Version Changes
Revised. Amendments from Version 1
This latest version of the review article "Calcium imaging analysis – how far have we come?" contains a clearer breakdown of the possible image analysis options for different experimental set-ups of calcium imaging experiments such as in vitro versus in vivo methods. We also expand on the 'Quantification' section as this is an important section that was previously very brief. In this section we discuss identification of action potentials from calcium signals as this can be a controversial topic.
Abstract
Techniques for calcium imaging were first demonstrated in the mid-1970s, whilst tools to analyse these markers of cellular activity are still being developed and improved today. For image analysis, custom tools were developed within labs and until relatively recently, software packages were not widely available between researchers. We will discuss some of the most popular methods for calcium imaging analysis that are now widely available and describe why these protocols are so effective. We will also describe some of the newest innovations in the field that are likely to benefit researchers, particularly as calcium imaging is often an inherently low signal-to-noise method. Although calcium imaging analysis has seen recent advances, particularly following the rise of machine learning, we will end by highlighting the outstanding requirements and questions that hinder further progress and pose the question of how far we have come in the past sixty years and what can be expected for future development in the field.
Keywords: Calcium Imaging, Denoising, Motion Correction, Classification, Quantification, Machine Learning, Neural Networks
Introduction
The ability to image calcium ion (Ca 2+) dynamics in cells has long been of interest, particularly in the neurosciences, where it can be used as a marker for neuronal excitability. The origins of calcium imaging began in the mid-1970s ( Blinks et al., 1976; Moisescu et al., 1975), however the most Ca 2+ specific BAPTA (1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid)-based dye was developed in 1980 by Roger Tsien, and its derivatives are still used today ( Tsien, 1980). In the past forty years, the methods available for measuring Ca 2+ fluxes in cells have expanded to include ratiometric, fluorescence lifetime, or fluorescence intensity-based dyes, and genetically-encoded calcium indicators (GECIs) ( Miyawaki et al., 1997; Ohkura et al., 2005). The use of microscopy modalities has also advanced to include light-sheet microscopy (LSM; Huisken et al., 2004) for long-term imaging, and 2-photon microscopy (2PM; Denk et al., 1990) for deep tissue and cell specific uncaging techniques. The combination of Ca 2+ indicator and imaging modality used will reflect the properties of the sample and the scientific question, as well as the methodologies available to the researcher. For example, in vitro imaging, or in vivo invertebrate imaging, may use exogenous or GECIs, imaged using LSM, epifluorescence, 2PM or other fluorescence microscopes depending on the temporal and spatial resolution, timescale of imaging, and thickness of the sample being taken into consideration. Other specialist options for in vivo imaging of GECIs are available for imaging in awake and behaving animals including miniaturized forms of 1- or 2- photon endoscopic fluorescence microscopes (miniscopes) for single-cell in vivo recordings ( Cai et al., 2016; Chen et al., 2013; Silva, 2017).
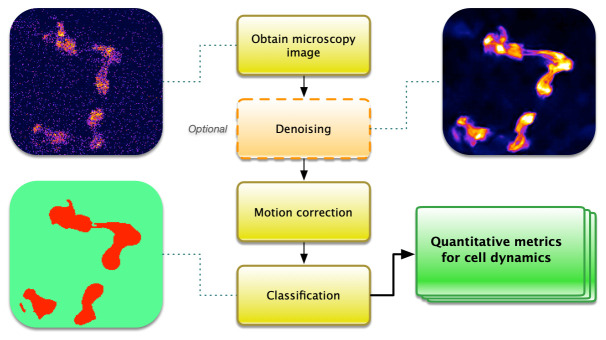
Calcium imaging is an inherently noisy method due to the high spatiotemporal information desired from a sample often showing low signal-to-noise alongside drift or cell movement, particularly for living organisms. In recent years, a number of software packages have been written for individual aspects of the commonly used pipeline in calcium imaging analysis ( Figure 1). This processing pipeline includes image denoising, motion correction, classification for cell identification, and quantification of calcium signals. As calcium imaging is used across a broad range of samples, from sub-cellular, cellular, networks, bulk tissue dynamics to whole organisms and behaving animals, aspects of this pipeline can vary substantially with no ‘one size fits all’ approach.
Figure 1. The steps of a common pipeline for calcium imaging analysis can be subdivided into three areas before quantitative analysis is performed.
Denoising is an optional step that can help to improve signal-to-noise and enhance features. Motion correction may be necessary in cases of drift or movement. Classification can select regions of interest for which quantitative analysis is performed.
Denoising
Live-cell imaging generally requires short exposure times and low excitation power to limit the effects of photo-toxicity and photo-bleaching. This leads to image degradation in the form of noise. In fluorescence microscopy the two prevalent noise sources are Poisson noise and Gaussian noise. Poisson noise is caused by the stochastic and discrete nature of photon emission and tends to be dominant at low light levels, whereas Gaussian noise describes the intrinsic thermal and electronic fluctuation in the image sensor ( Luisier et al., 2011)
Although denoising is not a required step in the pipeline, effective denoising can improve the subsequent steps by artificially enhancing signal-to-noise. Traditionally, image denoising has been based on local averaging approaches, such as the application of a Gaussian smoothing filter ( Buades et al., 2005; Lindenbaum et al., 1994). Alternative methods include a local filter method such as anisotropic filters ( Broser et al., 2004; Kitamura & Häusser, 2011) ( Perona & Malik, 1990), or in the frequency domain, Wiener filters ( Wiener, 1950) and wavelet thresholding methods ( Donoho, 1995)( Besbeas et al., 2004; Wegner et al., 2006).
Local methods are computationally light but have clear limitations. First, the averaging often involved in local methods introduces blur, causing features to appear less well defined. Second, they do not perform well for high noise levels, since the correlations between neighbouring pixels deteriorate ( Shao et al., 2014). In the context of calcium imaging, local methods have been shown to perform well ( Malik et al., 2011).
Non-local filters solve some of the problems by using self-similarity of natural images beyond neighbouring pixels, thus exploiting global information ( Shao et al., 2014). The first method to propose this is the non-local means method ( Buades et al., 2005), in which subregions of an image referred to as patches are restored by weighted averaging of all other patches in an image. Since then, there have been a number of improvements such as invariance to patches that are rotated or mirrored with respect to each other ( Grewenig et al., 2011), improved computational efficiency, and automated parameter tuning and extension to 3D image stacks ( Coupé et al., 2008). Although non-local filters are better at high noise levels, they will typically lead to artefacts like over-smoothing ( Shao et al., 2014). A modern, well-balanced and state-of-the-art non-local method is ND-SAFIR, which is specifically geared towards application in fluorescence microscopy imaging ( Boulanger et al., 2010). ND-SAFIR is a powerful method for removing Poisson-Gaussian noise. It is based on non-local means denoising using a variance stabilisation step, followed by calculating the spatial and temporal patch-based weighted averages for intensity values. The method is widely applicable between experimental samples and can be used directly for 2D+t and 3D+t datasets ( Buades et al., 2011).
In recent years, deep learning methods have become state-of-the-art for denoising. Methods such as DnCNN ( Zhang et al., 2017), FFDNet ( Zhang et al., 2018) and CARE ( Weigert et al., 2018) rely on convolutional neural networks that are trained in a supervised learning approach. However, this requires ground truths to be available for model training, which may be difficult to obtain in practice. A different approach was developed in noise2noise ( Lehtinen et al., 2018), where instead of learning the mapping from noisy images to clean targets, the model is trained with other noisy images as targets. The images must be corresponding pairs displaying the same objects but with independent noise. Assuming the noise sources underlying the images have zero-mean distributions, the weights of the network will then converge during training to the same values as a network trained with clean targets because the noise that manifests in the weights cancels out. A more recent method, noise2void ( Krull et al., 2019), aims to resolve this issue of needing ground truths, by using self-supervised learning. Here, the network is optimised to predict the value of each pixel from the values of neighbouring pixels in an image, thus requiring no separate ground truths. In another recent method, DeepInterpolation ( Lecoq et al., 2020), the need for ground truth training data is avoided by treating the denoising task as a nonlinear interpolation problem. This assumes that the data have a sequential component, such that spatiotemporally overlapping features can be exploited. DeepCAD is a new deep self-supervised denoising method that reduces detection noise and thereby improve the signal-to-noise more than tenfold, which it claims can improve the accuracy of neuron extraction and spike inference ( Li et al., 2021).
Motion correction
Motion correction is often required to ensure consistent image processing across a time stack. We distinguish between two types of motion: (a) drift occurring in the imaging system itself caused by thermal gradients in the microscope, vibrations and mechanical instability ( Kreft et al., 2005); (b) subject motion such as fluctuations in the immersion media or the movement of organisms ( Jenkinson et al., 2002). Drift will typically play a significant role when imaging the same field of view over multiple days, which can be rectified by using standard registration methods ( Dubbs et al., 2016)( Thévenaz et al., 1998).
More complex motion such as organism movement can be harder to correct as it is often non-uniform, over a large area, and causes movement in-and-out of the focal plane. These require non-rigid registration methods or motion tracking. A commonly used example available in Python and MATLAB is Non-Rigid Motion Correction, NoRMcorre ( Pnevmatikakis & Giovannucci, 2017), which uses patch-based field of view registration whereby separate images are then merged by smooth interpolation. The popularity of NoRMcorre may in part be due to its general applicability.
Two correction methods have been produced for in vivo imaging in awake rodents, one based on the rigid-transform-based Lucas–Kanade (gradient descent) ( Lucas & Kanade, 1981) image registration algorithm using MathWorks® MATLAB platform ( Greenberg & Kerr, 2009), the other using a Hidden Markov Model ( Dombeck et al., 2007). Although effective, these methods have not been packaged for easy implementation and are reliant on cells remaining in the x- and y- dimensions as it cannot track following movement between z-axes. In cases with z-axis movement, tracking-based methods may be more reliable, and specialist options exist using control theory and machine learning approaches for post-processing ( Nguyen et al., 2017), or applied to a motorised stage ( Cong et al., 2017; Kim et al., 2017). A MathWorks® MATLAB toolbox, miniscope 1-photon imaging pipeline (MIN1PIPE), has been developed to include denoising, motion correction and signal extraction ( Lu et al., 2018). MIN1PIPE motion correction includes several steps including the Lucas-Kanade and Kanade-Lucas-Tomasi ( Lucas & Kanade, 1981; Shi & Tomasi, 1994) trackers, and Log-Demons registration ( Vercauteren et al., 2009), and outperforms the Lucas-Kanade, Kanade-Lucas-Tomasi, and NoRMcorre for using sample 2-photon videos ( Lu et al., 2018).
Tracking methods specifically designed to be more basic to implement and widely available include plug-ins for image processing packages ( Abramoff et al., 2004) such as Trackmate ( Tinevez et al., 2017), or Time Series Analyzer ( Balaji, 2014).
Classification
Classification is required to ensure that the quantification can be performed over specific regions of interest, such as for subcellular area, specific cells, or tissue regions. Classification can be achieved through pixel- or object-based segmentation. Pixel-based methods map each pixel to a class according to the spectral similarities. Popular pixel-based methods for calcium image analysis include Maximum Likelihood Classification (MLC) ( Malik et al., 2011) or Otsu thresholding to separate ‘light’ and ‘dark’ clustered pixels ( Otsu, 1979) as used as part of the SIMA Python package ROI pipeline ( Kaifosh et al., 2014).
Object-based segmentation is a two-step process using both spectral and spatial/contextual information to group pixels into objects which are then classified. CaImAn is an open-source package with modules for classification, motion correction, source extraction, and spike deconvolution. The classification method is based on convolutional neural networks ( Giovannucci et al., 2019). It was packaged into EZcalcium in an effort to improve usability by providing a GUI in MathWorks® MATLAB ( Cantu et al., 2020). However, using limited CaImAn function in EZcalcium does not easily allow for segmentation of more complex structures or large organelles or clusters of cells and is better for somas or smaller, less complex areas. Cellpose is another generalist, deep learning-based segmentation method that uses entirely open source packages in Python with a GUI to aid implementation. There is also a web-based option for testing Cellpose, which makes it very easy to use ( Stringer et al., 2020), though it too can be limited at detecting more complex cell shapes such as dendrites and axons.
DenoiSeg is an extension of Noise2Void that offers an end-to-end neural network, which is jointly optimised to denoise and segment images. The denoising capability is learnt by the self-supervised learning principle that noise2void introduced ( Krull et al., 2019). By combining this with a supervised learning approach using a few annotated ground truths of segmentation maps, the final segmentation performance ends up performing better than without co-learning, i.e. having two separate networks perform the respective tasks ( Buchholz et al., 2020).
Cell classification methods have been discussed with the conclusion that ‘learning-based methods score among the best-performing methods, but well-optimized traditional methods can even surpass these approaches in a fraction of the time’ ( Vicar et al., 2019).
Quantification
The aim of each step is for signal extraction to allow a quantitative output from the images of calcium signals. The most commonly used measure is the relative fluorescence variation (ΔF/F0) for classified cells. Packages will therefore either provide this data of the baseline fluorescence (F0) and deviations from baseline (ΔF), for further analysis, or provide a direct plot. Background subtraction may need to be considered as not all packages will take this into account. Multiple methods can be used, including subtracting the intensity values from a region of the image that does not contain Ca 2+ indicator from the intensity values in regions of interest. However care should be taken using background subtraction with ratiometric indicators ( Shkryl, 2020). F0 baseline values can be calculated by averaging the values before the onset of stimulation in the same region ( Galizia et al., 1999), or by low-pass filtering the signal ( Balkenius et al., 2009) (For review ( Balkenius et al., 2015).
Signal extraction from single cells can be particularly difficult for in vivo recordings due to large background fluxes and high spatial overlaps of cells outside of the focus plane which is further increased in 1-photon compared to 2-photon imaging. Semi-automated ROI analysis ( Barbera et al., 2016; Klaus et al., 2017; Pinto & Dan, 2015), principal component analysis independent components analysis (PCA/ICA) ( Mukamel et al., 2009), clustering based approaches (like Suite2P; ( Pachitariu et al., 2017), and constrained nonnegative matrix factorization (CNMF) ( Pnevmatikakis et al., 2016) approaches are techniques that have been explored with different strengths for detecting background and spatial overlap. An ‘extended’ CNMF method (CNMF-E) has been developed with an adjusted spatiotemporal background model that outperformed PCA/ICA for the simulated and experimental datasets that were tested ( Zhou et al., 2018). For a package method, the toolbox MIN1PIPE combines a CNMF ( Pnevmatikakis et al., 2016) with additional steps to remove false positives ( Lu et al., 2018). CaImAn also builds upon the CNMF algorithm ( Pnevmatikakis et al., 2016) to allow it to be fully automated, and CNMF-E for 1-photon endoscopic data ( Zhou et al., 2018).
Another feature commonly needed by researchers is timing of neuronal action potentials (APs) or ‘spike detection’ through deconvolution of the extracted signal. A wide range of algorithms can be used as discussed in the results to the Spikefinder challenge ( Berens et al., 2018) as there are multiple methods of varying complexity that can be used. EZcalcium directly shows the raw fluorescence, inferred activity and deconvolved neural ‘spiking’, whereby the data can then be exported into file formats for proprietary (.mat, .xlsx) or open (.csv) software programmes for further analysis ( Cantu et al., 2020; Giovannucci et al., 2019). The ability to accurately detect spikes requires knowledge of ground truth, usually from electrophysiological recordings. Calcium imaging can be susceptible to variation between neuron type, calcium indicator and its concentration used, the optical resolution, the sampling rate and the noise level. Therefore, it is fundamental to understand how specific indicators react under the given imaging conditions, which cannot be readily generalized across protocols. To try and improve the accuracy of spike detection, a toolkit using a supervised algorithm of spike inference has been developed using a ‘ground truth database’ from a large number of sets of calcium imaging with corresponding electrophysiological measurements ( Rupprecht et al., 2021).
Conclusion
A great number of analysis advancements have been made since calcium imaging was first developed. Popular packages for various steps of the pipeline ( Figure 1) include CaImAn, SIMA, Suite2P, and EZcalcium ( Cantu et al., 2020; Giovannucci et al., 2019; Kaifosh et al., 2014; Pachitariu et al., 2017). Although these packages are great starting tools for the community, many require programming knowledge in Python or commercial packages such as MathWorks® MATLAB. Many of the available options are only semi-automated and the limited automated options available are often designed for a very limited experimental context and are not actively supported when problems are experienced, e.g. other than for cells of a specific size and shape imaged in vitro. EZcalcium is one of the most intuitive options, which has improved the usability of CaImAn, NoRMCorre, but again seems best suited to analyse cell bodies. Suite2P and EZcalcium both attempt to offer an automated pipeline from raw images to spike extraction with no prior programming knowledge required by the user ( Cantu et al., 2020; Pachitariu et al., 2017). As both packages are suited to similar experimental data, the choice may be based upon personal preference.
It therefore seems that perhaps some of the biggest advances could be made by designing packages for detecting neuritic structures or organelles and improving the spatial resolution of the analysis to be intracellular, such as has been used for calcium sparks ( Berens et al., 2018). Longitudinal tracking of specific cells across imaging sessions also remains a challenge so that individual cells can be identified between multiple imaging sessions. A MathWorks® MATLAB toolkit has been made with reported error rates of < 5 % ( Sheintuch et al., 2017); an alternative approach is also available using CaImAn ( Giovannucci et al., 2019) though direct comparisons between these methods is difficult without knowing ground truths. Calcium imaging for population activity has also been highlighted as an area that requires further research, particularly when imaging over larger fields of view. Using models specific for neuron types imaged may improve detection of APs, which are commonly under-represented in population activity measurements ( Huang et al., 2021). Recent toolboxes with large datasets containing ground-truths may reduce false negatives during analysis ( Rupprecht et al., 2021). On the other end of the scale, pipelines for functional imaging in organisms such as zebrafish, C. elegans and Drosophila, where motion correction is often required and improved analysis for connectomics purposes are much needed.
As the application of machine learning in calcium imaging analysis matures, a higher level of automation and throughput for analysis tasks can be expected to follow. This will be enabled by more generalised and robust machine learning models. The barrier to training and deploying these methods will also reduce as more research is made into few-shot learning (using small training datasets) in addition to training approaches such as self-supervised and unsupervised learning.
Data availability
No data are associated with this article.
Acknowledgments
This publication was supported by COST Action NEUBIAS (CA15124), funded by COST (European Cooperation in Science and Technology).
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 2; peer review: 3 approved]
References
- Abramoff MD, Magalhaes PJ, Ram SJ: Image Processing with ImageJ. Biophotonics International. 2004;11(7):36–42. Reference Source [Google Scholar]
- Balaji J: Time series analyzer.2014; (Accessed: 7 June 2021). Reference Source [Google Scholar]
- Balkenius A, Bisch-Knaden S, Hansson B: Interaction of visual and odour cues in the mushroom body of the hawkmoth Manduca sexta. J Exp Biol. 2009;212(Pt 4):535–541. 10.1242/jeb.021220 [DOI] [PubMed] [Google Scholar]
- Balkenius A, Johansson AJ, Balkenius C: Comparing Analysis Methods in Functional Calcium Imaging of the Insect Brain. PLoS One.Public Library of Science,2015;10(6):e0129614. 10.1371/journal.pone.0129614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera G, Liang B, Zhang L, et al. : Spatially Compact Neural Clusters in the Dorsal Striatum Encode Locomotion Relevant Information. Neuron.Cell Press,2016;92(1):202–213. 10.1016/j.neuron.2016.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens P, Freeman J, Deneux T, et al. : Community-based benchmarking improves spike rate inference from two-photon calcium imaging data. bioRxiv. 2018;177956. 10.1101/177956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besbeas P, De Feis I, Sapatinas T: A Comparative Simulation Study of Wavelet Shrinkage Estimators for Poisson Counts. Int Stat Rev.John Wiley & Sons, Ltd.2004;72(2):209–237. 10.1111/j.1751-5823.2004.tb00234.x [DOI] [Google Scholar]
- Blinks JR, Prendergast FG, Allen DG: Photoproteins as biological calcium indicators. Pharmacol Rev. 1976;28(1):1–93. [PubMed] [Google Scholar]
- Boulanger J, Kervrann C, Bouthemy P, et al. : Patch-based nonlocal functional for denoising fluorescence microscopy image sequences. IEEE Trans Med Imaging. 2010;29(2):442–454. 10.1109/TMI.2009.2033991 [DOI] [PubMed] [Google Scholar]
- Broser PJ, Schulte R, Lang S, et al. : Nonlinear anisotropic diffusion filtering of three-dimensional image data from two-photon microscopy. J Biomed Opt.International Society for Optics and Photonics,2004;9(6):1253–1264. 10.1117/1.1806832 [DOI] [PubMed] [Google Scholar]
- Buades A, Coll B, Morel JM: A non-local algorithm for image denoising. 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'05). 2005;2(0):60–65. 10.1109/CVPR.2005.38 [DOI] [Google Scholar]
- Buades A, Coll B, Morel JM: Non-Local Means Denoising. Image Processing On Line. 2011;1:208–212. 10.5201/ipol.2011.bcm_nlm [DOI] [Google Scholar]
- Buchholz TO, Prakash M, Schmidt D: DenoiSeg: joint denoising and segmentation. European Conference on Computer Vision. 2020;324–337. 10.1007/978-3-030-66415-2_21 [DOI] [Google Scholar]
- Cai DJ, Aharoni D, Shuman T, et al. : A shared neural ensemble links distinct contextual memories encoded close in time. Nature.Nature Publishing Group,2016;534(7605):115–118. 10.1038/nature17955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu DA, Wang B, Gongwer MW, et al. : EZcalcium: Open-Source Toolbox for Analysis of Calcium Imaging Data. Front Neural Circuits.Frontiers Media S.A.2020;14:25. 10.3389/fncir.2020.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, et al. : Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature.Nature,2013;499(7458):295–300. 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Wang Z, Chai Y, et al. : Rapid whole brain imaging of neural activity in freely behaving larval zebrafish ( Danio rerio). eLife. 2017;6:e28158. 10.7554/eLife.28158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupé P, Yger P, Prima S, et al. : An optimized blockwise nonlocal means denoising filter for 3-D magnetic resonance images. IEEE Trans Med Imaging. 2008;27(4):425–441. 10.1109/TMI.2007.906087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW: Two-photon laser scanning fluorescence microscopy. Science. 1990;248(4951):73–76. 10.1126/science.2321027 [DOI] [PubMed] [Google Scholar]
- Dombeck DA, Khabbaz AN, Collman F, et al. : Imaging Large-Scale Neural Activity with Cellular Resolution in Awake, Mobile Mice. Neuron.Elsevier.2007;56(1):43–57. 10.1016/j.neuron.2007.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho DL: De-noising by soft-thresholding. IEEE Trans Inf Theory. 1995;41(3):613–627. 10.1109/18.382009 [DOI] [Google Scholar]
- Dubbs A, Guevara J, Yuste R: moco: Fast Motion Correction for Calcium Imaging. Front Neuroinform.Frontiers,2016;10:6. 10.3389/fninf.2016.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galizia CG, Menzel R, Holldobler B: Optical imaging of odor-evoked glomerular activity patterns in the antennal lobes of the ant camponotus rufipes. Naturwissenschaften.Naturwissenschaften,1999;86(11):533–537. 10.1007/s001140050669 [DOI] [PubMed] [Google Scholar]
- Giovannucci A, Friedrich J, Gunn P, et al. : CaImAn an open source tool for scalable calcium imaging data analysis. eLife.NLM (Medline),2019;8:e38173. 10.7554/eLife.38173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DS, Kerr JN: Automated correction of fast motion artifacts for two-photon imaging of awake animals. J Neurosci Methods.Elsevier,2009;176(1):1–15. 10.1016/j.jneumeth.2008.08.020 [DOI] [PubMed] [Google Scholar]
- Grewenig S, Zimmer S, Weickert J: Rotationally invariant similarity measures for nonlocal image denoising. J Vis Commun Image Represent. 2011;22(2):117–130. 10.1016/j.jvcir.2010.11.001 [DOI] [Google Scholar]
- Haykin S, Widrow B: Least-Mean-Square Adaptive Filters. Wiley Online Library. 2003;31. 10.1002/0471461288 [DOI] [Google Scholar]
- Huang L, Ledochowitsch P, Knoblich U, et al. : Relationship between simultaneously recorded spiking activity and fluorescence signal in gcamp6 transgenic mice. eLife.eLife Sciences Publications Ltd,2021;10:e51675. 10.7554/eLife.51675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisken J, Swoger J, Del Bene F, et al. : Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305(5686):1007–1009. 10.1126/science.1100035 [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, et al. : Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17(2):825–841. 10.1016/s1053-8119(02)91132-8 [DOI] [PubMed] [Google Scholar]
- Kaifosh P, Zaremba JD, Danielson NB, et al. : SIMA: Python software for analysis of dynamic fluorescence imaging data. Front Neuroinform.Frontiers Research Foundation.2014;8:80. 10.3389/fninf.2014.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Kim J, Marques JC, et al. : Pan-neuronal calcium imaging with cellular resolution in freely swimming zebrafish. Nat Methods.Nature Publishing Group.2017;14(11):1107–1114. 10.1038/nmeth.4429 [DOI] [PubMed] [Google Scholar]
- Kitamura K, Häusser M: Dendritic Calcium Signaling Triggered by Spontaneous and Sensory-Evoked Climbing Fiber Input to Cerebellar Purkinje Cells In Vivo. J Neurosci.Society for Neuroscience,2011;31(30):10847–10858. 10.1523/JNEUROSCI.2525-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus A, Martins GJ, Paixao VB, et al. : The Spatiotemporal Organization of the Striatum Encodes Action Space. Neuron.Cell Press,2017;95(5):1171–1180.e7. 10.1016/j.neuron.2017.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft M, Stenovec M, Zorec R: Focus-drift correction in time-lapse confocal imaging. Ann N Y Acad Sci.Ann N Y Acad Sci,2005;1048:321–330. 10.1196/annals.1342.029 [DOI] [PubMed] [Google Scholar]
- Krull A, Buchholz TO, Jug F: Noise2void - learning denoising from single noisy images. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition.2019;2129–2137. 10.1109/CVPR.2019.00223 [DOI] [Google Scholar]
- Lecoq J, Oliver M, Siegle JH, et al. : Removing independent noise in systems neuroscience data using DeepInterpolation. bioRxiv.Cold Spring Harbor Laboratory,2020;2020.10.15.341602. 10.1101/2020.10.15.341602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen J, Munkberg J, Hasselgren J, et al. : Noise2Noise: Learning Image Restoration without Clean Data. arXiv:1803.04189v3.2018. Reference Source [Google Scholar]
- Li X, Zhang G, Wu J, et al. : Reinforcing neuron extraction and spike inference in calcium imaging using deep self-supervised denoising. Nat Methods. 2021. 10.1038/s41592-021-01225-0 [DOI] [PubMed] [Google Scholar]
- Lindenbaum M, Fischer M, Bruckstein A: On Gabor’s contribution to image enhancement. Pattern Recogn. 1994;27(1):1–8. 10.1016/0031-3203(94)90013-2 [DOI] [Google Scholar]
- Lu J, Li C, Singh-Alvarado J, et al. : MIN1PIPE: A Miniscope 1-Photon-Based Calcium Imaging Signal Extraction Pipeline. Cell Rep. 2018;23(12):3673–3684. 10.1016/j.celrep.2018.05.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas BD, Kanade T: Iterative image registation technqiue with an application to stereo vision.1981;2:674–679. Reference Source [Google Scholar]
- Luisier F, Blu T, Unser M: Image denoising in mixed poisson-gaussian noise. IEEE Trans Image Process. 2011;20(3):696–708. 10.1109/TIP.2010.2073477 [DOI] [PubMed] [Google Scholar]
- Malik WQ, Schummers J, Sur M, et al. : Denoising two-photon calcium imaging data. PLoS One.PLoS One,2011;6(6):e20490. 10.1371/journal.pone.0020490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, et al. : Fluorescent indicators for Ca 2+ based on green fluorescent proteins and calmodulin. Nature.Nature Publishing Group;1997;388(6645):882–887. 10.1038/42264 [DOI] [PubMed] [Google Scholar]
- Moisescu DG, Ashley CC, Campbell AK: Comparative aspects of the calcium-sensitive photoproteins aequorin and obelin. Biochim Biophys Acta. 1975;396(1):133–140. 10.1016/0005-2728(75)90196-6 [DOI] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, Schnitzer MJ: Automated Analysis of Cellular Signals from Large-Scale Calcium Imaging Data. Neuron.NIH Public Access,2009;63(6):747–760. 10.1016/j.neuron.2009.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen JP, Linder AN, Plummer GS, et al. : Automatically tracking neurons in a moving and deforming brain. PLoS Comput Biol.Edited by E. Dyer. Public Library of Science.2017;13(5):e1005517. 10.1371/journal.pcbi.1005517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura M, Matsuzaki M, Kasai H, et al. : Genetically encoded bright Ca 2+ probe applicable for dynamic Ca 2+ imaging of dendritic spines. Anal Chem.American Chemical Society.2005;77(18):5861–5869. 10.1021/ac0506837 [DOI] [PubMed] [Google Scholar]
- Otsu N: Threshold Selection Method From Gray-Level Histograms. IEEE Trans Syst Man Cybern. 1979;9(1):62–66. 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Pachitariu M, Stringer C, Dipoppa M, et al. : Suite2p: beyond 10,000 neurons with standard two-photon microscopy. bioRxiv.Cold Spring Harbor Laboratory.2017;061507. 10.1101/061507 [DOI] [Google Scholar]
- Perona P, Malik J: Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell. 1990;12(7):629–639. 10.1109/34.56205 [DOI] [Google Scholar]
- Pinto L, Dan Y: Cell-Type-Specific Activity in Prefrontal Cortex during Goal-Directed Behavior. Neuron.Cell Press,2015;87(2):437–450. 10.1016/j.neuron.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Giovannucci A: NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. J Neurosci Methods.Elsevier B.V.2017;291:83–94. 10.1016/j.jneumeth.2017.07.031 [DOI] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, et al. : Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron.Cell Press,2016;89(2):285–99. 10.1016/j.neuron.2015.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupprecht P, Carta S, Hoffmann A, et al. : A Database and deep learning toolbox for noise-optimized, generalized spike inference from calcium imaging. bioRxiv. 2021. 10.1101/2020.08.31.272450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Yan R, Li X, et al. : From Heuristic Optimization to Dictionary Learning: A Review and Comprehensive Comparison of Image Denoising Algorithms. IEEE Trans Cybern. 2014;44(7):1001–1013. 10.1109/TCYB.2013.2278548 [DOI] [PubMed] [Google Scholar]
- Sheintuch L, Rubin A, Brande-Eilat N, et al. : Tracking the Same Neurons across Multiple Days in Ca 2+ Imaging Data. Cell Rep.Elsevier B.V.,2017;21(4):1102–1115. 10.1016/j.celrep.2017.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Tomasi C: Good features to track. In 1994 Proceedings of IEEE Conference on Computer Vision and Pattern Recognition.Publ by IEEE,1994;593–600. 10.1109/CVPR.1994.323794 [DOI] [Google Scholar]
- Shkryl VM: Error correction due to background subtraction in ratiometric calcium measurements with CCD camera. Heliyon.Elsevier Ltd,2020;6(6):e04180. 10.1016/j.heliyon.2020.e04180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ: Miniaturized two-photon microscope: Seeing clearer and deeper into the brain. Light Sci Appl.Nature Publishing Group,2017;6(8):e17104. 10.1038/lsa.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C, Wang T, Michaelos M: Cellpose: A generalist algorithm for cellular segmentation. bioRxiv. 2020;2020.02.02.931238. 10.1101/2020.02.02.931238 [DOI] [PubMed] [Google Scholar]
- Thévenaz P, Ruttimann UE, Unser M: A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7(1):27–41. 10.1109/83.650848 [DOI] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, et al. : TrackMate: An open and extensible platform for single-particle tracking. Methods.Academic Press Inc.2017;115:80–90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Tsien RY: New Calcium Indicators and Buffers with High Selectivity Against Magnesium and Protons: Design, Synthesis, and Properties of Prototype Structures. Biochemistry. 1980;19(11):2396–2404. 10.1021/bi00552a018 [DOI] [PubMed] [Google Scholar]
- Vercauteren T, Pennec X, Perchant A, et al. : Diffeomorphic demons: efficient non-parametric image registration. NeuroImage.Academic Press,2009;45(1 Suppl):S61–S72. 10.1016/j.neuroimage.2008.10.040 [DOI] [PubMed] [Google Scholar]
- Vicar T, Balvan J, Jaros J, et al. : Cell segmentation methods for label-free contrast microscopy: Review and comprehensive comparison. BMC Bioinformatics.BioMed Central Ltd.2019;20(1):360. 10.1186/s12859-019-2880-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner FV, Both M, Fink RHA: Automated Detection of Elementary Calcium Release Events Using the á Trous Wavelet Transform. Biophys J.The Biophysical Society,2006;90(6):2151–63. 10.1529/biophysj.105.069930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert M, Schmidt U, Boothe T, et al. : Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat Methods. 2018;15(12):1090–1097. 10.1038/s41592-018-0216-7 [DOI] [PubMed] [Google Scholar]
- Wiener N: Extrapolation, interpolation, and smoothing of stationary time series: with engineering applications.Technology Press.1950. Reference Source [Google Scholar]
- Zhang K, Zuo W, Chen Y, et al. : Beyond a Gaussian denoiser: Residual learning of deep CNN for image denoising. IEEE Trans Image Process. 2017;26(7):3142–3155. 10.1109/TIP.2017.2662206 [DOI] [PubMed] [Google Scholar]
- Zhang K, Zuo W, Zhang L: FFDNet: Toward a fast and flexible solution for CNN Based image denoising. IEEE Trans Image Process. 2018;27(9):4608–4622. 10.1109/TIP.2018.2839891 [DOI] [PubMed] [Google Scholar]
- Zhou P, Resendez SL, Rodriguez-Romaguera J, et al. : Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife.NLM (Medline),2018;7:e28728. 10.7554/eLife.28728 [DOI] [PMC free article] [PubMed] [Google Scholar]