Abstract
The spindle assembly checkpoint (SAC) is a surveillance mechanism that promotes accurate chromosome segregation in mitosis. The checkpoint senses the attachment state of kinetochores, the proteinaceous structures that assemble onto chromosomes in mitosis in order to mediate their interaction with spindle microtubules. When unattached, kinetochores generate a diffusible inhibitor that blocks the activity of the anaphase-promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase required for sister chromatid separation and exit from mitosis. Work from the past decade has greatly illuminated our understanding of the mechanisms by which the diffusible inhibitor is assembled and how it inhibits the APC/C. However, less is understood about how SAC proteins are recruited to kinetochores in the absence of microtubule attachment, how the kinetochore catalyzes formation of the diffusible inhibitor, and how attachments silence the SAC at the kinetochore. Here, we summarize current understanding of the mechanisms that activate and silence the SAC at kinetochores and highlight open questions for future investigation.
Keywords: Checkpoint, Kinetochores, Chromosome segregation, Mitosis, Aneuploidy, Catalysis
1. Introduction
Accurate transfer of genetic information encoded in DNA across generations is a fundamental aspect of life. Eukaryotes ensure transfer of genetic information in each cell division by first replicating their chromosomes and then segregating them equally to daughter cells in mitosis. The segregation of chromosomes is mediated by kinetochores, the proteinaceous structures that assemble onto the centromeric region of chromosomes to mediate interaction with spindle microtubules [1]. The process of microtubule capture by kinetochores on mitotic chromosomes is at least partially stochastic and requires time. If there is insufficient time in mitosis, chromosomes do not connect correctly and segregate improperly, generating daughter cells that have either gained or lost a chromosome, a state referred to as aneuploidy. Depending on its degree, aneuploidy can lead to inviability, developmental defects or contribute to the genesis and progression of diseases such as cancer.
To ensure that there is sufficient time in mitosis for all chromosomes to connect to spindle microtubules, cells employ a mechanism known as the spindle assembly checkpoint (SAC) [2], [3], [4], [5] (Fig. 1). When kinetochores are unattached, they generate a checkpoint signal that delays the onset of anaphase. Once all kinetochores are attached, production of the checkpoint signal stops, thereby ensuring that anaphase only occurs after all chromosomes are bound to spindle microtubules. Classical genetic screens in the budding yeast S. cerevisiae identified the majority of SAC components, which include the Mitotic Arrest Deficient (MAD) and the Budding Uninhibited by Benzimidazoles (BUB) genes [6], [7]. Other components essential for the SAC include the Mps1 kinase, Cdc20 and, in metazoans, kinetochore receptors such as the Rod-Zwilch-ZW10 (RZZ) complex (Fig. 2).
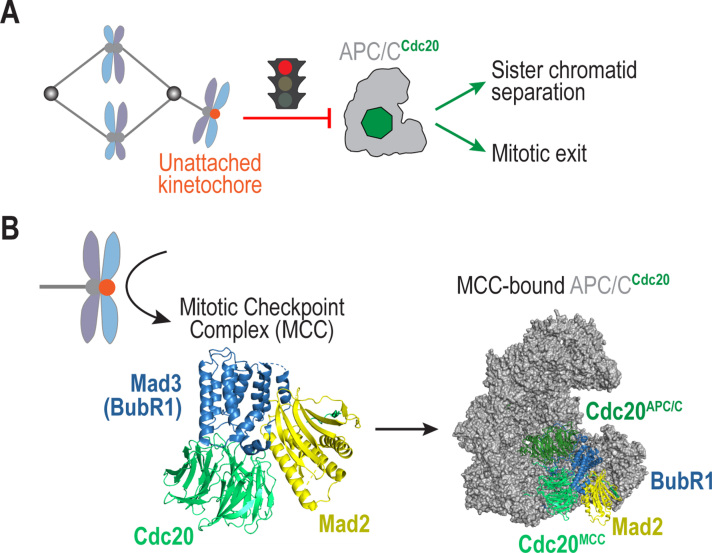
Fig. 1.
Schematic of Spindle Assembly Checkpoint (SAC) – dependent control of mitosis. (A) During mitosis, unattached kinetochores (orange) catalyze formation of an inhibitor that targets the APC/CCdc20, a ubiquitin ligase whose activity is essential for sister chromatid separation and mitotic exit. The SAC thereby ensures that the metaphase-anaphase transition occurs only after all kinetochores are attached to spindle microtubules. (B) The SAC functions by catalyzing the formation of the Mitotic Checkpoint Complex (MCC), which then binds APC/CCdc20 to block its activity. Shown here are the crystal structure of the MCC from S. pombe (PDB 4aez, left) [12] and the cryo-EM structure of human APC/CCdc20 bound to the MCC (PDB 5lcw, right) [15]. Note that Bub3, a component of the MCC in most species, is absent in these structures. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
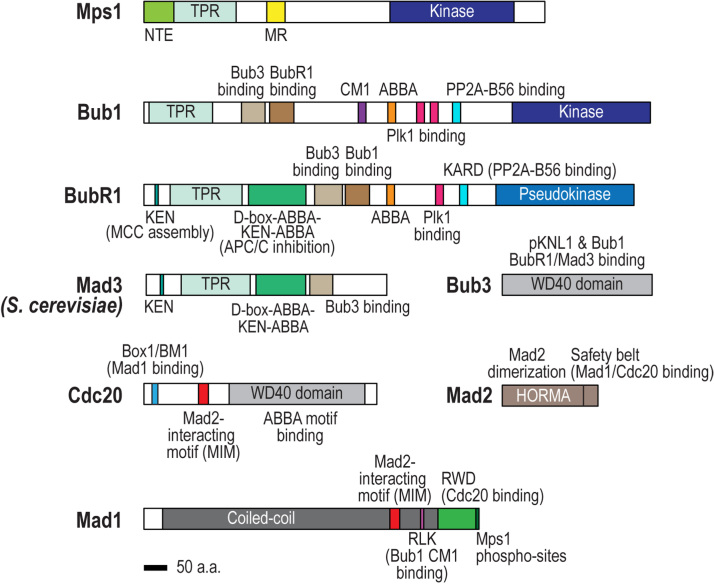
Fig. 2.
Domain organization of SAC proteins. The SAC-relevant motifs/domains discussed in the text of Mps1, Bub1, Mad1, Mad2, BubR1/Mad3, Bub3, and Cdc20 are shown. Cartoons are based on the human proteins except for Mad3, which was based on the S. cerevisiae protein. Scale bar represents 50 amino acids.
The efforts of many groups over the last decade have greatly advanced understanding of the downstream events in checkpoint signaling, namely how the SAC signal generated at kinetochores blocks anaphase onset [8], [9], [10], [11], [12], [13], [14], [15], [16]. A combination of cell biology, genetics and biochemistry and ultimately beautiful structures made possible by advances in molecular cryo-EM conducted on biochemically reconstituted complexes, showed that the SAC generates a diffusible inhibitor that binds and inhibits the APC/C [15], [16]. By comparison, how the SAC signal is generated specifically at unattached kinetochores is less well understood. Recent technical advances in cellular analysis in coordination with complex biochemical reconstitutions of kinetochore components are helping to fill this gap, by defining how kinetochores act as catalysts for production of a diffusible anaphase inhibitor [17]. In this review, we focus on recent advances in understanding checkpoint signal generation at unattached kinetochores and silencing of this signal following microtubule attachment.
2. Overview of checkpoint signaling
The SAC works by inhibiting the activity of an E3 ubiquitin ligase known as the APC/C [18], [19] (Fig. 1). During mitosis, the APC/C is activated by its co-activator protein Cdc20 and the resulting APC/CCdc20 complex ubiquitinates the anaphase inhibitors securin and cyclin B to promote sister chromatid separation and exit from mitosis [18], [19] (Fig. 1A). When kinetochores are unattached, they produce a diffusible signal that blocks APC/CCdc20 activity towards securin and cyclin B [20], [21], [22], [23], [24], [25], [26]. While a single unattached kinetochore is sufficient to elicit a SAC response [27], the strength of this diffusible SAC signal is proportional to the number of unattached kinetochores present, indicating that the SAC is not an all-or-none response but rather modulated by the number of active signaling centers [28], [29]. These properties of the SAC enable unattached kinetochores to limit the premature segregation of chromosomes, thereby protecting against the formation of aneuploid progeny.
The primary action of unattached kinetochores with respect to SAC signaling is to catalyze the formation of the Mitotic Checkpoint Complex (MCC), a potent inhibitor of APC/CCdc20 [2], [3], [4], [5] (Fig. 1B). The MCC is a tetrameric complex composed of Mad2, Cdc20, BubR1 (Mad3 in yeast and nematodes) and Bub3 [12], [30], [31], [32], [33]; Bub3 is not part of the MCC in organisms such as S. pombe but when present, it is reported to increase the affinity of the MCC for the APC/C [11], [34], [35]. One fascinating aspect of the SAC is that Cdc20 has dual roles, as the APC/C activator and as a subunit of the MCC [18], [19]. Thus, during a SAC-dependent mitotic arrest, the APC/C is bound by two molecules of Cdc20: an activating Cdc20 and a Cdc20 that is part of the inhibitory MCC [13] (Fig. 1B). The ability of the MCC to bind APC/CCdc20 explains how perturbing the spindle in metaphase to reactivate the SAC can rapidly inhibit active APC/C [23]. The activating and inhibitory functions of Cdc20 can be separated both in vivo and in vitro [13], [15], [16], [36]. Consistent with the MCC being a stoichiometric inhibitor of APC/CCdc20, partial reduction of MCC components is sufficient to cause a SAC defect, whereas partial reduction of SAC catalysts such as Mad1 has a less significant effect [37], [38].
The MCC is formed from two different subcomplexes, namely Mad3/BubR1-Bub3 and Mad2-Cdc20 [2], [3], [4], [5]. The Mad3/BubR1-Bub3 complex exists throughout the cell cycle and it is not thought to be regulated [30], [39]. By contrast, the formation of the Mad2-Cdc20 complex is catalyzed by unattached kinetochores and it is the rate-limiting reaction in checkpoint signaling [17], [40], [41]. Unattached kinetochores act in two steps to promote the formation of the Mad2-Cdc20 complex: (1) local concentration of SAC components and (2) catalysis of Mad2-Cdc20 complex formation.
According to the highly influential Mad2 template mechanism [42], formation of the Mad2-Cdc20 complex is promoted by the structurally related Mad1-Mad2 complex [43], [44]. Mad2 interacts with both Mad1 and Cdc20 in a similar manner through a domain in Mad2’s C-terminus known as the “safety belt” that wraps around a Mad2-interacting motif (MIM) in Mad1 and Cdc20 [43], [44]. Whereas the Mad1-Mad2 complex is stable and present throughout the cell cycle, formation of Mad2-Cdc20 complexes is catalyzed by unattached kinetochores during mitosis. The initiation step in spindle checkpoint signaling is recruitment of the Mad1-Mad2 complex to unattached kinetochores. In metazoans, this recruitment event is not well understood and involves multiple kinetochore receptors, including Bub1-Bub3, the RZZ (Rod, Zwilch, ZW10) complex and the Ndc80 complex. The activity of Mps1, an essential spindle checkpoint kinase, is also critical for the recruitment of Mad1-Mad2 complexes to kinetochores [45] (see below).
Once localized, the Mad1-Mad2 complex recruits free, cytosolic Mad2; subsequent events at the kinetochore convert this recruited Mad2 into a form that is bound to Cdc20 [43], [44]. The Mad2-Cdc20 dimer then rapidly interacts with the BubR1-Bub3 complex to assemble the full MCC, which diffuses away to bind APC/CCdc20 and inhibits its activity. The ability of MCC to inhibit the APC/CCdc20 is achieved by a region in BubR1/Mad3 that is composed of APC/C degrons: a D-box, a KEN box and two ABBA motifs. These degrons organize into a D-box – ABBA – KEN – ABBA cassette which wraps around the activating molecule of Cdc20 in the APC/C and functions as a pseudosubstrate inhibitor, preventing the APC/C from binding its substrates [13], [14], [15], [16], [36]. In addition, MCC-bound APC/CCdc20 is limited in its ability to engage with E2 ubiquitin conjugating enzymes [15], [16].
Microtubule attachment silences SAC signaling through multiple different mechanisms. First, microtubules promote the dynein motor-dependent “stripping” of SAC proteins such as Mad1-Mad2 from the kinetochore [46], [47], [48], [49]. Second, kinetochore-localized protein phosphatases dephosphorylate the scaffold protein Knl1, thereby removing Bub1-Bub3 from kinetochores [50], [51], [52], [53], [54], [55], [56], [57]. Third, microtubule binding by Ndc80 displaces Mps1 from kinetochores [58], [59]. In addition to the above, intra-kinetochore stretching after microtubule attachment contributes to SAC satisfaction [60], [61]. Aside from these kinetochore-dependent silencing mechanisms, two parallel cytosolic pathways - one involving the Mad2-related protein p31 associated with the AAA+ enzyme TRIP13, and one involving Cdc20 autoubiquitination - contribute to APC/CCdc20 activation by catalyzing MCC disassembly [62], [63], [64], [65], [66], [67], [68].
3. Recruitment of checkpoint components to kinetochores
The first step in spindle checkpoint signaling is the recruitment of checkpoint proteins to unattached kinetochores. SAC proteins localize to the outer kinetochore region, facilitated by the KMN (Knl1, Mis12 and Ndc80) network [1]. Understanding the molecular mechanisms that regulate the recruitment of SAC components to the kinetochore is critical to understand how the SAC is activated and how different mechanisms silence the SAC once kinetochores are attached.
3.1. Bub1, BubR1 and Bub3
Pioneering work in species such as yeast has shown that Bub1 is a critical upstream component of the SAC [69], [70]. While the kinase activity of Bub1 does not play an essential role in the SAC [71], [72], [73], [74], [75], [76], Bub1’s primary role appears to be to recruit other SAC components, such as Mad1, Mad2, Cdc20 and BubR1, and to catalyze local formation of the Mad2-Cdc20 complex.
In the current model for checkpoint signaling, Bub1 is co-recruited to kinetochores with its binding partner, the WD40 domain-containing protein Bub3 [77] (Figs. 2, 3A). During mitosis, the Bub1-Bub3 complex binds to the Knl1 component of the KMN network [78]. Knl1 possesses repetitive sequences in its N-terminus known as MELTs (named after the single amino acid code) that are phosphorylated in mitosis by Mps1 and, to a lesser extent, by Plk1 [75], [78], [79], [80], [81], [82] (Fig. 3A). Phosphorylation of the Knl1 MELTs creates binding sites for the Bub1-Bub3 complex [78], [79], [80]. Bub3 directly docks onto phosphorylated MELTs and its partner Bub1 increases the affinity of the Bub3–phospho-MELT interaction [83]. Depending on the organism, Knl1 has between 2 and 27 MELT repeats spread across its N-terminal region [84]. Other species-specific motifs adjacent to the MELTs likely also contribute to the affinity of Knl1 for Bub1-Bub3 [85]. Systematic analyses of the different MELTs in Knl1 suggest that they may have different affinities for the Bub1-Bub3 complex [75], [86], [87], [88], [89], but why a significant number of MELTs and related motifs are present in Knl1 is unclear. One attractive possibility is that this multiplicity of motifs enables tuning the stoichiometry of Bub1-Bub3 complexes relative to the Knl1 scaffold at kinetochores, in a manner dictated by the microtubule attachment status [90].
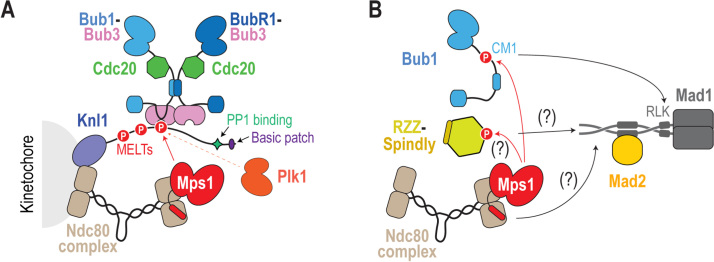
Fig. 3.
Mechanisms for SAC protein recruitment at kinetochores. (A) The Bub1-Bub3-BubR1-Cdc20 module docks onto repetitive motifs in the Knl1 N-terminus that are phosphorylated by the kinase Mps1, which is recruited to kinetochores by the Ndc80 complex; Plk1 kinase activity targeting Knl1 also contributes to a varying extent, depending on the species. (B) Localization of the Mad1-Mad2 complex to unattached kinetochores is mediated by Bub1, the RZZ complex and the Ndc80-Mps1 module. Mps1 phosphorylates Bub1 CM1 to promote its interaction with Mad1 and may also target the RZZ complex; the Ndc80 complex may also contribute independently of recruiting Mps1. While not depicted here, the RZZ complex promotes expansion of the fibrous corona, the outermost region of the kinetochore, into crescents and rings that do not contain Bub1; thus, Bub1 and RZZ may recruit spatially distinct pools of Mad1-Mad2 complexes.
Bub1 is structurally related to the MCC component BubR1, which also interacts with Bub3 [77] (Figs. 2, 3A). However BubR1 lacks the Bub1 loop that increases the affinity of the interaction between Bub3 and phospho-MELTs by an order of magnitude [35], [91]. Instead, BubR1-Bub3 recruitment primarily relies on formation of a heterotetrameric complex with the Bub1-Bub3 complex that depends on the presence of a region downstream of their Bub3 binding motifs [91], [92] (Fig. 3A). The interaction between Bub1-Bub3 and BubR1-Bub3 can be recapitulated in vitro, in the absence of post-translational modifications [91], but may be subjected to post-translational control in vivo [93]. Several species express a BubR1 orthologue known as Mad3, which lacks the C-terminal region downstream of the Bub3 binding domain and does not localize to kinetochores [94], [95]; an exception is S. pombe Mad3, which does not interact with Bub3 but still localizes to kinetochores through an interaction with Bub1 that depends on its N-terminal TPR region [96].
While the mechanism of BubR1 recruitment to kinetochores in humans is clear, preventing the Bub1-BubR1 interaction does not inhibit checkpoint signaling [91], [97]. Consistent with these perturbations of BubR1 localization in human cells, in organisms such as S. cerevisiae and C. elegans, the BubR1 orthologue Mad3 is essential for SAC signaling as integral component of the MCC but does not localize to unattached kinetochores [94], [95]. These observations support the notion that kinetochores act primarily to catalyze the formation of the Mad2-Cdc20 complex, which then interacts with BubR1/Mad3-Bub3 to form the MCC. While not critical for checkpoint signaling, kinetochore-localization of BubR1 is important for regulating kinetochore-microtubule attachments, at least in part by recruitment of the PP2A-B56 phosphatase [98], [99], [100], [101].
An interesting aspect of Bub1, BubR1 and Bub3 is that significant levels of these proteins remain at kinetochores upon microtubule attachment, unlike other SAC proteins such as Mad1 and Mad2 [90]. Because most of our understanding of how Bub1, BubR1 and Bub3 are recruited in human cells comes from studies where cells are treated with nocodazole to prevent kinetochore-microtubule attachments, it will be interesting to determine if these same mechanisms contribute to Bub1 localization at attached kinetochores. For instance, Mps1 is greatly reduced at attached kinetochores in human cells but because the Bub proteins persist, it is possible that the Plk1 kinase, which phosphorylates a similar primary sequence as Mps1 [102], is important to maintain Bub proteins following attachment [81], [82], [103]. The function of Bub proteins at attached kinetochores is not clear, but could include promoting chromosome alignment, mediating the recruitment of Aurora B to inner centromeres, and maintaining a foundational level of SAC components at the kinetochore in order to enable the SAC to respond rapidly if attachments are lost [104].
3.2. Mps1
The mitotic kinase Mps1 acts at multiple steps of the SAC. Among the known functions of Mps1 kinase are (1) phosphorylation of the Knl1 MELT repeats that mediate the recruitment of the Bub1-Bub3 complex to kinetochores (see above), (2) release of the Mad1-Mad2 complex from nuclear pores at the beginning of mitosis [105], [106], (3) recruitment of the Mad1-Mad2 complex to unattached kinetochores [107], [108], [109], [110], (4) catalysis of the formation of Mad2-Cdc20 complexes [17] and (5) promotion of the expansion of the outermost fibrous corona region of the kinetochore in the absence of microtubule attachments [111], [112], presumably a means to amplify the SAC signal emanating from individual kinetochores. Mps1 is notably absent in C. elegans, where many of its functions are compensated for by Plk1 [82].
Mps1 is recruited to kinetochores through a direct interaction with the Ndc80 complex [113], [114], [115], a component of the KMN network that is essential to form load-bearing microtubule attachments [1] (Fig. 3A). The Ndc80 complex is composed of Spc24, Spc25, Nuf2 and Ndc80 (Hec1 in humans); the latter binds microtubules directly through its N-terminal calponin-homology (CH) domain [1]. The N-terminus of Mps1, which possesses a TPR domain, an N-terminal extension (NTE) and a middle region (MR), is critical for kinetochore recruitment (Fig. 2). The NTE self-interacts with the TPR domain of Mps1; during mitosis the NTE is released from the TPR to allow for the NTE and the TPR domain to interact with the CH domain of Ndc80 [58], [59], [116]. Consistent with the Mps1-Ndc80 interaction being important, deletion of the Mps1 N-terminus prevents kinetochore targeting of Mps1 and abolishes SAC signaling [115]. The NTE itself is part of an Mps1 kinase autoinhibitory mechanism that is released by Mps1 autophosphorylation [117], but whether relief of autoinhibition relates to Mps1 kinetochore targeting is unclear. Interestingly, Mps1 kinase activity negatively regulates its own targeting to kinetochores; autophosphorylation of Mps1 causes its release from kinetochores whereas inhibition of Mps1 activity with small molecule inhibitors increases the amount of Mps1 at kinetochores [108], [118], [119], but the importance of this activity-dependent regulation of localization is not clear. In vitro, the MR of Mps1 interacts directly with the Nuf2 component of the Ndc80 complex [58]. While a fragment of Mps1 containing a repeat of the MR is sufficient for kinetochore targeting via Nuf2, deletion of the MR does not prevent Mps1 recruitment [58]. However, mutation of S281 within the MR, which is phosphorylated by Cdk1, prevents Mps1 recruitment [120]. Therefore, it is unclear how the MR contributes to Mps1 kinetochore recruitment. Interestingly in fission yeast, kinetochore recruitment of Mph1, the orthologue of Mps1, also requires an interaction with Ndc80 that depends on its N-terminus, even though Mph1 does not possess recognizable NTE, TPR or MR domains [121], [122].
Mps1 recruitment is strongly dependent on phosphorylation by Cdk1 and Aurora B [107], [123], [124]. While the targets of Aurora B that promote Mps1 recruitment have not been fully elucidated, in most organisms Aurora B inhibition weakens but does not abolish checkpoint signaling [125], [126], [127], an observation that is at odds with Mps1 kinetochore recruitment being essential for the SAC. In human cells, ARHGEF17 was also shown to contribute to Mps1 kinetochore recruitment and to spindle checkpoint signaling, but the mechanism by which it acts is not understood [128].
In humans, Mps1 recruitment is observed at unattached kinetochores. In vitro work showed that the binding of Ndc80 to microtubules displaces Mps1, suggesting a direct link between microtubule binding to kinetochores and Mps1 removal from the kinetochore to turn off checkpoint signaling [58], [59]. However, in systems such as S. cerevisiae, Mps1 recruitment to kinetochores is not attachment-regulated [119], [129], indicating that other mechanisms participate in shutting off Mps1 signaling. In budding yeast, displacement of substrates from Mps1 following attachment is the currently favored mechanism (see below).
3.3. Mad1-Mad2 complex
The recruitment of the Mad1-Mad2 complex to kinetochores is the most important event in SAC signaling and, at the same time, the least understood outside of yeast (Fig. 3B). Mad1 and Mad2 form a heterotetrameric complex throughout the cell cycle [33]. In interphase, the Mad1-Mad2 complex is localized at nuclear pore complexes, specifically at a region known as the basket, where it interacts with Tpr/Megator [130], [131], [132], [133]; Tpr/Megator promotes Mad1-Mad2 proteostasis in interphase [133], [134]. In yeast, the Mad1-Mad2 complex has a role in nuclear transport [135] but whether this function is conserved in metazoans is unclear. Nuclear pore-localized Mad1-Mad2 complexes may also catalyze formation of Mad2-Cdc20 complexes in interphase [68], [136]. During mitotic entry, the activities of Mps1 and Mad1-tethered Cdk1-cyclin B displace the Mad1-Mad2 complex from the nuclear pore to promote its recruitment to unattached kinetochores [105], [106]. In most systems, recruitment of the Mad1-Mad2 complex depends on Mad1 alone, although in S. cerevisiae Mad1 and Mad2 recruitment is co-dependent [94].
The Mad1-Mad2 complex directly interacts with Bub1, which possesses a conserved motif (CM1) that is phosphorylated by the sequential action of the mitotic kinases Cdk1 and Mps1 [73], [137], [138], [139], [140], [141] (Fig. 3B). Phosphorylated CM1 directly interacts with a C-terminal RLK motif of Mad1 [137], [138], [139], [142]. Recently, the crystal structure of the human Mad1 C-terminus – Bub1 CM1 complex was solved, revealing how phosphorylation of T461 in the Bub1 CM1 creates an interaction surface for the RLK motif of Mad1 [143]. In yeast, the Mad1-Bub1 interaction is sufficient for kinetochore recruitment of the Mad1-Mad2 complex [137], [144]. However, in metazoans, there are at least two other kinetochore components aside from Bub1 that contribute to the recruitment of the Mad1-Mad2 complex: the RZZ (Rod, Zwilch, ZW10) complex and Ndc80 [113], [145], [146] (Fig. 3B). Moreover, the N-terminus of Mad1 also contributes to the recruitment of Mad1-Mad2 to kinetochores [136], [147]. Therefore, the mechanisms that recruit Mad1-Mad2 to kinetochores in metazoans have proved challenging to elucidate. In C. elegans, mutating the Bub1 CM1 does not affect the recruitment of Mad1-Mad2 to kinetochores; instead, a different interaction between the Bub1 kinase domain and a region of the coiled-coil domain in Mad1 is necessary [75], [141].
Recently, the role of human Bub1 in Mad1-Mad2 complex recruitment and/or SAC signaling was called into question, which was surprising given that it is essential for SAC signaling in all other species tested, including mice [148]. While some groups reported a SAC defect upon Bub1 depletion [73], [112], [149], [150], [151], others suggested that Bub1 does not contribute to the SAC in humans [152], [153], [154]. However, recent efforts have clarified that, as Bub1 is a catalytic component of the SAC, residual levels of Bub1 that remain either upon incomplete RNAi experiments or through CRISPR-Cas9 edits that can be bypassed by the RNA splicing machinery, are sufficient to support checkpoint signaling [112], [151], [155]. Since other mammalian species such as mice rely on Bub1 for the SAC [148], it is also possible that human cell lines recovered after complete Bub1 gene knockout may have selected for compensatory mechanisms that bypass its requirement for the SAC [154], [156]. Thus, after some confusion, there is now a consensus that penetrant removal of Bub1 is required to observe a functional effect on the SAC in human cells, in agreement with its proposed role as a catalyst in SAC signaling.
In metazoans, the RZZ complex contributes to Mad1-Mad2 kinetochore localization [145], [146] (Fig. 3B). The RZZ complex recruits the coiled-coil dynein activator Spindly that in turn recruits the dynein-dynactin complex to kinetochores to promote chromosome capture and bi-orientation [49]. Interestingly, Spindly is required for Mad1-Mad2 recruitment and checkpoint signaling in C. elegans but not in human cells [157], [158]. Whether RZZ interacts directly with the Mad1-Mad2 complex remains to be clarified. In addition, the relationship between RZZ and Bub1-recruited pools of Mad1-Mad2 at the kinetochore has not been resolved, but interesting possibilities include recruitment to spatially distinct kinetochore regions or in temporally distinct mitotic stages [112], [140], [152]. In addition, Bub1 is also partially required for RZZ recruitment, suggesting an interplay between these two complexes [151]. The RZZ-Spindly complex is central to expansion of the outermost kinetochore region, the fibrous corona; in the absence of microtubule attachment, Mad1-Mad2 localize to this expanded corona region [159], [160]. In human cells deleted for the RZZ subunit Rod, corona expansion does not occur but Mad1 is still observed localizing to the outer kinetochore, albeit only transiently [112]. Thus, it is possible that the effect of RZZ or Spindly inhibitions on Mad1-Mad2 recruitment is at least in part due to their contribution to corona expansion.
Finally, the Ndc80 complex is essential for Mad1-Mad2 complex recruitment to unattached kinetochores [113] (Fig. 3B). As the Ndc80 complex localizes Mps1 (see above), whose activity is required to recruit the Mad1-Mad2 complex, the effect of Ndc80 depletions on Mad1-Mad2 recruitment may be indirect. However, we note that in C. elegans, where Mps1 is absent, Ndc80 is nonetheless required for Mad1-Mad2 complex kinetochore localization [95], suggesting a potential Mps1-independent role for the Ndc80 complex in Mad1-Mad2 recruitment.
3.4. Cdc20
Cdc20 is both an activator of the APC/C and a checkpoint component that promotes APC/C inhibition in the absence of kinetochore-microtubule interactions. Cdc20 is also recruited to kinetochores across species, from yeast to humans [161], [162], [163], [164]. Kinetochore recruitment of Cdc20 depends on Bub1 and BubR1, which possess a motif known as ABBA (also known as the Phe-box and the A motif) that binds between blades two and three of the C-terminal WD40 domain of Cdc20 [165], [166], [167], [168], [169] (Figs. 2, 3A). In C. elegans, where Bub1 is the sole Cdc20 kinetochore receptor, mutating the ABBA motif of Bub1 abolishes Cdc20 recruitment and SAC signaling, indicating that Cdc20 recruitment to kinetochores is essential for the SAC [164]. However in humans, both kinetochore-localized Bub1 and BubR1 contribute to Cdc20 recruitment to kinetochores [167]. Whether Bub1 or BubR1 is the primary Cdc20 receptor at kinetochores is a matter of controversy, although mutants in Bub1 or BubR1 that prevent Cdc20 binding abolish or weaken the checkpoint [138], [150], [167], [168], [170]. Elucidating the role of kinetochore-localized Cdc20 in the SAC in human cells will require mutation of Bub1 and BubR1, individually and in combination. In S. cerevisiae, mutating the putative ABBA motif in Bub1 only partially affected the SAC [171], although it is not clear whether S. cerevisiae Bub1 directly interacts with Cdc20. Interestingly, Cdc20 fluxes rapidly through the kinetochore, with a half-life of 0.5–2 s depending on the organism, which is consistent with Cdc20 transiently localizing to the kinetochore but acting in the cytoplasm, either to activate the APC/C or to form part of the MCC [164], [172]. By comparison, both Bub1 and BubR1 exhibit slower turnover at kinetochores [35], [173], indicating that the Bub1-Cdc20 and BubR1-Cdc20 interactions are highly dynamic.
The summary above indicates that many questions about how checkpoint proteins are recruited to kinetochores to initiate SAC signaling remain unresolved, especially in human cells. Of critical importance is to understand Mad1-Mad2 kinetochore recruitment at a mechanistic and structural level, which remains a major open question in the field. The dynamic control of Mps1 localization - including via its own activity - also merits further investigation. On a related note, the difference in SAC signaling following Mps1 versus Aurora B inhibition, despite the significant negative impact of Aurora B inhibition on Mps1 kinetochore localization, needs to be re-investigated using newer tools such as endogenously-tagged Mps1 [124]. It will also be important to continue efforts to define the molecular basis and significance of Cdc20 kinetochore localization across systems.
4. Catalysis of Mad2-Cdc20 complex formation
The function of kinetochores in the SAC is to catalyze the formation of Mad2-Cdc20 complexes [17], [41], [174] (Fig. 4). Both Mad2 and Cdc20 are expressed throughout the cell cycle, but their ability to form a complex is kinetically disfavored [40]. In addition, mechanisms involving factors such as TRIP13-p31comet actively disassemble Mad2-Cdc20 complexes throughout the cell cycle [175] (Fig. 4A), indicating that unattached kinetochores must overcome the inherent kinetic barrier to accelerate Mad2-Cdc20 complex formation and to win the tug-of-war against disassembly. The mechanisms described in the previous section to recruit Mad2 and Cdc20 to kinetochores concentrate these proteins at kinetochores; indeed, tethering of Mad1 to attached kinetochores is sufficient to trigger a constitutive SAC [176], [177], [178], [179]. But, in addition to recruiting checkpoint components, kinetochores facilitate the assembly of Mad2-Cdc20 complexes that then leave the kinetochore to form MCCs. In vitro, the addition of purified kinetochore-containing chromosomes or catalysts such as Bub1, Mps1 and the Mad1-Mad2 complex greatly accelerates the rate of Mad2-Cdc20 complex formation [17], [41] (Fig. 4A).
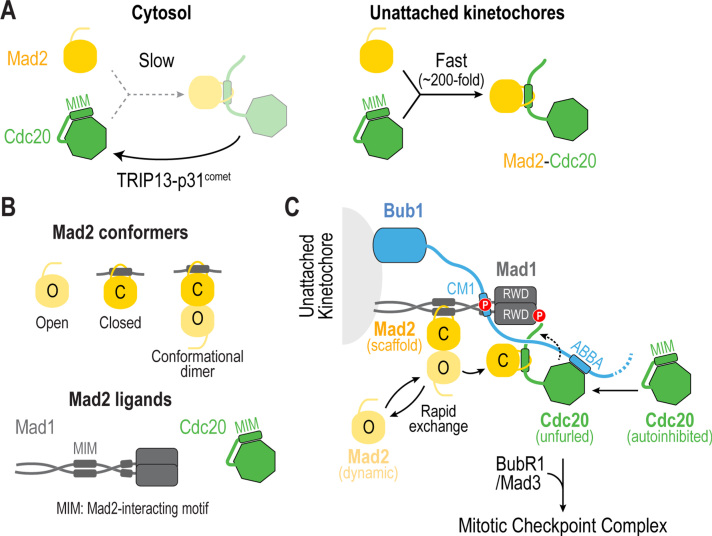
Fig. 4.
Catalysis of Mad2-Cdc20 complex formation at unattached kinetochores. (A) (left) Complex formation between Mad2 and Cdc20 is kinetically disfavored and subjected to disassembly by TRIP13-p31comet. (right) During mitosis, unattached kinetochores accelerate the rate of Mad2-Cdc20 complex formation by ~200 fold. (B) Mad2 has two different conformations, Open (O) and Closed (C), which interact with each other through dimerization. C-Mad2 is found bound to its ligands Mad1 and Mad2, which possess Mad2-interacting motifs (MIMs). (C) Proposed mechanism for how unattached kinetochores catalyze formation of the Mad2-Cdc20 complex. Mad2 is at two populations at the kinetochore: one that is stably bound to Mad1 (Mad2 scaffold) and a second that is cytosolic, transiently gets recruited to kinetochores through dimerization and becomes linked to Cdc20 (Mad2 dynamic). Kinetochores catalyze the transfer of Mad2 dynamic to Cdc20 by (1) recruiting Mad2 and Cdc20 to kinetochores, (2) positioning Cdc20 in close proximity to Mad2, and (3) unfurling Cdc20 to expose its MIM and promote the formation of the Mad2-Cdc20 complex.
Because the formation of the Mad2-Cdc20 complex formation is slow [40], this is a highly regulatable step. The exact nature of the kinetic barrier is unclear, but one proposed contributing factor is that Cdc20 is in a conformation in which its N-terminal Mad2-interacting motif is masked, thereby limiting interaction with Mad2 (Fig. 4A). In support of this view, the N-terminus of Cdc20 interacts with its C-terminal WD40 domain in vitro [174] and the Cdc20 N-terminus self-interacts [180], both of which may contribute to masking the Mad2-binding motif. In addition, truncations in Cdc20 that eliminate regions upstream or downstream of the Mad2-interacting motif make Cdc20 a better ligand for Mad2 [32], [181]. These observations suggest that catalyzing formation of the Mad2-Cdc20 complex requires exposing the Mad2-binding site on Cdc20 and placing it in a context that promotes association with Mad2.
Central to the catalysis of Mad2-Cdc20 complex formation is the ability of Mad2 to exist as two conformers: open (O) and closed (C) [43], [44] (Fig. 4B). O-Mad2 is free and cytosolic, whereas C-Mad2 is bound to its ligands Mad1 and Cdc20. In addition, the O-Mad2 and C-Mad2 conformers associate to form what is known as the conformational dimer (Fig. 4B). Conversion of Mad2 from an open to a closed state is also an inherently slow process that is accelerated by both the presence of a ligand and by dimerization [182], [183]. In the first step of catalysis, O-Mad2 is recruited from the cytosol to unattached kinetochores by the Mad1-Mad2 complex through O-Mad2–C-Mad2 dimerization [43], [44]. Kinetochore catalysts then assist in the transfer of O-Mad2 from Mad1-C-Mad2 to Cdc20 in order to form the Mad2-Cdc20 complex. In vitro, both the N-terminal tail and the C-terminal WD40 domain of Cdc20 are required to catalyze formation of the Mad2-Cdc20 complex [180]. As described below, the C-terminal WD40 domain of Cdc20 interacts with the ABBA motif of Bub1, whereas the N-terminal tail interacts with Mad1 (Fig. 4C). These two interactions likely underlie the mechanisms by which Bub1 and Mad1-Mad2 accelerate formation of Mad2-Cdc20 complexes. Below we discuss current thinking on the mechanisms promoting catalysis at kinetochores.
4.1. Bub1 positions Cdc20
Artificially targeting Mad1 to human kinetochores is sufficient to activate the checkpoint, even if all kinetochores are attached [176]. Interestingly, the arrest induced in this artificial condition is dependent on Bub1, even though Bub1 does not contribute to the localization of tethered Mad1 [112], [178]. These observations indicate that Bub1 has an essential role in the SAC that goes beyond contributing to Mad1-Mad2 complex recruitment to unattached kinetochores. Notably, data from S. pombe showed that even with kinetochore-tethered Mad1, the interaction between Mad1 and the Bub1 CM1 motif is necessary to generate a checkpoint signal [178]. An interesting natural parallel is observed in C. elegans, where the Bub1 CM1 does not contribute to Mad1 kinetochore recruitment but is nonetheless essential for the SAC [141]. These cell biological data are consistent with the observation that Bub1 accelerates the rate of Mad2-Cdc20 complex formation in vitro [17].
Following phosphorylation by mitotic kinases, the Bub1 CM1 motif interacts directly with the Mad1 C-terminus [73], [137], [138], [139], [140], [143], [178] (Fig. 4C). Phosphorylated CM1 interacts with the RLK motif, a basic patch in the coiled-coil of Mad1 that precedes its C-terminal RWD domain [142], [184] (Fig. 2). The affinity of phospho-CM1 for the Mad1 RLK is relatively low (2–16 µM); [138], [139], [143] but this low affinity may be compensated for by local concentration of both proteins on the kinetochore scaffold. Because the Bub1-Mad1 interaction contributes to the checkpoint beyond Mad1-Mad2 complex recruitment [141], [178], this interaction likely functions to constrain Bub1 and Mad1 to facilitate the formation of the Mad2-Cdc20 complex. Interestingly, the ABBA motif of Bub1, which is 30–50 amino acids downstream of the CM1 domain, recruits Cdc20 to kinetochores (Fig. 2). Because the Bub1–Cdc20 interaction promotes catalysis even in an in vitro context where kinetochore localization does not play a role [180], the simplest explanation is that the Bub1 CM1–Mad1 interaction positions Cdc20 in close proximity to the O-Mad2 associated with the Mad1-C-Mad2 scaffold [141], [180] (Fig. 4C). In support of this conclusion, the CM1 and the ABBA motif must both be present in the same Bub1 molecule to support Mad2-Cdc20 complex formation at kinetochores in C. elegans [141]. These observations suggest that Bub1 acts as a "matchmaker", with its CM1-ABBA motifs bringing together Cdc20 and O-Mad2. We note that the relative importance of Bub1 in catalyzing formation of the Mad2-Cdc20 complex in vitro depends on the concentrations of the two reactants [180], of which Cdc20 is limiting in vivo [185]. Thus, the relative “essentiality” of Bub1 in checkpoint signaling in vivo may depend on the precise cellular concentrations of Cdc20 and Mad2.
4.2. Mad1 unfurls the Cdc20 N-terminal tail to expose its Mad2-interacting motif
Aside from Bub1, the Mad1-Mad2 complex and the Mps1 kinase are critical catalysts accelerating formation of Mad2-Cdc20 complexes in vitro [17]. Mps1 phosphorylates the C-terminus of Mad1 that contains the conserved RWD domain [17], [138] (Fig. 2). Phosphorylation of the Mad1 C-terminus creates a binding surface for the Cdc20 N-terminus, at a region upstream of its Mad2-interacting motif that is known as BM1 or Box1 [138] (Fig. 2). While Mad1 and Cdc20 are not required for each other’s recruitment, both in vitro and in vivo data indicate that this interaction promotes the unfurling of the Cdc20 N-terminus to expose its Mad2 binding motif [141], [180] (Fig. 4C). In support of this view, mutating the Mad1 RWD domain or preventing phosphorylation of Mad1 by Mps1 abrogates catalysis of Mad2-Cdc20 complex formation [141], [180]. Therefore, the Mad1 RWD domain provides a critical role in promoting formation of the Mad2-Cdc20 complex. We note that, while the RWD domain is evolutionarily conserved, the BM1 of Cdc20 is not, which suggests existence of additional interaction surfaces in Cdc20 that mediate binding to the Mad1 RWD domain.
The studies described above have led to the model that kinetochores catalyze formation of Mad2-Cdc20 complexes by positioning Cdc20 in close proximity to O-Mad2 and unfurling the Mad2-binding site in Cdc20 to facilitate the coupling of O-Mad2, recruited onto the Mad1-Mad2 scaffold, with Cdc20 (Fig. 4C). While the molecular details of the Bub1 CM1-Mad1 interaction have recently been elucidated [143], biochemical and structural analysis of the Mad1-Cdc20 interaction and of Cdc20 autoinhibition will be critical to understand how kinetochores act as catalysts in the SAC. In addition to mechanistic work complemented by mutational analysis in vivo, a complementary approach being taken to understand SAC signal generation is to activate the SAC in the cytoplasm. Early work showed that overexpressing Mps1 in yeast was sufficient to activate the SAC independently of kinetochores [186], [187], a result that pre-staged the success of biochemical reconstitution efforts. Recently, artificial dimerization approaches have been developed to bring checkpoint scaffolds and catalysts, mutated to prevent their kinetochore localization, into close proximity [129], [188], [189]. These efforts indicate that SAC signal activation can be achieved in the cytoplasm independently of kinetochores and are making it feasible to probe the quantitative features of SAC signal generation.
5. Checkpoint silencing at the kinetochore
Once microtubules are attached to kinetochores, the SAC signal must be rapidly silenced to promote mitotic exit. Failure to silence the SAC leads to prolonged mitotic arrest, which can result in premature loss of sister chromatid cohesion, cell death or cell cycle arrest at the subsequent G1 [190], [191], [192], [193].
Mechanisms for checkpoint silencing can be classified into two groups: cytoplasmic and kinetochore-based. There are two cytoplasmic mechanisms: active disassembly of Mad2-Cdc20 complex by TRIP13-p31comet and Cdc20 ubiquitylation by the APC/C [62], [63], [64], [65], [66], [67], [68]. Here, we will focus on the mechanisms that silence the SAC at the kinetochore, which include dynein-mediated stripping of SAC components, intra-kinetochore stretching, and the actions of kinetochore-localized phosphatases (Fig. 5).
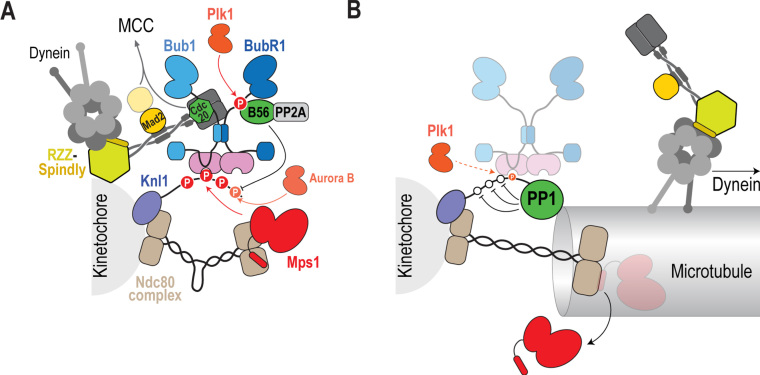
Fig. 5.
SAC silencing at kinetochores. (A) At unattached kinetochores Mps1, anchored onto the Ndc80 complex, phosphorylates the Knl1 MELT repeats, which recruit the Bub module (Bub1, BubR1, Bub3 and Cdc20). Bub1, along with the RZZ complex, recruits the Mad1-Mad2 complex. The RZZ complex also recruits Spindly-Dynein. Phosphorylation of BubR1 by Plk1 promotes recruitment of the PP2A-B56 phosphatase, which opposes phosphorylation of the PP1 binding motif on Knl1 by Aurora B. (B) Microtubule attachments trigger the poleward transport of the Mad1-Mad2 complex by RZZ-Spindly-Dynein – this is a critical event in checkpoint silencing as preventing this transport leads to an active checkpoint despite kinetochore-microtubule attachment. In addition, microtubule attachment by Ndc80 causes either the displacement of Mps1 from kinetochores or intra-kinetochore stretching (depicted here for illustrative purposes as extension of the Ndc80 complex; we emphasize however that the molecular effector whose stretch affects SAC signaling is not known) which physically separates Mps1 from Knl1. Finally, PP1 recruitment causes dephosphorylation of the Knl1 MELTs to remove the Bub module from kinetochores. A small pool of the Bub module remains at attached kinetochores, which we speculate requires Plk1’s ability to also phosphorylate Knl1 MELTs.
5.1. Dynein-mediated stripping of SAC components
Soon after their initial discovery it became evident that SAC proteins such as Mad1 and Mad2 localize to unattached but not to microtubule-attached and aligned kinetochores [94], [130]. Recent high resolution live-imaging experiments have revealed that, upon the establishment of end-on microtubule attachments, SAC components are rapidly removed from kinetochore [194]. Moreover, end-on microtubule attachments and not transient lateral attachments, remove SAC components from attached kinetochores [194]. The removal of SAC proteins upon microtubule attachment is a major mechanism for SAC silencing at the kinetochore, a process known as “stripping”. In effect, stripping removes Mad1-Mad2 complexes from the local environment that is required for them to catalyze Mad2-Cdc20 complex formation.
SAC protein stripping depends on the minus-end directed motor dynein that is recruited to kinetochores by the RZZ-Spindly complex [46], [47], [48], [49] (Fig. 5A). The RZZ complex is also required to promote the recruitment of the Mad1-Mad2 complex to unattached kinetochores (see above). The current view is that once attachments have occurred, kinetochore-localized dynein transports cargo consisting of SAC proteins such as Mad1-Mad2, Spindly and the RZZ complex, towards spindle poles (Fig. 5B). The fate of these proteins after they reach the spindle poles is unknown, but they likely disassemble and are released into the cytosol. The process of stripping can be difficult to study in vivo, but experiments where cells are depleted of ATP have helped visualize dynein-dependent accumulation of SAC components at spindle poles [46], [195], [196]. Consistent with kinetochore-localized dynein being required for stripping, a Spindly mutant that cannot recruit dynein retains the Mad1-Mad2 complex at attached kinetochores [196]. Surprisingly, however, depletion of Spindly does not prevent attachment-dependent removal of Mad1-Mad2 [195], [196], indicating the existence of another mechanism for SAC protein removal that is inhibited by Spindly at kinetochores and can only be revealed upon depletion of Spindly. Indeed, organisms such as yeast do not require kinetochore dynein to remove SAC proteins from attached kinetochores, indicating that a dynein-independent mechanism might be more ancient. Understanding the nature of this alternative pathway, how it is inhibited by Spindly and how end-on attachments activate dynein to transport SAC proteins towards spindle poles, are all important open questions. Moreover, it is unknown how the cargo for removal is formed at kinetochores because connections between RZZ-Spindly-dynein and Mad1-Mad2 have not been reported. Thus, an important focus for the future will be to overcome the lack of mechanistic understanding of Mad1-Mad2 kinetochore recruitment, especially of how the RZZ complex contributes to this recruitment, since this gap limits understanding of both checkpoint activation and microtubule attachment-regulated removal from kinetochores.
5.2. Intra-kinetochore stretching
Intra-kinetochore stretching following microtubule attachment is correlated with cessation of SAC signaling [60], [61]. Intra-kinetochore stretching occurs when kinetochores are end-on attached to microtubules and the distance between the inner and outer kinetochore regions increase [60], [61]. How intra-kinetochore stretching relates to SAC silencing is unclear, but a clue came from studies showing that artificial tethering of the Mps1 kinase to kinetochores can cause a constitutively active checkpoint when targeted to Knl1, but not when tethered to outer kinetochore components such as Ndc80 [129]. Thus, close proximity of Mps1 to Knl1 is critical to activate the SAC. Because microtubule engagement by the kinetochore may cause Ndc80 to move away from Knl1 [197], [198], an attractive model is that, upon attachment, Mps1 bound to Ndc80 is displaced from targets such as Knl1, which in turn contributes to checkpoint silencing (Fig. 5). Further analysis of the relationship between intra-kinetochore stretching and SAC silencing will benefit from perturbations designed specifically to disrupt stretching [199]. Such an effort will also require molecular definition of the mechanically compliant component(s) at kinetochores, whose extension under force is responsible for the relative displacement.
5.3. Kinetochore-localized phosphatases
Finally, kinetochore-localized phosphatases oppose mitotic kinases to promote SAC silencing [50], [51], [52], [53], [54], [55], [56], [57]. The two well-characterized kinetochore phosphatases are protein phosphatase 1 (PP1) and protein phosphatase 2 (PP2A)-B56, which are recruited by Knl1 and BubR1, respectively (Fig. 5). While PP2A-B56 is specifically recruited to unattached kinetochores [101], PP1 recruitment correlates with the establishment of kinetochore-microtubule attachments [52], [54], indicating that PP1 recruitment is a trigger to stop SAC signaling once attachments are made.
PP1 binds to Knl1 through canonical PP1-docking motifs known as SILK and RRVSF [52], [53], [54], [55] (Fig. 3A). The serine residues in these motifs are phosphorylated by Aurora kinases and phosphorylation is thought to reduce PP1 binding to Knl1 [52], [200] (Fig. 5A). Interestingly, Knl1 also possesses a basic patch motif that binds microtubules in vitro and also contributes to SAC silencing [55]. In yeast, mutating the basic patch motif prevents SAC silencing by impairing PP1 function at the kinetochore [201], although in C. elegans the basic patch appears to work independently of PP1 [55]. Surprisingly, microtubule binding and PP1 binding to Knl1 are mutually exclusive in vitro [202], which highlights the lack of consensus on whether microtubule binding to Knl1 contributes to SAC silencing at kinetochores.
PP2A-B56 is recruited to kinetochores through BubR1, whose ‘KARD’ domain contains the PP2A-B56 docking motif LxxIxE [100], [203] (Figs. 2, 5A). Recruitment of PP2A-B56 to BubR1 requires phosphorylation of the KARD domain by kinases such as Plk1 and Cdk1 [99], [100], [203]. Because PP2A-B56 is recruited specifically to unattached kinetochores, its major function might be to limit SAC signaling. BubR1-bound PP2A-B56 has also been implicated in controlling microtubule attachments to ensure proper chromosome alignment by opposing the activity of Aurora B [98], [99], [100], [125].
Preventing the recruitment of either PP1 or PP2A-B56 at the kinetochores causes a SAC silencing defect. Among the direct targets of kinetochore-localized phosphatases in the SAC are Knl1 itself, which is dephosphorylated at its MELT repeats to remove Bub1 and BubR1 from the kinetochore; the Mps1 activation loop, and Bub1/BubR1 at motifs that participate in Plk1 binding [56], [57], [103], [164], [204] (Fig. 5). Protein phosphatases are known to cross-talk; for instance, PP1 can interact with PP2A-B56 to promote its dephosphorylation and activation [205]. Conversely, PP2A-B56 dephosphorylates the PP1 docking site in Knl1 to promote PP1 kinetochore recruitment [57]. Therefore, it has been difficult to attribute specific phospho-substrates to specific phosphatases at the kinetochore. Indeed, phosphatases can substitute for each other at the kinetochore, suggesting that their roles are redundant and that they only differ in how they respond to kinase inputs during mitosis [206]. Moreover, recent work has suggested that one of the primary functions of kinetochore phosphatases is to oppose local Plk1 activity. Bub1 and BubR1 recruit Plk1 to kinetochores in a phospho-dependent manner (Fig. 2) and both PP1 and PP2A-B56 can dephosphorylate Bub1 and BubR1 to limit localized Plk1 kinase activity, which is known to phosphorylate the MELT repeats in Knl1 [103]. The complexity of protein phosphatase action at kinetochores is an exciting area of research with many open questions, with a major challenge being to deconvolve the action of specific phosphatase complexes and to understand the short linear motifs that regulate their targeting and local activities.
We note that the three mechanisms we have discussed for SAC silencing at the kinetochore may represent different facets of a common underlying mechanism. For instance, intra-kinetochore stretching may promote phosphatase recruitment that activates the dynein module [199]. Thus, in addition to work on each facet, there is also a need to develop a more holistic picture of SAC silencing at the kinetochore, and to account for features of the SAC such as its rapid reactivation when perturbations occur after full attachment [23].
6. Concluding remarks
In this review, we have discussed the events occurring at kinetochores that initiate SAC signaling and locally silence the SAC following microtubule attachment. The SAC represents a paradigm for highly localized signaling in a sub-cellular domain; thus, its mechanistic elucidation has implications not only for understanding how the SAC acts to ensure genome integrity during mitosis but also for generally understanding how highly localized events can control the physiological state of the entire cell. The SAC conceptual framework is also influencing analysis of HORMA domain proteins with a Mad2-like fold that are implicated in DNA repair, meiotic recombination, autophagy, and bacterial antiviral immunity [207].
Recent work has also revealed repurposing of mechanisms and components central to SAC signaling in other cellular processes such as mitotic entry [208], regulation of sister chromatid separation [209] and insulin signaling [210]. Moreover, mutations in SAC components such as BubR1 and TRIP13 are the cause of Mosaic Variegated Aneuploidy (MVA), a rare genetic disease associated with microcephaly, developmental delay and propensity to developing tumors [211], [212], [213], [214]. These findings highlighting that understanding of the SAC is likely to yield broader insights beyond unattached kinetochore-triggered control of mitosis. While the combination of genetics, cell biology, biochemistry, in vitro reconstitutions, and structural biology has led to an increasingly detailed view of how the SAC is wired, many key questions about local events that occur at kinetochores, highlighted in the sections above, remain unanswered. Addressing these open questions will continue to occupy SAC enthusiasts and draw in newcomers eager to understand how this ancient pathway ensures genome integrity during organismal development and protects against the chromosome missegregation-triggered genomic havoc that is a characteristic feature of cancers [215], [216].
Acknowledgements
We thank K. Corbett for many helpful discussions on the structural mechanisms underlying spindle checkpoint signaling. A.D. is funded by NIH R01 GM074215 and the Ludwig Institute for Cancer Research. J.P. is supported by CR UK Program grant C29/A24397 and an Investigator award from Wellcome.
Contributor Information
Pablo Lara-Gonzalez, Email: plgonzalez@ucsd.edu.
Jonathon Pines, Email: jon.pines@icr.ac.uk.
Arshad Desai, Email: abdesai@ucsd.edu.
References
- 1.Musacchio A., Desai A. A molecular view of kinetochore assembly and function. Biology. 2017;6(1) doi: 10.3390/biology6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lara-Gonzalez P., Westhorpe F.G., Taylor S.S. The spindle assembly checkpoint. Curr. Biol. 2012;22(22):R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Jia L., Kim S., Yu H. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem. Sci. 2013;38(6):302–311. doi: 10.1016/j.tibs.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Musacchio A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 2015;25(20):R1002–R1018. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 5.Corbett K.D. Molecular mechanisms of spindle assembly checkpoint activation and silencing. Prog. Mol. Subcell. Biol. 2017;56:429–455. doi: 10.1007/978-3-319-58592-5_18. [DOI] [PubMed] [Google Scholar]
- 6.Li R., Murray A.W. Feedback control of mitosis in budding yeast. Cell. 1991;66(3):519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 7.Hoyt M.A., Totis L., Roberts B.T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66(3):507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 8.King E.M., van der Sar S.J., Hardwick K.G. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS One. 2007;2(4):342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burton J.L., Solomon M.J. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21(6):655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sczaniecka M., Feoktistova A., May K.M., Chen J.S., Blyth J., Gould K.L., Hardwick K.G. The spindle checkpoint functions of Mad3 and Mad2 depend on a Mad3 KEN box-mediated interaction with Cdc20-anaphase-promoting complex (APC/C) J. Biol. Chem. 2008;283(34):23039–23047. doi: 10.1074/jbc.M803594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lara-Gonzalez P., Scott M.I., Diez M., Sen O., Taylor S.S. BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J. Cell Sci. 2011;124(Pt 24):4332–4345. doi: 10.1242/jcs.094763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao W.C., Kulkarni K., Zhang Z., Kong E.H., Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484(7393):208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 13.Izawa D., Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2015;517(7536):631–634. doi: 10.1038/nature13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Fiore B., Wurzenberger C., Davey N.E., Pines J. The mitotic checkpoint complex requires an evolutionary conserved cassette to bind and inhibit active APC/C. Mol. Cell. 2016;64(6):1144–1153. doi: 10.1016/j.molcel.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alfieri C., Chang L., Zhang Z., Yang J., Maslen S., Skehel M., Barford D. Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature. 2016;536(7617):431–436. doi: 10.1038/nature19083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi M., VanderLinden R., Weissmann F., Qiao R., Dube P., Brown N.G., Haselbach D., Zhang W., Sidhu S.S., Peters J.M., Stark H., Schulman B.A. Cryo-EM of mitotic checkpoint complex-bound APC/C reveals reciprocal and conformational regulation of ubiquitin ligation. Mol. Cell. 2016;63(4):593–607. doi: 10.1016/j.molcel.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faesen A.C., Thanasoula M., Maffini S., Breit C., Muller F., van Gerwen S., Bange T., Musacchio A. Basis of catalytic assembly of the mitotic checkpoint complex. Nature. 2017;542(7642):498–502. doi: 10.1038/nature21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson E.R., Brown N.G., Peters J.M., Stark H., Schulman B.A. Posing the APC/C E3 ubiquitin ligase to orchestrate cell division. Trends Cell Biol. 2019;29(2):117–134. doi: 10.1016/j.tcb.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barford D. Structural interconversions of the anaphase-promoting complex/cyclosome (APC/C) regulate cell cycle transitions. Curr. Opin. Struct. Biol. 2020;61:86–97. doi: 10.1016/j.sbi.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Rieder C.L., Khodjakov A., Paliulis L.V., Fortier T.M., Cole R.W., Sluder G. Mitosis in vertebrate somatic cells with two spindles: implications for the metaphase/anaphase transition checkpoint and cleavage. Proc. Natl. Acad. Sci. USA. 1997;94(10):5107–5112. doi: 10.1073/pnas.94.10.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S.H., Lin D.P., Matsumoto S., Kitazono A., Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279(5353):1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 22.Hwang L.H., Lau L.F., Smith D.L., Mistrot C.A., Hardwick K.G., Hwang E.S., Amon A., Murray A.W. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279(5353):1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 23.Clute P., Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1999;1(2):82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- 24.Hagting A., Den Elzen N., Vodermaier H.C., Waizenegger I.C., Peters J.M., Pines J. Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J. Cell Biol. 2002;157(7):1125–1137. doi: 10.1083/jcb.200111001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thornton B.R., Toczyski D.P. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell Biol. 2003;5(12):1090–1094. doi: 10.1038/ncb1066. [DOI] [PubMed] [Google Scholar]
- 26.Heasley L.R., Markus S.M., DeLuca J.G. “Wait anaphase” signals are not confined to the mitotic spindle. Mol. Biol. Cell. 2017;28(9):1186–1194. doi: 10.1091/mbc.E17-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rieder C.L., Cole R.W., Khodjakov A., Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 1995;130(4):941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dick A.E., Gerlich D.W. Kinetic framework of spindle assembly checkpoint signalling. Nat. Cell Biol. 2013;15(11):1370–1377. doi: 10.1038/ncb2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collin P., Nashchekina O., Walker R., Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat. Cell Biol. 2013;15(11):1378–1385. doi: 10.1038/ncb2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardwick K.G., Johnston R.C., Smith D.L., Murray A.W. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 2000;148(5):871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudakin V., Chan G.K., Yen T.J. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154(5):925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Z., Bharadwaj R., Li B., Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 2001;1(2):227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 33.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell. 2002;13(3):755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han J.S., Vitre B., Fachinetti D., Cleveland D.W. Bimodal activation of BubR1 by Bub3 sustains mitotic checkpoint signaling. Proc. Natl. Acad. Sci. USA. 2014;111(40):E4185–E4193. doi: 10.1073/pnas.1416277111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overlack K., Bange T., Weissmann F., Faesen A.C., Maffini S., Primorac I., Muller F., Peters J.M., Musacchio A. BubR1 promotes Bub3-dependent APC/C inhibition during spindle assembly checkpoint signaling. Curr. Biol. 2017;27(19):2915–2927. doi: 10.1016/j.cub.2017.08.033. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sewart K., Hauf S. Different functionality of Cdc20 binding sites within the mitotic checkpoint complex. Curr. Biol. 2017;27(8):1213–1220. doi: 10.1016/j.cub.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Heinrich S., Geissen E.M., Kamenz J., Trautmann S., Widmer C., Drewe P., Knop M., Radde N., Hasenauer J., Hauf S. Determinants of robustness in spindle assembly checkpoint signalling. Nat. Cell Biol. 2013;15(11):1328–1339. doi: 10.1038/ncb2864. [DOI] [PubMed] [Google Scholar]
- 38.Meraldi P., Draviam V.M., Sorger P.K. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7(1):45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Chen R.H. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 2002;158(3):487–496. doi: 10.1083/jcb.200204048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simonetta M., Manzoni R., Mosca R., Mapelli M., Massimiliano L., Vink M., Novak B., Musacchio A., Ciliberto A. The influence of catalysis on mad2 activation dynamics. PLoS Biol. 2009;7(1):10. doi: 10.1371/journal.pbio.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulukian A., Han J.S., Cleveland D.W. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell. 2009;16(1):105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., Musacchio A. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 2005;15(3):214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 43.Yu H. Structural activation of Mad2 in the mitotic spindle checkpoint: the two-state Mad2 model versus the Mad2 template model. J. Cell Biol. 2006;173(2):153–157. doi: 10.1083/jcb.200601172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mapelli M., Musacchio A. MAD contortions: conformational dimerization boosts spindle checkpoint signaling. Curr. Opin. Struct. Biol. 2007;17(6):716–725. doi: 10.1016/j.sbi.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 45.Pachis S.T., Kops G. Leader of the SAC: molecular mechanisms of Mps1/TTK regulation in mitosis. Open Biol. 2018;8(8):180109. doi: 10.1098/rsob.180109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., Salmon E.D. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001;155(7):1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wojcik E., Basto R., Serr M., Scaerou F., Karess R., Hays T. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 2001;3(11):1001–1007. doi: 10.1038/ncb1101-1001. [DOI] [PubMed] [Google Scholar]
- 48.Basto R., Scaerou F., Mische S., Wojcik E., Lefebvre C., Gomes R., Hays T., Karess R. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr. Biol. 2004;14(1):56–61. doi: 10.1016/j.cub.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 49.Griffis E.R., Stuurman N., Vale R.D. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 2007;177(6):1005–1015. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinsky B.A., Nelson C.R., Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr. Biol. 2009;19(14):1182–1187. doi: 10.1016/j.cub.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanoosthuyse V., Hardwick K.G. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 2009;19(14):1176–1181. doi: 10.1016/j.cub.2009.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu D., Vleugel M., Backer C.B., Hori T., Fukagawa T., Cheeseman I.M., Lampson M.A. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 2010;188(6):809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenberg J.S., Cross F.R., Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 2011;21(11):942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meadows J.C., Shepperd L.A., Vanoosthuyse V., Lancaster T.C., Sochaj A.M., Buttrick G.J., Hardwick K.G., Millar J.B. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev. Cell. 2011;20(6):739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espeut J., Cheerambathur D.K., Krenning L., Oegema K., Desai A. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol. 2012;196(4):469–482. doi: 10.1083/jcb.201111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Espert A., Uluocak P., Bastos R.N., Mangat D., Graab P., Gruneberg U. PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing. J. Cell Biol. 2014;206(7):833–842. doi: 10.1083/jcb.201406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nijenhuis W., Vallardi G., Teixeira A., Kops G.J., Saurin A.T. Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat. Cell Biol. 2014;16(12):1257–1264. doi: 10.1038/ncb3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ji Z., Gao H., Yu H. CELL DIVISION CYCLE. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science. 2015;348(6240):1260–1264. doi: 10.1126/science.aaa4029. [DOI] [PubMed] [Google Scholar]
- 59.Hiruma Y., Sacristan C., Pachis S.T., Adamopoulos A., Kuijt T., Ubbink M., von Castelmur E., Perrakis A., Kops G.J. CELL DIVISION CYCLE. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science. 2015;348(6240):1264–1267. doi: 10.1126/science.aaa4055. [DOI] [PubMed] [Google Scholar]
- 60.Uchida K.S., Takagaki K., Kumada K., Hirayama Y., Noda T., Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 2009;184(3):383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maresca T.J., Salmon E.D. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 2009;184(3):373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Habu T., Kim S.H., Weinstein J., Matsumoto T. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 2002;21(23):6419–6428. doi: 10.1093/emboj/cdf659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Westhorpe F.G., Tighe A., Lara-Gonzalez P., Taylor S.S. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J. Cell Sci. 2011;124(Pt 22):3905–3916. doi: 10.1242/jcs.093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mansfeld J., Collin P., Collins M.O., Choudhary J.S., Pines J. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat. Cell Biol. 2011;13(10):1234–1243. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foster S.A., Morgan D.O. The APC/C subunit Mnd2/Apc15 promotes Cdc20 autoubiquitination and spindle assembly checkpoint inactivation. Mol. Cell. 2012;47(6):921–932. doi: 10.1016/j.molcel.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uzunova K., Dye B.T., Schutz H., Ladurner R., Petzold G., Toyoda Y., Jarvis M.A., Brown N.G., Poser I., Novatchkova M., Mechtler K., Hyman A.A., Stark H., Schulman B.A., Peters J.M. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat. Struct. Mol. Biol. 2012;19(11):1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye Q., Rosenberg S.C., Moeller A., Speir J.A., Su T.Y., Corbett K.D. TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. Elife. 2015;4:07367. doi: 10.7554/eLife.07367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim D.H., Han J.S., Ly P., Ye Q., McMahon M.A., Myung K., Corbett K.D., Cleveland D.W. TRIP13 and APC15 drive mitotic exit by turnover of interphase- and unattached kinetochore-produced MCC. Nat. Commun. 2018;9(1):4354. doi: 10.1038/s41467-018-06774-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roberts B.T., Farr K.A., Hoyt M.A. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell Biol. 1994;14(12):8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hardwick K.G., Murray A.W. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 1995;131(3):709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharp-Baker H., Chen R.H. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 2001;153(6):1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fernius J., Hardwick K.G. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 2007;3(11):213. doi: 10.1371/journal.pgen.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klebig C., Korinth D., Meraldi P. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 2009;185(5):841–858. doi: 10.1083/jcb.200902128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perera D., Taylor S.S. Sgo1 establishes the centromeric cohesion protection mechanism in G2 before subsequent Bub1-dependent recruitment in mitosis. J. Cell Sci. 2010;123(Pt 5):653–659. doi: 10.1242/jcs.059501. [DOI] [PubMed] [Google Scholar]
- 75.Moyle M.W., Kim T., Hattersley N., Espeut J., Cheerambathur D.K., Oegema K., Desai A., Bub1-Mad1 A. interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J. Cell Biol. 2014;204(5):647–657. doi: 10.1083/jcb.201311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baron A.P., von Schubert C., Cubizolles F., Siemeister G., Hitchcock M., Mengel A., Schroder J., Fernandez-Montalvan A., von Nussbaum F., Mumberg D., Nigg E.A. Probing the catalytic functions of Bub1 kinase using the small molecule inhibitors BAY-320 and BAY-524. Elife. 2016;5:12187. doi: 10.7554/eLife.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor S.S., Ha E., McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 1998;142(1):1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.London N., Ceto S., Ranish J.A., Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 2012;22(10):900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., Millar J.B. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 2012;22(10):891–899. doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamagishi Y., Yang C.H., Tanno Y., Watanabe Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 2012;14(7):746–752. doi: 10.1038/ncb2515. [DOI] [PubMed] [Google Scholar]
- 81.von Schubert C., Cubizolles F., Bracher J.M., Sliedrecht T., Kops G., Nigg E.A. Plk1 and Mps1 cooperatively regulate the spindle assembly checkpoint in human cells. Cell Rep. 2015;12(1):66–78. doi: 10.1016/j.celrep.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 82.Espeut J., Lara-Gonzalez P., Sassine M., Shiau A.K., Desai A., Abrieu A. Natural loss of Mps1 kinase in nematodes uncovers a role for polo-like kinase 1 in spindle checkpoint initiation. Cell Rep. 2015;12(1):58–65. doi: 10.1016/j.celrep.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Primorac I., Weir J.R., Chiroli E., Gross F., Hoffmann I., van Gerwen S., Ciliberto A., Musacchio A. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. Elife. 2013;2:01030. doi: 10.7554/eLife.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vleugel M., Hoogendoorn E., Snel B., Kops G.J. Evolution and function of the mitotic checkpoint. Dev. Cell. 2012;23(2):239–250. doi: 10.1016/j.devcel.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 85.Vleugel M., Omerzu M., Groenewold V., Hadders M.A., Lens S.M.A., Kops G. Sequential multisite phospho-regulation of KNL1-BUB3 interfaces at mitotic kinetochores. Mol. Cell. 2015;57(5):824–835. doi: 10.1016/j.molcel.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 86.Vleugel M., Tromer E., Omerzu M., Groenewold V., Nijenhuis W., Snel B., Kops G.J. Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. J. Cell Biol. 2013;203(6):943–955. doi: 10.1083/jcb.201307016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang G., Lischetti T., Nilsson J. A minimal number of MELT repeats supports all the functions of KNL1 in chromosome segregation. J. Cell Sci. 2014;127(Pt 4):871–884. doi: 10.1242/jcs.139725. [DOI] [PubMed] [Google Scholar]
- 88.Krenn V., Overlack K., Primorac I., van Gerwen S., Musacchio A. KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT repeats. Curr. Biol. 2014;24(1):29–39. doi: 10.1016/j.cub.2013.11.046. [DOI] [PubMed] [Google Scholar]
- 89.Roy B., Han S.J., Fontan A.N., Joglekar A.P. The copy-number and varied strengths of MELT motifs in Spc105 balance the strength and responsiveness of the spindle assembly checkpoint. Elife. 2020;9:55096. doi: 10.7554/eLife.55096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Etemad B., Vertesy A., Kuijt T.E.F., Sacristan C., van Oudenaarden A., Kops G. Spindle checkpoint silencing at kinetochores with submaximal microtubule occupancy. J. Cell Sci. 2019;132(12):231589. doi: 10.1242/jcs.231589. [DOI] [PubMed] [Google Scholar]
- 91.Overlack K., Primorac I., Vleugel M., Krenn V., Maffini S., Hoffmann I., Kops G.J., Musacchio A. A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. Elife. 2015;4:05269. doi: 10.7554/eLife.05269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang G., Lischetti T., Hayward D.G., Nilsson J. Distinct domains in Bub1 localize RZZ and BubR1 to kinetochores to regulate the checkpoint. Nat. Commun. 2015;6:7162. doi: 10.1038/ncomms8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Taylor S.S., Hussein D., Wang Y., Elderkin S., Morrow C.J. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 2001;114(Pt 24):4385–4395. doi: 10.1242/jcs.114.24.4385. [DOI] [PubMed] [Google Scholar]
- 94.Gillett E.S., Espelin C.W., Sorger P.K. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 2004;164(4):535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Essex A., Dammermann A., Lewellyn L., Oegema K., Desai A. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol. Biol. Cell. 2009;20(4):1252–1267. doi: 10.1091/mbc.E08-10-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leontiou I., London N., May K.M., Ma Y., Grzesiak L., Medina-Pritchard B., Amin P., Jeyaprakash A.A., Biggins S., Hardwick K.G. The Bub1-TPR domain interacts directly with Mad3 to generate robust spindle checkpoint arrest. Curr. Biol. 2019;29(14):2407–2414. doi: 10.1016/j.cub.2019.06.011. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang G., Mendez B.L., Sedgwick G.G., Nilsson J. Two functionally distinct kinetochore pools of BubR1 ensure accurate chromosome segregation. Nat. Commun. 2016;7:12256. doi: 10.1038/ncomms12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lampson M.A., Kapoor T.M. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 2005;7(1):93–98. doi: 10.1038/ncb1208. [DOI] [PubMed] [Google Scholar]
- 99.Elowe S., Hummer S., Uldschmid A., Li X., Nigg E.A. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21(17):2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Suijkerbuijk S.J., Vleugel M., Teixeira A., Kops G.J. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell. 2012;23(4):745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 101.Foley E.A., Maldonado M., Kapoor T.M. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 2011;13(10):1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dou Z., von Schubert C., Korner R., Santamaria A., Elowe S., Nigg E.A. Quantitative mass spectrometry analysis reveals similar substrate consensus motif for human Mps1 kinase and Plk1. PLoS One. 2011;6(4):18793. doi: 10.1371/journal.pone.0018793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cordeiro M.H., Smith R.J., Saurin A.T. Kinetochore phosphatases suppress autonomous Polo-like kinase 1 activity to control the mitotic checkpoint. J. Cell Biol. 2020;219(12):202002020. doi: 10.1083/jcb.202002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim T., Gartner A. Bub1 kinase in the regulation of mitosis. Anim. Cells Syst. 2021;25(1):1–10. doi: 10.1080/19768354.2021.1884599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cunha-Silva S., Osswald M., Goemann J., Barbosa J., Santos L.M., Resende P., Bange T., Ferras C., Sunkel C.E., Conde C. Mps1-mediated release of Mad1 from nuclear pores ensures the fidelity of chromosome segregation. J. Cell Biol. 2020;219(3):201906039. doi: 10.1083/jcb.201906039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jackman M., Marcozzi C., Barbiero M., Pardo M., Yu L., Tyson A.L., Choudhary J.S., Pines J. Cyclin B1-Cdk1 facilitates MAD1 release from the nuclear pore to ensure a robust spindle checkpoint. J. Cell Biol. 2020;219(6):201907082. doi: 10.1083/jcb.201907082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Santaguida S., Tighe A., D’Alise A.M., Taylor S.S., Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 2010;190(1):73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hewitt L., Tighe A., Santaguida S., White A.M., Jones C.D., Musacchio A., Green S., Taylor S.S. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J. Cell Biol. 2010;190(1):25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maciejowski J., George K.A., Terret M.E., Zhang C., Shokat K.M., Jallepalli P.V. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J. Cell Biol. 2010;190(1):89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Abrieu A., Magnaghi-Jaulin L., Kahana J.A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D.W., Labbe J.C. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106(1):83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 111.Sacristan C., Ahmad M.U.D., Keller J., Fermie J., Groenewold V., Tromer E., Fish A., Melero R., Carazo J.M., Klumperman J., Musacchio A., Perrakis A., Kops G.J. Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis. Nat. Cell Biol. 2018;20(7):800–810. doi: 10.1038/s41556-018-0130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rodriguez-Rodriguez J.A., Lewis C., McKinley K.L., Sikirzhytski V., Corona J., Maciejowski J., Khodjakov A., Cheeseman I.M., Jallepalli P.V. Distinct roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr. Biol. 2018;28(21):3422–3429. doi: 10.1016/j.cub.2018.10.006. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Martin-Lluesma S., Stucke V.M., Nigg E.A. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297(5590):2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- 114.Kemmler S., Stach M., Knapp M., Ortiz J., Pfannstiel J., Ruppert T., Lechner J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28(8):1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nijenhuis W., von Castelmur E., Littler D., De Marco V., Tromer E., Vleugel M., van Osch M.H., Snel B., Perrakis A., Kops G.J. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J. Cell Biol. 2013;201(2):217–231. doi: 10.1083/jcb.201210033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pachis S.T., Hiruma Y., Tromer E.C., Perrakis A., Kops G. Interactions between N-terminal modules in MPS1 enable spindle checkpoint silencing. Cell Rep. 2019;26(8):2101–2112. doi: 10.1016/j.celrep.2019.01.017. e6. [DOI] [PubMed] [Google Scholar]
- 117.Combes G., Barysz H., Garand C., Gama Braga L., Alharbi I., Thebault P., Murakami L., Bryne D.P., Stankovic S., Eyers P.A., Bolanos-Garcia V.M., Earnshaw W.C., Maciejowski J., Jallepalli P.V., Elowe S. Mps1 phosphorylates Its N-terminal extension to relieve autoinhibition and activate the spindle assembly checkpoint. Curr. Biol. 2018;28(6):872–883. doi: 10.1016/j.cub.2018.02.002. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jelluma N., Dansen T.B., Sliedrecht T., Kwiatkowski N.P., Kops G.J. Release of Mps1 from kinetochores is crucial for timely anaphase onset. J. Cell Biol. 2010;191(2):281–290. doi: 10.1083/jcb.201003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Koch L.B., Opoku K.N., Deng Y., Barber A., Littleton A.J., London N., Biggins S., Asbury C.L. Autophosphorylation is sufficient to release Mps1 kinase from native kinetochores. Proc. Natl. Acad. Sci. USA. 2019;116(35):17355–17360. doi: 10.1073/pnas.1901653116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hayward D., Alfonso-Perez T., Cundell M.J., Hopkins M., Holder J., Bancroft J., Hutter L.H., Novak B., Barr F.A., Gruneberg U. CDK1-CCNB1 creates a spindle checkpoint-permissive state by enabling MPS1 kinetochore localization. J. Cell Biol. 2019;218(4):1182–1199. doi: 10.1083/jcb.201808014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Heinrich S., Windecker H., Hustedt N., Hauf S. Mph1 kinetochore localization is crucial and upstream in the hierarchy of spindle assembly checkpoint protein recruitment to kinetochores. J. Cell Sci. 2012;125(Pt 20):4720–4727. doi: 10.1242/jcs.110387. [DOI] [PubMed] [Google Scholar]
- 122.Chmielewska A.E., Tang N.H., Toda T. The hairpin region of Ndc80 is important for the kinetochore recruitment of Mph1/MPS1 in fission yeast. Cell Cycle. 2016;15(5):740–747. doi: 10.1080/15384101.2016.1148842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saurin A.T., van der Waal M.S., Medema R.H., Lens S.M., Kops G.J. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat. Commun. 2011;2:316. doi: 10.1038/ncomms1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alfonso-Perez T., Hayward D., Holder J., Gruneberg U., Barr F.A. MAD1-dependent recruitment of CDK1-CCNB1 to kinetochores promotes spindle checkpoint signaling. J. Cell Biol. 2019;218(4):1108–1117. doi: 10.1083/jcb.201808015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003;161(2):267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003;161(2):281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Santaguida S., Vernieri C., Villa F., Ciliberto A., Musacchio A. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 2011;30(8):1508–1519. doi: 10.1038/emboj.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Isokane M., Walter T., Mahen R., Nijmeijer B., Heriche J.K., Miura K., Maffini S., Ivanov M.P., Kitajima T.S., Peters J.M., Ellenberg J. ARHGEF17 is an essential spindle assembly checkpoint factor that targets Mps1 to kinetochores. J. Cell Biol. 2016;212(6):647–659. doi: 10.1083/jcb.201408089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aravamudhan P., Goldfarb A.A., Joglekar A.P. The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling. Nat. Cell Biol. 2015;17(7):868–879. doi: 10.1038/ncb3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen R.H., Shevchenko A., Mann M., Murray A.W. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 1998;143(2):283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Campbell M.S., Chan G.K., Yen T.J. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 2001;114(Pt 5):953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- 132.Lee S.H., Sterling H., Burlingame A., McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22(21):2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lince-Faria M., Maffini S., Orr B., Ding Y., Claudia F., Sunkel C.E., Tavares A., Johansen J., Johansen K.M., Maiato H. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 2009;184(5):647–657. doi: 10.1083/jcb.200811012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schweizer N., Ferras C., Kern D.M., Logarinho E., Cheeseman I.M., Maiato H. Spindle assembly checkpoint robustness requires Tpr-mediated regulation of Mad1/Mad2 proteostasis. J. Cell Biol. 2013;203(6):883–893. doi: 10.1083/jcb.201309076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cairo L.V., Ptak C., Wozniak R.W. Mitosis-specific regulation of nuclear transport by the spindle assembly checkpoint protein Mad1p. Mol. Cell. 2013;49(1):109–120. doi: 10.1016/j.molcel.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 136.Rodriguez-Bravo V., Maciejowski J., Corona J., Buch H.K., Collin P., Kanemaki M.T., Shah J.V., Jallepalli P.V. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell. 2014;156(5):1017–1031. doi: 10.1016/j.cell.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.London N., Biggins S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 2014;28(2):140–152. doi: 10.1101/gad.233700.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ji Z., Gao H., Jia L., Li B., Yu H. A sequential multi-target Mps1 phosphorylation cascade promotes spindle checkpoint signaling. Elife. 2017;6:22513. doi: 10.7554/eLife.22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang G., Kruse T., Lopez-Mendez B., Sylvestersen K.B., Garvanska D.H., Schopper S., Nielsen M.L., Nilsson J. Bub1 positions Mad1 close to KNL1 MELT repeats to promote checkpoint signalling. Nat. Commun. 2017;8:15822. doi: 10.1038/ncomms15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Qian J., Garcia-Gimeno M.A., Beullens M., Manzione M.G., Van der Hoeven G., Igual J.C., Heredia M., Sanz P., Gelens L., Bollen M. An attachment-independent biochemical timer of the spindle assembly checkpoint. Mol. Cell. 2017;68(4):715–730. doi: 10.1016/j.molcel.2017.10.011. e5. [DOI] [PubMed] [Google Scholar]
- 141.Lara-Gonzalez P., Kim T., Oegema K., Corbett K., Desai A. A tripartite mechanism catalyzes Mad2-Cdc20 assembly at unattached kinetochores. Science. 2021;371(6524):64–67. doi: 10.1126/science.abc1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Brady D.M., Hardwick K.G. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 2000;10(11):675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- 143.Fischer E.S., Yu C.W.H., Bellini D., McLaughlin S.H., Orr C.M., Wagner A., Freund S.M.V., Barford D. Molecular mechanism of Mad1 kinetochore targeting by phosphorylated Bub1. EMBO Rep. 2021:52242. doi: 10.15252/embr.202052242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Scott R.J., Lusk C.P., Dilworth D.J., Aitchison J.D., Wozniak R.W. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16(9):4362–4374. doi: 10.1091/mbc.E05-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Buffin E., Lefebvre C., Huang J., Gagou M.E., Karess R.E. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 2005;15(9):856–861. doi: 10.1016/j.cub.2005.03.052. [DOI] [PubMed] [Google Scholar]
- 146.Kops G.J., Kim Y., Weaver B.A., Mao Y., McLeod I., Yates J.R., 3rd, Tagaya M., Cleveland D.W. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 2005;169(1):49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chung E., Chen R.H. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol. Biol. Cell. 2002;13(5):1501–1511. doi: 10.1091/mbc.02-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Perera D., Tilston V., Hopwood J.A., Barchi M., Boot-Handford R.P., Taylor S.S. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev. Cell. 2007;13(4):566–579. doi: 10.1016/j.devcel.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 149.Meraldi P., Sorger P.K. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 2005;24(8):1621–1633. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vleugel M., Hoek T.A., Tromer E., Sliedrecht T., Groenewold V., Omerzu M., Kops G.J. Dissecting the roles of human BUB1 in the spindle assembly checkpoint. J. Cell Sci. 2015;128(16):2975–2982. doi: 10.1242/jcs.169821. [DOI] [PubMed] [Google Scholar]
- 151.Zhang G., Kruse T., Guasch Boldu C., Garvanska D.H., Coscia F., Mann M., Barisic M., Nilsson J. Efficient mitotic checkpoint signaling depends on integrated activities of Bub1 and the RZZ complex. EMBO J. 2019;38(7):100977. doi: 10.15252/embj.2018100977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Silio V., McAinsh A.D., Millar J.B. KNL1-Bubs and RZZ provide two separable pathways for checkpoint activation at human kinetochores. Dev. Cell. 2015;35(5):600–613. doi: 10.1016/j.devcel.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 153.Currie C.E., Mora-Santos M., Smith C.A., McAinsh A.D., Millar J.B.A. Bub1 is not essential for the checkpoint response to unattached kinetochores in diploid human cells. Curr. Biol. 2018;28(17):R929–R930. doi: 10.1016/j.cub.2018.07.040. [DOI] [PubMed] [Google Scholar]
- 154.Raaijmakers J.A., Medema R.H. Killing a zombie: a full deletion of the BUB1 gene in HAP1 cells. EMBO J. 2019;38(22) doi: 10.15252/embj.2019102423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Meraldi P. Bub1-the zombie protein that CRISPR cannot kill. EMBO J. 2019;38(7):101912. doi: 10.15252/embj.2019101912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang G., Kruse T., Boldu C.G., Garvanska D.H., Coscia F., Mann M., Barisic M., Nilsson J. Response to Raaijmakers & Medema. EMBO J. 2019;38(22) doi: 10.15252/embj.2019103547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gassmann R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R., 3rd, Oegema K., Cheeseman I.M., Desai A. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008;22(17):2385–2399. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yamamoto T.G., Watanabe S., Essex A., Kitagawa R. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J. Cell Biol. 2008;183(2):187–194. doi: 10.1083/jcb.200805185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kops G., Gassmann R. Crowning the kinetochore: the fibrous corona in chromosome segregation. Trends Cell Biol. 2020;30(8):653–667. doi: 10.1016/j.tcb.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 160.Allan L.A., Camacho Reis M., Ciossani G., Huis In ’t Veld P.J., Wohlgemuth S., Kops G.J., Musacchio A., Saurin A.T. Cyclin B1 scaffolds MAD1 at the kinetochore corona to activate the mitotic checkpoint. EMBO J. 2020;39(12) doi: 10.15252/embj.2019103180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kallio M.J., Beardmore V.A., Weinstein J., Gorbsky G.J. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 2002;158(5):841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li D., Morley G., Whitaker M., Huang J.Y. Recruitment of Cdc20 to the kinetochore requires BubR1 but not Mad2 in Drosophila melanogaster. Mol. Cell Biol. 2010;30(13):3384–3395. doi: 10.1128/MCB.00258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yang Y., Tsuchiya D., Lacefield S. Bub3 promotes Cdc20-dependent activation of the APC/C in S. cerevisiae. J. Cell Biol. 2015;209(4):519–527. doi: 10.1083/jcb.201412036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim T., Lara-Gonzalez P., Prevo B., Meitinger F., Cheerambathur D.K., Oegema K., Desai A. Kinetochores accelerate or delay APC/C activation by directing Cdc20 to opposing fates. Genes Dev. 2017;31(11):1089–1094. doi: 10.1101/gad.302067.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Burton J.L., Xiong Y., Solomon M.J. Mechanisms of pseudosubstrate inhibition of the anaphase promoting complex by Acm1. EMBO J. 2011;30(9):1818–1829. doi: 10.1038/emboj.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.He J., Chao W.C., Zhang Z., Yang J., Cronin N., Barford D. Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Mol. Cell. 2013;50(5):649–660. doi: 10.1016/j.molcel.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Di Fiore B., Davey N.E., Hagting A., Izawa D., Mansfeld J., Gibson T.J., Pines J. The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev. Cell. 2015;32(3):358–372. doi: 10.1016/j.devcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Diaz-Martinez L.A., Tian W., Li B., Warrington R., Jia L., Brautigam C.A., Luo X., Yu H. The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. J. Biol. Chem. 2015;290(4):2431–2443. doi: 10.1074/jbc.M114.616490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lu D., Hsiao J.Y., Davey N.E., Van Voorhis V.A., Foster S.A., Tang C., Morgan D.O. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J. Cell Biol. 2014;207(1):23–39. doi: 10.1083/jcb.201402041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lischetti T., Zhang G., Sedgwick G.G., Bolanos-Garcia V.M., Nilsson J. The internal Cdc20 binding site in BubR1 facilitates both spindle assembly checkpoint signalling and silencing. Nat. Commun. 2014;5:5563. doi: 10.1038/ncomms6563. [DOI] [PubMed] [Google Scholar]
- 171.Roy B., Han J.S.Y., Fontan A.N., Joglekar A.P. Aurora B phosphorylates Bub1 to promote spindle assembly checkpoint signaling. biorxiv. 2021 doi: 10.1101/2021.01.05.425459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Howell B.J., Moree B., Farrar E.M., Stewart S., Fang G., Salmon E.D. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 2004;14(11):953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 173.Asghar A., Lajeunesse A., Dulla K., Combes G., Thebault P., Nigg E.A., Elowe S. Bub1 autophosphorylation feeds back to regulate kinetochore docking and promote localized substrate phosphorylation. Nat. Commun. 2015;6:8364. doi: 10.1038/ncomms9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Han J.S., Holland A.J., Fachinetti D., Kulukian A., Cetin B., Cleveland D.W. Catalytic assembly of the mitotic checkpoint inhibitor BubR1-Cdc20 by a Mad2-induced functional switch in Cdc20. Mol. Cell. 2013;51(1):92–104. doi: 10.1016/j.molcel.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Vader G. Pch2(TRIP13): controlling cell division through regulation of HORMA domains. Chromosoma. 2015;124(3):333–339. doi: 10.1007/s00412-015-0516-y. [DOI] [PubMed] [Google Scholar]
- 176.Maldonado M., Kapoor T.M. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat. Cell Biol. 2011;13(4):475–482. doi: 10.1038/ncb2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ballister E.R., Riegman M., Lampson M.A. Recruitment of Mad1 to metaphase kinetochores is sufficient to reactivate the mitotic checkpoint. J. Cell Biol. 2014;204(6):901–908. doi: 10.1083/jcb.201311113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Heinrich S., Sewart K., Windecker H., Langegger M., Schmidt N., Hustedt N., Hauf S. Mad1contribution to spindle assembly checkpoint signalling goes beyond presenting Mad2 at kinetochores. EMBO Rep. 2014;15(3):291–298. doi: 10.1002/embr.201338114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kruse T., Larsen M.S., Sedgwick G.G., Sigurdsson J.O., Streicher W., Olsen J.V., Nilsson J. A direct role of Mad1 in the spindle assembly checkpoint beyond Mad2 kinetochore recruitment. EMBO Rep. 2014;15(3):282–290. doi: 10.1002/embr.201338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Piano V., Alex A., Stege P., Maffini S., Stoppiello G.A., Huis In ’t Veld P.J., Vetter I.R., Musacchio A. CDC20 assists its catalytic incorporation in the mitotic checkpoint complex. Science. 2021;371(6524):67–71. doi: 10.1126/science.abc1152. [DOI] [PubMed] [Google Scholar]
- 181.Zhang Y., Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol. Cell Biol. 2001;21(15):5190–5199. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Luo X., Tang Z., Xia G., Wassmann K., Matsumoto T., Rizo J., Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat. Struct. Mol. Biol. 2004;11(4):338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 183.Hara M., Ozkan E., Sun H., Yu H., Luo X. Structure of an intermediate conformer of the spindle checkpoint protein Mad2. Proc. Natl. Acad. Sci. USA. 2015;112(36):11252–11257. doi: 10.1073/pnas.1512197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kim S., Sun H., Tomchick D.R., Yu H., Luo X. Structure of human Mad1C-terminal domain reveals its involvement in kinetochore targeting. Proc. Natl. Acad. Sci. USA. 2012;109(17):6549–6554. doi: 10.1073/pnas.1118210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gross F., Bonaiuti P., Hauf S., Ciliberto A. Implications of alternative routes to APC/C inhibition by the mitotic checkpoint complex. PLoS Comput. Biol. 2018;14(9) doi: 10.1371/journal.pcbi.1006449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hardwick K.G., Weiss E., Luca F.C., Winey M., Murray A.W. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273(5277):953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 187.Poddar A., Daniel J.A., Daum J.R., Burke D.J. Differential kinetochore requirements for establishment and maintenance of the spindle checkpoint are dependent on the mechanism of checkpoint activation in Saccharomyces cerevisiae. Cell Cycle. 2004;3(2):197–204. [PubMed] [Google Scholar]
- 188.Chen C., Whitney I.P., Banerjee A., Sacristan C., Sekhri P., Kern D.M., Fontan A., Kops G., Tyson J.J., Cheeseman I.M., Joglekar A.P. Ectopic activation of the spindle assembly checkpoint signaling cascade reveals its biochemical design. Curr. Biol. 2019;29(1):104–119. doi: 10.1016/j.cub.2018.11.054. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yuan I., Leontiou I., Amin P., May K.M., Soper Ni Chafraidh S., Zlamalova E., Hardwick K.G. Generation of a spindle checkpoint arrest from synthetic signaling assemblies. Curr. Biol. 2017;27(1):137–143. doi: 10.1016/j.cub.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Brito D.A., Rieder C.L. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr. Biol. 2006;16(12):1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gascoigne K.E., Taylor S.S. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14(2):111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 192.Uetake Y., Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 2010;20(18):1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gorbsky G.J. Cohesion fatigue. Curr. Biol. 2013;23(22):R986–R988. doi: 10.1016/j.cub.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 194.Kuhn J., Dumont S. Spindle assembly checkpoint satisfaction occurs via end-on but not lateral attachments under tension. J. Cell Biol. 2017;216(6):1533–1542. doi: 10.1083/jcb.201611104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chan Y.W., Fava L.L., Uldschmid A., Schmitz M.H., Gerlich D.W., Nigg E.A., Santamaria A. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 2009;185(5):859–874. doi: 10.1083/jcb.200812167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gassmann R., Holland A.J., Varma D., Wan X., Civril F., Cleveland D.W., Oegema K., Salmon E.D., Desai A. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010;24(9):957–971. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F., Stukenberg P.T., Desai A., Salmon E.D. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137(4):672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Roscioli E., Germanova T.E., Smith C.A., Embacher P.A., Erent M., Thompson A.I., Burroughs N.J., McAinsh A.D. Ensemble-level organization of human kinetochores and evidence for distinct tension and attachment sensors. Cell Rep. 2020;31(4) doi: 10.1016/j.celrep.2020.107535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Uchida K.S.K., Jo M., Nagasaka K., Takahashi M., Shindo N., Shibata K., Tanaka K., Masumoto H., Fukagawa T., Hirota T. Kinetochore stretching-mediated rapid silencing of the spindle-assembly checkpoint required for failsafe chromosome segregation. Curr. Biol. 2021;31:1581–1591. doi: 10.1016/j.cub.2021.01.062. [DOI] [PubMed] [Google Scholar]
- 200.Nasa I., Rusin S.F., Kettenbach A.N., Moorhead G.B. Aurora B opposes PP1 function in mitosis by phosphorylating the conserved PP1-binding RVxF motif in PP1 regulatory proteins. Sci. Signal. 2018;11(530):eaai8669. doi: 10.1126/scisignal.aai8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Roy B., Verma V., Sim J., Fontan A., Joglekar A.P. Delineating the contribution of Spc105-bound PP1 to spindle checkpoint silencing and kinetochore microtubule attachment regulation. J. Cell Biol. 2019;218(12):3926–3942. doi: 10.1083/jcb.201810172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bajaj R., Bollen M., Peti W., Page R. KNL1 binding to PP1 and microtubules is mutually exclusive. Structure. 2018;26(10):1327–1336. doi: 10.1016/j.str.2018.06.013. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hertz E.P.T., Kruse T., Davey N.E., Lopez-Mendez B., Sigurethsson J.O., Montoya G., Olsen J.V., Nilsson J. A conserved motif provides binding specificity to the PP2A-B56 phosphatase. Mol. Cell. 2016;63(4):686–695. doi: 10.1016/j.molcel.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 204.Hayward D., Bancroft J., Mangat D., Alfonso-Perez T., Dugdale S., McCarthy J., Barr F.A., Gruneberg U. Checkpoint signaling and error correction require regulation of the MPS1 T-loop by PP2A-B56. J. Cell Biol. 2019;218(10):3188–3199. doi: 10.1083/jcb.201905026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Grallert A., Boke E., Hagting A., Hodgson B., Connolly Y., Griffiths J.R., Smith D.L., Pines J., Hagan I.M. PP1-PP2A phosphatase relay controls mitotic progression. Nature. 2015;517(7532):94–98. doi: 10.1038/nature14019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Smith R.J., Cordeiro M.H., Davey N.E., Vallardi G., Ciliberto A., Gross F., Saurin A.T. PP1 and PP2A use opposite phospho-dependencies to control distinct processes at the kinetochore. Cell Rep. 2019;28(8):2206–2219. doi: 10.1016/j.celrep.2019.07.067. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gu Y., Desai A., Corbett K.D. Evolutionary dynamics and molecular mechanisms of HORMA domain protein signaling. Annu. Rev. Biochem. 2021 doi: 10.1146/annurev-biochem-090920-103246. [DOI] [PubMed] [Google Scholar]
- 208.Lara-Gonzalez P., Moyle M.W., Budrewicz J., Mendoza-Lopez J., Oegema K., Desai A. The G2-to-M transition is ensured by a dual mechanism that protects cyclin B from degradation by Cdc20-Activated APC/C. Dev. Cell. 2019;51(3):313–325. doi: 10.1016/j.devcel.2019.09.005. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hellmuth S., Gomez H.L., Pendas A.M., Stemmann O. Securin-independent regulation of separase by checkpoint-induced shugoshin-MAD2. Nature. 2020;580(7804):536–541. doi: 10.1038/s41586-020-2182-3. [DOI] [PubMed] [Google Scholar]
- 210.Choi E., Zhang X., Xing C., Yu H. Mitotic checkpoint regulators control insulin signaling and metabolic homeostasis. Cell. 2016;166(3):567–581. doi: 10.1016/j.cell.2016.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Matsuura S., Matsumoto Y., Morishima K., Izumi H., Matsumoto H., Ito E., Tsutsui K., Kobayashi J., Tauchi H., Kajiwara Y., Hama S., Kurisu K., Tahara H., Oshimura M., Komatsu K., Ikeuchi T., Kajii T. Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am. J. Med. Genet. A. 2006;140(4):358–367. doi: 10.1002/ajmg.a.31069. [DOI] [PubMed] [Google Scholar]
- 212.Suijkerbuijk S.J., van Osch M.H., Bos F.L., Hanks S., Rahman N., Kops G.J. Molecular causes for BUBR1 dysfunction in the human cancer predisposition syndrome mosaic variegated aneuploidy. Cancer Res. 2010;70(12):4891–4900. doi: 10.1158/0008-5472.CAN-09-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yost S., de Wolf B., Hanks S., Zachariou A., Marcozzi C., Clarke M., de Voer R., Etemad B., Uijttewaal E., Ramsay E., Wylie H., Elliott A., Picton S., Smith A., Smithson S., Seal S., Ruark E., Houge G., Pines J., Kops G., Rahman N. Biallelic TRIP13 mutations predispose to Wilms tumor and chromosome missegregation. Nat. Genet. 2017;49(7):1148–1151. doi: 10.1038/ng.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bohers E., Sarafan-Vasseur N., Drouet A., Paresy M., Latouche J.B., Flaman J.M., Sesboue R., Frebourg T. Gradual reduction of BUBR1 protein levels results in premature sister-chromatid separation then in aneuploidy. Hum. Genet. 2008;124(5):473–478. doi: 10.1007/s00439-008-0572-y. [DOI] [PubMed] [Google Scholar]
- 215.Umbreit N.T., Zhang C.Z., Lynch L.D., Blaine L.J., Cheng A.M., Tourdot R., Sun L., Almubarak H.F., Judge K., Mitchell T.J., Spektor A., Pellman D. Mechanisms generating cancer genome complexity from a single cell division error. Science. 2020;368(6488):eaba0712. doi: 10.1126/science.aba0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shoshani O., Brunner S.F., Yaeger R., Ly P., Nechemia-Arbely Y., Kim D.H., Fang R., Castillon G.A., Yu M., Li J.S.Z., Sun Y., Ellisman M.H., Ren B., Campbell P.J., Cleveland D.W. Chromothripsis drives the evolution of gene amplification in cancer. Nature. 2021;591(7848):137–141. doi: 10.1038/s41586-020-03064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]