Abstract
Mitotic cell divisions ensure stable transmission of genetic information from a mother to daughter cells in a series of generations. To ensure this crucial task is accomplished, the cell forms a bipolar structure called the mitotic spindle that divides sister chromatids to the opposite sides of the dividing mother cell. After successful establishment of stable attachments of microtubules to chromosomes and inspection of connections between them, at the heart of mitosis, the cell starts the process of segregation. This spectacular moment in the life of a cell is termed anaphase, and it involves two distinct processes: depolymerization of microtubules bound to chromosomes, which is also known as anaphase A, and elongation of the spindle or anaphase B. Both processes ensure physical separation of disjointed sister chromatids. In this chapter, we review the mechanisms of anaphase B spindle elongation primarily in mammalian systems, combining different pioneering ideas and concepts with more recent findings that shed new light on the force generation and regulation of biochemical modules operating during spindle elongation. Finally, we present a comprehensive model of spindle elongation that includes structural, biophysical, and molecular aspects of anaphase B.
Keywords: Anaphase B, Spindle elongation, Motor proteins, Microtubule sliding, Chromosome segregation, Microtubule pushing, Microtubule pulling
Highlights
-
•
Overview of spindle elongation dynamics in the context of different mitotic phases.
-
•
Synopsis of pioneering ideas and recent structural literature on metaphase and anaphase spindles in human cells.
-
•
Dissection of force-generation mechanisms during anaphase B and experimental support for their existence in mammals.
-
•
Description of changes in protein localization after metaphase-to-anaphase transition for a large set of spindle proteins.
-
•
Detailed molecular models of key aspects of anaphase B and their temporal regulation.
1. Introduction to the world of anaphase B
At the very essence of cell division lies the mitotic spindle, a fascinating self-organizing structure that is composed primarily of tubulin and various associated proteins [1], [2]. Successful separation of sister chromatids requires their attachment to the spindle microtubules (MTs) on opposite sides of the bipolar spindle structure, through kinetochores, protein complexes on centromere of each chromatid [3]. Each kinetochore that is not attached to MTs emits an inhibitory signal through a series of complex events controlled by the spindle assembly checkpoint (SAC) [4]. Once all kinetochores are attached and the first levels of the inhibitory signal are removed, a cascade of events leads to a point of no return where the cell starts the process of chromosome segregation. This process is called anaphase (Greek ana, meaning going back) [5] and its mechanisms will form the basis of this review.
The process of chromosome segregation was depicted more than 150 years ago, starting an active anaphase fascination that spurred tremendous research in the following decades, but the mechanisms of this crucial process of chromosome movement are debatable even today [6], [7]. Anaphase involves two mechanistically distinct steps, shortening of kinetochore MTs that leads to the movement of each chromatid towards its respective pole, and spindle elongation that further separates the disjointed sister chromatids [8]. These distinct steps, which are in some organisms temporally divided while in others occur simultaneously, were classified as anaphase A and anaphase B, respectively [5], [9]. This review will focus on the mechanisms of anaphase B in human cells, with the contribution of data from other model organisms when introducing key concepts developed in a variety of model systems.
Anaphase B spindle elongation is broadly deployed through a highly diverse tree of life [7], [10]. In bacterial cells the mechanisms that separate the low-copy-number plasmids make simple DNA segregating machines that use elongation of ParM protein filaments between sister plasmids, resembling the elongation and bipolarity of the eukaryotic mitotic spindle [11]. In addition, the separation of sister chromatids in many other model systems is also highly dependent on anaphase B spindle elongation, like in yeasts and nematodes, whereas in other organisms the contribution varies [10]. In mammalian mitosis and meiosis, anaphase A and B equally contribute to net chromosome segregation movement [12], [13].
Anaphase spindle elongation is an important process because its failure can lead to chromosome mis-segregation [14], [15]. Accordingly, it has been shown that spindle elongation greatly contributes to the resolution of lagging chromosomes during anaphase in human cells and its absence can contribute to the generation of tetraploid cells [14], [16]. Finally, as spindle elongation precedes cytokinesis and telophase, and controls various crucial aspects of both processes [17], [18], its timely and correct execution is essential for cell survival.
2. Main constituents of mammalian anaphase B spindle – structural studies
Anaphase mitotic spindle is a highly complex structure made up of different MT subpopulations, all contributing to various extents to the active poleward movement of chromosomes, cell and spindle elongation: kinetochore MTs are attached to the kinetochore and form parallel bundles known as k-fibers, interpolar MTs extend from the opposite sides to the center of the spindle forming antiparallel overlapping bundles, termed collectively as the spindle midzone [19], and astral MTs grow from the spindle poles towards the cell cortex, and are not characterized by MT bundling [20], [21], [22], [23], [24], [25] (Fig. 1).
Fig. 1.
Structure of the anaphase mitotic spindle in human cells. Schematic representation of a mid-anaphase mitotic spindle in a human cell depicting in detail parts of the spindle (boxed regions 1–3 together with magnifications) characterized by extensive microtubule bundling. In all figures where spindles are depicted, MTs denotes microtubules; plus and minus signs denote the respective microtubule ends, blue circles represent kinetochores and gray circles centrosomes, light gray thick line is the plasma membrane and the reticulate structure below is the actin cortex, dark gray thick lines are kinetochore fibers, blue lines are bridging fibers and thin gray lines are astral microtubules. In all figures, please see text for details and references. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The possibility that k-fibers could be coupled to interpolar MTs around spindle poles has been envisioned in pioneering models of mitosis [26], [27], but the exact nature of this connection was tested only recently. In mammalian mitotic spindles, antiparallel midzone MTs termed bridging MTs were found to be highly crosslinked with sister k-fibers near kinetochores by usage of laser ablation and photoactivation approaches [12]. Additionally, a recent electron microscopy (EM) study reported midzone MT bundles that do not reach poles but are tightly connected to kinetochore MTs near chromosomes in HeLa cells [28]. Midzone MTs are classified by the location of their plus-ends, which can be either between the chromosomes, at the chromosome or between the chromosome and the pole [28] (Fig. 1, box 1).
Much more information about bridging MTs comes from studies of human metaphase spindles where each pair of sister k-fibers is linked with a bridging fiber [29], [30]. EM studies established that minus-ends of bridging MTs are localized along kinetochore MTs [23], [31], [32], in agreement with augmin-mediated nucleation of bridging MTs [33]. Imaging of live cells suggests that bridging midzone MTs during early anaphase are the same ones that were present in metaphase [12]. However, additional fusion of preexisting MTs [34] and nucleation of new MTs by augmin complex [35] take place in the midzone region later in anaphase. Thus, the spindle midzone during anaphase is probably composed of antiparallel MTs originating from metaphase, which may serve as substrates for de novo MT nucleation in this region during late anaphase.
Less information is available for k-fiber MTs during anaphase, but more was acquired during metaphase where the data on the average number of MTs varies from 8 to 17 per each fiber in human [36], [37] to around 20 in Ptk1 cells [38]. The percentage of kinetochore MTs that directly contact the structure of the centrosome reaches around 50% for HeLa and U2OS and around 72% for RPE1 cells [37] (Fig. 1, box 3), similar to number reported for Ptk1 cells [39], [40]. To conclude, the mitotic spindle during anaphase is composed of different subpopulations of MTs where kinetochore and interpolar MTs physically intermingle with each other.
3. Dynamics of spindle elongation – passing through phases
Spindle elongation in human mitotic cells in culture starts before nuclear envelope breakdown (NEBD) or during prometaphase, depending on the exact pathway, reaching a metaphase steady state length 5–8 min after NEBD [41] (Fig. 2). After metaphase, the spindle starts to elongate again during anaphase B, which has been studied in a variety of model systems where the characteristic elongation distances and rates were measured [7], [10]. In human mitotic cells, anaphase B usually starts with a short 30–50 s delay compared to the start of the anaphase A [6], [42], and the spindle elongates by a total of 8 µm during anaphase, and for 3 more microns after kinetochore-MT release in telophase [6] (Fig. 2).
Fig. 2.
Dynamics of spindle elongation, chromosome segregation, and overlap length in human mitosis. Typical time course in human cells for centrosome distance, chromatid distance and length of the overlap is plotted in the graph representing key cell and spindle morphological changes and velocity regimes that define transition phases. Note: Chromatid-to-chromatid movement during late anaphase/telophase is related to chromosome contraction that occurs after kinetochore release from microtubules during late anaphase. P, prophase; PM, prometaphase; Meta, metaphase; Ana, anaphase; T, telophase.
Post-metaphase spindle elongation in human cells can be described by distinct characteristic velocity regimes: early anaphase is characterized by the fast spindle elongation while middle and late anaphase are characterized by the gradual stall in spindle elongation, formation of the central spindle and actomyosin ring [42], [43], [44] (Fig. 2). During these last phases of anaphase, cytokinesis starts to cleave the mother cell into two daughter cells [18]. Anaphase is followed by telophase, characterized by the onset of nuclear envelope reformation and chromosome decondensation [17] (Fig. 2). Furthermore, the anaphase spindle elongation is accompanied by the plasma membrane elongation during mid-anaphase (Fig. 2) that increases the cell boundaries, making new space for the separating chromosomes [43].
4. Origin of forces required for spindle elongation – battle of concepts
The origin of the forces required for anaphase B spindle elongation is still debated between two views depending on where the force is generated, mainly pulling from the outside of the spindle or pushing from the inside of the spindle [5], [45] (Fig. 3). Approaches that tried to solve the anaphase B puzzle by introducing a universal mechanism were subsequently replaced by more organism-specific approaches as it became evident that different organisms perform anaphase B differently [1], and the contribution of different mechanisms to net chromosome separation also varies [10]. However, despite that, even within one organism, there is still no consensus on the dominant mechanism of anaphase B spindle elongation and this rivalry of concepts still endures [7]. Generally, it was envisioned that pulling, wherever it may happen, would be the preferred mechanism of moving objects for distances greater than 10 µm, since pushing MTs would experience high buckling due to resistance from the firm nonmoving objects [45], drastically reducing the effectiveness of such approach.
Fig. 3.
Two main views of force-generation during anaphase spindle elongation. Schematic representation of mid-anaphase spindle depicting the locations of two main views of force-generation during anaphase B. Pushing from the inside of the spindle (top scheme) is depicted by black arrows in spindle interior, in between separating kinetochores (blue dots) and pulling from the outside (bottom scheme) is depicted by black arrows on the sites distant from the kinetochores, on the other side of the centrosome structure (gray dots). (+) sign represents the plus-ends. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4.1. Pulling by astral MTs
Forces for spindle elongation may be generated by astral MTs pulling on the spindle poles from the cortex. The underlying mechanism of this pulling force can be explained by minus-end directed molecular motor walking and actively pulling astral MTs [46] (Fig. 4A) or a polymer ratchet mechanism where plus-ends of astral MTs anchor to a cortical adapter and depolymerize resulting in the pull of the poles [45], [47], [48], [49] (Fig. 4B). Cortical pulling forces require direct contact between astral MTs and the cell cortex, either via dynein, +TIP network proteins like CLASPs (CLIP-associated proteins) or kinesin-13 motors [48], [50], although with possible exceptions such as anchoring to the cytoplasmic actin filament network as shown in Drosophila cell-free embryo extract [51]. There is also a possibility that astral MTs depolymerize at the pole. However, this is not supported by the speckle microscopy experiments in newt lung cells, which showed no evidence for the poleward transport of astral MTs [52].
Fig. 4.
Proposed mechanisms of force-generation during anaphase B. Schematic representation of mid-anaphase spindle in human cells portraying all possible mechanistical models of force-generation for spindle elongation (A–E). Boxes 1–2 represent different locations of microtubule pushing that depends on the location of bridging MTs (blue thick lines) minus-ends. In boxes, black arrows represent the pushing force from bridging microtubules. Gray arrows represent the movement of the microtubule next to its arrow. Light orange structures represent adapter proteins, and dark orange structure represent motor proteins walking along astral microtubules (a small dark orange arrow represents the direction of movement of molecular motor). Small blue dots in D represent the polymerization of tubulin subunits into bridging microtubules at their plus-ends and small gray dots in B-C represent depolymerization of tubulin subunits from astral microtubule minus- (C) or plus-end (B). (?) sign depicts that this mechanism of astral microtubule pulling is not supported by current experimental data. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In this cortical pulling view, the spindle poles are pulled via astral MTs that are anchored both at the cortex and the spindle pole, and the force is transmitted to the poles and associated k-fibers towards the disjointed sister chromatid [53]. This view also implies that the same is happening on the other half of the bipolar spindle, and the collective action of these two force-generating sites results in the pull of the spindle poles towards the cortical region, thereby resulting in the elongation of the spindle, that is sometimes asymmetric regarding the net effect of the pull to each pole [54]. The phenomenon of asymmetric spindle elongation powered by cortical forces has been shown in unperturbed C. elegans mitosis [55] and in Ptk2 cells under confinement where forced spindle elongation depends on the presence of astral MTs and dynein activity [56].
Generally, spindle orientation and positioning are widely regarded as processes that are completely dependent on cortical force generators, in a series of different organisms from yeast to humans [57]. The evidence for external pulling forces that act on the spindle poles to elongate the spindle during anaphase has mainly been gathered directly by the usage of laser ablation approaches in the C. elegans embryos [55], mammalian Ptk1 cells [58], echinoderms and insect spermatocytes [59], where cutting of astral MTs drastically reduces the rate of spindle elongation during anaphase.
4.2. Pushing by midzone MTs
The other key model of spindle elongation revolves around midzone antiparallel MTs that could push into various structures and by various mechanistic methods [1]. First, antiparallel MTs could push with their growing plus ends by means of active MT polymerization. In this view, force-generating site would be at MT plus-ends [60] (Fig. 4D). Second, antiparallel adjacent MTs could slide apart from each other by the action of mitotic motors [61], in a manner analogous to class-II myosin filaments, that drive the sliding filament mechanism of muscle contraction [62]. This model postulates that if a MT crosslinker has plus-end directed activity, it would slide apart the adjacent antiparallel MTs, resulting in a decrease of the length of the antiparallel overlap and the increase in the length between their minus-ends [61]. These separating minus-ends could push into different structures within the cell. The presented mechanism was termed the mitotic sliding filament mechanism [61] and in this model the force-generating site would be within the MT antiparallel overlap (Fig. 4E).
The site at which minus- or plus-ends could push depends on the nature of MT organization within the spindle [6]. Consequently, if the antiparallel MTs are reaching the spindle poles, their minus-ends could push directly into the structure of the spindle pole excreting direct compressive forces on the poles (Fig. 4, box 1). This view is supported by the laser ablation experiments demonstrating pushing forces are responsible for spindle elongation in budding yeast [63], [64] and by the large set of EM data from various organisms [20], [22], [24], [65], where the minus ends of the overlapping midzone MTs physically interact directly with the spindle poles suggesting direct pole pushing. On the other hand, if the midzone MTs do not reach the spindle pole, their minus-ends could push into other MTs that are connected directly to the spindle poles, namely k-fibers [6], [66] (Fig. 4, box 2). Such MT coupling mechanism is supported by the EM studies of mammalian cells showing that antiparallel MTs do not directly interact with the pole during metaphase and anaphase but they interact with the adjacent MTs, mainly those that are part of the k-fiber [23], [28], [31].
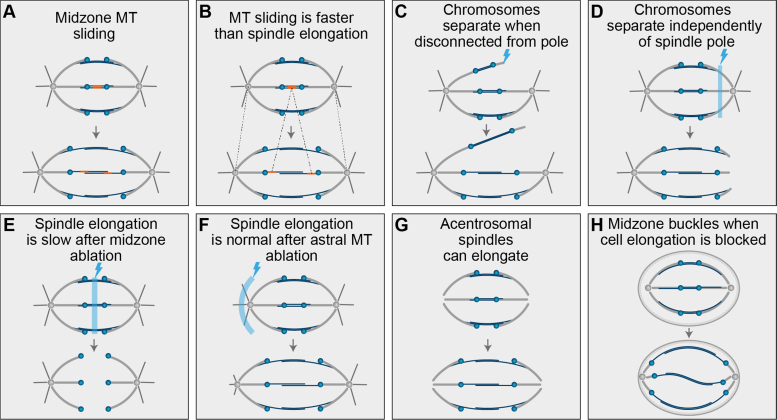
This notion of antiparallel MTs not interacting directly with poles has been used as an indication in favor of a cortical pulling anaphase B mechanism being more plausible in mammalian systems [7], but we argue that there is a lot of evidence against this hypothesis. First, early light microscopy observations of anaphase spindles marked by photo-bleaching and more recent photoactivation approaches in living cells both identified clear sliding of midzone MTs [12], [28], [67], [68] (Fig. 5A). Furthermore, it has been found recently that the velocity of MT sliding in human cells is much greater than the velocity of spindle elongation [68] (Fig. 5B), which would not be the case if the force generator site is at the cortex and the midzone sliding is just a response to the external pulling by astral MTs, as was suggested previously [69]. In that case one would expect the exact opposite, the velocity of spindle elongation would be equal or greater than the velocity of sliding, the situation not observed in human [68] or Drosophila embryo mitosis [70]. However, the observation of slower spindle elongation velocity when compared to midzone MT sliding could be as well attributed to ongoing minus-end depolymerization, although this process is downregulated following anaphase onset (121). Next, as shown by the laser ablation approach, a complete disconnection of a pair of sister k-fibers and their associated bridging MTs from one spindle pole, and with them astral MTs, led to chromosome segregation velocities similar to unperturbed chromosomes in human cells [12] (Fig. 5C). Similar observation was reported when the whole pole-proximal region of k-fibers on one side of the spindle was removed with a laser [28] (Fig. 5D). An analogous argument was raised after groundbreaking micromanipulation studies on grasshopper spindles [71]. Likewise, laser ablation of all midzone MTs abrogated spindle elongation in human spindles [12], [28] (Fig. 5E) and in some instances resulted in the movement of the poles towards the chromosomes [28], opposite of the direction that would be expected from cortical pulling forces [55], while laser perturbation of astral MTs did not perturb spindle elongation [12] (Fig. 5F). Furthermore, acentrosomal anastral spindles made artificially in human cells are capable of spindle elongation [72], [73] (Fig. 5G), and lastly, the buckled midzones observed in situations when cell length changes cannot follow the spindle elongation during anaphase [74] (Fig. 5H) all suggest that the force required for spindle elongation is predominantly generated internally in human spindles. Nevertheless, there is still a possibility that cortical pulling forces might play a role in the late anaphase spindle elongation as discussed later (see Section 7), and there is evidence that midzone MTs could also act as a brake to control the rate and extent of spindle elongation during late anaphase [34], [68].
Fig. 5.
Schematic representation of the experimental data that support internal force-generation in human spindles during anaphase B. Dark orange lines in A,B represent photoactivated tubulin subunits within midzone microtubules, light blue zones together with lightning sign in C–F represent the location where laser ablation was performed. Dashed gray lines in B represent the movement of spindle poles and photoactivated tubulin in the spindle midzone over time. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
5. Cell cycle regulation of anaphase B start and end – controlling the risk
During the metaphase-to-anaphase transition, the global protein phosphorylation state of the cell is reversed by the activation of E3 ubiquitin ligase APC/C which targets Cyclin B for degradation in proteasomes leading to downregulation of CDK1 activity and increase in PP1/PP2A phosphatase activity [75]. This allows the dephosphorylation of proteins that were previously phosphorylated by mitotic kinases, thus characterizing anaphase as a stage of net protein dephosphorylation [42]. The described phosphorylation-dephosphorylation switch at anaphase onset coincides with large changes in protein localization and activity (Fig. 6), ensuing profound changes in spindle behavior.
Fig. 6.
Patterns of localization of major motor proteins and non-motor regulators at the anaphase-to-metaphase transition. Stars in palette of blue colors represent schematical approximate locations of various motor proteins from kinesin-superfamily and dynein, as defined by the scheme on the right. Non-motor regulators are depicted in palette of orange colors as defined by the scheme on the right. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The coordination of anaphase exit depends on Aurora B activity gradient at the spindle midzone that monitors the extent of chromosome separation during anaphase. The aurora B activity ensures that completion of nuclear envelope reformation and chromosome decondensation occur only after sufficient sister chromatid separation [17], thus defining a chromosome segregation checkpoint in late anaphase.
5.1. Dynamics of changes in protein localization, function and activity
Here, we will briefly discuss the most dramatic changes happening at anaphase onset for some of the proteins of the mitotic spindle (Fig. 6). First, the major metaphase outward force generators, including EG5/kinesin-5 and KIF15/kinesin-12, do not change location drastically following anaphase onset [76]. Second, the kinetochore bound minus end-directed dynein is barely detectable or is completely removed from kinetochores at the metaphase-to-anaphase transition [77]. On the other hand, minus-end directed HSET/kinesin-14 seems to remain associated with minus-end of midzone MTs upon anaphase onset [78]. Third, levels of dynein and crosslinking proteins like NuMA increase at the cortical areas at anaphase onset, while they decrease at the poles and across the spindle [79], [80]. Furthermore, proteins that are involved in the control of kinetochore-derived polymerization forces, like KIF18A/kinesin-8 [81], and proteins that promote polar ejection forces (PEFs) like chromokinesins KID/kinesin-10 and KIF4A/kinesin-4 [82], are partially removed after anaphase onset from kinetochores and chromosomes, respectively. Interestingly, both KIF18A and KIF4A are transferred to the spindle midzone in human cells following anaphase onset [81], [83] and whereas KIF4A is directly dependent on PRC1 to localize to midzone MTs [84], for KIF18A this dependency has not been studied to date.
The localization of the major crosslinker of antiparallel MTs, PRC1, is controlled by its phosphorylation state, where phosphorylation by CDK1 inhibits its strong binding to MTs before anaphase [84]. However, a fraction of PRC1 associates with the antiparallel MTs even in the metaphase spindle [29], [30]. This transition pattern of localization to spindle midzone following anaphase onset is observed for most of PRC1 interaction partners during metaphase and anaphase counting various motors and MAPs including: regulators of MT dynamics CLASPs and kinesins-13, plus-end tracking proteins EB1, molecular motors KIF4A, MKLP1/kinesin-6, MKLP2/kinesin-6, CENP-E/kinesin-7, KIF14/kinesin-3, and kinases polo-like kinase 1 (PLK1), citron kinase 1 and Aurora kinase B, thus forming a main protein scaffold in antiparallel region of the spindle [19], [85] (Fig. 6). This regulatory PRC1 hub is involved in the control of anaphase B and cytokinesis in many model organisms [10].
6. Molecular modules and players involved in spindle elongation
Regarding spindle elongation in human cells, a lot of success has been achieved in identifying regulators of MT dynamics, proteins that bundle and stabilize MTs, kinases, and phosphatases that fine-tune the process [5], [42], [86], whereas the data on the force-generating proteins that elongate the spindle remained scarce. In other organisms, a force generating role has been described for the motors of the kinesin-5 and kinesin-6 families during anaphase in yeast cells [87], [88]. Also, other aspects of spindle elongation, like regulation of midzone stability, overlap length and the role of the midzone in braking spindle elongation, also lack details, which limits our understanding of these processes in human cells. In this part of the manuscript, we will assemble the gathered knowledge on different mechanisms of anaphase B in human cells, proposing molecular models behind each phenomenon involved in spindle elongation.
6.1. MT sliding motors and active force generation
Thus far, spindle elongation in human cells has been shown to almost completely stop after the addition of nocodazole [74], which depolymerizes MTs, after the addition of taxol [17], which stabilizes MTs against depolymerization and after perturbation of top signaling effectors that regulate multiple mitotic targets, such as aurora kinases, PLK1, CDK1 or Cyclin B [17], [89], [90]. Likewise, spindle elongation is abrogated after inhibition of topoisomerase-II [42], which resolves DNA supercoil threads and regulates activation of the midzone pool of Aurora B [91], and after inducing MT rigor-bound state of EG5, a motor that localizes to the antiparallel region [17], [92]. However, none of these studies reported a block in spindle elongation by specific removal of force-producing proteins responsible for elongation which created a puzzle because the prevailing view in the field is that anaphase B is ATP-dependent and motor driven [5], [7], [93], [94].
Nevertheless, as spindle elongation is drastically perturbed when EG5 is locked in a rigor-bound state to midzone MTs [17], this motor might have an active role during elongation. But, EG5 seems to be dispensable for spindle elongation in human cells as inhibition of its ATPase activity that induces a weak MT-binding state does not affect the rate of spindle elongation when compared to untreated cells [68], [74]. Similar findings were also reported in Drosophila embryo mitosis after injection of antibodies against kinesin-5 [70] and in pig epithelial cells where EG5 inhibition modestly increased the rate of elongation [92]. This is contrary to dominant role of the motor in generation of outward forces during pre-anaphase stages [95]. Besides, the lack of effect on elongation cannot be explained by the presence of KIF15 motor [68], [74], which acts redundantly with EG5 during metaphase [95]. Additionally, single depletion or inhibition of PRC1 or its interacting partners including MKLP1, MKLP2, KIF4A, CENP-E, KIF14 and additionally KIF18A and HSET did not stop spindle elongation during anaphase [68], [74], [96], although most of them reduced the rate of spindle elongation when compared to control cells [68]. This points out that plus-end directed motors might be operating redundantly within the antiparallel overlap, or at other sites within the spindle.
Interestingly, the fact that spindles shorten after EG5 inhibition during metaphase, but elongate soon after anaphase onset, points to the fact that additional outward force-generating mechanisms are turned on at this point. Our lab has recently reported that the protein, that is able to supplement for the lack of EG5 activity during anaphase spindle elongation, is PRC1-dependent [68]. After double perturbation of EG5 and PRC1, no spindle elongation is observed. As a result of a complete block in spindle elongation, the cells show either a delay or complete absence of cytokinesis, both leading to large defects in chromosome segregation [68]. The observed delay is consistent with the chromosome separation checkpoint mediated by Aurora B that delays telophase onset until sufficient chromosome segregation is achieved [17].
Interestingly, when KIF4A depletion is combined with EG5 inhibition, spindles also lose the capacity to elongate, in a phenotype that completely mimics the one seen after PRC1 depletion and EG5 inhibition [68]. This implies that PRC1 works as a scaffold to recruit KIF4A to the spindle midzone, as was reported previously [84], and force-generation by PRC1-KIF4A complexes is acting redundantly with EG5 activity during spindle elongation in human cells (Fig. 7, box 1). Furthermore, this phenotype is directly related to the impairment of the antiparallel MT sliding rather than disrupting global organization of midzone antiparallel MTs, suggesting that without force-generation in the overlap region, spindle elongation is blocked [68], corroborating the data gathered by laser ablation approaches [12], [28]. Both EG5 and KIF4A are reported to efficiently slide MTs in vitro [97], [98], but they use distinct mechanistical principles: EG5 is homotetramer that crosslinks two antiparallel MTs, creating a system in which MTs are both its cargo and its track [97] whereas KIF4A is a homodimer that requires PRC1 to perform efficient sliding [98].
Fig. 7.
Molecular mechanisms and major biochemical modules of anaphase B in human cells. Boxes 1–7 represent biochemical molecular modules involved in different aspects of spindle elongation and chromosome segregation during anaphase B in human cells. Box 1, midzone microtubule sliding by various redundant motor proteins (as depicted) where small arrows represent the movement of motor-heads and bigger blue arrows movement of bridging microtubules; Box 2, midzone braking due to the frictional action of PRC1 (blue arrows) opposing the outward movement of bridging microtubules; Box 3, protein components involved in overlap length regulation. (?) sign depicts probable contribution of the displayed protein. (˫) sign represents inhibition of microtubule polymerization (depicted as incorporation of blue dots at microtubule plus-ends) by a displayed protein; Box 4, probable molecular interface between bridging and kinetochore fibers including protein components as depicted, the blue thick arrow represents the movement of bridging microtubules. Box 5, factors involved in polar ejection forces. Gray dots represent the polymerization of plus-end of the astral microtubule. Box 6, minus-end depolymerization of the kinetochore fiber is depicted by dissociation of small gray dots depicting tubulin subunits. Box 7, main components of machinery responsible for cortical force-generation. Small gray dots represent depolymerization of tubulin subunits at minus-ends of astral microtubules and the gray arrow represents the movement of the dynein complex toward minus-end of astral microtubules. The text below and above each box represents the estimated activity of each module in depicted phases of mitosis. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Regarding other possible candidates that could act redundantly with EG5, a significant contribution has been shown for different sliding motors [68] including kinesins-6 MKLP1 [99] and MKLP2 [100] and kinesin-8 protein KIF18A [101] (Fig. 7, box 1), although not to the same extent as KIF4A [68]. This could imply that MT sliding during anaphase may be a result of massive redundancy between EG5-generated MT sliding that is teamed with the midzone localized and mostly PRC1-dependent sliding motors whose function in antiparallel overlap in upregulated following anaphase onset and a drop in the CDK1 activity [19]. The anaphase B redundancy in motor activity, which was foreseen earlier [102], could be dependent on the cellular context as well, including differences between spindles in the exact organization of MTs, levels of expression of genes encoding motor proteins, regulation of their activity during specific phases, or regulation of activity and localization of their regulators. These questions will surely merit future experimental work on protein redundancy during anaphase B spindle elongation.
6.2. MT crosslinkers - midzone stability and braking mechanisms
The MTs of the central spindle during late anaphase can resist high-pressure forces [103] and show increased resistance to nocodazole-induced depolymerization [74], when compared to MTs in early anaphase midzone in HeLa cells. This dramatic increase in MT stability following anaphase onset has been shown in yeast cells [104], [105], while in Drosophila embryos it was observed only in the pole-proximal region of the anaphase spindle [106]. However, it remained unclear if this increase in MT stability, mediated probably by a decrease in PRC1 phosphorylation state after anaphase onset [74], [107], is important for all stages during anaphase. Recent work indicates that this increase in central spindle MT stability is not essential for anaphase spindle elongation during early anaphase as reduced MT stability does not follow the trend of reduced spindle elongation velocities across different conditions [68].
This brings us to the question of how spindles know when to stop elongating. We argue that recent data points out that this is mediated by a temporally controlled counterforce to the active MT sliding in the midzone induced by the action of PRC1 during late anaphase in human cells [34], [108], [109]. Similar mechanism has been proposed to work during anaphase B in yeast spindles [110]. Experimental work on human cells showed that during late anaphase, cells with perturbed PRC1 do not stop spindle elongation at the same time as control spindles, and this led to spindle over-elongation, demonstrating that PRC1 is an important player in the control of extent of spindle elongation [34], [68]. One possibility is that PRC1 induces frictional forces within the spindle midzone to oppose cortical pulling by astral MTs mediated by cytoplasmic dynein on the cortex [34]. However, as mentioned previously, the cortical pulling mechanism does not seem to contribute greatly to the spindle elongation, meaning this mechanism of spindle elongation impeding is unlikely in human cells. We speculate this mechanism instead involves braking imposed to the sliding of midzone MTs [68].
It is possible that PRC1 is by itself taking part in this braking action (Fig. 7, box 2). Recent in vitro work supports this view as it showed that frictional forces induced by PRC1 scale with MT sliding velocity and the number of PRC1 crosslinks but do not depend on the overlap length or PRC1 density within overlaps [108], [111]. Interestingly, as PRC1 does not substantially resist the relative sliding of two MTs crosslinked by kinesin-5 motors [112], this suggests that resisting forces might depend on the nature and strength of the motor force production within the overlap. This PRC1 braking action is probably temporarily regulated in the cell. In that regard, the possibility that PRC1 could induce large friction to sliding during early anaphase is not supported by recent FRAP experiments demonstrating that turnover of PRC1 in the midzone of early anaphase spindles is relatively fast [34], [109], and the reported rates are not different from turnover rates during metaphase [109]. However, turnover rates drastically reduce as the cell approaches late anaphase and telophase, coinciding with increase in the signal of PRC1 within the midzone [109], and the drastic drop in spindle elongation velocity in human cells [42]. The notion that PRC1 residence time on antiparallel MTs is important for the extent of the force that resists MT sliding is reinforced by the recent in vitro work showing that greater retention of PRC1 molecules in overlaps leads to larger frictional forces [111]. The idea of temporal regulation of braking forces by increase in both midzone stability and PRC1 friction is nicely recapitulated by PRC1 over-expression [68] and PLK1 inhibition experiments [89], [107], which both induce premature and extensive PRC1 bundling of midzone MTs during metaphase, resulting in blocked spindle elongation during anaphase B.
6.3. Plus-end controlling proteins and overlap length regulation
On the other hand, regarding the importance of polymerization of interpolar MTs during anaphase, a recent study pointed out that the ends of MTs in overlap regions are nondynamic in human cells and supported a model in which the change in overlap length within MT bundles is directly related to the extent of MT sliding [34]. However, the peak-metaphase overlap length, measured to be around 5 µm in human cells inferred from PRC1 signal length [30], [34], cannot explain the extent of the spindle elongation since spindle elongates in anaphase by 6–9 µm [42], and if model of nondynamic ends is correct, the spindle could elongate to a maximum extent of 5 µm after which the midzone MTs would completely slide apart. This phenomenon was never observed in anaphase of normal human cells where overlap length in central spindle during late anaphase is around 2 µm [19].
We argue that interpolar MTs polymerize at the midzone as the anaphase B spindle elongates, as was reported in Ptk1 cells [67], [113], possibly assisted by the augmin-/γ-TuRC-dependent branching of midzone MT plus-ends in human cells [114]. The dynamics of midzone plus-ends is then regulated by PRC1-dependent protein KIF4A, a known regulator of interpolar MTs plus-ends [84], [115] and possibly also by PRC1-dependent CLASP proteins, known to localize to the spindle midzone, where they control plus-end dynamics [116], [117], MT-depolymerizing kinesin KIF18A that localizes to the central spindle in anaphase [81] and regulates overlap length in metaphase [115] (Fig. 7, box 3), and various members of kinesin-13 family [118]. However, the exact molecular mechanism of overlap length regulation during anaphase merits further experimental work.
Importance of overlap length for spindle elongation was substantiated by notion that spindle over-elongation is observed after depletion of PRC1-interacting partner KIF4A [74], which inhibits the dynamics of MT plus-end [119] (Fig. 7, box 3). Accordingly, it is known that PRC1-bound MT bundles increase drastically in length after KIF4A depletion [84], implying longer overlaps within those bundles. Thus, if KIF4A is depleted from the spindle midzone, MT dynamics is switched to favoring polymerization of interpolar MTs, leading to longer overlaps that could support binding of more motors that exert their forces even during late anaphase, leading to spindle over-elongation. Thus, KIF4A controls the length of antiparallel overlaps during anaphase by regulating polymerization at their plus-ends [74], defining the final length of the spindle. However, it is not clear if KIF4A requires PRC1 to localize to plus-ends of interpolar MTs during late anaphase in human cells, although current experimental data support a view in which KIF4A can perform this function independently of PRC1 [74]. To conclude, sudden increase in the stability of spindle MTs, an increase in the PRC1-mediated frictional forces and KIF4A activity at plus-ends [74] solidify and shorten the midzone overlap regions during late anaphase resulting in the slowdown of further sliding in the midzone [34], [108] (Fig. 7, box 2).
6.4. Transmission of the force from midzone to spindle poles
One interesting notion inferred from recent studies is that MT sliding velocities within the spindle midzone are almost equal to the chromosome segregation velocity [28], [68]. This implies that the force-transmission from the spindle midzone to the chromosome-proximal region is a very efficient process. It is interesting that the region between separating chromosomes and 1–2 µm from kinetochores is also defined to the same extent by the antiparallel overlaps measured by the extent of PRC1 signal [29], [30]. This suggests that transmission of force generated in the midzone is efficient in the region where bridging antiparallel MTs are present, and where they are strongly crosslinked to kinetochore MTs. On the other hand, how is this coordinated with the ongoing depolymerization of k-fiber MTs at kinetochores, which also contributes to chromosome segregation velocity during anaphase [12], remains unknown, and requires additional experimental data.
Interestingly, sliding velocities also tend to correlate with spindle elongation velocities, but sliding is always slower than spindle elongation within an individual cell [68]. This may be due to depolymerization of bridging MTs at their minus-ends, or an increased activity of minus-end directed motors near the pole [120] (Fig. 7, box 4). Observation of such phenomenon is similar in nature to the rate of transport of γTuRC-capped MTs, which is faster near the equator than the poles in the metaphase spindle of human cells [121] and to the flux velocity decrease near spindle poles in Xenopus [122] and human spindles [123]. However, the details of the molecular nature of the connections between k-fibers and bridging fibers during anaphase, and the dynamic nature of their interactions, awaits further studies, but probably includes the activity of MT-crosslinkers like NuMa, EG5, KIF15 and dynein (Fig. 7, box 4), similar to metaphase spindles [124].
6.5. Tubulin polymerization and active force-generation
Theoretically, it would be possible that spindles elongate as a result of pushing forces exerted directly by MT polymerization from a plus-ends of interpolar MTs. Such growth against the barriers is a well-known way of MT pushing [125]. This MT pushing mechanism has been described in yeasts and it is involved in the positioning of spindle pole bodies or the spindle, without contributing to anaphase B spindle elongation [63], [126]. In newt lung cells, pushing forces from growing MTs were demonstrated by a laser ablation approach where the removed chromosome fragment without kinetochore was pushed away from the proximal pole by the action of PEFs [127]. This force that ejects chromosome arms from the poles could result from the push of growing astral MTs directly into chromosome arm or from action of chromokinesins walking on MTs while they are connected to chromosome [128] (Fig. 7, box 5). Interestingly, it was shown that PEFs are inactivated by degradation of KID motor at anaphase onset in Xenopus egg extracts [129]. A similar reduction of PEFs at anaphase onset likely occurs also in human cells, though to a smaller extent given that after double-depletion of KID/KIF4A, chromosomes segregate more closer to the spindle poles [42]. Regarding direct pushing by growing plus-ends of interpolar MTs in the midzone, current experimental data does not support such possibility in human cells as KIF4A and EG5 promote MT depolymerization [119], [130], thus in their absence MTs are expected to polymerize more, and promote spindle elongation, which is the opposite from the block of spindle elongation that was observed after perturbation of these two motors in human cells [68]. Conversely, the CLASP-dependent polymerization of MTs between chromosomes is essential for anaphase B in acentrosomal spindles of C. elegans oocytes [131], [132]. For direct testing of the role of MT polymerization forces during anaphase spindle elongation in human cells, additional studies are needed.
6.6. Minus-end controlling proteins and poleward flux
The phenomenon of poleward flux represents the active movement of tubulin subunits within an individual spindle MT toward its respective spindle pole [133], and it was described in some systems that this process slows down as anaphase progresses [134]. A more contemporary view of flux sees it as the result of sliding forces generated by interpolar MTs that are coupled with k-fibers whose minus-ends depolymerize around poles [123], [135], [136], [137]. In human cells with inhibited flux during metaphase, following kinesins-13 depletion (Fig. 7, box 6), the velocity of spindle elongation was unchanged [138]. In Drosophila embryo mitosis and Xenopus egg extracts, a reduction in poleward flux rates after anaphase onset has been shown to act as a switch between steady-state metaphase length and anaphase spindle elongation [139], [140]. On the other hand, attenuation of MT‐flux in human cells either had no effect on spindle length [138], or resulted in shorter spindles [141]. Spindle shortening after flux perturbation could be explained by a situation in which plus-end polymerization is reduced, while minus-end depolymerization still continues. Recent data from human cells showed that MT flux driven by four kinesins regulates spindle length by specifically counteracting depolymerization of kinetochore MTs at their plus-ends driven primarily by MCAK/kinesin-13 [135]. This could explain why MT flux becomes dispensable to regulate spindle length in the absence of MCAK during metaphase [138]. In anaphase, when MCAK activity is downregulated [138], [142] and kinetochore MTs depolymerize at plus-ends, the spindle elongation may start after minus-end depolymerization slows down, as in Drosophila [140], but that notion requires direct testing. However, the minus-end depolymerization is likely more pronounced in human spindles during anaphase when compared to Drosophila embryo cells, as MT sliding rates in human cells are higher than the rates of spindle elongation during anaphase B [28], [68], [140].
6.7. Cortical force generators and spindle elongation
As already described, one way of cortical force generation revolves around motor-dependent mechanisms in which minus-end directed motor dynein in complex with dynactin-NuMA pulls the astral MTs toward the cortex [1] (Fig. 7, box 7). In human cells, the final spindle length at the end of anaphase is reduced after NuMA depletion [143]. However, NuMA is during anaphase and metaphase also located throughout the mitotic spindle where it mediates connections between parallel MTs [123], [124], [143] (Fig. 7, box 4), meaning conclusions based on depletion approaches are not straightforward. Furthermore, the effects of disruption of NuMa and cortical forces on cell elongation rather than spindle elongation are also possible [43], and would lead to the inability of the spindle to elongate when it reaches the cortical area [74]. Moreover, spindle elongation in human cells does not depend on the presence of dynein, since after total depletion of dynein, spindles are able to elongate similarly to controls [95], [96]. More specifically, removal of both dynein cortical adapters LGN and a 4.1 family protein does not affect rates of sister chromatid segregation in HeLa cells during early anaphase, but only during later stages [43]. In agreement, there was no acceleration of spindle pole separation after cutting of spindle midzone region during early anaphase [12], [28], similar to other systems where midzone forces are predominant [63]. In line with this, disruption of the actin cytoskeleton by actin-antagonizing drugs, which abolishes the astral MT link to the cortex and thus the forces they may impose on the spindle, does not affect spindle pole separation in human cells [144]. On the other hand, one could argue that astral MT connections to the actin cytoskeleton are not essential for spindle elongation [56]. In conclusion, cortical pulling forces mediated by dynein seem dispensable for fast spindle elongation and chromosome segregation during early anaphase in human cells.
Still, as astral MTs might be exerting pulling by depolymerization driven forces coupled to cortical or other adapters, which could be dynein and actin-independent, direct evidence for the absence of activity of cortical forces for spindle elongation in human cells is still lacking, contrary to evidence for their role in spindle positioning and orientation [43]. One would argue that normal mitosis in cells that lack most astral MTs observed in monoastral bipolar spindles after treatment with the plk4 inhibitor centrinone [56], [72], or in an anastral spindles observed after ablation of both centrosomes during prophase [73], suggests that astral force generation is not essential for anaphase B. Yet, as these experimental setups were not designed to study the anaphase B mechanisms directly, the conclusions in that part are challenging. One promising approach in that regard seems to be to control the growing ends of MTs by the localized control of EB1 activity using optogenetic approaches, similar to what has recently been done on metaphase spindles in human cells [145], where disruption of both cortical and midzone growing MTs had an impact on spindle length.
7. Temporal control of distinct mechanisms during anaphase B – maybe all models were right?
The different anaphase B mechanisms of inside pushing and outside pulling could operate in a coordinated manner during distinct phases of anaphase (Fig. 8). Thus, it is possible that spindle elongation might depend on midzone-generated forces during early anaphase B spindle elongation, as there is a lot of evidence supporting this [12], [28], [68], while during late anaphase, forces generated on the cortex may help to elongate the spindle. Although little experimental evidence currently support the letter possibility, like clear reduction in chromosome segregation velocity during late anaphase after depletion of dynein cortical adapters [43], the lack of experimental approaches directly addressing this problem does not firm the conclusion in which cortical forces are not contributing to spindle elongation.
Fig. 8.
Proposed scheme of temporally divided activities of different spindle elongation mechanisms. Typical time course in human cells for centrosome distance and length of overlap is plotted in the graph that further depicts molecular force-generating mechanisms which act in respective phases of mitosis and are presumably responsible for the observed dynamics of centrosome movement and overlap length changes. P, prophase; PM, prometaphase, Meta, metaphase; Ana, anaphase; T, telophase.
One observations however, argues against this temporal displacement possibility, at least before late anaphase, and that is the striking similarity between the rates of spindle elongation during both prometaphase and early anaphase [41], [68] (Fig. 8), implying similar mechanisms. However, the rates during early and late anaphase are clearly different, although the latter are probably controlled by the midzone braking mechanisms [34], [108] (Fig. 8), indicating there is a possibility that cortical forces might operate during late anaphase to elongate the spindle. In addition, since astral MTs might be exerting forces independently of dynein pulling by motor-activity, the experimental challenge in directly assessing this problem is to disrupt astral MTs without disrupting other MTs in the spindle, a challenge that seems to be improved after the development of optogenetic tools for fast control of protein localization and activity in specific space and time [115], [145]. Such localized approaches could finally settle the old debate, and contradictory experimental data gathered using similar techniques in various systems, that could end up in the situation in which all models were right, just at different time points.
8. Conclusions
Anaphase B is a very robust phase of mitosis that relies on the multiple distinct and coordinated mechanisms to accomplish the crucial task of chromosome segregation. We reason this is due to the fact that anaphase represents the final point in the mitotic process, where after satisfaction of complex survey mechanisms that ensure proper connections between MTs and chromosomes, the cell must achieve sufficient chromosome segregation before onset of chromosome decondensation, nuclear envelope reformation and furrow ingression. The final result is two daughter cells with an equal number of chromosomes. In that regard, anaphase is not only adapted to precision rather than to speed [146], but it is also highly robust and characterized by redundancy to achieve effective separation of sister chromatids to adapt to ever-changing environment. Finally, in many regards, early anaphase seems more similar to metaphase than to late anaphase when the central spindle solidifies in preparation for the late mitotic stages, with some notable differences including downregulation of minus-end directed motors and other mechanisms that might oppose anaphase movement, recruitment of additional plus-end generating motors to the midzone region and upregulation of other outward directed mechanisms that together help to elongate the spindle.
Acknowledgements
We thank Ivana Šarić for the drawings, Patrik Risteski and Juraj Simunić for the comments. The work in the Tolić group is funded by the European Research Council (ERC) (ERC Synergy Grant, GA Number 855158, European Union), Croatian Science Foundation Cooperation Programme with Croatian Scientists in Diaspora “Research Cooperability” (Project PZS-2019-02-7653, Croatia), the Science and Innovation Grant cofinanced by the European Structural and Investment Funds (ESIF) within the Operational Programme Competitiveness and Cohesion (OPCC) 2014–2020 (Grant KK.01.1.1.04.0057, Croatia and European Union), and QuantiXLie Centre of Excellence, a project cofinanced by the Croatian Government, Croatia and European Union through the European Regional Development Fund - the Competitiveness and Cohesion Operational Programme (Grant KK.01.1.1.01.0004). We also acknowledge earlier support from the ERC (Consolidator Grant, GA Number 647077, European Union). Finally, while we attempted a balanced literature review within the scope of this article, we apologize to all our colleagues whose work could not be cited due to space restrictions.
Contributor Information
Kruno Vukušić, Email: kvukusic@irb.hr.
Iva M. Tolić, Email: tolic@irb.hr.
References
- 1.McIntosh J.R., Molodtsov M.I., Ataullakhanov F.I. Biophysics of mitosis. Q. Rev. Biophys. 2012;45(2):147–207. doi: 10.1017/S0033583512000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavin N., Tolic I.M. Self-organization and forces in the mitotic spindle. Annu. Rev. Biophys. 2016;45:279–298. doi: 10.1146/annurev-biophys-062215-010934. [DOI] [PubMed] [Google Scholar]
- 3.Maiato H., Gomes A.M., Sousa F., Barisic M. Mechanisms of chromosome congression during mitosis. Biology. 2017;6(1):13. doi: 10.3390/biology6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kops G., Snel B., Tromer E.C. Evolutionary dynamics of the spindle assembly checkpoint in eukaryotes. Curr. Biol. 2020;30(10):R589–R602. doi: 10.1016/j.cub.2020.02.021. [DOI] [PubMed] [Google Scholar]
- 5.Maiato H., Lince-Faria M. The perpetual movements of anaphase. Cell Mol. Life Sci. 2010;67(13):2251–2269. doi: 10.1007/s00018-010-0327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vukušić K., Buđa R., Tolić I.M. Force-generating mechanisms of anaphase in human cells. J. Cell Sci. 2019;132(18) doi: 10.1242/jcs.231985. [DOI] [PubMed] [Google Scholar]
- 7.Scholey J.M., Civelekoglu-Scholey G., Brust-Mascher I. Anaphase B. Biology. 2016;5(4) doi: 10.3390/biology5040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ris H. The anaphase movement of chromosomes in the spermatocytes of the grasshopper. Biol. Bull. 1949;96(1):90–106. [PubMed] [Google Scholar]
- 9.Inoue S., Ritter H., Jr. Dynamics of mitotic spindle organization and function. Soc. Gen. Physiol. Ser. 1975;30:3–30. [PubMed] [Google Scholar]
- 10.Roostalu J., Schiebel E., Khmelinskii A. Cell cycle control of spindle elongation. Cell Cycle. 2010;9(6):1084–1090. doi: 10.4161/cc.9.6.11017. [DOI] [PubMed] [Google Scholar]
- 11.Gayathri P., Fujii T., Moller-Jensen J., van den Ent F., Namba K., Lowe J. A bipolar spindle of antiparallel ParM filaments drives bacterial plasmid segregation. Science. 2012;338(6112):1334–1337. doi: 10.1126/science.1229091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vukušić K., Buđa R., Bosilj A., Milas A., Pavin N., Tolić I.M. Microtubule sliding within the bridging fiber pushes kinetochore fibers apart to segregate chromosomes. Dev. Cell. 2017;43(1):11–23. doi: 10.1016/j.devcel.2017.09.010. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.FitzHarris G. Anaphase B precedes anaphase A in the mouse egg. Curr. Biol. 2012;22(5):437–444. doi: 10.1016/j.cub.2012.01.041. [DOI] [PubMed] [Google Scholar]
- 14.Lens S.M.A., Medema R.H. Cytokinesis defects and cancer. Nat. Rev. Cancer. 2019;19(1):32–45. doi: 10.1038/s41568-018-0084-6. [DOI] [PubMed] [Google Scholar]
- 15.Gordon D.J., Resio B., Pellman D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012;13(3):189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 16.Cimini D., Cameron L.A., Salmon E.D. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 2004;14(23):2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 17.Afonso O., Matos I., Pereira A.J., Aguiar P., Lampson M.A., Maiato H. Feedback control of chromosome separation by a midzone Aurora B gradient. Science. 2014;345(6194):332–336. doi: 10.1126/science.1251121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Avino P.P., Giansanti M.G., Petronczki M. Cytokinesis in animal cells. Cold Spring Harb. Perspect. Biol. 2015;7(4) doi: 10.1101/cshperspect.a015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee K.Y., Davies T., Mishima M. Cytokinesis microtubule organisers at a glance. J. Cell Sci. 2012;125(Pt 15):3495–3500. doi: 10.1242/jcs.094672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding R., McDonald K.L., McIntosh J.R. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 1993;120(1):141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIntosh J.R., Landis S.C. The distribution of spindle microtubules during mitosis in cultured human cells. J. Cell Biol. 1971;49(2):468–497. doi: 10.1083/jcb.49.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winey M., Mamay C.L., O’Toole E.T., Mastronarde D.N., Giddings T.H., Jr., McDonald K.L., McIntosh J.R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 1995;129(6):1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mastronarde D.N., McDonald K.L., Ding R., McIntosh J.R. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 1993;123(6 Pt 1):1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald K., Pickett-Heaps J.D., McIntosh J.R., Tippit D.H. On the mechanism of anaphase spindle elongation in Diatoma vulgare. J. Cell Biol. 1977;74(2):377–388. doi: 10.1083/jcb.74.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tolic I.M. Mitotic spindle: kinetochore fibers hold on tight to interpolar bundles. Eur. Biophys. J. 2018;47(3):191–203. doi: 10.1007/s00249-017-1244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goode D. Microtubule turnover as a mechanism of mitosis and its possible evolution. Biosystems. 1981;14:271–287. doi: 10.1016/0303-2647(81)90034-4. [DOI] [PubMed] [Google Scholar]
- 27.Margolis R.L., Wilson L., Keifer B.I. Mitotic mechanism based on intrinsic microtubule behaviour. Nature. 1978;272(5652):450–452. doi: 10.1038/272450a0. [DOI] [PubMed] [Google Scholar]
- 28.Yu C.H., Redemann S., Wu H.Y., Kiewisz R., Yoo T.Y., Conway W., Farhadifar R., Muller-Reichert T., Needleman D. Central-spindle microtubules are strongly coupled to chromosomes during both anaphase A and anaphase B. Mol. Biol. Cell. 2019;30(19):2503–2514. doi: 10.1091/mbc.E19-01-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajtez J., Solomatina A., Novak M., Polak B., Vukusic K., Rudiger J., Cojoc G., Milas A., Sumanovac Sestak I., Risteski P., Tavano F., Klemm A.H., Roscioli E., Welburn J., Cimini D., Gluncic M., Pavin N., Tolic I.M. Overlap microtubules link sister k-fibres and balance the forces on bi-oriented kinetochores. Nat. Commun. 2016;7:10298. doi: 10.1038/ncomms10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polak B., Risteski P., Lesjak S., Tolic I.M. PRC1-labeled microtubule bundles and kinetochore pairs show one-to-one association in metaphase. EMBO Rep. 2017;18(2):217–230. doi: 10.15252/embr.201642650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Toole E., Morphew M., McIntosh J.R. Electron tomography reveals aspects of spindle structure important for mechanical stability at metaphase. Mol. Biol. Cell. 2020;31(3):184–195. doi: 10.1091/mbc.E19-07-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nixon F.M., Honnor T.R., Clarke N.I., Starling G.P., Beckett A.J., Johansen A.M., Brettschneider J.A., Prior I.A., Royle S.J. Microtubule organization within mitotic spindles revealed by serial block face scanning electron microscopy and image analysis. J. Cell Sci. 2017;130(10):1845–1855. doi: 10.1242/jcs.203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. Manenica, V. Štimac, I. Koprivec, J. Simunić, I.M. Tolić, Augmin regulates kinetochore tension and spatial arrangement of spindle microtubules by nucleating bridging fibers, bioRxiv 2020.09.10.291740, 2020. doi: 10.1101/2020.09.10.291740. [DOI]
- 34.Pamula M.C., Carlini L., Forth S., Verma P., Suresh S., Legant W.R., Khodjakov A., Betzig E., Kapoor T.M. High-resolution imaging reveals how the spindle midzone impacts chromosome movement. J. Cell Biol. 2019;218(8):2529–2544. doi: 10.1083/jcb.201904169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uehara R., Nozawa R.S., Tomioka A., Petry S., Vale R.D., Obuse C., Goshima G. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 2009;106(17):6998–7003. doi: 10.1073/pnas.0901587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McEwen B.F., Chan G.K., Zubrowski B., Savoian M.S., Sauer M.T., Yen T.J. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell. 2001;12(9):2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R. Kiewisz, W. Conway, D. Needleman, T. Muller-Reichert, Ultrastructural characterization of k-fiber in mitotic spindles in human cells in culture, Mol. Biol. Cell 31 (26) (ASCB meeting abstract 1434).
- 38.McEwen B.F., Heagle A.B., Cassels G.O., Buttle K.F., Rieder C.L. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J. Cell Biol. 1997;137(7):1567–1580. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald K.L., O’Toole E.T., Mastronarde D.N., McIntosh J.R. Kinetochore microtubules in PTK cells. J. Cell Biol. 1992;118(2):369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rieder C.L. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 1981;84(1):145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- 41.Magidson V., O’Connell C.B., Loncarek J., Paul R., Mogilner A., Khodjakov A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146(4):555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su K.C., Barry Z., Schweizer N., Maiato H., Bathe M., Cheeseman I.M. A regulatory switch alters chromosome motions at the metaphase-to-anaphase transition. Cell Rep. 2016;17(7):1728–1738. doi: 10.1016/j.celrep.2016.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiyomitsu T., Cheeseman I.M. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 2013;154(2):391–402. doi: 10.1016/j.cell.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 2009;10(1):9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tolic-Norrelykke I.M. Push-me-pull-you: how microtubules organize the cell interior. Eur. Biophys. J. 2008;37(7):1271–1278. doi: 10.1007/s00249-008-0321-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pavin N., Tolic-Norrelykke I.M. Dynein, microtubule and cargo: a menage a trois. Biochem. Soc. Trans. 2013;41(6):1731–1735. doi: 10.1042/BST20130235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laan L., Pavin N., Husson J., Romet-Lemonne G., van Duijn M., Lopez M.P., Vale R.D., Julicher F., Reck-Peterson S.L., Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148(3):502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozlowski C., Srayko M., Nedelec F. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 2007;129(3):499–510. doi: 10.1016/j.cell.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 49.Grishchuk E.L., Molodtsov M.I., Ataullakhanov F.I., McIntosh J.R. Force production by disassembling microtubules. Nature. 2005;438(7066):384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- 50.Akhmanova A., Steinmetz M.O. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 2015;16(12):711–726. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- 51.Telley I.A., Gaspar I., Ephrussi A., Surrey T. Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. J. Cell Biol. 2012;197(7):887–895. doi: 10.1083/jcb.201204019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterman-Storer C.M., Desai A., Bulinski J.C., Salmon E.D. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 1998;8(22):1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- 53.Grill S.W., Hyman A.A. Spindle positioning by cortical pulling forces. Dev. Cell. 2005;8(4):461–465. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 54.Grill S.W., Howard J., Schaffer E., Stelzer E.H., Hyman A.A. The distribution of active force generators controls mitotic spindle position. Science. 2003;301(5632):518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- 55.Grill S.W., Gonczy P., Stelzer E.H., Hyman A.A. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409(6820):630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- 56.Guild J., Ginzberg M.B., Hueschen C.L., Mitchison T.J., Dumont S. Increased lateral microtubule contact at the cell cortex is sufficient to drive mammalian spindle elongation. Mol. Biol. Cell. 2017;28(14):1975–1983. doi: 10.1091/mbc.E17-03-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.di Pietro F., Echard A., Morin X. Regulation of mitotic spindle orientation: an integrated view. EMBO Rep. 2016;17(8):1106–1130. doi: 10.15252/embr.201642292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aist J.R., Bayles C.J., Tao W., Berns M.W. Direct experimental evidence for the existence, structural basis and function of astral forces during anaphase B in vivo. J. Cell Sci. 1991;100(Pt 2):279–288. doi: 10.1242/jcs.100.2.279. [DOI] [PubMed] [Google Scholar]
- 59.Hiramoto Y., Nakano Y. Micromanipulation studies of the mitotic apparatus in sand dollar eggs. Cell Motil. Cytoskelet. 1988;10:172–184. doi: 10.1002/cm.970100122. [DOI] [PubMed] [Google Scholar]
- 60.Dogterom M., Kerssemakers J.W., Romet-Lemonne G., Janson M.E. Force generation by dynamic microtubules. Curr. Opin. Cell Biol. 2005;17(1):67–74. doi: 10.1016/j.ceb.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 61.McIntosh J.R., Hepler P.K., Van Wie D.G. Model for mitosis. Nature. 1969;224:659–663. [Google Scholar]
- 62.Huxley H., Hanson J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 1954;173(4412):973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- 63.Tolic-Norrelykke I.M., Sacconi L., Thon G., Pavone F.S. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr. Biol. 2004;14(13):1181–1186. doi: 10.1016/j.cub.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 64.Khodjakov A., La Terra S., Chang F. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr. Biol. 2004;14(15):1330–1340. doi: 10.1016/j.cub.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 65.Ward J.J., Roque H., Antony C., Nedelec F. Mechanical design principles of a mitotic spindle. Elife. 2014;3 doi: 10.7554/eLife.03398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pavin N., Tolic I.M. Mechanobiology of the mitotic spindle. Dev. Cell. 2021;56(2):192–201. doi: 10.1016/j.devcel.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 67.Saxton W.M., McIntosh J.R. Interzone microtubule behavior in late anaphase and telophase spindles. J. Cell Biol. 1987;105(2):875–886. doi: 10.1083/jcb.105.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vukušić K., Ponjavić I., Buđa R., Risteski P., Tolić I.M. Microtubule sliding modules based on kinesins EG5 and PRC1-dependent KIF4A drive human spindle elongation. Dev. Cell. 2021 doi: 10.1016/j.devcel.2021.04.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aist J.R., Liang H., Berns M.W. Astral and spindle forces in PtK2 cells during anaphase B: a laser microbeam study. J. Cell Sci. 1993;104(Pt 4):1207–1216. doi: 10.1242/jcs.104.4.1207. [DOI] [PubMed] [Google Scholar]
- 70.Brust-Mascher I., Sommi P., Cheerambathur D.K., Scholey J.M. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol. Biol. Cell. 2009;20(6):1749–1762. doi: 10.1091/mbc.E08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nicklas R.B., Kubai D.F., Hays T.S. Spindle microtubules and their mechanical associations after micromanipulation in anaphase. J. Cell Biol. 1982;95(1):91–104. doi: 10.1083/jcb.95.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dudka D., Castrogiovanni C., Liaudet N., Vassal H., Meraldi P. Spindle-length-dependent HURP localization allows centrosomes to control kinetochore-fiber plus-end dynamics. Curr. Biol. 2019;29(21):3563–3578. doi: 10.1016/j.cub.2019.08.061. e6. [DOI] [PubMed] [Google Scholar]
- 73.Khodjakov A., Cole R.W., Oakley B.R., Rieder C.L. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 2000;10(2):59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 74.Hu C.K., Coughlin M., Field C.M., Mitchison T.J. KIF4 regulates midzone length during cytokinesis. Curr. Biol. 2011;21(10):815–824. doi: 10.1016/j.cub.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wurzenberger C., Gerlich D.W. Phosphatases: providing safe passage through mitotic exit. Nat. Rev. Mol. Cell Biol. 2011;12(8):469–482. doi: 10.1038/nrm3149. [DOI] [PubMed] [Google Scholar]
- 76.Gable A., Qiu M., Titus J., Balchand S., Ferenz N.P., Ma N., Collins E.S., Fagerstrom C., Ross J.L., Yang G., Wadsworth P. Dynamic reorganization of Eg5 in the mammalian spindle throughout mitosis requires dynein and TPX2. Mol. Biol. Cell. 2012;23(7):1254–1266. doi: 10.1091/mbc.E11-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pfarr C.M., Coue M., Grissom P.M., Hays T.S., Porter M.E., McIntosh J.R. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990;345(6272):263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- 78.Cai S., Weaver L.N., Ems-McClung S.C., Walczak C.E. Proper organization of microtubule minus ends is needed for midzone stability and cytokinesis. Curr. Biol. 2010;20(9):880–885. doi: 10.1016/j.cub.2010.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seldin L., Poulson N.D., Foote H.P., Lechler T. NuMA localization, stability, and function in spindle orientation involve 4.1 and Cdk1 interactions. Mol. Biol. Cell. 2013;24(23):3651–3662. doi: 10.1091/mbc.E13-05-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kotak S., Busso C., Gonczy P. NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J. 2014;33(16):1815–1830. doi: 10.15252/embj.201488147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stumpff J., von Dassow G., Wagenbach M., Asbury C., Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell. 2008;14(2):252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wandke C., Barisic M., Sigl R., Rauch V., Wolf F., Amaro A.C., Tan C.H., Pereira A.J., Kutay U., Maiato H., Meraldi P., Geley S. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J. Cell Biol. 2012;198(5):847–863. doi: 10.1083/jcb.201110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kurasawa Y., Earnshaw W.C., Mochizuki Y., Dohmae N., Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23(16):3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu C., Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA. 2005;102(2):343–348. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.She Z.Y., Wei Y.L., Lin Y., Li Y.L., Lu M.H. Mechanisms of the Ase1/PRC1/MAP65 family in central spindle assembly. Biol. Rev. Camb. Philos. Soc. 2019;94(6):2033–2048. doi: 10.1111/brv.12547. [DOI] [PubMed] [Google Scholar]
- 86.Afonso O., Figueiredo A.C., Maiato H. Late mitotic functions of Aurora kinases. Chromosoma. 2017;126(1):93–103. doi: 10.1007/s00412-016-0594-5. [DOI] [PubMed] [Google Scholar]
- 87.Straight A.F., Sedat J.W., Murray A.W. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 1998;143(3):687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fu C., Ward J.J., Loiodice I., Velve-Casquillas G., Nedelec F.J., Tran P.T. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev. Cell. 2009;17(2):257–267. doi: 10.1016/j.devcel.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brennan I.M., Peters U., Kapoor T.M., Straight A.F. Polo-like kinase controls vertebrate spindle elongation and cytokinesis. PLoS One. 2007;2(5) doi: 10.1371/journal.pone.0000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Afonso O., Castellani C.M., Cheeseman L.P., Ferreira J.G., Orr B., Ferreira L.T., Chambers J.J., Morais-de-Sa E., Maresca T.J., Maiato H. Spatiotemporal control of mitotic exit during anaphase by an aurora B-Cdk1 crosstalk. Elife. 2019;8 doi: 10.7554/eLife.47646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Coelho P.A., Queiroz-Machado J., Carmo A.M., Moutinho-Pereira S., Maiato H., Sunkel C.E. Dual role of topoisomerase II in centromere resolution and aurora B activity. PLoS Biol. 2008;6(8) doi: 10.1371/journal.pbio.0060207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Collins E., Mann B.J., Wadsworth P. Eg5 restricts anaphase B spindle elongation in mammalian cells. Cytoskeleton. 2014;71(2):136–144. doi: 10.1002/cm.21158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cande W.Z. Nucleotide requirements for anaphase chromosome movements in permeabilized mitotic cells: anaphase B but not anaphase A requires ATP. Cell. 1982;28(1):15–22. doi: 10.1016/0092-8674(82)90370-1. [DOI] [PubMed] [Google Scholar]
- 94.Cande W.Z., McDonald K.L. In vitro reactivation of anaphase spindle elongation using isolated diatom spindles. Nature. 1985;316(6024):168–170. doi: 10.1038/316168a0. [DOI] [PubMed] [Google Scholar]
- 95.van Heesbeen R.G., Tanenbaum M.E., Medema R.H. Balanced activity of three mitotic motors is required for bipolar spindle assembly and chromosome segregation. Cell Rep. 2014;8(4):948–956. doi: 10.1016/j.celrep.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 96.Zhu C., Zhao J., Bibikova M., Leverson J.D., Bossy-Wetzel E., Fan J.B., Abraham R.T., Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 2005;16(7):3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kapitein L.C., Peterman E.J., Kwok B.H., Kim J.H., Kapoor T.M., Schmidt C.F. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435(7038):114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 98.Wijeratne S., Subramanian R. Geometry of antiparallel microtubule bundles regulates relative sliding and stalling by PRC1 and Kif4A. Elife. 2018;7 doi: 10.7554/eLife.32595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nislow C., Lombillo V.A., Kuriyama R., McIntosh J.R. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359(6395):543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- 100.Adriaans I.E., Hooikaas P.J., Aher A., Vromans M.J.M., van Es R.M., Grigoriev I., Akhmanova A., Lens S.M.A. MKLP2 is a motile kinesin that transports the chromosomal passenger complex during anaphase. Curr. Biol. 2020;30(13):2628–2637. doi: 10.1016/j.cub.2020.04.081. e9. [DOI] [PubMed] [Google Scholar]
- 101.Su X., Arellano-Santoyo H., Portran D., Gaillard J., Vantard M., Thery M., Pellman D. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat. Cell Biol. 2013;15(8):948–957. doi: 10.1038/ncb2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goldstein L.S. Functional redundancy in mitotic force generation. J. Cell Biol. 1993;120(1):1–3. doi: 10.1083/jcb.120.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Salmon E.D., Goode D., Maugel T.K., Bonar D.B. Pressure-induced depolymerization of spindle microtubules. III. Differential stability in HeLa cells. J. Cell Biol. 1976;69(2):443–454. doi: 10.1083/jcb.69.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhai Y., Kronebusch P.J., Borisy G.G. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol. 1995;131(3):721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Higuchi T., Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433(7022):171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheerambathur D.K., Civelekoglu-Scholey G., Brust-Mascher I., Sommi P., Mogilner A., Scholey J.M. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J. Cell Biol. 2007;177(6):995–1004. doi: 10.1083/jcb.200611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu C.K., Ozlu N., Coughlin M., Steen J.J., Mitchison T.J. Plk1 negatively regulates PRC1 to prevent premature midzone formation before cytokinesis. Mol. Biol. Cell. 2012;23(14):2702–2711. doi: 10.1091/mbc.E12-01-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gaska I., Armstrong M.E., Alfieri A., Forth S. The mitotic crosslinking protein PRC1 acts like a mechanical dashpot to resist microtubule sliding. Dev. Cell. 2020;54(3):367–378. doi: 10.1016/j.devcel.2020.06.017. e5. [DOI] [PubMed] [Google Scholar]
- 109.J. Asthana, N.I. Cade, W.M. Lim, T. Surrey, PRC1 and EB1 binding dynamics reveal a solidifying central spindle during anaphase compaction in human cells, bioRxiv 195347, 2020. doi: 10.1101/2020.07.09.195347. [DOI]
- 110.Thomas E.C., Ismael A., Moore J.K. Ase1 domains dynamically slow anaphase spindle elongation and recruit Bim1 to the midzone. Mol. Biol. Cell. 2020;31(24):2733–2747. doi: 10.1091/mbc.E20-07-0493-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.A. Alfieri, I. Gaska, S. Forth, Two modes of PRC1-mediated mechanical resistance to kinesin-driven microtubule network disruption, bioRxiv 244491, 2020. doi: 10.1101/2020.08.10.244491. [DOI] [PubMed]
- 112.Subramanian R., Wilson-Kubalek E.M., Arthur C.P., Bick M.J., Campbell E.A., Darst S.A., Milligan R.A., Kapoor T.M. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 2010;142(3):433–443. doi: 10.1016/j.cell.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shelden E., Wadsworth P. Interzonal microtubules are dynamic during spindle elongation. J. Cell Sci. 1990;97(Pt 2):273–281. doi: 10.1242/jcs.97.2.273. [DOI] [PubMed] [Google Scholar]
- 114.Uehara R., Kamasaki T., Hiruma S., Poser I., Yoda K., Yajima J., Gerlich D.W., Goshima G. Augmin shapes the anaphase spindle for efficient cytokinetic furrow ingression and abscission. Mol. Biol. Cell. 2016;27(5):812–827. doi: 10.1091/mbc.E15-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jagric M., Risteski P., Martincic J., Milas A., Tolic I.M. Optogenetic control of PRC1 reveals its role in chromosome alignment on the spindle by overlap length-dependent forces. Elife. 2021;10 doi: 10.7554/eLife.61170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu J., Wang Z., Jiang K., Zhang L., Zhao L., Hua S., Yan F., Yang Y., Wang D., Fu C., Ding X., Guo Z., Yao X. PRC1 cooperates with CLASP1 to organize central spindle plasticity in mitosis. J. Biol. Chem. 2009;284(34):23059–23071. doi: 10.1074/jbc.M109.009670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pereira A.L., Pereira A.J., Maia A.R., Drabek K., Sayas C.L., Hergert P.J., Lince-Faria M., Matos I., Duque C., Stepanova T., Rieder C.L., Earnshaw W.C., Galjart N., Maiato H. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol. Biol. Cell. 2006;17(10):4526–4542. doi: 10.1091/mbc.E06-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Uehara R., Tsukada Y., Kamasaki T., Poser I., Yoda K., Gerlich D.W., Goshima G. Aurora B and Kif2A control microtubule length for assembly of a functional central spindle during anaphase. J. Cell Biol. 2013;202(4):623–636. doi: 10.1083/jcb.201302123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bringmann H., Skiniotis G., Spilker A., Kandels-Lewis S., Vernos I., Surrey T. A kinesin-like motor inhibits microtubule dynamic instability. Science. 2004;303(5663):1519–1522. doi: 10.1126/science.1094838. [DOI] [PubMed] [Google Scholar]
- 120.Brust-Mascher I., Civelekoglu-Scholey G., Scholey J.M. Mechanism for Anaphase B: evaluation of “Slide-and-Cluster” versus “Slide-and-Flux-or-Elongate” models. Biophys. J. 2015;108(8):2007–2018. doi: 10.1016/j.bpj.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lecland N., Luders J. The dynamics of microtubule minus ends in the human mitotic spindle. Nat. Cell Biol. 2014;16(8):770–778. doi: 10.1038/ncb2996. [DOI] [PubMed] [Google Scholar]
- 122.Yang G., Cameron L.A., Maddox P.S., Salmon E.D., Danuser G. Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindle. J. Cell Biol. 2008;182(4):631–639. doi: 10.1083/jcb.200801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.P. Risteski, M. Jagrić, I.M. Tolić, Sliding of kinetochore fibers along bridging fibers helps center the chromosomes on the spindle, bioRxiv 424837, 2020. doi: 10.1101/2020.12.30.424837. [DOI]
- 124.Elting M.W., Prakash M., Udy D.B., Dumont S. Mapping load-bearing in the mammalian spindle reveals local kinetochore fiber anchorage that provides mechanical isolation and redundancy. Curr. Biol. 2017;27(14):2112–2122. doi: 10.1016/j.cub.2017.06.018. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dogterom M., Yurke B. Measurement of the force-velocity relation for growing microtubules. Science. 1997;278(5339):856–860. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- 126.Vogel S.K., Raabe I., Dereli A., Maghelli N., Tolic-Norrelykke I. Interphase microtubules determine the initial alignment of the mitotic spindle. Curr. Biol. 2007;17(5):438–444. doi: 10.1016/j.cub.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 127.Rieder C.L., Davison E.A., Jensen L.C., Cassimeris L., Salmon E.D. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 1986;103(2):581–591. doi: 10.1083/jcb.103.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Almeida A.C., Maiato H. Chromokinesins. Curr. Biol. 2018;28(19):R1131–R1135. doi: 10.1016/j.cub.2018.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Funabiki H., Murray A.W. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102(4):411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- 130.Kim C.D., Kim E.D., Liu L., Buckley R.S., Parameswaran S., Kim S., Wojcik E.J. Small molecule allosteric uncoupling of microtubule depolymerase activity from motility in human Kinesin-5 during mitotic spindle assembly. Sci. Rep. 2019;9(1):19900. doi: 10.1038/s41598-019-56173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dumont J., Oegema K., Desai A. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 2010;12(9):894–901. doi: 10.1038/ncb2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Laband K., Le Borgne R., Edwards F., Stefanutti M., Canman J.C., Verbavatz J.M., Dumont J. Chromosome segregation occurs by microtubule pushing in oocytes. Nat. Commun. 2017;8(1):1499. doi: 10.1038/s41467-017-01539-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mitchison T., Evans L., Schulze E., Kirschner M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell. 1986;45(4):515–527. doi: 10.1016/0092-8674(86)90283-7. [DOI] [PubMed] [Google Scholar]
- 134.Mitchison T.J., Salmon E.D. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 1992;119(3):569–582. doi: 10.1083/jcb.119.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Steblyanko Y., Rajendraprasad G., Osswald M., Eibes S., Jacome A., Geley S., Pereira A.J., Maiato H., Barisic M. Microtubule poleward flux in human cells is driven by the coordinated action of four kinesins. EMBO J. 2020;39(23) doi: 10.15252/embj.2020105432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Matos I., Pereira A.J., Lince-Faria M., Cameron L.A., Salmon E.D., Maiato H. Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J. Cell Biol. 2009;186(1):11–26. doi: 10.1083/jcb.200904153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pereira A.J., Maiato H. Maturation of the kinetochore-microtubule interface and the meaning of metaphase. Chromosome Res. 2012;20(5):563–577. doi: 10.1007/s10577-012-9298-8. [DOI] [PubMed] [Google Scholar]
- 138.Ganem N.J., Upton K., Compton D.A. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 2005;15(20):1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 139.Rogers G.C., Rogers S.L., Schwimmer T.A., Ems-McClung S.C., Walczak C.E., Vale R.D., Scholey J.M., Sharp D.J. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427(6972):364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- 140.Brust-Mascher I., Civelekoglu-Scholey G., Kwon M., Mogilner A., Scholey J.M. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc. Natl. Acad. Sci. USA. 2004;101(45):15938–15943. doi: 10.1073/pnas.0407044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Maffini S., Maia A.R., Manning A.L., Maliga Z., Pereira A.L., Junqueira M., Shevchenko A., Hyman A., Yates J.R., 3rd, Galjart N., Compton D.A., Maiato H. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr. Biol. 2009;19(18):1566–1572. doi: 10.1016/j.cub.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ganguly A., Bhattacharya R., Cabral F. Cell cycle dependent degradation of MCAK: evidence against a role in anaphase chromosome movement. Cell Cycle. 2008;7(20):3187–3193. doi: 10.4161/cc.7.20.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kotak S., Busso C., Gonczy P. NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J. 2013;32(18):2517–2529. doi: 10.1038/emboj.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wheatley S.P., Hinchcliffe E.H., Glotzer M., Hyman A.A., Sluder G., Wang Y. CDK1 inactivation regulates anaphase spindle dynamics and cytokinesis in vivo. J. Cell Biol. 1997;138(2):385–393. doi: 10.1083/jcb.138.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dema A., van Harren J., Charafeddine R., Witmann T. EB1 photoinactivation reveals distinct roles of microtubule plus end dynamics in spindle size and orientation. Mol. Biol. Cell. 2020;31(26) (ASCB meeting abstract 1434) [Google Scholar]
- 146.Nicklas R.B. Chromosome movement: current models and experiments on living cells. Soc. Gen. Physiol. Ser. 1975;30:97–117. [PubMed] [Google Scholar]