ABSTRACT
Bacteriophage predation is an important factor in bacterial community dynamics and evolution. Phage-bacterium interaction has mainly been studied in lab cultures, while dynamics in natural habitats, and especially in the plant root niche, are underexplored. To better understand this process, we characterized infection of the soil bacterium Bacillus subtilis NCBI 3610 by the lytic phage SPO1 during growth in LB medium and compared it to root colonization. Resistance in vitro was primarily through modification of the phage receptor. However, this type of resistance reduced the ability to colonize the root. From a line that survived phage infection while retaining the ability to colonize the root, we identified a new phage resistance mechanism involving potassium (K+) ion influx modulation and enhanced biofilm formation. Furthermore, we show that potassium serves as a stimulator of root colonization among diverse growth-promoting bacilli species, with implications for plant health.
KEYWORDS: Bacillus subtilis; bacteriophage, evolution; biofilms; plant-microbe interactions
INTRODUCTION
Plant roots are associated with diverse bacteria in the soil (1), which can affect many aspects of plant life, including root architecture (2), nutrient acquisition (3), and disease state (2, 4). The root rhizosphere, i.e., the area close to the root surface, is enriched with specific bacterial taxa in comparison to that in bulk soil. This unique microbial composition is determined by the soil surrounding the plant root and its preexisting bacterial diversity (5, 6) and plant genotype (7) as well as the interaction with other bacteria (8) and with phage (9). Although a large body of research has been conducted to characterize each of these factors, the whole picture, and especially the role of phage in bacterial root colonization, is far from complete.
Phage are viruses that infect and kill bacteria (10, 11). As bacterial predators, they influence bacterial community dynamics through elimination of their sensitive hosts. Bacteria, in turn, respond by rapid evolution of phage resistance (12). In addition to influencing bacterial interaction with phage, newly acquired resistance (10, 13) can also cause changes in colony morphology (14), genome-wide mutation rate (15), and lateral spread of genetic material (16). In recent years, insights have been gained into how phage-bacterium interactions affect bacterial growth in ocean (17) and mammalian gut ecosystems (18). However, little is known about the effects of phage-bacterium interactions in other ecosystems, especially in soil and the root rhizosphere (19).
To address this question, we utilized Bacillus subtilis strain NCBI 3610 (henceforth, B. subtilis) (20) and its cognate lytic phage SPO1 (21) to explore their interaction during root colonization of the plant model system Arabidopsis thaliana. B. subtilis is a Gram-positive spore-forming bacterium isolated from soil and is able to colonize plant roots (22). Root colonization by B. subtilis is mediated by formation of a biofilm on the root (23). Biofilms are bacterial communities encased in an extracellular matrix. The B. subtilis matrix is mainly composed of sugar polymers, encoded by the eps operon, and protein fibers encoded by the tapA-sipW-tasA operon (24). B. subtilis defective in biofilm formation exhibits severe defects in root colonization (23). The phage SPO1 is a lytic phage, representing a large and diverse group of Myoviridae bacteriophages, harboring a long contractile tail. SPO1 phage exhibits a complex infection cycle, involving the subversion of the host transcription machinery for its own use (25). SPO1 utilizes the wall teichoic acid polymers (WTAs) as receptors to invade B. subtilis cells (26). WTAs are long sugar polymers that are incorporated into the cell wall and membrane of B. subtilis and other Gram-positive bacteria (27). SPO1 binds these polymers when they are decorated by glucose moieties (gWTAs). Comparison of phage-bacterium evolution upon infection in LB medium and during root colonization revealed that phage infection in vitro and in planta exhibits different evolutionary trajectories. Characterization of bacteria resistant to phage infection during root colonization led to the identification of a novel phage resistance mechanism through modulation of potassium (K+) ion influx and enhanced biofilm formation. Furthermore, we show that potassium serves as a stimulator of root colonization among diverse bacilli species.
RESULTS
Loss of phage receptor results in a fitness cost for root colonization.
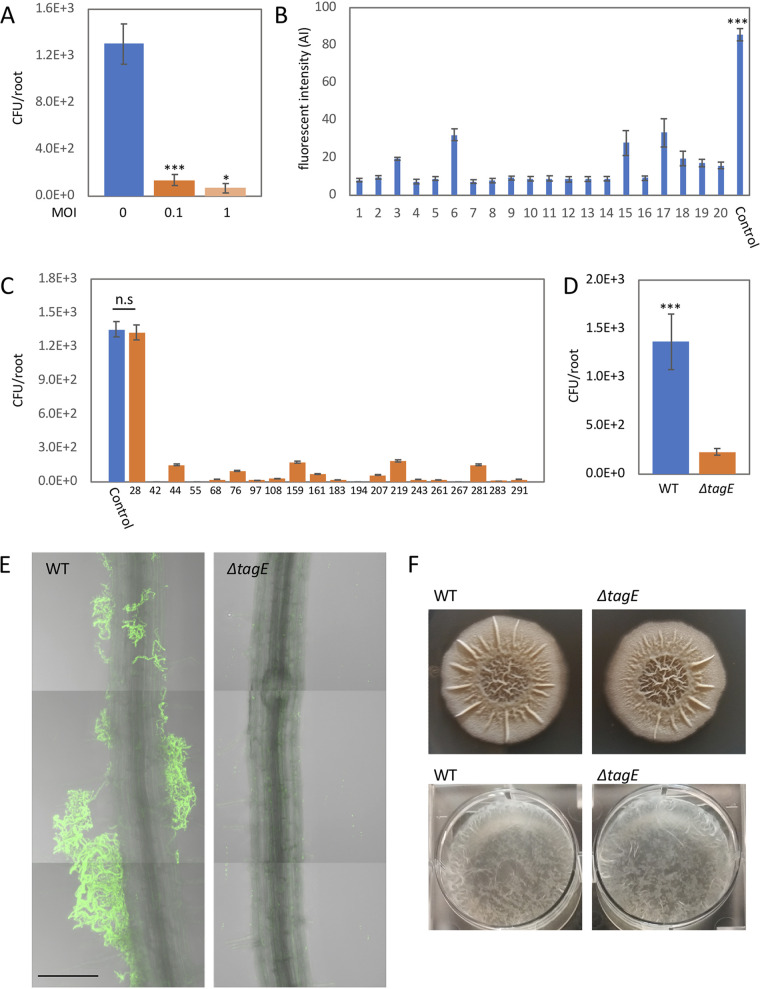
To explore phage-bacterium interactions during root colonization, we inoculated the roots of A. thaliana with B. subtilis 3610 bacteria together with SPO1 phage at either a high (phage/bacteria, 1:1), or low (phage/bacteria, 1:10) multiplicity of infection (MOI). Measuring bacterial colonization (CFU) after 48 h revealed a reduction in root colonization of ∼95% at an MOI of 1 and an ∼90% reduction at an MOI of 0.1 (Fig. 1A), indicating that SPO1 can efficiently infect and kill its host bacteria during root colonization. Significant levels of PFU were recovered from the root after SPO1 infection (5.56 × 104 ± 4.67 × 104, for MOI of 1, n = 6). However, no increase in the amount of the phage was observed in comparison to the input (PFU = 106), probably reflecting the fact that phage cannot adhere to the root without the presence of the host bacteria. Selection pressure by phage drives bacterial evolution of resistance mechanisms (28). To explore the evolution of phage resistance mechanisms in vitro versus those in planta, we isolated 300 bacteria (screen 1) that survived phage infection on the root and restreaked them on agar plates containing SPO1 phage, (maximum of 10 bacteria from each root). We found 20 in planta survivors that became resistant to phage infection in vitro. On the other hand, 100% of 70 bacteria isolated after surviving phage infection in LB medium (taken from 3 independent LB plates) became immune to further infection. Twenty randomly selected in vitro survivors became phage resistant through loss of the phage receptor, as judged by a lack of staining with concanavalin A, Alexa Fluor 488 conjugate (ConA488), a lectin that binds specifically glycosylated WTA (gWTA) (Fig. 1B) (see also reference 26). Of the 20 SPO1-resistant bacteria isolated from the plant all but one (m28) had lost their ability to recolonize the root (Fig. 1C). Nineteen of these isolates concomitantly lost their ConA488 staining (see Fig. S1A in the supplemental material). One of the isolates, m28, was completely phage resistant in vitro when infected in liquid culture (Fig. S1B) but still exhibited faint ConA488 staining (Fig. S1A and C) and partial sensitivity when infected on roots (Fig. 2A). The correlation between SPO1 sensitivity and root colonization ability suggests that gWTA is important for efficient root colonization. To test this hypothesis, we inoculated roots with ΔtagE bacteria, which lack gWTA (29), and found a significant reduction in root colonization by CFU measurement and fluorescent bacteria (Fig. 1D and E). Because biofilm formation has been shown to be necessary for root colonization by B. subtilis (30), we tested the ability of ΔtagE bacteria to form a biofilm in vitro. ΔtagE cells exhibit normal biofilm formation both on agar plates and in liquid medium (Fig. 1F). Of note, gWTA is required for nasal epithelium colonization by Staphylococcus aureus (31). Our results suggest that gWTA in B. subtilis is similarly important for plant surface adhesion, irrespective of biofilm formation.
FIG 1.
SPO1 phage receptor is necessary for bacterial root adhesion. (A) Seedlings were inoculated with B. subtilis 3610 together with SPO1 phage, at either a high (phage/bacteria, 1:1) or low (phage/bacteria, 1:10) multiplicity of infection (MOI) or with no phage (MOI of 0) for 48 h on agar plates, and the number of colonizing bacteria was counted. Shown are averages and standard deviations (SDs) from 2 independent experiments with an n of ≥3 for each (n throughout the paper is the number of roots sampled [biological replicates] with 3 technical replicates from each root). *, P < 0.05; ***, P < 0.005. (B) Twenty randomly selected bacterial colonies that survived SPO1 infection were stained with ConA488 and observed under a compound microscope. Shown are average and SD fluorescent intensity values of 10 bacteria from each of the colonies. ***, P < 0.005. (C) Twenty isolated SPO1 resistant bacteria were inoculated on seedlings and grown for 48 h, and the number of colonizing bacteria were counted. Shown are averages and SDs with an n of ≥3. (D) Seedlings were inoculated with either WT or ΔtagE bacteria for 48 h, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. ***, P < 0.005. (E) Seedlings were inoculated with either WT or ΔtagE bacteria expressing green fluorescent protein (GFP) (amyE::PrrnE-gfp) for 48 h on agar plates. Shown are representative overlay ×200 magnification confocal images of differential interference contrast (DIC) (root) and GFP fluorescence (bacteria). Scale bar, 50 μm. (F) WT and ΔtagE bacteria were inoculated onto MSgg agar plates (top) or liquid medium (bottom). Shown are representative biofilm images.
FIG 2.
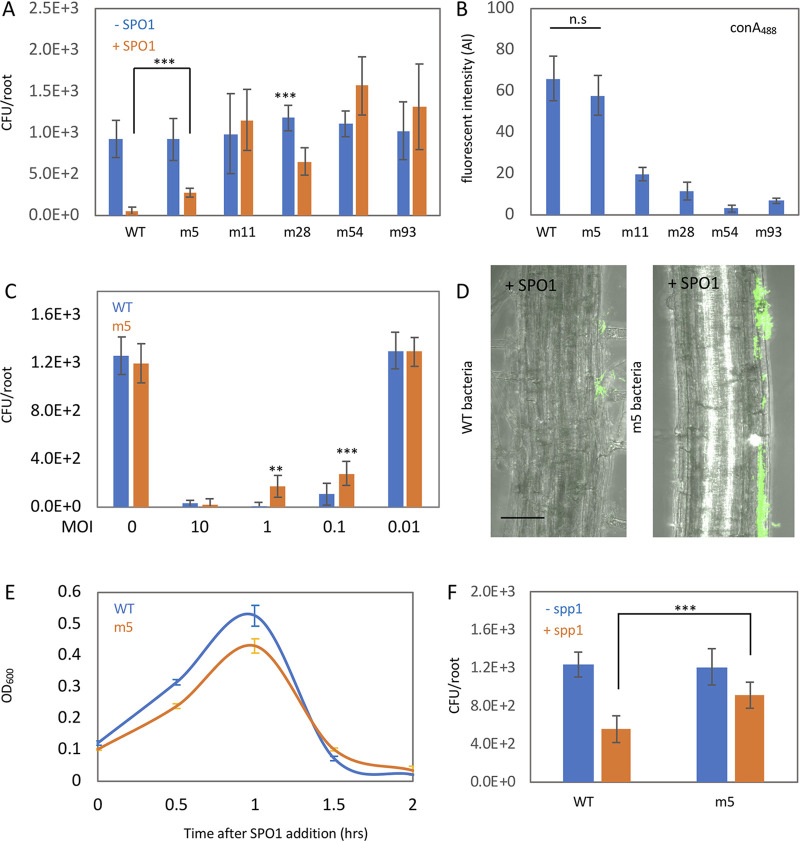
A novel SPO1 resistance mechanism. (A) Seedlings were inoculated with the indicated bacterial strains, with or without SPO1 addition, for 48 h on agar plates, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. ***, P < 0.005. (B) Bacterial strains were stained with ConA488 and observed under the microscope. Shown are average and SD of fluorescence intensity values of 10 bacteria from each colony. n.s., not significant. (C) Seedlings were inoculated with either WT or m5 bacteria, with the indicated phage MOI, for 48 h on agar plates, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. **, P < 0.01; ***, P < 0.005. (D) Seedlings were inoculated with either WT or m5 bacteria expressing GFP (amyE::PrrnE-gfp), in the presence of SPO1, for 48 h on agar plates. Shown are representative overlay ×200 magnification confocal images of DIC (root) and GFP fluorescence (bacteria). Scale bar, 20 μm. (E) WT and m5 bacteria were infected with SPO1 in LB medium, and OD600 was followed. Shown are averages and SDs, n = 3. (F) Seedlings were inoculated with either WT or m5 bacteria, with or without SPP1 phages, for 48 h on agar plates, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. ***, P < 0.005.
SPO1 resistance profile of phage survival isolates. (A) SPO1-resistant bacteria isolated from plants were stained with ConA488 and observed under the microscope. Shown are average and SD fluorescent intensity values from 10 bacteria of each of the surviving colonies. (B) The indicated bacterial strains were infected with SPO1 in LB medium, and OD600 was followed. Shown are averages and SDs with an n of 3. (C) The indicated bacterial strains were stained with ConA488. Shown are representative DIC (left) and fluorescence from ConA488 (right) images from each strain. Download FIG S1, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2021 Tzipilevich and Benfey.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacteria evolved a novel phage resistance mechanism.
Our results indicate that loss of the phage receptor results in a fitness cost to bacteria during root colonization. To identify alternative pathways utilized by bacteria to resist phage infection during root colonization, we utilized the bacteria that survived phage infection on the root (100 of the 300 bacteria from screen 1) and infected them with SPO1, this time during root colonization (screen 2). Most of the bacteria exhibited phage sensitivity similar to that of the parental bacteria. However, of 100 bacterial strains tested, we found 4 that survived SPO1 infection on roots better than wild-type (WT) cells (Fig. 2A). To explore the mechanism of bacterial survival, we sequenced the genomes of m5, m11, m54, and m93, the 4 bacteria isolated during screen 2, along with m28 isolated from screen 1. Table 1 presents the mutations in each of the bacteria.
TABLE 1.
Bacterial strains
| Bacterial strain | Mutation |
|---|---|
| m5 | Missense mutation in KtrC P189T |
| m11 | Mutations in YwbO promoter |
| m28 | Missense mutation GtaB Y170S |
| m54 | Missense mutations in YwrK T334S and CdaA F97L |
| m93 | Frame shift in tagE T1013 leading to premature stop codon |
m28 and m93 harbor mutations in gtaB and tagE, genes involved in the WTA glycosylation pathway (27). Interestingly, m11 and m54 do not have mutations in genes previously implicated in WTA glycosylation but nonetheless exhibited complete (m54) and partial (m11) loss of ConA488 staining (Fig. 2B and Fig. S1C). m11, m54, and m93 exhibited phage resistance in vitro (Fig. S1B).
One mutant, m5, exhibited enhanced phage resistance when infected in planta at a medium MOI (1 to 0.1) (Fig. 2C, CFU, and Fig. 2D, fluorescent bacteria), phage sensitivity in vitro, and ConA488 staining similar to that of WT (Fig. 2B and E and Fig. S2B), indicating that it harbors a novel phage resistance mechanism. The m5 mutant also exhibited enhanced resistance to infection by SPP1, a lytic phage from a different bacteriophage family (Siphoviridae) (Fig. 2F). SPP1 utilizes YueB, a membrane protein, as a receptor. However, gWTA also plays an important role for SPP1 infection as well (32). Nevertheless, m5 has a resistance mechanism that is not phage-family specific. m5 harbors a single point mutation in the ktrC gene, encoding a low-affinity potassium channel. This missense mutation changes proline 189 into threonine (KtrC P189T) and resides in a conserved RCK domain, known to bind the regulatory molecule ci-di-AMP (33, 34). ci-di-AMP negatively regulates potassium uptake (35). We hypothesized that KtrC P189T is a gain of function mutation, affecting the regulation of potassium uptake.
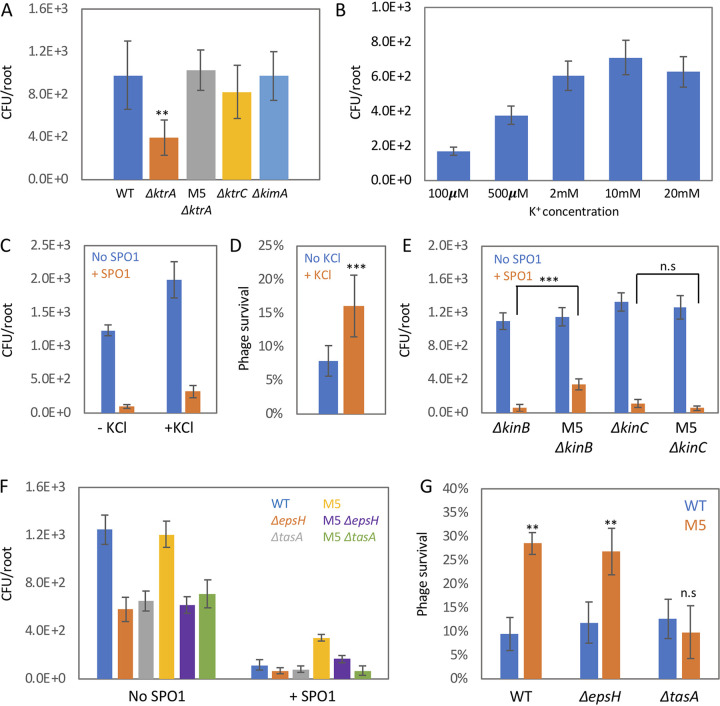
Potassium modulates B. subtilis root colonization and phage resistance. (A) Seedlings were inoculated with WT bacteria and grown for 48 h on 0.25× MS agar plates in the absence or presence of 5 mM of the indicated salts, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. ***, P < 0.005. (B) Seedlings were inoculated with ΔkinC bacteria, with or without SPO1 addition, and grown for 48 h on 0.25× MS agar plates in the presence or absence of 5 mM KCl; the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. (C) Seedlings were inoculated with the indicated bacterial strains and grown for 48 h on 0.25× MS agar plates, in the absence or presence of 5 mM KCl, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. ***, P < 0.005. Download FIG S2, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2021 Tzipilevich and Benfey.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Potassium enhances root colonization and phage resistance through modulation of biofilm formation.
Several potassium channels are encoded in the B. subtilis genome: ktrAB and kimA encode high-affinity channels (36, 37), while the ktrCD operon encodes a low-affinity channel (36). To understand the effect of the KtrC P189T mutation and potassium uptake on bacterial root colonization, we examined the colonization efficiency of bacteria lacking these channels. While ΔktrC and ΔkimA mutants colonized the root with similar efficiency to WT bacteria (Fig. 3A), ΔktrA bacteria exhibited reduced root colonization (Fig. 3A). The KtrC P189T mutation is able to restore the colonization of ΔktrA bacteria (Fig. 3A, M5 ΔktrA), suggesting that this mutation enhances potassium uptake through the KtrC channel. Growing plants on media with different levels of potassium revealed a positive correlation between potassium concentration and root colonization by WT bacteria (Fig. 3B), with the effect plateauing at 10 mM. Similarly, root colonization on 0.25 Murashige and Skoog (MS) plates, with addition of 5 mM potassium (KCl) (equal to total of ∼10 mM) indicated a positive effect of potassium on root colonization (Fig. 3C). Stimulation of root colonization was specific to potassium ions, as neither sodium nor nitrogen had a similar effect (Fig. S2A). Simply adding extra potassium to the plant growth medium was sufficient to enhance survival of WT cells upon phage infection (Fig. 3C and D), reinforcing the idea that the KtrC P189T mutation affects phage infection through increased potassium uptake.
FIG 3.
Potassium modulates B. subtilis root colonization and phage resistance through biofilm formation. (A) Seedlings were inoculated with the indicated bacterial strains for 48 h on agar plates, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. **, P < 0.01. (B) Seedlings were inoculated with WT B. subtilis on MS agar plates supplemented with the indicated potassium concentration for 48 h, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. (C and D) Seedlings were inoculated with WT B. subtilis, with or without SPO1 addition, for 48 h on 0.25× MS agar plates in the presence or absence of 5 mM KCl, and the number of colonizing bacteria was counted. Shown are averages and SDs of CFU values (C) and the percentages of SPO1 survival (D), from 2 independent experiments with an n of ≥3 for each. ***, P < 0.005. (E) Seedlings were inoculated with the indicated bacterial strains, with or without SPO1 addition, for 48 h on agar plates, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. ***, P < 0.005. (F and G) Seedlings were inoculated with the indicated bacterial strains, with or without SPO1 addition, for 48 h on agar plates in the presence or absence of 5 mM KCl, and the number of colonizing bacteria was counted. Shown are average and SD CFU values (F) and percentages of SPO1 survival (G) from 2 independent experiments with an n of ≥3 for each, **, P < 0.01.
It was previously shown that B. subtilis bacteria sense potassium influx through the cell membrane, utilizing KinC and KinB protein kinases to induce biofilm formation genes and sliding motility, respectively (38, 39). Both biofilm formation and sliding motility play important roles during root colonization (23, 40). We found that m5 ΔkinC bacteria, but not m5 ΔkinB bacteria, lost SPO1 resistance (Fig. 3E). Addition of potassium to the plant growth medium failed to stimulate phage resistance for ΔkinC bacteria (Fig. S2C). Thus, we conclude that KinC is involved in the effect of potassium on root colonization and phage resistance.
KinC activation induces a phosphorylation cascade that culminates in induction of biofilm matrix genes (38). Disruption of the biofilm matrix operons epsA-O and tapA-sipW-tasA abolished the effect of potassium on root colonization (Fig. S2C). Interestingly, disruption of the tapA-sipW-tasA operon, but not epsA-O, reduced the phage resistance phenotype of m5 cells (Fig. 3F and G), suggesting that an increase in the protein fiber component was responsible for the phage resistance effect of increased potassium influx. Thus, our analysis revealed a novel phage adaptation mechanism that works through enhanced potassium influx by stimulating biofilm matrix formation.
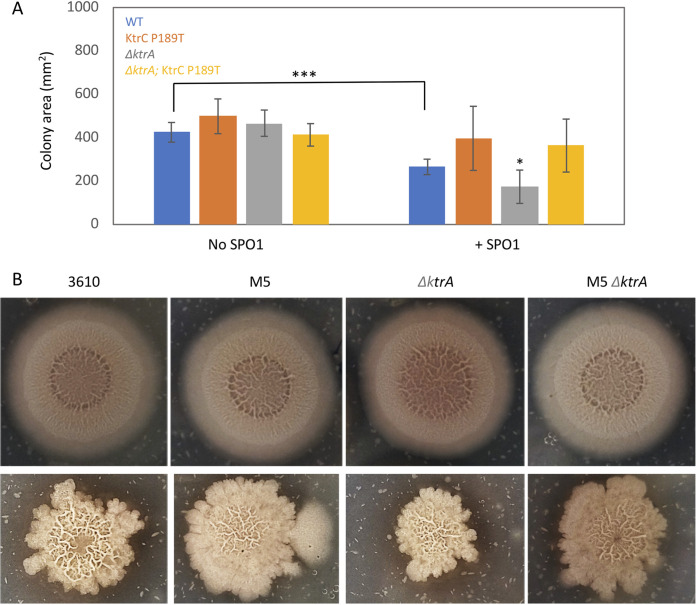
To further characterize the role of biofilm matrix formation in phage resistance, we monitored SPO1 infection during biofilm formation in vitro (20). Measurement of biofilm diameter on MSgg agar plates revealed significant reduction in colony diameter on plates containing SPO1 phage (Fig. 4A). Of note, ΔktrA cells, although able to form normal biofilm in the absence of phage, exhibited a significant decrease in biofilm diameter in the presence of phage in comparison to that of WT infected cells (Fig. 4A and B). Similar to what was observed in plants, the KtrC P189T mutation was able to compensate for the increased sensitivity of ΔktrA cells (Fig. 4A and B). Thus, our in vitro analysis provides further support for the hypothesis that potassium influx modulates biofilm formation to enhance phage resistance. Equivalent results were obtained for pellicle biofilm formation on liquid MSgg medium (Fig. S3), where enhanced potassium influx in m5 bacteria, or addition of extra potassium to the medium of WT bacteria, increased the chance of surviving SPO1 infection, while decreased influx, due to the ΔktrA mutation, increased phage sensitivity.
FIG 4.
Potassium modulates phage resistance during biofilm formation in vitro. (A) The indicated bacterial strains were inoculated onto MSgg agar plates with or without SPO1 addition for 48 h, and colony area was measured. Shown are averages and SDs from an n of 6. *, P < 0.05; ***, P < 0.005. (B) Shown are representative images for the experiment described for panel A with (bottom) or without (top) SPO1 addition.
Potassium modulates phage resistance during pellicle biofilm formation. (A) The indicated bacterial strains were inoculated in MSgg medium with or without SPO1 addition for 48 h, and the number of cultures exhibiting partial SPO1 survival (see panel C) were counted. Shown are the percentages of the cultures surviving SPO1, n ≥ 30. ***, P < 0.005. (B) WT bacteria were inoculated into MSgg medium with or without 5 mM KCl and infected with SPO1 for 48 h, as described for panel A. Shown are the percentages of the cultures surviving SPO1, n ≥ 30. ***, P < 0.005. (C) Shown are representative images for the experiment described in panels A and B, for an uninfected culture (1), a culture undergoing complete phage lysis (2), and a culture counted as partially surviving phage infection (3). Download FIG S3, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2021 Tzipilevich and Benfey.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
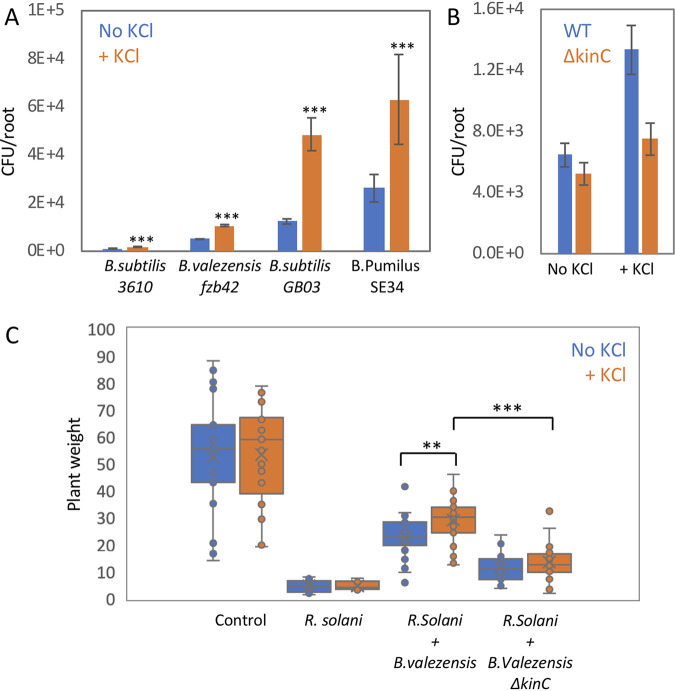
Potassium enhances root colonization by diverse bacilli species.
To determine if potassium is able to enhance colonization by other bacilli species, we analyzed its effect on Bacillus velezensis FZB42, a plant-associated bacterium (41), shown to enhance plant health through growth stimulation and pathogen inhibition. We found that potassium enhances root colonization of B. velezensis FZB42 (Fig. 5A), and this phenomenon was abolished in B. velezensis FZB42 ΔkinC cells (Fig. 5B). A similar phenomenon was observed for B. subtilis GB03 and B. pumilus SE34, two other plant growth-promoting bacteria (Fig. 5A). Thus, potassium serves as a wide-spread signal, stimulating root colonization by diverse bacilli species. B. velezensis FZB42 inhibits the growth of the plant fungal pathogen Rhizoctonia solani (42), raising the possibility that simply adding potassium to the growth medium could enhance B. velezensis FZB42 colonization and fungal protection. Indeed, potassium enhanced plant growth when inoculated with Rhizoctonia solani in the presence of B. velezensis FZB42 in comparison to growth of plants inoculated with fungi but no bacteria (Fig. 5C), and this effect of plant protection was abolished in ΔkinC cells, which are unable to sense potassium influx (Fig. 5C).
FIG 5.
Potassium modulates root colonization by diverse bacillus species. (A) Seedlings were inoculated with the indicated bacterial species for 48 h on 0.25× MS agar plates in the presence or absence of 5 mM KCl, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. ***, P < 0.005. (B) Seedlings were inoculated with either WT or ΔkinC B. velezensis FZB42 bacteria for 48 h on 0.25× MS agar plates in the presence or absence of 5 mM KCl, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with and n of ≥3 for each. (C) Seedlings were inoculated with either WT or ΔkinC B. velezensis FZB42 bacteria, for 48 h on 0.25× MS agar plates in the presence or absence of 5 mM KCl. Next, plates were inoculated with R. solani and incubated for an additional 7 days, and plant weight was measured. Untreated plants (neither bacteria nor fungi) were used as control. Shown are averages and SDs, n ≥ 20. **, P < 0.01; ***, P < 0.005.
DISCUSSION
Phage infection has been extensively characterized in lab cultures. Recent work revealed that phage-bacterium interactions in natural environments can differ significantly from those observed in the lab (43) due to the fitness constraints encountered by bacteria in their natural environment. This can impede the evolution of phage receptors, the main resistance pathway observed in the lab (44). We found that gWTA, the SPO1 phage receptor, is essential for root adhesion; thus, receptor modification has a fitness cost in the root environment. Of note, it has been shown that gWTA is essential for adhesion of Staphylococcus aureus to the nasal epithelium (31), while gWTA also serves as phage receptor for several S. aureus-infecting phages (45). It will be interesting to determine if phage-bacterium evolution is constrained in this niche as well.
While evolution of receptor modification is not favored, we found that bacteria in the root adapt to phage infection through evolution of enhanced biofilm formation. Consistent with previous in vitro work (38), we show that during plant colonization, altering potassium efflux through KtrCD channels enhances B. subtilis biofilm formation and thus promotes survival upon phage infection (Fig. 6). Our results indicate that protein fiber density, encoded by the tapA-sipW-tasA operon, is the main determinant of SPO1 resistance. Interestingly, it has been shown that curli protein fibers are important for T7 resistance in a biofilm of Escherichia coli bacteria (46). Phage-bacterium interaction in biofilm is an emerging area of research (47). Biofilms are an important mode of life for bacteria colonizing natural environments, such as plant roots, as well as for pathogenic bacteria (48). Thus, in order to understand bacterial community dynamics in nature, and to deal with biofilm pathogenicity, it is important to explore the interaction of phage and bacteria in this specific niche.
FIG 6.
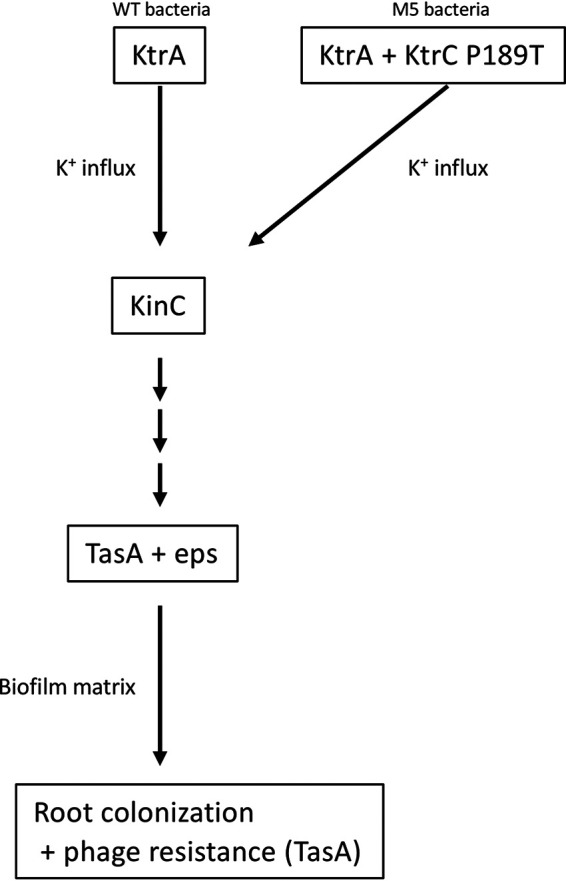
A model describing the effect potassium on root colonization and phage resistance.
Phage utilize bacterial appendages decorating the cell surface as receptors. Appendages such as flagella, type IV pili, exopolysaccharides, and transporters of nutrients, are important for bacterial survival in the natural environment and specifically, in the plant niche (9). Evolution of phage resistance has been associated with reduced host fitness on plants. An example is phage utilizing type IV pili (49), where the evolved mutant bacteria growly similar to the parental strain in planktonic culture but suffer from reduced biofilm formation and plant leaf colonization. Thus, it is important to explore phage-bacterium interactions in their natural context to gain insight into their coevolutionary dynamics.
Our genetic screen opens the way to discovering new phage resistance strategies that are not accessible during in vitro growth. An interesting future direction derives from the observation that among in planta phage-resistant bacteria, we found 4 of 100 with genetically inherited enhanced phage resistance. This suggests that 96% of the bacteria survived infection in a nongenetic manner. It would be interesting to determine how nongenetic mechanisms enable bacteria to survive phage infection. One possibility is by colonizing root niches that are less accessible to phage, a strategy that was recently demonstrated for phage infection of bacteria colonizing the gut ecosystem (50).
Potassium is an essential macronutrient for bacteria and plants. Unlike nitrogen and phosphate, it is not incorporated into cellular macromolecules but remains a soluble ion (51). Potassium is involved in the regulation of many processes, including osmotic adaptation, membrane potential, phloem transport, regulation of metabolic enzymes, and biotic and abiotic stress responses. Potassium, nitrogen, and phosphate, are the three main ions consumed by plants and are heavily utilized for fertilization in modern agriculture. (52). The effect of nitrogen and phosphorous on root microbial communities has been characterized (6, 53, 54). However, the role of potassium in bacterial colonization is less well explored. Our analysis suggests that addition of potassium is a simple method to enhance bacillus colonization. Many bacillus species exert positive effects on their plant host by enhancing plant growth and inhibiting pathogens. Our results suggest that adding potassium could enhance the ability of B. velezensis to protect seedlings from R. solani infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
WT B. subtilis NCBI 3610 and AR16 (55) (B. subtilis PY79) were kindly provided by Sigal Ben-Yehuda (Hebrew University); ΔtagE (BKK35730), ΔktrA (BKK31090), ΔktrC (BKK14510), and ΔkimA (BKK04320) strains (all in B. subtilis 168) were purchased from the Bacillus Genetic Stock Center (http://www.bgsc.org/). Genomic DNA was extracted from AR16 and mutant strains using a Wizard genomic DNA purification kit (Promega) and transformed into B. subtilis NCBI 3610. The media and growth conditions used for DNA transformation of B. subtilis NCBI 3610 are described at http://2013.igem.org/Team:Groningen/protocols/Transformation. The bacteria were cultivated routinely on Luria broth (LB) medium. When needed, the medium was solidified with 1.5% agar. For biofilm formation, bacteria grown overnight were inoculated into MSgg medium and incubated without shaking for 2 days at 30°C as described in reference 20. This medium was also solidified with 1.5% agar.
Phage strains and infection conditions.
SPO1 (1P4) and SPP1 (1P7) phage were purchased from the Bacillus Genetic Stock Center. Phage lysate was routinely prepared by adding approximately 109 phage to mid-log cells grown in LB medium supplemented with 10 mM MgSO4, until the culture was completely cleared. The lysate was then filtered through a 0.22-μm Millipore filter. For lysis dynamics in LB medium (Fig. 2E and Fig. S1B), approximately 109 phage were added to mid-log-phase growing cells at multiplicity of infection (MOI) of 1, and the optical density at 600 nm (OD600) was monitored at 30-min intervals. For SPO1 infection in pellicle biofilm (Fig. S3), bacteria grown overnight were inoculated into MSgg medium, and phage was added at a MOI of 1. Bacteria were incubated without shaking for 2 days at 30°C, and the number of cultures with surviving cells was counted by eye. For SPO1 infection in solid biofilm (Fig. 4), bacteria grown overnight were inoculated into MSgg medium supplemented with 105 PFU and solidified with 1.6% agar. A spot of 5 μl from the bacterial culture was inoculated onto the plate and incubated for 2 days at 30°C. Plates were scanned and colony diameter was determined using ImageJ.
Monitoring bacterial growth and phage infection on plant roots.
Plants were grown on 0.5 MS medium containing 1.1 g Murashige and Skoog basal salts (in 500 ml double-distilled water [ddH2O]), 1% sucrose, 1% agar, and 5 ml (in 500 ml ddH2O) morpholineethanesulfonic acid (MES; 50 g/liter, titrated to pH 5.8 with NaOH). Plants were stratified for 2 days in a 4°C dark room and grown vertically for 4 to 10 days under long-day light conditions. Bacteria from fresh colonies were grown in LB medium to an OD600 of 1.0 and then diluted 1:100 in 1× phosphate-buffered saline (PBS) for CFU measurements and microscopy, yielding approximately 1 × 106. Phage were added to the 1× PBS at an MOI of 0.1 unless otherwise indicated. Six-day-old seedlings were transferred onto square petri dishes containing 0.5 MS but without sucrose. Two microliters of bacterial dilution was deposited immediately above the root tip and left to dry for 2 min. The square plates were kept in a vertical position during the incubation time at 22°C under long-day light conditions (16 h light/8 h darkness) in a plant growth chamber. For bacterial CFU counting and microscopy, plants were incubated with bacteria for 48 h. Then, the inoculated plant roots were cut and washed three times in sterile water. For CFU counting, the seedlings were transferred to a tube with 1 ml of 1× PBS and vortexed vigorously for 20 s; then, a serial dilution was plated on LB plates. For PFU measurement, roots were treated as described for CFU, but without the washing step.
Microscopy.
Roots were observed using a Zeiss LSM 880 laser scanning confocal microscope with 20× lens objective. ConA488 (Thermo Fisher C11252) staining was performed as described previously (26). Bacteria were observed using a Zeiss LSM 880 laser scanning confocal microscope with a 40× (water immersion) lens objective. Fluorescence intensity was measured using ImageJ.
DNA sequence analysis.
DNA was extracted from an overnight culture of the mutant bacterial strain along with the WT strain using a DNeasy PowerSoil Pro kit (Qiagen), according to the manufacturer’s instructions. DNA sequencing (DNA-seq) libraries were prepared using a Nextera DNA Flex library preparation kit (Illumina), according to the manufacturer’s instructions. Illumina NextSeq 500 high-output 50-bp single reads were partially assembled into contigs using the SPAdes algorithm (56) with default parameters. The contigs were aligned to the genome of the WT bacteria from our lab stock.
Data analysis and statistics.
All graphs and statistical tests were performed in Excel. P values throughout the paper were determined using Student’s t test, except for Fig. S3, where a chi-square test was performed.
Data availability.
All the data utilized to generate the graph can be found online at https://www.doi.org/10.6084/m9.figshare.14925375. The DNA-seq data set is available in the SRA repository under accession number PRJNA729435.
ACKNOWLEDGMENTS
We thank I. Taylor and E. Pierre-Jerome from the Benfey lab for critical reading of the manuscript. We also thank S. Ben-Yehuda (Hebrew university) for providing bacterial strains.
This work was supported by a grant from the NSF (PHY-1915445) to P.N.B., by the Howard Hughes Medical Institute to P.N.B., and by a BARD Postdoctoral Fellowship (FI-574-2018) to E.T.
Footnotes
Citation Tzipilevich E, Benfey PN. 2021. Phage-resistant bacteria reveal a role for potassium in root colonization. mBio 12:e01403-21. https://doi.org/10.1128/mBio.01403-21.
Contributor Information
Philip N. Benfey, Email: philip.benfey@duke.edu.
Gustavo H. Goldman, Universidade de Sao Paulo
REFERENCES
- 1.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, Del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. 2012. Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verbon EH, Liberman LM. 2016. Beneficial microbes affect endogenous mechanisms controlling root development. Trends Plant Sci 21:218–229. doi: 10.1016/j.tplants.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hacquard S, Garrido-Oter R, González A, Spaepen S, Ackermann G, Lebeis S, McHardy AC, Dangl JL, Knight R, Ley R, Schulze-Lefert P. 2015. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17:603–616. doi: 10.1016/j.chom.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Lugtenberg B, Kamilova F. 2009. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 5.Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. 2013. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA 110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkel OM, Salas-González I, Castrillo G, Spaepen S, Law TF, Teixeira PJPL, Jones CD, Dangl JL. 2019. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol 17:e3000534. doi: 10.1371/journal.pbio.3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner MR, Lundberg DS, Del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T. 2016. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat Commun 7:12151. doi: 10.1038/ncomms12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finkel OM, Salas-González I, Castrillo G, Conway JM, Law TF, Teixeira PJPL, Wilson ED, Fitzpatrick CR, Jones CD, Dangl JL. 2020. A single bacterial genus maintains root growth in a complex microbiome. Nature 587:103–108. doi: 10.1038/s41586-020-2778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koskella B, Taylor TB. 2018. Multifaceted impacts of bacteriophages in the plant microbiome. Annu Rev Phytopathol 56:361–380. doi: 10.1146/annurev-phyto-080417-045858. [DOI] [PubMed] [Google Scholar]
- 10.Koskella B, Brockhurst MA. 2014. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev 38:916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salmond GP, Fineran PC. 2015. A century of the phage: past, present and future. Nat Rev Microbiol 13:777–786. doi: 10.1038/nrmicro3564. [DOI] [PubMed] [Google Scholar]
- 12.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Valera F, Martin-Cuadrado A-B, Rodriguez-Brito B, Pasić L, Thingstad TF, Rohwer F, Mira A. 2009. Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 14.Buckling A, Rainey PB. 2002. The role of parasites in sympatric and allopatric host diversification. Nature 420:496–499. doi: 10.1038/nature01164. [DOI] [PubMed] [Google Scholar]
- 15.Pal C, Macia MD, Oliver A, Schachar I, Buckling A. 2007. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 450:1079–1081. doi: 10.1038/nature06350. [DOI] [PubMed] [Google Scholar]
- 16.Haaber J, Leisner JJ, Cohn MT, Catalan-Moreno A, Nielsen JB, Westh H, Penadés JR, Ingmer H. 2016. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat Commun 7:13333. doi: 10.1038/ncomms13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul JH, Sullivan MB. 2005. Marine phage genomics: what have we learned? Curr Opin Biotechnol 16:299–307. doi: 10.1016/j.copbio.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Shkoporov AN, Hill C. 2019. Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 25:195–209. doi: 10.1016/j.chom.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Pratama AA, van Elsas JD. 2018. The 'neglected' soil virome - potential role and impact. Trends Microbiol 26:649–662. doi: 10.1016/j.tim.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart CR, Casjens SR, Cresawn SG, Houtz JM, Smith AL, Ford ME, Peebles CL, Hatfull GF, Hendrix RW, Huang WM, Pedulla ML. 2009. The genome of Bacillus subtilis bacteriophage SPO1. J Mol Biol 388:48–70. doi: 10.1016/j.jmb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Cao S, Chai Y, Clardy J, Kolter R, Guo J-h, Losick R. 2012. A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol Microbiol 85:418–430. doi: 10.1111/j.1365-2958.2012.08109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. 2013. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci USA 110:E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoet PP, Coene MM, Cocito CG. 1992. Replication cycle of Bacillus subtilis hydroxymethyluracil-containing phages. Annu Rev Microbiol 46:95–116. doi: 10.1146/annurev.mi.46.100192.000523. [DOI] [PubMed] [Google Scholar]
- 26.Habusha M, Tzipilevich E, Fiyaksel O, Ben-Yehuda S. 2019. A mutant bacteriophage evolved to infect resistant bacteria gained a broader host range. Mol Microbiol 111:1463–1475. doi: 10.1111/mmi.14231. [DOI] [PubMed] [Google Scholar]
- 27.Brown S, Santa Maria JP, Jr, Walker S. 2013. Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckling A, Brockhurst M. 2012. Bacteria-virus coevolution. Adv Exp Med Biol 751:347–370. doi: 10.1007/978-1-4614-3567-9_16. [DOI] [PubMed] [Google Scholar]
- 29.Allison SE, D'Elia MA, Arar S, Monteiro MA, Brown ED. 2011. Studies of the genetics, function, and kinetic mechanism of TagE, the wall teichoic acid glycosyltransferase in Bacillus subtilis 168. J Biol Chem 286:23708–23716. doi: 10.1074/jbc.M111.241265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banda J, Bellande K, von Wangenheim D, Goh T, Guyomarc'h S, Laplaze L, Bennett MJ. 2019. Lateral root formation in Arabidopsis: a well-ordered LRexit. Trends Plant Sci 24:826–839. doi: 10.1016/j.tplants.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 31.Winstel V, Kühner P, Salomon F, Larsen J, Skov R, Hoffmann W, Peschel A, Weidenmaier C. 2015. Wall teichoic acid glycosylation governs Staphylococcus aureus nasal colonization. mBio 6:e00632. doi: 10.1128/mBio.00632-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baptista C, Santos MA, Sao-Jose C. 2008. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. J Bacteriol 190:4989–4996. doi: 10.1128/JB.00349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rocha R, Teixeira-Duarte CM, Jorge JMP, Morais-Cabral JH. 2019. Characterization of the molecular properties of KtrC, a second RCK domain that regulates a Ktr channel in Bacillus subtilis. J Struct Biol 205:34–43. doi: 10.1016/j.jsb.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Grundling A. 2013. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci USA 110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gundlach J, Krüger L, Herzberg C, Turdiev A, Poehlein A, Tascón I, Weiss M, Hertel D, Daniel R, Hänelt I, Lee VT, Stülke J. 2019. Sustained sensing in potassium homeostasis: cyclic di-AMP controls potassium uptake by KimA at the levels of expression and activity. J Biol Chem 294:9605–9614. doi: 10.1074/jbc.RA119.008774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holtmann G, Bakker EP, Uozumi N, Bremer E. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J Bacteriol 185:1289–1298. doi: 10.1128/JB.185.4.1289-1298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gundlach J, Herzberg C, Kaever V, Gunka K, Hoffmann T, Weiß M, Gibhardt J, Thürmer A, Hertel D, Daniel R, Bremer E, Commichau FM, Stülke J. 2017. Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci Signal 10:eaal3011. doi: 10.1126/scisignal.aal3011. [DOI] [PubMed] [Google Scholar]
- 38.Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci USA 106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grau RR, de Oña P, Kunert M, Leñini C, Gallegos-Monterrosa R, Mhatre E, Vileta D, Donato V, Hölscher T, Boland W, Kuipers OP, Kovács ÁT. 2015. A duo of potassium-responsive histidine kinases govern the multicellular destiny of Bacillus subtilis. mBio 6:e00581. doi: 10.1128/mBio.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bais HP, Fall R, Vivanco JM. 2004. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol 134:307–319. doi: 10.1104/pp.103.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan B, Wang C, Song X, Ding X, Wu L, Wu H, Gao X, Borriss R. 2019. Bacillus velezensis FZB42 in 2018: the Gram-positive model strain for plant growth promotion and biocontrol. Front Microbiol 10:1279. doi: 10.3389/fmicb.2019.01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kröber M, Wibberg D, Grosch R, Eikmeyer F, Verwaaijen B, Chowdhury SP, Hartmann A, Pühler A, Schlüter A. 2014. Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing. Front Microbiol 5:252. doi: 10.3389/fmicb.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diaz-Munoz SL, Koskella B. 2014. Bacteria-phage interactions in natural environments. Adv Appl Microbiol 89:135–183. doi: 10.1016/B978-0-12-800259-9.00004-4. [DOI] [PubMed] [Google Scholar]
- 44.Westra ER, van Houte S, Oyesiku-Blakemore S, Makin B, Broniewski JM, Best A, Bondy-Denomy J, Davidson A, Boots M, Buckling A. 2015. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr Biol 25:1043–1049. doi: 10.1016/j.cub.2015.01.065. [DOI] [PubMed] [Google Scholar]
- 45.Moller AG, Lindsay JA, Read TD. 2019. Determinants of phage host range in staphylococcus species. Appl Environ Microbiol 85:e00209-19. doi: 10.1128/AEM.00209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vidakovic L, Singh PK, Hartmann R, Nadell CD, Drescher K. 2018. Dynamic biofilm architecture confers individual and collective mechanisms of viral protection. Nat Microbiol 3:26–31. doi: 10.1038/s41564-017-0050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen MF, Svenningsen SL, Roder HL, Middelboe M, Burmolle M. 2019. Big impact of the tiny: bacteriophage-bacteria interactions in biofilms. Trends Microbiol 27:739–752. doi: 10.1016/j.tim.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 48.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 49.Kim S, Rahman M, Seol SY, Yoon SS, Kim J. 2012. Pseudomonas aeruginosa bacteriophage PA1O requires type IV pili for infection and shows broad bactericidal and biofilm removal activities. Appl Environ Microbiol 78:6380–6385. doi: 10.1128/AEM.00648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lourenço M, Chaffringeon L, Lamy-Besnier Q, Pédron T, Campagne P, Eberl C, Bérard M, Stecher B, Debarbieux L, De Sordi L. 2020. The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28:390.e5–401.e5. doi: 10.1016/j.chom.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Amtmann A, Armengaud P. 2009. Effects of N, P, K and S on metabolism: new knowledge gained from multi-level analysis. Curr Opin Plant Biol 12:275–283. doi: 10.1016/j.pbi.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 52.Sustr M, Soukup A, Tylova E. 2019. Potassium in root growth and development. Plants (Basel) 8:435. doi: 10.3390/plants8100435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leff JW, Jones SE, Prober SM, Barberán A, Borer ET, Firn JL, Harpole WS, Hobbie SE, Hofmockel KS, Knops JMH, McCulley RL, La Pierre K, Risch AC, Seabloom EW, Schütz M, Steenbock C, Stevens CJ, Fierer N. 2015. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc Natl Acad Sci USA 112:10967–10972. doi: 10.1073/pnas.1508382112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castrillo G, Teixeira PJPL, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield NW, Mieczkowski P, Jones CD, Paz-Ares J, Dangl JL. 2017. Root microbiota drive direct integration of phosphate stress and immunity. Nature 543:513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenberg A, Sinai L, Smith Y, Ben-Yehuda S. 2012. Dynamic expression of the translational machinery during Bacillus subtilis life cycle at a single cell level. PLoS One 7:e41921. doi: 10.1371/journal.pone.0041921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SPO1 resistance profile of phage survival isolates. (A) SPO1-resistant bacteria isolated from plants were stained with ConA488 and observed under the microscope. Shown are average and SD fluorescent intensity values from 10 bacteria of each of the surviving colonies. (B) The indicated bacterial strains were infected with SPO1 in LB medium, and OD600 was followed. Shown are averages and SDs with an n of 3. (C) The indicated bacterial strains were stained with ConA488. Shown are representative DIC (left) and fluorescence from ConA488 (right) images from each strain. Download FIG S1, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2021 Tzipilevich and Benfey.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Potassium modulates B. subtilis root colonization and phage resistance. (A) Seedlings were inoculated with WT bacteria and grown for 48 h on 0.25× MS agar plates in the absence or presence of 5 mM of the indicated salts, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. ***, P < 0.005. (B) Seedlings were inoculated with ΔkinC bacteria, with or without SPO1 addition, and grown for 48 h on 0.25× MS agar plates in the presence or absence of 5 mM KCl; the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. (C) Seedlings were inoculated with the indicated bacterial strains and grown for 48 h on 0.25× MS agar plates, in the absence or presence of 5 mM KCl, and the number of colonizing bacteria was counted. Shown are averages and SDs from 2 independent experiments with an n of ≥3 for each. ***, P < 0.005. Download FIG S2, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2021 Tzipilevich and Benfey.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Potassium modulates phage resistance during pellicle biofilm formation. (A) The indicated bacterial strains were inoculated in MSgg medium with or without SPO1 addition for 48 h, and the number of cultures exhibiting partial SPO1 survival (see panel C) were counted. Shown are the percentages of the cultures surviving SPO1, n ≥ 30. ***, P < 0.005. (B) WT bacteria were inoculated into MSgg medium with or without 5 mM KCl and infected with SPO1 for 48 h, as described for panel A. Shown are the percentages of the cultures surviving SPO1, n ≥ 30. ***, P < 0.005. (C) Shown are representative images for the experiment described in panels A and B, for an uninfected culture (1), a culture undergoing complete phage lysis (2), and a culture counted as partially surviving phage infection (3). Download FIG S3, TIF file, 2.3 MB (2.3MB, tif) .
Copyright © 2021 Tzipilevich and Benfey.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All the data utilized to generate the graph can be found online at https://www.doi.org/10.6084/m9.figshare.14925375. The DNA-seq data set is available in the SRA repository under accession number PRJNA729435.