ABSTRACT
The gut microbiota affects the physiology and metabolism of animals and its alteration can lead to diseases such as gut dysplasia or metabolic disorders. Several reports have shown that the immune system plays an important role in shaping both bacterial community composition and abundance in Drosophila, and that immune deficit, especially during aging, negatively affects microbiota richness and diversity. However, there has been little study at the effector level to demonstrate how immune pathways regulate the microbiota. A key set of Drosophila immune effectors are the antimicrobial peptides (AMPs), which confer defense upon systemic infection. AMPs and lysozymes, a group of digestive enzymes with antimicrobial properties, are expressed in the gut and are good candidates for microbiota regulation. Here, we take advantage of the model organism Drosophila melanogaster to investigate the role of AMPs and lysozymes in regulation of gut microbiota structure and diversity. Using flies lacking AMPs and newly generated lysozyme mutants, we colonized gnotobiotic flies with a defined set of commensal bacteria and analyzed changes in microbiota composition and abundance in vertical transmission and aging contexts through 16S rRNA gene amplicon sequencing. Our study shows that AMPs and, to a lesser extent, lysozymes are necessary to regulate the total and relative abundance of bacteria in the gut microbiota. We also decouple the direct function of AMPs from the immune deficiency (IMD) signaling pathway that regulates AMPs but also many other processes, more narrowly defining the role of these effectors in the microbial dysbiosis observed in IMD-deficient flies upon aging.
KEYWORDS: microbiota, innate immunity, aging, gnotobiotic animals, immune effectors, gut
INTRODUCTION
The gut microbiota is the complex array of microbes commonly associated with the digestive tract of animals. This bacterial consortium greatly affects host physiology, for example by promoting immune function or intestinal homeostasis (1–4). Imbalance of the microbiota, called dysbiosis, has been identified as a cause of gut dysplasia and chronic inflammatory diseases, especially during aging (5).
The fruit fly Drosophila melanogaster is a powerful model to decipher host-microbe interactions (6–8). Its genetic tractability, the possibility to generate gnotobiotic animals, and the simplicity of its natural microbiota have made Drosophila melanogaster a convenient model to gain insight into host-microbiota relationships (7, 9, 10). Drosophila harbors a simple gut microbiota composed of only a few dominant species, mainly belonging to Acetobacteraceae and Lactobacillaceae, which influence multiple aspects of fly physiology, such as growth (11, 12), behavior (13), life span (14), and infection resistance (15, 16). In turn, the microbiota can be shaped by various host and environmental factors, such as food composition or age of the flies (17–21).
Innate immunity is a key regulator of microbial abundance in Drosophila (22–25). Upon acute bacterial infection, Drosophila immune responsive tissues (the fat body and hemocytes in systemic infection, and epithelium in local infection) sense microbe-associated molecular patterns (MAMPs) to activate signaling pathways. In Drosophila, two immune pathways, the immune deficiency (IMD) and Toll pathways, regulate the expression of genes encoding immune effectors that fight invading microbes (22, 26). Studies in Drosophila have revealed a key role of the IMD pathway in the gut to fight pathogens and keep symbiotic bacteria in check (27–29). It is, however, unclear how the IMD pathway can effectively combat pathogens but tolerate symbiotic microbiota members in the digestive tract. In fact, the microbiota induces a low level of activation of the IMD pathway (27). Several reports have demonstrated that immune tolerance toward the indigenous microbiota is sustained by several negative feedback loops that prevent hyperactivation of the IMD pathway by peptidoglycan (the bacterial elicitor recognized by the IMD pathway) released from commensal bacteria (30–32). Compartmentalization of the immune response to restricted areas can also favor microbiota growth and control (33, 34). However, IMD pathway activation is necessary to regulate both microbiota composition and proliferation, and dysregulation of this pathway leads to abnormal bacterial growth and premature death of the host (27, 31, 32, 35). Notably, mutations affecting the IMD transcription factor Relish lead to a higher gut microbiota load and a shifted bacterial composition compared to wild-type flies (27, 36). Moreover, aged Relish mutant flies display dysbiosis associated with a loss of gut epithelium integrity and premature death of the animals (21, 37). Collectively, these studies point to an important role of the IMD pathway in control of the microbiota, notably during aging. However, the IMD pathway regulates hundreds of immune effectors and affects numerous physiological processes, such as enterocyte delamination and digestion (38–42). As previous studies have used mutations that suppress the whole pathway (e.g Relish), the precise role of individual immune effectors downstream of the IMD pathway in shaping the gut microbial community has remained elusive.
Antimicrobial peptides (AMPs) are molecules that contribute to innate defenses by targeting the negatively charged membranes of microbes (43). These peptides are produced in large quantities by the fat body during systemic infection, but also in local epithelia such as the gut. Seven classic families of inducible AMPs with several isoforms have been identified in D. melanogaster (43, 44). Use of CRISPR/Cas9 has recently enabled the generation of individual and combined AMP mutants, allowing direct investigation of their role in host defense (45). Hanson et al. showed that Drosophila AMPs are essential for resisting infection by Gram-negative bacteria that trigger the IMD pathway, but appear to be less involved in defense against Gram-positive bacterial infection (45).
Another key group of effector proteins that are potential regulators of Gram-positive bacteria in the gut are the lysozymes (46, 47). Lysozymes specifically cleave peptidoglycan exposed on the cell wall of Gram-positive bacteria (48). The Drosophila genome encodes at least 17 putative lysozymes, whose functions have never been formally addressed. Among them, six lysozyme genes (LysB, D, E, P, S, and X) are clustered in the genome at cytogenetic map position 61F. This group of lysozyme genes, notably LysB, LysD, LysE, and LysP, is strongly expressed in the digestive tract (46), and may contribute to digestive activities of the gut by degrading peptidoglycan from dietary bacteria. Furthermore, lysozyme genes are expressed in the gut upon microbiota colonization in Drosophila, and these proteins have been proposed to modulate immune signaling (27, 30, 49). Lysozymes may contribute to gut immunity either as direct antimicrobials, or by cleaving peptidoglycan and modulating activation of the IMD pathway (30). As such, AMPs and lysozymes may shape microbiota composition by direct interactions with microbes.
In this study, we decipher the role of two classes of antimicrobial effectors of the Drosophila digestive tract, the antimicrobial peptides (AMPs) and the lysozymes, on the gut microbiota. We characterized the microbiota composition in mutant flies lacking either the 14 AMP genes from seven gene families or the four gut-specific lysozyme-encoding genes in a gnotobiotic setup using 16S rRNA gene amplicon sequencing (referred to as 16S sequencing). We also assessed the role of these effectors in controlling the abundance of individual microbiota members by performing mono-association experiments. Finally, we confirmed that certain immune effectors can directly control the proliferation of microbiota members by performing systemic infections. Our findings demonstrate a direct role for both AMPs and lysozymes in controlling both the composition and abundance of the microbiota in Drosophila melanogaster.
RESULTS
Impact of AMPs and lysozymes on microbiota composition.
To decipher the role of AMPs and lysozymes in the regulation of gut microbiota composition, we performed 16S sequencing on gnotobiotic flies. DrosDel isogenic flies with the following genotypes were used for all experiments: the wild-type strain w1118 (referred to as w), a compound mutant strain lacking Defensin, Cecropins (4 genes), Drosocin, Diptericins (2 genes), Attacins (4 genes), Metchnikowin, and Drosomycin, referred to as “ΔAMP14” (50), and a newly generated lysozyme-deficient mutant (referred to LysB-PΔ) (for details see the Extended Materials and Methods in Text S2 of the supplemental material). The LysB-PΔ mutation is an 11.5-kb deletion, removing LysC (a putative pseudogene) and the four lysozyme genes (i.e., Lys B, LysD, LysE, and Lys P) that are known to be strongly expressed in the digestive tract (46) (Fig. S1). As expected, gut extracts from LysB-PΔ flies have reduced lysozyme activity ex vivo, as monitored by their ability to digest peptidoglycan from Enterococcus faecalis (Fig. S1). We additionally included Relish (RelE20) flies lacking IMD signaling as a comparative control to determine to what extent AMPs contribute to the phenotype of IMD-deficient flies.
To avoid preexisting microbial community biases in different fly stocks, we performed this analysis in a gnotobiotic system with two different experimental designs. First, we analyzed the microbiota of 12-day-old flies with gut bacteria acquired through vertical transmission from gnotobiotic parental flies (i.e., germfree parents inoculated with a known community upon adult emergence) (Fig. 1A). Second, we analyzed aging-dependent changes in the adult microbiota. Here, we inoculated emerging germfree (GF) adults with a known microbiota and analyzed changes in the community structure 10 and 29 days after colonization (Fig. 2A). In this way, we uncoupled the effects of juvenile development and metamorphosis from the adult microbiota composition and abundance.
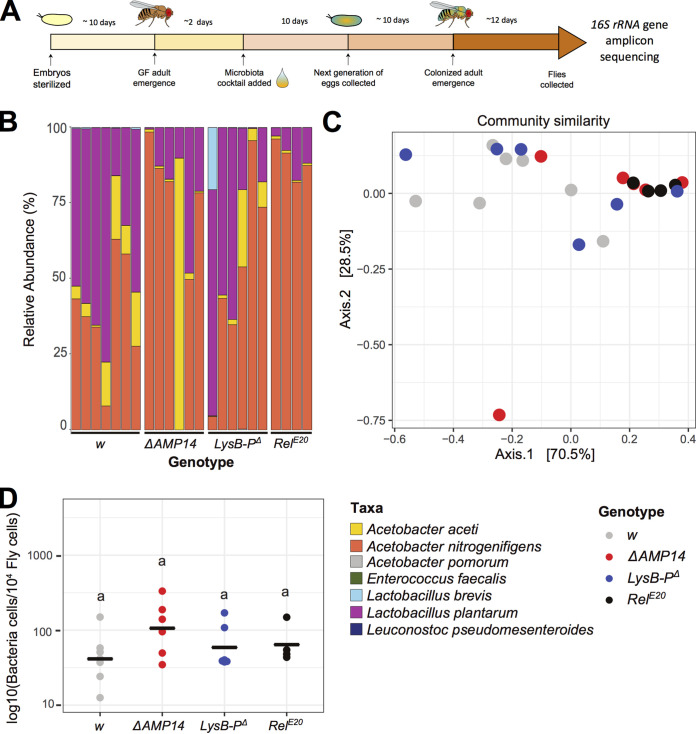
FIG 1.
The role of AMPs and lysozymes on microbiota composition and abundance in a gnotobiotic vertical transmission setup. (A) Scheme of the experimental procedure for fly colonization and collection for 16S rRNA gene amplicon sequencing. Parental embryos were collected, sterilized in 3% bleach, and kept on antibiotic food until the adult stage. Emerging GF flies were then associated with a bacterial cocktail (microbiota cocktail) containing six representative microbiota members. Their eggs were collected over 3 days, allowed to develop to adulthood, and finally the microbiota of their adult female progeny was analyzed at ∼12 days after emergence. (B) Relative community composition of the gut microbiota in wild-type iso w1118 (w) wild-type flies, Relish (RelE20), antimicrobial peptide (ΔAMP14), and gut lysozyme (LysB-PΔ) mutants as determined by 16S rRNA gene amplicon sequencing. Each bar represents a biological replicate of multiple pooled flies (see Table S1 in the supplemental material for the numbers of flies included in each sample). (C) Principal coordinate analysis (PCoA) of gut communities in w wild-type flies, RelE20, ΔAMP14, and LysB-PΔ, as determined by 16S rRNA gene amplicon sequencing. Overall colocalization of ΔAMP14 (red dots) and RelE20 (black dots) samples and separation of these from wild-type (gray dots) samples shows that ΔAMP14 and RelE20 samples are similar to each other and differ from wild-type samples. Stochastic distribution of LysB-PΔ samples shows high variability in community structures between samples. (D) Absolute quantification by qPCR of the total number of bacterial cells normalized to the host gene Actin5C. Horizontal black bars show mean values. Details of the statistical outcomes are provided in Table S2.
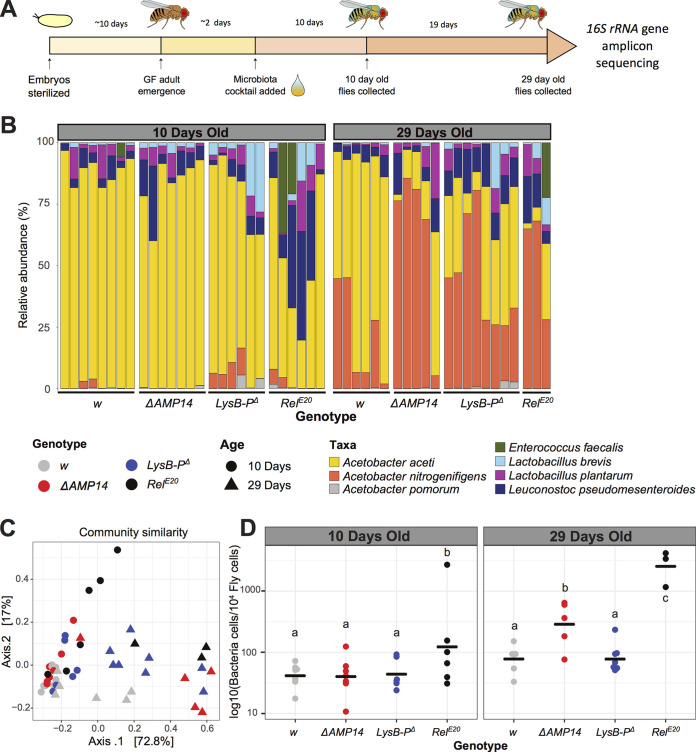
FIG 2.
The role of AMPs and lysozymes in microbiota composition and abundance on adult microbiota in a gnotobiotic setup of young and aged flies. (A) Scheme of the experimental procedure for fly colonization and collection for 16S rRNA gene amplicon sequencing. Embryos were collected, sterilized in 3% bleach, and kept on antibiotic food until the adult stage. Emerging GF flies were associated with a bacterial cocktail containing six representative microbiota members. Females were collected for DNA extraction and 16S rRNA gene amplicon sequencing at 10 and 29 days after colonization. See Table S1 for the number of flies included in each sample. (B) Relative community composition of the gut microbiota in iso w1118 (w) wild-type flies and Relish (RelE20), antimicrobial peptide (ΔAMP14), and gut lysozyme (LysB-PΔ) mutants at 10 days (left panel) and 29 days (right panel) after colonization. Each bar in the plot represents a biological replicate with a pool of 5 flies each. (C) Principal coordinate analysis based on Bray-Curtis dissimilarities on the gut communities of w control flies, RelE20, ΔAMP14, and LysB-PΔ mutants at 10 and 29 days after colonization, based on 16S rRNA gene amplicon sequencing. Separation of the 10-day-old (dots) and 29-day-old clusters on the first axis indicates that aging is the major factor defining bacterial community composition in adults. Separation of ΔAMP14 and RelE20 (red and black triangles) from wild-type and LysB-PΔ (gray and blue triangles) on the same axis in the 29-day samples indicates that aging and loss of immune effectors act on microbiota composition in similar directions. (D) Absolute quantification of the total number of bacterial cells by qPCR, normalized to the host gene Actin5C. Horizontal black bars show mean values. Details of the statistical outcomes are provided in Table S2.
We inoculated the flies with a cocktail of six bacterial isolates that were previously described as common Drosophila microbiota members (19), or that were associated with the food that was used in this study (see the Materials and Methods) (8, 10, 19). These included previously characterized bacterial species as members of the Drosophila gut microbiota: Acetobacter pomorum (51), Lactobacillus plantarum (11), and Enterococcus faecalis (52). Our cocktail also included some incompletely characterized bacterial strains: an Acetobacter sp. (53), an isolate of Lactobacillus brevis, and an isolate of Leuconostoc pseudomesenteroides (see Materials and Methods, Text S1). Acetobacter, a genus of Gram-negative bacteria, and Lactobacillus plantarum, a Gram-positive species, both have DAP-type peptidoglycan known to activate the IMD pathway (52, 54–56). In contrast, Leuconostoc pseudomesenteroides, Lactobacillus brevis (54, 57), and Enterococcus faecalis are Gram-positive bacteria with lysine-type peptidoglycan (58), which typically activates the Toll pathway during systemic infections. Although there is no evidence for a role of the Toll pathway in the midgut (22, 42, 55), Lys-type Gram-positive bacteria can induce a basal immune reaction in the gut through the release of the metabolite uracil, which activates reactive oxygen species (ROS) production through the Duox enzyme (59).
The 16S sequencing of the six-component cocktail yielded seven amplicon sequence variants (ASVs) (60), also referred to as zero-noise operational taxonomic units (OTUs) (61) or sub-OTUS (62), across the 72 samples, with a minimum of 42,596 reads per sample after quality and abundance filtering (see the Materials and Methods and Table S1 for details). These ASVs mapped to the known species in the inoculum cocktail. Sequencing showed the Acetobacter sp. (53) fraction mapped to two ASVs that were distinguishable by a single nucleotide difference in their 16S amplicon. These ASVs were associated with two closely related species, Acetobacter aceti and Acetobacter nitrogenifigens (63, 64), based on their highly similar sequence.
We first focused our analysis on flies with microbiota acquired through vertical transmission from parents raised in a gnotobiotic environment (Fig. 1A, see the Materials and Methods). We found that RelE20 and ΔAMP14 flies harbored communities dominated by A. nitrogenifigens, whereas the wild-type strain had a greater prevalence of La. plantarum (Fig. 1B). In contrast, LysB-PΔ flies had highly variable community compositions (see below), suggesting a different mode of action for these genes compared to the AMPs (Fig. 1B).
Similarities between bacterial communities were assessed using β-diversity analyses. Dissimilarities between all samples were calculated using Bray-Curtis distances plotted in a multidimensional space using principal component analysis (PCoA). This was complemented with an analysis of the dispersal (variability and spread) of the communities, and a permutation based, multivariate analysis of variance was applied to test statistical significance. These analyses showed that community compositions within LysB-PΔ and ΔAMP14 sample groups were more variable than the wild-type (Fig. S2A, 0.05 < P < 0.1), in that communities of some samples resembled wild-type flies while others resembled RelE20 flies (Fig. 1C). One LysB-PΔ sample had a completely different profile from all other samples, with higher abundance of La. brevis (Fig. 1B). This suggests that the loss of AMPs or lysozymes increases stochasticity in microbiota composition. Surprisingly, communities in RelE20 mutants were more consistent between replicates, which indicates either that the stochasticity is not due to perturbation of the immune response or that the communities in these mutants stabilize earlier than in other genotypes due to other factors regulated by the IMD pathway.
In terms of community composition, distribution of data in the PCoA shows that ΔAMP14 samples mimic RelE20 (pairwise ADONIS: p-adjustedΔAMPs vs Rel = 0.5), and both differ noticeably from the wild type, as demonstrated by general colocalization of ΔAMP14 and RelE20 samples, and separation from the wild-type samples (Fig. 1C, pairwise ADONIS: p-adjustedw vs Rel = 0.02; p-adjustedw vs ΔAMPs = 0.06). This suggests that loss of AMPs recapitulates the effect of a general loss of the IMD pathway on the microbiota structure. As expected from the variable community composition found in LysB-PΔ mutants (Fig. 1B), the PCoA did not reveal a distinct cluster for these samples (Fig. 1C).
Finally, we measured total bacterial loads in our samples using universal 16S rRNA gene primers (65) and Drosophila Actin 5C primers (53). We did not detect a statistically significant difference in total 16S rRNA gene copy numbers between the different genotypes, indicating that wild-type and mutant flies do not harbor different quantities of microbes under these conditions (Fig. 1D). Moreover, the change in microbiome variation between genotypes is not a statistical artifact of a change in absolute microbiome abundance.
Overall, our results show the microbiota composition in ΔAMP14 flies is similar to the microbiota of RelE20 mutants that completely lack IMD signaling, suggesting that the changes in community composition observed in IMD pathway mutants is at least partly due to the specific loss of AMP production.
Control of microbiota structure by AMPs and lysozymes during aging.
Next, we focused on the microbiota structure of adult flies that were raised in GF conditions throughout larval development and colonized only after emergence. We analyzed microbiota of these flies at both 10 and 29 days after colonization (Fig. 2A). Here, microbial communities were generally dominated by the two Acetobacter variants. At 10 days after colonization, A. aceti was the most abundant species, whereas by 29 days after colonization, A. nitrogenifigens was the dominant species, suggesting distinct competitive ability of the two bacteria tied to the 16S sequence variants detected in our Acetobacter sp. isolate.
As RelE20 mutants died earlier than other genotypes during the aging process, only three samples with fewer flies than other genotypes were included for the 29-day time point (Fig. 2B, Table S1). RelE20 mutants harbored elevated abundance of E. faecalis in 1/3 of the samples, which was not observed in other genotypes. Some samples in this genotype also had higher proportions of La. plantarum and Le. pseudomesenteroides (in two and three samples, respectively) at day 10, a trend that was not observed at day 29 (Fig. 2B). However, we cannot conclude whether this change in community structure is real or a consequence of high mortality in this genotype, leading to analysis of the survivors only. In contrast to the vertical transmission setup (Fig. S2A), RelE20 communities had high dispersion; the highest variation was observed at 10 days (Fig. S2B), and decreased at 29 days (Fig. S2C). This indicates that immunity mutations cause stochasticity in microbiota composition, but the communities are still capable of stabilizing over a long period of time.
β-Diversity and PCoA showed significant (ADONIS P = 0.001) separation of the 10-day-old and 29-day-old flies on the first axis, clearly pointing to aging as the major factor defining the community composition in adults (Fig. 2C). Interestingly, in 29-day-old flies, ΔAMP14 and RelE20 were separated from wild type and LysB-PΔ mutants on the same axis (Fig. 2C), indicating that aging and loss of immune effectors act on microbiota composition in similar directions. In 10-day-old flies, we did not see similar clustering of samples except for RelE20 mutants, which were more widely dispersed on the plot (Fig. 2C). This indicates that mutations in the IMD pathway act on microbiota composition differently in young versus old flies. A statistically significant genotype × age interaction (ADONIS P = 0.03) supports this interpretation.
Careful examination of the relative abundance of bacteria in wild-type and mutant flies reveals interesting trends (Fig. 2B). We found that wild-type flies maintained Acetobacter aceti as the dominant Acetobacter ASV even after 29 days, while the proportion of lactobacilli in the community remained small. However, although Acetobacter aceti was similarly abundant at 10 days in ΔAMP14 and LysB-PΔ flies, Acetobacter nitrogenifigens became predominant in 29-day samples, and the proportion of lactobacilli in some samples was higher than wild type, particularly in LysB-PΔ flies. This change in relative abundances was even more dramatic in RelE20 mutants, which were distinguished by disproportionate loads of Acetobacter nitrogenifigens and lactobacilli.
Investigation of each time point separately showed that loss of AMPs did not affect the community composition in 10-day-old flies (Fig. 2B, pairwise ADONIS, qw vs ΔAMPs = 0.1). However, loss of lysozymes had detectable effects (pairwise ADONIS, qw vs Lys = 0.02) on the abundance of Acetobacter pomorum, A. nitrogenifigens, or La. brevis depending on the samples, which further supports the idea of increased stochasticity in LysB-PΔ mutants compared to wild type. This stochasticity is clearly shown by the community dispersal (Fig. S2B).
At 29 days, microbial communities in the wild type differed from those of ΔAMP14, LysB-PΔ, and RelE20 genotypes (pairwise ADONIS p-adjustedw vs ΔAMPs = 0.04; p-adjustedw vs Lys = 0.04; p-adjustedw vs Rel = 0.04) (Fig. 2B and C). In the ΔAMP14 strain, the relative abundance of Gram-negative A. nitrogenifigens consistently increased, whereas in LysB-PΔ mutants the relative abundance of Gram-positive lactobacilli increased (Fig. 2B). This suggests that lysozymes act preferentially on Gram-positive bacteria, and the action of AMPs is limited to Acetobacteraceae. As all genotypes contain communities that are similarly variable (Fig. S2C), the observed differences in community composition at day 29 are unlikely to be an artifact of heterogeneity in variance among different groups.
Analysis of the total microbiota abundance showed that bacterial load differed between genotypes mainly in aged flies. At 10 days old, RelE20 flies harbored significantly larger amounts of total bacteria compared to the other genotypes, primarily due to one sample that had a high load typical in 29-day-old samples of this genotype (Fig. 2D). In 29-day-old flies, both ΔAMP14 and particularly RelE20 flies had higher bacterial loads (Fig. 2D). These data support the notion that the IMD pathway is crucial in regulating microbiota load as the flies age and that AMPs significantly contribute to this effect of the IMD pathway.
In agreement with previous reports, our data show that microbial community composition shifts and bacterial load increases with age (17, 18, 27), and that this effect is exacerbated by loss of antimicrobials.
Effect of AMPs and lysozymes on individual microbiota members.
The 16S sequencing provided us a first glimpse of how AMPs and gut lysozymes regulate microbiota structure at the community level. To further characterize the effect of these antimicrobials on individual microbiota members, we used a mono-association setup where we colonized flies with each bacterial isolate from the commensal cocktail used in the 16S sequencing experiment. GF adult flies were mono-associated with a single bacterial species and the bacterial load of females was measured 6 days after colonization by quantitative qPCR (Fig. 3A). We quantified 16S rRNA gene copies using primers that recognize Acetobacteraceae (66) and Firmicutes (including La. plantarum, Le. Pseudomesenteroides, and La. brevis) and normalized their abundance to host cells using primers for Actin 5C (53) (Fig. 3B).
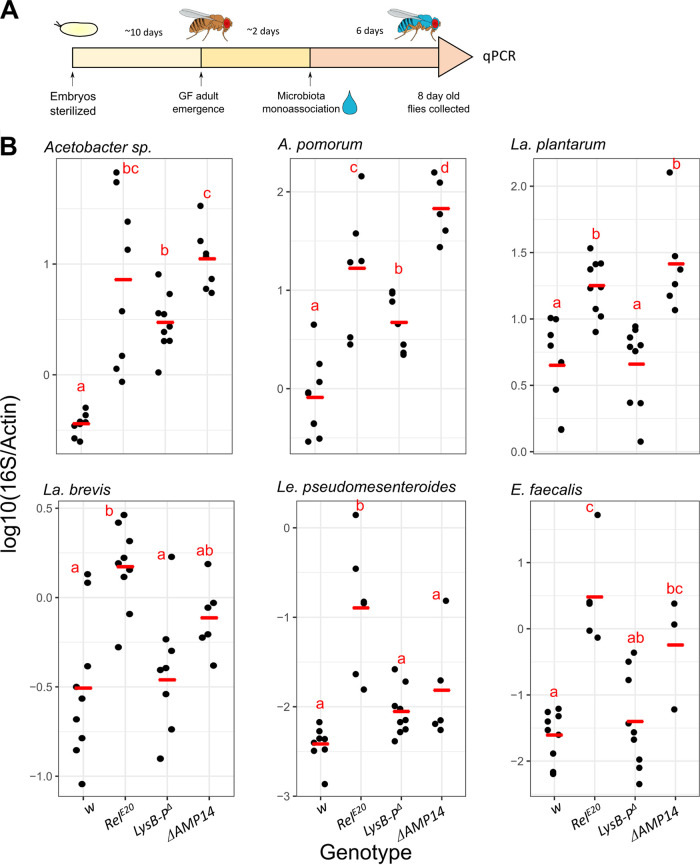
FIG 3.
Regulation of individual microbiota members in mono-association. (A) Scheme of the experimental procedure of the mono-association experiment. Embryos were collected, sterilized in 3% bleach, and kept on antibiotic food until the adult stage. Newly emerged GF flies were then mono-associated with a single bacterial isolate. Six days after colonization, the host and bacterial DNA was extracted and qPCR analysis of the microbial load was performed. (B) Total microbial load was determined by quantitative PCR (qPCR) in female flies at 6 days after mono-association with iso w1118 (w) wild-type flies versus Relish (RelE20), antimicrobial peptide (ΔAMP14), and gut lysozyme (LysB-PΔ) mutant flies. Bacterial loads were assessed by qPCR with family/phylum-specific 16S rRNA gene primers and normalized to the host gene Actin5C. Red horizontal bars show mean values. Each dot represents a sample containing five individuals. Letters represent statistical significance (P < 0.05) of adjusted P values (FDR) from pairwise contrasts obtained from a main general linear mixed model; samples with shared letters are not statistically different from each other.
As expected, all mono-associated taxa established a higher load in RelE20 flies compared to wild-type flies (Fig. 3B). Interestingly, the abundance of both Acetobacter sp. and A. pomorum isolates was high in LysB-PΔ but especially in ΔAMP14 mutants (Fig. 3B), indicating that AMPs most prominently control the proliferation of these Gram-negative microbiota members. Surprisingly, in contrast to shifts toward increased lactobacilli seen in the absence of lysozymes in gnotobiotic experiments (Fig. 1, Fig. 2), mono-associated La. plantarum increased in abundance in the absence of AMPs but not lysozymes (Fig. 3B). This was surprising considering that lysozymes are expected to digest Gram-positive bacteria. The differing trends resulting from these approaches may depend on bacterial community dynamics in gnotobiotic experiments, or age-related differences between the experimental setups. Interestingly, ΔAMP14 harbored significantly more E. faecalis compared to the wild-type w (Fig. 3B and p-adjusted = 0.032), and, indeed, ΔAMP14 had bacterial abundances equivalent or even greater than RelE20 flies for all bacterial taxa except Le. pseudomesenteroides (Fig. 3B).
Overall, our data indicate that in the absence of bacterial community dynamics, AMPs and, to a lesser extent, lysozymes are major effectors regulating gut microbiota abundance.
Systemic infection with microbiota members.
Previously, we showed that a lack of AMPs in the gut significantly affects the microbiota composition and growth. However, it is unclear whether AMPs have preferential antimicrobial activity that selects for core microbiota members, and to date it has not been demonstrated that AMPs directly control members of the microbiota community.
To address this, we used a systemic infection model to effectively “incubate” gut microbiota members in hemolymph with or without AMPs. Flies that fail to control bacterial proliferation ultimately die (67). We systemically infected flies with three representative bacteria that are normally present in the digestive tract and followed fly survival. We challenged wild-type, ΔAMP14, RelE20, and spzrm7 female flies by clean injury and with three different bacterial species: Acetobacter sp. and La. plantarum, which have DAP-type peptidoglycan, and E. faecalis which has Lys-type peptidoglycan. RelE20 flies lack a functional IMD response and are known to be very susceptible to systemic infection by most Gram-negative bacteria and certain classes of Gram-positive bacteria, while spätzlerm7 (spzrm7) mutants lack Toll immune signaling and are susceptible to Gram-positive bacteria and fungi. We observed that ΔAMP14 flies were more susceptible to Gram-negative Acetobacter sp., mimicking the susceptibility of RelE20 mutants (Fig. 4). As expected, spzrm7 flies were highly susceptible to Gram-positive La. plantarum and E. faecalis infection, dying completely within 1 week. However, ΔAMP14 flies did not have increased mortality when infected with these bacterial species. Flies did not die upon clean injury, indicating that the phenotype is specific to bacterial infection and is not due to a technical bias in the experiment (Fig. S3).
FIG 4.
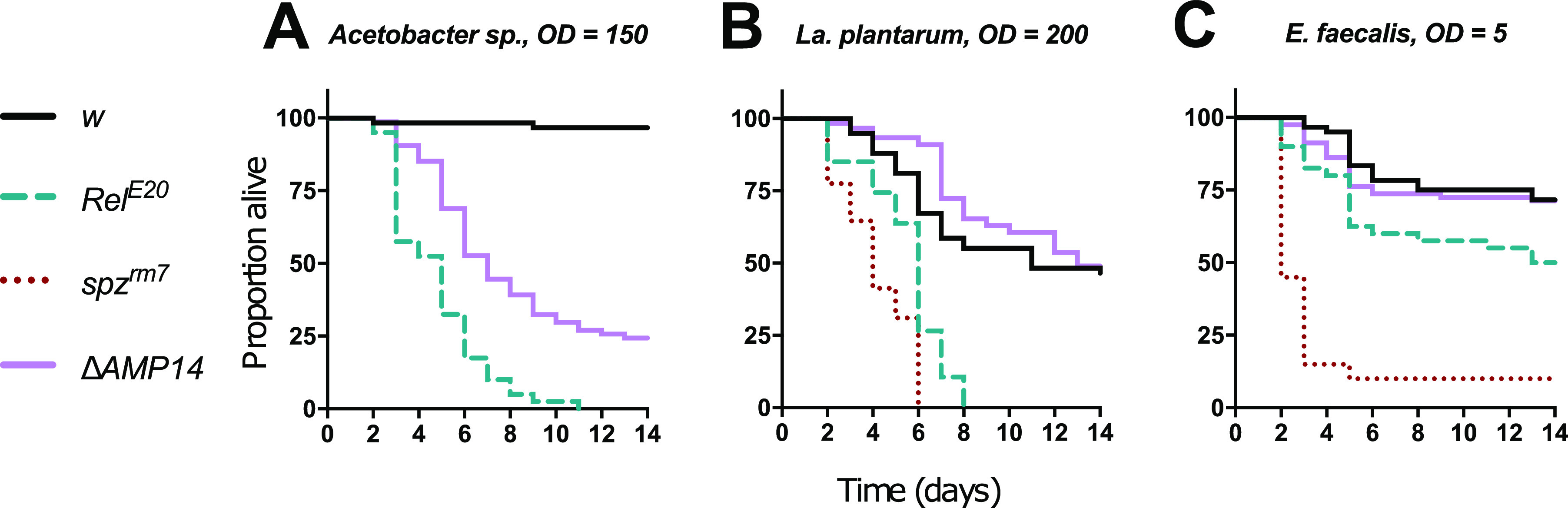
Survival upon systemic infection with microbiota bacteria. Female iso w1118 wild-type flies (w), and Relish (RelE20), antimicrobial peptide (ΔAMP14), and spaetzle (spzrm7) mutants were pricked in the thorax with three common microbiota bacteria: Gram-negative bacterium Acetobacter sp. (A) and two Gram-positive bacteria, La. plantarum (B) and E. faecalis (C). The ΔAMP14 mutants were significantly more susceptible than wild type only to Acetobacter sp. infection (P < 0.001), and otherwise resisted infection like wild type (P > 0.1). Pellet densities are reported for all systemic infections as the OD600 value.
These systemic infections confirm that AMPs can play a direct role in the control of Acetobacter sp., bacteria typically found in the gut, but have a lesser impact on La. plantarum and E. faecalis proliferation. This trend is consistent with the results above showing that AMPs most prominently contribute to Acetobacter control after gnotobiotic or mono-associative colonization.
DISCUSSION
In Drosophila, the immune system, and particularly the IMD pathway, has been robustly demonstrated to be an important regulator of the gut microbiota and intestinal homeostasis (21, 22, 27, 29, 32, 36). Several reports have indicated the importance of the IMD pathway in maintaining balanced microbiota during aging, and mutants for this pathway (e.g Relish) have atypical microbiota abundance and composition (27). While it is clear the IMD pathway is a regulator of the gut microbiota, little is known about the effectors mediating this regulation. In addition to regulating most AMP expression in the gut (21), the IMD pathway regulates other physiological aspects, including expression of digestive enzymes (41) and enterocyte delamination (39, 68, 69). The present work extends these studies by more narrowly defining the role of AMPs and lysozymes by comparing specific loss of these effectors to total loss of IMD signaling.
Our results confirm a prominent role for the IMD pathway in regulating both microbiota load and diversity, especially upon aging (21, 27, 36). The ΔAMP14 genotype mimics the Relish phenotype in many respects, showing that AMPs indeed contribute downstream of IMD to shape the microbiota composition. Our experiments consistently showed an increase in the load of Acetobacter species in both RelE20 and ΔAMP14 flies. The observation that both ΔAMP14 and RelE20 flies are also susceptible to Acetobacter systemic infection, together with previous studies showing that AMPs contribute to survival upon Gram-negative bacterial infection (45, 50), provides strong evidence that direct microbicidal activity of AMPs regulates these Gram-negative bacteria. Collectively, this indicates for the first time that the basal level of IMD pathway activity induced by the gut microbiota (27) leads to the production of AMPs that prevent overgrowth of Gram-negative commensals, such as Acetobacter. Future studies should clarify which AMP(s) among the 14 deleted in the ΔAMP14 flies regulate Acetobacter.
La. plantarum is an important member of Drosophila microbiota that is associated with the host, both in larval development and adulthood (11, 33, 70). As the DAP-type peptidoglycan found in the cell wall of these bacteria can activate the IMD pathway, we might expect to see an action of AMPs against them. However, the IMD pathway and AMP mutants are not very susceptible to DAP-type Gram-positive bacteria (45, 50). Moreover, d-alanylation of La. plantarum lipoteichoic acid has recently been proposed as a mechanism to protect against the action of AMPs and lysozymes (49). Here, we found that the AMPs play a role in controlling La. plantarum abundance in a mono-colonization setup but not when bacteria are in a community context. This might be due to the dynamics between microbiota members (e.g., competition between different species) or due to differential affinity of AMPs for the peptidoglycan of distinct species. It is possible that the abundance of La. plantarum is maintained at a threshold level in the gut and this is naturally achieved in a community through bacterial interactions. However, La. plantarum overgrowth can be inhibited by AMPs in a context where it becomes the only dominant member of the community.
Other factors that could influence the microbiota include developmental effects and sex-specific effects on immunity and nutrition (71, 72). Here, we only assessed the microbiota of adult females, but future studies should confirm if these AMP-microbiota interactions are consistent in other conditions. It should also be noted that we transferred the microbiota directly using beads, which could affect microbial community structure, e.g., by accruing microbial load over time or by avoiding bottleneck events that could have exacerbated stochastic microbe species takeover or drop out. We also used both propionic acid and methylparaben sodium salt (Moldex) in our food medium. These two preservative compounds are routinely used to prevent fly food spoilage, though it is known that they can affect microbial growth (73). Future investigations would benefit from monitoring microbial loads in the food while manipulating microbiota transfer techniques and fly diet.
The genome of Drosophila contains many genes encoding lysozymes, likely as a consequence of living in bacterially enriched habitats (46). Indeed, animals feeding on fermenting medium, such as ruminants or fruit flies, have a much higher number of lysozyme gene copies compared to animals feeding on “clean food” (74, 75). In many insects, lysozymes are induced upon systemic infection, pointing to a possible role as immune effectors. In contrast, Drosophila lysozymes are strongly expressed in the gut, indicating a specific role in the digestive process (46, 47). Of note, one uncharacterized gene annotated as encoding a putative lysozyme (CG6429) is strongly induced upon systemic infection and is partially regulated by the IMD pathway (38).
In this study, we generated a LysB-PΔ mutant deficient for four lysozyme genes strongly expressed in the gut. LysB-PΔ gut extracts have reduced lysozyme activity (Fig. S1), confirming that these four genes indeed contribute to gut lysozyme activity. As lysozymes are known to digest peptidoglycan and can exhibit bactericidal activity alone or in combination with AMPs (48), we were interested to monitor the impact of lysozymes on the gut microbiota. We expected that loss of lysozymes would have a greater effect on Gram-positive bacteria, as the thin peptidoglycan layer of Gram-negative bacteria is protected by their external lipopolysaccharide (LPS) membrane. Consistently, 16S sequencing revealed that LysB-PΔ mutants exhibited increased relative community Lactobacillus abundance. However, mono-association experiments revealed a role of lysozymes in suppressing growth of only Gram-negative Acetobacter species. This effect was less marked than that of AMP deficient mutants.
An interesting observation of our study is that flies lacking AMPs or lysozymes displayed greater community stochasticity, similar to the phenotype of RelE20 flies. This suggests that multiple factors, including AMPs, lysozymes, and bacteria-bacteria interactions, contribute to stability of the gut microbiota, and that loss of these factors increases stochasticity. We avoided complications of fly genetic backgrounds by using isogenic fly strains. While the isogenization process homogenizes the genetic background, it also increases the degree of homozygosity along the genome, with a possible increase in genetic interactions. Thus, our study on AMPs and lysozymes using the iso Drosdel background should be reinforced by other studies using other backgrounds or alternative approaches.
In Drosophila, the induction of genes for antibacterial peptides after infection is blocked in IMD pathway mutants, such as RelE20, resulting in high susceptibility. These flies also cannot control their microbiota load, especially during aging (27). As expected, we found similar gut microbiota structure in Relish and AMP mutants. Indeed, both genotypes were unable to control the microbiota load and composition, but RelE20 flies had a more severe phenotype, with 16S analysis showing atypical microbial composition at early life stages and marked inability to control all inoculated bacterial species in mono-association experiments. This is likely due to the multiple roles of the IMD pathway in gut physiology, apoptosis, nutrition, and metabolism (39, 76, 77), the loss of which, in addition to AMPs, may exacerbate gut dysbiosis or hasten the inability of the flies to control microbiota growth. This indicates that although AMPs play an important role in control of microbiota members, they contribute only partially to the dysbiosis of mutant flies with perturbed IMD pathways.
Collectively, our work is the first to show direct involvement of AMPs and lysozymes in the control of Drosophila gut microbiota. Consequences of the loss of these effectors are exacerbated during aging, and their loss contributes to increased microbiota abundance and shifted composition. This work shows that immune effectors typically associated with resistance to pathogenic infections also help shape the beneficial gut community, consistent with the idea that host-symbiont interactions use the same “language” typically associated with pathogenesis (78).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains used in this study and their origins are as follows: Acetobacter sp. (53), Acetobacter pomorum (51), Lactobacillus plantarum (11), and Enterococcus faecalis (52). Lactobacillus brevis and Leuconostoc pseudomesenteroides were isolated from the “Valais” population, collected in the Valais canton of Switzerland in 2007 (79). Briefly, homogenates from 20 flies were spread over Man, Rogosa and Sharpe (MRS)-d-Mannitol 2.5% plates. A single colony was used to prepare liquid cultures (described below) and establish glycerol stocks, as well as for 16S rRNA gene full-length amplification using universal primers. The PCR products were sequenced by Sanger sequencing and assigned to taxa based on a Microbial BLASTn search against the nucleotide database of NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC=MicrobialGenomes). See Text S1 in the supplemental material for full 16S rRNA gene sequences of these isolates and Acetobacter sp. (53).
A. pomorum and Acetobacter sp. were cultivated in (MRS)-d-Mannitol 2.5% medium in aerobic conditions at 29°C for at least 18 h with agitation. La. plantarum, La. brevis, and Le. pseudomesenteroides were cultivated in anaerobic conditions in (MRS)-d-Mannitol 2.5% medium at 29°C for at least 18 h standing. E. faecalis was cultured in brain heart infusion (BHI) broth medium in aerobic conditions at 37°C for at least 16 h with agitation.
Gnotobiotic fly cultivation and media.
All experiments were done at 25°C with a 12-h dark/light cycle. Embryos were collected and emerging adults were a mix of males and females. Only the microbiotas of females were assessed. Because antibiotic treatments over multiple generations may result in epigenetic effects that may interfere with phenotypes (80–84), GF flies were freshly generated for each experiment. Please see the Extended Methods for details.
To avoid sticky “biofilm-like” formation on the medium, flies were transferred to fresh medium every 3 to 4 days. To avoid a decrease in microbial loads, the microbiota of each tube was transferred to the next using glass beads (Fig. S4). Briefly, flies were anaesthetized on ice and removed on sterile caps. Then, 10 to 20 glass beads were transferred to the old tube and shaken for 10 s. The beads were then transferred to the new sterile tube and were shaken again for 10 s to spread the bacteria around the tubes. Adults were then added in the new tubes. For 16S RNA-seq experiments, flies were sampled at 10 days and 29 days after colonization taken at least 3 days after the last transfer to fresh food.
For vertically transmitted microbiota, we let gnotobiotic adults (colonized as described above 3 to 4 days previously) lay eggs on fresh medium for 3 days. We let larvae grow in this original medium and began transferring the emerging adults to new tubes 5 days after the first fly emerged. We collected adults 10 to 15 days after emergence and analyzed their associated bacterial community.
For mono-association experiments, we colonized flies with each isolate that was included in the commensal cocktail. Because during mono-association changes in community structure are not a concern, we maintained the flies in their original tube throughout the experiment. Flies were sampled 6 days after colonization.
DNA extraction and qPCR.
Surface-sterilized flies (washed in sterile water and EtOH) were mechanically lysed by bead beating and the DNA extraction was carried out on samples using a DNeasy blood and tissue kit (Qiagen).
The qPCRs for absolute quantification were carried out as previously described (85) and for relative quantification qPCRs as described in reference 66. The universal (85) and Acetobacter-specific 16S (66) and Actin 5C primers (66) were previously described. Firmicutes-specific primers (antisense primer 5′-AGCGTTGTCCGGATTTAT-3′, sense primer 5′-CATTTCACCGCTACACAT-3′) were designed by aligning the 16S rRNA gene sequences of the four Firmicutes species that were used in this study. Their specificity (lack of amplification in Acetobacteraceae) was determined on plasmid DNA containing specific 16S sequences, as well as on DNA extracted from flies mono-associated with microbiota members described in this study.
16S rRNA gene amplicon sequencing and data processing.
Amplification of the V4 region of the 16S rRNA gene and the library preparation protocol were done as previously described (85), except that the first cycle of PCR was performed with 30 cycles. Libraries were verified by Fragment analyzer, mixed with 10% PhiX library (Illumina number FC-110-3001), and subjected to Illumina MiSeq v3 paired-end sequencing in one lane, with all libraries multiplexed.
Sequencing data have been processed using Divisive Amplicon Denoising Algorithm 2 (DADA2) pipeline (“dada2” package version 1.14.1 in R) and “phyloseq” package version 1.30.0. Please see the Extended Methods for further details.
Diversity and statistical analysis.
Permutational multivariate analysis of variance (ADONIS, “adonis” function) based on Bray–Curtis distances (“vegdist” function) (86) was used to test the effects of age and genotype on community structure, and “metaMDS” function was used for plotting beta-diversity. For pairwise comparisons of ADONIS, “adonis.pair” function was used from the “EcolUtils” package. To test the dispersion of communities, we used the function “betadisper” (87, 88) and compared the distances of individual samples to group centroids in multidimensional space using “permutest.”
All statistical analyses were performed using R (version 3.6.3). We used general linear mixed models (“lme4” package version 1.1.23) to test for the effects of age, genotype, and their interaction (depending on the experimental design) on bacterial loads or dispersion of bacterial communities. Pairwise comparisons were performed using “emmeans” and “pairs” functions (“emmeans” package version 1.5.1). P values were adjusted using the FDR method.
Systemic infection and life span assay.
Systemic infections were performed by pricking 5- to 7-day-old conventionally reared adult females in the thorax with a 100-μm-thick insect pin dipped into a concentrated pellet of bacteria. The bacteria were grown to the following concentration values measured by the optical density at 600 nm (OD600) in (MRS)-d-Mannitol 2.5% broth (Acetobacter sp., OD600 = 150; La. plantarum, OD600 = 200) or BHI (E. faecalis, OD600 = 5). Infected flies were maintained at 25°C for experiments. At least three replicate survival experiments were performed for each infection, with 20 flies per vial on standard fly medium without yeast. Survivals were scored daily and flies were moved to fresh medium every 2 days.
Survival data were analyzed using the survival package in R 3.6.3 with a Cox proportional hazards model (coxph() function) including experiment as a covariate in the final model, when significant.
Data availability.
16S rRNA gene amplicon sequencing data have been submitted to the NCBI database under BioProject no. PRJNA742915.
Lysozyme mutant characterization. (A) Diagram of the LysB-PΔ mutation, which deletes 11.5 kb of nucleotide sequence from the Drosophila genome. LysB-PΔ flies lack the four lysozyme genes (LyB, LysD, LysE, and LysP) that are highly expressed in the gut, along with the LysC pseudogene. (B) LysB-PΔ mutants have a reduced capacity to degrade peptidoglycan in the gut compared to wild-type flies. Gut extracts from wild-type flies (w) and lysozyme mutants (LysB-PΔ) were incubated with E. faecalis peptidoglycan for 48 h. The lysozyme activity was monitored by change in optical density of the peptidoglycan solution. Commercial lysozyme was included as a positive control (See the Extended Methods for details). Download FIG S1, EPS file, 0.1 MB (106.3KB, eps) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Homogeneity of multivariate dispersals in microbial communities. Distance to the center of each group (genotype) represents the spread of the communities in the multivariate space (PCoA) in a vertical transmission setup (A), 10 days after colonization (B), and 29 days after colonization (C). Different letters indicate significant differences (P < 0.05) between P values upon pairwise comparison, calculated by permutations on multivariate equivalent of variances. A significant difference between groups indicates that samples belonging to one group are more variable in microbiota composition compared to the other group. Download FIG S2, EPS file, 0.2 MB (172.3KB, eps) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ΔAMP14 flies survive clean injury. Females from the indicated genotypes were pricked with a clean 100-μm insect pin. No genotype displayed pronounced susceptibility to clean injury. Download FIG S3, EPS file, 0.1 MB (61.5KB, eps) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbiota transfer with beads. See detailed protocol in the Materials and Methods in the section “Gnotobiotic fly cultivation and media.” FIG S4, TIF file, 0.4 MB (393.3KB, tif)
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA gene sequences of La. brevis, Le. pseudomesenteroides, and Acetobacter sp. (see Erkosar et al., 2017) used in this study. Download Text S1, DOCX file, 0.01 MB (13.6KB, docx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of flies used in each sample and number of reads that were retained after each step of the 16S rRNA gene analysis. Download Table S1, XLSX file, 0.02 MB (18.1KB, xlsx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of the statistics used for the data analyzed in Fig. 1 and Fig. 2. Download Table S2, XLSX file, 0.02 MB (18KB, xlsx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extended Materials and Methods. Supplementary information about Drosophila genetics, 16S data analysis, gnotobiotic fly cultivation, and lysozyme peptidoglycan digestion assay. Download Text S2, DOCX file, 0.03 MB (30.6KB, docx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We are grateful to Tadeusz J. Kawecki (University of Lausanne, Switzerland) for providing bacterial isolates, Xiaoxue Li for providing data on LysB-PΔ strain validation, Iatsenko Igor (Max Planck Institute, Germany) for LysB-PΔ strain isogenization, and to Hannah Westlake for critical reading of the manuscript.
This work was supported by the Swiss National Science Foundation grant no. 310030_185295. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
A.M., B.E., and B.L. conceived the study and designed the experiments; A.M., B.E., and M.A.H. collected the data; B.E. analyzed the data; S.K. provided critical reagents; A.M., B.E., M.A.H., and B.L. wrote the manuscript.
We declare no competing interests.
Footnotes
Citation Marra A, Hanson MA, Kondo S, Erkosar B, Lemaitre B. 2021. Drosophila antimicrobial peptides and lysozymes regulate gut microbiota composition and abundance. mBio 12:e00824-21. https://doi.org/10.1128/mBio.00824-21.
Contributor Information
B. Erkosar, Email: berra.erkosarcombe@unil.ch.
B. Lemaitre, Email: bruno.lemaitre@epfl.ch.
William W. Ja, The Scripps Research Institute
Margaret J. McFall-Ngai, University of Hawaii at Manoa
REFERENCES
- 1.Belkaid Y, Harrison OJ. 2017. Homeostatic immunity and the microbiota. Immunity 46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim S, Jazwinski SM. 2018. The gut microbiota and healthy aging: a mini-review. Gerontology 64:513–520. doi: 10.1159/000490615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fung TC, Olson CA, Hsiao EY. 2017. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci 20:145–155. doi: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. 2016. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med 8:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng MY, Inohara N, Nuñez G. 2017. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol 10:18–26. doi: 10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas AE. 2018. The Drosophila model for microbiome research. Lab Anim (NY) 47:157–164. doi: 10.1038/s41684-018-0065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma D, Storelli G, Mitchell M, Leulier F. 2015. Studying host-microbiota mutualism in Drosophila: harnessing the power of gnotobiotic flies. Biomed J 38:285–293. doi: 10.4103/2319-4170.158620. [DOI] [PubMed] [Google Scholar]
- 8.Broderick NA, Lemaitre B. 2012. Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3:307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludington WB, Ja WW. 2020. Drosophila as a model for the gut microbiome. PLoS Pathog 16:e1008398. doi: 10.1371/journal.ppat.1008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erkosar B, Storelli G, Defaye A, Leulier F. 2013. Host-intestinal microbiota mutualism: “learning on the fly”. Cell Host Microbe 13:8–14. doi: 10.1016/j.chom.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. 2011. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab 14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 12.Keebaugh ES, Yamada R, Obadia B, Ludington WB, Ja WW. 2018. Microbial quantity impacts Drosophila nutrition, development, and lifespan. iScience 4:247–259. doi: 10.1016/j.isci.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer C, Trautman EP, Crawford JM, Stabb EV, Handelsman J, Broderick NA. 2017. Metabolite exchange between microbiome members produces compounds that influence Drosophila behavior. Elife 6:1–25. doi: 10.7554/eLife.18855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HY, Lee SH, Lee JH, Lee WJ, Min KJ. 2019. The role of commensal microbes in the lifespan of Drosophila melanogaster. Aging (Albany NY) 11:4611–4640. doi: 10.18632/aging.102073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sansone CL, Cohen J, Yasunaga A, Xu J, Osborn G, Subramanian H, Gold B, Buchon N, Cherry S. 2015. Microbiota-dependent priming of antiviral intestinal immunity in Drosophila. Cell Host Microbe 18:571–581. doi: 10.1016/j.chom.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fast D, Petkau K, Ferguson M, Shin M, Galenza A, Kostiuk B, Pukatzki S, Foley E. 2020. Vibrio cholerae-symbiont interactions inhibit intestinal repair in Drosophila. Cell Rep 30:1088–1100.e5. doi: 10.1016/j.celrep.2019.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark RI, Salazar A, Yamada R, Fitz-Gibbon S, Morselli M, Alcaraz J, Rana A, Rera M, Pellegrini M, Ja WW, Walker DW. 2015. Distinct shifts in microbiota composition during Drosophila aging impair intestinal function and drive mortality. Cell Rep 12:1656–1667. doi: 10.1016/j.celrep.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Qi Y, Jasper H. 2016. Preventing age-related decline of gut compartmentalization limits microbiota dysbiosis and extends lifespan. Cell Host Microbe 19:240–253. doi: 10.1016/j.chom.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pais IS, Valente RS, Sporniak M, Teixeira L. 2018. Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol 16:e2005710. doi: 10.1371/journal.pbio.2005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandehoef C, Molaei M, Karpac J. 2020. Dietary adaptation of microbiota in Drosophila requires NF-κB-dependent control of the translational regulator 4E-BP. Cell Rep 31:107736. doi: 10.1016/j.celrep.2020.107736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. 2009. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev 23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchon N, Silverman N, Cherry S. 2014. Immunity in Drosophila melanogaster—from microbial recognition to whole-organism physiology. Nat Rev Immunol 14:796–810. doi: 10.1038/nri3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemaitre B, Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu Rev Immunol 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 24.Royet J, Gupta D, Dziarski R. 2011. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat Rev Immunol 11:837–851. doi: 10.1038/nri3089. [DOI] [PubMed] [Google Scholar]
- 25.Lesperance DNA, Broderick NA. 2020. Gut bacteria mediate nutrient availability in Drosophila diets. Appl Environ Microbiol 87:e01401-20. doi: 10.1128/AEM.01401-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin SJH, Cohen LB, Wasserman SA. 2020. Effector specificity and function in Drosophila innate immunity: getting AMPed and dropping Boms. PLoS Pathog 16:e1008480. doi: 10.1371/journal.ppat.1008480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broderick NA, Buchon N, Lemaitre B. 2014. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio 5:e01117–14–e01114. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liehl P, Blight M, Vodovar N, Boccard F, Lemaitre B. 2006. Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog 2:e56. doi: 10.1371/journal.ppat.0020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosco-Drayon V, Poidevin M, Gomperts Boneca I, Narbonne-Reveau K, Royet J, Charroux B. 2012. Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe 12:153–165. doi: 10.1016/j.chom.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Zaidman-Rémy A, Hervé M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B. 2006. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Paredes JC, Welchman DP, Poidevin M, Lemaitre B. 2011. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35:770–779. doi: 10.1016/j.immuni.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 32.Charroux B, Capo F, Kurz CL, Peslier S, Chaduli D, Viallat-Lieutaud A, Royet J. 2018. Cytosolic and secreted peptidoglycan-degrading enzymes in Drosophila respectively control local and systemic immune responses to microbiota. Cell Host Microbe 23:215–228.e4. doi: 10.1016/j.chom.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 33.Ryu J, Kim S, Lee H, Bai JY, Nam Y, Bae J, Lee DG, Shin SC, Ha E, Lee W. 2008. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 34.Neyen C, Runchel C, Schüpfer F, Meier P, Lemaitre B. 2016. The regulatory isoform rPGRP-LC resolves immune activation through ESCRT-mediated receptor clearance. Nat Immunol 17:1150–1158. doi: 10.1038/ni.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaidman-Rémy A, Poidevin M, Hervé M, Welchman DP, Paredes JC, Fahlander C, Steiner H, Mengin-Lecreulx D, Lemaitre B. 2011. Drosophila immunity: analysis of PGRP-SB1 expression, enzymatic activity and function. PLoS One 6:e17231. doi: 10.1371/journal.pone.0017231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo L, Karpac J, Tran SL, Jasper H. 2014. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell 156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iatsenko I, Boquete J, Lemaitre B. 2018. Microbiota-derived lactate activates production of reactive oxygen species by the intestinal NADPH oxidase Nox and shortens Drosophila lifespan. Immunity 49:929–942. doi: 10.1016/j.immuni.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 38.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. 2002. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J 21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhai Z, Boquete JP, Lemaitre B. 2018. Cell-specific Imd-NF-κB responses enable simultaneous antibacterial and intestinal epithelial cell shedding upon bacterial infection. Immunity 48:897–910. doi: 10.1016/j.immuni.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 40.Myllymäki H, Valanne S, Rämet M. 2014. The Drosophila Imd signaling pathway. J Immunol 192:3455–3462. doi: 10.4049/jimmunol.1303309. [DOI] [PubMed] [Google Scholar]
- 41.Erkosar B, Erkosar Combe B, Defaye A, Bozonnet N, Puthier D, Royet J, Leulier F. 2014. Drosophila microbiota modulates host metabolic gene expression via IMD/NF-κB signaling. PLoS One 9:e94729. doi: 10.1371/journal.pone.0094729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. 2009. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Hanson MA, Cohen LB, Marra A, Iatsenko I, Wasserman SA, Lemaitre B. 2020. The Drosophila Baramicin polypeptide gene protects against fungal infection. bioRxiv 10.1101/2020.11.23.394148. [DOI] [PMC free article] [PubMed]
- 44.Imler J, Bulet P. 2005. Antimicrobial peptides in Drosophila : structures, activities and gene regulation. Chem Imunol Allergy 86:1–21. doi: 10.1159/000086648. [DOI] [PubMed] [Google Scholar]
- 45.Hanson MA, Dostálová A, Ceroni C, Poidevin M, Kondo S, Lemaitre B. 2019. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. Elife 8:e44341. doi: 10.7554/eLife.44341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daffre S, Kylsten P, Samakovlis C, Hultmark D. 1994. The lysozyme locus in Drosophila melanogaster: an expanded gene family adapted for expression in the digestive tract. Mol Gen Genet 242:152–162. doi: 10.1007/BF00391008. [DOI] [PubMed] [Google Scholar]
- 47.Regel R, Matioli SR, Terra WR. 1998. Molecular adaptation of Drosophila melanogaster lysozymes to a digestive function. Insect Biochem Mol Biol 28:309–319. doi: 10.1016/s0965-1748(97)00108-2. [DOI] [PubMed] [Google Scholar]
- 48.Nash JA, Ballard TNS, Weaver TE, Akinbi HT. 2006. The peptidoglycan-degrading property of lysozyme is not required for bactericidal activity in vivo. J Immunol 177:519–526. doi: 10.4049/jimmunol.177.1.519. [DOI] [PubMed] [Google Scholar]
- 49.Zaynoun A, Kallassy Awad M, Rejasse A, Courtin P, Gomberts Boneca I, Chapot-Chartier M-P, Sanchis Borja V, El Chamy L. 2019. D-Alanylation of techoic acids in bacilli impedes the immune sensing of peptidoglycan in Drosophila. bioRxiv doi: 10.1101/631523. [DOI]
- 50.Carboni A, Hanson MA, Lindsay SA, Wasserman SA, Lemaître B. 2021. Cecropins contribute to Drosophila host defence against fungal and Gram-negative bacterial infection. bioRxiv 10.1101/2021.05.06.442783. [DOI] [PMC free article] [PubMed]
- 51.Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 52.Lemaitre B, Reichhart JM, Hoffmann JA. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci U S A 94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erkosar B, Kolly S, van der Meer JR, Kawecki TJ. 2017. Adaptation to chronic malnutrition leads to reduced dependence on microbiota in Drosophila. mBio 8:e01496. doi: 10.1128/mBio.01496-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schleifer KH, Kandler O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, Mengin-Lecreulx D, Lemaitre B. 2003. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol 4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 56.Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, Silverman N. 2004. Monomeric and polymeric Gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20:637–649. doi: 10.1016/S1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 57.Kleerebezem M, Hols P, Bernard E, Rolain T, Zhou M, Siezen RJ, Bron PA. 2010. The extracellular biology of the lactobacilli. FEMS Microbiol Rev 34:199–230. doi: 10.1111/j.1574-6976.2010.00208.x. [DOI] [PubMed] [Google Scholar]
- 58.Chang JD, Wallace AG, Foster EE, Kim SJ. 2018. Peptidoglycan compositional analysis of Enterococcus faecalis biofilm by stable isotope labeling by amino acids in a bacterial culture. Biochemistry 57:1274–1283. doi: 10.1021/acs.biochem.7b01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee KA, Kim SH, Kim EK, Ha EM, You H, Kim B, Kim MJ, Kwon Y, Ryu JH, Lee WJ. 2013. Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153:797–811. doi: 10.1016/j.cell.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 60.Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar R. 2016. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv doi: 10.1101/081257. [DOI]
- 62.Amnon A, Daniel M, Navas-Molina J, Kopylova E, Morton J, Xu ZZ, Eric K, Thompson L, Hyde E, Gonzalez A, Knight R. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2:e00191-16. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamada Y, Yukphan P. 2008. Genera and species in acetic acid bacteria. Int J Food Microbiol 125:15–24. doi: 10.1016/j.ijfoodmicro.2007.11.077. [DOI] [PubMed] [Google Scholar]
- 64.Dutta D, Gachhui R. 2006. Novel nitrogen-fixing Acetobacter nitrogenifigens sp. nov., isolated from Kombucha tea. Int J Syst Evol Microbiol 56:1899–1903. doi: 10.1099/ijs.0.64101-0. [DOI] [PubMed] [Google Scholar]
- 65.Kešnerová L, Mars RAT, Ellegaard KM, Troilo M, Sauer U, Engel P. 2017. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol 15:e2003467. doi: 10.1371/journal.pbio.2003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erkosar B, Yashiro E, Zajitschek F, Friberg U, Maklakov AA, van der Meer JR, Kawecki TJ. 2018. Host diet mediates a negative relationship between abundance and diversity of Drosophila gut microbiota. Ecol Evol 8:9491–9502. doi: 10.1002/ece3.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Duneau D, Ferdy J-B, Revah J, Kondolf H, Ortiz GA, Lazzaro BP, Buchon N. 2017. Stochastic variation in the initial phase of bacterial infection predicts the probability of survival in D. melanogaster. Elife 6:e28298. doi: 10.7554/eLife.28298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhai Z, Huang X, Yin Y. 2018. Beyond immunity: the Imd pathway as a coordinator of host defense, organismal physiology and behavior. Dev Comp Immunol 83:51–59. doi: 10.1016/j.dci.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 69.Petkau K, Ferguson M, Guntermann S, Foley E. 2017. Constitutive immune activity promotes tumorigenesis in Drosophila intestinal progenitor cells. Cell Rep 20:1784–1793. doi: 10.1016/j.celrep.2017.07.078. [DOI] [PubMed] [Google Scholar]
- 70.Cox CR, Gilmore MS. 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun 75:1565–1576. doi: 10.1128/IAI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Belmonte RL, Corbally MK, Duneau DF, Regan JC. 2020. Sexual dimorphisms in innate immunity and responses to infection in Drosophila melanogaster. Front Immunol 10:3075. doi: 10.3389/fimmu.2019.03075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wat LW, Chao C, Bartlett R, Buchanan JL, Millington JW, Chih HJ, Chowdhury ZS, Biswas P, Huang V, Shin LJ, Wang LC, Gauthier MPL, Barone MC, Montooth KL, Welte MA, Rideout EJ. 2020. A role for triglyceride lipase brummer in the regulation of sex differences in Drosophila fat storage and breakdown. PLoS Biol 18:e3000595. doi: 10.1371/journal.pbio.3000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tefit MA, Gillet B, Joncour P, Hughes S, Leulier F. 2018. Stable association of a Drosophila-derived microbiota with its animal partner and the nutritional environment throughout a fly population’s life cycle. J Insect Physiol 106:2–12. doi: 10.1016/j.jinsphys.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 74.Callewaert L, Michiels CW. 2010. Lysozymes in the animal kingdom. J Biosci 35:127–160. doi: 10.1007/s12038-010-0015-5. [DOI] [PubMed] [Google Scholar]
- 75.Ito Y, Nakamura M, Hotani T, Imoto T. 1995. Insect lysozyme from house fly (Musca domestica) larvae: possible digestive function based on sequence and enzymatic properties. J Biochem 118:546–551. doi: 10.1093/oxfordjournals.jbchem.a124943. [DOI] [PubMed] [Google Scholar]
- 76.Molaei M, Vandehoef C, Karpac J. 2019. NF-κB shapes metabolic adaptation by attenuating Foxo-mediated lipolysis in Drosophila. Dev Cell 49:802–810. doi: 10.1016/j.devcel.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nandy A, Lin L, Velentzas PD, Wu LP, Baehrecke EH, Silverman N. 2018. The NF-κB factor Relish regulates Atg1 expression and controls autophagy. Cell Rep 25:2110–2120.e3. doi: 10.1016/j.celrep.2018.10.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McFall-Ngai M, Heath-Heckman EAC, Gillette AA, Peyer SM, Harvie EA. 2012. The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol 24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Joye P, Kawecki TJ. 2019. Sexual selection favours good or bad genes for pathogen resistance depending on males’ pathogen exposure. Proc Biol Sci 286:20190226. doi: 10.1098/rspb.2019.0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qiu W, Fang M, Magnuson JT, Greer JB, Chen Q, Zheng Y, Xiong Y, Luo S, Zheng C, Schlenk D. 2020. Maternal exposure to environmental antibiotic mixture during gravid period predicts gastrointestinal effects in zebrafish offspring. J Hazard Mater 399:123009. doi: 10.1016/j.jhazmat.2020.123009. [DOI] [PubMed] [Google Scholar]
- 81.Hu Y, Peng J, Tai N, Hu C, Zhang X, Wong FS, Wen L. 2015. Maternal antibiotic treatment protects offspring from diabetes development in nonobese diabetic mice by generation of tolerogenic APCs. J Immunol 195:4176–4184. doi: 10.4049/jimmunol.1500884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim HY, Asselman J, Jeong TY, Yu S, De Schamphelaere KAC, Kim SD. 2017. Multigenerational effects of the antibiotic tetracycline on transcriptional responses of Daphnia magna and its relationship to higher levels of biological organizations. Environ Sci Technol 51:12898–12907. doi: 10.1021/acs.est.7b05050. [DOI] [PubMed] [Google Scholar]
- 83.Ghosh D, Veeraraghavan B, Elangovan R, Vivekananda P. 2020. Antibiotic resistance and epigenetics : more to it than meets the eye. Antimicrob Agents Chemother 64:e02225-19. doi: 10.1128/AAC.02225-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Badal S, Her YF, James Maher L. 2015. Nonantibiotic effects of fluoroquinolones in mammalian cells. J Biol Chem 290:22287–22297. doi: 10.1074/jbc.M115.671222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kešnerová L, Emery O, Troilo M, Liberti J, Erkosar B, Engel P. 2020. Gut microbiota structure differs between honeybees in winter and summer. ISME J 14:801–814. doi: 10.1038/s41396-019-0568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 87.Anderson MJ. 2006. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62:245–253. doi: 10.1111/j.1541-0420.2005.00440.x. [DOI] [PubMed] [Google Scholar]
- 88.Anderson MJ, Ellingsen KE, McArdle BH. 2006. Multivariate dispersion as a measure of beta diversity. Ecol Lett 9:683–693. doi: 10.1111/j.1461-0248.2006.00926.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lysozyme mutant characterization. (A) Diagram of the LysB-PΔ mutation, which deletes 11.5 kb of nucleotide sequence from the Drosophila genome. LysB-PΔ flies lack the four lysozyme genes (LyB, LysD, LysE, and LysP) that are highly expressed in the gut, along with the LysC pseudogene. (B) LysB-PΔ mutants have a reduced capacity to degrade peptidoglycan in the gut compared to wild-type flies. Gut extracts from wild-type flies (w) and lysozyme mutants (LysB-PΔ) were incubated with E. faecalis peptidoglycan for 48 h. The lysozyme activity was monitored by change in optical density of the peptidoglycan solution. Commercial lysozyme was included as a positive control (See the Extended Methods for details). Download FIG S1, EPS file, 0.1 MB (106.3KB, eps) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Homogeneity of multivariate dispersals in microbial communities. Distance to the center of each group (genotype) represents the spread of the communities in the multivariate space (PCoA) in a vertical transmission setup (A), 10 days after colonization (B), and 29 days after colonization (C). Different letters indicate significant differences (P < 0.05) between P values upon pairwise comparison, calculated by permutations on multivariate equivalent of variances. A significant difference between groups indicates that samples belonging to one group are more variable in microbiota composition compared to the other group. Download FIG S2, EPS file, 0.2 MB (172.3KB, eps) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ΔAMP14 flies survive clean injury. Females from the indicated genotypes were pricked with a clean 100-μm insect pin. No genotype displayed pronounced susceptibility to clean injury. Download FIG S3, EPS file, 0.1 MB (61.5KB, eps) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbiota transfer with beads. See detailed protocol in the Materials and Methods in the section “Gnotobiotic fly cultivation and media.” FIG S4, TIF file, 0.4 MB (393.3KB, tif)
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA gene sequences of La. brevis, Le. pseudomesenteroides, and Acetobacter sp. (see Erkosar et al., 2017) used in this study. Download Text S1, DOCX file, 0.01 MB (13.6KB, docx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of flies used in each sample and number of reads that were retained after each step of the 16S rRNA gene analysis. Download Table S1, XLSX file, 0.02 MB (18.1KB, xlsx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of the statistics used for the data analyzed in Fig. 1 and Fig. 2. Download Table S2, XLSX file, 0.02 MB (18KB, xlsx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extended Materials and Methods. Supplementary information about Drosophila genetics, 16S data analysis, gnotobiotic fly cultivation, and lysozyme peptidoglycan digestion assay. Download Text S2, DOCX file, 0.03 MB (30.6KB, docx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
16S rRNA gene amplicon sequencing data have been submitted to the NCBI database under BioProject no. PRJNA742915.
Lysozyme mutant characterization. (A) Diagram of the LysB-PΔ mutation, which deletes 11.5 kb of nucleotide sequence from the Drosophila genome. LysB-PΔ flies lack the four lysozyme genes (LyB, LysD, LysE, and LysP) that are highly expressed in the gut, along with the LysC pseudogene. (B) LysB-PΔ mutants have a reduced capacity to degrade peptidoglycan in the gut compared to wild-type flies. Gut extracts from wild-type flies (w) and lysozyme mutants (LysB-PΔ) were incubated with E. faecalis peptidoglycan for 48 h. The lysozyme activity was monitored by change in optical density of the peptidoglycan solution. Commercial lysozyme was included as a positive control (See the Extended Methods for details). Download FIG S1, EPS file, 0.1 MB (106.3KB, eps) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Homogeneity of multivariate dispersals in microbial communities. Distance to the center of each group (genotype) represents the spread of the communities in the multivariate space (PCoA) in a vertical transmission setup (A), 10 days after colonization (B), and 29 days after colonization (C). Different letters indicate significant differences (P < 0.05) between P values upon pairwise comparison, calculated by permutations on multivariate equivalent of variances. A significant difference between groups indicates that samples belonging to one group are more variable in microbiota composition compared to the other group. Download FIG S2, EPS file, 0.2 MB (172.3KB, eps) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ΔAMP14 flies survive clean injury. Females from the indicated genotypes were pricked with a clean 100-μm insect pin. No genotype displayed pronounced susceptibility to clean injury. Download FIG S3, EPS file, 0.1 MB (61.5KB, eps) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbiota transfer with beads. See detailed protocol in the Materials and Methods in the section “Gnotobiotic fly cultivation and media.” FIG S4, TIF file, 0.4 MB (393.3KB, tif)
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA gene sequences of La. brevis, Le. pseudomesenteroides, and Acetobacter sp. (see Erkosar et al., 2017) used in this study. Download Text S1, DOCX file, 0.01 MB (13.6KB, docx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of flies used in each sample and number of reads that were retained after each step of the 16S rRNA gene analysis. Download Table S1, XLSX file, 0.02 MB (18.1KB, xlsx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of the statistics used for the data analyzed in Fig. 1 and Fig. 2. Download Table S2, XLSX file, 0.02 MB (18KB, xlsx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extended Materials and Methods. Supplementary information about Drosophila genetics, 16S data analysis, gnotobiotic fly cultivation, and lysozyme peptidoglycan digestion assay. Download Text S2, DOCX file, 0.03 MB (30.6KB, docx) .
Copyright © 2021 Marra et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.