Abstract
The seasonal nature of outbreaks of respiratory viral infections with increased transmission during low temperatures has been well established. Accordingly, temperature has been suggested to play a role on the viability and transmissibility of SARS-CoV-2, the virus responsible for the COVID-19 pandemic. The receptor-binding domain (RBD) of the Spike glycoprotein is known to bind to its host receptor angiotensin-converting enzyme 2 (ACE2) to initiate viral fusion. Using biochemical, biophysical, and functional assays to dissect the effect of temperature on the receptor–Spike interaction, we observed a significant and stepwise increase in RBD-ACE2 affinity at low temperatures, resulting in slower dissociation kinetics. This translated into enhanced interaction of the full Spike glycoprotein with the ACE2 receptor and higher viral attachment at low temperatures. Interestingly, the RBD N501Y mutation, present in emerging variants of concern (VOCs) that are fueling the pandemic worldwide (including the B.1.1.7 (α) lineage), bypassed this requirement. This data suggests that the acquisition of N501Y reflects an adaptation to warmer climates, a hypothesis that remains to be tested.
Keywords: coronavirus, COVID-19, SARS-CoV-2, Spike glycoproteins, RBD, ACE2, temperature, N501Y, neutralization, variants of concern
Abbreviations: ACE2, angiotensin-converting enzyme 2; AEC, airway epithelial cell; BLI, biolayer interferometry; ITC, isothermal titration calorimetry; mAb, monoclonal antibody; RBD, receptor-binding domain; RBM, receptor-binding motif; VOC, variant of concern; VOI, variant of interest; WT, wild-type
The etiological agent of the COVID-19 pandemic is the SARS-CoV-2 virus (1). While it will likely take years to understand the spread of SARS-CoV-2 infection in the human population, several factors could be modulating transmission dynamics and are currently being heavily scrutinized. As for other respiratory viruses, host immunity, population density, human behavioral factors, humidity, and temperature likely modulate its transmission (2, 3, 4, 5, 6, 7). Different steps in the replication cycle of coronaviruses could be affected by such factors, particularly viral entry. This process is mediated by the viral Spike (S) glycoprotein. The Spike glycoprotein uses its receptor-binding domain (RBD) to interact with its host receptor angiotensin-converting enzyme 2 (ACE2) (8, 9, 10). Cleavage by cell surface proteases or endosomal cathepsins (8, 11, 12) releases the fusion peptide and triggers irreversible conformational changes in the Spike glycoprotein enabling membrane fusion and viral entry (13, 14).
Airway transmission of SARS-CoV-2 is confronted to the temperature gradient that exists in human airways. In the nasal mucosa it is around 30 to 32 °C, it moves up to 32 °C in the upper trachea, and around 36 °C in the bronchi (15, 16). Emerging results strongly suggest that the Spike glycoprotein of SARS-CoV-2 evolved to replicate in the upper airways (17). This was linked to Spike stability, which was enhanced by the introduction of the D614G mutation (18, 19, 20) but also enhanced its use of cell surface and endosomal proteases (17, 21). Additionally, recent studies have noticed an increase replication of SARS-CoV-2 in primary human airway epithelial cells at 33 °C compared with 37 °C (22), while higher temperatures (39–40 °C) decreased overall viral replication (23). Since it was previously documented that temperature modulates the affinity of another viral envelope glycoprotein (HIV-1 Env) for its receptor (24); herein, we evaluate to what extent temperature affects the interaction of SARS-CoV-2 Spike with ACE2 and concomitantly, its effect on viral attachment.
Results
Conservation of ACE2-interacting residues among SARS-CoV-2 Spike isolates
We first evaluated the conservation of the ACE2-binding site on the SARS-CoV-2 Spike. Based on previously published structural data (25), ACE2 interacts with 17 key residues on the RBD primarily located on the receptor-binding motif (RBM). To determine the degree of conservation of these residues, we used the COVID-19 CoV Genetics program (26) and analyzed SARS-CoV-2 sequences deposited in 2019 to 2020 or in 2021 (up to June 18th 2021). Over the 2019 to 2020 period, no major sequence variations were observed except for the N501Y mutation (4.4%), which started to arise at the end of the year in at least three independent lineages of interest (B.1.1.7, B.1.351, P.1) (27, 28, 29). In 2021 sequences, most residues were still found to be >99% conserved except for variations found at residues 417 (K417N, 1.5%; K417T, 2.6%) and 501 (N501Y, 65.2%), with the latter becoming predominant among all the deposited sequences in 2021 (Fig. 1A). These mutations are found in emergent variants of concern (VOCs), including the B.1.1.7 (N501Y), B.1.351 (K417N/N501Y), and P.1 lineages (K417T/N501Y) (30). Among them, the B.1.1.7 lineage (also known as alpha variant), which was first identified in the United Kingdom, was shown to have increased infectivity and transmissibility (31, 32). This lineage spread rapidly and is the major circulating strain in early 2021 worldwide, replacing the D614G strain, which was predominant in 2019 to 2020 (Fig. 1B) (33). Sequence variations were also found in other RBM residues (Fig. S1), notably mutations L452R (8.8%), E484K (7.7%), T478K (5.9%), S477N (2.2%), and N439K (1.2%), which are also found in other various VOCs (34, 35, 36, 37, 38) and were shown to either increase infectivity or promote the evasion of antibody responses (30, 34, 38, 39, 40, 41, 42).
Figure 1.
Enhanced binding of ACE2 to SARS-CoV-2Spike at low temperatures.A and B, LOGO depictions of the frequency of SARS-CoV-2 Spike residues known to (A) contact with ACE2 or (B) corresponding to B.1.1.7 defining mutations. Worldwide sequences deposited in the GISAID database in 2019 to 2020 and in 2021 (up to June 18th, 2021) were aligned using the COVID CoV Genetics program. The height of the letter indicates its frequency over total sequences. Residues corresponding to the WIV04 reference sequence are shown in black and residues corresponding to VOCs are shown in violet. A box with a cross inside (☒) indicates the presence of a residue deletion. C–E, cell-surface staining of transfected 293T cells expressing SARS-CoV-2 Spike (WT, D614G, Furin KO, D614G Furin KO, D614G N501Y, or B.1.1.7 variant) or SARS-CoV-1 Spike (WT) using (C) CV3-25 mAb or (D and E) ACE2-Fc. F, cell-surface staining of Vero E6 or primary human AECs from two healthy donors infected with authentic SARS-CoV-2 viruses (D614G or B.1.1.7 variant) using ACE2-Fc. C–F, the graphs shown represent the binding of primary antibodies performed at (C and D) 37 °C, 22 °C, and 4 °C or (E and F) at 37 °C and 4 °C. ACE2-Fc binding was normalized to CV3-25 binding in each experiment at each temperature. The graphs shown represent the median fluorescence intensities (MFI). Error bars indicate means ± SEM. These results were obtained in at least three independent experiments. Statistical significance was tested using (C–E) one-way ANOVA with a Holm–Sidak posttest or (F) a paired t test (∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, nonsignificant).
Low temperatures increase SARS-CoV-2 Spike–ACE2 interaction
To measure the effect of temperature on the Spike–ACE2 interaction, we use a system where we express the full-length native Spike at the surface of cells and measure its interaction with the ACE2 receptor using a recombinant ACE2-Fc chimeric protein. This recombinant protein is composed of an ACE2 ectodomain linked to a human IgG1 Fc (43). 293T cells were transfected with a plasmid encoding the SARS-CoV-2 wild-type (WT) Spike (Wuhan-Hu-1 strain). Forty-eight hours posttransfection, cells were incubated at different temperatures (37 °C, 22 °C, and 4 °C) before measuring ACE2-Fc binding by flow cytometry. To ensure that any differential recognition was not linked to a temperature-dependent variation in Spike levels, we used the conformational-independent S2-targeting monoclonal antibody (mAb) CV3-25 as an experimental control (44, 45). As shown in Figure 1C, temperature did not alter CV3-25 recognition, indicating that temperature does not affect the overall amount of Spike at the surface of these cells. Therefore, the CV3-25 mAb was used to normalize Spike expression levels among the different mutants or variants (Fig. 1, D–F).
Interestingly, we observed a gradual increase in ACE2-Fc recognition concomitant with the temperature decrease (Fig. 1D), suggesting a temperature-dependent interaction between Spike and ACE2. Since temperature was suggested to also affect Spike stability (17, 46, 47), which in turn could explain its decreased receptor binding at 37 °C, we introduced the D614G change, known to increase trimer stability (17, 19) in combination or not with furin cleavage site mutations (FKO), known to prevent Spike proteolytic cleavage (48). The same stepwise increase in ACE2-Fc binding at lower temperatures was observed with all Spike constructs (Fig. 1D), indicating that low temperature can enhance ACE2-Fc binding independently of the strength of association between the S1 and S2 subunits. To extend these results to the Spike of emergent circulating strains, we evaluated ACE2-Fc binding to the Spike N501Y mutant and the Spike from the B.1.1.7 lineage. The N501Y mutation is located at the RBD-ACE2 interface and has been previously shown to strengthen the interaction with ACE2 by inserting an aromatic ring into a cavity at the binding interface (47, 48). Despite significantly higher binding of ACE2-Fc at 37 °C (Fig. 1E), a similar enhancement was observed with both the N501Y mutant and the B.1.1.7 variant at low temperatures (Fig. 1D). To evaluate whether this phenotype was conserved among other Betacoronaviruses, we also performed the same experiment using the closely related SARS-CoV-1 Spike and similar changes were observed (Fig. 1D).
To confirm our observations in a more physiological model, we infected a highly permissive cell line (Vero E6) and primary airway epithelial cells (AECs) from two different healthy donors using authentic SARS-CoV-2 virus isolated from patients infected with SARS-CoV-2 D614G or B.1.1.7 (Fig. 1F). Using flow cytometry, we discriminated the infected cells using an antinucleocapsid (N) mAb and measured the binding of ACE2-Fc at the cell surface (Fig. S2, A and B). We first noticed that the expression of trimeric Spike, as quantified by CV3-25 binding, is specific for the N+ population (Fig. S2, A and B). In agreement with results from transfected cells, the binding of ACE2 to cell surface Spike was higher at cold temperature (4 °C) compared with 37 °C for both D614G- and B.1.1.7-infected cells (Fig. 1F and Fig. S2C). Importantly, ACE2 bound to the B.1.1.7 Spike about two times more than to the D614G Spike at 37 °C (Fig. 1F and Fig. S2C). Similar level of binding could only be achieved for the D614G Spike by decreasing the temperature to 4 °C (Fig. 1F). Overall, low temperatures appear to promote Spike–ACE2 interaction independently of Spike trimer stability and emerging mutations, although the B.1.1.7 variant exhibited a pronounced improvement in binding at warmer temperatures.
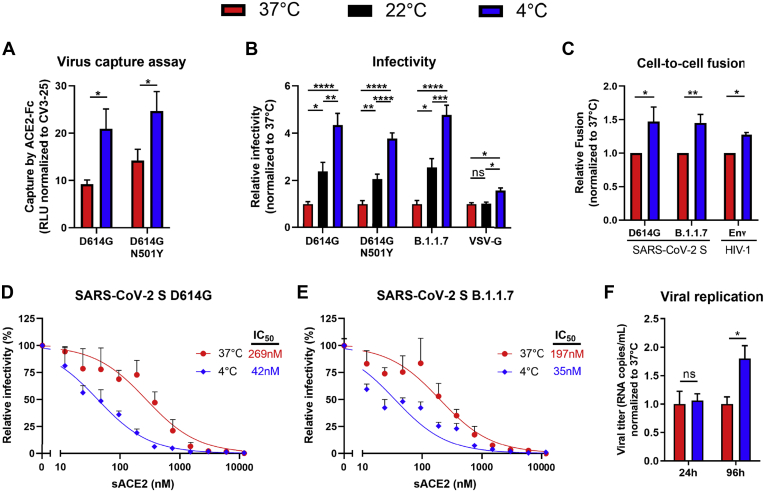
Low temperatures improve the viral attachment of SARS-CoV-2 virions
Next, we investigated the effect of enhanced ACE2 binding at low temperatures on SARS-CoV-2 Spike functional properties, including its ability to mediate viral attachment and fusion, and the subsequent consequences on early viral replication kinetics. To assess viral attachment, we adapted a previously described virus capture assay (49) where we generate lentiviral particles bearing SARS-CoV-2 Spike and look at their ability to interact with ACE2-Fc immobilized on ELISA plates. In agreement with a better affinity for ACE2 at lower temperatures, more SARS-CoV-2 D614G pseudoviral particles were captured at 4 °C compared with 37 °C (Fig. 2A). In line with these results, we also observed enhanced infectivity and cell-to-cell fusion mediated by SARS-CoV-2 Spike D614G at 4 °C compared with 37 °C, while a marginal increase was seen with an unrelated viral glycoprotein (VSV-G) (Fig. 2, B and C). In agreement with an enhanced affinity of HIV-1 Env for its receptor CD4 (24), HIV-1 Env-mediated fusion was also found to be temperature-dependent. Similarly, the capacity of soluble ACE2 (sACE2) to neutralize pseudovirions bearing SARS-CoV-2 Spike D614G was significantly improved when preincubating the virus with sACE2 at 4 °C when compared with 37 °C prior infection of 293T-ACE2 target cells (Fig. 2, D and E). Similar effects of temperature on Spike-mediated attachment and fusion and on sensitivity to sACE2 neutralization were observed when using the Spike N501Y mutant or B.1.1.7 variant (Fig. 2, A–E). To analyze the impact of temperature on viral replication in a more physiological model, we used authentic SARS-CoV-2 D614G viruses to infect reconstituted primary human airway epithelia (MucilAir). Infections were performed at 4 °C or 37 °C for 30 min, virus-containing medium was then discarded to remove any unbound virus before keeping the cells at 37 °C for 4 days. While no significant differences in viral titers were observed at 24 h postinfection, viral replication at 96 h postinfection was found to be significantly higher when the initial infection was performed at 4 °C versus 37 °C (Fig. 2F). Altogether, this suggests that lower temperatures improve the initial attachment of SARS-CoV-2, which in turn can alter the subsequent kinetics of viral replication.
Figure 2.
SARS-CoV-2 viral attachment and infectivity is higher at low temperatures.A, pseudoviruses encoding the luciferase gene (Luc+) and bearing SARS-CoV-2 Spike (D614G or D614G N501Y) were tested for virus capture by ACE2-Fc at 37 °C or 4 °C. RLU obtained using ACE2-Fc was normalized to the signal obtained with the CV3-25 mAb. B, pseudoviruses Luc+ bearing SARS-CoV-2 Spike (D614G, D614G N501Y or B.1.1.7), or VSV-G as a control, were used to infect 293T-ACE2 cells. Virions were incubated at 37 °C, 22 °C, or 4 °C for 1 h prior infection of 293T-ACE2 cells for 48 h at 37 °C. C, cell-to-cell fusion was measured between 293T effector cells expressing HIV-1 Tat and SARS-CoV-2 Spike (D614G or B.1.1.7), or HIV-1 EnvJRFL as a control, which were incubated at 37 °C or 4 °C for 1 h prior coculture with TZM-bl-ACE2 target cells. B and C, RLUs were normalized to the signal obtained with cells preincubated at 37 °C. D and E, pseudoviruses Luc+ bearing SARS-CoV-2 Spike (WT, D614G or B.1.1.7) were used to infect 293T-ACE2 cells in presence of increasing concentrations of sACE2 at 37 °C for 1 h prior infection of 293T-ACE2 cells. Fitted curves and IC50 values were determined using a normalized nonlinear regression. F, authentic SARS-CoV-2 D614G virus was used to infect reconstituted human airway epithelia. Viral attachment was performed at 37 °C or 4 °C and cells were further cultured at 37 °C for 96 h. Viral titers (RNA copies/ml) were monitored at 24 h and 96 h postinfection using one-step qRT-PCR. Viral titer values were normalized to the signal obtained with virions adsorbed to the cells at 37 °C. Error bars indicate means ± SEM. These results were obtained in at least three independent experiments. Statistical significance was tested using (A, C and F) an unpaired t test or (B) one-way ANOVA with a Holm–Sidak post-test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, nonsignificant).
Low temperatures enhance the affinity of SARS-CoV-2 RBD for ACE2
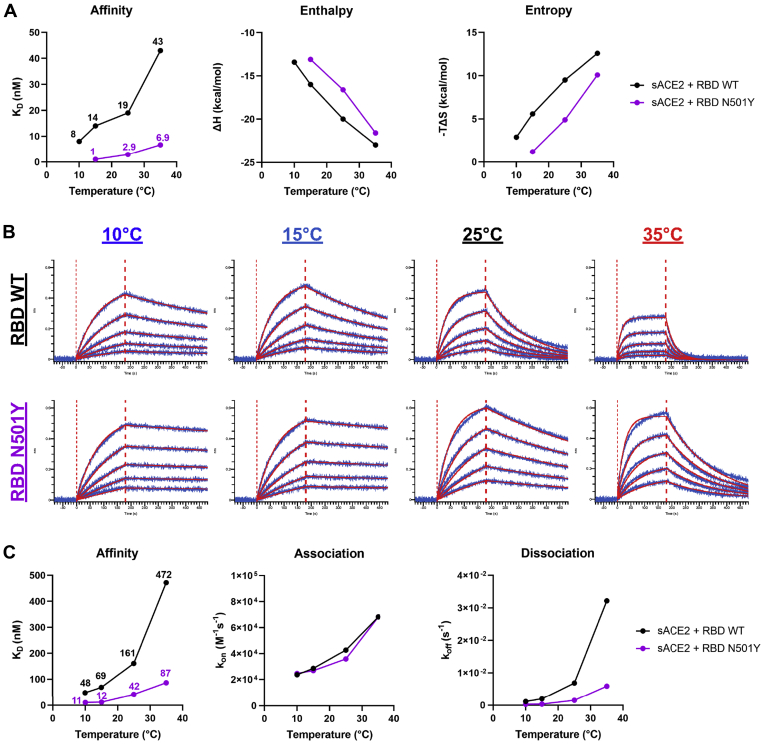
We then evaluated whether the impact of temperature on ACE2 interaction could be recapitulated by the RBD alone. Isothermal titration calorimetry (ITC) was used to measure the binding of ACE2 to RBD at different temperatures ranging from 10 to 35 °C (Fig. S3A). The binding of ACE2 to RBD WT at 25 °C is characterized by a dissociation constant (KD) of 19 nM in a process that is associated with a favorable change in enthalpy of −20 kcal/mol, which is partially compensated by an unfavorable entropy contribution of 9.5 kcal/mol (Fig. 3A and Fig. S3B). The data obtained at different temperatures reveal a threefold increase in binding affinity from 43 nM at 35 °C to 14 nM at 15 °C. The observed effect of the temperature on KD is expected based on the known temperature dependence of Gibbs energy of binding, ΔG(T), which allows calculation of the expected KD values at any temperature (Fig. S3C). Furthermore, as expected, the binding affinity of sACE2 to RBD N501Y was at least 6-fold higher than to RBD WT (Fig. 3A). The KD value for the N501Y mutant is 2.9 nM at 25 °C and the respective values for the enthalpy and entropy contributions are −16.6 and 4.9 kcal/mol. The gain in affinity is the result of a loss in unfavorable entropy, which is larger than and overcompensates the loss in favorable enthalpy. Compared with ACE2 binding to RBD WT, the increase in binding affinity upon a drop in temperature is larger for the N501Y mutant with the KD changing from 6.9 nM at 35 °C to 1 nM at 15 °C.
Figure 3.
Enhanced affinity of SARS-CoV-2 RBD for ACE2 at low temperatures.A, the thermodynamic parameters of sACE2 binding to SARS-CoV-2 RBD WT or N501Y measured by ITC at 10 °C, 15 °C, 25 °C, and 35 °C. The graphs shown represent the affinity (KD), enthalpy (ΔH), and entropy (−TΔS) values obtained at the different temperatures. All ITC titration curves and thermodynamics values are shown in Fig. S3. B and C, binding kinetics between SARS-CoV-2 RBD (WT or N501Y) and sACE2 assessed by BLI at different temperatures. B, biosensors loaded with RBD proteins were soaked in twofold dilution series of sACE2 (500 nM–31.25 nM) at different temperatures (10 °C, 15 °C, 25 °C, or 35 °C). Raw data are shown in blue and fitting model is shown in red. C, graphs represent the affinity constants (KD), on rates (Kon) and off rates (Koff) values obtained at different temperatures and calculated using a 1:1 binding model. All BLI data are summarized in Table S1.
To better characterize how the temperature affects the binding kinetics between RBD and ACE2, we used biolayer interferometry (BLI) at the same temperatures as for ITC. RBD proteins were immobilized on biosensors and were soaked in increasing concentrations of sACE2, ranging from 31.25 to 500 nM (Fig. 3B). Again, affinity between RBD WT and sACE2 was found to be higher at lower temperatures (∼10-fold increase between 35 °C and 10 °C). Changes in affinity were explained by a major decrease in the off rate kinetics at low temperatures, despite a concomitant decrease in on-rate kinetics (Fig. 3C and Table S1). Compared with its WT counterpart, introduction of the N501Y mutation significantly decreased the off rate resulting in a 4.6-fold increase in KD when performed at 25 °C. Remarkably, RBD WT reached a similar affinity for sACE2 at 10 °C than the one achieved by RBD N501Y at 25 °C (Fig. 3B). Altogether, this indicates that low temperatures or the N501Y mutation confers analogous affinity changes that are favorable for Spike RBD-ACE2 interaction.
Low temperatures modulate SARS-CoV-2 Spike trimer opening
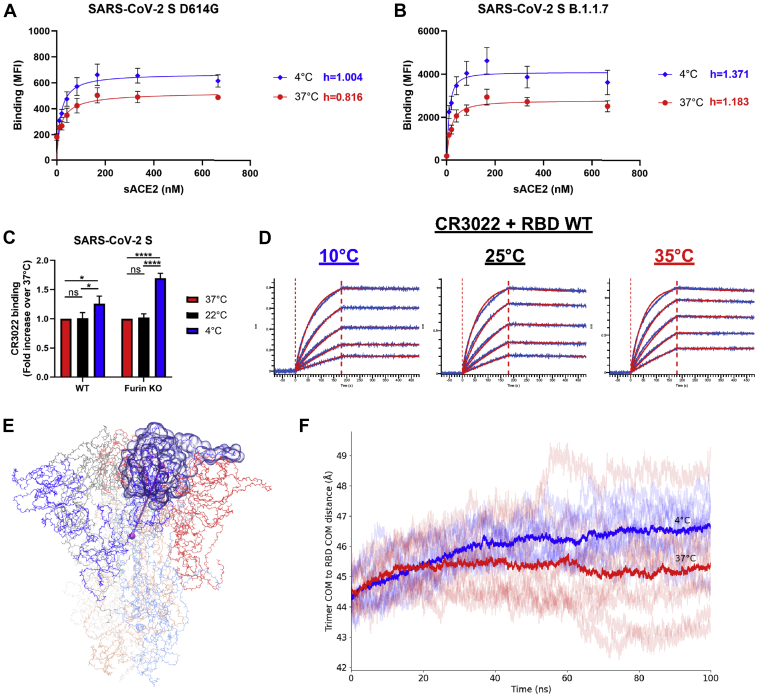
Since ACE2 interaction with Spike occurs when its RBD is in the “up” conformation (50, 51, 52), we sought to determine if temperature could also be modulating Spike trimer opening (i.e., RBD accessibility). To do so, we evaluated the degree of cooperativity between sACE2 monomer binding within the Spike trimers by calculating the Hill coefficient (h), since ACE2 is thought to interact with Spike RBDs in a sequential manner (51). The h values are calculated from the steepness of dose–response curves generated upon incubation of Spike-expressing cells with increasing concentrations of sACE2 as previously described (43). We observed that the binding cooperativity of ACE2 to Spike D614G was slightly negative at 37 °C (h = 0.816), while being neutral at 4 °C (h = 1.004) (Fig. 4A). On the contrary, the binding cooperativity to Spike B.1.1.7 was already slightly positive at 37 °C (h = 1.183) and was further improved at 4 °C (h = 1.371), suggesting that B.1.1.7 mutations could facilitate a coordinated Spike opening in addition to its increased ACE2-RBD interaction, thus fueling the viral entry process (Fig. 4B). Spike conformational changes induced by temperature variation were also investigated by measuring the binding of the CR3022 mAb, which specifically recognizes the RBD “up” conformation (53, 54). Despite no clear change in binding affinity to RBD at low temperatures, CR3022 bound better to the membrane-bound trimeric Spike at 4 °C compared with 37 °C (Fig. 4, C and D and Table S1). Since CR3022 is known to disrupt prefusion Spike trimer (RBD) (53, 54), we also confirmed this phenotype using an uncleaved Spike version (Furin KO) (Fig. 4C). However, the increase in binding by CR3022 at 4 °C was minor compared with the one observed with ACE2-Fc and no change was seen at 22 °C, whereas ACE2-Fc binding was significantly higher (Figs. 1D and 4C). This confirms that low temperatures facilitate the exposure of the RBD in the “up” conformation, but it is unlikely sufficient on its own to recapitulate the temperature-dependent modulation of ACE2 interaction described in Figures 1 and 2.
Figure 4.
SARS-CoV-2Spike trimer “opens” at low temperatures.A and B, binding of sACE2 to SARS-CoV-2 Spike (A) D614G or (B) B.1.1.7 expressed on 293T cells was measured at 37 °C or 4 °C by flow cytometry. Cells were preincubated with increasing amounts of sACE2 and its binding was detected using an anti-ACE2 staining. The Hill coefficients were determined using GraphPad software. C, cell-surface staining of transfected 293T cells expressing SARS-CoV-2 Spike (WT or Furin KO) using the CR3022 mAb when performed at 37 °C, 22 °C, or 4 °C. A–C, the graphs shown represent the median fluorescence intensities (MFI). Error bars indicate means ± SEM. These results were obtained in at least three independent experiments. Statistical significance was tested using (C) one-way ANOVA with a Holm–Sidak posttest (∗p < 0.05; ∗∗∗∗p < 0.0001; ns, nonsignificant). D, binding kinetics between RBD WT and CR3022 mAb assessed by BLI at 10 °C, 25 °C, or 35 °C. Biosensors loaded with RBD were soaked in twofold dilution series of CR3022 (100 nM–6.25 nM). Raw data are shown in blue and fitting model (1:1 binding model) is shown in red. All BLI data are summarized in Table S1. E, snapshot of SARS-CoV-2 Spike ectodomain (PDB 6VXX) (14) with one RBD indicated in transparent surface and one protomer’s RBD-to-trimer center-of-mass distance indicated with a cylinder. F, traces of the RBD-to-trimer distances from three replicas each of all-atom, fully glycosylated, and solvated MD simulations of the closed SARS-CoV-2 S trimer at 4 °C (blues) and 37 °C (reds) with dataset averages shown in heavy traces.
To better understand how low temperature affects the conformational dynamics of Spike and the propensity of RBD to sample the “up” conformation, we performed all-atom molecular dynamics (MD) simulations to measure the distance between the center of mass of the trimer and the center of mass of each RBD subunit using the structure of a fully glycosylated closed SARS-CoV-2 Spike ectodomain trimer as a model (Fig. 4E) (14). Shown in Figure 4F is the RBD-to-trimer center distances of all S1 subunits in three replicas for each temperature (37 °C or 4 °C). At 4 °C, this distance is on average about 1.5 Å longer than at 37 °C, suggesting that lower temperatures favor conformations that are, on average, closer to RBD opening than do higher temperatures. This quaternary structural sensitivity to temperature is consistent with the observation that CR3022 is more reactive against full Spike trimers, but not RBD alone, at lower temperatures (Fig. 4, C–F).
Discussion
In this study, we analyzed the role of temperature in modulating the affinity of SARS-CoV-2 Spike glycoprotein for its host receptor ACE2. We observed a significant enhancement in the affinity at low temperatures, which could be explained by favorable thermodynamics changes leading to a stabilization of the RBD-ACE2 interface and by the triggering of more “open” conformations of the Spike trimer. Consequently, SARS-CoV-2 entry events and early replication kinetics were found to be amplified by enhanced viral adsorption at cold temperatures. This could potentially lead to higher transmissibility and faster replication in upper airway tissues upon exposure to the virus at lower seasonal temperatures. Increasing evidence postulates for the establishment of a clear cycle of seasonal spread for SARS-CoV-2 infection once it reaches the endemic phase (55, 56), in a similar manner as other human coronaviruses and other respiratory viruses (57). Optimal air temperature for SARS-CoV-2 transmission has been suggested to range from 5 to 15 °C (7, 58). When entering the upper airways, such low temperature creates a gradient of temperature from the nasal cavity to the trachea, where it reaches around 33 °C (59, 60, 61). Combined with the fact that ACE2 is highly expressed in nasal epithelial cells (62), this makes the nasal cavity a remarkably favorable microenvironment for SARS-CoV-2 initial adsorption and early inoculum amplification (63), as previously observed (45, 64). While we did not explore this possibility, temperature could also be affecting viral replication kinetics postexposure and one could speculate that elevated body temperature resulting from SARS-CoV-2 infection (>38 °C) could participate in limiting virus replication in vivo by interfering with viral entry, as previously suggested (65).
In summary, our results suggest that the RBD from the original strain isolated in Wuhan requires lower temperature for optimal interaction with ACE2, whereas the N501Y mutation frees RBD from this requirement. A recent study compared a selection of SARS-CoV-2 Spike from emerging variants of concern (VOC) and variants of interest (VOI) for their sensitivity to cold temperature (66). The majority of them bound better to ACE2 at physiological temperature, notably for lineages harboring the N501Y mutation (B.1.1.7, B.1.351, and P.1) or the L452R mutation (B.1.617.2 and B.1.429). While all emerging variants bound better to ACE2 at low temperature, their sensitivity to cold temperature was less pronounced, especially for those harboring the N501Y mutation (66).
Whether this mechanism contributes to viral transmission and the apparent lack of seasonality for VOCs transmitted at warmer temperatures remains to be demonstrated. Our results indicate that the RBD-ACE2 affinity should be taken into consideration when evaluating the impact of the temperature on SARS-CoV-2 transmission.
Experimental procedures
Experimental procedures are provided as supporting information.
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information (8, 13, 14, 26, 43, 44, 45, 49, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank the CRCHUM BSL3 and Flow Cytometry Platforms for technical assistance. We thank Dr Stefan Pöhlmann and Dr Markus Hoffmann (Georg-August University) for the plasmids coding for SARS-CoV-2 and SARS-CoV-1 S, Dr M. Gordon Joyce (U.S. MHRP) for the CR3022 mAb, and Dr Jason McLellan for the plasmid coding for sACE2. The CV3-25 mAb was produced using the pTT vector kindly provided by the Canada Research Council.
Author contributions
J. P., W. M., A. S., and A. F. conceptualization; J. P. formal analysis; G. B. and A. F. funding acquisition; J. P, J. R., R. G., S. D., C. F., S. P. A., D. A., N. G. V., A. T., M. B., S. Y. G., G. G., A. P., E. B., G. B., C. F. A., A. S., and A. F. investigation; J. P., J. R., S. M., C. F. A., and A. S. methodology; H. C., M. R., M. P., and A. F. resources; A. F. supervision; J. P. validation; J. P., A. S., and A. F. writing—original draft; J. R., R. G., S. D., C. F., S. P. A., D. A., A. T., M. B., S. Y. G., W. M., M. P., E. B., G. B., and C. F. A. writing—review and editing.
Funding and additional information
This work was supported by le Ministère de l’Économie et de l’Innovation (MEI) du Québec, Programme de soutien aux organismes de recherche et d’innovation to A. F., the Fondation du CHUM, a CIHR foundation grant #352417, a CIHR stream 1 and 2 for SARS-CoV-2 Variant Research to A. F. and G. B., a CIHR operating grant Pandemic and Health Emergencies Research/Projet #177958 to A. F., an Exceptional Fund COVID-19 from the Canada Foundation for Innovation (CFI) #41027, the Sentinelle COVID Quebec network led by the Laboratoire de Santé Publique du Quebec (LSPQ) in collaboration with Fonds de Recherche du Québec-Santé (FRQS) and Genome Canada – Génome Québec, and by the Ministère de la Santé et des Services Sociaux (MSSS) and MEI. A. F. is the recipient of Canada Research Chair on Retroviral Entry no. RCHS0235 950-232424. J. P. and S. P. A. are supported by CIHR fellowships. R. G. is supported by an MITACS Accélération postdoctoral fellowship. This work used the Extreme Science and Engineering Discovery Environment (XSEDE) (78) resource Stampede2 at the Texas Advanced Computing Center through allocation TG-MCB070073N. XSEDE is supported by National Science Foundation grant number ACI-1548562. G. B. is the recipient of a CIHR foundation grant. The CRCHUM’s Respiratory Cell and Tissue Biobank (directed by E. B.) is supported by the Respiratory Health Research Network of Québec. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The views expressed in this presentation are those of the authors and do not reflect the official policy or position of the Uniformed Services University, US Army, the Department of Defense, or the US Government.
Biography

Jérémie Prévost is a PhD candidate in the Département de Microbiologie, Infectiologie et Immunologie at Université de Montréal. The focus of Jérémie’s thesis is on HIV-1 envelope glycoprotein and the impact of its conformation on antibody responses. During the COVID-19 pandemic, he applied his knowledge to study SARS-CoV-2 Spike glycoprotein. His current research aims to understand the impact of Spike conformation of emerging variants on viral replication and immune evasion.
Edited by Craig Cameron
Supporting information
References
- 1.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalziel B.D., Kissler S., Gog J.R., Viboud C., Bjornstad O.N., Metcalf C.J.E., Grenfell B.T. Urbanization and humidity shape the intensity of influenza epidemics in U.S. cities. Science. 2018;362:75–79. doi: 10.1126/science.aat6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowell S.F., Ho M.S. Seasonality of infectious diseases and severe acute respiratory syndrome-what we don't know can hurt us. Lancet Infect. Dis. 2004;4:704–708. doi: 10.1016/S1473-3099(04)01177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang H.Y., Lee J.K.W. The impact of temperature on the risk of COVID-19: A multinational study. Int. J. Environ. Res. Public Health. 2021;18:4052. doi: 10.3390/ijerph18084052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tosepu R., Gunawan J., Effendy D.S., Ahmad O.A.I., Lestari H., Bahar H., Asfian P. Correlation between weather and Covid-19 pandemic in Jakarta, Indonesia. Sci. Total Environ. 2020;725:138436. doi: 10.1016/j.scitotenv.2020.138436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mecenas P., Bastos R., Vallinoto A.C.R., Normando D. Effects of temperature and humidity on the spread of COVID-19: A systematic review. PLoS One. 2020;15 doi: 10.1371/journal.pone.0238339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sajadi M.M., Habibzadeh P., Vintzileos A., Shokouhi S., Miralles-Wilhelm F., Amoroso A. Temperature, humidity, and latitude analysis to estimate potential spread and seasonality of coronavirus disease 2019 (COVID-19) JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.11834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., Zambon M., Rey F.A., Corti D., Veesler D. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039.e1015. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., Diamond M.S., Ciorba M.A., Whelan S.P.J., Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindemann J., Leiacker R., Rettinger G., Keck T. Nasal mucosal temperature during respiration. Clin. Otolaryngol. Allied Sci. 2002;27:135–139. doi: 10.1046/j.1365-2273.2002.00544.x. [DOI] [PubMed] [Google Scholar]
- 16.McFadden E.R., Jr., Pichurko B.M., Bowman H.F., Ingenito E., Burns S., Dowling N., Solway J. Thermal mapping of the airways in humans. J. Appl. Physiol. (1985) 1985;58:564–570. doi: 10.1152/jappl.1985.58.2.564. [DOI] [PubMed] [Google Scholar]
- 17.Laporte M., Raeymaekers V., Van Berwaer R., Vandeput J., Marchand-Casas I., Thibaut H.J., Van Looveren D., Martens K., Hoffmann M., Maes P., Pohlmann S., Naesens L., Stevaert A. The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., Veinotte K., Egri S.B., Schaffner S.F., Lemieux J.E., Munro J.B. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e738. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., Li W., Izard T., Rader C., Farzan M., Choe H. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schafer A., Nakajima N., Takahashi K., Lee R.E., Mascenik T.M., Graham R., Edwards C.E., Tse L.V. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., Suzuki T., Tokunaga K. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12:848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.V'Kovski P., Gultom M., Kelly J.N., Steiner S., Russeil J., Mangeat B., Cora E., Pezoldt J., Holwerda M., Kratzel A., Laloli L., Wider M., Portmann J., Tran T., Ebert N. Disparate temperature-dependent virus-host dynamics for SARS-CoV-2 and SARS-CoV in the human respiratory epithelium. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herder V., Dee K., Wojtus J.K., Goldfarb D., Rozario C., Gu Q., Jarrett R.F., Epifano I., Stevenson A., McFarlane S., Stewart M.E., Szemiel A.M., Pinto R.M., Garriga A.M., Graham S.V. Elevated temperature inhibits SARS-CoV-2 replication in respiratory epithelium independently of the induction of IFN-mediated innate immune defences. bioRxiv. 2020 doi: 10.1101/2020.12.04.411389. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thali M., Olshevsky U., Furman C., Gabuzda D., Li J., Sodroski J. Effects of changes in gp120-CD4 binding affinity on human immunodeficiency virus type 1 envelope glycoprotein function and soluble CD4 sensitivity. J. Virol. 1991;65:5007–5012. doi: 10.1128/jvi.65.9.5007-5012.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 26.Chen A.T., Altschuler K., Zhan S.H., Chan Y.A., Deverman B.E. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. Elife. 2021;10 doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., Hulswit R.J.G., Franco L.A.M., Ramundo M.S., de Jesus J.G., Andrade P.S. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O'Toole A., Amato R., Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C.V. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 29.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann M., Arora P., Gross R., Seidel A., Hornich B.F., Hahn A.S., Kruger N., Graichen L., Hofmann-Winkler H., Kempf A., Winkler M.S., Schulz S., Jack H.M., Jahrsdorfer B., Schrezenmeier H. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e2312. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Washington N.L., Gangavarapu K., Zeller M., Bolze A., Cirulli E.T., Schiabor Barrett K.M., Larsen B.B., Anderson C., White S., Cassens T., Jacobs S., Levan G., Nguyen J., Ramirez J.M., 3rd, Rivera-Garcia C. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell. 2021;184:2587–2594.e7. doi: 10.1016/j.cell.2021.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26:2002106. doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e819. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., Lytras S., Czudnochowski N., Shah R., Meury M., Jesudason N. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e1120. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West A.P., Wertheim J.O., Wang J.C., Vasylyeva T.I., Havens J.L., Chowdhury M.A., Gonzalez E., Fang C.E., Di Lonardo S.S., Hughes S., Rakeman J.L., Lee H.H., Barnes C.O., Gnanapragasam P.N.P., Yang Z. Detection and characterization of the SARS-CoV-2 lineage B.1.526 in New York. Nat. Commun. 2021;12:4886. doi: 10.1038/s41467-021-25168-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozer E.A., Simons L.M., Adewumi O.M., Fowotade A.A., Omoruyi E.C., Adeniji J.A., Dean T.J., Taiwo B.O., Hultquist J.F., Lorenzo-Redondo R. High prevalence of SARS-CoV-2 B.1.1.7 (UK variant) and the novel B.1.5.2.5 lineage in Oyo State, Nigeria. medRxiv. 2021 doi: 10.1101/2021.04.09.21255206. [preprint] [DOI] [Google Scholar]
- 37.Mlcochova P., Kemp S., Dhar M.S., Papa G., Meng B., Mishra S., Whittaker C., Mellan T., Ferreira I., Datir R., Collier D.A., Singh S., Pandey R., Marwal R., Datta M. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021 doi: 10.1038/s41586-021-03944-y. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-Gonzalez A., Glasner D.R., Reyes K.R., Gliwa A.S., Reddy N.P., Sanchez San Martin C., Federman S., Cheng J., Balcerek J. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184:3426–3437.e3428. doi: 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prevost J., Finzi A. The great escape? SARS-CoV-2 variants evading neutralizing responses. Cell Host Microbe. 2021;29:322–324. doi: 10.1016/j.chom.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J. Reduced sensitivity of infectious SARS-CoV-2 variant B.1.617.2 to monoclonal antibodies and sera from convalescent and vaccinated individuals. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 41.Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., Alford B., Buchser W.J., Ellebedy A.H., Fremont D.H., Diamond M.S. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e474. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anand S.P., Chen Y., Prevost J., Gasser R., Beaudoin-Bussieres G., Abrams C.F., Pazgier M., Finzi A. Interaction of human ACE2 to membrane-bound SARS-CoV-1 and SARS-CoV-2 S glycoproteins. Viruses. 2020;12:1104. doi: 10.3390/v12101104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jennewein M.F., MacCamy A.J., Akins N.R., Feng J., Homad L.J., Hurlburt N.K., Seydoux E., Wan Y.H., Stuart A.B., Edara V.V., Floyd K., Vanderheiden A., Mascola J.R., Doria-Rose N., Wang L. Isolation and characterization of cross-neutralizing coronavirus antibodies from COVID-19+ subjects. Cell Rep. 2021;36:109353. doi: 10.1016/j.celrep.2021.109353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ullah I., Prevost J., Ladinsky M.S., Stone H., Lu M., Anand S.P., Beaudoin-Bussieres G., Benlarbi M., Ding S., Gasser R., Fink C., Chen Y., Tauzin A., Goyette G., Bourassa C. Live imaging of SARS-CoV-2 infection in mice reveals neutralizing antibodies require Fc function for optimal efficacy. Immunity. 2021;54:2143–2158. doi: 10.1016/j.immuni.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edwards R.J., Mansouri K., Stalls V., Manne K., Watts B., Parks R., Janowska K., Gobeil S.M.C., Kopp M., Li D., Lu X., Mu Z., Deyton M., Oguin T.H., 3rd, Sprenz J. Cold sensitivity of the SARS-CoV-2 spike ectodomain. Nat. Struct. Mol. Biol. 2021;28:128–131. doi: 10.1038/s41594-020-00547-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen H.T., Zhang S., Wang Q., Anang S., Wang J., Ding H., Kappes J.C., Sodroski J. Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects. J. Virol. 2020;95 doi: 10.1128/JVI.02304-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Target. Ther. 2020;5:92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding S., Laumaea A., Benlarbi M., Beaudoin-Bussieres G., Gasser R., Medjahed H., Pancera M., Stamatatos L., McGuire A.T., Bazin R., Finzi A. Antibody binding to SARS-CoV-2 S glycoprotein correlates with but does not predict neutralization. Viruses. 2020;12:1214. doi: 10.3390/v12111214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu C., Wang Y., Liu C., Zhang C., Han W., Hong X., Wang Y., Hong Q., Wang S., Zhao Q., Wang Y., Yang Y., Chen K., Zheng W., Kong L. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.Y., Katsamba P.S., Sampson J.M., Schon A., Bimela J., Boyington J.C., Nazzari A., Olia A.S., Shi W., Sastry M. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe. 2020;28:867–879.e865. doi: 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., Tan T.K., Rijal P., Coombes N., Bewley K.R., Tree J.A. Neutralization of SARS-CoV-2 by destruction of the prefusion spike. Cell Host Microbe. 2020;28:445–454.e446. doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lavine J.S., Bjornstad O.N., Antia R. Immunological characteristics govern the transition of COVID-19 to endemicity. Science. 2021;371:741–745. doi: 10.1126/science.abe6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X., Huang J., Li C., Zhao Y., Wang D., Huang Z., Yang K. The role of seasonality in the spread of COVID-19 pandemic. Environ. Res. 2021;195:110874. doi: 10.1016/j.envres.2021.110874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lagace-Wiens P., Bullard J., Cole R., Van Caeseele P. Seasonality of coronaviruses and other respiratory viruses in Canada: Implications for COVID-19. Can. Commun. Dis. Rep. 2021;47:132–138. doi: 10.14745/ccdr.v47i03a02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang Z., Huang J., Gu Q., Du P., Liang H., Dong Q. Optimal temperature zone for the dispersal of COVID-19. Sci. Total Environ. 2020;736:139487. doi: 10.1016/j.scitotenv.2020.139487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holden W.E., Wilkins J.P., Harris M., Milczuk H.A., Giraud G.D. Temperature conditioning of nasal air: Effects of vasoactive agents and involvement of nitric oxide. J. Appl. Physiol. (1985) 1999;87:1260–1265. doi: 10.1152/jappl.1999.87.4.1260. [DOI] [PubMed] [Google Scholar]
- 60.Kang D., Ellgen C., Kulstad E. Possible effects of air temperature on COVID-19 disease severity and transmission rates. J. Med. Virol. 2021;93:5358–5366. doi: 10.1002/jmv.27042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elad D., Wolf M., Keck T. Air-conditioning in the human nasal cavity. Respir. Physiol. Neurobiol. 2008;163:121–127. doi: 10.1016/j.resp.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 62.Sungnak W., Huang N., Becavin C., Berg M., Queen R., Litvinukova M., Talavera-Lopez C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L., Network H.C.A.L.B. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramasamy R. Perspective of the relationship between the susceptibility to initial SARS-CoV-2 infectivity and optimal nasal conditioning of inhaled air. Int. J. Mol. Sci. 2021;22:7919. doi: 10.3390/ijms22157919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., Chen G., Olivier K.N., Ghio A., Tse L.V., Leist S.R. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e414. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Z., Yang Z., Ou J., Zhang H., Zhang Q., Dong M., Zhang G. Temperature dependence of the SARS-CoV-2 affinity to human ACE2 determines COVID-19 progression and clinical outcome. Comput. Struct. Biotechnol. J. 2021;19:161–167. doi: 10.1016/j.csbj.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gong S.Y., Chatterjee D., Richard J., Prévost J., Tauzin A., Gasser R., Bo Y., Vézina D., Goyette G., Gendron-Lepage G., Medjahed H., Roger M., Côté M., Finzi A. Contribution of single mutations to selected SARS-CoV-2 emerging variants spike antigenicity. Virology. 2021;563:134–145. doi: 10.1016/j.virol.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prevost J., Gasser R., Beaudoin-Bussieres G., Richard J., Duerr R., Laumaea A., Anand S.P., Goyette G., Benlarbi M., Ding S., Medjahed H., Lewin A., Perreault J., Tremblay T., Gendron-Lepage G. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep. Med. 2020;1:100126. doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoffmann M., Muller M.A., Drexler J.F., Glende J., Erdt M., Gutzkow T., Losemann C., Binger T., Deng H., Schwegmann-Wessels C., Esser K.H., Drosten C., Herrler G. Differential sensitivity of bat cells to infection by enveloped RNA viruses: Coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beaudoin-Bussieres G., Laumaea A., Anand S.P., Prevost J., Gasser R., Goyette G., Medjahed H., Perreault J., Tremblay T., Lewin A., Gokool L., Morrisseau C., Begin P., Tremblay C., Martel-Laferriere V. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. mBio. 2020;11 doi: 10.1128/mBio.02590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussieres G., Brassard N., Laumaea A., Vezina D., Prevost J., Anand S.P., Bourassa C., Gendron-Lepage G., Medjahed H., Goyette G. A single BNT162b2 mRNA dose elicits antibodies with Fc-mediated effector functions and boost pre-existing humoral and T cell responses. Cell Host Microbe. 2021;29:1137–1150. doi: 10.1016/j.chom.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lodge R., Lalonde J.P., Lemay G., Cohen E.A. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., Preiser W., Doerr H.W., Chow V.T., de Kruif J., Peiris J.S. Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F.T., Mattos C. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 76.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mao Y., Wang L., Gu C., Herschhorn A., Xiang S.H., Haim H., Yang X., Sodroski J. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat. Struct. Mol. Biol. 2012;19:893–899. doi: 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Towns J.C.,T., Dahan M., Foster I., Gaither K., Grimshaw A., Hazlewood V., Lathrop S., Lifka D., Peterson G.D., Roskies R., Scott J.R., Wilkins-Diehr N. XSEDE: Accelerating scientific discovery. Comput. Sci. Eng. 2014;16:62–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.