Siu et al. review the cell biology of fertilization with a focus on the factors involved in gamete attachment and fusion.
Abstract
Fertilization is defined as the union of two gametes. During fertilization, sperm and egg fuse to form a diploid zygote to initiate prenatal development. In mammals, fertilization involves multiple ordered steps, including the acrosome reaction, zona pellucida penetration, sperm–egg attachment, and membrane fusion. Given the success of in vitro fertilization, one would think that the mechanisms of fertilization are understood; however, the precise details for many of the steps in fertilization remain a mystery. Recent studies using genetic knockout mouse models and structural biology are providing valuable insight into the molecular basis of sperm–egg attachment and fusion. Here, we review the cell biology of fertilization, specifically summarizing data from recent structural and functional studies that provide insights into the interactions involved in human gamete attachment and fusion.
Introduction
During sexual reproduction, the oocyte and sperm fuse to generate a new and unique embryo. The journey of a sperm to an egg ends in the ampulla of the female oviduct. From there, the sperm must overcome a number of physical and biochemical barriers. After undergoing the acrosome reaction and binding the ova, the sperm penetrates through the cumulus oophorus cells and the zona pellucida (ZP) to reach the perivitelline space (PVS) and oocyte membrane. Upon fusion of the sperm and egg membranes, the sperm nucleus and organelles are incorporated into the egg cytoplasm.
An understanding of the mechanisms of mammalian fertilization is crucial to treat infertility and develop new methods of birth control. Infertility affects 15% of couples globally, and in one third of these cases, the underlying cause is unknown (Gelbaya et al., 2014). Developments in assisted reproductive technologies have provided couples with new options to conceive but may have epigenetic side effects (Mani et al., 2020). Furthermore, only 40% of couples manage to have a child despite 2 yr of treatment. Safety, efficacy, and acceptability of contraceptives are also critically important, but many current female contraceptive methods have side effects that limit long-term use (Aitken et al., 2008), while male contraceptives are limited to condoms or vasectomy (Kanakis and Goulis, 2015). A better understanding of the molecular players involved in fertilization is necessary to drive innovation in both assisted reproductive technologies and contraception.
In this review, we will first briefly review the events that prepare the gametes for fertilization. We will then discuss how recent studies of genetically altered mice and structural biology efforts have shed light on the molecular mechanisms of sperm–egg attachment and fusion. We will also discuss the gaps in current knowledge and suggest new perspectives and future directions in the search for other protein factors involved at the gamete fusion synapse.
Cell biology of gametes
Fertilization requires proper gametogenesis (oogenesis in the female and spermatogenesis in the male), which produces haploid cells and introduces diversity. Primordial germ cells (PGC) are the embryonic precursors to spermatocytes and ova. The cells produced by the first few divisions of the fertilized egg are totipotent and capable of differentiating into any cell type, including germ cells. PGCs originate within the primary ectoderm of the embryo and then migrate into the yolk sac. Between weeks 4 and 6, the PGCs migrate back into the posterior body wall of the embryo, where they stimulate cells of the adjacent coelomic epithelium and mesonephros to form primitive sex cords and induce the formation of the genital ridges and gonads. The sex (gonadal) cords surround the PGCs and give rise to the tissue that will nourish and regulate the development of the maturing sex cells (ovarian follicles in the female and Sertoli cells in the male).
Egg
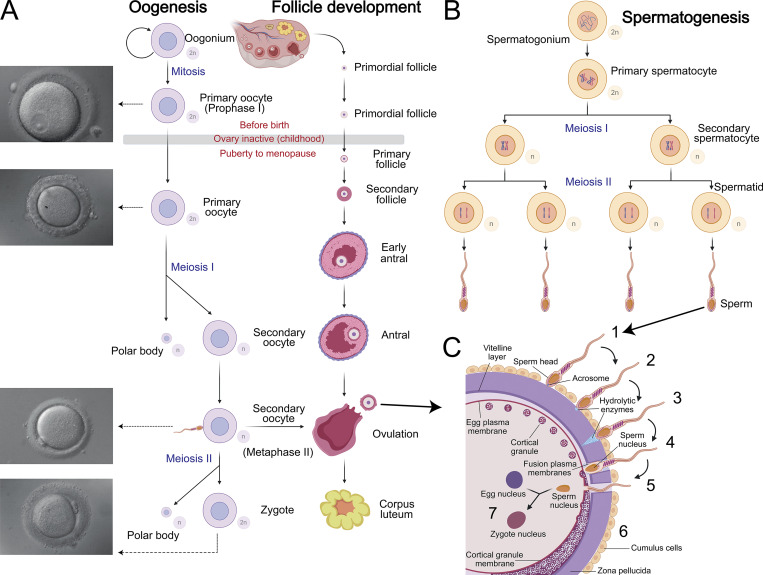
Oogenesis is a complex differentiation process by which mature functional ova develop from germ cells (Fig. 1 A; Edson et al., 2009). In humans, oogenesis begins in the ovary at 6–8 wk of fetus development, when PGCs differentiate into oogonia. By the 12th week, several million oogonia enter prophase, the first meiotic division and become dormant until shortly before ovulation (Hayashi et al., 2020). Due to their large and watery nuclei, these cells are referred to as germinal vesicles (Pan and Li, 2019). These primary oocytes become enclosed by follicle cells to form primordial follicles. The number of primordial follicles peaks at ∼7 million by the fifth month of fetal life, with ∼700,000 left at birth and 400,000 by puberty (Marcozzi et al., 2018). All of the egg cells that the ovaries will release are already present at birth.
Figure 1.
Gametogenesis and fertilization. (A–C) Illustration of oogenesis and follicle development (A), spermatogenesis (B), and the major steps in fertilization (C): (1) initial contact, (2) acrosome reaction, (3) ZP penetration, (4) sperm–egg fusion, (5) entry of sperm nucleus, (6) cortical reaction, and (7) fusion of the sperm and egg nuclei. The oocyte with its ZP measures 130 μm in diameter. Created with BioRender.
During each menstrual cycle, hormones from the hypothalamic–pituitary–gonadal axis restart the division of the primary oocytes in meiosis I and follicular development (Atwood and Vadakkadath Meethal, 2016). Primary follicles develop into secondary follicles, containing each growing oocyte surrounded by two or more layers of proliferating follicle cells. ZP glycoproteins are secreted by the oocyte of the primary follicle and possibly the follicular cells (Törmälä et al., 2008). Although these glycoproteins form a physical barrier between the follicle cells and the oocyte, follicle cells and the oocyte remain connected through transzonal cytoplasmic projections from the follicle cells until fertilization (Makabe et al., 2006). A reciprocal dialog between the oocyte and its surrounding follicular cells coordinates the different phases of follicular development and the maintenance of meiotic arrest (Dalbies-Tran et al., 2020). Oocyte-derived microvilli control female fertility by optimizing ovarian follicle selection in mice (Zhang et al., 2021). The epithelium of 5–12 primary follicles proliferates to form a multilayered capsule around the oocyte. A few of these growing follicles continue to enlarge in response to follicle-stimulating hormone (FSH; Visser and Themmen, 2014). A single follicle becomes dominant, and the others degenerate by atresia (Atwood and Vadakkadath Meethal, 2016). Meiosis of the oocyte in the mature preovulatory follicle is blocked until a surge in levels of FSH and luteinizing hormone that occurs midway through the menstrual cycle. The membrane of the germinal vesicle nucleus breaks down, the chromosomes align in metaphase, and the oocyte expels its first polar body. The secondary oocyte then begins the second meiotic division, which is arrested at the meiotic metaphase II stage until ovulation (Gougeon, 1996). Ovulation depends on the breakdown of the follicle wall and occurs ∼38 h after the increase in levels of FSH and luteinizing hormone (Holesh et al., 2021). The disruption of the follicle wall expulses the oocyte, which is captured by the fimbriated mouth of the oviduct and moved into the ampulla. The oocyte retains its ability to be fertilized for ∼24 h and completes meiosis only if it is fertilized.
Sperm
In contrast to oogenesis, which is complete before birth, spermatogenesis is a continuous process that begins at puberty (Fig. 1 B). In humans, spermatogenesis takes 74 d to complete; thus, multiple spermatogenesis events occur simultaneously to allow for continual sperm production. Spermatogenesis occurs in the testis in a stepwise manner, beginning with diploid spermatogonia at the basal surface of seminiferous tubules and ending with mature elongated spermatozoa that are released in tubule lumens in a process called spermiation (Clermont, 1972; Yang and Oatley, 2014). During spermatogenesis, mitosis results in gene amplification, meiosis results in genome reduction, and finally maturation occurs (Hess and Renato de Franca, 2008). At this stage, sperm are not motile and are fertilization incompetent. Two additional sperm maturational processes are required outside the testis. First, sperm undergo a maturation process during epididymal transit (Bedford et al., 1973) involving posttranslational modifications of previously synthesized proteins and acquisition of proteins from the epididymal epithelium (James et al., 2020; see text box). After ejaculation into the female reproductive tract, dilution triggers additional changes in sperm, collectively termed capacitation (see text box), that prepare the sperm for the acrosome reaction.
Epididymal maturation
Sperm exchange with the epididymal epithelium occurs by direct interaction with epithelial cells, by interaction with soluble proteins in the epididymal fluid or via extracellular exosome-like vesicles released by epithelial cells called epididymosomes (James et al., 2020). The purposes of this exchange are to redistribute sperm proteins and change the composition and lipid balance of the sperm membrane. These changes take place during the transit from the epididymis initial segment, through the caput and the corpus, to the cauda where sperm are stored (Cornwall, 2009). Epididymal transit lasts 10–12 d in mammals, but storage is dependent on sexual activity. Since fertilization is not immediate, fertilizing capacities of the spermatozoa are preserved by decapacitation factors that are active in the epididymis. An example of a decapacitation factor is SPINK3, which is secreted by seminal vesicles; it impairs sperm membrane hyperpolarization and calcium influx through CatSper (Zalazar et al., 2020). Epididymal plasma and sperm represent only a small fraction (5%) of semen in men (Batruch et al., 2011). Two thirds of the volume of semen comes from the seminal vesicles and the other third from the prostate. These secretions protect the sperm and prevent early maturation.
Sperm capacitation
More than 70 yr ago, Austin and Chang described capacitation as the changes required for sperm to fertilize oocytes in vivo (Austin, 1952; Chang, 1951). Once sperm enter the female reproductive tract, they undergo capacitation. Capacitation results in hyperactivation of sperm movement and initiation of the acrosome reaction (Saling et al., 1979; Florman and First, 1988). During capacitation, stabilizing or decapacitation factors that are adsorbed on the sperm plasma membrane are removed (Bedford and Chang, 1962). These agents that initiate removal of decapacitation factors are electrolytes, energy substrates, and proteins such as seminal plasma protein or albumin. Removal of decapacitation factors increases sperm plasma membrane fluidity, allowing an increase in the permeability to calcium, chloride, and bicarbonate ions (Gangwar and Atreja, 2015). Sperm motility depends on the membrane potential, intracellular pH, and balance of intracellular ions (reviewed in Nowicka-Bauer and Szymczak-Cendlak, 2021). The most important ion for this function is Ca2+ (Hwang et al., 2019). This secondary messenger is an important signaling pathways activator that regulates sperm motility (Finkelstein et al., 2020). The activation of soluble adenyl cyclases generates cyclic adenosine monophosphate that in turn activates serine/threonine protein kinase A, which induces a cascade of protein phosphorylation initiating the induction of sperm motility (Chen et al., 2000). Protein phosphorylation, sperm hyperactivation, and the acrosome reaction are used in vitro to evaluate capacitation. Capacitation can be induced in vitro by incubation in medium containing calcium, bicarbonate ions, and serum albumin (Touré, 2019).
Mammalian sperm capacitation occurs during sperm migration in the female tract. Mammalian males ejaculate millions of sperm cells into the female reproductive tract, but only a few hundred sperm at most reach the oocytes. This massive elimination process likely prevents polyspermy (reviewed in Kölle, 2015). Selection of human sperm during the journey begins in the acidic environment of the vagina. In the cervix, only morphologically normal sperm can migrate. Some sperm immediately pass into the cervical mucus, whereas the remaining sperm becomes a part of the coagulum. The next selection occurs at the uterus–tubal junction, the connection between the uterus and the oviduct that represents a major obstacle for sperm migration (Kölle, 2015). Experiments in mice indicate that sperm motility alone is insufficient for sperm migration through the uterus–tubal junction (Fujihara et al., 2018). Uterine contractions facilitate sperm transport as do molecular interactions. Several proteins, such as ADAM3 and other ADAM family members, are known to be involved in this step in mice (Yamaguchi et al., 2009; Xiong et al., 2019); most ADAM proteins have human orthologues.
Spermatogenesis takes place in a species-specific cycle called the seminiferous epithelial cycle and is regulated in particular through the hypothalamic–pituitary–testicular axis. Indeed, at puberty, the testes (interstitial steroidogenic Leydig cells) secrete an increased amount of testosterone, which triggers growth of the testes, maturation of the seminiferous tubules, and the commencement of spermatogenesis. The Sertoli cells are the major somatic cells present in the seminiferous tubules and are considered to be the main regulators of spermatogenesis. They orchestrate spermatogenesis by supporting spermatogonial stem cells, determining the testis size, organizing meiotic and postmeiotic development and sperm output, supporting androgen production by maintaining the development and function of Leydig cells, and regulating other aspects of testis function like peritubular myoid cells, immune cells, and the vasculature, which participate in the maintenance of the spermatogonial stem cell niche.
Acrosome reaction
The acrosome is a secretory vesicle located on the anterior region of sperm that originates from the spermatid Golgi apparatus. An acrosomal granule is formed by the fusion of proacrosomal vesicles in the vicinity of the nucleus. The region increases in size and spreads over the anterior part of the nucleus. The acrosome reaction is driven by SNARE complexes and results in the exocytosis of the contents of the acrosome upon fusion of the plasma membrane with the outer acrosomal membrane (Fig. 1 C; reviewed in Okabe, 2016; De Blas et al., 2005). The timing of the acrosome reaction is critical. Only sperm that have undergone this reaction are fertilization competent, but when a high proportion of sperm undergo the acrosome reaction prematurely, success of in vitro fertilization is low (Wiser et al., 2014). Several studies indicate that only a fraction of sperm is capable of undergoing spontaneous acrosome reaction. In human and mice sperm samples, 15–20% of cells undergo spontaneous acrosome reaction (Nakanishi et al., 2001), whereas only 20–30% undergo progesterone-induced acrosome reaction (Stival et al., 2016), suggesting physiological heterogeneity of sperm population. In addition, Inoue et al. demonstrated that acrosome-reacted mouse spermatozoa recovered from the PVS can fertilize other eggs (Inoue et al., 2011).
Based on in vitro data, it was thought that the acrosome reaction occurs when the sperm contacts the ZP, particularly the ZP3 protein (Litscher and Wassarman, 1996). Using transgenic mice that express fluorescent markers in the acrosome (Nakanishi et al., 1999) and the midpiece mitochondria (Hasuwa et al., 2010), real-time observation of acrosomal exocytosis was possible. These experiments showed that most mouse spermatozoa capable of fertilization had undergone the acrosome reaction before contact with the oocyte ZP (Jin et al., 2011). Most spermatozoa begin to react in the isthmus of the oviduct before reaching the ampulla (Hino et al., 2016; La Spina et al., 2016). Contact with the ZP in vitro probably makes it possible to complete a partial acrosome reaction. The most important function of the acrosome reaction is to induce changes in the sperm membrane (Okabe, 2016). The relocations of IZUMO1 and SPACA6, proteins essential for sperm–egg fusion, that occur after the acrosome reaction are illustrative examples of these changes (Sosnik et al., 2009; Barbaux et al., 2020; Satouh et al., 2012). The presence of these proteins on the sperm membrane, in addition to the classic markers Pisum sativum agglutinin, Peanut agglutinin lectins, or CD46, can be used as markers for the acrosome reaction (Ito and Toshimori, 2016). The acrosome and its disruption are both crucial for effective fertilization, as low fertilization rates are observed upon intracytoplasmic sperm injection of acrosome-intact sperm (Morozumi and Yanagimachi, 2005) or round spermatozoa lacking acrosomes (Dávila Garza and Patrizio, 2013).
ZP penetration
The ZP is a physical barrier between the oocyte and the follicular cells that forms from glycoproteins secreted from the primary follicles (Fig. 1 C). The human ZP consists of four glycoproteins (hZP1–hZP4; Harris et al., 1994). Mice, which have been used for most of the ZP studies in mammals, express only three ZP glycoproteins (mZP1–mZP3; Litscher and Wassarman, 2007). Analysis of mouse lines expressing human ZP proteins demonstrated that only hZP2 is important in human sperm–egg binding (Gupta, 2021). Experiments using purified native or recombinant human ZP proteins have shown that hZP1, hZP3, and hZP4 bind to the capacitated human spermatozoa and induce the acrosome reaction (Gupta, 2021). ZP1 is required for the structural integrity of the ZP (Chakravarty et al., 2008). To better understand the roles of ZP glycoproteins, further studies, particularly on ZP protein glycosylation, are needed. The species-specific binding of the ZP to sperm is presumably related to these carbohydrate moieties (Clark, 2014). The sialyl-Lewis(x) sequence is the major carbohydrate ligand for human sperm–egg binding (Pang et al., 2011). The current hypothesis that hZP1, hZP3, and hZP4 bind to capacitated sperm and hZP2 binds to sperm with intact acrosomes will need to be revisited due to the recent demonstration that the acrosome reaction takes place before ZP contact. Regardless, the role of the ZP in preventing polyspermy is clear. Indeed, ZP hardening is due to ZP2 cleavage by ovastacin, a protease released into the PVS by cortical granules after the first sperm–egg fusion (Burkart et al., 2012).
Sperm–egg attachment and membrane fusion
After penetration of the ZP, the sperm enters the PVS and can attach and fuse with the egg plasma membrane. The development of genetic knockout animal models has proven critical in determining the importance of various sperm and egg proteins in sperm–egg attachment and fusion. Surprisingly, genetic knockout studies revealed that many factors originally thought to be important for fertilization were in fact not necessary (reviewed in Okabe, 2018, 2015). The proteins from sperm and egg that are essential for sperm–egg membrane interaction and fusion are listed in Table 1 and are discussed individually from a structural and functional perspective in the sections below.
Table 1. Cellular protein factors involved in sperm–egg attachment or fusion.
| Protein | Year identified | Role in fertilization | Structural features | References |
|---|---|---|---|---|
| CD9 | 1999 | CD9 is expressed on the surface of the oocyte and accumulates during the attachment event; it may modulate the integrity of the oocyte membrane; its precise role in sperm–egg fusion remains unclear | CD9 is a tetraspanin with four transmembrane domains and two extracellular loops (short and long) | Miyado et al., 2000; Le Naour et al., 2000; Kaji et al., 2000; Chen et al., 1999; Umeda et al., 2020; Zimmerman et al., 2016; Zhang and Huang, 2012; Dahmane et al., 2019; Runge et al., 2007; Zhu et al., 2002; Chalbi et al., 2014; Rubinstein et al., 2006; Ziyyat et al., 2006 |
| IZUMO1 | 2005 | IZUMO1 relocates to the equatorial region of the sperm head after the acrosome reaction; high-affinity binding of IZUMO1 to JUNO results in initial attachment of sperm and egg in the PVS | The protein has an N-terminal 4HB, followed by a β-hinge and an IgSF domain; the structure is stabilized by five disulfide bonds | Inoue et al., 2005; Ellerman et al., 2009; Young et al., 2015; Satouh et al., 2012; Aydin et al., 2016; Ohto et al., 2016; Nishimura et al., 2016; Kato et al., 2016 |
| JUNO | 2014 | JUNO is expressed on the surface of the oocyte membrane and serves as the receptor of IZUMO1 | JUNO has structural similarity to folate receptors; it is a globular α/β protein composed of five α helices, three 310 helices, and four short β strands stabilized by eight disulfide bonds | Bianchi et al., 2014; Kato et al., 2016; Han et al., 2016; Jean et al., 2019; Yamaguchi et al., 2007; Aydin et al., 2016; Ohto et al., 2016 |
| SPACA6 | 2014 | SPACA6 is expressed in sperm and localized to the equatorial segment after the acrosome reaction, but its specific role in sperm–egg fusion remains unknown | The three-dimensional structure of SPACA6 is currently unknown; SPACA6 is similar in organization to IZUMO1 with a signal peptide, followed by an α-helical domain, an IgSF domain, a transmembrane helix, and a cytoplasmic tail | Lorenzetti et al., 2014; Noda et al., 2020; Barbaux et al., 2020 |
| TMEM95 | 2014 | TMEM95 is localized to the equatorial segment of sperm and is essential for sperm–egg fusion and male fertility in mice, but its specific role in sperm–egg fusion is currently unknown | The structure of TMEM95 is currently unknown; TMEM95 consists of a signal peptide, an N-terminal helix-rich region, a transmembrane helix, and a leucine-rich cytoplasmic domain | Pausch et al., 2014; Zhang et al., 2016; Noda et al., 2020; Fernandez-Fuertes et al., 2017; Lamas-Toranzo et al., 2020 |
| SOF1 | 2020 | SOF1 is predicted to be a secreted factor essential for fusion; its role is still not fully understood | No structural information to date; primary sequence analysis revealed the presence of conserved LLLL and CFNLAS motifs | Noda et al., 2020 |
| FIMP | 2020 | FIMP is involved in sperm–egg fusion; only the transmembrane form is important in fertilization, but its role is still not fully determined | No structural information to date | Fujihara et al., 2020 |
| DCST1/DCST2 | 2021 | DCST1 and DCST2 are involved in sperm–egg fusion; stability of SPACA6 is regulated by DCST1/2; DCST1/DCST2 are evolutionary conserved in vertebrates and invertebrates | No structural information to date; contains six putative transmembrane helices | Inoue et al., 2021 |
Sperm IZUMO1
In 2005, Inoue et al. discovered that homozygous Izumo1−/− mice are healthy and show normal mating behavior, but males are infertile. IZUMO1 is named after a shrine in Japan that honors the deity for marriage (Inoue et al., 2005). The spermatozoa of Izumo1−/− mice can undergo acrosomal reaction and penetrate the ZP but fail to fuse with oocytes. When the fusion step is bypassed using intracytoplasmic sperm injection, Izumo1−/− spermatozoa can fertilize oocytes, resulting in offspring; thus, IZUMO1 is only necessary at the adhesion/fusion stage of fertilization. An anti-IZUMO1 antibody, OBF13, completely abolishes gamete fusion by blocking IZUMO1 from binding to its receptor. There are four IZUMO family members (Ellerman et al., 2009), but in mice, IZUMO1 is the only paralog that is essential to fertilization (Inoue et al., 2005). IZUMO1 is a type I transmembrane protein consisting of 350 residues that is expressed exclusively in sperm (Inoue et al., 2005; Ellerman et al., 2009; Young et al., 2015). As sperm transit through the epididymis, IZUMO1 undergoes posttranslational modifications. In immature spermatozoa isolated from the proximal caput region of the epididymis, IZUMO1 is localized to both the acrosome and flagella of spermatozoa and is phosphorylated at two sites (S339 and S346; Young et al., 2015). In the cauda epididymis, IZUMO1 is found predominantly in the acrosome of spermatozoa and is phosphorylated at seven residues (S346, S352, S356, S366, T372, S374, and S375; Young et al., 2015). Cell-based fluorescence studies show that after the acrosomal reaction, IZUMO1 is relocated to the membrane surface in the equatorial segment of the acrosome (Satouh et al., 2012).
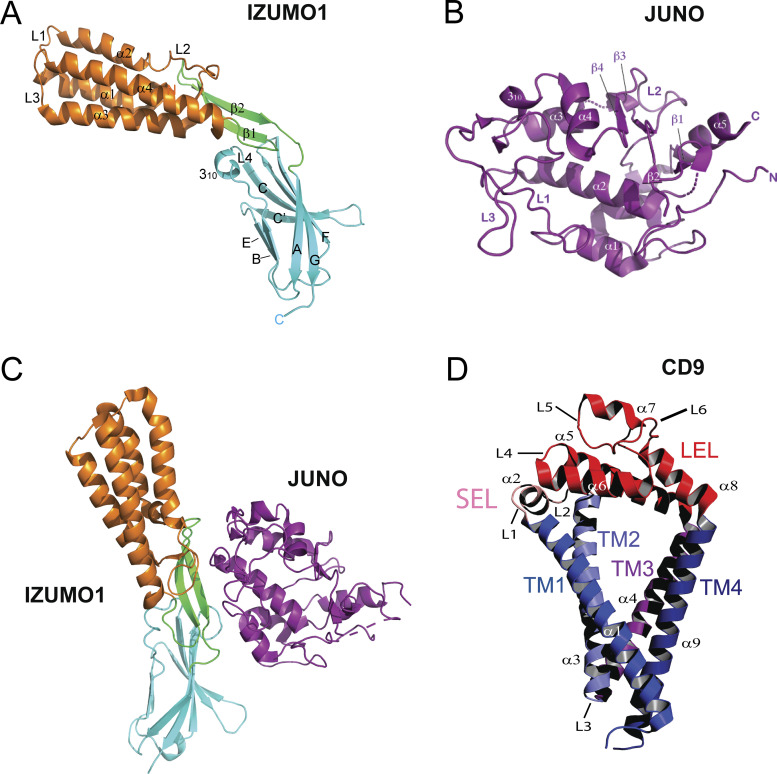
Three crystal structures of the human and mouse IZUMO1 ectodomain were recently published (Aydin et al., 2016; Ohto et al., 2016; Nishimura et al., 2016). In one structure, IZUMO1 is in an upright conformation; however, other crystallographic structures are angled at the hinge region in a “boomerang” shape, which is also observed in solution small-angle x-ray scattering studies (Aydin et al., 2016). The structural discrepancy is not unusual, because the crystal lattice can induce distortions. The crystal structures of the human and mouse IZUMO1 ectodomain show that the extracellular region is organized into two domains, an N-terminal four-helix bundle (4HB) and an Ig-superfamily (IgSF) domain (Fig. 2 A; Aydin et al., 2016; Ohto et al., 2016; Kato et al., 2016). The two domains are connected by a β-hairpin that serves as a flexible hinge. There are five disulfide bonds, one buried at the protein core and four others that are solvent exposed on the surface. Three disulfide bonds connect the N-terminal 4HB domain to the hinge region, and the fourth links the hinge region to the IgSF domain. Interestingly, IZUMO1 shows marked similarities to two protozoan Plasmodium sp. parasite proteins: TRAP, which plays a critical role in gliding motility and host invasion (Song et al., 2012), and SPECT1, which plays a role in host cell fusion and hepatocyte invasion (Ishino et al., 2004; see text box).
Figure 2.
Structures of cellular factors involved in sperm–egg attachment and fusion. (A) Human IZUMO1 is shaped like a boomerang in an unbound state (Protein Data Bank [PDB] accession no. 5F4T). The 4HB, hinge, and IgSF domains are shown in orange, green, and cyan, respectively. (B) Human JUNO (PDB accession no. 5F4Q) belongs to the folate receptor family. (C) The structure of human IZUMO1–JUNO complex (PDB accession no. 5F4E) reveals that JUNO binds to IZUMO1 via the β-hairpin hinge, four residues from the 4HB domain and two from the IgSF domain. (D) Human CD9 (PDB accession no. 6K4J) adopts a conical shape formed by four transmembrane helices (TM1–TM4; blue) and two extracellular loops (SEL, pink; and LEL, red).
Perspectives: Similarity to Plasmodium host invasion proteins
The β-hinge region of IZUMO1 is highly similar to an extensible β-ribbon region in TRAP (root-mean-square deviation [RMSD] 1.4 Å; Nishimura et al., 2016). The TRAP β-ribbon has been proposed to undergo conformational changes upon binding to a host cell to mediate sporozoite gliding and host cell invasion. The IZUMO1 4HB domain shows structural similarities to Plasmodium berghei SPECT1 (RMSD 3.3 Å; Nishimura et al., 2016; Aydin et al., 2016). SPECT1 is required for cell traversal of sporozoites. Both SPECT1 and IZUMO1 adopt 4HBs with the same connectivity. In SPECT1, the 4HB is proposed to be a metastable structure that transitions from a solvent-accessible to a membrane-associated state. It has also been proposed that SPECT1 interacts with SPECT2, which has a membrane-attack complex/perforin domain, to form a pore. How the two proteins cooperatively mediate pore formation remains to be determined, but the similarity of IZUMO1 to proteins involved in parasite entry is intriguing.
Oocyte JUNO
In 2014, Bianchi et al. made the groundbreaking discovery of the oocyte receptor for IZUMO1 (Bianchi et al., 2014). The group iteratively cloned and expressed the entire mouse oocyte cDNA library in mammalian cells and tested each clone for IZUMO1 binding using avidity-based extracellular interaction screening (AVEXIS; Kerr and Wright, 2012). Folate receptor δ (or folate receptor 4), which was aptly renamed JUNO, after the Roman goddess of marriage and fertility, was the only protein that bound to IZUMO1. Mouse JUNO shares 58% sequence identity with human folate receptors FOLR-α and FOLR-β but does not bind to folate (Kato et al., 2016; Han et al., 2016). Juno−/− mice show normal development and mating behaviors, but females are infertile, and eggs from Juno−/− mice are unable to fuse with wild-type sperm (Bianchi et al., 2014). Moreover, an anti-JUNO antibody incubated with human zona-free oocytes effectively blocks fertilization (Jean et al., 2019).
While JUNO is primarily expressed on the surface of oocytes, it is also expressed on CD4+ CD25+ regulatory T cells, albeit at a much lower level (Yamaguchi et al., 2007). JUNO is highly expressed in unfertilized eggs but upon fusion with sperm is rapidly shed from the cell surface into extracellular vesicles (Bianchi et al., 2014). By the anaphase II stage, which takes place 30–40 min after fertilization, JUNO is barely detectable at the cell surface (Bianchi et al., 2014). The rapid removal of JUNO from the egg surface may help prevent the entry of more than one sperm into an oocyte.
JUNO is a glycoprotein of 250 residues with a C-terminal glycosylphosphatidylinositol anchor. The crystal structures of human and mouse JUNO, both alone and in complex with human IZUMO1, were determined in 2016 (Aydin et al., 2016; Kato et al., 2016; Ohto et al., 2016; Han et al., 2016). The overall structure of human JUNO resembles structures of FOLR-α and FOLR-β with RMSDs of 1.1 Å and 1.0 Å, respectively. Like the folate receptors, JUNO has a compact, globular shape with five α helices, three 310 helices, and four short β strands stabilized by eight conserved disulfide bonds (Fig. 2 B). Despite its structural homology to folate receptors, five key residues in JUNO (A93, G121, Q122, R154, and G155) are not conserved compared with the folate-binding sites of FOLR-α and FOLR-β (Aydin et al., 2016). The aromatic and charged residues that in FOLR-α and FOLR-β anchor folate in the binding site through hydrogen bonds are replaced by alanine or glycine in JUNO, resulting in a larger cavity that cannot bind folate. Recombinant IZUMO1 binds to oocytes (and to nongamete human cells transfected with JUNO) but does not bind to oocytes that have been preincubated with an anti-JUNO antibody (Bianchi et al., 2014). The cocrystal structure of IZUMO1 in complex with JUNO reveals a 1:1 stoichiometry with a binding interface of ∼910 Å2 (Aydin et al., 2016). Biolayer interferometry, surface plasmon resonance, and isothermal titration calorimetry revealed that the complex of JUNO and IZUMO1 has a dissociation constant between 48 and 91 nM (Aydin et al., 2016; Ohto et al., 2016). The tight binding affinity results from an additive effect of extensive van der Waals, hydrophobic, and aromatic interactions, as well as two salt bridges. IZUMO1 binds to JUNO primarily via the β-hairpin hinge, with four residues from the 4HB domain and two from the IgSF domain also contributing to the binding (Fig. 2 C). In JUNO, the binding site is an elongated surface formed by the flanking regions of helices α1–α3 and loops L1–L3. The IZUMO1–JUNO interaction is not strictly species specific, as there is cross-species interaction between human IZUMO1 and hamster JUNO (see text box).
Perspectives: Cross-species interactions
Fertilization is a species-specific event, as sperm typically cannot fertilize eggs from a different species. The ZP provides an effective barrier against cross-species fertilization, but beyond this glycoprotein layer, IZUMO1-JUNO recognition is promiscuous. Human sperm cannot penetrate the hamster ZP, but they can fuse with zona-free hamster eggs (Inoue et al., 2005). Indeed, zona-free hamster eggs have been used to assess human sperm quality in fertility treatments. Using the ELISA-based AVEXIS platform, human IZUMO1 was confirmed to bind to hamster JUNO in solution (Bianchi and Wright, 2015). Like human IZUMO1, mouse and pig IZUMO1 also bind to hamster JUNO in solution (Bianchi and Wright, 2015). The results are consistent with the ability of human, mouse, and pig sperm to fuse with zona-free hamster eggs (Creighton and Houghton, 1987; Hanada and Chang, 1972).
Hamster and human JUNO are highly similar, with a sequence identity of 73%; however, eight residues at the IZUMO1–JUNO interface are not conserved. To understand the cross-species specificity, a homology model of hamster JUNO was generated based on the crystal structure of human JUNO. Despite key substitutions in hamster JUNO, the IZUMO1-binding site preserves the same structural architecture and physiochemical characteristics as human JUNO, with the exception of E45. In human JUNO, E45 forms a key salt bridge at the IZUMO1–JUNO interface. How an E45L substitution in hamster JUNO is able to maintain binding remains unclear. It was previously shown that E45 is critical for human JUNO recognition to IZUMO1, as E45A or E45K mutations severely reduced the interaction (Aydin et al., 2016). It may be possible that other interactions between hamster JUNO and human IZUMO1 compensate for the loss of the critical salt bridge. A crystal structure of hamster JUNO in complex with human IZUMO1 would provide important insight into the molecular basis of cross-species specificity in IZUMO1–JUNO recognition.
The binding sites on both IZUMO1 and JUNO have been verified by alanine-substitution experiments (Ohto et al., 2016). W62 and L81 in JUNO and W148 in IZUMO1 play critical roles at the interface, as substitution of these residues by alanine dramatically reduces binding affinity (Ohto et al., 2016). These residues are strictly conserved across mammalian species. To verify the biological relevance of the IZUMO1–JUNO interface, the binding of oocytes to COS-7 cells expressing wild-type IZUMO1 or to mutants with one or more mutations to residues proposed to be important in JUNO binding was tested. Mutating W148, K154, H157, I158, R160, or L163 in IZUMO1 significantly reduced oocyte binding. COS-7 cells that expressed IZUMO1 with multiple mutations at the JUNO-binding interface showed a complete lack of binding to oocytes (Ohto et al., 2016). These results confirmed the JUNO-binding residues identified in the crystal structures and biophysical studies (Ohto et al., 2016).
Oocyte CD9
The importance of CD9 in sperm–egg fusion was first described in 1999 (Chen et al., 1999) and confirmed in 2000 (Miyado et al., 2000; Le Naour et al., 2000; Kaji et al., 2000; Chen et al., 1999). CD9 is expressed on the plasma membrane of oocytes, and an anti-CD9 antibody inhibits sperm–egg fusion in a dose-dependent manner (Chen et al., 1999). Interestingly, anti-CD9 antibodies do not block sperm from binding to oocytes but instead prevent the fusion of sperm and egg membranes (Miyado et al., 2000; Le Naour et al., 2000). These findings are consistent with mouse studies, which showed that CD9−/− mice develop normally and that male mice are fertile but female mice have dramatically reduced fertility (Miyado et al., 2000; Le Naour et al., 2000; Kaji et al., 2000). When the sperm–egg fusion step is bypassed by injecting capacitated sperm into the cytoplasm of CD9−/− oocytes, the fertilized eggs show normal implantation efficiencies, and embryos develop normally.
CD9 belongs to the tetraspanin superfamily and is 228 amino acids long. It has four membrane-spanning domains (TM1–TM4) linked by a short extracellular loop (SEL) between TM1 and TM2 and a large extracellular loop (LEL) between TM3 and TM4 (Fig. 2 D). The transmembrane regions are highly conserved among tetraspanins, with sequence divergence only in the extracellular loops. The first structure of CD9 was recently determined to 2.7 Å resolution (Umeda et al., 2020). The four transmembrane helices tilt toward the cytoplasmic membrane interface to form a cone-shaped structure that creates a spacious cavity in the intramembranous region (Fig. 2 D). This is reminiscent of the CD81 structure; CD9 and CD81 have ∼60% sequence similarity and the same overall fold (RMSD of 1.9 Å; Umeda et al., 2020; Zimmerman et al., 2016). Previous studies have demonstrated the localization of tetraspanins in curved regions of cell membranes (Zhang and Huang, 2012; Dahmane et al., 2019). As CD9 clusters at the contact region of the egg and sperm membranes, the tight array of cone-shaped CD9 may increase the curvature of the oocyte membrane, effectively causing it to protrude. CD9-knockout oocytes produce short and sparse microvillus structures with a large radius of curvature of microvillar tips, which results in impaired fusion ability with spermatozoa (Runge et al., 2007).
The relative lengths of the SEL and LEL of tetraspanins control access to the intramembranous cavity. In silico analysis revealed that the LEL undergoes a conformational change between the open and closed states during binding partner recognition (Umeda et al., 2020). In the closed-state CD9 structure, the LEL is weakly associated with the SEL. In the open state, the LEL moves away from the SEL, thereby allowing access to the intramembranous cavity. The importance of the LEL in fertilization was demonstrated in a domain-swapping experiment, in which the LEL of a fertilization-incompetent tetraspanin CD53 was swapped with its equivalent section from CD9. The CD9–CD53LEL chimera had dramatically reduced fertilization competency, whereas the chimera with the LEL from CD9, CD53–CD9LEL, was ∼50% competent (Umeda et al., 2020). This suggests that additional regions such as the SEL and transmembrane domains are also important in fertilization. Alanine-substitution experiments on residues within the LEL have produced conflicting results. Mutation of the 173SFQ175 motif in the murine CD9 LEL suggests that these residues are essential for fertilization (Zhu et al., 2002). However, the murine 173SFQ175 LEL region is not conserved in human CD9 (175TFT177). A triple-alanine mutations of this region in both murine and human CD9 revealed that contrary to previous findings, both mutants rescued fertilization in CD9−/− oocytes (Umeda et al., 2020). Further studies are required to probe the roles of specific CD9 LEL, SEL, and transmembrane residues in fertilization.
Like other tetraspanins, CD9 can act as a scaffolding protein to bring together multiple protein partners to execute a biological function. For instance, CD9 associates with Igs, integrins, and other adhesion receptors and proteins (reviewed in Charrin et al., 2014). Recently, interaction studies using human sperm and mouse oocytes revealed that IZUMO1 and JUNO colocalize with CD9 on the surface of the egg during sperm–egg attachment. Along with sperm IZUMO1, egg CD9 accumulates at adhesion area surroundings, suggestive of a cis interaction with egg JUNO (Chalbi et al., 2014; Ravaux et al., 2018). In the same context, by measuring the force necessary to break contact between one sperm and an egg, it was suggested that CD9 induces the clustering of sperm receptors on the oocyte membrane, generating fusion-competent sites (Jégou et al., 2011).
Single-particle cryoelectron microscopy studies of CD9 in complex with EWI-2 provide insights into how CD9 engages with its targets. EWI-2 belongs to the IgSF, with four to eight predicted IgSF domains and a single-pass transmembrane anchor. EWI-2 is a major binding partner to both CD9 and CD81 (Runge et al., 2007; Umeda et al., 2020; Rubinstein et al., 2006). An anti-IgSF8 antibody had moderate inhibitory effects on sperm–egg binding, suggesting that mouse EWI-2 participates in gamete interactions (Glazar and Evans, 2009). Cryoelectron microscopy revealed that a 2:2 heterotetrameric arrangement of the extracellular domains of two EWI-2 molecules forms a tight dimer and that the EWI-2 transmembrane helix is sandwiched by two CD9 molecules. The transmembrane helix of EWI-2 interacts with TM3 and TM4 of CD9 via hydrophobic residues (Umeda et al., 2020). The nonspecific nature of the transmembrane hydrophobic interactions in the CD9–EWI-2 complex may explain the promiscuous nature of tetraspanins.
Newly identified players in mammalian fertilization
The use of CRISPR-Cas9 technology has led to the recent identification of six new factors essential for mammalian fertilization: SPACA6, TMEM95, SOF1, FIMP, and DCST1/DCST2.
Sperm SPACA6
In 2014, Lorenzetti et al. characterized a mutant mouse line that had a deletion removing Spaca6 (Lorenzetti et al., 2014). Male homozygous knockout mice were infertile with a phenotype that closely resembles that of Izumo1-deficient mice. Subsequent studies by two other groups confirmed that Spaca6 deletion in male mice results in infertility, although mating behavior is normal and sperm are motile and morphologically normal (Noda et al., 2020; Barbaux et al., 2020). Fertility could be restored by a transgene (Noda et al., 2020). In a human zona-free in vitro fertilization assay, an anti-SPACA6 antibody reduced fertilization rates by threefold (Barbaux et al., 2020).
Recovery of oocytes from female mice that were mated with Spaca6−/− male mice revealed that the spermatozoa were trapped in the PVS. This indicates that knockout spermatozoa migrate through the female genital tract to the oocyte and penetrate the ZP but fail to fuse with the oocyte membrane. When Spaca6−/− sperm was injected into the cytoplasm of oocytes to bypass the membrane fusion step, fertilization was successful, and the fertilized eggs showed normal embryonic development, suggesting that SPACA6 does not play a critical role downstream of sperm–egg fusion.
SPACA6 is primarily expressed in testis, with low levels of expression in the epididymis, seminal vesicle, and ovary (Noda et al., 2020; Lorenzetti et al., 2014). Orthologues of SPACA6 have been annotated in bull, hamster, human, mouse, rat, and zebrafish (Noda et al., 2020). In fresh spermatozoa, SPACA6 is not detected on the plasma membrane; rather, it is localized underneath the membrane of the sperm head. After the acrosomal reaction, SPACA6 relocates to the equatorial segment of the sperm head, with reduced levels detected in the midpiece, and completely diminishes from the neck region (Barbaux et al., 2020). Immunofluorescence staining revealed that the localization of IZUMO1 is unaffected in Spaca6−/− sperm before and after the acrosomal reaction (Barbaux et al., 2020). To verify this result, Spaca6−/− male mice were mated with female mice and oocytes were extracted and immunostained with an anti-IZUMO1 antibody revealing that IZUMO1 distribution in Spaca6−/− spermatozoa was identical to that in wild-type spermatozoa (Barbaux et al., 2020). This confirmed that SPACA6 does not affect IZUMO1 localization.
Both IZUMO1 and SPACA6 belong to the IgSF and are expressed in sperm and localized to the equatorial segment upon the acrosomal reaction (Noda et al., 2020). The phenotypes of knockout mutants are highly similar as well. The similarities also extend into their domain organization. Both proteins have an N-terminal signal peptide, followed by a helical domain, a single IgSF domain, a single N-linked glycosylation site, a monotopic transmembrane helix, and a short cytoplasmic tail (Noda et al., 2020). Despite these similarities, the proteins are not redundant, as both cell-based and mouse studies show that one cannot compensate for the lack of the other (Barbaux et al., 2020). Moreover, SPACA6 does not accumulate at the interface with IZUMO1, and SPACA6-expressing COS-7 or HEK293T cells do not bind to the surface of the oocyte (Inoue et al., 2015; Noda et al., 2020). No interaction was detected between SPACA6 and IZUMO1 by coimmunoprecipitation from testis extracts (Noda et al., 2020). How SPACA6 interacts with other sperm and oocyte proteins to mediate sperm–egg adhesion and fusion remains to be determined.
Sperm TMEM95
A genome-wide analysis designed to reveal genetic associations with infertility in bulls revealed the essential role of TMEM95 in fertility (Pausch et al., 2014). A nonsense mutation that introduces a premature stop codon in Tmem95 diminishes male fertility, although it does not significantly affect sperm morphology or motility (Pausch et al., 2014). TMEM95 is conserved in primary sequence among bull, hamster, mouse, rat, and humans (Zhang et al., 2016; Noda et al., 2020). In bulls, TMEM95 is expressed in spermatozoa and is localized on the acrosome, on the equatorial segment and on the connecting piece (Pausch et al., 2014). Bull spermatozoa with a knockout mutation in Tmem95 are unable to fuse with oocytes, suggesting that TMEM95 is required for sperm–oocyte fusion (Fernandez-Fuertes et al., 2017).
RT-PCR analysis revealed that in mice Tmem95 is expressed exclusively in testis. Expression begins on day 21 postpartum when spermiogenesis begins. Tmem95−/− mice that carry a 1,919-bp deletion in the Tmem95 locus have normal mating behavior but males are infertile (Noda et al., 2020). The Tmem95−/− spermatozoa have normal morphology and motility and bind to oocytes; however, the mutant sperm have impaired ability to fuse with oocytes and accumulate in the PVS. Expression of a Tmem95 transgene in Tmem95−/− male mice restored fertility (Noda et al., 2020). The fertility of Tmem95−/− female mice is unaffected (Noda et al., 2020). Consistent with the study by Noda et al., Lamas-Toranzo et al. also found that Tmem95−/− male mice were infertile (Lamas-Toranzo et al., 2020).
In silico analysis suggested that TMEM95 shares organizational similarities with IZUMO1 (Zhang et al., 2016). Like IZUMO1, it is a type I single-pass transmembrane protein with a signal peptide at the N terminus, a helix-rich N-terminal region, and a transmembrane helix. TMEM95 has an additional leucine-rich cytoplasmic domain compared with IZUMO1. Examination of the localization of IZUMO1 in acrosome-reacted wild-type and Tmem95−/− sperm revealed no difference in IZUMO1 translocation (Lamas-Toranzo et al., 2020). Moreover, TMEM95 disappears after the acrosomal reaction (Fernandez-Fuertes et al., 2017). Since IZUMO1 relocates to the equatorial segment only after the acrosomal reaction, this suggests that TMEM95 and IZUMO1 function independently. Experiments using the AVEXIS platform showed that TMEM95 does not bind to JUNO or IZUMO1 (Lamas-Toranzo et al., 2020). In contrast, coimmunoprecipitation studies using HEK293T cells coexpressing IZUMO1 and TMEM95 suggested that IZUMO1 does bind to TMEM95 (Noda et al., 2020). Further studies are required to verify whether or not IZUMO1 and TMEM95 interact.
Sperm SOF1
Another new molecular player important for male fertility identified by Noda and collaborators using CRISPR-Cas9–mediated gene knockout was a gene called 1700034O15Rik (also known as Llcfc1; Noda et al., 2020). The gene was aptly renamed SOF1 (sperm–oocyte fusion required 1). SOF1 is widely conserved in mammals and is highly expressed in the testis. SOF1 is predicted to be a 147-residue secreted protein with conserved LLLL and CFN(L/S)AS motifs. These motifs are observed in the DUF4717 family of proteins that have an unknown function but are exclusively found in eukaryotes. SOF1 reportedly undergoes posttranslational modifications during sperm maturation, and it was detected as a protein singlet in testicular germ cells but a doublet in acrosome-intact spermatozoa (Noda et al., 2020).
The sterility of Sof1−/− male mice is likely due to defective membrane fusion (Noda et al., 2020). The morphology and motility of Sof1−/− spermatozoa are similar to wild type; however, when used for in vitro fertilization with cumulus-intact oocytes, Sof1−/− spermatozoa do not fuse or fertilize oocytes and accumulate in the PVS. This inability to fuse was also observed using zona-free oocytes, but sperm continued to bind to the oocyte membrane, suggesting a role in either sperm–egg fusion or in the control of sperm–egg adhesion properties. Expression levels and localization of IZUMO1 are not affected in Sof1−/− spermatozoa before or after the acrosome reaction. Thus, sterility in Sof1−/− male mice is not due to a disruption of IZUMO1.
Sperm FIMP
CRISPR-Cas9–mediated deletion of Fimp (also known in mice as 4930451I11Rik) results in failure in sperm–egg fusion in mice (Fujihara et al., 2020). Similar to SOF1, FIMP is also a small protein of 132 amino acids that is highly expressed in the testis. The expression of this testis-specific gene is first observed 20 d after birth. The protein is detected in two distinct isoforms: membrane anchored and secreted. Only the transmembrane form appears to be critical for sperm-oocyte fusion in mice. Fimp−/− mice have normal testicular and sperm morphologies, and Fimp−/− spermatozoa penetrate the ZP but fail to fuse with oocytes (Fujihara et al., 2020). In an in vitro fertilization assay using zona-free oocytes, Fimp−/− sperm ability to fuse are severely reduced. IZUMO1 localization and expression levels in Fimp−/− mice are similar to the wild type. FIMP localizes to the equatorial segment membrane, but the FIMP-mCherry signal disappeared in 40% of the acrosome-reacted sperm (Fujihara et al., 2020). In contrast to IZUMO1, it does not appear FIMP is involved in the initial attachment step, as FIMP-expressing cells do not bind oocytes. The precise role of FIMP in sperm–egg fusion remains unclear.
Sperm DCST1/DCST2
As identified by gene disruption and complementation experiments, the evolutionarily conserved factors dendrocyte expressed seven transmembrane protein (DC-STAMP) domain-containing 1 and 2 (DCST1/DCST2) are required for gamete fusion (Inoue et al., 2021). Individual or double gene deletion results in male sterility with the same phenotype as that of Izumo1−/− or Spaca6−/− knockouts. Although their molecular mechanism of action is still unknown, DCST1 and DCST2 function might be intrinsically related to SPACA6. Surprisingly, while the rescued double transgenic males had normal fertility, SPACA6 was not detected. The protein stability of SPACA6 may be differently regulated by DCST1/DCST2 and IZUMO1 (Inoue et al., 2021).
Molecular mechanism of sperm–egg fusion
When the acrosome-reacted spermatozoon reaches the PVS, it is primed to interact and fuse with the egg. The molecular mechanism of mammalian sperm–egg attachment and fusion requires a complex sequence of events that for the most part remain a mystery (Fig. 3). However, new insights into some of the steps have now been obtained through structural, biophysical, and genetic analyses of the proteins essential to the adhesion fusion process.
Figure 3.
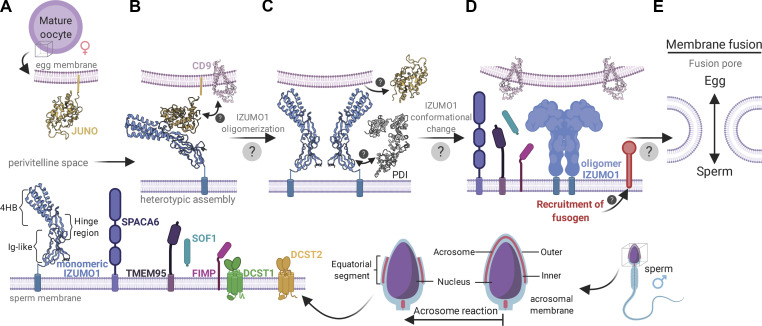
Current model of sperm–egg attachment and fusion. (A) Acrosome reaction. After the acrosome reaction, IZUMO1 (blue), SPACA6 (purple), and TMEM95 (violet) colocalize to the equatorial regions of sperm. FIMP (pink) appears to function before the acrosome reaction. There are conflicting data on whether or not TMEM95 interacts with IZUMO1. SOF1 (turquoise) is a secreted sperm protein. DCST1 (green) and DCST2 (orange) are transmembrane proteins implicated in regulating the protein stability of SPACA6. (B) Initial attachment. After the sperm reaches the PVS, it attaches to the egg. IZUMO1 is localized on the equatorial segment of acrosome-reacted sperm and its counterpart receptor, JUNO (yellow), on the oocyte membrane. JUNO specifically recognizes and binds to IZUMO1 in a monomeric conformation. IZUMO1 binding to JUNO drives the accumulation of CD9 (pink) at the sperm–egg interface to form a physical anchor that holds the sperm and oocyte membranes in proximity. (C) IZUMO1 multimerization. After the initial IZUMO1–JUNO attachment, the complex undergoes a dimerization event. The trigger for IZUMO1 oligomerization is not fully understood; however, colocalization analysis revealed the presence of PDI (gray) on the sperm surface. JUNO is thought to be shed from the oolemma and into the PVS after fertilization. (D) Fusogen recruitment. The bona fide sperm–egg fusogen remains a mystery. However, data suggest that IZUMO1 forms a scaffold to recruit the gamete fusion complex. The roles of SPACA6, TMEM95, and SOF1 remain unclear, but these proteins likely play roles in fusion. (E) Fusion pore formation. The merger of the egg and sperm membranes requires modulation of the membrane architecture. The fusogen is thought to catalyze the formation of a hemifusion intermediate, which is a stalk-like structure where the outer leaflets of the sperm and egg membrane bilayers mix. Subsequently, the inner bilayer leaflets mix to form the fusion pore. The precise mechanism of this step will require the identification of the sperm–egg fusogen. Created with BioRender.
Initial attachment
At the molecular level, the first step in the attachment of the sperm to the egg involves binding of IZUMO1, which is localized on the equatorial segment of acrosome-reacted sperm (Satouh et al., 2012), to its counterpart receptor JUNO on the oocyte membrane (Fig. 3 A). JUNO is monomeric when it binds to IZUMO1 (Fig. 3 B; Inoue et al., 2015). Comparison of the crystal structures of IZUMO1 with and without JUNO suggests that there is a binding-induced conformational change in IZUMO1 whereby the 4HB domain moves ∼20° to adopt an upright conformation (Aydin et al., 2016). IZUMO1 binding to JUNO drives the accumulation and local membrane organization of CD9. The accumulation of CD9 at the sperm–egg interface causes the egg membrane to protrude toward the sperm membrane (Chalbi et al., 2014).
IZUMO1 multimerization
After the initial IZUMO1–JUNO attachment, Inoue et al. suggested that the IZUMO1–JUNO complex undergoes a multimerization event that is critical for sperm–egg fusion (Fig. 3 C; Inoue et al., 2015). Bimolecular fluorescence complementation and photon-counting histogram analyses were used on a cultured cell–oocyte system to reveal that IZUMO1 forms a multimer at the cell–oocyte interface, but not on the rest of the cell surface. After IZUMO1 oligomerization, JUNO is not detected at the cell surface and presumably is shed (Inoue et al., 2015).
Inoue et al. suggested that the trigger for IZUMO1 multimerization involves a protein disulfide isomerase (PDI). Localization studies revealed the presence of PDIs on the sperm surface (Fig. 3 C; Ellerman et al., 2006). PDIs are responsible for proper folding of extracellular or membrane proteins during the maturation process in the endoplasmic reticulum. Western blot and proteomic analysis detected at least four PDI members on the sperm surface, PDI, ERp57, ERp72, and P5. Interestingly, PDI inhibitors reduce sperm–egg fusion in vitro in a dose-dependent manner (Ellerman et al., 2006), and a membrane-impermeable thiol-reactive reagent significantly reduces cell–oocyte binding (Inoue et al., 2015). To identify those PDIs that function in gamete fusion, sperm were preincubated with antibodies that specifically blocked each PDI member, and the ability of the spermatozoon to fuse with the oocyte was assessed (Ellerman et al., 2006). This revealed ERp57 is critical for gamete fusion. On the IZUMO1 ectodomain, 10 cysteines form five disulfide bonds. Four of the five disulfide bonds are located on the surface and are solvent accessible. The N-terminal helical domain of IZUMO1 was proposed to undergo a collapse or becomes buried at the oligomeric interface (Inoue et al., 2015). ERp57 and/or other PDIs may catalyze a thiol-disulfide exchange during this conformational rearrangement.
Fusogen recruitment
After IZUMO1 multimerization, the next step is thought to involve the recruitment of the bona fide human sperm–egg fusogen. Dimerized IZUMO1 was suggested to directly recruit a tight binding unidentified oocyte receptor (Fig. 3 D; Inoue et al., 2015). The identity of the gamete fusion complex remains unknown. SPACA6 was proposed to interact with IZUMO1 to mediate the binding of an oocyte receptor (Lorenzetti et al., 2014). However, coimmunoprecipitation studies of testis extracts did not show an interaction between SPACA6 and IZUMO1, whereas coimmunoprecipitation analysis using HEK293T cells showed interactions between IZUMO1 and SPACA6, FIMP, TMEM95, and SOF1 (Noda et al., 2020). Expression of all five proteins on HEK293T cells did not lead to fusion with zona-free oocytes, suggesting the recruitment of a yet-to-be discovered fusogen is required (Noda et al., 2020). Intriguingly, IZUMO1 and SPACA6 both contain an IgSF domain. While IgSF domains are known to facilitate protein–protein interactions, their role and importance in sperm–egg fusion is unknown (see text box). A complete understanding of the interplay of these proteins and the composition of the human gamete fusion machinery will require additional biochemical and functional experimentation.
Perspectives: Role of the IgSF in sperm–egg fusion
Many of the essential protein players involved in sperm–egg attachment and fusion contain IgSF domains (Table 1). The IgSFs belong to a large superfamily of proteins that have diverged in sequence and function; ∼500 nonimmunological proteins (nonantibody and non–T cell receptor) with IgSF domains are encoded in the human genome. The IgSF domain is ∼110 residues in size and is defined by two β sheets packed face to face (Bork et al., 1994; Harpaz and Chothia, 1994). The IgSF fold displays a common core composed of four anti-parallel β strands sandwiched by a second set of three to five β strands. Based on the number of strands and relative locations, several distinct subtypes have been defined. Most common are the variable (V) and constant (C) Ig domains, while a third type (I) is an intermediate structure between the V and C types.
Human IZUMO1, SPACA6, EWI-2, and EWI-F (CD9P-1) all contain at least one IgSF domain. The importance of IgSF domains in sperm–egg fusion is not limited to humans. In lower eukaryotes, the HAP2/GCS1 sperm–egg fusogen contains a C-type IgSF domain. Moreover, Caenorhabditis elegans encodes a transmembrane protein with an IgSF domain, termed SPE45, that is required for gamete fusion (Nishimura et al., 2015; Singaravelu et al., 2015). Interestingly, the IgSF domains of SPE45 and IZUMO1 share a common function, despite only 8.7% sequence identity (Nishimura et al., 2015, 2016). This was demonstrated by the finding that a chimeric SPE45 that contains the murine IZUMO1 IgSF domain had ∼77% of the activity of wild-type SPE45 (Nishimura et al., 2015). The IgSF domain may act as a scaffold to recruit binding partners in cis and/or in trans. Various organisms have IgSF proteins that act during gamete interactions, indicating the widespread utility of IgSF domains in fertilization.
The importance of the IgSF domains in human IZUMO1 and C. elegans SPE45 raises the question of whether there are special features unique to these IgSF domains. The crystal structure of human IZUMO1 revealed a novel Ig domain fold with a 2+5 β-sheet arrangement. The IZUMO1 IgSF domain is the only known member to adopt a 2+5 arrangement, thus suggesting a new IgSF subtype. It is not clear whether the IgSF domain in SPE45 adopts a similar structure to the IZUMO1 IgSF domain. The precise roles played by these IgSF domains in IZUMO1, SPACA6, EWI-2, and SPE45 are currently unknown. However, the IgSF domains in other cell surface proteins often form homo- or heterodimers. Structures of IgSF oligomers reveal that all regions of the domain surface can be used for interaction with other molecules. Noncovalent association of IgSF-type domains usually occurs through the exposed faces of the β sheets. Continued studies will undoubtedly reveal important functions of IgSF proteins in gamete fusion.
Fusion pore formation
The merger of the egg and sperm membranes is an energetically unfavorable process and must require modulation of the membrane architecture in order to form a fusion pore (Fig. 3 E). The formation of a fusion pore typically proceeds through one of two mechanisms, either via a hemifusion intermediate or via direct fusion (Chernomordik and Kozlov, 2005). In the case of hemifusion, the fusion of the two membranes occurs through the sequential mergers of each pair of bilayer leaflets. First, the outer membrane leaflets contact and mix to form the hemifusion stalk intermediate. This is followed by mixing of the inner leaflets to form the fusion pore. Enveloped viral-cell fusion proceeds through a hemifusion intermediate that is catalyzed by a viral fusion glycoprotein as previously discussed (Harrison, 2015; Sapir et al., 2008; White et al., 2008; Podbilewicz, 2014). The viral fusogens all contain distinctive hydrophobic fusion peptides that are inserted into the host target membrane when triggered. In direct fusion, proteins on both membranes arrange into complexes at the site of fusion and bind in trans to bring the two membranes together. This forms a continuous connection between the two protein-lined pores to allow for content mixing. In yeast vacuolar fusion, two proteolipid hexamers formed in trans by the V0 subunit of vacuolar H+-ATPase establishes a bridging channel/pore between the two membranes (Peters et al., 2001; Chernomordik and Kozlov, 2003). The pore is subsequently opened through a Ca2+-triggered conformational change that expands the proteolipid hexameric complex.
It is thought that sperm–egg fusion also proceeds via a membrane hemifusion intermediate, similar to viral-cell fusion. This is at least true in lower eukaryotic cells, in which the sperm–egg fusogen HAP2/GCS1 has a similar overall structure to the class II viral fusogens (Fédry et al., 2017), such as tick-borne encephalitis virus E glycoprotein. No evidence for HAP2/GCS1 orthologues has been found in vertebrates or mammals; thus, in an early vertebrate ancestor, a new fusogen likely replaced HAP2/GCS1 (Vance and Lee, 2020). The structures of CD9, IZUMO1, and JUNO lack characteristics common to viral fusogens such as the prototypical hydrophobic fusion peptide (Aydin et al., 2016). Moreover, no readily identifiable fusion peptides were detected upon sequence analysis of SOF1, DCST1/DCST2, TMEM95, FIMP, or SPACA6. Furthermore, cell fusion assay experiments show that the sperm proteins alone or together are not able to trigger cell–cell fusion (Fujihara et al., 2020; Noda et al., 2020; Barbaux et al., 2020; Lamas-Toranzo et al., 2020). Thus, additional factors remain to be identified that are essential for fusion pore formation.
Concluding remarks
While significant advances have been made to fully understand the molecular mechanism of fertilization, many questions are still outstanding. The identification of proteins involved in the sperm–egg fusion process remains the holy grail in reproductive biology. Understanding the interplay of all the partners involved has the potential to impact multiple areas of biology. Identifying the full complement of proteins involved in sperm–egg attachment and fusion will allow the mapping of genotype–phenotype correlations and improve diagnostic tests for people suffering from infertility. Understanding the mechanisms of sperm–egg fusion will also reveal ways to improve assisted reproductive technologies for humans and animals. High and predictable fertility rates for cows, pigs, chickens, and sheep are essential for efficient food animal production. Finally, although generally safe and effective, current hormone-based contraceptives may lead to adverse side effects that discourage many from long-term use. The safety and acceptability of contraceptives are particularly important for women, since they bear the greatest burden of contraceptive side effects. It is important to innovate and develop new alternative contraceptives that better meet the reproductive needs and desires of women and couples. Molecules that disrupt sperm–egg protein–protein interactions by binding to the sperm or egg protein side of the axis should result in a potent contraceptive. These reasons underscore why understanding the mechanisms of sexual fertilization is one of the most crucial biological questions.
Acknowledgments
The authors thank Pr. J.-P. Wolf (University of Paris-Cochin Hospital, Assistance Publique - Hôpitaux de Paris, Paris, France) for the gift of the human gamete images.
This work was supported by the Canadian Institutes of Health Research (grant PJT-153281 and Canada Research Chair in Structural Virology, J.E. Lee; and Banting Postdoctoral Fellowship, V.H.B. Serrao), the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Université de Paris, and the Agence Nationale pour la Recherche (grant FERTILIVE ANR-17-CE18-0005-02 to A. Ziyyat). In addition, this work was supported by the Bill and Melinda Gates Foundation (grant INV-024197 to J.E. Lee and A. Ziyyat) and the New Frontiers in Research Fund (grant NFRFE-2019-00230 to J.E. Lee and A. Ziyyat). This research was funded in whole or in part by the Bill and Melinda Gates Foundation. For the purpose of Open Access, the author has applied a CC-BY public copyright license to any Author Accepted Manuscript (AAM) version arising from this submission.
The authors declare no competing financial interests.
K.K. Siu, V.H.B. Serrao, A. Ziyyat, and J.E. Lee designed and wrote the manuscript. A. Ziyyat and J.E. Lee conceived and secured funding.
References
- Aitken, R.J., Baker M.A., Doncel G.F., Matzuk M.M., Mauck C.K., and Harper M.J.K.. 2008. As the world grows: contraception in the 21st century. J. Clin. Invest. 118:1330–1343. 10.1172/JCI33873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood, C.S., and Vadakkadath Meethal S.. 2016. The spatiotemporal hormonal orchestration of human folliculogenesis, early embryogenesis and blastocyst implantation. Mol. Cell. Endocrinol. 430:33–48. 10.1016/j.mce.2016.03.039 [DOI] [PubMed] [Google Scholar]
- Austin, C.R. 1952. The capacitation of the mammalian sperm. Nature. 170:326. 10.1038/170326a0 [DOI] [PubMed] [Google Scholar]
- Aydin, H., Sultana A., Li S., Thavalingam A., and Lee J.E.. 2016. Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex. Nature. 534:562–565. 10.1038/nature18595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaux, S., Ialy-Radio C., Chalbi M., Dybal E., Homps-Legrand M., Do Cruzeiro M., Vaiman D., Wolf J.-P., and Ziyyat A.. 2020. Sperm SPACA6 protein is required for mammalian Sperm-Egg Adhesion/Fusion. Sci. Rep. 10:5335. 10.1038/s41598-020-62091-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batruch, I., Lecker I., Kagedan D., Smith C.R., Mullen B.J., Grober E., Lo K.C., Diamandis E.P., and Jarvi K.A.. 2011. Proteomic analysis of seminal plasma from normal volunteers and post-vasectomy patients identifies over 2000 proteins and candidate biomarkers of the urogenital system. J. Proteome Res. 10:941–953. 10.1021/pr100745u [DOI] [PubMed] [Google Scholar]
- Bedford, J.M., and Chang M.C.. 1962. Removal of decapacitation factor from seminal plasma by high-speed centrifugation. Am. J. Physiol. 202:179–181. 10.1152/ajplegacy.1962.202.1.179 [DOI] [PubMed] [Google Scholar]
- Bedford, J.M., Calvin H., and Cooper G.W.. 1973. The maturation of spermatozoa in the human epididymis. J. Reprod. Fertil. Suppl. 18:199–213. [PubMed] [Google Scholar]
- Bianchi, E., and Wright G.J.. 2015. Cross-species fertilization: the hamster egg receptor, Juno, binds the human sperm ligand, Izumo1. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370:20140101. 10.1098/rstb.2014.0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, E., Doe B., Goulding D., and Wright G.J.. 2014. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature. 508:483–487. 10.1038/nature13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork, P., Holm L., and Sander C.. 1994. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 242:309–320. 10.1016/S0022-2836(84)71582-8 [DOI] [PubMed] [Google Scholar]
- Burkart, A.D., Xiong B., Baibakov B., Jiménez-Movilla M., and Dean J.. 2012. Ovastacin, a cortical granule protease, cleaves ZP2 in the zona pellucida to prevent polyspermy. J. Cell Biol. 197:37–44. 10.1083/jcb.201112094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty, S., Kadunganattil S., Bansal P., Sharma R.K., and Gupta S.K.. 2008. Relevance of glycosylation of human zona pellucida glycoproteins for their binding to capacitated human spermatozoa and subsequent induction of acrosomal exocytosis. Mol. Reprod. Dev. 75:75–88. 10.1002/mrd.20726 [DOI] [PubMed] [Google Scholar]
- Chalbi, M., Barraud-Lange V., Ravaux B., Howan K., Rodriguez N., Soule P., Ndzoudi A., Boucheix C., Rubinstein E., Wolf J.P., et al. 2014. Binding of sperm protein Izumo1 and its egg receptor Juno drives Cd9 accumulation in the intercellular contact area prior to fusion during mammalian fertilization. Development. 141:3732–3739. 10.1242/dev.111534 [DOI] [PubMed] [Google Scholar]
- Chang, M.C. 1951. Fertilizing capacity of spermatozoa deposited into the fallopian tubes. Nature. 168:697–698. 10.1038/168697b0 [DOI] [PubMed] [Google Scholar]
- Charrin, S., Jouannet S., Boucheix C., and Rubinstein E.. 2014. Tetraspanins at a glance. J. Cell Sci. 127:3641–3648. 10.1242/jcs.154906 [DOI] [PubMed] [Google Scholar]
- Chen, M.S., Tung K.S., Coonrod S.A., Takahashi Y., Bigler D., Chang A., Yamashita Y., Kincade P.W., Herr J.C., and White J.M.. 1999. Role of the integrin-associated protein CD9 in binding between sperm ADAM 2 and the egg integrin alpha6beta1: implications for murine fertilization. Proc. Natl. Acad. Sci. USA. 96:11830–11835. 10.1073/pnas.96.21.11830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Cann M.J., Litvin T.N., Iourgenko V., Sinclair M.L., Levin L.R., and Buck J.. 2000. Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science. 289:625–628. 10.1126/science.289.5479.625 [DOI] [PubMed] [Google Scholar]
- Chernomordik, L.V., and Kozlov M.M.. 2003. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 72:175–207. 10.1146/annurev.biochem.72.121801.161504 [DOI] [PubMed] [Google Scholar]
- Chernomordik, L.V., and Kozlov M.M.. 2005. Membrane hemifusion: crossing a chasm in two leaps. Cell. 123:375–382. 10.1016/j.cell.2005.10.015 [DOI] [PubMed] [Google Scholar]
- Clark, G.F. 2014. A role for carbohydrate recognition in mammalian sperm-egg binding. Biochem. Biophys. Res. Commun. 450:1195–1203. 10.1016/j.bbrc.2014.06.051 [DOI] [PubMed] [Google Scholar]
- Clermont, Y. 1972. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol. Rev. 52:198–236. 10.1152/physrev.1972.52.1.198 [DOI] [PubMed] [Google Scholar]
- Cornwall, G.A. 2009. New insights into epididymal biology and function. Hum. Reprod. Update. 15:213–227. 10.1093/humupd/dmn055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton, P., and Houghton J.A.. 1987. Visualization of pig sperm chromosomes by in-vitro penetration of zona-free hamster ova. J. Reprod. Fertil. 80:619–622. 10.1530/jrf.0.0800619 [DOI] [PubMed] [Google Scholar]
- Dahmane, S., Doucet C., Le Gall A., Chamontin C., Dosset P., Murcy F., Fernandez L., Salas D., Rubinstein E., Mougel M., et al. 2019. Nanoscale organization of tetraspanins during HIV-1 budding by correlative dSTORM/AFM. Nanoscale. 11:6036–6044. 10.1039/C8NR07269H [DOI] [PubMed] [Google Scholar]
- Dalbies-Tran, R., Cadoret V., Desmarchais A., Elis S., Maillard V., Monget P., Monniaux D., Reynaud K., Saint-Dizier M., and Uzbekova S.. 2020. A Comparative Analysis of Oocyte Development in Mammals. Cells. 9:E1002. 10.3390/cells9041002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávila Garza, S.A., and Patrizio P.. 2013. Reproductive outcomes in patients with male infertility because of Klinefelter’s syndrome, Kartagener’s syndrome, round-head sperm, dysplasia fibrous sheath, and ‘stump’ tail sperm: an updated literature review. Curr. Opin. Obstet. Gynecol. 25:229–246. 10.1097/GCO.0b013e32835faae5 [DOI] [PubMed] [Google Scholar]
- De Blas, G.A., Roggero C.M., Tomes C.N., and Mayorga L.S.. 2005. Dynamics of SNARE assembly and disassembly during sperm acrosomal exocytosis. PLoS Biol. 3:e323. 10.1371/journal.pbio.0030323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edson, M.A., Nagaraja A.K., and Matzuk M.M.. 2009. The mammalian ovary from genesis to revelation. Endocr. Rev. 30:624–712. 10.1210/er.2009-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerman, D.A., Myles D.G., and Primakoff P.. 2006. A role for sperm surface protein disulfide isomerase activity in gamete fusion: evidence for the participation of ERp57. Dev. Cell. 10:831–837. 10.1016/j.devcel.2006.03.011 [DOI] [PubMed] [Google Scholar]
- Ellerman, D.A., Pei J., Gupta S., Snell W.J., Myles D., and Primakoff P.. 2009. Izumo is part of a multiprotein family whose members form large complexes on mammalian sperm. Mol. Reprod. Dev. 76:1188–1199. 10.1002/mrd.21092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fédry, J., Liu Y., Péhau-Arnaudet G., Pei J., Li W., Tortorici M.A., Traincard F., Meola A., Bricogne G., Grishin N.V., et al. 2017. The Ancient Gamete Fusogen HAP2 Is a Eukaryotic Class II Fusion Protein. Cell. 168:904–915.e10. 10.1016/j.cell.2017.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fuertes, B., Laguna-Barraza R., Fernandez-Gonzalez R., Gutierrez-Adan A., Blanco-Fernandez A., O’Doherty A.M., Di Fenza M., Kelly A.K., Kölle S., and Lonergan P.. 2017. Subfertility in bulls carrying a nonsense mutation in transmembrane protein 95 is due to failure to interact with the oocyte vestments. Biol. Reprod. 97:50–60. 10.1093/biolre/iox065 [DOI] [PubMed] [Google Scholar]
- Finkelstein, M., Etkovitz N., and Breitbart H.. 2020. Ca2+ signaling in mammalian spermatozoa. Mol. Cell. Endocrinol. 516:110953. 10.1016/j.mce.2020.110953 [DOI] [PubMed] [Google Scholar]
- Florman, H.M., and First N.L.. 1988. The regulation of acrosomal exocytosis. I. Sperm capacitation is required for the induction of acrosome reactions by the bovine zona pellucida in vitro. Dev. Biol. 128:453–463. 10.1016/0012-1606(88)90307-7 [DOI] [PubMed] [Google Scholar]
- Fujihara, Y., Miyata H., and Ikawa M.. 2018. Factors controlling sperm migration through the oviduct revealed by gene-modified mouse models. Exp. Anim. 67:91–104. 10.1538/expanim.17-0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara, Y., Lu Y., Noda T., Oji A., Larasati T., Kojima-Kita K., Yu Z., Matzuk R.M., Matzuk M.M., and Ikawa M.. 2020. Spermatozoa lacking Fertilization Influencing Membrane Protein (FIMP) fail to fuse with oocytes in mice. Proc. Natl. Acad. Sci. USA. 117:9393–9400. 10.1073/pnas.1917060117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwar, D.K., and Atreja S.K.. 2015. Signalling Events and Associated Pathways Related to the Mammalian Sperm Capacitation. Reprod. Domest. Anim. 50:705–711. 10.1111/rda.12541 [DOI] [PubMed] [Google Scholar]
- Gelbaya, T.A., Potdar N., Jeve Y.B., and Nardo L.G.. 2014. Definition and epidemiology of unexplained infertility. Obstet. Gynecol. Surv. 69:109–115. 10.1097/OGX.0000000000000043 [DOI] [PubMed] [Google Scholar]
- Glazar, A.I., and Evans J.P.. 2009. Immunoglobulin superfamily member IgSF8 (EWI-2) and CD9 in fertilisation: evidence of distinct functions for CD9 and a CD9-associated protein in mammalian sperm-egg interaction. Reprod. Fertil. Dev. 21:293–303. 10.1071/RD08158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gougeon, A. 1996. Regulation of ovarian follicular development in primates: facts and hypotheses. Endocr. Rev. 17:121–155. 10.1210/edrv-17-2-121 [DOI] [PubMed] [Google Scholar]
- Gupta, S.K. 2021. Human Zona Pellucida Glycoproteins: Binding Characteristics With Human Spermatozoa and Induction of Acrosome Reaction. Front. Cell Dev. Biol. 9:619868. 10.3389/fcell.2021.619868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, L., Nishimura K., Sadat Al Hosseini H., Bianchi E., Wright G.J., and Jovine L.. 2016. Divergent evolution of vitamin B9 binding underlies Juno-mediated adhesion of mammalian gametes. Curr. Biol. 26:R100–R101. 10.1016/j.cub.2015.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada, A., and Chang M.C.. 1972. Penetration of zone-free eggs by spermatozoa of different species. Biol. Reprod. 6:300–309. 10.1093/biolreprod/6.2.300 [DOI] [PubMed] [Google Scholar]
- Harpaz, Y., and Chothia C.. 1994. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J. Mol. Biol. 238:528–539. 10.1006/jmbi.1994.1312 [DOI] [PubMed] [Google Scholar]
- Harris, J.D., Hibler D.W., Fontenot G.K., Hsu K.T., Yurewicz E.C., and Sacco A.G.. 1994. Cloning and characterization of zona pellucida genes and cDNAs from a variety of mammalian species: the ZPA, ZPB and ZPC gene families. DNA Seq. 4:361–393. 10.3109/10425179409010186 [DOI] [PubMed] [Google Scholar]
- Harrison, S.C. 2015. Viral membrane fusion. Virology. 479-480:498–507. 10.1016/j.virol.2015.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasuwa, H., Muro Y., Ikawa M., Kato N., Tsujimoto Y., and Okabe M.. 2010. Transgenic mouse sperm that have green acrosome and red mitochondria allow visualization of sperm and their acrosome reaction in vivo. Exp. Anim. 59:105–107. 10.1538/expanim.59.105 [DOI] [PubMed] [Google Scholar]
- Hayashi, K., Shimamoto S., and Nagamatsu G.. 2020. Environmental factors for establishment of the dormant state in oocytes. Dev. Growth Differ. 62:150–157. 10.1111/dgd.12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, R.A., and Renato de Franca L.. 2008. Spermatogenesis and cycle of the seminiferous epithelium. Adv. Exp. Med. Biol. 636:1–15. 10.1007/978-0-387-09597-4_1 [DOI] [PubMed] [Google Scholar]
- Hino, T., Muro Y., Tamura-Nakano M., Okabe M., Tateno H., and Yanagimachi R.. 2016. The Behavior and Acrosomal Status of Mouse Spermatozoa In Vitro, and Within the Oviduct During Fertilization after Natural Mating. Biol. Reprod. 95:50. 10.1095/biolreprod.116.140400 [DOI] [PubMed] [Google Scholar]
- Holesh, J.E., Bass A.N., and Lord M.. 2021. Physiology, Ovulation. In StatPearls. StatPearls Publishing, Treasure Island, FL. [PubMed] [Google Scholar]
- Hwang, J.Y., Mannowetz N., Zhang Y., Everley R.A., Gygi S.P., Bewersdorf J., Lishko P.V., and Chung J.-J.. 2019. Dual Sensing of Physiologic pH and Calcium by EFCAB9 Regulates Sperm Motility. Cell. 177:1480–1494.e19. 10.1016/j.cell.2019.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, N., Ikawa M., Isotani A., and Okabe M.. 2005. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 434:234–238. 10.1038/nature03362 [DOI] [PubMed] [Google Scholar]
- Inoue, N., Satouh Y., Ikawa M., Okabe M., and Yanagimachi R.. 2011. Acrosome-reacted mouse spermatozoa recovered from the perivitelline space can fertilize other eggs. Proc. Natl. Acad. Sci. USA. 108:20008–20011. 10.1073/pnas.1116965108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, N., Hagihara Y., Wright D., Suzuki T., and Wada I.. 2015. Oocyte-triggered dimerization of sperm IZUMO1 promotes sperm-egg fusion in mice. Nat. Commun. 6:8858. 10.1038/ncomms9858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, N., Hagihara Y., and Wada I.. 2021. Evolutionarily conserved sperm factors, DCST1 and DCST2, are required for gamete fusion. eLife. 10:e66313. 10.7554/eLife.66313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino, T., Yano K., Chinzei Y., and Yuda M.. 2004. Cell-passage activity is required for the malarial parasite to cross the liver sinusoidal cell layer. PLoS Biol. 2:E4. 10.1371/journal.pbio.0020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, C., and Toshimori K.. 2016. Acrosome markers of human sperm. Anat. Sci. Int. 91:128–142. 10.1007/s12565-015-0323-9 [DOI] [PubMed] [Google Scholar]
- James, E.R., Carrell D.T., Aston K.I., Jenkins T.G., Yeste M., and Salas-Huetos A.. 2020. The Role of the Epididymis and the Contribution of Epididymosomes to Mammalian Reproduction. Int. J. Mol. Sci. 21:E5377. 10.3390/ijms21155377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean, C., Haghighirad F., Zhu Y., Chalbi M., Ziyyat A., Rubinstein E., Gourier C., Yip P., Wolf J.P., Lee J.E., et al. 2019. JUNO, the receptor of sperm IZUMO1, is expressed by the human oocyte and is essential for human fertilisation. Hum. Reprod. 34:118–126. 10.1093/humrep/dey340 [DOI] [PubMed] [Google Scholar]
- Jégou, A., Ziyyat A., Barraud-Lange V., Perez E., Wolf J.P., Pincet F., and Gourier C.. 2011. CD9 tetraspanin generates fusion competent sites on the egg membrane for mammalian fertilization. Proc. Natl. Acad. Sci. USA. 108:10946–10951. 10.1073/pnas.1017400108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, M., Fujiwara E., Kakiuchi Y., Okabe M., Satouh Y., Baba S.A., Chiba K., and Hirohashi N.. 2011. Most fertilizing mouse spermatozoa begin their acrosome reaction before contact with the zona pellucida during in vitro fertilization. Proc. Natl. Acad. Sci. USA. 108:4892–4896. 10.1073/pnas.1018202108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji, K., Oda S., Shikano T., Ohnuki T., Uematsu Y., Sakagami J., Tada N., Miyazaki S., and Kudo A.. 2000. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat. Genet. 24:279–282. 10.1038/73502 [DOI] [PubMed] [Google Scholar]
- Kanakis, G.A., and Goulis D.G.. 2015. Male contraception: a clinically-oriented review. Hormones (Athens). 14:598–614. 10.14310/horm.2002.1623 [DOI] [PubMed] [Google Scholar]
- Kato, K., Satouh Y., Nishimasu H., Kurabayashi A., Morita J., Fujihara Y., Oji A., Ishitani R., Ikawa M., and Nureki O.. 2016. Structural and functional insights into IZUMO1 recognition by JUNO in mammalian fertilization. Nat. Commun. 7:12198. 10.1038/ncomms12198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr, J.S., and Wright G.J.. 2012. Avidity-based extracellular interaction screening (AVEXIS) for the scalable detection of low-affinity extracellular receptor-ligand interactions. J. Vis. Exp. 3881:e3881. 10.3791/3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölle, S. 2015. Transport, Distribution and Elimination of Mammalian Sperm Following Natural Mating and Insemination. Reprod. Domest. Anim. 50(Suppl 3):2–6. 10.1111/rda.12576 [DOI] [PubMed] [Google Scholar]
- La Spina, F.A., Puga Molina L.C., Romarowski A., Vitale A.M., Falzone T.L., Krapf D., Hirohashi N., and Buffone M.G.. 2016. Mouse sperm begin to undergo acrosomal exocytosis in the upper isthmus of the oviduct. Dev. Biol. 411:172–182. 10.1016/j.ydbio.2016.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas-Toranzo, I., Hamze J.G., Bianchi E., Fernández-Fuertes B., Pérez-Cerezales S., Laguna-Barraza R., Fernández-González R., Lonergan P., Gutiérrez-Adán A., Wright G.J., et al. 2020. TMEM95 is a sperm membrane protein essential for mammalian fertilization. eLife. 9:e53913. 10.7554/eLife.53913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Naour, F., Rubinstein E., Jasmin C., Prenant M., and Boucheix C.. 2000. Severely reduced female fertility in CD9-deficient mice. Science. 287:319–321. 10.1126/science.287.5451.319 [DOI] [PubMed] [Google Scholar]
- Litscher, E.S., and Wassarman P.M.. 1996. Characterization of mouse ZP3-derived glycopeptide, gp55, that exhibits sperm receptor and acrosome reaction-inducing activity in vitro. Biochemistry. 35:3980–3985. 10.1021/bi952722m [DOI] [PubMed] [Google Scholar]
- Litscher, E.S., and Wassarman P.M.. 2007. Egg extracellular coat proteins: from fish to mammals. Histol. Histopathol. 22:337–347. 10.14670/HH-22.337 [DOI] [PubMed] [Google Scholar]
- Lorenzetti, D., Poirier C., Zhao M., Overbeek P.A., Harrison W., and Bishop C.E.. 2014. A transgenic insertion on mouse chromosome 17 inactivates a novel immunoglobulin superfamily gene potentially involved in sperm-egg fusion. Mamm. Genome. 25:141–148. 10.1007/s00335-013-9491-x [DOI] [PubMed] [Google Scholar]
- Makabe, S., Naguro T., and Stallone T.. 2006. Oocyte-follicle cell interactions during ovarian follicle development, as seen by high resolution scanning and transmission electron microscopy in humans. Microsc. Res. Tech. 69:436–449. 10.1002/jemt.20303 [DOI] [PubMed] [Google Scholar]
- Mani, S., Ghosh J., Coutifaris C., Sapienza C., and Mainigi M.. 2020. Epigenetic changes and assisted reproductive technologies. Epigenetics. 15:12–25. 10.1080/15592294.2019.1646572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcozzi, S., Rossi V., Salustri A., De Felici M., and Klinger F.G.. 2018. Programmed cell death in the human ovary. Minerva Ginecol. 70:549–560. 10.23736/S0026-4784.18.04274-0 [DOI] [PubMed] [Google Scholar]
- Miyado, K., Yamada G., Yamada S., Hasuwa H., Nakamura Y., Ryu F., Suzuki K., Kosai K., Inoue K., Ogura A., et al. 2000. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 287:321–324. 10.1126/science.287.5451.321 [DOI] [PubMed] [Google Scholar]
- Morozumi, K., and Yanagimachi R.. 2005. Incorporation of the acrosome into the oocyte during intracytoplasmic sperm injection could be potentially hazardous to embryo development. Proc. Natl. Acad. Sci. USA. 102:14209–14214. 10.1073/pnas.0507005102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi, T., Ikawa M., Yamada S., Parvinen M., Baba T., Nishimune Y., and Okabe M.. 1999. Real-time observation of acrosomal dispersal from mouse sperm using GFP as a marker protein. FEBS Lett. 449:277–283. 10.1016/S0014-5793(99)00433-0 [DOI] [PubMed] [Google Scholar]
- Nakanishi, T., Ikawa M., Yamada S., Toshimori K., and Okabe M.. 2001. Alkalinization of acrosome measured by GFP as a pH indicator and its relation to sperm capacitation. Dev. Biol. 237:222–231. 10.1006/dbio.2001.0353 [DOI] [PubMed] [Google Scholar]
- Nishimura, H., Tajima T., Comstra H.S., Gleason E.J., and L’Hernault S.W.. 2015. The Immunoglobulin-like Gene spe-45 Acts during Fertilization in Caenorhabditis elegans like the Mouse Izumo1 Gene. Curr. Biol. 25:3225–3231. 10.1016/j.cub.2015.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, K., Han L., Bianchi E., Wright G.J., de Sanctis D., and Jovine L.. 2016. The structure of sperm Izumo1 reveals unexpected similarities with Plasmodium invasion proteins. Curr. Biol. 26:R661–R662. 10.1016/j.cub.2016.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Lu Y., Fujihara Y., Oura S., Koyano T., Kobayashi S., Matzuk M.M., and Ikawa M.. 2020. Sperm proteins SOF1, TMEM95, and SPACA6 are required for sperm-oocyte fusion in mice. Proc. Natl. Acad. Sci. USA. 117:11493–11502. 10.1073/pnas.1922650117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicka-Bauer, K., and Szymczak-Cendlak M.. 2021. Structure and Function of Ion Channels Regulating Sperm Motility-An Overview. Int. J. Mol. Sci. 22:3259. 10.3390/ijms22063259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto, U., Ishida H., Krayukhina E., Uchiyama S., Inoue N., and Shimizu T.. 2016. Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization. Nature. 534:566–569. 10.1038/nature18596 [DOI] [PubMed] [Google Scholar]
- Okabe, M. 2015. Mechanisms of fertilization elucidated by gene-manipulated animals. Asian J. Androl. 17:646–652. 10.4103/1008-682X.153299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe, M. 2016. The Acrosome Reaction: A Historical Perspective. Adv. Anat. Embryol. Cell Biol. 220:1–13. 10.1007/978-3-319-30567-7_1 [DOI] [PubMed] [Google Scholar]
- Okabe, M. 2018. Beware of memes in the interpretation of your results - lessons from gene-disrupted mice in fertilization research. FEBS Lett. 592:2673–2679. 10.1002/1873-3468.13101 [DOI] [PubMed] [Google Scholar]
- Pan, B., and Li J.. 2019. The art of oocyte meiotic arrest regulation. Reprod. Biol. Endocrinol. 17:8. 10.1186/s12958-018-0445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, P.-C., Chiu P.C.N., Lee C.-L., Chang L.-Y., Panico M., Morris H.R., Haslam S.M., Khoo K.-H., Clark G.F., Yeung W.S.B., and Dell A.. 2011. Human sperm binding is mediated by the sialyl-Lewis(x) oligosaccharide on the zona pellucida. Science. 333:1761–1764. 10.1126/science.1207438 [DOI] [PubMed] [Google Scholar]
- Pausch, H., Kölle S., Wurmser C., Schwarzenbacher H., Emmerling R., Jansen S., Trottmann M., Fuerst C., Götz K.-U., and Fries R.. 2014. A nonsense mutation in TMEM95 encoding a nondescript transmembrane protein causes idiopathic male subfertility in cattle. PLoS Genet. 10:e1004044. 10.1371/journal.pgen.1004044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, C., Bayer M.J., Bühler S., Andersen J.S., Mann M., and Mayer A.. 2001. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature. 409:581–588. 10.1038/35054500 [DOI] [PubMed] [Google Scholar]
- Podbilewicz, B. 2014. Virus and cell fusion mechanisms. Annu. Rev. Cell Dev. Biol. 30:111–139. 10.1146/annurev-cellbio-101512-122422 [DOI] [PubMed] [Google Scholar]
- Ravaux, B., Favier S., Perez E., and Gourier C.. 2018. Egg CD9 protein tides correlated with sperm oscillations tune the gamete fusion ability in mammal. J. Mol. Cell Biol. 10:494–502. 10.1093/jmcb/mjy005 [DOI] [PubMed] [Google Scholar]
- Rubinstein, E., Ziyyat A., Wolf J.-P., Le Naour F., and Boucheix C.. 2006. The molecular players of sperm-egg fusion in mammals. Semin. Cell Dev. Biol. 17:254–263. 10.1016/j.semcdb.2006.02.012 [DOI] [PubMed] [Google Scholar]
- Runge, K.E., Evans J.E., He Z.-Y., Gupta S., McDonald K.L., Stahlberg H., Primakoff P., and Myles D.G.. 2007. Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev. Biol. 304:317–325. 10.1016/j.ydbio.2006.12.041 [DOI] [PubMed] [Google Scholar]
- Saling, P.M., Sowinski J., and Storey B.T.. 1979. An ultrastructural study of epididymal mouse spermatozoa binding to zonae pellucidae in vitro: sequential relationship to the acrosome reaction. J. Exp. Zool. 209:229–238. 10.1002/jez.1402090205 [DOI] [PubMed] [Google Scholar]
- Sapir, A., Avinoam O., Podbilewicz B., and Chernomordik L.V.. 2008. Viral and developmental cell fusion mechanisms: conservation and divergence. Dev. Cell. 14:11–21. 10.1016/j.devcel.2007.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satouh, Y., Inoue N., Ikawa M., and Okabe M.. 2012. Visualization of the moment of mouse sperm-egg fusion and dynamic localization of IZUMO1. J. Cell Sci. 125:4985–4990. 10.1242/jcs.100867 [DOI] [PubMed] [Google Scholar]
- Singaravelu, G., Rahimi S., Krauchunas A., Rizvi A., Dharia S., Shakes D., Smith H., Golden A., and Singson A.. 2015. Forward Genetics Identifies a Requirement for the Izumo-like Immunoglobulin Superfamily spe-45 Gene in Caenorhabditis elegans Fertilization. Curr. Biol. 25:3220–3224. 10.1016/j.cub.2015.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, G., Koksal A.C., Lu C., and Springer T.A.. 2012. Shape change in the receptor for gliding motility in Plasmodium sporozoites. Proc. Natl. Acad. Sci. USA. 109:21420–21425. 10.1073/pnas.1218581109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnik, J., Miranda P.V., Spiridonov N.A., Yoon S.-Y., Fissore R.A., Johnson G.R., and Visconti P.E.. 2009. Tssk6 is required for Izumo relocalization and gamete fusion in the mouse. J. Cell Sci. 122:2741–2749. 10.1242/jcs.047225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stival, C., Puga Molina L.C., Paudel B., Buffone M.G., Visconti P.E., and Krapf D.. 2016. Sperm Capacitation and Acrosome Reaction in Mammalian Sperm. Adv. Anat. Embryol. Cell Biol. 220:93–106. 10.1007/978-3-319-30567-7_5 [DOI] [PubMed] [Google Scholar]
- Törmälä, R.-M., Jääskeläinen M., Lakkakorpi J., Liakka A., Tapanainen J.S., and Vaskivuo T.E.. 2008. Zona pellucida components are present in human fetal ovary before follicle formation. Mol. Cell. Endocrinol. 289:10–15. 10.1016/j.mce.2008.01.029 [DOI] [PubMed] [Google Scholar]
- Touré, A. 2019. Importance of SLC26 Transmembrane Anion Exchangers in Sperm Post-testicular Maturation and Fertilization Potential. Front. Cell Dev. Biol. 7:230. 10.3389/fcell.2019.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda, R., Satouh Y., Takemoto M., Nakada-Nakura Y., Liu K., Yokoyama T., Shirouzu M., Iwata S., Nomura N., Sato K., et al. 2020. Structural insights into tetraspanin CD9 function. Nat. Commun. 11:1606. 10.1038/s41467-020-15459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance, T.D.R., and Lee J.E.. 2020. Virus and eukaryote fusogen superfamilies. Curr. Biol. 30:R750–R754. 10.1016/j.cub.2020.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser, J.A., and Themmen A.P.N.. 2014. Role of anti-Müllerian hormone and bone morphogenetic proteins in the regulation of FSH sensitivity. Mol. Cell. Endocrinol. 382:460–465. 10.1016/j.mce.2013.08.012 [DOI] [PubMed] [Google Scholar]
- White, J.M., Delos S.E., Brecher M., and Schornberg K.. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189–219. 10.1080/10409230802058320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiser, A., Sachar S., Ghetler Y., Shulman A., and Breitbart H.. 2014. Assessment of sperm hyperactivated motility and acrosome reaction can discriminate the use of spermatozoa for conventional in vitro fertilisation or intracytoplasmic sperm injection: preliminary results. Andrologia. 46:313–315. 10.1111/and.12068 [DOI] [PubMed] [Google Scholar]
- Xiong, W., Wang Z., and Shen C.. 2019. An update of the regulatory factors of sperm migration from the uterus into the oviduct by genetically manipulated mice. Mol. Reprod. Dev. 86:935–955. 10.1002/mrd.23180 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T., Hirota K., Nagahama K., Ohkawa K., Takahashi T., Nomura T., and Sakaguchi S.. 2007. Control of immune responses by antigen-specific regulatory T cells expressing the folate receptor. Immunity. 27:145–159. 10.1016/j.immuni.2007.04.017 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, R., Muro Y., Isotani A., Tokuhiro K., Takumi K., Adham I., Ikawa M., and Okabe M.. 2009. Disruption of ADAM3 impairs the migration of sperm into oviduct in mouse. Biol. Reprod. 81:142–146. 10.1095/biolreprod.108.074021 [DOI] [PubMed] [Google Scholar]
- Yang, Q.-E., and Oatley J.M.. 2014. Spermatogonial stem cell functions in physiological and pathological conditions. Curr. Top. Dev. Biol. 107:235–267. 10.1016/B978-0-12-416022-4.00009-3 [DOI] [PubMed] [Google Scholar]
- Young, S.A.M., Aitken J., and Baker M.A.. 2015. Phosphorylation of Izumo1 and Its Role in Male Infertility. Asian J. Androl. 17:708–710. 10.4103/1008-682X.156119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalazar, L., Stival C., Nicolli A.R., De Blas G.A., Krapf D., and Cesari A.. 2020. Male Decapacitation Factor SPINK3 Blocks Membrane Hyperpolarization and Calcium Entry in Mouse Sperm. Front. Cell Dev. Biol. 8:575126. 10.3389/fcell.2020.575126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.A., and Huang C.. 2012. Tetraspanins and cell membrane tubular structures. Cell. Mol. Life Sci. 69:2843–2852. 10.1007/s00018-012-0954-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Cai H., Yang Q., Shi T., Pan C., Lei C., Dang R., Chen H., and Lan X.. 2016. Identification of novel alternative splicing transcript and expression analysis of bovine TMEM95 gene. Gene. 575:531–536. 10.1016/j.gene.2015.09.026 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Wang Y., Feng X., Zhang S., Xu X., Li L., Niu S., Bo Y., Wang C., Li Z., et al. 2021. Oocyte-derived microvilli control female fertility by optimizing ovarian follicle selection in mice. Nat. Commun. 12:2523. 10.1038/s41467-021-22829-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, G.-Z., Miller B.J., Boucheix C., Rubinstein E., Liu C.C., Hynes R.O., Myles D.G., and Primakoff P.. 2002. Residues SFQ (173-175) in the large extracellular loop of CD9 are required for gamete fusion. Development. 129:1995–2002. 10.1242/dev.129.8.1995 [DOI] [PubMed] [Google Scholar]
- Zimmerman, B., Kelly B., McMillan B.J., Seegar T.C.M., Dror R.O., Kruse A.C., and Blacklow S.C.. 2016. Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket. Cell. 167:1041–1051.e11. 10.1016/j.cell.2016.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziyyat, A., Rubinstein E., Monier-Gavelle F., Barraud V., Kulski O., Prenant M., Boucheix C., Bomsel M., and Wolf J.-P.. 2006. CD9 controls the formation of clusters that contain tetraspanins and the integrin alpha 6 beta 1, which are involved in human and mouse gamete fusion. J. Cell Sci. 119:416–424. 10.1242/jcs.02730 [DOI] [PubMed] [Google Scholar]