Abstract
Background
Here we report a case of para-Bombay phenotype due to a novel mutation FUT1 c.361G>A p.(Ala121Thr) and a nonfunctional allele FUT1*01N.13(c.881_882delTT) which showed a discrepancy in the routine ABO blood group typing.
Materials and Methods
The ABO phenotype and the Lewis blood group were typed with serological methods. The ABH antigens in saliva were determined by a hemagglutination inhibition test. The CDS region of ABO, FUT1and FUT2 were amplified with polymerase chain reaction and then directly sequenced. The novel mutation was confirmed by cloning and sequencing. Three-dimensional (3-D) structural analysis of the mutant and wild-type Fut1 were performed by the Chimera software.
Results
A, B and H antigens were not detected on the surface of red blood cells (RBCs) by the serological technique, and the B and H blood group substances were detected in the saliva, while the Lewis phenotype was Le(a–b+). Sequencing and cloning analysis showed the presence of a novel FUT1 mutation c.361G>A and a nonfunctional allele FUT1*01N.13(c.881_882delTT). The ABO genotype was ABO*B.01/ABO*O.01.01. The in silico analysis showed that the mutation p.(Ala121Thr) of FUT1did not change the 3-D structure of the whole enzyme but caused a certain amplitude of turnover in the loop region where Ala121 was located.
Conclusions
A novel FUT1 allele (FUT1*c.361G>A) was identified in a Chinese individual with para-Bombay B phenotype. The FUT1c.361G>A mutation may significantly downregulate the expression of H antigens on RBCs by damaging the enzyme conformation.
Keywords: FUT1gene, Para-Bombay phenotype, Mutation
Introduction
The Bombay and para-Bombay phenotypes are characterized by the deficiency of ABH antigens on the red blood cell (RBC) which are believed to result from a defect in the FUT1 (H)gene that encodes the 2α-fucosyltransferase responsible for the synthesis of the H antigen on RBCs [1]. The highly homologous FUT2 (secretor)genenormally encodes the very similar 2α-fucosyltransferase T2 (α2FucT2) that is closely linked to FUT1 and synthesizes H antigen in secretions [2]. In the Bombay phenotype, the FUT2gene is inactive together with FUT1. Since H antigen is the precursor of the A and B antigens, neither antigen can be synthesized. In the para-Bombay phenotype, FUT1 is inactive or mutated with an active FUT2 gene so that a trace of H/A/B antigen is present on RBCs, which is either adsorbed from the plasma or synthesized by mutated Fut1 enzyme. That is, the Bombay phenotypes are H-deficient nonsecretors whereas the para-Bombay phenotypes are H-deficient secretors. In both phenotypes, ABH antigen is very weakly expressed and is often only detected by adsorption-elution tests with the corresponding blood group antisera. The anti-H produced by individuals with the Bombay phenotype is usually strong whereas that from para-Bombay individuals is much weaker [3].
Till now, over 50 variants of the FUT1 gene are described, and most variants are identified in either the Bombay or the para-Bombay phenotype [4]. The first reported FUT1 mutation c.725T>G, is associated with the deletion of the FUT2gene and has been found only in persons of subcontinental Indian descent [5]. The most common FUT1 gene mutations for the para-Bombay phenotype in Chinese contain dinucleotide deletions and point mutation − c.547delAG, c.880delTT and c.658C>T [6, 7]. In East Asia, the synonymous single nucleotide variant c.357C>T in the FUT2gene is commonly found and does not influence the enzyme activity [8]. The incidence of FUT1mutations giving rise to Bombay and para-Bombay phenotypes varies in different populations from an estimated 1:1,000,000 to 1:1,000 [9]. Most para-Bombay phenotypes have been found in Eastern Asia [10, 11, 12, 13, 14, 15, 16]. Some estimated frequencies are one in 5,000 Thais [17], one in 8,000 Taiwanese [18] and one in 15,620 Hong Kong Chinese [12]. The para-Bombay phenotype has a high frequency of approximately 1:50 in the Lahu ethnic minority of China [8]. All were either homozygous or heterozygous for one or both of two FUT1 alleles: 328G>A, Ala110Thr and 658C>T, Arg220Cys. Here, we describe a novel mutation FUT1 c.361G>A, p.(Ala121Thr) in a Chinese individual with para-Bombay phenotype.
Materials and Methods
A 31-year-old male patient was admitted to the reproductive medicine department of Woman and Children's Hospital, with primary infertility. Venous blood specimens were collected into dried tubes containing 3.2% trisodium citrate. ABO blood group typing was firstly determined by an IH-1000 system (Bio-Rad, Roanne, France), then confirmed by manual tube methods. Absorption and elution test and Lewis blood group assessment were performed with standard serological techniques [19]. The following reagents were used in tube methods: monoclonal anti-Lea, anti-Leb (Sanquin, Amsterdam, the Netherlands), monoclonal anti-A, anti-B, anti-H and an ABO RBC kit (Shanghai Hemo-Pharmaceutical & Biological Co. Ltd., Shanghai, China). Genomic DNA was extracted from a peripheral blood sample using a blood DNA kit (Tiangen, Beijing, China). The presence or absence of ABH antigens in saliva was determined by a hemagglutination inhibition test [19]. The primer design and polymerase chain reaction (PCR) amplification of the CDS region of ABO, FUT1and FUT2 were performed as described in the literature [20, 21], and the haplotypes of FUT1were identified by TOPO TA cloning sequencing (Invitrogen, CA, USA). The novel mutation site was also amplified and then sequenced from 120 random, apparently healthy Chinese donors. The alleles were named according to the nomenclature used in the International Society of Blood Transfusion, and the nucleotide and amino acid mutations were named according to the Human Genome Variation Society. The unliganded X-ray structure of fucosyltransferase (PDB code, 2HLH) was used as the template to construct the initial molecular model. The 3-dimensional (3-D) model of wild-type Fut1 was built through homology modeling by the Phyre2 [8]. The in silico mutation was modeled by the Chimera software (version 1.11.2; University of California, San Francisco, CA, USA) [22]. The structural figures were also generated in Chimera or prepared by Pymol-2.3.2 (The PyMOL Molecular Graphics System, Version 2.0, Schrödinger LLC) [23]. The effect of the mutation on the enzyme function was predicted by PolyPhen2 (Polymorphism Phenotyping Version 2.2.2) [24] and PROVEAN (Protein Variation Effect Analyzer, Version 1.1.5, The J. Craig Venter Institute, Rockville, MD, USA) [25]. Based on pairs of false rate (FPR) thresholds by the PolyPhen2, a mutation is appraised qualitatively, as benign (FPR >0.902), possibly damaging (FPR <0.902) or probably damaging (FPR <0.493). To the PROVEAN, if the score is equal to or below a predefined threshold −2.5, the protein variant is predicted to have a “deleterious” effect, otherwise a “neutral” effect (threshold >–2.5). The number of sequences and clusters used for prediction were 212 and 30.
Results
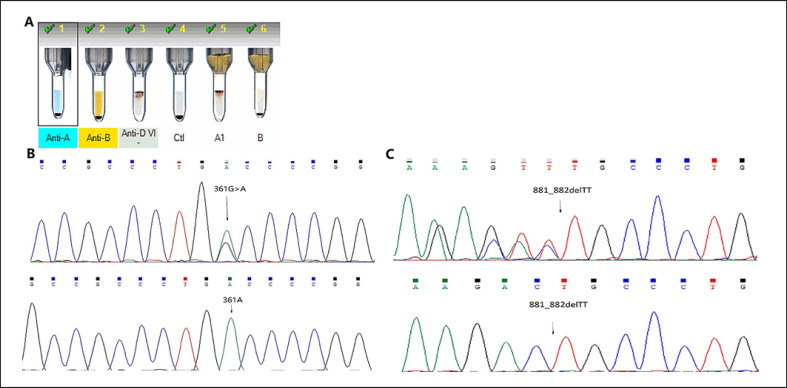
The serological results of ABO phenotype showed a discrepancy between the forward and reverse typing (Table 1, Fig. 1a). A, B and H antigens were not detected on the surface of RBCs both by routine methods and the absorption and elution test, but the B and H blood group substances were detected in the saliva. The Lewis phenotype was Le(a–b+) (Table 1). The serological characteristics showed that RJ204 had a rare H-deficient phenotype and was identified as the para-Bombay B phenotype.
Table 1.
Serological analysis of ABH and Lewis antigens1
| No. | Forward grouping | Reverse grouping | Antigens in saliva | Lewis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| anti-A | anti-B | anti-H | Ac | Bc | Oc | A | B | H | ||
| RJ204 | − | −2 | −2 | 4+ | − | 1+ | − | + | + | Le(a–b+) |
−, negative; 1+–4+, positive according to agglutination strength; Ac, A1 red blood cells; Bc, B red blood cells; Oc, O red blood cells.
The results shown in the table were obtained by tube methods.
B and H antigen was not detected by adsorption and elution test either.
Fig. 1.
The results of the serological analysis, DNA sequencing and TA cloning. A The ABO phenotype was determined by an IH-1000 system. B A heterozygous mutation (A/G) was detected at c.361 (upper panel), and the sequence of the isolated variant FUT1 allele after cloning was shown (lower panel). C TT deletion at position c.881_882 (upper panel) and haplotype of the FUT1c.881_882 delTT (lower panel).
Direct DNA sequencing and TOPO TA cloning of the whole FUT1 coding region revealed one novel mutation FUT1c.361G>A (GenBank No. MN938362) (Fig. 1b) and a reported nonfunctional allele FUT1*01N.13(c.881_882delTT) (Fig. 1c). A homozygous single nucleotide variant c.357C>T, p.(Asn119=) was identified in the FUT2 gene. The novel FUT1 mutation c.361G>A was not detected in 120 random Chinese individuals with the common ABO phenotype, and it was not in the SNP database (https://www.ncbi.nlm.nih.gov/snp/). The ABO genotype was ABO*B.01/ABO*O.01.01.
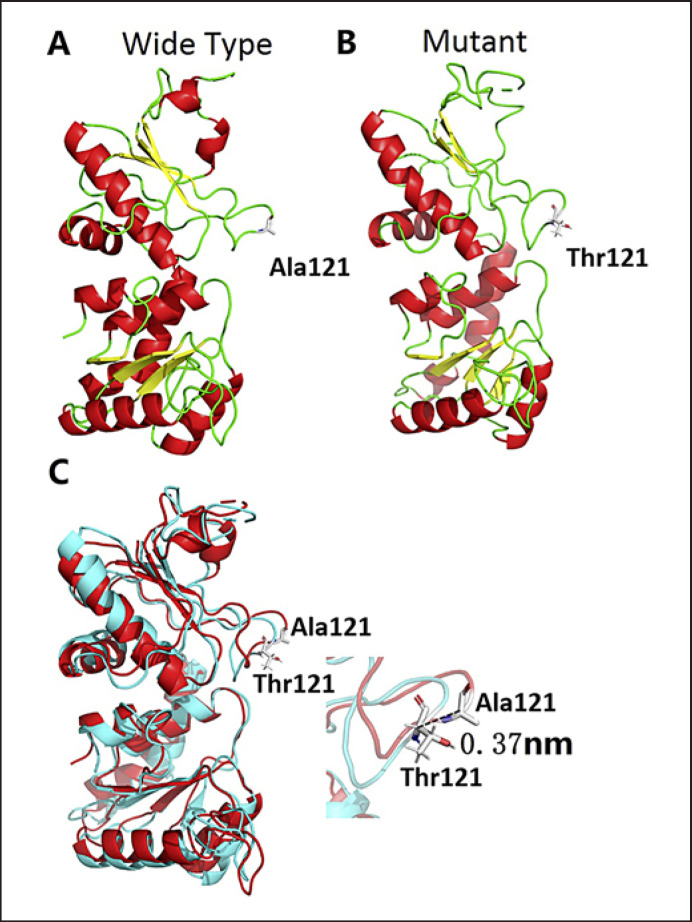
The complete structure of the unliganded wild-type and mutant Fut1 enzymes were mainly built from the crystal structure 2HLH (Fig. 2a, b). The overall structure of the mutant was predicted to be similar to that of the wild-type Fut1 enzyme. The results of the structural comparison of wild-type and mutant Fut1 enzymes showed that there was a displacement of amino acids caused by mutation. This mutation causes a dramatical turnover of the loop Ala115-Phe124, where Ala121 was located. The space displacement between Ala121 and Thr121 was 0.37 nm (based on the Cα of the Ala121 and Thr121, Fig. 2c).
Fig. 2.
The different conformations in wild-type and mutant Fut1 enzymes. The amino acids were colored by elements in a ball and sticks method. O, N, C and H atoms represented by red, blue, long silver and short silver sticks. A The overall structure of the wild-type Fut1 enzyme and the side of Ala121 is shown. B The overall structure of the mutant Fut1 enzyme and the side of Thr121 is shown. C The structural comparison of wild-type and mutant Fut1 enzymes. The wild-type enzyme is colored in red and the mutant enzyme in blue-green. The black dotted line indicates the distance between the Cα of the Ala121 and Thr121.
The effect of the mutation on the enzyme function was predicted by the algorithms of PolyPhen2, and a score of 0.849 was obtained (threshold <0.902) based on the HumVar indicating the mutation was possibly damaging. The PROVEAN score of mutation was −2.569 (threshold −2.5) indicating the mutation was deleterious.
Discussion
In this report, we identified a novel FUT1 mutation in para-Bombay B phenotype, which may impair the function and stability of Fut1 and significantly downregulate the expression of H antigens on the RBCs with a reported nonfunctional allele FUT1*01N.13.
According to the serological results, no A, B and H antigens on the surface of RBCs were found even by the absorption and elution test, while the B and H antigens could be detected in the saliva. These results suggested that RJ204 was an ABH secretor and belonged to the para-Bombay B phenotype. The undetectable B and H antigens on RBCs may be because of the limited sensitivity of the absorption and elution test in our laboratory. The Lewis phenotype was Le(a–b+), and the ABO genotype was ABO*B.01/ABO*O.01.01, consistent with the individual's secretor status and ABO reversing grouping, respectively.
A novel mutation FUT1 c.361G>A was detected, and the corresponding FUT1 allele was confirmed by direct DNA sequencing and sequencing after cloning. This mutation is predicted to cause the substitution p.(Ala121Thr). Alanine is a hydrophobic and nonpolar molecule, while threonine is a hydrophilic and polar molecule. The mutation introducing threonine may disturb the structure of the polypeptide by causing a difference in bulk and physical properties between the normal and the mutated amino acid. In the 3-D structural modeling studies, there is a 0.37-nm space displacement between Ala121 and Thr121 and a certain amplitude of turnover of loop Ala115-Phe124. These changes can influence the conformation of the enzyme. The score of the algorithms of PolyPhen2 was 0.849 which was below the threshold (0.902) showing the effect of the mutation on the enzyme function was harmful referring to HumVar which is the preferred model for diagnostics of Mendelian diseases to distinguish mutations with drastic effects from all the remaining human variations including abundant mildly deleterious alleles. The PROVEAN score was −2.569 (threshold −2.5), which is below the default threshold, and the variant is considered “deleterious.” Those results suggested the local conformation and function of the enzyme might be damaged by the mutation.
In conclusion, a novel FUT1 allele (FUT1*c.361G>A) was identified in a Chinese individual with para-Bombay B phenotype. The FUT1c.361G>A mutation, along with the other reported silenced FUT1 mutation, causes the rare para-Bombay phenotype by changing the conformation and then affecting the efficiency of the encoded 2α-fucosyltransferase through the amino acid substitution Ala121Thr.
Statement of Ethics
The study was approved by the ethics committee of Ruijin Hospital and Woman and Children's Hospital. Written informed consent was obtained from the patient. The patient identity is not disclosed in the paper.
Conflict of Interest Statement
The authors declare no conflict of interest.
Funding Sources
This work was supported by the Shanghai Municipal Natural Science Foundation (17ZR1417000), WeiGao Science Foundation of the Chinese Society of Blood Transfusion, WGSF-CSBT(CSBT-WG-2019-01), Young Scholars of Fujian Provincial Department of Health Office (2010-2-110) and the National Natural Science Foundation of China (82000183).
Author Contributions
L.H. and Y.S. contributed equally to this work.
Acknowledgments
The authors thank Dr. Fang Li from Shanghai Jiao Tong University School of Life Sciences and Biotechnology for her technical assistance.
References
- 1.Oriol R, Candelier JJ, Mollicone R. Molecular genetics of H. Vox Sang. 2000;78(Suppl 2):105–8. [PubMed] [Google Scholar]
- 2.Zhang A, Chi Q, Ren B. Genomic analysis of para-Bombay individuals in south-eastern China: the possibility of linkage and disequilibrium between FUT1 and FUT2. Blood Transfus. 2015 Jul;13((3)):472–7. doi: 10.2450/2015.0185-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storry JR, Johannesson JS, Poole J, Strindberg J, Rodrigues MJ, Yahalom V, et al. Identification of six new alleles at the FUT1 and FUT2 loci in ethnically diverse individuals with Bombay and Para-Bombay phenotypes. Transfusion. 2006 Dec;46((12)):2149–55. doi: 10.1111/j.1537-2995.2006.01045.x. [DOI] [PubMed] [Google Scholar]
- 4.Nykamp K, Anderson M, Powers M, Garcia J, Herrera B, Ho YY, et al. Invitae Clinical Genomics Group Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017 Oct;19((10)):1105–17. doi: 10.1038/gim.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koda Y, Soejima M, Johnson PH, Smart E, Kimura H. Missense mutation of FUT1 and deletion of FUT2 are responsible for Indian Bombay phenotype of ABO blood group system. Biochem Biophys Res Commun. 1997 Sep;238((1)):21–5. doi: 10.1006/bbrc.1997.7232. [DOI] [PubMed] [Google Scholar]
- 6.Yu LC, Yang YH, Broadberry RE, Chen YH, Lin M. Heterogeneity of the human H blood group alpha(1,2)fucosyltransferase gene among para-Bombay individuals. Vox Sang. 1997;72((1)):36–40. doi: 10.1046/j.1423-0410.1997.00036.x. [DOI] [PubMed] [Google Scholar]
- 7.Cai XH, Jin S, Liu X, Fan LF, Lu Q, Xiang D. Molecular genetic analysis for the para-Bombay blood group revealing two novel alleles in the FUT1 gene. Blood Transfus. 2011 Oct;9((4)):466–8. doi: 10.2450/2011.0115-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai XH, Fang CJ, Jin S, Liu X, Fan LF, Yang XP, et al. H blood-group deficiency has a high frequency in Lahu Chinese. Transfus Med. 2011 Jun;21((3)):209–10. doi: 10.1111/j.1365-3148.2010.01066.x. [DOI] [PubMed] [Google Scholar]
- 9.Daniels G. Human blood groups. 3rd ed. Oxford: Wiley Blackwell; 2013. [Google Scholar]
- 10.Reviron J, Jacquet A, Salmon C. [An example of chromosome cis A1B. Immunologic and genetic study of the induced phenotype] Nouv Rev Fr Hematol. 1968 May-Jun;8((3)):323–38. [PubMed] [Google Scholar]
- 11.Pacuszka T, Kościelak J, Seyfried H, Walewska I. Biochemical, serological and family studies in individuals with Cis AB phenotypes. Vox Sang. 1975;29((4)):292–300. doi: 10.1111/j.1423-0410.1975.tb00510.x. [DOI] [PubMed] [Google Scholar]
- 12.Takizawa H, Takizawa H. [Blood group substances decomposing enzymes. 31. Immunological properties of Bombay phenotype] Igaku To Seibutsugaku. 1970 Nov;81((5)):227–30. [PubMed] [Google Scholar]
- 13.Beranovц║ G, Prodanov P, Hrubisko M, et al. A new variant in the ABO blood group system: Bh. Vox Sang, 1969;16((6)):449–456. doi: 10.1111/j.1423-0410.1969.tb04771.x. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi HO, Hazama F. Another Japanese A2B3 blood-group family with the propositus having O-group father. Proe Ipn Acad. 1966;42:517–20. [Google Scholar]
- 15.Kogure T. A family with unusual inheritance of ABO blood groups. Jinrui Idengaku Zasshi. 1971 Oct;16((2)):69–78. [PubMed] [Google Scholar]
- 16.Salmon C, Rouger P, Rodier L, Liberge G, Juszczak G, Cartron JP, et al. Different H deficient phenotypes present in one kindred. Rev Fr Transfus Immunohematol. 1980;23((3)):251–8. doi: 10.1016/s0338-4535(80)80129-2. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko M, Nishihara S, Shinya N, Kudo T, Iwasaki H, Seno T, et al. Wide variety of point mutations in the H gene of Bombay and para-Bombay individuals that inactivate H enzyme. Blood. 1997 Jul;90((2)):839–49. [PubMed] [Google Scholar]
- 18.Prodanov P, Drazhev G. A A1 xh variant of Ah in a Bulgarian family. Vox Sang. 1978;34((3)):162–3. [PubMed] [Google Scholar]
- 19.Mark K, Fung AFE, Steven L., Spitalnik, Connie M., Westhoff . 19th ed. Bethesda (MD): American Association of Blood Banks; 2017. AABB Technical Manual. pp. pp. 777–778. [Google Scholar]
- 20.Yip SP, Chee KY, Chan PY, Chow EY, Wong HF. Molecular genetic analysis of para-Bombay phenotypes in Chinese: a novel non-functional FUT1 allele is identified. Vox Sang. 2002 Oct;83((3)):258–62. doi: 10.1046/j.1423-0410.2002.00184.x. [DOI] [PubMed] [Google Scholar]
- 21.Cai XH, Jin S, Liu X, Shen W, Lu Q, Wang JL, et al. Molecular genetic analysis for the B subgroup revealing two novel alleles in the ABO gene. Transfusion. 2008 Nov;48((11)):2442–7. doi: 10.1111/j.1537-2995.2008.01878.x. [DOI] [PubMed] [Google Scholar]
- 22.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25((13)):1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 23.William Humphrey AD, Schulten Klaus. VMD visual molecular dynamics. J Mol Graph. 1996;14((1)):33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 24.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010 Apr;7((4)):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7((10)):e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]