Abstract
Protein post-translational modifications regulate protein structure and function. Lysine benzoylation is a new type of histone mark with unique physiological relevance. To construct proteins with this modification site-specifically introduced, here we generated orthogonal tRNAPyl-MaBzKRS pairs by engineering Methanomethylophilus alvus pyrrolysyl-tRNA synthetase, allowing the genetic incorporation of ε-N-benzoyllysine (BzK) into proteins with high efficiency in E. coli and mammalian cells. Two types of MaBzKRS were identified to incorporate BzK using mutations located at different positions of the amino acid binding pocket. These MaBzKRS are small in size and highly expressed, which will afford broad utilities in studying the biological effects of lysine benzoylation.
Keywords: lysine benzoylation, posttranslational modification, genetic code expansion, pyrrolysyl-tRNA synthetase, noncanonical amino acid
Graphical Abstract
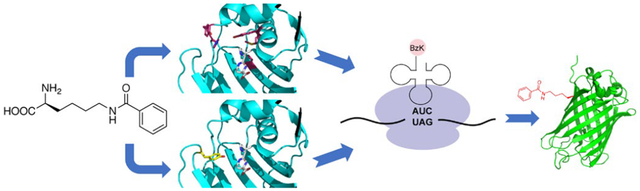
Two M. alvus PylRS mutants were engineered to genetically encode ε-N-benzoyllysine, a newly discovered histone posttranslational modification, into proteins in E. coli and mammalian cells with high efficiency. Mutations at different locations in M. alvus PylRS active site (pink and yellow) were able to achieve high specificity for the same amino acid substrate (grey).
Protein post-translational modifications (PTMs) expand the functional diversity of the proteome and play important roles in regulating protein activity, structure, and function. [1] The ability to prepare proteins with the desired PTM site-specifically introduced at the target sites is critical for the understanding of the principles, types, and effects of protein PTMs on biological processes as well as for the prevention and treatment of associated diseases. Aside from various chemical and semisynthetic methods, [2–5] genetically encoding noncanonical amino acids (ncAAs) using orthogonal tRNA-synthetase pairs in live cells [6,7] has been extensively employed to introduce various PTMs and mimetics into proteins. Representative examples include glycosylation, [8,9] phosphorylation, [10–12] alkylation or acylation, [13] [14–19] sulfation, [20,21] [22] and so on. Knockout of release factor further allows the simultaneous incorporation of PTM-bearing ncAAs at multiple sites into proteins. [23]
Lysine benzoylation has been discovered and characterized as a histone mark, [24] which is associated with gene expression and has physiological relevance distinct from lysine acetylation. More interestingly, sodium benzoate, a widely used food preservative, can generate benzoyl CoA and thus stimulate lysine benzoylation, raising concerns for its safety. Whether lysine benzoylation spreads throughout the proteome and how it exerts physiological effect awaits further studies. Recently, Ji et al. reported the genetic incorporation of ε-N-benzoyllysine (BzK) using a mutant synthetase derived from Methanosarcina barkeri pyrrolysyl-tRNA synthetase (MbPylRS), which contains a Y349W mutation and is poly-specific for multiple ncAAs. [25] Here we independently evolved BzK-specific synthetases from Methanomethylophilus alvus pyrrolysyl-tRNA synthetase (MaPylRS), which incorporate BzK into proteins in E. coli and mammalian cells in excellent fidelity and enhanced efficiency, as verified with mass spectrometry and BzK-specific antibodies. These enhanced BzK-specific MaPylRS mutants would facilitate the study of lysine benzoylation both in vitro and in live cells. In addition, our identification of two different types of BzK-specific MaPylRS mutants suggests that PylRS could be engineered to recognize a ncAA using potentially dissimilar mechanisms.
In comparison with MbPylRS and Methanosarcina mazei pyrrolysyl-tRNA synthetase (MmPylRS), the recently discovered MaPylRS is much smaller in size (MaPylRS 275 aa vs. MbPylRS 419 aa and MmPylRS 454 aa) due to the lack of the N-terminal noncatalytic domain. [26,27] The smaller size of MaPylRS makes it more amenable to delivery via viral vectors with limited capacity and easier to construct gene libraries for directed evolution. MaPylRS is also more stable and has higher expression level in E. coli cells. [28] Transplantation of mutations identified in the active sites of MmPylRS or MbPylRS into the MaPylRS often lead to improved ncAA incorporation efficiency in E. coli. [29,30] MaPylRS protein can also be concentrated several fold higher without aggregation in vitro, which greatly facilitates cell-free genetic code expansion as well. [28]
We therefore decided to use MaPylRS as the parental synthetase to engineer BzK-specific mutants. On the basis of the crystal structure of MaPylRS (Figure 1b), [28] we randomized Tyr126, Met129, and Val168 in the active site and fixed His227 to Ile and Tyr228 to Pro. The H227I/Y228P mutations are at the second layer of the amino acid-binding pocket and have been shown to improve ncAA incorporation efficiency in E. coli. [28]. The resultant mutant library JL1.3 was subjected to selections using procedures described previously. [31,32] After selection, hits exhibiting best BzK-dependent phenotype converged on the following mutations Y126T/M129R/V168H/H227I/Y228P, which was named as MaBzKRS.
Figure 1.
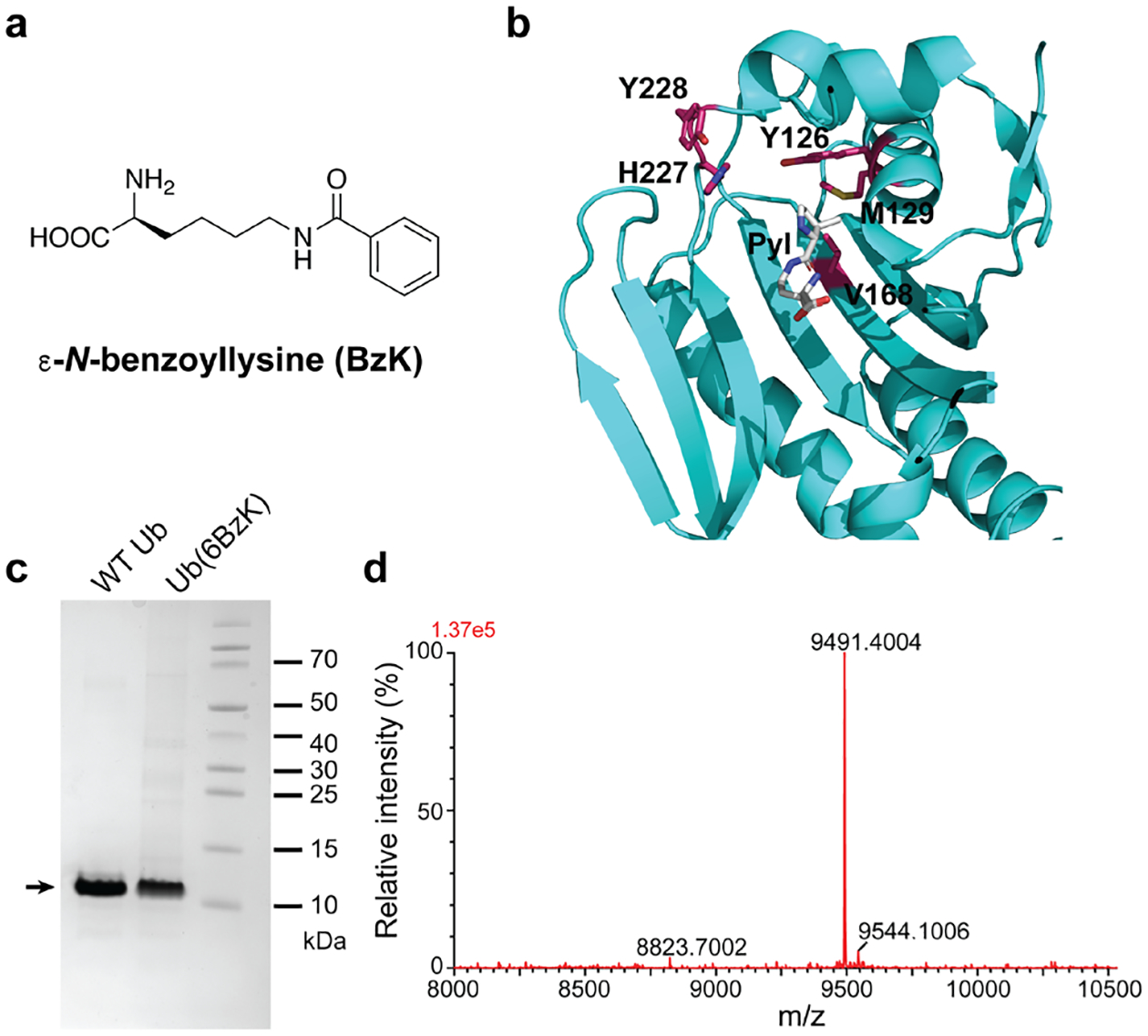
Genetic incorporation of BzK into proteins in E. coli using the evolved Ma-tRNAPyl/MaBzKRS. (a) Structure of BzK. (b) Structure of M. alvus PylRS amino acid binding pocket with residues mutated in the library shown in stick. To illustrate the position of the bound Pyl, M. alvus PylRS apo form (PDB 6JP2) was superimposed on M. mazei PylRSc bound with Pyl (PDB 2ZCE). (c) SDS-PAGE analysis of WT Ub and Ub(6BzK) expressed and purified from E. coli. (d) ESI-TOF MS spectrum of the intact Ub(6BzK).
To evaluate the incorporation of BzK into proteins in E. coli, we expressed the ubiquitin (Ub) gene containing a TAG codon at the permissive site 6 (Ub-6TAG) with the Ma-tRNAPyl/MaBzKRS pair (encoded by plasmid pEvol-MaBzKRS) in E. coli. The cells were grown with 2 mM of BzK added in growth media. After purification through nickel affinity chromatography, the protein was analyzed with SDS-PAGE and full-length Ub(6BzK) was produced in the yield of 4.5 mg/L (Fig. 1c). To determine the identity, the purified protein was then analyzed with electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS) (Fig. 1d). A peak was observed at 9491.4 Da, corresponding to intact Ub containing BzK at site 6 (expected 9491.8 Da). We manually searched for a peak at 9387.7 Da, corresponding to debenzoylation of BzK to Lys, and found that the peak was at the noise level, suggesting that the endogenous CobB, the only sirtuin identified in E. coli, has low debenzoylation activity in the DH10β strain used. No peaks corresponding to mis-incorporation of other natural amino acids at site 6 were detected. These results demonstrated that the evolved Ma-tRNAPyl/MaBzKRS pair was able to incorporate BzK into proteins in high fidelity in E. coli.
To further determine the incorporation specificity of BzK, we incorporated it into GFP and detected with an antibody specific for benzoyllysine. We first expressed the super-fold GFP (sfGFP) gene containing a TAG codon at permissive site 151 with the Ma-tRNAPyl/MaBzKRS pair in E. coli. After purification, full-length sfGFP(151BzK) protein was produced in the yield of 35 mg/L (Figure 2a), indicating that BzK was incorporated in sfGFP by the Ma-tRNAPyl/MaBzKRS pair in high efficiency. The purified proteins were immunoblotted with an anti-GFP mAb or an anti-benzoyllysine mAb (Figure 2b). Only sfGFP(151BzK) showed clear and strong signal for anti-benzoyllysine mAb. We also expressed the enhanced GFP gene containing a TAG codon at the permissive site 182 with the Ma-tRNAPyl/MaBzKRS pair in E. coli. The purified EGFP(182BzK) (yielding 6.6 mg/L) ran at the similar position with the WT EGFP on SDS-PAGE (Figure 2c), and only EGFP(182BzK) showed robust signal when immunoblotted by the anti-benzoyllysine mAb (Figure 2d). These results demonstrate the efficient incorporation of BzK by the Ma-tRNAPyl/MaBzKRS pair in E. coli and BzK specificity verification with the anti-benzoyllysine mAb.
Figure 2.
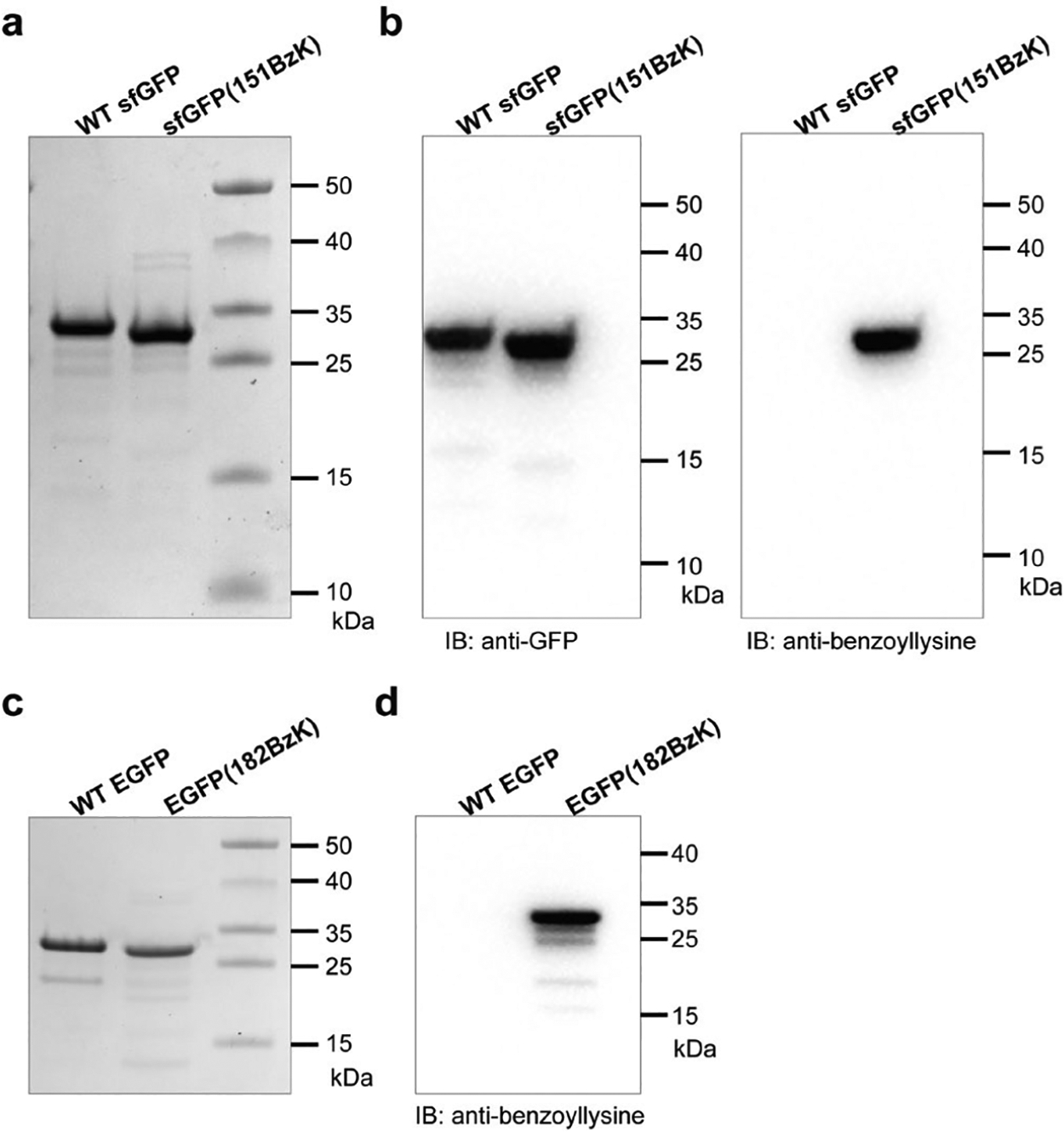
Genetic incorporation of BzK into GFP and verification with anti-benzoyllysine mAb. (a) SDS-PAGE analysis of WT sfGFP and sfGFP(151BzK). (b) Western blots of WT sfGFP and sfGFP(151BzK) detected by an anti-GFP antibody (left) and an anti-benzoyllysine antibody (right). (c) SDS-PAGE analysis of WT EGFP and EGFP(182BzK). (d) Western blot of WT EGFP and EGFP(182Bzk) detected by an anti-benzoyllysine antibody.
To enable the generation of BzK-containing proteins in mammalian cells, we tested BzK incorporation into EGFP in HEK-293T cells. We transfected HEK-293T cells with plasmid pcDNA3.1-EGFP(182TAG) expressing EGFP gene containing a TAG codon at permissive site 182 and plasmid pNEU-BzKRS expressing the Ma-tRNAPyl/MaBzKRS pair. Cells were cultured in the presence or absence of 2 mM BzK, followed with flow cytometric analysis. Suppression of the 182TAG codon of the EGFP gene would produce full-length EGFP, rendering cells fluorescent. As showed in Figure 3a, strong green fluorescence of EGFP were detected from cells cultured in the presence of BzK, and no green fluorescence was detected from cells without BzK added. Compared with 24 h incubation, more cells became green fluorescent after incubating for 48 h. Confocal fluorescence microscopy of these cells confirmed the above results (Figure 3b). In the presence of BzK, strong EGFP fluorescence was observed inside the cells, and cell morphology were normal, suggesting no obvious cytotoxicity of BzK to HEK-293T cells. No fluorescence signal was detected when BzK was not added. The successful incorporation of BzK into EGFP in HEK-293T cells was also demonstrated by Western blot analysis of cell lysate (Figure 3c). Immunoblot with an anti-GFP antibody showed that full-length EGFP was produced only when BzK was added. Immunoblot with the anti-benzoyllysine antibody confirmed that BzK was incorporated into the full-length EGFP, showing a clear band at the same position with the EGFP band. Together, these results indicate that the Ma-tRNAPyl/MaBzKRS pair was able to incorporate BzK into proteins in HEK-293T cells with high specificity.
Figure 3.
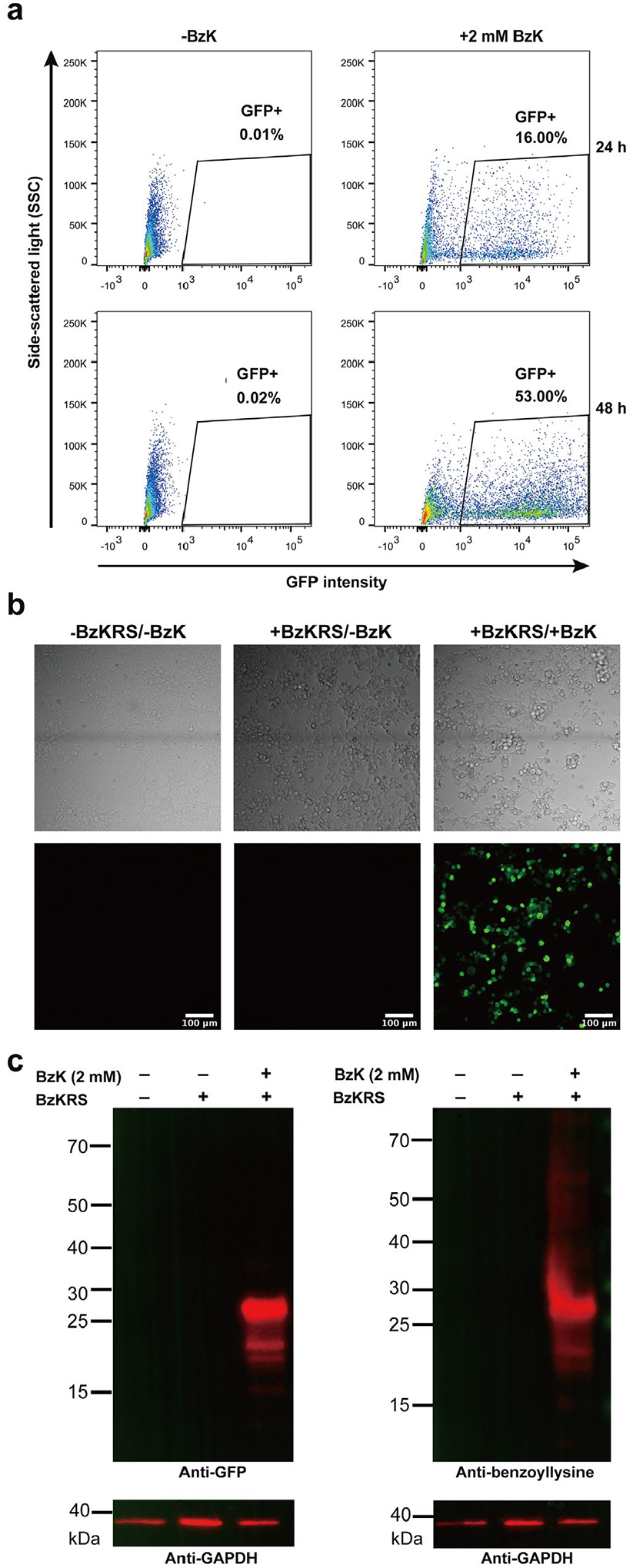
Genetic incorporation of BzK into proteins in mammalian cells. (a) Flow cytometric analysis of BzK incorporation into EGFP(182TAG) in HEK-293T cells. Cells were co-transfected with pNEU-BzKRS and pcDNA3.1-EGFP(182TAG) and cultured with or without BzK for 24–48 h. (b) Fluorescence confocal microscopic analysis of BzK incorporation into EGFP(182TAG) in HEK-293T cells. (c) Western blot analysis of BzK incorporation into EGFP(182TAG) in HEK-293T cells. An anti-GFP antibody was used to detect EGFP, and an anti-benzoyllysine antibody was used to detect BzK incorporated into EGFP protein.
During the preparation of this work, Ji et al. reported that MbPylRS(Y349W) incorporates BzK into proteins in E. coli and mammalian cells. [25] We thus compared MaBzKRS with MbPylRS(Y349W) using the same expression plasmid pEvol to express corresponding tRNAPyl/PylRS and the reporter plasmid pBAD-sfGFP(151TAG) to express EGFP gene containing a TAG codon at the permissive 151 site. The pEvol and pBAD-sfGFP(151TAG) plasmids were co-transformed in E. coli, and the cells were grown with or without 2 mM BzK. EGFP fluorescence intensity of cells were measured and normalized to cell optical density at 600 nm (OD600). Judging from normalized fluorescence intensity in the presence of BzK, MaBzKRS showed similar incorporation efficiency to MbPylRS(Y349W) (Figure 4a), but as for the normalized fluorescence intensity ratio of +BzK over −BzK, MaBzKRS was 2.2 fold of MbPylRS(Y349W), suggesting that MaBzKRS has a higher BzK fidelity. Since the active sites of MaPylRS, MbPylRS, and MmPylRS are well conserved, and transplanting mutations from MbPylRS or MmPylRS to MaPylRS often leads to higher incorporation efficiency, we therefore made the corresponding mutation in MaPylRS to generate MaPylRS(Y206W). Gratifyingly, MaPylRS(Y206W) was not only able to incorporate BzK, but also showed a 3-fold increase of efficiency over MbPylRS(Y349W) and MaBzKRS. We thus named MaPylRS(Y206W) as MaBzKRS2. The second layer mutation H227I/Y228P of MaPylRS has been shown to increase ncAA incorporation efficiency, [28] but adding these two mutations to MaBzkRS2 reduced the incorporation efficiency to 27%. We also wondered if mutations of MaBzKRS could synergize with that of MaBzKRS2. However, introducing of Y206W into MaBzKRS reduced the incorporation efficiency to 22% instead.
Figure 4.
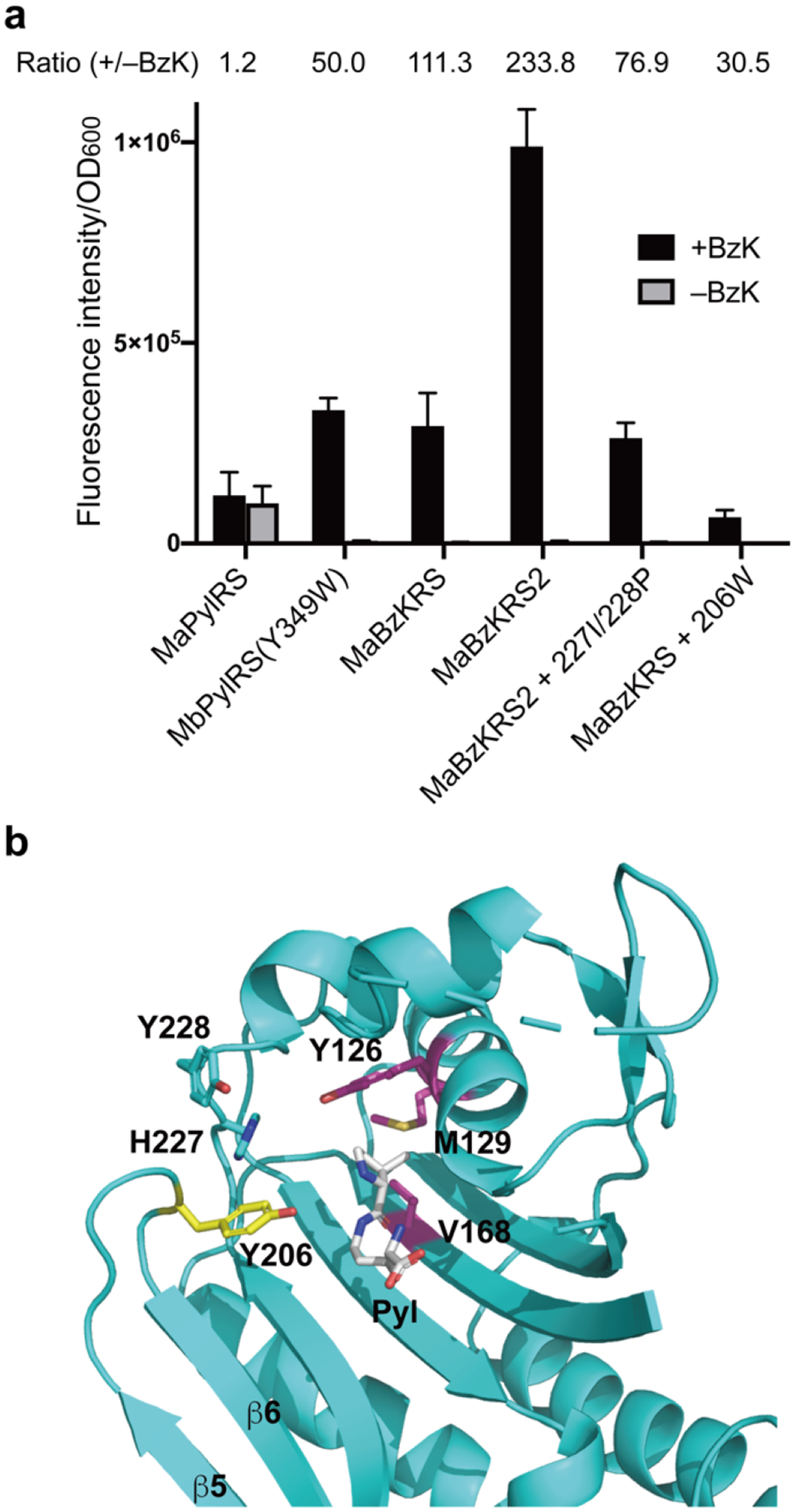
Two different types of MaBzKRS for BzK incorporation. (a) Comparison of the BzK incorporation efficiency of different MaBzKRS mutants. Measured sfGFP fluorescence intensity was normalized to cell optical density. Normalized fluorescence ratio for +/− BzK was indicated for each mutant. Error bars represent s.d., n = 3 independent experiments. (b) Structure of MaPylRS showing the different locations of mutated amino acid residues in MaBzKRS (pink and cyan) and in MaBzKRS2 (yellow). To illustrate the position of the bound Pyl, M. alvus PylRS apo form (PDB 6JP2) is superimposed on M. mazei PylRSc bound with Pyl (PDB 2ZCE).
These mutation results suggest that MaBzKRS2 and MaBzKRS may have dissimilar mechanism in recognizing BzK. In fact, the five mutations in MaBzKRS are located in the active site pocket, with Y126T/M129R/V168H in the first layer directly contacting BzK side chain, and H227I/Y228P in the second layer beyond the first layer residues (Figure 4b). That is, these mutations are more likely to shape the amino acid binding pocket to accommodate the ncAA side chain. The Y206W mutation in MaBzKRS2, in contrast, is located on the tip of the β5- β6 hairpin (Figure 4b), which is in a dynamic equilibrium of open and closed conformation, affecting the opening/closing of the amino acid binding tunnel. [28] It is intriguing to observe that two sets of mutations at different locations of the active site of MaPylRS can achieve the same specificity for BzK, but the detailed molecular mechanism awaits further studies.
In summary, we generated two different MaPylRS mutants for genetic incorporation of BzK into proteins with high efficiency and fidelity in E. coli and mammalian cells. Sharing the small size, excellent stability, and high expression level with the parental MaPylRS, these BzK-specific synthetases will prove useful in introducing lysine benzoylation site-specifically into proteins both in cell-free translation systems and in live cells, facilitating the research of this PTM. In addition, MbPylRS (Y349W) has been shown to incorporate many other ncAAs, and the transplanted MaPylRS (Y206W) may also enhance the incorporation efficiency of these ncAAs.
Supplementary Material
Acknowledgements
L.W. acknowledges the support of the NIH (R01GM118384). S.R. acknowledges the support of the NIH (GM121607).
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].Walsh CT, Garneau-Tsodikova S, Gatto GJJ, Angew. Chem. Int. Ed. Engl 2005, 44, 7342–7372. [DOI] [PubMed] [Google Scholar]
- [2].Muir TW, Annu. Rev. Biochem 2003, 72, 249–289. [DOI] [PubMed] [Google Scholar]
- [3].Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM, Cell 2007, 128, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Szewczuk LM, Tarrant MK, Cole PA, Methods Enzymol. 2009, 462, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wright TH, Bower BJ, Chalker JM, Bernardes GJL, Wiewiora R, Ng WL, Raj R, Faulkner S, Vallee MRJ, Phanumartwiwath A, et al. , Science 2016, 354, aag1465–aag1465. [DOI] [PubMed] [Google Scholar]
- [6].Wang L, Brock A, Herberich B, Schultz PG, Science 2001, 292, 498–500. [DOI] [PubMed] [Google Scholar]
- [7].Wang L, Schultz PG, Angew. Chem. Int. Ed 2004, 44, 34–66. [DOI] [PubMed] [Google Scholar]
- [8].Liu H, Wang L, Brock A, Wong C-H, Schultz PG, J. Am. Chem. Soc 2003, 125, 1702–1703. [DOI] [PubMed] [Google Scholar]
- [9].Yang B, Wang N, Schnier PD, Zheng F, Zhu H, Polizzi NF, Ittuveetil A, Saikam V, DeGrado WF, Wang Q, et al. , J. Am. Chem. Soc 2019, 141, 7698–7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, Benner J, Noren CJ, Rinehart J, Soll D, Science 2011, 333, 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hoppmann C, Wong A, Yang B, Li S, Hunter T, Shokat KM, Wang L, Nat. Chem. Biol 2017, 13, 842–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Luo X, Fu G, Wang RE, Zhu X, Zambaldo C, Liu R, Liu T, Lyu X, Du J, Xuan W, et al. , Nat. Chem. Biol 2017, 13, 845–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Guo J, Wang J, Lee JS, Schultz PG, Angew. Chem. Int. Ed. Engl 2008, 47, 6399–6401. [DOI] [PubMed] [Google Scholar]
- [14].Ai H-W, Lee JW, Schultz PG, Chemical communications 2010, 46, 5506–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nguyen DP, Garcia Alai MM, Virdee S, Chin JW, Chem. Biol 2010, 17, 1072–1076. [DOI] [PubMed] [Google Scholar]
- [16].Lee Y-J, Wu B, Raymond JE, Zeng Y, Fang X, Wooley KL, Liu WR, ACS Chem. Biol 2013, 8, 1664–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xiao H, Xuan W, Shao S, Liu T, Schultz PG, ACS Chem. Biol 2015, 10, 1599–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang A, Ha S, Ahn J, Kim R, Kim S, Lee Y, Kim J, Söll D, Lee H-Y, Park H-S, Science 2016, 354, 623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fu C, Chen Q, Zheng F, Yang L, Li H, Zhao Q, Wang X, Wang L, Wang Q, Angew. Chem. Int. Ed. Engl 2019, 58, 1392–1396. [DOI] [PubMed] [Google Scholar]
- [20].Liu CC, Schultz PG, Nat. Biotechnol 2006, 24, 1436–1440. [DOI] [PubMed] [Google Scholar]
- [21].He X, Chen Y, Beltran DG, Kelly M, Ma B, Lawrie J, Wang F, Dodds E, Zhang L, Guo J, et al. , Nat. Commun 2020, 11, 4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Italia JS, Peeler JC, Hillenbrand CM, Latour C, Weerapana E, Chatterjee A, Nat. Chem. Biol 2020, 16, 379–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Johnson DB, Xu J, Shen Z, Takimoto JK, Schultz MD, Schmitz RJ, Xiang Z, Ecker JR, Briggs SP, Wang L, Nat. Chem. Biol 2011, 7, 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Huang H, Zhang D, Wang Y, Perez-Neut M, Han Z, Zheng YG, Hao Q, Zhao Y, Nat. Commun 2018, 9, 3374–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ji Y, Ren C, Miao H, Pang Z, Xiao R, Yang X, Xuan W, Chemical communications 2021, 57, 1798–1801. [DOI] [PubMed] [Google Scholar]
- [26].Borrel G, Parisot N, Harris HMB, Peyretaillade E, Gaci N, Tottey W, Bardot O, Raymann K, Gribaldo S, Peyret P, et al. , BMC genomics 2014, 15, 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Meineke B, Heimgärtner J, Lafranchi L, Elsässer SJ, ACS Chem. Biol 2018, 13, 3087–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Seki E, Yanagisawa T, Kuratani M, Sakamoto K, Yokoyama S, ACS Synth Biol 2020, 9, 718–732. [DOI] [PubMed] [Google Scholar]
- [29].Yamaguchi A, Iraha F, Ohtake K, Sakamoto K, Molecules 2018, 23, 2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu J, Cheng R, Van Eps N, Wang N, Morizumi T, Ou W-L, Klauser PC, Rozovsky S, Ernst OP, Wang L, J. Am. Chem. Soc 2020, 142, 17057–17068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liu J, Zheng F, Cheng R, Li S, Rozovsky S, Wang Q, Wang L, J. Am. Chem. Soc 2018, 140, 8807–8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu J, Cheng R, Wu H, Li S, Wang PG, DeGrado WF, Rozovsky S, Wang L, Angew. Chem. Int. Ed. Engl 2018, 57, 12702–12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


