Mitophagy is regulated by multiple interconnected pathways, which remain poorly understood in physiological settings. Shen et al. describe a mechanism of Pink1-dependent mitophagy utilizing Vps13D that acts in parallel to Parkin-mediated mitophagy during development.
Abstract
Defects in autophagy cause problems in metabolism, development, and disease. The autophagic clearance of mitochondria, mitophagy, is impaired by the loss of Vps13D. Here, we discover that Vps13D regulates mitophagy in a pathway that depends on the core autophagy machinery by regulating Atg8a and ubiquitin localization. This process is Pink1 dependent, with loss of pink1 having similar autophagy and mitochondrial defects as loss of vps13d. The role of Pink1 has largely been studied in tandem with Park/Parkin, an E3 ubiquitin ligase that is widely considered to be crucial in Pink1-dependent mitophagy. Surprisingly, we find that loss of park does not exhibit the same autophagy and mitochondrial deficiencies as vps13d and pink1 mutant cells and contributes to mitochondrial clearance through a pathway that is parallel to vps13d. These findings provide a Park-independent pathway for Pink1-regulated mitophagy and help to explain how Vps13D regulates autophagy and mitochondrial morphology and contributes to neurodegenerative diseases.
Introduction
Autophagy, the delivery of intracellular cargo to lysosomes, plays an important role in cell homeostasis and health. Cells use autophagy as a way to recycle intracellular contents and waste for energy maintenance. Various human diseases, including neurodegenerative conditions and cancer, have been linked to defects in autophagy (Jiang and Mizushima, 2014). For example, a decrease in autophagic flux and failure to clear autophagy substrates contributes to Parkinson’s disease and amyotrophic lateral sclerosis pathogenesis (Hou et al., 2020; Vicencio et al., 2020). Knowledge of the role of autophagy in the development of these diseases, however, is limited by an incomplete understanding of the molecular mechanisms and factors that regulate autophagy.
Elegant studies in yeast characterized a core autophagy machinery that comprises the autophagy-related (atg) genes that are functionally conserved in higher eukaryotes. This autophagy machinery regulates the engulfment of intracellular cargoes, often marked by ubiquitination, through the formation of a double-membrane autophagosome structure. Autophagosomes fuse with lysosomes to degrade the engulfed cargoes, completing the autophagy process (Nakatogawa et al., 2009). While most studies have characterized the role of autophagy in starvation and cell stress, they are not able to account for the role of autophagy in other processes, such as development and cell death. For example, it has been reported that autophagy can occur without core autophagy machinery components, such as atg6/BECN1, atg3, and atg7 (Scarlatti et al., 2008; Chang et al., 2013; Tsuboyama et al., 2016). This is surprising because these genes are crucial for starvation-induced autophagy (Nakatogawa et al., 2009). Furthermore, selective forms of autophagy that recruit specific intracellular cargoes possess unique regulatory pathways that are unaccounted for in the current model of the core autophagy machinery (Lazarou et al., 2015; Rakovic et al., 2019; Chino and Mizushima, 2020).
Selective autophagy is important for the removal of specific organelles (Anding and Baehrecke, 2017). One such organelle that is removed by autophagy is the mitochondrion, which has crucial roles in cell metabolism and death (Friedman and Nunnari, 2014). The Pink1/Parkin pathway regulates mitophagy, the autophagic removal of mitochondria. Pink1 acts as a SerThr kinase that phosphorylates ubiquitin chains on the outer mitochondrial membrane and recruits the E3 ubiquitin ligase Park/Parkin to mitochondria, marking mitochondria for degradation through the autophagy pathway (Ordureau et al., 2014; Koyano et al., 2014; Wauer et al., 2015). Studies of this pathway in cell lines have expanded our understanding of mitophagy and how this process contributes to multiple diseases, such as Parkinson’s disease and Alzheimer’s disease (Palikaras et al., 2018, Wang et al., 2019). However, the initial mechanistic studies of Pink1/Parkin-dependent mitophagy were conducted in HeLa cells, which required exogenous expression of Parkin (Lazarou et al., 2015). Mitophagy under physiological conditions in animals is more complex and often utilizes different mechanistic pathways that cannot be explained by these existing models. For example, pink1- and parkin-independent mitophagy is known to occur in multiple cell lines and tissues (Lee et al., 2018; Bhujabal et al., 2017; McWilliams et al., 2018; Allen et al., 2013; von Stockum et al., 2018). These studies reveal that there is much to learn regarding the relationship among Pink1, Parkin, and mitophagy under physiological conditions.
The context-dependent removal of specific autophagic cargoes is important for animal development and health. For example, macrophages must specifically target foreign microbes for autophagy following phagocytosis to control infection and utilizes regulatory pathways not used in starvation-induced autophagy (Pilli et al., 2012). This example demonstrates the need to investigate selective autophagy in the context of appropriate physiological stimuli. The development of Drosophila larvae to prepupae involves the autophagic removal of larval organs (Chang et al., 2013). In larval intestine cells, mitochondria are rapidly cleared through mitophagy, and previous studies have used this system to screen for genes required for mitophagy under physiological conditions, including pink1 and park (Liu et al., 2016). One gene required for this process is vps13d, which is required for proper mitochondrial morphology and clearance (Anding et al., 2018). vps13d is an essential gene, with vps13d loss-of-function mutants being lethal during development in both flies and mammals (Anding et al., 2018). vps13d also ranks as one of the most important genes for survival in cells lines (Blomen et al., 2015; Wang et al., 2015; Dempster et al., 2019; Meyers et al., 2017). Furthermore, mutations in vps13d have been linked to several diseases, most notably a familial neurological movement disorder involving chorea, dystonia, and ataxia (Seong et al., 2018; Gauthier et al., 2018; Nakada et al., 2015). In contrast to other Vps13 family members, only Vps13D is required for autophagy in flies, is essential, and contains a ubiquitin binding (UBA) domain that is required for proper mitochondrial morphology and clearance (Anding et al., 2018). However, the functional role that Vps13D plays in the autophagic machinery and the relationship to any well-defined mitophagy pathways are currently unknown.
Here, we identify a context-dependent function of Vps13D in the core autophagy machinery as a regulator of ubiquitin and Atg8a localization. We find that in the developing Drosophila larval intestine, Pink1 regulates mitochondrial clearance upstream of Vps13D. Unlike Pink1, Parkin does not regulate mitophagy in a Vps13D-dependent manner and instead influences mitochondrial clearance through a parallel pathway that is required for the clearance of a smaller subset of mitochondria within the cell. Importantly, our findings characterize a new form of Pink1-dependent and Parkin-independent mitophagy regulated by Vps13D. These results establish a mechanistic model for the role that Vps13D plays in mitophagy and help to explain the function of vps13d in development and disease.
Results
Vps13D regulates Atg8a and ubiquitin localization during developmental autophagy
Autophagy genes are required for the clearance of intracellular cargoes, including mitochondria, in Drosophila larval intestine cells (Chang et al., 2013; Anding et al., 2018). The poorly understood role of Vps13D in autophagy prompted us to consider how Vps13D interacts with the autophagy machinery to influence this process in larval intestine cells.
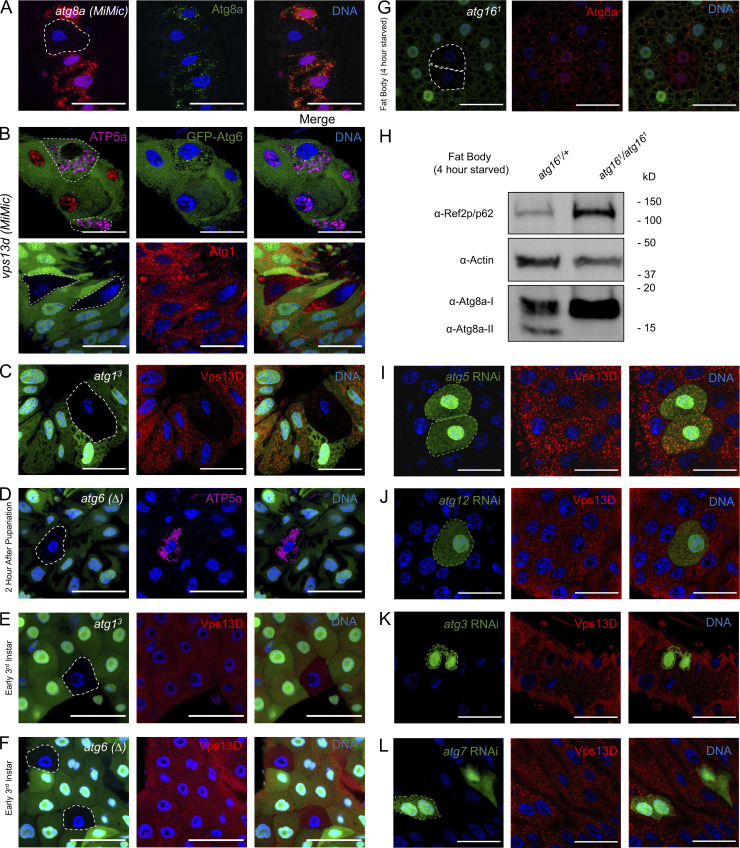
Atg8a (LC3/GABARAP in mammals) is an important regulator of autophagosome formation and mitophagy (Lazarou et al., 2015). We thus sought to examine the localization of Atg8a and mitochondria in vps13d mutants. vps13d (MiMic) loss-of-function intestine cells that lack GFP were generated through mosaic cell clone induction (GFP was pseudo-colored red to best visualize contrast in other color channels). Intestines that were isolated 2 h after prepupa formation were then stained with an antibody specific for endogenous Atg8a (Fig. S1 A). Compared with red-labeled heterozygous control cells, vps13d loss-of-function mutant intestine cells had an abnormal retention and localization of Atg8a. Atg8a puncta in these mutant cells were enlarged and seemed to localize around ATP5a-labeled mitochondria that failed to be degraded (Fig. 1, A and B).
Figure S1.
Vps13D puncta localization is not affected in early third instar larvae autophagy mutant cells or by loss of Atg8a conjugating pathway components. (A) atg8a (MiMic) intestine cells (nonred) stained with Atg8a-specific antibody have depleted Atg8a puncta (green) compared with heterozygous control cells (red) 2 h after pupariation. (B) Top: vps13d (MiMic) loss-of-function mutants (nonnuclear red) do not have bright, enlarged, Myo31DFNP0001-driven GFP-Atg6 puncta (green) that localize around ATP5a puncta (purple) compared with neighboring control cells (nuclear red). Bottom: vps13d (MiMic) loss-of-function mutants (nongreen) do not have altered Atg1 puncta (red) compared with neighboring control cells (green). (C) atg13 loss-of-function intestine cells (nongreen) have fewer Vps13D puncta (red) compared with neighboring heterozygous control cells (green) 2 h after pupariation. (D) atg6 (Δ) loss-of-function intestine cells (nongreen) retain ATP5a puncta (purple) compared with neighboring heterozygous control cells (green) 2 h after pupariation. (E) atg13 loss-of-function mutant intestine cells (nongreen) from early third instar larvae do not have altered Vps13D puncta (red) compared with neighboring heterozygous control cells (green). (F) atg6 (Δ) loss-of-function mutant intestine cells from early third instar larvae (nongreen) do not have altered Vps13D puncta (red) compared with neighboring heterozygous control cells (green). (G) atg161 fat body cells (nongreen) from early third instar larvae starved for 4 h have decreased Atg8a puncta (red) compared with neighboring heterozygous control cells (green). (H) Fat body dissected from homozygous atg161/atg161 mutant early third instar larvae starved for 4 h have increased Ref2p/p62, increased Atg8a-I, and decreased Atg8a-II compared with heterozygote atg161/+ controls. (I) atg5 RNAi-expressing intestine cells (green) do not have altered Vps13D puncta (red) compared with neighboring control cells (nongreen) 2 h after pupariation. (J) atg12 RNAi-expressing intestine cells (green) do not have altered Vps13D puncta (red) compared with neighboring control cells (nongreen) 2 h after pupariation. (K) atg3 RNAi-expressing intestine cells (green) do not have altered Vps13D puncta (red) compared with neighboring control cells (nongreen) 2 h after pupariation. (L) atg7 RNAi-expressing intestine cells (green) do not have altered Vps13D puncta (red) compared with neighboring control cells (nongreen) 2 h after pupariation. Scale bars in A–G and I–L are 40 μm. Representative of three or more independent biological experiments.
Figure 1.
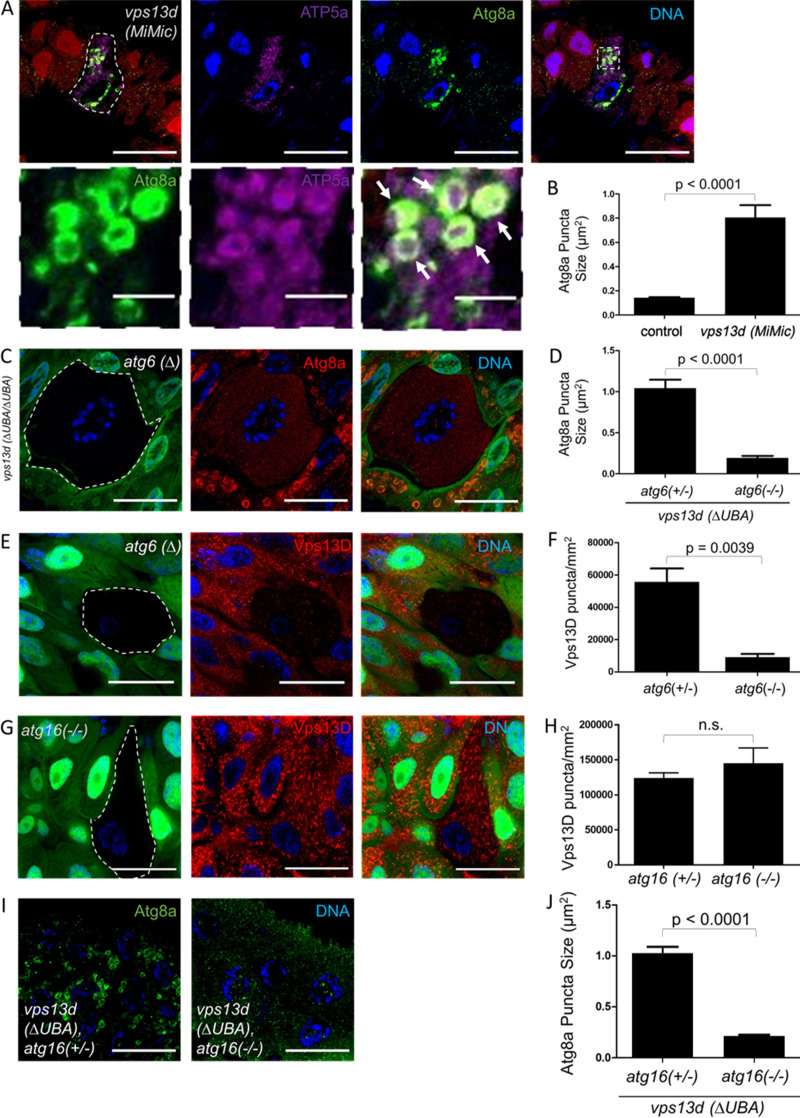
Vps13D regulates Atg8a localization around mitochondria during autophagy. (A) vps13d (MiMic) loss-of-function intestine cells (nonred) from 2-h-old prepupae have increased mitochondrial ATP5a puncta (purple) and enlarged Atg8a (green) puncta compared with neighboring control cells (red; top panels). Atg8a puncta tend to surround the ATP5a puncta in vps13d (MiMic) mutant cells (bottom panels). (B) Quantification of the size of Atg8a puncta surrounding ATP5a puncta in vps13d (MiMic) intestine cells (n = 6) compared with heterozygote controls (n = 22). (C) atg6 (Δ), vps13d (ΔUBA/ΔUBA) double-mutant intestine cells (nongreen) 2 h after pupariation have suppressed formation of enlarged Atg8a puncta (red) seen in control neighboring atg6 (Δ)/+, vps13d (ΔUBA/ΔUBA) single-mutant cells (green). (D) Quantification of Atg8a puncta size in atg6 (Δ), vps13d (ΔUBA/ΔUBA) double-mutant cells (n = 7) compared with atg6 (Δ)/+, vps13d (ΔUBA/ΔUBA) single-mutant cells (n = 20). (E) atg6 (Δ) loss-of-function intestine cells (nongreen) stained with antibody against Vps13D (red) were compared with neighboring heterozygous control cells (green) in 2-h-old prepupae. (F) Quantification of the amount of Vps13D puncta in atg6 (Δ) loss-of-function intestine cells (n = 5) compared with neighboring control cells (n = 13). (G) atg161 loss-of-function intestine cells (nongreen) have similar levels of Vps13D puncta (red) compared with neighboring heterozygote control cells (green) in 2-h-old prepupae. (H) Quantification of Vps13D puncta in atg161 cells (n = 9) compared with neighboring heterozygous control cells (n = 22). (I) atg161, vps13d (ΔUBA/ΔUBA) double-mutant intestines cells have suppressed formation of enlarged Atg8a puncta compared with atg161/+, vps13d (ΔUBA/ΔUBA) single-mutant cells 2 h after pupariation. (J) Quantification of Atg8a puncta size in atg161, vps13d (ΔUBA/ΔUBA) double-mutant intestines cells (n = 19) compared with atg161/+, vps13d (ΔUBA/ΔUBA) single-mutant cells (n = 14). Scale bars in A (top panel), C, E, G, and I are 40 μm. Scale bars in A (bottom panel) are 5 μm. Columns in B, D, F, H, and J were compared using two-tailed t test without Welch’s correlation, with error bars representing SEM. Representative of three or more independent biological experiments.
To determine whether the Atg8a localization phenotype in vps13d mutant cells was due to a defect in the autophagy machinery, we investigated whether this phenotype was influenced by loss of any core autophagy genes. atg6 loss-of-function mutants, atg6 (Δ), were generated in intestine cells that were homozygous for the vps13d (ΔUBA) mutant allele that lacks the UBA domain. Staining with the Atg8a antibody revealed that loss of atg6 in the cells lacking GFP (labeled green in all future instances unless indicated), which were therefore atg6 and vps13d double mutants, suppressed the altered Atg8a localization phenotype compared with neighboring vps13d (ΔUBA) control cells expressing GFP (Fig. 1, C and D). These data indicate that the unique Atg8a localization phenotype in vps13d mutant cells requires Atg6, an upstream regulator of autophagy, and suggest that a functional link exists between the core autophagy machinery and Vps13D. Consistent with this hypothesis, atg6 (Δ) mutant cells that lack GFP exhibit a significant decrease in Vps13D protein puncta in intestine cells 2 h after prepupa formation (Fig. 1, E and F). Like Atg6, Atg1/ULK acts as an initiator of autophagy early in the core autophagy machinery (Suzuki et al., 2007). In contrast to Atg8a, Atg6, and Atg1 localization were not affected by loss of vps13d (Fig. S1 B). Consistent with these findings, atg13 loss-of-function mutant cells lacking GFP also had decreased Vps13D protein puncta (Fig. S1 C).
Autophagy is required for removal of mitochondria from Drosophila larval intestine cells (Chang et al., 2013). Consistent with these previous findings, atg6 (Δ) mutant intestine cells lacking GFP were unable to clear mitochondria 2 h after prepupa formation (Fig. S1 D). Generation of atg13 and atg6 (Δ) mutants in intestine cells in early third instar larvae, before the onset of pupariation and autophagy in the larval intestine (Chang et al., 2013), did not affect Vps13D puncta levels (Fig. S1, E and F), suggesting a context- and stage-specific relationship between Vps13D and these core autophagy genes.
The Atg5/12/16 complex is required for lipidation of Atg8a and recruitment to the forming autophagosome (Shpilka et al., 2011). We used transcription activator-like effector nucleases to generate a frameshift mutation in all isoforms of Atg16, resulting in a strong loss-of-function mutant allele, atg161. Cells lacking atg161 accumulated the autophagy cargo receptor Ref2p and possessed decreased Atg8a lipidation and puncta formation in starved fat body cells (Fig. S1, G and H). Unlike atg1 and atg6 loss-of-function mutants, which act early in the core autophagy machinery (Suzuki et al., 2007), atg161 mutant cells lacking GFP did not affect Vps13D puncta 2 h after prepupa formation (Fig. 1, G and H). Knockdown of other components of the Atg8a conjugation machinery, including atg3, atg7, atg5, and atg12, also did not affect Vps13D localization 2 h after prepupa formation (Fig. S1, I–L). These findings suggest that while Vps13D puncta formation depends on Atg1 and Atg6, it either does not require or functions upstream of core autophagy machinery components required for Atg8a conjugation. To determine which of these possibilities was more likely, we conducted double-mutant analyses between the vps13d (ΔUBA) mutant and atg161. Compared with vps13d (ΔUBA) single-mutant cells that possess enlarged Atg8a puncta, loss of atg16 suppressed the Atg8a localization phenotype (Fig. 1, I and J). Taken together, these findings suggest that vps13d acts downstream of Atg1 and Atg6 but upstream of Atg8a conjugation pathways in Drosophila intestine cells 2 h after puparium formation.
Vps13D was previously found to localize to late endosomes and lysosomes (Anding et al., 2018). Therefore, we also considered the possibility that Vps13D could regulate autophagy at a later stage associated with autophagosome and lysosome fusion, a process that is mediated by Syx17 (Itakura et al., 2012; Takáts et al., 2013). To determine whether a relationship exists between Vps13D and Syx17, we analyzed Vps13D protein localization (purple) in intestine cells with reduced Syx17 function (green) and did not detect any change in Vps13D localization (Fig. S2 A). In addition, we analyzed Syx17 protein localization (red) in vps13d mutant intestine cells (nongreen) and did not detect a change in Syx17 localization (Fig. S2 A). These findings suggest that Vps13D does not regulate autophagy through Syx17-mediated autophagosome and lysosome fusion.
Figure S2.
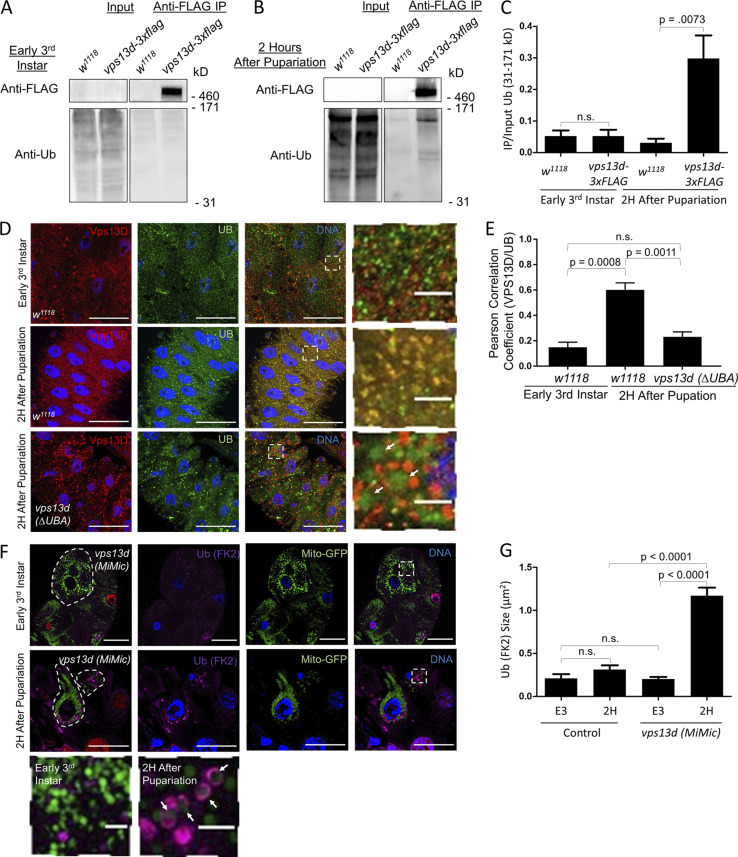
Proteins are ubiquitinated during pupariation in Drosophila and lysis of Vps13D-3×FLAG requires harsh conditions. (A) Top: syx17 RNAi-expressing larval intestine cells (GFP) did not have a difference in Vps13D puncta (purple) compared with control cells (non-GFP) 2 h after pupariation. Bottom: vps13d (MiMic) loss-of-function larval intestines cells (non-GFP) did not have a difference in Syx17 puncta (red) compared with control cells (green). (B) Whole-animal lysates from early third instar larval and 2 h after pupariation were lysed in harsh lysis conditions (1× Laemli buffer) and immunoblotted with antibody against ubiquitin and actin. (C) Control w1118 and vps13d-3×FLAG 2-h-old prepupae were lysed in harsh lysis conditions and immunoblotted with antibody against FLAG and actin. (D) vps13d (ΔUBA) intestines expressing mitochondrial localized mito-GFP (green) throughout the intestine 2 h after pupariation were stained with a pan-ubiquitin–specific antibody (red). Scale bars in A and D are 40 μm with the exception of enlarged images, which are 5 μm.
The functional requirements of both the ubiquitin E1–activating enzyme Uba1 and UBA domain encoding Vps13D protein for autophagy illustrate the importance of ubiquitin in Drosophila intestine cells (Chang et al., 2013; Anding et al., 2018). Therefore, we investigated the relationship between ubiquitin and Vps13D during autophagy in intestine cells. As expected, we detected an increase in the amount of ubiquitinated proteins in whole-animal lysates between early third instar larvae and 2 h after puparium formation (Fig. S2 B). The Vps13D UBA domain preferentially interacts with K63 ubiquitin in vitro (Anding et al., 2018). To verify that Vps13d interacts with ubiquitin in vivo, we used a Drosophila strain in which the vps13d gene was endogenously tagged with 3×FLAG on the C terminus (vps13d-3×FLAG). Unlike vps13d mutants, these flies are viable and fertile and complement the early lethal phenotype of vps13d (MiMic) flies. Importantly, immunoblotting revealed a distinct band in the vps13d-3×FLAG lysates at the predicted size of Vps13D-3×FLAG that was absent in the w1118 control lysates (Fig. S2 C). Immunoprecipitation of the FLAG epitope followed by Western blot analysis of the eluate using a ubiquitin antibody did not reveal ubiquitinated protein binding with Vps13D-3×FLAG in the early third instar stage in either the control w1118 or the vps13d-3×FLAG lysates (Fig. 2 A). By contrast, we detected a significant increase in ubiquitinated protein interacting with Vps13D-3×FLAG compared with the control w1118 eluate 2 h after pupariation (Fig. 2, B and C). Consistent with these biochemical results, Vps13D and ubiquitin colocalized to a greater extent 2 h after pupariation than in early third instar w1118 intestines (Fig. 2, D and E). Significantly, deletion of the UBA domain in vps13d suppressed the colocalization of Vps13D and ubiquitin (Fig. 2, D and E). Additionally, ubiquitin puncta in vps13d (ΔUBA) mutant cells were larger than ubiquitin puncta in control intestine cells under identical conditions (Fig. 2 D). These enlarged ubiquitin puncta colocalized with mito-GFP, a marker for mitochondria (Fig. S2 D).
Figure 2.
Vps13D interacts with and regulates ubiquitin localization. (A) Western blot of input and eluates from a FLAG immunoprecipitation (IP) of control w1118 and vps13d-3×flag early third instar larvae extracts that were probed with antibodies against ubiquitin. (B) Western blot of input and eluates from a FLAG IP of control w1118 and vps13d-3×flag 2-h-old prepupal extracts that were probed with antibodies against ubiquitin. Vps13D-3×FLAG levels were too low to be detected in the input with lysate conditions suitable for IP but were present in lysates from harsher lysis conditions (Fig. S2 B). (C) Quantification of the ratio of ubiquitinated proteins from 31 to 171 kD in the IP compared with input for w1118 and vps13d-3×FLAG lysates (n = 6 independent experiments). (D) w1118 and vps13d (ΔUBA) intestines were dissected from early third instar and 2 h after pupariation and costained with antibodies against Vps13D (red) and ubiquitin (green). vps13d (ΔUBA) intestines had larger and more spherical ubiquitin puncta (arrows). (E) Quantification of the Pearson correlation coefficient between Vps13D and ubiquitin in w1118 early third instar intestine cells (n = 25), 2 h after pupariation w1118 (n = 50), and vps13d (ΔUBA) (n = 44) intestine cells. (F) vps13d (MiMic) loss-of-function intestine cells (lacking red nuclei) have similar conjugated ubiquitin (purple) localization to neighboring heterozygote control cells (red nuclei; top panels) in early third instar larvae. By contrast, vps13d (MiMic) cells have enlarged conjugated ubiquitin (FK2) puncta compared with neighboring control cells (middle panels) in 2-h-old prepupae. A portion of these puncta tend to encircle the mitochondria (green) in vps13d (MiMic) mutant cells at 2 h after pupariation (bottom panels). (G) Quantification of the size of conjugated ubiquitin (FK2) puncta in vps13d (MiMic) intestine cells in early third instar and 2 h after pupariation (E3 and 2H, respectively, n = 6) intestine cells compared with control cells (E3, n = 12; 2H, n = 15). Scale bars in D and F are 40 μm with the exception of the enlarged images, which are 5 μm. Identical time point columns in C were compared using two-tailed t test without Welch’s correlation. Columns in E and G were compared using one-way ANOVA with Tukey’s post hoc analysis. Error bars are SEM. Representative of three or more independent biological experiments.
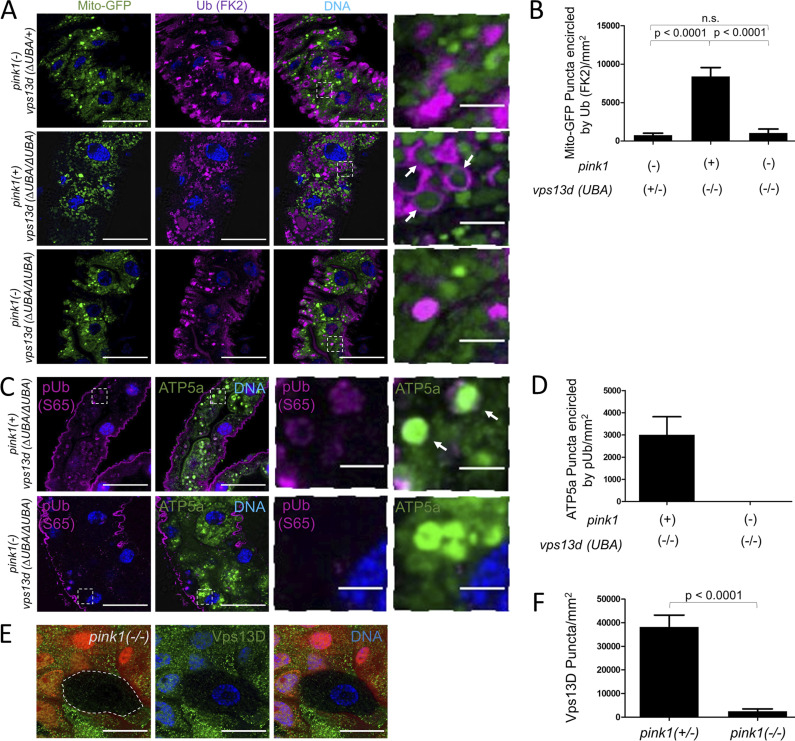
Ubiquitin is associated with cargoes that are targeted to autophagosomes for degradation, including mitochondria (Nakatogawa et al., 2009). Therefore, we hypothesized that the increased size of ubiquitin puncta in vps13d mutant cells could be explained by changes in protein bound to ubiquitin (conjugated ubiquitin) on mitochondria because of a failure in the clearance of these autophagic substrates. To test this possibility, we compared vps13d (MiMic) loss-of-function mutant intestine cells stained with an antibody specific for conjugated ubiquitin, the ubiquitin (FK2) antibody. In early third instar larvae, we did not detect a significant difference in conjugated ubiquitin between vps13d mutant cells (lacking nuclear RFP) and control cells (Fig. 2, F and G). By contrast, conjugated ubiquitin puncta were enlarged in vps13d mutant compared with control cells 2 h after puparium formation (Fig. 2, F and G). Interestingly, the colocalization of conjugated ubiquitin with mito-GFP revealed ubiquitin puncta that appeared to encircle mitochondria that failed to be cleared in vps13d mutant cells (Fig. 2, F and G), a localization pattern that is similar to Atg8a (Fig. 1 A). These results indicate that Vps13D influences the localization of Atg8 and conjugated ubiquitin that are associated with mitochondrial clearance.
Vps13D regulates mitophagy through a Pink1-dependent mechanism
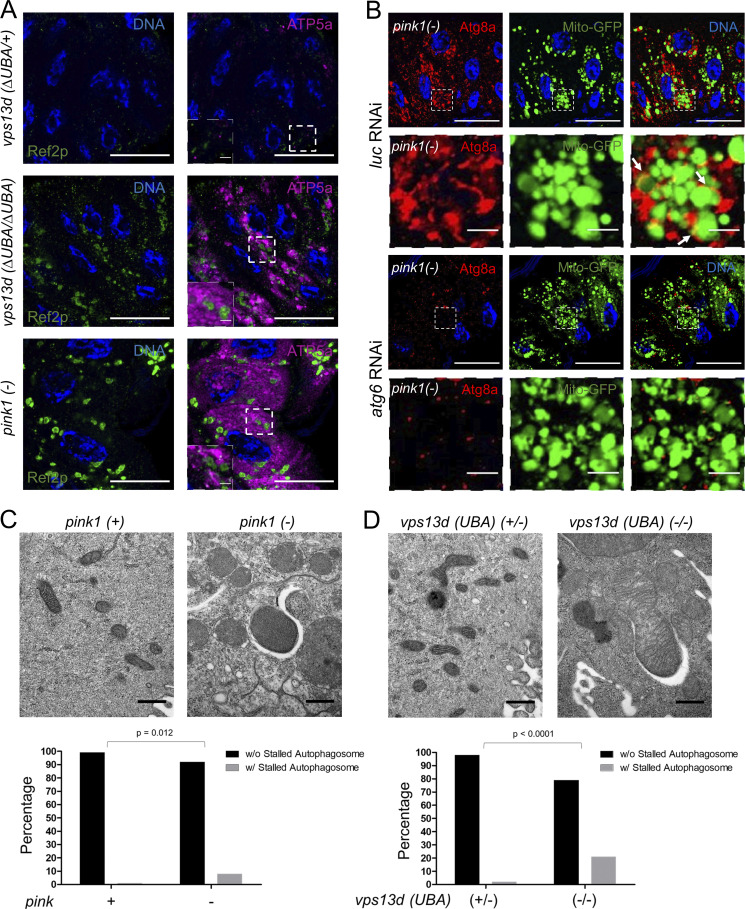
The influence of Vps13D on Atg8a, ubiquitin, and clearance of mitochondria prompted us to consider whether established mitophagy pathway genes have phenotypes that are similar to vps13d in intestine cells. Pink1 functions as a mitochondrial stress-sensing kinase that facilitates the targeting of mitochondrial-associated proteins for ubiquitination by Parkin and clearance during mitophagy (Lazarou et al., 2015). To determine whether Pink1 is required for mitochondrial clearance by autophagy in intestine cells, we generated pink1B9 loss-of-function mutant cell clones (lacking RFP) and stained with antibodies specific for the autophagy cargo receptor Ref2p/p62 and mitochondrial ATP5a. Cells lacking pink1 function had increased Ref2p/p62 and ATP5a puncta compared with neighboring control cells (Fig. 3, A, B, D, and E). In addition, pink1 mutant cells also failed to reduce in size (Fig. 3 C), an indicator of autophagy deficiency in intestine cells (Chang et al., 2013). Curiously, Ref2p puncta did not seem to localize around mitochondria in either pink1 or vps13d mutant cells (Fig. S3 A), suggesting that Pink1 and Vps13D may regulate clearance of other autophagic cargoes that are associated with Ref2p. Combined, these data indicate that pink1 influences markers of autophagy and mitochondrial clearance in a manner that is similar to the loss of vps13d.
Figure 3.
pink1 mutant cells have autophagy and mitochondrial phenotypes that are similar to vps13d mutant cells. (A) pink1B9 loss-of-function mutant (−/−) intestine cells (nonred) fail to reduce in size and have increased Ref2p puncta (green) compared with neighboring control pink1B9/+ heterozygous (+/−) cells (red) 2 h after pupariation. (B) Quantification of Ref2p puncta in pink1B9 mutant (−/−) cells (n = 6) compared with heterozygous control (+/−) cells (n = 15). (C) Quantification of cell size in pink1B9 mutant (−/−) cells (n = 9) compared with control (+/−) cells (n = 11). (D) pink1B9 loss-of-function mutant (−/−) intestine cells (nonred) have increased mitochondrial ATP5a (purple) puncta and exhibit enlarged Atg8a puncta (green) compared with heterozygous (+/−) control cells (red) 2 h after pupariation (top panels). Atg8a puncta in mutant cells tend to encircle mitochondrial ATP5a puncta (bottom panels). (E) Quantification of the amount of ATP5a puncta in pink1B9 mutant (−/−) cells (n = 9) compared with neighboring control (+/−) cells (n = 11). (F) Quantification of the size of Atg8a puncta in pink1B9 mutant (−/−) cells (n = 7) compared with neighboring control (+/−) cells (n = 13). (G) Quantification of the number of ATP5a puncta having at least 50% of the perimeter encircled by Atg8a in pink1B9/pink1B9 mutant (−/−) cells (n = 7) compared with vps13d (MiMic) mutant intestine cells (n = 6). (H) pink1B9/pink1B9 loss-of-function mutant (−/−) intestine cells (nonred) have enlarged conjugated ubiquitin (FK2) puncta (purple) compared with heterozygous neighboring control cells (red) 2 h after pupariation (top panels). These enlarged puncta do not seem to encircle mitochondria labeled by mito-GFP (green; bottom panels). (I) Quantification of conjugated ubiquitin (FK2) puncta size in pink1B9/pink1B9 mutant (−/−) cells (n = 9) compared with control (+/−) cells (n = 11). (J) Quantification of the number of mito-GFP puncta having a perimeter at least 50% encircled by conjugated ubiquitin in pink1B9/pink1B9 mutant (−/−) cells (n = 9) compared with vps13d (MiMic) mutant cells (n = 6). (K) Representative TEM images from sections of control w1118, pink1B9 (-), and vps13d (ΔUBA) male larval intestine cells 2 h after pupariation. Images on top and bottom are lower and higher resolution images, respectively. (L) Quantification of mitochondria area in w1118 (n = 202), pink1B9 (n = 188), and vps13d (ΔUBA) (n = 185) intestine cells 2 h after pupariation. (M) Quantification of the percentage of mitochondria <0.1 μm or ≥0.1 μm in w1118 (n = 202), pink1B9 (-) (n = 188), and vps13d (ΔUBA) (n = 185) intestine cells 2 h after pupariation. Fisher’s exact test values are as follows: w1118 vs. pink1B9, P < 0.0001; w1118 vs. vps13d (ΔUBA), P < 0.0001; and pink1B9 vs. vps13d (ΔUBA), P < 0.0001. Scale bars in A, D, and H are 40 μm with the exception of the enlarged images in D and H, which are 5 μm. Scale bars in K are 2.0 μm (top panels) and 0.5 μm (bottom panels). Columns in B, C, E–G, I, and J were compared using two-tailed t test without Welch’s correlation. Columns in L were compared using one-way ANOVA with Tukey’s post hoc analysis. Error bars are SEM. Representative of three or more independent biological experiments.
Figure S3.
Loss of atg6 suppresses pink1 loss-of-function Atg8a puncta phenotype. (A) Mutant vps13d (ΔUBA/ΔUBA) and pink1B9 (-) intestine cells have increased Ref2p puncta (green) that do not tend to localize around mitochondria labeled by ATP5a (purple) compared with control vps13d (ΔUBA/+) intestine cells 2 h after pupariation. (B) atg6 RNAi-expressing pink1B9 (-) intestine cells have suppression of the enlarged Atg8a puncta (red) that encircle mito-GFP (green) compared with control luc RNAi-expressing pink1B9 (-) mutant cells (arrows) 2 h after pupariation. Scale bars are 40 μm with the exception of those in enlarged figures and insets, which are 5 μm. (C) pink1B9 (−) loss-of-function mutant larval intestine cells (right) occasionally have stalled autophagosomes surrounding mitochondria (defined as an electron-lucent compartment surrounding at least 50% of the perimeter of a mitochondria) 2 h after pupariation. These structures are almost absent in control w1118 (+) intestine cells (left) 2 h after pupariation. The percentage of mitochondria with (w/) and without (w/o) stalled autophagosomes were quantified in w1118 (n = 118) and pink1B9 (n = 97) intestines 2 h after pupariation (bottom), and the P value was determined by Fisher’s exact test. (D) Representative TEM image of mitochondria with stalled autophagosome (defined as an electron-lucent compartment surrounding at least 50% of the perimeter of a mitochondria) in vps13d (ΔUBA/ΔUBA) (−/−) intestine cells 2 h after pupariation (right). Almost no mitochondria with stalled autophagosomes were seen in control vps13d (ΔUBA/+) (+/−) intestine cells 2 h after pupariation (left). The percentage of mitochondria with (w/) and without (w/o) stalled autophagosomes were quantified in vps13d (ΔUBA/+) (n = 83) and vps13d (ΔUBA/ΔUBA) (n = 91) intestines 2 h after pupariation (bottom), and the P value was determined by Fisher’s exact test. Representative of three or more independent biological experiments.
We next investigated the influence of pink1 function on the localization of Atg8a and ubiquitin with ATP5a as a marker of mitochondria. Antibody staining against Atg8a revealed enlarged puncta surrounding enlarged mitochondrial ATP5a puncta (Fig. 3, D–G), and these Atg8a puncta could be suppressed by knockdown of atg6 (Fig. S3 B), similar to vps13d mutant cell Atg8a puncta (Fig. 1, C and D). Antibody staining against conjugated ubiquitin also revealed that pink1 mutant cells had enlarged conjugated ubiquitin puncta (Fig. 3, H and I). In contrast to vps13d mutant cells, pink1 mutant cell ubiquitin puncta did not localize to the periphery of mitochondria (Fig. 3 J), suggesting potential differences between Pink1 and Vps13D.
Transmission electron microscopy (TEM) was used to investigate mitochondrial clearance and ultrastructure in control w1118, pink1 loss-of-function mutant, and vps13d (ΔUBA) mutant intestine cells 2 h after prepupa formation. Both pink1 and vps13d (ΔUBA) mutant cells possessed uncleared mitochondria that were larger than w1118 controls, which possess mitochondria with an area of ∼0.1 μm2 (Fig. 3, K and L). vps13d (ΔUBA) mutant mitochondria were on average larger than pink1 mutant mitochondria (Fig. 3, K and L). In addition, pink1 mutant cells possessed a larger percentage of mitochondria having an area <0.1 μm2 than vps13d (ΔUBA) mutants (Fig. 3 M). Both pink1 and vps13d mutant cells also had cup-shaped and electron-lucent structures adjacent to mitochondria, which appeared to be stalled phagophores of autophagosomes (Fig. S3, C and D). These data together with autophagy pathway genetic analyses and the enlarged Atg8a structures surrounding ATP5a in pink1 and vps13d loss-of-function mutants are consistent with Pink1 and Vps13D regulating autophagosome phagophore expansion.
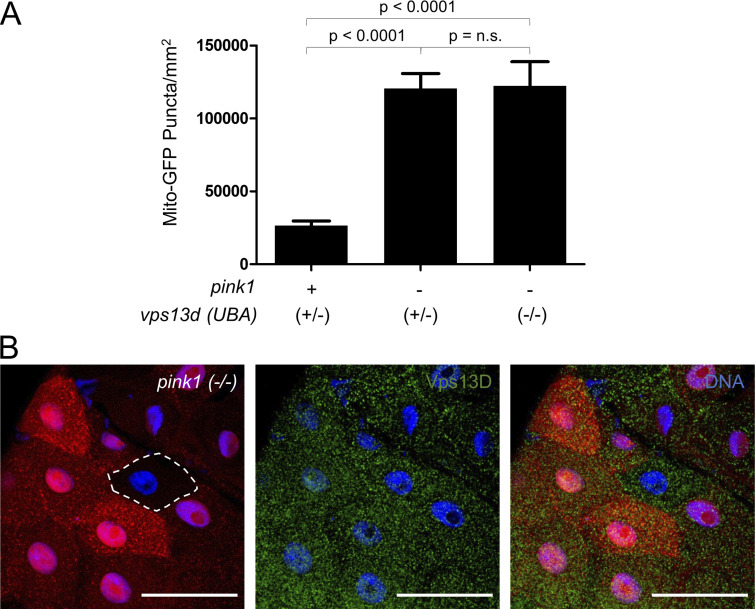
The similarities in vps13d and pink1 mutant cell phenotypes prompted us to investigate their functional relationship. Therefore, we conducted double-mutant genetic analyses to determine whether vps13d and pink1 are in a common pathway that regulates mitochondria structure and removal. We compared mitochondrial size in TEM sections of pink1 mutant with pink1;vps13d double-mutant intestine cells. These mutants had similar mitochondria area 2 h after puparium formation (Fig. 4, A and B). Furthermore, these single- and double-mutant genotypes had a similar proportion of remaining mitochondria that are <0.1 μm2 (Fig. 4 C), even though vps13d (ΔUBA) single mutants have larger mitochondria and fewer mitochondria that are <0.1 μm2 than control and pink1B9 single-mutant intestines (Fig. 3, K–M). These data indicate that the combined loss of both vps13d and pink1 fails to enhance single-mutant mitochondrial size phenotype, thus suggesting that Vps13D functions in the same pathway as Pink1 in the regulation of mitochondrial size.
Figure 4.
Vps13D functions in a mitophagy pathway with Pink1. (A) Representative TEM images of male pink1B9 (−);vps13d (ΔUBA/+) single-mutant and pink1B9 (−);vps13d (ΔUBA/ΔUBA) double-mutant intestine cells 2 h after pupariation. (B) Quantification of mitochondria area in pink1B9 (−);vps13d (ΔUBA/+) single-mutant (n = 187) and pink1B9;vps13d (ΔUBA/ΔUBA) double-mutant (n = 181) intestine cells 2 h after pupariation. (C) Quantification of the percentage of mitochondria <0.01 μm and ≥0.1 μm in pink1B9 (−);vps13d (ΔUBA/+) single-mutant (n = 187) and pink1B9 (−);vps13d (ΔUBA/ΔUBA) double-mutant (n = 181) intestine cells 2 h after pupariation using Fisher’s exact test (P = 0.7507). (D) Mito-QC was expressed using the Myo31DFNP0001 driver in pink1B9 (−);vps13d (ΔUBA) mutant intestine cells from 2-h-old male prepupae. pink1 (+);vps13d (ΔUBA)/+ heterozygote control cells possessed mostly red puncta without corresponding green puncta, reflecting mitochondria in autolysomes (mitolysosomes). By contrast, pink1B9 (−);vps13d (ΔUBA)/+ single-mutant, pink1 (+);vps13d (ΔUBA/ΔUBA) single-mutant, and pink1B9 (−);vps13d (ΔUBA)/ΔUBA) double-mutant intestine cells all exhibited red and green structures, reflecting mitochondria that failed to be cleared by mitophagy. These structures were either filamentous (arrowheads) or enlarged and punctate (arrows). Enlarged punctate structures were defined as having an area of at least 1.0 μm2. (E) Quantification of the amount of red and green structures that presented as enlarged puncta in pink1 (+);vps13d (ΔUBA)/+ heterozygote control (n = 17), pink1B9 (−);vps13d (ΔUBA)/+ single-mutant (n = 14), pink1 (+);vps13d (ΔUBA/ΔUBA) single-mutant, and pink1B9 (−);vps13d (ΔUBA)/ΔUBA) double-mutant (n = 14) cells 2 h after pupariation. (F) pink1B9/pink1B9 (−/−);vps13d (ΔUBA/ΔUBA) double-mutant intestine cells (nonred) have similar levels and size of Atg8a puncta (green) compared with pink1B9/+ (+/−);vps13d (ΔUBA/ΔUBA) single-mutant cells (red) 2 h after pupariation. (G) Quantification of Atg8a puncta number in pink1B9/pink1B9 (−/−);vps13d (ΔUBA/ΔUBA) double-mutant cells (n = 11) compared with neighboring pink1B9/+ (+/−);vps13d (ΔUBA/ΔUBA) single-mutant cells (n = 15). (H) Quantification of Atg8a puncta size in pink1B9/pink1B9 (−/−);vps13d (ΔUBA/ΔUBA) double-mutant cells (n = 11) compared with pink1B9/+ (+/−);vps13d (ΔUBA/ΔUBA) single-mutant cells (n = 15). Scale bars in A are 0.5 μm. Scale bars in D and F are 40 μm with the exception of the enlarged images in D, which are 5 μm. Columns in B, G, and H were compared using two-tailed t test without Welch’s correlation. Columns in E were compared using one-way ANOVA with Tukey’s post hoc analysis. Error bars are SEM. Representative of three or more independent biological experiments.
We sought to determine whether Vps13D and Pink1 act within the same pathway to regulate mitophagy. We used the Mito-QC system to analyze mitophagy, which utilizes mitochondria-localized tandem mCherry and GFP fluorescent tags, to label mitochondria outside of autolysosomes with both mCherry and GFP (yellow) and mitochondria inside autolysosomes with only red mCherry as the acidic environment of the autolysosome quenches GFP signal (McWilliams et al., 2016; Lee et al., 2018). In control cells that are wild type for pink1 and heterozygous for vps13d, most of the GFP signal from the Mito-QC was quenched, leaving only red mCherry puncta 2 h after prepupa formation (Fig. 4, D and E) and indicating that mitophagy was active. By contrast, pink1 mutants and pink1;vps13d double-mutant cells retained both GFP and mCherry (yellow) signal that was absent in the control (Fig. 4, D and E), indicating that mitophagy was impaired. However, it is worth noting that the morphology of the retained GFP and mCherry (yellow) signal differed between these single and double mutants. The pink1 mutants appeared as either filamentous structures or large, round, and punctate structures. By contrast, vps13d mutants only had the enlarged punctate structures (Fig. 4 D). Importantly, the distribution of the large and round yellow puncta were the same in pink1 and pink1;vps13d double-mutant cells (Fig. 4, D and E). Together with our TEM data (Fig. 4, A–C), these findings suggest that Vps13D and Pink1 function in a common pathway to regulate mitochondrial morphology and clearance.
To further investigate the relationship between Vps13D and Pink1, we determined how they influence Atg8a puncta in intestine cells 2 h after puparium formation. Like vps13d mutant intestine cells (Fig. 1, A and B), pink1 mutant intestine cells possess abnormal and enlarged Atg8a localization (Fig. 3, D, F, and G). Both vps13d mutant (labeled by nuclear RFP) and pink1;vps13d double-mutant (lacking nuclear RFP) intestine cells possessed similar Atg8a puncta size and amounts (Fig. 4, F–H). These findings suggest that pink1 and vps13d function in the same pathway to regulate Atg8a localization.
Pink1 senses mitochondrial stress and facilitates ubiquitination of mitochondria-associated proteins to facilitate mitophagy (Ordureau et al., 2014; Koyano et al., 2014; Wauer et al., 2015). In contrast to Atg8a localization, pink1 and vps13d mutant intestine cells differ in conjugated ubiquitin localization (Fig. 2, F and G; and Fig. 3, H and J). To further examine the relationship between these two regulators of mitochondrial clearance, we compared conjugated ubiquitin localization in either pink1 mutant, vps13d mutant, or pink1;vps13d double-mutant cells. In contrast to vps13d mutant cells, pink1;vps13d double-mutant cells exhibited the same pattern of conjugated ubiquitin localization as pink1 single-mutant cells that was not associated with the perimeter of mitochondria (Fig. 5, A and B). Furthermore, there was no additive increase in the remaining mitochondria in pink1 single-mutant cells compared with pink1;vps13d double-mutant cells 2 h after pupariation (Fig. S4 A), further suggesting that Vps13D regulates mitochondrial clearance in a Pink1-dependent manner.
Figure 5.
Loss of Pink1 suppresses ubiquitin localization to mitochondria in vps13d mutant cells. (A) pink1B9 (−);vps13d (ΔUBA/+) single-mutant intestine cells have conjugated ubiquitin puncta (purple) that do not encircle mitochondria labeled by mito-GFP (green) as frequently as intestines from pink1 (+);vps13d (ΔUBA/ΔUBA) single mutants 2 h after prepupae formation (top panels). Loss of Pink1 in a vps13d (ΔUBA/ΔUBA) background, resulting in a pink1B9 (−);vps13d (ΔUBA/ΔUBA) double mutant (bottom panels) suppresses the conjugated ubiquitin localization to mitochondria phenotype in vps13d mutant intestine cells (middle panels) 2 h after pupariation. (B) Quantification of the number of mito-GFP puncta with at least 50% of the perimeter encircled by conjugated ubiquitin in pink1B9 (−);vps13d (ΔUBA/+) (n = 15), pink1 (+);vps13d (ΔUBA/ΔUBA) (n = 11), and pink1B9 (−);vps13d (ΔUBA/ΔUBA) (n = 12) mutant intestines 2 h after pupariation. (C) pink1 (+);vps13d (ΔUBA/ΔUBA) single-mutant intestine cells have Ser65 phosphorylated ubiquitin (pUb) puncta (purple) surrounding mitochondria labeled by ATP5a (green) 2 h after puparium formation. pink1B9 (−); vps13d (ΔUBA/ΔUBA) double-mutant intestine cells do not have Ser65 pUb puncta surrounding mitochondria 2 h after prepupae formation. (D) Quantification of the number of ATP5a puncta with at least 50% of the perimeter encircled by Ser65 pUb in pink1 (+);vps13d (ΔUBA/ΔUBA) single-mutant (n = 15) and pink1B9;vps13d (ΔUBA/ΔUBA) double-mutant (n = 15) intestines 2 h after pupariation. (E) pink1B9/pink1B9 (−/−) mutant intestine cells (nonred) have decreased Vps13D puncta (green) compared with pink1B9/+ (+/−) heterozygous control neighboring cells (red). (F) Quantification of Vps13D puncta in pink1B9/pink1B9 (−/−) cells (n = 6) compared with control (+/−) cells (n = 14). Scale bars in A, C, and E are 40 μm with the exception of the enlarged images in A and C, which are 5 μm. Columns in B were compared using one-way ANOVA with Tukey’s post hoc analysis. Columns in F were compared using two-tailed t test without Welch’s correlation, with error bars representing SEM. Representative of three or more independent biological experiments.
Figure S4.
Loss of vps13d does not add to pink1 mitochondrial clearance phenotype, and loss of pink1 in early third instar intestine cells do not have altered Vps13D puncta. (A) Quantification of mito-GFP puncta remaining in pink1 (+);vps13d (ΔUBA/+) intestine cells (n = 12); pink1B9 (-);vps13d (ΔUBA/+) intestine cells (n = 12); and pink1B9 (-);vps13d (ΔUBA/ΔUBA) intestine cells (n = 11) 2 h after pupariation. Columns were compared using one-way ANOVA with Tukey’s post hoc analysis. (B) pink1B9/pink1B9 (−/−) loss-of-function mutant intestine cells (nonred) in early third instar larvae do not have altered Vps13D puncta (green) compared with heterozygous control (+/−) cells (red). Scale bars in B are 40 μm. Error bars are SEM. Representative of three or more independent biological experiments.
Pink1 can directly phosphorylate ubiquitin conjugated to proteins at the Ser65 residue, resulting in a conformation change that inhibits deubiquitination and can lead to further ubiquitination (Ordureau et al., 2014; Koyano et al., 2014; Wauer et al., 2015). vps13d mutant intestine cells were stained with an antibody specific for ubiquitin phosphorylated at Ser65. Like conjugated ubiquitin, phosphorylated ubiquitin localized around the periphery of mitochondria (labeled by ATP5a) in vps13d mutants but was absent in pink1;vps13d double mutants (Fig. 5, C and D). These data indicate that pink1 function is required for localization of conjugated and phosphorylated ubiquitin near the perimeter of mitochondria in vps13d mutant cells and suggests that pink1 is upstream of vps13d. To test this hypothesis, we analyzed Vps13D protein puncta localization in pink1 mutant intestine cells. pink1 mutant cells (lacking RFP) had reduced Vps13D protein puncta compared with neighboring RFP-labeled control cells (Fig. 5, E and F). Like with loss of core autophagy proteins, loss of Pink1 did not affect Vps13D puncta in early third instar larval intestine cells (Fig. S4 B), suggesting that the relationship between Pink1 and Vps13D is stage and context dependent. Taken together, these results suggest that Pink1 and Vps13D can function in a common pathway to regulate mitophagy, with Pink1 acting upstream of Vps13D.
Vps13D and Parkin function in parallel pathways downstream of Pink1
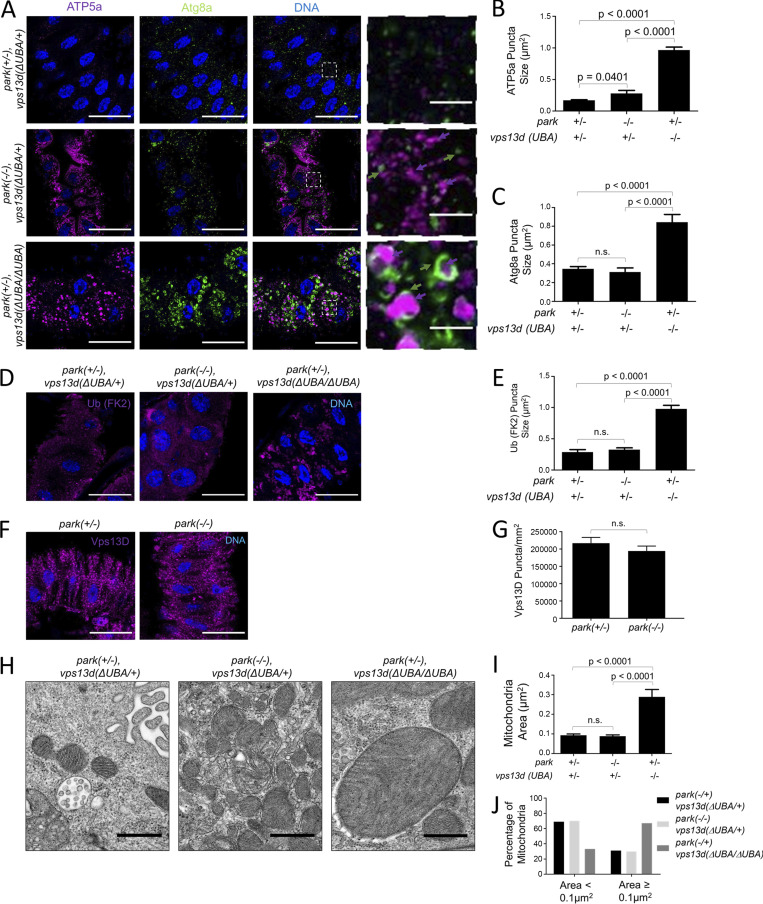
Pink1 phosphorylates both ubiquitin and Parkin, leading to ubiquitination of mitochondrial-associated proteins and recruitment of the core autophagy machinery for mitophagy (Ordureau et al., 2014; Koyano et al., 2014; Wauer et al., 2015; Lazarou et al., 2015). The relationship between Pink1 and Vps13D prompted us to hypothesize that Vps13D and Park, the Drosophila homologue of Parkin, function in the same pathway to regulate mitophagy. We expected park25 loss-of-function mutant intestine cells to have the same phenotype as pink1 and vps13d mutant cells 2 h after prepupa formation. While park25 mutant cells failed to clear mitochondria as indicated by abundant ATP5a staining compared with heterozygous control cells, homozygous park mutant cells had neither enlarged ATP5a puncta nor abnormal Atg8a as were detected in vps13d mutant cells (Fig. 6, A–C). In addition, park mutant cells lacked the enlarged conjugated ubiquitin puncta that were observed in homozygous vps13d mutant cells (Fig. 6, D and E). Furthermore, the ATP5a puncta size was smaller in park mutants than in pink1 mutants (P = 0.0019; Fig. 3 I and Fig. 6 E). Significantly, unlike pink1 mutants, park mutants did not affect Vps13D puncta in intestine cells (Fig. 6, F and G). Combined, these data suggest that Vps13D and Park act in parallel pathways to regulate mitophagy.
Figure 6.
Vps13D and Park function in separate mitochondrial clearance pathways. (A) park25/park25 (−/−);vps13d (ΔUBA/+) single-mutant intestine cells (middle panels) and park25/+ (+/−);vps13d (ΔUBA/ΔUBA) single-mutant intestine cells (bottom panels) 2 h after pupariation are unable to clear mitochondria as shown by ATP5a staining (purple) compared with park25/+ (+/−);vps13d (ΔUBA/+) heterozygous control cells (top panels). vps13d (ΔUBA) mutants have ATP5a that appear larger and less filamentous (purple arrows) and larger Atg8a puncta (green arrows) encircling the enlarged ATP5a puncta (purple arrows), which are not present in the park25 mutant cells. (B) Quantification of ATP5a puncta size in park25/+ (+/−);vps13d (ΔUBA/+) heterozygous control (n = 13), park25/park25 (−/−);vps13d (ΔUBA)/+ (n = 12), and park25/+ (+/−);vps13d (ΔUBA/ΔUBA) (n = 12) intestine cells. (C) Quantification of Atg8a puncta size in park25/+ (+/−);vps13d (ΔUBA/+) heterozygous control (n = 13), park25/park25 (−/−);vps13d (ΔUBA)/+ (n = 12), and park25/+ (+/−);vps13d (ΔUBA) (n = 12) intestine cells. (D) park25/+ (+/−);vps13d (ΔUBA/ΔUBA) single-mutant cells have larger conjugated ubiquitin (FK2) puncta (purple) than park25/park25 (−/−);vps13d (ΔUBA/+) single-mutant and park25/+ (+/−), vps13d (ΔUBA/+) heterozygous control intestine cells 2 h after pupariation. (E) Quantification of conjugated ubiquitin (FK2) size in park25/+ (+/−);vps13d (ΔUBA/+) heterozygous control (n = 11), park25/park25 (−/−);vps13d (ΔUBA)/+ (n = 12), and park25/+ (+/−);vps13d (ΔUBA/ΔUBA) (n = 11) intestine cells 2 h after pupariation. (F) park25/park25 (−/−) mutant intestine cells have similar levels of Vps13D puncta (purple) compared with heterozygous park25/+ (+/−) control intestine cells 2 h after pupariation. (G) Quantification of Vps13D puncta in park25/park25 (−/−) mutant intestine cells (n = 11) compared with park25/+ (+/−) control cells (n = 13). (H) Representative TEM images of park25/+ (+/−);vps13d (ΔUBA)/+ heterozygous control, park25/park25 (−/−);vps13d (ΔUBA/+) single-mutant, and park25/+ (+/−);vps13d (ΔUBA/ΔUBA) single-mutant intestine cells 2 h after pupariation. (I) Quantification of mitochondrial area from TEM images of park25/+ (+/−);vps13d (ΔUBA)/+ heterozygous control cells (n = 192), park25/park25 (−/−);vps13d (ΔUBA/+) single-mutant (n = 186), and park25/+ (+/−);vps13d (ΔUBA/ΔUBA) single-mutant (n = 184) intestine cells 2 h after pupariation. (J) Quantification of the percentage of mitochondria <0.1 μm and ≥0.1 μm from TEM images from park25/+ (+/−);vps13d (ΔUBA)/+ heterozygous control (n = 192), park25/park25 (−/−);vps13d (ΔUBA/+) single-mutant (n = 186), and park25/+ (+/−);vps13d (ΔUBA/ΔUBA) single-mutant (n = 184) intestine cells 2 h after pupariation. vps13d mutant intestine cells have larger mitochondria and fewer remaining mitochondria ≤0.1 μm2 than park mutant and control intestine cells. Columns were compared using Fisher’s exact test with the following values: park25/+;vps13d (ΔUBA)/+ vs. park25/park25;vps13d (ΔUBA/+), P = 0.9109, park25/+;vps13d (ΔUBA)/+ vs. park25/+;vps13d (ΔUBA/ΔUBA), P < 0.0001, and park25/park25;vps13d (ΔUBA/+) vs. park25/+;vps13d (ΔUBA/ΔUBA), P < 0.0001. Scale bars in A, D, and F are 40 μm with the exception of the enlarged images in A, which are 5 μm. Scale bars in H are 0.5 μm. Columns in B, C, E, and I were compared using one-way ANOVA with Tukey’s post hoc analysis. Columns in G were compared using two-tailed t test without Welch’s correlation. Error bars are SEM. Representative of three or more independent biological experiments.
The evaluation of mitochondrial size by immunofluorescence is difficult when puncta signals are saturated, as small puncta in close proximity may be interpreted as single large puncta. Thus, TEM was used to study the influence of Park on mitochondria size. vps13d mutant intestine cells had larger mitochondria and a higher proportion of mitochondria remaining with an area ≥0.1 μm2 compared with park mutant intestine cells 2 h after prepupa formation (Fig. 6, H–J). Significantly, park mutant intestine cells had smaller mitochondria than pink1 mutant cells (P < 0.0001) and had a smaller proportion of mitochondria with an area ≥0.1 μm2 (P < 0.0001; Fig. 3, K–M; and Fig. 6, H–J). While both Park and Vps13D regulate mitochondrial clearance, these data indicate that they function in parallel pathways.
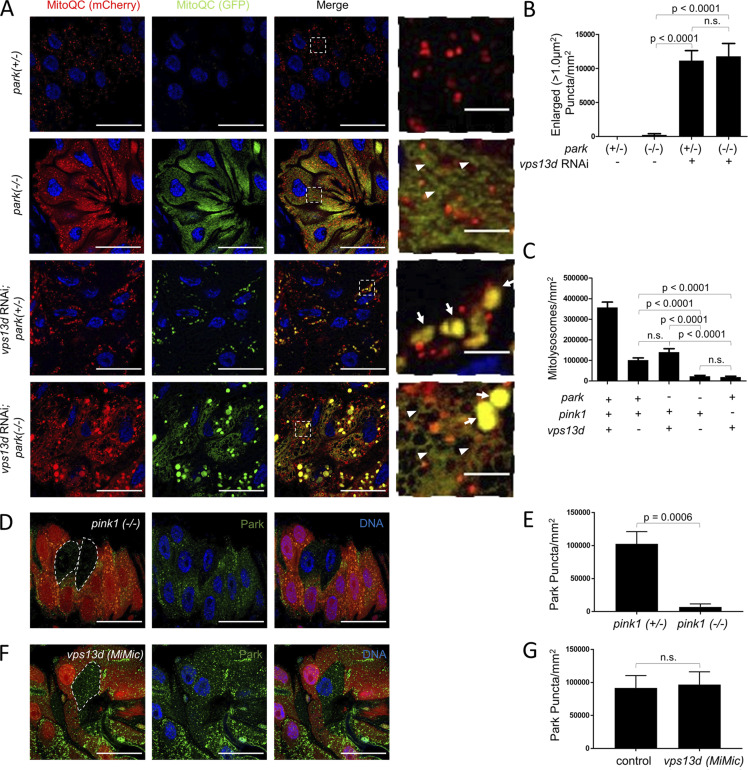
Park functions downstream of Pink1 (Narendra et al., 2010; Youle and Narendra, 2011). In addition, Vps13D appears to act downstream of Pink1 and in parallel to Park (Figs. 5 and 6). Thus, we hypothesized that Pink1 regulates both Vps13D- and Park-mediated clearance of mitochondria in intestine cells. We used Mito-QC to investigate the functions of vps13d and park in mitophagy. The early lethality of vps13d (ΔUBA) and park25 double mutants required the use of park mutants expressing vps13d RNAi in intestine cells for these analyses. While park heterozygous control intestine cells did not retain GFP, indicating active mitophagy 2 h after prepupa formation, park mutant intestine cells retained GFP signal in a filamentous distribution (Fig. 7 A). In contrast to park mutant cells, cells with reduced vps13d function retained GFP signal in enlarged, round puncta (Fig. 7 A), similar to the Mito-QC phenotype in vps13d mutant cells (Fig. 4 D). Importantly, intestine cells with reduced function of both park and vps13d had an additive phenotype with both filamentous and enlarged round puncta (Fig. 7, A and B). This phenotype is similar to the Mito-QC phenotype in pink1 mutant intestine cells (Fig. 4, C and D). To verify that the Mito-QC distribution we detected in different mutants were representative of mitochondria in intestine cells, we stained Mito-QC–expressing intestine cells with antibody against mitochondrial ATP5a protein, and detected Mito-QC and ATP5a colocalization (Fig. S5 A). Quantification of the number of mCherry-positive and GFP-negative puncta, which represent mitochondria in autolysosomes (mitolysosomes; Lee et al., 2018), revealed that deficiency in either Vps13D, Park, or Pink1 reduced mitolysosome formation in intestine cells 2 h after pupariation. Significantly, deficiencies in both Vps13D and Park resulted in a decrease in mitolysosomes with the same severity as deficiency in Pink1 alone (Fig. 7 C). These findings are consistent with Pink1 regulating mitophagy upstream of parallel Vps13D and Park pathways.
Figure 7.
Vps13D and Park function in parallel Pink1-regulated mitophagy pathways. (A) Mito-QC was expressed in intestine cells from park25/park25 (−/−) prepupae with and without expression of vps13d RNAi. park25/+ (+/−) heterozygous control cells expressing control uas-flp (to account for off-target effects of UAS expression) possessed mostly red puncta without corresponding green puncta, reflecting mitochondria in autolysomes (mitolysosomes). By contrast, park25/park25 (−/−) expressing flp, park25/+ (+/−) expressing vps13d RNAi, and park25/park25 (−/−) expressing vps13d RNAi intestine cells all exhibited red and green structures 2 h after pupariation, reflecting mitochondria that failed to be cleared by mitophagy. park25/park25 (−/−) cells had almost exclusively filamentous structures (arrowheads), park25/+ (+/−) expressing vps13d RNAi cells had almost exclusively enlarged punctate structures (arrows), and park25/park25 (−/−) expressing vps13d RNAi intestine cells had both filamentous and enlarged punctate structures. (B) Quantification of the number of enlarged (>1.0 μm2) punctate structures in (left to right) park25/+ (+/−) heterozygous control (n = 24), park25/park25 (−/−) expressing flp (n = 16), park25/+ (+/−) expressing vps13d RNAi (n = 15), and park25/park25 (−/−) expressing vps13d RNAi (n = 15) intestine cells. (C) Number of mitolysosomes (mCherry-only puncta) were quantified in the following intestine cell genotypes 2 h after pupariation (left to right): park25/+ (+), pink1 (+), luc RNA-expressing (+) intestine cells (n = 14); park25/+ (+), pink1 (+), vps13d RNAi–expressing (-) intestine cells (n = 13); park25/park25 (-), pink1 (+) mutant, luc RNAi–expressing (+) intestine cells (n = 13); park25/park25 (-), pink1 (+), vps13d RNAi–expressing (-) intestine cells (n = 13); and pink1 (-), luc RNAi–expressing (+) intestine cells (n = 11). P < 0.0001 for quantification of mitolysosomes in park25/+ (+), pink1 (+), luc RNAi–expressing (+) intestine cells with all other genotypes. (D) pink1B9/pink1B9 (−/−) loss-of-function mutant intestine cells (nonred) have reduced Park protein puncta (green) compared with neighboring pink1B9/+ (+/−) heterozygous control cells (red) 2 h after pupariation. (E) Quantification of Park protein puncta in pink1B9/pink1B9 (−/−) intestine cells (n = 9) compared with neighboring pink1B9/+ (+/−) control cells (n = 15). (F) vps13d (MiMic) loss-of-function mutant intestine cells (nonred) have similar levels of Park protein puncta (green) compared with neighboring heterozygous control (red) cells 2 h after pupariation. (G) Quantification of Park protein puncta in vps13d (MiMic) intestine cells (n = 9) compared with heterozygous control cells (n = 11). Scale bars in A, D, and F are 40 μm with the exception of the enlarged images in A, which are 5 μm. Columns in B and C were compared using one-sided ANOVA test with Tukey’s post hoc analysis, while E and G were compared using two-tailed t test without Welch’s correlation. Error bars are SEM. Representative of three or more independent biological experiments.
Figure S5.
Mito-QC represents retained mitochondria in vps13d and park mutants. (A) Colocalization of mitochondrial ATP5a (purple) with the Mito-QC GFP component (green), representing mitochondria that have not reached the autolysosome, expressed in vps13d (ΔUBA/ΔUBA) and park25/park25 (−/−) mutant intestine cells 2 h after pupariation. In both mutant intestines, ATP5a colocalized with nonautolysosomal Mito-QC puncta (GFP). (B) park25/park25 (−/−) loss-of-function mutant intestine cells stained with a Park-specific antibody had reduced Park protein puncta compared with heterozygous park25/+ (+/−) control cells 2 h after pupariation. Scale bars are 40 μm with the exception of those in enlarged figures and insets, which are 5 μm. Representative of three or more independent biological experiments. (C) vps13d (MiMic) loss-of-function larval intestine cells (nonnuclear RFP) have ATP5a (purple) and Park (green) puncta that colocalize (merge inset) 2 h after pupariation. Scale bars are 40 μm with the exception of those in insets, which are 5 μm.
To further explore the relationship among Pink1, Vps13D, and Park, we analyzed Park protein puncta localization using a Park-specific antibody (Fig. S5 B). While pink1 loss-of-function (lacking RFP) and park loss-of-function mutant cells each had a significant decrease in Park protein puncta (Fig. 7, D and E; and Fig. S5 B), vps13d (MiMic) loss-of-function mutant cells (lacking RFP) had no impact on Park puncta (Fig. 7, F and G). Furthermore, Park puncta localized to mitochondrial ATP5a puncta even in vps13d mutant intestine cells (Fig. S5 C). Since both Park and Vps13D protein puncta are reduced in pink1 mutant cells, these data are consistent with Pink1 regulation of mitophagy upstream of both Vps13D and Park.
Discussion
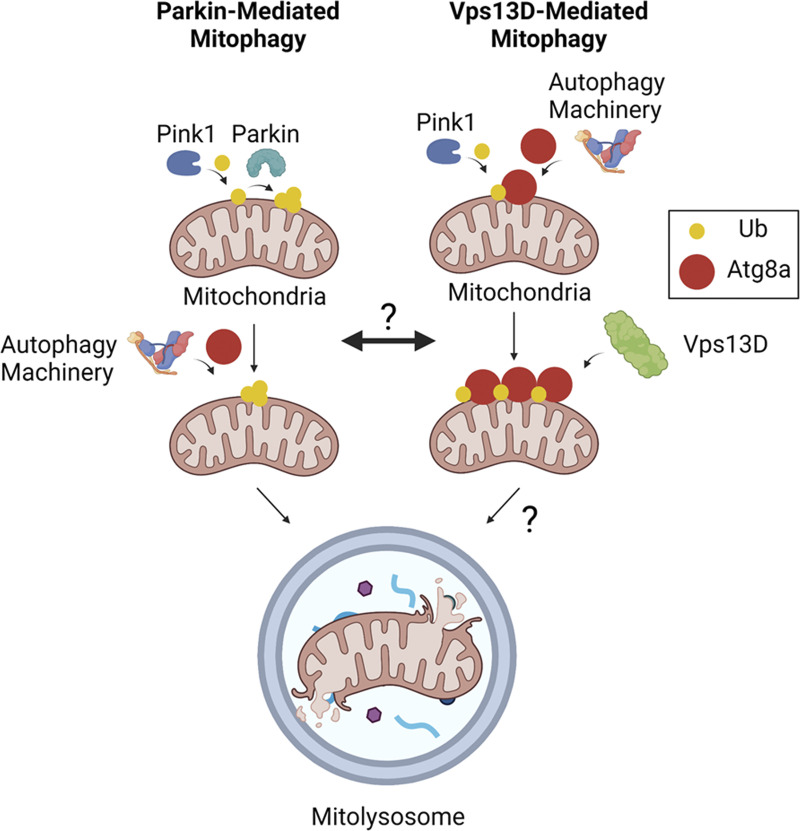
Vps13D plays an important role in mitochondrial morphology and mitophagy during development (Anding et al., 2018). Mutations in Vps13D have also been identified as causes of familial neurological movement disorders (Seong et al., 2018; Gauthier et al., 2018). However, little is known about the mechanistic relationship between Vps13D and the regulation of autophagy, mitophagy, and associated diseases. Here, we identify a role of Vps13D in the core autophagy machinery and Pink1-regulated mitophagy. Vps13D functions downstream of Pink1 and parallel to Park to regulate ubiquitin and Atg8a localization around mitochondria during mitophagy. Thus, we define a new form of Pink1-regulated mitophagy that acts through Vps13D. A hypothesized model for how Vps13D functions relative to Pink1, Park, and the core autophagy machinery is shown in Fig. 8.
Figure 8.
Hypothesized model for Parkin- and Vps13D-mediated mitophagy. Parkin-mediated mitophagy (left) involves a feed-forward ubiquitin loop mechanism initiated by Pink1 and amplified by Parkin, leading to downstream recruitment of the autophagy machinery (Ordureau et al., 2014; Lazarou et al., 2015). Vps13D-mediated mitophagy (right) involves upstream coordination of both Pink1 and the autophagy machinery in a Parkin-independent manner, leading to the recruitment of Vps13D. Created with BioRender.
Ubiquitin and regulators of ubiquitin localization and conjugation function in autophagy. In response to mitochondrial damage, Pink1 phosphorylates ubiquitin and Parkin, leading to expansion of ubiquitinated chains on the mitochondrial outer membrane (Ordureau et al., 2014; Koyano et al., 2014; Wauer et al., 2015). Previous studies have indicated that autophagy receptors, including p62 and OPTN, can interact with ubiquitinated chains on mitochondria and through physical interactions with Atg8a, function as bridges between mitochondria and the autophagic machinery to facilitate mitophagy (Lazarou et al., 2015; Kirkin and Rogov, 2019). We show that Vps13D interacts with ubiquitinated proteins and regulates ubiquitin localization in a context-dependent manner. In vps13d mutant cells, ubiquitin is able to localize to mitochondria. The accumulation of enlarged conjugated ubiquitin puncta around mitochondria suggests a problem with the processing of ubiquitin after localization to cargo. In addition, Pink1 is required for the localization of conjugated and phosphorylated ubiquitin around mitochondria in Vps13D mutants (Fig. 5, A–D). Thus, Vps13D seems to play a role that is downstream of Pink1 activity but upstream of the clearance of ubiquitinated mitochondria.
Previous work suggests that the recruitment of the core autophagy machinery to mitochondria during mitophagy depends on ubiquitination of mitochondrial outer membrane proteins following Pink1 recruitment (Lazarou et al., 2015). By contrast, we observed that recruitment of Atg8a to mitochondria is independent of Pink1 function (Fig. 3, D and G). These findings indicate that Pink1-regulated ubiquitination and Atg8a recruitment can act in separate pathways. Other groups have reported similar phenomena in human cell lines, where Atg8/LC3s can be recruited to mitochondria independent of ubiquitination events (Padman et al., 2019). In Drosophila, Vps13D appears to function downstream of these biochemical changes during mitophagy, as Vps13D mutant cells accumulate conjugated ubiquitin and Atg8a around the periphery of mitochondria (Fig. 1 A; Fig. 2 F; and Fig. 3, G and J). Thus, Vps13D could act as a mediator between ubiquitinated cargo and the core autophagy machinery once localized to mitochondria.
It remains unclear how loss of Vps13D results in accumulation of Atg8a and conjugated ubiquitin around mitochondria. Studies of yeast vps13, which is highly conserved with vps13d, suggest that the proteins in the vps13 family serve as lipid transporters between membranes at organelle contact sites (Lang et al., 2015). Furthermore, proteins in the Vps13 family all contain conserved Chorein_N domains with strong homology to the Chorein_N domain of Atg2 (Kumar et al., 2018), a core autophagy protein that acts as a tether between the ER and the forming autophagosome known as the phagophore. Atg2 facilitates the transfer of phospholipids to the expanding autophagosome membrane (Osawa and Noda, 2019). Replacement of the Atg2 Chorein_N domain with the Chorein_N domain of Vps13 resulted in the same autophagic activity as wild-type Atg2 (Osawa et al., 2019). However, the presence of this Chorein_N domain in all members of the Vps13 family is not sufficient to explain an autophagy-specific function, as Vps13D is the only member of the Vps13 family that is required for autophagy in flies. Rather, Vps13D is the only Vps13 family member with a UBA domain (Anding et al., 2018), and this UBA domain is required for clearance of mitochondria (Fig. 6, A and C). Although speculative, it is possible that Vps13D binds to ubiquitinated cargo, in this case Pink1-regulated ubiquitination of factors that are juxtaposed to mitochondria, and then this proximity could allow the transfer of lipids to the growing autophagosome membranes around mitochondrial cargoes. Without Vps13D, autophagosome components such as Atg8a simply accumulate in the vicinity of mitochondria.
Pink1-mediated phosphorylation of ubiquitin and Parkin leads to the rapid ubiquitination of outer mitochondrial membrane proteins, leading to local recruitment of the autophagy machinery (Ordureau et al., 2014; Koyano et al., 2014; Wauer et al., 2015). While phosphorylation of both ubiquitin and Parkin are required for Parkin-mediated mitophagy (Koyano et al., 2014), it is unknown whether this is the sole purpose of ubiquitin phosphorylation. The accumulation of phosphorylated ubiquitin around mitochondria in vps13d mutants is consistent with Pink1 regulating Vps13D-mediated mitophagy through phosphorylation of ubiquitin (Fig. 5, C and D). Structural studies of phosphorylated ubiquitin chains suggest that phosphorylated ubiquitin is more resistant to deubiquitinating enzymes, suggesting that Pink1 phosphorylation of ubiquitin chains on outer mitochondrial membrane proteins by Pink1 can enhance protein ubiquitination even without Parkin (Wauer et al., 2015). Thus, one possible model is that phosphorylation of outer mitochondrial membrane ubiquitin chains in a Pink1-dependent manner stabilizes these ubiquitin chains. Vps13D then preferentially interacts with these phosphorylated ubiquitin chains, potentially through the UBA domain, facilitating additional processing through the autophagic machinery. Without Vps13D, these conjugated ubiquitin chains accumulate on the outer mitochondrial membrane, consistent with the conjugated ubiquitin phenotype observed in Vps13D mutant cells (Fig. 2, F and G; and Fig. 5, A and B). Another potential Pink1 substrate could be Vps13D itself, as phosphorylation of Vps13D was significantly decreased in a pink1 mutant mouse model for Parkinson’s disease (Auburger et al., 2019).
Pink1 can recruit proteins to mitochondria in a Parkin-independent manner (Lazarou et al., 2015). Furthermore, Pink1 physically interacts with and recruits Atg6 to mitochondria and ER membrane contacts to mediate mitophagy (Michiorri et al., 2010). Significantly, this recruitment of Atg6 is independent of Parkin (Gelmetti et al., 2017). In addition, proteomic studies have suggested that Vps13D interacts with proteins present at the mitochondria and ER interface (Hung et al., 2017; Antonicka et al., 2020). Thus, Vps13D puncta could reflect recruitment to the mitochondria and ER interface that is driven by Pink1 and Atg6 without requiring Park. In support of this model, we have recently shown that Vps13D functions as a conserved regulator of mitochondria and ER contact (Shen et al., 2021). However, given that Park localizes to remaining mitochondria in vps13d-deficient cells (Fig. S5 C), we cannot rule out the possibility that there may be some crosstalk between Vps13d- and Park-mediated mitophagy pathways. While Park is required for ubiquitin accumulation, loss of Park does not have the same ubiquitin phenotypes as loss of Pink1 or Vps13D (Fig. 3 H and Fig. 6 D). These findings could be explained by contributions from other ubiquitin ligases involved in Vps13D-mediated mitophagy. Further studies that investigate the role of Park localization to mitochondria and other ubiquitin ligases in Vps13D-deficient genetic backgrounds will help to clarify this mechanism.
Mutations in pink1 and vps13d are both associated with neurological disease and abnormal mitochondrial morphology. These diseases also share some symptoms, such as tremors and dystonia (Seong et al., 2018; Gauthier et al., 2018; Armstrong and Okun, 2020). Our findings suggest that there may be a link between the pathology of Vps13D- and Pink1-related diseases. However, thorough comparisons are difficult since only 19 patients from 12 families with confirmed vps13d-associated neurological symptoms have been characterized (Seong et al., 2018; Gauthier et al., 2018). A more thorough characterization of vps13d patient symptoms and clinical findings will be required to fully understand the clinical implications of the relationship between Pink1 and Vps13D.
Materials and methods
Drosophila strains
Drosophila melanogaster strains used in this study are listed in Table S1. w1118 were used as controls. All flies were raised on standard cornmeal/molasses/agar media at 25°C.
Cell lines
All cells used in this study are listed in Table S2. Cells were cultured at 37°C in 5% CO2 in DMEM supplemented with 5% FBS and penicillin/streptomycin as previously described (Anding et al., 2018).
atg161 and vps13d-3×FLAG design
The atg161 loss-of-function mutant was designed using transcription activator-like effector nuclease (Katsuyama et al., 2013). The vps13d-3×FLAG strain was edited using CRISPR/Cas9 (Gratz et al., 2014). All germline injections were done by the University of Massachusetts Medical School CRISPR Core. Oligonucleotides used in this study are listed in Table S3. For atg161, the following template DNA sequence was used: 5′-TCGCCGATGAGGCCGCCAACGAACTGGGCGGGACTGGAGTTGCGCTGCAAGGA-3′ to produce a 5-bp deletion of CCCAG (2870–2874 in the genomic region), producing a frameshift in the 244th amino acid of Atg16 and affecting all isoforms of Atg16. A single female fly containing the insertion was collected and validated by DNA sequencing. For vps13d-3×flag, the following single guide RNA targeting sequence was used: 5′-TTTATAAAATGCAATAGGT-3′. A 2-kb region flanking the C terminus of genomic vps13d was amplified by PCR, and site-directed mutagenesis was used to insert the 3×flag sequence in frame immediately before the stop codon. This fragment was inserted into a TOPO vector via TOPO cloning and sequenced to ensure that no additional mutations were present and was used to tag the C terminus of vps13d with 3×flag. A single female fly containing the insertion was collected and validated by DNA sequencing.
Induction of mosaic RNAi and mutant cell clones
Mosaic fluorescent-negative mutant cell clones were induced as described (Anding et al., 2018). To induce mosaic pink1B9, atg13, atg6 (Δ), atg161, and vps13d (MiMic) loss-of-function mutant cell clones, we used the hsflp, FRT19A, mRFP; hsflp;;FRT2A, Ubi-nlsRFP, hsflp;;FRT82B, Ubi-nlsGFP, and hsflp;;FRT80B, ubi-nlsGFP flies and crossed them with the mutant alleles with the respective FRT site. 8-h egg lays were heat shocked for 90 min at 37°C. Mosaic analyses were conducted only where indicated; otherwise, whole mutant animal or midgut-specific RNAi was used. In experiments where entire pink1B9 mutant animals were used, only males were selected and compared to ensure no sex-specific differences. Unless otherwise stated, in all experiments where mutant or RNAi-expressing phenotypes were compared with other single- or double-mutant cell phenotypes, either a heterozygous mutant genetic background or background expressing a nontarget upstream activating sequence (UAS) element was used as a control.
Dissection and immunolabeling of Drosophila larval intestines
White prepupae were collected and allowed to develop on wet filter paper for 2 h before dissection. Intestines were immunostained, as previously described, with modifications (Anding et al., 2018). Intestines were removed in cold PBS before being placed in 4% paraformaldehyde solution for fixation at 4°C overnight. Where applicable, RNAi was expressed throughout the entire intestine using the Myo31DFNP0001 driver. For Atg8a/GABARAP staining, samples were placed in 1:1 4% paraformaldehyde/heptanes for 20 min before being incubated in methanol for 1 min at room temperature for fixation. After fixation, intestines were washed twice with PBS and then twice with 0.1% PBS with Triton X-100 (PBSTx) before blocking in 5% normal goat serum for 90 min and incubation with primary antibody in 0.1% PBSTx overnight. Intestines were then stained with secondary antibody for 3 h before nuclei staining and mounting. All antibodies used in this study are listed in Table S4. The following primary antibodies were used: rabbit anti-ref(2)p (1:1,000; from Gabor Juhasz, Biological Research Centre of the Hungarian Academy of Sciences, Budapest, Hungary), mouse anti-ATP synthase complex V (1:1,000, ab14748; Abcam), rabbit anti-ubiquitin (1:1,000, ab19247; Abcam), mouse anti-ubiquitin-FK2 (1:100, PW8810; Enzo Life Sciences), rabbit anti-Atg8a/GABARAP (1:100, #13733; Cell Signaling Technology), rabbit anti-Park (1:100, from Alex Whitworth, MRC Mitochondrial Biology Unit, University of Cambridge, Cambridge, UK), rabbit anti-pSer65 Ubiquitin (1:100, ABS1513-I; Millipore Sigma), mouse anti-Vps13D (1:50; Anding et al., 2018), and rat anti-Syx-17 (1:100, from Gabor Juhasz). The following secondary antibodies were used: anti-mouse Alexa Fluor 647 (A-21235; Invitrogen), anti-rabbit Alexa Fluor 546 (A-11035; Invitrogen), anti-rabbit Oregon Green 488 (O-6381; Molecular Probes), anti-rat Alexa Fluor 546 (A-11081; Thermo Fisher Scientific), and anti-mouse Alexa Fluor 546 (A-11030; Invitrogen). Nuclei were stained with Hoescht (Invitrogen), and samples were mounted with VECTASHIELD (Vector Laboratories). Only enterocytes in the anterior midgut and gastric caeca were imaged. Images were acquired using a Zeiss LSM 700 Axio Observer confocal microscope with Plan apochromat 63×/1.40 NA and EC Plan-Neofluar 40×/1.30 oil immersion objective at room temperature using Zen software (Zeiss). Signals in images were thresholded to only include distinct puncta.
TEM
TEM was conducted as previously described, with modifications (Anding et al., 2018). Intestines were dissected in PBS (GIBCO) 2 h after pupariation and fixed in a solution of 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.4 (Electron Microscopy Sciences) for 1 h at room temperature followed by overnight fixation at 4°C in fresh fix. Intestines were washed in 0.1 M sodium cacodylate buffer, pH 7.4; postfixed in 1% osmium tetroxide in distilled water for 1 h at room temperature; and washed in distilled water. Preparations were stained en bloc in 1% aqueous uranyl acetate overnight at 4°C in the dark, washed in distilled water, dehydrated through a graded ethanol series, treated with propylene oxide, and infiltrated in SPI-Pon Araldite for embedding. Ultrathin sections were cut on a Leica UC7 microtome. Sections were stained with uranyl acetate and lead citrate and examined on a Philips CM10 TEM. Images were taken down the length of the anterior region of the midgut to ensure an unbiased approach. For each genotype, at least three intestines were embedded and sectioned for analyses and quantification. We reviewed all images and selected representative images for analyses.
Western blot and immunoprecipitation
Tissue was lysed in 1× Laemli sample buffer diluted in radioimmunoprecipitation assay (RIPA) lysis buffer (10 mM Tris-Cl, pH 8.0; 1 mM EDTA, pH 8.0; 0.5 mM EGTA; 2.4 mM sodium deoxycholate; 140 mM sodium chloride) at a ratio of 10 μl lysis buffer per intestine and 30 μl per whole prepupa. Samples were homogenized in solution using a plastic pestle for 30 s before being boiled at 99°C for 6 min. Samples were run on 7.5% polyacrylamide gel, transferred onto 0.45-μm polyvinylidene fluoride membranes (Millipore Sigma), and probed with antibodies using standard protocols. Cells were cultured in 24-well plates and lysed at 80%–90% confluency using the same 1× Laemli sample buffer by removing media, washing with PBS, and incubating with 150 μl lysis buffer for 15 min with rotation. Primary antibodies used were mouse anti-FLAG (1:1,000; Millipore Sigma), anti-ubiquitin-P4D1 HRP (sc-8017; Santa Cruz Biotechnology), rabbit anti-Atg8a/GABARAP (1:1,000, #13733; Cell Signaling Technology), mouse anti-actin (1:1,000, Proteintech), and mouse anti-ATP5a (1:1,000; Abcam).
For immunoprecipitations, 2-h-old prepupae were lysed in RIPA lysis buffer supplemented with 1 mM N-ethylmaleimide, 1 mM PMSF, and Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific) at a ratio of 16 prepupae per 250 μl lysis buffer. Prepupae were crushed with a plastic pestle for 30 s and incubated on ice for 30 min before being centrifuged at 4°C at 13,000 rpm for 10 min. Supernatant was filtered through 0.45-μm cellulose acetate filters (Millipore Sigma). 30 μl of filtered supernatant was diluted in 10 μl of 4× Laemli sample buffer (Bio-Rad), boiled for 6 min at 99°C, and used as input. 200 μl of filtered supernatant (∼1 mg protein) was used for immunoprecipitation. 40 μl of anti-FLAG M2 magnetic bead slurry (Millipore Sigma) warmed to room temperature was washed twice with RIPA buffer before incubation with filtered supernatant for 2 h at 4°C on a rotator. For all control lysates, 1 μg of 3×FLAG peptide was added to account for nonspecific binding due to the 3×FLAG epitope. Following incubation, the supernatant was discarded, and the beads were washed four times with 1 mL 0.1% PBSTx. Beads were eluted with 20 μl 1× Laemli sample buffer diluted in RIPA lysis buffer and boiled for 6 min at 99°C. 20 μl of input and eluate was run on 7.5% polyacrylamide gel for Western blot analysis.
Quantification and statistical analyses
All quantification of cell size and puncta in immunofluorescence images were conducted using ImageJ software. Sample sizes (n) in immunofluorescence image quantifications represent number of cells. For quantification of puncta number, size, and number of puncta encircled, images were first thresholded to the brightest 2.5% of puncta with the exception of conjugated ubiquitin (FK2) puncta, which were thresholded to the brightest 5.0% of puncta due to dull puncta in control cells. Gaussian blur with 0.8 sigma was applied to reduce background noise. Quantification of mitolysosomes from Mito-QC was based on the approach described in Lee et al. (2018). Images of mCherry-positive puncta and mCherry and GFP–positive puncta for each sample were thresholded to the brightest 2.5% of puncta. The area of mCherry-positive puncta was then subtracted by the area of mCherry and GFP–positive puncta using the Image Calculator function in ImageJ. The remaining mCherry-only puncta, representative of mitolysosomes, were then quantified using the same criteria as previous quantifications of puncta number.
TEM analyses of mitochondria area was manually calculated by individually analyzing mitochondria using ImageJ. Regions for analyses were randomly selected from sections for each sample. For each analysis, at least 100 mitochondria from at least 20 sections from three individual animals were used for quantification. Sample sizes (n) in TEM quantifications represent number of mitochondria.
All experiments are representative of three independent replicates. No statistical methods were used to predetermine sample sizes. Preliminary experiments were conducted to achieve similar sample sizes as previously published studies using our model systems. Animals were not excluded for statistical analyses. Researchers were not blinded. Unless otherwise stated, two-tailed t test P values were calculated without Welch’s correlation. For comparisons among three or more genotypes, one-sided ANOVA tests were conducted with Tukey’s post hoc analysis. Data distribution was assumed to be normal, but this was not formally tested. P > 0.05 were considered nonsignificant. All bars are means, and error bars are SEM, unless otherwise stated.
Online supplemental material
Fig. S1 shows specificity of the Atg8a antibody, examines Atg1 and Atg6 localization in a vps13d-deficient background, compares Vps13D puncta formation in other core autophagy gene–deficient cells and time points, and demonstrates that the atg161 mutant is a strong loss-of-function mutant. Fig. S2 compares ubiquitinated protein levels in early third instar larvae compared with 2-h-old prepupae, shows that vps13d was successfully tagged with 3×FLAG, and shows pan-ubiquitin and mitochondria colocalization in a vps13d (ΔUBA) mutant background. Fig. S3 characterizes Ref2p localization, abnormal Atg8a puncta suppression by Atg6a depletion, and stalled autophagosome formation in pink1 and vps13d mutants. Fig. S4 quantifies mito-GFP levels in pink1 and pink1; vps13d mutants and shows that Vps13D puncta are not affected in pink1B9 loss-of-function mutant cells in early third instar larvae. Fig. S5 shows that ATP5a and the Mito-QC reporter colocalize in vps13d and park mutant cells, specificity of the Park antibody and localization in vps13d-deficient cells, and the relationship between Syx17 and Vps13D. Table S1 lists Drosophila lines used in this study. Table S2 lists the Drosophila lines used for each experiment. Table S3 lists primers and oligonucleotides used in this study. Table S4 lists antibodies used in this study.
Supplementary Material
lists Drosophila lines used in this study.
lists the Drosophila lines used for each experiment.
lists primers and oligonucleotides used in this study.
lists antibodies used in this study.
Acknowledgments
We thank the Baehrecke laboratory, G. Juhasz, L. Pallanck, T. Tomoda, A.J. Whitworth, the Vienna Drosophila Resource Center, the Bloomington Stock Center, the Kyoto Drosophila Genetic Resource Center, and the Electron Microscopy Core Facility at the University of Massachusetts Medical School for advice, flies, antibodies, and technical support.
This work was supported by National Institutes of Health grants R35GM131689 to E.H. Baehrecke and F30CA239374 to J.L. Shen.
The authors declare no competing financial interests.
Author contributions: J.L. Shen and E.H. Baehrecke designed experiments. J.L. Shen and T.M. Fortier performed experiments. R. Wang provided resources. J.L. Shen and E.H. Baehrecke wrote the manuscript, and all authors commented on it.
References
- Allen, G.F., Toth R., James J., and Ganley I.G.. 2013. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 14:1127–1135. 10.1038/embor.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anding, A.L., and Baehrecke E.H.. 2017. Cleaning house: Selective autophagy of organelles. Dev. Cell. 41:10–22. 10.1016/j.devcel.2017.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anding, A.L., Wang C., Chang T.-K., Sliter D.A., Powers C.M., Hofmann K., Youle R.J., and Baehrecke E.H.. 2018. Vps13D encodes a ubiquitin-binding protein that is required for the regulation of mitochondrial size and clearance. Curr. Biol. 28:287–295.e6. 10.1016/j.cub.2017.11.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonicka, H., Lin Z.-Y., Janer A., Aaltonen M.J., Weraarpachai W., Gingras A.-C., and Shoubridge E.A.. 2020. A high-density human mitochondrial proximity interaction network. Cell Metab. 32:479–497.e9. 10.1016/j.cmet.2020.07.017 [DOI] [PubMed] [Google Scholar]
- Armstrong, M.J., and Okun M.S.. 2020. Diagnosis and treatment of Parkinson disease: A review. JAMA. 323:548–560. 10.1001/jama.2019.22360 [DOI] [PubMed] [Google Scholar]
- Auburger, G., Gispert S., Torres-Odio S., Jendrach M., Brehm N., Canet-Pons J., Key J., and Sen N.-E.. 2019. SerThr-phosphoproteome of brain from aged PINK1-KO+A53T-SNCA mice reveals pT1928-MAP1B and pS3781-ANK2 deficits, as hub between autophagy and synapse changes. Int. J. Mol. Sci. 20:3284. 10.3390/ijms20133284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhujabal, Z., Birgisdottir Å.B., Sjøttem E., Brenne H.B., Øvervatn A., Habisov S., Kirkin V., Lamark T., and Johansen T.. 2017. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 18:947–961. 10.15252/embr.201643147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen, V.A., Májek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F.R., Olk N., Stukalov A., et al. 2015. Gene essentiality and synthetic lethality in haploid human cells. Science. 350:1092–1096. 10.1126/science.aac7557 [DOI] [PubMed] [Google Scholar]
- Chang, T.-K., Shravage B.V., Hayes S.D., Powers C.M., Simin R.T., Wade Harper J., and Baehrecke E.H.. 2013. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat. Cell Biol. 15:1067–1078. 10.1038/ncb2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chino, H., and Mizushima N.. 2020. ER-phagy: Quality control and turnover of endoplasmic reticulum. Trends Cell Biol. 30:384–398. 10.1016/j.tcb.2020.02.001 [DOI] [PubMed] [Google Scholar]
- Dempster, J.M., Rossen J., Kazachkova M., Pan J., Kugener G., Root D.E., and Tsherniak A.. 2019. Extracting biological insights from the Project Achilles Genome-Scale CRISPR screens in cancer cell lines. bioRxiv. (Preprint posted July 31, 2019). 10.1101/720243 [DOI]
- Friedman, J.R., and Nunnari J.. 2014. Mitochondrial form and function. Nature. 505:335–343. 10.1038/nature12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier, J., Meijer I.A., Lessel D., Mencacci N.E., Krainc D., Hempel M., Tsiakas K., Prokisch H., Rossignol E., Helm M.H., et al. 2018. Recessive mutations in VPS13D cause childhood onset movement disorders. Ann. Neurol. 83:1089–1095. 10.1002/ana.25204 [DOI] [PubMed] [Google Scholar]
- Gelmetti, V., De Rosa P., Torosantucci L., Marini E.S., Romagnoli A., Di Rienzo M., Arena G., Vignone D., Fimia G.M., and Valente E.M.. 2017. PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER-mitochondria tethering and autophagosome formation. Autophagy. 13:654–669. 10.1080/15548627.2016.1277309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz, S.J., Ukken F.P., Rubinstein C.D., Thiede G., Donohue L.K., Cummings A.M., and O’Connor-Giles K.M.. 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 196:961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, X., Watzlawik J.O., Fiesel F.C., and Springer W.. 2020. Autophagy in Parkinson’s disease. J. Mol. Biol. 432:2651–2672. 10.1016/j.jmb.2020.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, V., Lam S.S., Udeshi N.D., Svinkina T., Guzman G., Mootha V.K., Carr S.A., and Ting A.Y.. 2017. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife. 6:e24463. 10.7554/eLife.24463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura, E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Jiang, P., and Mizushima N.. 2014. Autophagy and human diseases. Cell Res. 24:69–79. 10.1038/cr.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuyama, T., Akmammedov A., Seimiya M., Hess S.C., Sievers C., and Paro R.. 2013. An efficient strategy for TALEN-mediated genome engineering in Drosophila. Nucleic Acids Res. 41:e163. 10.1093/nar/gkt638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin, V., and Rogov V.V.. 2019. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol. Cell. 76:268–285. 10.1016/j.molcel.2019.09.005 [DOI] [PubMed] [Google Scholar]
- Koyano, F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., et al. 2014. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 510:162–166. 10.1038/nature13392 [DOI] [PubMed] [Google Scholar]
- Kumar, N., Leonzino M., Hancock-Cerutti W., Horenkamp F.A., Li P., Lees J.A., Wheeler H., Reinisch K.M., and De Camilli P.. 2018. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217:3625–3639. 10.1083/jcb.201807019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, A.B., John Peter A.T., Walter P., and Kornmann B.. 2015. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 210:883–890. 10.1083/jcb.201502105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou, M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., and Youle R.J.. 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 524:309–314. 10.1038/nature14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.J., Sanchez-Martinez A., Martinez Zarate A., Benincá C., Mayor U., Clague M.J., and Whitworth A.J.. 2018. Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin. J. Cell Biol. 217:1613–1622. 10.1083/jcb.201801044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Lin J., Zhang M., Chen K., Yang S., Wang Q., Yang H., Xie S., Zhou Y., Zhang X., et al. 2016. PINK1 is required for timely cell-type specific mitochondrial clearance during Drosophila midgut metamorphosis. Dev. Biol. 419:357–372. 10.1016/j.ydbio.2016.08.028 [DOI] [PubMed] [Google Scholar]
- McWilliams, T.G., Prescott A.R., Allen G.F.G., Tamjar J., Munson M.J., Thomson C., Muqit M.M.K., and Ganley I.G.. 2016. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 214:333–345. 10.1083/jcb.201603039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams, T.G., Prescott A.R., Montava-Garriga L., Ball G., Singh F., Barini E., Muqit M.M.K., Brooks S.P., and Ganley I.G.. 2018. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 27:439–449.e5. 10.1016/j.cmet.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H., Dharia N.V., Montgomery P.G., Cowley G.S., Pantel S., et al. 2017. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49:1779–1784. 10.1038/ng.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiorri, S., Gelmetti V., Giarda E., Lombardi F., Romano F., Marongiu R., Nerini-Molteni S., Sale P., Vago R., Arena G., et al. 2010. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 17:962–974. 10.1038/cdd.2009.200 [DOI] [PubMed] [Google Scholar]
- Nakada, T.A., Boyd J.H., Russell J.A., Aguirre-Hernández R., Wilkinson M.D., Thair S.A., Nakada E., McConechy M.K., Fjell C.D., and Walley K.R.. 2015. VPS13D gene variant is associated with altered IL-6 production and mortality in septic shock. J. Innate Immun. 7:545–553. 10.1159/000381265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa, H., Suzuki K., Kamada Y., and Ohsumi Y.. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467. 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- Narendra, D.P., Jin S.M., Tanaka A., Suen D.-F., Gautier C.A., Shen J., Cookson M.R., and Youle R.J.. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298. 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau, A., Sarraf S.A., Duda D.M., Heo J.M., Jedrychowski M.P., Sviderskiy V.O., Olszewski J.L., Koerber J.T., Xie T., Beausoleil S.A., et al. 2014. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell. 56:360–375. 10.1016/j.molcel.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa, T., and Noda N.N.. 2019. Atg2: A novel phospholipid transfer protein that mediates de novo autophagosome biogenesis. Protein Sci. 28:1005–1012. 10.1002/pro.3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa, T., Kotani T., Kawaoka T., Hirata E., Suzuki K., Nakatogawa H., Ohsumi Y., and Noda N.N.. 2019. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26:281–288. 10.1038/s41594-019-0203-4 [DOI] [PubMed] [Google Scholar]
- Padman, B.S., Nguyen T.N., Uoselis L., Skulsuppaisarn M., Nguyen L.K., and Lazarou M.. 2019. LC3/GABARAPs drive ubiquitin-independent recruitment of optineurin and NDP52 to amplify mitophagy. Nat. Commun. 10:408. 10.1038/s41467-019-08335-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras, K., Lionaki E., and Tavernarakis N.. 2018. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20:1013–1022. 10.1038/s41556-018-0176-2 [DOI] [PubMed] [Google Scholar]
- Pilli, M., Arko-Mensah J., Ponpuak M., Roberts E., Master S., Mandell M.A., Dupont N., Ornatowski W., Jiang S., Bradfute S.B., et al. 2012. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 37:223–234. 10.1016/j.immuni.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovic, A., Ziegler J., Mårtensson C.U., Prasuhn J., Shurkewitsch K., König P., Paulson H.L., and Klein C.. 2019. PINK1-dependent mitophagy is driven by the UPS and can occur independently of LC3 conversion. Cell Death Differ. 26:1428–1441. 10.1038/s41418-018-0219-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlatti, F., Maffei R., Beau I., Codogno P., and Ghidoni R.. 2008. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 15:1318–1329. 10.1038/cdd.2008.51 [DOI] [PubMed] [Google Scholar]
- Seong, E., Insolera R., Dulovic M., Kamsteeg E.-J., Trinh J., Brüggemann N., Sandford E., Li S., Ozel A.B., Li J.Z., et al. 2018. Mutations in VPS13D lead to a new recessive ataxia with spasticity and mitochondrial defects. Ann. Neurol. 83:1075–1088. 10.1002/ana.25220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J.L., Fortier T.M., Zhao Y.G., Wang R., Burmeister M., and Baehrecke E.H.. 2021. Vmp1, Vps13D, and Marf/Mfn2 function in a conserved pathway to regulate mitochondria and ER contact in development and disease. Curr. Biol. 31:3028–3039.e7. 10.1016/j.cub.2021.04.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka, T., Weidberg H., Pietrokovski S., and Elazar Z.. 2011. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 12:226. 10.1186/gb-2011-12-7-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Kubota Y., Sekito T., and Ohsumi Y.. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12:209–218. 10.1111/j.1365-2443.2007.01050.x [DOI] [PubMed] [Google Scholar]
- Takáts, S., Nagy P., Varga Á., Pircs K., Kárpáti M., Varga K., Kovács A.L., Hegedűs K., and Juhász G.. 2013. Autophagosomal syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201:531–539. 10.1083/jcb.201211160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboyama, K., Koyama-Honda I., Sakamaki Y., Koike M., Morishita H., and Mizushima N.. 2016. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 354:1036–1041. 10.1126/science.aaf6136 [DOI] [PubMed] [Google Scholar]
- Vicencio, E., Beltrán S., Labrador L., Manque P., Nassif M., and Woehlbier U.. 2020. Implications of selective autophagy dysfunction for ALS pathology. Cells. 9:381. 10.3390/cells9020381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stockum, S., Marchesan E., and Ziviani E.. 2018. Mitochondrial quality control beyond PINK1/Parkin. Oncotarget. 9:12550–12551. 10.18632/oncotarget.23799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., and Sabatini D.M.. 2015. Identification and characterization of essential genes in the human genome. Science. 350:1096–1101. 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Liu N., and Lu B.. 2019. Mechanisms and roles of mitophagy in neurodegenerative diseases. CNS Neurosci. Ther. 25:859–875. 10.1111/cns.13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer, T., Swatek K.N., Wagstaff J.L., Gladkova C., Pruneda J.N., Michel M.A., Gersch M., Johnson C.M., Freund S.M.V., and Komander D.. 2015. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 34:307–325. 10.15252/embj.201489847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle, R.J., and Narendra D.P.. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12:9–14. 10.1038/nrm3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists Drosophila lines used in this study.
lists the Drosophila lines used for each experiment.
lists primers and oligonucleotides used in this study.
lists antibodies used in this study.