Abstract
Nuclear factor kappa B (NF-κB) has been implicated in the regulation of cell proliferation, transformation, and tumor development. We provide evidence for a direct link between NF-κB activity and cell cycle regulation. NF-κB was found to stimulate transcription of cyclin D1, a key regulator of G1 checkpoint control. Two NF-κB binding sites in the human cyclin D1 promoter conferred activation by NF-κB as well as by growth factors. Both levels and kinetics of cyclin D1 expression during G1 phase were controlled by NF-κB. Moreover, inhibition of NF-κB caused a pronounced reduction of serum-induced cyclin D1-associated kinase activity and resulted in delayed phosphorylation of the retinoblastoma protein. Furthermore, NF-κB promotes G1-to-S-phase transition in mouse embryonal fibroblasts and in T47D mammary carcinoma cells. Impaired cell cycle progression of T47D cells expressing an NF-κB superrepressor (IκBαΔN) could be rescued by ectopic expression of cyclin D1. Thus, NF-κB contributes to cell cycle progression, and one of its targets might be cyclin D1.
The inducible transcription factor NF-κB participates in the regulation of numerous genes, many of which are involved in inflammation and the immune response. The NF-κB/Rel family consists of five members (p50, p52, p65 [RelA], c-Rel, and RelB) which can form various homo- or heterodimeric complexes. NF-κB is activated by the release from cytoplasmic IκB proteins and subsequently translocates into the nucleus (3, 5, 34). Activation is triggered by signal-induced phosphorylation of IκB, which targets the inhibitor for rapid degradation by the proteasome (49).
Several observations have suggested a role of the NF-κB and IκB gene products in cell proliferation, transformation, and tumor development (47, 53). NF-κB controls the expression of a number of growth-promoting cytokines. In fact, a nuclear NF-κB-like DNA binding activity is induced during the G0-to-G1 transition after serum stimulation in mouse fibroblasts and in regenerating liver (6, 13–15, 18, 54). Interestingly, the NF-κB transactivation potential appears to be linked to signaling that controls cell cycle progression (9, 41).
The first evidence for a connection between NF-κB and cell death came from studies with mice lacking the RelA unit of NF-κB as a result of targeted mutation of the relA gene. These mice die before birth and show massive degeneration of liver cells caused by apoptosis (10). The antiapoptotic function of NF-κB is supported by several studies demonstrating that NF-κB activity prevents the induction of apoptosis by tumor necrosis factor alpha, ionizing radiation, and anticancer agents (4) and that c-Rel prevents spontaneous apoptosis of B cells (52). Recent data indicate that constitutive NF-κB activation is essential for apoptosis resistance of different types of tumor cells (7, 48). Interestingly, constitutive NF-κB is required for cell cycle progression of Hodgkin’s lymphoma cells (7). However, a direct link between NF-κB activity and cell cycle progression remains to be established.
The control of mammalian cell proliferation by extracellular signals takes place in mid- to late G1 phase of the cell cycle. D-type cyclins, in association with cyclin-dependent kinases CDK4 and CDK6, promote G1-to-S-phase transition by phosphorylating the retinoblastoma protein (pRB), thereby releasing the transcription factor E2F, which is required for the activation of S-phase-specific genes (8, 11, 21, 27, 39, 44, 46, 51).
The D-type cyclins are induced as part of the delayed early response to mitogenic stimulation by growth factors, form active holoenzymes with CDK4 or CDK6 by mid-G1, and are able to bind directly to pRB via their N-terminal L-X-C-X-E motifs. Furthermore, they have a substrate preference for pRB over histone H1, and they phosphorylate pRB in vitro on residues which are physiologically phosphorylated in G1 in vivo (44, 46, 51). Consistent with a major role in positive regulation of G1 progression, the D-type cyclins are required for S-phase entry, and their overexpression accelerates G1 and reduces dependency on exogenous growth factors (8). These data suggest that cyclin D-associated kinases and their pRB substrate are the central players of the G1 checkpoint control. In fact, it could be demonstrated that mitogenic signal transduction pathways from three classes of receptors converge and strictly require the cyclin D-CDK activity to induce S phase (31). In addition, members of different signal transduction pathways regulate cyclin D expression positively (e.g., the transforming mutant p21ras and p42/p44MAPK) or negatively (e.g., p38) (1, 2, 28, 40). However the transcriptional mechanisms that link mitogenic signal transduction to cyclin D expression are poorly understood.
Our data indicate that NF-κB transmits growth signals directly to key regulators of the cell cycle. NF-κB activates transcription of the cyclin D1 promoter primarily through a proximal binding site. The NF-κB binding sites that were identified are required for serum induction of cyclin D1 transcription. Inhibition of NF-κB activity in mouse embryo fibroblasts (MEF), T47D mammary carcinoma cells, or HeLa cells stably expressing a dominant negative IκBα mutant led to a delayed and reduced expression of cyclin D1 during G1 phase. Furthermore, inhibition of NF-κB resulted in retarded pRB phosphorylation and in impeded G1-to-S-phase transition. The impaired G1-to-S-phase transition caused by NF-κB inhibition could be overcome by ectopic expression of cyclin D1. These observations suggest that NF-κB directly contributes to stimulation of cell cycle progression by regulating the RB pathway.
MATERIALS AND METHODS
Cell culture.
Primary MEF (32), COS-7 cells, HeLa cells, and NIH 3T3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 U of penicillin-streptomycin per ml, and 1 mM sodium pyruvate. T47DΔMTcycD1 cells (37) (donated by E. Musgrove) were grown in RPMI 1640 phenol red-free medium supplemented with 10% FCS, 100 U of penicillin-streptomycin per ml, and 200 μg of G418 (Gibco BRL) per ml. These cells contain a stably integrated cyclin D1 expression vector driven by a zinc-inducible metallothionein promoter (37). Transient transfections into COS-7 and NIH 3T3 cells were carried out with Lipofectin (Gibco BRL). For stable transfections in MEF and T47DΔMTcycD1 cells, Superfect (Qiagen) was used. Stable clones were selected with G418 or hygromycin. All transfections were done according to the manufacturer’s protocols.
DNA constructs.
The cyclin D1 promoter-containing construct pD1luc was described previously (36). We generated mutant promoter constructs D1-κB1M and D1-κB2M, harboring two point mutations in the D1-κB1 (CGCGACCCCC) or the D1-κB2 (CGCGAGTTTT) binding site (introduced point mutations are underlined), respectively, by site-directed mutagenesis with the Clontech Transformer mutagenesis kit. For D1-κB1/2M, the mutant D1-κB1 sites were cloned as a SpeI/PmlI fragment into D1-κB2M.
The p50 and p65 expression plasmids pECEp50 and pECEp65 (38) and the IκBΔN expression plasmid (25) were described previously. For stable transfections of T47DΔMTcycD1 cells, IκBΔN was cloned as an NruI/EcoRV fragment in ppIXIhygM4. Plasmid c-jun-His6 was kindly provided by D. Bohmann; plasmid pBS-SP1 was provided by R. Tjian. Plasmid pGL2HIV was constructed by inserting the human immunodeficiency virus (HIV) core promoter as a BglII/HindIII fragment into pGL2 (Promega). pGL2HIVD1κB2 was constructed by cloning a double-stranded oligonucleotide containing cyclin D1 promoter sequence position from −37 to −19 into the MluI site of pGL2HIV.
EMSA.
For electrophoretic mobility shift assays (EMSA), cells were washed twice with phosphate-buffered saline (PBS), scraped off the plate, and lysed in EBL buffer (25) for 30 min at 4°C. The extracts were centrifuged at 15,000 rpm for 10 min at 4°C in a Sigma 2K15 centrifuge. The supernatant was used for further analysis. EMSA were performed as described previously (37). NF-κB-containing complexes were determined by adding p50 antibody (Rockland) or p65 antibody (no. sc109x; Santa Cruz) to the reaction mixture.
Western blotting.
Cells were washed twice with PBS, scraped off the plate, and lysed with extraction buffer (32) for 2 h at 4°C with occasional vortexing. The extracts were centrifuged at 15,000 rpm for 20 min at 4°C in a Sigma 2K15 centrifuge. The supernatant was used for further analysis. Extracted proteins (30 to 50 μg) were separated on sodium dodecyl sulfate-polyacrylamide gels. Gels were blotted onto nitrocellulose (Amersham) by a semidry method, and immunodetection was performed with the ECL enhanced chemiluminescence system. The primary antibodies used were mouse monoclonal antibodies against pRB (G3245; Pharmingen) or p16 (no. sc-1661; Santa Cruz), goat polyclonal antibody against p15 (no. sc-1429; Santa Cruz), and rabbit polyclonal antibodies against CDK4 (no. sc-260), CDK2 (no. sc-163), cyclin E (no. sc-481), p21 (no. sc-397), p27 (no. M-197), and IκBα (no. C-21) (all from Santa Cruz). Mouse monoclonal antibodies against cyclin D1 and cyclin D3 were donated by J. Bartek. Horseradish peroxidase-conjugated antimouse, antigoat, or antirabbit antibodies (Santa Cruz) were used for secondary detection.
pRB kinase assay.
For the pRB kinase assay, 200 μg of protein extracts (see “Western blotting” above) per immunoprecipitation was used with monoclonal anti-cyclin D1 antibody (5D4; donated by J. Bartek). Immunoprecipitation and pRB kinase assay were carried out as described previously (32), using as a substrate glutathione S-transferase–RB pocket (pRB amino acids 379 to 928).
Histone H1 kinase assay.
Cells were lysed in kinase extraction buffer (50 mM HEPES [pH 7.5], 250 mM NaCl, 5 mM EDTA, 0.1% Nonidet P-40, 1 mM dithiothreitol, 10 mM β-glycerophosphate, 1 mM NaF, 0.1 mM Na3VO4, 5 μg of leupeptin per ml, 2 μg of aprotinin per ml, and 0.1 M phenylmethylsulfonyl fluoride) and incubated for 30 min on ice with vigorous vortexing every 5 min. Two hundred micrograms of total extracted protein per assay was used. Following immunoprecipitation with polyclonal anti-CDK2 antibody (no. sc-163), the protein A-Sepharose beads were washed three times in kinase extraction buffer and twice in kinase assay buffer (50 mM HEPES [pH 7.5], 10 mM MgCl2, 1 mM dithiothreitol, 10 mM β-glycerophosphate, 1 mM NaF, 0.1 mM Na3VO4, 5 μg of leupeptin per ml, 2 μg of aprotinin per ml, and 0.1 M phenylmethylsulfonyl fluoride). The final pellet (15 μl of solid beads) was resuspended in 25 μl of kinase assay buffer with 2.5 μg of the histone H1 substrate (Boehringer), 50 μM ATP, and 5 μCi of [γ-32P]ATP and incubated at 30°C for 20 min. The reaction was stopped by adding 10 μl of 4× concentrated Laemmli sample buffer, and the products were separated on a sodium dodecyl sulfate–12% polyacrylamide gel.
Cell cycle analysis.
MEF cells were synchronized in G0 by serum starvation for 3 days (in DMEM without serum), followed by stimulation in DMEM supplemented with 10% FCS. T47DΔMTcycD1 cells were synchronized in early G1 by treatment with lovastatin (20 μM; Calbiochem) for 30 to 36 h and subsequent stimulation by removal of lovastatin and addition of mevalonate (2 mM; Sigma). For ectopic cyclin D1 expression, cells were additionally treated with 75 μM ZnSO4. Progression through the cell cycle was monitored by detection of the DNA content. Cells were washed twice with PBS, trypsinized, and fixed in 70% methanol for 2 h at −20°C. Subsequently, cells were precipitated (5 min of centrifugation at 500 × g at 4°C in a Sigma 6K15 centrifuge), washed with PBS, and resuspended in 1 ml of PBS containing 40 U of RNase A per ml and 40 μg of propidium iodide per ml. After incubation for 30 min at 37°C, DNA flow cytometric analysis was performed with an EPICS XL-MCL flow cytometer (Coulter). For quantification we used Multicycle AV software (Phoenix Flow Systems).
RESULTS
Transcription factor NF-κB activates the human cyclin D1 promoter.
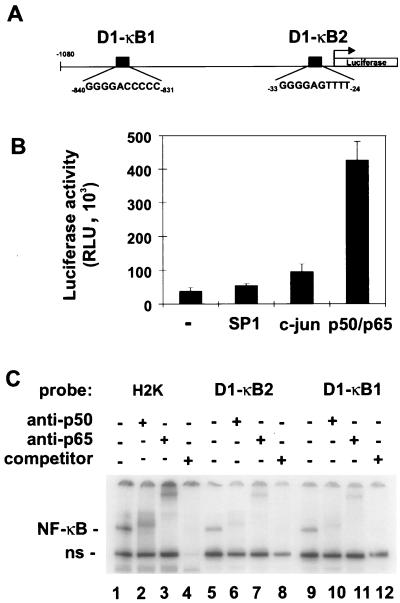
To investigate if NF-κB is directly connected with G1 checkpoint control, we searched for potential transcriptional targets involved in the regulation of the cell cycle. The human cyclin D1 promoter contains two putative NF-κB binding sites, termed D1-κB1 and D1-κB2 (Fig. 1A). Expression of p50 and p65 in COS-7 cells led to a 12-fold stimulation of a luciferase reporter gene under the control of the cyclin D1 promoter (Fig. 1B). Transcriptional activation of the cyclin D1 promoter by NF-κB was also observed in Huh-7 and C33A cells (data not shown). In contrast, transfection of either Sp1 or c-Jun, two other potential regulators of cyclin D1 expression (2, 22, 50), only weakly activated transcription in COS-7 cells.
FIG. 1.
Transcriptional regulation of the cyclin D1 promoter by NF-κB. (A) Schematic map of the cyclin D1 promoter. Sequences of NF-κB binding sites are shown and are designated D1-κB1 and D1-κB2. (B) NF-κB activates the cyclin D1 promoter. COS-7 cells were cotransfected with 200 ng of pD1luc and 100 ng of either SP1, c-Jun, p50/p65 expression constructs, or pUC18 to give a total of 400 ng. Luciferase activity was measured and standardized by cotransfection of β-galactosidase expression vectors. Results represent the means and standard deviations from three individual experiments. RLU, relative light units. (C) Specific binding of NF-κB to the cyclin D1 promoter. Protein extracts from COS-7 cells, transfected with p50/p65 expression constructs, were incubated either with a specific NF-κB oligonucleotide probe (H2K) or with oligonucleotide probes containing NF-κB binding sites of the cyclin D1 promoter (D1-κB1 and D1-κB2). The identity of NF-κB-containing complexes was determined by adding anti-p50 or anti-p65 antibodies to the reaction mixture, as indicated. In lanes 4, 8, and 12, a 50-fold excess of unlabeled H2K oligonucleotide was added to the reaction mixture. ns, nonspecific.
To test if the transcriptional activation of the cyclin D1 promoter by NF-κB was due to direct DNA binding, an EMSA was performed (Fig. 1C). Double-stranded oligonucleotides containing D1-κB1 and D1-κB2 sequences were generated and used to analyze DNA binding activity with protein extracts of COS-7 cells cotransfected with p50 and p65 expression constructs. As a control, a bona fide NF-κB binding site probe (H2K) was used. NF-κB binds to the D1-κB1, D1-κB2, and H2K sites with comparable efficiencies (Fig. 1C, lanes 1, 5, and 9), although D1-κB1 and D1-κB2 are not perfect NF-κB consensus sequences. The identity of the NF-κB–DNA complex was proven by reactivity towards antibodies against p50 and p65 (lanes 2, 3, 6, 7, 10, and 11). These results demonstrate that NF-κB can bind to the human cyclin D1 promoter and activate transcription.
The proximal NF-κB binding site D1-κB2 is necessary for NF-κB-dependent activation.
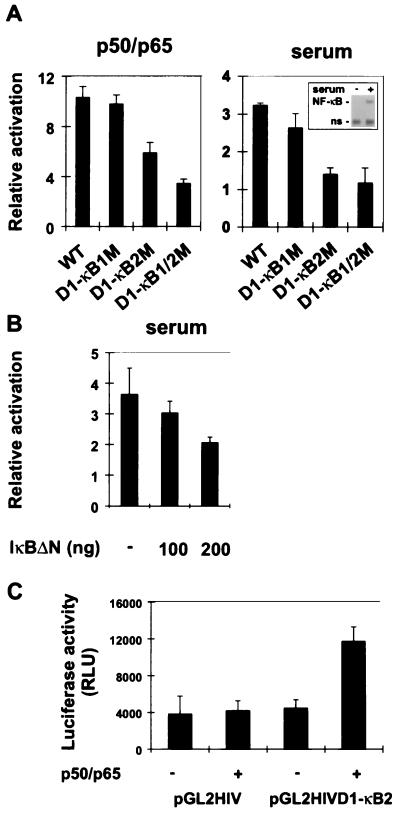
To investigate the transcriptional activation of the cyclin D1 promoter by NF-κB in more detail, we tested promoter constructs containing point mutations of the D1-κB1 and D1-κB2 sites. We cotransfected these constructs either with p50 and p65 expression constructs or with a control plasmid into COS-7 cells and measured luciferase expression (Fig. 2A, left panel). Mutation of the distal NF-κB binding site (D1-κB1M) did not interfere with NF-κB activation of the promoter. In contrast, mutation of the proximal NF-κB binding site (D1-κB2M), or of both, caused a significant reduction in the NF-κB responsiveness of the cyclin D1 promoter. The point mutations introduced into both sites prevented NF-κB binding to these sequences (data not shown). To test if NF-κB binding sites contribute to serum responsiveness of the cyclin D1 promoter (45, 51), we transfected wild-type and mutant promoters into NIH 3T3 cells and measured luciferase activity (Fig. 2A, right panel). Serum induction of the cyclin D1 promoter was completely dependent on a functional D1-κB2 site. By EMSA analysis (Fig. 2A, right panel, inset), we could demonstrate that growth factor addition activated NF-κB, confirming previous findings that NF-κB is induced upon G0-to-G1-phase transition (6, 15).
FIG. 2.
Functional characterization of NF-κB-responsive elements in the cyclin D1 promoter. (A) Reporter gene activation of wild-type (WT) and mutant constructs by p50/p65 was determined in COS-7 cells (left panel) as described for Fig. 1B. The graphs represent the means and standard deviations from three individual experiments. Serum induction of the cyclin D1 promoter was measured in NIH 3T3 cells as described previously (22) (right panel). Luciferase activity was measured as described for Fig. 1B. An EMSA was performed to show serum-induced NF-κB activation (inset). ns, nonspecific. (B) Induction of the cyclin D1 promoter by serum is reduced by IκBΔN expression. NIH 3T3 cells were transiently transfected with the cyclin D1 promoter-luciferase construct and increasing amounts of the IκBΔN expression vector, as indicated. Luciferase activity was measured after readdition of serum to the starved cells. (C) Activation of a heterologous promoter through the D1-κB2 sequence. A single copy of a cyclin D1 promoter fragment containing the D1-κB2 site was cloned in front of an HIV core promoter luciferase construct (pGL2HIV). The luciferase reporter gene assay was carried out as described for Fig. 1B. RLU, relative light units.
To confirm that growth factor induction of the cyclin D1 promoter depends on NF-κB activity, the cyclin D1 promoter construct was cotransfected with increasing amounts of IκBαΔN expression vector (Fig. 2B). In fact, repression of endogenous NF-κB activity caused a reduction in serum-induced promoter activity.
We next investigated the potential of the D1-κB2 sequence to mediate NF-κB-dependent transcriptional activation in a heterologous promoter context. The sequence was inserted into a luciferase expression construct which contains only the HIV core promoter (pGL2). pGL2 or pGL2D1κB2 was transfected together with p50 and p65 or with a control plasmid into COS-7 cells (Fig. 2C). A single copy of the D1-κB2 sequence could activate the core promoter threefold in the presence of NF-κB, demonstrating that this NF-κB element is functional in a different context. Taken together, these results suggest that NF-κB activates transcription of the cyclin D1 promoter primarily through the proximal binding site D1-κB2.
A stably expressed dominant negative IκB mutant abrogates serum-induced activation of NF-κB in MEF.
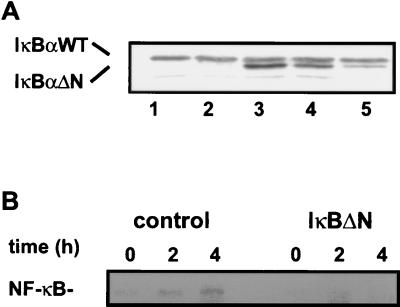
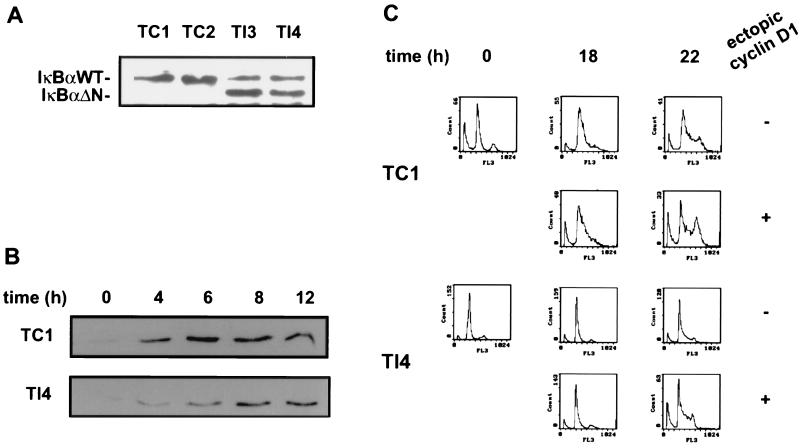
Primary MEF were used as a model system to investigate whether NF-κB regulates endogenous cyclin D1 expression and hence influences the RB pathway. To inhibit NF-κB activity, we stably expressed a superrepressor form of IκBα (IκBΔN) (7, 25). As a control, the empty expression plasmid was transfected. The expression level of IκBΔN was comparable to that of endogenous IκBα for clones I3 and I4 (Fig. 3A). In contrast, in clone I5 expression of IκBΔN was much lower.
FIG. 3.
IκBΔN expression reduces serum-induced NF-κB activity. (A) Expression of IκBΔN in primary MEF. MEF were transfected with an IκBΔN expression construct or an empty vector, and stable clones were selected with G418 (Gibco BRL). Control and IκBΔN-transfected MEF cells were lysed with extraction buffer and analyzed by Western blotting with anti-IκBα antibody. The positions of wild-type (WT) and mutant IκBα are indicated. Lanes 1 and 2, control clones (C1 and C2); lanes 3, 4, and 5, IκBΔN-expressing clones (I3, I4, and I5, respectively). (B) Serum induction of NF-κB DNA binding activity. MEF cells were synchronized in G0 by serum starvation for 3 days, followed by stimulation with DMEM plus 10% FCS. Serum-deprived and -stimulated C1 and I3 cells were extracted at the indicated time points and analyzed by EMSA. The NF-κB–DNA complex containing p50/p65 as determined by antibody supershifting and inhibition (data not shown) is indicated.
Serum-induced activation of NF-κB was analyzed by EMSA (Fig. 3B). Serum-deprived and -stimulated cells were extracted at the indicated time points and incubated with the H2K probe. NF-κB binding activity was induced in control cells but not in cells expressing the superrepressor (Fig. 3B). The NF-κB complex consisted of heterodimeric p50-p65 as determined by antibody supershifting and inhibition (data not shown).
NF-κB inactivation causes reduced and delayed cyclin D1 expression in G1 phase.
The effect of NF-κB inactivation on cyclin D1 expression during G1 phase was analyzed in synchronization experiments. Cells were starved of serum and then released from G0 by readdition of serum. Cells were extracted at the indicated time points, and protein extracts were analyzed by Western blotting (Fig. 4A). Cyclin D1 expression was delayed in cells expressing the NF-κB superrepressor compared to control cells (Fig. 4A, upper panel). The blots were stripped and analyzed for CDK4 expression. Consistent with the fact that CDK4 expression is not cell cycle dependent, CDK4 levels were not influenced by serum stimulation (Fig. 4A, upper panel). The difference in the kinetics of cyclin D1 induction was quantified by using multiple Western blots (Fig. 4A, bottom panel). To prove that the effect of NF-κB on cyclin D1 expression was not cell type specific, the same experiments were performed with synchronized HeLa cells either stably expressing the superrepressor or containing the empty vector (Fig. 4B). Again, inhibition of NF-κB caused delayed and reduced cyclin D1 expression.
FIG. 4.
NF-κB regulates serum-induced cyclin D1 expression. (A) Western blots of MEF protein extracts, prepared at the indicated times after serum induction of C1 and I3 cells (upper panel). Cyclin D1 was detected with the monoclonal antibody DCS-6. The blot was stripped and reprobed with polyclonal anti-CDK4 antibody. Three individual experiments were performed, and relative intensities of the cyclin D1 bands were quantified with Quantity One software (PDI Inc.) (lower panel). Error bars indicate standard deviations. Similar results were obtained with C2 and I4 cells (data not shown). (B) NF-κB-dependent cyclin D1 expression in HeLa cells. Protein extracts of control and IκBΔN-transfected cells were prepared at the indicated times after serum induction. Cyclin D1 expression was analyzed by Western blotting (upper panel). A nonspecific band which cross-reacts with the antibody served as loading control (data not shown). Serum-induced cyclin D1 expression was quantified as described for panel A, lower panel.
NF-κB inactivation leads to reduced cyclin D1-associated kinase activity and delayed pRB phosphorylation in mid- to late G1 phase.
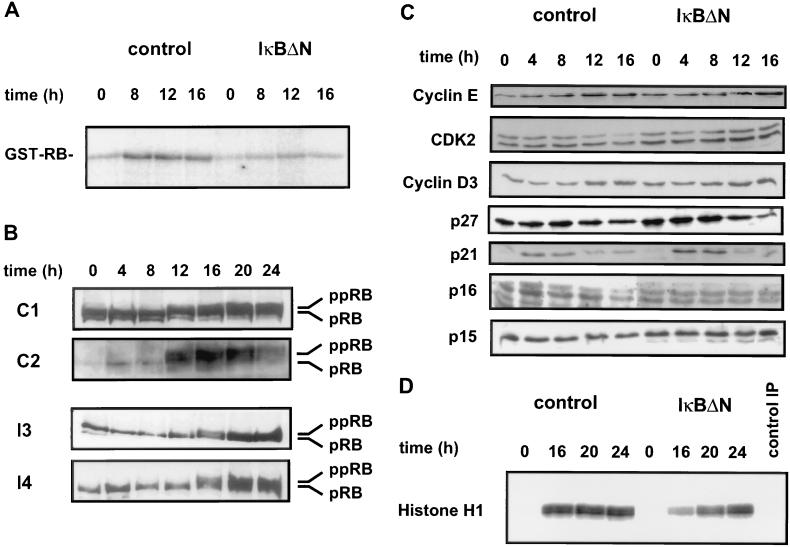
Defective cyclin D1 expression implies that NF-κB inactivation may have further consequences for the RB pathway, since cyclin D1 forms complexes with cyclin-dependent kinases (CDK4 and CDK6) which subsequently phosphorylate the retinoblastoma protein in mid- to late G1 phase. Hence, we assayed cyclin D1-associated pRB kinase activity at different times after release from serum starvation. In control cells, cyclin D1-associated kinase activity increased faster and reached a higher level than in cells expressing the NF-κB superrepressor (Fig. 5A). The specificity of the antibody was confirmed in a previous report (32). Surprisingly, cyclin D1-associated kinase activity was even more affected by IκBΔN than would be predicted from the more modest modulation of cyclin D1 expression (Fig. 4).
FIG. 5.
IκBΔN expression results in a delay of pRB phosphorylation (A) NF-κB inactivation affects cyclin D1-associated kinase activity. pRB phosphorylation by cyclin D1-associated kinase activity from synchronized C1 (control) and I4 (IκBΔN) cells was analyzed as described in Materials and Methods. Similar results were obtained with C2 and I3 cells (data not shown). (B) NF-κB inactivation causes a delay in phosphorylation of endogenous pRB. Western blots of protein extracts prepared after serum induction of either control (C1 and C2) or IκBΔN-expressing (I3 and I4) cells are shown. Hypophosphorylated (pRB) and hyperphosphorylated (ppRB) RB proteins were detected with the monoclonal antibody G3-245 (Pharmingen). (C) Expression of various cell cycle-regulatory proteins in either control (C1) or IκBΔN-expressing (I3) cells. Western blots of protein extracts prepared at the indicated times after serum induction are shown. Cyclin E, cyclin D3, CDK2, p27, p21, p16, and p15 were detected as described in Materials and Methods. Similar results were obtained with C2 and I4 cells (data not shown). (D) Histone H1 kinase activity of anti-CDK2 immunoprecipitates (IP) from synchronized C1 and I3 cells. Similar results were obtained with C2 and I4 cells (data not shown).
The subsequent analysis of the endogenous pRB phosphorylation status during G1 phase revealed that inactivation of NF-κB caused a delay in pRB phosphorylation (Fig. 5B). Whereas in control cells hyperphosphorylated RB (ppRB) was already observed at 12 h after serum stimulation, in IκBΔN-expressing cells pRB phosphorylation did not appear before 16 h.
To investigate if NF-κB would control expression of other components of the cell cycle machinery, we analyzed additional G1 cyclins, kinases, and inhibitors by Western blotting. NF-κB inactivation did not significantly interfere with the expression of cyclin E, cyclin D3, CDK2, p27, p21, p16, and p15 in synchronized cells (Fig. 5C). We also tested whether NF-κB inactivation would interfere with CDK2 kinase activity. Histone H1 phosphorylation by CDK2 was assayed at different times after release from serum starvation. In control cells CDK2 activity reached maximum levels after 16 h, while in IκBΔN-expressing cells full activity was observed only after 24 h (Fig. 5D). Thus, NF-κB inactivation interferes with both CDK4 and CDK2 kinase activities.
IκBΔN expression affects G1-to-S-phase transition.
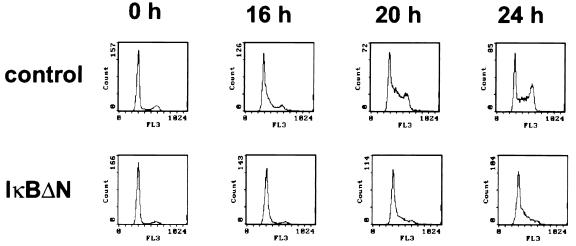
Since pRB phosphorylation is a prerequisite for G1-to-S-phase transition, NF-κB activity should influence progression of the cell cycle. The progression of synchronized MEF through the cell cycle was determined by fluorescence-activated cell sorter (FACS) analysis (Fig. 6) and quantified (Table 1). NF-κB inactivation indeed strongly delayed cell cycle progression. Control cells (C1) started to enter S phase at 16 h after serum stimulation, reached a maximum in S phase at 20 h, and passed into G2/M phase after 24 h. In contrast, in cells expressing IκBΔN (I1), significant S-phase entry was not observed before 20 h after serum readdition, and the maximum was still not reached after 24 h.
FIG. 6.
NF-κB inactivation causes retardation of G1-to-S-phase transition. FACS analysis of progression into S phase of C1 and I3 cells at 0, 16, 20, or 24 h after readdition of serum is shown. Progression through the cell cycle was monitored by detection of the DNA content by using propidium iodide staining. Similar results were obtained with C2 and I4 cells.
TABLE 1.
Cell cycle quantification for synchronized control (C1 and C2) and IκBΔN-expressing (I3, I4, and I5) MEF clones
| Time after serum stimulation (h) | Phase | % of cells
|
|||||
|---|---|---|---|---|---|---|---|
| Expt 1
|
Expt 2
|
Expt 3
|
|||||
| C1 | I3 | C2 | I4 | C1 | I5 | ||
| 0 | G1 | 75.0 | 85.9 | 92.0 | 88.0 | 73.3 | 83.8 |
| S | 11.4 | 5.1 | 5.5 | 4.7 | 19.8 | 13.8 | |
| G2/M | 13.5 | 9.0 | 2.5 | 7.3 | 6.9 | 2.4 | |
| 12 | G1 | 69.0 | 91.9 | 89.0 | 91.7 | 64.1 | 74.6 |
| S | 18.8 | 3.5 | 5.5 | 2.8 | 24.1 | 17.8 | |
| G2/M | 12.2 | 4.6 | 5.4 | 5.5 | 11.8 | 7.6 | |
| 16 | G1 | 40.9 | 73.7 | 74.3 | 82.4 | 39.6 | 50.9 |
| S | 45.3 | 17.9 | 16.1 | 10.4 | 56.9 | 43.2 | |
| G2/M | 13.7 | 8.5 | 9.7 | 7.2 | 3.5 | 5.9 | |
| 20 | G1 | 30.4 | 52.7 | 19.9 | 51.2 | 33.3 | 19.0 |
| S | 64.8 | 39.7 | 77.1 | 41.3 | 54.1 | 81.0 | |
| G2/M | 4.8 | 7.6 | 3.0 | 7.5 | 12.6 | 0.0 | |
| 24 | G1 | 27.8 | 49.3 | 16.3 | 42.0 | 40.9 | 27.2 |
| S | 50.3 | 48.3 | 60.0 | 51.6 | 33.3 | 34.3 | |
| G2/M | 21.9 | 2.5 | 23.7 | 6.4 | 25.7 | 38.5 | |
In another experiment, using different control and IκBΔN-expressing clones (Table 1, experiment 2), again at 20 h after serum stimulation more control cells (C2) than IκBΔN-expressing cells (I4) had entered S phase. While after 24 h control cells began to enter G2/M phase, IκBΔN-expressing cells still were undergoing G1-to-S-phase transition. Finally, in a third experiment (Table 1, experiment 3), control cells (C16) were compared with a third IκBΔN clone (I5), where only low IκBΔN expression was observed (Fig. 3A). In this case differences in cell cycle progression were weak. Only slightly more control cells than inhibitor-expressing cells had entered S phase at 16 h after serum stimulation. While most of the IκBΔN-expressing cells were in S phase at 20 h after serum stimulation, some of the control cells had reached G2/M phase or had even entered the next cell cycle. At 24 h after serum stimulation, synchronization in both populations was lost.
We also analyzed the effect of NF-κB inactivation on cell cycle progression in a T47D mammary carcinoma cell line modified to inducibly express cyclin D1 from a stably integrated zinc-responsive expression vector (37). These cells were stably transfected with IκBΔN or empty vector (Fig. 7A). As observed for MEF and HeLa cells, IκBΔN expression resulted in a pronounced delay of cyclin D1 induction during early to mid-G1 phase of the synchronized cells (Fig. 7B). Furthermore, as in MEF cells, inhibition of NF-κB activity in the T47D cells resulted in retarded G1-to-S-phase transition (Fig. 7C, compare first and third rows of panels). The IκBΔN-mediated delay of cell cycle progression could be overcome by zinc-induced ectopic cyclin D1 expression (Fig. 7C). However, ectopic cyclin D1 expression also led to somewhat accelerated cell cycle progression in control cells, perhaps due to the elevated cyclin D1 levels.
FIG. 7.
Retardation of G1-to-S-phase transition in T47DΔMTcycD1 cells, caused by NF-κB inactivation, was rescued by ectopic cyclin D1 expression. (A) Expression of IκBΔN in T47DΔMTcycD1 cells. Cells were transfected with an IκBΔN expression construct or an empty vector, and stable clones were selected with hygromycin (Sigma). Control and IκBΔN-transfected cells were lysed with extraction buffer and analyzed by Western blotting with anti-IκBα antibody. The positions of wild-type (WT) and mutant IκBα are indicated. Lanes 1 and 2, control clones (TC1 and TC2); lanes 3 and 4, IκBΔN-expressing clones (TI3 and TI4). (B) NF-κB-dependent cyclin D1 expression in T47DΔMTcycD1 cells. Western blots of protein extracts prepared from presynchronized TC1 and TI4 cells at the indicated time points after lovastatin removal and mevalonate addition are shown. Cyclin D1 was detected with the monoclonal antibody DCS-6. (C) FACS analysis of progression into S phase of TC1 and TI4 cells at 0, 18, or 22 h after lovastatin removal and mevalonate addition. Ectopic expression of cyclin D1 was induced by the addition of Zn2+ (+) or was not induced (−), as indicated. Progression through the cell cycle was monitored by detection of the DNA content. TC2 and TI3 cells (data not shown) gave similar results.
In summary, our data show that NF-κB inactivation led to delayed G1-to-S-phase transition in MEF and T47D cells. The observation that this can be rescued by ectopic cyclin D1 expression is consistent with the hypothesis that NF-κB regulates the RB pathway and that one of its targets could be cyclin D1, although other targets cannot be ruled out.
DISCUSSION
The present study provides evidence that the pleiotropic transcription factor NF-κB transmits growth signals directly to key regulators of the cell cycle. Our results suggest that NF-κB stimulates cyclin D1 transcription in G1 phase and thereby subsequently affects both pRB phosphorylation and G1-to-S-phase transition. The impaired cell cycle progression following NF-κB inhibition could be rescued by ectopic cyclin D1 expression, indicating that either cyclin D1 or another component of the RB pathway is controlled by NF-κB.
Interestingly, we observed that cyclin D1-associated kinase activity was even more affected upon NF-κB inactivation than would be expected from the modest modulation of cyclin D1 expression. However, narrow individual threshold levels are critical for key cell cycle regulators to exert their effects (46). Hence, subtle changes in their expression level can interfere with cell cycle progression. Alternatively, our results may indicate that NF-κB regulates cyclin D1-associated kinase activity through another pathway. So far we cannot identify any further cell cycle regulators whose expression levels were affected by NF-κB inactivation (Fig. 5C). However, we do not rule out that NF-κB could control expression or activity of additional components. The observation that ectopic expression of cyclin D1 can overcome the delay of G1-to-S-phase transition caused by NF-κB inactivation makes it unlikely that NF-κB controls an event late in G1 or S phase. In this respect, the observed delay in CDK2 activity (Fig. 5D) could be due to delayed pRB phosphorylation.
NF-κB activates transcription of the cyclin D1 promoter primarily through a proximal binding site. Weak residual NF-κB responsiveness after mutation of two identified major binding sites indicates the presence of further cryptic NF-κB elements. NF-κB binding sites as well as cellular NF-κB activation are required for serum induction of cyclin D1 transcription (Fig. 2). Even though the appropriate expression of cyclin D1 depended on NF-κB, activation of NF-κB, e.g., by tumor necrosis factor alpha, in serum-deprived cells was not sufficient to induce cyclin D1 (not shown). Thus, further growth factor-activated regulators must contribute in parallel to ensure efficient cyclin D1 induction in G1 phase. Recent data indicate that NF-κB can functionally interact with other transcription factors, such as c-Fos/c-Jun, SP1, or E2F-1 (26, 47). Since the cyclin D1 promoter has been shown to be regulated by these transcription factors (2, 22, 50), maximal activation might result from multiple functional interactions. The fact that the cyclin D1 promoter contains binding sites for all of these transcription factors indicates the possibility of multiple cooperative interactions. Such cooperativity could form the basis for the suggested function of cyclin D1 to integrate diverse mitogenic stimuli (31, 45, 51).
In agreement with previous findings, we observed growth factor activation of NF-κB DNA binding activity in early G1 phase (6, 15). The activation level varied between different cell types analyzed (Fig. 2A and 3B and data not shown). A relatively weak induction of NF-κB DNA binding activity in response to serum may indicate that growth factor signaling additionally leads to RelA phosphorylation, ultimately increasing the transactivation potential of NF-κB. Recently, it has been demonstrated that phosphorylation of the RelA subunit stimulates NF-κB transcriptional activity by promoting an interaction with CBP/p300 (55, 56). The observation that even cyclin-dependent kinases may regulate RelA through interaction with the coactivator CBP/p300 indicates a possible further link between NF-κB and cell cycle control (41).
The data presented here raise the question of how NF-κB is linked to mitogenic signal transduction. Activation of NF-κB involves the phosphorylation of IκBα at its regulatory N terminus, subsequent conjugation with ubiquitin, and degradation of the inhibitor mediated by the proteasome (3, 49). Recently an IκBα-specific kinase activity was identified as part of a 700-kDa complex (12), which can be activated by MEKK1 (29). Interestingly, MEKK1 can interact with Ras (42), a component of one major mitogenic signaling cascade, the Ras–Raf–mitogen-activated protein kinase (MAPK) pathway (20, 23, 33). Another interesting link is provided by the observation that the ribosomal S6 kinase pp90rsk, a downstream target of the Ras-Raf-MAPK pathway, phosphorylates IκBα (19, 43). Furthermore, it has been shown that the transforming mutant p21ras can activate cyclin D1 expression (1, 2). Consistently, Ras inactivation causes a decline in cyclin D1 protein levels, accumulation of hypophosphorylated pRB, and G1 arrest (1, 40). Finally, Raf kinase activates NF-κB (16, 24, 30), and NF-κB activity is required for Ras-mediated oncogenesis (17, 35). Taken together, these observations provide a connection between the mitogenic Ras-Raf-MAPK pathway, NF-κB activation, and cell cycle progression.
Recent data have indicated a role of the NF-κB and IκB gene products in cell proliferation, transformation, and tumor development (47, 53). Constitutive NF-κB activation is essential for survival and progression of Hodgkin’s lymphoma and breast cancer cells (7, 48). The direct link between NF-κB activity and the central pathway of G1 checkpoint control presented here provides a basis for understanding how NF-κB/Rel deregulation may result in tumorigenesis.
ACKNOWLEDGMENTS
We thank Alexandra Bohne, Uta Fischer, Heidi Riedel, and Heidrun Peter for excellent technical assistance.
This work was supported in part by grants from the Deutsche Forschungsgemeinschaft (SFB344) to C.S. and M.S. and by a grant from the Fond der Chemischen Industrie to M.S.
REFERENCES
- 1.Aktas H, Cai H, Cooper G M. Ras links growth factor signaling to the cell cycle machinery via regulation of cyclin D1 and the Cdk inhibitor p27KIP1. Mol Cell Biol. 1997;17:3850–3857. doi: 10.1128/mcb.17.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 3.Baeuerle P A, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 4.Baichwal V R, Baeuerle P A. Activate NF-κB or die? Curr Biol. 1997;7:R94–R96. doi: 10.1016/s0960-9822(06)00046-7. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin A S, Jr, Azizkhan J C, Jensen D E, Beg A A, Coodly L R. Induction of NF-κB DNA-binding activity during the G0-to-G1 transition in mouse fibroblasts. Mol Cell Biol. 1991;11:4943–4951. doi: 10.1128/mcb.11.10.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bargou R C, Emmerich F, Krappmann D, Bommert K, Mapara M Y, Arnold W, Royer H D, Grienstein E, Greiner A, Scheidereit C, Dörken B. Constitutive nuclear factor-κB-RelA activation is required for proliferation and survival of Hodgkin’s disease tumor cells. J Clin Invest. 1997;100:2961–2969. doi: 10.1172/JCI119849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartek J, Bartkova J, Lukas J. The retinoblastoma protein pathway and the restriction point. Curr Opin Cell Biol. 1996;8:805–814. doi: 10.1016/s0955-0674(96)80081-0. [DOI] [PubMed] [Google Scholar]
- 9.Bash J, Zong W X, Gelinas C. c-Rel arrests the proliferation of HeLa cells and affects critical regulators of the G1/S-phase transition. Mol Cell Biol. 1997;17:6526–6536. doi: 10.1128/mcb.17.11.6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beg A A, Sha W C, Bronson R T, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 11.Beijersbergen R L, Bernards R. Cell cycle regulation by the retinoblastoma family of growth inhibitory proteins. Biochim Biophys Acta. 1996;1287:103–120. doi: 10.1016/0304-419x(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z J, Parent L, Maniatis T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 13.Cressman D E, Greenbaum L E, Haber B A, Taub R. Rapid activation of post-hepatectomy factor/nuclear factor κB in hepatocytes, a primary response in the regenerating liver. J Biol Chem. 1994;269:30429–30435. [PubMed] [Google Scholar]
- 14.Cressman D E, Taub R. Physiologic turnover of nuclear factor κB by nuclear proteolysis. J Biol Chem. 1994;269:26594–26597. [PubMed] [Google Scholar]
- 15.Duckett C S, Perkins N D, Leung K, Agranoff A B, Nabel G J. Cytokine induction of nuclear factor κB in cycling and growth-arrested cells. Evidence for cell cycle-independent activation. J Biol Chem. 1995;270:18836–18840. doi: 10.1074/jbc.270.32.18836. [DOI] [PubMed] [Google Scholar]
- 16.Finco T S, Baldwin A S., Jr κB site-dependent induction of gene expression by diverse inducers of nuclear factor κB requires Raf-1. J Biol Chem. 1993;268:17676–17679. [PubMed] [Google Scholar]
- 17.Finco T S, Westwick J K, Norris J L, Beg A A, Der C J, Baldwin A S., Jr Oncogenic Ha-Ras-induced signaling activates NF-κB transcriptional activity, which is required for cellular transformation. J Biol Chem. 1997;272:24113–24116. doi: 10.1074/jbc.272.39.24113. [DOI] [PubMed] [Google Scholar]
- 18.FitzGerald M J, Webber E M, Donovan J R, Fausto N. Rapid DNA binding by nuclear factor κB in hepatocytes at the start of liver regeneration. Cell Growth Differ. 1995;6:417–427. [PubMed] [Google Scholar]
- 19.Ghoda L, Lin X, Greene W C. The 90-kDa ribosomal S6 kinase (pp90rsk) phosphorylates the N-terminal regulatory domain of IκBα and stimulates its degradation in vitro. J Biol Chem. 1997;272:21281–21288. doi: 10.1074/jbc.272.34.21281. [DOI] [PubMed] [Google Scholar]
- 20.Heldin C H. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 21.Helin K, Harlow E. The retinoblastoma protein as a transcriptional repressor. Trends Cell Biol. 1993;3:43–46. doi: 10.1016/0962-8924(93)90150-y. [DOI] [PubMed] [Google Scholar]
- 22.Herber B, Truss M, Beato M, Muller R. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene. 1994;9:2105–2107. [PubMed] [Google Scholar]
- 23.Hill C S, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 24.Kanno T, Siebenlist U. Activation of nuclear factor-κB via T cell receptor requires a Raf kinase and Ca2+ influx. Functional synergy between Raf and calcineurin. J Immunol. 1996;157:5277–5283. [PubMed] [Google Scholar]
- 25.Krappmann D, Wulczyn F G, Scheidereit C. Different mechanisms control signal-induced degradation and basal turnover of the NF-κB inhibitor IκB alpha in vivo. EMBO J. 1996;15:6716–6726. [PMC free article] [PubMed] [Google Scholar]
- 26.Kundu M, Guermah M, Roeder R G, Amini S, Khalili K. Interaction between cell cycle regulator, E2F-1, and NF-κB mediates repression of HIV-1 gene transcription. J Biol Chem. 1997;272:29468–29474. doi: 10.1074/jbc.272.47.29468. [DOI] [PubMed] [Google Scholar]
- 27.LaThangue N B. DRTF1/E2F: an expanding family of heterodimeric transcription factors implicated in cell-cycle control. Trends Biochem Sci. 1994;19:108–114. doi: 10.1016/0968-0004(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 28.Lavoie J N, L’Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 29.Lee F S, Hagler J, Chen Z J, Maniatis T. Activation of the IκBα kinase complex by MEKK1, a kinase of the JNK pathway. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Sedivy J M. Raf-1 protein kinase activates the NF-κB transcription factor by dissociating the cytoplasmic NF-κB-IκB complex. Proc Natl Acad Sci USA. 1993;90:9247–9251. doi: 10.1073/pnas.90.20.9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukas J, Bartkova J, Bartek J. Convergence of mitogenic signalling cascades from diverse classes of receptors at the cyclin D–cyclin-dependent kinase–pRb-controlled G1 checkpoint. Mol Cell Biol. 1996;16:6917–6925. doi: 10.1128/mcb.16.12.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall C J. Ras effectors. Curr Opin Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 34.May M J, Ghosh S. Rel/NF-κB and IκB proteins: an overview. Semin Cancer Biol. 1997;8:63–73. doi: 10.1006/scbi.1997.0057. [DOI] [PubMed] [Google Scholar]
- 35.Mayo M W, Wang C Y, Cogswell P C, Rogers Graham K S, Lowe S W, Der C J, Baldwin A S., Jr Requirement of NF-κB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1997;278:1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 36.Muller H, Lukas J, Schneider A, Warthoe P, Bartek J, Eilers M, Strauss M. Cyclin D1 expression is regulated by the retinoblastoma protein. Proc Natl Acad Sci USA. 1994;91:2945–2949. doi: 10.1073/pnas.91.8.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musgrove E A, Lee C S L, Buckley M F, Sutherland R L. Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc Natl Acad Sci USA. 1994;91:8022–8026. doi: 10.1073/pnas.91.17.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naumann M, Wulczyn F G, Scheidereit C. The NF-κB precursor p105 and the proto-oncogene product Bcl-3 are IκB molecules and control nuclear translocation of NF-κB. EMBO J. 1993;12:213–222. doi: 10.1002/j.1460-2075.1993.tb05647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 40.Peeper D S, Upton T M, Ladha M H, Neuman E, Zalvide J, Bernards R, DeCaprio J A, Ewen M E. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature. 1997;386:177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- 41.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 42.Russell M, Lange Carter C A, Johnson G L. Direct interaction between Ras and the kinase domain of mitogen-activated protein kinase kinase kinase (MEKK1) J Biol Chem. 1995;270:11757–11760. doi: 10.1074/jbc.270.20.11757. [DOI] [PubMed] [Google Scholar]
- 43.Schouten G J, Vertegaal A C, Whiteside S T, Israel A, Toebes M, Dorsman J C, van der Eb A J, Zantema A. IκBα is a target for the mitogen-activated 90 kDa ribosomal S6 kinase. EMBO J. 1997;16:3133–3144. doi: 10.1093/emboj/16.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherr C J. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 45.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 46.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 47.Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-κB. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 48.Sovak M A, Bellas R E, Kim D W, Zanieski G J, Rogers A E, Traish A M, Sonenshein G E. Aberrant nuclear factor-κB/Rel expression and the pathogenesis of breast cancer. J Clin Invest. 1997;100:2952–2960. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe G, Albanese C, Lee J, Reutens A, Vario G, Henglein B, Pestell R G. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol. 1998;18:3212–3222. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 52.Wu M, Lee H, Bellas R E, Schauer S L, Arsura M, Katz D, FitzGerald M J, Rothstein T L, Sherr D H, Sonenshein G E. Inhibition of NF-κB/Rel induces apoptosis of murine B cells. EMBO J. 1996;15:4682–4690. [PMC free article] [PubMed] [Google Scholar]
- 53.Wulczyn F G, Krappmann D, Scheidereit C. The NF-κB/Rel and IκB gene families: mediators of immune response and inflammation. J Mol Med. 1996;74:749–769. doi: 10.1007/s001090050078. [DOI] [PubMed] [Google Scholar]
- 54.Yamada Y, Kirillova I, Peschon J J, Fausto N. Initiation of liver growth by tumor necrosis factor: deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc Natl Acad Sci USA. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong H, SuYang H, Erdjument Bromage H, Tempst P, Ghosh S. The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 56.Zhong H, Voll R E, Ghosh S. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–672. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]