ABSTRACT
Various toxic compounds disrupt bacterial physiology. While bacteria harbor defense mechanisms to mitigate the toxicity, these mechanisms are often coupled to the physiological state of the cells and become ineffective when the physiology is severely disrupted. Here, we characterized such feedback by exposing Escherichia coli to protonophores. Protonophores dissipate the proton motive force (PMF), a fundamental force that drives physiological functions. We found that E. coli cells responded to protonophores heterogeneously, resulting in bimodal distributions of cell growth, substrate transport, and motility. Furthermore, we showed that this heterogeneous response required active efflux systems. The analysis of underlying interactions indicated the heterogeneous response results from efflux-mediated positive feedback between PMF and protonophores’ action. Our studies have broad implications for bacterial adaptation to stress, including antibiotics.
KEYWORDS: bacterial physiology, single-cell microscopy, cell-to-cell heterogeneity, efflux pumps, proton motive force, protonophore
INTRODUCTION
An electrochemical proton gradient across the cytoplasmic membrane, alternatively known as proton motive force (PMF), drives vital processes in cells. For example, PMF powers ATP synthesis (1, 2), transport of a wide range of substrates, including essential ions and metabolites (3–6), and motility (7–9). Furthermore, PMF plays an important role in cell division (10) and cell-to-cell signaling (11, 12). Due to its importance, PMF is a key target for chemical warfare between living organisms. For example, bacteria dissipate PMF of other species to increase their colonization (13–18). A host dissipates PMF of pathogens to slow or prevent their invasion (19, 20).
One common way to dissipate PMF is via protonophores. They are a class of ionophores that collapse the proton gradient across the cell membrane by shuffling protons (21–23). Protonophores have been extensively used in various research fields to perturb a wide range of cellular processes, particularly in the antibiotic research field, due to the critical role of PMF in antibiotic influx, efflux, and mechanism of action. For example, aminoglycoside influx is PMF driven (24), and the PMF dissipation by protonophores turns aminoglycoside from bactericidal to bacteriostatic (25) or generates antibiotic-tolerant persisters (26). Furthermore, efflux pumps, a main culprit for multidrug resistance, require PMF to pump antibiotic molecules out of cells (27). Because PMF dissipation has detrimental effects on cells, protonophores themselves work as antibiotic agents (17, 18, 28–33). Perhaps unsurprisingly, protonophores naturally produced by bacteria or hosts alter antibiotic efficacy (20, 34, 35).
The detailed action of protonophores has been extensively studied in vitro using reconstituted lipid-bilayer systems (for example, see references 36 and 37). However, in vivo effects of protonophores are less well understood, despite the fact that they are critical to bacterial physiology, microbial competition, host-pathogen interaction, and antibiotic action and resistance. In this study, we characterized the cellular response to protonophores.
RESULTS
Heterogeneous responses of bacteria to protonophores.
We measured the growth of E. coli treated with a common protonophore, carbonyl cyanide m-chlorophenyl hydrazine (CCCP). At increasing CCCP concentrations, the rate of population growth decreased gradually (see Fig. S1A in the supplemental material). We then monitored the growth of individual cells at 50 μM CCCP, an intermediate concentration at which a population exhibits a moderate growth reduction (Fig. S1A). We found an all-or-none effect of CCCP at the single-cell level (Fig. 1A); some cells did not grow, whereas other cells continued to grow at the same rate as untreated cells.
FIG 1.
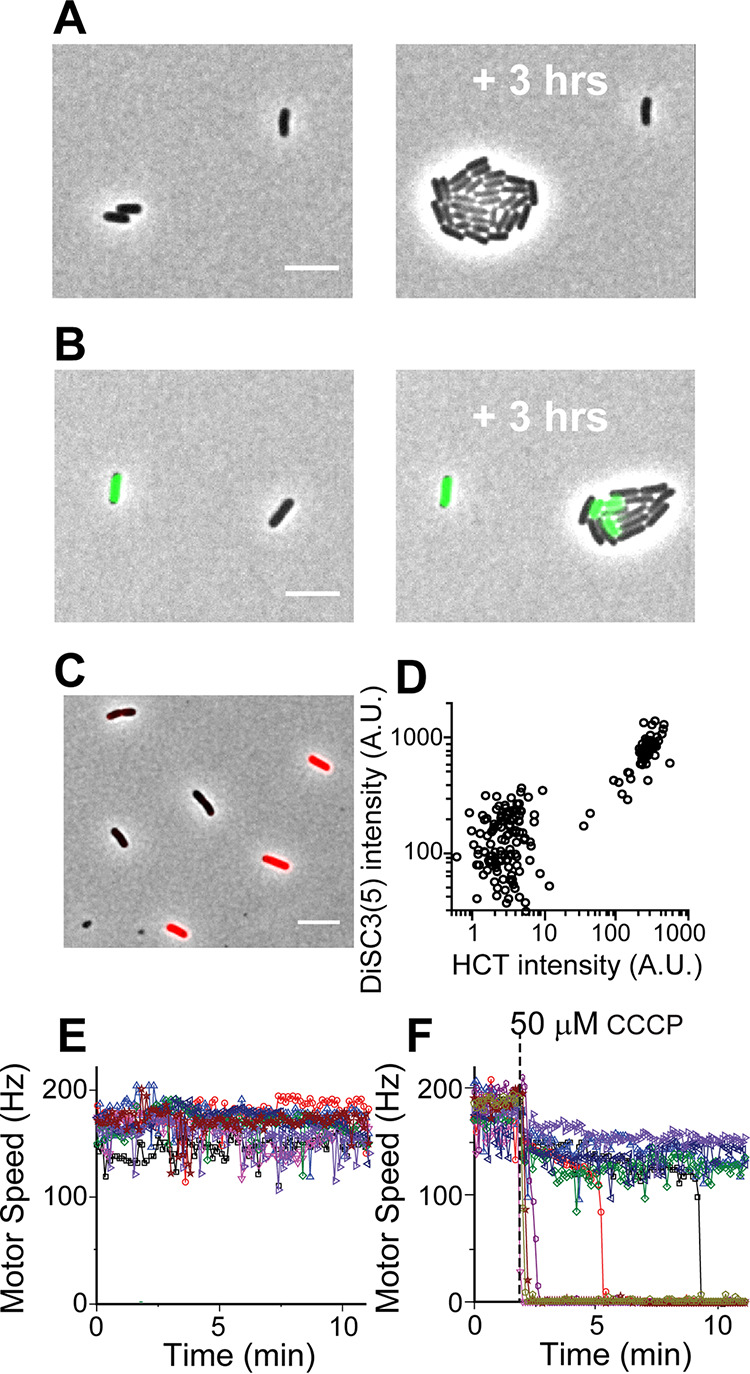
Heterogeneous responses of E. coli to 50 μM CCCP. (A) Some cells grew normally (growth rate of 0.87 ± 0.07/h, which is comparable to 0.83 ± 0.09/h for untreated cells). However, growth was completely inhibited in other cells. (B) Cells exhibited two distinct HCT levels, and HCT-bright cells did not grow. Out of 297 cells, 74 cells were HCT-bright and nongrowing. Cells transitioned from HCT-dim to HCT-bright during the experiment, as indicated by HCT-bright cells in the growing microcolony on the right. (C and D) Cells exhibited two distinct intracellular DiSC3(5) intensities, which are correlated with HCT intensities. The scale bar represents 5 μm. Note that HCT intensities are quantified in Fig. 2 and show the HCT intensity distribution differs between the ΔtolC and WT strains. Thus, we compared the DiSC3(5) intensity in the ΔtolC and WT strains. We found that DiSC3(5) intensity in the ΔtolC strain was moderately higher (∼50%). While this finding agrees with the previous finding that DiSC3(5) is a substrate of the efflux pumps (80), the efflux activity is only moderate and cannot explain the 10-fold difference in DiSC3(5) intensity between DiSC3(5)-bright and DiSC3(5)-weak cells in panel D. (E) In the absence of CCCP, the motor speeds were uniform across a population. (F) When exposed to CCCP, cells exhibited two distinct motor speeds.
Growth of E. coli under various conditions. (A) Growth curve of WT E. coli treated with various concentrations of CCCP. At 50 μM, the rate of population growth decreased moderately. (B) Growth curve of WT E. coli treated with various concentrations of HCT. At 0.1 μM (the concentration used in the present study), HCT has little effect on cell growth. (C) Growth of WT E. coli treated with 0.1 nM DiSC3(5). At this concentration, DiSC3(5) has little effect on cell growth. Download FIG S1, TIF file, 0.1 MB (154KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We then examined how CCCP affects substrate transport by using a fluorescent dye, Hoechst 33342 (HCT) (38). Intracellular HCT intensity was uniformly low in the absence of CCCP. At an intermediate CCCP concentration (50 μM), we observed coexistence of cells exhibiting two distinct HCT intensities (Fig. 1B). Importantly, HCT intensity was correlated with cell growth; cells with low intensity (HCT-dim) grew unperturbed, while those with high intensity (HCT-bright) exhibited no growth (Fig. 1B). Some cells transitioned from HCT-dim to HCT-bright during the experiment. After the transition, HCT-bright cells ceased to grow.
We then examined the mechanism for the coexistence of cells with the distinct cell growth and substrate transport states. Given that CCCP disrupts PMF (22), we hypothesized that the effect of CCCP on PMF is heterogeneous, i.e., it disrupts PMF severely in some cells but not in others. To examine this hypothesis, we evaluated PMF using two different approaches. First, we used a dye sensitive to membrane potential, DiSC3(5). When extracellular pH is comparable to that of intracellular pH (as is the case for our growth medium), membrane potential is primarily determined by PMF. DiSC3(5) accumulates in cells with strong PMF and self-quenches, resulting in low fluorescence (39, 40). We first confirmed that our working DiSC3(5) concentration (nanomolars) did not affect cell growth (Fig. S1C). We then exposed cells to 50 μM CCCP and found they exhibited two distinct DiSC3(5) intensities (Fig. 1C). Because DiSC3(5) and HCT fluorescence emission spectra are well separated, they can be used simultaneously. We found that DiSC3(5)-bright cells were also HCT-bright and did not grow (upper right group in Fig. 1D), whereas DiSC3(5)-dim cells were HCT-dim and grew (lower left group in Fig. 1D). This observation suggests that the coexistence of cells with two distinct HCT intensities and cell growth states is caused by the heterogeneous effect of CCCP on PMF.
To further test our hypothesis, we next measured the bacterial flagellar motor speed. Flagellar motor is powered by, and its speed is proportional to, PMF (7, 9). By measuring the motor rotation speed, we previously determined a relative change in PMF (41, 42). In the absence of CCCP, the motor speed remained high and constant (Fig. 1E). Treatment with 50 μM CCCP resulted in two subpopulations, one with slightly reduced rotation (high PMF) and the other with no rotation (zero PMF) (Fig. 1F). The time point at which each cell lost PMF varied, agreeing with our observation that cell growth stopped at various times during CCCP treatment (Fig. 1B). Our observation of two distinct motor speeds and DiSC3(5) intensities in a population supports our hypothesis that CCCP disrupts the PMF of cells heterogeneously.
We then quantified the degree of heterogeneity by characterizing HCT fluorescence intensity and motor speeds over a wide range of CCCP concentrations. At low CCCP concentrations (≤25 μM), HCT intensity in a population was low (HCT-dim) and its distribution was unimodal (Fig. 2A and B). The motor speed was barely affected (Fig. 3A). An increase in the CCCP concentration to 50 μM did not shift the center of the original peak in the HCT intensity distribution but led to the appearance of another peak on the right (HCT-bright cells), showing a bimodal distribution (Fig. 2C). This agrees with the motor speed data, which showed two distinct motor speeds at this concentration (Fig. 3B). At higher CCCP concentrations (≥70 μM), the original peak on the left in the HCT intensity distribution disappeared, indicating the enrichment of HCT-bright cells (Fig. 2D to E). This enrichment is accompanied by the complete collapse of PMFs, as indicated by motor speeds of zero in all cells (Fig. 3C to D).
FIG 2.
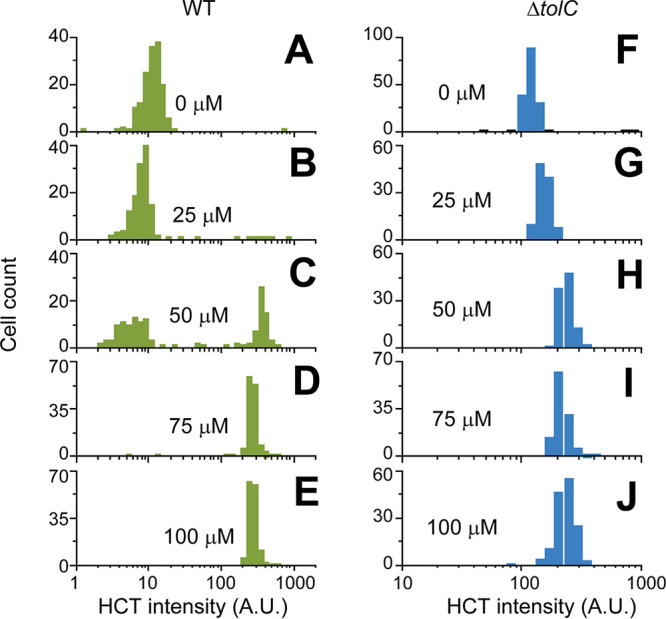
HCT fluorescence intensity distribution in cells treated with various CCCP concentrations. (A and B) At low CCCP concentrations (≤25 μM), HCT intensity was low across a population (HCT-dim), resulting in a unimodal distribution with the peak center near ∼10 AU. (C) Increasing the CCCP concentration to 50 μM did not shift the peak center but led to the appearance of another peak on the right (>100 AU), showing a bimodal distribution. At higher (≥75 μM) CCCP concentrations, the left low-intensity peak disappeared, showing the enrichment of HCT-bright cells. (F to J) The ΔtolC strain lacks a peak on the left, exhibiting a unimodal distribution. More than 200 cells were analyzed for each condition. We made a similar observation for two other protonophores, TCS and indole (Fig. S3 and S4).
FIG 3.
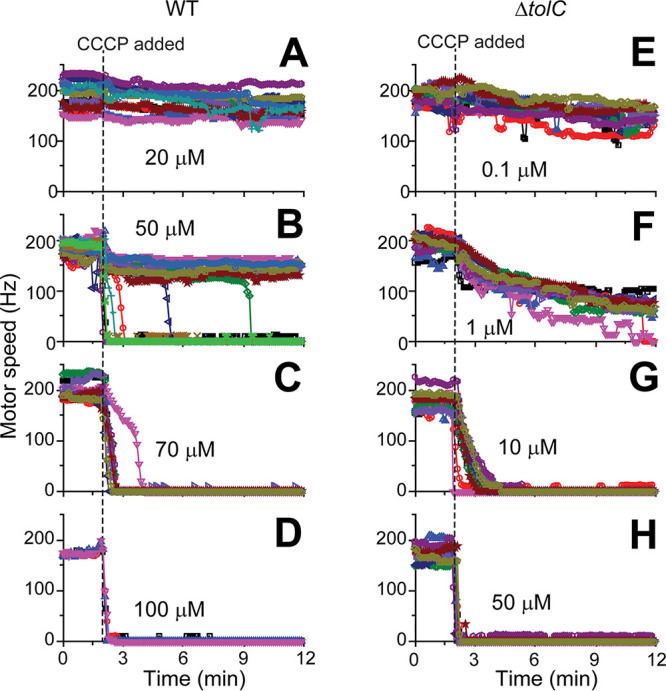
Flagellar motor speeds of cells treated with various CCCP concentrations. (A) At low CCCP concentrations, the motor speeds of WT cells were barely affected. (B) Cells exhibited two distinct motor speeds at 50 μM CCCP, one with a slightly reduced rotation (high PMF) and the other with no rotation (zero PMF). (C and D) At higher CCCP concentrations (≥70 μM), the motor speeds were zero in all cells. (E to H) The ΔtolC strain exhibited a uniform and gradual reduction in the motor speed at increasing CCCP concentrations. Note that, at very low PMF values where the bacterial flagellar motor operates with one stator unit, the motor can transiently stop in a step-like manner (81, 82), which can explain the purple trajectory in panel F. Motor speed measurements are laborious and time-consuming. The motor speeds of 10 to 15 cells were analyzed for each condition except for the experiment with the WT strain exposed to 100 μM CCCP (where 4 cells were analyzed).
HCT intensity distribution in WT (left) and ΔtolC (right) cells treated with TCS. (Left) WT strain. At low TCS concentrations (≤5 μM), the HCT intensity was uniformly low across a population, leading to a unimodal distribution (with the peak center around ∼10 AU). Increasing TCS concentrations led to the emergence of the second peak on the right (>100 AU, HCT-bright cells), showing a bimodal distribution. At higher concentrations (≥30 μM), the left low-intensity peak disappeared, showing the enrichment of HCT-bright cells. (Right) ΔtolC strain. For all TCS concentrations tested, the left low-intensity peak is absent in the ΔtolC strain, which resulted in a unimodal distribution. The center of the single peak shifted moderately at increasing TCS concentrations. This observation is consistent with that made for CCCP (Fig. 2). Download FIG S3, TIF file, 0.2 MB (173.8KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
HCT intensity distribution in WT (left) and ΔtolC (right) cells treated with indole. The result is similar to that from the TCS experiment in Fig. S3. (Left) WT strain. Increasing indole concentrations led to the emergence of the second peak on the right (>100 AU, HCT-bright cells) and a mild shift in the center of the left low-intensity peak. The two peaks merged at 7.5 mM indole concentration. (Right) ΔtolC strain. For all indole concentrations tested, a left low-intensity peak is absent in the ΔtolC strain. The center of the single peak shifted moderately at increasing indole concentrations. Of note, we previously measured the motor speeds of WT cells at low indole concentrations (≤2 mM), which showed a uniform reduction in the motor speeds (41). Here, we observed a bimodal distribution of the HCT intensity at higher concentrations (≥2.5 mM). At 2.5 mM, the fraction of cells exhibiting high HCT intensities was less than 5%, which is difficult to detect with motor speed measurements because motor speed measurements are laborious, and it is challenging to track more than 20 cells. Download FIG S4, TIF file, 0.2 MB (169.7KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The heterogeneous effect of a protonophore is mediated by the efflux pumps.
Our observations described above confirm that cells exposed to CCCP exhibit distinct PMF levels. In bacteria, positive feedback is required to stabilize distinct phenotypic states (43). Here, we investigated a feedback mechanism that stabilizes two distinct PMF levels in a CCCP-exposed population. Bacteria can mitigate harmful effects of protonophores and other toxic compounds by extruding them with efflux pumps (44–46) (green arm in Fig. 4). However, these pumps are powered by PMF (27) (blue arm in Fig. 4) and, thus, are subject to disruption by protonophores (red arm in Fig. 4), suggesting an efflux-mediated positive feedback between protonophores and PMF (Fig. 4).
FIG 4.

Model of efflux-mediated positive feedback. Efflux pumps transport protonophores out of cells (44, 45) (green arm). The pump is powered by PMF (27) (blue arm) and, thus, is subject to disruption by protonophores (red arm).
We experimentally tested this potential role of efflux activity by repeating our measurements using the ΔtolC strain. In many bacterial species, including E. coli, TolC is a major component of efflux pumps (47), and these pumps can be inactivated by the tolC knockout. We first confirmed that the tolC knockout itself had little effect on the PMF level in the absence of CCCP (Fig. S2). Our HCT measurements showed that the HCT intensity was uniform across a ΔtolC population, and the analysis showed the absence of a left, low-intensity peak, which results in a narrow unimodal distribution (Fig. 2F). Increasing CCCP concentrations moderately shifted the peak center, but the distribution remained unimodal (Fig. 2F to J), which is in contrast to a bimodal distribution in the wild-type (WT) strain (Fig. 2A to E). This observation with the ΔtolC strain is consistent with motor speed measurements, which showed that ΔtolC cells exhibited a uniform and gradual reduction in the motor speed at increasing CCCP concentrations (Fig. 3E to H). These data indicate that the efflux pumps indeed play a critical role in the heterogeneous effect of a protonophore.
Motor speeds of WT (green) and ΔtolC (blue) cells in the absence of protonophores. Here, we compared the motor speeds of WT and ΔtolC strains. Their motor speeds were comparable, indicating that ΔtolC has no noticeable effect on the PMF. Download FIG S2, TIF file, 0.1 MB (85KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We next examined the HCT intensity distribution in cells treated with other common protonophores, 3,3′,4′,5-tetrachlorosalicylanilide (TCS) (48, 49) and indole (37, 41). Similar to CCCP, the WT cells exhibited two distinct HCT intensities in intermediate concentrations of these protonophores, and the analysis confirmed a bimodal distribution (Fig. S3 and S4, left). The centers of the two peaks were comparable to those observed in a CCCP-treated population, the left peak at ∼10 arbitrary units (AU) and the right peak at >100 AU. In a ΔtolC population, however, the HCT intensity was uniform, and the analysis showed the absence of a left, low-intensity peak (Fig. S3 and S4, right), which is consistent with our finding from the experiment with CCCP.
DISCUSSION
PMF is at the basis of vital physiological functions in cells (1–11). Protonophores are synthesized for a research purpose or produced naturally by living organisms (13–20). For example, indole (a protonophore [37, 41] tested in the present study) is one of the most abundant compounds in a dense bacterial culture and present in high concentrations in gut microbiome (12, 50). Protonophores collapse the total PMF that acts on protons by allowing them to equilibrate (22, 37, 51). Our additional analysis of the functional dependence of the motor speed on the CCCP concentration (see Fig. S5 in the supplemental material) is consistent with this known mechanism of action.
Our results agree with the known CCCP action as a protonophore. Previous in vitro measurements have showed that CCCP collapses the total PMF that acts on protons by allowing them to equilibrate (22, 37). To confirm this mechanism of CCCP action, we analyzed a relative change in the motor speed as a function of a CCCP concentration, [CCCP], in the ΔtolC strain: ω/ω0 =PMF/PMF0 = f ([CCCP],t), as we have shown before (41). The mean speeds during the last 2 min of CCCP exposure (Fig. 3E to H) were obtained from Gaussian fits to probability densities of speeds left) and normalized to the pre-CCCP treatment speed. We then fit the normalized speeds with a hyperbolic function and obtained the following relationship, PMF/PMF0 = 1/(2.64 × [CCCP]+1) (right). This agrees with an inverse relationship between PMF and [CCCP] from in vitro studies (22, 37), supporting that CCCP acts as a protonophore. We note that, unlike some other ionophores, such as butanol and indole (41), the PMF reduction at a given CCCP concentration takes several minutes (Fig. 3E to H). Download FIG S5, TIF file, 0.4 MB (378.5KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Efflux pumps transport protonophores out of cells, protecting them from protonophores’ harmful effects (44, 45). The present study demonstrates that this protection is heterogeneous, protecting some cells but not all. Our findings indicate that this heterogeneity emerges because protonophores affect their own efflux transport. For example, if cells initially have a strong efflux activity, upon the exposure to protonophores, they extrude protonophores better and maintain their PMF, thereby continuing to support the strong efflux activity (opposite for cells with weak efflux activity).
PMF has important roles in antibiotic influx, efflux, and mechanism of action. As such, protonophores were extensively used in antibiotic research. Interestingly, heterogeneous responses were observed in these studies. For example, a subpopulation of cells can tolerate antibiotics by not growing: bacterial persisters (52, 53). The postantibiotic effect, continued growth suppression after antibiotic withdrawal, is strongly skewed by a small subpopulation that resumes growth earlier than others (54). Protonophores have strong effects on the emergence of a persister subpopulation (26, 55) and an early-grower subpopulation (56). Importantly, these heterogeneous effects of protonophores involve efflux pumps (26, 55–57). This is consistent with our observation of heterogeneous growth phenotypes (i.e., the coexistence of nongrowing and growing subpopulations), which is mediated by the feedback among protonophores, PMF, and efflux pumps. Our additional motor-speed data indicate that, upon protonophore washout, cells in the nongrowing subpopulation recover their PMFs heterogeneously (Fig. S6). We believe that our findings are useful for antibiotic research, given the important role of PMF and efflux pumps to antibiotic action and widespread use of protonophores in the field. Furthermore, some protonophores are used as antibiotic agents (17, 18, 28–33), for which our findings can be directly applicable.
Recovery of PMF after CCCP washout. We have discussed in the main text that the treatment with 50 μM CCCP resulted in two subpopulations, one with a moderate reduction in the motor speed and the other with a motor speed of zero. When CCCP was removed, the former recovered the motor speed immediately. Partial recovery was observed in some cells in the latter subpopulation (red arrow). Download FIG S6, TIF file, 0.1 MB (87.6KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Our results have broad implications for bacterial adaptation to stress. In natural environments, various toxic compounds negatively affect bacterial physiology. While bacteria harbor defense mechanisms to mitigate the toxicity, these mechanisms are often coupled to the physiological state of the cells and become ineffective when the physiology is severely disturbed. In our studies, this coupling is manifested as the feedback between protonophores and efflux pumps. Similar coupling could be realized through other mechanisms. For example, efflux pumps extrude biocides or other plant-derived disinfectants, but their expression is altered by these compounds, thereby forming feedback (58, 59). Cytoplasmic pH is an important determinant for antibiotic efflux, as it contributes to PMF and can affect the expression of some efflux systems (60, 61). However, pH can be altered by antibiotics (51, 62) (e.g., nigericin specifically targets the cytoplasmic pH), suggesting an additional feedback mediated by pH. Our studies provide insight into how such coupling could affect bacterial adaptation to these toxic compounds.
Lastly, protonophores have been extensively utilized as a powerful tool to perturb various physiological processes in cells, including cell division, motility, and antibiotic transport. It was commonly assumed that increasing protonophore concentrations lead to gradual disruption of these processes. However, our studies confirm that the disruption is heterogeneous at the single-cell level. Our data from the experiment with the ΔtolC strain show that gradual disruption can be achieved but requires the inactivation of efflux activities. Additionally, we provide a functional dependency of the PMF loss on a CCCP concentration (Fig. S5), which can facilitate the use of CCCP to perturb PMF in a quantitative manner. These results should be useful for experimental designs and data interpretation in future studies.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli K-12 NCM3722 (63–65) and Neidhart’s morpholine propanesulfonic acid (MOPS) minimal medium (66) with glucose and ammonium as the carbon and nitrogen sources were used, except for the motor speed measurement (see below). See Table S1 in the supplemental material for all the ingredients and their concentrations used in the media. The medium pH is 7.0, which allowed E. coli to keep neutral cytoplasmic pH (67–69) and ensured HCT fluorescence does not incur any pH-related intensity changes (70). To make the ΔtolC strain (NMK320), the tolC gene deletion allele from the Keio deletion collection (71, 72) was transferred to the NCM3722 strain using P1 transduction (73). The Kmr gene was flipped out as previously described (71, 72).
Chemicals used in our experiments. Download Table S1, DOCX file, 0.02 MB (20.6KB, docx) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cells were cultured at 37°C with constant agitation at 250 rpm in a water bath (New Brunswick Scientific). To monitor their growth, the optical density at 600 nm (OD600) of the culture was measured using a Genesys20 spectrophotometer (Thermo-Fisher) with a standard cuvette (16.100-Q-10/Z8.5; Starna Cells Inc.). To prepare experimental cultures, cells were taken from −80°C stocks and first grown in 5 ml LB medium at an OD600 of ∼0.001 (seed culture). At an OD600 of ∼0.5 (before cells entered stationary phase), we resuspended cells in 5 ml minimal growth medium at very low densities (typically lower than an OD600 of ∼0.0001) and cultured them overnight (preculture). The next morning, the preculture was diluted in prewarmed, 5 ml minimal growth medium (experimental culture) to an OD600 of ∼0.01 (20 to 50 times dilution) and allowed to grow exponentially to an OD600 of ∼0.1, at which measurements were performed.
Fluorescence microscopy.
To make fluorescence measurements, cells were treated with protonophores at an OD600 of 0.1 for 10 min and then with HCT and/or DiSC3(5) for 60 min at 37ºC in constant agitation in the dark; 0.1 μM HCT and 0.1 nM DiSC3(5) were sufficient to produce discernible intracellular fluorescence signals while not affecting cell growth (Fig. S1). The cells then were loaded onto no. 1.5 cover glasses; 1-mm-thick 1.5% agarose pads, made with the same MOPS growth medium [containing the same concentrations of HCT, DiSC3(5) and/or protonophores], were used to cover the cells.
Cells were imaged with a prewarmed (at 37°C) inverted microscope (Olympus IX83 P2Z) with a Neo 5.5 sCMOS camera (Andor Neo). Intracellular HCT and DiSC3(5) were imaged using 4′,6-diamidino-2-phenylindole (DAPI) and Cy5 fluorescence filter sets. Images were acquired with MetaMorph Microscopy Automation and Image Analysis Software and analyzed with the MicrobeJ 5.13m4 plug-in in ImageJ (74). MicrobeJ can automatically segment cell boundaries from phase-contrast microscope images and apply the binary masks from the segmentation to measure fluorescence intensities inside and outside the cells. The latter (background) is subtracted from the former to determine intracellular fluorescence signals.
Motor speed measurement.
E. coli K-12 MG1655 with genetically modified flagellar filaments (EK07 [41]) was used. It was cultured in lysogeny broth (LB) (10 g tryptone, 5 g yeast extract, 10 g NaCl per 1 liter) to an OD600 of ∼2. Cells were sheared to truncate flagellar filaments, washed from LB to modified minimal medium (MM9; 50 mM Na2HPO4, 25 mM NaH2PO4, 8.5 mM NaCl, 18.7 mM NH4Cl, 0.1 mM CaCl2, 1 mM KCl, 2 mM MgSO4, pH 7.5) supplemented with 0.3% d-glucose and attached to the cover glass surface of a tunnel slide via poly-l-lysine (41, 75, 76). We have shown that the cytoplasmic pH of the cells in this medium is ∼7.8 (41). Based on these values (medium pH 7.5 and cytoplasmic pH 7.8), we estimate that the maximum contribution of ΔpH to the PMF under our condition is <30mV (41). Polystyrene beads (0.5 μm) were attached to truncated flagellar filaments and placed into the focus of a heavily attenuated optical trap (855-nm laser) to detect the motor rotation (41). Time course of the bead rotation was recorded with the position-sensitive detector (model 2931; New Focus, Irvine, CA) at 10 kHz, and a 2.5-kHz cutoff antialiasing filter was applied before processing the signal. Next, a flat-top window discrete Fourier transform (window size, 16,384 data points with a step dt of 0.01 s) was applied to the acquired x and y coordinates of a bead position to obtain a time series motor speed recording. The speed traces were then median filtered with a 401-point moving window after manual removal of spurious zeroes caused by flowing CCCP/medium into the slide. The filtered speed trace was then resampled to 10 samples/min for plotting. Measurements were made with a microscope equipped with back focal plane interferometry capability (77, 78), as described previously (77–79).
Footnotes
Citation Le D, Krasnopeeva E, Sinjab F, Pilizota T, Kim M. 2021. Active efflux leads to heterogeneous dissipation of proton motive force by protonophores in bacteria. mBio 12:e00676-21. https://doi.org/10.1128/mBio.00676-21.
Contributor Information
Teuta Pilizota, Email: teuta.pilizota@ed.ac.uk.
Minsu Kim, Email: minsu.kim@emory.edu.
Vaughn S. Cooper, University of Pittsburgh
REFERENCES
- 1.Maloney PC, Kashket ER, Wilson TH. 1974. A protonmotive force drives ATP synthesis in bacteria. Proc Natl Acad Sci U S A 71:3896–3900. doi: 10.1073/pnas.71.10.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell P. 1966. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol Rev Camb Philos Soc 41:445–501. doi: 10.1111/j.1469-185x.1966.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 3.Harold FM, Van Brunt J. 1977. Circulation of H+ and K+ across the plasma membrane is not obligatory for bacterial growth. Science 197:372–373. doi: 10.1126/science.69317. [DOI] [PubMed] [Google Scholar]
- 4.Ohsumi Y, Anraku Y. 1981. Active transport of basic amino acids driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae. J Biol Chem 256:2079–2082. doi: 10.1016/S0021-9258(19)69736-X. [DOI] [PubMed] [Google Scholar]
- 5.Nelson N. 1994. Energizing porters by proton-motive force. J Exp Biol 196:7–13. doi: 10.1242/jeb.196.1.7. [DOI] [PubMed] [Google Scholar]
- 6.Kim BH, Gadd GM. 2019. Membrane transport–nutrient uptake and protein excretion, p 31–57. Prokaryotic metabolism and physiology, 2nd ed. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 7.Fung DC, Berg HC. 1995. Powering the flagellar motor of Escherichia coli with an external voltage source. Nature 375:809–812. doi: 10.1038/375809a0. [DOI] [PubMed] [Google Scholar]
- 8.Manson MD, Tedesco P, Berg HC, Harold FM, Van der Drift C. 1977. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci U S A 74:3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabel CV, Berg HC. 2003. The speed of the flagellar rotary motor of Escherichia coli varies linearly with protonmotive force. Proc Natl Acad Sci U S A 100:8748–8751. doi: 10.1073/pnas.1533395100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strahl H, Hamoen LW. 2010. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci U S A 107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prindle A, Liu J, Asally M, Ly S, Garcia-Ojalvo J, Süel GM. 2015. Ion channels enable electrical communication in bacterial communities. Nature 527:59–63. doi: 10.1038/nature15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J-H, Lee J. 2010. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 13.van Belkum MJ, Kok J, Venema G, Holo H, Nes IF, Konings WN, Abee T. 1991. The bacteriocin lactococcin A specifically increases permeability of lactococcal cytoplasmic membranes in a voltage-independent, protein-mediated manner. J Bacteriol 173:7934–7941. doi: 10.1128/jb.173.24.7934-7941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao FH, Abee T, Konings WN. 1991. Mechanism of action of the peptide antibiotic nisin in liposomes and cytochrome c oxidase-containing proteoliposomes. Appl Environ Microbiol 57:2164–2170. doi: 10.1128/aem.57.8.2164-2170.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montville TJ, Bruno MEC. 1994. Evidence that dissipation of proton motive force is a common mechanism of action for bacteriocins and other antimicrobial proteins. Int J Food Microbiol 24:53–74. doi: 10.1016/0168-1605(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 16.Lamsa A, Liu W-T, Dorrestein PC, Pogliano K. 2012. The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Mol Microbiol 84:486–500. doi: 10.1111/j.1365-2958.2012.08038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruno ME, Kaiser A, Montville TJ. 1992. Depletion of proton motive force by nisin in Listeria monocytogenes cells. Appl Environ Microbiol 58:2255–2259. doi: 10.1128/aem.58.7.2255-2259.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venema K, Abee T, Haandrikman AJ, Leenhouts KJ, Kok J, Konings WN, Venema G. 1993. Mode of action of lactococcin B, a thiol-activated bacteriocin from Lactococcus lactis. Appl Environ Microbiol 59:1041–1048. doi: 10.1128/aem.59.4.1041-1048.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dankert JR, Esser AF. 1987. Bacterial killing by complement. C9-mediated killing in the absence of C5b-8. Biochem J 244:393–399. doi: 10.1042/bj2440393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCollister BD, Hoffman M, Husain M, Vázquez-Torres A. 2011. Nitric oxide protects bacteria from aminoglycosides by blocking the energy-dependent phases of drug uptake. Antimicrob Agents Chemother 55:2189–2196. doi: 10.1128/AAC.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell P. 2011. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biochim Biophys Acta 1807:1507–1538. doi: 10.1016/j.bbabio.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Kasianowicz J, Benz R, McLaughlin S. 1984. The kinetic mechanism by which CCCP (carbonyl cyanidem-chlorophenylhydrazone) transports protons across membranes. J Membr Biol 82:179–190. doi: 10.1007/BF01868942. [DOI] [PubMed] [Google Scholar]
- 23.McLaughlin SG, Dilger JP. 1980. Transport of protons across membranes by weak acids. Physiol Rev 60:825–863. doi: 10.1152/physrev.1980.60.3.825. [DOI] [PubMed] [Google Scholar]
- 24.Taber HW, Mueller JP, Miller PF, Arrow AS. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev 51:439–457. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruni GN, Kralj JM. 2020. Membrane voltage dysregulation driven by metabolic dysfunction underlies bactericidal activity of aminoglycosides. Elife 9:e58706. doi: 10.7554/eLife.58706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulsen IT, Brown MH, Skurray RA. 1996. Proton-dependent multidrug efflux systems. Microbiol Rev 60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aowicki D, Huczyński A. 2013. Structure and antimicrobial properties of monensin A and its derivatives: summary of the achievements. Biomed Res Int 2013:742149. doi: 10.1155/2013/742149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farha MA, Verschoor CP, Bowdish D, Brown ED. 2013. Collapsing the proton motive force to identify synergistic combinations against Staphylococcus aureus. Chem Biol 20:1168–1178. doi: 10.1016/j.chembiol.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Ahmed S, Booth IR. 1983. The use of valinomycin, nigericin and trichlorocarbanilide in control of the protonmotive force in Escherichia coli cells. Biochem J 212:105–112. doi: 10.1042/bj2120105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng X, Zhu W, Schurig-Briccio LA, Lindert S, Shoen C, Hitchings R, Li J, Wang Y, Baig N, Zhou T, Kim BK, Crick DC, Cynamon M, McCammon JA, Gennis RB, Oldfield E. 2015. Antiinfectives targeting enzymes and the proton motive force. Proc Natl Acad Sci U S A 112:E7073–E7082. doi: 10.1073/pnas.1521988112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeon AB, Ackart DF, Li W, Jackson M, Melander RJ, Melander C, Abramovitch RB, Chicco AJ, Basaraba RJ, Obregón-Henao A. 2019. 2-Aminoimidazoles collapse mycobacterial proton motive force and block the electron transport chain. Sci Rep 9:1513. doi: 10.1038/s41598-018-38064-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valderrama K, Pradel E, Firsov AM, Drobecq H, Bauderlique-Le Roy H, Villemagne B, Antonenko YN, Hartkoorn RC. 2019. Pyrrolomycins are potent natural protonophores. Antimicrob Agents Chemother 63:e01450-19. doi: 10.1128/AAC.01450-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vega NM, Allison KR, Khalil AS, Collins JJ. 2012. Signaling-mediated bacterial persister formation. Nat Chem Biol 8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crabbé A, Ostyn L, Staelens S, Rigauts C, Risseeuw M, Dhaenens M, Daled S, Van Acker H, Deforce D, Van Calenbergh S, Coenye T. 2019. Host metabolites stimulate the bacterial proton motive force to enhance the activity of aminoglycoside antibiotics. PLoS Pathog 15:e1007697. doi: 10.1371/journal.ppat.1007697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leblanc OH, Jr. 1971. The effect of uncouplers of oxidative phosphorylation on lipid bilayer membranes: carbonylcyanidem-chlorophenylhydrazone. J Membr Biol 4:227–251. doi: 10.1007/BF02431973. [DOI] [PubMed] [Google Scholar]
- 37.Chimerel C, Field CM, Piñero-Fernandez S, Keyser UF, Summers DK. 2012. Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim Biophys Acta 1818:1590–1594. doi: 10.1016/j.bbamem.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Berg van Saparoea HB, Lubelski J, van Merkerk R, Mazurkiewicz PS, Driessen AJ. 2005. Proton motive force-dependent Hoechst 33342 transport by the ABC transporter LmrA of Lactococcus lactis. Biochemistry 44:16931–16938. doi: 10.1021/bi051497y. [DOI] [PubMed] [Google Scholar]
- 39.Winkel T, Gray JD, Seistrup DA, Hamoen KH, Strahl LW. 2016. Analysis of antimicrobial-triggered membrane depolarization using voltage sensitive dyes. Front Cell Dev Biol 4:29. doi: 10.3389/fcell.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Síp M, Herman P, Plásek J, Hrouda V. 1990. Transmembrane potential measurement with carbocyanine dye diS-C3-5: fast fluorescence decay studies. J Photochem Photobiol B 4:321–328. doi: 10.1016/1011-1344(90)85037-W. [DOI] [PubMed] [Google Scholar]
- 41.Krasnopeeva E, Lo C-J, Pilizota T. 2019. Single-cell bacterial electrophysiology reveals mechanisms of stress-induced damage. Biophys J 116:2390–2399. doi: 10.1016/j.bpj.2019.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mancini L, Terradot G, Tian T, Pu Y, Li Y, Lo C-J, Bai F, Pilizota T. 2020. A general workflow for characterization of nernstian dyes and their effects on bacterial physiology. Biophys J 118:4–14. doi: 10.1016/j.bpj.2019.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitrophanov AY, Groisman EA. 2008. Positive feedback in cellular control systems. Bioessays 30:542–555. doi: 10.1002/bies.20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lomovskaya O, Lewis K. 1992. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci U S A 89:8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikaido H. 1996. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol 178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griffith JM, Basting PJ, Bischof KM, Wrona EP, Kunka KS, Tancredi AC, Moore JP, Hyman MRL, Slonczewski JL. 2019. Experimental evolution of Escherichia coli K-12 in the presence of proton motive force (PMF) uncoupler carbonyl cyanide m-chlorophenylhydrazone selects for mutations affecting PMF-driven drug efflux pumps. Appl Environ Microbiol 85:e02792-18. doi: 10.1128/AEM.02792-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du D, Wang-Kan X, Neuberger A, van Veen HW, Pos KM, Piddock LJV, Luisi BF. 2018. Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 48.MacLeod RA, Wisse GA, Stejskal FL. 1988. Sensitivity of some marine bacteria, a moderate halophile, and Escherichia coli to uncouplers at alkaline pH. J Bacteriol 170:4330–4337. doi: 10.1128/jb.170.9.4330-4337.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng X-C, Guo W-Q, Zheng H-S, Wu Q-L, Luo H-C, Ren N-Q. 2018. Effect of metabolic uncoupler, 3,3′,4′,5-tetrachlorosalicylanilide (TCS) on Bacillus subtilis: biofilm formation, flocculability and surface characteristics. RSC Adv 8:16178–16186. doi: 10.1039/C8RA02315H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berstad A, Raa J, Valeur J. 2015. Indole–the scent of a healthy “inner soil.” Microb Ecol Health Dis 26:27997. doi: 10.3402/mehd.v26.27997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholls DG, Ferguson SJ. 2013. Ion transport across energy-conserving membranes, p 13–25. Bioenergetics, 4th ed. Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 52.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 53.Şimşek E, Kim M. 2019. Power-law tail in lag time distribution underlies bacterial persistence. Proc Natl Acad Sci U S A 116:17635–17640. doi: 10.1073/pnas.1903836116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacKenzie FM, Gould IM. 1993. The post-antibiotic effect. J Antimicrob Chemother 32:519–537. doi: 10.1093/jac/32.4.519. [DOI] [PubMed] [Google Scholar]
- 55.Kwan BW, Valenta JA, Benedik MJ, Wood TK. 2013. Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother 57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srimani JK, Huang S, Lopatkin AJ, You L. 2017. Drug detoxification dynamics explain the postantibiotic effect. Mol Syst Biol 13:948. doi: 10.15252/msb.20177723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pu Y, Zhao Z, Li Y, Zou J, Ma Q, Zhao Y, Ke Y, Zhu Y, Chen H, Baker Matthew AB, Ge H, Sun Y, Xie Xiaoliang S, Bai F. 2016. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol Cell 62:284–294. doi: 10.1016/j.molcel.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blanco P, Hernando-Amado S, Reales-Calderon JA, Corona F, Lira F, Alcalde-Rico M, Bernardini A, Sanchez MB, Martinez JL. 2016. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4:14. doi: 10.3390/microorganisms4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alcalde-Rico M, Hernando-Amado S, Blanco P, Martínez JL. 2016. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front Microbiol 7:1483. doi: 10.3389/fmicb.2016.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martins A, Spengler G, Rodrigues L, Viveiros M, Ramos J, Martins M, Couto I, Fanning S, Pagès J-M, Bolla JM, Molnar J, Amaral L. 2009. pH modulation of efflux pump activity of multi-drug resistant Escherichia coli: protection during its passage and eventual colonization of the colon. PLoS One 4:e6656-e. doi: 10.1371/journal.pone.0006656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amaral L, Fanning S, Pagès JM. 2011. Efflux pumps of gram-negative bacteria: genetic responses to stress and the modulation of their activity by pH, inhibitors, and phenothiazines. Adv Enzymol Rel Areas Mol Biol 77:61–108. doi: 10.1002/9780470920541.ch2. [DOI] [PubMed] [Google Scholar]
- 62.Hards K, McMillan DGG, Schurig-Briccio LA, Gennis RB, Lill H, Bald D, Cook GM. 2018. Ionophoric effects of the antitubercular drug bedaquiline. Proc Natl Acad Sci U S A 115:7326–7331. doi: 10.1073/pnas.1803723115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soupene E, van Heeswijk WC, Plumbridge J, Stewart V, Bertenthal D, Lee H, Prasad G, Paliy O, Charernnoppakul P, Kustu S. 2003. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J Bacteriol 185:5611–5626. doi: 10.1128/JB.185.18.5611-5626.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lyons E, Freeling M, Kustu S, Inwood W. 2011. Using genomic sequencing for classical genetics in E. coli K12. PLoS One 6:e16717. doi: 10.1371/journal.pone.0016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown SD, Jun S. 2015. Complete genome sequence of Escherichia coli NCM3722. Genome Announc 3:e00879-15. doi: 10.1128/genomeA.00879-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Slonczewski JL, Rosen BP, Alger JR, Macnab RM. 1981. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc Natl Acad Sci U S A 78:6271–6275. doi: 10.1073/pnas.78.10.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zilberstein D, Agmon V, Schuldiner S, Padan E. 1984. Escherichia coli intracellular pH, membrane potential, and cell growth. J Bacteriol 158:246–252. doi: 10.1128/jb.158.1.246-252.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilks JC, Slonczewski JL. 2007. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J Bacteriol 189:5601–5607. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Swain BM, Guo D, Singh H, Rawlins PB, McAlister M, van Veen HW. 2020. Complexities of a protonatable substrate in measurements of Hoechst 33342 transport by multidrug transporter LmrP. Sci Rep 10:20026. doi: 10.1038/s41598-020-76943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomason LC, Costantino N, Court DL. 2007. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol Chapter 1:Unit 1.17. [DOI] [PubMed] [Google Scholar]
- 74.Ducret A, Quardokus EM, Brun YV. 2016. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y-K, Krasnopeeva E, Lin S-Y, Bai F, Pilizota T, Lo C-J. 2019. Comparison of Escherichia coli surface attachment methods for single-cell microscopy. Sci Rep 9:19418. doi: 10.1038/s41598-019-55798-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krasnopeeva E, Barboza-Perez UE, Rosko J, Pilizota T, Lo CJ. 5 July 2020. Bacterial flagellar motor as a multimodal biosensor. Methods doi: 10.1016/j.ymeth.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 77.Denk W, Webb WW. 1990. Optical measurement of picometer displacements of transparent microscopic objects. Appl Opt 29:2382–2391. doi: 10.1364/AO.29.002382. [DOI] [PubMed] [Google Scholar]
- 78.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. 1993. Direct observation of kinesin stepping by optical trapping interferometry. Nature 365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 79.Rosko J, Martinez VA, Poon WCK, Pilizota T. 2017. Osmotaxis in Escherichia coli through changes in motor speed. Proc Natl Acad Sci U S A 114:E7969–E7976. doi: 10.1073/pnas.1620945114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Salcedo-Sora JE, Jindal S, Hagan S, Kell DB. 2021. A palette of fluorophores that are differentially accumulated by wild-type and mutant strains of Escherichia coli: surrogate ligands for profiling bacterial membrane transporters. Microbiology 167:001016. doi: 10.1099/mic.0.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 82.Wadhwa N, Phillips R, Berg HC. 2019. Torque-dependent remodeling of the bacterial flagellar motor. Proc Natl Acad Sci U S A 116:11764–11769. doi: 10.1073/pnas.1904577116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth of E. coli under various conditions. (A) Growth curve of WT E. coli treated with various concentrations of CCCP. At 50 μM, the rate of population growth decreased moderately. (B) Growth curve of WT E. coli treated with various concentrations of HCT. At 0.1 μM (the concentration used in the present study), HCT has little effect on cell growth. (C) Growth of WT E. coli treated with 0.1 nM DiSC3(5). At this concentration, DiSC3(5) has little effect on cell growth. Download FIG S1, TIF file, 0.1 MB (154KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
HCT intensity distribution in WT (left) and ΔtolC (right) cells treated with TCS. (Left) WT strain. At low TCS concentrations (≤5 μM), the HCT intensity was uniformly low across a population, leading to a unimodal distribution (with the peak center around ∼10 AU). Increasing TCS concentrations led to the emergence of the second peak on the right (>100 AU, HCT-bright cells), showing a bimodal distribution. At higher concentrations (≥30 μM), the left low-intensity peak disappeared, showing the enrichment of HCT-bright cells. (Right) ΔtolC strain. For all TCS concentrations tested, the left low-intensity peak is absent in the ΔtolC strain, which resulted in a unimodal distribution. The center of the single peak shifted moderately at increasing TCS concentrations. This observation is consistent with that made for CCCP (Fig. 2). Download FIG S3, TIF file, 0.2 MB (173.8KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
HCT intensity distribution in WT (left) and ΔtolC (right) cells treated with indole. The result is similar to that from the TCS experiment in Fig. S3. (Left) WT strain. Increasing indole concentrations led to the emergence of the second peak on the right (>100 AU, HCT-bright cells) and a mild shift in the center of the left low-intensity peak. The two peaks merged at 7.5 mM indole concentration. (Right) ΔtolC strain. For all indole concentrations tested, a left low-intensity peak is absent in the ΔtolC strain. The center of the single peak shifted moderately at increasing indole concentrations. Of note, we previously measured the motor speeds of WT cells at low indole concentrations (≤2 mM), which showed a uniform reduction in the motor speeds (41). Here, we observed a bimodal distribution of the HCT intensity at higher concentrations (≥2.5 mM). At 2.5 mM, the fraction of cells exhibiting high HCT intensities was less than 5%, which is difficult to detect with motor speed measurements because motor speed measurements are laborious, and it is challenging to track more than 20 cells. Download FIG S4, TIF file, 0.2 MB (169.7KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Motor speeds of WT (green) and ΔtolC (blue) cells in the absence of protonophores. Here, we compared the motor speeds of WT and ΔtolC strains. Their motor speeds were comparable, indicating that ΔtolC has no noticeable effect on the PMF. Download FIG S2, TIF file, 0.1 MB (85KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Our results agree with the known CCCP action as a protonophore. Previous in vitro measurements have showed that CCCP collapses the total PMF that acts on protons by allowing them to equilibrate (22, 37). To confirm this mechanism of CCCP action, we analyzed a relative change in the motor speed as a function of a CCCP concentration, [CCCP], in the ΔtolC strain: ω/ω0 =PMF/PMF0 = f ([CCCP],t), as we have shown before (41). The mean speeds during the last 2 min of CCCP exposure (Fig. 3E to H) were obtained from Gaussian fits to probability densities of speeds left) and normalized to the pre-CCCP treatment speed. We then fit the normalized speeds with a hyperbolic function and obtained the following relationship, PMF/PMF0 = 1/(2.64 × [CCCP]+1) (right). This agrees with an inverse relationship between PMF and [CCCP] from in vitro studies (22, 37), supporting that CCCP acts as a protonophore. We note that, unlike some other ionophores, such as butanol and indole (41), the PMF reduction at a given CCCP concentration takes several minutes (Fig. 3E to H). Download FIG S5, TIF file, 0.4 MB (378.5KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recovery of PMF after CCCP washout. We have discussed in the main text that the treatment with 50 μM CCCP resulted in two subpopulations, one with a moderate reduction in the motor speed and the other with a motor speed of zero. When CCCP was removed, the former recovered the motor speed immediately. Partial recovery was observed in some cells in the latter subpopulation (red arrow). Download FIG S6, TIF file, 0.1 MB (87.6KB, tif) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Chemicals used in our experiments. Download Table S1, DOCX file, 0.02 MB (20.6KB, docx) .
Copyright © 2021 Le et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.


