Abstract
Stem cells have the unique ability to undergo asymmetric division which produces two daughter cells that are genetically identical, but commit to different cell fates. The loss of this balanced asymmetric outcome can lead to many diseases, including cancer and tissue dystrophy. Understanding this tightly regulated process is crucial in developing methods to treat these abnormalities. Here, we report that during a Drosophila female germline stem cell asymmetric division, the two daughter cells differentially inherit histones at key genes related to either maintaining the stem cell state or promoting differentiation, but not at constitutively active or silenced genes. We combine histone labeling with DNA Oligopaints to distinguish old versus new histones and visualize their inheritance patterns at a single‐gene resolution in asymmetrically dividing cells in vivo. This strategy can be applied to other biological systems involving cell fate change during development or tissue homeostasis in multicellular organisms.
Keywords: asymmetric cell division, Drosophila, epigenetics, FISH, histone
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics
This study shows locally non‐overlapping old versus new histone H3 in the Drosophila female germline. The genomic regions with asymmetric H3 inheritance harbor differentially expressed genes between the two daughter cells derived from asymmetric stem cell divisions.
Introduction
Epigenetic modifications can be maintained through multiple cell divisions, and mis‐regulation can lead to many diseases including cancers, tissue dystrophy, and infertility (Baylin & Ohm, 2006; Feinberg et al, 2006; Chambers et al, 2007; Fredly et al, 2013; Fitzsimons et al, 2014). The known epigenetic mechanisms include DNA methylation, chromatin remodeling, post‐translational modifications (PTMs) of histones, histone variants, and non‐coding RNAs (Kouzarides, 2007; Jin et al, 2011; Lee, 2012). Histone proteins interact with DNA intimately: The (H3‐H4)2 tetramer combines with two H2A/H2B dimers in an octamer structure encircled by DNA to form the basic unit of chromatin. Notably, most histone PTMs that regulate gene expression are found on the N‐termini of H3 and H4 (Kouzarides, 2007; Young et al, 2010; Huang et al, 2014; Allis & Jenuwein, 2016; Janssen et al, 2017). The age of a histone protein also determines the specific PTMs it carries (Xu et al, 2011; Zee et al, 2012; Alabert et al, 2015). The molecular mechanisms by which newly synthesized histones are incorporated onto DNA have been well studied, but little is known about how pre‐existing old histones are re‐incorporated during DNA replication followed by segregation during cell division (Alabert & Groth, 2012; Ahmad & Henikoff, 2018; Serra‐Cardona & Zhang, 2018). Recent studies have shed light on both the molecular mechanisms (Gan et al, 2018; Petryk et al, 2018; Yu et al, 2018) and the cellular responses (Lin et al, 2016; Reveron‐Gomez et al, 2018; Escobar et al, 2019) for old histone recycling in symmetrically dividing cells [reviewed by Escobar et al (2021)]. However, it remains elusive how these processes are regulated in asymmetrically dividing cells, including many types of adult stem cells.
The Drosophila germline stem cell (GSC) systems permit the visualization of asymmetric cell division (ACD) at single‐cell resolution (Fuller & Spradling, 2007). Both male and female GSCs can divide asymmetrically to produce a self‐renewed GSC and a cystoblast (CB, in the female) or a gonialblast (GB, in the male), which subsequently undergoes transit amplification of four mitoses before entering meiosis and terminal differentiation. During ACD of male GSCs, the old H3 is selectively segregated to the renewed GSC, whereas the new H3 is enriched in the GB (Tran et al, 2012). This asymmetry is specific to asymmetrically dividing GSCs and occurs with H3 and H4, but not with H2A (Wooten et al, 2019). Drosophila ovaries consist of 16‐18 ovarioles, each containing a niche with 2‐3 GSCs that can undergo ACD (Deng & Lin, 1997; Lin & Spradling, 1997; Xie & Spradling, 2000). Recent studies revealed that asymmetric microtubules closely interact with asymmetric sister centromeres to establish biased attachment and segregation of sister chromatids in female GSCs (Dattoli et al, 2020), resembling the results in male GSCs (Ranjan et al, 2019). However, key differences exist between these two systems. For example, one GB divides four times to produce 16 spermatogonial cells, all of which subsequently undergo meiosis to produce a total of 64 sperm (Tokuyasu et al, 1977; Hardy et al, 1979; Hardy et al, 1981). By contrast, the CB undergoes four rounds of mitosis but produces two pre‐oocytes and 14 nurse cells. One of the pre‐oocytes is determined as the oocyte and undergoes meiosis, whereas the other pre‐oocyte takes the nurse cell fate. Together, all 15 nurse cells undergo endoreplication and produce mRNAs and proteins to support oogenesis and early embryogenesis (King, 1957; Smith & Orr‐Weaver, 1991). Interestingly, the initial CB division produces two pre‐oocytes, but only one becomes the oocyte (Lin & Spradling, 1995). It was suggested that the spectrosome inheritance may play a role in determining oocyte versus nurse cell fate (Deng & Lin, 1997; de Cuevas & Spradling, 1998).
Here, we show that in both Drosophila female GSC and CB divisions, large chromosomal regions carrying distinct old versus new H3 can be detected. This feature gradually diminishes throughout subsequent mitotic divisions with further cellular differentiation. Further, we combined DNA Oligopaints with a two‐color histone‐labeling system to visualize sequence‐specific histone inheritance patterns in mitotically dividing female germ cells. We found that the two daughter cells produced from a GSC ACD differentially inherit histones at genomic regions of key genes, such as daughters against dpp (dad) for maintaining the stem cell state (Xie & Spradling, 1998; Casanueva & Ferguson, 2004; Kirilly et al, 2011) and bag of marbles (bam) for promoting differentiation (McKearin & Spradling, 1990; McKearin & Ohlstein, 1995). Overall, this study reveals how epigenetic information is inherited through ACD and provides a new technique to simultaneously visualize genetic and epigenetic information.
Results and Discussion
Old versus new H3 displays non‐overlapping patterns in dividing female GSCs, but not in dividing cystocytes
To investigate histone inheritance patterns, we used a heat shock‐controlled, dual‐fluorescent tag‐switching histone transgene driven by GreenEye‐nanos‐Gal4 (Holtzman et al, 2010; Laws et al, 2015), which drives gene expression in early‐stage female germ cells including GSCs (Fig 1A). Using this system, old histones are tagged with eGFP (green fluorescent protein) and new histones with mCherry (red fluorescent protein) (Fig 1B). Since the cell cycle of female GSCs is ~24 h without synchrony, with G2 being the longest phase for 18–20 h (Hsu et al, 2008; Ables & Drummond‐Barbosa, 2013; Hinnant et al, 2017), it is very likely that the heat shock‐induced histone tag switch occurs during G2 phase. A 34‐ to 40‐h recovery after heat shock would ensure that the tag‐switched GSCs finish a complete round of S phase, when mCherry‐tagged new histones are incorporated genome wide. GSCs then advance into the subsequent M phase, when old versus new histone distribution is examined (Fig 1C). As a control for cell cycle‐dependent global incorporation of new canonical histones, similar experiments were performed with an 18‐ to 20‐h recovery (~one cell cycle) after the heat shock. The GSCs at this first M phase were enriched with old H3 but had low to undetectable new H3, confirming that global new histone incorporation requires one complete S phase (Fig EV1A and B).
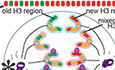
Figure 1. Non‐overlapping old versus new histone H3 distribution patterns in mitotic Drosophila female germline stem cells (GSCs), but not in cystocytes (CCs).
-
AA cartoon and corresponding immunofluorescence image depicting the Drosophila germarium. Terminal filament cells (TFC) and cap cells (CapC) create a niche for female germline stem cells (GSC), which divide asymmetrically to self‐renew and produce a cystoblast (CB). The CB undergoes four mitotic divisions to create differentiating cystocytes (CC). The germline (gray) is supported by somatic (green) escort cells (EC) and follicle stem cells (FSC) that produce follicle cells that surround developing egg chambers. The spectrosome (Spec, red) is a specialized organelle in the early‐stage germline, such as GSCs and CBs. The spectrosome structure is round while the fusome is branched, which occurs over the mitotic divisions in further differentiated germ cells, connecting the CCs within a cyst.
-
BA cartoon detailing how the heat shock‐controlled dual‐color system driven by GreenEye‐nanos‐Gal4 labels pre‐existing (old) histones with eGFP (green fluorescent protein) and newly synthesized (new) histones with mCherry (red fluorescent protein), respectively.
-
CA scheme of recovery time and histone incorporation after heat shock that induces an irreversible genetic switch in the histone transgene.
-
D–FOld (green) versus new (red) histone patterns for H3 (D), H4 (E), and H2A (F) in prometaphase female GSCs marked by anti‐H3S10p (gray).
-
GOld (red) versus new (green) histone patterns for H3Rev in prometaphase female GSCs marked by anti‐H3S10p (gray).
-
H–JOld (green) versus new (red) histone patterns for H3 (H), H4 (I), and H2A (J) in prometaphase female 4‐cell CCs marked by anti‐H3S10p (gray).
-
KOld (red) versus new (green) histone patterns for H3Rev in prometaphase female 4‐cell CC marked by anti‐H3S10p (gray).
-
LQuantification of overlap between old and new histones in late prophase and prometaphase GSCs and CCs using Spearman correlation: The measurement is from a single Z‐slice at the center of each mitotic nucleus, which shows results similar to analyzing every Z‐slice throughout the entire Z‐stack and averaging them (Fig EV2A, see Materials and Methods). Data were collected over 20, 16, 11, and 12 separate experiments for H3, H4, H2A, and H3Rev, respectively. Values are mean ± 95% CI. P‐value: pairwise ANOVA test with Bonferroni correction. ****: P < 0.0001, n.s.: not significant.
Data information: Asterisk: niche. Scale bars: 2 µm. See Dataset EV1 for individual data points for Fig 1L.
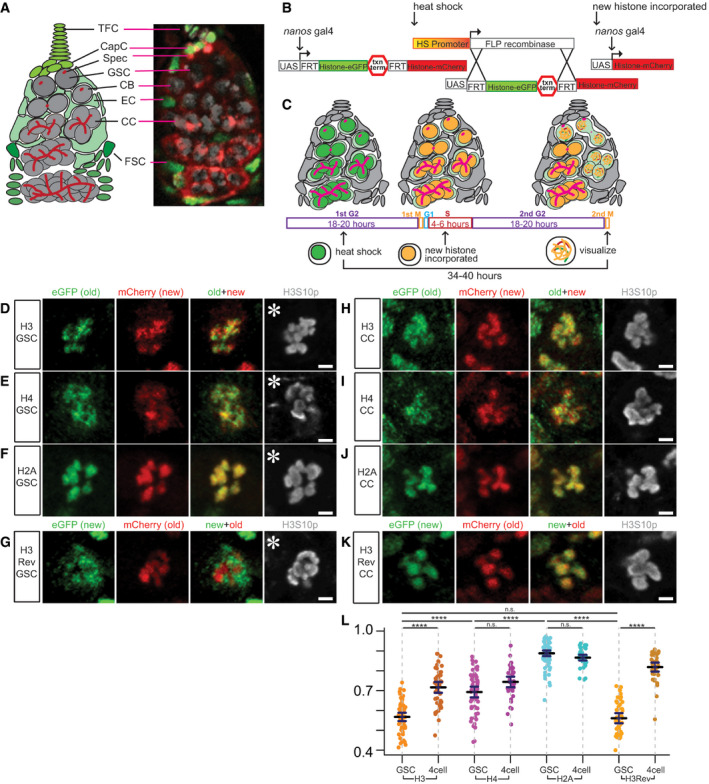
Figure EV1. Old versus new histone H3 distribution patterns in Drosophila female germline stem cells (GSCs) and cystocytes (CCs) during the first mitosis after heat shock‐induced genetic switch.
- Old (green, eGFP) versus new (red, mCherry) histone H3 patterns in a prometaphase female GSC marked by anti‐H3S10p (gray).
- Old (red, mCherry) versus new (green, eGFP) histone H3 patterns for H3Rev in a prophase female GSC marked by anti‐H3S10p (gray).
- Old (green, eGFP) versus new (red, mCherry) histone H3 patterns in a prophase female CC marked by anti‐H3S10p (gray).
- Old (red, mCherry) versus new (green, eGFP) histone H3 patterns for H3Rev in a prometaphase female CC marked by anti‐H3S10p (gray). During the first mitosis after heat shock, the new histone has not yet been globally incorporated into the chromatin, shown as low to undetectable fluorescent signals.
Data information: Scale bars: 2 µm.
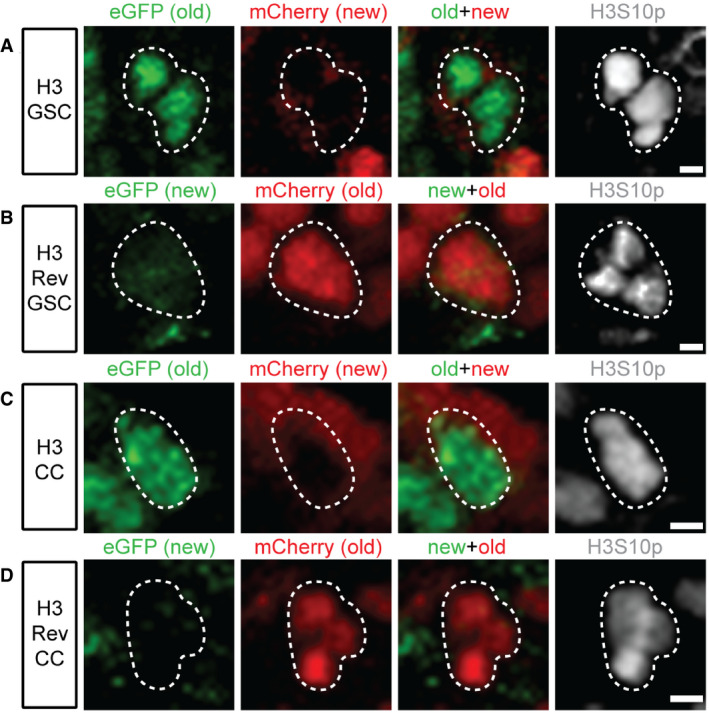
Figure EV2. Quantification of overlap between old and new histones, for H3, H3Rev, H4, H3.3, and H2A in different staged germ cells.
-
ASpearman values of single Z‐slice from 15 individual female GSCs re‐plotted from Fig 1L, compared to the average Spearman values obtained from the same 15 individual female GSCs by quantifying every single slice of the entire Z‐stack followed by averaging them. Data collected over 9 separate experiments. Values are mean ± 95% CI. P‐value: pairwise ANOVA test with Bonferroni correction. n.s.: not significant.
-
B, CQuantification of overlap between old and new H3 in late prophase and prometaphase germ cells at distinct differentiation stages. In these analyses, a slight difference between the H3 (B) and H3Rev (C): From the 4‐cell stage to 8‐cell stage, there is a slight decrease of correlation coefficient in the H3 line, but not the H3Rev line. This difference could arise from subtle protein stability differences between eGFP and mCherry used as the “old” histone tag and/or lower expression levels, as GreenEye‐nanos‐Gal4 expression tapers off in the later staged 4‐cell and 8‐cell. H3: GSC (n = 57), CB (n = 51), 2‐cell (n = 70), 4‐cell (n = 48), and 8‐cell (n = 35). Data collected from 20 separate experiments. H3Rev: GSC (n = 50), CB (n = 50), 2‐cell (n = 54), 4‐cell (n = 39), and 8‐cell (n = 37). Data collected from 12 separate experiments. Values are mean ± 95% CI. P‐value: pairwise ANOVA test with Bonferroni correction. ****: P < 0.0001. ***: P < 0.001. *: P < 0.05. n.s.: not significant.
-
D, EQuantification of overlap between old and new H4 (D) and H3.3 (E) in late prophase and prometaphase cells, which show no significant change among distinct differentiation stages. H4: GSC (n = 60), CB (n = 51), 2‐cell (n = 61), 4‐cell (n = 39), and 8‐cell (n = 42). Data collected from 16 separate experiments. H3.3: GSC (n = 55), CB (n = 46), 2‐cell (n = 52), 4‐cell (n = 51), and 8‐cell (n = 34). Data collected from 7 separate experiments. Values are mean ± 95% CI. P‐value: pairwise ANOVA test with Bonferroni correction. n.s.: not significant.
-
FQuantification of overlap between old and new H2A in late prophase and prometaphase cells at distinct differentiation stages. There is a slight but statistically significant decrease in overlap between GSCs and CBs for H2A, which could be due to many of the correlation values for the H2A in GSCs are compressed at or near the maximum possible correlation value at 1.0. This distribution of data points flattens the measurement variances within one sample and thus when comparing to another sample with more variance above and below the mean value, stringent statistical analysis could identify this as a significant difference between the two samples. H2A: GSC (n = 79), CB (n = 80), 2‐cell (n = 67), 4‐cell (n = 41), and 8‐cell (n = 38). Data collected from 11 separate experiments. Values are mean ± 95% CI. P‐value: pairwise ANOVA test with Bonferroni correction. ****: P < 0.0001. n.s.: not significant.
-
GDirect comparison of overlap between old and new H3 as well as old and new H2A at both GSC and CB stages. Data collected over 12 separate experiments. Values are mean ± 95% CI. P‐value: pairwise ANOVA test with Bonferroni correction. ****: P < 0.0001. n.s.: not significant.
Data information: See Dataset EV5 for individual data points for Fig EV2, EV3, EV4, EV5, EV6.
Using a mitotic marker (anti‐H3S10phos or H3S10p) and chromosomal morphology to identify late prophase and prometaphase GSCs, old and new H3 were found to occupy distinct chromosomal domains shown as separable signals on condensed chromosomes (Fig 1D). Furthermore, mitotic GSCs expressing old H4 (eGFP) and new H4 (mCherry) displayed a moderate separation (Fig 1E). Contrastingly, more overlapping signals were detected in mitotic GSCs where old H2A was labeled with eGFP and new H2A by mCherry (Fig 1F). As a control, we generated a transgenic line in which the old and new fluorescent tags were reversed and found that old H3 (mCherry) and new H3 (eGFP) displayed a similar separation pattern (Fig 1G).
Next, to determine whether the non‐overlapping old versus new H3 distribution patterns have cellular specificity, we imaged and quantified dividing cystocytes (CCs) at the 4‐cell stage (Figs 1A, C and H, and EV1C and D). For old and new H3, late prophase to prometaphase CCs showed a higher degree of overlap (Fig 1H versus Fig 1D). Old and new H4 displayed a pattern that was not obviously different from those in GSCs (Fig 1I versus Fig 1E). Moreover, the degree of overlap between old and new H2A was indistinguishable between GSCs and CCs (Fig 1J versus Fig 1F). Lastly, the H3Rev line also displayed more overlapping pattern between old and new H3 in late prophase to prometaphase CCs compared to GSCs (Fig 1K versus Fig 1G).
In order to quantify these imaging results, Spearman’s rank correlation coefficient (Schober et al, 2018) was measured for old versus new H3, H4, H2A, and H3Rev, respectively (Figs 1L and EV2A–G). In GSCs, old and new H3 consistently showed the lowest correlation coefficients of 0.567 (n = 57) in the H3 line and of 0.561 (n = 50) in the H3Rev line, indicative of the highest separation between these two signals. In contrast, old and new H2A displayed the highest correlation coefficient of 0.888 (n = 79), suggesting the highest overlap between these two signals. Consistent with the moderate separation between old and new H4, its average correlation coefficient of 0.693 (n = 60) is between that of H3 and H2A. Although canonical H3 and H4 form tetramers in vivo, their different behaviors could be due to H4 pairing with both H3 and the histone variant H3.3.
To directly test this, we examined the correlation coefficient of old versus new H3.3 using similar methods. Since new H3.3 is incorporated in a replication‐independent manner (Ahmad & Henikoff, 2002a; Ahmad & Henikoff, 2002b; Tagami et al, 2004), we used the 18‐ to 20‐h recovery after the heat shock induced H3.3‐eGFP to H3.3‐mCherry expression switch and examined the subsequent mitotic GSCs, when an average correlation coefficient of 0.752 was detected for old and new H3.3 (n = 55, Fig EV2E). The correlation coefficient of H4 (0.693) is actually very close to the average of the H3 and H3.3 correlation coefficients at 0.660.
In CCs, late prophase to prometaphase CCs showed an average Spearman correlation coefficient of 0.716 (n = 48) for old and new H3, indicating a significantly higher degree of overlap (Fig 1H versus Fig 1D and L). The H3Rev also had a significantly higher correlation coefficient of 0.818 (n = 39) in late prophase to prometaphase CCs compared to GSCs (Fig 1K versus Fig 1G and L). Old and new H4 displayed an average correlation coefficient of 0.744 (n = 39) in mitotic CCs, which was not significantly different from the correlation coefficient in mitotic GSCs (Fig 1I versus Fig 1E and L). Lastly, the degree of overlap between old and new H2A was indistinguishable between GSCs and CCs, with the latter having an average correlation coefficient of 0.866 (n = 41, Fig 1J versus Fig 1F and L). Together, these data demonstrate that the non‐overlapping old versus new H3 domains are more prominent in GSCs than in CCs.
Old versus new histone distribution patterns reveal CBs are more like GSCs than CCs
Given that old and new H3 appear in separable domains in GSCs but not in CCs (Fig 1), we further examined the histone distribution patterns for H3, H4, H2A, H3.3, and H3Rev in mitotic cells at each stage during transit amplification in the female germline and compared them with their distribution patterns in GSCs. Interestingly, we found that the separable old versus new H3 pattern gradually diminishes as germ cells differentiate (Fig EV2B for H3 and EV2C for H3Rev). Specifically, the old versus new H3 correlation coefficient does not significantly differ between GSCs and CBs in both H3 lines, suggesting that this separation is independent of the fluorescence tag. Additionally, both lines display a significantly increased correlation coefficient from the early‐staged GSC and CB cells to the subsequent 2‐cell and 4‐cell stages, suggesting increasing overlap between old and new H3 as germ cells differentiate. Furthermore, this gradual change of the old versus new histone correlation coefficient during germline differentiation is specific to H3 (Fig EV2B and C). Other histones, including H4, H3.3, and H2A (Fig EV2, EV3, EV4, EV5, EV6), showed no significant differences in the old versus new histone overlap between each consecutive stage, although there is a statistically significant decrease of H2A from GSCs to CBs (Fig EV2F and legend for possible reasons). However, the correlation coefficients for H2A in both GSCs and CBs were significantly higher than that for H3, respectively (Fig EV2G). We also found that in early prophase GSCs old H3 is more associated with H3S10p when compared to new H3, and this was not the case for association of old versus new H2A with H3S10p (Fig EV3, EV4, EV5, EV6). These imaging results are consistent with previous biochemistry work showing higher levels of H3S10 phosphorylation associated with old H3 in early mitotic cells (Lin et al, 2016).
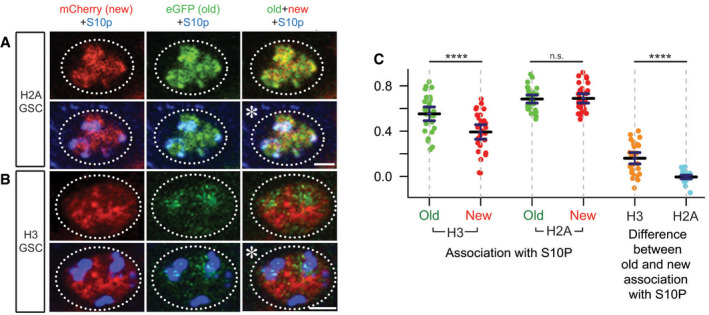
Figure EV3. H3S10p association with old versus new H3 and H2A histones in prophase GSCs.
-
A, BOld (green) versus new (red) histone patterns for H2A (A) and H3 (B) in early prophase female GSCs marked by anti‐H3S10p (blue). Asterisk: niche. Scale bars: 2 µm.
-
CSpearman correlation quantification of overlap between old histone signal and H3S10p (green dots) and new histone signal and H3S10p (red dots) shows a significant difference in H3 GSCs (n = 30, mean = 0.553 for old and 0.392 for new) but not H2A GSCs (n = 31, mean = 0.684 for old and 0.689 for new). For each GSC, the Spearman value for old histone associated with H3S10p was subtracted from the Spearman value for new histone associated with H3S10p to quantify the magnitude of difference for each individual cell (right side). There was a significant difference in the amount of old versus new H3 associated with H3S10p (orange dots, mean = 0.160) compared to the amount of old versus new H2A associated with H3S10p (blue dots, mean = −0.006). Data collected over 7 separate experiments. Values are mean ± 95% CI. P‐value: pairwise ANOVA test with Bonferroni correction. ****P < 0.0001. n.s.: not significant. See Dataset EV6 for individual data points.
Figure EV4. The inheritance patterns of old versus new histone variant H3.3 in GSCs and CBs.
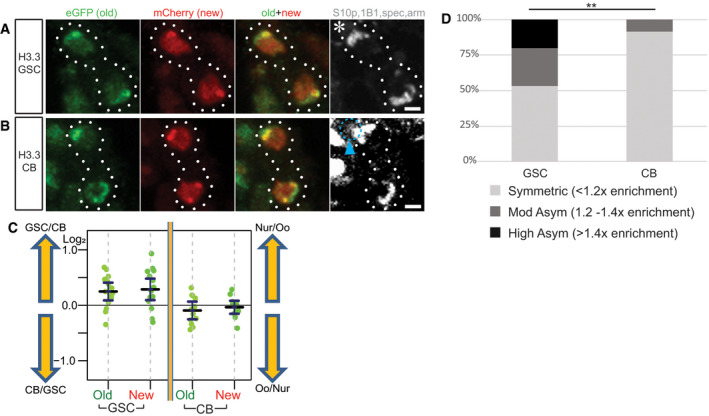
-
A, BTelophase cells marked by H3S10p (gray) show overlapping old (green) and new (red) histone variant H3.3 patterns during the GSC (A) and CB (B) divisions. Asterisk: niche. Arrowhead (cyan): biased spec inheritance. Scale bars: 2 µm.
-
CQuantification of log2 ratios of total old and new H3.3 histone inherited by each future daughter cell of the GSC and CB divisions, where a value of 0 is equal inheritance at exactly a 1:1 ratio. Data gathered over 7 separate experiments for GSCs and 9 separate experiments for CBs. Values are mean ± 95% CI. See Dataset EV7 for individual data points.
-
DSummary of total old histone variant H3.3 inherited in GSCs and CBs: < 1.2‐fold is symmetric, 1.2‐fold to 1.4‐fold is moderately asymmetric, and > 1.4‐fold is highly asymmetric, using previously established criteria for the degree of histone or histone variant asymmetry (Ranjan et al, 2019). **: P < 0.01, based on pairwise ANOVA test with Bonferroni correction.
Figure EV5. Histone transgene expression in bam mutant ovaries and RNA FISH results show bgcn gene is actively transcribed in the early‐stage female germline, including both GSCs and CBs.
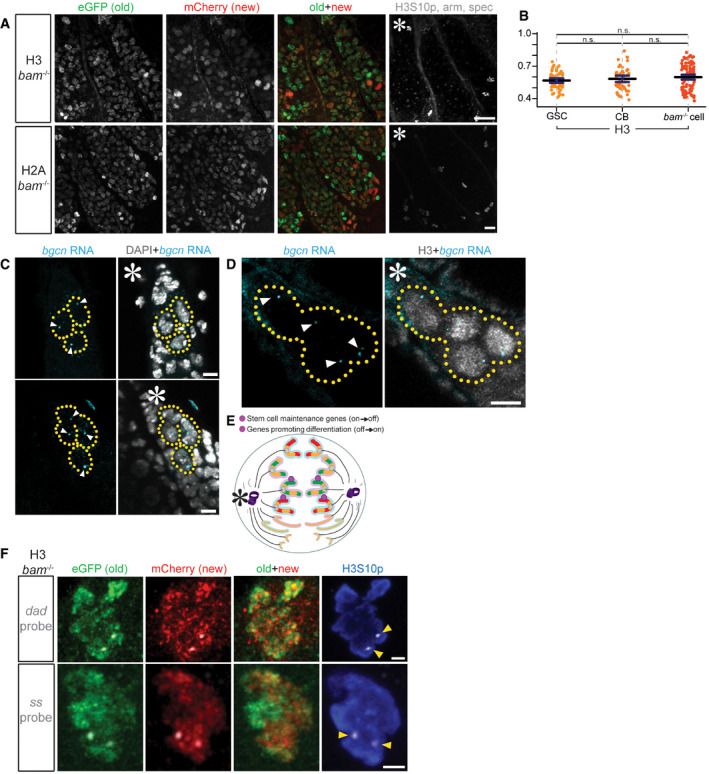
- Old (eGFP) and new (mCherry) H3 or H2A expression in bam mutant ovaries. Scale bars: 20 µm. Asterisk: niche.
- Spearman correlation coefficients are re‐plotted from Figs EV2B and 3D for direct comparison of the overlap between old and new H3 in WT GSCs, WT CBs, and bam mutant GSC‐like germ cells. Data collected over 12 separate experiments. Values are mean ± 95% CI. P‐value: pairwise ANOVA test with Bonferroni correction. n.s.: not significant. See Dataset EV8 for individual data points.
- RNA FISH using probes targeting nascent bgcn RNA (cyan) reveals active transcription in both GSCs and CBs, and nuclei are marked by DAPI (gray). Asterisk: niche. Scale bars: 5 µm.
- RNA IF‐FISH using probes targeting nascent bgcn RNA (cyan) reveals active transcription in both GSCs and CBs, and nuclei are marked by histone H3 (gray) driven by the GreenEye‐nanos‐Gal4 promoter. Asterisk: niche. Arrowheads: actively transcribed nascent bgcn. Scale bars: 5 µm. The bgcn transcript was detected in both GSCs and CBs in 100% of the germaria (n = 17).
- Probes recognize genes (magenta) that change their epigenetic state in a 2:2 ratio with biased old:new H3‐enriched regions. In this instance, both sister chromatids for maternal or paternal chromosome have an “agreement” on old (green at the maternal chromosomes indicated by the pink outline) versus new (red at the paternal chromosomes indicated by the blue outline) H3.
- Two examples showing probe signals condensed into 2 foci (gray) amid old (green) versus new (red) H3 distribution for dad gene (top) and ss gene (bottom) in bam mutant GSC‐like cells at prometaphase marked by anti‐H3S10p (blue). Yellow arrowheads: FISH probe signal. Scale bars: 1 µm.
Figure EV6. Oligopaint IF‐FISH reveals differential old versus new H3 inheritance at key genes for maintaining stem cell fate or promoting differentiation in WT female GSCs.
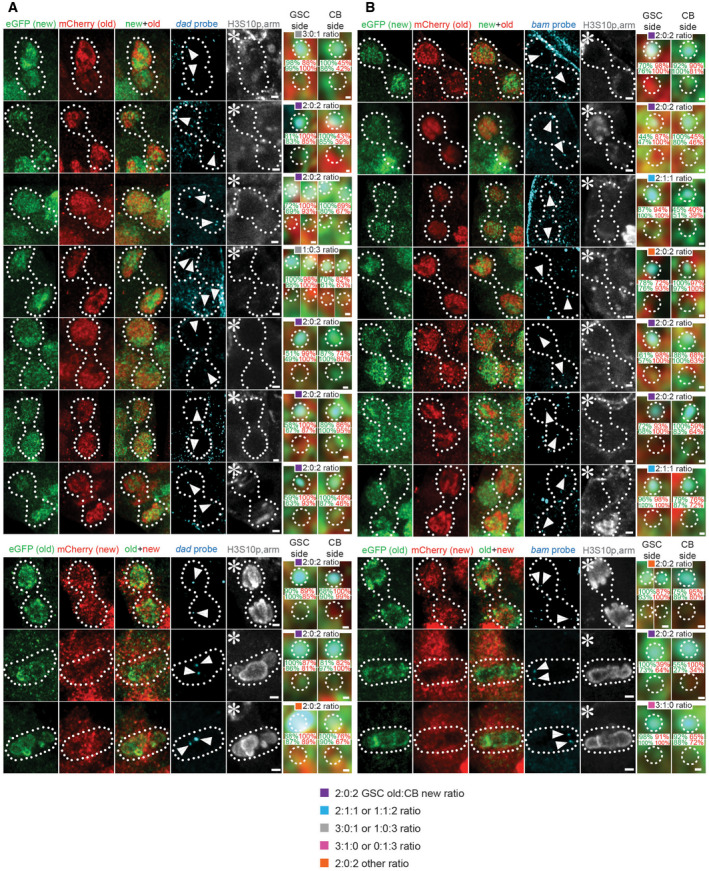
-
A, B, TopOld (red) versus new (green) histone H3Rev patterns with a single genomic locus labeled with fluorescent probes (cyan) for dad (A) and bam (B) in WT telophase female GSCs, marked by anti‐H3S10ph (gray).
-
A, B, bottomOld (green) versus new (red) histone H3 inheritance patterns with single genes labeled with fluorescent probes (cyan) for dad (A) and bam (B) in WT telophase and metaphase female GSCs, marked by anti‐H3S10ph (gray).
Data information: Zoomed‐in images of each probe are displayed to the right of each figure panel and include the ratio probe association with old versus new histone, and the percentage of red versus green signal strength within each probe signal. Asterisk: niche. Arrowheads: probe signal. Scale bars: 1 µm. Zoomed‐in scale bars: 0.1 µm. See Dataset EV4 telophase cells for individual data points.
Dividing female GSCs and CBs shows globally symmetric but locally asymmetric histone segregation patterns
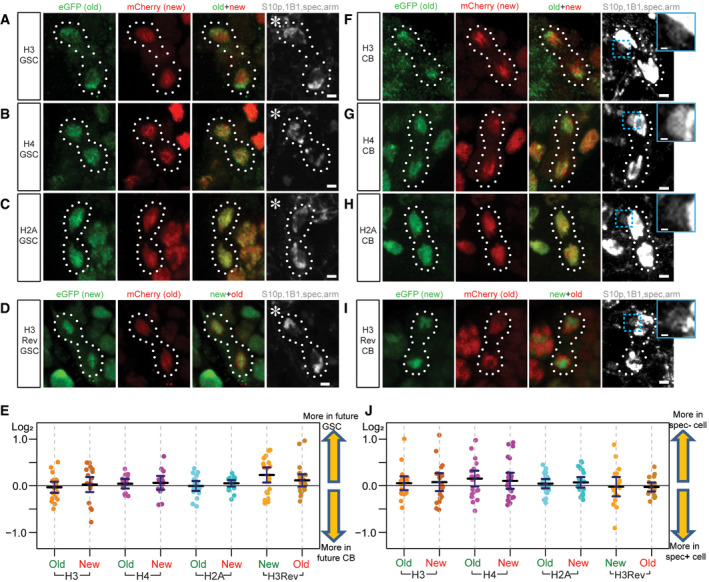
Next, we studied old versus new H3 segregation patterns in anaphase and telophase GSCs and CBs. Using the H3S10p marker and chromosomal morphology to identify anaphase and telophase GSCs, we found that the average ratios of old and new histone H3 (n = 21), H4 (n = 15), H2A (n = 19), and the H3Rev (n = 24) were nearly equal between the segregated sister chromatids in asymmetrically dividing GSCs (Fig 2A–D and E). Given the intriguing similarity between GSCs and CBs in old versus new H3 distribution patterns at prophase and prometaphase (Fig EV2B and C), we next examined old versus new H3 segregation patterns in anaphase and telophase CBs. It was speculated that the daughter cell resulting from CB division that inherits more spectrosome eventually differentiates into the oocyte, while the daughter cell inheriting less spectrosome commits to the nurse cell fate (Lin & Spradling, 1995; de Cuevas & Spradling, 1998). By using both anti‐α‐spectrin and anti‐1B1 to label spectrosome, we distinguished the putative pre‐oocyte versus pre‐nurse cell between the two daughter cells derived from CB division. The average ratios of old and new histone H3 (n = 19), H4 (n = 24), H2A (n = 23), and the H3Rev (n = 20) were nearly equal between the segregated sister chromatids in dividing CBs (Fig 2F–I and J). However, old and new H3 displayed non‐overlapping patterns in anaphase and telophase GSCs and CBs (Fig 2A, D, F and I), compared to the overlapping old and new H2A patterns (Fig 2C and H). Therefore, old and new H3 are inherited in a globally symmetric manner but show locally non‐overlapping patterns during both GSC and CB divisions.
Figure 2. Mitotic GSCs and CBs exhibit non‐overlapping old versus new histone H3 distribution patterns, but overall symmetric old and new histone segregation patterns.
-
A–CTelophase GSCs marked by H3S10p (gray) expressing old (green) and new (red) histone H3 show non‐overlapping patterns (A). Telophase GSCs expressing old (green) and new (red) histone H4 show moderate levels of overlap (B). GSCs expressing old (green) and new (red) histone H2A show high levels of overlap (C).
-
DA H3Rev control where the old and new fluorescent tags have been switched shows the non‐overlapping patterns of old (red) versus new (green) histone H3 in telophase GSCs marked by H3S10p (gray).
-
EQuantification of log2 ratios of total old and new histone inherited by each future daughter cell of the GSC division, where a value of 0 is equal inheritance at exactly a 1:1 ratio. Data gathered over 16 separate experiments for H3 GSCs. Data gathered over 9 separate experiments for H4 GSCs, H2A GSCs, and H3Rev GSCs. Values are mean ± 95% CI.
-
F–HTelophase CBs marked by H3S10p (gray) expressing old (green) and new (red) histone H3 also show non‐overlapping patterns (F). Telophase CBs expressing old (green) and new (red) histone H4 (G) and H2A (H), which depict more overlap like their patterns in GSCs.
-
IA H3Rev control with switched old and new fluorescent tags shows the non‐overlapping old (red) versus new (green) histone H3 patterns in telophase CBs marked by H3S10p (gray).
-
JQuantification of log2 ratios of total old and new histone inherited by each future daughter cell of the CB division, where a value of 0 is equal inheritance at exactly a 1:1 ratio. Data gathered over 12 separate experiments for H3 CBs and H4 CBs. Data gathered over 9 separate experiments for H2A CBs and H3Rev CBs. Values are mean ± 95% CI.
Data information: Asterisk: niche. Scale bars: 2 µm. Dashed box (cyan): biased spec inheritance. Zoomed‐in images of biased spectrosome inheritance (gray) are displayed to the right of panels (F‐I). Scale bars: 0.5 µm. See Dataset EV2 for individual data points for Fig 2E and J.
Next, we studied old and new H3.3 patterns (see previous discussion on H3.3, Fig EV4, EV5, EV6). Interestingly, old H3.3 showed significantly higher ratios of asymmetric inheritance patterns in GSC divisions (n = 15) compared to CB divisions (n = 12, Fig EV4D). During GSC ACD, the sister chromatids segregated toward the GSC side inherited more of both old and new H3.3 than the sister chromatids segregated toward the CB side. This biased segregation pattern was not detected in the CB division (Fig EV4C), indicating that the future GSC has more total H3.3 than the future CB during ACD of GSCs. Given the transcription‐dependent incorporation mode of H3.3 (Ahmad & Henikoff, 2002b; Tagami et al, 2004), it is possible that this biased H3.3 inheritance contributes to or is correlated with higher transcriptional activities in GSCs compared to CBs. These results are consistent with previous reports that stem cells might maintain an overall more open chromatin state. During differentiation, cells begin to “lock down” their chromatin toward a more restricted cell fate (Turner, 2008; Gaspar‐Maia et al, 2011; Golkaram et al, 2017; Yu et al, 2017).
GSC‐like bam mutant cells recapitulate the non‐overlapping old versus new H3 distribution patterns observed in wild‐type GSCs
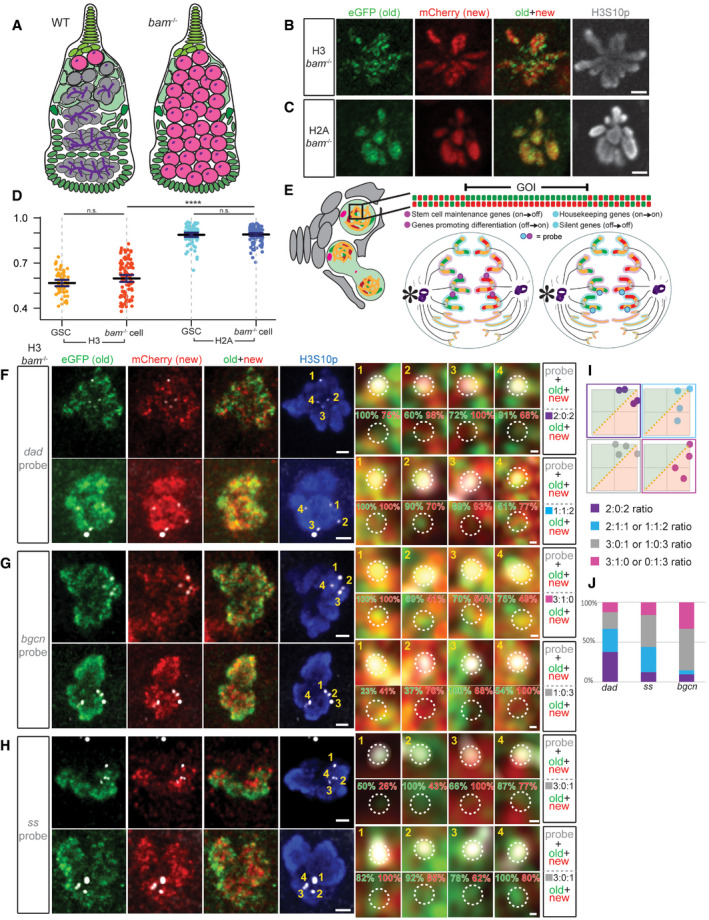
In wild‐type (WT) female ovaries, there are only 2–3 GSCs per ovariole with ~15‐min M phase in a ~24‐h cell cycle (Xie & Spradling, 2000; Fuller & Spradling, 2007; Ables & Drummond‐Barbosa, 2013; Hinnant et al, 2017). To obtain more GSC‐like cells, we used bag‐of‐marbles (bam) mutant ovaries. The bam gene is required for female GSC differentiation and bam mutant ovaries are enriched with undifferentiated GSC‐like cells [Fig 3A, (McKearin & Spradling, 1990; McKearin & Ohlstein, 1995; Chen & McKearin, 2003; Li et al, 2009)].
Figure 3. bam mutant germ cells recapitulate the separable old versus new H3 distribution in WT GSCs, and Oligopaint IF‐FISH reveals distinct old versus new H3 at key genes.
-
AA cartoon depicting a WT germarium with differentiating germline at different stages (gray) compared with a bam mutant ovariole filled with GSC‐like cells (pink) without further differentiated germ cells. Undifferentiated cells can be identified by the presence of dotted spectrosome versus branched fusome structure (purple).
-
B, CNon‐overlapping patterns of old (green) versus new (red) histone H3 (B) and overlapping patterns for H2A (C) in mitotic prometaphase bam mutant cells marked by anti‐H3S10p (gray), similar to WT GSCs. Scale bars: 2 µm.
-
DQuantification of the overlap between old and new histones in late prophase and prometaphase bam mutant cells using Spearman correlation, also plotted with and compared to the WT GSC values from Fig 1L. Data gathered over 4 separate experiments. Values are mean ± 95% CI. P‐value: pairwise ANOVA test with Bonferroni correction. ****: P < 0.0001. n.s.: not significant.
-
EA cartoon depicting the Oligopaint IF‐FISH scheme to identify old versus new histone inheritance at single genomic loci in mitotic cells. (Left) A germarium containing a GSC in prometaphase, where the 3D chromatin structure may disguise local asymmetries at genomic regions of interest. (Top) A linearized gene of interest (GOI) displays local asymmetries. (Bottom) Two anaphase cells depict potential Oligopaint IF‐FISH results. Maternal chromosomes are outlined in pink and paternal ones in blue. (Bottom Left) Probes recognize genes (magenta) that change their epigenetic state in a 2:2 ratio with biased old:new H3‐enriched regions. In this instance, each duplicated sister chromatid from either maternal or paternal chromosome has an “agreement” on old (green, inherited by GSC, niche is indicated by an asterisk) versus new (red, inherited by the CB) H3. (Bottom Right) Probes recognize genes (cyan) that maintain their epigenetic state associated with more symmetric regions (yellow) that do not have a strong bias for either old or new H3.
-
F–HExamples showing old (green) versus new (red) H3 distribution patterns and percentage of signal strength at a single genomic locus labeled with fluorescent probes (gray) for the dad gene (F), bgcn gene (G), and ss gene (H) in bam mutant GSC‐like cells at prometaphase marked by anti‐H3S10p (blue). Scale bars: 1 µm. Zoomed‐in images of each probe are displayed to the right of each figure panel. Scale bars: 0.1 µm.
-
IExamples of scatter plots showing probes' association with normalized old H3 (green, above X = Y line) versus new H3 (red, below X = Y line) and the (i):(ii):(iii) ratios that result from them, where (i) is the number of FISH signals more associated with old histone, (ii) is the number of FISH signals that had equal association with both old and new histones, and (iii) is the number of FISH signals more associated with new histone.
-
JQuantification of the probe association ratios with old versus new H3 for each candidate gene in bam mutant germ cells. Data gathered over 4 separate experiments for dad, 3 for bgcn, and 2 for ss.
Data information: See Dataset EV3a for individual data points for Fig 3D and Dataset EV3b for individual data points for Fig 3J.
We performed similar experiments using the dual‐color H3 and H2A transgenes in bam mutant ovaries [bam Δ86 /bam 1 (McKearin & Spradling, 1990; Bopp et al, 1993), Fig EV5A] and compared the results with those from WT GSCs. Similar to WT GSCs, GSC‐like bam cells displayed non‐overlapping old versus new H3 patterns (Fig 3B), but largely overlapping old versus new H2A patterns (Fig 3C), at late prophase to prometaphase. Quantification showed an average of 0.598 Spearman’s rank correlation coefficient for H3 (n = 98) and 0.890 for H2A (n = 116). Both coefficient values for H3 and H2A in GSC‐like bam cells were indistinguishable from those in WT GSCs (Fig 3D). Furthermore, GSC‐like bam cells showed significantly different correlation coefficients between old versus new H3 and old versus new H2A, similar to the significant difference detected in WT GSCs (Fig 3D). Together, these results suggest that the non‐overlapping old versus new H3 distribution in WT female GSCs are recapitulated in GSC‐like bam cells, which greatly facilitate higher throughput image acquisition and data analyses.
In summary, WT GSCs, WT CBs, and GSC‐like bam cells all displayed similar non‐overlapping old versus new H3 distribution patterns (Fig EV5B), which gradually diminished in correlation with germline differentiation after the CB stage (Fig EV2B and C). In accordance with this observation, it has been shown that other cellular features, such as cell cycle progression and spectrosome/fusome morphology, also change gradually during female germline differentiation (Lin & Spradling, 1995; Hinnant et al, 2017).
Differential H3 distribution at key genes for maintaining stem cell fate in bam cells
The non‐overlapping old versus new H3 distribution patterns in WT GSCs and GSC‐like bam cells introduce a possibility that old and new H3 could be differentially distributed at specific genomic regions. We hypothesize that these regions could harbor genes that change expression patterns during germline differentiation, such as genes important for maintaining the stem cell state (i.e., stemness genes) or for promoting cellular differentiation (i.e., differentiation genes). For example, the self‐renewed GSC could inherit old histones with PTMs to maintain active expression of the stemness genes, while the differentiating daughter cell could carry new histones without these PTMs (or with different PTMs) to turn off expression of the stemness genes. Other genes such as constitutively active genes or non‐germline lineage genes might display more symmetric histone inheritance patterns, which would allow both daughter cells to achieve similar gene expression patterns after ACD.
To visualize old versus new histones at specific genomic loci and between sister chromatids, we combined the dual‐color histone‐labeling scheme with DNA Oligopaint FISH technology [Fig 3E (Beliveau et al, 2012; Joyce et al, 2013)]. Our candidate target genes included the stemness gene daughters against dpp (dad) (Xie & Spradling, 1998; Casanueva & Ferguson, 2004; Kirilly et al, 2011), the germline differentiation gene bam, an actively transcribed gene in both GSCs and CBs called benign gonial cell neoplasm [bgcn, Fig EV5C and D (Gonczy et al, 1997; Lavoie et al, 1999)], and a neuronal gene that is completely silenced in the entire female germline lineage, spineless (ss) (Struhl, 1982; Wernet et al, 2006; Gan et al, 2010). Among these candidate genes, dad, bam, and ss are located at the right arm of the 3rd chromosome (3R), and bgcn is at the right arm of the 2nd chromosome (2R), all of which are autosomes.
Since the non‐overlapping old versus new H3 distribution pattern in GSC‐like bam mutant cells resembled that in WT GSCs (Fig 3B and D), we first used bam mutant ovaries to obtain a higher number of cells where histone distribution patterns at candidate genomic regions could be examined. We imaged mitotic bam cells using high‐resolution Airyscan microscopy (Sivaguru et al, 2018) to resolve the candidate genomic regions into four fluorescence in situ hybridization (FISH) “dots”, two of each corresponding to the replicated maternal and paternal chromosomes, respectively. We hypothesize if histones are differentially distributed at stemness and differentiation genes, we would detect two FISH signals associated with old H3 and two FISH signals associated with new H3 at those candidate gene loci. However, if the histones are symmetrically inherited, we would not be able to detect this pattern.
Using Oligopaint IF‐FISH, we first examined each candidate gene locus in GSC‐like bam cells at late prophase when the four distinct FISH dots were most likely detectable. The mitotic chromosomes were co‐immunostained with antibodies against H3S10p, GFP (old H3), and mCherry (new H3). As shown previously (Fig 3B), old and new H3 displayed non‐overlapping patterns in mitotic bam mutant cells (Fig 3F–H). We then asked whether the resolved four FISH signals show any distinct patterns associated with old versus new H3. We reasoned if two dots displayed an association with old H3 and the other two were enriched with new H3, there were two possibilities: (i) One sister chromatid from the maternal chromosome and one sister chromatid from the paternal chromosome are associated with old H3, while the others are associated with new H3 (left panel in Fig 3E); or (ii) both sister chromatids from either maternal or paternal chromosome are associated with old H3, and both of the remaining chromatids are associated with new H3 (Fig EV5E). It is worth noting that homologous chromosomes are not paired in the early‐stage female germ cells, unlike somatic cells in Drosophila (Joyce et al, 2013).
To examine the association of FISH signals with old versus new histones, we measured and normalized the mean intensity values for each channel (Materials and Methods). We then plotted the normalized mean intensity values in a two‐dimensional (2D) scatter plot and observed the resulting i:ii:iii ratios (Fig 3I). We classified all scatter plot patterns with (i) denoting the number of FISH signals more associated with the old histone (above the X = Y line), (ii) denoting the number of FISH signals that with an equal association between old and new histones (on the X = Y line), and (iii) denoting the number of FISH signals more associated with new histone (below the X = Y line). For the stemness dad gene (Fig 3F), ~38% of the bam germ cells showed two dots associated with the old histone and two with the new histone (a 2:0:2 ratio, n = 24, quantified in Fig 3J). In contrast, when probing for bgcn (Fig 3G, n = 21) and ss (Fig 3H, n = 25), fewer cells (9.5 and 12%, respectively) displayed this 2:0:2 ratio (Fig 3J). These results suggest that the 2:0:2 histone association pattern is more frequently detected for the gene that must change expression between the two daughter cells, such as dad. For genes that do not change expression between the two daughter cells, such as ss (“off” in both cells) and bgcn (“on” in both cells), this ratio is less frequent (Fig 3J).
Notably, a caveat for bam mutant cells is that the genetic lesions at the bam genomic locus might complicate FISH probing at this region. Moreover, in bam mutant ovaries, no ACD occurs because all germ cells overproliferate as GSC‐like cells. Therefore, to study histone inheritance patterns during ACD, we must examine Oligopaint IF‐FISH signals with old versus new H3 segregation patterns in asymmetrically dividing WT GSCs. Because very few GSC‐like bam mutant cells displayed the 2:0:2 ratio for the constitutively active bgcn gene and the silent ss gene, we next focused on differentially expressed dad gene and bam gene in WT GSCs.
Asymmetric H3 segregation at key genes for maintaining stem cell fate or for promoting differentiation during ACD of WT GSCs
In mitotic WT GSCs, we detected non‐overlapping old versus new H3 distribution patterns (Figs 1D, G and L, and 2A and D). Here, using Oligopaint IF‐FISH we visualized candidate genes with old versus new histone signals. Notably, FISH signals from duplicated sister chromatids of the maternal and paternal chromosomes often result in two adjacent dots. In prophase GSCs, this is likely due to the action of cohesin proteins between sister chromatids (Brooker & Berkowitz, 2014), and this phenomenon was also observed in bam mutant cells (Fig EV5F). In anaphase and telophase WT GSCs, the adjacent two dots result from co‐segregating maternal and paternal chromosomes via microtubule activities from the mitotic spindle. Co‐segregation of maternal and paternal autosomes has been previously reported in ACD of Drosophila male GSCs (Yadlapalli & Yamashita, 2013). In cases where the dots were adjacent with each other, two measurements were taken in the two Z planes with the highest FISH signals, for a total of four data points with two at each set of segregated sister chromatids.
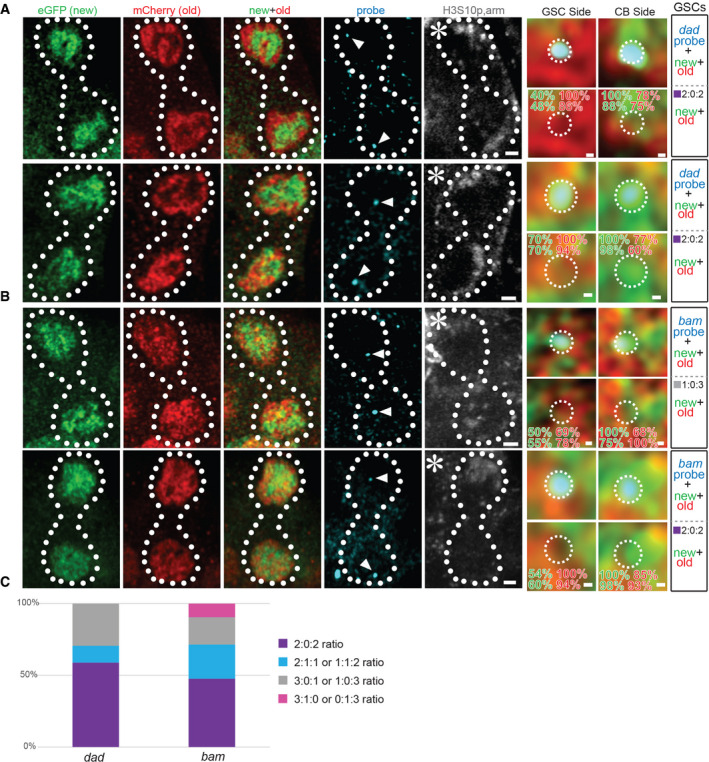
Next, we examined the segregation patterns of old versus new H3 at the genomic loci of the stemness dad gene and the differentiation bam gene in anaphase or telophase WT GSCs, when segregated sister chromatids can be readily distinguished (Fig 4A: dad gene locus, Fig 4B: bam gene locus). When probing the dad locus, the future GSC side had predominantly old H3, while the CB side mainly inherited new H3 at ~75% of the time (n = 12). A similar pattern was observed in ~67% of GSCs at the bam locus (n = 12). Specifically, out of the 10 telophase cells displaying a 2:0:2 ratio at the dad locus (n = 12), 90% showed old H3 on GSC side and new H3 on CB side. Additionally, out of the 10 telophase cells displaying a 2:0:2 ratio at the bam locus (n = 12), 80% displayed old H3 on GSC side and new H3 on CB side. Considering the total examined WT mitotic GSCs (including prometphase GSCs), ~59% of the dad gene locus (n = 17) and ~48% of the bam gene locus (n = 21) displayed two FISH dots associated with old H3 and two FISH dots with new H3, respectively (Fig 4C). If considering both the 2:0:2 and the 2:1:1 patterns, approximately 71% of the dad and 71% of the bam FISH signals fell in these two categories. Collectively, these data indicate a differential association of old versus new H3 at specific gene loci, followed by the preferential segregation of old H3‐enriched sister chromatids toward the GSC side and new H3‐enriched ones toward the CB side.
Figure 4. Oligopaint IF‐FISH reveals differential old versus new H3 inheritance at key genes for maintaining stem cell fate or promoting differentiation in WT female GSCs.
-
A, BOld (red) versus new (green) histone H3 distribution patterns and percentage of signal strength with a single genomic locus labeled with fluorescent probes (cyan) for dad (A) and bam (B) in WT telophase female GSCs, marked by anti‐H3S10p (gray).
-
CQuantification of the probe association ratios with old versus new H3 for each candidate gene in WT female GSCs. Data gathered over 8 separate experiments.
Data information: Asterisk: niche. Arrowheads: probe signal. Scale bars: 1 µm. Zoomed‐in images of each probe are displayed to the right of each figure panel. Scale bars: 0.1 µm. See Dataset EV4 for individual data points for Fig 4C.
In summary, in this study we detected differentially distributed old versus new H3 in prophase and prometaphase early‐stage female germ cells. This differential distribution has molecular specificity for H3 but for H2A, as well as cellular specificity for GSCs and CBs but not for further differentiated CCs. Using Oligopaint IF‐FISH, this differential distribution is likely associated with the key genes either for maintaining the stem cell state or for promoting differentiation. Overall, we have shown that old versus new H3 asymmetry could be global as in the Drosophila male GSCs (Tran et al, 2012) and Drosophila intestinal stem cells (preprint: Zion & Chen, 2020) or local as in the Drosophila female GSCs and Wnt3a‐induced mouse embryonic stem cells (Ma et al, 2020). Different stem cell systems have distinct cellular differentiation programs and varied amount of changes in gene expression profiles. Perhaps these differences could be related to the degree of histone asymmetry displayed during ACD in different systems. In the future, the combination of differential labeling of histones with Oligopaint IF‐FISH, which can be adapted and applied to other systems, will provide valuable insight into how ACD regulates cell fate decisions in multicellular organisms.
Materials and Methods
Reagents and Tools table
| Product name | Vendor | Catalog Number / Size |
|---|---|---|
| Bovine serum albumin (BSA) | Cell Signaling Technology | 9998S / 50 g |
| Dextran Sulfate sodium salt (from Leuconostoc spp.) | Sigma‐Aldrich | D8787 / 5 g |
| Formaldehyde, 16% Solution (w/v), Methanol‐free | ThermoFisher Scientific | 28906 / 10 × 1 ml ampules |
| **Formamide, ReagentPlus, ≥ 99.0% (GC) | Sigma‐Aldrich | F7503 / 250 ml |
| Heptane, ReagentPlus, 99% | Sigma‐Aldrich | H2198 / 1 l |
| Normal Goat Serum (NGS) | Cell Signaling Technology | 5425S / 10 ml |
| NP‐40 (Tergitol), 70% in H2O | Sigma‐Aldrich | NP40S / 100 ml |
| Phosphate‐buffered saline (PBS) | Quality Biological | 119‐069‐131 / 1000 ml |
| Polyethylene glycol (PEG‐8000) | Promega | V3011 / 500 g |
| Potassium chloride | Sigma‐Aldrich | PHR1329 / 5 g |
| Potassium phosphate monobasic | Sigma‐Aldrich | P0662 / 25 g |
| Schneider’s Insect Medium, 1× [+] L‐Glutamine | Life Technologies (Gibco) | 21720‐024 / 500 ml |
| Sodium chloride | ThermoFisher Scientific | BP358‐1 / 1 kg |
| Sodium phosphate monobasic monohydrate BioXtra for molecular biology, ≥ 99.5% (T) | Sigma‐Aldrich | 71507‐250G / 250 g |
| SSC, 20× | Quality Biological | 351‐003‐101 / 500 ml |
| TritonX‐100 | Sigma‐Aldrich | T8787 / 100 ml |
| Tween20 | Sigma‐Aldrich | P9416 / 50 ml |
Methods and Protocols
Fly strains and husbandry
Fly stocks were raised using standard Bloomington medium at 25°C. The following fly stocks were used: Heatshock‐flippase on the X chromosome (Bloomington Stock Center BL‐26902), GreenEye‐nanos‐Gal4 on the 2nd chromosome [Bloomington Stock Center BL‐32179, from Dr. Daniela Drummond‐Barbosa, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA (Holtzman et al, 2010)], UASp‐FRT‐H2A‐eGFP‐PolyA‐FRT‐H2A‐mCherry on the 3rd chromosome, UASp‐FRT‐H2A‐eGFP‐PolyA‐FRT‐H2A‐mCherry on the 2nd chromosome, UASp‐FRT‐H3‐eGFP‐PolyA‐FRT‐H3‐mCherry on the 2nd chromosome, UASp‐FRT‐H4‐eGFP‐PolyA‐FRT‐H4‐mCherry (2nd chromosome) (Wooten et al, 2019), UASp‐FRT‐H3‐mCherry‐PolyA‐FRT‐H3‐eGFP on the 3rd chromosome (Ranjan et al, 2019), nanos‐Gal4 (Van Doren et al, 1998) and bamΔ86 (Bopp et al, 1993) combined on the 3rd chromosome (from Dr. Mark Van Doren, Johns Hopkins University, Baltimore, Maryland, USA), and bam1 on the 3rd chromosome (McKearin & Spradling, 1990).
Generation of fly strains with different switchable dual‐color transgenes
Standard procedures were used for all molecular cloning experiments. Enzymes used for plasmid construction were obtained from New England Biolabs. The new histone sequence, histone‐mCherry, was recovered as an XbaI‐flanked fragment and was subsequently inserted into the XbaI site of the UASp plasmid to construct the UASp‐histone‐mCherry plasmid. The old histone sequence, histone‐eGFP, was inserted into a pBluescript‐FRT‐NheI‐SV40 PolyA‐FRT plasmid at the unique NheI site. The entire NotI‐FRT‐histone‐eGFP‐SV40 PolyA‐FRT‐EcoRI sequences were then subcloned into the UASp‐histone‐mCherry plasmid digested by NotI and EcoRI. The final UASp‐FRT‐histone‐eGFP‐PolyA‐FRT‐histone‐mCherry plasmids were introduced into w1118 flies by P‐element‐mediated germline transformation (BestGene Inc.). Transgenic flies with the following transgenes were newly generated in studies reported here:
UASp‐FRT‐H3.3‐eGFP‐PolyA‐FRT‐H3.3‐mCherry on the 2nd chromosome.
Heat shock scheme
Male flies carrying the UASp‐dual‐color histone transgenes were crossed with female virgins containing the GreenEye‐nanos‐Gal4 driver at 25°C. Once progenies were produced, vials with darkened pupae were submerged in a circulating 37°C water bath for 90 min and recovered in a 29°C incubator for 34–40 h for studies on canonical histones H3, H4, and H2A or for 17–20 h for studies on histone H3.3, before dissection for immunostaining and Oligopaint IF‐FISH experiments.
Immunostaining
Ovaries were dissected from female flies within 2 days post‐eclosion in room temperature Schneider’s Insect Medium. Ovaries were then fixed for 5–10 min in 4% formaldehyde in 1× PBS + 0.1% Triton. After fixation, ovaries were rinsed three times and underwent three washes, 5‐min/each in 1× PBS + 0.1% Triton. Primary antibodies were diluted in 1× PBS + 0.1% Triton + 3% BSA and incubated with ovaries overnight at 4°C on a nutator. The primary antibodies used were mouse anti α‐Spectrin (1:50, DSHB, Cat# 3A9), mouse anti hu‐li tai shao (1:50, DSHB, Cat# 1B1), mouse anti‐Armadillo (1:100, DSHB, Cat# N27A1), mouse anti‐H3S10ph (1:5000, Abcam, Cat# ab14955), rabbit anti‐GFP (1:200, Abcam, Cat# ab290), rabbit anti‐GFP (1:400, Invitrogen, Cat# A‐11122), and Chicken anti‐mCherry (1:1,000, Novus Biologicals, Cat#NBP2‐25158). After incubation with primary antibodies, gonads were rinsed three times and underwent three washes, 5‐min/each in 1× PBS + 0.1% Triton. Secondary antibodies were diluted in 1× PBS + 0.1% Triton + 5% NGS and incubated with ovaries for 2–4 h rotating at room temperature or at 4°C overnight in the dark. Secondary antibodies were the Alexa Fluor‐conjugated series used at 1:1,000 (goat anti‐mouse 405, goat anti‐rabbit 488, goat anti‐chicken 568; Molecular Probes). After incubation with secondary antibodies, ovaries were rinsed three times and underwent three washes, 5‐min/each in 1× PBS + 0.1% Triton at room temperature in the dark. Ovaries were placed on a glass slide and ovarioles were separated, followed by mounting with Vectashield Antifade Mounting Medium (Vector Laboratories, Cat# H‐1000). Images were taken using Zeiss LSM 700 confocal microscope with a 63× oil immersion objective. Images were processed and analyzed using Zen Black, FIJI software, and/or R Studio.
Quantification of old versus new histone overlap
Open images in Zen, identify the Z‐slice containing the most chromatin information (Z‐slices typically 0.9 µm), use slide bar to adjust signal so that 1 pixel in the cell was saturated in both the red channel and the green channel. Adjust background from 0 to 154 (this should not affect any signal in the cells, if it does the image capture settings might have been wrong). Zoom to 500 for prometaphase cells and add a scale bar. Minimize left and bottom side bars and export contents of window as a (.tif) file. Open exported image in FIJI. Draw a region of interest (ROI) around the cell to be quantified using the circle tool. Duplicate the ROI and split color channels. Subtract background from each channel using 120 rolling ball radius. For old versus new histone overlap analysis, use the FIJI Coloc2 plugin to analyze overlap between red and green channels with settings PSF (3) and Costes iterations (10). Values for Spearman correlation and Pearson correlation (both with and without Costes threshold) for each histone protein are saved and significance is quantified using a pairwise ANOVA test in R Studio.
Quantification of old versus new total histone protein
Open image in Zen, use slide bar to adjust signal so that 1 pixel in the telophase pair is saturated in both the red channel and the green channel. Adjust background from 0 to 154 (this should not affect any signal in the cells, if it does the image capture settings might have been wrong). Zoom to 800 for telophase nuclei and add a scale bar. Minimize left and bottom side bars and export contents of window as a (.tif) file. Open exported image in FIJI. Draw a region of interest (ROI) around the cell to be quantified using the circle tool. Duplicate the ROI and split color channels. Subtract background from each channel using 300 rolling ball radius. For total histone protein analysis, used Analyze>Measure tool to get Raw Integrated Density (RawIntDen, total signal) values for each channel. Repeated for each Z‐slice and summed RawIntDen from all Z‐slices for each channel to get total signal for each nucleus. Then, used summed RawIntDen values to create the ratio of total histone protein for either GSC‐CB or CC‐CC pairs.
Oligopaint probe preparation
Probe preparation is done on ice and protected from light exposure. Prepare a 10 ml PCR consisting of 1 ml 10× Taq buffer (NEB, Cat#B9014S), 200 µl 10 mM dNTPs (Promega, Cat# U151B), 100 µl 200 µM 5’fluor‐labeled forward primer, 100 µl 200 µM 5’phosphorylated reverse primer, 10 µl 1 ng/µl gene library template, 100 µl Taq (NEB, 5,000 U/ml, Cat# M0273L), 8490 µl MilliQ purified H2O. Aliquot PCR into 100 µl reactions (or 50 µl depending on thermocycler capacity) in strip tubes or plates (makes 100 rxns = 12.5 tube strips). PCR Program: 95°C‐5 min, (95°C‐30 s, 55°C‐30 s, 72°C‐15 s) ×40, 72°C‐5 min. Purify PCR using Zymo DNA clean & Concentrator‐500 columns (Cat# D4031/D4032) by pooling the PCR products in a reagent reservoir and mixing in 50 ml DNA Binding Buffer. Use two Zymo columns in two 50‐ml conical tubes to transfer 20 ml of PCR to columns (10 ml each) and spin at 4°C for 5 min at 3,000× g. Repeat until all PCR product is purified and then add 10 ml of DNA wash buffer to each column, and spin at 4°C for 5 min at 3,000× g. Transfer columns to new 50‐ml conical tubes and spin again for 5 min to get rid of any residual ethanol. Transfer columns to 50‐ml conical tubes and elute each by adding 2,800 µl Elution Buffer to the column, incubate at room temperature for 1 min, then spin for at 4°C for 3 min at 3,000× g. Combine DNA Eluant and add 500 µl 10× Lambda buffer (NEB, provided with Lambda exonuclease, Lot# 00371474), 100 µl Lambda exonuclease 5,000 U/ml (NEB, Cat# M0262L). Aliquot 100 µl (or 50 µl) into strip tubes or plates and set thermocycler to 37°C‐30 min, 75°C‐10 min to create single‐stranded probe. Combine the Lambda Exonuclease reactions into a reagent reservoir and mix in 1/50 (100 µl) glycogen (Thermo Scientific, 20 mg/ml Cat#R0561). Then, add 1/10 (500 µl) 4 M NH4OAc and mix well. Finally, add 2.5× (12.5 mls) ice‐cold 100% EtOH and mix well. Aliquot into 1.5‐ml tubes and place at −80°C for at least 30 min. Spin at maximum speed at 4°C for 1 h. Remove ALL EtOH and add 8 µl MilliQ purified H2O to each pellet. Incubate for 15 min at 42°C before combing pellets and analyzing probe concentration using fluorescent value measured by NanoDrop ND1000 Spectrophotometer.
DNA IF‐FISH
Ovaries were dissected from 10 or fewer female flies within 2 days post‐eclosion in room temperature Schneider’s Insect Medium. Ovaries were then transferred to an Eppendorf tube containing 200 µl 4% FA/1× PBS/0.5% NP‐40 plus 600 µl heptane (prepared on ice), shaken vigorously by hand, and then fixed for 5 min while rotating. After fixation, tissues were rinsed three times and underwent three washes, 5‐min/each in 1× PBS + 0.1% Triton. Tissues were blocked for 1 h at room temperature while rotating in 1× PBS + 0.3% Tween 20 + 1.5% BSA. Primary antibodies were diluted in 1× PBS + 0.3% Tween 20 + 1.5% BSA and incubated with ovaries overnight at 4°C. The primary antibodies used were α‐Spectrin (1:50, DSHB, Cat#3A9), mouse anti hu‐li tai shao (1:50, DSHB, Cat#1B1), mouse anti‐Armadillo (1:100, DSHB, Cat#N27A1), mouse anti‐H3S10ph (1:5,000, Abcam, Cat# ab14955), rabbit anti‐GFP (1:200, Abcam, Cat#ab290), rabbit anti‐GFP (1:400, Invitrogen, Cat#A‐11122), and Chicken anti‐mCherry (1:1,000, Novus Biologicals, Cat#NBP2‐25158). After incubation with primary antibodies, ovaries were washed three times, 20 min each time in 1× PBS + 0.1% Triton. Secondary antibodies were diluted in 1× PBS + 0.1% Triton + 5% NGS and incubated with ovaries for 2 h rotating at room temperature in the dark. Secondary antibodies were the Alexa Fluor‐conjugated series used at 1:1,000 (goat anti‐mouse 405, goat anti‐rabbit 488, goat anti‐chicken 568; Molecular Probes). After incubation with secondary antibodies, ovaries were washed twice, 20 min each time in 1× PBS + 0.1% Triton, and then washed once for 20 min with 1× PBS. During the 1× PBS wash, 200 µl 4% FA/1× PBS/0.5% NP‐40 plus 600 µl heptane fixative was prepared for each sample in a 1.5‐ml Eppendorf tube on ice. Using a P200 pipette with the tip removed and coated in 1× PBS + 0.1% Triton, transfer the ovaries to the cap of the Eppendorf tube containing fixative, bringing as little excess 1× PBS as possible. Close the tube and shake vigorously by hand, then leave on rotator at room temperature for no more than 5 min. Ovaries were then washed quickly four times using 2× SSC + 0.1% Tween 20 (2× SSCT) followed by one 10‐min wash in 2× SSCT + 2% PEG + 20% formamide rotating at room temperature, one 10‐min wash in 2× SSCT + 4% PEG + 40% formamide rotating at room temperature, and two 10‐min washes in 2× SSCT + 5% PEG + 50% formamide rotating at room temperature. Then, the tubes were moved from rotating at room temperature to shaking at 37°C for 3–5 h for pre‐denaturation/pre‐hybridization. The ovaries were then transferred to a PCR tube, excess buffer was removed, and hybridization buffer was added (36 µl 2×SSCT/5% PEG/10% dextran sulfate/50% formamide + 1 µl RNAse A (Thermo Scientific, Cat #R1253), + 4‐µl probe (from Oligopaint Probe Preparation)). Typically, 100–200 pmol of heterochromatic labeled oligo probes or 200–300 pmol of euchromatic Oligopaint probes are added per reaction (with a maximum of 4‐µl total probe) for a final volume of 40 µl. The tubes were gently mixed and then transferred to a thermocycler. Tubes were heated to 91°C for 2 min followed by a return to 37°C. The tubes were then transferred to a 37°C shaker and allowed to incubate/hybridize in the dark overnight for 18–20 h. Hybridization buffer was removed and tissues were washed once with 2× SSCT + 5% PEG + 50% formamide for 1 h at 37°C with shaking. Then, all but 50 µl of the supernatant was removed from the tube, 100 pmol of secondary probe was added to the wash, and tubes were incubated for 1 h at 37°C with shaking. Tissues were then washed twice with 2× SSCT + 5% PEG + 50% formamide for 30 min each time at 37°C with shaking. Then, tissues were washed with 2× SSCT + 2% PEG + 20% formamide for 10 min at room temperature with rotation on a nutator. Tissues were washed in 2× SSCT for 10 min at room temperature with rotation. Finally, tissues were rinsed three times with 2× SSCT at room temperature. Ovaries were placed on a glass slide and ovarioles were spread before mounting using Vectashield Antifade Mounting Medium (Cat# H‐1000). Images were taken using Zeiss LSM 800 or 980 confocal microscope with AiryScan processing using a 63× oil immersion objective. Images were processed and analyzed using Zen Black, FIJI software, and R Studio.
Quantification of FISH probe association with old versus new histone
Open image in Zen, use the slide bar to adjust signal so that 1 pixel in the entire cell is saturated for each channel. Adjust background from 0 to 154 (this should not affect any signal in the cells, if it does the image capture settings might have been wrong). Identify the Z‐slices containing the probe signal (Z‐slices taken at 0.13–0.15 µm, typically probe signal spans three Z‐slices). Select a single Z‐slice containing the most probe signal, and create a subset image containing the red, green, and probe channels but omitting the 4th channel containing H3S10ph signal. Zoom to 1,000, minimize left and bottom side bars and export the 3‐color contents of window as a (.tif) file. Open exported image in FIJI. Draw a region of interest (ROI) of 10 × 10 pixels around the probe signal to be quantified using the circle tool. Duplicate the ROI and split color channels. To examine whether the probe ROI is more associated with green (old) or red (new) histone, use Analyze>Measure tool to get mean intensity values for each channel. Then, normalize both red and green channel values by making the highest mean intensity value for each channel equal to 1 and dividing the other three signal values accordingly to create a percentage. Plot red versus green (0 to 1) normalized mean intensity values for each probe signal and, using a line at X = Y (or red = green), determined whether each probe signal was more associate with green (old) or red (new) histone. For each individual cell, take the ratio of green (old) versus red (new) association to examine frequency of a 2:0:2 ratio using different probe targets (for details see Fig 3I). For telophase cells where the two FISH signals from homologous chromosomes are very close to each other due to the pulling force of the mitotic spindle, we selected the two Z‐slices containing the most probe signal and measure both to achieve four total data points with two at each set of segregated sister chromatids.
RNA IF‐FISH
All reagents were prepared under RNase‐free conditions using RNase‐free solutions. All surfaces and equipment were cleaned with RNaseZAP (Sigma‐Aldrich, Cat#R2020), and experiments were performed separately at a bench designed for RNA work. Ovaries were dissected from 10 or fewer female adult flies within 5 days post‐eclosion in room temperature Schneider’s Insect Medium. Ovaries were then transferred to an Eppendorf tube containing 200 µl 4% Formaldehyde/1× PBS/0.5% NP‐40 plus 600 µl heptane (prepared on ice), shaken vigorously by hand, and then fixed for 10 min while rotating. After fixation, tissues were rinsed three times and underwent three 5‐min washes in 1× PBS + 0.3% Triton. Tissues were blocked for 1 h at room temperature while rotating in 1× PBS + 0.3% Tween 20 + 1.5% BSA. Primary antibodies were diluted in 1× PBS + 0.3% Tween 20 + 1.5% BSA + 1 µl RNase Inhibitor (Promega, Cat#N2111) and incubated with ovaries overnight at 4°C. The primary antibodies used were anti laminB (1:100, DSHB, Cat#ADL167.10), rabbit anti‐GFP (1:200, Abcam, Cat#ab290), and Chicken anti‐mCherry (1:1,000, Novus Biologicals, Cat#NBP2‐25158). After incubation with primary antibodies, ovaries were washed three times, each time for 20 min, in 1× PBS + 0.3% Triton. Secondary antibodies were diluted in 1× PBS + 0.3% Triton + 5% NGS + 1 µl RNase Inhibitor and incubated with ovaries for 2 h rotating in the dark at room temperature. Secondary antibodies were the Alexa Fluor‐conjugated series used at 1:1,000 (goat anti‐mouse 405, goat anti‐rabbit 488, goat anti‐chicken 568; Molecular Probes). After incubation with secondary antibodies, ovaries were washed twice, each time for 20 min in the dark, in 1× PBS + 0.3% Triton, and then washed once for 20 min with 1× PBS in the dark. During the 1× PBS wash, 200 µl 4% FA/1× PBS/0.5% NP‐40 plus 600 µl heptane fixative was prepared for each sample in 1.5‐ml Eppendorf tubes on ice. Using a P200 pipette with the tip removed and coated in 1× PBS + 0.3% Triton, transferred the ovaries to the cap of the Eppendorf tube containing fixative, bringing as little excess 1× PBS as possible. The tube was closed and shaken vigorously by hand and then left on rotator in the dark at room temperature for 5 min. Ovaries were then washed quickly four times using 2× SSCT followed by one 10‐min wash in 2× SSCT + 20% formamide rotating at room temperature in the dark, one 10‐min wash in 2× SSCT + 40% formamide rotating at room temperature in the dark, and two 10‐min washes in 2× SSCT + 50% formamide rotating at room temperature in the dark. The tubes were then moved from rotating at room temperature to shaking at 37°C in the dark for 3–5 h for pre‐denaturation/pre‐hybridization. Wash buffer was removed and hybridization buffer was added (36 µl 2xSSCT/10% dextran sulfate/50% formamide + 1 µl RNase Inhibitor + 4‐µl probe) for a final volume of 40 µl. The tubes were gently mixed and incubated/hybridized overnight in the dark for 18–20 h at 37°C with shaking. Then, hybridization buffer was removed and ovaries were washed once with 2× SSCT + 50% formamide for 1 h at 37°C with shaking in the dark. All but 50 µl of the supernatant was removed from the tube, 1 µl RNase Inhibitor + 1 µl (100 pmol) of secondary probe was added to the wash, and tubes were incubated for 1 h at 37°C with shaking in the dark. Tissues were washed twice with 2× SSCT + 50% formamide, each time for 30 min, at 37°C with shaking in the dark. Then, tissues were washed with 2× SSCT + 20% formamide for 10 min at room temperature with rotation in the dark. Tissues were washed in 2× SSCT for 10 min at room temperature with rotation in the dark. Finally, tissues were rinsed three times with 2× SSCT at room temperature. Ovaries were placed on a glass slide and ovarioles were spread before mounting using Vectashield Antifade Mounting Medium (Cat# H‐1000) or Vectashield Antifade Mounting Medium with DAPI (Cat# H‐1200). Images were taken using Zeiss LSM 700 confocal microscope using a 63× oil immersion objective. Images were processed and analyzed using Zen Black, FIJI software, and R Studio (Fig EV6).
Author contributions
EWK and XC conceptualized the study. KV‐L and RJ provided Oligopaint training and some relevant reagents. EWK, EHZ, and LS performed all the experiments and data analyses. EWK and XC wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Dataset EV7
Dataset EV8
Review Process File
Acknowledgements
We thank Drs. M. Van Doren, J. Gall, D. Drummond‐Barbosa, A. Spradling, and X.C. laboratory members for insightful suggestions. We also thank T. Wu (Harvard University) and E. Joyce (University of Pennsylvania) and their laboratory members for thoughtful discussions and reagents. We thank Johns Hopkins Integrated Imaging Center for imaging. This work was supported by F31GM122339 (E.W.K.), F31DK122702 (E.H.Z.), F31EY026786 (K.V.L.), R01EY025598 (R.J.), R01GM112008, R35GM127075, and the Howard Hughes Medical Institute (Grant# 55108512) (X.C.).
EMBO reports (2021) 22: e51530.
Data availability
No data were deposited in public databases.
References
- Ables ET, Drummond‐Barbosa D (2013) Cyclin E controls Drosophila female germline stem cell maintenance independently of its role in proliferation by modulating responsiveness to niche signals. Development 140: 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S (2002a) Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci USA 99(Suppl 4): 16477–16484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S (2002b) The histone variant H3.3 marks active chromatin by replication‐independent nucleosome assembly. Mol Cell 9: 1191–1200 [DOI] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S (2018) No strand left behind. Science 361: 1311–1312 [DOI] [PubMed] [Google Scholar]
- Alabert C, Barth TK, Reverón‐Gómez N, Sidoli S, Schmidt A, Jensen ON, Imhof A, Groth A (2015) Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev 29: 585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C, Groth A (2012) Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 13: 153–167 [DOI] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T (2016) The molecular hallmarks of epigenetic control. Nat Rev Genet 17: 487–500 [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE (2006) Epigenetic gene silencing in cancer ‐ a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6: 107–116 [DOI] [PubMed] [Google Scholar]
- Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Chang Y, Li JB, Senaratne TN, Williams BRet al (2012) Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci USA 109: 21301–21306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp D, Horabin JI, Lersch RA, Cline TW, Schedl P (1993) Expression of the Sex‐lethal gene is controlled at multiple levels during Drosophila oogenesis. Development 118: 797–812 [DOI] [PubMed] [Google Scholar]
- Brooker AS, Berkowitz KM (2014) The roles of cohesins in mitosis, meiosis, and human health and disease. Methods Mol Biol 1170: 229–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanueva MO, Ferguson EL (2004) Germline stem cell number in the Drosophila ovary is regulated by redundant mechanisms that control Dpp signaling. Development 131: 1881–1890 [DOI] [PubMed] [Google Scholar]
- Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA (2007) Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol 5: e201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, McKearin DM (2003) A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130: 1159–1170 [DOI] [PubMed] [Google Scholar]
- de Cuevas M, Spradling AC (1998) Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development 125: 2781–2789 [DOI] [PubMed] [Google Scholar]
- Dattoli AA, Carty BL, Kochendoerfer AM, Morgan C, Walshe AE, Dunleavy EM (2020) Asymmetric assembly of centromeres epigenetically regulates stem cell fate. J Cell Biol 219: e201910084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H (1997) Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila . Dev Biol 189: 79–94 [DOI] [PubMed] [Google Scholar]
- Escobar TM, Loyola A, Reinberg D (2021) Parental nucleosome segregation and the inheritance of cellular identity. Nat Rev Genet. 10.1038/s41576-020-00312-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar TM, Oksuz O, Saldana‐Meyer R, Descostes N, Bonasio R, Reinberg D (2019) Active and repressed chromatin domains exhibit distinct nucleosome segregation during DNA Replication. Cell 179: 953–963 e911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S (2006) The epigenetic progenitor origin of human cancer. Nat Rev Genet 7: 21–33 [DOI] [PubMed] [Google Scholar]
- Fitzsimons CP, van Bodegraven E, Schouten M, Lardenoije R, Kompotis K, Kenis G, van den Hurk M, Boks MP, Biojone C, Joca Set al (2014) Epigenetic regulation of adult neural stem cells: implications for Alzheimer's disease. Mol Neurodegener 9: 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredly H, Gjertsen BT, Bruserud O (2013) Histone deacetylase inhibition in the treatment of acute myeloid leukemia: the effects of valproic acid on leukemic cells, and the clinical and experimental evidence for combining valproic acid with other antileukemic agents. Clin Epigenetics 5: 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT, Spradling AC (2007) Male and female Drosophila germline stem cells: two versions of immortality. Science 316: 402–404 [DOI] [PubMed] [Google Scholar]
- Gan H, Serra‐Cardona A, Hua X, Zhou H, Labib K, Yu C, Zhang Z (2018) The Mcm2‐Ctf4‐polalpha axis facilitates parental histone H3–H4 transfer to lagging strands. Mol Cell 72: 140–151 e143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q, Chepelev I, Wei G, Tarayrah L, Cui K, Zhao K, Chen X (2010) Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA‐seq. Cell Res 20: 763–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar‐Maia A, Alajem A, Meshorer E, Ramalho‐Santos M (2011) Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol 12: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golkaram M, Jang J, Hellander S, Kosik KS, Petzold LR (2017) The role of chromatin density in cell population heterogeneity during stem cell differentiation. Sci Rep 7: 13307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Matunis E, DiNardo S (1997) bag‐of‐marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development 124: 4361–4371 [DOI] [PubMed] [Google Scholar]
- Hardy RW, Tokuyasu KT, Lindsley DL, Garavito M (1979) The germinal proliferation center in the testis of Drosophila melanogaster . J Ultrastruct Res 69: 180–190 [DOI] [PubMed] [Google Scholar]
- Hardy RW, Tokuyasu KT, Lindsley DL (1981) Analysis of spermatogenesis in Drosophila melanogaster bearing deletions for Y‐chromosome fertility genes. Chromosoma 83: 593–617 [DOI] [PubMed] [Google Scholar]
- Hinnant TD, Alvarez AA, Ables ET (2017) Temporal remodeling of the cell cycle accompanies differentiation in the Drosophila germline. Dev Biol 429: 118–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman S, Miller D, Eisman R, Kuwayama H, Niimi T, Kaufman T (2010) Transgenic tools for members of the genus Drosophila with sequenced genomes. Fly 4: 349–362 [DOI] [PubMed] [Google Scholar]
- Hsu HJ, LaFever L, Drummond‐Barbosa D (2008) Diet controls normal and tumorous germline stem cells via insulin‐dependent and ‐independent mechanisms in Drosophila . Dev Biol 313: 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y (2014) SnapShot: histone modifications. Cell 159: 458–458.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen KA, Sidoli S, Garcia BA (2017) Recent achievements in characterizing the histone code and approaches to integrating epigenomics and systems biology. Methods Enzymol 586: 359–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Li Y, Robertson KD (2011) DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer 2: 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce EF, Apostolopoulos N, Beliveau BJ, Wu CT (2013) Germline progenitors escape the widespread phenomenon of homolog pairing during Drosophila development. PLoS Genet 9: e1004013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RC (1957) Oogenesis in adult Drosophila melanogaster. II. Stage distribution as a function of age. Growth 21: 95–102 [PubMed] [Google Scholar]
- Kirilly D, Wang S, Xie T (2011) Self‐maintained escort cells form a germline stem cell differentiation niche. Development 138: 5087–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lavoie CA, Ohlstein B, McKearin DM (1999) Localization and function of Bam protein require the benign gonial cell neoplasm gene product. Dev Biol 212: 405–413 [DOI] [PubMed] [Google Scholar]
- Laws KM, Sampson LL, Drummond‐Barbosa D (2015) Insulin‐independent role of adiponectin receptor signaling in Drosophila germline stem cell maintenance. Dev Biol 399: 226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT (2012) Epigenetic regulation by long noncoding RNAs. Science 338: 1435–1439 [DOI] [PubMed] [Google Scholar]
- Li Y, Minor NT, Park JK, McKearin DM, Maines JZ (2009) Bam and Bgcn antagonize Nanos‐dependent germ‐line stem cell maintenance. Proc Natl Acad Sci USA 106: 9304–9309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC (1995) Fusome asymmetry and oocyte determination in Drosophila . Dev Genet 16: 6–12 [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC (1997) A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124: 2463–2476 [DOI] [PubMed] [Google Scholar]
- Lin S, Yuan ZF, Han Y, Marchione DM, Garcia BA (2016) Preferential phosphorylation on old histones during early mitosis in human cells. J Biol Chem 291: 15342–15357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Trieu TJ, Cheng J, Zhou S, Tang Q, Xie J, Liu JL, Zhao K, Habib SJ, Chen X (2020) Differential histone distribution patterns in induced asymmetrically dividing mouse embryonic stem cells. Cell Rep 32: 108003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKearin D, Ohlstein B (1995) A role for the Drosophila bag‐of‐marbles protein in the differentiation of cystoblasts from germline stem cells. Development 121: 2937–2947 [DOI] [PubMed] [Google Scholar]
- McKearin DM, Spradling AC (1990) bag‐of‐marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev 4: 2242–2251 [DOI] [PubMed] [Google Scholar]
- Petryk N, Dalby M, Wenger A, Stromme CB, Strandsby A, Andersson R, Groth A (2018) MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361: 1389–1392 [DOI] [PubMed] [Google Scholar]
- Ranjan R, Snedeker J, Chen X (2019) Asymmetric centromeres differentially coordinate with mitotic machinery to ensure biased sister chromatid segregation in germline stem cells. Cell Stem Cell 25: 666–681.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveron‐Gomez N, Gonzalez‐Aguilera C, Stewart‐Morgan KR, Petryk N, Flury V, Graziano S, Johansen JV, Jakobsen JS, Alabert C, Groth A (2018) Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication. Mol Cell 72: 239–249.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober P, Boer C, Schwarte LA (2018) Correlation coefficients: appropriate use and interpretation. Anesth Analg 126: 1763–1768 [DOI] [PubMed] [Google Scholar]
- Serra‐Cardona A, Zhang Z (2018) Replication‐coupled nucleosome assembly in the passage of epigenetic information and cell identity. Trends Biochem Sci 43: 136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M, Urban MA, Fried G, Wesseln CJ, Mander L, Punyasena SW (2018) Comparative performance of airyscan and structured illumination superresolution microscopy in the study of the surface texture and 3D shape of pollen. Microsc Res Tech 81: 101–114 [DOI] [PubMed] [Google Scholar]
- Smith AV, Orr‐Weaver TL (1991) The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development 112: 997–1008 [DOI] [PubMed] [Google Scholar]
- Struhl G (1982) Spineless‐aristapedia: a homeotic gene that does not control the development of specific compartments in Drosophila . Genetics 102: 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray‐Gallet D, Almouzni G, Nakatani Y (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61 [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT, Peacock WJ, Hardy RW (1977) Dynamics of spermiogenesis in Drosophila melanogaster. VII. Effects of segregation distorter (SD) chromosome. J Ultrastruct Res 58: 96–107 [DOI] [PubMed] [Google Scholar]
- Tran V, Lim C, Xie J, Chen X (2012) Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science 338: 679–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM (2008) Open chromatin and hypertranscription in embryonic stem cells. Cell Stem Cell 2: 408–410 [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R (1998) Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol 8: 243–246 [DOI] [PubMed] [Google Scholar]
- Wernet MF, Mazzoni EO, Çelik A, Duncan DM, Duncan I, Desplan C (2006) Stochastic spineless expression creates the retinal mosaic for colour vision. Nature 440: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten M, Snedeker J, Nizami ZF, Yang X, Ranjan R, Urban E, Kim JM, Gall J, Xiao J, Chen X (2019) Asymmetric histone inheritance via strand‐specific incorporation and biased replication fork movement. Nat Struct Mol Biol 26: 732–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Spradling AC (1998) decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 94: 251–260 [DOI] [PubMed] [Google Scholar]
- Xie T, Spradling AC (2000) A niche maintaining germ line stem cells in the Drosophila ovary. Science 290: 328–330 [DOI] [PubMed] [Google Scholar]
- Xu M, Wang W, Chen S, Zhu B (2011) A model for mitotic inheritance of histone lysine methylation. EMBO Rep 13: 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadlapalli S, Yamashita YM (2013) Chromosome‐specific nonrandom sister chromatid segregation during stem‐cell division. Nature 498: 251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NL, DiMaggio PA, Garcia BA (2010) The significance, development and progress of high‐throughput combinatorial histone code analysis. Cell Mol Life Sci 67: 3983–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Gan H, Serra‐Cardona A, Zhang L, Gan S, Sharma S, Johansson E, Chabes A, Xu RM, Zhang Z (2018) A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361: 1386–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wu C, Bhavanasi D, Wang H, Gregory BD, Huang J (2017) Chromatin dynamics during the differentiation of long‐term hematopoietic stem cells to multipotent progenitors. Blood Adv 1: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee BM, Britton LM, Wolle D, Haberman DM, Garcia BA (2012) Origins and formation of histone methylation across the human cell cycle. Mol Cell Biol 32: 2503–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zion E, Chen X (2020) Asymmetric histone inheritance regulates stem cell fate in Drosophila midgut. BioRxiv. 10.1101/2020.08.15.252403 [PREPRINT] [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Dataset EV7
Dataset EV8
Review Process File
Data Availability Statement
No data were deposited in public databases.


