Abstract
Cross communication between regulatory proteins is an important event in the control of eukaryotic gene transcription. Here we have examined the structural and functional interaction between two cellular regulatory proteins, YB-1 and Purα, on the 23-bp sequence element derived from the enhancer-promoter of the human polyomavirus JCV. YB-1 and Purα are single-stranded DNA binding proteins which recognize C/T- and GC/GA-rich sequences, respectively. Results from band shift studies demonstrated that while both proteins interact directly with their DNA target sequences within the 23-bp motif, each protein can regulate the association of the other one with the DNA. Affinity chromatography and coimmunoprecipitation provide evidence for a direct interaction between Purα and YB-1 in the absence of the DNA sequence. Ectopic expression of YB-1 and Purα in glial cells synergistically stimulated viral promoter activity via the 23-bp sequence element. Results from mutational studies revealed that residues between amino acids 75 and 203 of YB-1 and between amino acids 85 and 215 of Purα are important for the interaction between these two proteins. Functional studies with glial cells indicated that the region within Purα which mediates its association with YB-1 and binding to the 23-bp sequence is important for the observed activation of the JCV promoter by the Purα and YB-1 proteins. The results of this study suggest that the cooperative interaction between YB-1 and Purα mediates the synergistic activation of the human polyomavirus JCV genome by these cellular proteins. The importance of these findings for cellular and viral genes which are regulated by Purα and YB-1 is discussed.
The study of transcriptional regulation of viral genes by cellular proteins provides important information on the interaction between the virus and host cell during steps involved in reactivation of the virus and during the course of its lytic infection. Previously, we and others demonstrated that the human neurotropic JC virus (JCV) contains an enhancer-promoter region with the ability to interact with several regulatory proteins from glial and nonglial cells (38). JCV is a polyomavirus which infects greater than 70% of the human population during childhood, and its reactivation in immunocompromised individuals causes the fatal demyelinating disease progressive multifocal leukoencephalopathy (PML) (8, 17, 32). Although the JCV genome has been detected in several tissues (14, 15, 22, 33), replication of the viral genome is seen predominantly in glial cells of the central nervous system (CNS) (8). Several lines of studies, including in vitro transcription of the viral genome (1, 2), transfection of various cells with recombinant plasmids containing the viral regulatory sequence (16, 25, 46), and creation of transgenic animals containing the JCV promoter sequence (16, 23, 41), have established that the tissue-specific replication of the viral genome is due to the transcriptional activity of the viral promoter, which is more efficient in CNS cells. Therefore, much effort has been directed toward analysis of the JCV regulatory sequence and identification of host proteins which, upon interaction with their target sequence within the promoter, confer specificity to viral gene transcription. The typical regulatory sequence of JCV is comprised of two 98-bp tandem repeats, each containing a TATA box in juxtaposition with a pentanucleotide repeat, AGGGAAGGGA, and an NF-1 motif. This strain of JCV is called JCVMad-1 (18). Results from earlier studies led us to believe that the pentanucleotide repeat sequence, also referred to as the lytic control element (LCE), has the ability to modulate JCV early and late promoters (44, 45) and contributes to viral DNA replication (10, 30). In subsequent studies, two cellular single-stranded DNA binding proteins named Purα and YB-1, which recognize purine-rich and C/T-rich sequences, respectively, of the LCE, were identified (12, 26, 44). Results from subsequent studies indicated that in JCVMad-1, binding of YB-1 to its DNA target within the LCE is increased by Purα. In contrast, interaction of Purα with the LCE motif is diminished once YB-1 is included in the reaction mixture (12). Functionally, Purα was able to induce JCV early gene transcription, whereas YB-1 was the activator for the JCV late promoter (11). Results from DNA binding and transfection studies suggested a functional role for these proteins via the LCE motif in transcription of the JCV promoters in glial cells (11, 12).
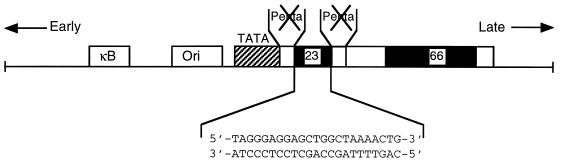
Another peculiarity in the JCV genome is the hypervariability of the structural organization of the viral control sequences (17). For example, the control region of a newly isolated JCV from brain tissue of a PML patient (51) contains a 23-bp sequence element within the pentanucleotide repeat, disrupting this important regulatory motif. A similar DNA sequence with an identical nucleotide composition is present within the JCV regulatory region of the JCV archetype, JCVCY (Fig. 1). JCVCY was first isolated from the urine of nonimmunosuppressed patients as well as healthy individuals (49). According to one model, JCVMad-1, which is routinely detected in brains of PML patients, may evolve from JCVCY by DNA rearrangements whereby the 23-bp sequence element is deleted. Although the importance of the 23-bp sequence in the regulation of the JCV genome remains to be investigated, early studies have indicated that despite disruption of the LCE motif, the presence of the 23-bp sequence element does not alter the level of viral gene transcription (51). These observations suggest (i) that the presence of the 23-bp sequence element within the JCV enhancer-promoter region does not interfere with the ability of the LCE binding proteins, i.e., YB-1 and Purα, to modulate viral gene transcription and/or (ii) that the 23-bp DNA sequence provides a new target for interaction of Purα and YB-1, thus ensuring efficient transcription of the viral genome in the infected cells. Here, we have performed a series of DNA binding and protein-protein interaction experiments to investigate the association between Purα, YB-1, and the 23-bp DNA sequence and their effect on transcription of the viral promoter in glial cells.
FIG. 1.
Structural organization of the regulatory region of JCVCY. NF-κB and ori, regulatory sequences for binding of the NF-κB transcription protein and the origin of viral DNA replication, respectively. The regulatory region of JCVCY contains one 98-bp repeat with two additional motifs of 23 and 66 bp. The 23-bp sequence element disrupts the pentanucleotide repeat sequence (AGGGAAGGGA). The nucleotide composition of the 23-bp insertion is shown at the bottom. The directions of transcription of the viral early and late promoters are indicated by the arrows.
We demonstrate that Purα and YB-1 have the ability to interact with the 23-bp sequence element. Evidently, the interplay between YB-1 and Purα determines their ability to bind to this element. Moreover, we demonstrate that ectopic expression of YB-1 and Purα causes synergistic activation of the JCV minimal promoter and that this event requires an intact 23-bp sequence element within the viral genome.
MATERIALS AND METHODS
Cell culture.
U-87MG cells were maintained in Dulbecco’s modified Eagle medium (GIBCO, Grand Island, N.Y.) supplemented with 10% fetal calf serum and penicillin-streptomycin. Cells were grown at 37°C in 7% CO2.
Construction of plasmids.
Reporter plasmids, designated pBLCAT3-CYE and pBLCAT3-CYL, contain the minimal promoter region from JCVCY, which spans nucleotides 5077 to 63, in the early and late gene orientations, respectively. These constructs were created by PCR amplification of the JCVCY regulatory region with the MS-1 (5′-TGAGCTCATGCTTGGCTGGCAA-3′) and MS-2 (5′-CCTAAAAAGCCTCCACGCCCT-3′) primers. The PCR-amplified DNA fragments were first cloned into the SmaI site of Bluescript SK (Stratagene, La Jolla, Calif.) and then transferred into the BamHI/PstI site of pBLCAT3 (29). Each clone was examined for the orientations of their inserts by direct DNA sequencing. pGEX2T-YB-1 was provided by Gene MacDonald (31). Glutathione S-transferase (GST)–YB-1 C-terminal deletion mutants were created as follows. pGEX2T-YB-1 was first digested with SalI/EcoRI, AlwNI/EcoRI, StyI/EcoRI, and BglI/EcoRI enzymes, and the appropriate DNA fragments were removed by gel purification. The 5′ and 3′ ends of the DNA fragments were blunt ended upon treatment with Klenow enzyme and religated with T4 DNA ligase. The plasmids obtained from these manipulations were designated pGEX2T-YB-1(1-203), -YB-1(1-125), -YB-1(1-75), and -YB-1(1-37).
Constructs containing YB-1 N-terminal deletion mutants were created by PCR amplification of the coding region of YB-1 with the 5′ primers YB-1 501 (5′-ACTGGATCCGCCGCCATGGCAGGTCCTGGTGGTGTT-3′), YB-1 737 (5′-ACTGGATCCGCCGCCATGGGTCGACCACAGTATTCC-3′), and YB-1 877 (5′-ACTGGATCCGCCGCCATGGGCCAAAGACAGCCTAGA-3′) and the 3′ primer 5′-GCGGGAATTCTCAGCTGGTGGATCGGCTGCTTTTGTCTC-3′. The amplified DNA fragments after treatment with EcoRI restriction enzyme were cloned into the EcoRI site of the pCDNA3 expression vector (Invitrogen, Carlsbad, Calif.). Constructs for N-terminal deletion mutants of YB-1 were designated cytomegalovirus (CMV)-YB-1(125-318), -YB-1(204-318), and -YB-1(250-318). Construction of GST-Purα and GST-Purα mutants is detailed elsewhere (19, 24). YB-1 was tagged in frame with a histidine (His) tag at the 5′ end by subcloning it into the Epstein-Barr virus (EBV)-His A expression vector (Invitrogen) at the BamHI/XhoI site. Purα was tagged at the 5′ end in frame with an influenza virus hemagglutinin (HA) tag by subcloning it into the CMV-pCDNA3 expression vector (Invitrogen) at the BamHI/XhoI site and was designated CMV-HA-Purα. The expression vector pEBV-His B Purα, containing the coding region of the Purα gene fused 3′ to a histidine epitope tag under the control of the Rous sarcoma virus (RSV) promoter, has been previously described (19). pEBV-His-Purα(216-322) has also been previously described (19). pEBV-His B Purα(85-322) was constructed by first subcloning the EcoRI fragment containing the coding region encompassing amino acids 85 to 322 from pGEX-1λT-Purα (24) into EcoRI-digested pCDNA3, generating pCDNA3-Purα. pCDNA-Purα was subsequently digested with BamHI and XhoI, and the BamHI/XhoI fragment containing the Purα-coding sequence was ligated in frame into BamHI/XhoI-cut pEBV-His B. Both pMAL-MBP-YB-1 and pMAL-MBP-Purα have been described previously (12). All plasmids were verified by DNA sequencing by using Sequenase (U.S. Biochemical Corp., Cleveland, Ohio).
EMSA.
For electrophoretic mobility shift assays (EMSAs), single-stranded DNA fragments representing the early strand (3′-ATCCCTCCTCGACCGATTTTGAC-5′) or late strand (5′-TAGGGAGGAGCTGGCTAAAACTG-3′) of the JCVCY 23-bp sequence were 5′ end labeled with [γ-32P]ATP by using T4 polynucleotide kinase. Bacterially expressed highly purified fusion proteins, i.e., maltose binding protein (MBP) fused to YB-1 (MBP–YB-1) or to Purα (MBP-Purα) (12), were mixed with the single-stranded probes (30,000 cpm/reaction) in a binding buffer containing 0.1 μg of poly(dI-dC)/μl, 12 mM HEPES (pH 7.9), 4 mM Tris-HCl (pH 7.5), 60 mM KCl, 5 mM MgCl2, and 0.1 mM dithiothreitol and incubated for 30 min at 4°C. The DNA-protein complexes were resolved on 6% polyacrylamide gels in 0.5× TBE (1× TBE is 89 mM Tris-HCl [pH 8.0], 89 mM boric acid, and 2 mM EDTA [pH 8.0]). For Fig. 2F and G, EMSAs were performed as described above with GST-Purα and GST–YB-1. The gels were dried, and complexes were detected by autoradiography at −70°C with an intensifying screen.
FIG. 2.
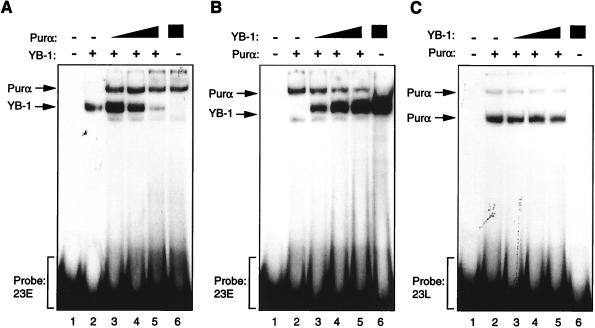
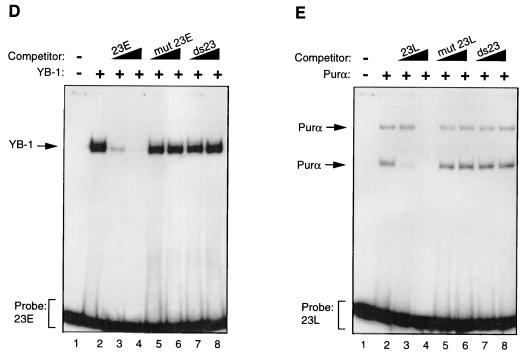
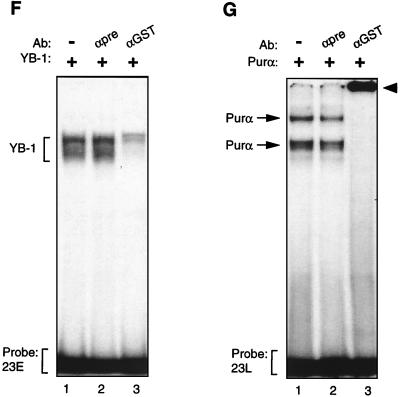
Purα and YB-1 modulate each other’s binding to 23-bp single strands. (A to C) Band shift assay. (A) A 23-bp single-stranded DNA probe from the early strand (23E) was incubated with 30 ng of MBP–YB-1 alone (lane 2) or with 200, 400, or 600 ng of MBP-Purα (lanes 3 to 5, respectively). Lanes 1 and 6 contain 600 ng of MBP and 600 ng of Purα, respectively. The positions of YB-1 and Purα complexes are shown by the arrows. (B) The 23-bp early DNA probe (23E) was incubated with 200 ng of Purα alone (lane 2) or with 30, 60, or 120 ng of YB-1 (lanes 3 to 5, respectively). (C) The 23-bp single-stranded DNA probe for the late strand (23L) was incubated with YB-1 and Purα fusion proteins as described for panel B. (D and E) Competitive band shift assays. (D) The 23E probe was mixed with YB-1 protein in the absence or presence of 50- and 250-fold molar excesses of unlabeled competitor DNA as indicated. (E) The 23L probe was incubated with Purα protein alone (lane 2) or in the presence of 50- and 250-fold molar excesses of competitor DNAs. The positions of the Purα and YB-1 complexes are shown by the arrows. (F and G) Antibody (Ab) supershift assays. (F) GST–YB-1 fusion protein (150 ng) was incubated with the 23E probe (lanes 1 to 3), and the binding reaction mixture included either 1 μg of preimmune (αpre) (lane 2) or 1 μg of anti-GST (lane 3) antibodies. The positions of the GST–YB-1:23E complexes are indicated by a bracket. (G) Two hundred nanograms of GST-Purα fusion protein was incubated with the 23L probe. The reaction mixture included preimmune (lane 2) and anti-GST (lane 3) antibodies as described for panel F. The positions of the GST-Purα:23L complexes are indicated by arrows. Supershifted complexes are depicted by an arrowhead.
In vitro transcription and translation assay.
To synthesize YB-1 and Purα proteins in vitro, we utilized a TNT coupled in vitro transcription-translation system (Promega, Madison, Wis.). The proteins were labeled with [35S]methionine as recommended by the supplier.
GST fusion protein affinity chromatography.
U-87MG cells expressing Purα or YB-1 were lysed in LB150 buffer, containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 0.1% Nonidet P-40, 50 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, and 1 μg of aprotinin per ml, for 30 min on ice. Lysates were scraped, collected into microcentrifuge tubes, vortexed briefly, and microcentrifuged for 30 min at 4°C. Five hundred micrograms of whole-cell extract prepared from U-87MG cells either untransfected or transfected with HA-tagged Purα (HA-Purα) was incubated with 1 μg of GST or GST–YB-1 fusion protein coupled to glutathione-Sepharose beads in 300 μl of LB 150 for 2 h at 4°C with continuous rocking. Alternatively, 500 μg of whole-cell extract prepared from U-87MG cells either untransfected or transfected with histidine-tagged YB-1 (His–YB-1) was incubated with 1 μg of GST or GST-Purα under similar conditions. After incubation, the beads were pelleted and washed five times with LB 150 buffer. Bound proteins were eluted with Laemmli sample buffer and heated at 95°C for 10 min. Complexes were resolved on sodium dodecyl sulfate (SDS)–10% polyacrylamide gel and analyzed for HA-Purα or His–YB-1 with either anti-HA or anti-T7 antibodies, respectively. Thirty micrograms of crude extracts was loaded on the gels as migration controls.
For mapping studies, GST pull-down experiments were performed with 35S-labeled in vitro-translated proteins and various GST fusion proteins. More specifically, 4 μl of 35S-labeled in vitro-translated YB-1 was incubated with 1 μg of GST, GST Purα, or GST-Purα amino- and carboxy-terminal deletion mutants immobilized on glutathione-Sepharose beads. Alternatively, 4 μl of 35S-labeled in vitro-translated Purα was incubated with 1 μg of GST, GST–YB-1, or carboxy-terminal GST-YB-1 deletion mutants. Additionally, 4 μl of 35S-labeled in vitro-translated amino-terminal YB-1 deletion mutants was incubated with full-length GST-Purα fusion protein immobilized on glutathione-Sepharose beads. All reactions were preformed in 300 μl of LB 150 plus 1 μg of BSA per μl for 2 h at 4°C with continuous rocking. After incubation, the beads were pelleted and washed five times with LB 150 buffer. Bound proteins were eluted with Laemmli sample buffer and heated at 95°C for 10 min. Complexes were resolved by SDS–10% polyacrylamide gel electrophoresis (SDS–10% PAGE), and proteins were detected by fluorography for the presence of Purα and YB-1. One-tenth of the input reaction mixture was loaded as a migration control.
Expression and purification of recombinant fusion proteins.
YB-1 and Purα fusion proteins used in band shift studies were prepared by methods described previously (12). Expression of GST–YB-1, GST-Purα, and the mutant variants was carried out by the procedure described previously (19). Briefly, 100 ml of overnight cultures of Escherichia coli DH5α, transformed with either pGEX2T-YB-1, pGEX2T-Purα, or their respective deletion mutant plasmids, was diluted 1:10 in fresh Luria-Bertani medium supplemented with ampicillin (100 μg/ml). Cultures were induced with 0.4 M isopropyl-β-d thiogalactopyranoside (IPTG) at an optical density of 0.6 and incubated for an additional 1.5 h at 37°C. Cells were collected by centrifugation and resuspended in 10 ml of lysis buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40 supplemented with 1 mM phenylmethylsulfonyl fluoride, 2 μM pepstatin A, 0.6 μM leupeptin, and 2 mM benzamidine. After sonication, clear cell lysates were prepared and incubated with 100 μl of 50% GST-Sepharose beads (Pharmacia, Piscataway, N.J.) overnight at 4°C. GST fusion proteins were washed three times with 50 ml of lysis buffer, and protein quality was analyzed by SDS-PAGE followed by Coomassie blue staining. Additionally, GST-Purα and GST–YB-1 proteins were eluted from the Sepharose beads with 10 mM glutathione in elution buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 5 mM dithiothreitol and subsequently dialyzed in 20 mM Tris-HCl (pH 7.4)–150 mM NaCl–10% glycerol.
Transient-transfection assays.
U-87MG cells were seeded in 60-mm-diameter culture dishes the day before transfection. Cells were transfected with plasmid DNA by calcium phosphate coprecipitation (21). The total amounts of DNA in the transfection mixtures were equalized to 30 μg with empty vector DNA. At 48 h posttransfection, cells were lysed by three freeze-thaw cycles, and the protein contents were determined by the method of Bradford (9). Chloramphenicol acetyltransferase (CAT) activity was determined by a previously described method (20). Results are expressed as fold activation. Transfections were performed at least three times with different plasmid preparations. In all transfection studies, 1 μg of plasmid expressing β-galactosidase (β-gal) (RSV-β-gal) was included in the transfection mixture, and the level of β-gal activity was used as the baseline to normalize the CAT levels from the various experiments.
Western blot analysis.
Whole-cell extracts were prepared from either untransfected or transfected U-87MG cells (4), and 30 μg of protein extract was separated by SDS–10% PAGE prior to transfer to nitrocellulose membranes (Schleicher and Schuell, Keene, N.H.). The filters were probed with specific antibodies (1:200 dilution) and developed with an ECL detection kit according to the recommendations of the manufacturer (Amersham, Arlington Heights, Ill.).
Coimmunoprecipitation assay.
EBV-His-YB-1 and CMV-HA-Purα expression plasmids were introduced into U-87MG cells by the calcium phosphate precipitation method (21). At 48 h posttransfection, cells were lysed in LB 150 buffer, and approximately 0.5 mg of whole-cell extract in a total volume of 0.5 ml of lysis buffer was incubated with 0.5 μg of either anti-His (Novagen, Madison, Wis.) or anti-HA (Berkeley Antibody, Richmond, Calif.) antibodies for 16 h at 4°C. Immunocomplexes were precipitated with the addition of protein A-Sepharose beads (Pharmacia) for an additional 2 h at 4°C. After four washes in lysis buffer, immunocomplexes were resolved by SDS–10% PAGE and analyzed by Western blotting with an ECL detection kit.
RESULTS
Interaction of Purα and YB-1 with the JCVCY 23-bp sequence.
To examine the ability of Purα and YB-1 to interact with the 23-bp DNA sequence, synthetic single-stranded DNA probes representing the early (23E) and late (23L) strands of the 23-bp sequence element were incubated with highly purified bacterially produced Purα and YB-1. The protein-DNA complexes were analyzed by native PAGE. As shown in Fig. 2A, lane 2, a major band corresponding to the YB-1:23E complex was observed. Inclusion of Purα in the binding reaction mixture at a low concentration enhanced the intensity of the band corresponding to the YB-1:23E complex and at higher concentrations showed an inhibitory effect on the association of YB-1 with the 23E probe (Fig. 2A, compare lane 2 to lanes 3, 4, and 5). The band corresponding to the Purα:23E complex showed slower electrophoretic mobility and migrated above the YB-1:23E complex. Of interest is that the intensity of the Purα:23E band was not enhanced upon increasing the concentration of Purα in the binding reaction mixture. These experiments were performed under conditions where the probe was in excess, indicating that the observed inhibition of YB-1:23E assembly by Purα may not be attributed to the limitation of the DNA probe.
In the reciprocal experiment, the 23E probe was incubated with purified Purα in the absence and presence of increasing amounts of YB-1. As shown in Fig. 2B, the addition of YB-1 to the binding reaction mixture enhanced the intensity of the band corresponding to YB-1:23E and gradually decreased the formation of the Purα:23E complex. These data suggest that cross communication between YB-1 and Purα may dictate the level of their association with the 23-bp sequence of the JCV regulatory region. A similar study was performed with the 23L DNA probe. As illustrated in Fig. 2C, while YB-1 showed no affinity for binding to the 23L probe, inclusion of YB-1 in the binding reaction mixture modestly decreased the intensities of the bands corresponding to the Purα:23L complex. Of note is that the ability of Purα to form two distinct bands on the gel is due to its capacity to interact with its target sequence as a monomer or multimer (24, 27, 36). Additionally, the 23L DNA is enriched in GA and contains the perfect binding site for Purα, GGAGGA. This may allow the formation of a more stable Purα:23L complex which remains intact, albeit partially, in the presence of YB-1.
To examine the specificity of YB-1 and Purα association with the 23E and 23L probes, respectively, we performed competitive band shift assays. As shown in Fig. 2D, the unlabeled 23E oligonucleotide inhibits binding of YB-1 to the 23E probe, whereas mutant 23E DNA with no binding site for YB-1, or the double-stranded 23-bp oligonucleotide (ds23), had no effect on binding of YB-1 to the 23E probe. Similarly, binding of Purα to the 23L probe was blocked by unlabeled 23L DNA but not by the mutant 23L, which lacks the Purα binding sites, or the ds23 DNA (Fig. 2E). These observations indicated that association of the DNA probes with YB-1 and Purα are specific and mediated through specific nucleotide sequences within the 23-bp sequence. We also performed antibody supershift experiments to further investigate the specificity of the interaction between the YB-1 and Purα proteins with their respective target sequences on 23-bp single strands. The addition of the preimmune antibody to the binding reaction mixture showed no effect on complex formation as evidenced by the electrophoretic mobility of the YB-1:23E complex (Fig. 2F, compare lane 1 to lane 2). However, the addition of anti-GST antibody to the binding mixture resulted in a substantial decrease in the intensity of the band corresponding to the YB-1:23E complex (Fig. 2F, lane 3), indicating disruption of the complex formed between YB-1 and the 23-bp early single strand. Similar experiments were also carried out to further investigate the interaction between Purα and the 23L probe (Fig. 2G). Although the preimmune antibody did not show any effect on the migration pattern of the GST-Purα:23L complexes (Fig. 2G, lane 2), anti-GST antibody completely supershifted the Purα:23L complexes (Fig. 2G, lane 3). Of note is that no complex was observed upon incubation of the anti-GST antibody with either of the 23-bp single-stranded probes (data not shown).
In conclusion, antibody supershift experiments further indicated the specificity of the interaction between YB-1 and Purα with their respective targets on the 23-bp single strands.
Interaction of YB-1 and Purα in the absence of the 23-bp sequence.
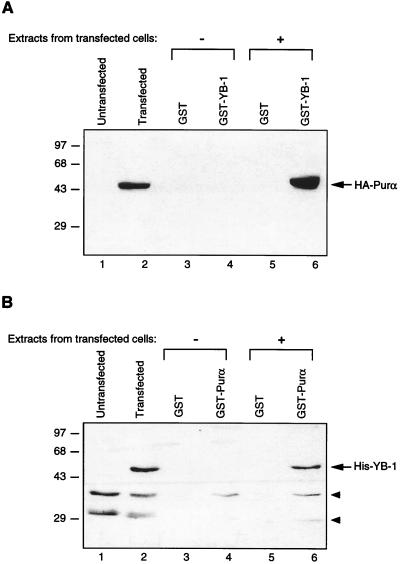
Results from DNA binding studies implied that Purα and YB-1 may interact with each other and that this interaction may determine the levels of their binding to the 23-bp motif. To investigate the direct interaction of YB-1 and Purα in glial cells, in vitro GST pull-down experiments were performed. To this end, U-87MG cells were transfected with an HA-tagged Purα expression plasmid (pCMV-HA-Purα) or with the histidine-tagged YB-1 expression plasmid (pEBV-His-YB-1). Forty-eight hours after transfection, protein extracts were prepared and incubated with bacterially produced GST and GST-Purα and GST–YB-1 fusion proteins immobilized on glutathione-Sepharose beads. After washing, proteins retained on the beads were analyzed by immunoblot analysis with an HA antibody which recognizes the HA tag on Purα or an anti-His antibody which recognizes the histidine tag on YB-1. As shown in Fig. 3A, incubation of extract derived from cells transfected with HA-Purα and Sepharose columns containing either GST or GST–YB-1 resulted in the specific retention of HA-Purα on the column containing GST–YB-1 (lane 6) but not on the column containing GST alone (lane 5). As expected, no band corresponding to the HA-Purα protein was detected in lanes corresponding to protein from untransfected cells (Fig. 3A, lane 1) or in the lanes containing eluates obtained after incubation with GST or GST–YB-1 (Fig. 3A, lanes 3 and 4). A band corresponding to HA-Purα was also detected in the unfractionated protein extract from the pCMV-HA-Purα-transfected cells (Fig. 3A, lane 2). In reciprocal experiments, protein extracts from the control untransfected cells and the cells transfected with pEBV-His-YB-1 were examined for production of His–YB-1 and for the ability of the ectopically produced YB-1 to bind to Purα. As shown in Fig. 3B, anti-His antibody was able to detect the YB-1 fusion protein in the extract from transfected cells (lane 2) and in the fractions that bound to GST-Purα but not GST alone (compare lane 6 to lane 5). Together, these observations point to the in vitro association of Purα and YB-1.
FIG. 3.
In vitro interaction of Purα and YB-1. (A) Bacterially produced GST or GST–YB-1 was immobilized on GST-Sepharose beads and incubated with whole-cell extracts prepared from untransfected U-87MG cells or U-87MG cells transfected with CMV-HA-Purα expression plasmid for 2 h at 4°C. The Sepharose beads were washed extensively with lysis buffer, and proteins interacting with GST or GST–YB-1 were analyzed by SDS-PAGE followed by immunoblotting with anti-HA antibody, which detected the HA-Purα fusion protein. (B) Whole-cell extracts prepared from untransfected U-87MG cells or cells transfected with EBV-His-YB-1 expression plasmid were incubated with either GST alone or GST-Purα. Proteins interacting with GST or GST-Purα were harvested and analyzed by immunoblotting with anti-His antibody for detection of His-tagged YB-1. Arrowheads indicate nonspecific bands detected with the anti-His antibody. Numbers on the left are molecular masses in kilodaltons.
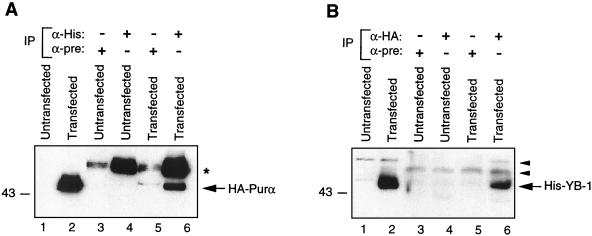
To examine the association of Purα and YB-1 in cellular extracts, U-87MG cells were cotransfected with pCMV-HA-Purα plus pEBV-His-YB-1, which allows production of HA-Purα and His–YB-1 fusion proteins, respectively, in these cells. Protein extracts from the transfected and the untransfected control cells were prepared and examined for the presence of a YB-1:Purα complex by coimmunoprecipitation and Western blotting techniques. As shown in Fig. 4A, the immunocomplexes obtained upon incubation of the protein extracts with anti-His antibody, which detects YB-1 fusion protein, contained HA-Purα fusion protein (lane 6). The HA-Purα, which was not detected in complexes from the control preimmune sera in either transfected or untransfected cells, comigrated with the HA-Purα detected in the total protein extract from the transfected cells (Fig. 4A, lane 2). In reciprocal experiments, we were able to detect His–YB-1 in the immunocomplexes that were pulled down by anti-HA antibody and contained HA-Purα fusion protein (Fig. 4B, lane 6). A band corresponding to His–YB-1 was also detected in total protein extract from the transfected cells. These results along with those of the previous in vitro binding studies strongly suggest that Purα and YB-1 can form heterocomplexes in the absence of their DNA target sequence within the 23-bp motif.
FIG. 4.
Coimmunoprecipitation of YB-1 and Purα. (A) Approximately 0.5 mg of whole-cell extracts prepared from untransfected U-87MG cells or cells transfected with EBV-His-YB-1 and CMV-HA-Purα expression plasmids were immunoprecipitated (IP) with 0.5 μg of monoclonal anti-His (α-His) or preimmune (mouse) serum (α-pre). Immunocomplexes were analyzed by SDS-PAGE followed by immunoblotting with an antibody directed against the HA tag for detection of HA-Purα fusion protein. An asterisk indicates the position of immunoglobulin G detected by the secondary antibody. (B) Whole-cell extracts (0.5 mg) were immunoprecipitated with an antibody directed against HA tag or preimmune mouse serum. The immunocomplexes were analyzed by SDS-PAGE followed by immunoblotting with anti-His antibody for detection of His–YB-1. Arrowheads indicate nonspecific bands.
Functional interaction of Purα and YB-1 on the JCVCY minimal promoter containing the 23-bp sequence.
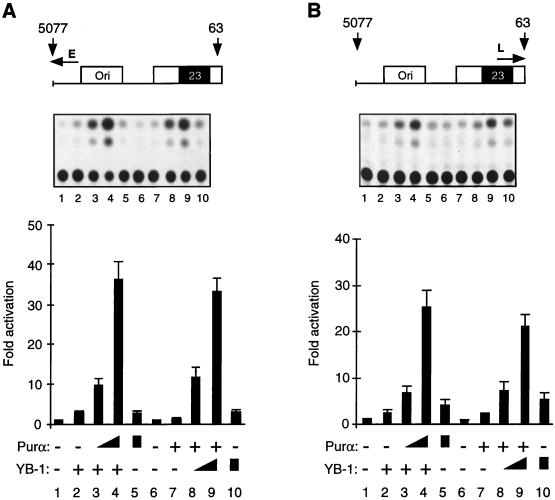
To investigate the functional importance of the interplay between Purα and YB-1 for the transcriptional activity of the 23-bp sequence, we performed cotransfection experiments. In these studies the human glial cell line U-87MG was transfected with the JCV minimal promoter construct containing the 23-bp motif, alone or together with the recombinants expressing Purα and YB-1. As shown in Fig. 5A, in the presence of a small amount of YB-1, no significant changes in the activity of the JCVE promoter were detected (compare lane 1 to lane 2). Under similar experimental conditions, the addition of Purα expression plasmids to the transfection mixture, in particular at the higher concentration, significantly increased the activity of the JCVE promoter in these cells (compare lane 2 to lane 4). The level of activation was substantially higher than the sum of the transcriptional activities obtained with YB-1 (lane 2) and Purα (lane 5) alone, suggesting that YB-1 and Purα may function synergistically to stimulate the JCV early promoter. Figure 5A also shows that at a lower concentration, Purα has no effect on the JCVE promoter (lane 7), whereas in the presence of YB-1, a drastic increase in viral promoter activity is observed (compare lane 7 with lanes 8 and 9). Again, the level of activation mediated by Purα plus YB-1 was much higher than the activity observed for the sum of YB-1 and Purα, pointing to the synergistic activity of these two proteins on the JCVE promoter.
FIG. 5.
Functional cooperation between Purα and YB-1 in transcriptional activation of the JCVCY minimal promoter in U-87MG cells. (A) pBLCAT3-CYE reporter plasmid containing the JCVCY minimal promoter (7.5 μg), as shown (top), was introduced into U-87MG cells alone or together with YB-1 and Purα expression plasmids. In cotransfections, as the plasmid concentration for one transactivator was kept constant at 10 μg for each lane (lanes 2 to 4 for YB-1 and lanes 7 to 9 for Purα), the plasmid concentration for the other transactivator was varied (5, 10, and 10 μg of plasmid DNA of Purα for lanes 3 to 5 respectively, and 5, 10, and 10 μg of plasmid DNA of YB-1 for lanes 8 to 10, respectively). One microgram of RSV-β-gal plasmid was added to each transfection mixture. The DNA concentrations for each lane were normalized with the addition of empty expression vector DNA. CAT activity was measured and normalized for β-gal activity. The transfection experiments were repeated at least three times. The data obtained from CAT assays were quantitated and are presented as fold activation relative to the basal expression of the minimal promoter (bottom). Error bars indicate standard deviations. Results of a representative CAT assay are shown in the middle. (B) Experiments similar to those detailed for panel A were performed with the JCV late promoter, pBLCAT3-CYL.
A similar set of cotransfection experiments was carried out to evaluate the cooperative action of YB-1 and Purα on the JCV late minimal promoter containing the 23-bp motif. As shown in Fig. 5B, consistent with their effect on the JCV early promoter, overexpression of YB-1 and Purα resulted in a synergistic activation of the viral late promoter in glial cells. Taken together, these results demonstrate that YB-1 and Purα enhance each other’s effect on both the JCVE and JCVL promoters.
We have also examined the transcriptional activities of the JCV minimal promoters in response to overexpression of YB-1 and Purα in kidney epithelial cells (CV-1). In contrast to the activity observed in glial cells, we did not observe any synergistic activity in these cells (data not shown).
Protein domains involved in formation of the Purα:YB-1 complex.
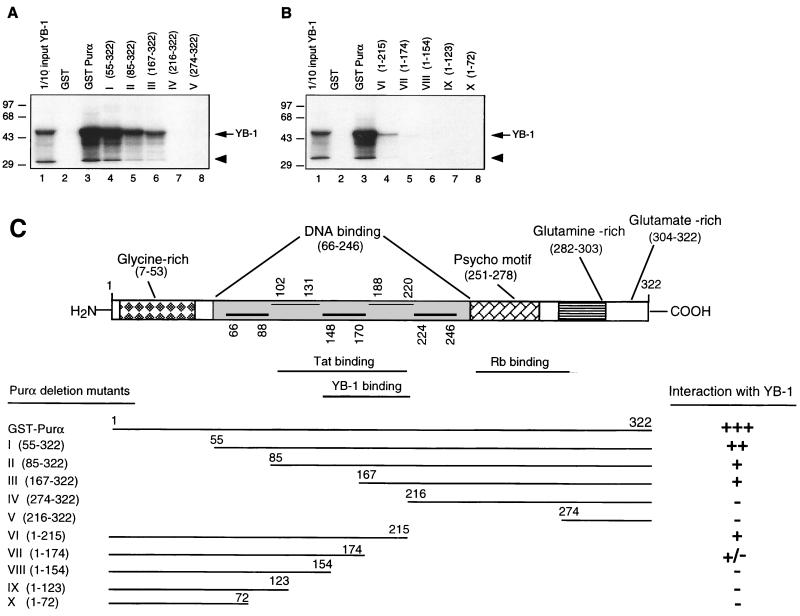
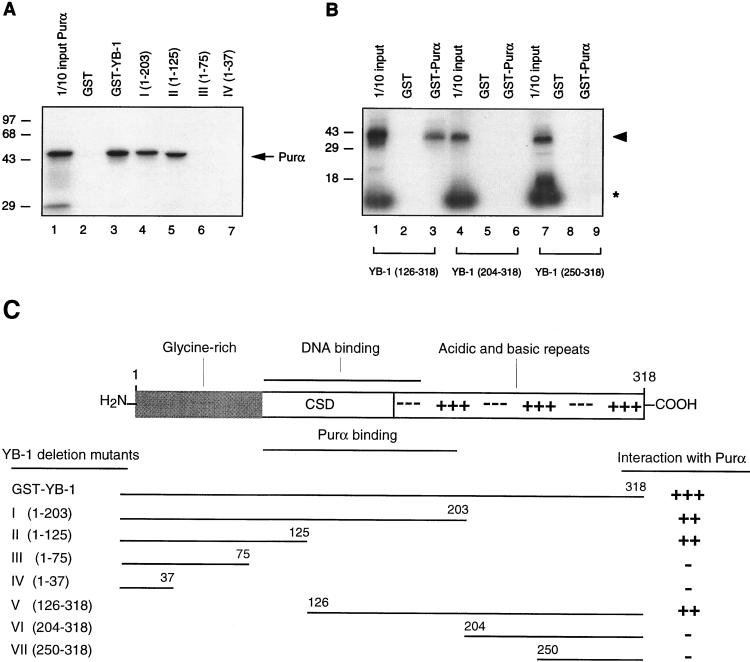
In the next series of experiments, we attempted to identify the region(s) within Purα which is important for its interaction with YB-1. In this study, in vitro-translated 35S-labeled YB-1 protein was prepared and used in GST pull-down assays with GST-Purα deletions encompassing various regions of Purα. As shown in Fig. 6A, binding of YB-1 to Purα was gradually decreased upon removal of amino acid residues 54 to 166 and was completely abrogated by further amino-terminal deletions. These data suggest that the region located between residues 167 and 216 is the minimal sequence which is important for Purα interaction with YB-1. Results with C-terminal deletion mutants further indicated that residues 174 to 215 are critical for complex formation between YB-1 and Purα. These results are summarized in Fig. 6C.
FIG. 6.
Mapping of the domain(s) of Purα involved in interaction with YB-1. (A and B) Mapping of the interaction domain of Purα with YB-1. GST-Purα or its N-terminal (A) or its C-terminal (B) deletion mutants were incubated with in vitro-translated [35S]methionine-labeled YB-1. Sepharose beads were washed three times with lysis buffer, and bound proteins were resolved by SDS–10% PAGE. One-tenth of the input YB-1 used in each reaction was loaded for migration controls (lane 1 in each panel). The labeled arrow marks the position of in vitro-translated [35S]methionine-labeled YB-1, and the arrowhead indicates a degradation product of YB-1. Numbers on the left are molecular masses in kilodaltons. (C) Summary of the results obtained from in vitro mapping assays. A schematic representation of the Purα protein is shown at the top (not shown to scale). The relative strengths of the interactions between GST-Purα and its deletion mutants with in vitro-translated [35S]methionine-labeled YB-1 are depicted.
To identify the region(s) of YB-1 necessary for interaction with Purα, in vitro-translated 35S-labeled Purα was incubated with full-length GST–YB-1 or various GST-YB-1 mutants, and their association with each other was examined by GST pull-down assays. As shown in Fig. 7A, the C-terminal deletion mutant of YB-1 which retains residues 1 to 125 is able to form a complex with Purα, whereas removal of residues 76 to 125 inhibits its association with Purα. In a separate series of experiments, three in vitro-translated amino-terminal YB-1 deletion mutants, YB-1(126-318), YB-1(204-318), and YB-1(250-318), were incubated with GST or GST-Purα. As shown in Fig. 7B, YB-1(126-318), interacted with GST-Purα (lane 3) but not with GST (lane 2). Neither YB-1(204-318) nor YB-1(250-318) was able to interact with GST-Purα (lanes 6 and 9, respectively). Taken together, these results suggest that the region spanning residues 76 to 204 of YB-1 is important for its association with Purα. These results are summarized in Fig. 7C. These results are interesting in light of our previous data (12). In that work we demonstrated that Purα mutants Purα(216-322) and Purα(274-322) enhanced the ability of YB-1 to interact with DNA. Those studies were conducted in the presence of the DNA probe, and the results were analyzed by band shift assay. Here, we evaluated the level of YB-1 and Purα interaction directly, in the absence of their target DNAs, by a combination of column chromatography and Western blot analysis. Therefore, it is likely that while stable association of Purα and YB-1 may require residues 167 to 216, their communication which results in enhancement of YB-1 DNA binding activity is mediated through residues which are located at the C terminus of Purα. On the other hand, it is possible that the residues 85 to 215 of Purα which binds to YB-1 are insufficient to alter YB-1 DNA binding activity.
FIG. 7.
Mapping the domain(s) of YB-1 involved in the interaction with Purα. (A) In vitro-translated 35S-labeled Purα was incubated with either GST alone or C-terminal deletion mutants of GST–YB-1 fusion proteins. Bound proteins were analyzed as described for Fig. 6. The labeled arrow marks the position of in vitro-translated 35S-labeled Purα. (B) Three different 35S-labeled in vitro-translated amino-terminal mutants of YB-1, i.e., YB-1(126-318), YB-1(204-318), and YB-1(250-318), were incubated with GST (lanes 2, 5, and 8) or GST-Purα (lanes 3, 6, and 9). Bound proteins were analyzed as described for Fig. 6. Lanes 1, 4, and 7 contain 1/10 of the amount used in the pull-down experiments with YB-1(126-318), YB-1(204-318), and YB-1(250-318), respectively. The arrowhead designates the position of the in vitro-translated amino-terminal deletion mutants. The asterisk denotes a nonspecific product present in the in vitro transcription-translation reactions. (C) A schematic representation of full-length YB-1 is shown at the top. CSD, the cold shock domain of YB-1. The relative strengths of the interactions observed between YB-1 and Purα are depicted on the right.
Interaction of Purα and YB-1 is important for their regulatory action on the JCVCY promoter.
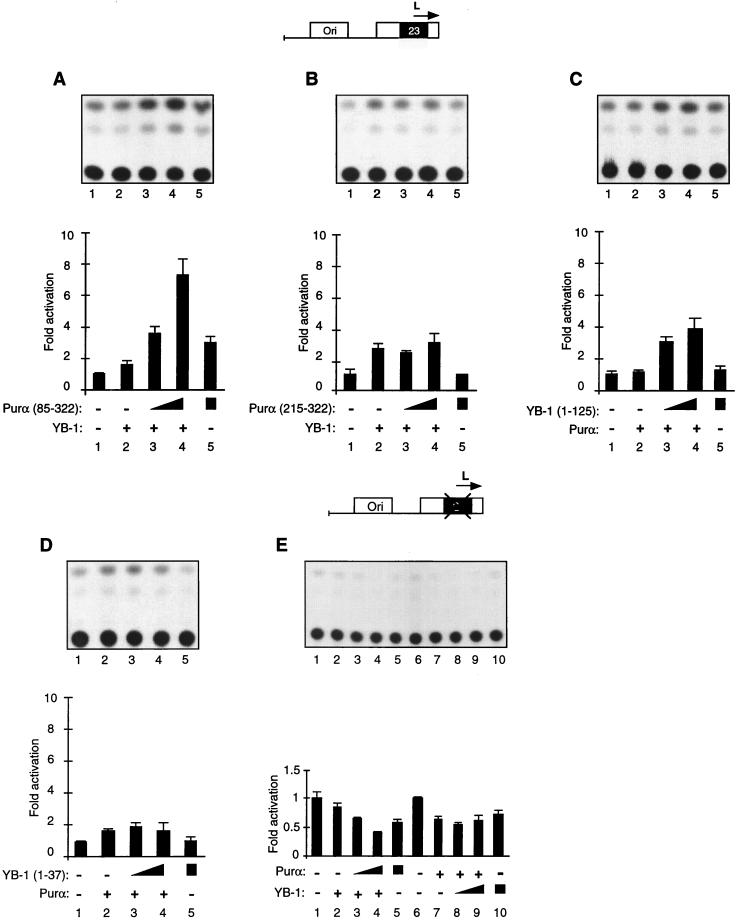
To further assess the functional interaction between YB-1 and Purα, transient-transfection studies utilizing deletion mutants of each protein were performed. Expression of these mutants was verified by Western blot analysis (data not shown). In these studies, U-87MG cells were transfected with the minimal JCVL promoter containing the 23-bp motif along with a small amount of YB-1 in the presence of the Purα mutant which binds YB-1 [Purα(85-322)] or its variant with no ability to bind YB-1 [Purα(216-322)]. As shown in Fig. 8A, ectopic expression of YB-1 and Purα(85-322) resulted in a synergistic activation of the JCV promoter. This is similar to the activity of full-length Purα and YB-1 (Fig. 5). Under similar conditions, no synergism between YB-1 and Purα(216-322) was detected (Fig. 8B). These observations suggest that the region within Purα which is important for its association with YB-1 is required for its functional interaction with YB-1 with the JCV promoter sequence. In a different series of experiments, cells were transfected with the JCVL promoter together with a plasmid expressing a mutant YB-1, YB-1(1-125), which binds to Purα, or its variant, YB-1(1-37), with no ability to bind Purα. As shown in Fig. 8C, while Purα or YB-1(1-125) alone did not show a significant stimulatory effect on the JCVL promoter (lanes 2 and 5, respectively), coproduction of these two proteins enhanced the activity of the JCVL promoter in the transfected cells. Under similar conditions, coproduction of the full-length Purα with YB-1(1-37) had no drastic effect on the transcription of the JCV promoter (Fig. 8D). These observations along with binding results indicate that a cooperative interaction between YB-1 and Purα may determine their transcriptional ability with the JCV promoter. In the next series of transfection experiments, we utilized a JCV promoter construct with a cluster mutation within the 23-bp sequence that converts GGA to AAA. As shown in Fig. 8E, ectopic expression of YB-1 in the absence and presence of Purα had no stimulatory effect on the mutant viral promoter (compare lane 1 to lanes 2 to 5). In fact, these proteins, alone or in combination, exert a negative effect on the basal promoter activity and decrease the level of transcription from the JCV minimal promoter. Figure 8E (bottom) shows a summary of the results from these experiments. These observations together suggest that association of Purα and YB-1 with each other and the viral 23-bp regulatory sequence is functionally important for transcription of the viral genome.
FIG. 8.
Functional interaction of YB-1 and Purα deletion mutants on the JCVCY late minimal promoter. (A and B). A 7.5-μg amount of JCVL minimal promoter reporter construct (shown above the panels) was introduced into U-87MG cells alone (lanes 1) or in combination with YB-1 and Purα deletion mutants. In cotransfections, as the plasmid concentration for YB-1 was kept constant at 10 μg (lanes 2 to 4), the plasmid concentrations for Purα deletion mutants were varied (5, 10, and 10 μg of plasmid in lanes 3 to 5, respectively). CAT activity was measured and normalized as described for Fig. 5. The lower panels show the quantitative analysis of the results from transfection experiments. Error bars indicate standard deviations. (C and D) Transfections were performed with 7.5 μg of JCVL promoter as described above and with 10 μg of Purα-expressing plasmid alone or with 5 or 10 μg of YB-1 mutants as indicated. (E) Approximately 7.5 μg of the JCVL minimal promoter reporter construct with a mutation in the 23-bp motif (shown at the top) was introduced into U-87MG cells alone or together with YB-1 and Purα expression plasmids. The experimental design was virtually identical to that described in the legend to Fig. 5. The bottom panel shows the quantitative analysis of representative experiments.
DISCUSSION
A mechanism, so-called cross-family interaction, in which one transcription factor modulates the function of another one is an important event in the control of eukaryotic gene transcription. Examples of this cross-family interaction include association of NF-κB family members with Jun/Fos or C/EBP family members (42, 43) and the interaction of Jun/Fos with MyoD (7), which can determine the regulatory activity of these factors. We have utilized the human neurotropic virus JCV as a model to unravel the regulatory pathways which include DNA-protein and protein-protein interactions in mediating transcription of eukaryotic genes in CNS cells. In earlier studies, we demonstrated that YB-1, a DNA binding protein which recognizes the inverted CCAAT sequence, may exert its regulatory action upon communication with other cellular proteins, including NF-κB subunits via the D-regulatory motif (39). Moreover, we demonstrated that YB-1 could bind to another viral regulatory motif, the LCE (26). As the 23-bp sequence, by disrupting the LCE motif, may affect its regulatory action on the JCV genome, we performed a series of structural and functional studies to analyze the functional importance of the 23-bp sequence for JCV gene transcription. Here, we demonstrate that while YB-1 binds to the 23-bp DNA sequence of JCV, its interaction with the DNA can be regulated by Purα, a single-stranded DNA binding protein that recognizes the GC/GA-rich motif. This regulatory event is mutual, as YB-1 was able to influence the association of Purα with its target GC/GA nucleotide within the 23-bp sequence. Of interest is that results from our band shift studies showed no evidence for formation of a ternary complex indicative of simultaneous association of the 23-bp sequence with YB-1 and Purα. Thus, according to one model, Purα, by transient interaction with YB-1, may induce a conformational change in YB-1, which inhibits its association with the DNA molecule. This event could be bidirectional, as interaction of Purα with the DNA is also affected. Such a mechanism has been previously reported, where the activity of MyoD1 is enhanced in the presence of heat shock protein 90 (40).
Alternatively, it is possible that Purα, by binding to the YB-1:23-bp complex, forms an unstable ternary complex, YB-1:23-bp:Purα, which may dissociate during gel electrophoresis. Results from our in vitro and in vivo protein-protein interaction studies indicated that these two proteins can form a heterodimer in the absence of DNA and that certain domains within each protein are critical for their heterodimerization. Results from functional studies demonstrated that overexpression of YB-1 and Purα synergistically stimulate transcription of the JCV genome. It was evident that the 23-bp sequence is important for such a regulatory effect. Thus, while Purα and YB-1 can form a complex off their target DNA sequences, their association with the DNA sequence is critical for their transcriptional activity. Taken together, these observations demonstrate that cooperative interaction of two cellular proteins with distinct DNA recognition sites positioned within the 23-bp motif modulates viral gene transcription. Of note is that in earlier studies we demonstrated that in the absence of the 23-bp sequence, the intact LCE may provide a target for the interplay of YB-1 and Purα and for their regulatory action on JCV early and late gene transcription (11, 12). While the 23-bp motif interrupts the pentanucleotide AGGGAAGGGA repeat of the LCE motif, it contains nucleotide sequences which provide targets for YB-1 and Purα binding. However, the important difference is that the early strand of LCE bound exclusively to YB-1, whereas its late strand interacts only with Purα. Here, we demonstrate that while YB-1 does not interact with the late 23-bp sequence element, both YB-1 and Purα form complexes with the early strand of the 23-bp sequence element. Interestingly, results from functional studies revealed that while both Purα and YB-1 alone can stimulate viral early and late gene transcription, together their activities are significantly increased. This observation differs from previous results for JCVMad-1, containing an intact LCE but not the 23-bp sequence element, in which Purα and YB-1 had more effect on early and late gene transcription, respectively. Thus, while the 23-bp sequence element provides an alternative site for binding of YB-1 and Purα, differential activities of these regulatory proteins on the viral genome may exist, suggesting that the structural organization of the viral promoter can dictate the activities of the participant regulatory proteins.
Both YB-1 and Purα represent cellular regulatory proteins with the ability to control expression of a broad range of cellular and viral genes. For example, earlier studies demonstrated that YB-1 modulates transcription of the human MDR1 gene (3), the chicken α-2(1) collagen gene (6), the grp 78 gene (28), the matrix metalloproteinase 2 gene (34), the major histocompatibility complex class II HLA-DR-α gene (35), the thyrotropin receptor gene (37), the gamma interferon gene (48), and the myosin light chain 2 V gene (52). On the other hand, earlier reports demonstrated that Purα stimulates expression of the myelin basic protein gene (23), human immunodeficiency virus type 1 (13), the transforming growth factor β1 gene (47), and the neuron-specific FE65 gene (50). As our results presented here suggest that the activities of YB-1 and Purα on the JCV genome could be regulated by their mutual interaction, one can postulate a similar mechanism for control of the other responsive viral and cellular genes. The common feature of YB-1 and Purα is their ability to interact in a sequence-specific manner with the single-stranded DNAs. Interestingly, while Purα recognizes the purine-rich sequences, YB-1 binds to the C/T-rich DNA elements. The ability of these two proteins to recognize complementary single-stranded elements leads to interesting speculation about their control over transcription. Furthermore, our data presented here demonstrate the physical interaction of these two proteins off the DNA molecule. This information, along with the earlier observation pointing to the ability of YB-1 to promote single-stranded DNA regions (31), suggests that YB-1 may function as a chaperone and, by promoting the single-stranded region within the duplex DNA, facilitate binding of Purα to its purine-rich target. Furthermore, in recent studies it has been shown that expression of YB-1 may be regulated by environmental stress, such as UV-irradiation or drug treatment, etc. (5). Thus, the functional role of cooperative interaction of YB-1 and Purα in stimulation of cellular genes whose products are important for cell survival during stress may become an important regulatory event. Moreover, as both Purα and YB-1 are expressed in a variety of cells and tissues, future studies will be directed toward the understanding of the interaction with the JCV genome in other tissues, such as B cells, where JCV has been detected.
ACKNOWLEDGMENTS
We thank G. MacDonald for providing the full-length YB-1 DNA and past and present members of the Center for NeuroVirology and NeuroOncology for sharing ideas, insightful discussion, and helpful discussion. We thank Cynthia Schriver for editorial assistance.
This work was made possible by grants awarded by NIH to K.K.
REFERENCES
- 1.Ahmed S, Chowdhury M, Khalili K. Regulation of the human neurotropic virus promoter, JCVE: identification of a novel activator domain located upstream from the 98 bp enhancer region. Nucleic Acids Res. 1990;18:7417–7423. doi: 10.1093/nar/18.24.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed S, Rappaport J, Tada H, Kerr D, Khalili K. A nuclear protein derived from brain cells stimulates transcription of the human neurotropic virus promoter, JCVE, in vitro. J Biol Chem. 1990;265:13899–13905. [PubMed] [Google Scholar]
- 3.Asakuno K, Kohno K, Uchiumi T, Kubo T, Sato S, Isono M, Kuwano M. Involvement of a DNA binding protein, MDR-NF1/YB-1, in human MDR1 gene expression by actinomycin D. Biochem Biophys Res Commun. 1994;199:1428–1435. doi: 10.1006/bbrc.1994.1390. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F, Brent R, Kingston R E, Moar D D, Siedman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 5.Bargou R C, Jurchott K, Wagener C, Bergmann S, Metzner S, Bommert K, Mapara M Y, Winzer K J, Dietel M, Dorken B, Royer H D. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med. 1997;3:447–450. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- 6.Bayarsaihan D, Enkhmandakh B, Lukens L N. Y-box proteins interact with the S1 nuclease-sensitive site in the chicken alpha 2(I) collagen gene promoter. Biochem J. 1996;319:203–207. doi: 10.1042/bj3190203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bengal E, Ransone L, Scharfmann R, Dwarki V J, Tapscott S J, Weintraub H, Verma I M. Functional antagonism between c-Jun and MyoD proteins: a direct physical association. Cell. 1992;68:507–519. doi: 10.1016/0092-8674(92)90187-h. [DOI] [PubMed] [Google Scholar]
- 8.Berger J R, Concha M. Progressive multifocal leukoencephalopathy: the evolution of a disease once considered rare. J Neurovirol. 1995;1:5–18. doi: 10.3109/13550289509111006. [DOI] [PubMed] [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Chang C-F, Tada H, Khalili K. The role of a pentanucleotide repeat sequence, AGGGAAGGGA, in the regulation of JC virus DNA replication. Gene. 1994;148:309–314. doi: 10.1016/0378-1119(94)90704-8. [DOI] [PubMed] [Google Scholar]
- 11.Chen N N, Khalili K. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Purα in glial cells. J Virol. 1995;69:5843–5848. doi: 10.1128/jvi.69.9.5843-5848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen N N, Chang C-F, Gallia G L, Kerr D A, Johnson E M, Krachmarov C P, Barr S M, Frisque R J, Bollag B, Khalili K. Cooperative action of cellular proteins YB-1 and Purα with the tumor antigen of the human JC polyomavirus determines their interaction with the viral lytic control element. Proc Natl Acad Sci USA. 1995;92:1087–1091. doi: 10.1073/pnas.92.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chepenik L G, Tretiakova A P, Krachmarov C P, Johnson E M, Khalili K. The single-stranded DNA binding protein, Purα, binds HIV-1 TAR RNA and activates HIV-1 transcription. Gene. 1998;210:37–44. doi: 10.1016/s0378-1119(98)00033-x. [DOI] [PubMed] [Google Scholar]
- 14.Chesters P M, Heritage J, McCance D J. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissue. J Infect Dis. 1983;147:676–684. doi: 10.1093/infdis/147.4.676. [DOI] [PubMed] [Google Scholar]
- 15.Dörries K. Progressive multifocal leukoencephalopathy: analysis of JC virus DNA from brain and kidney tissue. Virus Res. 1984;1:25–38. doi: 10.1016/0168-1702(84)90032-7. [DOI] [PubMed] [Google Scholar]
- 16.Feigenbaum L, Hinrichs S H, Jay G. JC virus and simian virus 40 enhancers and transforming proteins: role in determining tissue specificity and pathogenicity in transgenic mice. J Virol. 1992;66:1176–1182. doi: 10.1128/jvi.66.2.1176-1182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frisque R J, White F A., III . The molecular biology of JC virus, causative agent of progressive multifocal leukoencephalopathy. In: Roos R P, editor. Molecular neurovirology. Totowa, N.J: Humana Press; 1992. pp. 25–158. [Google Scholar]
- 18.Frisque R J, Bream G L, Cannella M T. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallia G L, Safak M, Khalili K. Interaction of single-stranded DNA binding protein, Purα, with human polyomavirus, JCV, early protein, T-antigen. J Biol Chem. 1998;273:32662–32669. doi: 10.1074/jbc.273.49.32662. [DOI] [PubMed] [Google Scholar]
- 20.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham F L, van der Eb A. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–457. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 22.Grinnell B W, Padgett B L, Walker D L. Distribution of nonintegrated DNA from JC papovavirus in organs of patients with progressive multifocal leukoencephalopathy. J Infect Dis. 1983;147:669–675. doi: 10.1093/infdis/147.4.669. [DOI] [PubMed] [Google Scholar]
- 23.Haas S, Haque N S, Beggs A H, Khalili K, Knobler R L, Small J A. Expression of the myelin basic protein gene in transgenic mice expressing human neurotropic virus, JCV, early protein. Virology. 1995;202:89–96. doi: 10.1006/viro.1994.1325. [DOI] [PubMed] [Google Scholar]
- 24.Johnson E M, Chen P L, Krachmarov C P, Barr S M, Kanovsky M, Ma Z W, Lee W H. Association of human Purα with the retinoblastoma protein, Rb, regulates binding to the single-stranded DNA Purα recognition element. J Biol Chem. 1995;41:24352–24360. doi: 10.1074/jbc.270.41.24352. [DOI] [PubMed] [Google Scholar]
- 25.Kenney S, Natarajan V, Strika V, Khoury G, Salzman N P. JC virus enhancer-promoter active in human brain cells. Science. 1984;226:1337–1339. doi: 10.1126/science.6095453. [DOI] [PubMed] [Google Scholar]
- 26.Kerr D, Chang C-F, Chen N N, Gallia G, Raj G, Schwartz B, Khalili K. Transcription of a human neurotropic virus promoter in glial cells: effect of YB-1 on expression of JC viral late gene. J Virol. 1994;68:7637–7643. doi: 10.1128/jvi.68.11.7637-7643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krachmarov C P, Chepenik L G, Barr S M, Khalili K, Johnson E M. Association of the HIV-1 Tat protein with human cellular protein, Purα, enhances binding of Purα to the JC virus Tat-responsive transcriptional control element. Proc Natl Acad Sci USA. 1996;93:14112–14117. doi: 10.1073/pnas.93.24.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W W, Hsiung Y, Wong V, Galvin K, Zhou Y, Shi Y, Lee A S. Suppression of grp78 core promoter element-mediated stress induction by the dbpA and dbpB (YB-1) cold shock domain proteins. Mol Cell Biol. 1997;17:61–68. doi: 10.1128/mcb.17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luckow B, Schultz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490–5498. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch K J, Frisque R J. Identification of critical elements within the JC virus DNA replication origin. J Virol. 1990;64:5812–5822. doi: 10.1128/jvi.64.12.5812-5822.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacDonald G H, Itoh-Lindstrom Y, Ting J P. The transcriptional regulatory protein, YB-1, promotes single-stranded regions in the DRA promoter. J Biol Chem. 1995;270:3527–3533. doi: 10.1074/jbc.270.8.3527. [DOI] [PubMed] [Google Scholar]
- 32.Major E O, Amemiya K, Tornatore C, Houff S A, Berger J R. Pathogenesis and molecular biology of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev. 1992;5:49–73. doi: 10.1128/cmr.5.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCance D J. Persistence of animal and human papovaviruses in renal and nervous tissues. In: Sever J L, Madden D L, editors. Polyomaviruses and human neurological diseases. New York, N.Y: Alan R. Liss; 1983. pp. 343–357. [PubMed] [Google Scholar]
- 34.Mertens P R, Harendza S, Pollock A S, Lovett D H. Glomerular mesangial cell-specific transactivation of matrix metalloproteinase 2 transcription is mediated by YB-1. J Biol Chem. 1997;272:22905–22912. doi: 10.1074/jbc.272.36.22905. [DOI] [PubMed] [Google Scholar]
- 35.Montani V, Taniguchi S I, Shong M, Suzuki K, Ohmori M, Giuliani C, Napolitano G, Saji M, Fiorentino B, Reimold A M, Ting J P, Kohn L D, Singer D S. Major histocompatibility class II HLA-DR alpha gene expression in thyrocytes: counter regulation by the class II transactivator and the thyroid Y box protein. Endocrinology. 1998;139:280–289. doi: 10.1210/endo.139.1.5673. [DOI] [PubMed] [Google Scholar]
- 36.Muralidharan V, Tretiakova A, Steplewski A, Haas S, Johnson E, Khalili K. Evidence for inhibition of MyEF-2 binding to MBP promoter by MEF-1/Pur α. J Cell Biochem. 1997;66:524–531. doi: 10.1002/(sici)1097-4644(19970915)66:4<524::aid-jcb11>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 37.Ohmori M, Shimura H, Shimura Y, Kohn L D. A Y-box protein is a suppressor factor that decreases thyrotropin receptor gene expression. Mol Endocrinol. 1996;10:76–89. doi: 10.1210/mend.10.1.8838147. [DOI] [PubMed] [Google Scholar]
- 38.Raj G, Khalili K. Transcriptional regulation: lessons from the human neurotropic polyomavirus, JCV. Virology. 1995;213:283–291. doi: 10.1006/viro.1995.0001. [DOI] [PubMed] [Google Scholar]
- 39.Raj G, Safak M, MacDonald G, Khalili K. Transcriptional regulation of human polyomavirus JCV: evidence for a functional interaction between RelA (p65) and the Y-box binding protein. J Virol. 1996;70:5944–5953. doi: 10.1128/jvi.70.9.5944-5953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaknovich R, Shue G, Kohtz D S. Conformational activation of a basic helix-loop-helix protein (MyoD1) by the C-terminal region of murine HSP90 (HSP84) Mol Cell Biol. 1992;12:5059–5068. doi: 10.1128/mcb.12.11.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Small J A, Scangos G A, Cork L, Jay G, Khoury G. The early region of human papovaviruses JC induces dysmyelination in transgenic mice. Cell. 1986;46:13–18. doi: 10.1016/0092-8674(86)90855-x. [DOI] [PubMed] [Google Scholar]
- 42.Stein B, Baldwin A S, Jr, Ballard D W, Greene W C, Angel P, Herrlich P. Cross-coupling of the NF-kappa B p65 and Fos/Jun transcription factors produces potentiated function. EMBO J. 1993;12:3879–3891. doi: 10.1002/j.1460-2075.1993.tb06066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein B, Cogswell P C, Baldwin A S., Jr Functional and physical association between NF-kappa B and C/EBP family member: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tada H, Khalili K. A novel sequence-specific DNA-binding protein, LCP-1, interacts with single-stranded DNA and differentially regulates early gene expression of the human neurotropic JC virus. J Virol. 1992;66:6885–6892. doi: 10.1128/jvi.66.12.6885-6892.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tada H, Lashgari M, Khalili K. Regulation of JCVL promoter function: evidence that a pentanucleotide “silencer” repeat sequence, AGGGAAGGGA, down-regulates transcription of the JC virus late promoter. Virology. 1991;180:327–338. doi: 10.1016/0042-6822(91)90037-c. [DOI] [PubMed] [Google Scholar]
- 46.Tada H, Lashgari M, Rappaport J, Khalili K. Cell-type specific expression of JC virus early promoter is determined by positive and negative regulation. J Virol. 1989;63:463–466. doi: 10.1128/jvi.63.1.463-466.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thatikunta P, Sawaya B E, Denisova L, Cole C, Yusibova G, Johnson E M, Khalili K, Amini S. Identification of a cellular protein that binds to Tat-responsive element of TGFβ-1 promoter in glial cells. J Cell Biochem. 1997;67:466–477. [PubMed] [Google Scholar]
- 48.Ting J P, Painter A, Zeleznik-Le N J, MacDonald G, Moore T M, Brown A, Schwartz B D. YB-1 DNA-binding protein represses interferon gamma activation of class II major histocompatibility complex gene. J Exp Med. 1994;179:1605–1611. doi: 10.1084/jem.179.5.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso Y, Hara K, Taguchi F. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocomprised individuals. J Virol. 1990;64:3139–3143. doi: 10.1128/jvi.64.6.3139-3143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zambrano N, DeRenzis S, Minopoli G, Faraonio R, Donini V, Scaloni A, Cimino F, Russo T. DNA-binding protein Purα and transcription factor YY1 function as transcription activators of the neuron-specific FE65 gene promoter. Biochem J. 1997;328:293–300. doi: 10.1042/bj3280293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zoltick P W, Reddy R, Chang C-F, Northrup B, Khalili K, Schwartzman R J. Isolation and characterization of a type II human neurotropic JC virus from a brain biopsy of a patient with progressive multifocal leukoencephalopathy. J Neurovirol. 1995;1:307–315. doi: 10.3109/13550289509114027. [DOI] [PubMed] [Google Scholar]
- 52.Zou Y, Chien K R. EF1A/YB-1 is a component of cardiac HF-1A binding activity and positively regulates transcription of the myosin light-chain 2v gene. Mol Cell Biol. 1995;15:2972–2982. doi: 10.1128/mcb.15.6.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]