Abstract
The formation of various tissues requires close communication between two groups of cells, epithelial and mesenchymal cells. COUP-TFs are transcription factors which have been shown to have functions in embryonic development. COUP-TFI is expressed mainly in the nervous system, and its targeted deletion leads to defects in the central and peripheral nervous systems. COUP-TFII is highly expressed in the mesenchymal component of the developing organs. A null mutation of COUP-TFII results in the malformation of the heart and blood vessels. From their expression pattern, we proposed that COUP-TFs regulate paracrine signals important for mesenchymal cell-epithelial cell interactions. In order to identify genes regulated by COUP-TF in this process, a rat urogenital mesenchymal cell line was stably transfected with a COUP-TFI expression vector. We found that NGFI-A, a gene with important functions in brain, organ, and vasculature development, has elevated mRNA and protein levels upon overexpression of COUP-TFI in these cells. A study of the promoter region of this gene identified a COUP-TF-responsive element between positions −64 and −46. Surprisingly, this region includes binding sites for members of the Sp1 family of transcription factors but no COUP-TF binding site. Mutations that abolish the Sp1 binding activity also impair the transactivation of the NGFI-A promoter by COUP-TF. Two regions of the COUP-TF molecule are shown to be important for NGFI-A activation: the DNA binding domain and the extreme C terminus of the putative ligand binding domain. The C-terminal region is likely to be important for interaction with coactivators. In fact, the coactivators p300 and steroid receptor activator 1 can enhance the transactivation of the NGFI-A promoter induced by COUP-TFI. Finally, we demonstrated that COUP-TF can directly interact with Sp1. Taken together, these results suggest that NGFI-A is a target gene for COUP-TFs and that the Sp1 family of transcription factors mediates its regulation by COUP-TFs.
COUP-TFs are transcription factors structurally related to the nuclear receptor superfamily (60), which embraces a growing number of hormone receptors (retinoic acid, thyroid hormone, vitamin D, and steroid hormones, among others) as well as many other proteins, such as COUP-TFs themselves, with unknown ligands (48). The structure of the members of this nuclear receptor superfamily consists of an N-terminal ligand-independent activation domain (activation function 1 [AF-1]), a highly conserved DNA binding domain (DBD), a hinge region, and a ligand binding domain (LBD). The LBD contains a ligand-dependent activation function (AF-2) (57).
There are two COUP-TF genes, COUP-TFI and COUP-TFII (for a review, see reference 58), which share a high degree of homology in their DBDs and putative LBDs. In addition, COUP-TFI and COUP-TFII are highly conserved among different species, suggesting that they play important roles in development. Although the expression of COUP-TFI and COUP-TFII is highly overlapping in mice (24, 43), COUP-TFI is abundantly expressed in the central and peripheral nervous system, while COUP-TFII is highly expressed in the mesenchymal component of the developing organs (46). Based on this expression pattern, we have proposed that COUP-TFI is important for neural development and that COUP-TFII is important in organogenesis through the regulation of mesenchymal cell-epithelial cell interactions. Indeed, the targeted disruption of either of these two genes in mice results in a lethal phenotype. COUP-TFI-deficient mice show defects in the development of the central and peripheral nervous systems (47; unpublished observations), whereas mutation of the COUP-TFII gene results in defects in the heart and vasculature formation (unpublished observations).
Vasculature and organ formation requires very tight communication between epithelial cell and mesenchymal cell populations (7). This communication is essential for the differentiation of these two cell types (14). Since COUP-TFII is highly expressed in the mesenchyme but is undetectable in the epithelium of most organs, it is likely that COUP-TFII plays an important role in mesenchymal cell-epithelial cell interactions during organogenesis. This process has been well studied for the prostate, where several growth factors and the extracellular matrix are involved in this communication between the mesenchyme and the epithelium (11, 19).
COUP-TFs are able to bind to a variety of dispositions of the basic AGGTCA motif, including those recognized by the receptors of retinoic acid (RAR), thyroid hormone, and vitamin D (13). This ability makes COUP-TFs capable of competing for the response elements of these receptors, thus acting as passive repressors of the transcriptional activation induced by them (12, 13, 56). Another mechanism of passive repression by COUP-TFs involves their ability to heterodimerize with the 9-cis retinoic acid receptor (RXR), reducing its availability for other nuclear receptors that use it as a partner. In addition, COUP-TFs contain an active repression domain within their putative LBDs (1, 31). This repression domain is capable of interacting with corepressors such as SMRT and N-CoR (50), molecules that can recruit histone deacetylase activities to the DNA to suppress transcription (2, 23, 39).
NGFI-A (38), also known as Egr-1 (53), Krox-24 (3), Zif268 (9, 29), TIS8 (32), CEF5 (51), or zfp-6 (15), is an early response gene that has also been shown to be expressed in prostate cancer. In a prostatic cancer cell line (LNCaP), NGFI-A regulates the expression of the retinoblastoma (Rb) gene and induces apoptosis (16, 17). In addition, several other key genes involved in vasculature and organ formation, e.g., those for transforming growth factor β1 (TGFβ1), platelet-derived growth factor A chain (PDGF-A), and basic fibroblast growth factor (FGF), are also transactivated by NGFI-A (6, 26, 27, 35). Furthermore, mice lacking both NGFI-A and NGFI-C, another member of this family of transcription factors, have severe defects in the development of all the accessory glands of the male reproductive tract (38a). All of this evidence suggests that NGFI-A may serve as a mediator for COUP-TFs during organogenesis.
In order to study the role of COUP-TFs in prostate development, we generated several rat urogenital mesenchymal (rUGM) cell lines that overexpress COUP-TFI. Using these cell lines, we identified NGFI-A as one of its target genes but, surprisingly, the regulation was positive. In addition, we identified a sequence of 19 bp responsible for COUP-TF induction. This sequence contains two imperfect Sp1 and Sp3 binding sites, and band shift analysis demonstrated that both Sp1 and Sp3 can indeed bind to this sequence. Using truncation mutants, we demonstrated that the DBD and the extreme C terminus of COUP-TF are necessary for the activation of the NGFI-A promoter. Finally, we showed that the coactivators p300 and steroid receptor coactivator 1 (SRC-1) can enhance the positive action of COUP-TF on the NGFI-A promoter. These results demonstrate that NGFI-A is a target gene for COUP-TF in urogenital mesenchymal cells and can be a mediator of the possible functions of COUP-TF in prostate development.
MATERIALS AND METHODS
Tissue culturing and generation of the stably transfected cell lines.
The rUGM cell line (62) was kindly provided by Leland Chung (University of West Virginia). These cells were grown in T medium (10) supplemented with 5% fetal bovine serum and 1% penicillin-streptomycin (Life Sciences-Gibco BRL). The stable cell lines were generated as follows. Ten-centimeter dishes of rUGM cells were transfected by the calcium phosphate method with plasmid pCNX containing the COUP-TFI open reading frame and the neomycin resistance gene. Six hours after transfection, the cells were washed and grown in fresh medium for 2 days. Cells were then split into 10 10-cm plates with regular medium containing 300 μg of Geneticin (Life Sciences-Gibco BRL) per ml, determined earlier to be the effective dose of antibiotic capable of killing untransfected cells. Cells were grown for 2 weeks, and colonies were picked and expanded.
Western and Northern blotting.
The clones obtained were checked for COUP-TF mRNA and protein expression levels. For protein analyses, nuclear extracts were obtained as previously described (4). Twenty micrograms of nuclear extract was run on sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to nitrocellulose membranes. After visualization of the transferred proteins by Ponceau staining, the membranes were blocked in 3% milk for 1 h and incubated with COUP-TF antiserum diluted 1:1,000 in 0.3% milk for 2 h. Membranes were washed, incubated with a horseradish peroxidase-conjugated antibody diluted 1:10,000 in 0.3% milk for 45 min, and washed again. Proteins were detected with a Renaissance chemiluminescence kit (NEN) by following the manufacturer’s directions. The process was repeated with an anti-NGFI-A polyclonal antibody (Santa Cruz) under the same conditions.
For Northern analyses, total RNA was prepared by the protocol of Chomczynski and Sacchi (8). Twenty micrograms of total RNA was denatured, electrophoresed in 2.2 M formaldehyde–1% agarose gels in morpholinepropanesulfonic acid (MOPS) buffer at 120 V for 3 to 4 h, and transferred to nylon membranes (Zeta-probe; Bio-Rad) overnight. RNA was cross-linked to the membranes by UV irradiation. A reverse transcription-PCR-generated clone of the NGFI-A mRNA open reading frame was used as a template to synthesize [α-32P]dCTP-labeled probes by random priming. The radiolabeled probes were hybridized to the blots in 50% formamide–3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.2% SDS at 42°C for 20 h. Unhybridized probes were removed by washing once in 2× SSC–0.5% SDS for 40 min at 65°C and three times in 0.2× SSC–0.5% SDS for 15 min at 65°C. Filters were then processed for autoradiography.
Promoter deletion analysis and transient transfections.
Three constructs containing fragments of the rat NGFI-A promoter between 5′ positions −1386, −807, and −389 and 3′ position +43 in plasmid pXP1 were kindly provided by Ana Pérez Castillo. Further deletions of the promoter were generated by PCR. The PCR products were cloned into vector pCR3.1 (Invitrogen) with a TA cloning kit. After determination of the orientation, the clones were inserted into vector pXP2. An oligonucleotide containing the sequence between positions −64 and −46 of the NGFI-A promoter (TCACGGCGGAGGCGGGCCC) as well as the sequence of a consensus TATA box (GGGTATATAA) was cloned into plasmid pXP2 to generate the pXP2−64/−46TATA construct. To generate the COUP-TF-responsive element mutant reporter constructs, oligonucleotides containing the mutations fused to a TATA box were cloned into vector pXP2. The mutant oligonucleotides were as follows: mutation 1, TCACTTCGGAGGCGGGCCC; mutation 2, TCACGGCGGATTCGGGCCC; mutation 3, TCACTTCGGATTCGGGCCC; mutation 4, TCACGGCTTAGGCGGGCCC; and mutation 5, TCACGGCGGAGGCTTGCCC (underlining indicates mutated nucleotides).
For transfection experiments, HeLa cells grown in six-well plates at a density of 3 × 105 cells/well were treated with Lipofectin (Life Sciences-Gibco BRL) in the presence of 250 ng of reporter construct and various amounts of expression plasmids. Cells were maintained in the presence of the Lipofectin complexes for 10 h, washed with Hanks’ buffered solution, and then grown in Dulbecco modified Eagle medium containing 10% fetal bovine serum. After 36 h of incubation, the cells were harvested. For transfection of rUGM cells, the cells were grown in six-well plates to a density of 105 cells/well in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Reporter plasmid (250 ng) and various amounts of expression plasmids were transfected into the cells with FuGene 6 (Boehringer Mannheim Biochemicals). Cells were harvested 36 h after transfection.
Band shift analysis.
Nuclear protein extracts were obtained from various cell lines as previously described (4). A double-stranded oligonucleotide containing the sequence between positions −64 and −46 of the NGFI-A promoter flanked by two unannealed restriction sites (Acc 65I and XhoI) was labeled by filling in of the uncoupled ends with Sequenase (Amersham) in the presence of [α-32P]dATP and [α-32P]dCTP. The nuclear extracts were incubated on ice for 15 min with 1 μg of poly(dI-dC) (Pharmacia) in 2.5% glycerol–10 mM Tris (pH 7.5)–50 mM NaCl–1 mM EDTA–1 mM dithiothreitol–1 mM phenylmethylsulfonyl fluoride–0.5 μg of bovine serum albumin per ml. Antibodies or competitor oligonucleotides were added during this incubation. Upon addition of 30,000 cpm of the oligonucleotide probe, the reaction mixture was incubated for 15 min on ice and then resolved on a 5% polyacrylamide gel run at 25 mA for 2.5 h. The gel was dried and exposed to X-ray film.
GST pull-down assays.
Glutathione S-transferase (GST) or GST–COUP-TFI proteins were expressed in Escherichia coli, and whole-cell extracts were prepared by sonication and centrifugation. Extracts from bacteria expressing GST alone or GST–COUP-TFI were run on a polyacrylamide gel to determine the concentrations of these proteins in the extracts. Equal amounts of GST or GST–COUP-TFI were incubated for 1 h at room temperature with glutathione-Sepharose 4B beads (Pharmacia) in NETN buffer (20 mM Tris [pH 8], 50 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40). Subsequently, the beads were washed twice with NETN buffer and incubated overnight with 35S-labeled Sp1 that had been produced in a rabbit reticulocyte lysate system (TNT; Promega) in NETN buffer. Finally, the beads were washed five times with NETN buffer, dried, resuspended in 30 μl of loading buffer (41), and boiled to release the interacting proteins. The released proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and the gel was dried and exposed to X-ray film.
RESULTS
rUGM cell lines overexpressing COUP-TFI have elevated levels of NGFI-A mRNA and protein.
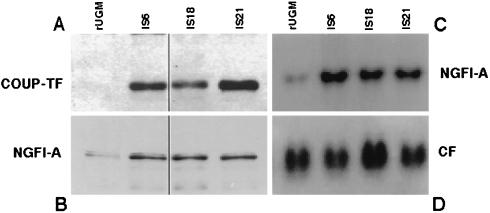
In order to study the role of COUP-TFs in prostate development, rUGM cell lines overexpressing COUP-TFI were established. Nuclear protein extracts from the overexpressing clones were analyzed by Western blotting and incubated with antibodies against COUP-TF. Figure 1A shows the levels of COUP-TFs in several overexpressing clones (IS6, IS18, and IS21) compared to the levels in the original cell line (rUGM). Clearly, the selected clones contain elevated levels of COUP-TFs. It is known that NGFI-A affects the proliferation and differentiation of cells (44) and is overexpressed in prostate cancer and in the prostatic cancer cell line LNCaP (17). It activates the expression of the Rb gene in these cells (16) as well as other genes (those for TGFβ1 [33], basic FGF [6], PDGF-A [26], and PDGF-B [25]) considered to be important for organogenesis. To assess whether NGFI-A is upregulated in the COUP-TF-overexpressing cell lines, the same blots were incubated with antibodies against proteins with known roles in organogenesis. As shown in Fig. 1B, the COUP-TFI-overexpressing cell lines (IS6, IS18, and IS21) contain three to five times more NGFI-A transcription factor than control untransfected cells. This effect occurs at the mRNA level, since the accumulation of the NGFI-A protein correlates with the accumulation of its mRNA, as assessed by Northern blotting (Fig. 1C). The blots were also hybridized with a cyclophilin probe to indicate equal loading in the filters (Fig. 1D). Thus, the overexpression of COUP-TFI in rUGM cells leads to an increased accumulation of NGFI-A mRNA and elevated levels of NGFI-A protein in these cells.
FIG. 1.
Elevated expression of NGFI-A in COUP-TFI-overexpressing rUGM cells. rUGM cells were stably transfected with a cytomegalovirus-driven COUP-TFI expression plasmid, and transfectants were selected in medium containing 300 μg of neomycin per ml. (A and B) Twenty micrograms of nuclear protein extract from three surviving clones (IS6, IS18, and IS21) was analyzed by Western blotting (A and B). The same membrane was incubated with an anti-COUP-TF antibody (A) and with an anti-NGFI-A antibody (B). The levels of COUP-TFs and NGFI-A proteins in the transfected clones and in the original cell line, rUGM, are shown. (C and D) Twenty micrograms of total RNA from the same clones (IS6, IS18, and IS21) was analyzed by Northern blotting. The amounts of NGFI-A mRNA in these clones and in the original cell line, rUGM, are shown (C). As a loading control, the filter was also hybridized with a cyclophilin probe (D).
Both COUP-TFI and COUP-TFII are able to activate NGFI-A promoter activity.
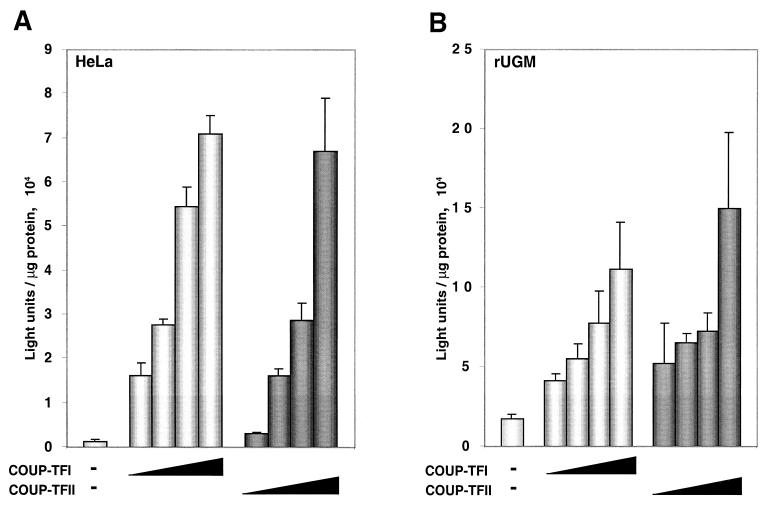
The elevated levels of NGFI-A mRNA in stably transfected cell lines overexpressing COUP-TFI suggested possible upregulation of the NGFI-A promoter by COUP-TFs. In order to determine whether the increase in the levels of NGFI-A mRNA caused by COUP-TFI is due to transcriptional regulation, we carried out transfection experiments to determine whether COUP-TF can upregulate NGFI-A promoter activity. For this purpose, we used a reporter construct containing the region spanning positions −1386 to +43 of the NGFI-A promoter. This construct was cotransfected with expression vectors for either COUP-TFI or COUP-TFII in HeLa cells. Figure 2A shows that both COUP-TFI and COUP-TFII can induce the expression of the NGFI-A promoter-driven luciferase reporter. The enhancement of reporter activity is proportional to the quantity of the cotransfected COUP-TF expression vector. Similar experiments were carried out with rUGM cells. Figure 2B clearly shows that COUP-TFs augment NGFI-A promoter activity in a dose-dependent manner. These results show that both COUP-TFI and COUP-TFII have similar effects on NGFI-A promoter activity in different cell lines and suggest that the effect of COUP-TF on NGFI-A mRNA levels is mediated by direct or indirect activation of a COUP-TF-responsive element in the NGFI-A promoter region.
FIG. 2.
Dose-dependent activation of the NGFI-A promoter by COUP-TFs. (A) A luciferase reporter plasmid (250 ng) containing 1.4 kb of the rat NGFI-A promoter region was transfected along with increasing amounts of COUP-TFI or COUP-TFII expression vector (0.1, 0.25, 0.5, and 0.75 μg). HeLa cells were lysed in 200 μl of lysis buffer (41), and 20-μl volumes of the extracts were assayed for luciferase activity. (B) The same reporter vector (125 ng) was transfected along with increasing amounts of COUP-TFI or COUP-TFII expression vector (0.05, 0.125, 0.25, and 0.375 μg). rUGM cells were lysed in 200 μl of lysis buffer, and 20-μl volumes of the lysates were assayed for luciferase activity. The values presented are the means of a representative experiment done in triplicate, and the error bars correspond to the standard deviations.
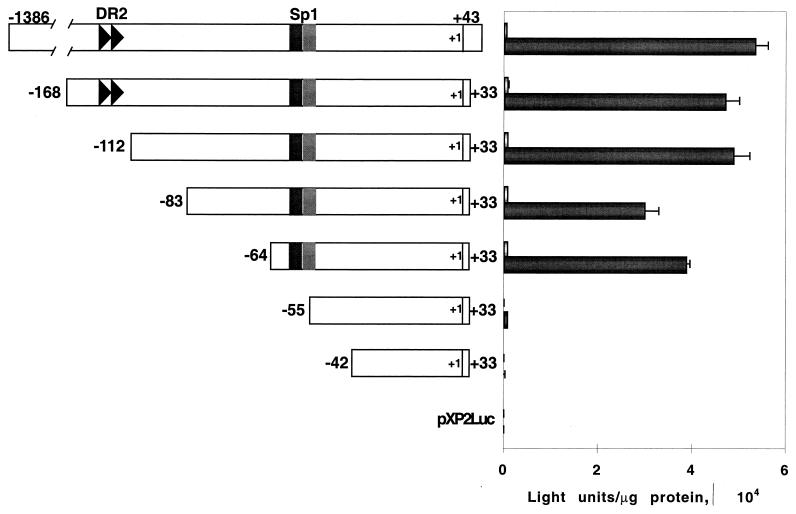
To define the region of the NGFI-A promoter that responds to COUP-TF activation, we generated a series of 5′ deletion mutants of the NGFI-A promoter construct. Figure 3 shows that deletion from positions −1386 to −64 of the NGFI-A promoter region has no significant impact on its response to COUP-TF. However, sequence analysis revealed a direct repeat of basic motif AGGTCA with a spacing of 2 nucleotides (DR2) between positions −144 and −130. DR2 is a binding site for COUP-TFs, as confirmed by band shift analysis (data not shown). Thus, it is surprising that the deletion of this region does not affect the activation of the promoter by COUP-TFs (Fig. 3). Further deletion from positions −64 to −55 resulted in a complete loss of COUP-TF responsiveness. Therefore, the COUP-TF-responsive element is localized between positions −64 and −55.
FIG. 3.
Deletion analysis of the NGFI-A promoter and delimitation of the COUP-TF-responsive element. The deletion mutants were generated by PCR and finally cloned in vector pXP2. Reporter constructs (250 ng) were transfected with 750 ng of an expression vector for COUP-TFI or the empty vector. On the right, the amount of luciferase activity in the cells transfected with the adjacent construct is shown. The dark bars correspond to the cells cotransfected with the COUP-TFI expression vector. The light bars correspond to the cells cotransfected with the empty vector. The values represent the means for three different cell culture wells, and the error bars represent the standard deviations.
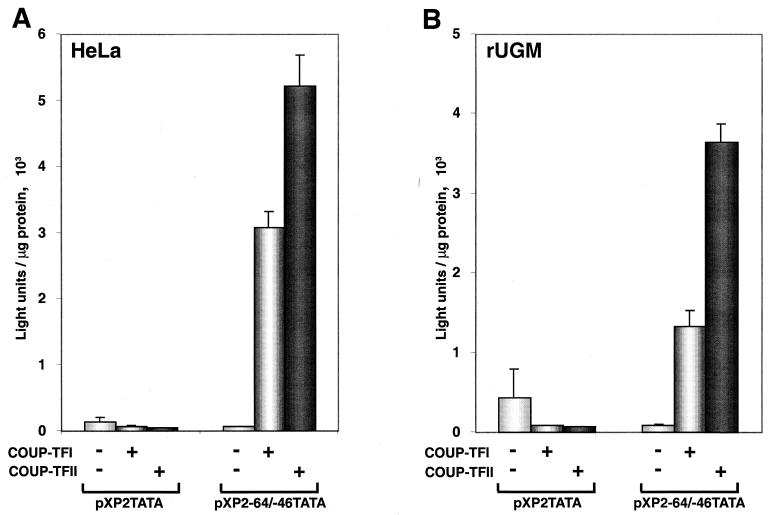
In order to confirm that the COUP-TF-responsive element is located in this region, we tested if a 19-bp oligonucleotide containing the sequence between positions −64 and −46 could confer COUP-TF inducibility to a heterologous promoter composed of a TATA box. As shown in Fig. 4, a reporter construct containing a single copy of the nucleotide sequence between positions −64 and −46 of the NGFI-A promoter was highly activated when cotransfected with either a COUP-TFI or a COUP-TFII expression vector in both HeLa and rUGM cells. In contrast, the basic promoter control construct was not activated by either COUP-TF vector. Taken together, these results demonstrate the existence in the NGFI-A promoter of a discrete COUP-TF-responsive element that localizes to the region between positions −64 and −46.
FIG. 4.
The COUP-TF-responsive element is located between positions −64 and −46 of the NGFI-A promoter. Oligonucleotides containing the sequence of the NGFI-A promoter between positions −64 and −46 were cloned into vector pXP2 containing a minimal promoter (TATA box). Either pXP2TATA or pXP2−64/−46TATA (250 ng) was transfected along with 750 ng of an expression plasmid for COUP-TFI or COUP-TFII or the empty vector as a control in HeLa (A) and rUGM (B) cells. The experiment was performed in triplicate. Error bars indicate standard deviations.
The COUP-TF-responsive element of the NGFI-A promoter region contains Sp1 binding sites.
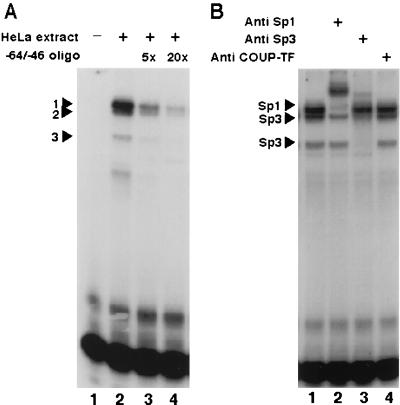
Close examination of the nucleotide sequence of the COUP-TF-responsive element on the NGFI-A promoter revealed several potential Sp1 binding sites. Sp1 and COUP-TFs have been shown to interact with and activate the human immunodeficiency virus (HIV) long terminal repeat (49). In order to identify the proteins that bind to the COUP-TF-responsive element of the NGFI-A promoter, we performed band shift analysis of this sequence. Incubation of a nuclear protein extract from HeLa cells with a labeled oligonucleotide probe containing the COUP-TF-responsive element of the NGFI-A promoter resulted in the formation of three major complexes that we could resolve by electrophoresis (Fig. 5A, lane 2). These complexes are specific, since the addition of an excess of unlabeled probe disrupted their formation (Fig. 5A, lanes 3 and 4). Preincubation of the nuclear protein extract with antibodies against the Sp1 and Sp3 transcription factors prior to the addition of the probe resulted in a reduction in the mobilities of band 1 and bands 2 and 3, respectively (Fig. 5B, compare lane 1 to lane 2 or 3). This result demonstrates that both factors can bind to this sequence. In addition, incubation of purified Sp1 protein with this element yields a complex with the same mobility properties as complex 1 (data not shown). In contrast, incubation of the nuclear protein extract with antibodies against COUP-TF had no effect on the mobilities of any of the complexes (Fig. 5B, lane 4). Band shift analysis of nuclear protein extracts from rUGM cells yielded similar results (data not shown).
FIG. 5.
The COUP-TF-responsive element of the NGFI-A gene is an Sp1 family binding site. (A) When 5 μg of a nuclear protein extract from HeLa cells was incubated with the 32P-labeled oligonucleotide containing the sequence between positions −64 and −46 of the NGFI-A promoter, three major complexes were observed (lane 2, complexes 1, 2, and 3 denoted by arrowheads). The formation of these complexes was inhibited by a 5- or 20-fold molar excess of the unlabeled probe in order to assess their specificity (lane 3 or 4, respectively). (B) Preincubation of the protein extract with antibodies against Sp1 and Sp3 altered the mobilities of specific bands (lanes 2 and 3, respectively), denoted here with arrowheads and the names of the proteins recognized in the complexes. Preincubation with an anti-COUP-TF antibody did not have any effect on the mobilities of these three complexes (lane 4).
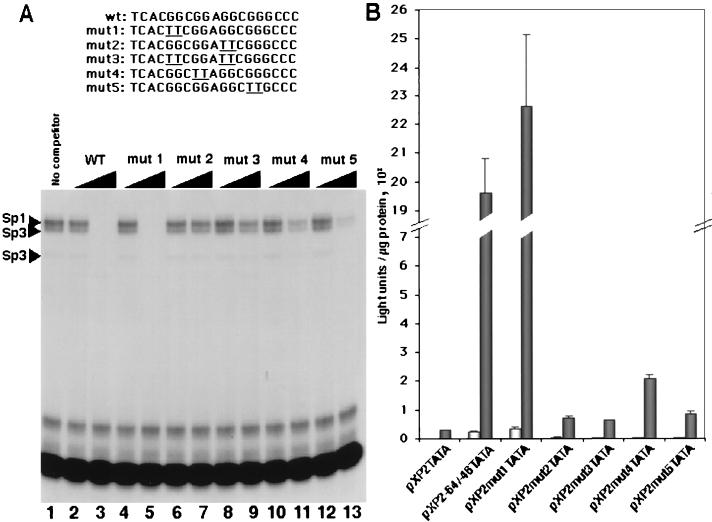
To determine which base pairs are important for the binding of these complexes, we performed mutational analysis of the COUP-TF-responsive element. The mutations that we generated changed selected pairs of guanidine residues to pairs of thymidine residues throughout the entire COUP-TF-responsive element region, as shown in Fig. 6. First, we tested the ability of oligonucleotides containing the mutations to compete for the formation of the complexes detected in the band shift analysis. An oligonucleotide containing mutation 2 or 3 was unable to compete for the binding of any of the complexes, even at a 50-fold molar excess (Fig. 6A, compare lanes 2 and 3 to lanes 6 and 7 and lanes 8 and 9). An oligonucleotide carrying mutation 4 or 5 was able to compete for complex formation, but with a lower efficiency (Fig. 6A, compare lanes 2 and 3 to lanes 10 and 11 and lanes 12 and 13). In contrast, mutation 1 had no effect on the ability of the oligonucleotide to compete for Sp1 or Sp3 binding to the sequence from positions −64 to −46 (Fig. 6A, compare lanes 2 and 3 to lanes 4 and 5).
FIG. 6.
The disruption of the binding site for Sp1 in the COUP-TF-responsive element impairs transactivation by COUP-TF. (A) Gel shift analysis of the complexes formed after incubation of the COUP-TF-responsive element with HeLa cell nuclear extracts and their inhibition by various mutant oligonucleotides. A HeLa cell nuclear protein extract (5 μg per lane) was incubated with a 5- or 50-fold molar excess of the competitor oligonucleotide prior to incubation with a 32P-labeled oligonucleotide containing the wild-type (wt or WT) COUP-TF-responsive element. (B) The same oligonucleotides as in panel A, cloned next to a minimal TATA box promoter in plasmid pXP2, were transfected into HeLa cells in the presence of a COUP-TFII expression vector (dark bars) or the empty vector (light bars). The graph shows the average luciferase activities of a representative experiment done in triplicate. Error bars represent standard deviations.
To assess the effect of these mutations on the enhancement of NGFI-A promoter activity by COUP-TF, the oligonucleotides carrying the mutations or the wild-type COUP-TF-responsive element were inserted 5′ to a TATA box minimal promoter. These constructs were then cotransfected with expression plasmids for COUP-TFI or COUP-TFII. Figure 6B shows that mutations affecting the binding of the Sp1 or Sp3 protein to the COUP-TF-responsive element also impair the ability of the COUP-TF-responsive element to mediate transactivation by COUP-TFI. Moreover, a tight correlation exists between the level of competition in the band shift assays and the level of transactivation mediated by the mutated oligonucleotides in the transfection experiments. Similar results were obtained when a COUP-TFII expression vector was cotransfected with the reporter constructs (data not shown). These results indicate that the central GC-rich Sp1 binding site in the region from positions −64 to −46 is important for COUP-TF activation of NGFI-A promoter activity.
COUP-TFI and Sp1 physically interact in vitro.
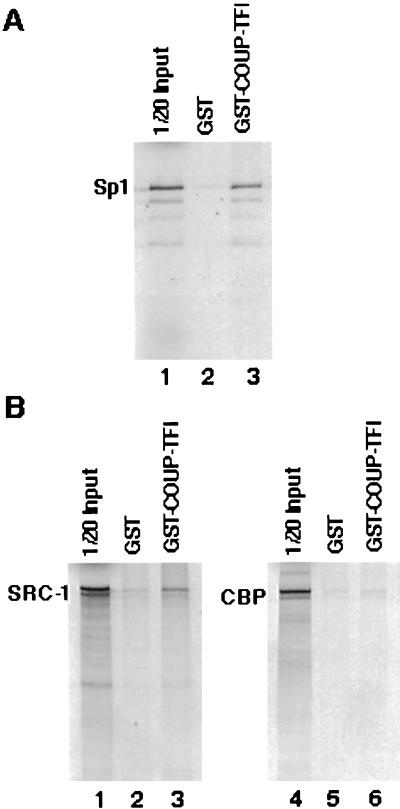
Gel shift analysis of the COUP-TF-responsive element incubated with either HeLa cell or COUP-TF-overexpressing rUGM cell nuclear extracts (Fig. 5 and data not shown) failed to show a direct interaction of COUP-TF with its responsive element in the NGFI-A promoter. In addition, the lack of a consensus COUP-TF binding site within the COUP-TF-responsive element suggests that COUP-TFs may transactivate the NGFI-A promoter through an interaction with Sp1. In order to test this possibility, we performed a GST pull-down assay. A GST–COUP-TFI fusion protein was incubated with a 35S-labeled Sp1 protein, and the complex formed was retained on glutathione-Sepharose beads and analyzed by SDS-PAGE. Figure 7A shows that a GST–COUP-TFI fusion protein can effectively retain Sp1 on a glutathione-Sepharose resin. In contrast, Sp1 fails to interact with GST alone. This result clearly indicates that COUP-TFI interacts specifically with Sp1 in vitro and suggests that Sp1 can serve as a docking protein for recruiting COUP-TF to the NGFI-A promoter and for mediating COUP-TF transactivation.
FIG. 7.
COUP-TFI interacts with Sp1 and SRC-1 in vitro. (A) Equal amounts of GST protein (lane 2) or GST–COUP-TFI fusion protein (lane 3) were incubated with 35S-labeled Sp1 protein. The complexes were retained on glutathione-Sepharose 4B beads and then analyzed by SDS-PAGE. A 1/20 quantity of the Sp1 protein input was run in parallel as a reference (lane 1). (B) Equal amounts of GST protein (lanes 2 and 5) or GST–COUP-TFI fusion protein (lanes 3 and 6) were incubated with radiolabeled coactivator SRC-1 (lanes 2 and 3) or CBP (lanes 5 and 6). The complexes were retained on a glutathione-Sepharose 4B matrix and resolved by SDS-PAGE. A 1/20 quantity of the SRC-1 (lane 1) or CBP (lane 4) protein input was run in parallel as a reference.
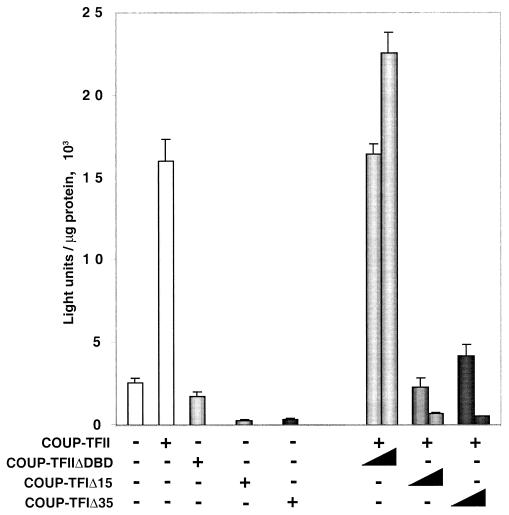
Both the DBD and the extreme C terminus of COUP-TFs are required for activation of the NGFI-A promoter.
COUP-TFs have been traditionally known as active or passive transcriptional repressors. There is evidence that transrepression and active repression are the predominant mechanisms of COUP-TF repressor activity (31). A recent report shows that the transrepression domain of the COUP-TFs includes their DBD as well as a region within their LBD (1). Leng et al. (31) demonstrated that a 35-amino-acid truncation of the C terminus of COUP-TFI is enough to eliminate its active repression activity. Furthermore, Shibata et al. (50) showed that this same truncation eliminates interactions with the corepressors SMRT and N-CoR. Thus, we tested whether deletion of the DBD or truncation of the C terminus of COUP-TF affected its NGFI-A promoter activation potential. As shown in Fig. 8, a COUP-TFII construct lacking its DBD is unable to activate the NGFI-A promoter. In addition, a truncation of as little as 15 amino acids from the C terminus of COUP-TFI is sufficient to completely abolish its ability to activate the NGFI-A promoter (Fig. 8). Moreover, this truncated protein acts as a dominant negative inhibitor of the wild-type protein. As expected, a larger truncation of 35 amino acids from the C terminus of COUP-TFI has a similar effect on the activation of the NGFI-A promoter. In contrast, DBD-lacking COUP-TFII did not affect the transactivation mediated by wild-type COUP-TF (Fig. 8). These results suggest the existence of two interfaces in the COUP-TF activating protein. The DBD appears to be necessary for direct or indirect targeting of COUP-TF to the DNA element, and the extreme C terminus appears to contain the interface needed to interact with coactivators or the basal transcriptional machinery.
FIG. 8.
Both the DBD and the 15 most C-terminal amino acids of COUP-TF are required for NGFI-A promoter activation. A reporter plasmid (250 ng) containing the sequence between positions −64 and +33 of the NGFI-A gene was transfected into HeLa cells along with a DBD deletion COUP-TFII mutant (COUP-TFIIΔDBD), COUP-TFI with a 15-amino-acid truncation of the C terminus (COUP-TFIΔ15), COUP-TFI with a 35-amino-acid or truncation of the C terminus (COUP-TFIΔ35) in the presence or absence of wild-type COUP-TFII. The experiment was performed in triplicate, and the graph shows the average luciferase activities and standard deviations.
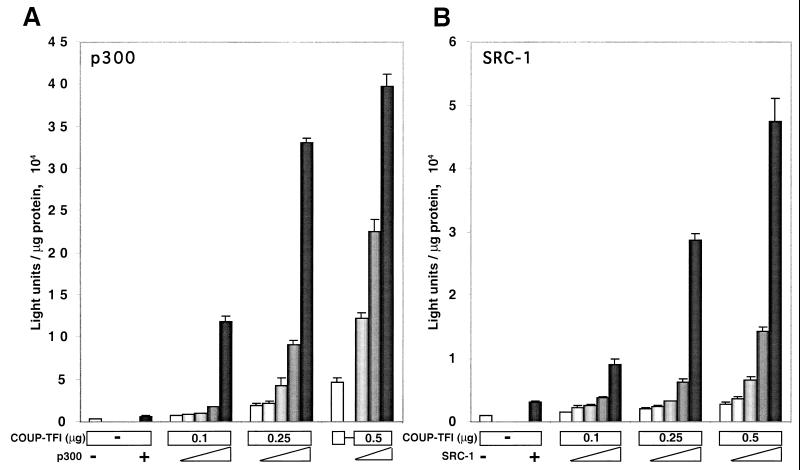
The coactivators p300 and SRC-1 further increase the activation of the NGFI-A promoter by COUP-TFs.
Coactivators have been shown to be important proteins for the action of nuclear receptors (41, 59), serving as bridges between them and the basal transcriptional machinery and recruiting histone acetylase activity to the chromatin of the receptor target genes (22, 40, 52). In our system, COUP-TFs function as transcriptional activators, so we wanted to test if coactivators could further enhance COUP-TF upregulation of the NGFI-A promoter. To this end, we cotransfected increasing amounts of expression vectors for two known coactivators, p300 and SRC-1, with a COUP-TFI expression vector. Figure 9A shows that COUP-TFI can transactivate the region from positions +33 to −64 of the NGFI-A promoter in a dose-dependent manner. An increase in the concentration of p300 enhances this transactivation of the NGFI-A promoter by COUP-TFI in a dose-dependent manner. In addition, the coactivation effect of p300 is proportional to the concentration of COUP-TFI. A similar result was observed when SRC-1 was cotransfected with a COUP-TFI expression vector (Fig. 9B). SRC-1 enhanced COUP-TFI transactivation of the region from positions −64 and +33 of the NGFI-A promoter in a dose-dependent manner and in proportion to the amount of COUP-TFI. Taken together, these results suggest that coactivators, such as p300 and SRC-1, play an important role in the activation of the NGFI-A promoter by COUP-TFI.
FIG. 9.
Coactivators p300 and SRC-1 can enhance the transactivation of the NGFI-A promoter by COUP-TFI. A reporter luciferase construct (250 ng) containing the sequence between positions −64 and +33 of the NGFI-A promoter was cotransfected into HeLa cells with the indicated amounts of a COUP-TFI expression plasmid and increasing amounts of expression vectors (0.25, 0.5, 0.75, and 1 μg) for either p300 (A) or SRC-1 (B). The experiment was performed in triplicate, and the graph shows the average luciferase activities and standard deviations.
COUP-TFs interact with SRC-1 in vitro.
Since the coactivators SRC-1 and CREB binding protein (CBP)/p300 are important for COUP-TF activation of the NGFI-A promoter, a possible mechanism of this activation would be the recruitment of these molecules to the NGFI-A promoter by either Sp1, COUP-TF, or both. We have previously demonstrated that SRC-1 can stimulate nuclear receptors and Sp1-mediated target gene expression (41). To verify that COUP-TF is also able to interact with the coactivators SRC-1 and CBP/p300, we performed a GST pull-down assay with a GST–COUP-TFI fusion protein and radiolabeled SRC-1 or CBP (Fig. 7B). We found that SRC-1 interacts with COUP-TFI specifically, since it is retained by GST–COUP-TFI but not by GST alone (Fig. 7B, compare lanes 3 and 2) on a glutathione-Sepharose matrix. In contrast, we failed to observe any specific interaction of CBP with COUP-TFI in a similar GST pull-down assay (Fig. 7B, compare lanes 6 and 5). These results suggest a direct interaction of SRC-1 with COUP-TF and/or Sp1 as the mechanism for the activation of the NGFI-A promoter by SRC-1. In contrast, coactivation of COUP-TF by CBP/p300 may be exerted through other, indirect interaction mechanisms.
DISCUSSION
COUP-TF was first described as an essential factor for the transcription of the chicken ovalbumin gene (42) and was subsequently cloned from many other organisms (for a review, see reference 58). The amino acid sequence conservation of COUP-TFs in different species suggests that COUP-TFs play important roles in vivo. Indeed, the targeted disruption of either COUP-TFI or COUP-TFII results in perinatal or embryonic lethality in mice, respectively. Mice lacking COUP-TFI display abnormal development of the central and peripheral nervous systems (47) and abnormal bone formation, whereas COUP-TFII-deficient mutant mice are defective in heart formation and angiogenesis (unpublished results). In the developing organs, COUP-TFII is expressed primarily in the mesenchymal compartment but is absent from epithelial cells. It has been demonstrated that the communication between the mesenchyme and the epithelium is a central process necessary for proper organogenesis and blood vessel formation (7, 11, 14). These mesenchymal cell-epithelial cell interactions are mediated by growth factors, which include members of the FGF family and PDGF, early in the process (7). At later stages, new factors, which include TGFβ1, among others, and molecules of the cell extracellular matrix, act as short-range signals that inhibit the proliferation of the epithelium and induce the differentiation of both cell groups into their final phenotypes (20). The prostate has been commonly used as a system to study mesenchymal cell-epithelial cell interactions and their role in organogenesis (10). Since COUP-TFII is expressed in the urogenital mesenchyme and the timing of its expression indicates its importance in prostate development, we decided to use a rat mesenchymal cell line to study the role of COUP-TFs in organogenesis. Our approach was first to produce cell lines that over- or underexpress COUP-TFs and then to compare the levels of expression of candidate genes regulated by these factors.
In the present study, we have demonstrated that COUP-TFs positively regulate the expression of the NGFI-A gene in prostate mesenchymal cells. Although COUP-TFs largely act as negative regulators of transcription, we show that they are also able to transactivate the endogenous gene as well as the NGFI-A promoter in a dose-dependent manner. The enhancement of transcription is mediated by a 19-bp sequence located at position −56 upstream of the transcriptional start site. This positive action on the transcription of the target gene is enhanced with increasing concentrations of intracellular levels of coactivators, such as p300 and SRC-1.
Only a few promoters have been reported to be transactivated by COUP-TFs; these include the arrestin promoter (36), the phosphoenolpyruvate carboxykinase promoter (21), the trout estrogen receptor gene (30), the vHNF1 promoter (45), and the HIV long terminal repeat (49). Transactivation by COUP-TFs can also be observed in an in vitro system (37). In most cases, the positive actions of COUP-TFs in transcription are mediated by their interaction with some other transcription factor. For example, COUP-TFs can interact with the octamer binding proteins to transactivate the vHNF1 gene (45). Furthermore, COUP-TFs interact with Sp1 and synergistically transactivate HIV gene expression (49). This interaction is mediated by the DBD of COUP-TFs, and it has been shown to occur in vitro and in cells. Another member of the Sp1 family of transcription factors, Sp3, can also bind to the same element and plays the role of a repressor of Sp1- and COUP-TF-induced activation (49).
There is growing evidence for functional and physical interactions between members of the nuclear receptor family and Sp1. It has been recently reported that the estrogen-induced expression of the c-fos and retinoic acid receptor α1 genes in MCF-7 cells depends on the formation of a complex between Sp1 and the estrogen receptor (18, 54). Similarly, COUP-TF and Sp1 interact and cooperate in the transactivation of the HIV long terminal repeat (49). In the present work, we demonstrate that COUP-TFs can activate the transcription of the NGFI-A gene through an element in its promoter that contains two imperfect Sp1 binding sites. Furthermore, the major complexes formed by this element when incubated with HeLa cell nuclear extracts contain Sp1 and Sp3. Moreover, mutational analysis showed that the abrogation of Sp1 or Sp3 binding to this element reduces the COUP-TF-induced transactivation of the NGFI-A promoter. This observation suggests that the activation of the NGFI-A promoter by COUP-TF could be mediated by the interaction of COUP-TF with Sp1. In fact, we demonstrated that these proteins interact strongly in vitro in a GST pull-down assay (Fig. 7A). In agreement with previously published data (49), this interaction would be mediated by the DBD of COUP-TF. Indeed, DBD-defective COUP-TFII is unable to abolish the activation mediated by the wild-type molecule, presumably because of its inability to bind to Sp1. The effect of Sp3 on the NGFI-A promoter is not clear. However, Sp3 may act as a repressor in the system, since it can impair transactivation by COUP-TFII in a dose-dependent manner (data not shown). This result is consistent with previously published observations (49).
We have previously shown that COUP-TFs serve as repressors. There are three mechanisms by which COUP-TFs can negatively affect transcription. First, their striking capacity to bind to the AGGTCA basic motif either in a direct or in a palindromic arrangement with different spacing makes them able to compete with most type II nuclear receptors (RAR, thyroid hormone receptor, RXR, vitamin D receptor, and peroxisomal proliferative factor-activated receptor) for binding sites (13). Second, COUP-TFs are able to heterodimerize with RXR, which acts as a heterodimerization partner and cofactor for most type II nuclear receptors. Third, COUP-TFs contain a repression domain within their LBDs (31) that has been shown to interact with corepressors, such as N-CoR and SMRT (50), which in turn recruit histone deacetylase activity to the promoter region to disrupt the active chromatin conformation, thus silencing promoter activity (23, 39). Our data demonstrate that COUP-TFs also contain an activation domain, the function of which is impaired by deletion of the 15 amino acids closest to the C terminus of the protein. The fact that p300 and SRC-1 can function as coactivators of COUP-TF-induced activation further supports the idea that they might be the factors that interact with such an activation domain. These data correlate with the existence of an AF-2 ligand-activated domain within the putative LBD of COUP-TFs (1). The AF-2 activation domain has been shown to be involved in transactivation and release of corepressors in a ligand-dependent manner for the thyroid hormone receptor and RAR (5, 61). Curiously, our 15-amino-acid deletion from the C terminus of COUP-TFI does not include the core of the AF-2 activation domain, indicating that residues located closer to the C terminus than this core are also important for the transactivation of the NGFI-A gene.
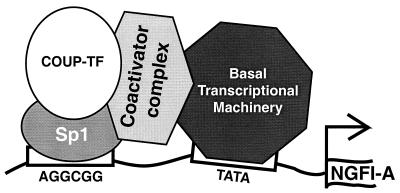
Considering all these results, we propose that Sp1 serves as a docking protein for COUP-TF in the transactivation of the NGFI-A promoter (Fig. 10). This notion is consistent with our observation that COUP-TFs and Sp1 directly interact in vitro (Fig. 7). Since neither Sp1 alone nor a Gal DBD–COUP-TF fusion protein bound directly to a 17-mer Gal4 binding site (data not shown) can fully activate the promoter, it is most likely that the Sp1–COUP-TF complex is responsible for the recruitment of coactivators to the DNA. The AF-2 activation domain of COUP-TF is probably responsible for the interaction with coactivators. SRC-1 has been shown to enhance Sp1 transactivation potential (41). In fact, a detectable increase in the basal activity of the NGFI-A promoter was observed upon expression of exogenous SRC-1 in HeLa cells (Fig. 9B), possibly due to the enhancement of Sp1 activation. However, this enhancement is minimal compared to the coactivation effect of SRC-1 or p300 when COUP-TF is present, suggesting an important role of COUP-TF in coactivator recruitment. Indeed, we found that SRC-1 interacts with COUP-TFI specifically (Fig. 7B). It is likely, then, that SRC-1 activates the NGFI-A promoter by interacting with either COUP-TF or Sp1 or both. In contrast, CBP does not seem to interact directly with COUP-TFI (Fig. 7B). Thus, CBP/p300 activation of the NGFI-A promoter may occur through a direct interaction of Sp1 with CBP/p300. However, we cannot exclude the possibility that CBP/p300 works through COUP-TF indirectly. In experiments with the progesterone receptor (PR), we found that CBP interacts weakly with PR but strongly with SRC-1. Thus, CBP/p300 activation of PR-mediated target gene expression is likely due to the recruitment of SRC-1 by PR and the subsequent recruitment of CBP/p300 by SRC-1. This scenario can also take place for CBP/p300 activation of the COUP-TF target gene. Sp3 could modulate the activation of the promoter through this element by competing for binding with the Sp1–COUP-TF complex. However, the data presented here are insufficient to eliminate the possibility that COUP-TFs enhance Sp1-mediated transactivation of the NGFI-A promoter. We are currently performing new experiments to achieve a better understanding of the mechanism.
FIG. 10.
Model for the transactivation of the NGFI-A promoter by COUP-TFs. We propose (see the text) that COUP-TF interacts with Sp1 through its DBD. The COUP-TF–Sp1 complex bound to DNA would then recruit coactivators that would facilitate the formation of the basal transcriptional complex to initiate RNA synthesis. This interaction would require at least in part the AF-2 activation domain of COUP-TF located in its C terminus.
Although NGFI-A was first cloned as an immediate-early gene (38), recent studies show that NGFI-A affects growth regulation and suppresses transformation by transactivation of genes important for these processes. These genes include those for TGFβ1 (34), basic FGF (6), PDGF-A and -B chains (25, 26), and Rb (16). In addition, NGFI-A is able to cooperate with other important factors, such as Sp1, p21/WAF1/Cip1, and Jun-B. It also stimulates apoptosis by transactivation of the p53 gene (for a review, see reference 35). Furthermore, NGFI-A is highly expressed in prostate tumors and in tumoral prostate cell lines and plays a role in their growth regulation. Although there is no direct in vivo evidence to suggest that COUP-TF activation of NGFI-A is physiologically important, the colocalization of NGFI-A and COUP-TFs in various regions during development and the fact that both factors are reported to be important for hippocampus development (55) imply that NGFI-A might be regulated by COUP-TFs in vivo. The regulation of NGFI-A by COUP-TFs provides support for a role of COUP-TFs in organogenesis and angiogenesis through their regulation of important genes, such as those for TGFβ1, basic FGF, PDGF, and Rb. In fact, we have determined the levels of Rb protein in our rUGM cell lines and have observed increased expression in COUP-TF-overexpressing cells (data not shown). It has been previously demonstrated that Rb is a potent regulator of TGFβ1 and TGFβ2 gene expression (28). Thus, COUP-TFs may carry out their role in embryogenesis and organogenesis through the regulation of developmentally important transcription factors, such as NGFI-A, which regulate the paracrine signals that control these processes. The position of COUP-TFs at the beginning of these cascades of transactivation may explain the lethal phenotype caused by their disruption in mice.
In summary, the present work demonstrates that NGFI-A is positively regulated by COUP-TFs, both in urogenital mesenchymal cells and in HeLa cells. The COUP-TF-responsive element on the NGFI-A promoter contains an Sp1 binding site. The activation requires the DBD and the extreme C-terminal end of COUP-TF and is probably mediated by interactions with coactivators. The interaction between COUP-TFs and NGFI-A and their coexpression in certain tissues strengthen the idea of a role for COUP-TFs in a regulatory cascade leading to important events during embryogenesis, such as organ and vasculature formation.
ACKNOWLEDGMENTS
We thank L. Chung for providing the rUGM cell line and A. Pérez Castillo for providing the full-length NGFI-A promoter. We also thank Z. Nawaz, S. Chua, D. Bramlett, C. Zhou, and N. Barron for suggestions on the paper.
C.P. was the recipient of a fellowship from the Ministerio de Educación y Cultura of Spain during 1996 and 1997. This work was supported by NIH grants.
REFERENCES
- 1.Achatz G, Holzl B, Speckmayer R, Hauser C, Sandhofer F, Paulweber B. Functional domains of the human orphan receptor ARP-1/COUP-TFII involved in active repression and transrepression. Mol Cell Biol. 1997;17:4914–4932. doi: 10.1128/mcb.17.9.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 3.Almendral J M, Sommer D, Macdonald-Bravo H, Burckhardt J, Perera J, Bravo R. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol. 1988;8:2140–2148. doi: 10.1128/mcb.8.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baniahmad A, Leng X, Burris T P, Tsai S Y, Tsai M-J, O’Malley B W. The tau 4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biesiada E, Razandi M, Levin E R. Egr-1 activates basic fibroblast growth factor transcription. Mechanistic implications for astrocyte proliferation. J Biol Chem. 1996;271:18576–18581. doi: 10.1074/jbc.271.31.18576. [DOI] [PubMed] [Google Scholar]
- 7.Carmeliet P, Collen D. Vascular development and disorders: molecular analysis and pathogenic insights. Kidney Int. 1998;53:1519–1549. doi: 10.1046/j.1523-1755.1998.00936.x. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1986;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Christy B A, Lau L F, Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with “zinc finger” sequences. Proc Natl Acad Sci USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung L W, Chang S M, Bell C, Zhau H E, Ro J Y, von Eschenbach A C. Co-inoculation of tumorigenic rat prostate mesenchymal cells with non-tumorigenic epithelial cells results in the development of carcinosarcoma in syngeneic and athymic animals. Int J Cancer. 1989;43:1179–1187. doi: 10.1002/ijc.2910430636. [DOI] [PubMed] [Google Scholar]
- 11.Chung L W, Cunha G R. Stromal-epithelial interactions. II. Regulation of prostatic growth by embryonic urogenital sinus mesenchyme. Prostate. 1983;4:503–511. doi: 10.1002/pros.2990040509. [DOI] [PubMed] [Google Scholar]
- 12.Cooney A J, Leng X, Tsai S Y, O’Malley B W, Tsai M J. Multiple mechanisms of chicken ovalbumin upstream promoter transcription factor-dependent repression of transactivation by the vitamin D, thyroid hormone, and retinoic acid receptors. J Biol Chem. 1993;268:4152–4160. [PubMed] [Google Scholar]
- 13.Cooney A J, Tsai S Y, O’Malley B W, Tsai M-J. Chicken ovalbumin upstream promoter transcription factor (COUP-TF) dimers bind to different GGTCA response elements, allowing COUP-TF to repress hormonal induction of the vitamin D3, thyroid hormone, and retinoic acid receptors. Mol Cell Biol. 1992;12:4153–4163. doi: 10.1128/mcb.12.9.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunha G R, Fujii H, Neubauer B L, Shannon J M, Sawyer L, Reese B A. Epithelial-mesenchymal interactions in prostatic development. I. Morphological observations of prostatic induction by urogenital sinus mesenchyme in epithelium of the adult rodent urinary bladder. J Cell Biol. 1983;96:1662–1676. doi: 10.1083/jcb.96.6.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darland T, Samuels M, Edwards S A, Sukhatme V P, Adamson E D. Regulation of Egr-1 (Zfp-6) and c-fos expression in differentiating embryonal carcinoma cells. Oncogene. 1991;6:1367–1376. [PubMed] [Google Scholar]
- 16.Day M L, Wu S, Basler J W. Prostatic nerve growth factor inducible A gene binds a novel element in the retinoblastoma gene promoter. Cancer Res. 1993;53:5597–5599. [PubMed] [Google Scholar]
- 17.Day M L, Zhao X, Wu S, Swanson P E, Humphrey P A. Phorbol ester-induced apoptosis is accompanied by NGFI-A and c-fos activation in androgen-sensitive prostate cancer cells. Cell Growth Differ. 1994;5:735–741. [PubMed] [Google Scholar]
- 18.Duan R, Porter W, Safe S. Estrogen-induced c-fos protooncogene expression in MCF-7 human breast cancer cells: role of estrogen receptor Sp1 complex formation. Endocrinology. 1998;139:1981–1990. doi: 10.1210/endo.139.4.5870. [DOI] [PubMed] [Google Scholar]
- 19.Fong C J, Sherwood E R, Braun E J, Berg L A, Lee C, Kozlowski J M. Regulation of prostatic carcinoma cell proliferation and secretory activity by extracellular matrix and stromal secretions. Prostate. 1992;21:121–131. doi: 10.1002/pros.2990210205. [DOI] [PubMed] [Google Scholar]
- 20.Gittes G K, Galante P E, Hanahan D, Rutter W J, Debase H T. Lineage-specific morphogenesis in the developing pancreas: role of mesenchymal factors. Development. 1996;122:439–447. doi: 10.1242/dev.122.2.439. [DOI] [PubMed] [Google Scholar]
- 21.Hall R K, Sladek F M, Granner D K. The orphan receptors COUP-TF and HNF-4 serve as accessory factors required for induction of phosphoenolpyruvate carboxykinase gene transcription by glucocorticoids. Proc Natl Acad Sci USA. 1995;92:412–416. doi: 10.1073/pnas.92.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassig C A, Schreiber S L. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 23.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 24.Jonk L J, de Jonge M E, Pals C E, Wissink S, Vervaart J M, Schoorlemmer J, Kruijer W. Cloning and expression during development of three murine members of the COUP family of nuclear orphan receptors. Mech Dev. 1994;47:81–97. doi: 10.1016/0925-4773(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 25.Khachigian L M, Lindner V, Williams A J, Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 26.Khachigian L M, Williams A J, Collins T. Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth factor A-chain promoter in cultured vascular endothelial cells. J Biol Chem. 1995;270:27679–27686. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 27.Kim S J, Park K, Rudkin B B, Dey B R, Sporn M B, Roberts A B. Nerve growth factor induces transcription of transforming growth factor-beta 1 through a specific promoter element in PC12 cells. J Biol Chem. 1994;269:3739–3744. [PubMed] [Google Scholar]
- 28.Kim S J, Wagner S, Liu F, O’Reilly M A, Robbins P D, Green M R. Retinoblastoma gene product activates expression of the human TGF-beta 2 gene through transcription factor ATF-2. Nature. 1992;358:331–334. doi: 10.1038/358331a0. [DOI] [PubMed] [Google Scholar]
- 29.Lau L F, Nathans D. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: coordinate regulation with c-fos or c-myc. Proc Natl Acad Sci USA. 1987;84:1182–1186. doi: 10.1073/pnas.84.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazennec G, Kern L, Valotaire Y, Salbert G. The nuclear orphan receptors COUP-TF and ARP-1 positively regulate the trout estrogen receptor gene through enhancing autoregulation. Mol Cell Biol. 1997;17:5053–5066. doi: 10.1128/mcb.17.9.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leng X, Cooney A J, Tsai S Y, Tsai M-J. Molecular mechanisms of COUP-TF-mediated transcriptional repression: evidence for transrepression and active repression. Mol Cell Biol. 1996;16:2332–2340. doi: 10.1128/mcb.16.5.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim R W, Varnum B C, Herschman H R. Cloning of tetradecanoyl phorbol ester-induced ’primary response’ sequences and their expression in density-arrested Swiss 3T3 cells and a TPA non-proliferative variant. Oncogene. 1987;1:263–270. [PubMed] [Google Scholar]
- 33.Liu C, Adamson E, Mercola D. Transcription factor EGR-1 suppresses the growth and transformation of human HT-1080 fibrosarcoma cells by induction of transforming growth factor beta 1. Proc Natl Acad Sci USA. 1996;93:11831–11836. doi: 10.1073/pnas.93.21.11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C, Calogero A, Ragona G, Adamson E, Mercola D. EGR-1, the reluctant suppression factor: EGR-1 is known to function in the regulation of growth, differentiation, and also has significant tumor suppressor activity and a mechanism involving the induction of TGF-beta1 is postulated to account for this suppressor activity. Crit Rev Oncogenesis. 1996;7:101–125. [PubMed] [Google Scholar]
- 35.Liu C, Rangnekar V M, Adamson E, Mercola D. Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene Ther. 1998;5:3–28. [PubMed] [Google Scholar]
- 36.Lu X P, Salbert G, Pfahl M. An evolutionary conserved COUP-TF binding element in a neural-specific gene and COUP-TF expression patterns support a major role for COUP-TF in neural development. Mol Endocrinol. 1994;8:1774–1788. doi: 10.1210/mend.8.12.7708064. [DOI] [PubMed] [Google Scholar]
- 37.Malik S, Karathanasis S. Transcriptional activation by the orphan nuclear receptor ARP-1. Nucleic Acids Res. 1995;23:1536–1543. doi: 10.1093/nar/23.9.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 38a.Milbrandt, J. Personal communication.
- 39.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 40.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 41.Oñate S A, Tsai S Y, Tsai M-J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 42.Pastorcic M, Wang H, Elbrecht A, Tsai S Y, Tsai M-J, O’Malley B W. Control of transcription initiation in vitro requires binding of a transcription factor to the distal promoter of the ovalbumin gene. Mol Cell Biol. 1986;6:2784–2791. doi: 10.1128/mcb.6.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pereira F A, Qiu Y, Tsai M-J, Tsai S Y. Chicken ovalbumin upstream promoter transcription factor (COUP-TF): expression during mouse embryogenesis. J Steroid Biochem Mol Biol. 1995;53:503–508. doi: 10.1016/0960-0760(95)00097-j. [DOI] [PubMed] [Google Scholar]
- 44.Pérez Castillo A, Pipaón C, García I, Alemany S. NGFI-A gene expression is necessary for T lymphocyte proliferation. J Biol Chem. 1993;268:19445–19450. [PubMed] [Google Scholar]
- 45.Power S C, Cereghini S. Positive regulation of the vHNF1 promoter by the orphan receptors COUP-TF1/Ear3 and COUP-TFII/Arp1. Mol Cell Biol. 1996;16:778–791. doi: 10.1128/mcb.16.3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu Y, Cooney A J, Kuratani S, DeMayo F J, Tsai S Y, Tsai M-J. Spatiotemporal expression patterns of chicken ovalbumin upstream promoter-transcription factors in the developing mouse central nervous system: evidence for a role in segmental patterning of the diencephalon. Proc Natl Acad Sci USA. 1994;91:4451–4455. doi: 10.1073/pnas.91.10.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu Y, Pereira F A, DeMayo F J, Lydon J P, Tsai S Y, Tsai M-J. Null mutation of mCOUP-TFI results in defects in morphogenesis of the glossopharyngeal ganglion, axonal projection, and arborization. Genes Dev. 1997;11:1925–1937. doi: 10.1101/gad.11.15.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu Y, Tsai S Y, Tsai M-J. COUP-TF. An orphan member of the steroid/thyroid hormone receptor superfamily. Trends Endocrinol Metab. 1994;5:234–239. doi: 10.1016/1043-2760(94)p3081-h. [DOI] [PubMed] [Google Scholar]
- 49.Rohr O, Aunis D, Schaeffer E. COUP-TF and Sp1 interact and cooperate in the transcriptional activation of the human immunodeficiency virus type 1 long terminal repeat in human microglial cells. J Biol Chem. 1997;272:31149–31155. doi: 10.1074/jbc.272.49.31149. [DOI] [PubMed] [Google Scholar]
- 50.Shibata H, Nawaz Z, Tsai S Y, O’Malley B W, Tsai M-J. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT) Mol Endocrinol. 1997;11:714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- 51.Simmons D L, Levy D B, Yannoni Y, Erikson R L. Identification of a phorbol ester-repressible v-src-inducible gene. Proc Natl Acad Sci USA. 1989;86:1178–1182. doi: 10.1073/pnas.86.4.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Oñate S A, Tsai S Y, Tsai M-J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 53.Sukhatme V P, Kartha S, Toback F G, Taub R, Hoover R G, Tsai-Morris C H. A novel early growth response gene rapidly induced by fibroblast, epithelial cell and lymphocyte mitogens. Oncogene Res. 1987;1:343–355. [PubMed] [Google Scholar]
- 54.Sun G, Porter W, Safe S. Estrogen-induced retinoic acid receptor alpha 1 gene expression: role of estrogen receptor-Sp1 complex. Mol Endocrinol. 1998;12:882–890. doi: 10.1210/mend.12.6.0125. [DOI] [PubMed] [Google Scholar]
- 55.Tole S, Christian C, Grove E A. Early specification and autonomous development of cortical fields in the mouse hippocampus. Development. 1997;124:4959–4970. doi: 10.1242/dev.124.24.4959. [DOI] [PubMed] [Google Scholar]
- 56.Tran P, Zhang X K, Salbert G, Hermann T, Lehmann J M, Pfahl M. COUP orphan receptors are negative regulators of retinoic acid response pathways. Mol Cell Biol. 1992;12:4666–4676. doi: 10.1128/mcb.12.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai M-J, O’Malley B W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 58.Tsai S Y, Tsai M-J. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocrinol Rev. 1997;18:229–240. doi: 10.1210/edrv.18.2.0294. [DOI] [PubMed] [Google Scholar]
- 59.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L H, Tsai S Y, Cook R G, Beattie W G, Tsai M-J, O’Malley B W. COUP transcription factor is a member of the steroid receptor superfamily. Nature. 1989;340:163–166. doi: 10.1038/340163a0. [DOI] [PubMed] [Google Scholar]
- 61.Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:206. doi: 10.1038/nsb0296-206. [DOI] [PubMed] [Google Scholar]
- 62.Zhau H E, Hong S J, Chung L W. A fetal rat urogenital sinus mesenchymal cell line (rUGM): accelerated growth and conferral of androgen-induced growth responsiveness upon a human bladder cancer epithelial cell line in vivo. Int J Cancer. 1994;56:706–714. doi: 10.1002/ijc.2910560516. [DOI] [PubMed] [Google Scholar]