Abstract
As the SARS-CoV-2 pandemic continues to spread, the necessity for rapid, easy diagnostic capabilities could never have been more crucial. With this aim in mind, we have developed a cost-effective and time-saving testing methodology/strategy that implements a sensitive reverse transcriptase loop-mediated amplification (RT-LAMP) assay within narrow, commercially available and cheap, glass capillaries for detection of the SARS-CoV-2 viral RNA. The methodology is compatible with widely used laboratory-based molecular testing protocols and currently available infrastructure. It employs a simple rapid extraction protocol that lyses the virus, releasing sufficient genetic material for amplification. This extracted viral RNA is then amplified using a SARS-CoV-2 RT-LAMP kit, at a constant temperature and the resulting amplified product produces a colour change which can be visually interpreted. This testing protocol, in conjunction with the RT-LAMP assay, has a sensitivity of ∼100 viral copies per reaction of a sample and provides results in a little over 30 min. As the assay is carried out in a water bath, commonly available within most testing laboratories, it eliminates the need for specialised instruments and associated skills. In addition, our testing pathway requires a significantly reduced quantity of reagents per test while providing comparable sensitivity and specificity to the RT-LAMP kit used in this study. While the conventional technique requires 25 μl of reagent, our test only utilises less than half the quantity (10 μl). Thus, with its minimalistic approach, this capillary-based assay could be a promising alternative to the conventional testing, owing to the fact that it can be performed in resource-limited settings, using readily available apparatus, and has the potential of increasing the overall testing capacity, while also reducing the burden on supply chains for mass testing.
Keywords: COVID-19, Capillary polymerase chain reaction (PCR), Loop-mediated isothermal amplification (LAMP), Molecular diagnostic
Graphical abstract
1. Introduction
The worldwide demand for reagents for real-time reverse transcriptase PCR (RT-qPCR) based diagnostic testing for COVID-19 has caused a bottleneck across the UK in its efforts to follow the World Health Organisation's advice to “Test, Test, Test” [1]. Polymerase chain reaction (PCR) testing has been critical in curbing the spread of the COVID-19 pandemic and the worldwide shortage of reagents has been understood to be a rate-limiting step [2]. Testing plays a crucial and powerful role in the public health arsenal [3] as these screening tests will detect the underlying illnesses in apparently healthy or asymptomatic persons [4]. RT-LAMP is another nucleic acid amplification-based molecular diagnostic technique that instead utilises an isothermal amplification route. It has existed for over 20 years and has been employed in diverse and even inaccessible settings [5,6]. RT-LAMP uses loop-forming primers and strand-displacement polymerases to achieve the isothermal amplification of a target RNA template. Hence, unlike an RT-PCR reaction, it does not require a thermal cycler, providing a crucial advantage by keeping the screening/testing strategy relatively simple, scalable, and not requiring expensive and sophisticated equipment that usually burdens diagnostic testing pipelines. Successful amplification in RT-LAMP reactions can be directly visualized via a variety of visual indicators, including phenol red dye, which changes colour upon successful amplification [5,7]. RT-LAMP assays are: 1. Specific because they employ 4 to 6 primers for every amplification reaction, which in theory reduces off-target effects in comparison to PCR, 2. Robust and thereby they will produce on the order of 109 copies of the target in an hour-long reaction [2], and 3. Performed almost anywhere because they simply require pipettes and a heating source (e.g., water baths, heat blocks) as the necessary equipment. Additionally, with regards to SARS-CoV-2 testing, recent studies have shown that saliva has a high diagnostic value for rapid screening tests [8] and when combined with the simplicity of an RT-LAMP and the non-invasive nature of the saliva collection approach this would satisfy the criteria for an efficient screening test [3].
Though RT-PCR continues to be the gold standard for the diagnosis of SARS-CoV-2, RT-LAMP assays have been shown to produce diagnostic results with high sensitivity and specificity [9,10]. Furthermore, the RT-LAMP assay's ability to tolerate PCR inhibitors eliminates the necessity for laborious RNA extraction and purification methodologies [11,12]. Several modified methods capable of performing LAMP assays in remote settings have also been documented [13]. However, for interpreting the associated colour-change, most of these colorimetric LAMP assays employ fully integrated real-time fluorescence or smartphone camera detection systems, which are either expensive or require the use of a specific phone model [14].
In normal times (pre-and post-pandemic), hospital laboratories concentrate on the diagnosis of patients. However, as the COVID-19 pandemic continued to spread, there is particular concern over the unexpected supply shortages of reagents and consumables (PCR tubes, pipette tips, microtiter plates) needed for the diagnosis of the SARS-CoV-2 virus [15]. These consumables and their related costs are not significant issues for well-funded laboratories, but they can be expensive and sometimes may not be available or affordable in resource-limited settings (15). A viable alternative is to therefore explore the use of low-cost consumables such as sterile transparent containers currently used for storage of samples and reagents in laboratories, as the cuvettes required for performing colorimetric assays [16]. Now, more than ever, there is a far greater need for the scientific community to innovate and find solutions, as even large international commercial companies struggle to cope with the demand for supplies (16).
To fulfil the need for a simplistic stand-alone (i.e. reader-free) methodology that can also cost-effectively execute a colorimetric isothermal nucleic acid amplification assay, we present here an (innovative molecular diagnostic testing procedure (that implements a LAMP assay) for the detection of SARS-CoV-2. This involves the use of a capillary to perform a LAMP assay, using only half the quantity of reagents usually required for the detection of the viral genome from a saliva sample. Furthermore, as would be required for any mass-testing in a pandemic, we have developed the technique to align itself with the standard operating processes and instrumentation of similar laboratory-based molecular testing methods, in an attempt to minimise disruption if adopted in a large-scale testing pathway. We validated our capillary-based RT-LAMP testing strategy using saliva samples spiked with different concentrations of deactivated virus and further compared it to the SARS-CoV-2 results performed in a PCR tube. We found our optimized RT-LAMP capillary-based procedure produces consistent results when compared to conventional RT-LAMP performed in a PCR-tube.
2. Materials and methods
This study was approved by the Ethics and Research board at the University of Southampton (62930).
2.1. Study design
This study intended to develop an experimental method for detecting SARS-CoV-2 RNA using a thin glass capillary capable of holding a maximum of 20 μl of liquid. We used SARS-CoV-2 plasmid control (Integrated DNA Technologies, Inc., USA) and inactivated SARS-CoV-2 to standardise the technique. Our study was designed to investigate the sensitivity and specificity of the colorimetric RT-LAMP assay and to evaluate its suitability as an alternative to RT-PCR testing for detecting SARS-CoV-2 viral RNA, isolated from saliva samples. This study was conducted at the University of Southampton, United Kingdom, in January of 2021.
2.2. Sample preparation and inactivation of virus
A saliva sample was collected in a tube from a COVID-19 negative individual, which was confirmed from an independent laboratory and then spiked with inactivated SARS-CoV-2 (BetaCoV/Australia/VIC01/2020). The following describes the preparation of viral culture and the inactivation process. SARS-CoV-2 and the Vero E6 kidney cell line from an African green monkey were obtained from Public Health England (PHE), UK. Cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen; cat no 11995065) supplemented with 10% (v/v) foetal bovine serum (FBS), glutamine and 50 u/ml penicillin-streptomycin at 37 °C in 5% (v/v) CO2. For viral cultures, cells were infected at an multiplicity of infection (MOI) of 0.005 and then maintained for 3 days in DMEM supplemented with 4% (v/v) FBS, 50 u/ml penicillin-streptomycin and 25 mM HEPES. The supernatant of the infected cell culture was clarified by centrifugation at 2000 g for 10 min and frozen as aliquots at −80 °C. Viral titres were determined by plaque assay on Vero E6 cell cultures. Inactivation of viral supernatants was achieved by UV-irradiation @ 260 nm for 30 min. Full inactivation of the virus was validated by plaque assay. All handling of live SARS-CoV-2 was performed in containment level 3 laboratories.
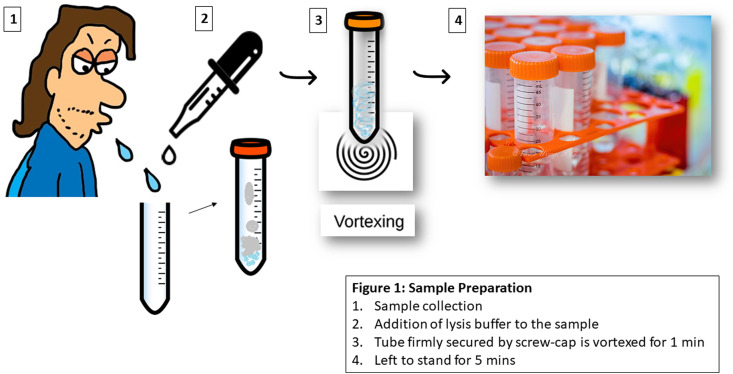
The saliva samples were spiked with inactivated virus that was diluted serially (5 × 105 - 5 × 10°) to identify the limit of detection (LoD) for the capillary. A 0.25% (v/v) solution of lysis buffer, Triton X 100 was prepared in PBS and then added to the spiked sample for disruption/lysis of the virion, and this was followed by vortexing for a minute and left to stand at room temperature for 5 min (Fig. 1 ). This latter step minimises the associated chance of contamination due to any aerosol formation. The lysis step is crucial to the release of viral RNA, which is used subsequently for amplification. The volume of this lysis buffer was kept at twice the volume of the sample. We also evaluated the efficiency of the lysis step and identified 0.25% (v/v) Triton X 100 in PBS as our lysis buffer of choice with a lysis time of 5 min for optimal test results.
Fig. 1.
Schematic for a simple rapid extraction protocol for virus lysis.
2.3. Colorimetric RT-LAMP
2.3.1. RT-LAMP assay in a PCR tube
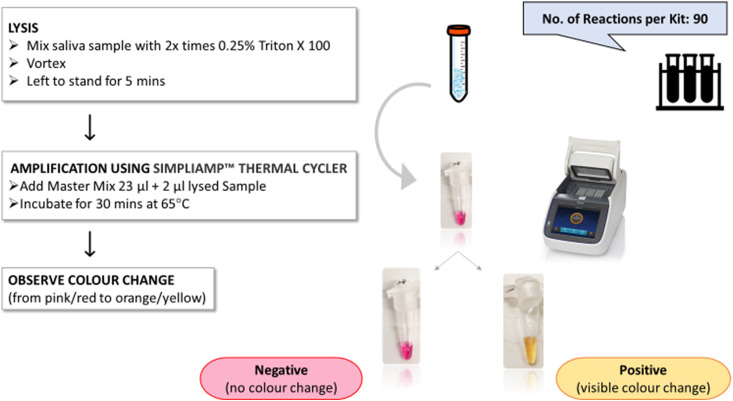
A RT-LAMP assay was performed in a biosafety level-2 certified laboratory set-up. The LAMP mix was first prepared on ice in a PCR-tube, without the addition of DNA templates (Table 1 ). We set up an RT-LAMP assay using the SARS-CoV-2 rapid colorimetric LAMP assay Kit commercialised by New England Biolabs (NEB), for a 25 μl reaction mixture (12.5 μL 2X Master Mix; 2.5 μl primer mix; 2.5 μl Guanidine hydrochloride and 5.5 μL DNase & RNase-free molecular grade water) mixed homogeneously and centrifuged. 2 μl of the lysed sample (RNA template) was then added to 23 μl of the LAMP mix, bringing the total volume to 25 μl. The samples were run on a SimpliAmp™ Thermal Cycler PCR machine at 65 °C for 30 min, following the manufacturer's specification. Each assay was performed with positive and negative controls for consistency. Positive reactions were identified through a visible change in colour from dark pink to orange/yellow. A negative test result was indicated by no colour change, due to the absence of SARS-CoV-2 viral RNA and hence lack of amplified product (Fig. 2 ). After amplification and interpretation of the results, care was taken in discarding the PCR-tubes in sealed bags without re-opening their lids, to prevent post-amplification contamination of the workspace.
Table 1.
Composition of the LAMP reaction mix.
| Components | WarmStart Colorimetric LAMP 2X Master Mix with UDG | SARS-Cov-2 Positive control | SARS-CoV-2 LAMP Primer Mix (N/E) | Nuclease-free Water | Guanidine Hydrochloride | Sample Nucleic Acid | Total Reaction volume |
|---|---|---|---|---|---|---|---|
| No Template control | 12.5 μl | – | 2.5 μl | 7.5 μl | 2.5 μl | – | 25 μl |
| Positive control | 12.5 μl | 2.0 μl | 2.5 μl | 5.5 μl | 2.5 μl | – | 25 μl |
| SARS-Cov-2 Test sample | 12.5 μl | – | 2.5 μl | 5.5 μl | 2.5 μl | 2 μl | 25 μl |
Fig. 2.
Schematic for a 30-min protocol illustrating sample preparation (including lysis using Triton X 100 followed by vortexing, incubation for 5 min) and the introduction of the RT-LAMP reaction mix and thereafter the sample to produce the colorimetric result.
2.3.2. RT-LAMP in a glass capillary
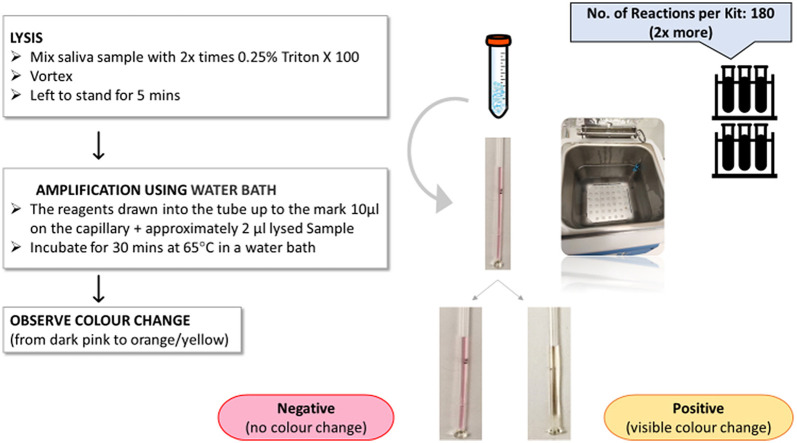
The capillary-based LAMP was performed using SARS-CoV-2 Rapid Colorimetric LAMP commercialised assay Kit (New England Biolabs UK, E2019S). The master mix for multiple reactions was prepared in a PCR tube and consisted of the following reagents: WarmStart® Colorimetric LAMP 2X Master Mix with UDG (2X), SARS-CoV-2 LAMP Primer Mix (N/E) (10X), Nuclease-free Water, Guanidine Hydrochloride (0.4 M) (10X). The master mix was introduced into a sterile thin Hirschmann ring caps disposable glass capillary with a capacity to hold 20 μL. Due to capillary action, the master mix was quickly drawn into the tube and was allowed to travel up to the mark on the capillary corresponding to a volume of 10 μl. This capillary was then introduced into the tube containing the lysed saliva sample allowing it to be drawn up into the capillary. Approximately 2 μl of the sample was drawn up by the capillary. The mixing of the master mix and saliva would happen as follows - due to the capillary action the sample is first transported through the empty capillary section towards the master mix at the top end of the capillary. Once the two liquids come in contact, a simple liquid-liquid in-diffusion process induces the mixing between the two liquids. This mixing progresses with time (a 5-min time interval was maintained between loading the capillary with sample and placing it in the water bath) and is further assisted by the elevated temperature in the water bath, ensuring proper mixing of the sample with the master mix. This is evidenced by the consistent colour change observed across the entire section of the capillary which holds the mix.
The inlet was immediately sealed using readily available superglue (The Box Every day, U.K). Multiple capillaries were prepared using the above method and five of them were stacked (held together with an elastic band) and placed in a water bath for 30 min at 65 °C. A visible colour change was observed between 15 to 45 min after incubation in the water bath, depending on the viral concentration present in the sample. The contents of the capillaries that turned yellow indicated positive detection of SARS-CoV-2 confirming successful amplification of the viral RNA present in them. Dark pink indicated a negative test result due to the lack of any amplified viral RNA product and hence the absence of SARS-CoV-2 (Fig. 3 ). After interpretation, capillaries were discarded into a carton box designated for glass disposal, without re-opening to prevent post-amplification contamination of workspaces.
Fig. 3.
Schematic for a 30-min protocol illustrating sample preparation (including lysis using Triton X 100 followed by vortexing, incubation for 5 min) and finally the introduction of the RT-LAMP reaction mix and thereafter the sample to produce the colorimetric result.
2.4. Limit of detection
Study of the LoD of our capillary platform was performed by spiking serially diluted SARS-CoV-2 plasmid control (from Integrated DNA Technologies, Inc., USA) into COVID-negative saliva. 2 μl of the saliva sample was used for the testing at final concentrations of 20,000, 2,000, 200, 100, and 50 copies/μl. Also, the LoD study was tested using inactivated SARS-CoV-2 virus (BetaCoV/Australia/VIC01/2020) which was spiked again in a saliva sample of a COVID-negative individual; 2 μl of the sample were tested at final concentrations of 5 × 105, 5 × 104, 5 × 103, 5 × 102, 5 × 101 and 5 × 10° pfu/ml. Each assay was repeated 5 times to ascertain the LoD for the above variable plasmid and inactivated virus concentrations.
3. Results and discussions
One of the objectives of this study was to minimise the sample processing associated with the RT-LAMP reaction (Fig. 1). To achieve this, we first optimized and validated the process of viral lysis of the inactivated SARS-CoV-2 spiked within the saliva samples using multiple buffers with varied concentrations. Also, the buffer's compatibility was checked with the RT-LAMP assay kit specifications. For the colorimetric RT-LAMP, phenol red was used as the visual indicator for the nucleic acid amplification process. When an RT-LAMP assay amplifies a target, it causes a drop in the pH within the tube from a pH value of 6.0–6.58, triggering a colour change from dark pink to orange/yellow. Table 3 shows the results for four different buffers IGEPAL, Tween 80, Tween 20, and Triton X 100 at four different concentrations, 0.25%, 0.50%, 0.75%, 1%. Spiked saliva samples (5 × 105 pfu/ml) as well as the positive and negative control samples treated with IGEPAL, Tween 80, and Tween 20 changed colour immediately on their addition to the master mix from dark pink to orange/yellow. This undesired colour change prior to amplification indicates a false positive, and these results were therefore labelled as invalid. As shown in Table 3, correct assay operation i.e. a colour change after amplification was observed only for Triton X 100 and only at concentrations of 0.25% and 0.5%. For consistency and comparative analysis, all the RT-LAMP assays were performed in parallel using the capillary method.
Table 3.
Shows comparative lysis efficiencies of four different buffers at four different concentrations in the extraction of viral RNA from the saliva sample spiked with inactivated virus (5 × 105 pfu/ml), the positive control, and the negative control.
| Lysis Buffer | Sample type | Concentrations |
|||
|---|---|---|---|---|---|
| 0.25% | 0.50% | 0.75% | 1% | ||
| IGEPAL | Spiked saliva sample | Invalid | Invalid | Invalid | Invalid |
| Positive control | Invalid | Invalid | Invalid | Invalid | |
| Negative control | Not applicable | Not applicable | Not applicable | Not applicable | |
| Tween 80 | Spiked saliva sample | Invalid | Invalid | Invalid | Invalid |
| Positive control | Invalid | Invalid | Invalid | Invalid | |
| Negative control | Not applicable | Not applicable | Not applicable | Not applicable | |
| Tween 20 | Spiked saliva sample | Invalid | Invalid | Invalid | Invalid |
| Positive control | Invalid | Invalid | Invalid | Invalid | |
| Negative control | Not applicable | Not applicable | Not applicable | Not applicable | |
| Triton X 100 | Spiked saliva sample | valid | valid | Invalid | Invalid |
| Positive control | valid | valid | Invalid | Invalid | |
| Negative control | Not applicable | Not applicable | Not applicable | Not applicable | |
Invalid: sample changes colour from dark pink to orange/yellow before amplification.
Valid: sample changes colour from dark pink to orange/yellow after amplification.
Not applicable: sample does not change colour after amplification.
3.1. SARS-CoV-2 RT-LAMP using PCR tubes
The workflow used to conduct the RT-LAMP is depicted in Fig. 2. The primer sequences used to target the nucleocapsid (N) gene and envelope (E) gene of SARS-CoV-2 is listed in Table 2. The compatibility of the buffer was tested with the LAMP assay kit by spiking serially diluted SARS-CoV-2 plasmid control and inactivated virus. Use of a suitable buffer was important to ensure the amplification process occurred without any unintended inhibition or interference. The test was able to amplify the samples from 40,000 copies per reaction to 100 copies per reaction of plasmid and 5 × 105 to 5 × 101 (pfu/ml) of inactivated virus per reaction. To obtain a more precise LoD, the experiments were run in replicates and we found consistent amplification of the sample with 100 copies per reaction and 5 × 101 (pfu/ml) per reaction as shown in Fig. 4 A/B.
Table 2.
SARS-CoV-2 LAMP Primer Sequences 5′ to 3’ end (New England Biolabs NEB #E2019S manual).
| Gene E Primer Set Sequence |
Primer Sequences (5’ 3′ end) 3′ end)
|
Source |
|---|---|---|
| E1-F3 | TGAGTACGAACTTATGTACTCAT | NEB #E2019S manual 2020 |
| E1-B3 | TTCAGATTTTTAACACGAGAGT | |
| E1-FIP | ACCACGAAAGCAAGAAAAAGAAGTTCGTTTCGGAAGAGACAG | |
| E1-BIP | TTGCTAGTTACACTAGCCATCCTTAGGTTTTACAAGACTCACGT | |
| E1-LF | CGCTATTAACTATTAACG | |
| E1-LB | GCGCTTCGATTGTGTGCGT | |
| Gene N2 Primer Set Sequence | ||
| N2–F3 | ACCAGGAACTAATCAGACAAG | |
| N2–B3 | GAC CTTCGGGAACGTGGTTGACC | |
| N2-LB | TTGATCTTTGAAATTTGGATCT | |
| N2-FIP | TTCCGAAGAACGCTGAAGCGGAACTGATTACAAACATTGGCC | |
| N2-BIP | CGCATTGGCATGGAAGTCACAATTTGATGGCACCTGTGTA | |
| N2-LF | GGGGGCAAATTGTGCAATTTG |
Fig. 4.
A/B: Detection of different concentrations of inactivated SARS-CoV-2 and SARS-CoV-2 plasmid spiked in saliva samples using PCR tubes. Colour change from dark pink to orange/yellow indicates a positive result. The negative sample remains dark pink. The results show that the LoD for the RT-LAMP in a PCR-tube (as seen in 4A) for the inactivated virus is 5 × 101 (pfu/ml) and (as seen in 4B) for the SARS-CoV-2 plasmid is 100 copies per reaction. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. SARS-CoV-2 RT-LAMP using capillary method
For quicker and easier detection of SARS-CoV-2 in patient samples, it would be advantageous to avoid the highly specialised and time-consuming RNA extraction and purification steps that are usually required in PCR techniques. High purity of RNA is crucial to all common detection systems like RT-PCR. However, we found that this step was not essential to perform accurate and sensitive RT-LAMP for detecting SARS-CoV-2. A simple sample preparation step with a buffer worked sufficiently well and thereby avoided the laborious RNA extraction method. To optimise detection results and get the fastest and most accurate outcome, we tested the capillary technique with both the inactivated virus and SARS-CoV-2 plasmid diluted at varied concentrations as mentioned in the methods section. The capillary with the master mix was introduced into the sample preparation tube and ∼2 μl of the sample was drawn into the capillary due to capillary action. The capillaries were sealed at both ends with superglue and then stacked and finally placed into the water bath for 30 min at 65 °C for amplification (Fig. 5 ). Positive results were indicated via the visible colour change in samples from dark pink to orange/yellow. Negative samples remained dark pink.
Fig. 5.
Capillaries placed together in a water bath for amplification at 65 °C.
The compatibility of the lysis buffer was also checked by the capillary method and the results were found to be comparable with the RT-LAMP PCR tube method. The test was able to amplify the samples spiked with either the plasmid or the inactivated virus in the buffer (Fig. 6 ). The LoD for the inactivated SARS-CoV-2 virus was found to be 5 × 101 (pfu/ml) per reaction and this is shown in Fig. 6A. As shown in Fig. 6B, the capillary method was able to amplify the plasmid from the lowest concentration of 100 copies per reaction. Thus, the capillary method showed a LoD that is comparable to that of the RT-LAMP PCR tube method.
Fig. 6.
(A & B): Detection of different concentrations of SARS-CoV-2 plasmid and inactivated SARS-CoV-2 virus spiked in saliva samples. Colour change from dark pink to orange/yellow indicates a positive result. The negative sample remains dark pink. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Cost-effectiveness and benefit analysis
There has been increased focus on the strategy of test and trace by the governments of all nations worldwide as the COVID-19 pandemic continues to pose a grave economic crisis due to the burden of lockdowns and restrictions. The UK alone has estimated the cost of test and trace to rise as high as £22 billion [17], and hence the cost is an important driver at this point. We have undertaken a cost-based analysis of our technique against an RT-LAMP assay (New England Biolabs UK kit E2019S) and found a per-test consumable cost of a conventional RT-LAMP in a PCR tube to be £10.40 excluding running costs and manpower, while a capillary-based RT-LAMP costs only 50% of this. This significant reduction in cost was primarily achieved by a lower volume of reagents used per test while maintaining sensitivity and specificity at the same levels as conventional methods. The capillary-based method showed an outcome of comparable accuracy with reduced quantities of master mix and employing a simple heating device such as a water bath versus the PCR method which uses an expensive thermal cycler. In the estimated cost-analysis above, pricing was quoted per kit and further prospects of reduction seems likely if kits are ordered in bulk.
3.4. Cross-reactivity validation
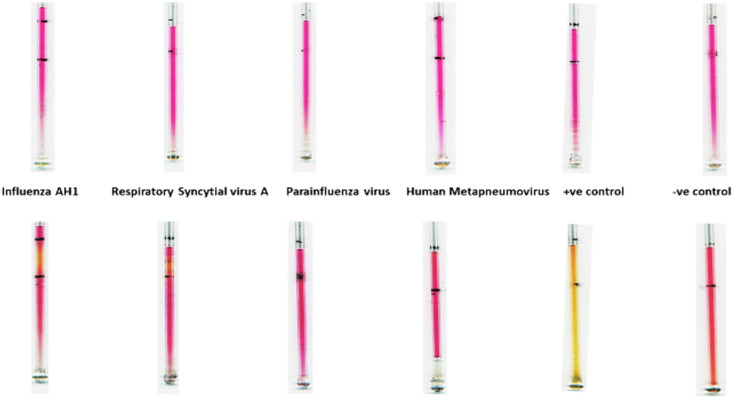
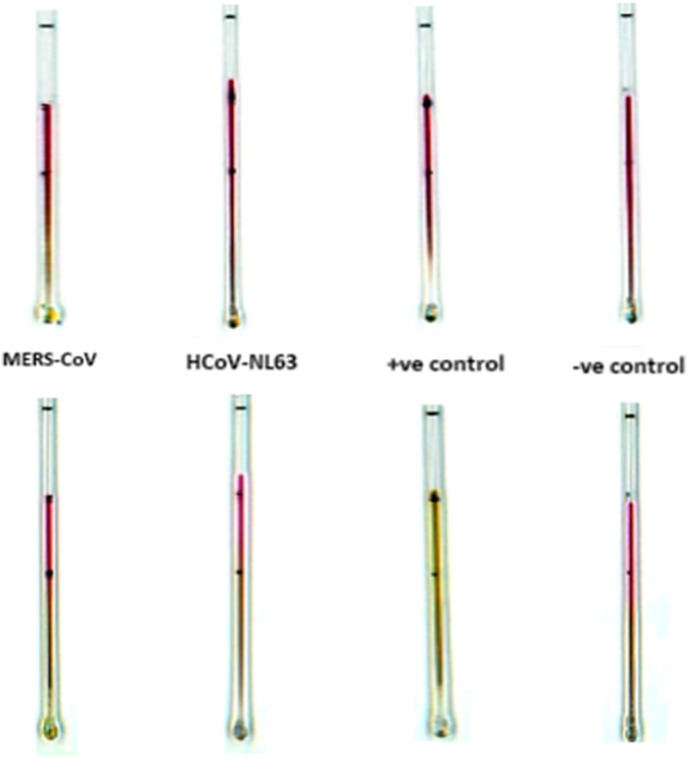
To investigate the cross-reactivity or specificity of the assay, we have tested (multiple times) our capillary-based method with six other relevant respiratory RNA/coronaviruses, Influenza AH1, Respiratory syncytial virus A, Parainfluenza virus, Human metapneumovirus, MERS, and HCoV-NL63 procured from ZeptoMetrix, which is widely used as a ‘standard’ for regulatory approval of commercial diagnostic tests. The samples from the kit were lysed in 0.25% Triton X 100 in PBS and tested with the capillary-based method. Results in Fig. 7, Fig. 8 show that there is no change in colour for the negative control and all the six tested RNA/coronaviruses, therefore validating the specificity of the method.
Fig. 7.
Image showing the results for the cross-reactivity testing with four RNA viruses, Influenza AH1, Respiratory syncytial virus A, Parainfluenza virus and Human metapneumovirus along with the positive and negative controls using the capillary method. Images in A and B are for the capillaries before and after performing the LAMP assay. A colour change from dark pink to orange/yellow indicates the presence of the COVID-19 virus, whereas no change in colour indicates the absence of the virus. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 8.
Image showing the results for the cross-reactivity testing with related coronaviruses, MERS-CoV and HCoV-NL63 along with positive and negative controls using the capillary method. Images in A and B are for the capillaries before and after performing the LAMP assay. A colour change from dark pink to orange/yellow indicates the presence of the SARS-CoV-2 virus, whereas no change in colour indicates the absence of the virus. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Enhancements of the LAMP technique have been constantly made ever since its inception in 2000 [18]. Here we have demonstrated a rapid and inexpensive loop-mediated isothermal amplification assay for the detection of SARS-CoV-2 that uses a simple 5-min sample inactivation step. This assay can be performed in any clinical laboratory or field setup, such as mobile labs, as it does not require any specialised equipment or skilled laboratory personnel. There are several advantages to the SARS-CoV-2 saliva-based RT-LAMP testing method described here: 1) We can eliminate the uncomfortable invasive nasal swab-based sampling method. 2) We developed and used a simple RNA extraction method from saliva samples while maintaining compatibility with a colorimetric RT-LAMP assay. 3) Overall, with a simplified two-step process of saliva preparation and virus detection, the test has a rapid sample-to-result turnaround time of 45 min 4) This test is cost-effective, which enhances its merit for mass testing.
Previous studies have shown that the RT-LAMP assays can detect SARS-CoV-2 with a high degree of sensitivity. These studies report very high sensitivities starting from 100 copies [19,20] of viral RNA per reaction down to 10–20 copies [21,22], detecting even as low as 2 or 3 copies [23,24] of viral RNA per reaction. The novel capillary-based method for detection of SARS-CoV-2 developed in this study has comparable analytical sensitivity (or limit of detection), specificity, precision, and achieved excellent agreement compared to the reference RT-LAMP performed instead in a PCR tube. This method was able to achieve a LoD of 100 copies per reaction and 5 × 101 pfu/ml respectively, which is the minimum detectable limit specified by the manufacturer of the assay, New England Biolabs, and further this is in accord with other studies [19,20,25].
The initial hypothesis was to develop a rapid, cost-effective, and simple method that is not necessarily a quantitative measure of infection but could be applied within any other application areas at short notice and utilised by users with even limited training this has been demonstarted in our capillary-based assay. All that is required to conduct the test is a heating-block or heater capable of providing a constant temperature of 60–65 °C. From the standpoint of reagents, half the quantity is used per test, meaning with the same volume of reagents, double the number of tests can be conducted. In addition, the advantage of a capillary-based RT-LAMP test is its rapidity in detection, yielding results within 20 min for samples with 40,000 copies per reaction and between 35 and 45 min for a lesser copy load of around 100 copies per reaction.
4. Conclusion
Based on our findings, we conclude that our novel way of using capillaries in LAMP reactions is a highly promising way to further increase testing capacity, potentially serving as a tool for effective SARS-CoV-2 surveillance at the community level. This novel RT-LAMP testing strategy can offer solutions to a nationwide shortage of COVID-19 mass testing not only in hospital settings but also in major public hotspots, gatherings and concert venues, sporting events, transport hubs, to name but a few. The possibilities accompanying a minimalistic approach like this in scaling-up the testing capacity while being cost-efficient are indeed exciting, especially for developing and low-income countries. The protocol presented here using non-invasive saliva samples in rapid community testing would provide more frequent tests to those who face the risk of daily transmission, but who would not have the time or resources to seek tests in a clinical setting. With minimal set-up, capillary-based RT-LAMP testing could be performed in essential workplaces such as factories, construction sites, and schools.
At the outset, the impetus was to develop an assay to contribute to the diagnostic challenge posed by the growing COVID-19 pandemic situation and its adverse impact on almost every person. Evidence of lesser cross-reactivity observed in RT-LAMP testing made it the assay of choice. Having used the capillary as the platform for quick and accurate SARS-CoV-2 diagnostic testing, we have recognised the potential which we believe can be further explored for detection of different infectious agents. The results are promising for Influenza-type viruses which are under study at the moment and could be used for other future microbiological outbreaks where a rapid response is required.
CRediT authorship contribution statement
Anto J.U.K. John: performed - writing, conceptualization, investigation, writing – original draft, formal analysis. Peijun J.W. He: formal analysis, conceptualization, writing – review & editing. Ioannis N. Katis: writing – review & editing. P.P. Galanis: writing – review & editing. Alice H. Iles: writing – review & editing. Robert W. Eason: writing – review & editing, supervision, funding acquisition. Collin L. Sones: formal analysis, conceptualization, writing – review & editing, supervision, funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge the funding received via an Impact Acceleration Award (IAA) from the University of Southampton and funding via the Engineering and Physical Sciences Research Council (EPSRC) Grant Nos. EP/R511766/1, EP/J008052/1, EP/K023454/1, EP/N004388/1, EP/M027260/1 and EP/P025757/1. The underpinning RDM data for this paper can be found at 10.5258/SOTON/D1766.
References
- 1.Scallan M.F., Dempsey C., MacSharry J., O'Callaghan I., O'Connor P.M., Horgan C.P., et al. bioRxiv; 2020. Validation of a Lysis Buffer Containing 4 M Guanidinium Thiocyanate (GITC)/Triton X-100 for Extraction of SARS-CoV-2 RNA for COVID-19 Testing: Comparison of Formulated Lysis Buffers Containing 4 to 6 M GITC, Roche External Lysis Buffer and Qiagen RTL Lysis Buffer. 2020.04.05.026435. [Google Scholar]
- 2.Bruce E.A., Huang M.-L., Perchetti G.A., Tighe S., Laaguiby P., Hoffman J.J., et al. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Q., Meyerson N.R., Clark S.K., Paige C.L., Fattor W.T., Gilchrist A.R., et al. medRxiv; 2020. Saliva TwoStep: an RT-LAMP Saliva Test for SARS-CoV-2 and its Assessment in a Large Population. 2020.07.16.20150250. [Google Scholar]
- 4.Anahtar M.N., McGrath G.E.G., Rabe B.A., Tanner N.A., White B.A., Lennerz J.K.M., et al. medRxiv; 2020. Clinical Assessment and Validation of a Rapid and Sensitive SARS-CoV-2 Test Using Reverse-Transcription Loop-Mediated Isothermal Amplification. 2020.05.12.20095638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chotiwan N., Brewster C.D., Magalhaes T., Weger-Lucarelli J., Duggal N.K., Ruckert C., et al. Rapid and specific detection of Asian- and African-lineage Zika viruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aag0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohaim M.A., Clayton E., Sahin I., Vilela J., Khalifa M.E., Al-Natour M.Q., et al. Artificial intelligence-assisted loop mediated isothermal amplification (AI-LAMP) for rapid detection of SARS-CoV-2. Viruses. 2020:12. doi: 10.3390/v12090972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., et al. medRxiv; 2020. Analytical Sensitivity and Efficiency Comparisons of SARS-COV-2 qRT-PCR Assays. 2020.03.30.20048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashir J., Yaqinuddin A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med. Hypotheses. 2020;141:109786. doi: 10.1016/j.mehy.2020.109786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): expansion of its practical application as a tool to achieve universal health coverage. J. Infect. Chemother. 2020;26:13–17. doi: 10.1016/j.jiac.2019.07.020. [DOI] [PubMed] [Google Scholar]
- 14.Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020;66:975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu K.J. The New York Times; New York: 2020. It's like Groundhog Day’: Coronavirus Testing Labs Again Lack Key Supplies. [Google Scholar]
- 16.Bwambok D.K., Christodouleas D.C., Morin S.A., Lange H., Phillips S.T., Whitesides G.M. Adaptive use of bubble wrap for storing liquid samples and performing analytical assays. Anal. Chem. 2014;86:7478–7485. doi: 10.1021/ac501206m. [DOI] [PubMed] [Google Scholar]
- 17.James W., UK Says COVID-19 “Test and Trace” System Cost to Rise to 22 Billion Pounds, Reuters, United Kingdom.
- 18.Liu M.-L., Xia Y., Wu X.-Z., Huang J.-Q., Guo X.-G. Loop-mediated isothermal amplification of Neisseria gonorrhoeae porA pseudogene: a rapid and reliable method to detect gonorrhea. Amb. Express. 2017;7:48. doi: 10.1186/s13568-017-0349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T., et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe Acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baek Y.H., Um J., Antigua K.J.C., Park J.H., Kim Y., Oh S., et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microb. Infect. 2020;9:998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu L., Wu S., Hao X., Li X., Liu X., Ye S., et al. medRxiv; 2020. Rapid Colorimetric Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform: iLACO. 2020.02.20.20025874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu R., Wu X., Wan Z., Li Y., Zuo L., Qin J., et al. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virol. Sin. 2020;35:344–347. doi: 10.1007/s12250-020-00218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang W.E., Lim B., Hsu C.C., Xiong D., Wu W., Yu Y., et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byrne R.L., Kay G.A., Kontogianni K., Brown L., Collins A.M., Cuevas L.E., et al. medRxiv; 2020. Saliva Offers a Sensitive, Specific and Non-invasive Alternative to Upper Respiratory Swabs for SARS-CoV-2 Diagnosis. 2020.07.09.20149534. [DOI] [PMC free article] [PubMed] [Google Scholar]