Abstract
SUMOylation of proteins regulates cell behaviors and is reversibly removed by small ubiquitin-like modifier (SUMO)-specific proteases (SENPs). The SENP family member SENP3 is involved in SUMO2/3 deconjugation and has been reported to sense cell stress and accumulate in several human cancer cells and macrophages. We previously reported that Senp3-knockout heterozygous mice showed smaller liver, but the pertinent mechanisms of SENP3 and SUMOylated substrates remain unclear. Thus, in this study, we investigated the interacting proteins with SENP3 and the alteration in hepatocytes treated with the xenobiotic diethylnitrosamine (DEN), which is specifically transformed in the liver and induces DNA double-strand breaks. Our data revealed that a certain amount of SENP3 was present in normal, untreated hepatocytes; however, DEN treatment promoted rapid SENP3 accumulation. SENP3 was mainly localized in the nuclei, and its level was significantly increased in the cytoplasm after 2 h of DEN treatment. The application of the recent proximity-dependent biotinylation (BioID) method led to the identification of 310 SENP3-interacting proteins that were involved in not only gene transcription but also RNA splicing, protein folding, and metabolism. Furthermore, after DEN exposure for a short duration, ribosomal proteins as well as proteins associated with mitochondrial ATP synthesis, membrane transport, and bile acid synthesis, rather than DNA repair proteins, were identified. This study provides insights into the diverse regulatory roles of SENP3, and the BioID method seems to be efficient for identifying physiologically relevant insoluble proteins.
Keywords: SUMO specific peptidase 3, SUMOylation, BioID, hepatocytes, diethylnitrosamine
Introduction
Small ubiquitin-like modifier (SUMO)-specific peptidase 3 (SENP3) regulates cellular activities by removing the conjugation of SUMO proteins from substrate proteins [1]. In recent years, SUMOylated proteins have been demonstrated to be involved in almost all important cell behaviors, and hundreds of SUMO2-modified proteins have been identified in different cells using proteomics approaches [2–5]. We previously reported that SENP3 plays regulatory roles in tumor cell proliferation and migration and in macrophage inflammatory response by targeting several SUMO2/3-modified proteins [6–9]. An important feature of SENP3 is its ability to rapidly accumulate and expand its substrates, from the nucleoli to nucleoplasm and even the cytoplasm, in response to a wide variety of stresses [10]. According to an earlier study, embryonic death at 8.5 days was observed upon Senp3 knockout; furthermore, Senp3 knockout heterozygous mice showed smaller liver [11], suggesting an important role of SENP3 in the liver. Moreover, a recent proteomics study of SUMO modification sites identified that among all organs, SUMO target proteins were the most diverse in the liver [12]. The liver is particularly susceptible to damage from biotransformation of most xenobiotics [13]; however, the pattern of SUMO2/3 modification and global regulation by SENP3 remains unclear. Thus, in this study, we investigated SENP3-interacting proteins and potential SUMO2/3 modification using a proteomics approach and further explored alterations in the SENP3-interacting protein profile in hepatocytes treated with the xenobiotic diethylnitrosamine (DEN).
Co-immunoprecipitation (co-IP) is commonly used to identify protein interaction partners. In recent years, the BirA–biotinylation–affinity system has been reported to overcome the limitations associated with co-IP. BirA, a biotin ligase from Escherichia coli, is fused with the protein of interest, and it mediates biotinylation of proximal interacting proteins in the presence of biotin; an affinity system eventually precipitates these modified proteins. This type of physical interaction and subsequent irreversible biotinylation has been reported to markedly increase the detection sensitivity and efficiency in vivo [14]. In addition, this system was found to be effective when applied to insoluble and membrane-associated proteins, emerging as a unique method to screen physiologically relevant protein–protein interactions (PPIs) [15]. As the interaction of SENP3 with SUMO2/3-modified target proteins is very dynamic and difficult to capture, we herein used the BirA–biotinylation–affinity method to precipitate SENP3-interacting proteins so as to explore the role of SENP3 in hepatocytes and the global SUMO2/3 modification dynamics.
Materials and Methods
Antibodies and reagents
Antibodies against SENP3 (D20A10), biotin (7075), and p62 (5114S) were obtained from Cell Signaling Technology (Boston, USA) Antibodies against FLAG M2 (F1804) and β-actin (A5441) were from Sigma (San Francisco, USA). Antibodies against nucleophosmin (NPM1, 60096-1-Ig) and ATP5A1 (60371) were from Proteintech Group (Chicago, USA). Antibody against Transketolase (sc-390179) was from Santa Cruz Biotechnology (Santa Cruz, USA). The antibody against HA was obtained from Affinity (Shanghai, China). Biotin, anti-FLAG M2 affinity gel (A2220), and DEN were purchased from Sigma (St Louis, USA), and NeutrAvidin™ agarose (29200) was obtained from Thermo Fisher Scientific (Pittsburgh, USA). Alexa Fluor 488-labeled goat anti-rabbit antibodies were from Molecular Probes (Eugene, USA), and Alexa Fluor 555-labeled goat anti-mouse antibodies were obtained from Invitrogen (Carlsbad, USA).
Plasmids
pCDH–BirA was kindly provided by Dr. Zhaoyuan Hou of the Department of Biochemistry and Molecular Cell Biology, Shanghai Jiao Tong University School of Medicine (Shanghai, China). Beneficial mutation of this plasmid had been performed from arginine 118 to glycine (R118G) according to the work reported by Roux in the Journal of Cell Biology in 2012. Here, it was simplified as pCDH–BirA. Based on pCDH–BirA, we generated pCDH–SENP3–BirA via subcloning. The primer sequences were as follows: forward, 5′-TGCTCTAGAATGAAAGAGACTATACAAGGGAC-3′ and reverse, 5′-CCGCTCGAGCACAGTGAGTTTGCAGTGACAC-3′. Based on FLAG–TKT that was kindly provided by Dr. Xuemei Tong of the Department of Biochemistry and Molecular Cell Biology, Shanghai Jiao Tong University School of Medicine (Shanghai, China), we generated GST–TKT plasmid by subcloning. The primer sequences were as follows: forward, 5′-CCGCAATTCGAGAGCTACCACAAGCCTG-3′; reverse, 5′-CCGCTCGAGCTAGGCCTTGGTGATGAGG-3′. The plasmids of Flag–SENP3 were generated as previously described [16].
Cell culture and transfection
HEK293T and human hepatocytes (LO2 cell line) were obtained from the American Type Culture Collection (Manassas, USA) and Cell Bank of the Chinese Academy of Sciences (Shanghai, China), respectively. The cell lines were maintained in Dulbecco’s modified Eagle’s medium and RPMI1640 (GIBCO, Carlsbad, USA), respectively, at 37°C in a humidified atmosphere of 5% CO2. All media were supplemented with 10% fetal bovine serum (GIBCO). For HEK293T cells, transfection with plasmid DNA and small interfering RNAs was performed using Lipofectamine 2000 (Invitrogen), whereas for LO2 cells, transfection was performed using FuGENE (Promega, Madison, USA). For the establishment of stable SENP3–BirA cell line, construction of the lentiviral plasmid pCDH–SENP3–BirA and virus packaging were conducted by GeneChem (Shanghai, China), and subsequently, LO2 cells were infected with the viral supernatant as per manufacturer’s instructions.
Western blot analysis
Western blot analysis was performed as previously described [11]. Briefly, proteins were separated by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE) and then transferred onto a nitrocellulose membrane. Specific proteins were detected using antibodies against SENP3 (1:1000), β-actin (1:2000), FLAG (1:1000), and voltage-dependent anion channel (VDAC, 1:1000) (overnight at 4°C). Secondary antibodies (1:5000) were allowed to react for 1 h at 37°C, and protein bands were visualized using an ECL system (EMD Millipore, Billerica, USA). Images scanned with ImageQuant™ LAS 4000 mini (GE Healthcare) were exported as TIFF in RGB mode.
Immunofluorescence assay
Immunofluorescence assay was performed as previously described [11]. Briefly, after being fixed with cold 4% paraformaldehyde, the cells or frozen tissue sections were permeabilized with 0.2% Triton for 5 min at room temperature, blocked with 10% goat serum for 1 h at room temperature, and incubated with primary antibodies overnight at 4°C. Immunofluorescence staining was performed, and images were obtained with a ZEISS 710 confocal microscope (Jena, Germany). The cells were counterstained with DAPI. To visualize the cytoplasmic staining of SENP3, which was predominantly in the nucleus, a mild permeabilization approach was used with 50 μg/ml digitonin [11].
Proximity-dependent biotinylation (BioID) pull-down assay
Prior to harvesting, transfected cells were incubated with 50 μM biotin for 24 h. The cells were then lysed in a lysis buffer [50 mM Tris-HCl, pH 7.4, 500 mM NaCl, 0.2% SDS (w/v), and 1× protease inhibitor]. Triton X-100 (2% final concentration) was then added and mixed by trituration. The lysates were sonicated (two sessions, 30 pulses, 4 times) and centrifuged at 16,500 g for 10 min at 4°C. Subsequently, streptavidin beads were added to the supernatant, followed by incubation on a rotator overnight at 4°C. The beads were washed successively with wash buffer 1 [2% (w/v) SDS], wash buffer 2 [0.1% (w/v) deoxycholic acid, 1% (w/v) Triton X-100, 1 mM ethylenediaminetetraacetic acid (EDTA), 500 mM NaCl, and 50 mM HEPES, pH 7.5], and wash buffer 3 [0.5% (w/v) deoxycholic acid, 0.5% (w/v) NP-40, 1 mM EDTA, 250 mM LiCl, and 10 mM Tris-HCl, pH 7.4] for 8 min each, and then mixed with a loading buffer. The supernatant was collected for western blot analysis.
FLAG IP assay
Briefly, the transfected cells were lysed in RIPA (50 mM Tris, 150 mM NaCl, 1% Triton, 0.5% sodium deoxycholate, and 0.1% SDS, pH 8.0) for 30 min at 4°C and centrifuged at 12,000 g at 4°C for 30 min. Anti-FLAG M2 affinity gel was added to the cell lysate, followed by incubation overnight at 4°C. After the last wash, FLAG-tagged proteins were eluted, and the supernatant was collected for western blot analysis [11].
Mass spectrometry
Once the BioID pull-down assay had been performed, the precipitations were subject to SDS–PAGE and silver staining of the gel according to the kit instruction (Sangon Biotech, Shanghai, China). After verification, washed beads were sent to Shanghai Institute of Materia Medica, Chinese Academy of Sciences (Shanghai, China), for analysis. The samples were digested with trypsin, and peptides were then re-dissolved in 15 μl 0.1% formic acid for liquid chromatography tandem mass spectrometry (MS) analysis (Orbitrap Fusion; Thermo Scientific, Waltham, USA). The raw files were analyzed using MaxQuant v1.6.0.1.
Co-immunoprecipitation
Routine co-immunoprecipitation (co-IP) was performed using a previously reported method [11]. Briefly, the cells were lysed in RIPA buffer (Thermo Fisher Scientific) for 30 min at 4°C and then centrifuged at 13,800 g at 4°C for 30 min. Cell lysates were incubated with specific antibodies overnight at 4°C, followed by incubation with protein-A/G agarose beads (Pierce, Rockford, USA) for 4 h at 4°C. The beads were then washed six times, mixed with a loading buffer, and finally subject to western blot analysis.
Glutathione S-transferase pull-down assay
Transfected cells with Flag-SENP3 were lysed in a lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100). Anti-Flag M2 affinity gel was added to the cell lysates and incubated overnight at 4°C. In the meantime, GST and GST–TKT fusion proteins were expressed in BL21 bacteria by induction with 0.5 mM IPTG in LB at 16°C overnight and lysed with lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1 mg/ml lysozyme, and protease inhibitor). The bacteria lysis freeze/thaw cycle was repeated 10 times and then incubated with anti-Flag M2 affinity gel overnight at 4°C. For the binding assay, the anti-Flag M2 affinity gel was washed five times with IP buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100). After the final wash, the Flag-tagged protein complex was eluted, and the supernatant was collected for western blot analysis.
FRET imaging
FRET was performed as previously described [7]. Briefly, LO2 cells were loaded on coverslips, followed by treatment with 15 mM DEN for 2 h. Then, cells were fixed in 4% paraformaldehyde for 10 min before permeabilization with 0.01% digitonin phosphate. Cells were incubated with primary antibodies against SENP3, TKT, and ATP5F1A in 3% bovine serum albumin at 4°C overnight. The monolayers were then washed with phosphate buffered saline (PBS) and incubated with appropriate secondary antibodies in PBS at 37°C for 2 h. Cells were counterstained with DAPI (Beyotime, Shanghai, China). FRET was performed on a laser scanning confocal microscope (Leica TCS SP8 STED; Leica, Nussloch, Germany) with a Plan Apochromat 63.0 × oil immersion. Images were scanned in 1024×1024 format, 1.58 μs pixel dwell, 12-bit depth, 4× line average and 3.0-μm pinhole. Acceptor photobleaching method was performed, in which the quenching of fluorescence emission of acceptor in the presence of donor could lead to the enhancement of the fluorescence emission of donor when they are interacted. Individual donor and acceptor images were analyzed. Five regions of interest were selected in the cytoplasm to be bleached. FRET efficiency was calculated using the software LAS-X (Leica). Ten cells were chosen to be repeated for FRET. Images for pre-bleach and post-bleach were acquired. Donor image and acceptor image were shown. FRET images were shown with pseudo-colored heat map according to the average FRET efficiency.
Bioinformatics
Proteomics data were analyzed by PTM BioLabs, Inc. (Hangzhou, China). WoLF PSORT was used to analyze the subcellular localization and classification of proteins. The interaction prediction database STRING v.11.0 was used to construct PPI networks and elucidate related biological processes. A medium confidence score of 0.7 was used to identify more potential interactions. PPI networks were reconstructed with Cytoscape v3.6.1.
Statistics analysis
The GraphPad Prism 5 software was used to analyze the data. Differences between individual groups were compared by using Student’s t-test. P<0.05 indicates a statistically significant difference.
Results and Discussion
SENP3 localization in the nucleus and cytoplasm and accumulation within the cells upon DEN exposure
DEN is a xenobiotic and is specifically transformed in the liver, disrupting double-stranded DNA and eventually causing hepatocellular carcinoma [17]; we herein used DEN to subject hepatocytes to stress. γ-H2AX level was used to assess the extent of DNA double-strand breaks so as to determine the effects of DEN in the normal liver cell line LO2. We found that the γ-H2AX level was increased in a time-dependent manner in LO2 cells after treatment with 15 mM DEN (Fig. 1A). SENP3 levels also showed an increase, as early as from 15 min of DEN treatment to 12 h, without any obvious cell death (Fig. 1B). In untreated hepatocytes, SENP3 was mainly present in the nuclei, but its level showed a considerable increase in the nuclei after DEN exposure (Fig. 1C). To demonstrate whether SENP3 could interact with cytoplasmic proteins, digitonin, a mild detergent with a low ability to permeabilize the nuclear membrane, was used to emphasize cytoplasmic proteins. The results showed that small amounts of SENP3 were found in the cytoplasm in untreated cells and that its level was significantly increased upon DEN treatment for 2 h (Fig. 1C). Cytoplasmic localization of SENP3 implies more substrates of de-SUMOylation in untreated or DEN-treated hepatocytes.
Figure 1.
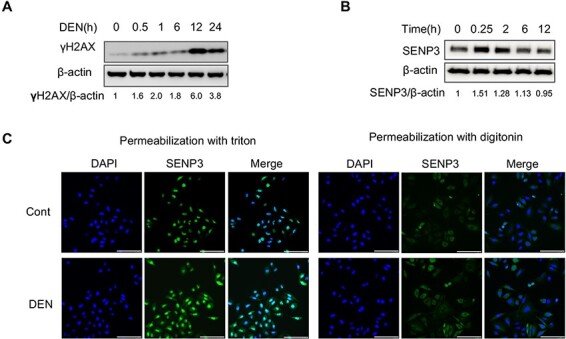
SENP3 localization in the nucleus and cytoplasm and accumulation within the cells upon DEN exposure (A,B) LO2 cells were treated with DEN for an indicated time period, and then DN damage and SENP3 level were analyzed by western blot analysis. (C) LO2 cells were treated with DEN for 2 h, and then SENP3 was visualized by immunofluorescence and distinguished between nuclear localization and cytoplasmic localization with different methods of permeabilization. Scale bar: 100 μm.
Proximity-dependent biotinylation of SENP3-interacting proteins by SENP3–BirA
The workflow for proximity-dependent biotinylation of SENP3-interacting proteins by SENP3–BirA is shown in Fig. 2A. SENP3–BirA was expressed at levels comparable to those of endogenous SENP3. The levels of biotinylated proteins were higher in biotin-treated hepatocytes as compared with those in untreated hepatocytes. Furthermore, cells transfected with pCDH–SENP3–BirA showed fewer biotinylated protein bands than pCDH–BirA-transfected cells, suggesting that BirA–SENP3 possibly caused the proximal-dependent biotinylation of SENP3-interacting proteins (Fig. 2B).
Figure 2.
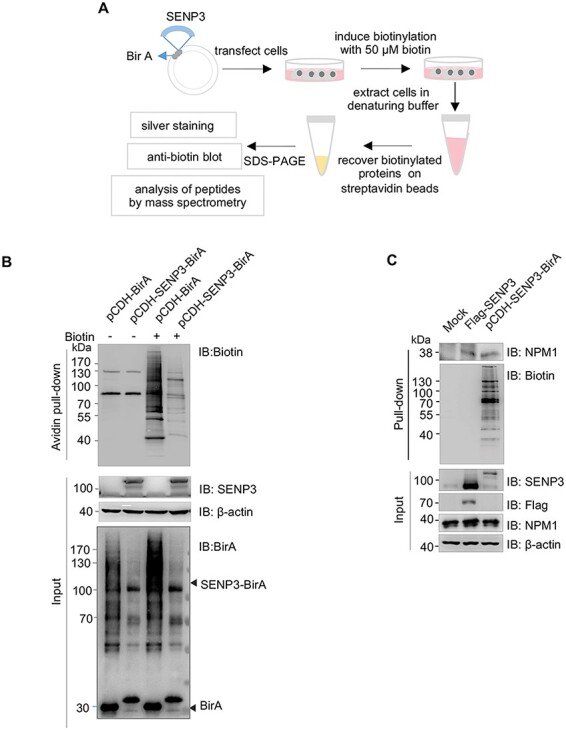
BioID of SENP3-interacting proteins by SENP3–BirA (A) Schematic workflow of the BioID method with SENP3–BirA. (B) HEK293T cells were transfected with the indicated plasmids. At 24 h after transfection, the expression levels of SENP3–BirA and control BirA were determined. Biotinylation of SENP3-interacting proteins was determined in the presence or absence of biotin pretreatment for 24 h. (C) LO2 cells were transfected with pCDH–SENP3–BirA or FLAG–SENP3. SENP3 was precipitated with indicating methods and then nucleophosmin (NPM1) was detected.
By measuring NPM1, a known SENP3-interacting protein [18], we further assessed whether an interaction exists between NPM1 and SENP3 using the BioID method and compared it with the traditional Co-IP method. After LO2 cells were transfected with pCDH–SENP3–BirA or FLAG–SENP3, SENP3 was precipitated using strepavidin and highly specific anti-Flag M2 beads, respectively. As shown in Fig. 2C, an equivalent amount of NPM1 was precipitated with both methods, whereas much less SENP3–BirA was needed than FLAG–SENP3, suggesting that the BioID method was more efficient than the co-IP method for precipitating SENP3-interacting proteins.
Identification and verification of SENP3-interacting proteins using the BioID method with SENP3–BirA
To identify the SENP3-interacting proteins in untreated and DEN-treated hepatocytes, LO2 cells stably transfected with BirA–SENP3 were analyzed using the BioID method and MS. Silver staining of the precipitates by avidin was performed after SDS–PAGE to show all biotinylated proteins mediated by SENP3–BirA (Fig. 3A). Then, the precipitates were identified, and a total of 310 proteins in untreated hepatocytes and 208 proteins in DEN-treated hepatocytes were detected as the SENP3-interacting proteins, in which >0.3% of sequence coverage was set as a cutoff value for MS data. Furthermore, 95 out of the 208 proteins were exclusive to DEN-treated hepatocytes, i.e. novel SENP3-interacting were identified in response to DEN treatment (Fig. 3B). The sequence coverage of SENP3 in the untreated and DEN-treated groups was 31.4% and 16.6%, respectively, and the intensity was 4.6×109 and 4.4×108, respectively (Supplementary Table S1). Three hits of SENP3-interacting proteins, i.e. nuclear protein NPM1, cytoplasmic protein transketolase (TKT), and mitochondrial inner membrane protein ATP synthase subunit (ATP5F1A), were chosen and validated. NPM1 and TKT were present in both DEN-treated hepatocytes and untreated hepatocytes, while ATP5F1A was exclusively present in DEN-treated hepatocytes (Fig. 3C), as revealed by MS data.
Figure 3.
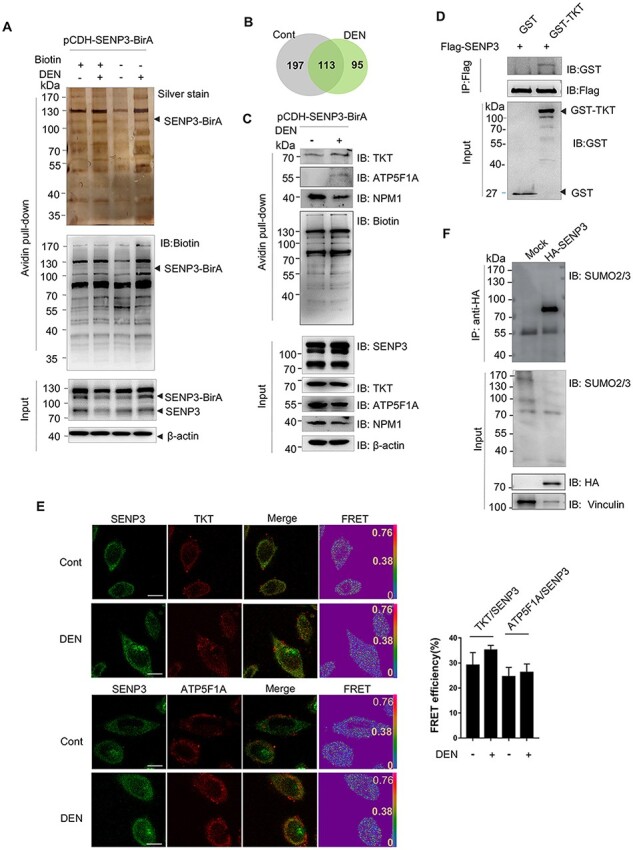
Identification and verification of SENP3-interacting proteins with SENP3–BirA (A) Silver staining of precipitations by BioID. LO2 cells were stably transfected with pCDH–SENP3–BirA (named LO2–SENP3–BirA). LO2–SENP3–BirA cells were untreated or treated with DEN. (B) A pie chart showing the number of proteins identified by MS. (C) The interaction of SENP3 with TKT, NPM1, and ATP5F1A was verified by co-IP. LO2 cells were transfected with pCDH–SENP3–BirA and treated with DEN. (D) The in vitro interaction of SENP3 with TKT was validated by GST pull-down assay. Flag-SENP3 was transfected into HEK293T cells, and SENP3 was purified from cell lysates with anti-Flag M2 gels after 24 h of transfection. GST–TKT was obtained from prokaryotic expression system. (E) The physical interaction of SENP3 with TKT or ATP5F1A was measured by FRET with acceptor photobleaching method. After treatment with DEN for 2 h, LO2 cells were fixed and stained with primary antibodies and fluorescence-conjugated secondary antibodies. Scale bar: 10 μm. The efficiency of FRET from 10 cells are shown. (F) Transient SUMOylated proteins that interact with SENP3 were evaluated. co-IP was performed in HEK293T cells transfected with HA–SENP3.
Alternatively, the interaction of SENP3 with TKT was validated by GST pull-down assay. SENP3 was extracted from HEK293T cells and then incubated with GSH-bound GST–TKT. SENP3 was indeed pulled down by TKT (Fig. 3D), which provided the evidence of direct interaction in vitro. In addition, these physical interactions were evaluated by fluorescence resonance energy transfer (FRET) assay. The efficiency of FRET of SENP3 with TKT was around 30% in untreated cells and increased to 35% in DEN-treated cells. Similarly, the efficiency of FRET of SENP3 and ATP5F1A was around 25%. Since an efficiency of >20% is the evidence of interaction [19], TKT and ATP5F1A were proved to interact with SENP3. There was some difference in FRET efficiency between the untreated group and the treated group, but no statistical significance was found (Fig. 3E). These results independently and mutually validated SENP3-interacting proteins revealed by MS.
To gain insight into the specificity of BioID approach, we compared our data with known SENP3 substrates and other endogenous SUMO2/3 proteomics studies. There are 13 substrates of SENP3 in cultured cell lines, 59 mouse liver-specific SUMOylated proteins, and 955 tissue-specific SUMOylated proteins reported by our lab and other labs [8,10,20–26], while 1, 3, and 93 proteins within individual datasets were identified in the present BioID assay, fold of enrichment achieving 3.7, 3.2, and 4.1 (log2), respectively (Supplementary Table S2). Compared with these proteomics studies, our data also provided more information about SENP3-associated dynamic SUMOylation.
Additionally, we also explored whether SENP3-interacting proteins are mainly SUMOylated substrates. HEK293 cells were transfected with HA–SENP3, followed by the precipitation process using the antibody against HA to harvest SENP3-interacting proteins, and the complex was recognized by SUMO2/3 antibodies. The results showed that SENP3-interacting proteins showed smear bands of conjugated proteins (Fig. 3F, upper panel), although deconjugation of substrate proteins within whole cell lysate was detected in SENP3-overexpressing cells (Fig. 3F, bottom panel). These data suggested that transient and dynamic interactions exist between SENP3 and SUMO2/3-conjugated substrates. Therefore, we believe that assessing SENP3-interacting proteins should provide insights into SUMOylated proteomics.
Analysis of SENP3-interacting proteins in untreated and DEN-treated hepatocytes
We analyzed the biological functions of SENP3-interacting proteins, particularly of novel SENP3 partners, using an integrated bioinformatics approach, including subcellular localization determination, Kyoto Encyclopedia of Genes and Genomes signaling pathway analysis, and PPI network analysis.
A total of 310 SENP3-interacting proteins were identified, with 44.2%, 33.9%, and 6.45% mainly localized in the nucleus, cytoplasm, and plasma membrane (or part of a secretory pathway), respectively. In the cytoplasm, 5.5% SENP3 partners were distributed within the mitochondria (Fig. 4A), and notably, the proportion of cytoskeletal and related proteins accounted for 2.26% SENP3 partners, such as filamin A, actin, keratin-2, keratin-18, keratin-9, and keratin-1. Unexpectedly, secreted, membrane, and endoplasmic reticulum proteins, such as albumin, fibrinogen, desmocollin, and the endoplasmic reticulum chaperone BiP, were also identified. These proteins are present in intracellular vesicles or the endoplasmic reticulum and Golgi bodies, making their identification challenging using the traditional co-IP method. Our findings thus highlight the advantages of using the BirA system for precipitating and consequently identifying proteins present in these compartments.
Figure 4.
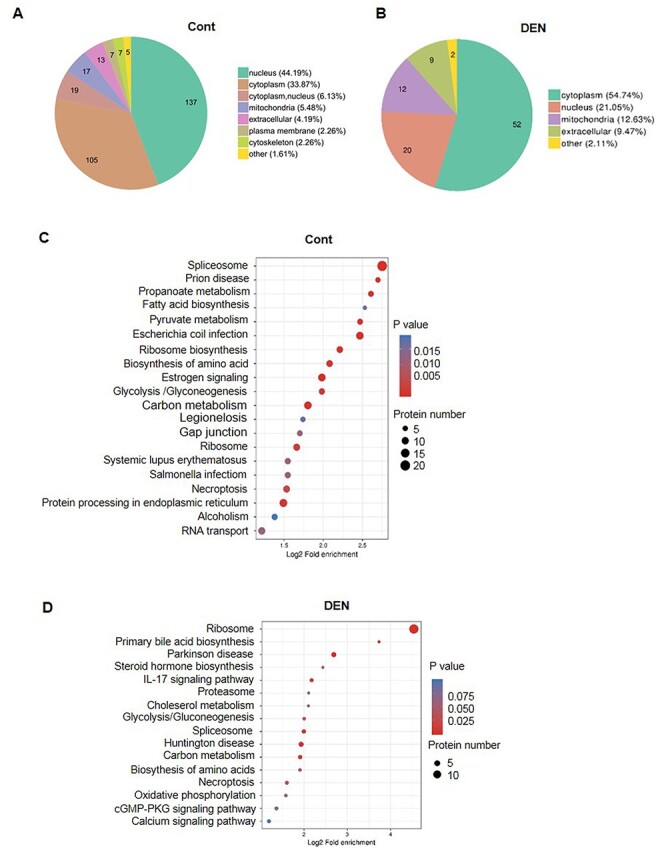
Analysis of SENP3-interacting proteins in untreated and DEN-treated hepatocytes Integrated bioinformatics analyses of 310 proteins from untreated LO2–SENP3–BirA cells and 95 novel proteins from DEN-treated LO2–SENP3–BirA cells were performed. (A,B) Localization analysis. (C,D) Kyoto Encyclopedia of Genes and Genomes signaling pathway analysis. (A,C) Untreated hepatocytes. (B,D) DEN-treated hepatocytes.
Of the 95 proteins that were exclusive to DEN-treated hepatocytes, 54.74% were cytoplasmic, 12.63% were mitochondrial, and 21.05% were nuclear proteins (Fig. 4B). The cytoplasmic proteins were mainly ribosomal proteins, indicating their involvement in nucleolus stress response or protein synthesis. Changes in SUMOylation of the cytoplasmic proteins argininosuccinate synthase and aldo–keto reductase family 1 member C1 and C3 were indicative of changes in metabolism. Furthermore, the mitochondrial proteome was composed of membrane proteins VDAC1, VDAC2, ATP/ADP translocase, SLA25A5, SLA25A6, and SLA25C and the inner membrane proteins ATP5F1A and ATP5F1B. Only a few studies have focused on the SUMOylation of mitochondrial proteins, and very little is known about the outer membrane protein Drp1 [9]. Our data expand the list of mitochondrial proteins that potentially act as reversible SUMO2/3-conjugated substrates.
In normal hepatocytes, SENP3-interacting proteins were assigned to 20 Kyoto Encyclopedia of Genes and Genomes signaling pathways, with the most being involved in spliceosome (ID: 03040) and prion disease (ID: 05020) (Supplementary Table S3). Most spliceosome-related SENP3-interacting proteins were either RNA editing-related proteins, including hnRNPs U, M, K, and A1, or RNA splicing proteins such as PRPF3 (encoding U4/U6 small nuclear ribonucleoprotein) and serine/arginine-rich splicing factors 3 and 7. Prion disease–related proteins, including BiP and heat shock 70-kDa proteins 6 and 1B, were associated with protein folding signaling pathways. In addition, our analysis revealed several metabolism-related signaling pathways that influence fatty acid synthesis, pyruvate metabolism, amino acid synthesis, and so on. The proteins involved in such pathways included fatty acid synthase and acetyl-CoA carboxylases 1 and 2 (Fig. 4C).
Among the newly identified SENP3-interacting proteins in DEN-treated hepatocytes, changes in ribosome-related pathways were remarkable (ID: 03010). In particular, there were 15 and 13 more new proteins in the 60S large and 40S small subunits, respectively (Fig. 4D, Supplementary Table S4). Due to the special localization of SENP3 in the nucleolus and its close relation with ribosome biogenesis, it becomes more difficult to distinguish whether the ribosomal proteins are specific SENP3-interacted proteins or non-specific pollutant in the MS studies. In the hit list from SENP3-BioID, compared to the identification of 10 kinds of 60S ribosomal proteins in the untreated hepatocytes, 28 kinds of ribosomal proteins as new hits were found in DEN-treated cells. It might suggest relatively the specific interaction of SENP3 with ribosomal proteins. In addition, one established SENP3-interacted protein TEX10 was identified in this study in untreated cells, which had been reported to be required for the maturation of the 60S preribosomal subunit [27,28], and importantly, the interaction disappeared after DEN treatment, supporting the notion of specific interaction of SENP3 with some proteins associated with ribosomal biogenesis as well.
SENP3-interacting proteins are also associated with other pathways involved in primary bile acid biosynthesis, Parkinson’s disease, interleukin-17 signaling, glycolysis/gluconeogenesis, and spliceosome. The pathways involved in bile acid biosynthesis and gluconeogenesis are hepatocyte-specific metabolic pathways. However, the SENP3-interacting fatty acid synthesis signaling pathway is not involved upon DEN treatment. Because both fatty acid and bile acid levels are closely associated with hepatocellular carcinoma [29], their de-SUMOylation may be considered as pathological factors.
We herein used STRING to evaluate the interactive (PPI) relationships. Consistent with the well-established role of SENP3 in ribosome biosynthesis [18,28], we found that SENP3 had a node that links nucleolar and ribosomal proteins in untreated hepatocytes (Fig. 5A). Interestingly, upon DEN treatment, SENP3 was associated with both a new series of ribosomal network and a mitochondrial membrane protein-related network (Fig. 5B). DEN evidently acutely triggers biotransformation, oxidative damage to macromolecules, and DNA damage in hepatocytes [30]. However, our data suggested that an additional rapid response to DEN treatment was the alterations in SENP3-regulated ribosomal proteins rather than in DNA repair proteins.
Figure 5.
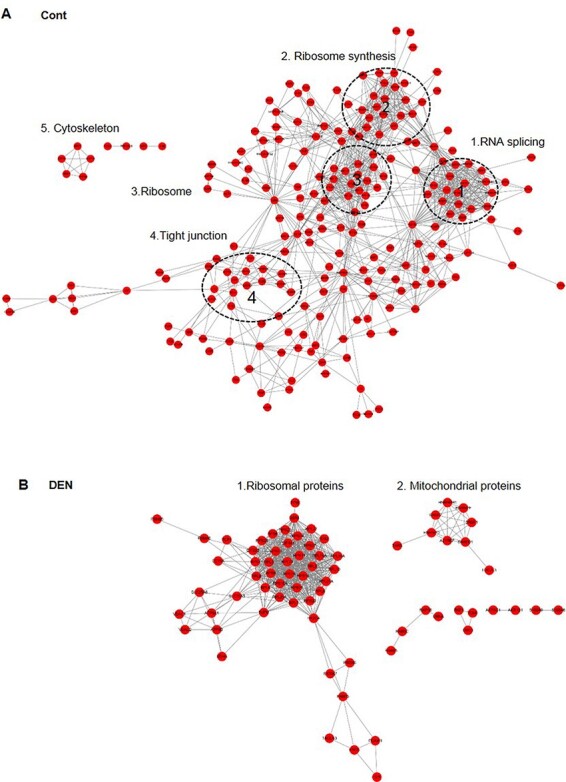
STRING network maps of SENP3-interacting proteins in untreated and DEN-treated hepatocytes (A) Untreated and (B) DEN-treated LO2–SENP3–BirA cells.
Conclusion
In recent years, the presence of 751–4300 SUMO modification sites and 395–1600 proteins has been reported in HeLa, U2OS, and HEK293 cells, with most of them being localized in the nucleus and involved in the majority of nuclear processes such as gene transcription, DNA repair, chromatin remodeling, pre-mRNA splicing, and ribosome assembly [2,31,32]. Our data added cytoplasmic proteins such as metabolic enzymes, mitochondrial proteins, and the cytoskeleton as potential SUMO2/3-modified proteins in hepatocytes. More importantly, these proteins were found to undergo dynamic changes (SUMO2/3 modification). To the best of our knowledge, this study for the first time reports proteomics data pertaining to the dynamic alteration of endogenous SUMO2/3 modifications in DEN-treated hepatocytes, which could be a potential undervalued regulatory mechanism. The BioID method is an effective system for identifying physiologically relevant proteins.
Supplementary Material
Acknowledgement
We thank Drs Zhaoyuan Hou and Xuemei Tong (Shanghai Jiao Tong University School of medicine) for kindly providing pCDH-BirA, Flag-TKT plasmids, and helpful comments.
Contributor Information
Fei Chen, Department of Biochemistry and Molecular Cell Biology, Shanghai Key Laboratory for Tumor Microenvironment and Inflammation, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Hongyu Yan, Department of Biochemistry and Molecular Cell Biology, Shanghai Key Laboratory for Tumor Microenvironment and Inflammation, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Chu Guo, Department of Biochemistry and Molecular Cell Biology, Shanghai Key Laboratory for Tumor Microenvironment and Inflammation, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Huiqin Zhu, Department of Biochemistry and Molecular Cell Biology, Shanghai Key Laboratory for Tumor Microenvironment and Inflammation, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Jing Yi, Department of Biochemistry and Molecular Cell Biology, Shanghai Key Laboratory for Tumor Microenvironment and Inflammation, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Xuxu Sun, Department of Biochemistry and Molecular Cell Biology, Shanghai Key Laboratory for Tumor Microenvironment and Inflammation, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Jie Yang, Department of Biochemistry and Molecular Cell Biology, Shanghai Key Laboratory for Tumor Microenvironment and Inflammation, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Supplementary Data
Supplementary Data is available at Acta Biochimica et Biophysica Sinica online.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (No. 31771522), Shanghai Municipal Science and Technology Commission (No. 16ZR1418400), the Innovative Research Team of High-level Local Universities in Shanghai, the National Natural Science Foundation of China (No. 31970587 to Dr Xuxu Sun), and the Foundation of Shanghai Oriental Scholar (No. TP2018045).
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Han Y, Huang C, Sun X, Xiang B, Wang M, Yeh ET, Chen Y, et al. SENP3-mediated De-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. J Biol Chem 2010, 285: 12906–12915. doi: 10.1074/jbc.M109.071431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendriks IA, Vertegaal ACO. A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Bio 2016, 17: 581–595. doi: 10.1038/nrm.2016.81 [DOI] [PubMed] [Google Scholar]
- 3.Lamoliatte F, Caron D, Durette C, Mahrouche L, Maroui MA, Caron-Lizotte O, Bonneil E, et al. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat Commun 2014, 5: 5409. doi: 10.1038/ncomms6409 [DOI] [PubMed] [Google Scholar]
- 4.Lamoliatte F, McManus FP, Maarifi G, Chelbi-Alix MK, Thibault P. Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat Commun 2017, 8: 14109. doi: 10.1038/ncomms14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin B, Wang J, Liu X, Fang S, Jiang B, Hofmann K, Yin J, et al. Ubiquitin-mimicking peptides transfer differentiates by E1 and E2 enzymes. BioMed Res Int 2018, 2018: 6062520. doi: 10.1155/2018/6062520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M, Sang J, Ren Y, Liu K, Liu X, Zhang J, Wang H, et al. SENP3 regulates the global protein turnover and the Sp1 level via antagonizing SUMO2/3-targeted ubiquitination and degradation. Protein Cell 2016, 7: 63–77. doi: 10.1007/s13238-015-0216-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Z, Wang M, Li J, Xiao M, Chin YE, Cheng J, Yeh ETH, et al. SUMOylation and SENP3 regulate STAT3 activation in head and neck cancer. Oncogene 2016, 35: 5826–5838. doi: 10.1038/onc.2016.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lao Y, Yang K, Wang Z, Sun X, Zou Q, Yu X, Cheng J, et al. DeSUMOylation of MKK7 kinase by the SUMO2/3 protease SENP3 potentiates lipopolysaccharide-induced inflammatory signaling in macrophages. J Biol Chem 2018, 293: 3965–3980. doi: 10.1074/jbc.M117.816769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, Han Y, Wang Y, Sun X, Yan S, Yeh ET, Chen Y, et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. Embo J 2009, 28: 2748–2762. doi: 10.1038/emboj.2009.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Yang J, Yi J. Redox sensing by proteins: oxidative modifications on cysteines and the consequent events. Antioxid Redox Signal 2012, 16: 649–657. doi: 10.1089/ars.2011.4313 [DOI] [PubMed] [Google Scholar]
- 11.Liu K, Guo C, Lao Y, Yang J, Chen F, Zhao Y, Yang Y, et al. A fine-tuning mechanism underlying self-control for autophagy: deSUMOylation of BECN1 by SENP3. Autophagy 2019, 16: 975–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendriks IA, Lyon D, Su D, Skotte NH, Daniel JA. Site-specific characterization of endogenous SUMOylation across species and organs. Nat Commun 2018, 9: 2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sunny NE, Parks EJ, Browning JD, Burgess SC. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Meta 2011, 14: 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta GD, Coyaud E, Goncalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, et al. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell 2015, 163: 1484–1499. doi: 10.1016/j.cell.2015.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roux KJ, Kim DI, Burke B. BioID: a screen for protein-protein interactions. Curr Protoc Protein Sci 2013, 74: 19.23.1–19.23.14. doi: 10.1002/0471140864.ps1923s74 [DOI] [PubMed] [Google Scholar]
- 16.Yan S, Sun X, Xiang B, Cang H, Kang X, Chen Y, Li H, et al. Redox regulation of the stability of the SUMO protease SENP3 via interactions with CHIP and Hsp90. Embo J 2010, 29: 3773–3786. doi: 10.1038/emboj.2010.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tolba R, Kraus T, Liedtke C, Schwarz M, Weiskirchen R. Diethylnitrosamine (DEN)-induced carcinogenic liver injury in mice. Lab Anim 2015, 49: 59–69. doi: 10.1177/0023677215570086 [DOI] [PubMed] [Google Scholar]
- 18.Haindl M, Harasim T, Eick D, Muller S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. Embo Rep 2008, 9: 273–279. doi: 10.1038/embor.2008.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roszik J, Tóth G, Szöll˝osi J, Vereb G. Validating pharmacological disruption of protein–protein interactions by acceptor photobleaching FRET imaging. Methods Mol Biol 2013, 986: 165–178. [DOI] [PubMed] [Google Scholar]
- 20.Kejia L, Chu G, Yimin L, Jie Y, Fei C, Yuzheng Z, Yi Y, et al. A fine-tuning mechanism underlying self-control for autophagy: deSUMOylation of BECN1 by SENP3. Autophagy 2020, 16: 975–990. doi: 10.1080/15548627.2019.1647944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nayak A, Lopez-Davila AJ, Kefalakes E, Holler T, Kraft T, Amrute-Nayak M. Regulation of SETD7 methyltransferase by SENP3 is crucial for sarcomere organization and cachexia. Cell Rep 2019, 27: 2725–2736. doi: 10.1016/j.celrep.2019.04.107 [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharjee J, Alahari S, Sallais J, Tagliaferro A, Post M, Caniggia I. Dynamic regulation of HIF1Alpha stability by SUMO2/3 and SENP3 in the human placenta. Placenta 2016, 40: 8–17. doi: 10.1016/j.placenta.2016.02.002 [DOI] [PubMed] [Google Scholar]
- 23.Guo C, Hildick KL, Luo J, Dearden L, Wilkinson KA, Henley JM. SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia. EMBO J 2013, 32: 1514–1528. doi: 10.1038/emboj.2013.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nayak A, Viale-Bouroncle S, Morsczeck C, Muller S. The SUMO-specific isopeptidase SENP3 regulates MLL1/MLL2 methyltransferase complexes and controls osteogenic differentiation. Mol Cell 2014, 55: 47–58. doi: 10.1016/j.molcel.2014.05.011 [DOI] [PubMed] [Google Scholar]
- 25.Hendriks IA, Lyon D, Su D, Skotte NH, Daniel JA, Jensen LJ, Nielsen ML. Site-specific characterization of endogenous SUMOylation across species and organs. Nat Commun 2018, 9: 2456. doi: 10.1038/s41467-018-04957-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lumpkin RJ, Gu H, Zhu Y, Leonard M, Ahmad AS, Clauser KR, Meyer JG, et al. Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat Commun 2017, 8: 1171. doi: 10.1038/s41467-017-01271-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castle CD, Cassimere EK, Denicourt C. LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol Biol Cell 2012, 23: 716. doi: 10.1091/mbc.e11-06-0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J 2011, 30: 1067–1078. doi: 10.1038/emboj.2011.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun L, Beggs K, Borude P, Edwards G, Bhushan B, Walesky C, Roy N, et al. Bile acids promote diethylnitrosamine-induced hepatocellular carcinoma via increased inflammatory signaling. Am J Physiol Gastrointest Liver Physiol 2016, 311: G91–G104. doi: 10.1152/ajpgi.00027.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arboatti AS, Lambertucci F, Sedlmeier MG, Pisani G, Monti J, Alvarez ML, Frances DEA, et al. Diethylnitrosamine increases proliferation in early stages of hepatic carcinogenesis in insulin-treated type 1 diabetic mice. Biomed Res Int 2018. doi: 10.1155/2018/9472939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendriks IA, Vertegaal ACO. SUMO in the DNA damage response. Oncotarget 2015, 6: 15734. doi: 10.18632/oncotarget.4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hang J, Wan R, Yan C, Shi Y. Structural basis of pre-mRNA splicing. Science 2015, 349: 1191–1198. doi: 10.1126/science.aac8159 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


