ABSTRACT
Azole resistance in pathogenic Aspergillus fumigatus has become a global public health issue threatening the use of medical azoles. The environmentally occurring resistance mutations, TR34/L98H (TR34) and TR46/Y121F/T289A (TR46), are widespread across multiple continents and emerging in the United States. We used whole-genome single nucleotide polymorphism (SNP) analysis on 179 nationally represented clinical and environmental A. fumigatus genomes from the United States along with 18 non-U.S. genomes to evaluate the genetic diversity and foundation of the emergence of azole resistance in the United States. We demonstrated the presence of clades of A. fumigatus isolates: clade A (17%) comprised a global collection of clinical and environmental azole-resistant strains, including all strains with the TR34/L98H allele from India, The Netherlands, the United Kingdom, and the United States, and clade B (83%) consisted of isolates without this marker mainly from the United States. The TR34/L98H polymorphism was shared among azole-resistant A. fumigatus strains from India, The Netherlands, the United Kingdom, and the United States, suggesting the common origin of this resistance mechanism. Six percent of azole-resistant A. fumigatus isolates from the United States with the TR34 resistance marker had a mixture of clade A and clade B alleles, suggestive of recombination. Additionally, the presence of equal proportions of both mating types further suggests the ongoing presence of recombination. This study demonstrates the genetic background for the emergence of azole resistance in the United States, supporting a single introduction and subsequent propagation, possibly through recombination of environmentally driven resistance mutations.
KEYWORDS: azole resistance, Aspergillus fumigatus, whole-genome sequencing, population genomics, population structure, TR34/L98H, drug resistance mechanisms, population genetics
INTRODUCTION
Aspergillus fumigatus is a globally distributed environmental fungus and the primary cause of aspergillosis, a serious and often fatal infection that mainly affects patients with impaired immune systems or lung function but can also occur in people without these underlying conditions (1, 2). Recent studies demonstrate that invasive pulmonary aspergillosis is a lethal and frequently misdiagnosed complication of severe influenza and severe acute respiratory syndrome coronavirus 2 (SARs-CoV-2) infections that can cause illness and death in previously healthy individuals (2–9). Medical azoles, such as itraconazole, posaconazole, and voriconazole, are the first line of therapy against aspergillosis. However, numerous clinical reports and surveillance studies document the emergence of azole-resistant A. fumigatus (ARAf) over the past decade (10).
Azole-resistant A. fumigatus isolates are characterized by a wide variety of genomic resistance mechanisms. However, the predominant azole resistance mechanism found among ARAf isolates is one or more substitutions in the sterol 14α-demethylase gene (cyp51A) involved in the biosynthesis of ergosterol, the main component of the fungal cell membrane; medical azoles target the product of this gene. Numerous reports document a variety of single nucleotide substitutions (SNPs) in the cyp51A gene that arise in patients that undergo prolonged antifungal therapy, such as patients with cystic fibrosis and chronic pulmonary aspergillosis (11–13). In contrast, ARAf isolates with duplications in the cyp51A gene promoter coupled with specific amino acid substitutions, known as TR34/L98H and TR46/Y121F/T289A, are becoming widespread in the environment and are frequently isolated from patients without previous exposure to antifungal drugs (14–19). Clinical ARAf isolates harboring these environmentally derived mutations are emerging as the predominant cause of resistance in patients without azole exposure (20).
It is now widely accepted that the emergence of TR34/L98H and TR46/Y121F/T289A substitutions in A. fumigatus is associated with selection pressure from azole fungicides that target the cyp51A gene product. In recent years, the use of these fungicides in crop agriculture in the United States has increased; therefore, it is not surprising that reports of ARAf isolates in patients and the environment harboring these substitutions has also increased (20, 21). The presence of clinical or environmental ARAf with TR34/L98H and TR46/Y121F/T289A substitutions has now been documented on six continents (14, 17, 22–26). Up to 15% of clinical isolates in Europe carry these substitutions, which dramatically decrease treatment options for patients infected with ARAf strains (27, 28). Recent genomic analysis demonstrated that azole-resistant isolates from patients and the environment are genetically related (29).
Although still uncommon in the United States, ARAf with environmentally derived substitutions in cyp51A have emerged in at least 3 states (30). While the initial survey of 1,026 A. fumigatus isolates conducted by the CDC from 2011 to 2013 did not identify isolates with TR34/L98H or TR46/Y121F/T289A substitutions, subsequent surveillance reported the emergence of these polymorphisms among clinical and environmental ARAf isolates in the United States (15, 31–35). However, it remains unknown whether isolates with these mutations were introduced into the United States or emerged independently. To address this question and to gain deeper insight into the genetic diversity and the emergence of ARAf isolates in the United States, we applied whole-genome sequencing on a subset of A. fumigatus isolates from a collection of 1,736 clinical and environmental isolates from various regions in the United States (15, 31). The aim was to (i) characterize the genetic relationship among ARAf isolates from the United States and global isolates, (ii) identify the genetic substitutions in the cyp51A gene that may influence antifungal susceptibility, and (iii) evaluate the extent of genetic recombination in the A. fumigatus population to test the hypothesis that recombination may have contributed to the distribution of the TR34/L98H allele in the United States.
RESULTS
Azole resistance and substitutions in cyp51A.
A total of 179 A. fumigatus isolates from 36 states were included in the analysis (see Table S1 in the supplemental material). This sample included (i) 164 representative clinical isolates selected from 1,736 A. fumigatus isolates submitted to the CDC as part of the ongoing passive surveillance for azole resistance from 2015 to 2017, (ii) 13 previously reported environmental isolates from a peanut farm in the state of Georgia, and (iii) two historical type isolates from the CDC strain collection (15). We included all isolates with elevated MICs to itraconazole and voriconazole available at the time as well as susceptible isolates from seven geographic areas in the United States (West, Midwest, Mountain, Central, Northeast, Mid-Atlantic, and Southeast) (Fig. 1). In addition, publicly available genomes of 18 isolates from The Netherlands, India, and the United Kingdom were included for comparison. Of the 179 U.S. samples, 46 (26%, 33 clinical and all 13 environmental) were deemed resistant to triazoles using an MIC cutoff value of 2 μg/ml for either itraconazole or voriconazole (Table S1).
FIG 1.
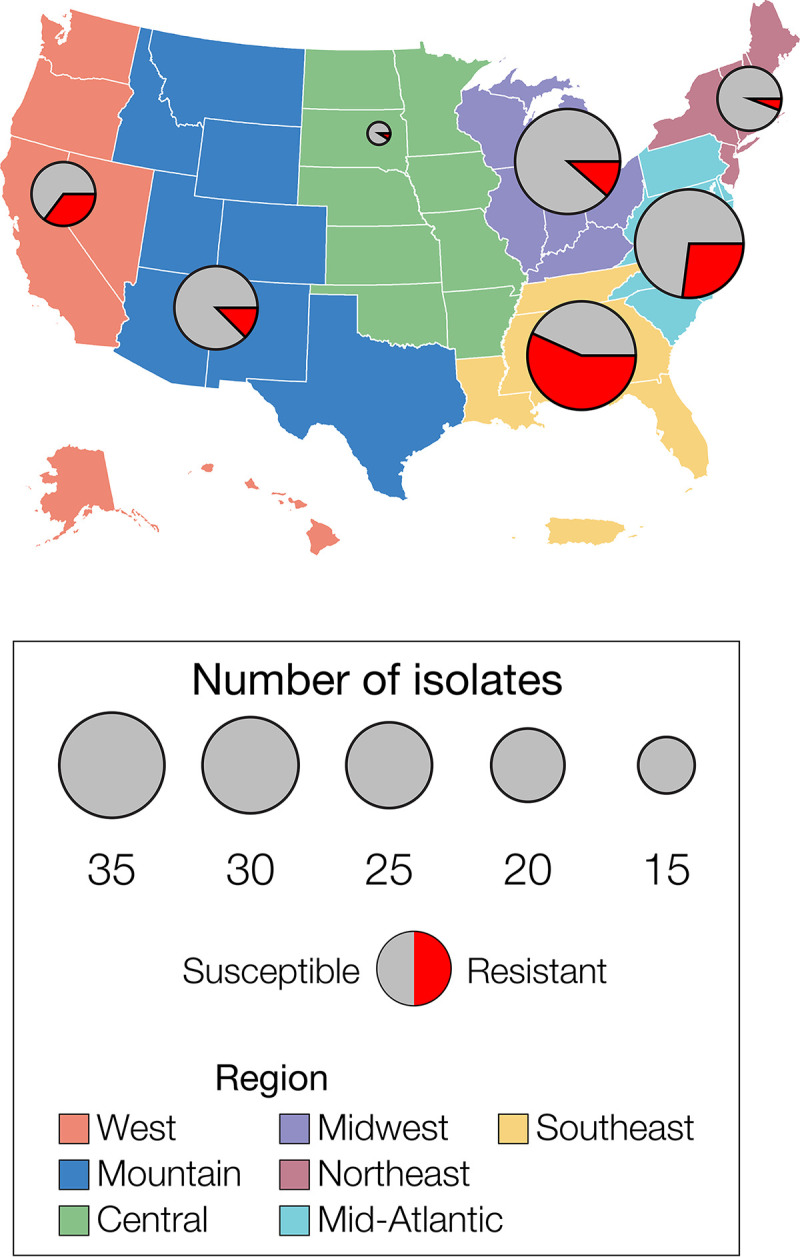
A. fumigatus isolates in the United States. Azole-resistant and -susceptible Aspergillus fumigatus isolates sampled from 7 Antimicrobial Resistance Laboratory Network (ARLN) regions in the United States. The total number of isolates and the number of resistant isolates are represented by circles and pie pieces, respectively.
Isolates. Aspergillus fumigatus isolates in this study. Download Table S1, PDF file, 0.1 MB (143.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To investigate the potential mechanisms of resistance, we downloaded cyp51A sequences and searched for nonsynonymous substitutions in this region. Forty-four isolates (25%) had nonsynonymous cyp51A substitutions, three of which, G448S, TR34/L98H, and TR34/L98H/S297T/F495I, are known to confer resistance to one or more triazole (Table 1; Table S1). Three clinical isolates with TR34/L98H substitution were identified in the Mid-Atlantic region. In addition, eight isolates with TR34/L98H/S297T/F495I were identified: two clinical (one from the Mid-Atlantic region one from the West), and six environmental isolates from a peanut farm in the Southeast. Other substitutions in cyp51A were also identified, but they were present in both resistant and susceptible isolates and were not linked to resistance (Table 1). An identical wild-type cyp51A allele was found in 135 isolates; of those, 120 were susceptible and 15 were resistant to azoles (Table S1). Additionally, we also searched for variants within the 3-hydroxy-3-methyl-glutaryl-coenzyme A (HMG-CoA) reductase-encoding gene, hmg1: all sequences were wild type.
TABLE 1.
cyp51A substitutions and in vitro antifungal susceptibility ranges of A. fumigatus isolates from the United States
| cyp51A substitution(s) | No. of isolates | Origin type | MIC range (μg/ml) |
|
|---|---|---|---|---|
| Itraconazole | Voriconazole | |||
| A9T | 4 | Clinical | 0.5 to 1.5 | 0.38 to 1 |
| D262Y | 1 | Clinical | 8 | 2 |
| F46Y/M172V/N248K/D255E/E427K | 8 | Both | 0.19 to >16 | 0.094 to 4 |
| G254V | 1 | Environmental | >16 | 4 |
| G448S | 2 | Clinical | 16 | 8 |
| I242V | 15 | Both | 0.25 to >16 | 0.25 to 2 |
| N248K | 1 | Clinical | 1 | 0.25 |
| P216L | 1 | Clinical | >32 | 0.047 |
| TR34/L98H | 14 | Clinical | 4 to 16 | 0.5 to 2 |
| TR34/L98H/S297T/F495I | 10 | Both | 8 to >16 | 0.25 to 4 |
| Wild type | 124 | Both | 0 to 2 | 0 to 1 |
Phylogenetic analysis using the neighbor-joining algorithm showed little genetic variation in the cyp51A locus, a total of 27 SNPs were identified in a 2,049-bp region. All sequences from ARAf isolates with the TR34/L98H substitution from U.S. and non-U.S. isolates formed a single monophyletic clade on the phylogenetic tree (see Fig. S1).
Genetic relationship in the cyp51A gene among A. fumigatus isolates. Analysis of the cyp51A gene using neighbor-joining algorithm identified multiple types of mutations among U.S. and non-U.S. Aspergillus fumigatus isolates: the environmentally derived mutation, TR34/L98H is seen among U.S. and non-U.S. azole-resistant A. fumigatus isolates. A small set of clinical and environmental isolates from the United States includes this environmental mutation coupled with two other substitutions, S297T and F495I. Substitutions such as A9T, G254V, I242V, D262Y, and N248K were commonly found among azole-resistant and azole-susceptible clinical isolates from the United States. Isolates with the wild-type allele were commonly found among the U.S. collection. Download FIG S1, PDF file, 0.4 MB (449.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genetic diversity of A. fumigatus isolates.
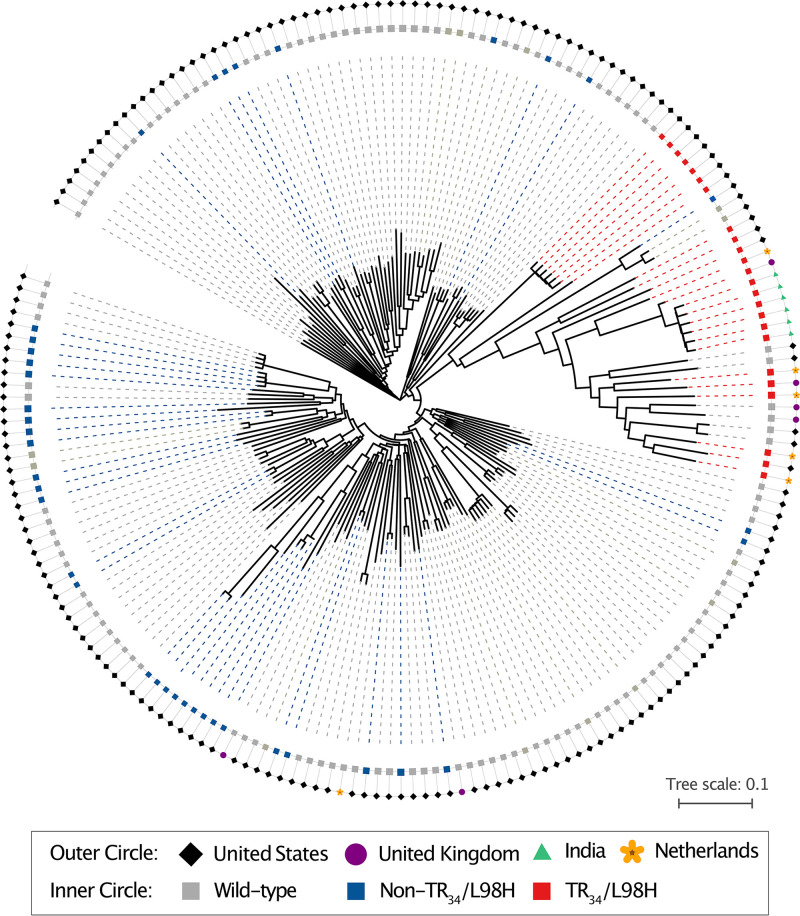
We performed whole-genome sequencing to investigate genetic diversity among isolates. All genomes had greater than 100× sequencing coverage and mapped to >95% of the publicly available reference genome AF293 (Table S1). Genomes representing the United States and the non-U.S. genomes shared a total of 228,546 SNPs. Phylogenetic analysis using the neighbor-joining algorithm showed the presence of two major clades (Fig. 2). Clade B represented azole-susceptible and -resistant isolates without the TR34/L98H resistance marker (N = 164 [83%]); resistant isolates belonging to this clade were geographically spread across the United States and can be further divided into two subclades. Clade A comprised 24 (12%) isolates with the TR34/L98H resistance marker from the United States, India, The Netherlands, and United Kingdom as well as 9 isolates without this marker from the United States, The Netherlands, and United Kingdom (Fig. 2). Notably, 7 of the 18 U.S. isolates clustering with clade A were located on the long branches at the base of clade A, suggestive of their intermediate position between the two clades (Fig. 2). Six environmental isolates with TR34/L98H/S297T/F495I from a peanut farm in Georgia were genetically related and likely came from a clonal population.
FIG 2.
A. fumigatus triazole resistance. Phylogenomic analysis, using the neighbor-joining (NJ) algorithm, of azole-resistant and -susceptible isolates from the United States, the United Kingdom, India, and The Netherlands suggests the present of two clades. Clade A consists mainly of azole-resistant A. fumigatus isolates carrying the TR34/L98H resistance marker from all geographic areas. Clade B consists of azole-resistant and -susceptible A. fumigatus isolates carrying non-TR34/L98H and wild-type alleles mainly from the United States.
Population structure of A. fumigatus.
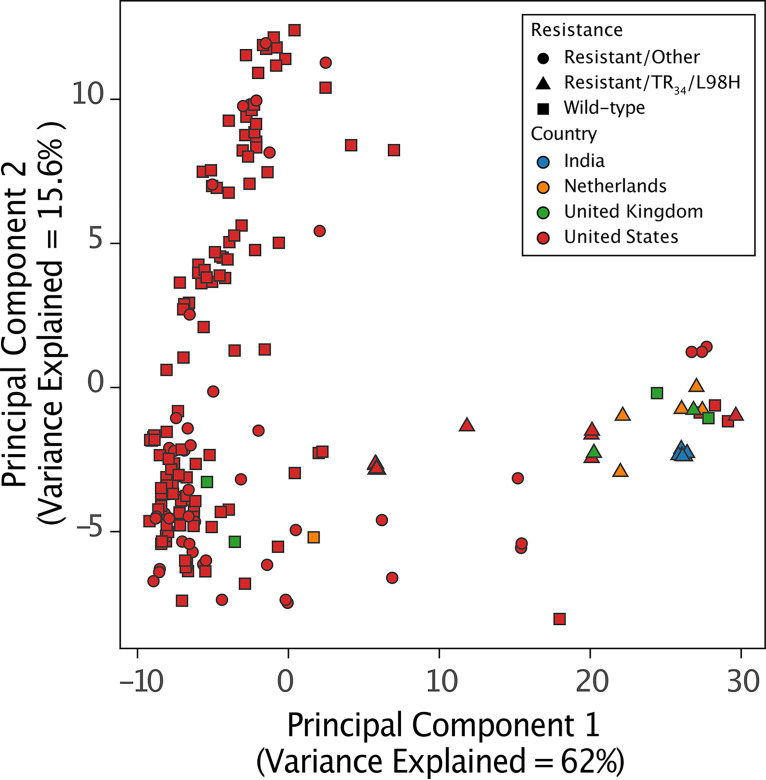
To explore the genetic relationships among isolates, we performed principal-component analysis. Analysis of the first two principal components, representing the majority of the variation (77.6% of the total variance) in this data set, demonstrated the presence of two populations, A and B, that corresponded to the A and B clades on the phylogenetic tree; in addition, 14 isolates were located between the two populations (Fig. 2 and 3) (17). A collection of globally represented A. fumigatus isolates harboring the TR34/L98H resistance marker clustered along the first principal component, capturing much of the uniqueness in this data set (62% variation) (Fig. 3). The majority of U.S. and non-U.S. isolates with the TR34/L98H resistance marker were found along this axis (population A). Isolates mainly representing azole-susceptible and ARAf isolates without the TR34/L98H marker clustered along the second principal component (population B) (Fig. 3). Less genomic variation was observed among A. fumigatus isolates within the United States (15.6%); however, two subpopulations within population B which correspond to the two subclades in clade B can be differentiated. In addition, 3 genotypes occupied an intermediate position between the two populations (Fig. 3).
FIG 3.
Global diversity of A. fumigatus isolates. Principal-component analysis indicated that most A. fumigatus isolates from the United States cluster can be found along the second principal component separately from non-U.S. A. fumigatus isolates. Azole-resistant A. fumigatus isolates without the TR34/L98H resistance marker and isolates carrying the wild-type allele can also be found along this axis (population B). A subset of A. fumigatus isolates harboring the TR34/L98H resistance marker from India, Netherlands, United States, and United Kingdom clustered along the first principal component (population A). An intermediate population of U.S. azole-resistant A. fumigatus isolates with the TR34/L98H resistance marker was observed (red triangles).
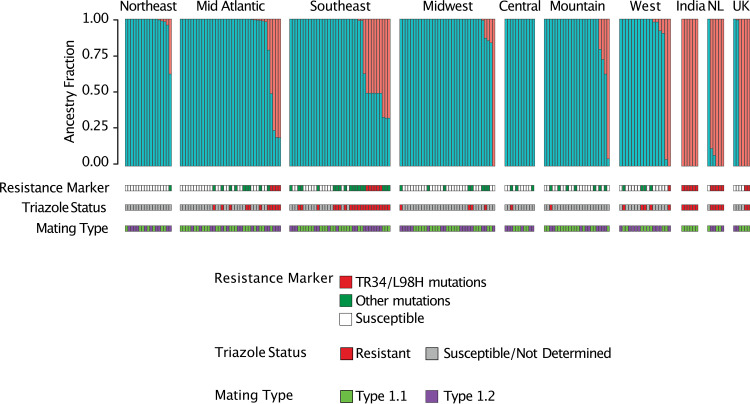
We used ADMIXTURE to further investigate the population structure and assess the extent of genetic recombination between A and B populations as well as between isolates with and without the TR34/L98H resistance marker (36). When criteria were set to two populations (K = 2), ADMIXTURE assigned 164 (83%) isolates from the United States to population B, while isolates with TR34/L98H markers from India, The Netherlands, and the United Kingdom were assigned to population A. In addition, six isolates (18%), representing the Midwest, Mountain, Southeast, and West regions of the United States were also assigned to population A (Fig. 4). Furthermore, 12 (6%) isolates from the United States, including those with the TR34/L98H markers, had a mixture of population A and population B alleles, indicative of recombination between the populations. Notably, all isolates with the TR34/L98H resistance marker shared more than 50% of their alleles with the isolates from population A, which suggests they may be a product of recombination between the two populations (Fig. 4). Similar results were observed when the ADMIXTURE criteria were set to 3 populations (K = 3) (see Fig. S2), in which case, the majority of isolates from the United States contained a mixture of alleles from two subpopulations corresponding to the two subclades within the B clade on the phylogenetic tree, while isolates with TR34/L98H marker were assigned to population A and possessed more than 50% of their alleles with the isolates from population A. Although a K value of >3 was not supported by principal-component analysis (PCA) or phylogenetic analysis, ADMIXTURE results for K values of 4 to 10 are shown in Fig. S3.
FIG 4.
Population structure of A. fumigatus isolates. ADMIXTURE analysis (K = 2) identified two populations of A. fumigatus isolates. A. fumigatus resistant isolates with TR34/L98H resistance marker from different areas of the United States share alleles with A. fumigatus isolates from India, The Netherlands, and United Kingdom. Most A. fumigatus isolates without the TR34/L98H were widespread across multiple regions of the United States and shared less than 50% of their alleles with non-U.S. isolates. Azole resistant A. fumigatus isolates with the TR34/L98H carrying the mat1-2 idiomorph were commonly seen in U.S. isolates, while mat1-1 was seen in non-U.S. isolates.
Population structure of A. fumigatus isolates. ADMIXTURE (K = 3) analysis demonstrates that the majority of isolates from the United States contained a mixture of alleles from two subpopulations. A. fumigatus resistant isolates with the TR34/L98H resistance marker from different areas of the United States share alleles with A. fumigatus isolates from India, The Netherlands, and the United Kingdom. Download FIG S2, PDF file, 0.4 MB (424.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ADMIXTURE results (for K = 2 through 10). Results for K values of 2 to 4 indicate that the U.S. isolates with the TR34/L98H marker share alleles with A. fumigatus isolates from India, The Netherlands, and the United Kingdom. No evidence for K value of >3 was detected by PCA or phylogenetic analysis. Download FIG S3, PDF file, 0.3 MB (309.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Distribution of mating types.
Aspergillus fumigatus isolates carrying a mixture of alleles from the two different populations, which suggests the presence of recombination and sexual reproduction. To further test whether A. fumigatus in the United States is capable of sexual reproduction, we mined genome assemblies for mat1-1 and mat1-2 genes. The mat1-1 gene was present in 55% of genomes, while the mat1-2 gene was found in 45% of A. fumigatus genomes (Fig. 4; see also Fig. S4). While the mat1-2 gene was more frequent among ARAf isolates, the difference was not significant (P > 0.05). This analysis demonstrates a nearly equal proportion of both mating types present in this sample and allele sharing between different mating types (Fig. 4).
Distribution of mating types among resistant and susceptible A. fumigatus isolates in the United States. A chi-square statistical analysis with 179 isolates from the United States suggests mating type does not influence antifungal susceptibility. Download FIG S4, PDF file, 0.4 MB (392.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
The environmentally associated polymorphism TR34/L98H occurs globally but only recently emerged in the United States among clinical and environmental ARAf. Azole resistance in A. fumigatus is primarily linked to substitutions in the cyp51A gene. Of the 46 resistant isolates from the United States in our collection, nearly two-thirds had nonsynonymous substitutions in the cyp51A gene, three of which, including the environmentally derived TR34/L98H mutation, are known to cause resistance. Other substitutions in cyp51A were also observed in both resistant and susceptible isolates; however, their role in conferring resistance to azoles requires additional investigation. In addition, we identified 16 resistant isolates with a wild-type copy of cyp51A, indicating the presence of other mechanisms of resistance in the U.S. population of A. fumigatus. No isolates with TR46/Y121F/T289A resistance polymorphisms were identified in our collection, although detection of isolates with these alleles in the United States was reported by others (33, 34).
Recent studies utilizing high-resolution whole-genome sequencing and microsatellite typing have identified two genetically distinct populations of A. fumigatus (17, 19, 29, 34). These studies indicate that most ARAf isolates harboring TR34/L98H form a single clade that is genetically distinct from other clades and includes isolates with other resistance mechanisms and wild-type genotypes. Consistent with these observations, phylogenetic analysis using whole-genome sequencing data and PCA identified the presence of two major populations in the United States, which, according to the nomenclature used by Sewell et al., were designated clade A and clade B (17).
Phylogenetic analysis demonstrated 83% of U.S. isolates clustered with clade B, all of which were either susceptible to azoles or had other mechanisms of azole resistance; no isolates with the TR34/L98H marker were detected in this population. Conversely, all U.S. isolates with the TR34/L98H marker clustered with clade A; however, most of them occupied an intermediate position between the two populations on the PCA plot and were placed on long branches at the base of clade A that suggested that they might have been products of recombination between the two populations. However, the main limitation of these analyses is that neither PCA nor phylogenetic analysis considers a possibility of recombination between isolates.
We used ADMIXTURE that considers recombination and estimates the contributions of alleles from different populations in a recombining population (36). This analysis confirmed that azole-resistant isolates with the TR34/L98H resistance marker from The Netherlands, India, and the United Kingdom belonged to population A. In addition, two isolates from the United States, one with TR34/L98H from the West and another susceptible isolate from the Midwest, possessed 100% of the alleles assigned to population A. Furthermore, 7% of the isolates from the United States contained a mixture of alleles from both populations, and all U.S. isolates with the TR34/L98H marker had at least 50% of the alleles from population A, suggesting ongoing recombination between the populations and a possible spread of TR34/L98H into population B in the United States (Fig. 3). Sexual reproduction was previously reported in A. fumigatus (37–40). Our analysis shows the nearly 1:1 distribution of the mating types in our sample that is consistent with a recombining population. Isolates with both mating types were identified among TR34/L98H strains in the United States.
To further investigate the origin of the TR34/L98H marker in the United States, we interrogated cyp51A gene genealogy, a much smaller region that is less likely to be affected by recombination than the entire genome. Unfortunately, only a small number of single nucleotide polymorphisms were detected in this locus, limiting the extent of phylogenetic inference that could be conducted with this locus. However, these limited data are consistent with a single monophyletic origin of TR34/L98H sequences. Interestingly, isolates with TR34/L98H/S297T/F495I were closely related to TR34/L98H sequences. Consistent with phylogenetic analysis, isolates with this marker also formed a separate phylogenetic subgroup on the genome phylogeny within clade A and consisted mainly of isolates from the United States, which was indicative of the genetic relatedness among these isolates.
Because of the environmental occurrence and the rapid spread in Europe and Asia, the emergence of TR34/L98H and TR46/Y121F/T289A mechanisms of resistance is a serious concern to public health. One of the major questions about the emergence of these isolates in the environment is whether these markers can arise independently in multiple populations or whether these substitutions have arisen once and are spreading through migration and recombination. In addition to an academic interest, the answer to this question has important public health implications. Although they do not provide direct proof, the combined results of PCA, ADMIXTURE, whole-genome, and single-locus phylogenetic analysis are consistent with the hypothesis of a single introduction of the TR34/L98H mutations in population A and the ongoing spread of this marker throughout clade A possibly through recombination. Since the majority of the U.S. isolates of A. fumigatus are represented in population B, the current low prevalence of the TR34/L98H mutations among the U.S. isolates is not surprising. However, the ability of isolates from two populations to recombine is suggestive of future spread of this mutation among U.S. A. fumigatus populations.
Our results demonstrate that isolates with this resistance mechanism have been introduced into the United States (21). The use of azole antifungals in agriculture in the United States is rapidly increasing; therefore, the proportion of resistance isolates in the environment and clinics is also expected to increase, underscoring the importance of public health, environmental surveillance, and the development of prevention measures (21). Therefore, the CDC and AR Laboratory network have launched surveillance for ARAf in the United States (https://www.cdc.gov/drugresistance/laboratories/AR-lab-network-testing-details.html).
MATERIALS AND METHODS
Isolates.
Species identification and antifungal susceptibility testing were previously reported on a combined collection of 1,736 clinical and environmental isolates in the United States (15, 31). A total of 179 clinical isolates, 46 azole resistant and 133 randomly selected azole susceptible, collected in 2015 to 2017 from 7 geographic areas (36 states) were included (see Table S1 in the supplemental material); the origin of the isolates was assigned based on the submitter address. Thirteen azole-resistant environmental isolates were also included. Two historical isolates, which later were identified as different aliquots of the same NRRL 163 (ATCC 1022) strain, were obtained from the Mycotic Diseases culture collection, Centers for Disease Control and Prevention, Atlanta, Georgia.
Growth, DNA extraction, and whole-genome sequencing.
All isolates were grown on Sabouraud dextrose agar (SDA) slants at 37°C for 24 h; conidia were harvested and preserved in 20% Tween 80-sterile water. Conidial suspensions were serially diluted; 1 μl of the suspension was placed in a 25-ml canonical tube containing yeast-peptone-dextrose (YPD) broth. This mixture was grown for 12 to 18 h in a 37°C centrifugal shaker at 300 × g. Fungal hyphae were transferred to a 2-ml centrifuge tube and spun at 13,000 × g to remove residual broth. For DNA extraction, a Zymo research fungal/bacterial miniprep kit (Zymo Research, USA) coupled with a Zymo research large-fragment DNA recovery kit was used per manufacturer’s guidelines. Genomic library preparation and sequencing were performed as previously described (41).
Gene analysis.
All sequence data were assembled using default parameters in SPAdes version 3.9 (42). Assemblies were evaluated using QUAST version 2.3 (43). Nucleotide BLAST databases were generated using the cyp51A gene (accession AF338569) and mating type genes mat1-1 and mat1-2 (44). De novo genome assemblies were compared to the cy51A gene using the blastn algorithm of BLAST+ (45). This process was repeated for the mating type genes. Genomic regions that covered the entire length of reference sequences and produced greater the 98% sequence similarity were designated that particular gene. Genes were analyzed using the MUSCLE algorithm in MEGA version 7, and neighbor-joining trees were plotted using iTOL and Microreact (46–48).
Variant calling and phylogenetic analysis.
Raw Illumina reads from U.S. and non-U.S. Aspergillus fumigatus genomes (BioProject PRJEB8623) were evaluated for quality using FastQC version 0.11.2 (49). All sequence data were mapped to AF293 (GCF_000002655.1) using BWA version 0.7.7 (50). Postalignment processing was performed using SAMtools version 1.7 and Picard tools version 2.5 (51). Single nucleotide polymorphisms were identified using the Genome Analysis Toolkit Haplotype Caller (GATK) version 4.0 (52). Variant filtering was performed by removing variants according to extracted values from Quality Depth (QD < 2) Fisher Strand (FS > 50), Strand Odds Ratio (SOR < 3), Mapping Quality (MQ < 50), MappingQualityRankSum (−10.5 < MQ > 10.5), and ReadPosRankSum (−5 < ReadPosRankSum > 5). Filtered variants were converted to allele sharing distances in TASSLE version 5.2.1.6 (53). Allele sharing distance is defined as 1 − identity by state (IBS), where IBS is defined as the probability of drawing the same allele from two different individuals. The neighbor-joining algorithm, as implemented in MEGA-CC, was used to construct an unrooted phylogenetic tree and visualized with the Interactive Tree Of Life (iTOL) and Microreact (46–48).
Population analysis.
Pairwise average nucleotide identities were estimated using a publicly available script (54). Principal-component analysis for the first two components was performed with resulting pairwise distance matrix using the prcomp function in the R programming language. To evaluate the structure of A. fumigatus isolates, an unsupervised analysis was executed for a K of 2 to a K of 10 populations in ADMIXTURE (36). All graphs shown in this paper were plotted using the ggplot2 library in R.
Data availability.
All raw Illumina reads in this study have been submitted to NCBI under BioProject accession number PRJNA632561.
ACKNOWLEDGMENT
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
This article is a direct contribution from Shawn R. Lockhart, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Matthew Fisher, Imperial College London, and John Perfect, Duke University.
Citation Etienne KA, Berkow EL, Gade L, Nunnally N, Lockhart SR, Beer K, Jordan IK, Rishishwar L, Litvintseva AP. 2021. Genomic diversity of azole-resistant Aspergillus fumigatus in the United States. mBio 12:e01803-21. https://doi.org/10.1128/mBio.01803-21.
Contributor Information
Anastasia P. Litvintseva, Email: frq8@cdc.gov.
Leah E. Cowen, University of Toronto
REFERENCES
- 1.Meis JF, Chowdhary A, Rhodes JL, Fisher MC, Verweij PE. 2016. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos Trans R Soc Lond B Biol Sci 371:20150460. doi: 10.1098/rstb.2015.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crum-Cianflone NF. 2016. Invasive aspergillosis associated with severe influenza infections. Open Forum Infect Dis 3 Suppl 1:1272. doi: 10.1093/ofid/ofw172.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meersseman W, Vandecasteele SJ, Wilmer A, Verbeken E, Peetermans WE, Van Wijngaerden E. 2004. Invasive aspergillosis in critically ill patients without malignancy. Am J Respir Crit Care Med 170:621–625. doi: 10.1164/rccm.200401-093OC. [DOI] [PubMed] [Google Scholar]
- 4.Wauters J, Baar I, Meersseman P, Meersseman W, Dams K, De Paep R, Lagrou K, Wilmer A, Jorens P, Hermans G. 2012. Invasive pulmonary aspergillosis is a frequent complication of critically ill H1N1 patients: a retrospective study. Intensive Care Med 38:1761–1768. doi: 10.1007/s00134-012-2673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taccone FS, Van den Abeele AM, Bulpa P, Misset B, Meersseman W, Cardoso T, Paiva JA, Blasco-Navalpotro M, De Laere E, Dimopoulos G, Rello J, Vogelaers D, Blot SI, AspICU Study Investigators. 2015. Epidemiology of invasive aspergillosis in critically ill patients: clinical presentation, underlying conditions, and outcomes. Crit Care 19:7. doi: 10.1186/s13054-014-0722-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nulens EF, Bourgeois MJ, Reynders MB. 2017. Post-influenza aspergillosis, do not underestimate influenza B. Infect Drug Resist 10:61–67. doi: 10.2147/IDR.S122390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah MM, Hsiao EI, Kirsch CM, Gohil A, Narasimhan S, Stevens DA. 2018. Invasive pulmonary aspergillosis and influenza co-infection in immunocompetent hosts: case reports and review of the literature. Diagn Microbiol Infect Dis 91:147–152. doi: 10.1016/j.diagmicrobio.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koehler P, Bassetti M, Chakrabarti A, Chen SCA, Colombo AL, Hoenigl M, Klimko N, Lass-Florl C, Oladele RO, Vinh DC, Zhu LP, Boll B, Bruggemann R, Gangneux JP, Perfect JR, Patterson TF, Persigehl T, Meis JF, Ostrosky-Zeichner L, White PL, Verweij PE, Cornely OA, European Confederation of Medical Mycology, International Society for Human Animal Mycology, Asia Fungal Working Group, INFOCUS LATAM/ISHAM Working Group, SHAM Pan Africa Mycology Working Group, European Society for Clinical Microbiology, Infectious Diseases Fungal Infection Study Group, ESCMID Study Group for Infections in Critically Ill Patients, Interregional Association of Clinical Microbiology and Antimicrobial Chemotherapy, Medical Mycology Society of Nigeria, Medical Mycology Society of China Medicine Education Association, Infectious Diseases Working Party of the German Society for Haematology and Medical Oncology, Association of Medical Microbiology, Infectious Disease Canada. 2021. Defining and managing COVID-19-associated pulmonary aspergillosis: the 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect Dis 21:e149–e162. doi: 10.1016/S1473-3099(20)30847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salmanton-Garcia J, Sprute R, Stemler J, Bartoletti M, Dupont D, Valerio M, Garcia-Vidal C, Falces-Romero I, Machado M, de la Villa S, Schroeder M, Hoyo I, Hanses F, Ferreira-Paim K, Giacobbe DR, Meis JF, Gangneux JP, Rodriguez-Guardado A, Antinori S, Sal E, Malaj X, Seidel D, Cornely OA, Koehler P, FungiScope European Confederation of Medical Mycology/The International Society for Human and Animal Mycology Working Group. 2021. COVID-19-associated pulmonary aspergillosis, March-August 2020. Emerg Infect Dis 27:1077–1086. doi: 10.3201/eid2704.204895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lestrade PPA, Meis JF, Melchers WJG, Verweij PE. 2019. Triazole resistance in Aspergillus fumigatus: recent insights and challenges for patient management. Clin Microbiol Infect 25:799–806. doi: 10.1016/j.cmi.2018.11.027. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhary A, Sharma C, Hagen F, Meis JF. 2014. Exploring azole antifungal drug resistance in Aspergillus fumigatus with special reference to resistance mechanisms 9:697–711. doi: 10.2217/fmb.14.27. [DOI] [PubMed] [Google Scholar]
- 12.Dudakova A, Spiess B, Tangwattanachuleeporn M, Sasse C, Buchheidt D, Weig M, Groß U, Bader O. 2017. Molecular tools for the detection and deduction of azole antifungal drug resistance phenotypes in Aspergillus species. Clin Microbiol Rev 30:1065–1091. doi: 10.1128/CMR.00095-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma C, Chowdhary A. 2017. Molecular bases of antifungal resistance in filamentous fungi. Int J Antimicrob Agents 50:607–616. doi: 10.1016/j.ijantimicag.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Alvarez-Moreno C, Lavergne RA, Hagen F, Morio F, Meis JF, Le Pape P. 2017. Azole-resistant Aspergillus fumigatus harboring TR34/L98H, TR46/Y121F/T289A and TR53 mutations related to flower fields in Colombia. Sci Rep 7:45631. doi: 10.1038/srep45631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurst SF, Berkow EL, Stevenson KL, Litvintseva AP, Lockhart SR. 2017. Isolation of azole-resistant Aspergillus fumigatus from the environment in the south-eastern USA. J Antimicrob Chemother 72:2443–2446. doi: 10.1093/jac/dkx168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoustra SE, Debets AJM, Rijs A, Zhang J, Snelders E, Leendertse PC, Melchers WJG, Rietveld AG, Zwaan BJ, Verweij PE. 2019. Environmental hotspots for azole resistance selection of Aspergillus fumigatus, the Netherlands. Emerg Infect Dis 25:1347–1353. doi: 10.3201/eid2507.181625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sewell TR, Zhu J, Rhodes J, Hagen F, Meis JF, Fisher MC, Jombart T. 2019. Nonrandom distribution of azole resistance across the global population of Aspergillus fumigatus. mBio 10:e00392-19. doi: 10.1128/mBio.00392-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Snelders E, Zwaan BJ, Schoustra SE, Meis JF, Van Dijk K, Hagen F, Van Der Beek MT, Kampinga GA, Zoll J, Melchers WJG, Verweij PE, Debets AJM. 2017. A novel environmental azole resistance mutation in Aspergillus fumigatus and a possible role of sexual reproduction in its emergence. mBio 8:e00791-17. doi: 10.1128/mBio.00791-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdolrasouli A, Rhodes J, Beale MA, Hagen F, Rogers TR, Chowdhary A, Meis JF, Armstrong-James D, Fisher MC. 2015. Genomic context of azole resistance mutations in Aspergillus fumigatus determined using whole-genome sequencing. mBio 6:e00536. doi: 10.1128/mBio.00536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lockhart SR, Beer K, Toda M. 2020. Azole-resistant Aspergillus fumigatus: what you need to know. Clin Microbiol Newsl 42:1–6. doi: 10.1016/j.clinmicnews.2019.12.003. [DOI] [Google Scholar]
- 21.Toda M, Beer KD, Kuivila KM, Chiller TM, Jackson BR. 2021. Trends in agricultural triazole fungicide use in the United States, 1992–2016 and possible implications for antifungal-resistant fungi in human disease. Environ Health Perspect 129:55001. doi: 10.1289/EHP7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdolrasouli A, Petrou MA, Park H, Rhodes JL, Rawson TM, Moore LSP, Donaldson H, Holmes AH, Fisher MC, Armstrong-James D. 2018. Surveillance for azole-resistant Aspergillus fumigatus in a centralized diagnostic mycology service, London, United Kingdom, 1998–2017. Front Microbiol 9:2234–2234. doi: 10.3389/fmicb.2018.02234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhary A, Kathuria S, Xu J, Sharma C, Sundar G, Singh PK, Gaur SN, Hagen F, Klaassen CH, Meis JF. 2012. Clonal expansion and emergence of environmental multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR(3)(4)/L98H mutations in the cyp51A gene in India. PLoS One 7:e52871. doi: 10.1371/journal.pone.0052871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagiwara D, Watanabe A, Kamei K, Goldman GH. 2016. Epidemiological and genomic landscape of azole resistance mechanisms in Aspergillus fungi. Front Microbiol 7:1382. doi: 10.3389/fmicb.2016.01382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buil JB, Snelders E, Denardi LB, Melchers WJG, Verweij PE. 2019. Trends in azole resistance in Aspergillus fumigatus, the Netherlands, 1994–2016. Emerg Infect Dis 25:176–178. doi: 10.3201/eid2501.171925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vermeulen E, Lagrou K, Verweij PE. 2013. Azole resistance in Aspergillus fumigatus: a growing public health concern. Curr Opin Infect Dis 26:493–500. doi: 10.1097/QCO.0000000000000005. [DOI] [PubMed] [Google Scholar]
- 27.Camps SM, Rijs AJ, Klaassen CH, Meis JF, O'Gorman CM, Dyer PS, Melchers WJ, Verweij PE. 2012. Molecular epidemiology of Aspergillus fumigatus isolates harboring the TR34/L98H azole resistance mechanism. J Clin Microbiol 50:2674–2680. doi: 10.1128/JCM.00335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lestrade PPA, Buil JB, van der Beek MT, Kuijper EJ, van Dijk K, Kampinga GA, Rijnders BJA, Vonk AG, de Greeff SC, Schoffelen AF, van Dissel J, Meis JF, Melchers WJG, Verweij PE. 2020. Paradoxal trends in azole-resistant Aspergillus fumigatus in a national multicenter surveillance program, the Netherlands, 2013–2018. Emerg Infect Dis 26:1447–1455. doi: 10.3201/eid2607.200088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhodes J, Abdolrasouli A, Dunne K, Sewell TR, Zhang Y, Ballard E, Brackin AP, van Rhijn N, Tsitsopoulou A, Posso RB, Chotirmall SH, McElvaney NG, Murphy PG, Talento AF, Renwick J, Dyer PS, Szekely A, Bromley MJ, Johnson EM, White PL, Warris A, Barton RC, Schelenz S, Rogers TR, Armstrong-James D, Fisher MC. 8 April 2021. Tracing patterns of evolution and acquisition of drug resistant Aspergillus fumigatus infection from the environment using population genomics. bioRxiv doi: 10.1101/2021.04.07.438821. [DOI]
- 30.Beer KD, Farnon EC, Jain S, Jamerson C, Lineberger S, Miller J, Berkow EL, Lockhart SR, Chiller T, Jackson BR. 2018. Multidrug-resistant Aspergillus fumigatus carrying mutations linked to environmental fungicide exposure — three states, 2010–2017. MMWR Morb Mortal Wkly Rep 67:1064–1067. doi: 10.15585/mmwr.mm6738a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berkow EL, Nunnally NS, Bandea A, Kuykendall R, Beer K, Lockhart SR. 2018. Detection of TR34/L98H CYP51A mutation through passive surveillance for azole-resistant Aspergillus fumigatus in the United States from 2015 to 2017. Antimicrob Agents Chemother 62:e02240-17. doi: 10.1128/AAC.02240-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vazquez JA, Manavathu EK. 2016. Molecular characterization of a voriconazole-resistant, posaconazole-susceptible Aspergillus fumigatus isolate in a lung transplant recipient in the United States. Antimicrob Agents Chemother 60:1129–1133. doi: 10.1128/AAC.01130-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiederhold NP, Gil VG, Gutierrez F, Lindner JR, Albataineh MT, McCarthy DI, Sanders C, Fan H, Fothergill AW, Sutton DA. 2016. First detection of TR34 L98H and TR46 Y121F T289A Cyp51 mutations in Aspergillus fumigatus isolates in the United States. J Clin Microbiol 54:168–171. doi: 10.1128/JCM.02478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang SE, Sumabat LG, Melie T, Mangum B, Momany M, Brewer MT. 25 May 2020. Evidence for the agricultural origin of antimicrobial resistance in a fungal pathogen of humans. bioRxiv doi: 10.1101/2020.05.24.113787. [DOI] [PMC free article] [PubMed]
- 35.Pham CD, Reiss E, Hagen F, Meis JF, Lockhart SR. 2014. Passive surveillance for azole-resistant Aspergillus fumigatus, United States. Emerg Infect Dis 20:1498–2013. doi: 10.3201/eid2009.140142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander DH, Lange K. 2011. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12:246. doi: 10.1186/1471-2105-12-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Gorman CM, Fuller H, Dyer PS. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- 38.Kwon-Chung KJ, Sugui JA. 2009. Sexual reproduction in Aspergillus species of medical or economical importance: why so fastidious? Trends Microbiol 17:481–487. doi: 10.1016/j.tim.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dyer PS, Paoletti M. 2005. Reproduction in Aspergillus fumigatus: sexuality in a supposedly asexual species? Med Mycol 43 Suppl 1:S7–S14. doi: 10.1080/13693780400029015. [DOI] [PubMed] [Google Scholar]
- 40.Paoletti M, Rydholm C, Schwier EU, Anderson MJ, Szakacs G, Lutzoni F, Debeaupuis JP, Latge JP, Denning DW, Dyer PS. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Biol 15:1242–1248. doi: 10.1016/j.cub.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 41.Oltean HN, Etienne KA, Roe CC, Gade L, McCotter OZ, Engelthaler DM, Litvintseva AP. 2019. Utility of whole-genome sequencing to ascertain locally acquired cases of coccidioidomycosis, Washington, USA. Emerg Infect Dis 25:501–506. doi: 10.3201/eid2503.181155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, García JL, García MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jiménez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Lafton A, Latgé J-P, Li W, Lord A, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 45.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Argimon S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, Feil EJ, Holden MTG, Yeats CA, Grundmann H, Spratt BG, Aanensen DM. 2016. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom 2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom. [Google Scholar]
- 50.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 54.Jain C, Rodriguez RL, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Isolates. Aspergillus fumigatus isolates in this study. Download Table S1, PDF file, 0.1 MB (143.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genetic relationship in the cyp51A gene among A. fumigatus isolates. Analysis of the cyp51A gene using neighbor-joining algorithm identified multiple types of mutations among U.S. and non-U.S. Aspergillus fumigatus isolates: the environmentally derived mutation, TR34/L98H is seen among U.S. and non-U.S. azole-resistant A. fumigatus isolates. A small set of clinical and environmental isolates from the United States includes this environmental mutation coupled with two other substitutions, S297T and F495I. Substitutions such as A9T, G254V, I242V, D262Y, and N248K were commonly found among azole-resistant and azole-susceptible clinical isolates from the United States. Isolates with the wild-type allele were commonly found among the U.S. collection. Download FIG S1, PDF file, 0.4 MB (449.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Population structure of A. fumigatus isolates. ADMIXTURE (K = 3) analysis demonstrates that the majority of isolates from the United States contained a mixture of alleles from two subpopulations. A. fumigatus resistant isolates with the TR34/L98H resistance marker from different areas of the United States share alleles with A. fumigatus isolates from India, The Netherlands, and the United Kingdom. Download FIG S2, PDF file, 0.4 MB (424.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ADMIXTURE results (for K = 2 through 10). Results for K values of 2 to 4 indicate that the U.S. isolates with the TR34/L98H marker share alleles with A. fumigatus isolates from India, The Netherlands, and the United Kingdom. No evidence for K value of >3 was detected by PCA or phylogenetic analysis. Download FIG S3, PDF file, 0.3 MB (309.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Distribution of mating types among resistant and susceptible A. fumigatus isolates in the United States. A chi-square statistical analysis with 179 isolates from the United States suggests mating type does not influence antifungal susceptibility. Download FIG S4, PDF file, 0.4 MB (392.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
All raw Illumina reads in this study have been submitted to NCBI under BioProject accession number PRJNA632561.