ABSTRACT
The closest relative of human pathogen Leishmania, the trypanosomatid Novymonas esmeraldas, harbors a bacterial endosymbiont “Candidatus Pandoraea novymonadis.” Based on genomic data, we performed a detailed characterization of the metabolic interactions of both partners. While in many respects the metabolism of N. esmeraldas resembles that of other Leishmaniinae, the endosymbiont provides the trypanosomatid with heme, essential amino acids, purines, some coenzymes, and vitamins. In return, N. esmeraldas shares with the bacterium several nonessential amino acids and phospholipids. Moreover, it complements its carbohydrate metabolism and urea cycle with enzymes missing from the “Ca. Pandoraea novymonadis” genome. The removal of the endosymbiont from N. esmeraldas results in a significant reduction of the overall translation rate, reduced expression of genes involved in lipid metabolism and mitochondrial respiratory activity, and downregulation of several aminoacyl-tRNA synthetases, enzymes involved in the synthesis of some amino acids, as well as proteins associated with autophagy. At the same time, the genes responsible for protection against reactive oxygen species and DNA repair become significantly upregulated in the aposymbiotic strain of this trypanosomatid. By knocking out a component of its flagellum, we turned N. esmeraldas into a new model trypanosomatid that is amenable to genetic manipulation using both conventional and CRISPR-Cas9-mediated approaches.
KEYWORDS: Trypanosomatidae, Leishmaniinae, bacterial endosymbiont, genomics, metabolism
INTRODUCTION
Trypanosomatidae is a family of obligate parasitic flagellates that includes the causative agents of some severe human diseases (genera Leishmania and Trypanosoma), as well as those affecting plants (genus Phytomonas) (1, 2). The vast majority of trypanosomatids parasitize insects (3), using them as sole either hosts (monoxenous species) or vectors, transmitting the flagellates to vertebrates or plants (dixenous species) (4). Trypanosomatids and related lineages, represented mostly by free-living forms, constitute the class Kinetoplastea, which, together with marine diplonemids, freshwater euglenids, and anoxic environment-dwelling symbiontids, belong to the phylum Euglenozoa (5, 6). Some members of this extremely diverse protist group harbor either extracellular or intracellular bacterial symbionts of varied provenience, as they include alphaproteobacteria, betaproteobacteria, gammaproteobacteria, and cyanobacteria, the latter confined to euglenids (6). From trypanosomatids, only betaproteobacteria of the order Burkholderiales have been described so far; they are products of at least two independent acquisitions. The common ancestor of the cosmopolitan genera Angomonas, Strigomonas, and Kentomonas was invaded by “Candidatus Kinetoplastibacterium” of the family Alcaligenaceae (7, 8), whereas the monospecific Novymonas harbors “Ca. Pandoraea novymonadis” of the family Burkholderiaceae (9). From several available genomes of “Ca. Kinetoplastibacterium,” one can predict that these are well-established symbionts that achieved a high level of mutual adaptation following long-term coevolution. In contrast, the endosymbiotic relationship between N. esmeraldas and “Ca. Pandoraea novymonadis” appears to be much less stable, as the trypanosomatid host does not strictly control bacterial replication, and therefore, the number of endosymbionts significantly varies from cell to cell (9). Compared to its free-living relatives, the genome of “Ca. Pandoraea novymonadis” is substantially reduced in size, exhibiting extensive gene loss, low GC content, and numerous gene rearrangements, revealing that bacteria depend heavily on the host for survival. The recently described unidentified endosymbiont of the plant pathogen Phytomonas borealis also shows signs of an unstable relationship (10).
How do the partners benefit from each other? In the above-mentioned symbiosis between the trypanosomatids of the subfamily Strigomonadinae and “Ca. Kinetoplastibacterium,” the long-term coevolution triggered significant changes in morphology, biochemistry, and physiology of both bacteria and their flagellate host. Each protist harbors a single bacterium, which replicates in a synchronous manner with its host, ensuring well-coordinated vertical transmission (11, 12). Compared to other trypanosomatids, Strigomonadinae are endowed with an enlarged mitochondrion, presumably because of their increased energy consumption (8, 13). It was proposed that the lack of cell wall in these bacteria facilitates exchange of metabolites (14), which likely include ATP and phosphatidylcholine from the trypanosomatid side (15), while the bacterium reciprocates by providing purines, polyamines, heme, some vitamins, lipids, and amino acids (16–18). Moreover, the synthesis of certain amino acids requires enzymes of both partners, making their metabolic pathways intricately intertwined (19). In addition to metabolites, some host-encoded proteins are targeted from the host cell cytoplasm into the endosymbiont (20). This is a hallmark of a rather stable and intimate relationship between both partners.
It is now generally accepted that endosymbiotic relationships not only were critical for the emergence of the complexity of eukaryotic cell (21, 22) but also have enormous importance in extant ecosystems (23, 24). Our current understanding of the intricacies of symbiotic relationships between eukaryotes on one side and bacteria or archaea on the other is mostly based on the knowledge derived from a variety of animals and plants (25–27). However, this does not align well with the fact that the diversity of protists that exceeds that of multicellular eukaryotes (28, 29) most likely reflects the diversity of respective endosymbiotic relationships.
One way of changing this situation is to introduce and characterize new protist-endosymbiont systems, which inevitably has to be accompanied by their amenability to reverse genetics. Indeed, there is a scarcity of refined molecular genetic tools that currently significantly limits the dissection of the protist-endosymbiotic relationships. However, once a basic tool kit becomes available for these organisms, they may provide outsized insight into symbiosis, as in the case of the cercozoan Paulinella chromatophora (30, 31). Thanks to the wide range of genetic manipulation methods available for the medically important dixenous genera Leishmania and Trypanosoma, as well as for some of their monoxenous kin (Leptomonas seymouri [32], Crithidia fasciculata [33], Phytomonas sp. [34], Herpetomonas muscarum [35], and Lotmaria passim [36]), we believe that the endosymbiont-bearing trypanosomatids can be turned into model systems. This is already exemplified by one species, Angomonas deanei, which was successfully genetically modified (20).
In this work, we describe the genome and transcriptome analysis of N. esmeraldas and characterize the metabolic interactions with its endosymbiont. Moreover, we report the development of a molecular toolbox, both conventional and CRISPR-Cas9 based, which will make it possible to tackle the symbiotic relationships between the flagellate host and its cytoplasmic bacteria.
RESULTS
Genome of N. esmeraldas.
The N. esmeraldas genome assembly contains 1,429 scaffolds with an N50 of 197,811 nucleotides (nt), a total length of approximately 32 Mbp, and a GC content of 62.7%. The genome annotation yielded 9,299 predicted proteins. These values are comparable to those from other available trypanosomatid genomes. The genome and annotation completeness was assessed by BUSCO, and 73% of the universal eukaryotic genes were identified in the assembly. This score is high compared to those of the reference trypanosomatid genomes, which are between 62.4% in the plant pathogen Phytomonas sp. strain Hart1 and 74.9% in the model human pathogen Trypanosoma brucei TREU927.
Metabolic potential of N. esmeraldas and its endosymbiont.
We compared the predicted metabolic capabilities of N. esmeraldas with those of the model human pathogen Leishmania major. Both trypanosomatids belong to the subfamily Leishmaniinae (37, 38) and are very similar with respect to 486 sequences of selected core metabolic enzymes (Table S1A). Only a few genes present in L. major, such as those encoding NAD-dependent glutamate dehydrogenase, transketolase, and cytosolic malate dehydrogenase, were not found in N. esmeraldas. The absence of the latter two enzymes may be compensated for by their respective orthologues in the endosymbiont. We also noted a large number of genes in the genome of N. esmeraldas encoding GP63 proteases and the pteridin/biopterin transporters, which are known virulence factors in Leishmania species responsible for human leishmaniases (39, 40).
Analyses of Novymonas esmeraldas. (A) List of 486 metabolic enzymes of L. major and their orthologues in N. esmeraldas. (B) Novymonas esmeraldas metabolic enzymes not present in L. major. (C) Endosymbiont-specific proteins of “Ca. Pandoraea novymonadis.” (D) Proteins of N. esmeraldas with a putative glycosomal/peroxisomal targeting signal. (E) List of enzymes of N. esmeraldas involved in sugar metabolism. (F) Enzymes of gluconeogenesis. (G) Enzymes of the pyrimidine and purine biosynthesis and purine salvage. (H) Enzymes of the FoF1-ATP synthase and the mitochondrial electron transport chain and mitochondrial contact site and crista organization system (MICOS). (I) Enzymes of amino acid metabolism. (J) Enzymes of the urea cycle. (K) Enzymes involved in synthesis of vitamins and cofactors. (L) Enzymes involved in lipid metabolism. (M) Enzymes involved in phosphor-lipid metabolism. (N) Genes upregulated in the endosymbiont-free N. esmeraldas (ES) compared to the wild-type cells (WT). Criteria: BH-adjusted P value of <0.001 and fold change of ≥1.5. (O) Genes downregulated in the endosymbiont-free N. esmeraldas (ES) compared to the wild-type cells (WT). Criteria: BH-adjusted P value of <0.001 and fold change of ≥1.5. (P) Top 10 GO terms most significantly enriched among genes upregulated in aposymbiotic N. esmeraldas. The terms are sorted according to the Fisher's test P values; those significantly enriched are in bold (P < 0.01). Only GO terms of the three main categories (biological process, molecular function, and cellular component) are included. Annotated, all genes are annotated with the respective GO term in the whole analyzed genome; Significant, a number of genes in the list of the up-/down-regulated ones are annotated with the respective GO term; Expected, an estimate of the number of genes a node of size “Annotated” would have if the significant genes were to be randomly selected from the gene pool. (R) Top 10 GO terms most significantly enriched among genes downregulated in aposymbiotic N. esmeraldas. The terms are sorted according to the Fisher's test P values; those significantly enriched are in bold (P value < 0.01). Only GO terms of the three main categories (biological process, molecular function, and cellular component) are included. Annotated, a total number of genes annotated with the respective GO term in the whole analyzed genome; Significant, a number of genes in the list of the up-/down-regulated ones are annotated with the respective GO-term; Expected, an estimate of the number of genes a node of size “Annotated” would have if the significant genes were to be randomly selected from the gene pool. (S) Data sets used in phylogenomic analysis. (T) Primers used in this study. Download Table S1, XLSX file, 0.7 MB (705.4KB, xlsx) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Of 9,299 predicted protein-coding genes in the N. esmeraldas genome, 871 were not found in L. major. The majority of them are hypothetical, 202 are unique to N. esmeraldas (Table S1B), and, curiously, seven are also present in the “Ca. Pandoraea novymonadis” genome. Three proteins in the latter category are involved in the synthesis of lysine from diaminopimelate, and one is an enzyme of the urea cycle. Another representative example is a pair of argininosuccinate lyases in both the host and endosymbiont genomes, which despite 50% sequence identity do not have a common origin (data not shown). From the perspective of the “Ca. Pandoraea novymonadis” genome, 815 of its protein-coding genes (Table S1C) are endosymbiont specific (not present in the host genome). Below, we discuss several aspects of the N. esmeraldas and “Ca. Pandoraea novymonadis” metabolisms in detail.
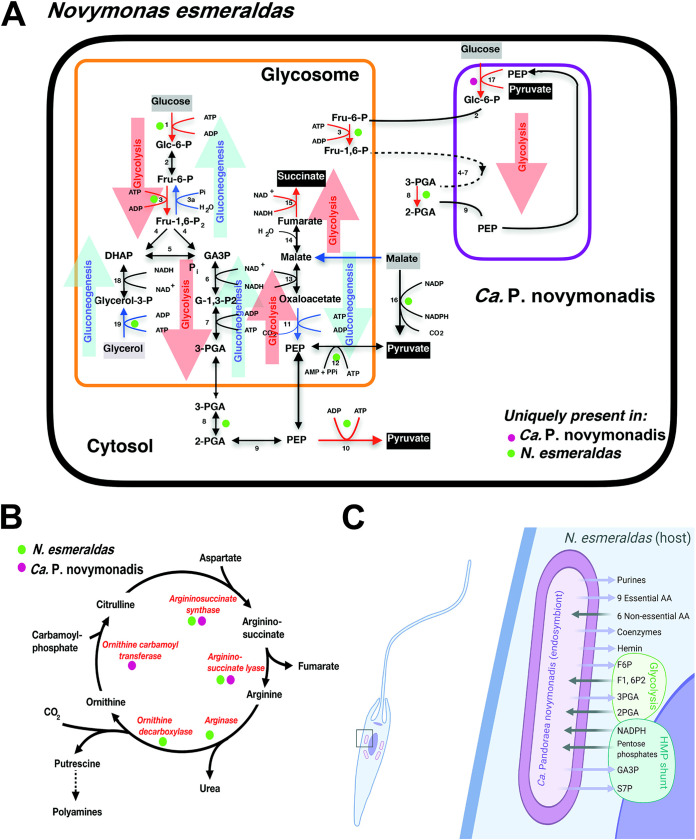
(i) Carbohydrate metabolism. Novymonas esmeraldas has a complete set of the glycolytic pathway enzymes. Several of them carry the peroxisomal/glycosomal targeting signal and, therefore, presumably function in the parasite’s glycosomes (Table S1D). Like most other trypanosomatids, N. esmeraldas is capable of using sugars other than glucose, such as galactose, fructose, xylulose, mannose, glucosamine and N-acetylglucosamine, as judged by the presence of all enzymes needed for their conversion to fructose-6-phosphate, an intermediate of glycolysis (41) (Table S1E; Fig. 1A). The gluconeogenic pathway, driven by pyruvate-phosphate dikinase, phosphoenolpyruvate carboxykinase, fructose bisphosphatase, and glucose-6-phosphate isomerase, is operational and localized in glycosomes. All genes encoding enzymes of the mitochondrial Krebs cycle were identified, and the classical hexose-monophosphate pathway (HMP), with the exception of transketolase, is present.
FIG 1.
Metabolic exchange between N. esmeraldas and its endosymbiont “Ca. Pandoraea novymonadis.” (A) Glycolysis and gluconeogenesis. Boxed metabolites are nutrients (in gray) or end products (in black). Glycolysis (red arrows) takes place both in glycosomes and in the endosymbiont. In the latter, exchange of intermediates with the host organism occurs. Gluconeogenesis (blue arrows) takes place exclusively in the host, where malate (resulting from mitochondrial amino acid metabolism) and glycerol (formed from lipid hydrolysis) are converted to glucose 6-phosphate. Enzymes: 1, hexokinase; 2, phosphoglucose isomerase; 3, phosphofructokinase; 3a, fructose-bisphosphatase; 4, fructose-bisphosphate aldolase; 5, triosephosphate isomerase; 6, glyceraldehyde-3-phosphate dehydrogenase; 7, phosphoglycerate kinase; 8, phosphoglycerate mutase; 9, enolase; 10, pyruvate kinase; 11, phosphoenolpyruvate carboxykinase; 12, pyruvate phosphate di-kinase; 13, malate dehydrogenase; 14, fumarate hydratase; 15, NADH-dependent fumarate reductase; 16, malic enzyme; 17, phosphoenolpyruvate-protein phosphotransferase. 18, glycerol-3-phosphate dehydrogenase (NAD); 19, glycerol kinase. Enzyme contributions by host and endosymbiont are indicated by dots of different colors. (B) The urea cycle of N. esmeraldas. The urea cycle is divided over host and endosymbiont. Enzyme contributions by host and endosymbiont are indicated by dots of different colors. (C) Summarized scheme of metabolic exchange between N. esmeraldas and its endosymbiont “Ca. Pandoraea novymonadis.” Abbreviations: F6P, fructose 6-phosphate; F1,6P2, fructose 1,6-bisphosphate; 3PGA, 3-phosphoglycerate; 2PGA, 2-phosphoglycerate; S7P, sedoheptulose 7-phosphate; GA3P, glyceraldehyde 3-phosphate; HMP, hexose-monophosphate shunt, or pentose-phosphate pathway.
The genome of “Ca. Pandoraea novymonadis” does not contain hexokinase and pyruvate kinase of the classical glycolytic pathway. Instead, the endosymbiont uses a typical bacterial-type phosphotransferase system (42) that couples the transfer of a high-energy phosphate from phosphoenolpyruvate to the internalization and phosphorylation of hexoses (Fig. 1A). Moreover, since the genes for phosphofructokinase and phosphoglycerate mutase were not found in its genome, “Ca. Pandoraea novymonadis” relies on the presence of these three host enzymes to convert hexoses to pyruvate, since the HMP shunt cannot compensate for this gene loss (Fig. 1A). In addition, the endosymbiont’s genome lacks three genes of the first oxidative part of the HMP, namely, glucose-6-phosphate dehydrogenase, 6-phospho-gluconolactonase, and 6-phospho-gluconate dehydrogenase. Interestingly, the transketolase gene is missing from the N. esmeraldas genome, making the host and its endosymbiont interdependent in carrying out both the HMP and glycolysis reactions (Table S1F). The presence of genes for fructose-1,6-bisphosphatase in the N. esmeraldas and “Ca. Pandoraea novymonadis” genomes suggests that gluconeogenesis plays an essential role in both partners, while the bacterium acquires sugar phosphates from the host cell via HMP. Moreover, the trypanosomatid fructose-bisphosphate aldolase and glucose-6-phosphate isomerase allow synthesis of sugars essential for both the bacterium and its eukaryotic host (Fig. 1A).
(ii) Pyrimidine biosynthesis and purine salvage. Trypanosomatids (including N. esmeraldas) are unable to synthesize their own purines but can salvage them and can synthesize pyrimidines (43, 44) (Table S1G). In contrast, “Ca. Pandoraea novymonadis” can synthesize its own pyrimidines and purines, yet it has a limited capacity for the salvage of purine bases and nucleosides. The biochemical pathways for the synthesis of pyrimidines differ in the host and the endosymbiont. While N. esmeraldas is endowed with a typical trypanosomatid-like pathway composed of soluble dihydroorotate dehydrogenase and bifunctional orotate phosphoribosyltransferase/orotidine-5-phosphate decarboxylase (45, 46), its endosymbiont harbors a typical bacterial-type pathway (ubiquinone-dependent dihydroorotate dehydrogenase, orotate phosphoribosyltransferase, and orotidine-5-phosphate decarboxylase) (Table S1G).
(iii) Oxidative phosphorylation and MICOS. All subunits of FoF1-ATP synthase and mitochondrial respiratory complexes I through IV were identified in the N. esmeraldas genome (Table S1H). Like some other trypanosomatids, N. esmeraldas possesses a functional complex I (NADH-ubiquinone oxidoreductase) (47). All subunits of complexes II (succinate dehydrogenase), III (cytochrome c reductase), and IV (cytochrome c oxidase) are present, although one of the auxiliary subunits of complex II appears to be missing. Its endosymbiont has genes for several subunits of complex I, three catalytic subunits of complex II, and three subunits of complex III (ubiquinol cytochrome c reductase, cytochrome b, and cytochrome c1), along with a gene encoding cytochrome c. No catalytic subunit and only a single associated subunit of complex IV were identified in “Ca. Pandoraea novymonadis.” Nevertheless, the finding of subunits of two ubiquinol oxidase complexes (cytochrome bo3 and cytochrome bd, expressed under high- and low-oxygen conditions, respectively [48]) in its genome implies that at least three of the respiratory complexes contribute to the endosymbiont’s oxidative phosphorylation. A similar situation has been reported for two other symbiont-bearing trypanosomatids, Angomonas deanei and Strigomonas culicis (49).
The mitochondrial contact site and crista organization system (MICOS) is a multiprotein complex responsible for crista formation. It is an ancestral eukaryotic protein complex of alphaproteobacterial origin (50). Trypanosomatids are endowed with its divergent form, which was recently carefully characterized in T. brucei (51). All the MICOS proteins were found in the N. esmeraldas (but not its bacterial symbiont) genome (Table S1H).
(iv) Metabolism of amino acids. Novymonas esmeraldas synthesizes nonessential amino acids, along with Thr and Met, and catabolizes amino acids into intermediates of the tricarboxylic acid (TCA) cycle (Table S1I). A few selected examples are described below. Like other Leishmaniinae (but not other trypanosomatids), N. esmeraldas synthesizes Arg from citrulline and aspartate, while Gly is split into CO2 and formic acid by the mitochondrial glycine-cleavage system, which is lacking in the endosymbiont. The trypanosomatid can salvage Met, as genes encoding all enzymes of the salvage cycle were found in its genome, which, however, does not contain enzymes of the aerobic aromatic amino acid metabolism pathway that convert Phe to fumarate and acetoacetate. Moreover, the flagellate under investigation can neither degrade nor synthesize Lys de novo. Among trypanosomatids, only Leptomonas and Crithidia spp. can convert the bacterial amino acid diaminopimelate to Lys (52). The detection of several diaminopimelate-metabolizing enzymes in both N. esmeraldas and its endosymbiont suggests that they have the capacity to perform this biochemical reaction.
(v) Urea cycle. Functionality of the urea cycle depends on both symbiotic partners (Fig. 1B). Novymonas esmeraldas and its endosymbiont contribute enzymes to the pathway as follows: ornithine carbamoyltransferase is supplied by the endosymbiont, argininosuccinate synthase and argininosuccinate lyase are provided by both host and bacterium, and arginase is supplied exclusively by the host. As a result, the urea cycle metabolites have to shuttle between the host and endosymbiont (Table S1J).
(vi) Vitamins and cofactors. Trypanosomatids are obligatory auxotrophs for a number of exogenous cofactors and/or vitamins. They can synthesize neither thiamine (vitamin B1) and riboflavin (vitamin B2) nor biotin (vitamin B7) and cobalamin (vitamin B12) (44). However, thanks to the presence of endosymbiont, N. esmeraldas can synthesize vitamins B1, B2, B6, and B7 but not B12 (Table S1K; Fig. 1C). Synthesis of tetrahydrofolic acid (vitamin B9) is of special importance for trypanosomatids, since pteridine and folate derivatives are essential cofactors in one-carbon transfer. Leishmania is a pteridine auxotroph and has evolved an elaborate and versatile pteridine salvage network capable of accumulating and reducing pteridines (53), which includes biopterin and folate transporters, pteridine reductase, and dihydrofolate reductase-thymidylate synthase (54). Novymonas esmeraldas also encodes a battery of enzymes responsible for the uptake of pteridine, its conversion into the dihydrofolic acid, and further utilization in the form of the various folate cofactors.
(vii) Heme biosynthesis, lipid metabolism, and ROS protection. Unlike most other trypanosomatids (55), N. esmeraldas is able to synthesize its own heme (Table S1K). The components of the heme biosynthetic pathway shared between the host and the endosymbiont are similar to what has been described for two other endosymbiont-harboring trypanosomatids, A. deanei and S. culicis (49).
Lipid metabolism is virtually the same in N. esmeraldas and L. major (Table S1L) (56, 57). The endosymbiont’s capacity for phospholipid metabolism appears to be limited, although, like its host, it can synthesize the ubiquinone required for its respiration (Table S1M).
Novymonas esmeraldas encodes catalase, a key component of the reactive oxygen species (ROS) pathway, which is related to its orthologues in other Leishmaniinae (58, 59). No catalase gene was found in the endosymbiont, while both host and endosymbiont have genes for superoxide dismutase, the presence of which renders “Ca. Pandoraea novymonadis” aerobic.
Gene expression changes in the aposymbiotic N. esmeraldas.
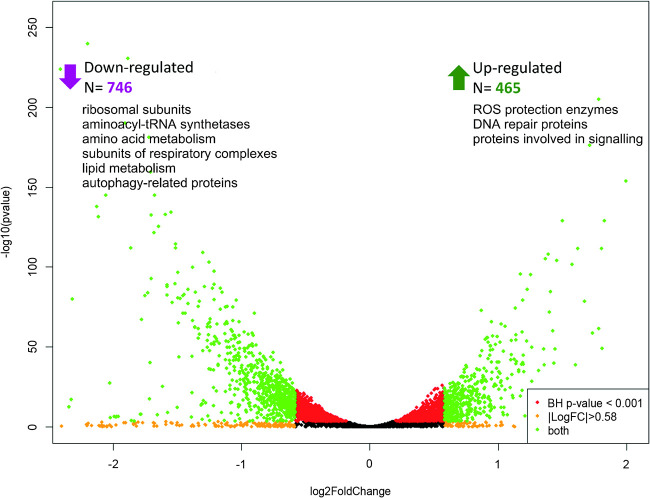
Using the aposymbiotic strain described earlier (42), we identified 1,211 differentially expressed genes (DEGs), of which 465 and 746 were up- and downregulated, respectively, in the aposymbiotic N. esmeraldas strain E262-AZI, compared to the wild-type endosymbiont-carrying isolate E262 (1.5-fold change; Benjamini-Hochberg [BH]-adjusted P value, <0.001) (Fig. 2; Table S1N and O). The analysis of DEGs was complicated by the lack of functional annotation for ∼42% and ∼34% of up- and downregulated genes, respectively. Nevertheless, as detailed below, the gene ontology (GO) term enrichment analysis sheds light on several aspects of the interactions between N. esmeraldas and “Ca. Pandoraea novymonadis” (Table S1P and R; Fig. S1 to S7).
FIG 2.
Volcano plot of differentially expressed genes between the E262 and E262-AZI N. esmeraldas. The green dots denote significantly differentially expressed genes [BH-adjusted P value of <0.001 and fold changes of ≥1.5, or log2(fold change) of ≥0.58].
A heat map for 50 genes with the highest variance across samples based on log-transformed expression values. Download FIG S1, PDF file, 0.08 MB (86.7KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The gene cohort downregulated in the aposymbiotic strain E262-AZI displays a significant enrichment in the GO terms “tRNA aminoacylation for protein translation” and “aminoacyl-tRNA ligase activity,” as well as the presence of a number of ribosomal proteins, multiple aminoacyl-tRNA synthetases, and rRNAs. Thus, the loss of endosymbiont apparently leads to a general decrease of protein biosynthesis in the trypanosomatid host. This may be accompanied by the reduced translation of proteins involved in the interactions at the trypanosomatid-bacterium interface, and/or proteins produced by the host and subsequently imported into the endosymbiont. The aminoacyl-tRNA synthetases downregulated in the endosymbiont-free N. esmeraldas include alanyl-, aspartyl-, cysteinyl-, glycyl-, leucyl-, lysyl-, prolyl-, seryl-, and valyl-tRNA charging enzymes. The downregulation of glycyl-, leucyl-, lysyl-, seryl-, and valyl-tRNA may be directly connected to the loss of endosymbionts, which synthetize the corresponding amino acids and provide them to the host (42).
The amino acid metabolism of N. esmeraldas also appears to be substantially impacted by the loss of bacteria. This is manifested in a significant decrease in the expression of genes of Gly metabolism (GO term “glycine metabolic process”), including two subunits of the Gly cleavage system (H and P proteins), as well as both cytosolic (NESM_000295500) and mitochondrial (NESM_000597600) hydroxymethyltransferases catalyzing Gly biosynthesis from Ser. The latter observation correlates with the downregulation of the glycyl-tRNA synthetase gene, which facilitates the incorporation of Gly. Moreover, Asp/Asn metabolism is downregulated in the endosymbiont-free N. esmeraldas, as reflected by the levels of Asn synthetase, which generates Asn from Asp, and three aspartate aminotransferases, catalyzing the interconversion of Asp and α-ketoglutarate to oxaloacetate and glutamate. In contrast, asparaginase, hydrolyzing Asn to aspartic acid, is upregulated in the endosymbiont-free cells, as is also the case for cysteine desulfurase, which catalyzes Ala biosynthesis from Cys and is involved in Fe-S cluster synthesis. However, other enzymes of the Cys metabolism, such as cystathionine γ-lyase and cystathionine β-lyase, responsible for the breakdown of cystathionine into Cys, α-ketobutyrate, ammonia, and homocysteine and pyruvate, respectively, are downregulated in the aposymbiotic strain. Similarly, the transcription of two genes for branched-chain amino acid aminotransferases, participating in the degradation of Leu, Ile, and Val provided by the endosymbiont (42), and Glu dehydrogenase, an enzyme involved in the metabolism of Glu, is also decreased in these cells. Furthermore, the absence of the endosymbionts causes the downregulation of Phe hydroxylase, catalyzing the conversion of Phe to Tyr, and of S-adenosylmethionine synthetase, responsible for the formation of S-adenosylmethionine (AdoMet) from Met and ATP. It appears that the processes of trans-methylation and polyamine biosynthesis are affected by the loss of endosymbiont, since AdoMet plays an important role in the respective reactions.
When metabolism is concerned, the loss of endosymbiont leads to a decrease in the mitochondrial respiratory activity. This is reflected by the downregulation of genes falling into the GO categories “ATP synthesis coupled proton transport,” “NAD(P)H dehydrogenase (quinone) activity,” and “mitochondrial inner membrane,” which include multiple subunits of respiratory complexes III, IV, and V (ATP synthase, cytochrome c reductase, and cytochrome c oxidase). Of note, a decreased expression of several subunits of respiratory complexes was recently reported for aposymbiotic cells of Strigomonas culicis (60). However, the situation with glycolysis and gluconeogenesis is less clear. On the one hand, the gene encoding 6-phosphofructo-2-kinase, responsible for the biosynthesis of fructose 2,6-bisphosphate, one of the major regulators of trypanosomatid energy metabolism (61), is significantly upregulated in the aposymbiotic N. esmeraldas, similarly to the endosymbiont-free S. culicis (60). On the other hand, two other glycolytic enzymes, hexokinase and enolase, along with the gluconeogenic enzyme phosphoenolpyruvate carboxykinase are significantly downregulated.
Lipid metabolism is also repressed in the endosymbiont-free N. esmeraldas, as suggested by the downregulation of genes within the GO categories “fatty acid biosynthetic process,” “3-oxo-arachidoyl-CoA synthase activity,” and “very-long-chain 3-ketoacyl-CoA synthase activity,” incorporating multiple fatty acid elongases, malonyl coenzyme A (malonyl-CoA) decarboxylase, β-ketoacyl synthase family protein, delta-5 fatty acid desaturase, 3-ketoacyl-CoA thiolase, isovaleryl-CoA dehydrogenase, 3-ketoacyl-CoA reductase, and enoyl-CoA hydratase.
The loss of the endosymbiont leads to increased ROS production, illustrated by the significant upregulation of genes encoding the enzymes involved in ROS protection, such as trypanothione synthetase, tryparedoxin, tryparedoxin-like proteins, tryparedoxin peroxidase, and glutathione peroxidase (Table S1N). In agreement with this observation, several genes encoding proteins of DNA repair (e.g., mismatch repair proteins MSH3 and MSH6) are also found to be upregulated, probably indicating DNA damage resulting from increased ROS production (Table S1O and R). Similar changes, namely, upregulation of tryparedoxin and genes involved in DNA repair, were reported in Angomonas deanei upon removal of the endosymbiont (62).
Sixteen genes encoding proteins associated with autophagy (e.g., autophagy-related protein 8 and ubiquitin-like modifier-activating enzyme ATG7) are significantly downregulated in the aposymbiotic cells. They fall into the “cellular response to nitrogen starvation” and “autophagy” categories and are among genes with the highest variance across samples (Fig. S1). This indicates a reduced requirement for the proteins involved in autophagy, which are likely used for controlling the number of bacteria in the wild-type cells (63). In contrast, genes related to autophagy are upregulated in the aposymbiotic strain of A. deanei, compared to the wild type (62).
The GO terms “cAMP biosynthetic process” and “intracellular signal transduction,” assigned to receptor adenylate cyclases, kinases, and a putative methylase, are significantly enriched in the cohort of genes upregulated in the aposymbiotic N. esmeraldas. Thus, the loss of the endosymbiont leads to a reduced amino acid, lipid, and carbohydrate metabolism, likely reflecting a significant contribution of “Ca. Pandoraea novymonadis” to these processes, as well as reduced needs of the bacterium-deprived trypanosomatid cells (of note, the trend is opposite in A. deanei, in which amino acid, lipid, and carbohydrate metabolism are all elevated upon bacterium elimination [62]). On the other hand, upregulation of genes whose products are involved in signaling cascades, ROS protection systems, and DNA repair also accompanies the loss of the bacterial participant of the Novymonas-“Ca. Pandoraea novymonadis” system.
Novymonas esmeraldas as a new model trypanosomatid.
We performed a phylogenomic analysis on a set of 640 proteins encoded by single-copy genes in all analyzed kinetoplastid genomes (Table S1S). Both maximum-likelihood and Bayesian trees had the same topology, which is consistent with the results of similar analyses performed earlier (64), and all branches but one displayed maximal statistical support (Fig. 3). Novymonas esmeraldas revealed an unambiguous sister relationship with the clade of dixenous Leishmaniinae (Leishmania, Porcisia, and Endotrypanum), confirming the inference based on the sequences of several genes made in the original description of this taxon (9).
FIG 3.
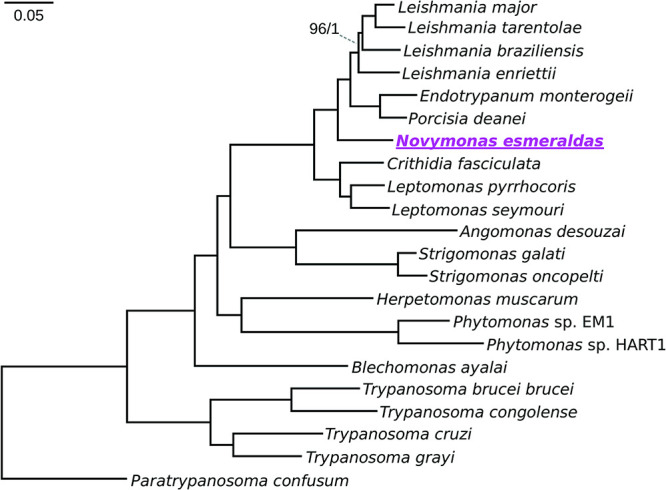
Phylogenetic position of N. esmeraldas. Maximum-likelihood phylogenomic tree based on the alignment of 359 proteins encoded by single-copy genes demonstrating the phylogenetic position of N. esmeraldas (underlined) as the closest described relative of Leishmania. All branches have maximal bootstrap support and posterior probability values (except the branch, where support values are indicated). The bar indicates number of substitutions per site.
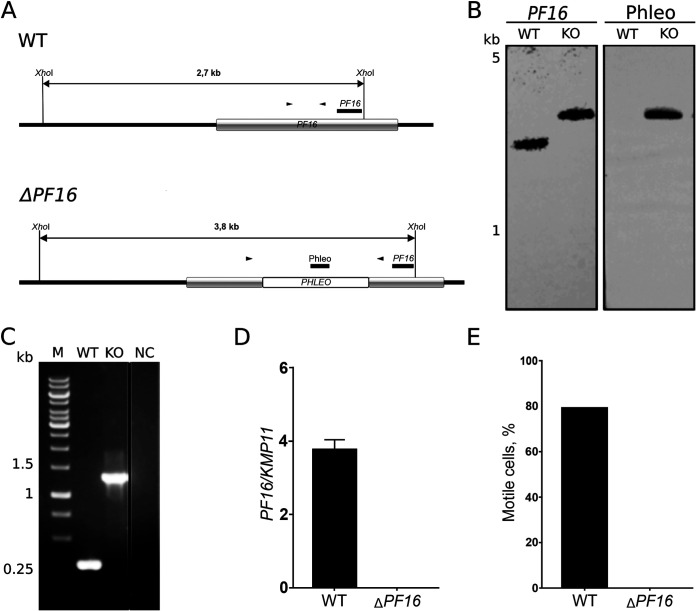
The available genomic and transcriptomic data for N. esmeraldas, combined with its capacity for genetic modifications, may provide novel insight into the establishment of an endosymbiotic relationship with a bacterium. As a proof of principle that an exogenous DNA can be integrated into the nucleus of N. esmeraldas, we first integrated the mCherry-expressing construct into its 18S rRNA locus. The resultant cells exhibited a bright red fluorescence (Fig. S8), indicative of a successful mCherry integration. Next, we set up a CRISPR-Cas9-dependent system for genetic manipulations of this protist. As a template, we used a system previously established in Leishmania mexicana (65), which relies on FLAG-Cas9 integrated into the same (i.e., the 18S rRNA) locus. Successful integration and expression of Cas9 were confirmed by PCR (Fig. S9A), reverse transcription-quantitative PCR (RT-qPCR) (Fig. S9B), and Western blotting with anti-FLAG antibodies (Fig. S9C). Of note, the expression of Cas9 varied significantly between the drug-selected populations, so for downstream experiments, we generated an N. esmeraldas population displaying a Cas9 expression level comparable to that in the L. mexicana transgenic strain (65) (Fig. S9B and C). We noted that Cas9 expression affects neither cell division (Fig. S9D) nor bacterial load (Fig. S9E). The resultant cells were used to ablate a single-copy gene, PF16, encoding a central pair protein of the flagellar axoneme (66). Ablation of this gene in other trypanosomatids makes them invariably immotile (67, 68). The PF16 homolog of N. esmeraldas was disrupted by insertion of the phleomycin resistance gene, and successful integration into both alleles was confirmed by Southern blotting, PCR, and RT-qPCR (Fig. 4A to D). All selected cells were immotile (Fig. 4E), proving that we have established two complementary systems for genetic manipulations in N. esmeraldas based on conventional and Cas9-mediated recombination.
FIG 4.
Genetic ablation of PF16 using CRISPR-Cas9-mediated approach in N. esmeraldas. (A) Schematic representation of the wild-type (top) and recombined (bottom) PF16 alleles of N. esmeraldas. Expected sizes of DNA fragments after XhoI digestion and positions of the annealed probes and primers for diagnostic PCR are indicated. (B) Southern blot analysis of the XhoI-digested N. esmeraldas genomic DNA of the WT and PF16-ablated strains with PF16 and Phleo probes. (C) Diagnostic PCR for the wild-type and recombined (KO) PF16 alleles of N. esmeraldas. NC, negative control. (D) RT-qPCR control for the wild-type and ΔPF16 alleles of N. esmeraldas. (E) Percent motile cells in the wild type and ΔPF16 N. esmeraldas.
Subgraph representing the 5 most significant GO terms of the category “Molecular Function” (in rectangles) enriched in the group of genes upregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S2, PDF file, 0.02 MB (22.8KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subgraph representing the 5 most significant GO terms of the category “Biological Process” (in rectangles) enriched in the group of genes upregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S3, PDF file, 0.08 MB (80.1KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subgraph representing the 5 most significant GO terms of the category “Cellular component” (in rectangles) enriched in the group of genes upregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S4, PDF file, 0.02 MB (25.8KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subgraph representing the 5 most significant GO terms of the category “Molecular Function” (in rectangles) enriched in the group of genes downregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S5, PDF file, 0.03 MB (28.9KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subgraph representing the 5 most significant GO terms of the category “Biological Process” (in rectangles) enriched in the group of genes downregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S6, PDF file, 0.08 MB (87.6KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subgraph representing the 5 most significant GO terms of the category “Cellular component” (in rectangles) enriched in the group of genes downregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S7, PDF file, 0.04 MB (39.7KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Live Novymonas esmeraldas cells expressing mCherry (fluorescence microscopy). Bright field, SYTO 24, mCherry and the merged image are presented in the corresponding columns. The mCherry-expressing and wild-type N. esmeraldas cells are shown in the upper and lower panels, respectively. Bar, 5 μm. Download FIG S8, PDF file, 0.05 MB (54.1KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Establishment of Cas9-expressing N. esmeraldas. (A) PCR confirmation of successful integration. Populations 1 to 4 are shown along with the wild type. (B) RT-qPCR of Cas9 expression in populations 1 to 4. Wild-type N. esmeraldas and previously established Cas9-expressing L. mexicana were used as negative and positive controls, respectively. (C) Western blotting confirmation of Cas9 expression using anti-FLAG antibodies. Labels are as in panel B. (D) Growth curves of WT and Cas9-expressing N. esmeraldas; (E) Bacterial load in WT and Cas9-expressing N. esmeraldas. Sizes of DNA (A) and protein (C) fragments are in kilobases and kilodaltons, respectively. Data in panels B and E are summarized from three independent biological replicates. Download FIG S9, PDF file, 0.1 MB (115.9KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Trypanosomatid flagellates proved instrumental in dissecting numerous features of the eukaryotic cell, such as the replication of mitochondrial DNA, RNA editing, polycistronic transcription, and trans-splicing, to name just the best-known examples (1). By turning the easily and affordably cultivable N. esmeraldas into a genetically tractable system, we extend its potential to addressing important questions related to endosymbiosis.
There are two endosymbiotic system known in trypanosomatids. The more extensively studied endosymbiosis is the one in the subfamily Strigomonadinae that coevolved for a long time with “Ca. Kinetoplastibacterium” This has significantly influenced several morphological features of their hosts, such as the packaging of kinetoplast DNA, the length of the paraflagellar rod, and the extent to which the mitochondrion penetrates the layer of subpellicular microtubules (8, 18, 69). These endosymbionts have a reduced peptidoglycan layer, coordinate their division with that of the host, and are transmitted vertically (70). The presence of these bacteria makes their hosts less fastidious with regard to nutrient intake, relieving the requirement for heme, several amino acids, and some vitamins, which are synthesized by the bacterium (18, 19). The observed close association of endosymbionts with the host’s glycosomes suggests that bacteria may fuel the energy metabolism of the host (71). While the acquisition of bacteria by the ancestor of Strigomonadinae was considered a singular event in the family Trypanosomatidae (72), the N. esmeraldas-“Ca. Pandoraea novymonadis” system (9) revealed that such a relationship is not exclusive. Less extensive genome reduction and the presence of a complete TCA cycle and amino acid synthesis pathways in the bacterium, combined with a variable number of bacteria per host cell and the use of lysosomes presumably to control their growth, suggested a relatively recent origin of this endosymbiosis. While the genome of “Ca. Pandoraea novymonadis” has already passed the period of an intense adaptation to endosymbiosis, the relationships of this bacterium with its host are apparently not as fine-tuned as in the case of “Ca. Kinetoplastibacterium” (42, 73).
In this work, we provide a detailed characterization of the metabolic cooperation between N. esmeraldas and “Ca. Pandoraea novymonadis” (Fig. 1). The bacterium has apparently lost the ability to synthesize polyamines and six amino acids (versus 13 in “Ca. Kinetoplastibacterium”), whereas the synthesis of heme, phospholipids, and all vitamins is still preserved (42). In this respect, it is less auxotrophic than the endosymbionts of Strigomonadinae, which lost the ability to synthesize some vitamins and phospholipids (16, 74). In line with this, the aposymbiotic N. esmeraldas strain exhibited significant deceleration of growth in the RPMI medium, which was almost fully recovered in the medium supplemented with amino acids, vitamins, and nucleic acid bases (supplemented M199), confirming the nutritional role of the endosymbiont (42). In the Strigomonadinae-“Ca. Kinetoplastibacterium” system, the bacteria influence the ability of flagellates to colonize insect hosts (75), regulate the oxygen consumption (76), affect mitochondrial functionality (77), facilitate control of protein synthesis and folding (78), and impact the activity of metalloproteinases (79, 80). Compared to “Ca. Kinetoplastibacterium,” “Ca. Pandoraea novymonadis” appears to be more self-sufficient for energy generation (42). It also retains more genes involved in cell division, which does not seem to be directly controlled by the host (9).
Previously, we sequenced the genome of “Ca. Pandoraea novymonadis” and compared it with those of free-living Pandoraea spp. Despite a significant reduction in size and numerous gene losses, it preserves a number of main metabolic pathways, all essential for the trypanosomatid host (42). In contrast to “Ca. Pandoraea novymonadis,” the genome of N. esmeraldas, analyzed here for the first time, did not experience a substantial reduction and is very similar in size to those of available endosymbiont-lacking trypanosomatids. Indeed, despite the presence of the endosymbiont, almost all genes of the typical trypanosomatid core metabolism have been retained.
The carbohydrate metabolism of N. esmeraldas resembles that of L. major and other Leishmaniinae. A complete glycolytic pathway, with part of its enzymes enclosed inside glycosomes (81), is operational. Meanwhile, relying on its own phosphotransferase system to phosphorylate hexose sugars (42), the endosymbiont became dependent on two host enzymes of the pathway, phosphofructokinase and phosphoglycerate mutase, to compensate for the absence of its own corresponding genes (Fig. 1). The same applies to the HMP shunt, interconnecting the host and its endosymbiont. “Ca. Pandoraea novymonadis” lacks the first three enzymes (glucose-6-phosphate dehydrogenase, 6-phosphogluconolactonase, and 6-phosphogluconate dehydrogenase) of this pathway, whereas N. esmeraldas has lost one of the shunt’s interconverting enzymes, transketolase. Moreover, with the loss of the first part of the HMP shunt, the endosymbiont became dependent on its host for the production of NADPH, required for ROS protection, anabolic reactions, and pentose sugars for the synthesis of nucleotides. Novymonas esmeraldas is unable to synthesize its own purines, while its endosymbiont has retained this capacity, with all the genes of the purine de novo biosynthetic pathway present in its genome, thus rendering its host completely independent of external purines. Moreover, N. esmeraldas can synthetize pyrimidines using trypanosomatid-specific enzymes (46), while its endosymbiont relies on its own set of bacterial-type enzymes to produce these compounds.
The metabolism of amino acids in N. esmeraldas is very similar to that described for the other Leishmaniinae (44, 54, 82). Pro, Thr, and Glu serve as important energy substrates which, together with Asp, feed directly into the Krebs cycle, the components of which have been retained in both partner cells. Met is converted via 2-ketobutyrate to succinyl-CoA, and the branched-chain amino acids Ile and Val are oxidized in the mitochondrion to succinyl-CoA and acetyl-CoA. Similarly to the related trypanosomatids, N. esmeraldas lacks most of the classical pathway of aromatic amino acid oxidation. Some genes of the anaerobic Phe degradation are present, and aromatic amino acids may be converted to their corresponding aromatic carboxylic acids and alcohols via this pathway. All members of the subfamily Leishmaniinae are auxotrophic for Arg, His, Ile, Leu, Phe, Ser, Trp, Tyr, and Val (44), and the present study confirmed the same feature in N. esmeraldas. In order to compensate for this shortcoming, “Ca. Pandoraea novymonadis” has retained the ability to synthesize all these amino acids. Conversely, N. esmeraldas synthesizes six nonessential amino acids (Ala, Asn, Asp, Cys, Met, and Pro), which its endosymbiont is unable to produce (42).
In order to further dissect the intricately intertwined metabolisms of these endosymbiotic partners and analyze the likely complex targeting of bacterial and host proteins, we have turned N. esmeraldas into a novel model trypanosomatid amenable to genetic manipulations by either conventional or CRISPR-Cas9-mediated technology. So far, a similar toolbox developed for the unrelated Angomonas-“Ca. Kinetoplastibacterium” system was instrumental in tracking the intracellular trafficking of the bacterium-targeted protein (20). Hence, following P. chromatophora, Angomonas deanei was only the second protist model (and the first one amendable to genetic manipulations) with the capacity to document the emergence of a nonorganellar protein-import machinery.
The important differences documented between the Angomonas-“Ca. Kinetoplastibacterium” and Novymonas-“Ca. Pandoraea novymonadis” systems warrant further comparative analyses of their genomes, metabolic pathways, and relationships with bacterial endosymbionts, which will shed light on the evolution of endosymbiosis. The development, refinement, and application of new genetic modification methods will facilitate achieving these goals.
MATERIALS AND METHODS
Cultivation, DNA and RNA isolation, and sequencing.
Novymonas esmeraldas wild-type strain E262 and its aposymbiotic derivate E262-AZI (42) were cultivated in M199 (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 2 μg/ml hemin (Jena Bioscience, Jena, Germany), 10% heat-inactivated fetal bovine serum (FBS; BioSera Europe, Nuaillé, France), 2 μg/ml biopterin, 100 U/ml of penicillin, and 100 μg/ml of streptomycin (all from Life Technologies/Thermo Fisher Scientific, Carlsbad, CA, USA) at 23°C. Total genomic DNA isolation and sequencing for the E262 cells were reported previously (42). Total RNA was isolated from 5 × 107 cells in three biological replicates using the RNeasy minikit (Qiagen, Hilden, Germany). The cDNA libraries were made and sequenced with 100-nt paired-end reads on the Illumina HiSeq 2000 platform (Macrogen, Seoul, Republic of Korea) as described elsewhere (83).
For genetic manipulations, wild-type cells were cultured at 23°C in brain heart infusion medium (BHI; VWR, Radnor, PA, USA), supplemented with 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 10% heat-inactivated FBS.
Genome assembly and annotation.
DNA sequencing reads were processed using the BBtools package, v. 36.021 (84). The reads were merged and quality-trimmed using BBmerge with the quality threshold of 20. Nonmerged reads were quality-trimmed using BBduk with the same parameters. The quality of raw and trimmed reads was assessed using FASTQC v.0.11.52 (85). The genome assembly was performed using the SPAdes genome assembler, v. 3.9.0, with default options (86), resulting in 1,429 scaffolds with N50 of 197,811 nt. The endosymbiont genome (consisting of 6 scaffolds; GenBank accession number MUHY00000000) was extracted from the assembly. The scaffolds shorter than 200 nt and/or with coverage less than 5 were filtered out. The resulting genome assembly contained 1,423 scaffolds with a total length of 32 Mbp.
Prior to assembly, transcriptome sequencing (RNA-seq) reads were subjected to adapter and quality trimming using Trimmomatic v. 0.32 (87) with the following parameters: illuminaclip: TruSeq3-PE-2.fa:2:20:10:8:true; leading: 3; trailing: 3; slidingwindow: 4:15; minlen: 75. All other parameters were left at the default settings. The reads were mapped on the genome using Bowtie 2 (88) with the “–very-sensitive” preset. The transcripts were predicted using StringTie (89) with default parameters. The genome was annotated with Companion (90) using the predicted transcripts, resulting in 8,638 predicted genes and 9,299 predicted proteins. Its completeness was assessed using BUSCO v. 4.1.4 (91).
Differential gene expression analysis.
The transcriptomes of the endosymbiont-free and wild-type N. esmeraldas strains (three independent biological replicates each) were sequenced using the Illumina NovaSeq platform, yielding ∼31 million 150-nt paired-end reads for each of the replicates. The raw reads were adapter and quality trimmed using Trimmomatic v. 0.39 with the following options: TruSeq3-PE-2.fa:2:20: 10; leading: 3; trailing: 3; slidingwindow: 4:15; minlen: 75 (87). The quality control before and after trimming was performed with FastQC v. 0.11.9 (85). Bowtie2 v. 2.4.1 (88) with the “–very-sensitive” option and other parameters left on the default settings was employed for read mapping, with the subsequent read sorting performed using SAMtools v. 1.1 (92). The mapped read counts for each gene were obtained using the featureCounts program with default settings (93). The differential expression analysis was performed using the DESeq2 R package (94). The genes showing over 1.5-fold expression change between E262 and E262-AZI cells with Benjamini-Hochberg-adjusted P values below 0.001 were selected for further analyses. Read counts were log-normalized with the DESeq2 rlog function and used for producing a heat map for 50 genes with the highest variance in expression according to the DESeq2 vignette instructions (94).
Gene ontology (GO) terms were assigned to N. esmeraldas proteins using PANNZER2 with a PPV threshold of 0.5 (95). GO term enrichment analysis was performed with the topGO R package, v. 2.42.0 (96). Additional functional annotation of differentially expressed genes was performed using BlastKOALA (97). Mitochondrial targeting sequence prediction was carried out with the TargetP-2.0 server (98).
Phylogenomic and metabolic analyses.
For phylogenomic data set construction, annotated proteins of the kinetoplastid species from Table S1S were clustered using OrthoFinder v. 1.6.12 (99) with default settings. The orthologous groups (OGs) containing proteins encoded by single-copy genes (1,175 OGs in total) were selected for further analysis. The respective protein sequences were aligned using MAFFT v. 7.402 with L-INS-i algorithm (100). At this step, the alignments with <65% average identity were filtered out, resulting in the decrease of the total number of proteins to 359. The remaining alignments were subsequently trimmed using trimAL v. 1.2rev59 (101) with the -strict option and concatenated into a supermatrix of 120,063 amino acid positions used for phylogenomic inferences. A maximum-likelihood phylogenomic tree was inferred with IQ-TREE software v. 1.6.12 (102), automatically selected LG + F + I + G4 model and 1,000 standard bootstrap replicates. A Bayesian tree was constructed using PhyloBayes-MPI v. 1.8 (103) with the LG + CAT model, four discrete Γ rate categories, and removal of invariant sites. Two independent chains ran for 30,000 generations, resulting in good mixing and convergence of all parameters (maxdiff < 0.1 and effective size > 300, estimated after discarding 20% burn-in). The tree topology shown in Fig. 3 became invariant after the first 15 iterations. It was visualized in Mega X v. 10.1.5 (104).
Metabolic pathways were analyzed as in references 44 and 105, using “all against all” BLASTp searches with an E value cutoff 10−20. This E value was chosen to discriminate between truly orthologous proteins and more distant homologues, which are not necessarily functional orthologues.
Genetic manipulations by homologous recombination.
For transfection, E262 cells were grown to a density of 2 × 107 cells/ml. In total, 5 × 108 cells were pelleted at 1,000 × g for 15 min and 4°C. Six to eight micrograms of linearized DNA construct was electroporated as described previously with three pulses of 25 μF at 1,500 V (3.75 kV/cm) with a 10-s pause between them (106). Cells were incubated in supplemented BHI without antibiotic for 16 h at 23°C and then under antibiotic selection for 2 to 4 weeks.
To generate a line of N. esmeraldas expressing mCherry, this gene was PCR amplified from the p2686 plasmid (107) using the primers mCherry_BglII_F and mCherry_NotI_R (all primers are listed in Table S1T) and cloned into the pF4X1.4neo vector (Jena Bioscience). The resultant plasmid was linearized with SwaI, gel purified, and used for transfection. The transformants were selected in liquid BHI medium supplemented with 200 μg/ml of neomycin (VWR) for 3 weeks.
Genetic manipulations by CRISPR/Cas9.
The same strategy, previously employed for L. mexicana (65), was adapted for N. esmeraldas. The trypanosomatids were transfected with 7 μg of the linearized Cas9/pLEXSYhyg2 plasmid, and the resulting populations were selected in liquid BHI medium supplemented with 200 μg/ml hygromycin. The integration was checked by PCR with the primers SSU_f and Cas9_13_r. Expression of Cas9 was monitored by Western blotting and RT-qPCR with the primers Cas9_10_f and Cas9_5_r. One population with the highest expression of Cas9 was chosen for subsequent experiments.
The U6 promoter, single guide RNA (sgRNA) with trans-activating CRISPR RNA (tracrRNA), and the U6 terminator were amplified using the following primers: PF16KO_Seed_f/Novy_U6_prom_r, Novy _U6_scaffold_f/PF16KO_Seed_r, Novy_U6_term_f/Novy_U6_scaffold_r, respectively. Three fragments were fused using the primers Novy_U6_nested_f and Novy_U6_nested_r and cloned into pLEXSY-neo2.1 (Jena Bioscience). The phleomycin resistance gene was amplified using the primers Spe-Phleo_f and BamHI-Phleo_r and cloned into pLS6-PFR2 (108). The donor DNA sequence was amplified from this plasmid using the primers Donor_PF16KO_f and Donor_PF16KO_r with 30-bp sequences of PF16 flanking the double-strand DNA break site. The linearized constructs containing the sgRNA under U6 and the PCR product of donor DNA were transfected into N. esmeraldas_Cas9 in two transfections, performed one by one. Populations were selected on liquid BHI medium supplemented with an additional 150 μg/ml hygromycin, 200 μg/ml neomycin, and 650 μg/ml phleomycin. Deletion of PF16 was confirmed by PCR with the primers PF16KO_int_check_f and PF16KO_int_check_r (expected fragment sizes for the wild type [WT] and mutant are 311 and 1,336 bp, respectively), by RT-qPCR with the primers PF16_qPCR_f and PF16_qPCR_r and by Southern blotting with probes for PF16 and the phleomycin resistance gene. The bacterial load was analyzed by qPCR with the primers GyrA_Pnovy_F and GyrA_Pnovy_R for the bacterial gyrase gene and HKG11_Novy_f and HKG11_Novy_r for a housekeeping gene (KMP11) used for normalization.
Fluorescence microscopy.
Parasites were washed with 1× phosphate-buffered saline (PBS; VWR) and mixed with 40% glycerol solution (vol/vol) in 1× PBS on poly-l-Lys-coated coverslips (Thermo Fisher Scientific). Slides were stained with SYTO 24 green fluorescent nucleic acid stain (Thermo Fisher Scientific), and cells were observed with an Olympus BX-53 microscope (Olympus, Tokyo, Japan) equipped with an Olympus DP73 digital camera. Images were taken with Olympus cellSens v.1.6 software, and merging was done in ImageJ v.1.51n (109).
Reverse transcription-quantitative PCR.
Cas9 expression was measured by RT-qPCR in the LightCycler480 (Roche Life Science, Penzberg, Germany) as described previously (110, 111) using the following primer pairs: 18S_for/18S_rev, Cas9_10_f/Cas9_5_r, HKG11_Novy_f/HKG11_Novy_r, and PF16_qPCR_f/PF16_qPCR_r. All experiments were performed in biological and technical triplicates, using 18S rRNA (18S) and kinetoplast membrane protein-11 (KMP11) expression for normalization.
DNA preparation and Southern blot analysis.
Probes for PF16 and phleomycin resistance gene were amplified with primers PF16_SB-probe_f/PF16_SB-probe_r and Phleo_SB-probe_f/Phleo_SB-probe_r, respectively, using a PCR digoxigenin (DIG) probe synthesis kit (Roche Life Science). For Southern blotting, 20 μg of DNA was digested with XhoI, separated on a 0.75% agarose gel, and probed as described previously (65, 112).
Data availability.
The whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JAECZO000000000. The version described in this paper is version JAECZO010000000. The data set used for phylogenomic inferences is available at https://figshare.com/articles/dataset/Novymonas_esmeraldas_phylogenomic_dataset/14618193.
ACKNOWLEDGMENTS
We thank members of our laboratories for stimulating discussions.
This work was supported by the Grant Agency of Czech Republic (20-07186S and 21-09283S) and the European Regional Funds (CZ.02.1.01/16_019/0000759) to V.Y. and J.L., grant SGS/PrF/2021 from the University of Ostrava and Moravskoslezský kraj research initiative (RRC/10/2019) to A.Z., State Assignment АААА-А19-119031390116-9 for ZIN RAS to A.Y.K., and ERC CZ (LL1601) to J.L. F.R.O. was supported by a grant from the de Duve Institute. A part of this work on comparative analysis of Novymonas esmeraldas and Leishmania spp. was funded by the Russian Science Foundation (grant 19-15-00054) to V.Y. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare no conflict of interest.
Footnotes
This article is a direct contribution from Julius Lukeš, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Eva Nowack, Heinrich Heine University Duesseldorf, and Sergio Munoz Gomez, Universite Paris-Saclay.
Citation Zakharova A, Saura A, Butenko A, Podešvová L, Warmusová S, Kostygov AY, Nenarokova A, Lukeš J, Opperdoes FR, Yurchenko V. 2021. A new model trypanosomatid, Novymonas esmeraldas: genomic perception of its “Candidatus Pandoraea novymonadis” endosymbiont. mBio 12:e01606-21. https://doi.org/10.1128/mBio.01606-21.
Contributor Information
Vyacheslav Yurchenko, Email: vyacheslav.yurchenko@osu.cz.
L. David Sibley, Washington University School of Medicine.
REFERENCES
- 1.Maslov DA, Opperdoes FR, Kostygov AY, Hashimi H, Lukeš J, Yurchenko V. 2019. Recent advances in trypanosomatid research: genome organization, expression, metabolism, taxonomy and evolution. Parasitology 146:1–27. doi: 10.1017/S0031182018000951. [DOI] [PubMed] [Google Scholar]
- 2.Lukeš J, Butenko A, Hashimi H, Maslov DA, Votýpka J, Yurchenko V. 2018. Trypanosomatids are much more than just trypanosomes: clues from the expanded family tree. Trends Parasitol 34:466–480. doi: 10.1016/j.pt.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Maslov DA, Votýpka J, Yurchenko V, Lukeš J. 2013. Diversity and phylogeny of insect trypanosomatids: all that is hidden shall be revealed. Trends Parasitol 29:43–52. doi: 10.1016/j.pt.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Lukeš J, Skalický T, Týč J, Votýpka J, Yurchenko V. 2014. Evolution of parasitism in kinetoplastid flagellates. Mol Biochem Parasitol 195:115–122. doi: 10.1016/j.molbiopara.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Butenko A, Hammond M, Field MC, Ginger ML, Yurchenko V, Lukeš J. 2021. Reductionist pathways for parasitism in euglenozoans? Expanded datasets provide new insights. Trends Parasitol 37:100–116. doi: 10.1016/j.pt.2020.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Kostygov AY, Karnkowska A, Votýpka J, Tashyreva D, Maciszewski K, Yurchenko V, Lukeš J. 2021. Euglenozoa: taxonomy, diversity and ecology, symbioses and viruses. Open Biol 11:200407. doi: 10.1098/rsob.200407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Souza W, Motta MC. 1999. Endosymbiosis in protozoa of the Trypanosomatidae family. FEMS Microbiol Lett 173:1–8. doi: 10.1111/j.1574-6968.1999.tb13477.x. [DOI] [PubMed] [Google Scholar]
- 8.Votýpka J, Kostygov AY, Kraeva N, Grybchuk-Ieremenko A, Tesařová M, Grybchuk D, Lukeš J, Yurchenko V. 2014. Kentomonas gen. n., a new genus of endosymbiont-containing trypanosomatids of Strigomonadinae subfam. n. Protist 165:825–838. doi: 10.1016/j.protis.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Kostygov A, Dobáková E, Grybchuk-Ieremenko A, Váhala D, Maslov DA, Votýpka J, Lukeš J, Yurchenko V. 2016. Novel trypanosomatid-bacterium association: evolution of endosymbiosis in action. mBio 7:e01985-15. doi: 10.1128/mBio.01985-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganyukova AI, Frolov AO, Malysheva MN, Spodareva VV, Yurchenko V, Kostygov AY. 2020. A novel endosymbiont-containing trypanosomatid Phytomonas borealis sp. n. from the predatory bug Picromerus bidens (Heteroptera: Pentatomidae). Folia Parasitol 67. doi: 10.14411/fp.2020.004 [DOI] [PubMed] [Google Scholar]
- 11.Catta-Preta CM, Nascimento MT, Garcia MC, Saraiva EM, Motta MC, Meyer-Fernandes JR. 2013. The presence of a symbiotic bacterium in Strigomonas culicis is related to differential ecto-phosphatase activity and influences the mosquito-protozoa interaction. Int J Parasitol 43:571–577. doi: 10.1016/j.ijpara.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Motta MC, Catta-Preta CM, Schenkman S, de Azevedo Martins AC, Miranda K, de Souza W, Elias MC. 2010. The bacterium endosymbiont of Crithidia deanei undergoes coordinated division with the host cell nucleus. PLoS One 5:e12415. doi: 10.1371/journal.pone.0012415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freymuller E, Camargo EP. 1981. Ultrastructural differences between species of trypanosomatids with and without endosymbionts. J Protozool 28:175–182. doi: 10.1111/j.1550-7408.1981.tb02829.x. [DOI] [PubMed] [Google Scholar]
- 14.Chang KP. 1974. Ultrastructure of symbiotic bacteria in normal and antibiotic-treated Blastocrithidia culicis and Crithidia oncopelti. J Protozool 21:699–707. doi: 10.1111/j.1550-7408.1974.tb03733.x. [DOI] [PubMed] [Google Scholar]
- 15.Motta MC, Soares MJ, Attias M, Morgado J, Lemos AP, Saad-Nehme J, Meyer-Fernandes JR, De Souza W. 1997. Ultrastructural and biochemical analysis of the relationship of Crithidia deanei with its endosymbiont. Eur J Cell Biol 72:370–377. [PubMed] [Google Scholar]
- 16.Klein CC, Alves JM, Serrano MG, Buck GA, Vasconcelos AT, Sagot MF, Teixeira MM, Camargo EP, Motta MC. 2013. Biosynthesis of vitamins and cofactors in bacterium-harbouring trypanosomatids depends on the symbiotic association as revealed by genomic analyses. PLoS One 8:e79786. doi: 10.1371/journal.pone.0079786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alves JM, Serrano MG, Maia da Silva F, Voegtly LJ, Matveyev AV, Teixeira MM, Camargo EP, Buck GA. 2013. Genome evolution and phylogenomic analysis of Candidatus Kinetoplastibacterium, the beta-proteobacterial endosymbionts of Strigomonas and Angomonas. Genome Biol Evol 5:338–350. doi: 10.1093/gbe/evt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alves JM, Klein CC, da Silva FM, Costa-Martins AG, Serrano MG, Buck GA, Vasconcelos AT, Sagot MF, Teixeira MM, Motta MC, Camargo EP. 2013. Endosymbiosis in trypanosomatids: the genomic cooperation between bacterium and host in the synthesis of essential amino acids is heavily influenced by multiple horizontal gene transfers. BMC Evol Biol 13:190. doi: 10.1186/1471-2148-13-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alves JMP. 2017. Working together: amino acid biosynthesis in endosymbiont-harbouring Trypanosomatidae, p 371–383. In D'Mello JPF (ed), The handbook of microbial metabolism of amino acids. CAB International, Wallingford, UK Boston, USA. [Google Scholar]
- 20.Morales J, Kokkori S, Weidauer D, Chapman J, Goltsman E, Rokhsar D, Grossman AR, Nowack EC. 2016. Development of a toolbox to dissect host-endosymbiont interactions and protein trafficking in the trypanosomatid Angomonas deanei. BMC Evol Biol 16:247. doi: 10.1186/s12862-016-0820-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Embley TM, Martin W. 2006. Eukaryotic evolution, changes and challenges. Nature 440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 22.Keeling PJ, Koonin EV. 2014. The origin and evolution of eukaryotes: a subject collection from Cold Spring Harbor Perspectives in Biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 24.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubilier N, Bergin C, Lott C. 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol 6:725–740. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- 26.McCutcheon JP, Moran NA. 2011. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 27.Müller DB, Vogel C, Bai Y, Vorholt JA. 2016. The plant microbiota: systems-level insights and perspectives. Annu Rev Genet 50:211–234. doi: 10.1146/annurev-genet-120215-034952. [DOI] [PubMed] [Google Scholar]
- 28.Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cárdenas P, Čepička I, Chistyakova L, Del Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karnkowska A, Karpov S, Kim E, Kolisko M, Kudryavtsev A, Lahr DJG, Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, Mitchell EAD, Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Spiegel FW, Torruella G, Youssef N, Zlatogursky V, Zhang Q. 2019. Revisions to the classification, nomenclature, and diversity of eukaryotes. J Eukaryot Microbiol 66:4–119. doi: 10.1111/jeu.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burki F, Roger AJ, Brown MW, Simpson AGB. 2020. The new tree of eukaryotes. Trends Ecol Evol 35:43–55. doi: 10.1016/j.tree.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Nowack EC, Price DC, Bhattacharya D, Singer A, Melkonian M, Grossman AR. 2016. Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora. Proc Natl Acad Sci USA 113:12214–12219. doi: 10.1073/pnas.1608016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer A, Poschmann G, Muhlich C, Valadez-Cano C, Hansch S, Huren V, Rensing SA, Stuhler K, Nowack ECM. 2017. Massive protein import into the early-evolutionary-stage photosynthetic organelle of the amoeba Paulinella chromatophora. Curr Biol 27:2763–2773.e5. doi: 10.1016/j.cub.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Kraeva N, Butenko A, Hlaváčová J, Kostygov A, Myškova J, Grybchuk D, Leštinová T, Votýpka J, Volf P, Opperdoes F, Flegontov P, Lukeš J, Yurchenko V. 2015. Leptomonas seymouri: adaptations to the dixenous life cycle analyzed by genome sequencing, transcriptome profiling and co-infection with Leishmania donovani. PLoS Pathog 11:e1005127. doi: 10.1371/journal.ppat.1005127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coburn CM, Otteman KM, McNeely T, Turco SJ, Beverley SM. 1991. Stable DNA transfection of a wide range of trypanosomatids. Mol Biochem Parasitol 46:169–179. doi: 10.1016/0166-6851(91)90210-w. [DOI] [PubMed] [Google Scholar]
- 34.Miranda MR, Saye M, Reigada C, Carrillo C, Pereira CA. 2015. Phytomonas: a non-pathogenic trypanosomatid model for functional expression of proteins. Protein Expr Purif 114:44–47. doi: 10.1016/j.pep.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 35.Sloan MA, Ligoxygakis P. 2020. Tools for the genetic manipulation of Herpetomonas muscarum. G3 (Bethesda) 10:1613–1616. doi: 10.1534/g3.120.401048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q, Lei J, Kadowaki T. 2019. Gene disruption of honey bee trypanosomatid parasite, Lotmaria passim, by CRISPR/Cas9 system. Front Cell Infect Microbiol 9:126. doi: 10.3389/fcimb.2019.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jirků M, Yurchenko V, Lukeš J, Maslov DA. 2012. New species of insect trypanosomatids from Costa Rica and the proposal for a new subfamily within the Trypanosomatidae. J Eukaryot Microbiol 59:537–547. doi: 10.1111/j.1550-7408.2012.00636.x. [DOI] [PubMed] [Google Scholar]
- 38.Kostygov AY, Yurchenko V. 2017. Revised classification of the subfamily Leishmaniinae (Trypanosomatidae). Folia Parasitol 64:20. doi: 10.14411/fp.2017.020. [DOI] [PubMed] [Google Scholar]
- 39.Olivier M, Atayde VD, Isnard A, Hassani K, Shio MT. 2012. Leishmania virulence factors: focus on the metalloprotease GP63. Microbes Infect 14:1377–1389. doi: 10.1016/j.micinf.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Jain M, Dole VS, Myler PJ, Stuart KD, Madhubala R. 2007. Role of biopterin transporter (BT1) gene on growth and infectivity of Leishmania. Am J Biochem Biotechnol 3:199–206. doi: 10.3844/ajbbsp.2007.199.206. [DOI] [Google Scholar]
- 41.Naderer T, Heng J, McConville MJ. 2010. Evidence that intracellular stages of Leishmania major utilize amino sugars as a major carbon source. PLoS Pathog 6:e1001245. doi: 10.1371/journal.ppat.1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kostygov AY, Butenko A, Nenarokova A, Tashyreva D, Flegontov P, Lukeš J, Yurchenko V. 2017. Genome of “Ca. Pandoraea novymonadis,” an endosymbiotic bacterium of the trypanosomatid Novymonas esmeraldas. Front Microbiol 8:1940. doi: 10.3389/fmicb.2017.01940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fish WR, Looker DL, Marr JJ, Berens RL. 1982. Purine metabolism in the bloodstream forms of Trypanosoma gambiense and Trypanosoma rhodesiense. Biochim Biophys Acta 719:223–231. doi: 10.1016/0304-4165(82)90092-7. [DOI] [PubMed] [Google Scholar]
- 44.Opperdoes FR, Butenko A, Flegontov P, Yurchenko V, Lukeš J. 2016. Comparative metabolism of free-living Bodo saltans and parasitic trypanosomatids. J Eukaryot Microbiol 63:657–678. doi: 10.1111/jeu.12315. [DOI] [PubMed] [Google Scholar]
- 45.Nara T, Aoki T. 2002. The pyrimidine-biosynthetic (pyr) gene cluster in trypanosomes. Tanpakushitsu Kakusan Koso 47:13–20. [PubMed] [Google Scholar]
- 46.Opperdoes FR, Michels PA. 2007. Horizontal gene transfer in trypanosomatids. Trends Parasitol 23:470–476. doi: 10.1016/j.pt.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Čermáková P, Maďarová A, Baráth P, Bellová J, Yurchenko V, Horváth A. 2021. Differences in mitochondrial NADH dehydrogenase activities in trypanosomatids. Parasitology 148:1161–1170. doi: 10.1017/S0031182020002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puustinen A, Finel M, Haltia T, Gennis RB, Wikström M. 1991. Properties of the two terminal oxidases of Escherichia coli. Biochemistry 30:3936–3942. doi: 10.1021/bi00230a019. [DOI] [PubMed] [Google Scholar]
- 49.Motta MC, Martins AC, de Souza SS, Catta-Preta CM, Silva R, Klein CC, de Almeida LG, de Lima Cunha O, Ciapina LP, Brocchi M, Colabardini AC, de Araujo Lima B, Machado CR, de Almeida Soares CM, Probst CM, de Menezes CB, Thompson CE, Bartholomeu DC, Gradia DF, Pavoni DP, Grisard EC, Fantinatti-Garboggini F, Marchini FK, Rodrigues-Luiz GF, Wagner G, Goldman GH, Fietto JL, Elias MC, Goldman MH, Sagot MF, Pereira M, Stoco PH, de Mendonca-Neto RP, Teixeira SM, Maciel TE, de Oliveira Mendes TA, Urmenyi TP, de Souza W, Schenkman S, de Vasconcelos AT. 2013. Predicting the proteins of Angomonas deanei, Strigomonas culicis and their respective endosymbionts reveals new aspects of the trypanosomatidae family. PLoS One 8:e60209. doi: 10.1371/journal.pone.0060209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muñoz-Gómez SA, Slamovits CH, Dacks JB, Baier KA, Spencer KD, Wideman JG. 2015. Ancient homology of the mitochondrial contact site and cristae organizing system points to an endosymbiotic origin of mitochondrial cristae. Curr Biol 25:1489–1495. doi: 10.1016/j.cub.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Kaurov I, Vancová M, Schimanski B, Cadena LR, Heller J, Bíly T, Potěšíl D, Eichenberger C, Bruce H, Oeljeklaus S, Warscheid B, Zdráhal Z, Schneider A, Lukeš J, Hashimi H. 2018. The diverged trypanosome MICOS complex as a hub for mitochondrial cristae shaping and protein import. Curr Biol 28:3393–3407.e5. doi: 10.1016/j.cub.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Flegontov P, Butenko A, Firsov S, Kraeva N, Eliáš M, Field MC, Filatov D, Flegontova O, Gerasimov ES, Hlaváčová J, Ishemgulova A, Jackson AP, Kelly S, Kostygov A, Logacheva MD, Maslov DA, Opperdoes FR, O'Reilly A, Sádlová J, Ševčíková T, Venkatesh D, Vlček Č, Volf P, Votýpka J, Záhonová K, Yurchenko V, Lukeš J. 2016. Genome of Leptomonas pyrrhocoris: a high-quality reference for monoxenous trypanosomatids and new insights into evolution of Leishmania. Sci Rep 6:23704. doi: 10.1038/srep23704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cunningham ML, Beverley SM. 2001. Pteridine salvage throughout the Leishmania infectious cycle: implications for antifolate chemotherapy. Mol Biochem Parasitol 113:199–213. doi: 10.1016/S0166-6851(01)00213-4. [DOI] [PubMed] [Google Scholar]
- 54.Opperdoes FR, Coombs GH. 2007. Metabolism of Leishmania: proven and predicted. Trends Parasitol 23:149–158. doi: 10.1016/j.pt.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Kořený L, Lukeš J, Oborník M. 2010. Evolution of the haem synthetic pathway in kinetoplastid flagellates: an essential pathway that is not essential after all? Int J Parasitol 40:149–156. doi: 10.1016/j.ijpara.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Lee SH, Stephens JL, Englund PT. 2007. A fatty-acid synthesis mechanism specialized for parasitism. Nat Rev Microbiol 5:287–297. doi: 10.1038/nrmicro1617. [DOI] [PubMed] [Google Scholar]
- 57.Uttaro AD. 2014. Acquisition and biosynthesis of saturated and unsaturated fatty acids by trypanosomatids. Mol Biochem Parasitol 196:61–70. doi: 10.1016/j.molbiopara.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 58.Kraeva N, Horáková E, Kostygov A, Kořený L, Butenko A, Yurchenko V, Lukeš J. 2017. Catalase in Leishmaniinae: with me or against me? Infect Genet Evol 50:121–127. doi: 10.1016/j.meegid.2016.06.054. [DOI] [PubMed] [Google Scholar]
- 59.Škodová-Sveráková I, Záhonová K, Bučková B, Füssy Z, Yurchenko V, Lukeš J. 2020. Catalase and ascorbate peroxidase in euglenozoan protists. Pathogens 9:317. doi: 10.3390/pathogens9040317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bombaça ACS, Brunoro GVF, Dias-Lopes G, Ennes-Vidal V, Carvalho PC, Perales J, d'Avila-Levy CM, Valente RH, Menna-Barreto RFS. 2020. Glycolytic profile shift and antioxidant triggering in symbiont-free and H2O2-resistant Strigomonas culicis. Free Radic Biol Med 146:392–401. doi: 10.1016/j.freeradbiomed.2019.11.025. [DOI] [PubMed] [Google Scholar]
- 61.Chevalier N, Bertrand L, Rider MH, Opperdoes FR, Rigden DJ, Michels PA. 2005. 6-Phosphofructo-2-kinase and fructose-2,6-bisphosphatase in Trypanosomatidae. Molecular characterization, database searches, modelling studies and evolutionary analysis. FEBS J 272:3542–3560. doi: 10.1111/j.1742-4658.2005.04774.x. [DOI] [PubMed] [Google Scholar]
- 62.Penha LL, Hoffmann L, Souza SS, Martins AC, Bottaro T, Prosdocimi F, Faffe DS, Motta MC, Ürményi TP, Silva R. 2016. Symbiont modulates expression of specific gene categories in Angomonas deanei. Mem Inst Oswaldo Cruz 111:686–691. doi: 10.1590/0074-02760160228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Randow F, Youle RJ. 2014. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe 15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Durante IM, Butenko A, Rašková V, Charyyeva A, Svobodová M, Yurchenko V, Hashimi H, Lukeš J. 2020. Large-scale phylogenetic analysis of trypanosomatid adenylate cyclases reveals associations with extracellular lifestyle and host-pathogen interplay. Genome Biol Evol 12:2403–2416. doi: 10.1093/gbe/evaa226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishemgulova A, Hlaváčová J, Majerová K, Butenko A, Lukeš J, Votýpka J, Volf P, Yurchenko V. 2018. CRISPR/Cas9 in Leishmania mexicana: a case study of LmxBTN1. PLoS One 13:e0192723. doi: 10.1371/journal.pone.0192723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loreng TD, Smith EF. 2017. The central apparatus of cilia and eukaryotic flagella. Cold Spring Harb Perspect Biol 9:a028118. doi: 10.1101/cshperspect.a028118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ralston KS, Lerner AG, Diener DR, Hill KL. 2006. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot Cell 5:696–711. doi: 10.1128/EC.5.4.696-711.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beneke T, Madden R, Makin L, Valli J, Sunter J, Gluenz E. 2017. A CRISPR Cas9 high-throughput genome editing toolkit for kinetoplastids. R Soc Open Sci 4:170095. doi: 10.1098/rsos.170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yurchenko V, Lukeš J. 2018. Parasites and their (endo)symbiotic microbes. Parasitology 145:1261–1264. doi: 10.1017/S0031182018001257. [DOI] [PubMed] [Google Scholar]
- 70.Catta-Preta CM, Brum FL, da Silva CC, Zuma AA, Elias MC, de Souza W, Schenkman S, Motta MC. 2015. Endosymbiosis in trypanosomatid protozoa: the bacterium division is controlled during the host cell cycle. Front Microbiol 6:520. doi: 10.3389/fmicb.2015.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loyola-Machado AC, Azevedo-Martins AC, Catta-Preta CMC, de Souza W, Galina A, Motta MCM. 2017. The symbiotic bacterium fuels the energy metabolism of the host trypanosomatid Strigomonas culicis. Protist 168:253–269. doi: 10.1016/j.protis.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 72.Hollar L, Lukeš J, Maslov DA. 1998. Monophyly of endosymbiont containing trypanosomatids: phylogeny versus taxonomy. J Eukaryot Microbiol 45:293–297. doi: 10.1111/j.1550-7408.1998.tb04539.x. [DOI] [PubMed] [Google Scholar]
- 73.Harmer J, Yurchenko V, Nenarokova A, Lukeš J, Ginger ML. 2018. Farming, slaving and enslavement: histories of endosymbiosis during kinetoplastid evolution. Parasitology 145:1311–1323. doi: 10.1017/S0031182018000781. [DOI] [PubMed] [Google Scholar]
- 74.Silva FM, Kostygov AY, Spodareva VV, Butenko A, Tossou R, Lukes J, Yurchenko V, Alves JMP. 2018. The reduced genome of Candidatus Kinetoplastibacterium sorsogonicusi, the endosymbiont of Kentomonas sorsogonicus (Trypanosomatidae): loss of the haem-synthesis pathway. Parasitology 145:1287–1293. doi: 10.1017/S003118201800046X. [DOI] [PubMed] [Google Scholar]
- 75.Fampa P, Correa-da-Silva MS, Lima DC, Oliveira SM, Motta MC, Saraiva EM. 2003. Interaction of insect trypanosomatids with mosquitoes, sand fly and the respective insect cell lines. Int J Parasitol 33:1019–1026. doi: 10.1016/s0020-7519(03)00124-3. [DOI] [PubMed] [Google Scholar]
- 76.de Azevedo-Martins AC, Alves JM, de Mello FG, Vasconcelos AT, de Souza W, Einicker-Lamas M, Motta MC. 2015. Biochemical and phylogenetic analyses of phosphatidylinositol production in Angomonas deanei, an endosymbiont-harboring trypanosomatid. Parasit Vectors 8:247. doi: 10.1186/s13071-015-0854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bombaça ACS, Dias FA, Ennes-Vidal V, Garcia-Gomes ADS, Sorgine MHF, d'Avila-Levy CM, Menna-Barreto RFS. 2017. Hydrogen peroxide resistance in Strigomonas culicis: effects on mitochondrial functionality and Aedes aegypti interaction. Free Radic Biol Med 113:255–266. doi: 10.1016/j.freeradbiomed.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 78.Brunoro GVF, Menna-Barreto RFS, Garcia-Gomes AS, Boucinha C, Lima DB, Carvalho PC, Teixeira-Ferreira A, Trugilho MRO, Perales J, Schwammle V, Catanho M, de Vasconcelos ATR, Motta MCM, d'Avila-Levy CM, Valente RH. 2019. Quantitative proteomic map of the trypanosomatid Strigomonas culicis: the biological contribution of its endosymbiotic bacterium. Protist 170:125698. doi: 10.1016/j.protis.2019.125698. [DOI] [PubMed] [Google Scholar]
- 79.d’Avila-Levy CM, Santos LO, Marinho FA, Matteoli FP, Lopes AHCS, Motta MCM, Santos ALS, Branquinha MH. 2008. Crithidia deanei: influence of parasite gp63 homologue on the interaction of endosymbiont-harboring and aposymbiotic strains with Aedes aegypti midgut. Exp Parasitol 118:345–353. doi: 10.1016/j.exppara.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 80.d'Avila-Levy CM, Souza RF, Gomes RC, Vermelho AB, Branquinha MH. 2003. A metalloproteinase extracellularly released by Crithidia deanei. Can J Microbiol 49:625–632. doi: 10.1139/w03-081. [DOI] [PubMed] [Google Scholar]
- 81.Opperdoes FR, Borst P. 1977. Localization of nine glycolytic enzymes in a microbody-like organelle in Trypanosoma brucei: the glycosome. FEBS Lett 80:360–364. doi: 10.1016/0014-5793(77)80476-6. [DOI] [PubMed] [Google Scholar]
- 82.Opperdoes F, Michels PA. 2008. The metabolic repertoire of Leishmania and implications for drug discovery, p 123–158. In Myler P, Fasel N (ed), Leishmania: after the genome. Caister Academic Press, Norfolk, UK. [Google Scholar]
- 83.Ishemgulova A, Butenko A, Kortišová L, Boucinha C, Grybchuk-Ieremenko A, Morelli KA, Tesařová M, Kraeva N, Grybchuk D, Pánek T, Flegontov P, Lukeš J, Votýpka J, Pavan MG, Opperdoes FR, Spodareva V, d'Avila-Levy CM, Kostygov AY, Yurchenko V. 2017. Molecular mechanisms of thermal resistance of the insect trypanosomatid Crithidia thermophila. PLoS One 12:e0174165. doi: 10.1371/journal.pone.0174165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bushnell B, Rood J, Singer E. 2017. BBMerge—accurate paired shotgun read merging via overlap. PLoS One 12:e0185056. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andrews S. 2019. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 8 March 2021.
- 86.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Steinbiss S, Silva-Franco F, Brunk B, Foth B, Hertz-Fowler C, Berriman M, Otto TD. 2016. Companion: a web server for annotation and analysis of parasite genomes. Nucleic Acids Res 44:W29–W34. doi: 10.1093/nar/gkw292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol 35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 94.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Törönen P, Medlar A, Holm L. 2018. PANNZER2: a rapid functional annotation web server. Nucleic Acids Res 46:W84–W88. doi: 10.1093/nar/gky350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alexa A, Rahnenfuhrer J. 2020. topGO: enrichment analysis for Gene Ontology, v2.42.0.
- 97.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 98.Almagro Armenteros JJ, Salvatore M, Emanuelsson O, Winther O, von Heijne G, Elofsson A, Nielsen H. 2019. Detecting sequence signals in targeting peptides using deep learning. Life Sci Alliance 2:e201900429. doi: 10.26508/lsa.201900429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Capella-Gutiérrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol 62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- 104.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Butenko A, Kostygov AY, Sádlová J, Kleschenko Y, Bečvář T, Podešvová L, Macedo DH, Žihala D, Lukeš J, Bates PA, Volf P, Opperdoes FR, Yurchenko V. 2019. Comparative genomics of Leishmania (Mundinia). BMC Genomics 20:726. doi: 10.1186/s12864-019-6126-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Robinson KA, Beverley SM. 2003. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol 128:217–228. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 107.Kelly S, Reed J, Kramer S, Ellis L, Webb H, Sunter J, Salje J, Marinsek N, Gull K, Wickstead B, Carrington M. 2007. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol Biochem Parasitol 154:103–109. doi: 10.1016/j.molbiopara.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sollelis L, Ghorbal M, MacPherson CR, Martins RM, Kuk N, Crobu L, Bastien P, Scherf A, Lopez-Rubio JJ, Sterkers Y. 2015. First efficient CRISPR-Cas9-mediated genome editing in Leishmania parasites. Cell Microbiol 17:1405–1412. doi: 10.1111/cmi.12456. [DOI] [PubMed] [Google Scholar]
- 109.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Záhonová K, Hadariová L, Vacula R, Yurchenko V, Eliáš M, Krajčovič J, Vesteg M. 2014. A small portion of plastid transcripts is polyadenylated in the flagellate Euglena gracilis. FEBS Lett 588:783–788. doi: 10.1016/j.febslet.2014.01.034. [DOI] [PubMed] [Google Scholar]
- 111.Grybchuk D, Macedo DH, Kleschenko Y, Kraeva N, Lukashev AN, Bates PA, Kulich P, Leštinová T, Volf P, Kostygov AY, Yurchenko V. 2020. The first non-LRV RNA virus in Leishmania. Viruses 12:168. doi: 10.3390/v12020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ishemgulova A, Kraeva N, Hlaváčová J, Zimmer SL, Butenko A, Podešvová L, Leštinová T, Lukeš J, Kostygov A, Votýpka J, Volf P, Yurchenko V. 2017. A putative ATP/GTP binding protein affects Leishmania mexicana growth in insect vectors and vertebrate hosts. PLoS Negl Trop Dis 11:e0005782. doi: 10.1371/journal.pntd.0005782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analyses of Novymonas esmeraldas. (A) List of 486 metabolic enzymes of L. major and their orthologues in N. esmeraldas. (B) Novymonas esmeraldas metabolic enzymes not present in L. major. (C) Endosymbiont-specific proteins of “Ca. Pandoraea novymonadis.” (D) Proteins of N. esmeraldas with a putative glycosomal/peroxisomal targeting signal. (E) List of enzymes of N. esmeraldas involved in sugar metabolism. (F) Enzymes of gluconeogenesis. (G) Enzymes of the pyrimidine and purine biosynthesis and purine salvage. (H) Enzymes of the FoF1-ATP synthase and the mitochondrial electron transport chain and mitochondrial contact site and crista organization system (MICOS). (I) Enzymes of amino acid metabolism. (J) Enzymes of the urea cycle. (K) Enzymes involved in synthesis of vitamins and cofactors. (L) Enzymes involved in lipid metabolism. (M) Enzymes involved in phosphor-lipid metabolism. (N) Genes upregulated in the endosymbiont-free N. esmeraldas (ES) compared to the wild-type cells (WT). Criteria: BH-adjusted P value of <0.001 and fold change of ≥1.5. (O) Genes downregulated in the endosymbiont-free N. esmeraldas (ES) compared to the wild-type cells (WT). Criteria: BH-adjusted P value of <0.001 and fold change of ≥1.5. (P) Top 10 GO terms most significantly enriched among genes upregulated in aposymbiotic N. esmeraldas. The terms are sorted according to the Fisher's test P values; those significantly enriched are in bold (P < 0.01). Only GO terms of the three main categories (biological process, molecular function, and cellular component) are included. Annotated, all genes are annotated with the respective GO term in the whole analyzed genome; Significant, a number of genes in the list of the up-/down-regulated ones are annotated with the respective GO term; Expected, an estimate of the number of genes a node of size “Annotated” would have if the significant genes were to be randomly selected from the gene pool. (R) Top 10 GO terms most significantly enriched among genes downregulated in aposymbiotic N. esmeraldas. The terms are sorted according to the Fisher's test P values; those significantly enriched are in bold (P value < 0.01). Only GO terms of the three main categories (biological process, molecular function, and cellular component) are included. Annotated, a total number of genes annotated with the respective GO term in the whole analyzed genome; Significant, a number of genes in the list of the up-/down-regulated ones are annotated with the respective GO-term; Expected, an estimate of the number of genes a node of size “Annotated” would have if the significant genes were to be randomly selected from the gene pool. (S) Data sets used in phylogenomic analysis. (T) Primers used in this study. Download Table S1, XLSX file, 0.7 MB (705.4KB, xlsx) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A heat map for 50 genes with the highest variance across samples based on log-transformed expression values. Download FIG S1, PDF file, 0.08 MB (86.7KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subgraph representing the 5 most significant GO terms of the category “Molecular Function” (in rectangles) enriched in the group of genes upregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S2, PDF file, 0.02 MB (22.8KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subgraph representing the 5 most significant GO terms of the category “Biological Process” (in rectangles) enriched in the group of genes upregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S3, PDF file, 0.08 MB (80.1KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subgraph representing the 5 most significant GO terms of the category “Cellular component” (in rectangles) enriched in the group of genes upregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S4, PDF file, 0.02 MB (25.8KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subgraph representing the 5 most significant GO terms of the category “Molecular Function” (in rectangles) enriched in the group of genes downregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S5, PDF file, 0.03 MB (28.9KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subgraph representing the 5 most significant GO terms of the category “Biological Process” (in rectangles) enriched in the group of genes downregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S6, PDF file, 0.08 MB (87.6KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subgraph representing the 5 most significant GO terms of the category “Cellular component” (in rectangles) enriched in the group of genes downregulated in aposymbiotic N. esmeraldas. Rectangle color represents the relative significance (dark red, most significant). Download FIG S7, PDF file, 0.04 MB (39.7KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Live Novymonas esmeraldas cells expressing mCherry (fluorescence microscopy). Bright field, SYTO 24, mCherry and the merged image are presented in the corresponding columns. The mCherry-expressing and wild-type N. esmeraldas cells are shown in the upper and lower panels, respectively. Bar, 5 μm. Download FIG S8, PDF file, 0.05 MB (54.1KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Establishment of Cas9-expressing N. esmeraldas. (A) PCR confirmation of successful integration. Populations 1 to 4 are shown along with the wild type. (B) RT-qPCR of Cas9 expression in populations 1 to 4. Wild-type N. esmeraldas and previously established Cas9-expressing L. mexicana were used as negative and positive controls, respectively. (C) Western blotting confirmation of Cas9 expression using anti-FLAG antibodies. Labels are as in panel B. (D) Growth curves of WT and Cas9-expressing N. esmeraldas; (E) Bacterial load in WT and Cas9-expressing N. esmeraldas. Sizes of DNA (A) and protein (C) fragments are in kilobases and kilodaltons, respectively. Data in panels B and E are summarized from three independent biological replicates. Download FIG S9, PDF file, 0.1 MB (115.9KB, pdf) .
Copyright © 2021 Zakharova et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JAECZO000000000. The version described in this paper is version JAECZO010000000. The data set used for phylogenomic inferences is available at https://figshare.com/articles/dataset/Novymonas_esmeraldas_phylogenomic_dataset/14618193.