ABSTRACT
Peptidoglycan (PG) is a highly cross-linked peptide-glycan mesh that confers structural rigidity and shape to most bacterial cells. Polymerization of new PG is usually achieved by the concerted activity of two membrane-bound machineries, class-A penicillin binding proteins (aPBPs) and class-B penicillin binding proteins (bPBPs) in complex with shape, elongation, division, and sporulation (SEDS) proteins. Here, we have identified four phylogenetically distinct groups of bacteria that lack any identifiable aPBPs. We performed experiments on a panel of species within one of these groups, the Rickettsiales, and found that bacteria lacking aPBPs build a PG-like cell wall with minimal abundance and rigidity relative to cell walls of aPBP-containing bacteria. This reduced cell wall may have evolved to minimize the activation of host responses to pathogens and endosymbionts while retaining the minimal PG-biosynthesis machinery required for cell elongation and division. We term these “peptidoglycan-intermediate” bacteria, a cohort of host-associated species that includes some human pathogens.
KEYWORDS: peptidoglycan, penicillin binding proteins, Rickettsiales, obligate intracellular bacteria, endosymbionts, orthogonal chemical probes, Gram-negative bacteria, host-pathogen interactions
INTRODUCTION
The peptidoglycan (PG) cell wall is the load-bearing structure of most bacterial cells, conferring stiffness and osmotic protection. It comprises a mesh-like sacculus that usually covers the entirety of the cell membrane and is composed of long glycan strands cross-linked by short peptide side chains (1, 2). In Gram-positive bacteria this can be many layers thick, while in Gram-negative bacteria it is much thinner and surrounded by a second lipid membrane. As a highly conserved and essential constituent of almost all bacterial cells, PG is recognized by the eukaryotic immune system as a pathogen-associated molecular pattern and is a potent activator of innate immunity.
Polymerization of the PG precursor, lipid II, into growing PG strands has two enzymatic requirements: glycosyltransferase (GTase) activity to form β1-4-glycosidic bonds between disaccharide residues, and transpeptidase (TPase) activity to cross-link the peptide side chains on adjacent strands. Class A penicillin binding proteins (aPBPs) are large transmembrane proteins that possess both activities (3, 4). In contrast, class B PBPs (bPBPs) are transmembrane proteins that only possess TPase activity. It was recently shown that shape, elongation, division, and sporulation (SEDS) proteins, which form a complex with bPBPs, possess GTase activity in some organisms (5–9). Thus, it is possible that bPBP/SEDS and aPBPs form two complementary cell wall synthetic motors that both possess full TPase and GTase activity.The aPBPs are widespread and essential in most bacterial species and were long thought to be the primary drivers of PG polymerization; however, several lines of recent evidence suggest that most nascent PG growth is, in fact, driven by the newly described bPBP/SEDS activity, with aPBPs playing a supportive role in repairing cell wall defects. First, the subcellular localization of aPBP within the cytoplasmic membrane is distinct from foci formed by bPBP/SEDS pairs in the Gram-positive bacterium Bacillus subtilis and the Gram-negative bacterium Escherichia coli, indicating distinct activities (10). Second, it has been shown that all four aPBP genes can be deleted in B. subtilis to generate slow-growing but viable bacteria (11). Third, decreased expression of aPBPs in E. coli results in cells that remain rod shaped, but with decreased levels of PG cross-linking and less ability to sustain cell wall damage (12). These data suggest distinct, though partly complementary, roles for aPBPs and bPBPs in PG polymerization.
Here, we identify a group of bacteria that lack any identifiable aPBP genes but can synthesize a PG-like structure that is required for viability, demonstrating that aPBPs are dispensable for PG polymerization. Compared with closely related species that have aPBPs, the PG in these “PG-intermediate” (PGi) organisms is less abundant, and the cells do not have a uniform rod shape. These PGi organisms all exhibit an obligate intracellular or endosymbiotic lifestyle, and the osmotic protection and avoidance of innate immune receptors may have driven the selection for a minimal PG cell wall.
RESULTS
The class A penicillin binding protein gene has been lost at least four times during evolution.
We previously used a comparative genomics approach to characterize the distribution of cell wall biosynthesis genes in obligate intracellular bacteria, which identified species that possessed almost all genes in the classical PG biosynthesis pathway but specifically lacked any identifiable aPBPs (13). Meeske et al. also performed a phylogenetic analysis showing that SEDS/bPBPs are more widely conserved than aPBPs and that some organisms lack aPBPs but retain a SEDS/bPBP synthase (6). Here, we extended this analysis to include 119 different strains across 9 major bacterial groups, including the identifiable obligate intracellular bacteria, facultative intracellular bacteria, and endosymbionts with complete genomes available in KEGG, as well as related free-living bacteria (Fig. 1A and Fig. S1). We identified clusters of bacteria that lack aPBPs in four unrelated groups, the Rickettsiales, the Actinobacteria, the Gammaproteobacteria, and the Planctomycetes Verrucomicrobia Chlamydiae (PVC) superphylum. These organisms were classified as PGi, with the criterion that they possess most lipid II biosynthesis genes, at least one SEDS and bPBP gene, but no identifiable aPBP gene. We also identified bacteria in five diverse groups that had lost the majority of their PG biosynthesis genes and would be predicted to lack PG synthesis capability (PG-negative [PGn]). In contrast, we term species that encode aPBPs, bPBPs/SEDS, and other genes in the PG biosynthesis “PG-classical” (PGc).
FIG 1.
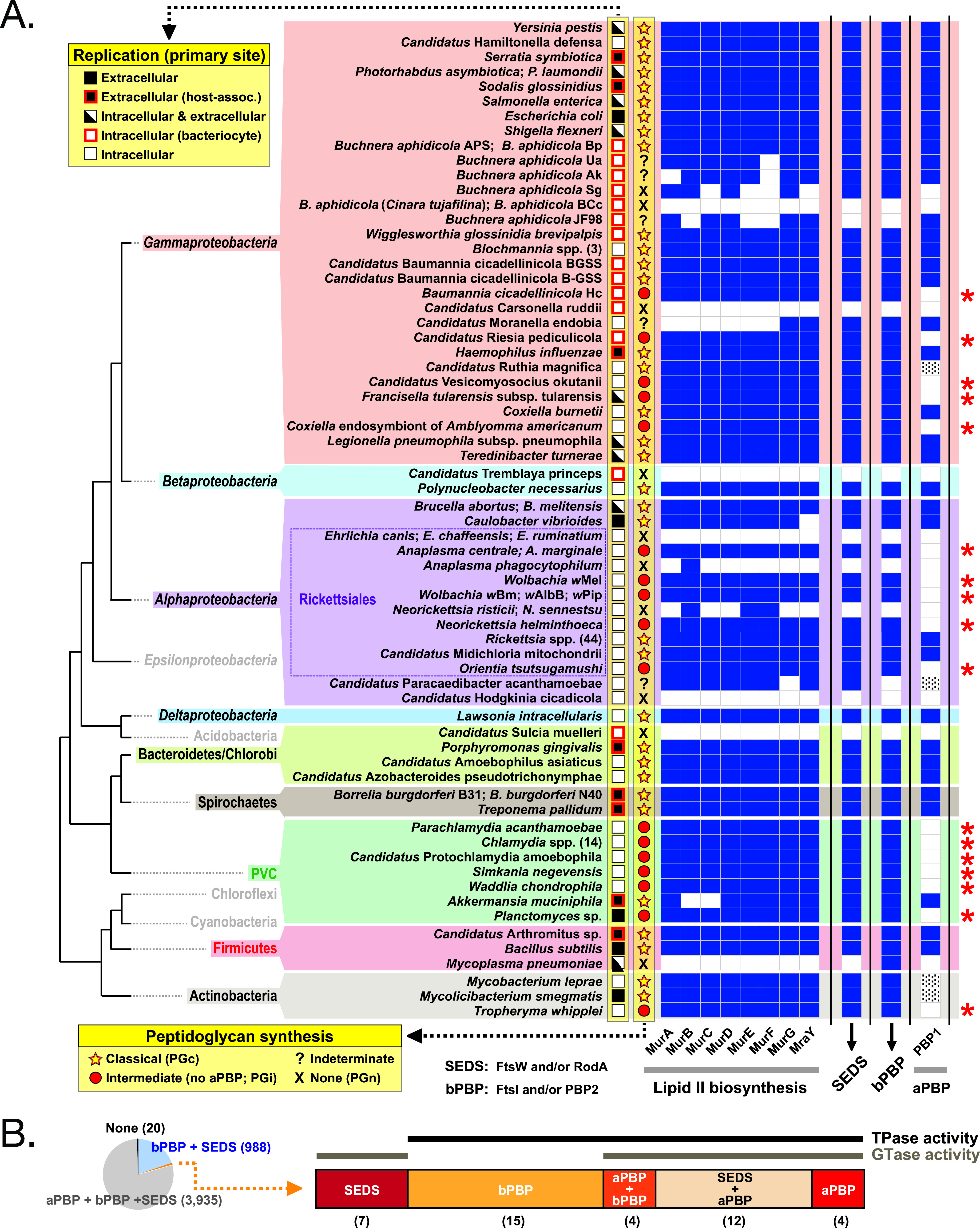
The class A PBP gene has been lost at least four times during evolution. (A) Analysis of key genes in the peptidoglycan biosynthesis pathway showing their presence or absence in obligate intracellular, facultative intracellular, host-associated, and free-living bacteria across 9 major bacterial groups. The predicted peptidoglycan (PG) status is shown as well as the primary site of replication of the bacteria. Genes involved in lipid II biosynthesis, SEDS family, class B PBPs, and class A PBPs were analyzed. The presence of a gene is shown by block color, the absence in white, and genes identified manually (not annotated as aPBP in KEGG) are shown with dashes. Bacterial organisms in the Gammaproteobacteria (Baumannia cicadellinicola Hc, “Candidatus Vesicomyosocius okutanii,” Francisella tularensis subsp. tularensis, Coxiella endosymbiont of Amblyomma americanum), Alphaproteobacteria (Anaplasma spp., Wolbachia spp., Orientia tsutsugamushi), PVC (Parachlamydia acanthamoebae, Chlamydia spp., “Candidatus Protochlamydia amoebophila,” Simkania negevensis, Waddlia chondrophila, Planctomyces sp.), and Actinobacteria (Tropheryma whipplei) all lack class A PBP homologs and are predicted to build intermediate PG-like structures (PGi) as highlighted by a red asterisk. PVC, Planctomycetes, Verrucomicrobia, and Chlamydiae; SEDS, shape, elongation, division, and sporulation; PBP, penicillin binding protein. Details of SEDS and bPBPs are shown in Fig. S1. (B) Relative quantification of bacterial strains containing different combinations of aPBP, bPBP, and SEDS proteins. This analysis was carried out on the 4,985 closed bacterial genomes available on KEGG in February 2020. Most bacteria (n = 4,923) either harbor all PG polymerization machines (80%) or qualify as PGi species (20%); a minority of species with other distributions (n = 42) are shown at the right as follows: SEDS only (n = 7), bPBP only (n = 15), bPBP and aPBP (n = 4), SEDS and aPBP (n = 12), and aPBP only (n = 4). Numbers of species in each category are shown in brackets. Bacteria binned to each category are listed in Table S1.
The class A PBP gene has been lost at least four times during evolution (extended version). Analysis of key genes in the peptidoglycan biosynthesis pathway showing their presence or absence in obligate intracellular, facultative intracellular, host-associated, and free-living bacteria across 9 phyla. The predicted peptidoglycan (PG) status is shown as well as the primary site of replication of the bacteria. Genes involved in lipid II biosynthesis, SEDS family, class B PBPs, and class A PBPs were analyzed. The presence of a gene is shown by block color, the absence in white, and genes identified manually (not annotated as aPBP in KEGG) are shown with dashes. Bacterial organisms in Gammaproteobacteria (Baumannia cicadellinicola Hc, “Candidatus Vesicomyosocius okutanii,” Francisella tularensis subsp. tularensis, Coxiella endosymbiont of Amblyomma americanum), Alphaproteobacteria (Anaplasma spp., Wolbachia spp., Orientia tsutsugamushi), PVC (Parachlamydia acanthamoebae, Chlamydia spp., “Candidatus Protochlamydia amoebophila,” Simkania negevensis, Waddlia chondrophila, Planctomyces sp.), and Actinobacteria (Tropheryma whipplei) all lack class A PBP homologs and are predicted to build intermediate PG-like structures (PGi). PVC, Planctomycetes, Verrucomicrobia, and Chlamydiae; SEDS, shape, elongation, division, and sporulation; PBP, peptidoglycan binding protein. Download FIG S1, PDF file, 0.1 MB (111.5KB, pdf) .
Copyright © 2021 Atwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
KEGGerator output indicating presence (1) or absence (0) of each KEGG ortholog involved in peptidoglycan synthesis, across all 4,989 bacterial genomes in KEGG. Each row contains a single taxon, including the KEGG group, genus, and KEGG organism identifier. Each column is a distinct ortholog identified with the KEGG identifier and, where applicable, gene locus. Columns Z, AA, and AB sum the number of different orthologs within the SEDS, bPBP, and aPBP families, respectively. Each sheet represents a different subset of bacteria. (A) All (n = 4,989) bacteria. (B) Bacteria (n = 3,935) containing at least one ortholog from all three families (SEDS, aPBP, bPBP). (C) Bacteria (n = 987) containing SEDS and bPBP but no aPBP. (D) Bacteria (n = 20) lacking SEDS, aPBP, and bPBP. (E) Bacteria (n = 7) containing SEDS but not aPBP or bPBP. (F) Bacteria (n = 19) containing bPBP but not SEDS or aPBP. (G) Bacteria (n = 4) containing aPBP and bPBP but not SEDS. (H) Bacteria (n = 12) containing SEDS and aPBP but not bPBP. (I) Bacteria (n = 4) containing aPBP but not SEDS or bPBP. Download Table S1, XLSX file, 1.0 MB (1MB, xlsx) .
Copyright © 2021 Atwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Taxonomic groups included in the current study. Each entry is identified by its KEGG organism identifier, genus, taxon name, and lifestyle (extracellular, extracellular-attached, intracellular-obligate, intracellular-facultative, bacteriocytic, bacteriocytic; extracellular, or free-living). (A) All 117 taxa in the analysis. (B) All taxa binned into 65 groups, based on taxonomic relatedness and PGP profile similarity. Download Table S2, XLSX file, 0.01 MB (17.2KB, xlsx) .
Copyright © 2021 Atwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
KEGG identifiers for all orthologs included in the current study, grouped by broad biochemical pathway. Download Table S3, XLSX file, 0.01 MB (9.6KB, xlsx) .
Copyright © 2021 Atwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
This result showed that aPBPs are dispensable for growth in host-associated bacteria. We extended this analysis to all 4,985 closed bacterial genomes available on KEGG without distinguishing organisms by lifestyle (Fig. 1B and Table S1). We found that the majority (3,935) encoded aPBPs, bPBPs, and SEDS (PGc), a smaller fraction (988) had specifically lost aPBPs (PGi), and a small number (20) had lost all three classes of genes (PGn). In contrast, we only identified 20 instances of strains that had retained an aPBP but lost SEDS and/or bPBP. This analysis builds on previous reports of organisms lacking aPBPs (6, 13) and demonstrates that while aPBPs are dispensable for growth, SEDS/bPBPs have been almost universally retained.
The Rickettsiales are an order of obligate intracellular bacteria that are dominated in nature by arthropod endosymbionts (Wolbachia strains and other species/strains) but also include a number of important arthropod-transmitted animal pathogens (species of Neorickettsia, Anaplasma, Ehrlichia, Orientia, Rickettsia). The pathogens Ehrlichia spp., Anaplasma phagocytophilum, Neorickettsia sennetsu, and Neorickettsia risticii were all classified as PGn, while the closely related pathogens Anaplasma marginale, Anaplasma centrale, Neorickettsia helminthoeca, Orientia tsutsugamushi, and Wolbachia strains were all classified as PGi. Remarkably, despite significant genome reduction, all 44 Rickettsia species/strains contain a full complement of PG synthesis genes and were classified as PGc.
The Gammaproteobacteria include obligate intracellular, bacteriocyte-associated intracellular, facultative intracellular, host-associated extracellular, and free-living extracellular species. Out of 30 Gammaproteobacteria strains, we identified 7 PGn/unclassified, 6 PGi and 17 PGc. We found three instances where closely related strains had distinct classifications: (i) Buchnera aphidicola, where five strains were predicted to be PGn (or unclassified) and two other strains of B. aphidicola (APS, Bp) were predicted to be PGc. B. aphidicola strain JF98 was the only strain in our data set that retained aPBP but lost bPBP/SEDS; however, this also lacks murB, murE, and murF and is likely in the process of pathway degradation. (ii) Baumannia cicadellinicola, where one strain (“Candidatus Baumannia cicadellinicola” BGSS) was classified as PGc while another strain (B. cicadellinicola Hc) lacked aPBP and was classified as PGi. (iii) Coxielliae, where Coxiella burnettii was classified as PGc and a Coxiella endosymbiont of Amblyomma americanum was classified as PGi.
The PVC group includes obligate intracellular pathogens (species of Chlamydia, Waddlia), amoeba endosymbionts (Simkania spp.), and extracellular bacterial species (Akkermansia muciniphila, Planctomyces spp.). We analyzed seven distinct species, and all except Akkermansia were classified as PGi. Akkermansia, which is a host-associated extracellular bacterium, retained aPBP and was classified as PGc.
The Actinobacteria are a group of Gram-positive bacteria that includes Mycobacterium species. We analyzed three species and found that Tropheryma whipplei, which is a human pathogen generally thought to have an obligate intracellular lifestyle, was classified as PGi, while the other species were classified as PGc.
Together, this analysis led us to the hypothesis that the PGi bacteria produce a PG wall that is different from that produced by PGc bacteria and that this would confer an advantage within the obligate intracellular/endosymbiotic life cycle. In order to test this hypothesis, we selected a group of six related organisms from within one of these groups, the Rickettsiales, and set out to determine whether they synthesized PG walls with different characteristics. For this analysis we selected representative PGc (Rickettsia canadensis), PGi (Orientia tsutsugamushi, Wolbachia endosymbiont of melanogaster [Wm], and Anaplasma marginale), and PGn (Anaplasma phagocytophilum, and Ehrlichia chaffeensis) species.
PG-intermediate bacterial species build a PG-like structure.
We used a NOD1 reporter assay to assess whether the PGc and PGi species generated detectable PG-like structures. NOD1 is a mammalian innate immune receptor that detects PG fragments containing the PG-specific amino acid meso-DAP that is commonly found in Gram-negative bacterial PG (14). The NOD1 reporter assay measures the response of human embryonic kidney (Hek1) reporter cells to meso-DAP-containing PG fragments (15). We purified 6 Rickettsiales species from host cells and added them to NOD1 reporter cells at a constant bacterial concentration (Fig. 2A and B). While PGn species did not stimulate a NOD1 response, both PGc and PGi induced NOD1 activation. This demonstrated the presence of meso-DAP-containing PG in R. canadensis, A. marginale, O. tsutsugamushi, and Wolbachia pipientis (Wp). The magnitude of this activation was higher in the PGc species R. canadensis than in PGi organisms, suggesting a higher abundance of PG in this species.
FIG 2.
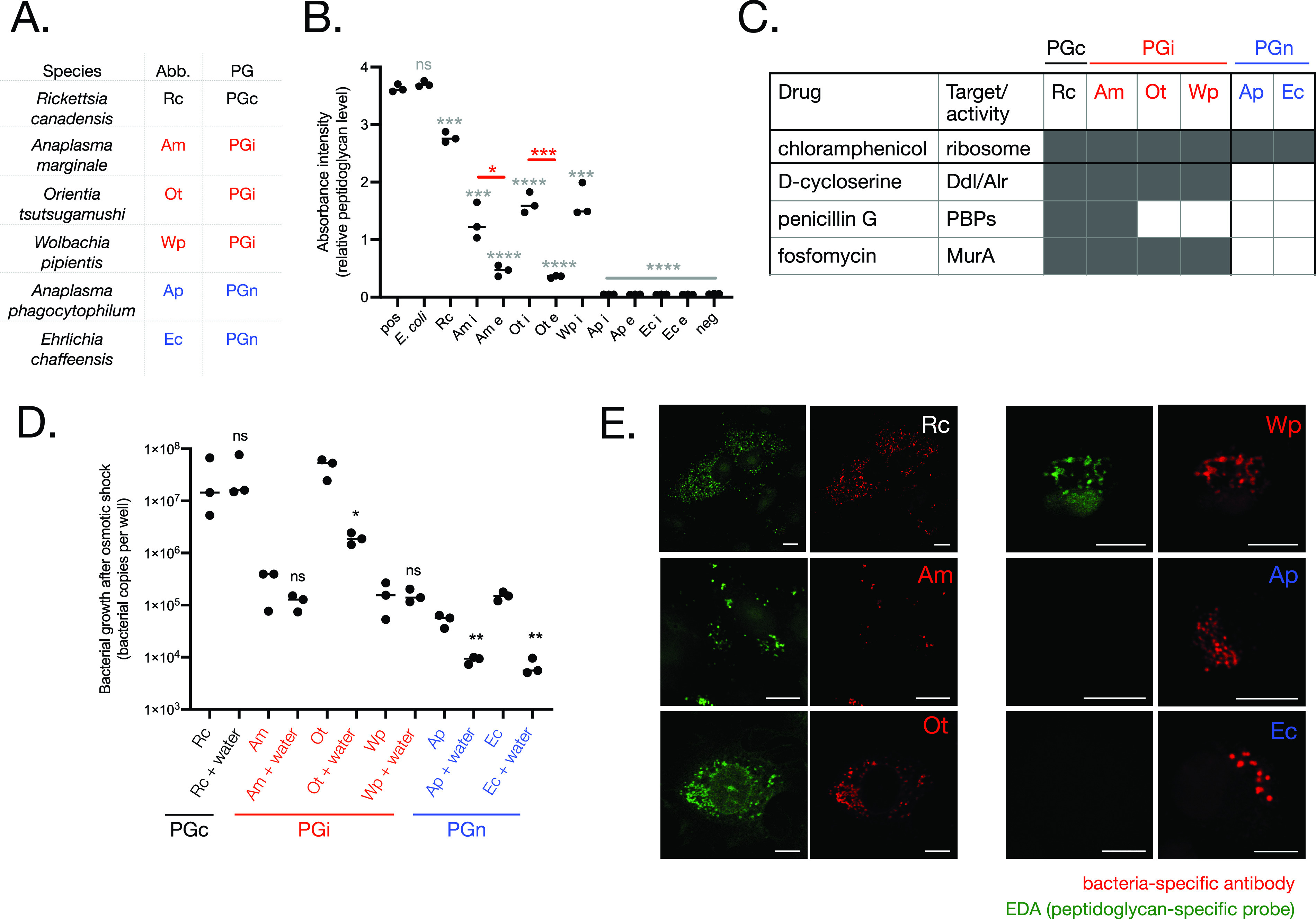
PG-intermediate Rickettsiales species build a PG-like structure that confers osmotic protection. (A) Overview of species in this figure. (B) Quantification of relative PG levels using a NOD1 reporter cell line. Extracellular (e) and intracellular (i) bacteria were isolated from host cells and quantified by qPCR, and 1 × 10E4 bacteria were added to HEK-Blue NOD1 reporter cells. Activation of NOD1 in response to peptidoglycan was measured by secreted alkaline phosphatase activity. Positive control (pos) was 10 μg/ml iE-DAP. Individual values and median are shown, and statistical analyses show the difference between two groups as measured by an unpaired t test using the software GraphPad Prism. Gray asterisks show the results of statistical comparisons between each group and the positive control. Red asterisks show the difference between the i and e population of a particular bacterial species. (C) Sensitivity of Rickettsiales to cell wall-targeting drugs. Bacteria were grown in the presence of drugs at the following concentrations: chloramphenicol, 100 μg/ml; penicillin G, 150 μg/ml; d-cycloserine 250 μg/ml; and phosphomycin, 40 μg/ml. The bacterial copy number after growth was compared with that in the absence of drugs. A bacterial species was scored as susceptible to that drug if the bacterial copy number was reduced in a statistically significant manner as measured by an unpaired t test. Gray cell, bacteria susceptible to drug; white cell, bacteria resistant to drug. Raw data are given in Fig. S1. (D) Quantification of bacterial regrowth after 10 min of incubation in sucrose phosphate buffer (SPG) or water. Bacteria were isolated from host cells, exposed to osmotic shock (pure water) or an osmotically protective buffer (SPG) for 10 min, and then grown in host cells for 7 days. Bacterial growth was measured by qPCR. Individual values and median are shown, and an unpaired t test was used to compare the water-treated and untreated groups within each bacterial species using the software GraphPad Prism. (E) Visualization of a PG-like structure in R. canadensis and PGi species. Bacteria were grown in the presence of a d-alanine analog containing an alkyne group (EDA). After fixation, incorporation of probes into nascent PG was detected by labeling with azide-alexa488 using a copper catalyzed click reaction. Bacteria were counterstained using bacterium-specific antibodies. PGc (R. canadensis) and PGi (A. marginale/Wp/O. tsutsugamushi) species could be labeled with EDA, while PGn (E. chaffeensis/A. phagocytophilum) species could not. Throughout the figure, PGc (R. canadensis) is indicated in black, PGi (A. marginale, O. tsutsugamushi, Wp) in red, and PGn (A. phagocytophilum, E. chaffeensis) in blue. P values throughout: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
We analyzed PG levels at different stages of the bacterial life cycle. The Anaplasmataceae A. marginale, A. phagocytophilum, and E. chaffeensis undergo a biphasic life cycle in which they differentiate between a replicative intracellular form and an infectious extracellular form (16). We recently showed that O. tsutsugamushi also differentiates into distinct intracellular and extracellular populations (manuscript in preparation). In contrast, differentiation has not been described in R. canadensis and Wp. We isolated bacteria from intracellular and extracellular populations in A. marginale, O. tsutsugamushi, A. phagocytophilum, and E. chaffeensis and found that in the PGi species A. marginale and O. tsutsugamushi, the PG levels were significantly higher in the intracellular population (Fig. 2B), at which time the bacteria are actively undergoing growth and division. PG could not be detected in either intracellular or extracellular populations of A. phagocytophilum or E. chaffeensis.
Next, we determined the susceptibility of the same six species to PG-targeting drugs, in order to determine whether they made a PG-like structure that was required for their growth (Fig. 2C and Fig. S2). We used the bacterial ribosomal inhibitor chloramphenicol as a positive control, because this is known to be effective against Rickettsiales. We tested the susceptibility to d-cycloserine (targets proteins involved in the isomerization of l-Ala to d-Ala and its dimerization into d-Ala-d-Ala); penicillin G (targets TPase activity of aPBP and bPBP), and fosfomycin (targets MurA, a cytoplasmic protein required for early stages of synthesis of the PG precursor lipid II). The PGc species R. canadensis was sensitive to all cell wall-targeting drugs that we tested. The PGi organisms were all sensitive to d-cycloserine and fosfomycin, consistent with the presence of an essential PG-like structure and as shown previously for some of these (17–19). A. marginale was susceptible to penicillin G; both O. tsutsugamushi and Wp were not. The susceptibility of PGi organisms to d-cycloserine and fosfomycin demonstrates that they build a PG-like structure that is required for their growth; however, the differential susceptibility of PGi organisms to penicillin G cannot currently be explained and may reflect different PBP modifications or differences in membrane structure or permeability. PGn organisms were insensitive to all the cell wall-targeting drugs that we tested, consistent with a lack of a functional PG cell wall.
Drug sensitivity assays. Bacterial copy number after 5 days (7 days for Wolbachia) of growth in the presence or absence of drugs was quantified by qPCR. Growth in the presence of a drug was normalized by growth in the absence of drug within the same experiment. RC, Rickettsia canadensis, grown for 5 days in Vero cells; AM, Anaplasma marginale grown for 5 days in Vero cells; OT, Orientia tsutsugamushi grown for 5 days in L929 cells; WP, Wolbachia pipientis grown for 7 days in JW18 cells; EC, Ehrlichia chaffeensis grown for 5 days in DH82 cells; AP, Anaplasma phagocytophilum grown for 5 days in HL60 cells. Drug concentrations were 100 μg/ml chloramphenicol, 150 μg/ml penicillin G, 250 μg/ml d-cycloserine, and 40 μg/ml phosphomycin. Statistical analysis between the untreated and drug-treated groups was carried out using an unpaired t test with GraphPad Prism software. Individual values and median are shown. P values: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. Download FIG S2, PDF file, 0.1 MB (64.1KB, pdf) .
Copyright © 2021 Atwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, we tested whether the PG-like structure of PGi species conferred osmotic protection. We isolated bacteria from their host cells, exposed them to hypoosmotic shock (water), and assessed the effect on subsequent bacterial growth (Fig. 2D). We found that the PGn species were highly sensitive to hypoosmotic shock, whereas growth of the PGi species A. marginale and Wp as well as the PGc R. canadensis was not affected. Growth of the PGi O. tsutsugamushi was affected by hypoosmotic shock, demonstrating that the amount of PG in this organism was not sufficient to confer complete protection. While the mechanistic basis of this difference is unknown, it may reflect the fact that A. marginale and Wp replicate within a membrane-bound vacuole and may have evolved additional rigidity in their membranes to survive vacuole-associated hypoosmolarity compared with the cytoplasm-residing PGi species O. tsutsugamushi.
Visualization of a PG-like structure in PG-intermediate species using a PG-specific metabolic probe.
We used a clickable d-amino acid analog, ethynyl-d-alanine (EDA), to determine the spatial localization of PG in PGc and PGi Rickettsiales species (Fig. 2E). This is an orthogonal chemical probe that incorporates into the PG of growing bacterial cells and can be conjugated to a fluorophore after fixation for fluorescence microscopy analysis. We have previously used this to label the PGi O. tsutsugamushi (17). This probe labeled both PGc and PGi species in our study, indicating the presence of some form of PG sacculus. However, we found that the labeling was frequently unsuccessful despite carefully controlling for variability. This may be due to batch-dependent differences in permeability of host cells or differences in the activity of PG synthesis machinery at different stages of bacterial growth. The PGn species A. phagocytophilum and E. chaffeensis could never be labeled with EDA, consistent with our hypothesis that these lack any PG-like structure and demonstrating that the probes do not bind nonspecifically to bacterial cells under these experimental conditions. It has been shown that some PGi Chlamydiales species only synthesize PG at their septum (20). Such localization was never observed in any of the species studied here.
Analysis of the cell shape of PGc, PGi, and PGn Rickettsiales.
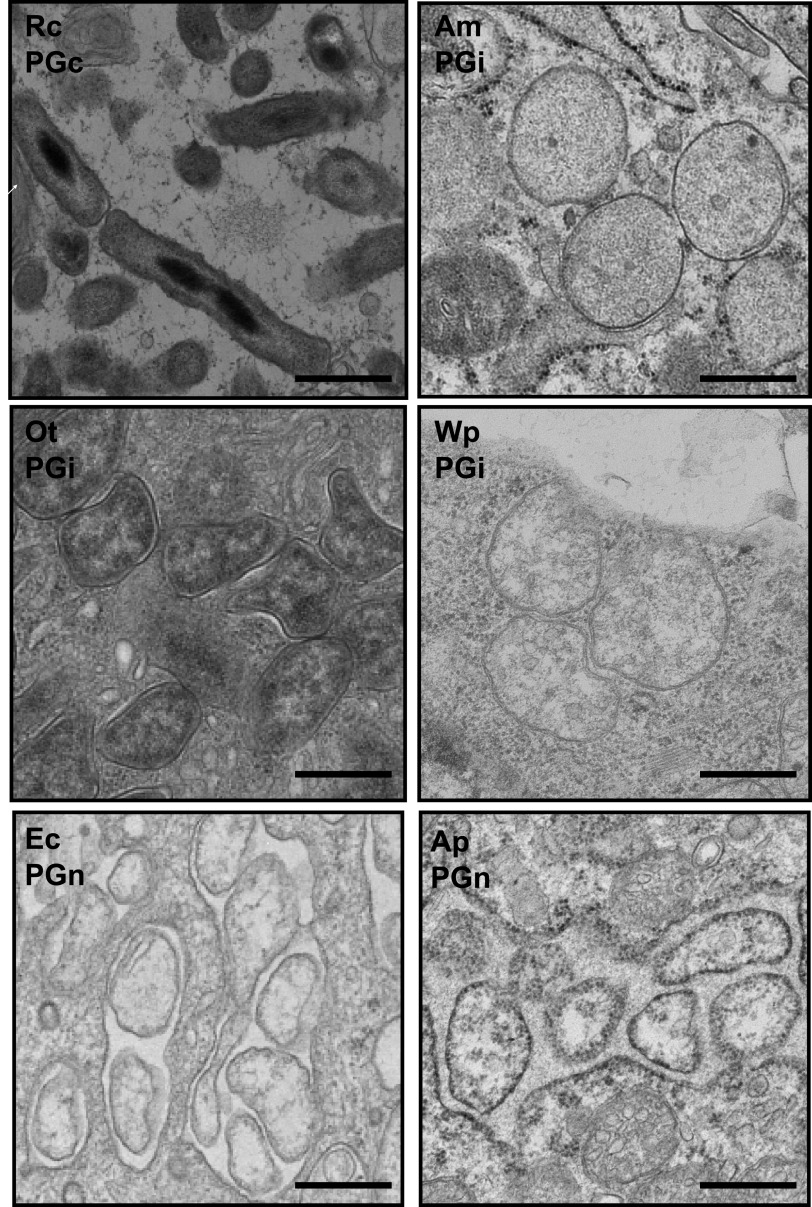
The low levels of NOD1 activation in PGi species (Fig. 2A), combined with the fact that PG has been difficult to detect in both O. tsutsugamushi and Wp, suggests that the absence of aPBP leads to a reduced amount of PG in the cell wall. We reasoned that this may result in reduced structural rigidity and that this would affect bacterial shape. Infected cells were examined by transmission electron microscopy to document the shape of the six Rickettsiales species within host cells (Fig. 3). We found that PGc R. canadensis forms regular, rod-shaped cells, while PGi and PGn species adopt irregular and/or round cells. This finding is consistent with an electron microscopy analysis showing that the Gammaproteobacteria “Candidatus Baumannia cicadellinicola” strain BGSS (endosymbiont of blue-green sharpshooter), classified here as PGc, is rod shaped while the closely related Baumannia cicadellinicola strain Hc (endosymbiont of glassy-winged sharpshooter) classified here as PGi, is irregular and non-rod shaped (21).
FIG 3.
Transmission electron microscopy analysis of PGc, PGi, and PGn Rickettsiales. Bacteria were grown in host cells and prepared for fixed thin-section transmission electron microscopy. Micrographs show that the PGi R. canadensis is rod-shaped compared with PGi and PGn Rickettsiales. PGi and PGn bacteria are round and/or pleiomorphic, reflecting the low level or absence of PG in their cell walls.
DISCUSSION
Here, we used comparative genomics to identify 16 species across 4 major bacterial groups that have lost aPBPs but retained genes for synthesis of the PG precursor lipid II as well as at least one bPBP and one SEDS gene. We characterized three of these species from within the order Rickettsiales and showed that they generate a PG-like structure that is required for bacterial growth. It has already been shown that multiple chlamydial species, which are not closely related to Rickettsiales, generate minimal PG-like structures (20, 22), and we hypothesize that an absence of aPBPs in Gammaproteobacteria and Actinobacteria will also generate bacteria with similar cell wall characteristics.
The species that lack aPBP were almost all associated with an intracellular lifestyle (15 out of 16). There are two possible reasons why adaptation to an intracellular lifestyle would confer selective pressure to reduce the amount of PG in the bacterial cell wall. First, PG is a strong stimulator of the host immune system via peptidoglycan recognition proteins such as NOD1/NOD2 and PG recognition proteins in mammals and peptidoglycan recognition proteins (PGRPs) in invertebrates. Activation of these systems leads to induction of antibacterial mechanisms that intracellular bacteria aim to evade, and we show here that both PGc and PGi Rickettsiales are able to activate NOD1. There is substantial variation in the abundance and distribution of PGRPs in invertebrate hosts (23), and differences in the magnitude of PG-sensing between hosts may underpin why some host-associated bacteria were able to retain a classical cell wall (PGc) while others were under strong selective pressure to reduce or remove it (PGi/PGn). Second, the intracellular niche (whether cytosolic or vacuolar) offers an osmotically protective environment, and therefore the bacteria may not require cell walls with the same rigidity as their free-living counterparts. Rickettsia species counter this argument yet are the only analyzed Rickettsiales species that synthesize lipopolysaccharide (24), which may require a more rigid cell wall for scaffolding this large glycoconjugate.
How do PGi bacteria build a PG wall in the absence of aPBPs, a bifunctional TPase and GTase normally considered a major driver of PG polymerization? All the PGi strains identified in our analysis retained at least one copy of the two known bPBP and SEDS genes. bPBPs have TPase activity, and SEDS from E. coli and B. subtilis are known to have GTase activity (5, 6, 8, 9). Thus, it is plausible that bPBP/SEDS complexes are the major drivers of PG polymerization in aPBP-negative PGi organisms. This is supported by the observation that aPBPs in B. subtilis are dispensable for growth (11), resulting in a reliance on bPBP/SEDS-driven PG polymerization analogous to the PGi species in our study. Our model for PG polymerization in PGc, PGi, and PGn bacteria is shown in Fig. 4.
FIG 4.
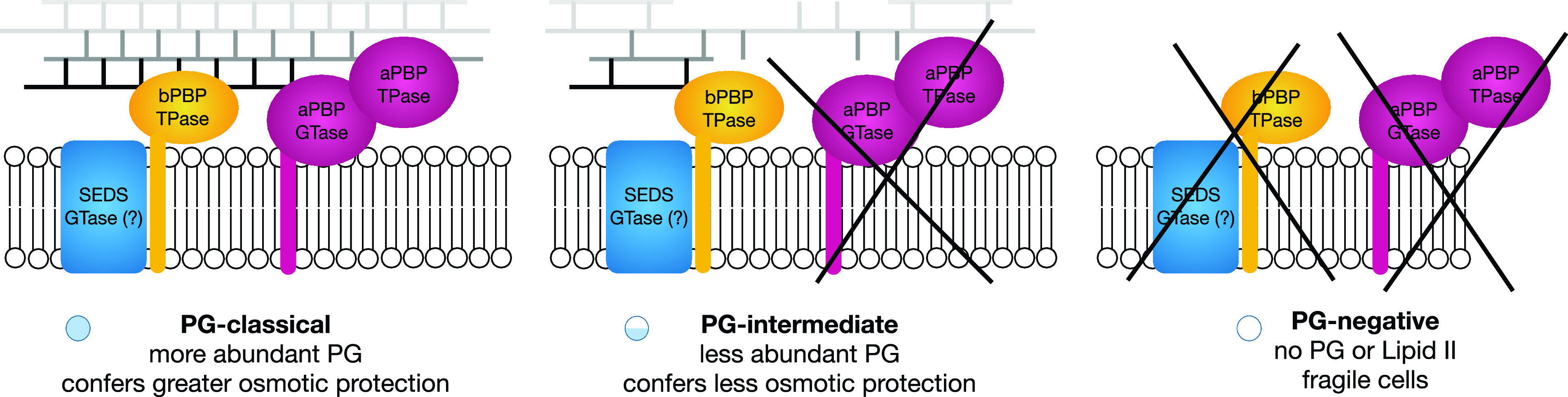
Predicted PG polymerization in PG-classic, PG-intermediate, and PG-negative organisms. SEDS, shape, elongation, division, and sporulation proteins (FtsW, RodA); bPBP, class B penicillin binding protein (PBP2, FtsI); aPBP, class A penicillin binding protein; TPase, transpeptidase activity; GTase, glycosyltransferase activity.
These results also raise the following question: what is the structure of the bacterial cell wall in PGi organisms? The reduced level of NOD1 activation shown in this study compared with E. coli and the PGc R. canadensis, the difficulty in d-alanine orthogonal probe labeling, and the historical inability to detect PG in PGi organisms (18, 25–28) suggest that the PG in these bacteria is not abundant, and in organisms with biphasic lifestyles this structure is further depleted during some stages of the growth cycle. Since PG in Gram-negative bacteria is only one or a few layers thick (1), the observed low abundance is likely to result from PG consisting of shorter glycan strands and/or less peptide cross-linking. Diminished cross-linking within the PG sacculus would result in a larger mesh size and lower structural support to the underlying membrane (Fig. 4). This would explain the reduced osmotic protection of PGi Rickettsiales compared with the closely related PGc relatives, as well as the loss of regularity in cell shape that would result from a less extensive—and therefore less rigid—PG sacculus. This interpretation of the PG structure in PGi organisms is consistent with a role for aPBP in increasing the cross-linking and repairing damage (12) rather than being the major driver of PG polymerization.
Together, these data show that aPBP is dispensable for PG polymerization and raises questions about the structure and synthesis of PG in organisms that naturally lack this important component of the PG biosynthesis machinery.
MATERIALS AND METHODS
Comparative genomics analyses.
A total of 119 bacterial taxa (Table S2) were selected for comparative analysis based on their lifestyle, phylogenomic relationship, and genome status (closed genomes only). Then, 32 genes of interest were chosen (Table S3), 10 lipid II biosynthesis genes, 2 SEDS family protein genes, 13 bPBP genes, and 7 aPBP genes. Identifiers for all taxa and genes were located in the Kyoto Encyclopedia of Genes and Genomes (KEGG) and used as input to kegghole, part of the keggerator package (24). This software queries KEGG for the presence of the input genes in all input taxa and generates a presence/absence matrix. Genomes that appeared to lack an input gene according to KEGG were manually queried using the protein sequence from a related organism and NCBI’s blastp program (default settings except for an E value cutoff of 1). Genes absent from all 117 query taxa were removed. Finally, phylogenetically related taxa with identical patterns of gene presence/absence were compressed into taxon groups for simplicity.
Bacterial propagation and quantification.
The cell lines and bacteria listed in Table 1 were used.
TABLE 1.
Cell lines and bacteria used in the study
| Bacterial species | Strain | Source of strain | Host cell type | Source of cell line |
|---|---|---|---|---|
| Rickettsia canadensis | CA410 | Stuart Blacksell | L929 (mouse fibroblast)HeLa (human epithelial) | ATCC CCL-1ATCC CCL-2 |
| Anaplasma marginale | 291 | ATCC (VR-1444) | Vero (monkey epithelial) | ATCC CRL-1586 |
| Orientia tsutsugamushi | UT76 | Kelly Brayton | Vero (monkey epithelial) | ATCC CRL-1586 |
| Wolbachia pipientis | wMel | Irene Newton | JW18 (Drosophila) | Irene Newton |
| Anaplasma phagocytophilum | HGE1 | Kelly Brayton | DH82 (dog macrophage) | ATCC CRL-10389 |
| Ehrlichia chaffeensis | Arkansas | Jere Mcbride | DH82 (dog macrophage) | ATCC CRL-10389 |
Cell culture.
L929 cells were grown in RPMI 1640 medium with HEPES (Thermo Fisher Scientific, USA; 22-400-071) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific; 16140071) in 25-cm2 flasks at 37°C and 5% CO2. HeLa cells were grown in 25-cm2 flasks with growth medium (Dulbecco’s modified Eagle’s medium [DMEM]; Thermo Fisher Scientific; 21013) supplemented with 10% heat-inactivated FBS (Thermo Fisher Scientific; 16140071) at 37°C and 5% CO2. Vero cells were grown in RPMI 1640 medium with HEPES, supplemented with 10% heat-inactivated FBS in 25-cm2 flasks at 37°C and 5% CO2. DH82 cells were grown in 25-cm2 flasks with Eagle’s minimum essential medium (EMEM; Sigma, USA; M0325) with 10% heat-inactivated FBS at 37°C and 5% CO2. JW18 cells were grown in Schneider’s insect medium (Sigma; S0146) with 10% heat-inactivated FBS at 25°C.
HEK-Blue hNOD1 (InvivoGen, USA; chkb-hnod1) cells were used for NOD1 reporter assays and were grown in 25-cm2 flasks at 37°C and 5% CO2 with growth medium DMEM with 4.5 g/liter glucose, 10% heat-inactivated FBS, 100 U/ml penicillin, 100 mg/ml streptomycin, 100 mg/ml Normocin, and 2 mM l-glutamine. Selective antibiotics, 30 μg/ml of blasticidin and 100 μg/ml of Zeocin, were added after passage 2 to maintain the cell line.
Bacterial copy number was determined by extracting DNA using alkaline lysis treatment and then performing quantitative PCR (qPCR) relative to known standards (29) (Table 2).
Table 2.
Primers and probes used in the study
| Primer/probe name | Target organism(s) | Target gene | Sequence |
|---|---|---|---|
| tsa47f | Orientia tsutsugamushi | tsa47 | TCCAGAATTAAATGAGAATTTAGGAC |
| tsa47r | TTAGTAATTACATCTCCAGGAGCAA | ||
| tsa47probe | [6FAM]TTCCACATTGTGCTGCAGATCCTTC[TAM] | ||
| Rick16Sf | Rickettsiaceae: Orientia tsutsugamushi/ Rickettsia canadensis | 16S | GCTACACGCGTGCTACAATGG |
| Rick16SRr | TGTGTACAAGGCCCGAGAACG | ||
| Rick16Sprobe | [6FAM] ATCGCTAGTAATCGCGGATCAGCATGCC [TAM] | ||
| Ana16Sf | Anaplasmataceae: Wolbachia/Anaplasma/Ehrlichia | 16S | ACTGGAGGAAGGTGGGGATG |
| Ana16Sr | TGATCCACGATTACTAGCGATTCC | ||
| Ana16Sprobe | [6FAM] TGGGCTACACACGTGCTACAATGG [TAM] |
Quantification of relative peptidoglycan levels using NOD1 reporter assay.
HEK-Blue hNod1 cells (Invitrogen, USA; hkb-hnod1) were grown in DMEM growth medium with 4.5 g/liter glucose, 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, 100 mg/ml streptomycin, 100 mg/ml Normocin, and 2 mM l-glutamine plus selective antibiotics (30 μg/ml of blasticidin and 100 μg/ml of Zeocin). Cells were seeded on clear-bottom black 96-well plates (Corning, USA; 29444-008) 2 days before infection. Bacteria were taken from prepared aliquots kept at −80°C in sucrose phosphate glutamine (SPG). All bacteria were heat-inactivated at 90°C for 30 min before being added to host cells. All infections were carried out in triplicate. After 2 days of infection, growth medium was replaced with HEK-Blue detection medium (InvivoGen, USA; hb-det2) for secreted embryonic alkaline phosphatase (SEAP) detection. The plate was further incubated at 37°C and 5% CO2 and quantified by spectrophotometry (Synergy H1; BioTek) at 640 nm from 6 h of the addition of detection media. Results were added to Prism (GraphPad Software, San Diego, CA, USA), and an unpaired t test was performed to compare bacterial SEAP levels to positive-control iE-DAP (γ-d-Glu-mDAP) at 10 μg/ml, as well as to perform pairwise comparisons between intracellular and extracellular bacteria in the case of Wolbachia and Orientia tsutsugamushi species.
Growth inhibition experiments.
Bacteria were grown in 12-well plates in their respective cell lines in the presence of drugs at the following concentrations: 100 μg/ml chloramphenicol, 150 μg/ml penicillin G, 250 μg/ml d-cycloserine, and 40 μg/ml phosphomycin. After 5 days of growth (7 days for Wolbachia) bacterial DNA was extracted and quantified by qPCR as described above. For quantifying the effect of osmotic shock, bacteria were isolated from host cells, resuspended in pure water for 10 min, and then grown in fresh host cells. Bacterial growth after 5 days was quantified by qPCR. Results were added to Prism (GraphPad Software, San Diego, CA, USA), and an unpaired t test was performed to compare growth in the presence or absence of drugs or growth in the presence or absence of osmotic shock.
Immunofluorescence labeling, click labeling, and confocal microscopy.
All fixed cells were permeabilized in 0.5% Triton X for 30 min, 100% ethanol for 1 h on ice, and 1 mg/ml lysozyme in sterile tris-EDTA for 1 h at room temperature. Primary antibodies were added for 1 h at 37°C (TSA56, 13-6, ANAF16C1, FtsZ, dog serum, and anti-p44; see Table 3). Samples were washed 3 times with phosphate-buffered saline-bovine serum albumin (PBS-BSA), and then the appropriate secondary antibodies were diluted 1:500 and incubated for 30 min at 37°C in the dark (goat anti-rat IgG Alexafluor 555 conjugate [Thermo Fisher A-21434], goat anti-rabbit, Alex Fluor 594 [Thermo Fisher A-11012], goat anti-mouse IgG superclonal Alexa Fluor 555 [Thermo Fisher A28180], and goat anti-canine IgG Texas red [Novus Biologicals NBP173511]). The nuclear stain Hoechst was diluted to 1:1,000 and included with the secondary antibody incubation. Before addition of the mounting medium, cells were washed with 1× PBS again.
Table 3.
Primary antibodies used in the study
| Species | Antibody name | Source |
|---|---|---|
| O. tsutsugamushi | TSA56 | In-house |
| R. canadensis | 13-6 | Ted Hackstadt, Rocky Mountain Laboratories |
| R. canadensis | 4994 | Ted Hackstadt, Rocky Mountain Laboratories |
| A. marginale | ANAF16C1 (Msp5) | Kelly Brayton, Washington State University |
| A. phagocytophilum | FtsZ | Jason Carlyon, Virginia Commonwealth University |
| Erlichia canis | Serum | Roman Ganta, Kansas State University |
| Wolbachia | Anti-p44 | Irene Newton, Indiana University |
The metabolic click-labeling is based on the Click-iT l-homopropargylglycine (HPG) Alexa Fluor protein synthesis assay kits (molecular probe by Life Technologies). To incorporate HPG at each time point, infected cells were incubated in the minimal medium without l-methionine (Dulbecco’s modified Eagle’s medium [DMEM], catalog [cat.] no. 21013) containing 25 μM HPG for 30 min at 37°C. Labeled bacteria were washed twice in PBS plus 1 mg/ml BSA before fixing with 1% formaldehyde or methanol (antibody 13-6 only) and subsequently permeabilized with 0.5% Triton X for 30 min, 100% ethanol for 1 h on ice, and 1 mg/ml lysozyme in sterile tris-EDTA for 1 h at room temperature. After washing with PBS plus 1 mg/ml BSA, Click-iT reaction cocktail was incubated with cells for 30 min at room temperature protected from light. The component of Click-iT reaction cocktail is based on Click-iT HPG Alexa Fluor protein synthesis assay kits [cat.] no. C10428. The azide dye (Alexa Fluor 488; Invitrogen A10266) was used at final concentration of 5 μM. After the click reaction, the cells were ready for immunofluorescent fluorescence labeling and imaging as described above.
Imaging was performed using an Observer Z1 LSM700 confocal microscope with an HBO 100 illuminating system equipped with a ×63/1.4 Plan-APOCHROMAT objective lens (Carl Zeiss, Germany) and 405-nm, 488-nm, and 555-nm laser lines. In some cases, we used a TCS SP8 confocal microscope (Leica Microsystems, Germany) equipped with a ×63/1.4 Plan-APOCHROMAT oil objective lens with a 1.4-mm working distance and 405-nm, 488-nm, 552- nm, and 638-nm laser lines.
Transmission electron microscopy.
Bacteria were grown in their respective cell lines, harvested by trypsinization, and pelleted at 1,000 × g in a microcentrifuge. Culture medium supernatants were removed, and the resultant cell pellets were fixed using 2.5% formaldehyde, 2.5% glutaraldehyde, and 0.1 M sodium cacodylate buffer, pH 7.4 (EMS catalog no. 15949). After overnight fixation at 4°C, the pellets were gently rinsed and floated in 0.1 M sodium cacodylate buffer, pH 7.4 and postfixed with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer, followed by en-bloc staining in 1% aqueous uranyl acetate. Pellets were dehydrated through a graded series of ethanol and propylene oxide up to 100% propylene oxide and then incubated in a 1:1 mixture of propylene oxide and EMBed 812 (Electron Microscopy Sciences; 14120). Pellets were equilibrated in 100% EMBed 812 overnight, placed into fresh EMBed812 in flat embedding molds, and cured at 60°C. Ultrathin sections (∼70 nm) were cut, and grids were stained with uranyl acetate and lead citrate. Sections were imaged using a Thermo Fisher FEI Tecnai 12 transmission electron microscope, and micrographs were recorded using a Gatan OneView 16-megapixel camera.
Data availability.
All data are included in the figures and supplementary information.
ACKNOWLEDGMENTS
We thank all the members of the lab for discussions and support. We thank the following people for providing strains and support in propagating them: Amelia Lindsey and Irene Newton, Ulrike Munderloh, and Kelly Brayton. We thank Raj Patel for preparing thin sections, and we acknowledge use of the Advanced Imaging Core Facility at Rutgers-Newark. We thank Arkady Mustaev for making the ethynyl-d-alanine. We thank the following colleagues for sharing antibodies with us: Roman Ganta, Ted Hackstadt, Kelly Brayton, Irene Newton, and Jason Carlyon.
This work was funded by a Royal Society Dorothy Hodgkin Research Fellowship (J.S.) (DH140154), a Rutgers-Newark, SASN Dean’s Research Award (E.M.B.), and funds from National Institutes of Health/National Institute of Allergy and Infectious Diseases grants (R21AI26108 and R21AI146773 to J.J.G., R21AI144385 and R56AI148645 to J.S., and R01AI152219 to T.D.).
This research was funded in whole, or in part, by the Wellcome Trust (220211). For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission. (This comment applies to author J.S.)
We declare no conflicts of interest.
Footnotes
Citation Atwal S, Chuenklin S, Bonder EM, Flores J, Gillespie JJ, Driscoll TP, Salje J. 2021. Discovery of a diverse set of bacteria that build their cell walls without the canonical peptidoglycan polymerase aPBP. mBio 12:e01342-21. https://doi.org/10.1128/mBio.01342-21.
Contributor Information
Jeanne Salje, Email: js2522@rutgers.edu.
Lotte Søgaard-Andersen, Max Planck Institute for Terrestrial Microbiology.
REFERENCES
- 1.Vollmer W, Bertsche U. 2008. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta 1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Vollmer W, Blanot D, de Pedro M. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 3.Sauvage E, Kerff F, Terrak M, Ayala J, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 4.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. 2006. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol Rev 30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 5.Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PDA, Suh H, Marto JA, Garner EC, Bernhardt TG. 2016. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol 1:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, Rudner DZ. 2016. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrichfreise B, Brunke M, Viollier P. 2016. Bacterial surfaces: the wall that SEDS built. Curr Biol 26:R1158–R1160. doi: 10.1016/j.cub.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 8.Emami K, Guyet A, Kawai Y, Devi J, Wu LJ, Allenby N, Daniel RA, Errington J. 2017. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol 2:16253. doi: 10.1038/nmicrobiol.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leclercq S, Derouaux A, Olatunji S, Fraipont C, Egan AJF, Vollmer W, Breukink E, Terrak M. 2017. Interplay between penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci Rep 7:43306. doi: 10.1038/srep43306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dion MF, Kapoor M, Sun Y, Wilson S, Ryan J, Vigouroux A, van Teeffelen S, Oldenbourg R, Garner EC. 2019. Bacillus subtilis cell diameter is determined by the opposing actions of two distinct cell wall synthetic systems. Nat Microbiol 4:1294–1305. doi: 10.1038/s41564-019-0439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McPherson D, Popham D. 2003. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J Bacteriol 185:1423–1431. doi: 10.1128/JB.185.4.1423-1431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vigouroux A, Cordier B, Aristov A, Alvarez L, Özbaykal G, Chaze T, Oldewurtel ER, Matondo M, Cava F, Bikard D, van Teeffelen S. 2020. Class-A penicillin binding proteins do not contribute to cell shape but repair cell-wall defects. Elife 9:e51998. doi: 10.7554/eLife.51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otten C, Brilli M, Vollmer W, Viollier P, Salje J. 2018. Peptidoglycan in obligate intracellular bacteria. Mol Microbiol 107:142–163. doi: 10.1111/mmi.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caruso R, Warner N, Inohara N, Núñez G. 2014. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity 41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Packiam M, Weinrick B, Jacobs WJ, Maurelli A. 2015. Structural characterization of muropeptides from Chlamydia trachomatis peptidoglycan by mass spectrometry resolves “chlamydial anomaly”. Proc Natl Acad Sci U S A 112:11660–11665. doi: 10.1073/pnas.1514026112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salje J. 2021. Cells within cells: Rickettsiales and the obligate intracellular bacterial lifestyle. Nat Rev Microbiol 19:375–390. doi: 10.1038/s41579-020-00507-2. [DOI] [PubMed] [Google Scholar]
- 17.Atwal S, Giengkam S, Chaemchuen S, Dorling J, Kosaisawe N, VanNieuwenhze M, Sampattavanich S, Schumann P, Salje J. 2017. Evidence for a peptidoglycan-like structure in Orientia tsutsugamushi. Mol Microbiol 105:440–452. doi: 10.1111/mmi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vollmer J, Schiefer A, Schneider T, Jülicher K, Johnston KL, Taylor MJ, Sahl H-G, Hoerauf A, Pfarr K. 2013. Requirement of lipid II biosynthesis for cell division in cell wall-less Wolbachia, endobacteria of arthropods and filarial nematodes. Int J Med Microbiol 303:140–149. doi: 10.1016/j.ijmm.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Wisseman CJ, Silverman D, Waddell A, Brown D. 1982. Penicillin-induced unstable intracellular formation of spheroplasts by Rickettsiae. J Infect Dis 146:147–158. doi: 10.1093/infdis/146.2.147. [DOI] [PubMed] [Google Scholar]
- 20.Liechti G, Kuru E, Packiam M, Hsu Y-P, Tekkam S, Hall E, Rittichier JT, VanNieuwenhze M, Brun YV, Maurelli AT. 2016. Pathogenic chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog 12:e1005590. doi: 10.1371/journal.ppat.1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett GM, McCutcheon JP, MacDonald BR, Romanovicz D, Moran NA. 2014. Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. mBio 5:e01697-14. doi: 10.1128/mBio.01697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, Hall E, Brun YV, VanNieuwenhze MS, Vollmer W, Horn M, Jensen GJ. 2013. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun 4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer W, Jiggins F. 2015. Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol Biol Evol 32:2111–2129. doi: 10.1093/molbev/msv093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Driscoll TP, Verhoeve VI, Guillotte ML, Lehman SS, Rennoll SA, Beier-Sexton M, Rahman MS, Azad AF, Gillespie JJ. 2017. Wholly Rickettsia! Reconstructed metabolic profile of the quintessential bacterial parasite of eukaryotic cells. mBio 8:e00859-17. doi: 10.1128/mBio.00859-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amano K, Tamura A, Ohashi N, Urakami H, Kaya S, Fukushi K. 1987. Deficiency of peptidoglycan and lipopolysaccharide components in Rickettsia tsutsugamushi. Infect Immun 55:2290–2292. doi: 10.1128/iai.55.9.2290-2292.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moulder J. 1993. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect Agents Dis 2:87–99. [PubMed] [Google Scholar]
- 27.Ghuysen J, Goffin C. 1999. Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob Agents Chemother 43:2339–2344. doi: 10.1128/AAC.43.10.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henrichfreise B, Schiefer A, Schneider T, Nzukou E, Poellinger C, Hoffmann T-J, Johnston KL, Moelleken K, Wiedemann I, Pfarr K, Hoerauf A, Sahl HG. 2009. Functional conservation of the lipid II biosynthesis pathway in the cell wall-less bacteria Chlamydia and Wolbachia: why is lipid II needed? Mol Microbiol 73:913–923. doi: 10.1111/j.1365-2958.2009.06815.x. [DOI] [PubMed] [Google Scholar]
- 29.Giengkam S, Blakes A, Utsahajit P, Chaemchuen S, Atwal S, Blacksell SD, Paris DH, Day NPJ, Salje J. 2015. Improved quantification, propagation, purification and storage of the obligate intracellular human pathogen Orientia tsutsugamushi. PLoS Negl Trop Dis 9:e0004009. doi: 10.1371/journal.pntd.0004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The class A PBP gene has been lost at least four times during evolution (extended version). Analysis of key genes in the peptidoglycan biosynthesis pathway showing their presence or absence in obligate intracellular, facultative intracellular, host-associated, and free-living bacteria across 9 phyla. The predicted peptidoglycan (PG) status is shown as well as the primary site of replication of the bacteria. Genes involved in lipid II biosynthesis, SEDS family, class B PBPs, and class A PBPs were analyzed. The presence of a gene is shown by block color, the absence in white, and genes identified manually (not annotated as aPBP in KEGG) are shown with dashes. Bacterial organisms in Gammaproteobacteria (Baumannia cicadellinicola Hc, “Candidatus Vesicomyosocius okutanii,” Francisella tularensis subsp. tularensis, Coxiella endosymbiont of Amblyomma americanum), Alphaproteobacteria (Anaplasma spp., Wolbachia spp., Orientia tsutsugamushi), PVC (Parachlamydia acanthamoebae, Chlamydia spp., “Candidatus Protochlamydia amoebophila,” Simkania negevensis, Waddlia chondrophila, Planctomyces sp.), and Actinobacteria (Tropheryma whipplei) all lack class A PBP homologs and are predicted to build intermediate PG-like structures (PGi). PVC, Planctomycetes, Verrucomicrobia, and Chlamydiae; SEDS, shape, elongation, division, and sporulation; PBP, peptidoglycan binding protein. Download FIG S1, PDF file, 0.1 MB (111.5KB, pdf) .
Copyright © 2021 Atwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
KEGGerator output indicating presence (1) or absence (0) of each KEGG ortholog involved in peptidoglycan synthesis, across all 4,989 bacterial genomes in KEGG. Each row contains a single taxon, including the KEGG group, genus, and KEGG organism identifier. Each column is a distinct ortholog identified with the KEGG identifier and, where applicable, gene locus. Columns Z, AA, and AB sum the number of different orthologs within the SEDS, bPBP, and aPBP families, respectively. Each sheet represents a different subset of bacteria. (A) All (n = 4,989) bacteria. (B) Bacteria (n = 3,935) containing at least one ortholog from all three families (SEDS, aPBP, bPBP). (C) Bacteria (n = 987) containing SEDS and bPBP but no aPBP. (D) Bacteria (n = 20) lacking SEDS, aPBP, and bPBP. (E) Bacteria (n = 7) containing SEDS but not aPBP or bPBP. (F) Bacteria (n = 19) containing bPBP but not SEDS or aPBP. (G) Bacteria (n = 4) containing aPBP and bPBP but not SEDS. (H) Bacteria (n = 12) containing SEDS and aPBP but not bPBP. (I) Bacteria (n = 4) containing aPBP but not SEDS or bPBP. Download Table S1, XLSX file, 1.0 MB (1MB, xlsx) .
Copyright © 2021 Atwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Taxonomic groups included in the current study. Each entry is identified by its KEGG organism identifier, genus, taxon name, and lifestyle (extracellular, extracellular-attached, intracellular-obligate, intracellular-facultative, bacteriocytic, bacteriocytic; extracellular, or free-living). (A) All 117 taxa in the analysis. (B) All taxa binned into 65 groups, based on taxonomic relatedness and PGP profile similarity. Download Table S2, XLSX file, 0.01 MB (17.2KB, xlsx) .
Copyright © 2021 Atwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
KEGG identifiers for all orthologs included in the current study, grouped by broad biochemical pathway. Download Table S3, XLSX file, 0.01 MB (9.6KB, xlsx) .
Copyright © 2021 Atwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Drug sensitivity assays. Bacterial copy number after 5 days (7 days for Wolbachia) of growth in the presence or absence of drugs was quantified by qPCR. Growth in the presence of a drug was normalized by growth in the absence of drug within the same experiment. RC, Rickettsia canadensis, grown for 5 days in Vero cells; AM, Anaplasma marginale grown for 5 days in Vero cells; OT, Orientia tsutsugamushi grown for 5 days in L929 cells; WP, Wolbachia pipientis grown for 7 days in JW18 cells; EC, Ehrlichia chaffeensis grown for 5 days in DH82 cells; AP, Anaplasma phagocytophilum grown for 5 days in HL60 cells. Drug concentrations were 100 μg/ml chloramphenicol, 150 μg/ml penicillin G, 250 μg/ml d-cycloserine, and 40 μg/ml phosphomycin. Statistical analysis between the untreated and drug-treated groups was carried out using an unpaired t test with GraphPad Prism software. Individual values and median are shown. P values: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. Download FIG S2, PDF file, 0.1 MB (64.1KB, pdf) .
Copyright © 2021 Atwal et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All data are included in the figures and supplementary information.