ABSTRACT
Marine phytoplankton and heterotrophic bacteria share a very close but usually changeable relationship. However, the ultimate fate of their unstable relationship on a long-term scale is unclear. Here, the relationship between Synechococcus and heterotrophic bacterial communities underwent a dramatic shift from antagonism to commensalism and eventually to mutualism during long-term cocultivation. The relationship change is attributed to the different (even opposite) effects of diverse bacterial members on Synechococcus and the ratio of beneficial to harmful bacteria. Different bacterial members also interact with each other (e.g., quorum-sensing communication, hostility, or mutual promotion) and drive a dynamic succession in the entire community structure that corresponds exactly to the shift in its relationship with Synechococcus. In the final mutualism stage, a self-sufficient nitrogen cycle, including nitrogen fixation, denitrification, and organic nitrogen degradation, contributed to the healthy survival of Synechococcus for 2 years without an exogenous nutrient supply. This natural selective trait of Synechococcus and heterotrophic bacteria toward mutualism under long-term coexistence provides a novel clue for understanding the ubiquity and competitive advantage of Synechococcus in global oceans.
KEYWORDS: Synechococcus, heterotrophic bacterial community, algae-bacteria interaction, mutualism, nitrogen cycle
INTRODUCTION
Phytoplankton (including cyanobacteria) and heterotrophic bacteria are closely related and are important regulators of the ocean ecosystem (1). As the dominant primary producers (phytoplankton) and drivers of biogeochemical cycles (bacteria) in the ocean (2, 3), they interact with each other and underpin most functions of marine ecological processes (4–6). The interactions between phytoplankton and heterotrophic bacteria are multifarious, spanning from mutualism (7) to antagonism (8) or from cooperation (9) to allelopathy (10). These interactions have recently been thoroughly examined in several excellent reviews (1, 11, 12). However, despite being scrutinized for at least half a century, the “black box” of the highly complex relationship between phytoplankton and heterotrophic bacteria is not well uncovered (12, 13).
Indeed, the relationship between phytoplankton and heterotrophic bacteria is usually unstable and can change under the influence of environmental and/or biological factors. For example, during algal blooms, the effect of the bacterial community on phytoplankton can shift from promoting algal growth or symbiosis with algae to antagonizing algae (14–17). In the long-term cocultivation of a heterotrophic bacterium, Tropicibacter sp., and Synechococcus sp. strain WH7803, the bacterial effect on Synechococcus shifted from weakly maintaining the growth of Synechococcus to promoting its high-density growth, which was likely driven by nutrient recycling (18). In contrast, a mutualist-to-parasite shift occurred in the cocultivation of phytoplankton and Roseobacter, which depended on the growth state of the phytoplankton and the concomitant changes in substances released from the bacterium (19, 20). Although these two cases, to a certain extent, reflect the complex and dynamic phytoplankton-bacterium interactions, they focused on a single bacterium’s effect on phytoplankton. In the natural environment, complex multispecies communities rather than a single bacterium interact with the phytoplankton. In this sense, it is more important to consider the role of bacterial communities, not just individual bacterial species, while investigating the relationship between phytoplankton and heterotrophic bacteria.
The most abundant constituents of phytoplankton are the picocyanobacteria, dominated by the genera Synechococcus and Prochlorococcus. Synechococcus is widely distributed in various marine environments, including coastal, pelagic, and polar oceans, etc. (21, 22). Despite their small size (diameter of <2 μm), over 40% of Synechococcus cells were found to be conjoint with other bacteria under in situ observation (23). Moreover, the metagenomic information collected during the Tara Oceans expedition indicated that the close relationship between heterotrophic bacteria and Synechococcus in oligotrophic oceans has an important role in carbon export processes (24). However, to date, the relationship between Synechococcus and heterotrophic bacteria at the community level has been poorly understood. Here, the dynamic relationship between Synechococcus sp. strain PCC7002 and a natural seawater bacterial community during a long-term cocultivation process was investigated. Intriguingly, we observed that their relationship changed dramatically, after which Synechococcus could survive permanently without added nutrients. Here, we unveil the underlying drivers of this phenomenon, which provides novel insight into the interactions between phytoplankton and heterotrophic bacteria in the ocean.
RESULTS
Dynamic relationships between Synechococcus and the bacterial community during long-term cocultivation.
During long-term cocultivation, the relationship between Synechococcus sp. PCC7002 and the coexisting heterotrophic bacterial community changed dramatically from antagonism to commensalism and finally to mutualism.
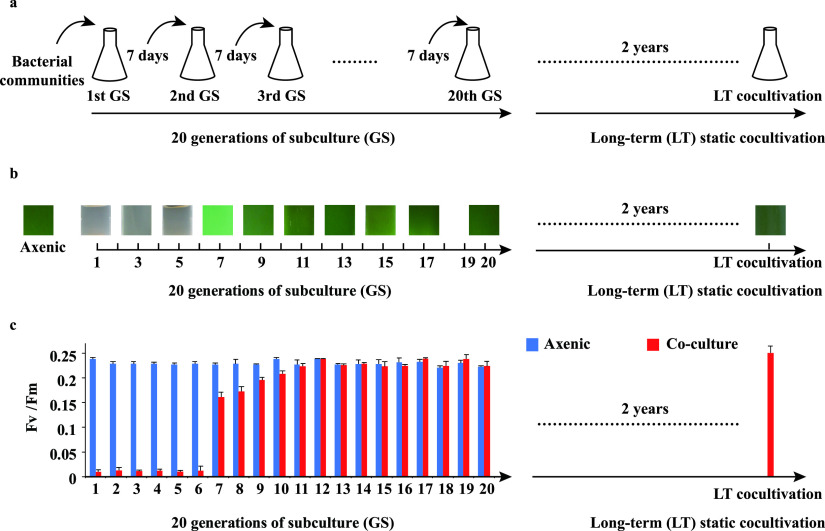
Upon inoculation of a natural bacterial community to an exponentially growing Synechococcus culture (i.e., the first generation of the subculture [1st GS]), the bacterial community showed a strong growth-inhibitory or algicidal effect on Synechococcus, which was reflected by the obvious color change in the cocultivation system from healthy green to colorless with a white deposition (Fig. 1b). The Synechococcus abundance in the cocultivation system also decreased significantly from 1.7 × 105 ± 0.14 × 105 to 5.0 × 104 ± 0.36 × 104 cells ml−1, while that in the bacterium-free control set increased from 1.7 × 105 ± 0.02 × 105 to 2.5 × 105 ± 0.01 × 105 cells ml−1. This inhibitory effect persisted until the 6th GS. We have also confirmed that no viral infection was involved in causing the decrease in the abundance of Synechococcus (see Text S1 and Fig. S1 in the supplemental material). The photosystem II maximum quantum yield (Fv/Fm) of Synechococcus remained as low as ca. 0.01 during the first 6 generations, while that in the control set was far higher (ca. 0.22) (Fig. 1c). Therefore, the first 6 GSs were called the antagonism stage of the relationship.
FIG 1.
Serial subculture and long-term static cocultivation of Synechococcus and the natural bacterial community. (a) The Synechococcus-bacteria cocultivation system was initially established by adding seawater bacterial communities to an axenic culture of Synechococcus sp. PCC7002 to be cocultivated for 7 days (i.e., the 1st GS). Next, the bacterial community from the 1st GS was added to another fresh axenic Synechococcus culture to generate the 2nd GS over another 7 days. Subculturing was carried out based on the above-described steps for a total of 20 generations. Thereafter, the 20th GS was transferred to large-volume fresh axenic Synechococcus cultures for 2 years of static cocultivation. (b) Color changes of the cocultivation system of Synechococcus and the natural bacterial community in different GSs. (c) Responses of the Fv/Fm of Synechococcus to the coexisting bacterial community during serial subculture and the long-term static cocultivation process.
Supplemental materials and methods, results, figure legends, and references. Download Text S1, DOC file, 0.06 MB (64KB, doc) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Time course of virus and Synechococcus abundances in the first generation of the subculture. Download FIG S1, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Thereafter, from the 7th to the 11th GSs, the bacterial community’s inhibitory effect on Synechococcus gradually weakened, and the cocultivation system was no longer clear but light green. The Fv/Fm also gradually increased from ca. 0.15 to 0.2 and reached the same level as that of axenic Synechococcus in the 11th GS. From the 11th GS onwards, there was little difference between the colors of the cocultivation system and the control set. The bacterial community no longer suppressed Synechococcus growth, and the Fv/Fm value was almost equal to that of the control set. This behavior continued until the end of the serial subculturing process (the 20th GS). Therefore, the stage from the 7th to the 20th GSs was called the commensalism stage. In the last four subcultures (i.e., the 17th to the 20th GSs), the Fv/Fm of Synechococcus was even slightly higher than that of the control set.
From then on, the coexisting bacterial community was transferred to a culture of exponentially growing Synechococcus for long-term observations. Interestingly, even after 2 years without any additional supply of nutrients, Synechococcus continued to grow healthily in the static cocultivation system, maintaining a green color. The Fv/Fm of Synechococcus was ca. 0.25, comparable to the Fv/Fm values of healthy axenic Synechococcus PCC7002 (25) and other cyanobacteria (26). In contrast, axenic Synechococcus in the control set (without additional N source replenishment) collapsed and turned colorless with a white deposition within 3 months. It should be mentioned that during the long-term cocultivation process, the coculture was not contaminated by any other phytoplankton, and Synechococcus was confirmed to be identical to the original Synechococcus sp. PCC7002 (Text S1). This indicated that a mutually beneficial interaction persisted between Synechococcus and the coexisting bacterial community, and this stage was called the mutualism stage.
Succession of bacterial community structure during long-term cocultivation.
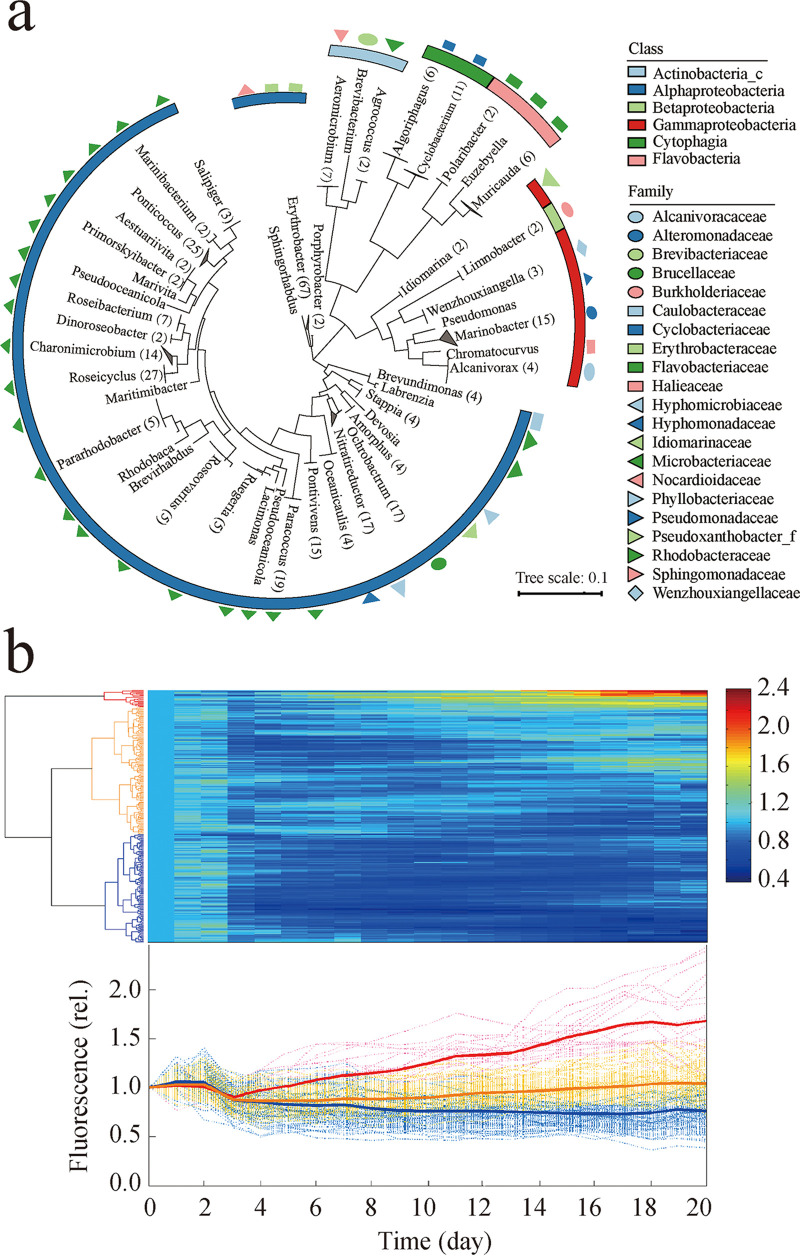
Throughout the cocultivation process, the bacterial communities changed dynamically, showing obvious succession characteristics. The changing bacterial communities can be clustered into four distinct groups (i.e., clusters I to IV) according to principal-coordinate analysis (PCoA) and hierarchical clustering analysis (Fig. 2a and b). Cluster I included the bacterial community profiles from the 1st, 3rd, and 5th GSs (Fig. 2b), all of which had a strong inhibitory effect on Synechococcus growth and corresponded exactly to the antagonism stage of their relationship with Synechococcus. Notably, in the antagonism stage, Planktosalinus and Marinicella were the most dominant genera, with relative abundances accounting for 18.9 to 53.6% and 13.2 to 27.5% of the whole bacterial community, respectively (Fig. 2c and d). The 6th GS, a transitional phase after which the growth-inhibitory effect of the bacterial community started to diminish, was in cluster II (Fig. 2b), with Polycyclovorans taking up the position of Marinicella as the second most abundant group. All the bacterial community profiles from the 7th to the 20th GSs were in cluster III (Fig. 2b), which corresponds exactly to the commensalism stage of the relationship with Synechococcus. Here, Pseudomonas became the most dominant bacterial group, with a relative abundance of 63.8 to 89.1% (Fig. 2c). Concurrently, the relative abundance of the genera Planktosalinus, Marinicella, and Polycyclovorans decreased to less than 1%. The bacterial community from the long-term static cocultivation system was found to be highly curated in cluster IV (Fig. 2b), corresponding exactly to the mutualism stage of its relationship with Synechococcus. In the mutualism stage, the genera Erythrobacter and Hyphobacterium predominated the bacterial profiles, with a total relative abundance of >80%.
FIG 2.

Dynamic changes in the bacterial community structure during long-term cocultivation with Synechococcus. Constrained principal-coordinate analysis (PCoA) based on the Bray-Curtis distance matrix (a), hierarchical clustering based on the Bray-Curtis distance matrix (b), bacterial community structure at the genus level (c), and dominant bacterial taxa with significant differences in different stages/clusters based on LEfSe analysis (LDA score of >2; P < 0.05) (d) are shown.
Effect of the bacterial individuals from the cocultivation system on Synechococcus growth.
Totals of 326 and 938 bacterial strains were isolated from the antagonism stage (represented by the 3rd GS) and the mutualism stage (i.e., long-term static cocultivation), respectively. The 326 strains from the antagonism stage belonged to 45 genera and 56 species (Fig. 3a; see also Table S1 at https://doi.org/10.6084/m9.figshare.15059517.v3), while the 938 strains from the mutualism stage had relatively low diversity, belonging to 25 genera and 34 species (see Table S2 at the URL mentioned above).
FIG 3.
Classification of 326 bacterial individuals from the antagonism stage and their effect on the growth of Synechococcus sp. PCC7002. (a) Maximum likelihood tree based on 16S rRNA genes. The strains belonging to the same genus were merged into one branch, where the number of strains is shown in parentheses. (b) Effect of each bacterial individual on the growth (using fluorescence as an indicator) of Synechococcus sp. PCC7002. Hierarchical clustering and a heat map were established based on the normalized fluorescence values and normalized fluorescence curves of Synechococcus-bacterium cocultures. These values were normalized against those of the axenic control. A total of 46.0% (150 out of 326) of the bacterial individuals had remarkable negative effects on Synechococcus growth (blue), 26.1% (85 out of 326) promoted the growth of Synechococcus (red), and the others had no obvious effects (yellow).
Interestingly, although the bacterial community in the antagonism stage showed a strong growth-inhibitory or algicidal effect on Synechococcus, among the 326 isolated bacterial strains, only 150 (46.0%) suppressed the growth of Synechococcus. In contrast, 85 out of these 326 bacterial strains (i.e., 26.1%) promoted the growth of Synechococcus. The remaining 91 strains (27.9%) had no obvious effects on Synechococcus (Fig. 3b; see also Table S3 at https://doi.org/10.6084/m9.figshare.15059517.v3). Similarly, although all bacterial communities in the mutualism stage showed a mutualistic relationship with Synechococcus, among the representative bacterial strains of all 34 different species, 1 bacterial strain (i.e., Microbacterium oxydans SN037) inhibited the growth of Synechococcus, and 14 strains (41.2%) promoted the growth of Synechococcus (see Table S3 at the URL mentioned above). Overall, the isolated bacterial community from the antagonism stage was composed of a higher proportion of inhibitory bacteria (46.0%) than beneficial bacteria (26.1%), while that isolated from the mutualism stage had more beneficial bacteria (ca. 41.2%) (see Table S3 at the URL mentioned above).
Interactions between different members of the bacterial community.
Forty-two and 34 bacterial strains, each as a representative strain of a different genus or species from the antagonism and mutualism stages, respectively, were tested for their quorum-sensing (QS) activity. Approximately 24% (10 of 42) of the representative strains from the antagonism stage and 20% (7 of 34) from the mutualism stage showed the ability to produce the QS signal molecule autoinducer-2 (AI-2) (Fig. 4), but the effect of different bacteria on Synechococcus growth was independent of their AI-2-producing capability. In addition, the bacterial communities coexisting with Synechococcus during the serial subculturing process were predicted to be involved in various quorum-sensing processes (Fig. S2; see also Table S4 at https://doi.org/10.6084/m9.figshare.15059517.v3), e.g., competence, motility, and iron uptake regulated by acylated homoserine lactones (AI-1), aromatic signaling molecules, or diffusible signal factors.
FIG 4.

Interactions between different members of the bacterial community. The ability of 42 (a) and 34 (b) representative bacterial individuals from the antagonism and mutualism stages, respectively, to produce autoinducer-2 and the impact of the two tested strains (Erythrobacter sp. SN021 and Pseudomonas sp. syn326) on these representative bacteria are shown.
Distribution of quorum-sensing genes in each cocultivation system. PICRUSt2 was applied for the predictions of microbial functional genes. Download FIG S2, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We also tested the effects of two representative strains, i.e., Pseudomonas sp. strain syn326 and Erythrobacter sp. strain SN021, on other isolates via plate assays. These two strains were phylogenetically similar to the most-abundant amplicon sequence variants (ASVs) in the commensalism and mutualism stages, i.e., Pseudomonas-like ASV_1 and Erythrobacter-like ASV_3 (see Table S5 at the URL mentioned above). Although Pseudomonas sp. syn326 did not influence the growth of Synechococcus, it inhibited the bacterium Algoriphagus sp. syn149, a strain with an inhibitory effect on Synechococcus, and promoted the growth of Nitratireductor sp. syn063 and Algoriphagus sp. syn216, which were both beneficial to Synechococcus (Fig. 4 and Fig. S3). Erythrobacter sp. SN021 was beneficial to Synechococcus growth and exhibited a strong ability to regulate the growth of other bacteria (Fig. 4 and Fig. S3). For instance, it inhibited the growth of Marinobacter sp. SN030 but promoted that of Marinobacter sp. SN009.
Bacterium-bacterium interactions. The inhibition zone around the agar slab containing Pseudomonas sp. syn326 (blue arrow) or Erythrobacter sp. SN021 (black arrow) indicates their inhibitory effect on the growth of the tested strains. The promotion zone indicates the beneficial effect. Download FIG S3, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rich inorganic nitrogen in the mutualism stage and nitrogen metabolism genes in the bacterial communities.
We determined the concentrations of different inorganic nitrogen compounds that may significantly affect the growth of Synechococcus in the mutualism stage and in the axenic control set (see Table S6 at https://doi.org/10.6084/m9.figshare.15059517.v3). In the control set containing only exponentially growing axenic Synechococcus cells, the concentrations of nitrate, nitrite, and ammonium were 8,015.3 ± 808.2 μmol/liter, 2.5 ± 0.4 μmol/liter, and 2,682.1 ± 236.4 μmol/liter, respectively, which were mainly from the fresh A+ medium. Surprisingly, in the mutualism stage, although no nutrients had been artificially supplemented for 2 years, the inorganic nitrogen in the cocultivation system remained at high levels (i.e., nitrate at 4,558.2 ± 677.2 μmol/liter, nitrite at 72.5 ± 12.4 μmol/liter, and ammonium at 2,277.5 ± 153.2 μmol/liter). As the host Synechococcus sp. PCC7002 cannot fix nitrogen (see Tables S7 and S8 at the URL mentioned above) (27, 28), and we did not artificially supplement N nutrients, the rich nitrogen source in the mutualism stage is a mystery, and it is speculated to come from the contribution of coexisting bacterial communities. To verify this hypothesis, we tested the 34 representative bacterial strains isolated from the mutualism stage to determine whether they contain the nitrogenase gene nifH. As a result, four strains (i.e., Rhizobium sp. SN019, Georhizobium sp. SN035, Mesorhizobium sp. SN007, and Bacillus sp. SN014) were found to be positive for the nifH gene, among which Mesorhizobium sp. SN007 and Bacillus sp. SN014 also showed actual nitrogenase activity in the acetylene reduction assay. Meanwhile, in the metagenome of the bacterial community at the mutualism stage, in addition to the nitrogen-fixing gene (nifH) that catalyzes nitrogen fixation, there are many other key genes involved in different nitrogen metabolism processes, i.e., organic nitrogen degradation, dissimilatory/assimilatory nitrate reduction, and denitrification (Fig. S4; see also Table S9 at the URL mentioned above). Among them, the most abundant genes were gdh (glutamate dehydrogenase) and gs (glutamate synthase), which were related to the conversion of Synechococcus-derived organic nitrogen to inorganic nitrogen (i.e., ammonia production). The narGHIJ genes, encoding nitrate reductases involved in dissimilatory nitrate reduction and nitrate-to-nitrite conversion, were also abundant in the mutualism stage. Nitrite can be further reduced to ammonium by the nitrite reductase encoded by the nrfA gene in the metagenome, which would support the growth of Synechococcus (Fig. S4). In addition, there was also a relatively high proportion of genes in the metagenome participating in denitrification, including the nirSK (nitrite reductase), norBC (nitric oxide reductase), and nosZ (nitrous oxide reductase) genes, which potentially induced the loss of nitrogen. Therefore, nitrogen input (i.e., nitrogen fixation) was particularly important to offset the possible nitrogen loss and maintain the high nitrogen concentration in the long-term mutualism stage of the coculture system.
Genes involved in the nitrogen cycle in the heterotrophic bacterial community cocultured with Synechococcus during the mutualism stage. The numbers in brackets beside each gene represent the abundances for the gene or gene family. Download FIG S4, TIF file, 1.0 MB (1,020.6KB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Phytoplankton and heterotrophic bacterium relationships are changeable and depend highly on many aspects, including external environmental and biotic factors. However, the underlying mechanisms of the relationship variability between phytoplankton and heterotrophic bacteria remain unclear. Here, we experimentally cocultivated a natural bacterial community with Synechococcus sp. PCC7002 and tracked the dynamics of their relationship on a long-term scale. We found that the relationship between the Synechococcus and heterotrophic bacterial communities underwent a dramatic shift from antagonism to commensalism and finally to mutualism (Fig. 1b and c). Concurrently, during this process, the bacterial communities also showed succession characteristics (clusters I to IV) (Fig. 2a and b) that were mapped to the timeline of the shift of the Synechococcus-bacterium relationship from antagonism to mutualism. This suggested that the succession of bacterial communities might be an important factor boosting the transition of the relationship between Synechococcus and the bacterial community.
Cluster I (Fig. 2a and b), representing the antagonistic stage (i.e., 1st through 5th GSs), was dominated by the genera Planktosalinus and Marinicella (Fig. 2c and d). These two bacterial genera are affiliated with Bacteroidetes and Gammaproteobacteria, respectively, which are frequent inhabitants of the algal phycosphere (12, 29, 30) and are known to include many algicidal bacteria (8, 31). In addition, Bacteroidetes or Gammaproteobacteria often increase significantly in the late algal bloom period (32–34). Similarly, the bacterial community in cluster I showed high relative abundances of Gammaproteobacteria (27 to 35%) and Bacteroidetes (25 to 57%) (Fig. 2c; see also Fig. S5 in the supplemental material). In this sense, the relatively high proportion of inhibitory bacteria may be the driving force behind the decline in Synechococcus at this stage. This was validated by the effects of bacterial individuals isolated from the cocultivation system on the growth of Synechococcus. As expected, nearly half of the bacterial individuals (46.0%) from the antagonism stage inhibited the growth of Synechococcus.
Dynamic changes in the bacterial community structure at the phylum level during long-term cocultivation with Synechococcus. Download FIG S5, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
From the 7th GS until the last (20th) GS of the serial subculturing process, no sign of Synechococcus growth inhibition was observed, suggesting that the antagonistic effect of the bacterial community diminished, giving rise to the emergence of the commensalism stage. The bacterial community structure profiles significantly differed from those of the previous stage (Fig. 2d), with a surge in Pseudomonas accounting for 60 to 90% of the total bacterial community (Fig. 2c). The ability of Pseudomonas species to promote the growth of host algae by itself or by regulating the growth of other bacteria has been reported. For example, cocultivation of the alga Chlorella vulgaris with a Pseudomonas strain yielded a 1.4-times-higher algal cell concentration than with axenic C. vulgaris (35). Additionally, some Pseudomonas spp. can also produce bacteriocins (peptidic toxins with antibacterial activity) and antibiotics (such as pyrrolnitrin, pyoluteorin, and 2,4-diacetylphloroglucinol) to inhibit other bacteria (29). Two representative strains of Pseudomonas (i.e., Pseudomonas species strains syn326 and SN018) isolated in this study (Fig. 4; see also Table S5 at https://doi.org/10.6084/m9.figshare.15059517.v3) had no inhibitory effect on the growth of Synechococcus, and in contrast, Pseudomonas sp. SN018 significantly promoted the growth of Synechococcus (Fig. 4). Although Pseudomonas sp. syn326 did not have a direct growth-promoting effect on Synechococcus, it inhibited the growth of some inhibitory bacteria (e.g., Algoriphagus sp. syn149) and promoted the growth of some beneficial bacteria (e.g., Nitratireductor sp. syn063 and Algoriphagus sp. syn216) (Fig. 4a). These abilities of Pseudomonas might have favored its proliferation, directly or indirectly benefiting Synechococcus growth and driving the establishment of a commensalism relationship between Synechococcus and the heterotrophic bacterial community.
The improved Fv/Fm of Synechococcus in the cocultivation system, especially in the last few generations (from the 17th to the 20th GSs) (Fig. 1c), suggested a positive interaction between Synechococcus and the heterotrophic bacterial community. Similar phenomena have also been observed by Motone et al. (36). Here, we asked how long this relationship could be maintained if there were no further subculture steps. Therefore, we conducted an additional static cocultivation experiment for 2 years. Interestingly, without any additional supplement of nutrients, Synechococcus maintained a healthy growth state for 2 years. In contrast, the axenic Synechococcus culture in the control set collapsed within 3 months. This indicates that a mutualistic relationship between Synechococcus and the bacterial community was formed in the cocultivation system. During the long-term mutualism stage, Hyphomonadaceae (comprising Hyphobacterium) and Erythrobacter were the most abundant taxa in the bacterial community. Most species of the Hyphomonadaceae can attach to or form biofilms and have the potential to degrade complex organic compounds (37). These characteristics may help them become the dominant bacterial group coexisting with the host Synechococcus. Moreover, Hyphomonadaceae have often been isolated from dinoflagellates and diatoms (38), suggesting an unknown close relationship between phytoplankton and Hyphomonadaceae. Erythrobacter also tends to inhabit the phycosphere environment (1, 39, 40) and can affect the growth of other bacteria. For instance, upon exposure to polyunsaturated aldehydes (PUAs), algal cell exudates with antimicrobial activity, most bacteria, but not Erythrobacter, were inhibited (41). Members of the Erythrobacter genus also have strong signaling molecule-producing capabilities and can regulate other bacterial interactions by quorum quenching (42, 43). Here, it was verified that the isolated representative strain of the genus Erythrobacter in the mutualism stage, namely, Erythrobacter SN021, can promote the growth of Synechococcus (Fig. 4b). In addition, 16 metagenome-assembled bacterial genomes (bin.1 to -16) (see Table S10 at https://doi.org/10.6084/m9.figshare.15059517.v3) were obtained from the mutualism stage, including a metagenome-assembled genome classified as Erythrobacter (i.e., bin.3). Among them, 12 strains possessed 4 to 7 auxin synthesis genes (see Table S10 at the URL mentioned above). Auxins are a class of phototrophic hormones, and it has been reported that auxins (especially indole-3-acetic acid, the most abundant auxin) have a growth-stimulating effect on phytoplankton (44–46). Furthermore, among all 34 different bacterial species from the mutualism stage, more than 40% significantly promoted the growth of Synechococcus, and only 1 had an inhibitory effect on the growth of Synechococcus (see Table S3 at the URL mentioned above). Overall, we speculated that the rise of beneficial bacteria in the bacterial community aids in maintaining the long-term mutualistic relationship between Synechococcus and the bacterial community at this stage.
We recognized that the final effect of the bacterial community on the host Synechococcus may not be the result of simply the accumulation or offset of the effects of different individual bacteria but could also be concomitant with close interactions among bacteria. It was roughly estimated that more than 20% of the strains isolated from the antagonism or mutualism stage had the ability to produce autoinducer-2 (Fig. 4). It was also predicted that bacterial communities may have additional forms of quorum-sensing communication throughout the serial subculture and static cocultivation processes, which would indicate that bacterium-bacterium interactions were common in these bacterial communities (Fig. S2). Besides, Pseudomonas sp. syn326 can differentially affect the growth of two strains with quorum-sensing activity but with different effects on Synechococcus growth (Fig. 4a), indicating that the bacterial interaction can spread through a cascade effect, forming a potential interaction network and likely driving the succession of the entire bacterial community structure during long-term cocultivation.
Nitrogen availability is an important growth-limiting factor for picocyanobacteria (47). Synechococcus sp. PCC7002 cannot fix nitrogen by itself (see Tables S7 and S8 at https://doi.org/10.6084/m9.figshare.15059517.v3) (27, 28); the unexpectedly high concentration of inorganic nitrogen (e.g., NO3− and NH4+) in the mutualistic cocultivation system implies that the bacterial community likely played a crucial role in providing available inorganic nutrients. Here, we did find evidence of the nitrogen-cycling genetic potential of the bacterial community via metagenomic analysis (Fig. S4; see also Table S9 at the URL mentioned above), including organic nitrogen mineralization, nitrogen fixation, and dissimilatory nitrate reduction. Previously, it was reported that a single bacterium can maintain the growth of Synechococcus for 200 days through nutrient recycling (i.e., mineralization) (18). Here, the abundant organic nitrogen degradation genes (e.g., gdh) found in the metagenomes of the long-term cocultivation system suggest that the bacterial community can drive the transformation of nitrogen from organic to inorganic forms during growth depending on the organic matter (containing organic nitrogen) released by Synechococcus (see Table S9 at the URL mentioned above) (48, 49), which in turn can support the growth of Synechococcus. Moreover, the abundant denitrification genes (nirS, nirK, norC, norB, and nosZ) found in the metagenomes implicate the potential of the bacterial community to cause nitrogen loss (Fig. S4). This hints that there must be a nitrogen replenishment process (e.g., nitrogen fixation) to compensate for the loss of nitrogen caused by denitrification during the mutualism stage and balancing nitrogen cycling. As expected, we validated the nitrogen-fixing gene (nifH) and nitrogenase activity of the bacterial individuals isolated from the mutualism stage and observed the potential nitrogen-fixing activity of four strains among them, further supporting our speculation that nitrogen-fixing activities may be a driving force for nitrogen supply in the mutualism stage. In addition, it has been reported that the growth of phytoplankton can be inhibited by high concentrations of NO2− (50, 51). The comparatively high abundance of nitrate reductase genes (narGHIJ genes), whose products can reduce NO3− to NO2−, may have been compensated for by the products of abundant nitrite reductase genes (nirK and nirS), nullifying the possibility of NO2− overproduction (Fig. S4; see also Table S9 at the URL mentioned above). Thus, without artificial supplementation of any nitrogen nutrients, a self-sufficient nitrogen cycle (especially including nitrogen fixation) within the cocultivation system during the long-term mutualism stage might have supported the healthy survival of the host Synechococcus, and in return, the organic carbon substances synthesized by Synechococcus photosynthesis met the nutritional needs of the heterotrophic bacterial community. However, no genes responsible for ammonia oxidation (e.g., amoA) were found in the metagenomes, so how nitrate was produced and maintained at a high concentration in the long-term mutualism stage is still unclear and requires further research. In addition to the nitrogen cycle, genomic capability for the metabolic generation of phosphorus, iron, and vitamin B12 was also observed in the metagenome; e.g., the bacterial community may contribute to the phosphorus cycle by the mineralization of organic phosphorus and the solubilization of recalcitrant phosphorus (see Table S11 at the URL mentioned above) and to the iron cycle through siderophore production and iron acquisition and transport (see Table S12 at the URL mentioned above). Besides, the bacterial community can also synthesize vitamin B12 necessary for the growth of Synechococcus sp. PCC7002 (52) (see Table S13 at the URL mentioned above). This indicates the complexity of nutritional self-sufficiency in the cocultivation system. In addition, we admit that the bacterial community cocultured with Synechococcus in the laboratory will be different from that of in situ seawater. Meanwhile, in the in situ environment, the relationship between Synechococcus and bacterial communities will be much more complex (such as grazing or viral lysis, etc.) than what we observed in the laboratory. These need more in-depth study in the future.
Taken together, the interactions among different bacterial members with significantly distinct effects (inhibitory or promotive) on Synechococcus growth may determine the overall relationship of the whole heterotrophic bacterial community with Synechococcus; the succession of the bacterial community structure during the long-term cocultivation process drove the dynamic changes of its relationship with Synechococcus from antagonism to commensalism and then to mutualism; the final mutualistic relationship between Synechococcus and the heterotrophic bacterial community allowed them to meet each other’s nutritional needs through the internal self-sufficient nutrient cycle (e.g., the nitrogen cycle), thereby supporting their healthy survival for up to 2 years without any exogenous nutrient supply (Fig. 5). This hints that the ubiquity and competitive advantage of Synechococcus in global oceans might also be partially attributed to the contribution of heterotrophic bacterial communities.
FIG 5.

Schematic of the succession of bacterial community structure and function during long-term cocultivation with Synechococcus. The initial bacterial community with a high proportion of inhibitory bacteria inhibited the growth of Synechococcus in the antagonism stage. Under the interaction between different members of the bacterial community during serial subculture, the inhibitory bacteria were suppressed to a point that they could not completely inhibit the growth of Synechococcus. The relationship between Synechococcus and the heterotrophic bacterial community gradually entered the commensalism stage. Thereafter, as the proportion of beneficial bacteria continued to increase, a mutually beneficial relationship was established between Synechococcus and the heterotrophic bacterial community. During the mutualism stage, a self-sufficient nitrogen cycle might contribute to the healthy survival of both Synechococcus and heterotrophic bacterial communities in 2 years without any exogenous nutrition supplementation.
MATERIALS AND METHODS
Establishment of the cocultivation system of Synechococcus and the natural bacterial community.
A model marine cyanobacterial strain, Synechococcus sp. PCC7002, was acquired from the Pasteur Culture Collection (53), which was isolated from the coast of the North Atlantic and belongs to subcluster 5.2 of the genus Synechococcus (see Fig. S6 in the supplemental material). The strain was streaked onto an A+ agar plate (54) and purified repeatedly in solid and liquid media. The sterility of the Synechococcus sp. PCC7002 culture was verified routinely and before all experiments by fluorescence microscopy, flow cytometry, agar media, and 16S rRNA gene sequencing (55). Axenic Synechococcus was inoculated into fresh A+ medium at a 1% volume ratio and cultivated to the exponential growth phase in 14 days at 26°C at 50 μmol m−2 s−1 under a 12-h-light/12-h-dark (12L:12D) photoperiod. Exponentially growing axenic Synechococcus cells were later employed for the experiments described below. For the natural bacterial community, three replicates of surface seawater samples (∼200 ml each) collected from the coast of Qingdao, China (36.06°N, 120.32°E), in summer (August) were incubated in the dark for 72 h (to suppress algal growth) and subjected to sequential filtration through 3.0- and 0.22-μm-pore-size polycarbonate filters (Millipore, Ireland). The 0.22-μm-pore-size filters, which retained the bacterial communities, were dispersed in 20 ml of an exponentially growing axenic Synechococcus culture (1.7 × 105 cells ml−1). The culture was gently shaken to allow the bacterial assemblage to separate from the filters. Like this, triplicate Synechococcus-bacterium cocultures were established along with a control set of triplicate axenic Synechococcus cultures in A+ medium.
Phylogenetic tree of 16S rRNA gene sequences of Synechococcus strains. Clade and subcluster designations follow the nomenclature of S. Bemal and A. C. Anil (FEMS Microbiol Ecol 92:fiw162, 2016, https://doi.org/10.1093/femsec/fiw162). The tree was constructed by the neighbor-joining method with Muscle, aligning 59 16S rRNA nucleotide sequences of Synechococcus strains with 1,000 bootstrap replicates. Download FIG S6, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Serial subculturing of the Synechococcus-bacterium coculture and subsequent long-term static cocultivation.
The above-described cocultivation systems were incubated at 26°C at 50 μmol m−2 s−1 under a 12L:12D photoperiod for 7 days (i.e., the first generation of the subculture [1st GS]). Following filtration through a 3.0-μm-pore-size polycarbonate membrane to remove large particles or biological aggregates, 1 ml of the filtrate containing the bacterial assemblage (and also Synechococcus cells due to the similar size range, i.e., <3 μm) was transferred to another exponentially growing axenic Synechococcus culture (20 ml) to initiate the “second generation” of the subculture (for another 7 days) (2nd GS). The remaining filtrate from the three parallel experimental groups (replicates) was mixed, and the bacteria in the filtrate were retained on 0.22-μm-pore-size membranes, flash-frozen, and preserved at −80°C for bacterial community analysis. Thereafter, subsequent subcultures (generations) were obtained according to the above-described steps until the end of the 20th generation (in total, 140 days of serial subculturing) (Fig. 1a). In addition, considering that viruses (especially cyanophage) and cyanobacteria from the natural seawater may be introduced into the experimental system, the abundance of viruses and the composition of cyanobacteria in the system were tested, and their interference was subsequently excluded (Text S1; see also Table S14 at https://doi.org/10.6084/m9.figshare.15059517.v3).
At the end of the 20th generation (20th GS), the final three replicates of the cocultivation system were mixed and filtered, and the <3-μm filtrate was inoculated into two new culture flasks (at a 1% ratio) containing 3 liters of an exponentially growing Synechococcus culture in A+ medium. The system was incubated at room temperature at 20°C to 25°C under natural light/dark conditions for more than 2 years (i.e., long-term static cocultivation).
Response of Synechococcus to bacterial communities during serial subculture and subsequent long-term static cocultivation.
To reveal the different responses of Synechococcus to bacterial communities during serial subculture and long-term static cocultivation, at the end of each subculture (generation) and 2-year static cocultivation, the maximum quantum yield of photosystem II (Fv/Fm) of Synechococcus in the cocultivation systems was measured and compared with that in the axenic control groups as an indicator of the autotroph’s photosynthetic performance. The samples were dark acclimated for 15 min, and the Fv/Fm was analyzed using a dual-wavelength pulse-amplitude-modulated fluorescence monitoring system (Dual-PAM; Heinz Walz, Germany) at a wavelength of 620 nm (56).
Dynamic changes in bacterial community structure during serial subculture and subsequent long-term static cocultivation.
To understand whether the bacterial community changed dynamically during long-term cocultivation, the bacterial community structure was analyzed according to the method of Picazo et al. (57) in each of the serial subcultures and in the 2-year static cocultivation system. Genomic DNA was extracted using the FastDNA Spin kit (MP Biomedicals). Detailed procedures for sequence analysis are provided in Text S1. Constrained principal-coordinate analysis (PCoA) and hierarchical clustering based on the Bray-Curtis distance matrix were employed to explore the similarities between different bacterial communities. The linear discriminant analysis (LDA) effect size (LEfSe) method (http://huttenhower.sph.harvard.edu/lefse/) was used to identify the most differentially abundant taxa between different groups. The raw reads were deposited in the Genome Sequence Archive of the National Genomics Data Center (NGDC) (58).
Isolation of bacterial individuals from the cocultivation system.
Individual bacteria were isolated from the cocultivation system using solid marine agar 2216E medium from the 3rd generation of the subculture (3rd GS), which represented the antagonism stage of the relationship, and during long-term static cocultivation, which represented the mutualism stage. In addition, potential nitrogen-fixing bacteria were also isolated using nitrogen-free media (i.e., Jensen’s medium and New Fabian broth [NFB] medium) under oxic and partially oxic conditions (59, 60). A total of 1,264 bacterial colonies were purified and identified (see Tables S1 and S2 at https://doi.org/10.6084/m9.figshare.15059517.v3) via 16S rRNA gene analysis (61). The detailed procedure for bacterial identification is provided in Text S1. The 16S rRNA gene sequences were deposited in GenBank.
Impacts of the representative bacterial individuals on the growth of Synechococcus.
The effects of all 326 different bacterial individuals from the 3rd GS and 34 out of 938 strains representing different species from the long-term static cocultivation system on Synechococcus growth were analyzed. Exponentially growing bacteria (with 3 parallel replicates) were washed and resuspended in A+ medium to a final concentration of ca. 106 cells ml−1. One milliliter of these bacterial suspensions was added to a 10-ml axenic Synechococcus (106 cells ml−1) culture and incubated at 26°C at 50 μmol m−2 s−1 under a 12L:12D photoperiod for 20 days. The chlorophyll fluorescence was measured at a fixed time every day using a BioTek Synergy HT plate reader with excitation/emission wavelengths of 440/680 nm to determine the growth status of Synechococcus. The following formula was used for the calculation of average changes in florescence values (μ): μ = [ln(Nt) − ln(N0)]/Δt, where Nt and N0 are the Synechococcus fluorescence values at t and 0 days, respectively, and Δt is the incubation time. Significant effects (promotion and inhibition) of bacteria on the growth of Synechococcus were determined by a t test (P < 0.05), using μ values between the cocultivation group and the axenic Synechococcus group.
Interactions among members of the heterotrophic bacterial community.
Forty-two representative bacterial strains belonging to different genera or having different impacts on Synechococcus from the 3rd GS and 34 strains representing different species from the long-term static cocultivation system were tested for the production of autoinducer-2, a QS molecule, as described previously by Bassler et al. (62). In addition, a plate assay was used to test the actual occurrence of interactions between different bacterial individuals. Pseudomonas sp. syn326 and Erythrobacter sp. SN021, representing the most abundant bacterial species from the 3rd GS and the long-term static cocultivation system, respectively, were selected to test their effects on other bacterial strains. The detailed procedures are included in Text S1.
Determination of inorganic nitrogen in the cocultivation system and the genes related to the nitrogen cycle in the bacterial community.
The inorganic nitrogen (i.e., nitrate, nitrite, and ammonium) concentrations in the cocultivation systems were measured using an AutoAnalyzer (AA3; Bran and Luebbe, Germany).
Metagenomic analysis was employed to detect the genes related to the nitrogen cycle in the 2-year static cocultivation system (see Text S1 for detailed procedures for sequence analysis). Sequence reads were deposited in the NGDC database. Additionally, 34 representative bacterial strains from the 2-year static cocultivation system were screened for the presence of the nitrogenase gene nifH using primers PolF and PolR (63, 64). Meanwhile, their potential for nitrogen fixation was further determined by the acetylene reduction assay (Text S1). Moreover, the 34 representative strains from the 2-year static cocultivation system and 42 representative strains from the 3rd GS were also tested for nitrate-reducing ability according to standard approaches (Text S1 and Fig. S7) (65).
Nitrate-reducing bacteria in different cocultivation systems. The abilities to reduce nitrate to nitrite or nitrogen by the 42 (a) and 34 (b) representative bacterial individuals from the antagonism and mutualism stages, respectively, were tested. Beneficial bacteria for the growth of Synechococcus are marked in red, and inhibitory bacteria are in blue. Download FIG S7, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data availability.
The data that support the findings of this study are available from the corresponding author upon reasonable request. Sequence data have been deposited in the NGDC database under accession no. CRA003602 and CRA003605 and in GenBank under accession no. MW409752 to MW410111.
ACKNOWLEDGMENTS
We appreciate Xiangan Han (from the Shanghai Veterinary Research Institute, the Chinese Academy of Agricultural Sciences) for providing Vibrio harveyi BB150 and BB170 and Dalin Shi (from the State Key Laboratory of Marine Environmental Science, Xiamen University) for providing the gas chromatograph for the acetylene reduction assay.
This work was funded by the Senior User Project of RV KEXUE (KEXUE2019GZ03) supported by the Center for Ocean Mega-Science, Chinese Academy of Sciences; National Natural Science Foundation of China (no. U1906216 and 31700104); the National Key Research and Development Programs of China (2016YFA0601402); the Key R&D Project in Shandong Province (2019GHY112037); the DICP and QIBEBT (DICP&QIBEBT UN201803); and the QIBEBT (QIBEBT ZZBS 201805), Dalian National Laboratory for Clean Energy (DNL), CAS.
Y.Z. conceived the study and designed the experiments; Z.Z., S.N., and L.T. performed experiments and data analyses; H.Z. and Z.H. undertook bacterium-bacterium interaction experiments; M.C. undertook nitrogen fixation activity experiments; Y.Z., S.-J.K., and N.J. contributed to the final version of the manuscript; and Z.Z., S.N., and Y.Z. contributed to the writing and discussion of the paper.
We declare no competing interests.
Footnotes
Citation Zhang Z, Nair S, Tang L, Zhao H, Hu Z, Chen M, Zhang Y, Kao S-J, Jiao N, Zhang Y. 2021. Long-term survival of Synechococcus and heterotrophic bacteria without external nutrient supply after changes in their relationship from antagonism to mutualism. mBio 12:e01614-21. https://doi.org/10.1128/mBio.01614-21.
Contributor Information
Yongyu Zhang, Email: zhangyy@qibebt.ac.cn.
Mary Ann Moran, University of Georgia.
REFERENCES
- 1.Amin SA, Parker MS, Armbrust EV. 2012. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 76:667–684. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falkowski PG, Fenchel T, Delong EF. 2008. The microbial engines that drive Earth’s biogeochemical cycles. Science 320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Meng Z, Liu X, Zhang X-H. 2019. Microbial assembly, interaction, functioning, activity and diversification: a review derived from community compositional data. Mar Life Sci Technol 1:112–128. doi: 10.1007/s42995-019-00004-3. [DOI] [Google Scholar]
- 4.Azam F, Fenchel T, Field JG, Gray JS, Meyer-Reil LA, Thingstad F. 1983. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 10:257–263. doi: 10.3354/meps010257. [DOI] [Google Scholar]
- 5.Chisholm SW. 2000. Oceanography: stirring times in the Southern Ocean. Nature 407:685–687. doi: 10.1038/35037696. [DOI] [PubMed] [Google Scholar]
- 6.Jiao N, Herndl G, Hansell D, Benner R, Kattner G, Wilhelm S, Kirchman D, Weinbauer M, Luo T, Chen F, Azam F. 2010. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat Rev Microbiol 8:593–599. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- 7.Xie B, Bishop S, Stessman D, Wright D, Spalding MH, Halverson LJ. 2013. Chlamydomonas reinhardtii thermal tolerance enhancement mediated by a mutualistic interaction with vitamin B12-producing bacteria. ISME J 7:1544–1555. doi: 10.1038/ismej.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su J, Yang X, Zhou Y, Zheng T. 2011. Marine bacteria antagonistic to the harmful algal bloom species Alexandrium tamarense (Dinophyceae). Biol Control 56:132–138. doi: 10.1016/j.biocontrol.2010.10.004. [DOI] [Google Scholar]
- 9.Zehr JP. 2015. How single cells work together. Science 349:1163–1164. doi: 10.1126/science.aac9752. [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Liu J, Yang L, Chen H, Zhang S, Zhao H, Zhang N. 2011. Allelopathic control of cyanobacterial blooms by periphyton biofilms. Environ Microbiol 13:604–615. doi: 10.1111/j.1462-2920.2010.02363.x. [DOI] [PubMed] [Google Scholar]
- 11.Ramanan R, Kim B-H, Cho D-H, Oh H-M, Kim H-S. 2016. Algae-bacteria interactions: evolution, ecology and emerging applications. Biotechnol Adv 34:14–29. doi: 10.1016/j.biotechadv.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Seymour JR, Amin SA, Raina J-B, Stocker R. 2017. Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat Microbiol 2:17065. doi: 10.1038/nmicrobiol.2017.65. [DOI] [PubMed] [Google Scholar]
- 13.Bell W, Mitchell R. 1972. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol Bull 143:265–277. doi: 10.2307/1540052. [DOI] [Google Scholar]
- 14.Cruz-López R, Maske H. 2016. The vitamin B1 and B12 required by the marine dinoflagellate Lingulodinium polyedrum can be provided by its associated bacterial community in culture. Front Microbiol 7:560. doi: 10.3389/fmicb.2016.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park BS, Joo J-H, Baek K-D, Han M-S. 2016. A mutualistic interaction between the bacterium Pseudomonas asplenii and the harmful algal species Chattonella marina (Raphidophyceae). Harmful Algae 56:29–36. doi: 10.1016/j.hal.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Tan S, Zhou J, Zhu X, Yu S, Zhan W, Wang B, Cai Z, Lindell D. 2015. An association network analysis among microeukaryotes and bacterioplankton reveals algal bloom dynamics. J Phycol 51:120–132. doi: 10.1111/jpy.12259. [DOI] [PubMed] [Google Scholar]
- 17.Mayali X, Azam F. 2004. Algicidal bacteria in the sea and their impact on algal blooms. J Eukaryot Microbiol 51:139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x. [DOI] [PubMed] [Google Scholar]
- 18.Christie-Oleza JA, Sousoni D, Lloyd M, Armengaud J, Scanlan DJ. 2017. Nutrient recycling facilitates long-term stability of marine microbial phototroph-heterotroph interactions. Nat Microbiol 2:17100. doi: 10.1038/nmicrobiol.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seyedsayamdost MR, Wang R, Kolter R, Clardy J. 2014. Hybrid biosynthesis of roseobacticides from algal and bacterial precursor molecules. J Am Chem Soc 136:15150–15153. doi: 10.1021/ja508782y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Tomasch J, Michael V, Bhuju S, Jarek M, Petersen J, Wagner-Dobler I. 2015. Identification of genetic modules mediating the Jekyll and Hyde interaction of Dinoroseobacter shibae with the dinoflagellate Prorocentrum minimum. Front Microbiol 6:1262. doi: 10.3389/fmicb.2015.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee MD, Ahlgren NA, Kling JD, Walworth NG, Rocap G, Saito MA, Hutchins DA, Webb EA. 2019. Marine Synechococcus isolates representing globally abundant genomic lineages demonstrate a unique evolutionary path of genome reduction without a decrease in GC content. Environ Microbiol 21:1677–1686. doi: 10.1111/1462-2920.14552. [DOI] [PubMed] [Google Scholar]
- 22.Sohm JA, Ahlgren NA, Thomson ZJ, Williams C, Moffett JW, Saito MA, Webb EA, Rocap G. 2016. Co-occurring Synechococcus ecotypes occupy four major oceanic regimes defined by temperature, macronutrients and iron. ISME J 10:333–345. doi: 10.1038/ismej.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malfatti F, Azam F. 2009. Atomic force microscopy reveals microscale networks and possible symbioses among pelagic marine bacteria. Aquat Microb Ecol 58:1–14. doi: 10.3354/ame01355. [DOI] [Google Scholar]
- 24.Guidi L, Chaffron S, Bittner L, Eveillard D, Larhlimi A, Roux S, Darzi Y, Audic S, Berline L, Brum J, Coelho LP, Espinoza JCI, Malviya S, Sunagawa S, Dimier C, Kandels-Lewis S, Picheral M, Poulain J, Searson S, Tara Oceans Coordinators, Stemmann L, Not F, Hingamp P, Speich S, Follows M, Karp-Boss L, Boss E, Ogata H, Pesant S, Weissenbach J, Wincker P, Acinas SG, Bork P, de Vargas C, Iudicone D, Sullivan MB, Raes J, Karsenti E, Bowler C, Gorsky G. 2016. Plankton networks driving carbon export in the oligotrophic ocean. Nature 532:465–470. doi: 10.1038/nature16942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu D, Zhang J, Lü C, Xia Y, Liu H, Jiao N, Xun L, Liu J. 2020. Synechococcus sp. strain PCC7002 uses sulfide:quinone oxidoreductase to detoxify exogenous sulfide and to convert endogenous sulfide to cellular sulfane sulfur. mBio 11:e03420-19. doi: 10.1128/mBio.03420-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santabarbara S, Villafiorita Monteleone F, Remelli W, Rizzo F, Menin B, Casazza AP. 2019. Comparative excitation-emission dependence of the Fv/Fm ratio in model green algae and cyanobacterial strains. Physiol Plant 166:351–364. doi: 10.1111/ppl.12931. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig M, Bryant D. 2012. Acclimation of the global transcriptome of the cyanobacterium Synechococcus sp. strain PCC 7002 to nutrient limitations and different nitrogen sources. Front Microbiol 3:145. doi: 10.3389/fmicb.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimmori Y, Kanesaki Y, Nozawa M, Yoshikawa H, Ehira S. 2018. Transcriptional activation of glycogen catabolism and the oxidative pentose phosphate pathway by NrrA facilitates cell survival under nitrogen starvation in the cyanobacterium Synechococcus sp. strain PCC 7002. Plant Cell Physiol 59:1225–1233. doi: 10.1093/pcp/pcy059. [DOI] [PubMed] [Google Scholar]
- 29.Palleroni NJ. 2015. Pseudomonas, p 323–379. In Whitman WB, Rainey F, Kämpfer P, Trujillo M, Chun J, DeVos P, Hedlund B, Dedysh S (ed), Bergey’s manual of systematics of archaea and bacteria. Wiley, Hoboken, NJ. [Google Scholar]
- 30.Zheng Q, Wang Y, Xie R, Lang AS, Liu Y, Lu J, Zhang X, Sun J, Suttle CA, Jiao N. 2018. Dynamics of heterotrophic bacterial assemblages within Synechococcus cultures. Appl Environ Microbiol 84:e01517-17. doi: 10.1128/AEM.01517-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer N, Bigalke A, Kaulfuß A, Pohnert G. 2017. Strategies and ecological roles of algicidal bacteria. FEMS Microbiol Rev 41:880–899. doi: 10.1093/femsre/fux029. [DOI] [PubMed] [Google Scholar]
- 32.Yang C, Li Y, Zhou Y, Zheng W, Tian Y, Zheng T. 2012. Bacterial community dynamics during a bloom caused by Akashiwo sanguinea in the Xiamen Sea area, China. Harmful Algae 20:132–141. doi: 10.1016/j.hal.2012.09.002. [DOI] [Google Scholar]
- 33.Landa M, Blain S, Christaki U, Monchy S, Obernosterer I. 2016. Shifts in bacterial community composition associated with increased carbon cycling in a mosaic of phytoplankton blooms. ISME J 10:39–50. doi: 10.1038/ismej.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Wang K, Shen L, Chen H, Hou F, Zhou X, Zhang D, Zhu X. 2018. Microbial community dynamics and assembly follow trajectories of an early-spring diatom bloom in a semienclosed bay. Appl Environ Microbiol 84:e01000-18. doi: 10.1128/AEM.01000-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Z, Tong YW. 2014. The interactions between Chlorella vulgaris and algal symbiotic bacteria under photoautotrophic and photoheterotrophic conditions. J Appl Phycol 26:1483–1492. doi: 10.1007/s10811-013-0186-1. [DOI] [Google Scholar]
- 36.Motone K, Takagi T, Aburaya S, Miura N, Aoki W, Ueda M. 2020. A zeaxanthin-producing bacterium isolated from the algal phycosphere protects coral endosymbionts from environmental stress. mBio 11:e01019-19. doi: 10.1128/mBio.01019-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abraham W-R, Rohd M. 2014. The family Hyphomonadaceae, p 283–297. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes. Springer, Berlin, Germany. [Google Scholar]
- 38.Strömpl C, Hold GL, Lünsdorf H, Graham J, Gallacher S, Abraham W-R, Moore ERB, Timmis KN. 2003. Oceanicaulis alexandrii gen. nov., sp. nov., a novel stalked bacterium isolated from a culture of the dinoflagellate Alexandrium tamarense (Lebour) Balech. Int J Syst Evol Microbiol 53:1901–1906. doi: 10.1099/ijs.0.02635-0. [DOI] [PubMed] [Google Scholar]
- 39.Li S, Chen M, Chen Y, Tong J, Wang L, Xu Y, Hu Z, Chen H. 2019. Epibiotic bacterial community composition in red-tide dinoflagellate Akashiwo sanguinea culture under various growth conditions. FEMS Microbiol Ecol 95:fiz057. doi: 10.1093/femsec/fiz057. [DOI] [PubMed] [Google Scholar]
- 40.Shibl AA, Isaac A, Ochsenkühn MA, Cárdenas A, Fei C, Behringer G, Arnoux M, Drou N, Santos MP, Gunsalus KC, Voolstra CR, Amin SA. 2020. Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc Natl Acad Sci USA 117:27445–27455. doi: 10.1073/pnas.2012088117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribalet F, Intertaglia L, Lebaron P, Casotti R. 2008. Differential effect of three polyunsaturated aldehydes on marine bacterial isolates. Aquat Toxicol 86:249–255. doi: 10.1016/j.aquatox.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 42.Muras A, López-Pérez M, Mayer C, Parga A, Amaro-Blanco J, Otero A. 2018. High prevalence of quorum-sensing and quorum-quenching activity among cultivable bacteria and metagenomic sequences in the Mediterranean Sea. Genes 9:100. doi: 10.3390/genes9020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elodie B, Soizic P, Didier S, Jocivania ODS, Yoan F, Nicole B, Laurent I, Alexandre E, Rapha LL. 2017. Quorum sensing and quorum quenching in the Mediterranean seagrass Posidonia oceanica microbiota. Front Mar Sci 4:218. [Google Scholar]
- 44.Buchan A, LeCleir GR, Gulvik CA, González JM. 2014. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol 12:686–698. doi: 10.1038/nrmicro3326. [DOI] [PubMed] [Google Scholar]
- 45.Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR, Morales RL, Berthiaume CT, Parker MS, Djunaedi B, Ingalls AE, Parsek MR, Moran MA, Armbrust EV. 2015. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522:98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 46.Segev E, Wyche TP, Kim KH, Petersen J, Ellebrandt C, Vlamakis H, Barteneva N, Paulson JN, Chai L, Clardy J, Kolter R. 2016. Dynamic metabolic exchange governs a marine algal-bacterial interaction. Elife 5:e17473. doi: 10.7554/eLife.17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aldunate M, Henríquez‐Castillo C, Ji Q, Lueders‐Dumont J, Mulholland MR, Ward BB, von Dassow P, Ulloa O. 2020. Nitrogen assimilation in picocyanobacteria inhabiting the oxygen-deficient waters of the eastern tropical North and South Pacific. Limnol Oceanogr 65:437–453. doi: 10.1002/lno.11315. [DOI] [Google Scholar]
- 48.Tu Q, Lin L, Cheng L, Deng Y, He Z. 2019. NCycDB: a curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes. Bioinformatics 35:1040–1048. doi: 10.1093/bioinformatics/bty741. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Z, Gonsior M, Schmitt-Kopplin P, Zhan Y, Zhang R, Jiao N, Chen F. 2019. Microbial transformation of virus-induced dissolved organic matter from picocyanobacteria: coupling of bacterial diversity and DOM chemodiversity. ISME J 13:2551–2565. doi: 10.1038/s41396-019-0449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang S, Wang J, Cong W, Cai Z, Ouyang F. 2004. Utilization of nitrite as a nitrogen source by Botryococcus braunii. Biotechnol Lett 26:239–243. doi: 10.1023/b:bile.0000013722.45527.18. [DOI] [PubMed] [Google Scholar]
- 51.Chen W, Liu H, Zhang Q, Dai S. 2011. Effect of nitrite on growth and microcystins production of Microcystis aeruginosa PCC7806. J Appl Phycol 23:665–671. doi: 10.1007/s10811-010-9558-y. [DOI] [Google Scholar]
- 52.Pérez AA, Rodionov DA, Bryant DA. 2016. Identification and regulation of genes for cobalamin transport in the cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol 198:2753–2761. doi: 10.1128/JB.00476-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- 54.Mou S, Zhang Y, Li G, Li H, Liang Y, Tang L, Tao J, Xu J, Li J, Zhang C, Jiao N. 2017. Effects of elevated CO2 and nitrogen supply on the growth and photosynthetic physiology of a marine cyanobacterium, Synechococcus sp. PCC7002. J Appl Phycol 29:1755–1763. doi: 10.1007/s10811-017-1089-3. [DOI] [Google Scholar]
- 55.Zhao X, Liu J, Zhou S, Zheng Y, Wu Y, Kogure K, Zhang X-H. 2020. Diversity of culturable heterotrophic bacteria from the Mariana Trench and their ability to degrade macromolecules. Mar Life Sci Technol 2:181–193. doi: 10.1007/s42995-020-00027-1. [DOI] [Google Scholar]
- 56.Mou S, Li G, Li H, Li F, Shao Z, Li J, Qu C, Zhang Y. 2018. Differential physiological responses of the coastal cyanobacterium Synechococcus sp. PCC7002 to elevated pCO2 at lag, exponential, and stationary growth phases. Sci China Earth Sci 61:1397–1405. doi: 10.1007/s11430-017-9206-5. [DOI] [Google Scholar]
- 57.Picazo A, Rochera C, Villaescusa JA, Miralles-Lorenzo J, Velázquez D, Quesada A, Camacho A. 2019. Bacterioplankton community composition along environmental gradients in lakes from Byers Peninsula (Maritime Antarctica) as determined by next-generation sequencing. Front Microbiol 10:908. doi: 10.3389/fmicb.2019.00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.National Genomics Data Center Members and Partners. 2020. Database resources of the National Genomics Data Center in 2020. Nucleic Acids Res 48:D24–D33. doi: 10.1093/nar/gkz913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subba Rao NS. 1977. Soil microorganisms and plant growth. Oxford & IBH Publishing, New Delhi, India. [Google Scholar]
- 60.Baldani VLD, Döbereiner J. 1980. Host-plant specificity in the infection of cereals with Azospirillum spp. Soil Biol Biochem 12:433–439. doi: 10.1016/0038-0717(80)90021-8. [DOI] [Google Scholar]
- 61.Tang L, Zhang Z, Zhou C, Cui R, Tian Y, Zhang Y. 2018. Roseicyclus marinus sp. nov., isolated from a Synechococcus culture, and emended description of the genus Roseicyclus. Int J Syst Evol Microbiol 68:1781–1786. doi: 10.1099/ijsem.0.002752. [DOI] [PubMed] [Google Scholar]
- 62.Bassler BL, Greenberg EP, Stevens AM. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol 179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poly F, Monrozier LJ, Bally R. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152:95–103. doi: 10.1016/s0923-2508(00)01172-4. [DOI] [PubMed] [Google Scholar]
- 64.Yang Q-S, Dong J-D, Ahmad M, Ling J, Zhou W-G, Tan Y-H, Zhang Y-Z, Shen D-D, Zhang Y-Y. 2019. Analysis of nifH DNA and RNA reveals a disproportionate contribution to nitrogenase activities by rare plankton-associated diazotrophs. BMC Microbiol 19:188. doi: 10.1186/s12866-019-1565-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tindall BJ, Sikorski J, Smibert RA, Krieg NR. 2007. Phenotypic characterization and the principles of comparative systematics, p 330–393. In Reddy CA, Beveridge TJ, Breznak JA, Marzluf GA, Schmidt TM, Snyder LR (ed), Methods for general and molecular microbiology, 3rd ed. ASM Press, Washington, DC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods, results, figure legends, and references. Download Text S1, DOC file, 0.06 MB (64KB, doc) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Time course of virus and Synechococcus abundances in the first generation of the subculture. Download FIG S1, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Distribution of quorum-sensing genes in each cocultivation system. PICRUSt2 was applied for the predictions of microbial functional genes. Download FIG S2, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterium-bacterium interactions. The inhibition zone around the agar slab containing Pseudomonas sp. syn326 (blue arrow) or Erythrobacter sp. SN021 (black arrow) indicates their inhibitory effect on the growth of the tested strains. The promotion zone indicates the beneficial effect. Download FIG S3, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes involved in the nitrogen cycle in the heterotrophic bacterial community cocultured with Synechococcus during the mutualism stage. The numbers in brackets beside each gene represent the abundances for the gene or gene family. Download FIG S4, TIF file, 1.0 MB (1,020.6KB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Dynamic changes in the bacterial community structure at the phylum level during long-term cocultivation with Synechococcus. Download FIG S5, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic tree of 16S rRNA gene sequences of Synechococcus strains. Clade and subcluster designations follow the nomenclature of S. Bemal and A. C. Anil (FEMS Microbiol Ecol 92:fiw162, 2016, https://doi.org/10.1093/femsec/fiw162). The tree was constructed by the neighbor-joining method with Muscle, aligning 59 16S rRNA nucleotide sequences of Synechococcus strains with 1,000 bootstrap replicates. Download FIG S6, TIF file, 2.1 MB (2.1MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nitrate-reducing bacteria in different cocultivation systems. The abilities to reduce nitrate to nitrite or nitrogen by the 42 (a) and 34 (b) representative bacterial individuals from the antagonism and mutualism stages, respectively, were tested. Beneficial bacteria for the growth of Synechococcus are marked in red, and inhibitory bacteria are in blue. Download FIG S7, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Sequence data have been deposited in the NGDC database under accession no. CRA003602 and CRA003605 and in GenBank under accession no. MW409752 to MW410111.