Abstract
Yeast and mammalian SWI-SNF complexes regulate transcription through active modification of chromatin structure. Human SW-13 adenocarcinoma cells lack BRG1 protein, a component of SWI-SNF that has a DNA-dependent ATPase activity essential for SWI-SNF function. Expression of BRG1 in SW-13 cells potentiated transcriptional activation by the glucocorticoid receptor, which is known to require SWI-SNF function. BRG1 also specifically repressed transcription from a transfected c-fos promoter and correspondingly blocked transcriptional activation of the endogenous c-fos gene. Mutation of lysine residue 798 in the DNA-dependent ATPase domain of BRG1 significantly reduced its ability to repress c-fos transcription. Repression by BRG1 required the cyclic AMP response element of the c-fos promoter but not nearby binding sites for Sp1, YY1, or TFII-I. Using human C33A cervical carcinoma cells, which lack BRG1 and also express a nonfunctional Rb protein, transcriptional repression by BRG1 was weak unless wild-type Rb was also supplied. Interestingly, Rb-dependent repression by BRG1 was found to take place through a pathway that is independent of transcription factor E2F.
The modification of chromatin structure is increasingly recognized to be an important facet of transcriptional regulation. Such alterations likely occur in concert with the actions of the general transcription factors and promoter-specific activators and repressors in order to allow regulatory changes to take place. Several multiprotein complexes with roles in this kind of regulation have been identified, including the yeast SAGA (23, 72) and RSC (9) complexes, the Drosophila ISWI-containing complexes (16, 29, 67, 69), and the yeast and human SWI-SNF complexes (ySWI-SNF and hSWI-SNF) (8, 14, 28, 32, 35, 37, 50, 55, 73, 74). The regulation of chromatin structure is likely to influence specific cellular processes that rely heavily on transcriptional events, including the control of cellular proliferation.
Several studies have suggested a role for hSWI-SNF components BRG1 (32), hBRM (50), and hSNF5-INI1 (49) in the control of cellular proliferation. BRG1 and hBRM, human homologues of yeast SNF2, bind to members of the retinoblastoma (Rb) tumor suppressor protein family and can trigger cellular growth arrest (15, 32, 61, 62). Recently, the hSNF5-INI1-encoding gene was found to be mutated in multiple malignant rhabdoid tumors, strongly suggesting that hSWI-SNF has a tumor suppressor function (71). In addition, we have reported that the N-terminal domain of the adenovirus E1A oncoprotein specifically blocks SWI-SNF-dependent transcription in budding yeast, suggesting that disruption of human and mouse SWI-SNF function may be important in oncogenic transformation by E1A (46).
The ySWI-SNF and hSWI-SNF complexes have been studied extensively in vitro and have been demonstrated to trigger chromatin remodeling and facilitation of sequence-specific DNA binding (reviewed in references 7, 31, and 35). In addition, genetic approaches have resulted in the identification of several yeast cellular genes whose transcriptional regulation requires a functional SWI-SNF complex (10, 38, 39, 41, 56). In higher eukaryotic systems, transfected promoters under the control of the glucocorticoid receptor (GR) (17, 50, 61) or transcription factor E2F (66) have been shown to be regulated by BRG1 or the related hBRM protein, but regulation of the corresponding endogenous genes has not yet been reported.
The c-fos proto-oncogene is regulated at the transcriptional level during mitogenesis and other cellular processes and is subject to both activation and repression of transcription. Rapid activation of c-fos transcription can be followed closely by transcriptional inactivation, resulting in transient expression of the c-fos mRNA (5, 12, 57, 70). Based on the importance of c-fos in cellular proliferation (36, 53) and the pivotal role of transcriptional regulatory mechanisms in c-fos expression, we tested the hypothesis that the BRG1 protein (and hSWI-SNF activity) regulates transcription of c-fos in vivo. Here we report that BRG1 specifically represses transcription of a transfected c-fos promoter construct and correspondingly inhibits transcriptional induction of the endogenous c-fos mRNA. Repression by BRG1 correlates with its activity as a functional component of the hSWI-SNF complex. Interestingly, this repression takes place through an Rb-mediated pathway that is independent of transcription factors E2F and GR.
MATERIALS AND METHODS
Plasmids.
The following plasmids were described previously: hBRG1 and hBRG1K798R (32); pCMVRb and E2 CAT (27); pGRneo (J. Mymryk); pMSG-CAT (Pharmacia); pCMV-HA-DP-1, pCMV-HA-DP-1127–410, and pCMV-HA-DP-1Δ103–126 (75); −76/+10 fos-CAT and pm87 (19); pm27 and pm28 (77); pm82 (18); EF-1α luciferase (47); pSV2CAT (21); pTKM (65); pCMV VP16-E2 (42); pCMV5 (1); and 3 × 22 E1BCAT, 3 × 22 mutCRE, 3 × 22 mutYY1, and E1BCAT (19).
To make pCMV-BRG1 and pCMV-K798R, we first constructed pCMV5-BRG1 and pCMV5-HA. For pCMV5-BRG1 (or pCMV5-K798R), plasmid hBRG1 (or hBRG1K798R) was digested with NarI and BamHI, and the two fragments containing BRG1 (or K798R) sequences were ligated to pCMV5 that had been digested with ClaI and BamHI. For pCMV5-HA, a double-stranded oligonucleotide (coding strand, CGACGCGTGCCATGGCCTACCCATATGATGTTCCAGATTACGCAAGCTTGTAGGATCCGC) was multimerized by incubation with deoxynucleoside triphosphates, T4 ligase, T4 polynucleotide kinase, and T4 DNA polymerase; digested with MluI and BamHI; and ligated with pCMV5 that had been digested with MluI and BamHI. BRG1 (or K798R) from pCMV5-BRG1 (or pCMV5K798R) was inserted into pCMV5-HA in two cloning steps, as follows. The 3′ end of BRG1 (or K798R) was amplified by PCR with the oligonucleotides 5′-ACCCCAGCCGCGACAA and 5′-TTTACGCGTCTCTTGTTCCTCCTCACTG, resulting in the insertion of a downstream MluI site and the ablation of the native stop codon. The products were multimerized as described above and then digested with NotI and MluI. The purified NotI-MluI fragment was ligated to the BglII-NotI fragment from pCMV5-BRG1 plus pCMV-HA that had been digested with BglII and MluI. The resulting clone (pBRG1-C) was digested with BglII and ligated to the BglII fragment from pCMV5-BRG1 (or K798R) containing the 5′ two-thirds of the cDNA. All oligonucleotides were from Oligos, etc.
To make pB1cfos, a 2.1-kb ApaI fragment, including all of exons 2 and 3, and the most 5′ 600 bp of exon 4, was excised from pc-fos-1 (American Type Culture Collection catalog no. 4102) and ligated into the ApaI site of pBluescript KS+ (Stratagene).
To make pAdloxtetBRG1 and pAdloxtetK798R, we first constructed pAdloxBRG1 and pAdloxK798R; pCMV5-BRG1 or pCMV5-K798R was digested with NdeI (complete) and BamHI (partial). The resulting 5.5-kb fragment containing BRG1 or K798R was purified and ligated into pAdlox (24) that had been digested with NdeI and BamHI. pAdloxBRG1 (or pAdloxK798R) was digested with MluI, blunt ended with Klenow polymerase, and then digested with ClaI to liberate a 5.4-kb fragment containing the BRG1 (or K798R) cDNA. This was ligated into pAdloxtet (23a) that had been digested with EcoRV and ClaI.
Cell culture and transfections.
SW-13 and C33A cells were grown in Dulbecco’s modified Eagle medium containing 10% fetal calf serum, and HeLa cells were grown in Dulbecco’s modified Eagle medium containing 10% newborn calf serum. All cultures were supplemented with penicillin and streptomycin. All cell culture materials were from GIBCO/BRL. Twenty hours prior to transfection, trypsinized cells from confluent cultures were pooled, diluted 1 to 20, and replated. Transfections were performed by calcium phosphate coprecipitation as previously described (2). They contained 2 μg of promoter plasmid with various amounts of expression constructs plus empty pCMV5 vector to bring the total amount of DNA to 25 μg per dish. All transfections were performed in duplicate.
CAT and luciferase assays.
For chloramphenicol acetyltransferase (CAT) assays, cells were washed twice in phosphate-buffered saline, harvested by scraping in 1 ml of phosphate-buffered saline, pelleted at 700 × g in a microcentrifuge, and resuspended in 100 μl of 250 mM Tris (pH 7.8). The cells were lysed and assayed as previously described (19), except that half of each extract was assayed. For some assays, less extract was added in order to preserve linearity. For luciferase assays, lysates were prepared as directed by Analytical Luminescence Laboratory and assayed directly in a Monolight 2010 luminometer (Analytical Luminescence Laboratory).
Viruses.
AdtTa was previously described (52). AdBRG1 and AdK798R were constructed as previously described (24), by using plasmids pAdloxtetBRG1 and pAdloxtetR798 (see above) and by using helper virus φ9 (23a) instead of φ5. φ9 is the same as φ5, except that it contains a larger deletion in the E3 region from nucleotide 28133 to nucleotide 33818 (4). Viruses were propagated and purified as already described (24).
Infections and RNase protection assay.
Cells (6 × 105 to 10 × 105 per 10-cm-diameter dish) were infected with virus in 1 ml of medium (see figure legends for multiplicities of infection [MOIs]) for 1 h at 37°C with intermittent rocking, after which 9 ml of medium was added. After 48 h, the cells were treated with 5 μM forskolin or 2 nM interleukin-6 (IL-6). Cells were harvested by mild trypsinization and gentle scraping, washed repeatedly, pelleted, and frozen. Preparation of total cytoplasmic RNA and RNase protection assays were performed as previously described (19). c-fos probe was transcribed from NarI-digested pB1cfos, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was transcribed from HindIII-digested HHCMC32 (American Type Culture Collection catalog no. 78105). Probe (10,000 to 15,000 cpm) and 10 μg of RNA were added to the hybridization mixtures. For infections followed by transfections, 3 × 105 to 4 × 105 cells were infected, transfected 20 h later, and harvested after an additional 40 h. Alternatively, nuclei were separated from cytoplasmic extract by centrifugation at 575 × g and suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer.
Western blotting.
Lysates (50 μg of protein) prepared as for CAT assay were mixed with sample buffer, boiled and subjected to SDS-PAGE. Separated proteins were transferred to nitrocellulose and assayed for the presence of BRG1 with antibody J1 (from Gerald Crabtree) or for hemagglutinin (HA)-tagged BRG1 with antibody 12CA5 (from BABCO). Immunoreactivity was determined by using horseradish peroxidase-conjugated protein A or 125I-labeled protein A and detected by enhanced chemiluminescence assay or autoradiography, respectively.
RESULTS
BRG1 specifically represses c-fos promoter activity.
SW-13 cells, derived from a human small-cell carcinoma of the adrenal cortex, were used to study the effect of BRG1 on c-fos transcription. These cells lack endogenous BRG1 protein as well as the related protein hBRM but express other members of the hSWI-SNF complex (15, 73). Expression of BRG1 in SW-13 cells results in a flat cell morphology following prolonged incubation (15 and data not shown).
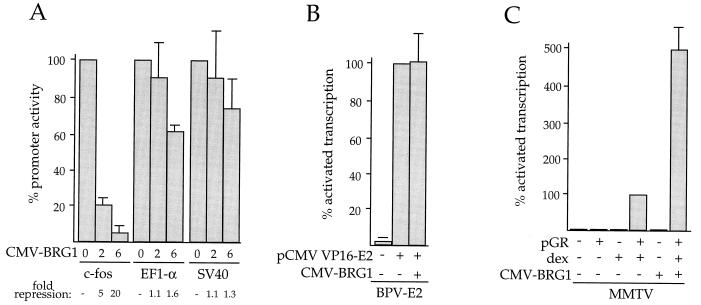
The reporter plasmid −76/+10 fos-CAT (hereafter referred to as pfos-CAT), containing nucleotides −76 to +10 of the mouse c-fos promoter linked to the CAT gene, was transfected into SW-13 cells along with a cytomegalovirus (CMV) promoter-driven, HA-tagged BRG1 cDNA construct, pCMV-BRG1. The parental pCMV5 vector was transfected as a control for pCMV-BRG1. pfos-CAT was transcriptionally active in the absence of BRG1 expression, routinely producing approximately 80,000 cpm during a standard 1-h CAT assay incubation. As shown in Fig. 1A, BRG1 expression caused 5- to 20-fold repression of CAT activity. The extent of the repression depended on the input DNA concentration and correlated with increasing amounts of detectable HA-tagged BRG1 protein by Western blot analysis (data not shown). In addition, transfection of a construct containing the BRG1 cDNA in the reverse orientation failed to significantly repress transcription (data not shown).
FIG. 1.
Transcriptional repression by BRG1. SW-13 cells were transfected by the calcium phosphate method with pCMV-BRG1 and either pfos-CAT, EF-1α-luciferase, or pSV2-CAT (A); pCMV VP16-E2, pCMV-BRG1, and pTKM (indicated as BPV-E2 (B); pGR, pCMV-BRG1, and pMSG-CAT (indicated as MMTV) (C). Cells were harvested 48 h after transfection and assayed for CAT or luciferase activity as described in Materials and Methods. Two micrograms of each reporter plasmid and 4 μg of each expression plasmid were used, except in panel B, where 2 μg of pCMV-BRG1 was used. (A) Average assay values for the promoter plasmids in the absence of BRG1 were 80,000 cpm (pfos-CAT), 400,000 light units (EF-1α-luciferase), and 100,000 cpm (pSV2-CAT). Data from BRG1-expressing cells are plotted as a percentage of these averages. The amount of pCMV-BRG1 transfected is indicated below each bar (in micrograms). The data are averages of three to six (A), three (B), or four (C) independent experiments, each performed in duplicate. dex, 1 μM dexamethasone. SV40, simian virus 40; CMV, cytomegalovirus.
To explore the specificity of this effect, three other promoter constructs were tested for an effect of BRG1. The human EF-1α promoter and the simian virus 40 enhancer-promoter were both found to be active in SW-13 cells but were only weakly affected by BRG1 expression, and this weak effect occurred only when the highest input plasmid amount was used (Fig. 1A). In addition, a fusion protein consisting of the DNA-binding domain of the bovine papillomavirus E2 protein (42) and the transcriptional activation domain of the herpes simplex virus VP-16 protein was used to activate transcription of a minimal thymidine kinase promoter containing bovine papillomavirus E2 binding sites (65). As shown in Fig. 1B, transcription activated by the VP16-E2 fusion protein was likewise unaffected by BRG1. In addition to these experiments with the mouse c-fos promoter, transcription of a similar construct containing the proximal human c-fos promoter was also repressed by BRG1, as was a longer murine c-fos promoter construct containing nucleotides −356 to +109 (data not shown). We conclude that the c-fos promoter is specifically repressed by BRG1 in SW-13 cells.
Expression of BRG1 reconstitutes hSWI-SNF activity in SW-13 cells.
Previously, ySWI-SNF and hSWI-SNF have been implicated in transcriptional activation by GR (17, 32, 50, 61, 76). To test if expression of BRG1 in SW-13 cells results in a functionally active hSWI-SNF complex, we examined the effect of BRG1 on GR-dependent transcription. Cells were transiently cotransfected with pCMV-BRG1, the GR expression construct pGRneo, and the reporter plasmid pMSG-CAT, which contains the GR-responsive mouse mammary tumor virus (MMTV) promoter. Figure 1C shows that GR-dependent transcription was potentiated fivefold by pCMV-BRG1, confirming that hSWI-SNF function was indeed reconstituted in SW-13 cells.
Since BRG1 expression alone (i.e., in the absence of GR and dexamethasone) had no effect on MMTV promoter activity (Fig. 1C), we can rule out the possibility that expression of BRG1 alone in SW-13 cells somehow results in activation of GR-dependent transcription and that such activation could lead indirectly to repression of c-fos transcription through unknown intermediate events.
The DNA-dependent ATPase domain of BRG1 is involved in repression.
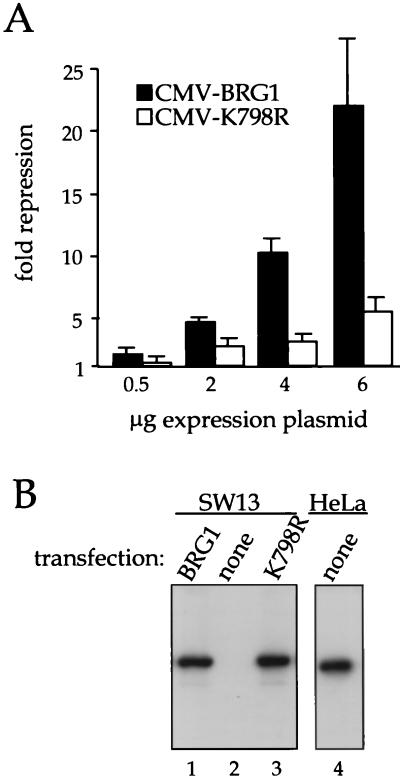
Chromatin remodeling by the hSWI-SNF complex requires a DNA-dependent ATPase activity supplied by BRG1 (28, 32, 37). If transcriptional repression by BRG1 involves the function of the hSWI-SNF complex, then mutation of the ATPase domain would be expected to reduce this activity. To test this possibility, we constructed plasmid pCMV-K798R, which encodes BRG1 with a lysine-to-arginine mutation in the ATPase domain that has been shown previously to impair BRG1 function and also that of ySWI-SNF and hSWI-SNF (32, 40). SW-13 cells were cotransfected with pfos-CAT and increasing amounts of either the wild-type or mutant BRG1 expression constructs. As shown in Fig. 2A, expression of mutant K798R had a much reduced effect on pfos-CAT compared with wild-type BRG1. Figure 2B shows the results of a Western blot analysis of a representative transfection experiment. No endogenous BRG1 was detected in untransfected cells (lane 2), and BRG1 and the K798R mutant were expressed at comparable levels (lanes 1 and 3). Also shown in Fig. 2B is a Western blot of an equivalent amount of extract from uninfected HeLa cells. We conclude that the DNA-dependent ATPase activity of BRG1 is required for transcriptional repression of c-fos. These data, together with those in Fig. 1C, strongly argue that BRG1 is functioning here as a component of the hSWI-SNF complex.
FIG. 2.
Repression requires the DNA-dependent ATPase domain of BRG1. (A) SW-13 cells were transfected with the indicated expression plasmids along with 2 μg of pfos-CAT. Cells were harvested after 48 h and assayed for CAT activity as described in Materials and Methods. Data (averages of six independent experiments) are plotted as fold repression calculated against pfos-CAT cotransfected with the parental pCMV5 expression plasmid. (B) SW-13 cells were transfected with 6 μg of pCMV-BRG1 or pCMV-K798R or left untransfected. After 48 h, they were harvested and subjected to Western blot analysis using anti-BRG1 antibody J1 (lanes 1 to 3) as described in Materials and Methods. HeLa cells were assayed similarly for endogenous BRG1 protein (lane 4).
BRG1 blocks transcriptional activation of the endogenous c-fos gene.
Because of the potential physical differences between transiently transfected DNA and normal chromatin, it was unclear if the physical configuration of the pfos-CAT plasmid represented a physiologically relevant substrate for BRG1 and hSWI-SNF in the cell. We therefore wanted to test if the endogenous c-fos gene could also be repressed by BRG1. To address this issue, we constructed a replication-deficient adenovirus vector, AdBRG1, capable of expressing the BRG1 protein in all cells of an experimental population. This reagent allowed us to measure the effect of BRG1 on the endogenous c-fos gene.
AdBRG1 consists of a full-length BRG1 cDNA placed in the E1 region of adenovirus vector φ9 (23a, 24) under the control of a minimal CMV promoter containing binding sites for the tet repressor (TetR) protein (22). In our experiments, cells were coinfected with this vector along with AdtTa, an adenovirus vector expressing a tetR-VP16 fusion protein which activates transcription from the promoter containing TetR binding sites on the coinfected vector (22). In control experiments with an adenovirus vector expressing green fluorescent protein, we noted that 100% of the cells in an SW-13 population could be infected (data not shown).
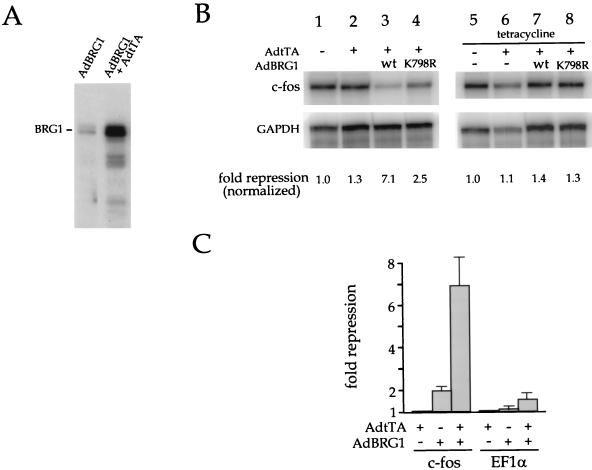
To test for an effect on endogenous c-fos transcription, cells were infected for 48 h with AdtTa alone or with AdtTA in combination with AdBRG1. Tetracycline, which inhibits the activity of the TetR-VP16 activator (22), was added to some of the cultures to block the expression of BRG1. At 45 min prior to harvesting, the cells were treated with 5 μM forskolin, which activates transcription of the c-fos gene via the cyclic AMP (cAMP) pathway. Total cytoplasmic RNA was isolated and assayed for c-fos mRNA by the RNase protection method. To accurately quantify the results, the RNA samples were also assayed for GAPDH mRNA as an internal control. As shown in Fig. 3A, coinfection of AdtTa and AdBRG1 resulted in induced expression of BRG1 protein. Figure 3B shows that expression of BRG1 caused sevenfold inhibition of cAMP-induced c-fos transcription (compare lanes 1 and 3). This effect required expression of BRG1, since addition of tetracycline to the culture medium completely abolished the effect (compare lanes 3 and 7) and efficiently blocked BRG1 expression, as judged by Western blot analysis (see Fig. 4A). Infection with AdtTa alone had no effect on endogenous c-fos expression (compare lanes 1 and 2), even at an MOI of 300 (data not shown).
FIG. 3.
BRG1 blocks induction of the endogenous c-fos gene. (A) tTa-induced expression of BRG1. SW-13 cells were infected with the indicated viruses and analyzed by Western blot analysis using anti-BRG1 antibody J1. (B) SW-13 cells were uninfected (lanes 1 and 5), infected with AdtTa alone (lanes 2 and 6), or infected with AdtTa in combination with either AdBRG1 (lanes 3 and 7) or AdK798R (lanes 4 and 8). Tetracycline (2.5 μg/ml), which abolishes tTA activator activity, was added 4 h p.i. (lanes 5 to 8). The MOI was 60 for AdtTa and 240 for AdBRG1 and AdK798R. Infection with AdtTa alone at an MOI of 300 produced no effect on c-fos induction (data not shown). After 48 h, cells were harvested for RNase protection assay with c-fos and GAPDH probes as described in Materials and Methods. RNase-resistant products were separated by electrophoresis through urea-polyacrylamide. They were visualized and quantified by using a PhosphorImager (Molecular Dynamics). The data shown are representative of three independent experiments (two in the case of the tetracycline addition experiments). (C) Cells were infected with the indicated viruses (same MOI as in panel A) and transfected on the following day with pfos-CAT or EF-1α-luciferase. After an additional 40 h, the cells were harvested and assayed for CAT or luciferase activity. Data are plotted as fold repression averaged from three independent experiments. wt, wild type.
FIG. 4.
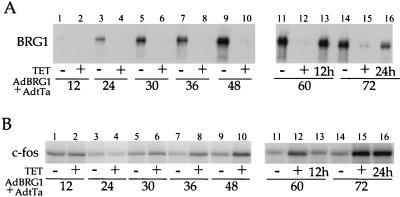
Time course of BRG1 protein expression and BRG1-induced repression. SW-13 cells were infected with AdBRG1 plus AdtTa as described in the legend to Fig. 3 and incubated for the indicated times in the presence or absence of 2.5-μg/ml tetracycline (TET). Cells were stimulated with forskolin for 45 min prior to harvesting. Harvested cells were processed for Western blot analysis of BRG1 (A) and RNase protection assay of c-fos mRNA (B). For lanes 13 of A and B, cells were first incubated for 48 h in the absence of tetracycline, and then tetracycline was added for the remaining 12 h of the 60-h period. For lanes 16 of A and B, cells were first incubated for 48 h in the absence of tetracycline, and then tetracycline was added for the remaining 24 h of the 72-h period. The designations 12h and 24h indicate this.
We also constructed AdK798R, expressing the corresponding ATPase-deficient mutant. Consistent with our transfection results, AdK798R showed a diminished ability to repress endogenous c-fos transcription compared with wild-type AdBRG1. No effect of BRG1 on the endogenous GAPDH mRNA was observed, consistent with our earlier observation that the effect on the c-fos promoter is specific. In other experiments, we observed that virally expressed BRG1 also blocked IL-6-induced transcriptional activation of the endogenous c-fos gene to an extent similar to that observed for forskolin-induced transcription (data not shown).
We broadened the analysis of virally expressed BRG1 to compare this system more fully with our earlier transfection results. SW-13 cells were infected with AdBRG1 and AdtTa and transfected on the following day with either pfos-CAT or EF-1α-luciferase. The cells were harvested 40 h later and assayed for CAT or luciferase activity. As shown in Fig. 3C, infection with the combination of AdBRG1 and AdtTa resulted in significant repression of transcription from pfos-CAT, with only a marginal effect on the EF-1α promoter. Thus, the promoter specificity observed in our earlier transfection experiments (Fig. 1) was reproduced with virally expressed BRG1. The experiments shown in Fig. 3 demonstrate that the endogenous c-fos gene is indeed regulated by BRG1 and that this process is similar or identical to that underlying repression of the transfected c-fos promoter.
Additionally, we compared the appearance of the BRG1 protein with the onset of repression of endogenous c-fos. SW-13 cells were coinfected with AdBRG1 and AdtTa and maintained in either the presence or the absence of 2.5-μg/ml tetracycline. The cells were stimulated with 5 μM forskolin for 45 min prior to harvesting at 12, 24, 30, 36, 48, 60, and 72 h postinfection (p.i.) and then analyzed for c-fos mRNA and BRG1 protein as described above. As shown in Fig. 4A, expression of BRG1 was barely detectable at 12 h p.i., and continued to increase until 48 h p.i., after which it remained high. Weak repression was reproducibly observed beginning at 30 h p.i., and repression increased thereafter in a manner that correlated with increased BRG1 expression (Fig. 4B). Importantly, cells that were treated with tetracycline starting at 48 h p.i. showed complete loss of repression within the next 24 h (Fig. 4B, compare lane 15 with lane 16). Interestingly, this corresponded to a decrease in BRG1 protein expression down to the level seen at 30 h p.i. in the absence of tetracycline (Fig. 4A, compare lane 16 with lane 5). These data clearly indicate that a threshold amount of BRG1 protein is required to cause repression and that this amount is achieved at around 30 to 36 h p.i.
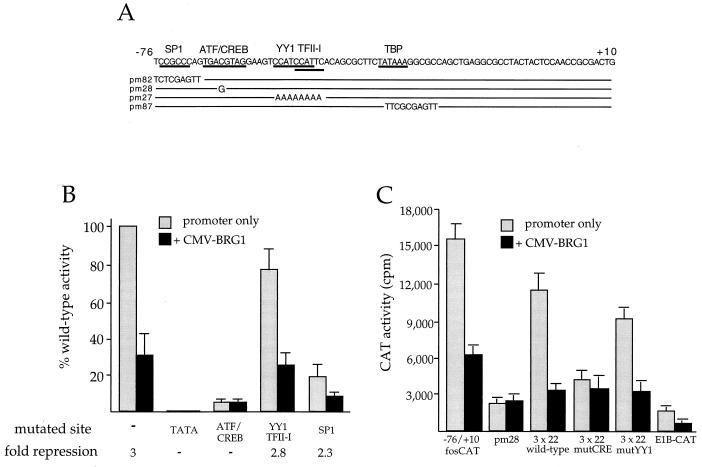
Repression by BRG1 requires the −67 ATF/CREB binding site in the c-fos promoter.
The −76 to +10 region of the c-fos promoter contains binding sites for transcription factors ATF/CREB (3), YY1 (51), and TFII-I (33) and also contains a TATA box and a consensus SP1 (20) binding site (Fig. 5A). We employed a series of linker substitution and point mutants to test the possible involvement of these factors in the observed response to BRG1. As shown in Fig. 5B, mutation of the TATA box resulted in complete loss of transcriptional activity in the absence of BRG1, and mutation of any of the other binding sites resulted in various decreases in transcription from pfos-CAT. Interestingly, whereas the YY1/TFII-I and SP-1 site mutants (pm27 and pm82, respectively) were sensitive to repression by BRG1, the ATF/CREB site mutant (pm28) was unaffected. We note that the overall activity of the pm28 mutant was low in the absence of BRG1. However, this activity was readily detectable (approximately 3,000 cpm compared with 50 to 100 cpm for the TATA mutant). We conclude that ATF/CREB-dependent transcription is necessary for BRG1 to repress transcription at the c-fos promoter. These data suggest that BRG1 specifically targets ATF/CREB-dependent transcription, although they do not rule out other possibilities (see Discussion).
FIG. 5.
Mutational analysis of the c-fos promoter. (A) The −76 to +10 sequence of the mouse c-fos promoter region is shown. Bars below the sequence indicate the binding sequences of the indicated transcription factors. There are two adjacent sites for YY1, whose core CCAT sequences are indicated. Except for SP1, all of the indicated factors have been demonstrated to bind to the mouse c-fos promoter. The SP1 site is a perfect match to the consensus SP1 binding site, and as shown here, mutation of this site results in a significant decrease in promoter activity. Sequences of pfos-CAT derivatives are also shown, with a solid line representing the wild-type sequence. (B) SW-13 cells were cotransfected with pCMV-BRG1 (3 μg) and pfos-CAT (2 μg) derivatives with mutations in the TATA box (pm87), ATF/CREB site (pm28), overlapping YY1 and TFII-I sites (pm27), or consensus SP1 site (pm 82). Cells were harvested 48 h after transfection and assayed for CAT activity. Data are plotted as percent wild-type promoter activity to show the effect of each mutation in the absence and the presence of BRG1. Each mutant was tested along with the wild-type promoter three to six independent times, and the percent wild-type activities were averaged. A dash for fold repression indicates that the promoter-only value divided by CMV-BRG1 equals 1. (C) SW-13 cells were transfected with the indicated fosCAT derivatives along with CMV-BRG1 and assayed as described above. This experiment was performed four times, each time in duplicate.
To further assess this issue, we employed a second set of CAT reporter constructs, each containing three copies of a 22-bp region of the c-fos promoter (positions −72 to −51), encompassing the cAMP response element (CRE) and YY1 sites, placed upstream of a minimal adenovirus E1B promoter sequence (19). Plasmid 3 × 22-E1BCAT contains the wild-type c-fos sequence. Plasmids 3 × 22 mutCRE and 3 × 22 mutYY1 contain inactivating mutations in the CRE and YY1 DNA-binding sites, respectively, as described previously (19). Each construct was cotransfected into SW-13 cells with 2 μg of a BRG1 expression construct. As shown in Fig. 5C, all constructs produced readily detectable amounts of CAT activity, albeit much lower than those observed with the −76 to +10 wild-type construct. Again, the activity of each construct, with the exception of plasmid 3 × 22mutCRE, was repressed by BRG1 expression. These data are consistent with involvement of the CRE in BRG1-mediated repression.
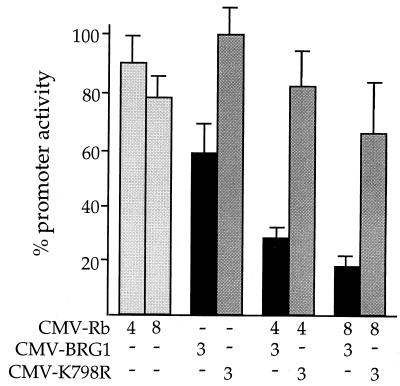
Repression of c-fos by BRG1 depends on Rb.
BRG1 and the related hBRM protein have been shown to interact directly with the Rb gene product pRb (15, 61). Interaction with Rb has been implicated in the ability of BRG1 to activate GR-dependent transcription (61) and in the ability of hBRM to repress E2F-dependent transcription (66). We therefore wanted to test the hypothesis that repression of c-fos transcription by BRG1 is an Rb-mediated event. To do this, we performed transfection experiments with the human cervical carcinoma line C33A, which encodes a mutant, nonfunctional Rb protein and also fails to produce the BRG1 protein (50). C33A cells were transiently transfected with pfos-CAT, along with either pCMV-BRG1, pCMV-Rb, or the two expression constructs combined. The cells were harvested 48 h later for CAT assay.
As in SW-13 cells, pfos-CAT was active in C33A cells, routinely producing at least 25,000 cpm during a 1-h CAT assay. As shown in Fig. 6, expression of BRG1 alone resulted in only a modest decrease in pfos-CAT activity, consistent with the idea that full transcriptional repression of the c-fos promoter requires the combination of BRG1 and Rb. Indeed, cotransfection of increasing amounts of the Rb expression construct significantly potentiated the effect of BRG1 on pfos-CAT activity. The combined effect on transcription was 5-fold, compared with 1.7-fold repression in the absence of pCMV-Rb. The effect of Rb on the c-fos promoter was clearly dependent on the presence of BRG1, since transfection of pCMV-Rb alone had no effect on pfos-CAT activity, at either 4 or 8 μg of input DNA. Again, mutant K798R produced only a slight effect on c-fos promoter activity, in either the presence or the absence of Rb. Western blot analysis demonstrated that expression of Rb had no effect on the level of BRG1 expression in these experiments (data not shown). We concluded that Rb acts together with BRG1 to repress transcription of the fos promoter. Recently, gel supershift experiments with an anti-Rb antibody have provided evidence that Rb from a HeLa cell extract can interact with the −123 to +45 region of the c-fos promoter (54). These and other aspects of c-fos regulation make it likely that the combined action of BRG1 and Rb is directed to the c-fos promoter (see Discussion).
FIG. 6.
Repression of the c-fos promoter by BRG1 depends on Rb. C33A cells were transfected with the indicated expression constructs, along with 2 μg of pfos-CAT. Cells were harvested after 48 h and assayed for CAT activity. The amount of input DNA (in micrograms) for each expression construct is given below each bar. The activity of pfos-CAT transfected along with the pCMV5 parental expression vector served as 100% activity for calculation of percent promoter activity. The data are presented as averages from five independent experiments. CMV, cytomegalovirus.
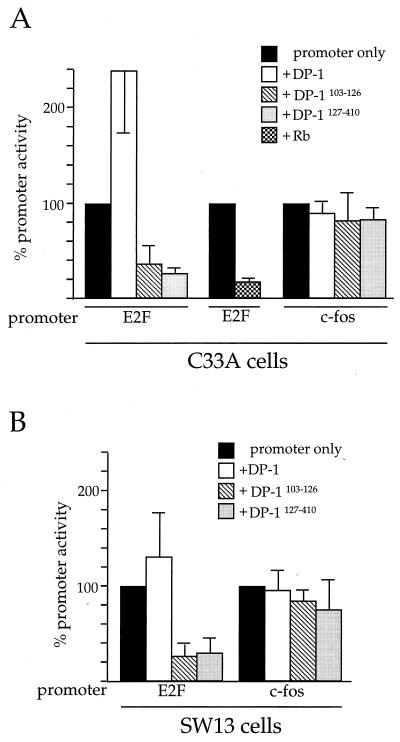
Repression of c-fos by BRG1 is independent of transcription factor E2F.
As noted above, the BRG1-related protein hBRM has been shown to block E2F-stimulated transcription through interaction with Rb (66). We therefore considered the hypothesis that repression of c-fos transcription by BRG1 is an indirect consequence of the combined action of BRG1 and Rb on E2F-dependent transcription. If the c-fos promoter were under the direct or indirect control of E2F, then repression of E2F-dependent transcription by Rb and BRG1 would lead to the observed decrease in c-fos transcription.
To test this hypothesis, we first employed two dominant-negative DP-1 expression constructs in order to abolish E2F-dependent transcription independently of the action of Rb or BRG1. DP-1 protein normally heterodimerizes with E2F to activate transcription at E2F DNA-binding sites (25). If the c-fos promoter is controlled by E2F, then expression of the dominant-negative DP-1 protein would be expected to result in loss of c-fos promoter activity.
To perform this experiment, we employed two different dominant-negative DP-1 expression clones. Each clone was cotransfected along with pfos-CAT into C33A cells, and the cells were harvested after 48 h for assay of CAT activity. To demonstrate that dominant-negative DP-1 protein can, in fact, abolish cellular E2F activity, clones pCMV-HA-DP-1127–410 and pCMV-HA-DP-1Δ103–126 (75) were first cotransfected with the E2-CAT reporter, containing the adenovirus E2 gene promoter under the control of E2F/DP-1. As shown in Fig. 7A, E2-CAT activity was significantly blocked when either of the dominant-negative DP-1 proteins was expressed, demonstrating that E2F activity was effectively blocked during the experiment. Overexpression of wild-type DP-1 caused a greater-than-twofold increase in E2-CAT activity. Importantly, Fig. 7A illustrates that dominant-negative DP-1 had absolutely no effect on transcription from the pfos-CAT construct, and the wild-type DP-1 protein also produced no effect. This indicates that the activity of the c-fos promoter is not controlled by E2F, either directly or indirectly.
FIG. 7.
Repression of c-fos by BRG1 is E2F independent. C33A (A) or SW-13 (B) cells were transfected with 5 μg of the indicated expression plasmids along with the indicated promoter constructs. Cells were harvested 48 h after transfection and assayed for CAT activity. The activity of pfos-CAT or E2-CAT transfected along with the pCMV5 parental expression vector served as 100% activity for calculation of percent promoter activity. The data are presented as averages from three independent experiments.
We also blocked cellular E2F activity by expressing Rb in C33A cells, which normally express a mutant, nonfunctional Rb protein. As shown in Fig. 7A, expression of Rb severely diminished the activity of the transfected E2-CAT construct in these cells, again demonstrating inhibition of cellular E2F activity. In contrast, as already presented in Fig. 6, expression of Rb alone in C33A cells had no effect on the c-fos promoter.
The DP1 dominant-negative mutants were also used to transfect SW-13 cells, along with either E2-CAT or pfos-CAT (Fig. 7B). Again, there was a clear effect of both proteins on E2F-dependent transcription but no effect on the c-fos promoter.
We conclude, therefore, that repression of the c-fos promoter by the combined action of BRG1 and Rb clearly does not take place through an E2F-mediated mechanism in C33A cells or SW-13 cells.
DISCUSSION
The complexity of transcriptional regulation at the c-fos locus is well established. The complete transcriptional control region of c-fos contains numerous cis-acting elements that respond rapidly to serum, growth factors, cAMP, and other signals (reviewed in reference 30). In addition to the binding of sequence-specific transcription factors controlled by these signaling pathways, there is in vivo and in vitro evidence for precisely positioned nucleosomes in the proximal promoter region of c-fos, including the region of the CRE (26, 60). Chromatin remodeling could therefore have a role in establishing or altering the physical state of the gene in relation to the transcriptional apparatus. We have presented evidence that BRG1, functioning as a component of the hSWI-SNF complex, specifically represses transcription directed by a transiently transfected mouse c-fos promoter and also of that directed by the endogenous human c-fos gene. A human c-fos proximal promoter construct was also repressed by BRG1 in SW-13 cells (data not shown). This is the first report documenting regulation of an endogenous gene by BRG1 and hSWI-SNF. Using transfected promoters, we also demonstrated that repression requires Rb, which is known to bind directly to BRG1. It remains to be determined if repression of the endogenous (as opposed to transfected) c-fos promoter also depends on Rb. We have shown that endogenous GAPDH mRNA is unaffected by BRG1, consistent with the data presented in Fig. 1 demonstrating that BRG1 acts specifically. It will be interesting to determine if the effect of BRG1 on endogenous c-fos is highly selective or if additional genes are regulated similarly.
We demonstrated that ectopic expression of BRG1 results in potentiation of GR-dependent transcription and that repression of the c-fos promoter involves the DNA-dependent ATPase domain of BRG1. These results strongly suggest that BRG1 acted in our experiments as a component of the SWI-SNF complex. It should be pointed out, however, that under our transfection conditions (approximately 30% of the cells are transfected), there could be an excess of free BRG1 in the transfected cells and that this population of BRG1 could function to repress transcription. It remains to be determined if c-fos transcription is repressed by intact SWI-SNF or by free BRG1.
Rb has previously been implicated in two types of BRG1 (or hBRM)-mediated transcriptional regulation: activation of GR-dependent transcription and repression of E2F-dependent transcription. Despite the involvement of Rb, we found that GR-dependent transcription and E2F-dependent transcription do not have a role in the repression of c-fos. For instance, expression of BRG1 alone (i.e., in the absence of GR and dexamethasone) had no effect on transcription of the GR-dependent MMTV promoter (Fig. 1). This indicates that repression of c-fos (which only requires BRG1) is not mediated through a GR-dependent process. Similarly, we found that ablation of E2F-dependent transcription using dominant-negative DP-1 mutants had no effect on c-fos transcription (Fig. 7), demonstrating that BRG1 cannot repress c-fos simply through inhibition of E2F activity. These results strongly suggest that the BRG1-Rb interaction leads to repression of c-fos by way of a distinct mechanism, possibly through interactions with other Rb-binding proteins. Since Rb is itself a highly regulated protein, our data also suggest that the Rb-BRG1 complex dictates a number of important cell cycle-related transcriptional changes.
Considering the −76 to +10 region of the c-fos promoter in isolation, mutation of the CRE (in pm28) resulted in complete loss of sensitivity to BRG1 in our assays. A CRE is also located in the corresponding location of the human promoter. Thus, a possible model for the effect of BRG1 on c-fos could involve nucleosomal rearrangement of the c-fos promoter (already occupied by ATF/CREB) and repression of ATF/CREB-dependent transcription by an Rb-associated histone deacetylase (6, 44, 45). Interestingly, the CRE is known to bind transcription factor ATF-2, which was reported to interact directly with Rb (34) (in that case, the ATF-2–Rb interaction correlated with transcriptional activation, not repression). We have detected ATF-2 in extracts of SW-13 cells by Western blot analysis using a specific antibody against ATF-2 (data not shown), and experiments are now in progress to examine its role in BRG1-mediated repression. However, the lack of responsiveness of pm28 to BRG1 could simply be due to the fact that the transcriptional activity of this mutant is very low, thus preventing the observation of an effect of BRG1 acting through some other component of the transcriptional machinery.
In transgenic mice carrying fos-lacZ fusion genes, transcription was found to be critically dependent on several distinct regulatory elements, suggesting that they act in an interdependent manner. This interdependency is consistent with our observation that BRG1 can affect both cAMP, and IL-6-stimulated transcription, even though the signaling pathways involved appear quite distinct (59, 63). Therefore, hSWI-SNF, possibly acting through the CRE, may act on the endogenous gene in a broader context. These questions are currently under investigation, as is the precise role of Rb in this process.
Transcriptional repression of c-fos by an Rb control element (RCE), located between positions −102 and −71 of the human c-fos promoter, has been previously described (11, 58, 68). The RCE contains an SP1-like binding site which is critical for its function, and a consensus SP-1 site is found in the −76 to +10 region of the mouse promoter studied here. In our experiments, the SP1 site played little or no role in BRG1-mediated repression. Replacement of this site with a heterologous sequence resulted in a significant drop in c-fos promoter activity, but the remaining activity was still largely sensitive to repression by BRG1 (Fig. 5). Consistent with this, the simian virus 40 enhancer-promoter, which contains several binding sites for SP1 (20), was unaffected by BRG1 expression (Fig. 1). However, it is intriguing that Rb can trigger repression through a homologous sequence in the human promoter, and we do not know if BRG1 is involved mechanistically with the RCE.
A recent report has implicated hSWI-SNF in a tumor supressor function (71), and earlier it was shown that expression of BRG1 in SW-13 cells results in growth arrest with a flat cell morphology after a prolonged 10-day incubation (15). It is interesting that growth arrest does not typically result in inhibition of c-fos transcription. For instance, cells arrested in G0 or other phases of the cell cycle are able to support rapid transcriptional activation of the c-fos gene by various inducers, including serum, growth factors, and cAMP (5, 13, 43, 48, 64). This feature of c-fos regulation is inconsistent with the idea that repression by BRG1 is a simple consequence of growth arrest. In fact, since c-fos is required for normal mitotic growth, as well as the G0-to-G1 transition (36), it is likely that repression of c-fos plays an important role in subsequent growth arrest. As noted above, the findings that Rb can associate with the c-fos promoter in vitro and that BRG1 can bind to Rb make it highly likely that repression of c-fos transcription by BRG1 is due to direct action at this promoter. In addition, data presented in Fig. 4 support this idea. These data establish that a threshold level of BRG1 is required for repression to occur. Specifically, we observed that when ongoing BRG1 synthesis was limited by addition of tetracycline, repression was completely abolished even though the level of BRG1 protein only decreased to about a third of its maximal level. This level of BRG1 protein corresponds to that seen 30 to 36 h p.i. in the absence of tetracycline. Since this time period is precisely when repression is initially observed, there seems to be a close link between BRG1 protein and transcriptional repression of c-fos.
As noted by Trouche et al., repression by BRG1 or hBRM may be considered problematic in that these proteins are usually credited with a coactivator function (66). Along with their results, ours suggest that the nucleosome remodeling activity of SWI-SNF can favor either activation or repression. As such, the effect of nucleosome remodeling on transcription might depend on whether binding by an activator or a repressor is facilitated by this activity at any particular locus.
ACKNOWLEDGMENTS
We thank David Allis and Peter Lewis for reading the manuscript critically.
This work was supported by Public Health Service grant CA60675 from the National Cancer Institute.
REFERENCES
- 1.Andersson S, Davis D L, Dahlback H, Jornvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 2.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, and Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 3.Berkowitz L A, Riabowol K T, Gilman M Z. Multiple sequence elements of a single functional class are required for cyclic AMP responsiveness of the mouse c-fos promoter. Mol Cell Biol. 1989;9:4272–4281. doi: 10.1128/mcb.9.10.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bett A J, Haddara W, Prevec L, Graham F L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci USA. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bravo R. Genes induced during the G0/G1 transition in mouse fibroblasts. Semin Cancer Biol. 1990;1:37–46. [PubMed] [Google Scholar]
- 6.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 7.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 8.Cairns B R, Kim Y J, Sayre M H, Laurent B C, Kornberg R D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 10.Carlson M, Laurent B C. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 11.Chen L I, Nishinaka T, Kwan K, Kitabayashi I, Yokoyama K, Fu Y H, Grunwald S, Chiu R. The retinoblastoma gene product RB stimulates Sp1-mediated transcription by liberating Sp1 from a negative regulator. Mol Cell Biol. 1994;14:4380–4389. doi: 10.1128/mcb.14.7.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen T A, Allfrey V G. Rapid and reversible changes in nucleosome structure accompany the activation, repression, and superinduction of murine fibroblast protooncogenes c-fos and c-myc. Proc Natl Acad Sci USA. 1987;84:5252–5256. doi: 10.1073/pnas.84.15.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa J J, Keffer J M, Goff J P, Metcalfe D D. Aphidicolin-induced proliferative arrest of murine mast cells: morphological and biochemical changes are not accompanied by alterations in cytokine gene induction. Immunology. 1992;76:413–421. [PMC free article] [PubMed] [Google Scholar]
- 14.Cote J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 15.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 16.Elfring L K, Deuring R, McCallum C M, Peterson C L, Tamkun J W. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fryer C J, Archer T K. Chromatin remodelling By the glucocorticoid receptor requires the Brg1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 18.Gedrich R W. Ph.D. thesis. Charlottesville: University of Virginia; 1995. [Google Scholar]
- 19.Gedrich R W, Engel D A. Identification of a novel E1A response element in the mouse c-fos promoter. J Virol. 1995;69:2333–2340. doi: 10.1128/jvi.69.4.2333-2340.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gidoni D, Dynan W S, Tjian R. Multiple specific contacts between a mammalian transcription factor and its cognate promoters. Nature. 1984;312:409–413. doi: 10.1038/312409a0. [DOI] [PubMed] [Google Scholar]
- 21.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 23a.Hardy, S. Unpublished data.
- 24.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps M L. Construction of adenovirus vectors through Cre-lox recombination. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helin K, Wu C L, Fattaey A R, Lees J A, Dynlacht B D, Ngwu C, Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 26.Herrera R E, Nordheim A, Stewart A F. Chromatin structure analysis of the human c-fos promoter reveals a centrally positioned nucleosome. Chromosoma. 1997;106:284–292. doi: 10.1007/s004120050249. [DOI] [PubMed] [Google Scholar]
- 27.Hiebert S W, Chellappan S P, Horowitz J M, Nevins J R. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- 28.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 29.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 30.Janknecht R, Cahill M A, Nordheim A. Signal integration at the c-fos promoter. Carcinogenesis. 1995;16:443–450. doi: 10.1093/carcin/16.3.443. [DOI] [PubMed] [Google Scholar]
- 31.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 32.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 33.Kim D W, Cheriyath V, Roy A L, Cochran B H. TFII-I enhances activation of the c-fos promoter through interactions with upstream elements. Mol Cell Biol. 1998;18:3310–3320. doi: 10.1128/mcb.18.6.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S J, Wagner S, Liu F, O’Reilly M A, Robbins P D, Green M R. Retinoblastoma gene product activates expression of the human TGF-beta 2 gene through transcription factor ATF-2. Nature. 1992;358:331–334. doi: 10.1038/358331a0. [DOI] [PubMed] [Google Scholar]
- 35.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 36.Kovary K, Bravo R. The Jun and Fos protein families are both required for cell cycle progression in fibroblasts. Mol Cell Biol. 1991;11:4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 38.Laurent B C, Carlson M. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes Dev. 1992;6:1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- 39.Laurent B C, Treich I, Carlson M. Role of yeast SNF and SWI proteins in transcriptional activation. Cold Spring Harbor Symp Quant Biol. 1993;58:257–263. doi: 10.1101/sqb.1993.058.01.030. [DOI] [PubMed] [Google Scholar]
- 40.Laurent B C, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 41.Laurent B C, Treitel M A, Carlson M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol Cell Biol. 1990;10:5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li R, Knight J D, Jackson S P, Tjian R, Botchan M R. Direct interaction between Sp1 and the BPV enhancer E2 protein mediates synergistic activation of transcription. Cell. 1991;65:493–505. doi: 10.1016/0092-8674(91)90467-d. [DOI] [PubMed] [Google Scholar]
- 43.Lin D, Shields M T, Ullrich S J, Appella E, Mercer W E. Growth arrest induced by wild-type p53 protein blocks cells prior to or near the restriction point in late G1 phase. Proc Natl Acad Sci USA. 1992;89:9210–9214. doi: 10.1073/pnas.89.19.9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 45.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 46.Miller M E, Cairns B R, Levinson R S, Yamamoto K R, Engel D A, Smith M M. Adenovirus E1A specifically blocks SWI/SNF-dependent transcriptional activation. Mol Cell Biol. 1996;16:5737–5743. doi: 10.1128/mcb.16.10.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan C J, Pledger W J. Cell cycle dependent growth factor regulation of gene expression. J Cell Physiol. 1989;141:535–542. doi: 10.1002/jcp.1041410312. [DOI] [PubMed] [Google Scholar]
- 49.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein with homology to Saccharomyces cerevisiae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Natesan S, Gilman M Z. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 1993;7:2497–2509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- 52.Neering S J, Hardy S F, Minamoto D, Spratt S K, Jordan C T. Transduction of primitive human hematopoietic cells with recombinant adenovirus vectors. Blood. 1996;88:1147–1155. [PubMed] [Google Scholar]
- 53.Nishikura K, Murray J M. Antisense RNA of proto-oncogene c-fos blocks renewed growth of quiescent 3T3 cells. Mol Cell Biol. 1987;7:639–649. doi: 10.1128/mcb.7.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pai S R, Bird R C. Interaction of the Rb tumor suppressor protein with the c-fos promoter in c-fos transfected cells overexpressing c-fos and Rb. Anticancer Res. 1997;17:3265–3272. [PubMed] [Google Scholar]
- 55.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 57.Rivera V M, Greenberg M E. Growth factor-induced gene expression: the ups and downs of c-fos regulation. New Biol. 1990;2:751–758. [PubMed] [Google Scholar]
- 58.Robbins P D, Horowitz J M, Mulligan R C. Negative regulation of human c-fos expression by the retinoblastoma gene product. Nature. 1990;346:668–671. doi: 10.1038/346668a0. [DOI] [PubMed] [Google Scholar]
- 59.Sassone-Corsi P. Transcription factors responsive to cAMP. Annu Rev Cell Dev Biol. 1995;11:355–377. doi: 10.1146/annurev.cb.11.110195.002035. [DOI] [PubMed] [Google Scholar]
- 60.Schild-Poulter C, Sassone-Corsi P, Granger-Schnarr M, Schnarr M. Nucleosome assembly on the human c-fos promoter interferes with transcription factor binding. Nucleic Acids Res. 1996;24:4751–4758. doi: 10.1093/nar/24.23.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 62.Strober B E, Dunaief J L, Guha S, Goff S P. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- 64.Takasuka T, Ishibashi S, Ide T. Expression of cell-cycle-dependent genes in serum stimulated cells whose entry into S phase is blocked by cytochalasin D. Biochim Biophys Acta. 1987;909:161–164. doi: 10.1016/0167-4781(87)90038-8. [DOI] [PubMed] [Google Scholar]
- 65.Thierry F, Dostatni N, Arnos F, Yaniv M. Cooperative activation of transcription by bovine papillomavirus type 1 E2 can occur over a large distance. Mol Cell Biol. 1990;10:4431–4437. doi: 10.1128/mcb.10.8.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 68.Udvadia A J, Rogers K T, Higgins P D, Murata Y, Martin K H, Humphrey P A, Horowitz J M. Sp-1 binds promoter elements regulated by the RB protein and Sp-1-mediated transcription is stimulated by RB coexpression. Proc Natl Acad Sci USA. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 70.Verma I M, Mitchell R L, Sassone-Corsi P. Proto-oncogene fos: an inducible gene. Princess Takamatsu Symp. 1986;17:279–290. [PubMed] [Google Scholar]
- 71.Versteege I, Sevenet N, Lange J, Rousseau-Merck M-F, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 72.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 74.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 75.Wu C L, Classon M, Dyson N, Harlow E. Expression of dominant-negative mutant DP-1 blocks cell cycle progression in G1. Mol Cell Biol. 1996;16:3698–3706. doi: 10.1128/mcb.16.7.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 77.Zhou Q, Gedrich R W, Engel D A. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J Virol. 1995;69:4323–4330. doi: 10.1128/jvi.69.7.4323-4330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]