Abstract
Simian virus 40 (SV40) large tumor antigen (T antigen) has been shown to inhibit p53-dependent transcription by preventing p53 from binding to its cognate cis element. Data presented in this report provide the first direct functional evidence that T antigen, under certain conditions, may also repress p53-dependent transcription by a mechanism in which the transactivation domain of p53 is abrogated while DNA binding is unaffected. Specifically, p53 purified as a complex with T antigen from mouse cells was found to bind DNA as a transcriptionally inactive intact complex, while that purified from human cells was found to bind DNA independently of T antigen and could activate p53-dependent transcription. This difference in activity may be dependent on a different interaction of T antigen with mouse and human p53 and, in addition, on the presence of super T, which is found only in transformed rodent cells. These results suggest that subtle yet important differences exist between the inhibition of p53 by T antigen in mouse and human cells. The implications of this finding with respect to SV40-associated malignancies are discussed.
p53 is an important tumor suppressor gene, found to be mutated or absent in over 50% of all cancers studied (23). It functions as a sequence-specific DNA-binding transcription factor (21, 24). In response to double-stranded DNA breaks, p53 is converted from a latent to an active form (17). This results in increased expression of p53-responsive proteins such as p21 which are required for growth arrest at the G1-to-S phase transition (12). It also mediates apoptosis via the increased expression of proteins such as Bax (30). Inactivation of p53, therefore, results in the loss of a cell cycle checkpoint required for repair of damaged DNA and prevents apoptosis in response to severe DNA damage. In the absence of these responses, oncogenic mutations which may result in tumor progression can accumulate. From the above, it is clear that the transcriptional activation function of p53 is critical to its role as a tumor suppressor.
A number of proteins bind to p53 and negatively affect its transcriptional activity. The cellular oncoprotein MDM2 has been shown to inhibit p53 via three different mechanisms. First, when bound to p53, MDM2 conceals the activation domain of p53 from the transcription machinery, thereby indirectly repressing p53-dependent transcription (32). Second, it has been found to promote the rapid degradation of p53 via a ubiquitin-proteosome pathway, resulting in decreased levels of p53 available to activate transcription (15, 22). Finally, MDM2 may itself function as an active repressor of transcription, which, via its interaction with p53, represses p53-responsive genes (39).
Proteins encoded by DNA tumor viruses also inhibit p53 activity in similar ways. The human papillomavirus type 16 E6 protein forms a complex with p53, thus promoting its polyubiquitination and subsequent degradation (35, 38). The adenovirus early 1B (E1B) 55K protein, a transcriptional repressor, binds to p53 and is thereby targeted to p53-responsive genes (41). The large tumor antigen (T antigen) of simian virus 40 (SV40) also forms with p53 a complex that inhibits p53 function in SV40-infected and SV40-transformed cells. Experiments performed with baculovirus-expressed human p53 and T antigen led Bargonetti et al. (1) to propose that T antigen inhibits p53 function by preventing it from binding to its cognate cis element. Furthermore, Segawa et al. (36) reported similar results when they examined the DNA-binding activity of p53 in crude nuclear extracts isolated from a human p53 null cell line transiently transfected with plasmids expressing p53 and T antigen; in addition, when baculovirus-expressed T antigen was added to a mouse cell lysate containing wild-type p53, DNA binding was abolished. Taken together, data derived from these experiments support the model in which T antigen inhibits p53 function by preventing it from binding to DNA.
T antigen is often found as a 90-kDa protein in the nuclei of SV40-infected and/or -transformed cells. However, higher-molecular-weight forms of T antigen, designated super T, have also been detected in SV40-transformed rodent cell lines (37). Forms of super T are reported to arise from internal in-phase duplications in the coding region of the T-antigen gene (28, 29) or, as is the case for a commonly occurring 100-kDa form, by differential splicing between two integrated partial copies of the T-antigen gene (25). The duplication which forms the 100-kDa protein includes the first exon, the intron, and part of the second exon upstream of the complete coding sequence for the T-antigen gene. It is proposed that transcription starts at the upstream copy of T antigen and continues through the host DNA and into the full-length copy of the gene. The long primary transcript is spliced, but a short region of extra RNA, possibly from the first copy of the duplicated control region, is retained and encodes the extra amino acids present in the 100-kDa super-T protein (25). The presence of super T was found to correlate with anchorage-independent growth in mouse cell lines (2, 6). Despite the compelling effect of super T in transformation, the molecular mechanism by which super T functions to inhibit p53-dependent transcription remains to be elucidated.
In the present study, we show that p53 immunopurified from a human cell line and a monkey cell line copurifies with T antigen, while that purified from two mouse cell lines copurifies with a 100-kDa form of super T. When purified from mouse cells, the p53-T antigen complex can bind specifically to DNA in electrophoretic mobility shift analysis (EMSA). However, despite this DNA-binding activity, the complex is not capable of activating p53-dependent transcription in vitro. Therefore, our data suggest that in mouse cell lines, T antigen/super T abrogates the transactivation domain of p53 and does not affect DNA binding. When purified from either a human or a monkey cell line, the p53-T antigen complex also binds specifically to DNA in EMSA, but surprisingly T antigen is not present in the resulting shifted complex and hence this complex can support p53-dependent transcription in vitro. This finding suggests that in human and monkey cells, p53 may not be inhibited by T antigen. The different activities of the p53-T antigen complex purified from mouse cell lines and from human and monkey cell lines were found to be dependent on a different interaction of T antigen with mouse and human p53 and, in addition, possibly on the presence of super T. These results suggest for the first time the existence of subtle differences between the inhibition of p53 by T antigen in human and mouse cells that may have physiologically significant consequences.
MATERIALS AND METHODS
Protein purification.
p53-T antigen complex was immunopurified from nuclear extracts prepared by the method of Dignam et al. (11). One milliliter of nuclear extract (7 mg of protein/ml) was incubated for 3 h at 4°C with 100 μl of packed protein A-Sepharose beads to which Pab 421, a monoclonal antibody specific for p53, was covalently linked. Beads were washed twice with 0.5 M KCl D buffer (20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) and once with 0.1 M KCl D buffer. p53 was eluted from the washed beads with 100 μl of 421 epitope oligopeptide (KKGQSTSRHKK) at 1 mg/ml in 0.1 M KCl D buffer. p53-T antigen complex was also purified by using beads to which Pab 108, a monoclonal antibody specific for T antigen, was covalently linked. In this case the complex was eluted with EG (ethylene glycol) buffer (50% EG, 0.5 M NaCl, 10% glycerol, 20 mM Tris HCl [pH 8.5], 1 mM EDTA) and dialyzed overnight against 0.1 M KCl D buffer. To purify p53 in the absence of T antigen, Pab 421 anti-p53 beads were incubated with nuclear extract and washed as described above. T antigen was eluted from the bound p53 by two 10-min washes in 0.1 ml of 2 M urea in 0.1 M KCl D buffer. The T-antigen-containing supernatant was dialyzed overnight against 0.1 M KCl D buffer. The remaining beads were washed overnight with 0.1 M KCl D buffer, after which p53 was eluted as described above. To immunodeplete the Pab 421-purified complex, a volume of covalently linked Pab 108 beads equal to one-fifth of the protein sample volume was added, and incubation was carried out for 3 h at 4°C with gentle rotation. The supernatant was retained and incubated with fresh Pab 108 beads for a further 3 h, after which the supernatant was collected. A control human p53 (no T antigen present) was prepared from HeLa cells infected with recombinant vaccinia virus expressing p53 as described previously (26) and purified with Pab 421 anti-p53 beads as described above. Recombinant T antigen was prepared from Spodoptera frugiperda SF21 insect cells infected with recombinant baculovirus (a gift from C. Prives, Columbia University) as described by Bargonetti et al. (1). Proteins were analyzed by electrophoresis on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels which were subjected to Western blotting or were silver stained to visualize bands.
EMSA.
The sequence of the oligonucleotide probe containing the ribosomal gene cluster (RGC) p53-binding site is 5′-AGCTTGCCTCGAGCTTGCCTGGACTTGCCTGGTCGACGC-3′; the sequence of that containing the p53-binding site from the p21 promoter is 5′-AGCTTAATTCTCGAGGAACATGTCCCAACATGTTGCTCGAGG-3′. Probes were labeled with the Klenow fragment of Escherichia coli DNA polymerase. When required, preincubation reactions were performed for 20 min at 4°C prior to EMSA. Binding reaction mixtures contained 60 mM KCl, 12% glycerol, 5 mM MgCl2, 1 mM EDTA, 0.1 μg of bovine serum albumin, 0.5 μg of poly(dG-dC), 200 pg of 32P-labeled probe, proteins and antibodies as indicated, and water in a total volume of 12.5 μl. Antibody N-19 (Santa Cruz Biotechnology Inc.), recognizing an amino-terminal epitope mapping to within residues 2 to 20, was used against p53. Pab 108, recognizing an amino-terminal epitope mapping to within residues 1 to 82, was used against T antigen. Reaction mixtures were incubated for 30 min at 30°C and then analyzed on a 3% polyacrylamide gel containing 0.5× TBE (0.045 mM Tris-borate, 0.045 mM sodium borate, 0.001 mM EDTA [pH 8.0]). Electrophoresis was carried out in 0.5× TBE. The gel was dried, and DNA-protein complexes were visualized with a PhosphorImager using Adobe Photoshop software. Densitometry was performed with ImageQuant software.
In vitro transcription.
Reactions were performed as described previously (26). Briefly, 70 μg of HeLa cell nuclear extract (7 mg of protein/ml) and 150 ng of a synthetic target promoter containing five p53-responsive sites immediately upstream of the adenovirus E4 TATA box and chloramphenicol acetyltransferase (CAT) reporter gene (5RGCE4CAT) were mixed in a final volume of 50 μl with 60 mM KCl, 12 mM HEPES (pH 7.9), 12% glycerol, 6 mM MgCl2, 0.4 mM ribonucleoside triphosphates, 7 mM β-mercaptoethanol, p53, and T antigen as indicated and incubated at 30°C for 60 min. Control reactions were performed with 150 ng of synthetic promoter containing five GAL4-binding sites immediately upstream of the adenovirus E4 TATA box fused to CAT (5GAL4E4CAT) (5) in the presence of 100 ng of bacterially expressed GAL4-VP16 protein. Reactions were stopped by the addition of 50 μl of stop buffer (2% SDS, 200 mM NaCl, 20 mM EDTA, 20 μg of tRNA per ml, 100 μg of proteinase K per ml) followed by incubation at 39°C for 10 min. After phenol-chloroform extraction, the RNA was ethanol precipitated. Primer extension was performed by resuspending the RNA pellet in 10 μl of annealing buffer (125 mM KCl, 25 mM Tris-HCl [pH 8.3], 1,000 cpm of radiolabeled primer) and incubating it at 70°C for 10 min and then at 39°C for 30 min. Twenty-four microliters of extension buffer (5 mM MgCl2, 50 mM KCl, 20 mM Tris HCl [pH 8.3], 0.3 mM deoxynucleoside triphosphates, 10 mM dithiothreitol, 10 U of Moloney murine leukemia virus reverse transcriptase) was then added, and the mixture was incubated for 30 min at 39°C. The reaction was stopped by the addition of 35 μl of stop buffer; primer extension products were ethanol precipitated and resuspended in 4 μl of formamide sequencing dye prior to electrophoresis on a 10% acrylamide-urea gel. The gel was dried, and primer extension products were visualized with a PhosphorImager using Adobe Photoshop software. Densitometry was performed with ImageQuant software.
RESULTS
p53-T antigen complex can specifically bind to DNA.
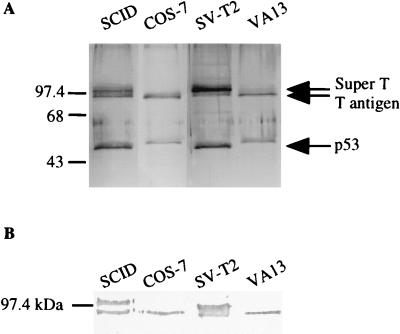
To define how T antigen inhibits p53-mediated transcription, p53 was purified from nuclear extracts prepared from two mouse (SCID and SVT2), one monkey (COS-7), and one human (WI38 VA13) cell line, all of which are stably transformed with SV40, using anti-p53 antibody Pab 421 (Fig. 1A). From the mouse cell lines, two proteins with apparent molecular masses of 90 and 100 kDa copurified with p53; from the human and monkey cell lines, only one protein of 90 kDa was copurified. All of the copurified proteins were identified as SV40 T antigen by Western blot analysis using anti-T-antigen antibody Pab 108 (Fig. 1B). From the SCID mouse cell line, the 100-kDa T antigen copurified in stoichiometric amounts with the 90-kDa form; from the SV-T2 mouse cell line, the 100-kDa protein was the major form of purified T antigen. Two lines of evidence suggest that the 100-kDa protein is a previously identified form of super T. First, the identification of this band as T antigen was further confirmed by sequence analysis of peptides resulting from the digestion of this protein (data not shown). Second, a form of super T with the same molecular mass as the 100-kDa T antigen that we observed has been identified in SV-T2 cells (40).
FIG. 1.
p53 immunopurified from SV40-transformed cells copurifies with T antigen. (A) Silver-stained SDS-polyacrylamide gel of the p53-T antigen complex purified from four SV40-transformed cell lines derived from mouse (SCID and SV-T2), human (WI38 VA13), and monkey (COS-7) cells. (B) Western blot of the p53-T antigen complex, using anti-T antigen antibody Pab 108, indicating that two forms of T antigen (regular T antigen and super T) are present in the mouse cell lines.
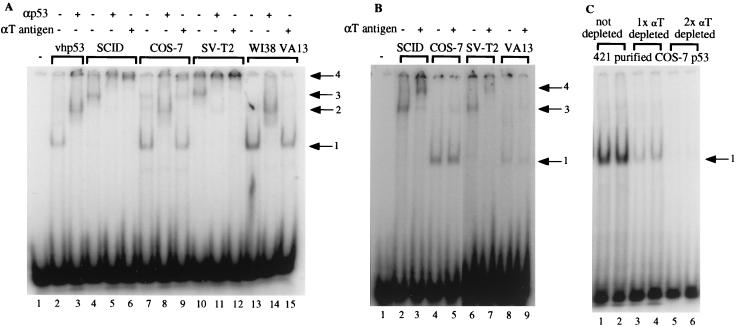
As stated above, T antigen interacts with p53 and is thought to inhibit its binding to DNA. Therefore, when the p53 samples shown in Fig. 1A were used in EMSA with a probe containing the p53 cis element identified in the RGC, the results were unexpected (Fig. 2A). Vaccinia virus-expressed human p53 (vhp53) purified from HeLa cells was used as a T-antigen-minus control. It produced a retarded p53-DNA complex (lane 2) which was supershifted by the addition of anti-p53 antibody (lane 3). p53 purified from the monkey cell line COS-7 and the human cell line WI38 VA13 formed complexes similar in mobility to those formed by vhp53 (compare lane 2 to lanes 7 and 13). These complexes could be supershifted by the addition of anti-p53 antibody but not by the addition of anti-T-antigen antibody (cf. lanes 8, 9, 14, and 15). Western blot analysis was performed to ensure that equivalent amounts of p53 were present in all samples (data not shown). This result suggested that although p53 was purified in a complex with T antigen, it could bind DNA and that when bound to DNA, the p53 was no longer complexed with T antigen. p53 purified from the mouse cell lines SCID and SVT2, however, formed a complex with the DNA probe that migrated more slowly than the control p53-DNA complex (compare lane 2 to lanes 4 and 10). The addition of both anti-p53 antibody and anti-T-antigen antibody supershifted this complex (lanes 5, 6, 11, and 12), suggesting that the mouse p53-T antigen complex remained intact during the gel electrophoresis. This result indicated that the DNA-binding ability of mouse p53 was also unaffected by T antigen, but when purified from mouse cells, the p53-T antigen complex remained intact when bound to DNA.
FIG. 2.
p53 copurified with T antigen can bind to a DNA probe in EMSA. (A) A 200-pg aliquot of radiolabeled probe containing the p53-binding site from RGC was incubated with approximately 50 ng of vhp53 purified from HeLa cells or 50 ng of each of the protein samples shown in Fig. 1A and 100 ng of antibody as indicated for 30 min at 30°C prior to electrophoresis on a 3% polyacrylamide gel. Arrows indicate positions of retarded complexes: 1, DNA-p53; 2, DNA-p53-N19 (αp53); 3, DNA-p53-T antigen; 4, DNA-p53-T antigen-N19 or Pab 108 (αT antigen). (B) Like panel A but with a probe containing the p53-binding site identified in the p21 promoter. The gels shown in panels A and B were subjected to electrophoresis for 3 and 4 h, respectively, which accounts for the more advanced migration of the bands in panel B. (C) Like panel A but with increasing amounts (50 and 100 ng) or equivalent volumes of p53-T antigen complex purified from COS-7 cells with Pab 421 either before (lanes 1 and 2) or after one (lanes 3 and 4) or two (lanes 5 and 6) rounds of immunodepletion with anti-T-antigen antibody Pab 108.
To determine if these results were peculiar to the cis element identified in the RGC, further EMSA was performed with a probe containing the p53 element identified in the p21 promoter. Similar results were obtained (Fig. 2B), again demonstrating that p53-T antigen complexes purified from the mouse cell lines SCID and SV-T2 can bind DNA in the presence of T antigen (lanes 2, 3, 6, and 7) while those purified from the monkey and human cell lines bind DNA in the absence of T antigen (lanes 4, 5, 8, and 9). It was possible that excess p53 not bound to T antigen present in the p53-T antigen preparation from the human and monkey cell lines was responsible for the p53-DNA shift observed in EMSA, even though visualization by silver staining indicated that the ratio of p53 to T antigen was approximately 1:1 (Fig. 1A). To test this possibility, Pab 421-purified p53-T antigen complex from COS-7 cells was immunodepleted with anti-T-antigen antibody. Analysis by SDS-PAGE followed by silver staining indicated that immunodepletion resulted in the equal loss of both p53 and T antigen from the sample (data not shown). Importantly, EMSA revealed no DNA binding with the twice-immunodepleted sample (Fig. 2C; compare lanes 1 and 2 to lanes 5 and 6). This result strongly argues that the p53 DNA binding observed was not due to the presence of excess unbound p53. Taken together, these results suggest the existence of alternative mechanisms for the inhibition of p53-dependent transcription by T antigen. In addition, they suggest that the mechanism for inhibition of p53 may differ between mouse cells and human and monkey cells.
The p53-T antigen complex purified from mouse cells is transcriptionally inactive.
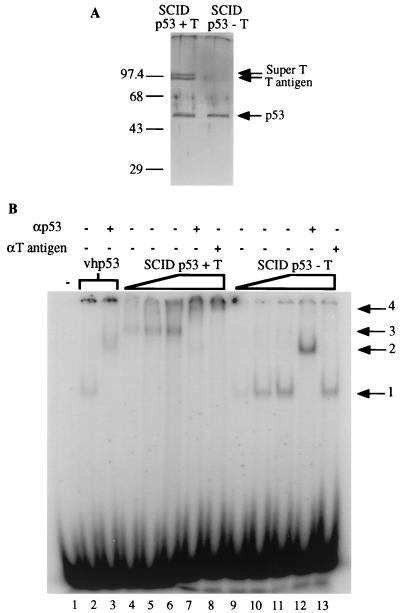
We next wanted to determine whether the mouse p53-T antigen complex, which could bind to DNA, could also support p53-dependent transcription. The complex was immunopurified from SCID cells; as a control, SCID p53 was also purified in the presence of 2 M urea in order to dissociate T antigen. The resulting p53 samples are shown in Fig. 3A. When tested by EMSA, the p53-T antigen complex resulted in the slowly migrating band in which both p53 and T antigen were present (Fig. 3B, lanes 4 to 8). In contrast, the p53 protein purified in the presence of 2 M urea produced a retarded complex that was similar in mobility to that produced by the control vhp53 (compare lanes 9 to 12 to lanes 2 and 3) and did not react with the anti-T-antigen antibody Pab 108 (lane 13).
FIG. 3.
Mouse p53 binds to DNA as a complex with T antigen in EMSA. (A) Silver-stained SDS-polyacrylamide gel of p53 purified from SCID cells with (+ T) or without (− T) T antigen. p53 was purified in the absence of T antigen by washing the p53-T antigen complex when bound to Pab 421 antibody linked to protein A-Sepharose beads with a 2 M urea solution. Sizes are indicated in kilodaltons. (B) Approximately 50 ng of vhp53 or increasing amounts (25, 50, and 75 ng) of the proteins used for panel A were incubated for 30 min at 30°C with 200 pg of radiolabeled probe containing the p53-binding site from RGC with 100 ng of antibody as indicated prior to electrophoresis on a 3% polyacrylamide gel. Arrows indicate positions of retarded complexes: 1, DNA-p53; 2, DNA-p53-N19 (αp53); 3, DNA-p53-T antigen; 4, DNA-p53-T antigen-N19 (αp53) or Pab 108 (αT antigen).
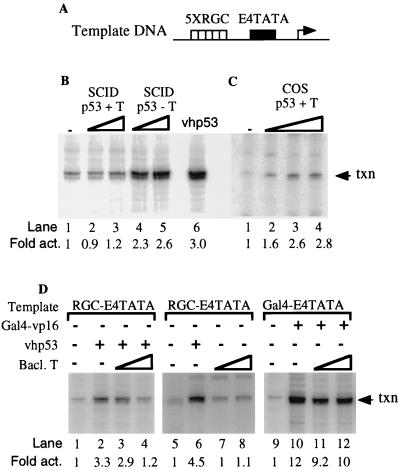
The ability of these p53 samples to stimulate transcription was next tested in an in vitro transcription assay as described previously (26). Unfractionated HeLa cell nuclear extract and a promoter template containing five p53 DNA-binding sites positioned immediately upstream of the adenovirus E4 TATA box (Fig. 4A) were incubated in the absence and presence of increasing amounts of p53. In the absence of exogenously added p53, the nuclear extract supported a low level of basal transcription (Fig. 4B, lane 1). The addition of vhp53 resulted in a threefold stimulation of transcription above this basal level (lane 6). By comparison to the control vhp53, addition of the mouse p53-T antigen complex isolated from SCID cells did not enhance transcription over basal levels (compare lane 6 to lanes 2 and 3) even though it bound DNA in EMSA. When the T-antigen proteins were removed, however, mouse p53 activated transcription 2.6-fold over basal levels (compare lane 1 to lanes 4 and 5). This indicates that loss of p53 activity was due to inhibition by T antigen and that the p53 was fully active on its own. These results were reproducible and therefore strongly suggest that in mouse cells, T antigen may inhibit p53-dependent transcription by blocking the transactivation domain of p53 and not by preventing it from binding to its cognate cis element.
FIG. 4.
Mouse p53-T antigen complex is transcriptionally inactive, while human p53-T antigen complex retains activity. (A) Schematic diagram of template DNA used in in vitro transcription reactions. (B) In vitro transcription reaction using HeLa nuclear extract (lane 1) with increasing amounts (300 and 600 ng) of SCID p53 purified with (lanes 2 and 3) or without (lanes 4 and 5) T antigen. vhp53 (200 ng) was used as a positive control (lane 6). Transcription (txn) products were subjected to electrophoresis on a 10% acrylamide-urea gel and visualized with a PhosphoImager using Adobe Photoshop software. (C) Like panel B but with increasing amounts (250, 500, and 750 ng) of COS-7 p53-T antigen complex (lanes 2 to 4). (D) Like panel B but with 100 ng of vhp53 (lanes 2 to 4 and 6) and increasing amounts (100 [lanes 3 and 7] and 400 [lanes 4 and 8] ng) of baculovirus-expressed T antigen (Bacl. T). In lanes 9 to 12, in vitro transcription was performed with a promoter similar to that used for panel A but with the RGC elements replaced with GAL4-binding sites. GAL4-VP16 (100 ng) was added to activate transcription (lanes 10 to 12) in the presence of increasing amounts (100 and 400 ng) of baculovirus-expressed T antigen (lanes 11 and 12).
Interestingly, COS-7 p53, which also purified in a complex with T antigen (Fig. 1) but bound DNA independently of T antigen (Fig. 2), retained the ability to activate p53-dependent transcription 2.8-fold over basal levels (Fig. 4C; compare lane 1 to lanes 2 to 4). This surprising result suggests that in monkey cells, T antigen may not necessarily inhibit p53 function despite its association with p53. T antigen can itself function as an activator of transcription from a simple promoter consisting of certain TATA elements and one upstream transcription factor-binding site (13, 33). Therefore, we performed assays in the presence of baculovirus-expressed purified T antigen to determine if T antigen alone could activate transcription in this system (Fig. 4D). p53-dependent transcription was inhibited when a fourfold excess of baculovirus-expressed T antigen was added to vhp53 (cf. lanes 2 and 4). The effect of T antigen on basal transcription was tested in assays using the amount of baculovirus T antigen sufficient to inhibit p53-dependent transcription. Transcription was not affected by the addition of T antigen alone (cf. lanes 7 and 8), suggesting that the transcription observed in Fig. 3C is p53 dependent and not T antigen dependent. Finally, the effect of T antigen on activated transcription driven by bacterially expressed GAL4-VP16 was also tested in assays using template DNA containing five upstream GAL4-binding sites fused to the adenovirus TATA box. The addition of GAL4-VP16 to these reactions resulted in a 12-fold increase in transcription levels. This transactivation was minimally affected by the addition of baculovirus T antigen (compare lane 10 to lanes 11 and 12). Therefore, unbound T antigen does not affect transcription in these assays.
DNA binding by a p53-T antigen complex is species dependent.
Next we wanted to determine what contributed to the different activities of the p53-T antigen complexes purified from either the mouse or the human and monkey cell lines. The difference may depend on the p53 present in these cell lines. At the amino acid level, monkey p53 is 96% identical to human p53, while mouse p53 is only 79% identical to human p53. Therefore, differences at the amino acid level or in protein modification may account for distinct interactions of mouse and human or monkey p53 with T antigen, with subsequent effects on p53-T antigen complex activity. Alternatively, the difference may be due to the presence of super T in the mouse cell lines but not in the human and monkey cell lines.
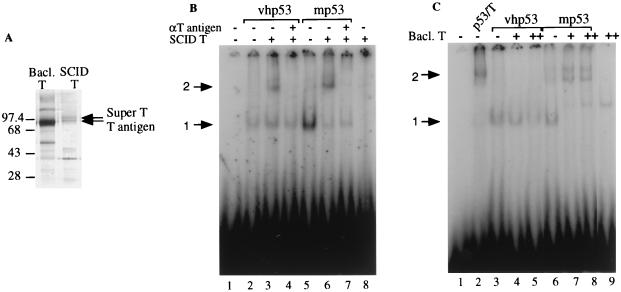
To address this issue, we performed EMSA with vhp53 and mouse p53 purified from SCID cells in the absence of T antigen. These samples were incubated with either a baculovirus-expressed T antigen (a 90-kDa protein) or T antigen purified from mouse SCID cells (90- and 100-kDa proteins [Fig. 5A]). Ideally, a preparation of the 100-kDa form of super T alone would have been used in these experiments, but to our knowledge no clone that only makes super T is available. Results are shown in Fig. 5B and C. The addition of T antigen/super T purified from SCID cells to mouse p53 resulted in the generation of a supershifted complex (Fig. 5B, lane 6). This complex was abolished by the addition of anti-T-antigen antibody (lane 7). T antigen/super T alone did not bind to DNA (lane 8). The addition of the same amount of T antigen/super T to human p53 also resulted in the generation of a supershifted complex (lane 3). However, we consistently observed that less human p53 than mouse p53 was supershifted under these conditions (cf. lanes 3 and 6), suggesting that T antigen/super T can form a DNA-binding complex more efficiently with mouse p53 than with human p53.
FIG. 5.
Both mouse p53 and mouse T/super T contribute to the formation of a p53-T antigen complex that can bind to DNA. (A) Silver-stained SDS-polyacrylamide gel of baculovirus-expressed T antigen purified with anti-T-antigen antibody Pab 108 from SF21 insect cells (Bacl. T) and T antigen purified from stably transformed mouse SCID cells in the absence of p53 as described in Materials and Methods (SCID T). Sizes are indicated in kilodaltons. (B) Approximately 50 ng of vhp53 or 100 ng of SCID p53 (no T antigen present; mp53) was incubated for 30 min at 30°C with 200 pg of radiolabeled probe containing the p53 binding site from RGC, 50 ng of T antigen purified from SCID cells, and 100 ng of antibody (αT) as indicated prior to electrophoresis on a 3% polyacrylamide gel. DNA-protein complexes were visualized with a PhosphoImager using Adobe Photoshop software. Arrows indicate positions of retarded complexes: 1, DNA-p53; 2, DNA-p53-T antigen. (C) Like panel B but with 100 ng of SCID p53-T antigen complex (lane 2), 100 ng of vhp53, or 100 ng of SCID p53 (no T antigen present; mp53) incubated with 50 or 100 ng of baculovirus-expressed T antigen as indicated. Arrows indicate positions of retarded complexes: 1, DNA-p53; 2, DNA-p53-T antigen.
When baculovirus T antigen was used in these experiments, a supershifted complex was formed only with mouse p53 (Fig. 5C; compare lane 6 to lanes 7 and 8). Two slowly migrating complexes, possibly due to incomplete renaturation of the mouse p53 after the harsh conditions of the purification procedure, were observed. The slower-migrating complex migrated at a position similar to that of the complex formed by the mouse p53-T antigen complex immunopurified from the SCID cell line (lane 2), and addition of both anti-p53 antibody and anti-T-antigen antibody supershifted this slower-migrating complex (data not shown). In comparison to mouse p53, DNA binding of human p53 was inhibited by baculovirus T antigen (compare lane 3 to lanes 4 and 5 [1.4- and 1.9-fold reduction in binding, respectively]). Baculovirus T antigen alone did not bind specifically to DNA (lane 9). Therefore, in these in vitro assays, the DNA-binding ability of human p53 was inhibited by the addition of baculovirus-expressed T antigen, in accordance with previous observations (1). This result concurs with the data from in vitro transcription assays using vhp53, where transcription was inhibited by the addition of baculovirus-expressed T antigen (Fig. 4D, lanes 2 to 4). The fact that transcription and DNA binding are observed with p53-T antigen complex purified from COS-7 monkey cells suggests that there may be an important difference between this in vivo complex and that prepared in vitro by using virally expressed proteins. In addition, the molar ratio of T antigen to p53 may be higher in the in vitro complex, therefore affecting its activity.
The EMSA results presented in Fig. 5 suggest that mouse p53 is different from human p53 in its ability to interact with T antigen. Indeed, this difference was also noted when we attempted to purify p53-T antigen complex from monkey and mouse cells by using anti-T-antigen antibody. The complex remained intact when purified under stringent conditions from the SCID mouse cell line but dissociated when purified from the monkey COS-7 cell line (data not shown), suggesting that T antigen binds strongly to mouse p53 and weakly to human p53. Therefore, mouse p53, in addition to mouse T/super T, may contribute to the formation of a DNA-binding p53-T antigen complex.
An active conformation of p53 is required for p53-T antigen complex DNA binding.
Wild-type p53 can be converted from an inactive or latent state to an active state that binds DNA (17). The latent state is thought to be dependent on a C-terminal negative regulatory domain within p53 that is proposed to interact with a motif in the core of the p53 tetramer, thereby forming a conformationally inactive complex (18). A number of conditions that modify the C terminus of p53 which convert p53 from a latent to an active state have been described. These include anti-p53 antibody Pab 421 (17, 18), phosphorylation (16, 17), acetylation (14), short single strands of DNA (19), and the redox/repair protein Ref-1 (20). The biological relevance of this model of activation is demonstrated by the fact that UV-induced activation of the transcriptional function of p53 does not require an increase in p53 protein levels (18).
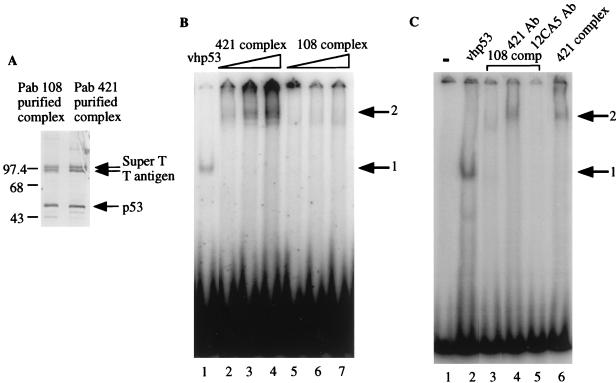
The p53-T antigen complexes used in the previous experiments were purified by using Pab 421 antibody. This antibody recognizes an epitope in the C terminus of p53 and is thought to convert p53 from its latent to its active state, thereby significantly increasing its DNA-binding activity (18, 31). Consequently, we wanted to determine if this method of purification affected the DNA-binding ability of the complex. First, the mouse p53-T antigen complexes eluted from Pab 421 beads by using a 50% EG solution and 421 peptide were compared. In both cases, similar binding affinities were observed in EMSA (data not shown), indicating that binding was not dependent on the presence of 421 peptide. Next, we attempted to purify the complex by using p53-specific antibodies D0-1 and Pab 1801 (Santa Cruz), both of which recognize amino-terminus epitopes in human p53. However, in each case control experiments indicated that the stringent conditions required to elute p53 from these antibodies resulted in DNA-binding-deficient protein. Therefore, the complex was purified by using anti-T-antigen antibody Pab 108, which is not known to activate p53, and eluted with 50% EG solution, which does not affect the DNA-binding ability of p53 (36a). This method of purification resulted in a complex similar to that purified with Pab 421 when analyzed by SDS-PAGE followed by silver staining (Fig. 6A). However, the complex purified with Pab 108 had a significantly reduced affinity for DNA compared to that purified with Pab 421 (Fig. 6B; compare lanes 2 to 4 to lanes 5 to 7), suggesting that an active conformation of p53 may be required for p53-T antigen complex DNA-binding ability. To test if this was the case, we incubated the Pab 108-purified complex with Pab 421 prior to EMSA and then observed DNA binding (Fig. 6C; cf. lanes 3 and 4). The resulting shifted band migrated parallel with that produced by the Pab 421-purified complex (cf. lanes 4 and 6) and could be supershifted by the addition of anti-p53 and anti-T-antigen antibodies (data not shown). The activation of binding was not observed following incubation with an unrelated antibody (lane 5) or with bovine serum albumin (data not shown). These results strongly argue that the DNA-binding ability of the complex is dependent on the active conformation of p53.
FIG. 6.
Purification of p53-T antigen complex by Pab 421 is required for DNA-binding ability. (A) Silver-stained SDS-polyacrylamide gel of p53-T antigen complex purified from mouse SCID cells by using Pab 108 (anti-T antigen) or Pab 421 (anti-p53). Sizes are indicated in kilodaltons. (B) vhp53 (50 ng), increasing amounts (20, 40, and 60 ng) of Pab 421-purified SCID p53-T antigen complex, or increasing amounts (50, 75, and 100 ng) of Pab 108-purified complex were incubated for 30 min at 30°C with 200 pg of radiolabeled probe containing the p53-binding site from RGC prior to electrophoresis on a 3% polyacrylamide gel. Arrows indicate positions of retarded complexes: 1, DNA-p53; 2, DNA-p53-T antigen. (C) Like panel B but with 50 ng of vhp53, 100 ng of Pab 108-purified SCID p53-T antigen complex, or 50 ng of Pab 421-purified complex as indicated. In lanes 4 and 5, Pab 108-purified complex (108 comp) was preincubated for 20 min at 4°C with 5 μg of Pab 421 or 5 μg of antibody 12CA5 in 0.1 M D buffer, as indicated.
DISCUSSION
In this paper we present biochemical evidence that p53 isolated in a complex with T antigen can bind to DNA, provided that p53 is in an active conformation. We also demonstrate that p53 exhibits species-specific interactions with T antigen. In vitro, T antigen apparently dissociates from human p53 in the presence of DNA, resulting in a transcriptionally active form of p53 bound to DNA. In contrast, mouse p53 binds DNA in a complex with T antigen, resulting in transcriptionally inactive p53. Our results indicate that this difference may be attributable to a difference between mouse and human p53 and, in addition, possibly to the presence of super T in the mouse cell lines. On the basis of our results, we conclude that T antigen, under certain conditions, may repress p53-dependent transcription by a mechanism in which the transactivation domain of p53 is inhibited while DNA binding is unaffected. This mechanism is different from the previous proposed mechanism in which T antigen inhibits p53 by preventing it from binding DNA (1, 36). Therefore, similar to the inhibition of p53 by MDM2, it seems that T antigen may inhibit p53 by more than one mechanism.
Long et al. (27) have shown, using nuclear extracts in EMSA, that a small fraction of p53 from SV40-transformed monkey and rat cells specifically binds to DNA as a complex with T antigen. Our results, however, differ from theirs in that we observe significant DNA binding by p53-T antigen complex purified from mouse cells. The reason for this discrepancy may be that we used purified p53 activated by Pab 421 antibody as opposed to crude nuclear extracts. However, we did not observe DNA binding by p53-T antigen complex when using crude nuclear extracts in EMSA, either in the presence or in the absence of Pab 421 (data not shown). This may have been because the concentration of p53 in the crude extract was too low for Pab 421 activation to occur or for DNA binding to be observed.
The observation that the transactivation domain of mouse p53 may be inhibited by T antigen while DNA binding is unaffected has several implications. First, the surface of p53 required for DNA binding must be accessible in the mouse p53-T antigen complex. The region of p53 that is required for T-antigen binding has been mapped to residues 126 to 218, a region within the core domain required for DNA binding (34). However, resolution of the crystal structure of a human p53 core domain-DNA complex has localized the region of p53 directly contacted by DNA to three structural elements which include the H2 helix (amino acids [aa] 278 to 286), the L1 loop (aa 112 to 124), and the L3 loop (aa 236 to 251) (7). None of these elements overlap with the region required by p53 to interact with T antigen. Therefore, it is possible that p53 can concurrently form a complex with T antigen and bind DNA. Second, interaction of T antigen with the DNA-binding domain of p53 must either alter or block the activation domain of p53. Thus, it appears that the alteration of one domain on p53 may affect the function of the others.
Our results suggest that T antigen can form a transcriptionally inactive DNA-binding complex with mouse p53. The adenovirus E1B 55K protein also forms an inhibitory complex with p53 without displacing p53 from its cognate cis element (41). p53-mediated transcription is prevented because E1B 55K is a repressor of transcription which, via its interaction with p53, is targeted to p53-responsive genes. The mechanism of repression adopted by T antigen, however, is unlikely to be identical to that adopted by E1B 55K, as T antigen can function as a transcriptional activator (13, 33). T antigen is thought to activate transcription through direct interactions with both the basal transcription complex and upstream-bound transcription factors and may act like a component of TFIID by augmenting, or replacing, a function of TAFII250 (9). Additionally, T antigen has been shown to enhance formation of TATA-binding protein–TFIIA complex on certain TATA elements (10). The fact that T antigen does not activate transcription when tethered to promoter DNA by p53 indicates that its activation function, as previously suggested (42), may be conformation dependent and that binding to p53 may alter its conformation. Consistent with this view, GAL4-T antigen fusion protein has been shown to be transcriptionally inactive when brought to a target promoter bearing GAL4 DNA-binding sites (13). When bound as a complex with p53 to DNA, T antigen may inhibit p53-dependent transcription by steric hindrance of the activation domain of p53 or by deleteriously affecting the assembly or conformation of the basal transcription machinery. In addition, T antigen may interfere with the interaction of p53 with coactivators of transcription such as p300 (14).
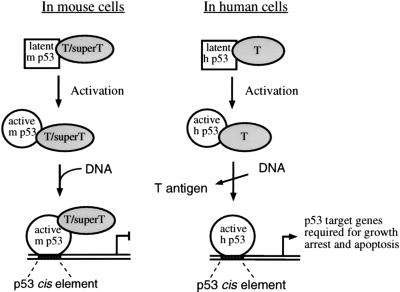
Mouse cells are more readily transformed with SV40 than are human cells. Using the data presented in this paper, we propose a model which may explain the molecular mechanism of this difference (Fig. 7). In mouse cell lines, latent p53 and T antigen/super T form a complex that cannot bind DNA. Upon activation of p53, a conformational change in p53 may alter the p53-T antigen-super T complex such that DNA binding occurs. p53-responsive promoters would therefore be blocked by the transcriptionally inactive complex, and the tumor suppressor function of p53 would be reduced or lost. In human cell lines, latent p53 and T antigen also form a complex that is unable to bind to DNA. Again, upon activation of p53, a conformational change in p53 may alter the p53-T antigen complex such that DNA binding occurs. However, because human p53 appears to bind to DNA independently of T antigen and to retain the ability to activate transcription, p53 function would not be completely lost. Therefore, p53 target genes that are required for growth arrest and apoptosis would be activated in human cells containing T antigen and an active form of p53. This model relies on the fact that p53 must adopt an active conformation. Significantly, cellular factors that may activate p53 similarly to Pab 421 antibody, including protein kinases (16, 17), coactivator p300 (14), and the redox/repair protein Ref-1 (20), are found in the cells.
FIG. 7.
Proposed model of p53 inhibition by T antigen in mouse (m) and human (h) cells.
It is tempting to speculate that our model may also explain why SV40 is known to cause tumors in rodents (3, 8) but has not proven to do so in humans. Although recently this virus has been linked to some human cancers, a cause-and-effect relationship has not been established (4). Finally, it will be of interest to determine whether the differences reported here between species-specific forms of p53 are also applicable to interactions with other viral and cellular proteins. If so, such a distinction between rodent and primate p53 could have significant ramifications for the use of rodent models of transformation.
ACKNOWLEDGMENTS
We thank Arnold Berk and Carol Prives for providing cells and viruses, and we thank Francisco Renteria for help with cell culture. We also thank Frances Sladek, Noelle L’Etoile, Renee Yew, and members of the Liu laboratory for many helpful discussions and valuable comments on the manuscript.
This work was supported by grants CA75180-01 (X.L.) from the National Cancer Institute and DAMD17-96-1-6076 (X.L.) from U.S. Army Breast Cancer Research Program.
REFERENCES
- 1.Bargonetti J, Reynisdottir I, Friedman P, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 1992;6:1886–1898. doi: 10.1101/gad.6.10.1886. [DOI] [PubMed] [Google Scholar]
- 2.Butel J S, Wong C, Evans B K. Fluctuation of simian virus 40 (SV40) super T-antigen expression in tumors induced by SV40-transformed mouse mammary epithelial cells. J Virol. 1986;60:817–821. doi: 10.1128/jvi.60.2.817-821.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carbone M, Rizzo P, Pass H I. Simian virus 40, poliovaccines and human tumors: a review of recent developments. Oncogene. 1997;15:1877–1888. doi: 10.1038/sj.onc.1201375. [DOI] [PubMed] [Google Scholar]
- 4.Carbone M, Rizzo P, Grimley P M, Procopio A, Mew D J Y, Shridhar V, de Bartolomeis A, Esposito V, Giuliano M T, Steinberg S M, Levine A, Giordano A, Pass H I. Simian virus-40 large T-antigen binds p53 in human mesotheliomas. Nat Med. 1997;3:908–912. doi: 10.1038/nm0897-908. [DOI] [PubMed] [Google Scholar]
- 5.Carey M, Leatherwood J, Ptashne M. A potent Gal4 derivative activates transcription at a distance in vitro. Science. 1990;247:710–712. doi: 10.1126/science.2405489. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Verderame M, Lo A, Pollack R. Nonlytic simian virus 40-specific 100K phosphoprotein is associated with anchorage-independent growth in simian virus 40-transformed and revertant mouse cell lines. Mol Cell Biol. 1981;1:994–1006. doi: 10.1128/mcb.1.11.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 8.Cicala C, Pompetti F, Carbone M. SV40 induces mesotheliomas in hamsters. Am J Pathol. 1993;142:1524–1533. [PMC free article] [PubMed] [Google Scholar]
- 9.Damania B, Alwine J C. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 10.Damania B, Lieberman P L, Alwine J C. Simian virus 40 large T antigen stabilizes the TATA-binding protein–TFIIA complex on the TATA element. Mol Cell Biol. 1998;18:3926–3935. doi: 10.1128/mcb.18.7.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcript initiation by RNA Pol II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Diery W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 13.Gruda M C, Zabolotny J M, Xiao J H, Davidson I, Alwine J C. Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol Cell Biol. 1993;13:961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 15.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 16.Hupp T R, Lane D P. Regulation of the cryptic sequence-specific DNA-binding function of p53 by protein kinases. Cold Spring Harbor Symp Quant Biol. 1994;59:195–206. doi: 10.1101/sqb.1994.059.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Hupp T R, Meek D W, Midgley C A, Lane D P. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 18.Hupp T R, Sparks A, Lane D P. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 19.Jayaraman L, Prives C. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 20.Jayaraman L, Murthy K G K, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 21.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 22.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 23.Levine A J, Momand J, Finlay C A. The p53 tumor suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 24.Levine A J. p53, the cellular gatekeeper for the growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 25.Levitt A, Chen S, Blanck G, George D, Pollack R E. Two integrated partial repeats of simian virus 40 together code for a super-T antigen. Mol Cell Biol. 1985;5:742–750. doi: 10.1128/mcb.5.4.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Berk A J. Reversal of in vitro p53 squelching by both TFIIB and TFIID. Mol Cell Biol. 1995;15:6474–6478. doi: 10.1128/mcb.15.11.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long S-B, Ho H-Y, Chen C-L, Lai M-D. Complex of simian virus large T antigen and p53 can bind DNA specifically. Anticancer Res. 1995;15:1375–1380. [PubMed] [Google Scholar]
- 28.Lovett M, Clayton C E, Murphy D, Rigby P W J, Smith A E, Chaudry F. Structure and synthesis of a simian virus 40 super-T antigen. J Virol. 1982;44:963–973. doi: 10.1128/jvi.44.3.963-973.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.May E, Kress M, Daya-Grosjean L, Monier R, May P. Mapping of the viral mRNA encoding a super-T antigen of 115,000 daltons expressed in simian virus 40-transformed rat cell lines. J Virol. 1981;37:24–35. doi: 10.1128/jvi.37.1.24-35.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 31.Mundt M, Hupp T, Fritsche M, Merkle C, Hansen S, Lane D, Groner B. Protein interactions at the carboxyl terminus of p53 result in the induction of its in vitro transactivation potential. Oncogene. 1997;15:237–244. doi: 10.1038/sj.onc.1201174. [DOI] [PubMed] [Google Scholar]
- 32.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 33.Rice P W, Cole C N. Efficient transcriptional activation of many simple modular promoters by simian virus 40 large T antigen. J Virol. 1993;67:6689–6697. doi: 10.1128/jvi.67.11.6689-6697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruppert J M, Stillman B. Analysis of a protein-binding domain of p53. Mol Cell Biol. 1993;13:3811–3820. doi: 10.1128/mcb.13.6.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 36.Segawa K, Minowa A, Sugasawa K, Takano T, Hanaoka F. Abrogation of p53-mediated transactivation by SV40 large T antigen. Oncogene. 1992;8:543–548. [PubMed] [Google Scholar]
- 36a.Sheppard, H. M., and X. Liu. Unpublished results.
- 37.Smith A F, Smith R, Pouche E. Characterization of different tumor antigens present in cells transformed by simian virus 40. Cell. 1979;18:335–346. doi: 10.1016/0092-8674(79)90053-9. [DOI] [PubMed] [Google Scholar]
- 38.Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh I M, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 39.Thut C J, Goodrich J A, Tijan R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson V G, Williams R L. Origin binding by a 100,000-dalton super-T antigen from SVT2 cells. J Virol. 1985;56:102–109. doi: 10.1128/jvi.56.1.102-109.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 42.Zhu J, Abate M, Rice P W, Cole C. The ability of simian virus 40 large T antigen to immortalize primary mouse embryo fibroblasts cosegregates with its ability to bind p53. J Virol. 1991;65:6872–6880. doi: 10.1128/jvi.65.12.6872-6880.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]