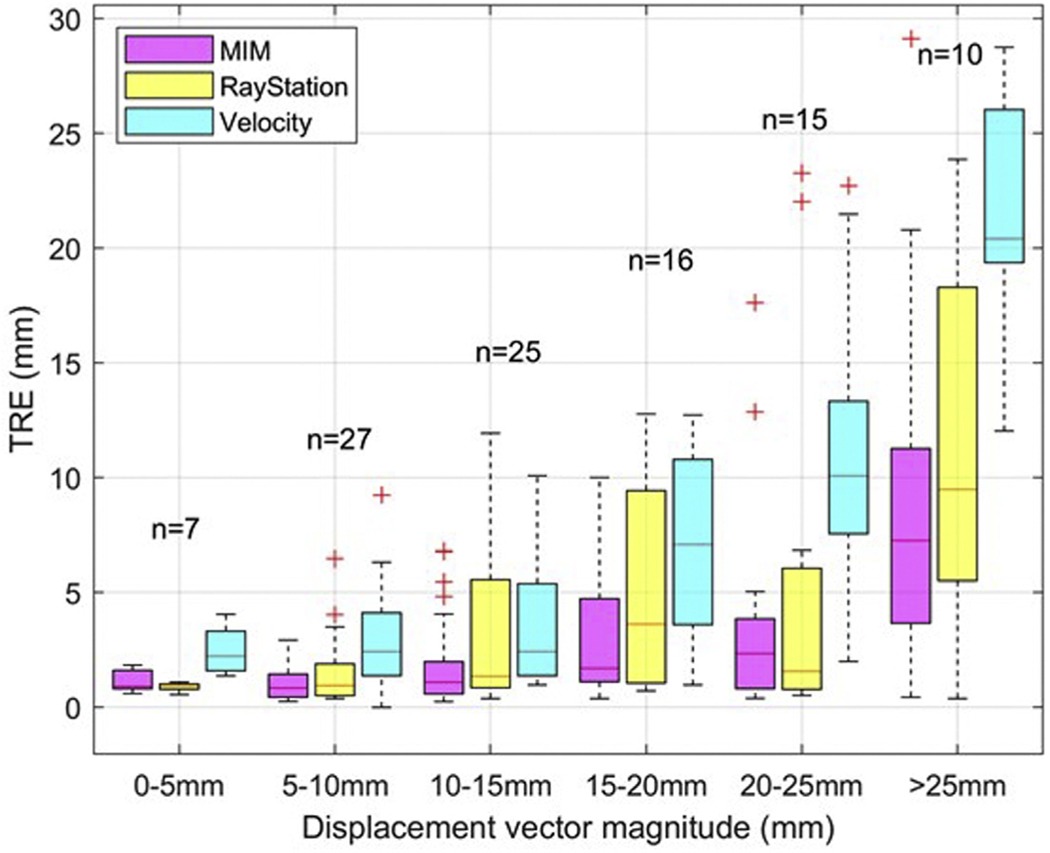
Figure 4 presents a box-plot summary of TRE analysis for the 3 tested systems that provided the DICOM export option for extracting all 100 pairs of homologous points. In all cases, the entire data sets were registered. The points were grouped in 5-mm bins based on their displacements between the 00% and 50% phases. All registration platforms performed better for points that exhibited less movement. Registration performance was fairly consistent for points with less than 10-mm displacements between 00% and 50% phases. The median TRE was below 5 mm for points moving up to 15 mm for Velocity and up to 25 mm for MIM and Raystation. Often, it is possible to improve the registration performance locally, and this aspect may have to be given consideration, depending on the endpoint of any particular study. The current version of Mirada does not allow point propagation or DICOM export, thus was excluded from this analysis. For Mirada users, Latifi et al12 provided a method to analyze TRE based on exported DVF files.
Figure 4.
Target registration error (TRE) boxplot for different ranges of displacement vectors of 2 matching points in the 0% and 50% phases (5 mm bins).
TG132-recommended tolerance for TRE is “maximum voxel dimension (2–3 mm).” Based on our results, this tolerance can hardly be met when the entire data sets are used for DIR. The results may improve if the deformable registration is performed over a smaller ROI. We recommend that clinical trial principal investigators (PIs) provide a test patient data set with identified homologous landmarks for the anatomic sites and propose pertinent tolerances for TRE values (eg, mean, max, or percentage), with consideration of the study goals. We also encourage users to modify the registration results based on the matching landmark points if the software/algorithm allows manual adjustments.
For contour-based metrics, TG132 recommended the tolerance for both DSC and MDA as “within contouring uncertainty of the structure” and set an example value of 2 to 3 mm for MDA and 0.8 to 0.9 for DSC. Our test with POPI data set showed that none of the commercial DIR software met the TG132 recommended values for every structure. Table 2 shows TG132-recommended analysis metrics based on the DIR of the POPI data set using MIM with forward (a) and inverse (b) deformation directions, with average values over all tested MIM systems and their corresponding standard deviations. Table 3a shows all DIR evaluation metrics calculated with our in-house software for the 6 structures in the POPI model. The use of in-house software rules out any variations associated with the implementation of metrics calculation algorithms in different systems. Note that contouring uncertainty differs with structures, and the effect of the contouring uncertainty on the evaluation metrics also differs. Yang et al19 reported DSC, HD95, and MDA values from interrater variabilities based on 3 human experts on 3 cases in the 2017 AAPM Thoracic auto-segmentation grand challenge, as shown in Table 3b. Table 3 illustrated that the interrater variability is different for each structure. It would be inappropriate to use 2 to 3 mm for MDA and 0.8 to 0.9 for DSC for all structures. The values in Table 3a that exceed Table 3b are highlighted in bold. As shown, this provides a more consistent highlight of problematic structures from multiple metrics.
Table 2.
(a) DIR analysis metrics for structures of the lung data set using MIM. (b) Analysis metrics for structures of the lung data set using MIM in inversed direction (with average values over 6 institutions and standard deviation)
| (a) | HD (mm) | MDA (mm) | DSC | Jaccard |
|---|---|---|---|---|
| Esophagus | 15.63 ± 0.1 | 1.39 ± 0.1 | 0.75 ± 0.003 | 0.61 ± 0.004 |
| Left ventricle | 21.38 ± 0.9 | 2.18 ± 0.1 | 0.91 ± 0.002 | 0.84 ± 0.003 |
| Left lung | 29.40 ± 0.9 | 0.81 ± 0.1 | 0.98 ± 0.000 | 0.96 ± 0.001 |
| Right lung | 33.67 ± 0.4 | 1.38 ± 0.1 | 0.97 ± 0.000 | 0.95 ± 0.001 |
| Spinal cord | 3.91 ± 0.1 | 0.43 ± 0.6 | 0.94 ± 0.001 | 0.88 ± 0.002 |
| Trachea | 6.02 ± 1.1 | 0.72 ± 0.1 | 0.92 ± 0.004 | 0.85 ± 0.006 |
| (b) | HD (mm) | MDA (mm) | DSC | Jaccard |
|
| ||||
| Esophagus | 15.46 ± 0.6 | 1.48 ± 0.1 | 0.75 ± 0.002 | 0.60 ± 0.003 |
| Left ventricle | 20.50 ± 0.2 | 2.12 ± 0.1 | 0.91 ± 0.001 | 0.84 ± 0.002 |
| Left lung | 25.64 ± 1.0 | 0.65 ± 0.1 | 0.98 ± 0.001 | 0.96 ± 0.001 |
| Right lung | 32.76 ± 0.4 | 1.17 ± 0.1 | 0.98 ± 0.000 | 0.95 ± 0.001 |
| Spinal cord | 4.71 ± 0.9 | 0.42 ± 0.1 | 0.94 ± 0.001 | 0.88 ± 0.001 |
| Trachea | 6.61 ± 0.4 | 0.80 ± 0.1 | 0.90 ± 0.001 | 0.82 ± 0.002 |
Abbreviations: DIR = deformable image registration; DSC = Dice similarity coefficient; HD = Hausdorff distance.
Table 3.
(a) DIR evaluation metrics of the 6 structures in the POPI model for 4 tested systems. (b) Interuser variations reported for the same structure by Yang et al.19 Bolded text in (a) indicate values that are inferior to the reported interuser variation baseline
| (a) | DSC |
HD (mm) |
HD95 (mm) |
MDA (mm) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIM | Velocity | RS | Mirada | MIM | Velocity | RS | Mirada | MIM | Velocity | RS | Mirada | MIM | Velocity | RS | Mirada | |
| Esophagus | 0.75 | 0.71 | 0.73 | 0.77 | 15.7 | 17.5 | 15.9 | 13.9 | 5.2 | 6.2 | 7.4 | 5.4 | 1.6 | 2.0 | 1.9 | 1.6 |
| Left ventricle | 0.91 | 0.90 | 0.89 | 0.92 | 21.0 | 22.7 | 23.1 | 20.4 | 9.9 | 10.5 | 10.3 | 9.9 | 2.6 | 2.8 | 3.3 | 2.5 |
| Leftt lung | 0.98 | 0.96 | 0.98 | 0.98 | 29.4 | 22.4 | 29.6 | 20.0 | 4.2 | 5.3 | 5.9 | 2.4 | 0.9 | 1.3 | 1.0 | 0.6 |
| Right lung | 0.97 | 0.95 | 0.98 | 0.98 | 33.5 | 44.0 | 40.0 | 28.8 | 7.4 | 13.8 | 6.6 | 3.9 | 1.5 | 2.7 | 1.4 | 1.0 |
| Spinal cord | 0.94 | 0.89 | 0.94 | 0.94 | 3.9 | 4.9 | 3.9 | 3.7 | 1.7 | 2.1 | 1.7 | 1.4 | 0.5 | 0.9 | 0.6 | 0.5 |
| Trachea | 0.92 | 0.87 | 0.91 | 0.92 | 5.3 | 7.1 | 5.0 | 4.4 | 2.3 | 3.5 | 2.1 | 2.0 | 0.8 | 1.1 | 0.7 | 0.7 |
| (b) | DSC | HD95 (mm) | MDA | |||||||||||||
|
| ||||||||||||||||
| Esophagus | 0.82 ± 0.04 | 3.33 ± 0.90 | 1.07 ± 0.25 | |||||||||||||
| Heart | 0.93 ± 0.02 | 6.42 ± 1.82 | 2.21 ± 0.59 | |||||||||||||
| Left lung | 0.96 ± 0.02 | 5.17 ± 2.73 | 1.51 ± 0.67 | |||||||||||||
| Right lung | 0.96 ± 0.02 | 6.71 ± 3.91 | 1.87 ± 0.87 | |||||||||||||
| Spinal cord (thoracic) | 0.86 ± 0.04 | 2.38 ± 0.39 | 0.88 ± 0.23 | |||||||||||||
Abbreviations: DIR = deformable image registration; DSC = Dice similarity coefficient; HD = Hausdorff distance.
The interrater variability for different organs can be obtained from literature.19–21 However, interrater variability levels could change with imaging modalities and contouring guidelines adopted.19,22,23 Clinical trial PIs are advised to establish baseline contour-based metrics for their study site by pooling a group of manual contours on the same image data set from multiple trial participants. Each trial participant should also test their DIR software on those image data sets and compare with the ground truth contours provided by trial PIs. This helps trial participants understand the performance and the limitations of their DIR for the study disease site. In addition, some DIR programs provide multiple algorithms as well as methods to manually correct errors and improve DIR results. These tests provide guidance to trial participants for determining the settings and procedures to obtain the most accurate DIR.
As illustrated in our POPI data set analysis, it is possible that the commercial software cannot meet the tolerance of contouring uncertainty for certain structures. However, depending on the structure’s distance to high-dose gradient regions as well as its tolerance dose, it is possible that slightly greater errors in contouring accuracy would not have appreciable effect on the treatment plan. Clinical trial PIs could relax contouring uncertainty tolerance for individual structures based on dosimetric effect analysis.
Based on the DIR tests exercised by our working group, the following suggestions are provided for trial-specific image registration QA:
Clinical trial PIs should provide test patient data sets for participants to manually contour and collect contouring uncertainty information for each organ.
Clinical trial PIs may relax contouring uncertainty tolerances based on the expected potential dosimetric effect. PI is advised to be cautious in relaxing tolerances in the areas that are in close proximity to the target region.
Each trial PI and participants should test their DIR program on the test patient provided to observe/understand the limitation of their DIR tools.
Trial PI and participants are encouraged to establish procedures to use features provided by DIR software to minimize registration inaccuracies.
Self-Evaluation and Patient-Specific QA Recommendations
The validation of an image registration for the purpose of clinical trials is a 2-part process. Assuming that the imaging systems are properly tested and their use for clinical trials has already been credentialed, the image registration validation adds the requirement to credential the performance of the system (software) used to register the images, as well as for the specific treatment site relevant to the clinical trial. The assessment will consist of aligning (rigidly and nonrigidly) image sets for which the exact transformation that brings them in congruence is known.
System evaluation
Institutions participating in clinical trials that require image registration and deformation should seek self-study evaluation for each system involved. Users are allowed to use manual and/or automated registrations to create the best match using the data set provided. Ideally, the registration software must be able to generate a “transformation” file in a format that can be further processed for analyses. For rigid registration, the reported values should include 3 Cartesian displacements and 3 rotation angles. The image sets used for the rigid image registration provided by TG132 have been evaluated by our working group and can be used based on our analysis elaborated in the section Commercial Systems and User Testing. For deformable registration, DVF comparison is most desirable, but there are various reasons that make this approach not practical at the current stage (see the section Current System Limitations, Recommendations to Vendors, and Future Work for Phantoms for details). As such, in the development of the registration software output, sampling the DVF by reporting landmark displacements is a more practical alternative. In addition, contour-based metrics are also recommended. The working group has validated an in-house designed code for calculating required and optional (Fig 1) quantitative evaluation metrics. Detailed information on user-prepared input files and step-by-step instruction is provided on the website where the in-house metrics calculator can be downloaded (https://carinaai.bitbucket.io/nrg-analyzer/). Institutions seeking self-study evaluation would provide the required data sets, which will be evaluated for compliance, and the calculated results for the DIR evaluation metrics will be sent back to the institution for reference. This working group used the POPI data set as an example for testing DIR accuracy between 2 images (phase 00% and 50%), and results are elaborated on in the section Commercial Systems and User Testing. Note that the DIR accuracy is highly site-dependent and deformation-level dependent, and it is impractical to cover all clinical scenarios in this current report. Therefore, the POPI data set can be used as part of the initial system evaluation, and users should go through a clinical trial self-evaluation using a trial-provided data set that is more relevant to the studied anatomic site and possible deformation levels.
Clinical trial self-evaluation
AAPM task group reports, TG179 and TG66,24,25 provide certain guidelines in initial and continuing QA of systems involving image guidance and imaging fusion, and provide the format requirement of input data for imaging fusion software. Depending on the specific protocol, the input data can range from CT or MR simulation scans to daily on-board images including on-board CT, CBCT, and on-board volumetric MR images. The clinical trial self-study evaluation process must ensure that the institution follows these guidelines by using site information questionnaires similar to those provided by more formal credentialing (http://rpc.mdanderson.org/RPC/credentialing/IGRT/IGRTCredentailing.aspx). Based on the image guided radiotherapy (IGRT) credentialing form, an updated image registration questionnaire is provided in Appendix E2 for rigid and deformable image registration self-study evaluation. This questionnaire includes items specific to the requirement of TG132, effectively requiring attestation from the institution that the system has been commissioned for clinical use. It is important to point out that the ultimate responsibility of the system commissioning falls on the end user of the system, as with any other software used in clinical trials (ie, treatment planning systems). The main goal of the self-study evaluation process is to ensure that all institutions use their registration software and clinical processes in accordance with the requirements or expectations of the trial. The main components of the self-study evaluation process are:
Site information questionnaire: This questionnaire serves as a basic attestation by the institution that they follow the standard guidelines for initial and continuing QA of their imaging systems (input data) as well as their image registration software.
Completion of the IROC IGRT credentialing (http://rpc.mdanderson.org/RPC/credentialing/IGRT/IGRTCredentailing.aspx).
Completion of the rigid and deformable image registration tests in compliance with the present credentialing guidelines.
Completion of site-specific deformable image registration tests, using the recommended data set provided by clinical trial PIs. It is the trial PIs’ prerogative and responsibility to recommend specific workflows and metrics for the self-study evaluation. If the ground-truth DVF is available, institutions are expected to submit the DVF in the registration software’s native format or in DICOM format.
Each individual clinical trial can benefit from using a specific data set from their trial study for best evaluation of image registration. This report only means to provide general recommendations in image registration evaluation, using the POPI data set as 1 case example. Site specific data sets (brain/head and neck, abdomen/thorax, or pelvis) should be provided, by the clinical trial PIs, to the institution. The provided data set should include the primary and secondary images, DVF if available, along with the contours of the ROIs and references points in DICOM coordinates on the primary and the secondary data sets. Ideally, institutions should be blinded to the true deformation between these data sets when performing their image registration tests. Resulting registration accuracy should be compared with the provided ground truth, and the evaluation metrics can be calculated using the tools provided by the commercial systems, as well as the previously mentioned calculator.
Recommendations for system- and patient-specific QA
The site information questionnaire should be updated on an annual basis, and the self-study evaluation steps must be repeated when new DIR software is introduced to the clinic or when major upgrades occur that can affect the performance of the DIR.
The patient specific validation is hampered by lack of availability of the ground truth. Therefore, it will be the responsibility of the protocol team to determine the nature and level of patient specific validation required. The most difficult cases to evaluate are, nonetheless, those involving deformable registration. Unless advances in registration assessment will make big strides in DVF evaluation in the absence of a ground truth, it is all but certain that the expert visual evaluation will remain the ultimate reliable tool at hand. At best, one can investigate the accuracy of contour and landmark transformations and deem as acceptable any errors that are no larger than inter- and intrauser variability. Most likely a decision will need to be made as to where high local accuracy (vs global) is most desirable because a global deformable registration of acceptable performance is likely not achievable in many cases. It is intuitive, for example, that high-dose gradient regions overlapping low-contrast regions may be among the most unfavorable scenarios and most susceptible to inaccuracies. This leads to another important aspect, which is the propagation of the registration error when other treatment plan metrics are analyzed. For the purpose of patient/site specific validation, a comprehensive literature review follows, to provide our current knowledge on evaluation metrics and expected results/challenges with respect to the anatomic site of interest.
Site-Specific Discussion and Example Cases
DIR algorithms use a model to describe the deformation that inevitably will have limitations. Success or failure of the DIR application depends on multiple variables, such as algorithm, metrics, site, image quality, and clinical goals. Various studies have shown that DIR results are site specific.12,26 For example, an algorithm that performs well for head and neck applications may not be suited for abdomen or thorax; however, generally speaking, the anatomic differences in individual cases had a greater effect on the DIR performance than the algorithm used.16,27 We believe it is important to review literature in those relevant site-specific studies in this report, so that clinical trial PIs can make an informative decision when they are creating their self-study evaluation data set.
Thorax
DIR in the thorax has various applications that present unique challenges. DIR has been applied in lung radiation therapy for contour propagation and auto-segmentation,28 intra- and interfraction dose accumulation,29 4-dimensional image analysis, and other applications such as ventilation mapping13,29–32 and radiomics.33 Adaptive radiation therapy and the use of DIR in lung radiation therapy is of great interest due to the changes encountered during the course of treatment such as tumor regression, tumor displacement, pleural effusion, and/or atelectasis (collapsed lung), which result in a deviation from the planned dose delivered.34
Challenges of DIR application in lung RT include breathing motion, sliding tissue effect, and large geometric changes due to atelectasis. The sliding tissue effect occurring at the wall of the pleura for lung and liver can result in large discontinuities in the DVF.33,35Unless specifically considered in the DIR method, the DVF may under- or overestimate the amount of deformation at the sliding interface. Large geometric changes during the course of lung radiation therapy can result in difficulties for DIR algorithms in detecting anatomic correspondence, decreasing DIR accuracy.36 It has been shown that DIR methods in lung perform well for contour propagation; however, the still need to be verified carefully for dose accumulation. Latifi et al12 showed the mean and max TRE values of individual points within the lung far exceeding the 2 and 5 mm criteria respectively proposed in the AAPM TG132 report. The results of DIR must be inspected carefully for DVFs that are nonphysical (eg, negative spatial Jacobian), which could lead to errors in dose propagation and accumulation,37 and careful attention must be applied to areas where significant anatomic changes occur, which produce highly localized deformations that are not handled well in typical DIR algorithms.26 Additionally, the DIR procedure used has an effect on the results, as shown by Kadoya et al,38 who observed both variation between institutions using different software, but also, to a lesser extent, variation between institutions using the same software. Table 4 presents a quantitative summary of DIR studies in the thorax region.
Table 4.
Summary of the results in the lung for common commercial algorithms in use currently for CT-CT (or 4DCT) registration
| Data set | # Cases | Software [# institutions] | Results | |
|---|---|---|---|---|
| Sarrudis et al23 | Patient — lung 4DCT | 6 | Raystation (Hybrid Intensity) [1] SmartAdapt [1] Velocity [1] |
DSC OARs > 0.83 (3 structures, all software) DSC GTV mean [min, max]: 0.65 [0–0.92] 0.62–072 [0–0.91]‡ 0.39 [0–0.9] |
| Loi et al26 | Patient — synthetic deformation | 1 × (2 deformations) | ABAS [1] MIM [2] MIRADA [1] Raystation (Hybrid intensity) [8] SmartAdapt [2] Velocity [1] |
DSC CTVs & OARs < 55c: 0.94* 0.91*† 0.98 0.95*† 0.92*†,‡ 0.98* |
| Latifi et al12 | Patient — POPI lung 4DCT set | 1 (TG132 4DCT set, inhale, exhale) 6 (POPI inhale, exhale pairs, 100 points/ each) |
MIRADA [1] | TRE avg. = [3.49–8.9] mm (min-max for 7 cases) DSC avg. = 0.92 (9 structures, 3 cases, forward/reverse DIR) |
| Velec et al39 | Patient — 4DCT | 16 | Raystation (Biomechanical) [1] Raystation (Hybrid intensity) [1] Raystation (Biomechanical) [1] Raystation (Hybrid intensity) [1] |
Mean DTA for:*
GTV, heart, bronchus, ribs, lungs§ 1.3, 1.8, 1.1,1.0, 0.8 mm 0.9,1.7,0.8,0.6,0.3 Mean TRE for lung:* 2.9 2.7 |
| Kadoya et al38 | Patient — DIR laboratory Lung 4DCT set | 10 (inhale, exhale pairs) | MIM [5] Raystation (Hybrid Intensity)[4] Velocity [3] |
Mean 3D TRE [min- max]: 3.29 [2.17–3.16] mm† 3.28 [1.26–3.91] mm† 5.01 [4.02–6.20] mm† |
Abbreviations: 4D = 4-dimensional; CT = computed tomography; CTV = clinical target volume; DIR = deformable image registration; DSC = Dice similarity coefficient; DTA = distance to agreement; GTV = gross tumor volume; OAR = organs at risk; TG132 = task group 132; TRE = target registration error.
Value estimated from figure.
Average over multiple centers.
SmartAdapt gives different results when repeated max/min are from repetitive DIRs done 10x.
Used as a controlling structure in DIR.
Head and neck
DIR combined with adaptive radiation therapy in head and neck has the potential to improve treatment outcomes.40 Difficulties lie in determining optimal strategies for when to apply ART.41,42 Primary challenges for the accurate use of DIR during the course of treatment include tumor response changes, parotid gland shrinkage, neck flexion changes, and weight loss.43 Similar to the results found in lung, DIR for dose accumulation in head and neck patients must be done carefully due to errors resulting from organ shrinkage and mass changes44; however, contour propagation was found to behave reasonably well across multiple algorithms.15 Certain organs at risk remain a challenge, however, such as the pharyngeal constrictors, owing to their small size, low contrast, and proximity to air cavities.12 Pukala et al16 evaluated 5 commercial algorithms using 10 virtual head and neck phantoms and showed little difference between the various algorithms. However, differences observed between phantoms emphasize the need to assess DIR accuracy on multiple cases. Additionally, dosimetric results (ie, dose volume histogram differences) illustrated the difficulty in generalizing any correlation between TRE and dose volume histogram errors. Table 5 presents a quantitative summary of DIR studies in the head and neck region.
Table 5.
Summary of a sample of results for HN and common commercial algorithms in use currently for CT-CT (or 4DCT) registration
| Data set | No. of cases | Software [no. of institutions] | Results | |
|---|---|---|---|---|
| Loi et al26 | Patient — synthetic deformation | 1 × (2 deformations) | ABAS [1] MIM [2] MIRADA [1] Raystation (Hybrid intensity) [8] SmartAdapt [2] Velocity [1] |
DSC CTVs and OARs < 55c: 0.81* 0.88*† 0.91 0.89*† 0.78*,† 0.86* |
| Latifi et al12 | Patient (diagnostic, planning CT) | 3 | MIRADA [1] | DSC OARs with/ without pharyngeal constrictors 0.74, 0.81 |
| Pukala et al16 | Patient — synthetic deformation (DIREP Library) | 10 | MIM [1] Pinnacle [1] Raystation (Hybrid Intensity) [1] SmartAdapt [1] Velocity [1] |
Mean TRE for: brainstem, cord, mandible, L/R parotid: 0.5, 0.5, 0.9, 1.2, 1.5 1.0,1.2, 1.9, 1.7 1.0, 1.6, 2.0, 2.4 1.1, 1.1, 2.1, 2.1, 1.8 1.2, 1.8, 1.5, 2.2, 1.6 |
| Nie et al15 | Patient — synthetic deformation | 1 | MIM [1] OnQ rts [1] Velocity [1] |
DSC for target, OARs 0.94, 0.9 0.92, 0.88 0.79, 0.73 |
Abbreviations: 4D = 4-dimensional; CT = computed tomography; CTV = clinical target volume; DSC = Dice similarity coefficient; OAR = organs at risk; TRE = target registration error.
Value estimated from figure.
Average over multiple centers.
Abdomen
DIR has also been applied in the abdomen for liver and pancreas. For liver, it has been used in modeling liver motion for targeting accuracy45,46 and modeling of dose-dependent anatomic changes in liver that may improve correlation of functional liver imaging with radiation dose.47 For pancreas, DIR can be used to aid in motion assessment,48 segmentation of internal target volumes,49 and adaptive radiation therapy (online or offline) positioning correction.50 Obstacles for accurate DIR in the abdomen include low-contrast features in abdominal CT, sliding effect for liver, and significant inter- and intra-fractional motion. Additionally, as found by Fukumitsu et al,27 fiducial registration error is also dependent on initial rigid registration, tumor diameter, and the use of CT contrast. Results, although similar for both algorithms used (MIM and Velocity), were again found to be case dependent. Velec et al39 evaluated the use of RayStation’s biomechanical algorithm on liver images. For liver 4-dimensional CT, compared with the existing RayStation Hybrid-Intensity algorithm used in geometry-based mode (ROI driven only), the biomechanical implementation improves internal ROI accuracy significantly. Compared to hybrid intensity with controlling ROIs, the biomechanical implementation performs comparably. However, the biomechanical algorithm outperforms the intensity-driven DIR algorithm in multimodality applications. Table 6 presents a quantitative summary of DIR studies in the abdomen.
Table 6.
Summary of some of the limited reported commercial DIR results in the abdomen/pelvis (liver) for common commercial algorithms for CT-CT (or 4DCT) registration
| Data set | No. of cases | Software [no. of institutions] | Results | |
|---|---|---|---|---|
| Fukumitsu et al27 | Patient — pre/post treatment CT | 24 | MIM [1] Velocity [1] |
TRE mean (fiducials): (noncontrast/contrast CT) 9.3 ± 9.9 mm/7.4 ± 7.7 mm 11.0 ± 10.0 mm/8.9 ± 7.2 mm |
| Velec et al39 | Patient — 4DCT | 10 | Raystation (Biomechanical) [1] Raystation (Hybrid intensity) [1] Raystation (Biomechanical) [1] Raystation (Hybrid intensity) [1] |
Mean DTA for:* kidneys, stomach, spleen†, liver† 1.5, 1.9, 0.7, 0.8 mm 0.7, 1.3, 0.1, 0.3 mm Mean TRE for liver (vessels POI)*: 2.6 mm 3.3 mm |
| Ribeiro et al51 | Patient — synthetic 4DCT (derived from 4D MRI DVF and exhale CT) | 9 (3 patients × 3 DVFs) | Raystation (Hybrid Intensity) [1] Raystation (Biomechanical) [1] Mirada |
Mean proton field-specific geometric error (mm) for 3 DVFs: 1.05, 2.44, 2.84 1.01, 2.34, 2.66 0.74, 1.93, 2.34 |
Abbreviations: 4D = 4-dimensional; CT = computed tomography; DIR = deformable image registration; DVF = deformation vector field; MRI = magnetic resonance imaging; TRE = target registration error.
Value estimated from figure.
Used as a controlling structure in DIR.
Current System Limitations, Recommendations to Vendors, and Future Work for Phantoms
Limitations of commercial systems and recommendations for vendors
From results shown in the section Commercial Systems and User Testing, all commercial registration software evaluated by this working group was found to be partially compliant with TG132 recommendations (Table 7). One common deficiency among all vendors is the format of the exported DVF files, which has not been standardized throughout vendors. MIM, Velocity, and Mirada (through scripting) can export the DVF in DICOM format, whereas Raystation exports in a text format. The lack of proper DICOM standard definitions for this registration type limits the direct comparison between user-provided DVF and the gold-standard DVF and further limits the interpretability and usability outside the original system. Also, DVF files may be too large, thus making their display, transfer, and postprocessing cumbersome with the current means. The AAPM working group on Integration of Health Care Enterprise in Radiation Oncology recently formed a subgroup comprised of clinical users and industry partners focused on creating proper DICOM definitions for deformable image registration (Integration of Health Care Enterprise in Radiation Oncology Deformable Image Registration [UN45]). This should help standardize the transfer of these files between systems.
Table 7.
Compliance status of commercial software for TG132-recommended tests
| MIM | MIRADA | Raystation | Velocity | |
|---|---|---|---|---|
| Version | 6.8 | RTX1.8 | 6.0 | 3.2 |
| TG132 recommended metrics for evaluation | ||||
| TRE | 3 | 0 | 2 | 2* |
| MDA | 3 | 1 | 2 | 2 |
| DSC | 3 | 1 | 2 | 2 |
| Jacobian determinant | 3 | 1 | 2 | 2 |
| Consistency | 3 | 1 | 2 | 2 |
| TG132 vendor recommendations (7.B) | ||||
| Disclose basic components of their registration algorithm | Yes | Yes | Yes | Yes |
| Provide the ability to export the registration matrix or deformation vector field | Yes | Yes† | Yes† | Yes† |
| Provide tools to qualitatively evaluate the image registration | Yes | Yes | Yes | Yes |
| Provide the ability to identify landmarks on 2 images and calculate TRE | See row 1 | No | See row 1 | See row 1 |
| Provide the ability to calculate the DSC and MDA | See row 2–3 | No | See row 2–3 | See row 2–3 |
| Support the integration of a request and report system | Yes‡ | Yes‡ | Yes | Yes‡ |
Abbreviations: DSC = Dice similarity coefficient; MDA = mean distance to agreement; TG132 = task group 132; TRE = target registration error.
Level 3: Compliant with direct display of the results.
Level 2: Compliant with using scripting provided by the system.
Level 1: Support Digital Imaging and Communications in Medicine export, and users can calculate metrics using a third party software.
Level 0: Not compliant, not able to export.
Velocity has a separate workspace for defining TRE points. Importing points as part of the structure set allows for calculation of TRE but through using surface distance metrics, which does not provide the error in each direction.
DVF files exported from the tested systems are not in a standard Digital Imaging and Communications in Medicine format.
Most systems support the integration of a report generation but do not fully support the integration of the request.
There are other issues encountered with some systems in their current versions at the time of publication of this report, including difficulty with importing reference points for TRE calculations. The recommendation from TG132 is for the vendor to provide the ability to identify landmarks and calculate TRE. In the context of self-study evaluation, the user must be able to import predefined points into their system, calculate the transformed location of those points, and calculate TRE. Several systems have limitations in how they handle the TRE calculations for points defined outside of the system or require extensive workarounds to be able to perform these calculations properly. Admittedly, TRE calculation based on a set of 20 to 100 points may not be sufficient to properly evaluate DIR performance. Yet, it is still a reasonable approach when direct DVF comparison is not yet available for those commercial systems. Table 7 provides a list of the vendor recommended, the user tests from TG132, and the compliance status of each registration software (current version at the time of publication) with each test.
Aside from system compliance with TG132 tests, it should be noted that the TG132 itself has specific limitations when used as a guideline for self-study evaluation. One limitation is that the recommended site-specific testing is fully qualitative, which may be helpful for QA within a given institution but would not be sufficient for self-study evaluation among all clinical trial participants. Clinical trials aiming to use deformable registration should provide a set of site-specific data sets that are ideally designed to be sensitive to the inherent differences in various deformable registration techniques with recommendations on quantitative tests and requiring users to provide their testing results as part of the self-study evaluation process (Appendix E2). Additionally, TG132 does not make any direct recommendations on testing deformable registration for dose accumulation, which is expected to be the main reason for using deformable registration in clinical trials. This topic is addressed in our sister NRG Oncology publication.52 It is worth noting that there are discrepancies in the reported rigid registration results for the CBCT image in phantom data set 1 provided by TG132. Two systems (MIM and Velocity) reported 3.0 cm shift and the other 2 systems (Raystation and Mirada) reported 1.5 cm shift, while the expected shift should be 3.0 cm as updated by TG132 (Fig E1). However, the phantom displacement between the 2 image sets (CT and CBCT) in Raystation and Mirada is indeed 1.5 cm. We suspected that the discrepancies are from the header information of the created CBCT image using ImSimQA; therefore, the results for the CBCT image tests were discarded.
Recommendation for future end-to-end phantom development
Appropriate phantoms are needed for commissioning and QA of deformable image registration in a commercial system. Although phantom tests cannot replace patient-specific QA, they are an effective tool in the commissioning to (1) validate the functionality of the software, (2) evaluate the deformable registration QA tools provided in the software, (3) establish the workflow of DIR and perform end-to-end test, and (4) verify the accuracy of image registration. To cover different scenarios of deformable registration in a clinical setting, a set of ideal phantoms should include the following features: (1) be available for different imaging modalities (CT, MR, or ultrasound), (2) represent real patient anatomies as closely as possible, (3) represent a range of deformations that are clinically relevant, and (4) have a known ground truth deformation information. The phantom sets could be a number of fabricated physical phantoms, digital phantoms derived from physical phantom scans, computer-created synthetic digital phantoms, or the combination of these.
Multimodality deformation phantoms
Multimodality image fusion is common in radiation therapy. The algorithm used for multimodality DIR is different from that for mono-modality DIR in most commercial software systems and thus should be commissioned using multimodality phantoms. Examples of such phantoms include a tissue-like prostate phantom imaged using CT, MRI, ultrasound, and CBCT53; a plastic anthropomorphic head phantom that is CT/MR/PET compatible54; and a digital full-body 4D phantom that can be combined with simulation packages (PET, single-photon emission computed tomography, CT, MRI, Ultrasound) to generate realistic imaging data.55 A single phantom that is compatible with all imaging modalities is desirable, but it may not be achievable; thus, the clinical trials involving multimodality image deformation should consider multiple data sets for the DIR self-study evaluation.
Site-specific deformation phantoms
Different human anatomic regions have different natures of deformation, as explained in the section Site Specific Discussion and Example Cases. Site-specific deformation phantoms are needed for comprehensive tests of various clinical scenarios. Most anthropomorphic phantoms published in the literature are specific to an anatomic site, for example, head and neck16,54,56 thorax,17,30,57–59 abdomen,14 or prostate.53,59 A phantom that includes full-body anatomy can be achieved with a synthetic digital phantom.55 Site-specific phantoms usually include a typical deformation pattern for their corresponding anatomic site, but more patterns could be associated with a single site in real clinical settings. Phantoms are also needed for other clinical cases, for example, fusing arms-up scan with arms-down scan for an area of clinical interest in shoulder or axilla region.
Physical versus digital phantoms
Both physical and digital phantoms are capable of verification of image registration accuracy and validation of software system functionality. Physical phantoms are more suitable for system end-to-end tests during the image registration commissioning process, along with other test items such as image integrity and data transfer. Physical phantoms can be imaged by different scanners to generate realistic multimodality images. The ground truth deformation information is usually available only for a set of landmark points but not for every voxel in the image of physical phantoms. Physical phantoms are also limited in the freedom of deformation, depending on the actual design and material of the phantom. A physical phantom that can be deformed voxel by voxel is yet to be constructed. On the other hand, digital phantoms are more capable of mimicking different clinical deformations and are thus more suitable for software functionality validation. A digital phantom can be created from clinical scans, so it can represent more closely the anatomy of a real human body. A synthetic digital phantom that is created by applying DVF to a reference image has full ground truth deformation information for every voxel in the image. It should be noted though that the predefined DVF may be physically inappropriate in a digital phantom. Digital phantoms can test the image registration process, but they are not suitable for an end-to-end test of a software system. Digital phantoms are easy to be distributed so they are particularly useful for multi-institutional or multisystem comparison study.16,38
Conclusions
TG132 is an instructive reference for systems that perform rigid and deformation image registration, and it includes important tests to analyze the accuracy of these registrations. However, it does not provide a practical clinical guideline on implementing those recommended tests with current commercial systems. The NRG Oncology working group on image registration is a more practical guideline that developed and made available practical data sets and analytical tools for clinics to qualitatively and quantitatively assess their workflow in employing commercial rigid and deformation registration applications for radiation therapy treatments. The testing data presented in our study are provided by the users of various commercial systems, thus representing more realistic and clinically achievable scenarios. Further investigations with different digital phantoms and physical phantoms may be considered and made available to those needing to assess their imaging coregistration capability. The NRG Oncology will continue to provide resources to ensure the highest quality care for patients enrolled in clinical trials and include other clinics that may not be enrolled in trials but wish to undergo quality improvement of their services.
Supplementary Material
Acknowledgments
Sources of support: This project was supported by grant U24CA180803-06 (IROC) and 2U10CA180868-06 (NRG) from the National Cancer Institute (NCI).
Disclosures: Y.R. discloses support from NIH R44CA254844, outside of the submitted work; Q.C. discloses funding support from NIH R43EB027523, R44CA254844 and Varian Research Grant, outside the submitted work; R.K. reports personal fees from ViewRay Inc, outside the submitted work.
Footnotes
All data generated and analyzed during this study are included in this published article (and its supplementary information files). Phantoms used in this study are available for download at https://www.aapm.org/pubs/reports/report132.aspand https://www.creatis.insa-lyon.fr/rio/popi-model?action=show&redirect=popi.
Supplementary Materials
Supplementary material for this article can be found at https://doi.org/10.1016/j.prro.2021.02.007.
References
- 1.Brock KK, Mutic S, McNutt TR, Li H, Kessler ML. Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the AAPM Radiation Therapy Committee Task. Med Phys. 2017;44:E43–E76. [DOI] [PubMed] [Google Scholar]
- 2.West JB, Fitzpatrick JM, Toms SA, Maurer CR, Maciunas RJ. Fiducial point placement and the accuracy of point-based, rigid body registration. Neurosurgery. 2001;48:810–816. [DOI] [PubMed] [Google Scholar]
- 3.Fitzpatrick JM, West JB, Maurer CR. Predicting error in rigid-body point-based registration. IEEE Trans Med Imag. 1998;17:694–702. [DOI] [PubMed] [Google Scholar]
- 4.Takacs B. Comparing face images using the modified Hausdorff distance. Patt Recog. 1998;31:1873–1881. [Google Scholar]
- 5.Huttenlocher DP, Klanderman GA, Rucklidge WJ. Comparing images using the Hausdorff Distance. IEEE Trans Pattern Analy Mach Intell. 1993;15:850–863. [Google Scholar]
- 6.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 7.Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med Imaging. 2015;15:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalana V, Kim YM. A methodology for evaluation of boundary detection algorithms on medical images. IEEE Trans Med. Imag. 1997;16:642–652. [DOI] [PubMed] [Google Scholar]
- 9.Cross VV, Sudkamp TA. Similarity and Compatibility in Fuzzy Set Theory: Assessment and Application. Heidelberg, Germany: Physica-Verlag; 2002. [Google Scholar]
- 10.Sulaiman NH, Mohamad D. A Jaccard-based similarity measure for soft sets. IEEE Symp Hum Sci Eng Res. 2012:659–663. [Google Scholar]
- 11.Hoffmann C, Krause S, Stoiber EM, et al. Accuracy quantification of a deformable image registration tool applied in a clinical setting. J Appl Clin Med Phys. 2014;15:4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latifi K, Caudell J, Zhang G, Hunt D, Moros EG, Feygelman V. Practical quantification of image registration accuracy following the AAPM TG-132 report framework. J Appl Clin Med Phys. 2018;19: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latifi K, Zhang G, Stawicki M, van Elmpt W, Dekker A, Forster K. Validation of three deformable image registration algorithms for the thorax. J Appl Clin Med Phys. 2013;14:3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao Y, Wang L, Xu X, et al. An anthropomorphic abdominal phantom for deformable image registration accuracy validation in adaptive radiation therapy. Med Phys. 2017;44:2369–2378. [DOI] [PubMed] [Google Scholar]
- 15.Nie K, Pouliot J, Smith E, Chuang C. Performance variations among clinically available deformable image registration tools in adaptive radiotherapy - how should we evaluate and interpret the result? J Appl Clin Med Phys. 2016;17:328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pukala J, Johnson PB, Shah AP, et al. Benchmarking of five commercial deformable image registration algorithms for head and neck patients. J Appl Clin Med Phys. 2016;17:25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandemeulebroucke J, Sarrut D, Clarysse P. The POPI-model, a point-validated pixel-based breathing thorax model. XVth International Conference on the Use of Computers in Radiation Therapy (ICCR). 2007; Toronto, Canada. [Google Scholar]
- 18.Vaman C, Staub D, Williamson J, Murphy MJ. A method to map errors in the deformable registration of 4DCT images. Med Phys. 2010;37:5765–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang JZ, Veeraraghavan H, Armato SG, et al. Auto-segmentation for thoracic radiation treatment planning: A grand challenge. Med Phys. 2017;44:3297–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelms BE, Tome WA, Robinson G, Wheeler J. Variations in the contouring of organs at risk: test case from a patient with oropharyngeal cancer. Int J Radiat Oncol Biol Phys. 2012;82: 368–378. [DOI] [PubMed] [Google Scholar]
- 21.Fiorino C, Reni M, Bolognesi A, Cattaneo GM, Calandrino R. Intra-and inter-observer variability in contouring prostate and seminal vesicles: Implications for conformal treatment planning. Radiother Oncol. 1998;47:285–292. [DOI] [PubMed] [Google Scholar]
- 22.Hardcastle N, Tome WA, Cannon DM, et al. A multi-institution evaluation of deformable image registration algorithms for automatic organ delineation in adaptive head and neck radiotherapy. Radiat Oncol. 2012;7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarudis S, Karlsson A, Bibac D, Nyman J, Back A. Evaluation of deformable image registration accuracy for CT images of the thorax region. Phys Med. 2019;57:191–199. [DOI] [PubMed] [Google Scholar]
- 24.Bissonnette JP, Balter PA, Dong L, et al. Quality assurance for image-guided radiation therapy utilizing CT-based technologies: A report of the AAPM TG-179. Med Phys. 2012;39:1946–1963. [DOI] [PubMed] [Google Scholar]
- 25.Mutic S, Palta JR, Butker EK, et al. Quality assurance for computed-tomography simulators and the computedtomography-simulation process: Report of the AAPM radiation therapy committee task group no. 66. Med Phys. 2003;30:2762–2792. [DOI] [PubMed] [Google Scholar]
- 26.Loi G, Fusella M, Lanza E, et al. Performance of commercially available deformable image registration platforms for contour propagation using patient-based computational phantoms: A multi-institutional study. Med Phys. 2018;45:748–757. [DOI] [PubMed] [Google Scholar]
- 27.Fukumitsu N, Nitta K, Terunuma T, et al. Registration error of the liver CT using deformable image registration of MIM Maestro and Velocity AI. BMC Med Imaging. 2017;17:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang G, Huang TC, Guerrero T, et al. Use of three-dimensional (3D) optical flow method in mapping 3D anatomic structure and tumor contours across four-dimensional computed tomography data. J Appl Clin Med Phys. 2008;9:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssens G, Orban de Xivry J, Fekkes S, Dekker A, et al. Evaluation of nonrigid registration models for interfraction dose accumulation in radiotherapy. Med Phys. 2009;36:4268–4276. [DOI] [PubMed] [Google Scholar]
- 30.Castillo R, Castillo E, Guerra R, et al. A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets. Phys Med Biol. 2009;54:1849–1870. [DOI] [PubMed] [Google Scholar]
- 31.Guerrero T, Zhang G, Huang TC, Lin KP. Intrathoracic tumour motion estimation from CT imaging using the 3D optical flow method. Phys Med Biol. 2004;49:4147–4161. [DOI] [PubMed] [Google Scholar]
- 32.Zhang GG, Huang TC, Forster KM, et al. Dose mapping: Validation in 4D dosimetry with measurements and application in radiotherapy follow-up evaluation. Comput Methods Programs Biomed. 2008;90: 25–37. [DOI] [PubMed] [Google Scholar]
- 33.Sarrut D, Baudier T, Ayadi M, Tanguy R, Rit S. Deformable image registration applied to lung SBRT: Usefulness and limitations. Phys Med. 2017;44:108–112. [DOI] [PubMed] [Google Scholar]
- 34.Kavanaugh J, Hugo G, Robinson CG, Roach MC. Anatomical adaptation-early clinical evidence of benefit and future needs in lung cancer. Semin Radiat Oncol. 2019;29:274–283. [DOI] [PubMed] [Google Scholar]
- 35.Al-Mayah A, Moseley J, Velec M, Brock KK. Sliding characteristic and material compressibility of human lung: Parametric study and verification. Med Phys. 2009;36:4625–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guy CL, Weiss E, Christensen GE, Jan N, Hugo GD. CALIPER: A deformable image registration algorithm for large geometric changes during radiotherapy for locally advanced non-small cell lung cancer. Med Phys. 2018;45:2498–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guy CL, Weiss E, Che S, Jan N, Zhao S, Rosu-Bubulac M. Evaluation of image registration accuracy for tumor and organs at risk in the thorax for compliance with TG 132 recommendations. Adv Radiat Oncol. 2019;4:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadoya N, Nakajima Y, Saito M, et al. Multi-institutional validation study of commercially available deformable image registration software for thoracic images. Int J Radiat Oncol Biol Phys. 2016;96: 422–431. [DOI] [PubMed] [Google Scholar]
- 39.Velec M, Moseley JL, Svensson S, Hardemark B, Jaffray DA, Brock KK. Validation of biomechanical deformable image registration in the abdomen, thorax, and pelvis in a commercial radiotherapy treatment planning system. Med Phys. 2017;44:3407–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heukelom J, Fuller CD. Head and neck cancer adaptive radiation therapy (ART): Conceptual considerations for the informed clinician. Semin Radiat Oncol. 2019;29:258–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Q, Chi Y, Chen PY, Krauss DJ, Yan D, Martinex A. Adaptive replanning strategies accounting for shrinkage in head and neck IMRT. Int J Radiat Oncol Biol Phys. 2009;75:924–932. [DOI] [PubMed] [Google Scholar]
- 42.McCulloch MM, Lee C, Rosen BS, et al. Predictive models to determine clinically relevant deviations in delivered dose for head and neck cancer. Pract Radiat Oncol. 2019;9:E422–E431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castadot P, Lee JA, Parraga A, Geets X, Macq B, Gregoire V. Comparison of 12 deformable registration strategies in adaptive radiation therapy for the treatment of head and neck tumors. Radiother Oncol. 2008;89:1–12. [DOI] [PubMed] [Google Scholar]
- 44.Zhong HL, Chetty IJ. Caution must be exercised when performing deformable dose accumulation for tumors undergoing mass changes during fractionated radiation therapy. Int J Radiat Oncol Biol Phys. 2017;97:182–183. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen TN, Moseley JL, Dawson LA, Jaffray DA, Brock KK. Adapting liver motion models using a navigator channel technique. Med Phys. 2009;36:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehrbar S, Johl A, Kuhni M, et al. ELPHA: Dynamically deformable liver phantom for real-time motion-adaptive radiotherapy treatments. Med Phys. 2019;46:839–850. [DOI] [PubMed] [Google Scholar]
- 47.Polan DF, Feng M, Lawrence TS, Ten Haken RK, Brock KK. Implementing radiation dose-volume liver response in biomechanical deformable image registration. Int J Radiat Oncol Biol Phys. 2017;99:1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reese AS, Yang X, Lu W, et al. Deformable image registration as a method to assess motion for pancreatic cancer using 4D computed tomography (CT) scans. Int J Radiat Oncol Biol Phys. 2012;84: S771. [Google Scholar]
- 49.Tai A, Liang Z, Erikson B, Li XA. Management of respiration-induced motion with 4-dimensional computed tomography (4DCT) for pancreas irradiation. Int J Radiat Oncol Biol Phys. 2013; 86:908–913. [DOI] [PubMed] [Google Scholar]
- 50.Ahunbay EE, Kimura B, Liu F, Erickson BA, Li XA. Comparison of various online strategies to account for interfractional variations for pancreatic cancer. Int J Radiat Oncol Biol Phys. 2013;86:914–921. [DOI] [PubMed] [Google Scholar]
- 51.Ribeiro CO, Knopf A, Langendijk JA, Weber DC, Lomax AJ, Zhang Y. Assessment of dosimetric errors induced by deformable image registration methods in 4D pencil beam scanned proton treatment planning for liver tumours. Radiother Oncol. 2018;128: 174–181. [DOI] [PubMed] [Google Scholar]
- 52.Glide-Hurst C, Lee P, Yock AD, et al. Adaptive radiation therapy (ART) strategies and technical considerations: A state of the ART review from NRG Oncology. Int J Radiat Oncol Biol Phys. 2021; 109:1054–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ionascu D, Castillo E, Qin A, et al. Performance of cross-modality DIR algorithms using images computed from a novel, tissue-like phantom capable of reproducible degrees of deformation. Med Phys. 2016;43:3738. [Google Scholar]
- 54.Mutic S, Dempsey JF, Bosch WR, et al. Multimodality image registration quality assurance for conformal three-dimensional treatment planning. Int J Radiat Oncol Biol Phys. 2001;51: 255–260. [DOI] [PubMed] [Google Scholar]
- 55.Segars WP, Bond J, Frush J, et al. Population of anatomically variable 4D XCAT adult phantoms for imaging research and optimization. Med Phys. 2013;40:043701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singhrao K, Kirby N, Pouliot J. A three-dimensional head-and-neck phantom for validation of multimodality deformable image registration for adaptive radiotherapy. Med Phys. 2014;41:121709. [DOI] [PubMed] [Google Scholar]
- 57.Castillo R, Castillo E, Fuentes D, et al. A reference dataset for deformable image registration spatial accuracy evaluation using the COPDgene study archive. Phys Med Biol. 2013;58:2861–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kashani R, Hub M, Kessler ML, Balter JM. Technical note: A physical phantom for assessment of accuracy of deformable alignment algorithms. Med Phys. 2007;34:2785–2788. [DOI] [PubMed] [Google Scholar]
- 59.Stanley N, Glide-Hurst C, Kim J, et al. Using patient-specific phantoms to evaluate deformable image registration algorithms for adaptive radiation therapy. J Appl Clin Med Phys. 2013;14: 177–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.