Abstract
The rapid spread of the SARS-CoV-2 virus that caused the COVID-19 disease, has highlighted our urgent need for sensitive, fast and accurate diagnostic technologies. In fact, one of the main challenges for flatting COVID-19 spread charts is the ability to accurately and rapidly identify asymptomatic cases that result in spreading the virus to close contacts. SARS-CoV-2 virus mutation is also relatively rapid, which makes the detection of COVID-19 diseases still crucial even after the vaccination. Conventional techniques, which are commercially available have focused on clinical manifestation, along with molecular and serological detection tools that can identify the SARS-CoV-2 virus however, owing to various disadvantages including low specificity and sensitivity, a quick, low cost and easy approach is needed for diagnosis of COVID-19. Scientists are now showing extensive interest in an effective portable and simple detection method to diagnose COVID-19. There are several novel methods and approaches that are considered viable advanced systems that can meet the demands. This study reviews the new approaches and sensing technologies that work on COVID-19 diagnosis for easy and successful detection of SARS-CoV-2 virus.
Keywords: SARS-CoV-2, COVID-19, Biosensor, Immunoassay, Bioassay, PCR
Graphical abstract

1. Introduction
The fight against infectious diseases caused by viruses is still challenging and regarded as an endless task for public health care [1]. Coronaviruses are the members of Coronaviridae family and can cause respiratory and neurological diseases [2,3]. Six human coronaviruses (HCoVs) have been detected, including, HCoV-229E, HCoV-HKU1, HCoV-OC43, HCoV NL63 to date, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV). Among them SARS -CoV and MERS-CoV caused pandemic in the past [[2], [3], [4], [5]]. The last and newest coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a novel infectious disease, called coronavirus disease 2019 (COVID-19). COVID-19 was started from Wuhan, China in the late 2019 and is considered as the most widespread pandemic of recent 100 years [6,7]. The pandemic declaration was performed by world health organization (WHO) in March 2019. The symptoms of COVID-19 are not specific and they are similar to the common flu. The preliminary symptoms of coronavirus disease are related to respiratory illness including cough, fever, and difficulty in breathing [8]. Due to the wide range in severity of the disease many individuals remain asymptomatic or have mild symptoms defining a population that is not tested at the time of acute infection [9]. The worldwide confirmed cases of COVID-19 are more than 94 million cases and confirmed deaths are more than 2 million cases in the world until March 2021. The living and working conditions of billions of people, have been significantly disrupted owing to different forms of social distancing and city lockdowns. Newly, the COVID-19 vaccines are produced and applied for high risk groups of people but still the best way for avoiding of COVID-19 spreading is social distancing, and continuous, inclusive and universal testing [10] specially because the vaccination rate is very slow comparing with virus spreading. Since the transmission rate of SARS-CoV-2 virus is very high, and novel in many aspects and produced a lot of difficulties in all countries around the world to get control over it, thus to break the chain of transmission a uniform diagnostic approach is still important. The rapid and accurate detections of SARS-CoV-2 virus is useful for the early diagnosis of COVID-19, which would be important for slowing down the spread of virus and save life of sensitive groups, especially because based on WHO results more than 30% of virus carriers, show no symptoms [11]. To this date, there are a number of reports about the new coronavirus detection (SARS-CoV-2) using wide range of methods. Several methods are used for other Coronaviridae family members previously and some novel methods are specifically designed for SARS-CoV-2 recently [12].
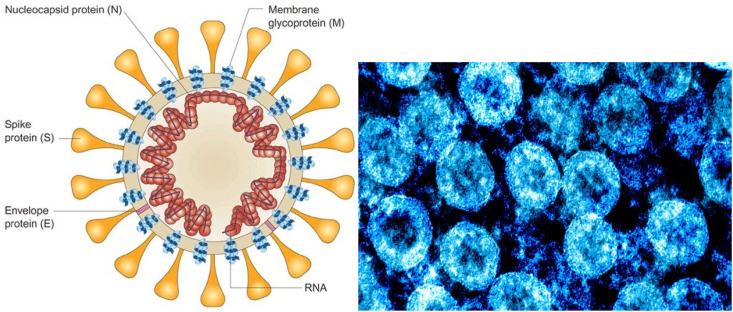
The studies on the SARS-CoV-2 were started in the first days of pandemic in China and virus structure was detected. As can be seen in Fig. 1 (a), the SARS-CoV-2 has circular shape (60–140 nm) and is containing a genetic material and spike proteins on the surface and its structure is comparable to the viruses of the Coronaviridae family. SARS-CoV-2 contains four structural proteins on the surface including spike glycoprotein (S), small envelope protein (E), matrix protein (M), and nucleocapsid protein (N). The spike protein is responsible for the virus infection property [13]. Fig. 1(b) shows the TEM image of SARS-CoV-2 that is identified in the Broncho alveolar lavages of patients after cell culture [14]. Now it is well-known that SARS-CoV-2 enters the cells through interaction with the angiotensin-converting enzyme 2 (ACE2) receptor and it could be found in most human organs including epithelial cells (lung alveolar and intestines cells) suggesting them being the primary region for infection onset while oral, nasal mucosa, and nasopharynx has been shown to lack ACE2 expression [15].
Fig. 1.
(a): Schematic structure and TEM image of the SARS-CoV-2 virus and 1(b): TEM photo of the SARS-CoV-2 [14,16].
Biosensors are analytical devices including a biological sensing element and they are using for detecting a wide range of biological molecules, bacteria and viruses in very different methods [[17], [18], [19]]. In this article we review all biomolecules that are used as analyte and also all methods that are applied for SARS-CoV-2 detection in literatures.
The present manuscript reviews all of the methods that are applied to detect SARS-CoV-2 virus. In addition, the analytes that are used as target for detection are classified and the advantages and disadvantages of different detection methods are compared. The purpose of present manuscript is to help researchers to find the novel detection methods and analytes, and develop the methods for improving specificity, sensitivity, selectivity and detection limit in future. The papers that are focused on SARS-CoV-2 detection methods and published after new pandemic starting in late 2019 are selected for this review.
2. Methods of SARS-CoV-2 virus detection
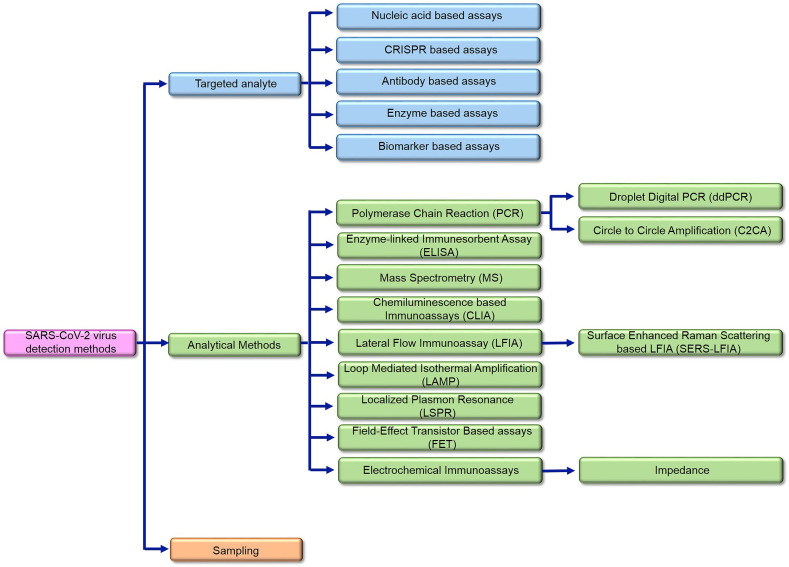
Whereas, COVID-19 is emerged as a worldwide pandemic recently, several detection methods are reported for that in literatures. The reviewed COVID-19 detection articles are classified into three main groups. Fig. 2 shows the classification diagram of approaches and applied methods. As Fig. 2 shows, in first part, the utilized biomolecules that are targeted as analyte are reviewed (section 3). In the second part, the methods and technologies that are used as sensing methods are reviewed (section 4) and in the third part the sampling methods are presented (section 5). Different types of nanoparticles are used in wide range of detection methods and applied for all types of target analytes.
Fig. 2.
The infograph of the reviewed approaches for SARS-CoV-2 detection methods.
3. SARS-CoV-2 virus detection based on targeted analyte
The biosensors use several types of biomolecules as targeted analyte for decays. The common biomolecules as targeted analyte are biomarkers, DNA, RNA, enzymes and antigen in different types of bioassays. In this section the most utilized targeted analytes in COVID-19 detection assays are classified and explained.
3.1. Nucleic acid based assays
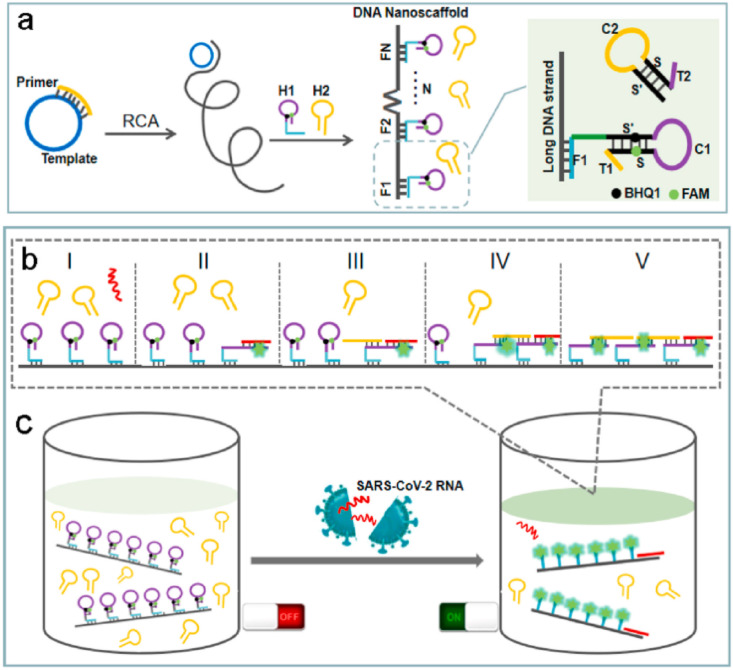
The genome sequence length of SARS-CoV-2 is about 30 Kb with a 5′ cap structure and 3′- poly-(A) tail enveloped by a complex of structural protein to form a crown like enveloped virus. The first method that was used for SARS-CoV-2 virus in early period of COVID-19 outbreak was DNA sequence detection [20]. DNA sequencing technology can be used to explore novel mutations and the evolution of SARS-CoV-2 isolates [21]. However DNA sequencing assays, require highly professional technicians to operate expensive instruments under stringent laboratory conditions. DNA sequencing is designed based on hybridization between the primer and target strands of DNA and a detection marker shows that the hybridization is processed and the target strand is detected. Among other studies which use DNA as assay analyte, Jiao et al. [22] successfully designed a DNA nanoscaffold hybrid chain reaction (DNHCR) method for rapid detection of COVID-19 based on SARS-CoV-2 RNA triggered isothermal amplification. In this method, as can be seen in Fig. 3 , the DNA nanoscaffolds have been first constructed by the self-assembly of long DNA strands and self-quenching probes (H1). Then, the SARS-CoV-2 RNA will initiate the hybridization of H1 and free H2 DNA probes along the nanoscaffold, and an illuminated DNA nanostring is instantly obtained. By taking advantages of the localization design of the H1 probes and the temperature tolerance of the isothermal amplification, the reaction time reduced to within about 10 min and high signal gain is obtained in wide temperature range of 15 °C −35 °C. Additionally, the reliability of DNHCR method in serum and saliva samples have been validated as well (Fig. 3). Therefore, comparing with previous reported biosensors for SARS-CoV-2 RNA detection, DNHCR-based method is expected to provide a simple and faster alternative to the traditional SARS-CoV-2 qRT-PCR assay, significantly simplified and affordable operation [22].
Fig. 3.
Schematic diagram for detection of SARS-CoV-2 RNA based on LAMP method. (a) The synthesis of DNA nanoscaffolds produced by RCA; (b) The procedure of DNHCR triggered by the SARS-CoV-2 RNA; (c) The detection of the target SARS-CoV-2 RNA [22].
3.2. CRISPR based assays
Clustered regularly interspaced short palindromic repeats (CRISPR) is a family of DNA sequences found in the genomes of prokaryotic organisms like bacteria and archaea. These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote. CRISPR is used in COVID-19 detection assays as analyte in some published studies. For instance, Hou et al. [23], have developed an isothermal CRISPR-based COVID-19 assay for SARS-CoV-2 detection based on Cas13a ORF with near single-copy sensitivity and compared their diagnostic performance with other technological platforms, including metagenomic sequencing and RT-PCR. The authors have claimed that their method demonstrated a sensitivity of 100% by detecting all 62 COVID-19 cases and no false positives were found in all 62 negative cases, including all the hCoV-infected ones [23]. In another study, broughton et al. [24] have reported the development and initial validation of a CRISPR–Cas12-based assay for detection of SARS-CoV-2 from extracted patient sample RNA, called SARS-CoV-2 DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR). This assay performs simultaneous RT–LAMP for RNA extracted from nasopharyngeal or oropharyngeal swabs, followed by Cas12 detection of predefined coronavirus sequences, after which cleavage of a reporter molecule confirms detection of the virus. They have joined isothermal amplification with CRISPR–Cas12 DETECTR technology to develop a rapid (30–40 min) test to detect of SARS-CoV-2 in clinical samples. For the method testing, the researchers have used contrived reference samples and clinical samples from patients in the United States, including 36 patients with COVID-19 infection and 42 patients with other viral respiratory infections. The results have showed the CRISPR-based DETECTR assay provides a visual and faster alternative to the US Centers for Disease Control and Prevention SARS-CoV-2 real-time RT–PCR assay, with 95% positive predictive agreement and 100% negative predictive agreement [24]. Ding et al. [25] developed a novel assay method for simple, rapid, ultrasensitive and visual detection of coronavirus SARS-CoV-2 and HIV virus and called that All In One Dual CRISPR (AIOD-CRISPR). In this assay, a pair of crRNAs was introduced to initiate dual CRISPR-Cas12a detection and improve detection sensitivity. The AIOD-CRISPR assay system was successfully utilized to detect nucleic acids (DNA and RNA) of SARS-CoV-2 and HIV with a sensitivity of few copies. The authors were engineered AIOD-CRISPR without pre-amplification could detect as low as 1.2 copies of DNA targets and 4.6 copies of RNA targets in 40 min incubation. The distinctive advantages listed for the assay are (i) system is a true single reaction system; (ii) AIOD-CRISPR is an isothermal nucleic acid detection method; (iii) AIOD-CRISPR-based detection is very fast, robust, highly specific, and nearly single-molecule sensitive and (iv) AIOD-CRISPR enables one-step CRISPR-Cas12a-based RNA detection. Therefore the developed method possesses a high potential for developing next-generation point-of-care molecular diagnostics compared to real time PCR [25].
3.3. Antibody based assays
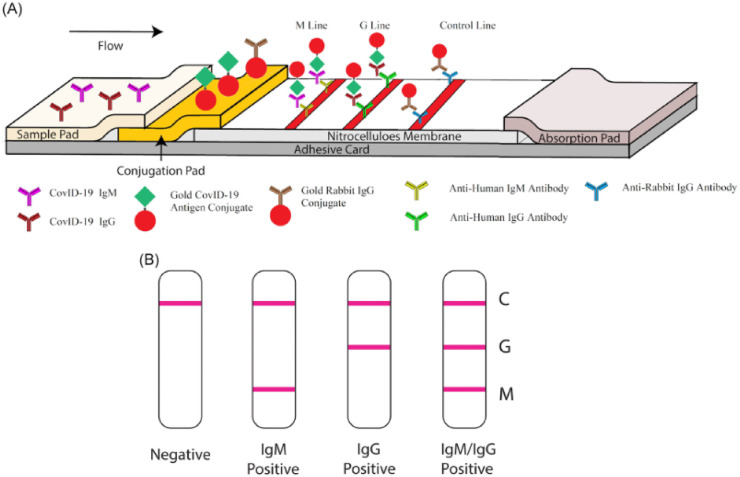
Nucleic acid testing, considered as the current primary method for diagnosing COVID-19, might lead to false negatives and is difficult to be applied for every suspected patient because of the existence of asymptomatic [26]. In addition, detecting specific antibodies in blood, like the immunoglobulin M (IgM) antibody, against the SARS-CoV-2 is another choice for COVID-19 diagnosis, as it is widely accepted that IgM is an important indicator in the acute infection period [26]. It is highly accepted that the IgM provides the first line of defense during viral infections, before the generation of adaptive, high affinity immunoglobulin G (IgG) responses that are important for long term immunity and immunological memory. The main idea for antibody based assay design is that IgM initially raises after the first contact of virus with body and rapidly decreases after short time, while IgG produces in a second time and remains in the blood even after recovery. Therefore the ability of recognition of the anti-SARS-CoV-2 immunoglobulins that also named SARS-CoV-2 antibodies can guide the researcher to identify infected patients and to recognize those who surpassed the illness and can safely end isolation [27]. The drawback of this approach is that the sequential production of IgM and IgG is questionable as reported for previous SARS and MERS virus, when the immune system encounter SARS CoV-2. In another word, IgM responses were either found earlier than IgG or together with IgG, later than IgG or were missing [28]. Furthermore, Detection of immunoglobulin A (IgA) and IgM has been suggested by some researcher as a more accurate diagnostic tool. The obtained results for SARS infection outbreak have shown that, just like the MERS, IgG can be detected as early as 2 days after the onset of fever and IgM can be detected at the same time or after the IgG [29]. There are several well-known methods for IgG and IgM detection [30,31], for instance, Li et al. [27], reported a strategy for COVID-19 detection based on IgM and IgG antibodies measurements. In this study, the commercial IgG-IgM combined antibody kit to detection of suffered SARS‐CoV‐2 cases from healthy cases was used. The samples obtained from 58 patients in Wuhan were tested and it was found that 94.83% of the positive patients had both IgM‐ and IgG‐positive test lines, and 1.72%, 3.45% had only IgM or only IgG‐positive lines, respectively. The test time was from day 8 to day 33 after infection symptoms appeared and the test time was less than 15 min. Fig. 4 , Shows the schematic illustration of IgG-IgM combined antibody testing kit that used in this study. As can be seen in Fig. 4(A) the test trip has two Mouse antihuman monoclonal antibodies (anti-IgG and anti-IgM) stripped on two separated test lines. A mixture of SARS‐CoV‐2 antibodies (including IgM and IgG) and rabbit IgG conjugated to colloidal gold nanoparticles (AuNP) are sprayed on the pad as sample and controlling solution respectively. The AuNP- rabbit IgG conjugates immobilize on the control line while AuNP- SARS‐CoV‐2 IgM conjugated SARS‐CoV‐2 and AuNP- SARS‐CoV‐2 IgG conjugated on M and G line respectively. If the sample does not contain SARS‐CoV‐2 antibodies, no labeled complexes bind at the test zone and no line could be observed. Fig. 4(B) shows the testing results for negative, IgM positive, IgG positive and IgM/IgG positive samples [27]. Additionally, Wang et al. [32], have developed a SARS-CoV-2 proteome peptide microarray to analyze antibody interactions at the amino acid resolution. Using the previous fabricated microarray, researcher showed the possibility of employing SARS-CoV-1 antibodies to detect the SARS-CoV-2 nucleocapsid phosphoprotein. For fabrication of SARS-CoV-2 proteome microarray, in the first step, the reference sequences of 10 proteins were encoded by the SARS-CoV-2 coronavirus genome and prepared the peptide library and then, all the peptides were labeled with a C-terminal biotin group and printed onto a three-dimensional (3D) modified microscope slide using biotin−streptavidin chemistry. After optimization the microarray was used for antibodies detection. The final results show that the designed SARS-CoV-2 peptide-based microarray is enable to detect the humoral antibody response in COVID-19 patients and animal models and can help to find the potential targets for COVID-19 diagnosis and treatments [32]. In another work, Steiner at al. [33], described an arrayed imaging reflectometry (AIR) platform for studying IgG and IgM in different types of Influenza viruses. The array was exposed fetal bovine serum (FBS) and pooled normal human serum (PNHS) and after preparation the diluted samples were added in the arrays and used for IgG and IgM antibodies detection and AIR images were analyzed and the results were compared with Enzyme-linked immunosorbent assay (ELISA) assay results. The obtained results show the array was initially acceptable for using commercial mono- and poly-clonal antibodies (Sino Biological) doped in PNHS produced strong signals to circulating different types of Influenza viruses and Responses to SARS-CoV-2 antigens effectively differentiated between serum samples from positive and negative COVID-19 cases, with generally good correlation to ELISA data [33]. Furthermore, Wu et al. [34], have demonstrated a new method for isolation fully human single-domain antibodies and their application for screening of antibodies against SARS-CoV-2. They explained development of a phage-displayed single-domain antibody library by grafting naive complementarity-determining regions (CDRs) into framework regions of a human germline immunoglobulin heavy chain variable region (IGHV) allele. Panning this library against SARS-CoV-2 RBD and S1 subunit identified fully human single-domain antibodies targeting five distinct epitopes on SARS-CoV-2 RBD with subnanomolar to low nanomolar affinities. Some of these antibodies neutralize SARS-CoV-2 by targeting a cryptic epitope located in the spike trimeric interface [34].
Fig. 4.
Schematic performance of rapid SARS-CoV-2 IgM-IgG combined antibody detection biosensor. M line detect the IgM and G line detect IgG in samples [27].
3.4. Enzyme based assays
Enzymes, as a biological molecule have the potential to be used for COVID-19 detection in different roles like analyte or detection label. Several different types of methods can be used to fabricate of biosensor assays using enzymes. A novel enzyme-based immunodetection assay is introduced by Conzelmann et al. [35], directly quantifies the amount of de novo synthesized viral spike protein within fixed and permeabilized cells. This method is an in-cell ELISA method that provides a rapid and quantitative detection of SARS-CoV-2 infection in microtiter format, regardless of the virus isolate or target cell culture. It follows the established method of performing ELISA assays and requires low-cost instrumentation. Using the in-cell ELISA has some advantages, including the determination of TCID50 of virus stocks, antiviral efficiencies (IC50 values) of drugs or neutralizing activity of sera and is a promising alternative to study SARS-CoV-2 infection and inhibition and may facilitate future research. In this method, SARS-CoV-2 infected cells were prepared and after washing cells once with buffer, cells are stained with 1:5000 diluted mouse anti-SARS-CoV-2 S protein antibody in specific buffer for 1 h. After sample preparation, the ELISA detection was done and the results shows that S protein specific in-cell ELISA quantifies SARS-CoV-2 infection rates of different cell lines and allows to rapidly screen and determine the potency of antiviral compounds. Thus, this method represents a trustable, rapid, accessible and convenient alternative to the current laboratory techniques studying SARS-CoV-2 and will facilitate the future research and drug development on COVID-19 [35]. In another study, Xiang et al. [36], have compared two methods for SARS-CoV-2 detection. In the first method, they collected 63 samples and used ELISA for IgG and IgM antibodies detection. In the second method, they have collected 91 plasma samples and used the colloidal gold-immunochromatographic assay (GICA) for detection. They used the IgG/IMG antibody ELISA kits and IgG/IgM antibody GICA kits for first and second method respectively. All the serum samples used for this study were obtained from confirmed COVID-19 patients (diagnosed by qRT-PCR). The results showed the sensitivity of the combined ELISA IgM and IgG detection was 55/63 (87.3%), The sensitivity of the combined GICA IgM and IgG detection was 75/91 (82.4%), and the healthy controls were negative. The authors have mentioned that the healthy controls are all negative and the specificity is very good. The results showed that ELISA and GICA for specific IgM and IgG antibodies are serological assays and they can offer a high-throughput alternative that enables for uniform tests for all suspected patients, and can facilitate more complete identification of infected cases and avoidance of unnecessary cross infection among unselected patients. The applied method is fast and simple and does not need huge clinical diagnosis instruments [36].
3.5. Biomarker based assays
A biological marker (Biomarker) is a characteristic that is objectively measured and evaluated as an indicator of normal biological process, pathogenic process or pharmacologic responses to a therapeutic intervention [37]. The published reports show that biomarkers can be used in biosensor assays for COVID-19 detection. A device that is fabricated with Shan et al. [38] is focused on nonmaterial-based sensor array with multiplexed capabilities for detection and monitoring of VOC biomarkers mixture from exhaled breath. As VOC is not a specific COVID-19 biomarker and it can be detect in all types of lung infection, the designed array sensor is not specifically designed for COVID-19 and can detect all type of lung infections including COVID-19. They used a developed breath device composed of a nanomaterial-based hybrid sensor array with multiplexed detection capabilities that can detect disease specific biomarkers from exhaled breath. The array is consisted of eight gold nanoparticles working modified on substrate and the sampling is doing with blowing into the device for 2–3 s. The tests set and training data indicate 76% and 94% accuracy in control group respectively and 95% and 90% accuracy COVID-19 suffering patients and patients with other lung infections [38].
4. SARS-CoV-2 detection based on applied analytical methods
Another classification for coronavirus detection is based on the tools and methods that are used for detection part. There are a wide range of detection methods that are used in published reports in different fields including spectroscopic and electrochemical methods. The main methods that are used for SARS-CoV-2 virus detection are reviewed as follows.
4.1. Polymerase chain reaction (PCR)
DNA sequencing methods are slow and relatively expensive methods. Therefore, these make significant limitation for extensive application for identifying SARS-CoV-2 specially during a worldwide pandemic that requires rapid and low cost disease diagnosis. On the other hand, the polymerase chain reaction (PCR) detection method is cheaper, easier and has a short turnaround time. Therefore, many institutes and companies have developed SARS-CoV-2 PCR detection for virus detection and controlling further spreading PCR [[39], [40], [41], [42]].
The PCR technology has made a revolution in nucleic acids analysis, allowing it on large laboratories scale. Kary B Mullis who designed and developed PCR technology was awarded by the Nobel Prize in Chemistry in 1993 [43]. PCR is an enzymatic method to produce numerous copies of a gene by separating the two strands of the DNA containing the gene segment, marking its location with a primer, and using a DNA polymerase to assemble a copy alongside each segment and continuously copy the copies [44]. During past years, this technology has been extensively expanded with new variants such as isothermal PCR, real time PCR and so forth [45]. Recently, PCR is applicable to highly sensitive detection of various pathogens in clinical and environmental samples such as food authenticity and food control, being routinely applied to the detection/identification of allergens, microorganisms, pathogen and disease detection and control [[46], [47], [48]]. One of the most useful applications of PCR is virus detection and this method is the most applied method for the SARS-CoV-2 virus detection. One of the methods that is commercially available for COVID-19 diagnosis is real-time reverse transcriptase-PCR (RT-PCR) that is including two methods: one step or two step assays. The difference between one step and two step assay is in number of tubes and buffers. One-step RT-PCR kit utilizes a single tube and buffer for RT and PCR steps, but two-step RT-PCR assay use two tube to perform RT and PCR separately in different tubes with independently optimized buffers. One-step RT-PCR assays is rapid, reproducible, and therefor works better for COVID-19 diagnosis during the present pandemic because they require limited sample management and reduce the risk of cross-contamination and human errors [49]. Two-step RT-PCR assays, on the other hand, are generally more flexible and tunable and offer superior sensitivity and low detection limit [50]. There are some studies with both methods for SARS-CoV-2 detection. At present, the novel coronavirus is mostly detected using the PCR technology [[51], [52], [53], [54], [55]]. Chu et al. [56] reported two one-step quantitative real time RT-PCR assays to detect two different region (ORF1b and N) of the viral genome. They have designed the primer and probe sets to react with SARS-CoV-2 and its closed related family, SARS coronavirus to avoid the possible scenario in which the genetic diversity of SARS-CoV-2 is much more diverse than expected. The assays were evaluated using a panel of positive and negative control samples. The obtained results in this study, showed using RNA extracted from cells infected by SARS coronavirus as a positive control, the designed assays were illustrated a dynamic range of at least seven orders of magnitude (2 × 10−4 to 2000 TCID50/reaction) and the detection limit of these assays were shown to be below 10 copies per reaction using DNA plasmids as positive standards. The accuracy of method is 100% and all the negative and positive obtained results are correct [56]. In another study, Azzi et al. [57] have found that saliva is a reliable tool to detect SARS-CoV-2. They have analyzed salivary samples of COVID-19 patients by RT-PCR and compared the results with their clinical and laboratory data. Twenty-five subjects were recruited in this study, 17 males and 8 females. Cardiovascular and/or dysmetabolic disorders were observed in 65.22% of cases. They collected the data including age, sex, comorbidities and drugs, lactate dehydrogenase (LDH) and ultrasensitive reactive C protein values in the same day that a salivary swab was collected. All the samples tested positive for the presence of SARS-CoV-2, while there was an inverse association between LDH and Ct values. Two patients showed positive salivary results on the same days when their pharyngeal or respiratory swabs showed conversion [57].
4.1.1. Droplet Digital PCR (ddPCR)
Whereas the sensitivity of RT-qPCR assays are reported to vary from 30% to 60%, some newly developed PCR methods with better performance are reported in the literature. One of them is Droplet Digital PCR (ddPCR) method which is a method for performing digital PCR based on water-in-oil (W/O) emulsion droplet technology. Digital PCR was conceptualized in 1990s and is based on dilation and fractionation of a sample in to hundreds to millions of separate reaction chambers that each chamber may contain a single or more copies of target sequence while others contain no target sequence and counting the positive partitions versus negative partitions. Positive droplets are read as droplets that contain at least a single copy or more copies of the target gene exhibiting increased fluorescence, while negative droplets have no copies of the target gene and exhibit no fluorescence. The ddPCR technology uses reagents and workflows similar to those used for most standard TaqMan probe-based assays. The sample is subsequently divided into thousands of single nanoliter-sized droplets (20,000 or more based on system) in a combination of water-in-oil emulsion droplet technology and microfluidics. Each Single droplet may or may not contain one or more targets sequence. Each droplet is amplified using a thermal cycler to end-point and then the droplets are placed in a droplet reader that analyzes the droplets individually based on a two-color detection system depending on the used fluorescent dye. In the next step, the fluorescent signals are measured for individual droplets in various channels to differentiate positive droplets from negative droplets. The massive sample partitioning is a key aspect of the ddPCR technique [58,59]. Both dPCR and ddPCR techniques are used for SARS-CoV-2 detection recently. For instance, Dong et al. [60], have analyzed a total of 196 clinical pharyngeal swab samples from 103 suspected patients, 77 close contacts and 16 supposed convalescents by RT-dPCR and compared their results with RT-qPCR. They are applied one step RT-dPCR method for detection open reading frame 1 ab (ORF1ab), nucleocapsid protein (N) and envelope protein (E) gene of SARS-CoV-2. They found that 39.4 °C fever suspected patients, 19 (19/25) negative and 42 (42/49) equivocal tested by RT-qPCR were positive according to RT-dPCR. The sensitivity of SARS-CoV-2 detection was significantly improved from 28.2% by RT-qPCR to 87.4% by RT-dPCR. They also compared 29 close contacts samples and the results showed 16 (16/17) equivocal and 1 negative tested by RT-qPCR were positive according to RT-dPCR, that applied the RT-qPCR is missing a lot of asymptomatic patients. The overall sensitivity, specificity and diagnostic accuracy of RT-dPCR were 91%, 100% and 93%, respectively. The final results showed that the dPCR is more sensitive and has better detection limit than RT-PCR [60]. Furthermore, Yu et al. [61], have reported that show the reverse transcription–PCR is sensitive and reliable, but ddPCR performed better in detecting low-viral-load samples. They used 323 samples from 76 COVID-19 confirmed patients were analyzed with ddPCR and RT-PCR based 2 target genes (ORF1ab and N). The samples were collected from Nasal swabs, throat swabs, sputum, blood, and urine samples. The obtained results showed the 95 samples that tested positive by both methods and the detailed results showed that sputum is a better indicator of viral replication in the body than throat and nasal swabs, and the viral load of sputum samples tends to increase and then decrease during the course of the disease. In addition, quantitative monitoring of viral load in lower respiratory tract samples could help to evaluated disease progression, especially in case of low viral loads [61].
4.1.2. Circle-to-circle amplification (C2CA) sensor
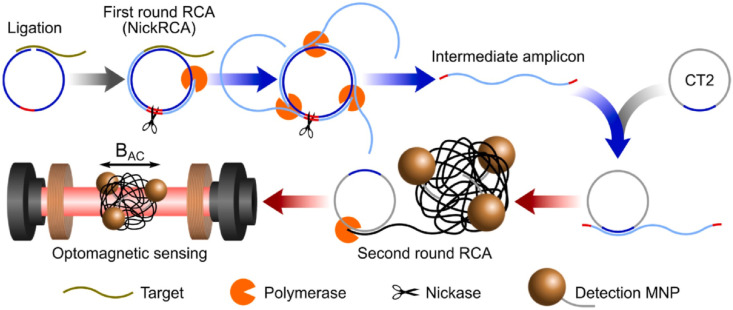
Circle-to-circle amplification (C2CA) is a specific and precise nucleic acid amplification method that is consisted of more than one round of padlock probe ligation and rolling circle amplification (RCA). Although this method contains several step by step operation processes, C2CA shows high amplification efficiency and very negligible increase of false-positive risk. The homogeneous C2CA amplification eliminates the additional monomerization and ligation steps after the first round that is required in normal C2CA of RCA and makes one step process. Tian et al. [62] illustrated a homogeneous and isothermal nucleic acid quantification strategy based on C2CA and optomagnetic analysis of magnetic nanoparticle (MNP) assembly. Fig. 5 shows the proposed methodology after the first round. As can be seen in this figure, the second round of RCA produces amplicon coils that anneal to detection probes grafted onto MNPs, resulting in MNP assembly that can be detected in real-time using an optomagnetic sensor. The new applied method for detection of synthetic complementary DNA of SARS-CoV-2 and RNA-dependent RNA polymerase (RdRpa) coding sequence, achieving a subfemto molar (0.4 fM) detection limit with a dynamic detection range of 3 orders of magnitude and 100% sensitivity. A mathematical model was set up and validated to predict the assay performance. Moreover, the proposed method was specific to distinguish SARS-CoV and SARS-CoV-2 sequences with high similarity [62].
Fig. 5.
Schematic performance of homogeneous C2CA. In the first round of RCA, polymerases act tandemly to generate intermediate amplicons. Intermediate amplicons anneal to CT2 for the second round of RCA, generating amplicon coils that lead to the assembly of MNPs. Sequence to a ligation step, all processes of amplification, hybridization, and detection take place simultaneously on-chip at 37 °C [62].
4.2. Enzyme-linked immunosorbent assay (ELISA)
It is well-known that immunological assays can be used for biological detections owing to the capability reducing the time and cost of analysis. The enzyme-linked immunesorbent assay (ELISA), stands out among immunological assays because of its specificity, simplicity and sensitivity, and other advantages. Basically, ELISA is an immunological technique that involves an enzyme that is a protein for catalyzing a biochemical reaction, to detect the presence of an antibody or an antigen in a sample [63]. There are two major methods for ELISA detection that are called, indirect and the sandwich ELISA. The indirect ELISA uses two antibodies that one of them is specific to the antigen and the other one is coupled to an enzyme and this second antibody gives the assay its ‘‘enzyme-linked’’ name [64]. In the sandwich ELISA the antigen is bound between two antibodies named the capture antibody and the detection antibody respectively. The detection antibody can be coupled to an enzyme or can bind the conjugate (enzyme-linked antibody) that will produce the biochemical reaction [65]. Among other applications, ELISA method has found application for detecting of COVID-19 during the current pandemic. For example, Zhao et al. [29], have collected the IgM and IgG antibodies form the plasma samples of 173 patients with positive SARS-CoV-2 infection and tested them using ELISA kits. In summary, the ELISA for total antibody detection was developed based on a double-antigen sandwich immunoassay (Ab-ELISA), using mammalian cell–expressed recombinant antigens containing the receptor binding domain (RBD) of the spike protein of SARS-CoV-2 as the immobilized and horseradish peroxidase–conjugated antigen. The IgM μ-chain capture method (IgM-ELISA) was used to detect the IgM antibodies, using the same HRP-conjugate RBD antigen as the Ab-ELISA. The IgG antibodies were tested using an indirect ELISA kit (IgG-ELISA) based on a recombinant nucleoprotein. The specificity of the assays for Ab, IgM, and IgG was determined to be 99.1% (211/213), 98.6% (210/213), and 99.0% (195/197), respectively, by testing the samples collected from healthy individuals before the SARS-CoV-2 outbreak. The seroconversion rates for Ab, IgM, and IgG were 93.1%, 82.7%, and 64.7%, respectively for 173 positive SARS-CoV-2 infection patients. The obtained results showed that even in the early stage of disease (first week), the combining RNA and antibody detection significantly improved the sensitivity of pathogenic diagnosis for COVID-19 (P = 0.007). They believe the antibody tests have improved the diagnosis value in combined with RNS routine tests and provide strong empirical support for serological testing of COVID-19 patients [29]. Furthermore, Guo et al. [66] have applied antibody, as analyte for detecting of COVID-19. To achieve this goal, an indirect ELISA protocol was developed for detecting IgM, IgA and IgG antibodies against SARS-CoV-2 using purified rNPs as coating antigens, and finally the concentration of the coated rNPs and plasma dilutions are optimized. 208 plasma samples were collected from 82 confirmed and 58 probable cases (qPCR negative, but with typical manifestation) and the average duration detection for IgM, IgA and IgG was 5, 5 and 14 days after symptom onset, respectively. The positive rates of IgM antibodies were 75.6% and 93.1%, respectively and the positive detection rate was significantly increased (98.6%) by conjugating the PCR and ELISA method. The final results showed the developed method can be used as a decision aid method for COVID-19 diagnosis [66]. In another approach, Li et al. [67] have used the sandwich ELISA kit to help the vaccine developers with monitoring the virus expression level in the cultures during the optimization process of manufacturing parameters. They have generated specific antibodies that are essential for accurate serological diagnostic tool of COVID-19. Li and coworkers have also reported that the polyclonal and monoclonal antibodies were generated by immunizing animals with synthetic peptides corresponding to the different areas of Nucleoprotein (N) of COVID-19 [67].
4.3. Mass spectrometry (MS)
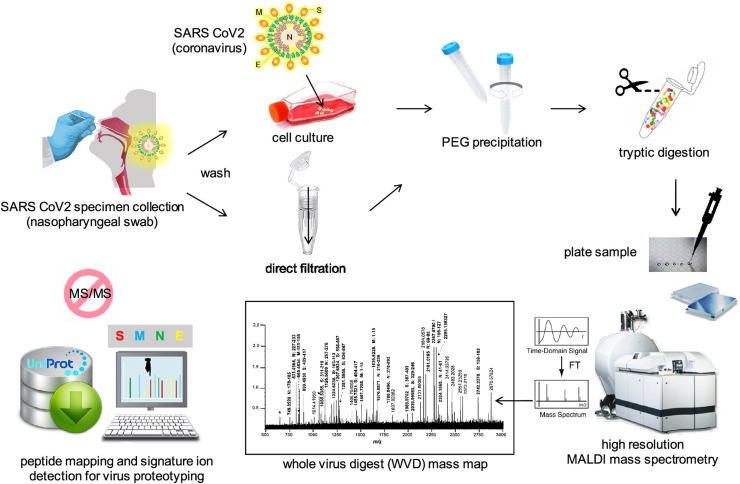
Approaches conjugating immunoassay methodology with mass spectrometric (MS) detection have been developed to cover some of the important limitations of these techniques. However, the high molecular weight and substantial heterogeneity create some additional challenges for the MS analysis of biological molecules. Therefore, some conjugation reagents were designed to improve the ionization and detection of biological molecules [68]. It seems MS method is a suitable alternative for PCR detection method during the pandemic. This method has been reported as a diagnostic method for SARS-CoV-2 detection that complement genomic information and increase our current knowledge of the COVID-19 disease using gargle solution samples of COVID-19 patients. Ihling el al., identified peptides originating from SARS-CoV-2 nucleoprotein in gargle solution samples. They planned to use this method for providing a robust, sensitive and reliable MS-based method for COVID-19 detection [69]. In another study, Gouveia and coworkers proposed the use of tandem mass spectrometry to detect SARS-CoV-2 marker peptides in nasopharyngeal swabs [70]. They showed that the signal from the viruses in some samples is low and can be overlooked when interpreting shotgun proteomic data acquired on a restricted window of the peptidome landscape. For solving this problem, Gouveia et al. [70], simulated the detection process via spiking the nasopharyngeal swabs with different quantities of purified SARS-CoV-2 viral material that were used to develop a nano LC−MS/MS acquisition method and then the results were then successfully applied on COVID-19 clinical samples. The authors argued that peptides named ADETQALPQR and GFYAQGSR from the nucleocapsid protein show the maximum increase of signal intensity and their elution can be obtained within a 3 min time period in the tested conditions. Additionally, Dollman et al. [71] have applied a high resolution MS approach to detect and characterize SARS-CoV-2 in cell cultured and nasopharyngeal swab specimens. The applied strategy is shown in Fig. 6 which is based on their previous works for influenza virus detection. The researchers have collected SARS-CoV-2 virus specimens by swabs and after washing and growing in cell culture and other preparation methods the virus could be detected via MS. They have detected and assigned Peptide ions for three of the most abundant structural viral proteins, membrane, including nucleocapid, and spike by virtue of the high resolution and mass accuracy within the mass maps of whole virus digests, without the need for tandem mass spectrometry (MS/MS). The MALDI-MS based approaches offer high sample throughput and speed, compared with those of LC−MS strategies, and detection limits at some 105 copies, or orders of magnitude less with selected ion monitoring that compete favorably with conventional RT-PCR strategies. The detection of signature peptides unique to SARS-CoV-2 enables its unambiguous detection from the influenza virus [71].
Fig. 6.
Schematic of the approach used to detect SARS-CoV-2 virus using high resolution mass spectrometry [71].
4.4. Chemiluminescence based immunoassay (CLIA)
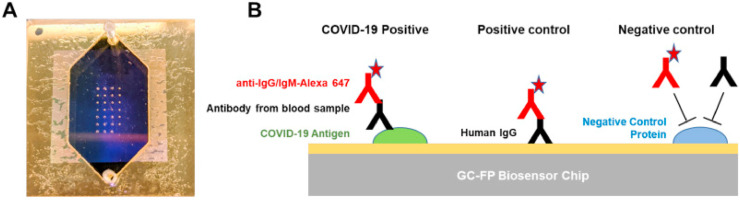
Chemiluminescence based immunoassay (CLIA) has been greatly developed in the past decades due to its good sensitivity and specificity and conjugated with newly-developed technologies. CLIA has been used for detecting several biomolecules, including antibody, aptamer, DNA and some other biomolecules [72]. In this method, the light emission arises during the course of a chemical reaction. When molecules formed in an electronically excited state and then decay to the ground state, light emitted. Recently, this method is using for successful detection of COVID-19 [73]. For instance, Cai et al. [74] have developed a peptide-based magnetic chemiluminescence enzyme immunoassay (MCLIA) could detect IgG and IgM antibodies by evaluating the given serum from healthy and infected patients for pathogens other than SARS-CoV-2. They synthesized twenty peptides deduced from the genomic sequence as candidate antigens from the orf1a/b, S, and N proteins. Each kind of peptide was labeled with biotin, and purified and subsequently bound to streptavidin-coated magnetic beads. At the same time, serum samples were mixed with the beads carrying corresponding peptides and then, the beads were subjected to antibody conjugation and reacted with the substrate to form the antibody assay. To evaluate assay performance, they detected IgG and IgM antibodies in the confirmed patient's serum. The positive rate of IgG and IgM was 71.4% and 57.2%, respectively. The researcher concluded that combination of immunoassay and RT-PCR might enhance the diagnostic accuracy of COVID-19 [74]. Furthermore, Diao et al. [75] developed an accurate, rapid and simple antibody assay method using the viral antigen for diagnosis of SARS-CoV-2 infection. They designed a fluorescence immunochromatographic assay for detecting nucleocapsid protein of SARS-CoV-2 in nasopharyngeal swab sample and urine. The experiments were completed in the double-blind evaluation clinical trial with the nucleic acid test as the golden standard during 10 min. They have found the nucleic acid test positive rate was 87% and 64%, depending on the CT value, the positive rate of N antigen detection of SARS-CoV-2 was 59% and 63% separately and 100% of nucleocapsid protein positive and negative participants accord with nucleic acid test. Moreover, the detection sensitivity of N antigen is 100%, which greatly reduced the false positive rate of nucleic acid detection. As an advantage of this method, the earliest patient after 3 days of fever can be identified by this method. The obtained results showed that this N antigen detection method not only guarantee early diagnosis in hospitals, but also can be used for large-scale screening in community [75]. Additionally, Zhang et al. [76], have used an automated CLIA to evaluate serum IgM and IgG antibodies of SARS-CoV-2 for monitoring the dynamic process of antibody production during disease progression and the value of antibody detection for the laboratory diagnosis of COVID-19. Using the CLIA Detection Kit, IgM and IgG antibodies were detected within 30 min after that the sample is loaded. The magnetic particle-coated antigens were used, including the mixed recombinant SARS-CoV-2 full length spike protein S1 and the full length nucleocapsid protein N, for better sensitivity achievement. The results showed a positive correlation between the amount of detected SARS-CoV-2 IgM or IgG antibody in a sample, and the concentration of the SARS-CoV-2 IgM or IgG antibody (AU/mL) was automatically calculated according to RLU and built-in calibration curve. A concentration of 10.0 AU/mL was regarded as positive. Their results showed the detection of specific SARS-CoV-2 antibodies, as a useful complement to nucleic acid detection, will present a rapid, comprehensive and accurate diagnostic approach to effectively distinguish between COVID-19 and other types of influenza virus disease [76]. Furthermore, a new multiplexed grating-coupled fluorescent plasmonics (GC-FP) platform was designed and fabricated by Cady et al. [77] for antibodies detection and quantification against COVID-19 in human blood serum and dried blood spot samples as shown in Fig. 7 . As seen in this figure, GC-FP chips were printed of spots of target and control antigens/proteins using microarray printer. The GC-FP platform measures antibody-antigen binding interactions for multiple targets in a single sample, and has 100% selectivity and sensitivity (n = 23) when measuring serum IgG levels against three COVID-19 antigens (spike S1, spike S1S2, and the nucleocapsid protein). The designed biosensor platform illustrates response for diluted serum samples as low as 1:1600 and results were highly correlated with two commercial COVID-19 antibody detection methods, including ELISA and a Luminex-based microsphere immunoassay. The GC-FP method was also used for more complex samples like dried blood spot samples (n = 63) and showed 100% selectivity and 87.6% sensitivity for diagnosing prior COVID-19 infection. The test was also evaluated for detection of multiple immunoglobulin isotypes with successful detection of IgM, IgG and IgA antibody-antigen interactions. Finally, a machine learning approach was developed to accurately score patient samples for prior COVID-19 infection, using antibody binding data for all three COVID-19 antigens, which were used in the test [77].
Fig. 7.
A) GC-FP biosensor chip shown with gasket and fluidic cover attached. B) Schematic illustration of GC-FP biosensor chip including Anti-IgG as sample capture (first line), IgG as positive control (second line) and controlling protein as negative control (for third line) [77].
4.5. Lateral flow immunoassay (LFIA)
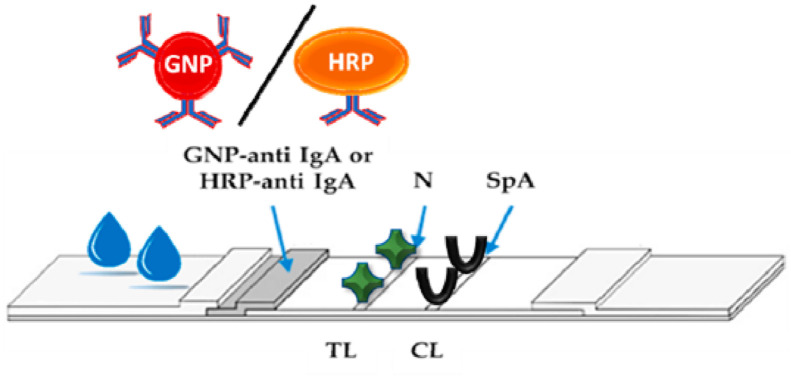
The lateral flow immunoassay (LFIA) is an immune-chromatographic assay (ICA) that is designed to detect the presence of an analyte by a specific labeled-antigen or labeled-antibody that is commercially available for a wide array of targets, including infectious agents, hormones, drugs, pesticides and mycotoxins [78]. The best known and first commercial developed LFIA was the pregnancy test for the detection of human chorionic gonadotropin (HCG) hormone in the urine sample. This method is rapid, simple, low cost, portable, highly sensitive and specific [79]. Point-of-care devices based on LFIA principle have been recently aimed at detecting the serologic response to the infection by specifically and separately targeting immunoglobulins belonging to the M and G classes in the blood serums [80]. The recent reports have shown that the multi-target LFIA can provide the specific and sensitive detection of total antibodies to SARS-COV-2 [81]. For instance, shown in Fig. 8 , a dual optical/chemiluminescence format of a LFIA immunosensor for IgA detection in serum and saliva was prepared by Roda et al. [82]. As seen in this figure, a labeled anti-human IgA determined the bound IgA fraction and a dual colorimetric and chemiluminescence could detect the IgA antibody. The researchers claimed that the applied method is affordable and ultrasensitive for IgA detection in COVID-19 patients. A simple smartphone-camera-based device was used to measure the color signal provided by nanogold-labeled anti-human IgA. For chemiluminescence transduction, a contact imaging portable device based on cooled CCD was applied and measured the light signal resulting from the reaction of the HRP-labeled anti-human IgA with a H2O2/luminol/enhancers substrate. Totally, 25 serum and 9 saliva samples from infected and/or recovered individuals were analyzed by the colorimetric LFIA. By switching to CL detection, the same immunosensor exhibited higher detection capability, revealing the presence of salivary IgA in infected individuals [82].
Fig. 8.
Schematic illustration of LFIA strip IgA antibody detection biosensor [82].
A research work focused on LFIA-based biosensor for COVID-19 detection was conducted by Kim et al. [83]. They utilized phage technology to produce four SARS-CoV-2 nucleocapsid protein (NP)-specific single-chain variable fragment-crystallizable fragment (scFv-Fc) fusion antibodies. It is evident that scFv-Fc antibodies can specifically and efficiently bind to SARS-CoV-2 NP antigen, but not to NPs of other coronaviruses and based on this fact, they monitored three diagnostic antibody pairs for use on a cellulose nanobead based LFIA platform. The results after optimization showed that the prepared biosensor can detect SARS-CoV-2 virus at levels as low as 1 × 104 pfu/reaction. Moreover, there was no cross-reactivity with NPs of SARS-Co-V, MERS-CoV, influenza virus, or negative control nasal swab specimens. In addition, the results indicated that the LFIA biosensor could successfully distinguish between SARS-CoV-2 positive and negative samples. The detection limit of LFIA biosensor would meet the conditions for the clinical use through optimization [83]. The effect of applying nanoparticles on sensitivity and selectivity of LFIA detection is investigated by Huang et al. [26]. They have introduced a simple and time consuming method for antibody measurement using a small amount of sample (10–20 μL serum for each test). The SARS-CoV-2 nucleoprotein was coated on an analytical membrane for preparing colloidal gold nanoparticle-based lateral-flow assay (AuNP-LFA) strips, as sample capture, and antihuman IgM was conjugated with AuNPs as detecting reporter. To achieve the best performance of AuNP-LFA, the pH value and the amount of antihuman IgM was optimized, and the performance of AuNP-LFA was evaluated by testing serum samples of COVID-19 patients and healthy individuals. The results showed the sensitivity and specificity of AuNP-LFA are 100% and 93.3%, respectively and have good agreement with PCR (R = 0.872). In addition there was no interference from other viruses thrombocytopenia syndrome virus (SFTSV) and dengue virus (DFV). The results indicated that the applied method is rapid and can be used for portable detection of the IgM antibody for diagnosing the SARS-CoV-2 virus [26].
4.5.1. Surface-enhanced Raman scattering-based lateral flow immunoassay (SERS-LFIA)
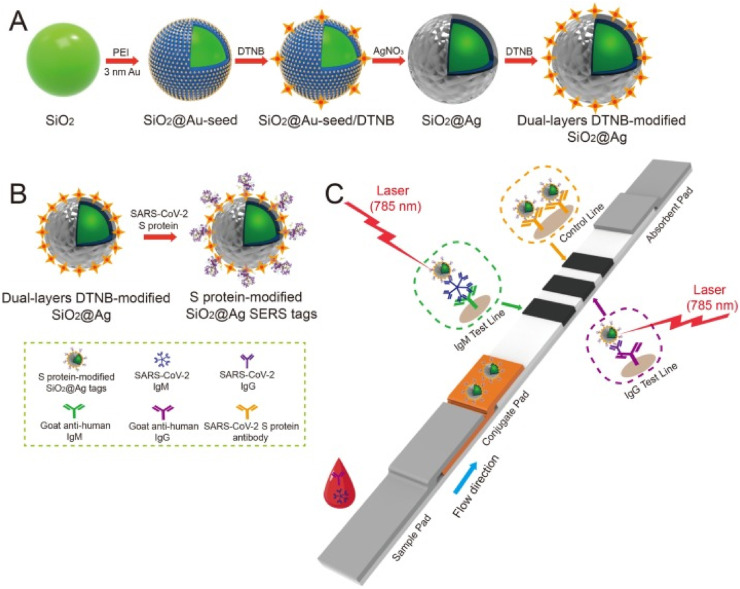
Surface-enhanced Raman Scattering (SERS)-based immunoassay is a novel rapid detection technology that combines SERS labelling and antigen-antibody immune interaction and typically uses gold or silver nanoparticles to enhance the Raman scattering signals of active molecules [84]. They have been combined with an immunoassay and have found extensive applications in biomedical diagnostics. The SERS is a sensitive method but required multiple washing and incubation steps and cannot be used for on-site detection. In contrast, lateral flow assay configurations are simple, rapid, and low cost. Thus, they are suitable for rapid on-site detection of targeted molecules [85]. Inasmuch as the SERS-based lateral flow immunoassays (SERS-LFIA) provide the combination of high sensitivity of SERS with the facile of lateral flow assay, a wide range of SERS-LFIA was described recently [86]. This method is used for diagnosis of COVID-19 via IgG/IgM antibodies detection. Liu et al. [28] have designed and illustrated a SERS-LFIA for simultaneous high sensitive detection of IgM/IgG antibodies. As can be seen in Fig. 9 , the SERS tags were labeled with dual layers of Raman dye and they were fabricated by coating a complete Ag shell onto the SiO2 core. Sequence to Ag coating, anti-human IgM and IgG were immobilized onto the two test lines of the strip to capture the formed SiO2@Ag-spike (S) protein-anti-SARS-CoV-2 IgM/IgG immunocomplexes. They used the mentioned method for 19 negative serum samples from COVID-19 patients and 49 negative serum samples from healthy people to show the capability of proposed immunoassay. Their final results showed the limit of detection (LOD) of SERS-LFIA is 800 times higher than that of standard Au nanoparticle-based LFIA for target IgM and IgG [28].
Fig. 9.
Schematic diagram of the preparation of the dual-layers DTNB-modified SiO2@Ag NPs. (B) Preparation of SARS-CoV-2 S protein-modified SiO2@Ag SERS tags. (C) Operating principle of the high-sensitivity and simultaneous analysis of anti-SARS-CoV-2 IgM/IgG via the SERS-LFIA strip [28].
4.6. Loop mediated isothermal amplification (LAMP)
Loop-mediated isothermal amplification (LAMP) was designed and fabricated for DNA detection by Notomi in 2000 [87]. LAMP is simple, still fast, selective and efficient virus detection method that amplifies DNA using DNA polymerase being carried out in isothermal conditions without complex lab equipment requirement [88]. The LAMP reaction generally proceeds in a constant temperature, and the target DNA can be amplified in 30 min [89]. In this method, four sets of primers including two forward inner primers and two backward inner primers are used to recognize six distinct sequences on the target DNA of virus and designed specifically for virus detection [87]. The two forward inner primers and two backward inner primers are selected for the first and second stages of the process respectively. In novel approaches, five or six primer sets are used for achieving more selective and efficient virus detection and shorter detection time [90,91]. Recently, this method is used for COVID-19 detection in several studies [92]. For example, Zhang et al. [93] have reported a method to identify SARS-CoV-2 virus RNA from purified RNA or cell lysis by LAMP method, using a visual, colorimetric detection. Sequent optimizing the assay design, the experiments used for real samples of COVID-19 positive patients in Wuhan. Additionally, the experiments were verified using RNA samples purified from respiratory swabs collected from COVID-19 patients with equivalent performance to a commercial RT-qPCR test that only heating and visual inspection. Researchers believe that combination between a quick sample preparation method and an easy detection process may realize the development of portable, field detection in addition to a rapid screening for point-of-need testing applications [93]. In another study, Huang et al. [89] have prepared a COVID-19 detection kit based on the RT-LAMP technology to achieve the detection of SARS-CoV-2 in 30 min. They prepared 4 different sets of LAMP primers consisted of 6 primers in each set, for targeting the viral RNA of SARS-CoV-2 in the regions of orf1ab, S gene and N gene. Huang and coworkers have used a colorimetric change at a constant 65 °C to report the results that enables the outcome of viral RNA amplification to be read by the naked eyes without the need for a dedicated instrument with an extra cost. The sensitivity was 80 copies of viral RNA per ml in a sample. All the results were obtained form 16 clinical samples with 8 positives and 8 negatives in China and results were compared with the conventional RT-qPCR. The researcher pointed out the developed method could provide a rapid and large scale screening medical test and patients could be diagnosed in the early stages [89]. Additionally, Yu et al. [94], introduced a rapid detection method for COVID-19 coronavirus using a reverse transcriptional-LAMP diagnostic platform. They have developed an isothermal RT-LAMP method for COVID-19 to amplify a fragment of the ORF1ab gene using 6 primers. They have certified the species specificity by comparing the target sequences with other viral genomes including 9 corona and 2 influenza viruses using NCBI BLAST tool. They established and optimized the method and after obtaining the results, compared the sensitivity of results with RT-qPCR results. The 43 samples of their method initially detected with RT-qPCR and the results revealed that 97.6% (42/43) of collected samples showed consistent signal after incubation with RNA (0.2–47 ng/μL) during 40 min. They repeated the experiments multiple times with random positive signal, to confirm the results [94]. Yan et al. [95], have developed a RT-LAMP assay to detect SARS-CoV-2 in individuals with COVID-19 for monitoring suspected patients, close contacts and high-risk groups. They designed five sets of primers that target the orf1ab gene and the spike gene for optimization of the assay. The evaluation for sensitivity and specificity of detection was applied using real time turbidity monitoring and visual observation. They used a commercial real time RT-PCR kit as the reference standard for the RT-LAMP. The primer sets orf1ab-4 and S-123 amplified the genes in the shortest times. The mean (±SD) times were 18 ± 1.32 min and 20 ± 1.80 min, respectively, and the optimum reaction temperature was 63 °C. The sensitivities were 20 and 200 copies per reaction with primer sets orf1ab-4 and S-123, respectively. This assay showed no cross-reactivity with 60 other respiratory pathogens [95]. In another approach, a method based on LAMP with two stage isothermal amplification, that called Penn-RAMP, is introduced by El-Tholoth et al. [96]. Two different tests were carried out in closed tubes with either fluorescence or colorimetric detection. The incubation was done without any thermal cycling and amplification process was monitored in real time with fluorescent dye. Two type of targets were used in this study. The first one obtained and purified form complete genome sequences of various COVID-19 and the other one was synthesized DNA containing the targeted sequence to mimic the COVID-19 target. Their results showed that RAMP has 10 times higher sensitivity than LAMP and RT-PCR if the purified targets are tested and 100 times better sensitivity than LAMP and RT-PCR if rapidly prepared sample mimics are tested. However, the researcher mentioned due to fortunate scarcity of COVID-19 infections in the USA, they were not able to test the assays and methods with patient real samples. Thus, they tested the spiked sample collection with inactivated virus particles [96]. Furthermore, Baek et al. [97] were developed a reverse transcription LAMP (RT-LAMP) assay for the specific detection of SARS-CoV-2. The primer sets for RT-LAMP assay were designed to target the nucleocapsid gene of the viral RNA, and showed a detection limit of 100 RNA copies close to that of qRT-PCR. The assay showed the capability of a rapid detection span of 30 min combined with the colorimetric visualization. The baek and coworkers indicated that there was no interference for the other type of coronaviruses, such as HCoV-229E, HCoV-NL63, HCoV-OC43, and MERS-CoV as well as human infectious influenza viruses (type B, H1N1pdm, H3N2, H5N1, H5N6, H5N8, and H7N9), and other respiratory disease-causing viruses (RSVA, RSVB, ADV, PIV, MPV, and HRV) in offered method and the obtained results have high agreement to the qRT-PCR results [97].
4.7. Localized plasmon resonance (LSPR)
Plasmonic biosensors have shown excellent performance for bio-detection because they use an enhanced electric field in the penetration depth and they are extremely sensitive to the changes in the refractive index of the medium [98]. Plasmonic biosensors are typically worked by amplifying the analyte signal, including Plasmonic enhanced fluorescence (PEF) [99], surface enhanced Raman Scattering (SERS) [84], surface-enhanced infrared absorption (SEIRA) [100], and plasmonic-based colorimetric analysis [101]. In addition, the operation of plasmonic biosensors can be based on detecting changes in the relevant frequency of the surface plasmon resonance in the presence of target analyte like surface plasmon resonance (SPR) and localized plasmon resonance (LSPR) biosensors, which are commercially available these days. LSPR is an optical phenomenon generated by a light wave trapped within conductive nanoparticles (NPs) smaller than the wavelength of light because of interactions between the incident light and surface electrons in a conduction band. The noble metal nanostructure absorbs photon energy to produce obvious extinction characteristics and near-field enhancement characteristics. The resonance wavelength is sensitive to the refractive index change at the interface between the nanostructure and surrounding media when LSPR occurs [102]. Currently, this method is widely used in biosensors for virus detection studies like Corona virus detection. Funari et al. [103], have developed an opto-microfluidic sensing platform with gold nanospikes immobilized by electrodeposition based on the principle of LSPR, to detect the specific antibodies for the SARS-CoV-2. Researchers tested the antibodies via spiking of antibodies protein in 1 μL of human plasma diluted in 1 mL of buffer solution, within approximately 30 min. The results showed the target antibody concentration could be correlated with the LSPR wavelength peak shift of gold nanospikes caused by the local refractive index change owing to the antigen–antibody binding and achieved a limit of detection of approximately 0.08 ng/mL 0.5 pM), falling under the clinical relevant concentration range. The explained opto-microfluidic platform offers a promising point-of-care testing tool to complement standard serological assays and makes SARS-CoV-2 quantitative diagnostics easier, cheaper, and faster [103]. Qiu et al. [104], prepared a dual-functional plasmonic biosensor combining the plasmonic photothermal (PPT) effect and LSPR sensing transduction that could provide an alternative and promising solution for the clinical COVID-19 diagnosis. The results of this study showed, the LSPR sensing unit attained a real-time and label-free detection of viral sequences including RdRp-COVID, ORF1ab-COVID, and E genes from SARS-Cov-2 with this configuration. They functionalized the two-dimensional gold nanoislands (AuNIs) with complementary DNA receptors and performed a sensitive detection of the selected sequences from SARS-CoV-2 through nucleic acid hybridization. Two different angles of incidence were applied and therefore the plasmonic resonances of PPT and LSPR can be excited at two different wavelengths that significantly enhanced the sensing stability, sensitivity, and reliability. Based on the obtained results, using the AuNI chips dramatically improved the hybridization kinetics and the specificity of nucleic acid detection. For obtaining the better results, they generated the thermoplasmonic heat on the same AuNIs chip when illuminated at their plasmonic resonance frequency. The researchers have showed that applied heat can elevate the in situ hybridization temperature and increase the discrimination accuracy of two similar gene sequences. The fabricated dual-functional LSPR biosensor exhibited a high sensitivity toward the selected SARS-CoV-2 sequences with a lower detection limit down to the concentration of 0.22 pM and enabled the precise detection of the specific target in a multigene mixture [104].
4.8. Field-effect transistor-based assay (FET)
Among a wide range of biosensors, field effect transistor (FET) based biosensors are one of the established approaches owing to their advantages, including, rapid, low cost and simple detection. Generally, FET is a solid-state device that the electroconductivity of the semiconductor between the source and drain terminals is regulated by a third gate electrode through an insulator [105]. FET based biosensors are classified into different groups according to the technique of gate voltage application, design, gate material, and the channel region. FET based biosensors have found an extensive application in biomolecules detection [106]. Recently, this technique coupled with graphene advantages is used for COVID-19 detection. Seo et al. [107], developed a new graphene based biosensing device functionalized with SARS-CoV-2 spike antibody, called COVID-19 FET sensor, for Coronavirus detection. These researchers pointed out that their FET sensor does not require any sample pretreatment or labeling. As can be seen in Fig. 10 , in the first step, for sensor fabrication, the graphene was transferred to a SiO2/Si substrate using conventional wet-transfer method. In the next step, the transferred graphene was patterned into linear shapes by photolithography and etched by reactive ion-etching method. Then the SARS-CoV-2 spike antibody was immobilized onto the fabricated device via banding 1-pyrenebutyric acid N-hydroxysuccinimide ester as interface coupling agent. The antigen protein was used as target species for sensor design and calibration. The sensor performance was determined using antigen protein, cultured virus, and nasopharyngeal swab specimens from COVID-19 patients, and the electrical performance was evaluated on semiconductor analyzer. The final results showed that the FET device could detect the SARS-CoV-2 spike protein at the concentrations of 1 fg/mL in phosphate-buffered saline and 100 fg/mL clinical transport medium. In addition, the FET sensor successfully detected SARS-CoV-2 in culture medium (limit of detection [LOD]: 16 pfu/mL) and clinical samples (LOD: 242 copies/mL) [107].
Fig. 10.
Schematic illustration of COVID-19 sensor based on FET. The sensing material is Graphene and SARS-CoV-2 antibody is conjugated onto the graphene sheet via 1-pyrenebutyric acid N-hydroxysuccinimide ester, which is probe linker [107].
4.9. Electrochemical immunoassays
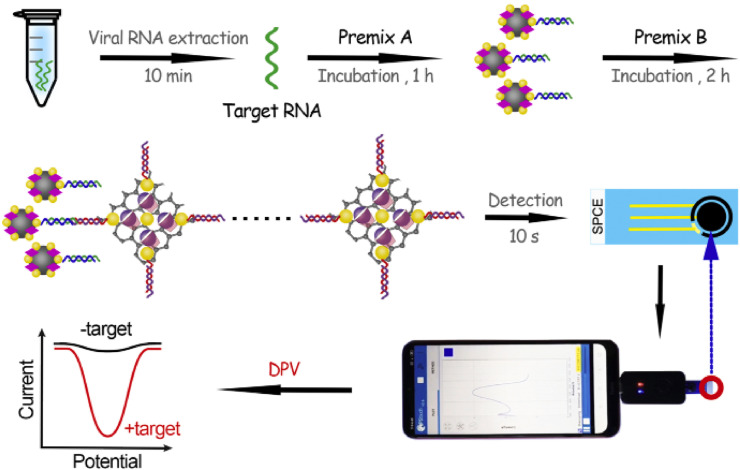
Electrochemical biosensors are analytical devices including a biological sensing element and a transducer that converts a chemical reaction to an electronic response [[108], [109], [110], [111]]. The reaction under investigation would either generate a measurable current (amperometiric) [112], potential or charge accumulation (poteomtiometric) [84] or conductive properties of a medium (conductometric [113] or Impedance [114,115]) between the electrodes. The electrochemical biosensors have a wide range of application for detecting several types of biological elements including DNA [84,116], RNA [117,118], antibody [119], biomarker [120], proteins [121,122], viruses [17,123] and bacteria [124]. The DNA/RNA biosensors are well known as simple, sensitive and powerful tools for detection of biomolecules in the early clinical diagnosis and monitoring of disease. Several types of biosensors have been introduced for detecting viruses, such as old types of Coronavirus [125,126]. The electrochemical biosensors also used for detection of COVID-19 disease in different methods and with different analyte such as DNA and protein [[127], [128], [129]]. Zhoa et al. [130], developed a new super sandwich-type electrochemical biosensor based on p-sulfocalixarene (SCX8) functionalized graphene (SCX8-RGO) to enrich TB for SARS-CoV-2 RNA detection using a portable electrochemical smartphone. They prepared two premixes A and B for SARS-CoV-2 detection. For premix A, Fe3O4 NPs were applied for synthesis of Au@Fe3O2 nanocomposite and for the premix B, HAuCl4 and TB were used for preparing Au@SCX8-RGO-TB nanocomposite. Fig. 11 illustrate the schematic process of SARS-CoV-2 detection electrochemical biosensor fabrication. As seen in this figure, the CPs labeled with thiol and immobilized on the surfaces of the Au@Fe3O4 nanoparticles and formed CP/Au@Fe3O4 nanocomposites and then the host-guest complexes (SCX8-TB) were immobilized on RGO to form Au@SCX8-TB-RGO-TB nanocomposite and the sandwich structure produced. The researcher used 88 samples from confirmed patients and 8 samples from recovered patients infected by SARS-CoV-2. Electrochemical measurement was used as detection method and the results were compared with results from RT-qPCR. The prepared biosensor showed high specificity and selectivity during silico analysis and actual testing, and its detectable ratios (85.5% and 46.2%) were higher than those obtained using RT-qPCR (56.5% and 7.7%). The LOD for the clinical specimen was 200 copies/mL, and very low volume of samples, only two copies (10 μL) of SARS-CoV-2 were required per assay. Additionally, the mentioned method does not require nucleic acid amplification and reverse transcription, and is a plug-and-play diagnostic system [130].
Fig. 11.
Schematic representation of SARS-CoV-2 detection using the electrochemical biosensor. (A) Preparation of premix A and B; (B) Process of electrochemical detection using a smartphone [130].
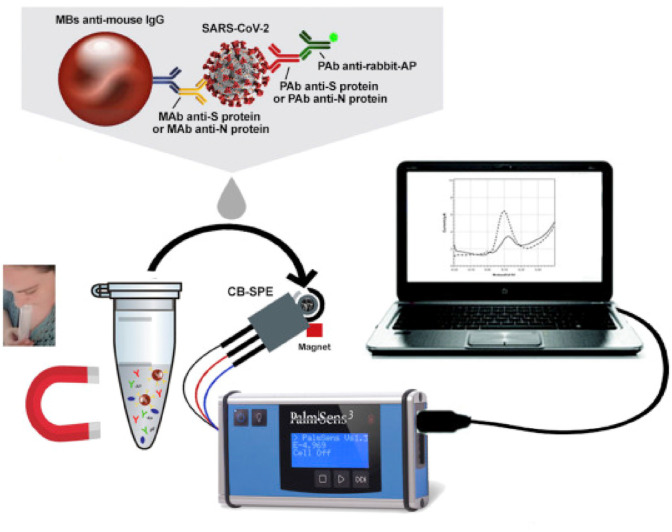
In another study, a label free paper-based electrochemical platform as a screening tool to detect IgG and IgM antibodies related to SARS-CoV-2 virus is suggested by Yakoh et al. [131]. The fabricated sensor was consisted of three parts called counter, closing part, and support material. The square-wave voltammetry (SWV) technique was used for obtaining the electrochemical response. In the working part, the SARS-CoV-2 spike protein containing receptor-binding domain (SP RBD) is immobilized to capture incoming SARS-CoV-2 antibodies. A series of assays were applied for both SARS-CoV-2-infected and -uninfected patients samples, and the results were compared with a commercial standard ELISA method. The experimental time, sensitivity, selectivity and LOD were reported 30 min, 100%, 90% and 1 ng/mL, respectively [131]. Hashemi et al. [132], fabricated a rapid electrochemical diagnostic kit, composed of fixed/screen printed electrodes that could detect pathogenic viruses such as SARS-CoV-2 and/or animal viruses through the differentiable fingerprint of their viral glycoproteins at different voltage. The sensor was comprised of a working electrode that was activated upon coating a layer of combination between graphene oxide (GO) and sensitive chemical compounds with gold nanostars (Au NS). This layer was capable of detecting the trace amount of viruses in any aquatic biological media, for instance, blood, saliva and oropharyngeal/nasopharyngeal swab, via interaction with active functional groups of their glycoproteins around 1 min and any extra extraction and/or biomarkers for detection of targeted viruses were not required. The LOD and sensitivity of sensor were calculated as 1.68 × 10−22 μgmL−1 and 0.0048 μAμgmL−1cm−2, respectively, toward detection of SARS-CoV-2 in biological media, while blind clinical evaluations of 100 suspected samples furtherly confirmed the superior sensitivity/specificity of developed nanosystem toward the rapid identification of patients even at incubation and prodromal periods of disease [132]. In another study, a ROS detection system was used for direct detection of COVID-19 in sputum. The authors applied the ROS/H2O2 system includes an integrated portable automatic electrochemical readout device and a sensor as the main diagnostic part of the system. The sensor was a three electrode system and the working electrode was made on steel needles coated with Multi-Wall Carbon Nanotubes (MWCNTs). The test was conducted in 30s and results show the ROS level of sputum with the calibrated response in correlation with the probability of COVID-19 involvement [133]. In the other study, as seen in Fig. 12 , Fabiani et al. [134] designed and fabricated a sensitive electrochemical immunoassay for rapid and smart detection of SARS-CoV-2 in untreated saliva. The electrochemical assay was fabricated on magnetic beads as support. The Spike (S) protein or Nucleocapsid (N) protein was immobilized on magnetic beads as immunological chain and secondary antibody with alkaline phosphatase as immunological label. After the enzymatic reaction of 1-naphthol on immobilized magnetic bead the electrochemical detection was carried out using screen-printed electrodes modified with carbon black nanomaterial. The analytical features of the electrochemical immunoassay were evaluated using the standard solution of S and N protein in buffer solution and untreated saliva with a LOD equal to 19 ng/mL and 8 ng/mL in untreated saliva, respectively for S and N protein. The fabricated immunoassay was tested using cultured virus in biosafety level 3 and saliva clinical samples were used as real sample and the results were compared with the results of nasopharyngeal swab specimens tested with Real-Time PCR. The researchers have reported that their test is rapid and the data has good agreement with Standard methods as well as the LOD is low and the miniaturization and portability is achievable for their method [134].
Fig. 12.
The MBs-based assay for SARS-CoV-2 detection in untreated saliva [134].
Alafeef et al. [135] developed a graphene based electrochemical DNA biosensor to detect the SARS-CoV-2 thiol-modified nucleocapsid phosphoprotein (N-gene) capped by gold nanoparticles (AuNPs). The sensor response was obtained from RNA samples collected from Vero cells infected with SARS-CoV-2, while SARS-CoV and MERS-CoV RNA were used as negative controls. The capability of the sensor chip to differentiate the positive COVID-19 samples from the negative ones was investigated using 48 clinical samples and compared with commercial RT-PCR diagnostic kit. The researcher argued the required time for process was less than 5 min, the sensitivity was 231 (copies μL−1)−1 and LOD was 6.9 copies/μL without the need for any further amplification. They tested their sensor with clinical samples from 22 COVID-19 positive patients and 26 healthy asymptomatic subjects confirmed using the FDA-approved RT-PCR COVID-19 diagnostic kit, and prepared sensor could distinguished the positive COVID-19 samples from the negative ones with 100% accuracy, sensitivity, and specificity [135].
4.9.1. Impedance
Impedance measurements involve the application of a small sinusoidal AC voltage probe and the determination of current response. Impedance sensors detect a change in one of these equivalent circuit parameters upon analyte binding. There are several impedance applications in biosensing detections. This technique has recently used for diagnosis of COVID-19 as detection method of biosensor. For example, a rapid detection of SAES-CoV-2 antibodies was explained using impedance based immunosensor by Rashed et al. [136]. In this study, a non-faradic capacitive immunosensing assay was used with a commercial impedance detector. The immunosensor was fabricated using pre-coated receptor binding domain (RBD) of SARS-CoV-2 spike protein on 16-well plate containing sensing electrodes with samples of anti-SARS-CoV-2 monoclonal antibody CR3022 (0.1 μg/mL, 1.0 μg/mL, 10 μg/mL) and subsequent blinded testing was performed on six serum specimens taken from COVID-19 and non-COVID-19 patients. The obtained impedance results show a strong correlation with standard ELISA test results (R2 = 0.9) [136].
5. Sampling
There are some studies that shows the sampling method is effective on COVID-19 detection results. This field is still under study. Wyllie et al. [137], reported a research that shows the sampling method is effective on sensitivity. They compared two different sampling methods, nasopharyngeal and saliva samples obtained from confirmed COVID-19 patients and self-collected samples from healthcare workers at moderated to high risk of COVID-19 exposure, and detect the samples with RT-PCR. The final results demonstrated that saliva is a viable and preferable alternative to nasopharyngeal swabs for SARS-CoV-2 detection. The obtained results indicated that not only saliva samples results are comparable to nasopharyngeal swabs in early hospitalization, but also is more consistent during extended hospitalization and recovery. Additionally, COVID-19 detection from the saliva sampling may also it be a useable alternative for identifying mild or subclinical infections for example asymptomatic healthcare workers. Additionally, the saliva self-sampling does not require direct healthcare worker-patient interaction, a source of several major testing bottlenecks and overall nosocomial infection risk, and alleviates supply demands on swabs and personal protective equipment [137].
6. Summary and concluding remarks
In this article, the literature on COVID-19 detection methods is reviewed in different categories, including the biomolecules used as analyte, the detection, and sampling methods. This review compares the new reported technologies with commercially available ones that are used in these days. Disease diagnosis is an efficient and valuable tool for controlling the spread of COVID-19 during the pandemic. It is noteworthy that the SARS-CoV-2 virus transmutations and spreading are very rapid, and we are still in the middle of the pandemic. Thus, finding a simple, rapid, low-cost, accurate and sensitive detection method of SARS-CoV-2 virus currently possesses the highest priority.
Table 1 presents the current methods and targeted analyte that are used for COVID-19 detection commercially and in the lab and their advantages. Among the different methods developed so far for detecting COVID-19, the PCR and antibody-based diagnostics are widely acclaimed for their high accuracy and sensitivity. However, the mentioned methods require pretreatment steps, large instruments, and are expensive that makes them inappropriate for applications that require on-site analysis. Alternative technologies such as LAMP, FET, and electrochemical methods are growing rapidly worldwide that can help to develop novel detection methods with high performance. The obtained results have shown that the novel methods have acceptable sensitivity, accuracy, and detection limit. In addition, these alternative methods possess a high potential to decrease the waiting-time for test results. It is apparent from Table 1 that the targeted analyte type is not an influential factor on bioassay sensitivity, and all types of analytes could serve properly as bioassay target.
Table 1.
Applied methods and targeted analytes for the detection of COVID-19.
| Methods | Analyte | Detection time | Sensitivity | Accuracy | DOL | Advantages | Reference |
|---|---|---|---|---|---|---|---|
| RT-PCR | RNA | 75 min | – | – | 10 copies/reaction | Specific | [56] |
| PCR | DNA | – | 91% | 93% | 2 copies/reaction | specific | [60] |
| RT-PCR | C2CA | 100 min | 100% | 0.4 fM | specific | [62] | |
| ELISA | IgG/IgM | – | 89.6% | – | – | Portable, fast | [29] |
| ELISA | IgG/IgM | 30 min | 85.4% | 98.6% | 1 copies/μL | Portable, fast | [66] |
| MS | DNA | 3 h | – | – | 105 copies | specific | [69] |
| MS | DNA | 24–48 h | – | – | 155–106 copies/μL | Specific | [71] |
| CLIA | IgG/IgM | – | – | 81.5% | – | Fast, portable | [74] |
| CLIA | DNA | – | 100% | 87% | – | Specific, fast, portable | [75] |
| LFIA | IgG/IgM | – | – | – | 1 × 104 Pfu | Sensitive | [83] |
| LFIA | IgG/IgM | – | 100% | – | – | sensitive | [26] |
| LAMP | DNA | – | 97.6% | – | 0.2–47 ng/μL | Specific, portable | [94] |
| LSPR | IgG/IgM | 2 h | – | – | 0.08 ng/mL | sensitive | [103] |
| LSPR | RNA | – | 3.2 copies | – | 0.22 pM | Specific, sensitive | [104] |
| FET | antibody | – | – | – | 16 Pfu/μL | Specific, sensitive | [107] |
| electrochemical | DNA | – | – | – | 200 copies/mL | Sensitive, fast, portable, specific | [101] |
| electrochemical | IgG/IgM | – | 100% | – | 1 ng/mL | Sensitive, fast | [131] |
| electrochemical | IgG/IgM | – | 0.0048 μAμgmL−1cm−2 | – | 1.68 × 10−22 μg/mL | Sensitive, fast | [132] |
| electrochemical | IgG/IgM | – | – | – | 19 ng/mL | Sensitive, fast, portable | [134] |
The recently developed strategies and methods and lessons learned can be deployed for facing future virus outbreaks and new generations of SARS-CoV-2. Using gold nanoparticles has greatly enhanced the performance metrics such as sensitivity, specifity, and accuracy of the COVID-19 sensors owing to their electron transfer improvement, electrocatalytic and bio-labelling properties, and high surface to volume ratio. The cost-effective manufacturing of lab-on-chip sensors with paper based platform can make possible the low-cost and disposable detection of COVID-19 without needing skilled operators. However, this novel field is still at its infancy with the capability of shortening the total-analytical-time from hours to minutes. The overall progress is promising, but a tangible success on detecting the SARS-CoV-2 is yet to be obtained for developing a commercially viable lab-on-chip based COVID-19 sensor.
References
- 1.Ribeiro B.V., Cordeiro T.A.R., Oliveira e Freitas G.R., Ferreira L.F., Franco D.L. Biosensors for the detection of respiratory viruses: a review. Talanta Open. 2020;2:100007. doi: 10.1016/j.talo.2020.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma L., Zeng F., Cong F., Huang B., Huang R., Ma J., Guo P. Development of a SYBR green-based real-time RT-PCR assay for rapid detection of the emerging swine acute diarrhea syndrome coronavirus. J. Virol Methods. 2019;265:66–70. doi: 10.1016/j.jviromet.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang R., Li J.M. [The way to reduce the false negative results of 2019 novel coronavirus nucleic acid detection] Zhonghua Yixue Zazhi. 2020;100:801–804. doi: 10.3760/cma.j.cn112137-20200215-00288. [DOI] [PubMed] [Google Scholar]
- 4.Xie C., Jiang L., Huang G., Pu H., Gong B., Lin H., Ma S., Chen X., Long B., Si G., Yu H., Jiang L., Yang X., Shi Y., Yang Z. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int. J. Infect. Dis. 2020;93:264–267. doi: 10.1016/j.ijid.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou L., Sun Y., Lan T., Wu R., Chen J., Wu Z., Xie Q., Zhang X., Ma J. Retrospective detection and phylogenetic analysis of swine acute diarrhoea syndrome coronavirus in pigs in southern China. Transbound. Emerg. Dis. 2019;66:687–695. doi: 10.1111/tbed.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang S., Peng W., Zhu Y., Lu S., Zhou M., Lin W., Wu W., Huang S., Jiang L., Luo X., Deng M. Recent progress in understanding 2019 novel coronavirus (SARS-CoV-2) associated with human respiratory disease: detection, mechanisms and treatment. Int. J. Antimicrob. Agents. 2020;55:105950. doi: 10.1016/j.ijantimicag.2020.105950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao C., Zhu L., Jin C.C., Tong Y.X., Xiao A.T., Zhang S. Prevalence and impact factors of recurrent positive SARS-CoV-2 detection in 599 hospitalized COVID-19 patients. Clin. Microbiol. Infect. 2021 doi: 10.1016/j.cmi.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krähling V., Halwe S., Rohde C., Becker D., Berghöfer S., Dahlke C., Eickmann M., Ercanoglu M.S., Gieselmann L., Herwig A., Kupke A., Müller H., Neubauer-Rädel P., Klein F., Keller C., Becker S. Development and characterization of an indirect ELISA to detect SARS-CoV-2 spike protein-specific antibodies. J. Immunol. Methods. 2021;490:112958. doi: 10.1016/j.jim.2021.112958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi L., Sun Q., He J.a., Xu H., Liu C., Zhao C., Xu Y., Wu C., Xiang J., Gu D., Long J., Lan H. Development of SPR biosensor for simultaneous detection of multiplex respiratory viruses. Bio Med. Mater. Eng. 2015;26(Suppl 1):S2207–S2216. doi: 10.3233/BME-151526. [DOI] [PubMed] [Google Scholar]
- 11.Chen K., Wu L., Jiang X., Lu Z., Han H. Target triggered self-assembly of Au nanoparticles for amplified detection of Bacillus thuringiensis transgenic sequence using SERS. Biosens. Bioelectron. 2014;62:196–200. doi: 10.1016/j.bios.2014.06.046. [DOI] [PubMed] [Google Scholar]
- 12.Antiochia R. Nanobiosensors as new diagnostic tools for SARS, MERS and COVID-19: from past to perspectives. Mikrochim. Acta. 2020;187:639. doi: 10.1007/s00604-020-04615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moulahoum H., Ghorbanizamani F., Zihnioglu F., Turhan K., Timur S. How should diagnostic kits development adapt quickly in COVID 19-like pandemic models? Pros and cons of sensory platforms used in COVID-19 sensing. Talanta. 2021;222:121534. doi: 10.1016/j.talanta.2020.121534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Nair M.S., Huang Y., Ho D.D. bioRxiv; 2021. Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. 2021.2001.2025.428137. [DOI] [PubMed] [Google Scholar]
- 17.Ilkhani H., Farhad S. A novel electrochemical DNA biosensor for Ebola virus detection. Anal. Biochem. 2018 doi: 10.1016/j.ab.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Pilevar M., Kim K.T., Lee W.H. Recent advances in biosensors for detecting viruses in water and wastewater. J. Hazard Mater. 2020:124656. doi: 10.1016/j.jhazmat.2020.124656. [DOI] [PubMed] [Google Scholar]
- 19.Bhatta D., Stadden E., Hashem E., Sparrow I.J.G., Emmerson G.D. Multi-purpose optical biosensors for real-time detection of bacteria, viruses and toxins. Sensor. Actuator. B Chem. 2010;149:233–238. [Google Scholar]
- 20.Ji T., Liu Z., Wang G., Guo X., Akbar Khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Detection of COVID-19: a review of the current literature and future perspectives. Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen M., Zhou Y., Ye J., Abdullah Al-Maskri A.A., Kang Y., Zeng S., Cai S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020;10:97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao J., Duan C., Xue L., Liu Y., Sun W., Xiang Y. DNA nanoscaffold-based SARS-CoV-2 detection for COVID-19 diagnosis. Biosens. Bioelectron. 2020;167:112479. doi: 10.1016/j.bios.2020.112479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., Wu J., Liao Y., Gou X., Li Y., Wang X., Su H., Gu B., Wang J., Xu T. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding X., Yin K., Li Z., Liu C. bioRxiv; 2020. All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) Assay: A Case for Rapid, Ultrasensitive and Visual Detection of Novel Coronavirus SARS-CoV-2 and HIV Virus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C., Wen T., Shi F.J., Zeng X.Y., Jiao Y.J. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 2020;5:12550–12556. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J. Med. Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H., Dai E., Xiao R., Zhou Z., Zhang M., Bai Z., Shao Y., Qi K., Tu J., Wang C., Wang S. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sensor. Actuator. B Chem. 2020:129196. doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montes R., Céspedes F., Baeza M. Highly sensitive electrochemical immunosensor for IgG detection based on optimized rigid biocomposites. Biosens. Bioelectron. 2016;78:505–512. doi: 10.1016/j.bios.2015.11.081. [DOI] [PubMed] [Google Scholar]
- 31.Bayart J.-L., Gusbin C., Lardinois B., Scohy A., Kabamba-Mukadi B. Analytical and clinical evaluation of new automated chemiluminescent immunoassays for the detection of IgG and IgM anti-Bartonella henselae antibodies. Diagn. Microbiol. Infect. Dis. 2020;98:115203. doi: 10.1016/j.diagmicrobio.2020.115203. [DOI] [PubMed] [Google Scholar]
- 32.Wang H., Wu X., Zhang X., Hou X., Liang T., Wang D., Teng F., Dai J., Duan H., Guo S., Li Y., Yu X. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. ACS Cent. Sci. 2020 doi: 10.1021/acscentsci.0c00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steiner D.J., Cognetti J.S., Luta E.P., Klose A.M., Bucukovski J., Bryan M.R., Schmuke J.J., Nguyen-Contant P., Sangster M.Y., Topham D.J., Miller B.L. Array-based analysis of SARS-CoV-2, other coronaviruses, and influenza antibodies in convalescent COVID-19 patients. Biosens. Bioelectron. 2020;169:112643. doi: 10.1016/j.bios.2020.112643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y., Li C., Xia S., Tian X., Kong Y., Wang Z., Gu C., Zhang R., Tu C., Xie Y., Yang Z., Lu L., Jiang S., Ying T. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898. doi: 10.1016/j.chom.2020.04.023. e895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conzelmann C., Gilg A., Gross R., Schutz D., Preising N., Standker L., Jahrsdorfer B., Schrezenmeier H., Sparrer K.M.J., Stamminger T., Stenger S., Munch J., Muller J.A. An enzyme-based immunodetection assay to quantify SARS-CoV-2 infection. Antivir. Res. 2020;181:104882. doi: 10.1016/j.antiviral.2020.104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang J., Yan M., Li H., Liu T., Lin C., Huang S., Shen C. medRxiv; 2020. Evaluation of Enzyme-Linked Immunoassay and Colloidal GoldImmunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19) [Google Scholar]
- 37.Rama E.C., Costa-García A. Screen-printed electrochemical immunosensors for the detection of cancer and cardiovascular biomarkers. Electroanalysis. 2016;28:1700–1715. [Google Scholar]
- 38.Shan B., Broza Y.Y., Li W., Wang Y., Wu S., Liu Z., Wang J., Gui S., Wang L., Zhang Z., Liu W., Zhou S., Jin W., Zhang Q., Hu D., Lin L., Zhang Q., Li W., Wang J., Liu H., Pan Y., Haick H. Multiplexed nanomaterial-based sensor array for detection of COVID-19 in exhaled breath. ACS Nano. 2020;14:12125–12132. doi: 10.1021/acsnano.0c05657. [DOI] [PubMed] [Google Scholar]
- 39.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;vol. 25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Cai K., Zhang R., He X., Shen X., Liu J., Xu J., Qiu F., Lei W., Wang J., Li X., Gao Y., Jiang Y., Xu W., Ma X. Novel one-step single-tube nested quantitative real-time PCR assay for highly sensitive detection of SARS-CoV-2. Anal. Chem. 2020;92:9399–9404. doi: 10.1021/acs.analchem.0c01884. [DOI] [PubMed] [Google Scholar]
- 41.Xiao A.T., Tong Y.X., Gao C., Zhu L., Zhang Y.J., Zhang S. Dynamic profile of RT-PCR findings from 301 COVID-19 patients in Wuhan, China: a descriptive study. J. Clin. Virol. 2020;127:104346. doi: 10.1016/j.jcv.2020.104346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yip C.C., Ho C.C., Chan J.F., To K.K., Chan H.S., Wong S.C., Leung K.H., Fung A.Y., Ng A.C., Zou Z., Tam A.R., Chung T.W., Chan K.H., Hung I.F., Cheng V.C., Tsang O.T., Tsui S.K.W., Yuen K.Y. Development of a novel, genome subtraction-derived, SARS-CoV-2-specific COVID-19-nsp2 real-time RT-PCR assay and its evaluation using clinical specimens. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21072574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petralia S., Conoci S. PCR technologies for point of care testing: progress and perspectives. ACS Sens. 2017;2:876–891. doi: 10.1021/acssensors.7b00299. [DOI] [PubMed] [Google Scholar]
- 44.Faria H.A.M., Zucolotto V. Label-free electrochemical DNA biosensor for zika virus identification. Biosens. Bioelectron. 2019;131:149–155. doi: 10.1016/j.bios.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 45.Poon L.L., Wong B.W., Chan K.H., Ng S.S., Yuen K.Y., Guan Y., Peiris J.S. Evaluation of real-time reverse transcriptase PCR and real-time loop-mediated amplification assays for severe acute respiratory syndrome coronavirus detection. J. Clin. Microbiol. 2005;43:3457–3459. doi: 10.1128/JCM.43.7.3457-3459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenkins M., O'Brien C., Fetterer R., Santin M. RT-PCR specific for Cryspovirus is a highly sensitive method for detecting Cryptosporidium parvum oocysts. Food Waterborne Parasitol. 2016;5:14–20. [Google Scholar]
- 47.Laude A., Valot S., Desoubeaux G., Argy N., Nourrisson C., Pomares C., Machouart M., Le Govic Y., Dalle F., Botterel F., Bourgeois N., Cateau E., Leterrier M., Le Pape P., Morio F. Is real-time PCR-based diagnosis similar in performance to routine parasitological examination for the identification of Giardia intestinalis, Cryptosporidium parvum/Cryptosporidium hominis and Entamoeba histolytica from stool samples? Evaluation of a new commercial multiplex PCR assay and literature review. Clin. Microbiol. Infect. 2016;22 doi: 10.1016/j.cmi.2015.10.019. 190.e191-190.e198. [DOI] [PubMed] [Google Scholar]
- 48.Tachibana H., Saito M., Tsuji K., Yamanaka K., Hoa L.Q., Tamiya E. Self-propelled continuous-flow PCR in capillary-driven microfluidic device: microfluidic behavior and DNA amplification. Sensor. Actuator. B Chem. 2015;206:303–310. [Google Scholar]
- 49.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Shanti N., Saini A., Stewart C.E. Two-Step versus One-Step RNA-to-CT 2-Step and One-Step RNA-to-CT 1-Step: validity, sensitivity, and efficiency. J. Biomol. Tech. 2009;20:172–179. [PMC free article] [PubMed] [Google Scholar]
- 51.Jin Y., Wang M., Zuo Z., Fan C., Ye F., Cai Z., Wang Y., Cui H., Pan K., Xu A. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int. J. Infect. Dis. 2020;94:49–52. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen T., Duong Bang D., Wolff A. Novel coronavirus disease (COVID-19): paving the road for rapid detection and point-of-care diagnostics. Micromachines. 2019;11 doi: 10.3390/mi11030306. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sah R., Rodriguez-Morales A.J., Jha R., Chu D.K.W., Gu H., Peiris M., Bastola A., Lal B.K., Ojha H.C., Rabaan A.A., Zambrano L.I., Costello A., Morita K., Pandey B.D., Poon L.L.M. Complete genome sequence of a 2019 novel coronavirus (SARS-CoV-2) strain isolated in Nepal. Microbiol. Resour. Announc. 2020;9 doi: 10.1128/MRA.00169-20. e169-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.To K.K., Tsang O.T., Yip C.C., Chan K.H., Wu T.C., Chan J.M., Leung W.S., Chik T.S., Choi C.Y., Kandamby D.H., Lung D.C., Tam A.R., Poon R.W., Fung A.Y., Hung I.F., Cheng V.C., Chan J.F., Yuen K.Y. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valitutto M.T., Aung O., Tun K.Y.N., Vodzak M.E., Zimmerman D., Yu J.H., Win Y.T., Maw M.T., Thein W.Z., Win H.H., Dhanota J., Ontiveros V., Smith B., Tremeau-Brevard A., Goldstein T., Johnson C.K., Murray S., Mazet J. Detection of novel coronaviruses in bats in Myanmar. PloS One. 2020;15 doi: 10.1371/journal.pone.0230802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A., Fasano M., Sessa F., Tettamanti L., Carinci F., Maurino V., Rossi A., Tagliabue A., Baj A. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pomari E., Piubelli C., Perandin F., Bisoffi Z. Digital PCR: a new technology for diagnosis of parasitic infections. Clin. Microbiol. Infect. 2019;25:1510–1516. doi: 10.1016/j.cmi.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 59.Nyaruaba R., Mwaliko C., Kering K.K., Wei H. Droplet digital PCR applications in the tuberculosis world. Tuberculosis. 2019;117:85–92. doi: 10.1016/j.tube.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Dong L., Zhou J., Niu C., Wang Q., Pan Y., Sheng S., Wang X., Zhang Y., Yang J., Liu M., Zhao Y., Zhang X., Zhu T., Peng T., Xie J., Gao Y., Wang D., Dai X., Fang X. Talanta; 2020. Highly Accurate and Sensitive Diagnostic Detection of SARS-CoV-2 by Digital PCR; p. 121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y., Gao G., Wang S., Ma C., Xie R., Wang F., Tan C., Zhu L., Guo Y., Zhang F. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 2020;71:793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020;165:112356. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- 63.Asensio L., González I., García T., Martín R. Determination of food authenticity by enzyme-linked immunosorbent assay (ELISA) Food Contr. 2008;19:1–8. [Google Scholar]
- 64.Servat A., Feyssaguet M., Blanchard I., Morize J.L., Schereffer J.L., Boue F., Cliquet F. A quantitative indirect ELISA to monitor the effectiveness of rabies vaccination in domestic and wild carnivores. J. Immunol. Methods. 2007;318:1–10. doi: 10.1016/j.jim.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 65.Costantini F., Sberna C., Petrucci G., Reverberi M., Domenici F., Fanelli C., Manetti C., de Cesare G., DeRosa M., Nascetti A., Caputo D. Aptamer-based sandwich assay for on chip detection of Ochratoxin A by an array of amorphous silicon photosensors. Sensor. Actuator. B Chem. 2016;230:31–39. [Google Scholar]
- 66.Guo L., Ren L., Yang S., Xiao M., Chang, Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q.W., Xu S.Y., Zhu H.D., Xu Y.C., Jin Q., Sharma L., Wang L., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu D., Ju C., Han C., Shi R., Chen X., Duan D., Yan J., Yan X. Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2020;173:112817. doi: 10.1016/j.bios.2020.112817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stevens K.G., Pukala T.L. Conjugating immunoassays to mass spectrometry: solutions to contemporary challenges in clinical diagnostics. Trac. Trends Anal. Chem. 2020;132:116064. doi: 10.1016/j.trac.2020.116064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ihling C., Tanzler D., Hagemann S., Kehlen A., Huttelmaier S., Arlt C., Sinz A. Mass spectrometric identification of SARS-CoV-2 proteins from gargle solution samples of COVID-19 patients. J. Proteome Res. 2020;19:4389–4392. doi: 10.1021/acs.jproteome.0c00280. [DOI] [PubMed] [Google Scholar]
- 70.Gouveia D., Miotello G., Gallais F., Gaillard J.C., Debroas S., Bellanger L., Lavigne J.P., Sotto A., Grenga L., Pible O., Armengaud J. Proteotyping SARS-CoV-2 virus from nasopharyngeal swabs: a proof-of-concept focused on a 3 min mass spectrometry window. J. Proteome Res. 2020;19:4407–4416. doi: 10.1021/acs.jproteome.0c00535. [DOI] [PubMed] [Google Scholar]
- 71.Dollman N.L., Griffin J.H., Downard K.M. Detection, mapping, and proteotyping of SARS-CoV-2 coronavirus with high resolution mass spectrometry. ACS Infect. Dis. 2020 doi: 10.1021/acsinfecdis.0c00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao Q., Xu C. Research progress on chemiluminescence immunoassay combined with novel technologies. Trac. Trends Anal. Chem. 2020;124:115780. [Google Scholar]
- 73.Kricka L., Thorpe G., Stott R. In: Principles and Practice of Immunoassay. Price C.P., Newman D.J., editors. Palgrave Macmillan UK; London: 1991. Luminescence immunoassay; pp. 417–445. [Google Scholar]
- 74.Cai X.F., Chen J., Li Hu J., Long Q.X., Deng H.J., Liu P., Fan K., Liao P., Liu B.Z., Wu G.C., Chen Y.K., Li Z.J., Wang K., Zhang X.L., Tian W.G., Xiang J.L., Du H.X., Wang J., Hu Y., Tang N., Lin Y., Ren J.H., Huang L.Y., Wei J., Gan C.Y., Chen Y.M., Gao Q.Z., Chen A.M., He C.L., Wang D.X., Hu P., Zhou F.C., Huang A.L., Wang D.Q. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of coronavirus disease 2019. J. Infect. Dis. 2020;222:189–193. doi: 10.1093/infdis/jiaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Diao B., Wen K., Chen J., Liu Y., Yuan Z., Han C., Chen J., Pan Y., Chen L., Dan Y., Wang J., Chen Y., Deng G., Zhou H., Wu Y. medRxiv; 2020. Diagnosis of Acute Respiratory Syndrome Coronavirus 2 Infection by Detection of Nucleocapsid Protein. 2020.2003.2007.20032524. [Google Scholar]
- 76.Zhang J., Zhang X., Liu J., Ban Y., Li N., Wu Y., Liu Y., Ye R., Liu J., Li X., Li L., Qin X., Zheng R. Serological detection of 2019-nCoV respond to the epidemic: a useful complement to nucleic acid testing. Int. Immunopharm. 2020;88:106861. doi: 10.1016/j.intimp.2020.106861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cady N.C., Tokranova N., Minor A., Nikvand N., Strle K., Lee W.T., Page W., Guignon E., Pilar A., Gibson G.N. Multiplexed detection and quantification of human antibody response to COVID-19 infection using a plasmon enhanced biosensor platform. Biosens. Bioelectron. 2021;171:112679. doi: 10.1016/j.bios.2020.112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakasone N., Toma C., Lu Y., Iwanaga M. Development of a rapid immunochromatographic test to identify enteropathogenic and enterohemorrhagic Escherichia coli by detecting EspB. Diagn. Microbiol. Infect. Dis. 2007;57:21–25. doi: 10.1016/j.diagmicrobio.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 79.Leuvering J.H.W., Thal P.J.H.M., Waart M.v.d., Schuurs A.H.W.M. Sol particle immunoassay (SPIA) J. Immunoassay. 1980;1:77–91. doi: 10.1080/01971528008055777. [DOI] [PubMed] [Google Scholar]
- 80.Karakus C., Salih B.A. Comparison of the lateral flow immunoassays (LFIA) for the diagnosis of Helicobacter pylori infection. J. Immunol. Methods. 2013;396:8–14. doi: 10.1016/j.jim.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 81.Cavalera S., Colitti B., Rosati S., Ferrara G., Bertolotti L., Nogarol C., Guiotto C., Cagnazzo C., Denina M., Fagioli F., Di Nardo F., Chiarello M., Baggiani C., Anfossi L. A multi-target lateral flow immunoassay enabling the specific and sensitive detection of total antibodies to SARS COV-2. Talanta. 2021;223:121737. doi: 10.1016/j.talanta.2020.121737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roda A., Cavalera S., Di Nardo F., Calabria D., Rosati S., Simoni P., Colitti B., Baggiani C., Roda M., Anfossi L. Dual lateral flow optical/chemiluminescence immunosensors for the rapid detection of salivary and serum IgA in patients with COVID-19 disease. Biosens. Bioelectron. 2021;172:112765. doi: 10.1016/j.bios.2020.112765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim H.Y., Lee J.H., Kim M.J., Park S.C., Choi M., Lee W., Ku K.B., Kim B.T., Changkyun Park E., Kim H.G., Kim S.I. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens. Bioelectron. 2020:112868. doi: 10.1016/j.bios.2020.112868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ilkhani H., Hughes T., Li J., Zhong C.J., Hepel M. Nanostructured SERS-electrochemical biosensors for testing of anticancer drug interactions with DNA. Biosens. Bioelectron. 2016;80:257–264. doi: 10.1016/j.bios.2016.01.068. [DOI] [PubMed] [Google Scholar]
- 85.Xi J., Yu Q. The development of lateral flow immunoassay strip tests based on surface enhanced Raman spectroscopy coupled with gold nanoparticles for the rapid detection of soybean allergen β-conglycinin. Spectrochim. Acta Mol. Biomol. Spectrosc. 2020;241:118640. doi: 10.1016/j.saa.2020.118640. [DOI] [PubMed] [Google Scholar]
- 86.Wang J., Chen Q., Jin Y., Zhang X., He L., Zhang W., Chen Y. Surface enhanced Raman scattering-based lateral flow immunosensor for sensitive detection of aflatoxin M1 in urine. Anal. Chim. Acta. 2020;1128:184–192. doi: 10.1016/j.aca.2020.06.076. [DOI] [PubMed] [Google Scholar]
- 87.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao V.X.T., Wong T.I., Zheng X.T., Tan Y.N., Zhou X. Colorimetric biosensors for point-of-care virus detections. Mater. Sci. Energy Technol. 2020;3:237–249. doi: 10.1016/j.mset.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang W.E., Lim B., Hsu C.C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hayasaka D., Aoki K., Morita K. Development of simple and rapid assay to detect viral RNA of tick-borne encephalitis virus by reverse transcription-loop-mediated isothermal amplification. Virol. J. 2013;10:68. doi: 10.1186/1743-422X-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Soliman H., El-Matbouli M. An inexpensive and rapid diagnostic method of Koi Herpesvirus (KHV) infection by loop-mediated isothermal amplification. Virol. J. 2005;2:83. doi: 10.1186/1743-422X-2-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Freire-Paspuel B., Garcia-Bereguiain M.A. Low clinical performance of the Isopollo COVID-19 detection kit (M Monitor, South Korea) for RT-LAMP SARS-CoV-2 diagnosis: a call for action against low quality products for developing countries. Int. J. Infect. Dis. 2021;104:303–305. doi: 10.1016/j.ijid.2020.12.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. medRxiv; 2020. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. 2020.2002.2026.20028373. [Google Scholar]
- 94.Yu L., Wu S., Hao X., Li X., Liu X., Ye S., Han H., Dong X., Li X., Li J., Liu N., Liu J., Zhang W., Pelechano V., Chen W.-H., Yin X. medRxiv; 2020. Rapid Colorimetric Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform: iLACO. 2020.2002.2020.20025874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.El-Tholoth M., Bau H.H., Song J. 2020. A Single and Two-Stage, Closed-Tube, Molecular Test for the 2019 Novel Coronavirus (COVID-19) at Home, Clinic, and Points of Entry, ChemRxiv : the Preprint Server for Chemistry. [Google Scholar]
- 97.Baek Y.H., Um J., Antigua K.J.C., Park J.H., Kim Y., Oh S., Kim Y.I., Choi W.S., Kim S.G., Jeong J.H., Chin B.S., Nicolas H.D.G., Ahn J.Y., Shin K.S., Choi Y.K., Park J.S., Song M.S. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microb. Infect. 2020;9:998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anker Jeffrey N., Paige Hall W., lyandres Olga, Shah Nilam C., Zhao J., duyne R.P.V. Biosensing with plasmonic nanosensors. Nat. Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 99.He H., Muhammad P., Guo Z., Peng Q., Lu H., Liu Z. Controllably prepared molecularly imprinted core-shell plasmonic nanostructure for plasmon-enhanced fluorescence assay. Biosens. Bioelectron. 2019;146:111733. doi: 10.1016/j.bios.2019.111733. [DOI] [PubMed] [Google Scholar]
- 100.Wang L.-X., Jiang X.-E. Bioanalytical applications of surface-enhanced infrared absorption spectroscopy. Chin. J. Anal. Chem. 2012;40:975–982. [Google Scholar]
- 101.Zhang Z., Wang H., Chen Z., Wang X., Choo J., Chen L. Plasmonic colorimetric sensors based on etching and growth of noble metal nanoparticles: strategies and applications. Biosens. Bioelectron. 2018;114:52–65. doi: 10.1016/j.bios.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 102.Xu T., Geng Z. Strategies to improve performances of LSPR biosensing: structure, materials, and interface modification. Biosens. Bioelectron. 2021;174:112850. doi: 10.1016/j.bios.2020.112850. [DOI] [PubMed] [Google Scholar]
- 103.Funari R., Chu K.Y., Shen A.Q. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. 2020;169:112578. doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 105.Sadighbayan D., Hasanzadeh M., Ghafar-Zadeh E. Biosensing based on field-effect transistors (FET): recent progress and challenges. Trac. Trends Anal. Chem. 2020;133:116067. doi: 10.1016/j.trac.2020.116067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sengupta J., Hussain C.M. Graphene-based field-effect transistor biosensors for the rapid detection and analysis of viruses: a perspective in view of COVID-19. Carbon Trends. 2021;2:100011. doi: 10.1016/j.cartre.2020.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 108.Grieshaber D., MacKenzie R., Vörös J., Reimhult E. Electrochemical biosensors - sensor principles and architectures. Sensors. 2008;8:1400–1458. doi: 10.3390/s80314000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ilkhani H., Ganjali M.R., Arvand M., Hejazi M.S., Azimi F., Norouzi P. Electrochemical spectroscopic investigations on the interaction of an ytterbium complex with DNA and their analytical applications such as biosensor. Int. J. Biol. Macromol. 2011;49:1117–1123. doi: 10.1016/j.ijbiomac.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 110.Ilkhani H., Ganjali M.R., Arvand M., Norouzi P. Electrochemical and spectroscopic study of samarium ion interaction with DNA in different pHs. Int. J. Electrochem. Sci. 2010;5:168–176. [Google Scholar]
- 111.Ilkhani H., Ganjali M.R., Arvand M., Norouzi P. Interaction study of ss-DNA and Yb 3+ ions in aqueous solutions by electrochemical and spectroscopic techniques. J. Mol. Liq. 2012;165:119–124. [Google Scholar]
- 112.Bollella P., Gorton L. Enzyme based amperometric biosensors. Curr. Opin. Electrochem. 2018;10:157–173. [Google Scholar]
- 113.Kolahchi N., Braiek M., Ebrahimipour G., Ranaei-Siadat S.O., Lagarde F., Jaffrezic-Renault N. Direct detection of phenol using a new bacterial strain-based conductometric biosensor. J. Environ. Chem. Eng. 2018;6:478–484. [Google Scholar]
- 114.Ilkhani H., Arvand M., Ganjali M.R., Marrazza G., Mascini M. Nanostructured screen printed graphite electrode for the development of a novel electrochemical genosensor. Electroanalysis. 2013;25:507–514. [Google Scholar]
- 115.Raoof J.B., Hejazi M., Ojani R., Asl E. A comparative study of carbon nanotube paste electrode for development of indicator-free DNA sensors using DPV and EIS: human interleukin-2. Int. J. Electrochem. Sci. 2009;4:1436–1451. [Google Scholar]
- 116.Ilkhani H., Zhang H., Zhou A. A novel three-dimensional microTAS chip for ultra-selective single base mismatched Cryptosporidium DNA biosensor. Sensor. Actuator. B Chem. 2019;282:675–683. [Google Scholar]
- 117.Zhang Y., Yan Y., Chen W., Cheng W., Li S., Ding X., Li D., Wang H., Ju H., Ding S. A simple electrochemical biosensor for highly sensitive and specific detection of microRNA based on mismatched catalytic hairpin assembly. Biosens. Bioelectron. 2015;68:343–349. doi: 10.1016/j.bios.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 118.Bettazzi F., Hamid-Asl E., Esposito C.L., Quintavalle C., Formisano N., Laschi S., Catuogno S., Iaboni M., Marrazza G., Mascini M., Cerchia L., De Franciscis V., Condorelli G., Palchetti I. Electrochemical detection of miRNA-222 by use of a magnetic bead-based bioassay. Anal. Bioanal. Chem. 2013;405:1025–1034. doi: 10.1007/s00216-012-6476-7. [DOI] [PubMed] [Google Scholar]
- 119.Ilkhani H., Sarparast M., Noori A., Bathaie S.Z., Mousavi M.F. Electrochemical aptamer/antibody based sandwich immunosensor for the detection of EGFR, a cancer biomarker, using gold nanoparticles as a signaling probe. Biosens. Bioelectron. 2015;74:491–497. doi: 10.1016/j.bios.2015.06.063. [DOI] [PubMed] [Google Scholar]
- 120.Ilkhani H., Ravalli A., Marrazza G. Design of an affibody-based recognition strategy for human epidermal growth factor receptor 2 (HER2) detection by electrochemical biosensors. Chemosensors. 2016;4:23. [Google Scholar]
- 121.Sun L., Chen Y., Chen F., Ma F. Peptide-based electrochemical biosensor for matrix metalloproteinase-14 and protein-overexpressing cancer cells based on analyte-induced cleavage of peptide. Microchem. J. 2020;157:105103. [Google Scholar]
- 122.Tanak A.S., Jagannath B., Tamrakar Y., Muthukumar S., Prasad S. Non-faradaic electrochemical impedimetric profiling of procalcitonin and C-reactive protein as a dual marker biosensor for early sepsis detection. Anal. Chim. Acta X. 2019;3:100029. doi: 10.1016/j.acax.2019.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu F., Choi K.S., Park T.J., Lee S.Y., Seo T.S. Graphene-based electrochemical biosensor for pathogenic virus detection. BioChip J. 2011;5:123–128. [Google Scholar]
- 124.Riu J., Giussani B. Electrochemical biosensors for the detection of pathogenic bacteria in food. Trac. Trends Anal. Chem. 2020;126:115863. [Google Scholar]
- 125.Subak H., Ozkan-Ariksoysal D. Label-free electrochemical biosensor for the detection of Influenza genes and the solution of guanine-based displaying problem of DNA hybridization. Sensor. Actuator. B Chem. 2018;263:196–207. [Google Scholar]
- 126.Lee T., Park S.Y., Jang H., Kim G.-H., Lee Y., Park C., Mohammadniaei M., Lee M.-H., Min J. Fabrication of electrochemical biosensor consisted of multi-functional DNA structure/porous au nanoparticle for avian influenza virus (H5N1) in chicken serum. Mater. Sci. Eng. C. 2019;99:511–519. doi: 10.1016/j.msec.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 127.Farzin L., Sadjadi S., Sheini A., Mohagheghpour E. A nanoscale genosensor for early detection of COVID-19 by voltammetric determination of RNA-dependent RNA polymerase (RdRP) sequence of SARS-CoV-2 virus. Microchimica Acta. 2021;188:121. doi: 10.1007/s00604-021-04773-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kudr J., Michalek P., Ilieva L., Adam V., Zitka O. COVID-19: a challenge for electrochemical biosensors. Trac. Trends Anal. Chem. 2021;136:116192. doi: 10.1016/j.trac.2021.116192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Imran S., Ahmadi S., Kerman K. Electrochemical biosensors for the detection of SARS-CoV-2 and other viruses. Micromachines. 2021;12:174. doi: 10.3390/mi12020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao H., Liu F., Xie W., Zhou T.C., OuYang J., Jin L., Li H., Zhao C.Y., Zhang L., Wei J., Zhang Y.P., Li C.P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hashemi S.A., Golab Behbahan N.G., Bahrani S., Mousavi S.M., Gholami A., Ramakrishna S., Firoozsani M., Moghadami M., Lankarani K.B., Omidifar N. Ultra-sensitive viral glycoprotein detection NanoSystem toward accurate tracing SARS-CoV-2 in biological/non-biological media. Biosens. Bioelectron. 2021;171:112731. doi: 10.1016/j.bios.2020.112731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Miripour Z.S., Sarrami-Forooshani R., Sanati H., Makarem J., Taheri M.S., Shojaeian F., Eskafi A.H., Abbasvandi F., Namdar N., Ghafari H., Aghaee P., Zandi A., Faramarzpour M., Hoseinyazdi M., Tayebi M., Abdolahad M. Real-time diagnosis of reactive oxygen species (ROS) in fresh sputum by electrochemical tracing; correlation between COVID-19 and viral-induced ROS in lung/respiratory epithelium during this pandemic. Biosens. Bioelectron. 2020;165:112435. doi: 10.1016/j.bios.2020.112435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fabiani L., Saroglia M., Galata G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020 doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171:112709. doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Geng B., Muenker M.C., Moore A.J., Vogels C.B.F., Petrone M.E., Ott I.M., Lu P., Venkataraman A., Lu-Culligan A., Klein J., Earnest R., Simonov M., Datta R., Handoko R., Naushad N., Sewanan L.R., Valdez J., White E.B., Lapidus S., Kalinich C.C., Jiang X., Kim D.J., Kudo E., Linehan M., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Weizman O.-E., Wong P., Yang Y., Bermejo S., Odio C., Omer S.B., Dela Cruz C.S., Farhadian S., Martinello R.A., Iwasaki A., Grubaugh N.D., Ko A.I. medRxiv; 2020. Saliva Is More Sensitive for SARS-CoV-2 Detection in COVID-19 Patients than Nasopharyngeal Swabs. 2020.2004.2016.20067835. [Google Scholar]