Abstract
Hematopoietic cells require cytokine-initiated signals for survival as well as proliferation. The pathways that transduce these signals, ensuring timely regulation of cell fate genes, remain largely undefined. The NFIL3 (E4BP4) transcription factor, Bcl-xL, and constitutively active mutants of components in Ras signal transduction pathways have been identified as key regulation proteins affecting murine interleukin-3 (IL-3)-dependent cell survival. Here we show that expression of NFIL3 is regulated by oncogenic Ras mutants through both the Raf–mitogen-activated protein kinase and phosphatidylinositol 3-kinase pathways. NFIL3 inhibits apoptosis without affecting Bcl-xL expression. By contrast, Bcl-xL levels are regulated through the membrane proximal portion in the cytoplasmic domain of the receptor (βc chain), which is shared by IL-3 and granulocyte-macrophage colony-stimulating factor. Activation of either pathway alone is insufficient to ensure cell survival, indicating that multiple independent signal transduction pathways mediate the survival of developing B-lymphoid cells.
Genes that regulate programmed cell death are highly conserved among organisms as diverse as insects, nematodes, and mammals. In the nematode Caenorhabditis elegans, each cell death during development is controlled through the coordinated action of three gene products, CED-3, CED-4, and CED-9 (13), while in mammalian cells, this function is performed by a group of CED-related proteins such as the caspases, Apaf-1, and Bcl-2 family members (25, 29, 40). Indeed, the activation of caspase-3 (CPP32) is essential for programmed cell death induced by growth factor deprivation in several cytokine-dependent hematopoietic cells, including murine interleukin-3 (IL-3)-dependent cells such as Baf-3 pro-B lymphocytes (30). Moreover, enforced expression of the IL-3-regulated Bcl-xL gene protects Baf-3 cells from apoptosis caused by IL-3 depletion (27), suggesting that Bcl-xL regulation of caspase-3 activity is a major mechanism contributing to programmed cell death in this cell system.
Genetic programs controlling apoptosis also share upstream elements that determine the fate of specific cell types. For instance, the fates of the sister cells of specific neurons (NSM neurons) in C. elegans are regulated by two transcription factors, CES-1 and CES-2 (12). CES-2 is a member of the basic region/leucine zipper (bZIP) transcription factor superfamily and is thought to act through its consensus binding sequence to negatively regulate expression of the ces-1 gene, causing cells to undergo apoptosis (28). Recently, we demonstrated that two CES-2-related transcription factors, E2A-HLF and NFIL3 (also called E4BP4), play critical roles in the regulation of apoptosis in mammalian pro-B lymphocytes.
The E2A-HLF chimeric transcription factor is expressed by human pro-B-cell leukemias harboring the t(17;19)(q22;p13) translocation (14, 17). In this chimera, the transactivation domain of E2A (also called E12/E47) is fused to the bZIP domain of hepatic leukemia factor, which has high homology with CES-2. E2A-HLF binds to the same DNA sequence as does CES-2 and forms homodimers that trans activate gene expression (15, 17, 18, 20, 21). A dominant-negative suppressor of E2A-HLF induces apoptotic cell death in leukemic cells with the t(17;19) translocation. Moreover, E2A-HLF markedly delays apoptosis caused by growth factor deprivation in murine IL-3-dependent pro-B lymphocytes including Baf-3 and FL5.12 cells (19), suggesting that the chimeric protein contributes to leukemogenesis by subverting the transcriptional regulation system of programmed cell death in mammalian B-cell precursors. Recently, we showed that a related bZIP protein, NFIL3 (also called E4BP4), is a cytokine-regulated mammalian transcription factor highly homologous to CES-2 (16). Originally isolated as a transcription factor binding to the adenovirus E4 promoter sequence with trans-repressor activity (9), NFIL3 was later identified as a trans activator of IL-3 gene expression in T cells (39). This protein also has a high degree of homology with CES-2 and avidly binds to the same DNA sequence recognized by CES-2 and E2A-HLF (9, 16). In Baf-3 and FL5.12 cells, NFIL3 is a delayed early transcription factor induced by IL-3 stimulation. Moreover, enforced expression of NFIL3 in FL5.12 cells delays apoptosis caused by IL-3 deprivation without promoting cell division (16), indicating that induction of NFIL3 is one of the mechanisms through which IL-3 suppresses apoptosis.
Previous studies have demonstrated that mutations leading to constitutive Ras activation promote the development of hematopoietic malignancies (reviewed in reference 22). We have also demonstrated that constitutively active Ras proteins block apoptosis induced by IL-3 deprivation in murine IL-3-dependent pro-B cells (33). Moreover, the membrane distal region of the cytoplasmic domain of the signaling receptor (common β chain or βc) shared by IL-3 and the granulocyte-macrophage colony-stimulating factor (GM-CSF) is required for both the survival and the activation of Ras in IL-3-dependent hematopoietic cells (23). Most recently, we identified the Raf–mitogen-activated protein kinase (MAPK) and rapamycin-wortmannin-sensitive pathways (most likely phosphatidylinositol 3-kinase [PI3-K] dependent) as pathways downstream of oncogenic Ras in the prevention of apoptosis in cytokine-deprived hematopoietic cells (24).
Here, we delineate two apparently independent pathways that regulate apoptosis in cytokine-deprived Baf-3 cells. One emanates from the membrane proximal portion of the βc chain and is involved in the rapid regulation of Bcl-xL expression; the other involves the NFIL3 transcription factor downstream of oncogenic Ras mutants. These data indicate that multiple pathways are involved in the cytokine-mediated survival of pro-B lymphocytes.
MATERIALS AND METHODS
Cell culture and cell growth assay.
Mouse IL-3-dependent Baf-3 pro-B lymphocytes were cultured in RPMI 1640 medium containing 10% fetal calf serum, 20 mM HEPES, 50 μM 2-mercaptoethanol, and 0.5% 10T1/2 conditioned medium as a source of murine IL-3. In some experiments, recombinant mouse IL-3 (Wako Pure Chemicals, Osaka, Japan) was used at concentrations described in the figure legends. Stable transfectants of truncated forms of the human GM-CSF receptor and Ras mutants were established in Baf-3 cells as described previously (23, 33). Transfectants were maintained in medium containing either 1 mg of G418 per ml of 200 μg of hygromycin per ml. To deplete IL-3, cells were washed twice with either IL-3-free growth medium or complete serum-free AIM-V medium (Gibco-BRL). Viable cell counts were determined by trypan blue dye exclusion in triplicate assays. BOSC23 cells, an ecotropic retrovirus packaging cell line, were purchased from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum.
Retrovirus-mediated gene expression in Baf-3 cells.
To construct a control CD8-expressing vector plasmid (pMX/IRES-CD8) for retroviral gene transfer from the pMX retroviral vector (a gift of T. Kitamura) (31), we inserted an IRES-CD8 cassette in which the mouse CD8 (lyt-2) cDNA was fused in frame to the internal ribosomal entry site sequence. Particular genes were expressed by inserting their cDNAs immediately after the 5′ long terminal repeat sequence (Fig. 1A). The dominant-negative NFIL3 mutant contains a defective NFIL3 basic domain characterized by substitution of nine of the amino acid residues critical for DNA binding (YWEKRRKNNEAAKRSRE mutated to YWEQSQQYSEPPQRSRE; single-letter code). Retrovirus was made by the method of Kitamura and coworkers (31) with minor modifications. Briefly, 3 × 106 BOSC23 cells were transfected with 6 μg of plasmid vector DNA with Lipofectamine (Gibco-BRL) according to the manufacturer’s instructions. Two days later, 2 × 105 Baf-3 cells were infected with retrovirus by adding the conditioned medium from transfected BOSC23 cells. The infection was performed in dishes coated with recombinant human fibronectin fragment (Retronectin; TaKaRa, Tokyo, Japan) at a concentration of 20 μg/cm2. After 24 h, the cells were separated from the dish with cell dissociation buffer (Gibco-BRL) and transferred to a new flask. Cells were cultured until the total number reached more than 3 × 106 (usually after 2 days), and the percentage of CD8-positive cells was determined by flow cytometry to monitor infection efficiency. CD8-positive cells were labeled with magnetic microbeads conjugated with the CD8 monoclonal antibody and isolated on MACS separation columns (Miltenyi Biotec, Gladbach, Germany) according to the manufacturer’s instructions. The selection procedure was repeated until more than 95% of cells were shown to be positive for CD8 by flow cytometry.
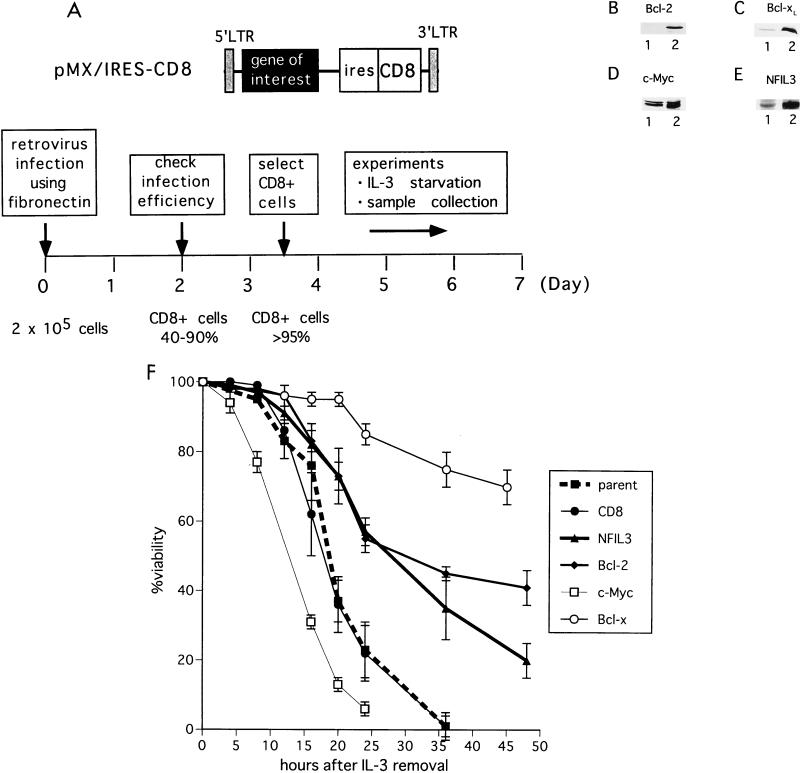
FIG. 1.
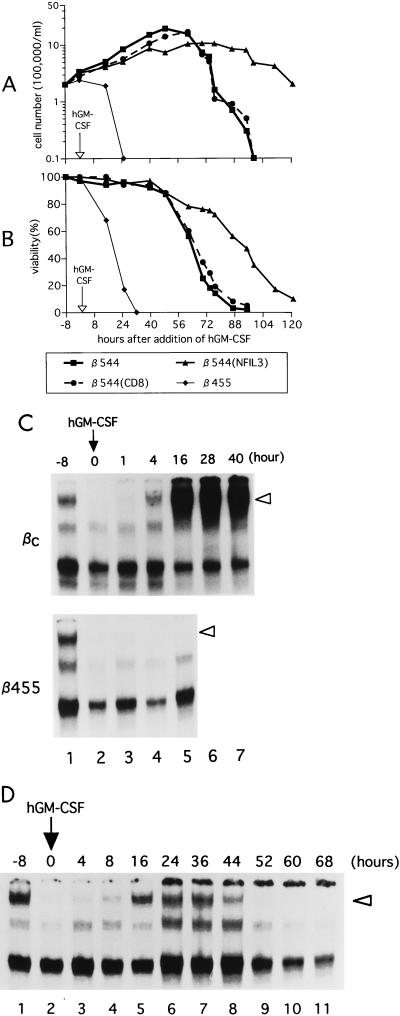
Schematic representation of retrovirus transduction experiments. (A) Structure of the retroviral vector used in this study (top); (ires, internal ribosomal entry site). Retrovirus-mediated gene transfer was followed by cell selection with CD8 magnetic beads (bottom) (also see Materials and Methods). LTR, long terminal repeat. (B to E) Immunoblot analysis. Cell lysates from parental Baf-3 cells (lanes 1 [B to E]) and cells infected with retrovirus expressing Bcl-2 (B), Bcl-xL (C), c-Myc (D), and NFIL3 (E) (each in lane 2) were analyzed with specific antibodies recognizing each protein. (F) Parental Baf-3 cells and cells expressing CD8 only (control), NFIL3, Bcl-2, c-Myc, or Bcl-xL were cultured in IL-3-free medium; cell viability was determined at each time point by trypan blue dye exclusion. Each value is the mean (± standard deviation) of three independent experiments.
Electrophoretic mobility shift analysis.
Binding reactions in the electrophoretic mobility shift assay (EMSA) were performed by incubating 12 μg of nuclear protein lysate at 30°C for 15 min with a 32P-end-labeled DNA oligonucleotide probe (2 × 104 cpm) containing the hepatic leukemia factor consensus binding site sequence (5′-GCTACATATTACGTAACAAGCGTT-3′) in 12% glycerol–12 mM HEPES (pH 7.9)–4 mM Tris (pH 7.9)–133 mM KCl–1.5 μg of sheared calf thymus DNA–300 μg of bovine serum albumin per ml as previously described (37). Complexes were tested for binding to antisera by incubating 1 μl of polyvalent immune rabbit antiserum to the nuclear protein lysates at 4°C for 30 min prior to the DNA binding reaction. Nondenaturing polyacrylamide gels containing 4% acrylamide and 2.5% glycerol were prerun at 4°C in a high-ionic-strength Tris-glycine buffer for 30 min and run at 50 mA for approximately 45 min. The gel was then dried under vacuum and analyzed by autoradiography.
Immunoblot analysis and antibodies.
Cells were solubilized in Nonidet P-40 lysis buffer (150 mM NaCl, 1.0% Nonidet P-40, 50 mM Tris, pH 8.0), and total cellular proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After wet electrotransfer of the proteins onto polyvinylidene difluoride membranes, proteins were detected by appropriate antibodies according to standard procedures. The blots were then stained with primary antibodies followed by horseradish peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin secondary antibodies and subjected to chemiluminescence detection according to the manufacturer’s instructions (Amersham). The following antibodies were used for immunodetection. Anti-Bcl-x polyclonal antibodies were purchased from Transduction Laboratories (Lexington, Ky.), anti-Bcl-2 (ΔC21) and anti-c-Myc (N-262) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.), and anti-Akt and anti-phosphorylated (Ser-473) Akt-specific antibodies were purchased from New England BioLabs (Beverly, Mass.). Anti-NFIL3 antibodies were previously described (16).
RNA extraction and Northern blot analysis.
Total cellular RNA was isolated with the RNeasy kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). RNA samples (20 μg each) were separated by electrophoresis in 1% agarose gels containing 2.2 M formaldehyde, transferred to nylon membranes, and hybridized with appropriate probes according to standard procedure.
RESULTS
NFIL3 delays apoptosis in Baf-3 cells deprived of IL-3.
Clonal variation and difficulties in isolating stable clones have impeded studies in cytokine-dependent cells deprived of growth factors. Thus, we established a retrovirus-mediated gene expression system that allows mass transfer of genes followed by rapid selection without prolonged culture in antibiotics (see Materials and Methods) (Fig. 1A). With this procedure, more than 95% of infected Baf-3 cells expressed high levels of CD8 proved by flow cytometry (data not shown). Immunoblot analysis documented protein expression in these cells (Fig. 1B to E).
Removal of IL-3 from the culture medium induced apoptosis in CD8-expressing Baf-3 cells, which died at essentially the same rate as Baf-3 cells that did not receive the retrovirus (Fig. 1F). Cells programmed to express Bcl-xL, Bcl-2, or NFIL3 were protected from apoptosis, with Bcl-xL exerting the most profound antiapoptotic effect, while, as expected (3), those expressing the c-Myc protein died more rapidly than controls. These results indicate that, in addition to Bcl-2 family members, NFIL3 can inhibit apoptosis in IL-3-deprived Baf-3 cells in agreement with previous findings in FL5.12 cells (16).
Higher IL-3 concentrations are required for survival of Baf-3 cells expressing a dominant-negative NFIL3 protein.
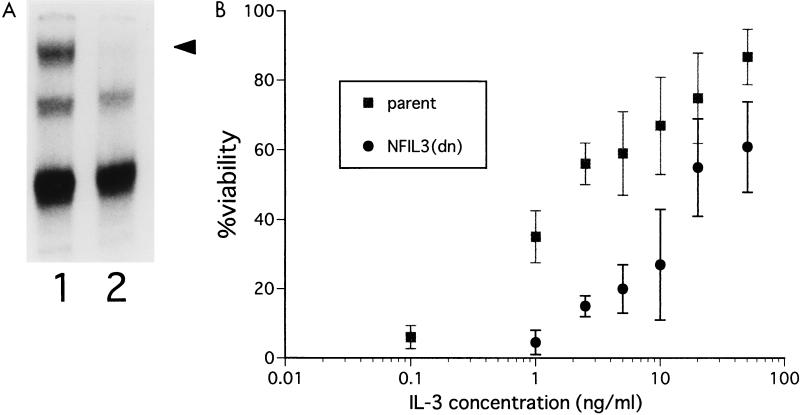
To confirm the antiapoptotic role of NFIL3 in Baf-3 cells grown in IL-3-containing medium, we attempted to suppress NFIL3 function by using a dominant-negative mutant [NFIL3(dn)] containing a defective basic region but an intact leucine zipper domain, which resulted in the formation of nonfunctional heterodimers with wild-type NFIL3 (see Materials and Methods). By EMSA, Baf-3 cells expressing NFIL3(dn) lacked functional NFIL3 protein detectable with an oligonucleotide probe containing the NFIL3 binding site (TTACGTAA) (Fig. 2A), indicating that the mutant had suppressed DNA binding by NFIL3 in a dominant-negative fashion. These cells required a 10-fold-greater concentration of IL-3 to survive than did parental Baf-3 cells or control CD8-expressing cells (Fig. 2B). The cell cycle distribution of NFIL3(dn)-positive cells was unchanged as shown by flow cytometric analysis (data not shown). Thus, the effect of NFIL3 on cytokine-mediated cell survival was independent of an effect on cell proliferation.
FIG. 2.
Baf-3 cells expressing dominant-negative NFIL3 require higher concentrations of IL-3 to survive. (A) Antibody-perturbed EMSA with lysates extracted from parental Baf-3 cells grown in 25 ng of IL-3 per ml (lane 1) and Baf-3 cells grown under the same conditions that express the dominant-negative NFIL3 protein (lane 2). The reaction mixture includes a 32P-labeled oligonucleotide probe containing the NFIL3 binding sequence and anti-NFIL3 antiserum. The arrowhead indicates a supershifted complex containing NFIL3. (B) Parental Baf-3 cells and cells expressing the dominant-negative NFIL3 protein were cultured in serum-free AIM-V medium with recombinant murine IL-3 at the indicated concentrations for 36 h, after which the percentage of surviving cells was determined. Each value is the mean (± standard deviation) of three independent experiments.
Oncogenic Ras induces NFIL3 expression.
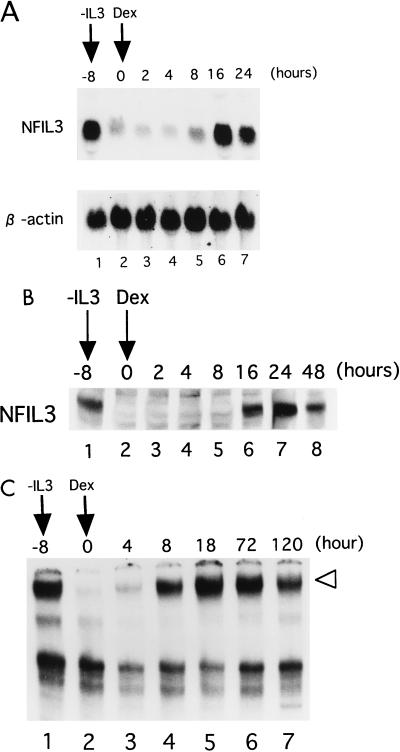
We previously reported that NFIL3 is an IL-3-responsive gene in Baf-3 cells (16). Since pathways induced by oncogenic Ras are required for the optimal survival of these cells (23, 24, 33), we considered that NFIL3 may be regulated through downstream pathways of oncogenic Ras mutants. To test this possibility, we used Baf-3 cells expressing a constitutively active Ras protein [Ras(G12V)] under the regulation of dexamethasone (Dex) (23). When cells were cultured in IL-3-free medium for 8 h, the mRNA expression levels of endogenous NFIL3 were down-regulated (lane 2, Fig. 3A). By 16 h after the addition of Dex, however, NFIL3 transcripts were induced to levels similar to those induced by IL-3 (lane 6). In accord with mRNA expression, NFIL3 protein expression was also induced by Dex as shown by immunoblot analysis (Fig. 3B). EMSA revealed that NFIL3 protein bound to DNA recovered by 8 h after addition of Dex (Fig. 3C), when protein expression was barely detectable by immunoblot analysis (Fig. 3B). This apparent discrepancy may be explained by the different sensitivities of the two assays, because, in general, EMSA is more sensitive than immunoblotting. In addition, alteration of the DNA binding ability of NFIL3 may contribute to the discordance (see Discussion). These effects appeared to be induced by Ras(G12V), not by Dex itself, since neither mRNA nor protein expression of NFIL3 was induced in wild-type Baf-3 cells after the addition of Dex (data not shown), suggesting that the oncogenic Ras protein had induced NFIL3 expression.
FIG. 3.
Induction of NFIL3 by an oncogenic Ras mutant [Ras(G12V)]. Baf-3 cells containing Dex-inducible Ras(G12V) cDNA were cultured in IL-3-free medium. Dex (10−7 M) was added to the culture medium 8 h after IL-3 removal, and cells were cultured for the indicated period (hours). (A) Northern blot analysis of total RNA. The blot was first hybridized with a mouse NFIL3 cDNA probe and then rehybridized with a human β-actin probe. (B) Immunoblot analysis with whole-cell lysates. NFIL3 protein was detected with the polyclonal antibodies. (C) Antibody-perturbed EMSA with nuclear extracts. The arrowhead indicates a supershifted complex containing NFIL3.
Downstream pathways of Ras mediating NFIL3 expression.
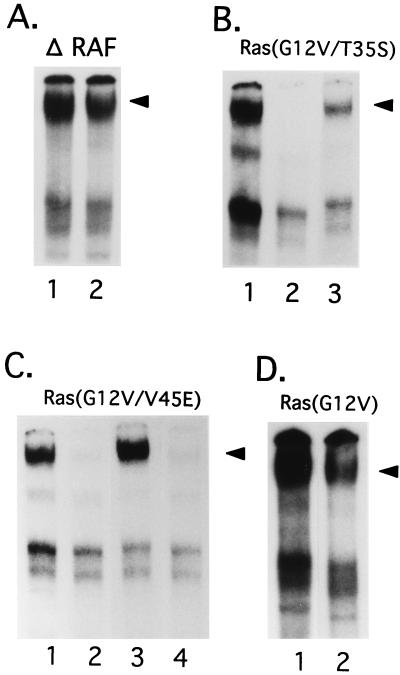
To identify downstream targets of Ras that could mediate NFIL3 expression in Baf-3 cells, we tested NFIL3 induction by various Ras or Raf mutants, which were shown to prevent apoptosis without obvious signs of cell division in a previous study (33). The ability of these mutants to prevent apoptosis and phosphorylate p42/MAPK, p70/S6 kinase, and Akt is summarized in Table 1. Cells expressing the ΔRaf mutant (truncated N terminus), which constitutively activates MAPK, were highly resistant to apoptosis. As expected, the cells retained their capacity to express NFIL3 even when cultured in IL-3-free medium (Fig. 4A, lane 2), indicating that activation of the Raf-MAPK pathway is sufficient for the induction of this transcription factor. In experiments to test this hypothesis, the Ras(G12V/T35S) mutant, which specifically but weakly activates the Raf-MAPK pathways (2), had less ability than ΔRaf to induce NFIL3 (Fig. 4B, lane 3) and only weakly influenced cell survival (Table 1). The Ras(G12V/V45E) mutant, which activates S6K and Akt but not Raf-MAPK pathways (34) (see also Fig. 8), prevented apoptosis efficiently and induced appreciable levels of NFIL3 (Fig. 4C, lane 3), suggesting that pathways downstream of Ras other than Raf-MAPK pathways contribute to NFIL3 regulation. This interpretation was confirmed by results with rapamycin, an inhibitor of the PI3-K–S6K pathway (26, 36), which dramatically reduced the levels of NFIL3 induced by Ras(G12V/V45E) (lane 4) and compromised cell survival (Table 1). By contrast, substantial amounts of NFIL3 protein were detected in Baf-3 cells expressing Ras(G12V) and cultured in rapamycin-containing medium (Fig. 4D, lane 2), as would be expected with the use of a reagent that does not affect the Raf-MAPK pathways. In summary, the Ras-Raf and rapamycin-sensitive pathways independently induce NFIL3 expression, apparently through effects on the phosphorylation of p42/MAPK or p70/S6K.
TABLE 1.
Summary of activation of Ras-Raf mutants
| Mutant | Phosphorylation
|
% Viability 48 h after IL-3 star-vation | ||
|---|---|---|---|---|
| p42/MAPK | p70/S6K | Akt | ||
| ΔRaf | + | − | NDa | 60 |
| Ras(G12V) | + | + | + | >90 |
| Ras(G12V/T35S) | Weak | − | ND | 10 |
| Ras(G12V/V45E) | − | + | + | 75 |
| Ras(G12V) + rapamycin | + | − | ND | 80 |
| Ras(G12V/V45E) + rapamycin | − | − | ND | <10 |
| Ras(G12V) + wortmannin | + | ND | − | 80 |
| Ras(G12V/V45E) + wortmannin | − | ND | − | <10 |
ND, not done.
FIG. 4.
Antibody-perturbed EMSA with nuclear lysates extracted from Baf-3 cells expressing various mutants of Ras and Raf. (A) Baf-3 cells expressing ΔRaf were cultured in IL-3-containing medium (lane 1) or IL-3-free medium for 18 h (lane 2). (B) Baf-3 cells expressing Dex-regulated Ras(G12V/T35S) (lane 1) were washed to remove IL-3 and then cultured in IL-3-free medium for 8 h (lane 2). Cells were then cultured in IL-3-free medium containing 10−7 M Dex to induce Ras(G12V/T35S) for another 18 h (lane 3). (C) Baf-3 cells expressing Ras(G12V/V45E) were cultured in IL-3-containing medium (lane 1) and then washed to remove IL-3 and cultured in IL-3-free medium for 8 h (lane 2). Cells were then cultured without (lane 3) or with (lane 4) rapamycin (10 ng/ml) and maintained in IL-3-free medium containing 10−7 M Dex to induce Ras(G12V/V45E). (D) Baf-3 cells inducibly expressing Ras(G12V) were cultured in IL-3-free medium for 8 h, then treated without (lane 1) or with (lane 2) rapamycin (10 ng/ml), and then cultured in IL-3-free medium containing 10−7 M Dex to induce Ras(G12V). Arrowheads indicate supershifted DNA-protein complexes containing NFIL3.
FIG. 8.
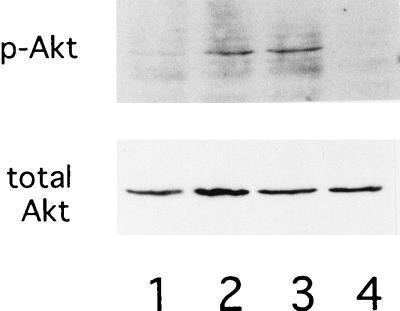
Expression and phosphorylation of Akt in Baf-3 cells expressing Ras(G12V/V45E). Baf-3 cells containing Dex-inducible Ras(G12V/V45E) were cultured in IL-3-containing medium without Dex (lane 1) or with 10−7 M Dex for 16 h (lane 2). The Dex-treated cells were then cultured in IL-3-free, Dex-containing medium for 12 h without wortmannin (lane 3) or with 100 nM wortmannin (lane 4). Phosphorylated Akt at Ser-473 (upper panel) or total Akt (lower panel) protein was detected with antibodies specific for each protein.
Ras-independent pathways mediating NFIL3 expression.
To further analyze pathways regulating the expression of NFIL3, we used Baf-3 cells expressing the full-length or truncated form of the βc human shared cytokine receptor chain. Because the membrane distal region of the cytoplasmic βc chain activates Ras signaling pathways, β544 cells that express the βc chain truncated at amino acid 544 do not activate the Ras pathways after addition of hGM-CSF to the culture medium (33). As controls, we used β455 cells expressing a βc chain lacking all of its cytoplasmic portion, as well as βc cells expressing the full-length βc chain.
As in an earlier study (23), the βc cells grew as well in hGM-CSF-containing medium as in medium that contained IL-3, while the β455 cells could not divide and rapidly underwent apoptotic cell death. The β544 cells proliferated well for approximately 60 h with no evidence of loss of viability but thereafter underwent apoptosis at a rapid rate (Fig. 5A and B). Flow cytometry analysis with propidium iodide staining demonstrated that the β544 cells had accumulated in G0/G1 phase prior to the onset of apoptosis at 60 h following the addition of hGM-CSF (data not shown).
FIG. 5.
Ras-independent pathways also mediate NFIL3 expression. (A and B) β544 cells, β544 cells expressing NFIL3, and β544 cells expressing CD8 as a control were cultured in cytokine-free medium for 8 h (−8 to 0); then hGM-CSF was added to the medium at a concentration of 5 ng/ml. Cell number (A) and percent viability (B) were determined by trypan blue dye exclusion. A representative experiment is shown; similar results were obtained in two other independent experiments. (C and D) EMSA with nuclear lysates extracted from βc or β455 (C) or β544 (D) cells expressing the full-length or a truncated form of the human common β chain. Cells were cultured in cytokine-free medium for 8 h (−8 to 0); then hGM-CSF was added to the medium at a concentration of 5 ng/ml, and the cells were cultured for the indicated period (hours). Open arrowheads indicate supershifted DNA-protein complexes containing NFIL3.
hGM-CSF induced NFIL3 expression in βc cells but not in β455 cells (Fig. 5C). In β544 cells exposed to hGM-CSF, NFIL3 expression was transient and much less pronounced than in βc cells (Fig. 5D), suggesting that Ras is required for the stable expression of NFIL3. Interestingly, as long as β544 cells expressed NFIL3, they remained alive, but when NFIL3 began to fall, the cells underwent apoptosis. To confirm the apparent role of NFIL3 in the survival of these cells, we infected β544 cells with a retrovirus made from the pMX-NFIL3/IRES-CD8 vector and selected CD8-positive cells. As demonstrated in Fig. 5A and B, β544 cells constitutively expressing NFIL3 were more resistant to apoptosis, indicating that NFIL3 at least partially substitutes for Ras signals in preventing the suicide. These results therefore suggest a role for Ras-mediated pathways in the maintenance of both NFIL3 levels and cell survival in response to cytokines.
Induction of Bcl-xL by oncogenic Ras.
Cell survival signals originating from cytokine receptors also regulate proteins involved in the general apoptosis program, including members of the Bcl-2 and caspase families. In fact, we previously reported that caspase-3 activity is elevated in cytokine-dependent cells that undergo apoptosis caused by cytokine deprivation (30). Because caspase activity is regulated by some of the Bcl-2 family members, we compared expression levels of the members of the Bcl-2 family (Bcl-2, Bcl-xL, Bax, Bad, and Bak) in Baf-3 cells cultured in medium with or without IL-3. The results indicated that, of the proteins analyzed, only Bcl-xL (mRNA and protein) was regulated by IL-3 (Fig. 6A and 7A), in agreement with findings of Leverrier et al. (27). Moreover, enforced expression of this protein protected cells from apoptosis caused by IL-3 starvation (Fig. 1F), underscoring its major role in the general apoptosis program regulating the cytokine-mediated survival of Baf-3 cells.
FIG. 6.
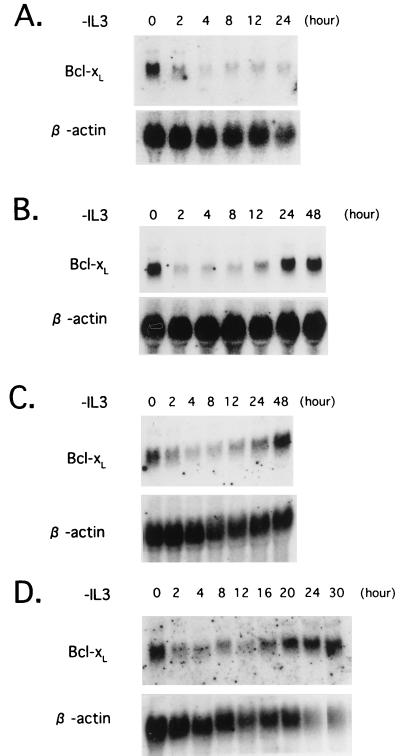
Bcl-xL mRNA expression in Baf-3 cells. The blots were first hybridized with a mouse Bcl-x cDNA probe and then rehybridized with a human β-actin probe. (A) Parental Baf-3 cells were cultured in IL-3-free medium for the indicated periods. (B and C) Baf-3 cells containing Dex-inducible Ras(G12V) (B) or Ras(G12V/V45E) (C) were cultured in medium containing 10−7 M Dex for 16 h; the cells were then cultured in IL-3-free, Dex-containing medium for the indicated period. (D) Baf-3 cells containing Dex-inducible Ras(G12V/V45E) were cultured in medium containing 10−7 M Dex for 16 h; the cells were then cultured in IL-3-free, Dex-containing medium with 100 nM wortmannin for the indicated period.
FIG. 7.
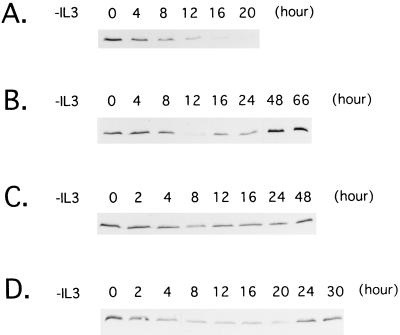
Expression of Bcl-xL protein in Baf-3 cells. (A) Parental Baf-3 cells were cultured in IL-3-free medium for the indicated periods. (B and C) Baf-3 cells containing Dex-inducible Ras(G12V) (B) or Ras(G12V/V45E) (C) were cultured in medium containing 10−7 M Dex for 16 h; the cells were then cultured in IL-3-free, Dex-containing medium for the indicated period. (D) Baf-3 cells containing Dex-inducible Ras(G12V/V45E) were cultured in medium containing 10−7 M Dex for 16 h; the cells were then cultured in IL-3-free, Dex-containing medium with 100 nM wortmannin for the indicated period.
To determine whether oncogenic Ras blocks apoptosis through Bcl-xL gene regulation, we performed Northern blot and immunoblot analysis with RNA and total cell lysates extracted from Baf-3 cells expressing the Ras(G12V) mutant. Bcl-xL mRNA and protein levels were down-regulated in IL-3-deprived Baf-3 cells expressing the oncogenic Ras mutant, as previously observed in parental Baf-3 cells, with a return to baseline levels by 24 h (Fig. 6B and 7B). Both Ras(G12V/V45E) and ΔRaf induced Bcl-xL expression in the same manner as Ras(G12V) did (Fig. 6C and 7C and data not shown), indicating that both the Ras–PI3-K and Raf-MAPK pathways are involved in the slow induction of Bcl-xL.
Since involvement of the PI3-K–Akt pathways in cytokine-mediated cell survival has been reported recently (1, 35), we used wortmannin to test whether the pathways contribute to inhibition of apoptosis through induction of Bcl-xL. Baf-3 cells expressing Ras(G12V) or Ras(G12V/V45E) by Dex were cultured without IL-3. As we previously reported (24), cells expressing Ras(G12V/V45E), but not those expressing Ras(G12V), underwent apoptosis by addition of 100 nM wortmannin in culture medium (Table 1), suggesting that signals mediated by the Ras–PI3-K pathways play central roles in inhibiting apoptosis in cells expressing Ras(G12V/V45E) cultured in IL-3-free medium. As expected, levels of phosphorylated Akt were down-regulated by the treatment of wortmannin (Fig. 8). However, expression levels of Bcl-xL mRNA and protein were not affected (Fig. 6D and 7D), suggesting that signals mediated by the PI3-K–Akt pathways protect cells from apoptosis through mechanisms independent of Bcl-xL.
Ras-independent induction of Bcl-xL.
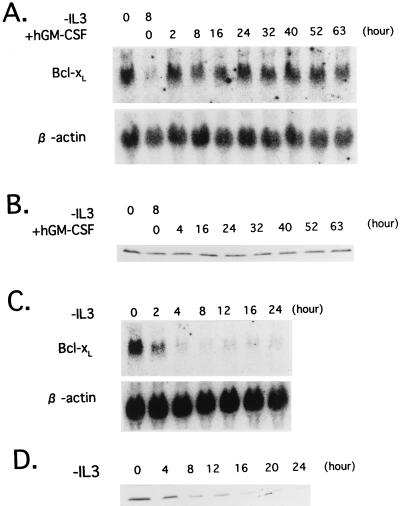
The results described above raised the possibility that the regulation of the Bcl-xL gene expression involves Ras-independent pathways. To test the possibility, we cultured β544 cells in hGM-CSF-containing medium, which does not activate the Ras pathways, and analyzed the expression of Bcl-xL. Bcl-xL mRNA was barely detectable 8 h after IL-3 removal, returning rapidly to its original level after the addition of hGM-CSF (Fig. 9A). Moreover, Bcl-xL protein levels were maintained for more than 63 h (Fig. 9B), when the cells underwent apoptosis (Fig. 5B), indicating that Ras-independent pathways can induce the expression of Bcl-xL but are unable to maintain the survival of Baf-3 cells.
FIG. 9.
Expression of Bcl-xL mRNA (A and C) and protein (B and D) in Baf-3 cells. The Northern blots were first hybridized with a mouse Bcl-x cDNA probe and then rehybridized with a human β-actin probe. (A and B) β544 cells were cultured in medium lacking either IL-3 or hGM-CSF for 8 h (lanes 2). Then cells were cultured in medium containing hGM-CSF for the indicated periods. (C and D) Baf-3 cells expressing NFIL3 were cultured in IL-3-free medium for the indicated periods.
We next tested whether NFIL3 induces expression of Bcl-xL. Baf-3 cells expressing NFIL3 were deprived of IL-3, and total RNA and cell lysates were extracted. Northern blot and immunoblot analysis revealed that both mRNA and protein expression of Bcl-xL were quickly down-regulated by IL-3 starvation (Fig. 9C and D), suggesting that NFIL3 delays apoptosis through mechanisms other than the induction of Bcl-xL.
DISCUSSION
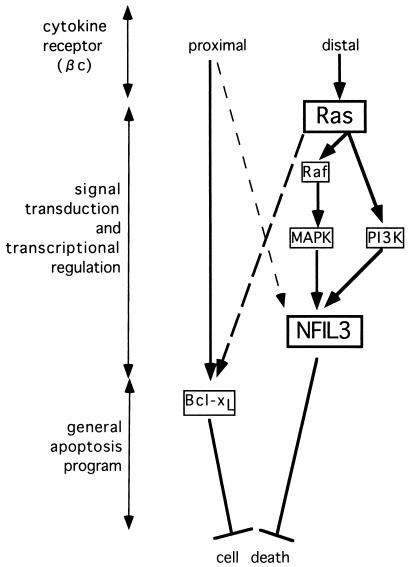
In earlier studies, we established that constitutively active Ras proteins and NFIL3 block apoptosis in cytokine-starved murine IL-3-dependent pro-B lymphocytes (16, 23, 24). We and others also demonstrated that the general cell death machinery, including Bcl-xL and caspase-3, ultimately controls the death of cytokine-deprived Baf-3 lymphocytes (27, 30), but the pathways regulating the expression and activity of these proteins remained unclear. Here we describe two distinct signaling pathways that appear to regulate cell survival (summarized in Fig. 10); one originates from the membrane proximal portion of the βc chain and is involved in the rapid and stable regulation of Bcl-xL, while the other pathway involves the NFIL3 transcription factor downstream of oncogenic Ras proteins.
FIG. 10.
Proposed signaling pathways regulating cell survival in Baf-3 cells that originate from segments of the cytokine-activated βc chain and proceed to the general apoptosis program through the signal transduction and transcriptional regulatory system. Signals arising from the membrane proximal portion of the βc chain contribute to the rapid and stable expression of Bcl-xL but induce NFIL3 only transiently (lightly dashed line). By contrast, signals from the membrane distal domain of the βc chain activate Ras-mediated pathways. A constitutively active Ras protein [Ras(G12V)] regulates the stable expression of the NFIL3 transcription factor through both the Raf-MAPK and PI3-K pathways. Oncogenic Ras is also involved in the slow but stable induction of Bcl-xL (heavily dashed line).
Signals originating from the membrane proximal domain of the βc chain likely regulate the expression of Bcl-xL, as shown by experiments using β544 cells stimulated with hGM-CSF, in which Bcl-xL expression levels were demonstrated to be similar to those in Baf-3 cells cultured with IL-3 (Fig. 9A and B). Oncogenic Ras did not block the rapid decline of Bcl-xL mRNA and protein levels after IL-3 deprivation. In addition, wortmannin-sensitive pathways including pathways involving PI3-K and Akt did not regulate Bcl-xL expression (Fig. 6 and 7). These data agree with the results of experiments by Packham et al. using the 32D, IL-3-dependent murine myeloid cell line, which indicate that the regulation of Bcl-xL protein levels is mediated by the Jak kinase pathway and is independent of other signaling effectors including PI3-K and Ras (32). Nevertheless, β544 cells cultured with hGM-CSF underwent apoptosis, indicating that signals from the membrane distal domain of the βc chain are also required to protect cells from apoptosis. Because Ras is activated through signals mediated by the βc chain distal portion, and because an oncogenic Ras mutant [Ras(G12V)] rescued β544 cells from apoptosis completely (23), it appears likely that one or more genes downstream of Ras(G12V) are required for cell survival in addition to Bcl-xL. NFIL3 appears to be a signaling component in Ras-mediated survival pathways, because this transcription factor is induced by IL-3 (16) and Ras(G12V) (Fig. 3) in Baf-3 cells, but only transiently by hGM-CSF in β544 cells (Fig. 5D), and the enforced expression of NFIL3 in β544 cells delays apoptosis (Fig. 5A and B).
We previously reported that DNA binding ability, as well as the expression levels of NFIL3, is regulated by IL-3 (16). DNA binding of this transcription factor may also be up-regulated by Ras-mediated signals (Fig. 3B and C). Evidence has been published that in vitro-phosphorylated NFIL3 is able to bind its consensus sequence more avidly than nonphosphorylated protein (5). Thus, oncogenic Ras may regulate NFIL3 not only by up-regulating its expression levels but also through posttranslational effects on DNA binding.
The incomplete ability of NFIL3 to rescue β544 cells cultured in medium containing hGM-CSF indicates that this transcription factor is not the only downstream target of Ras(G12V) that is required to prevent cell death. Candidates for additional components of Ras-mediated survival pathways include other transcription factors, members of the antiapoptotic Bcl-2 superfamily, or members of the BH3 domain-containing group of cell death activators, such as Bad, Bax, Bak, etc. Recently, IL-3-dependent phosphorylation of Bad overexpressed in FL5.12 cells was reported to mediate cell survival (38). Moreover, Bad phosphorylation in this cell system is mediated at least in part by Akt, which is downstream of PI3-K (10, 11). However, because endogenous Bad proteins in Baf-3 cells cultured in IL-3-containing medium are not phosphorylated at detectable levels (data not shown), it is difficult to evaluate the importance of this mechanism.
The downstream factors through which NFIL3 delays apoptosis in IL-3-deprived Baf-3 cells have as yet not been identified. One possibility is that NFIL3 induces the expression of cytokines in Baf-3 cells and blocks apoptosis through an autocrine mechanism, because this transcription factor has been reported to trans activate the IL-3 promoter in T cells (39). However, we have been unable to detect the IL-3 mRNA or protein in Baf-3 cells overexpressing NFIL3 and in conditioned medium from these cells (16), although the involvement of another cytokine(s) has not been ruled out. Another possibility is that NFIL3 induces antiapoptotic Bcl-2 family members; however, we have not been able to detect the induction of a known antiapoptotic Bcl-2 family member (Fig. 9C and D and data not shown). Recent findings on the regulation of programmed cell death in C. elegans may provide insight into this issue. The EGL-1 protein, which was recently identified as an upstream component of the general cell death machinery in the worm, contains a Bcl-2 homology region 3 (BH3)-like domain but does not contain a BH1, BH2, or BH4 domain (6). EGL-1 directly binds to CED-9 and is considered to inhibit its function. Because Egl-1 likely acts downstream of CES-1 and CES-2 (6), mammalian members of the BH3-only-containing group of cell death activators, such as Bid, Bik, or Hrk, would appear to be candidate target proteins of NFIL3.
Factors involved in the control of apoptosis have been implicated in oncogenic signaling cascades. NFIL3, for example, may act in some cells as a physiological counterpart of the leukemogenic E2A-HLF fusion transcription factor (16), by transmitting survival signals emanating from oncogenic Ras proteins. Ras pathways are activated in leukemogenesis not only by activating point mutations, but also by loss-of-function mutations of the NF1 tumor suppressor gene in juvenile-type chronic myelogenous leukemia (4). In addition, the BCR-ABL chimeric tyrosine kinase, implicated in the pathogenesis of Philadelphia chromosome-positive leukemias, acts in part through Ras-mediated signals (7, 8). Not surprisingly, we have shown that NFIL3 expression is induced by the BCR-ABL chimera in Baf-3 cells (data not shown), suggesting that Ras-NFIL3 pathways may be common targets for a variety of oncogenes. The dissection of transcriptional networks through which NFIL3 affects cell survival may ultimately provide insight into the role of Ras gene activation and its contribution to malignant transformation.
ACKNOWLEDGMENTS
We are indebted to T. Kitamura for providing the pMX retrovirus vector, John Gilbert for editorial review, and John L. Cleveland for critical comments.
This research was supported by grants-in-aid from the Ministry of Education, Science and Culture of Japan; by the Senri Life Science Foundation; by grants from the National Cancer Institute (CA 59571 and Cancer Center Core CA 21765); and by the American Lebanese Syrian Associated Charities (ALSAC), St. Jude Children’s Research Hospital.
REFERENCES
- 1.Ahmed N N, Grimes H L, Bellacosa A, Chan T O, Tsichlis P N. Transduction of interleukin-2 antiapoptotic and proliferative signals via Akt protein kinase. Proc Natl Acad Sci USA. 1997;94:3627–3632. doi: 10.1073/pnas.94.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akasaka K, Tamada M, Wang F, Kariya K, Shima F, Kikuchi A, Yamamoto M, Shirouzu M, Yokoyama S, Kataoka T. Differential structural requirements for interaction of Ras protein with its distinct downstream effectors. J Biol Chem. 1996;271:5353–5360. doi: 10.1074/jbc.271.10.5353. [DOI] [PubMed] [Google Scholar]
- 3.Askew D S, Ashmun R A, Simmons B C, Cleveland J L. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 4.Bollag G, Clapp D W, Shih S, Adler F, Zhang Y Y, Thompson P, Lange B J, Freedman M H, McCormick F, Jacks T, Shannon K. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12:144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 5.Chen W-J, Lewis K S, Chandra G, Cogswell J P, Stinnett S W, Kadwell S H, Gray J G. Characterization of human E4BP4, a phosphorylated bZIP factor. Biochim Biophys Acta. 1995;1264:388–396. doi: 10.1016/0167-4781(95)00182-4. [DOI] [PubMed] [Google Scholar]
- 6.Conradt B, Horvitz H R. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 7.Cortez D, Kadlec L, Pendergast A M. Structural and signaling requirements for BCR-ABL-mediated transformation and inhibition of apoptosis. Mol Cell Biol. 1995;15:5531–5541. doi: 10.1128/mcb.15.10.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortez D, Stoica G, Pierce J H, Pendergast A M. The BCR-ABL tyrosine kinase inhibits apoptosis by activating a Ras-dependent signaling pathway. Oncogene. 1996;13:2589–2594. [PubMed] [Google Scholar]
- 9.Cowell I G, Skinner A, Hurst H C. Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol Cell Biol. 1992;12:3070–3077. doi: 10.1128/mcb.12.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 11.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 12.Ellis R E, Horvitz H R. Two C. elegans genes control the programmed deaths of specific cells in the pharynx. Development. 1991;112:591–603. doi: 10.1242/dev.112.2.591. [DOI] [PubMed] [Google Scholar]
- 13.Ellis R E, Yuan J Y, Horvitz H R. Mechanisms and functions of cell death. Annu Rev Cell Biol. 1991;7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- 14.Hunger S P, Ohyashiki K, Toyama K, Cleary M L. HLF, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 1992;6:1608–1620. doi: 10.1101/gad.6.9.1608. [DOI] [PubMed] [Google Scholar]
- 15.Hunger S P, Brown R, Cleary M L. DNA-binding and transcriptional regulatory properties of hepatic leukemia factor (HLF) and the t(17;19) acute lymphoblastic leukemia chimera E2A-HLF. Mol Cell Biol. 1994;14:5986–5996. doi: 10.1128/mcb.14.9.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikushima S, Inukai T, Inaba T, Nimer S D, Cleveland J L, Look A T. Pivotal role for the NFIL3/E4BP4 transcription factor in IL-3-mediated survival of pro-B lymphocytes. Proc Natl Acad Sci USA. 1997;94:2609–2614. doi: 10.1073/pnas.94.6.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inaba T, Roberts W M, Shapiro L H, Jolly K W, Raimondi S C, Smith S D, Look A T. Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia. Science. 1992;257:531–534. doi: 10.1126/science.1386162. [DOI] [PubMed] [Google Scholar]
- 18.Inaba T, Shapiro L H, Funabiki T, Sinclair A E, Jones B G, Ashmun R A, Look A T. DNA-binding specificity and trans-activating potential of the leukemia-associated E2A-hepatic leukemia factor fusion protein. Mol Cell Biol. 1994;14:3403–3413. doi: 10.1128/mcb.14.5.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inaba T, Inukai T, Yoshihara T, Seyschab H, Ashmun R A, Canman C E, Laken S J, Kastan M B, Look A T. Reversal of apoptosis by the leukemia-associated E2A-HLF chimeric transcription factor. Nature. 1996;382:541–544. doi: 10.1038/382541a0. [DOI] [PubMed] [Google Scholar]
- 20.Inukai T, Inaba T, Yoshihara T, Look A T. Cell transformation mediated by homodimeric E2A-HLF transcription factors. Mol Cell Biol. 1997;17:1417–1424. doi: 10.1128/mcb.17.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inukai T, Inaba T, Ikushima S, Look A T. The AD1 and AD2 transactivation domains of E2A are essential for the antiapoptotic activity of the E2A-HLF chimeric oncoprotein. Mol Cell Biol. 1998;18:6035–6043. doi: 10.1128/mcb.18.10.6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joneson T, Bar Sagi D. Ras effectors and their role in mitogenesis and oncogenesis. J Mol Med. 1997;75:587–593. doi: 10.1007/s001090050143. [DOI] [PubMed] [Google Scholar]
- 23.Kinoshita T, Yokota T, Arai K, Miyajima A. Suppression of apoptotic death in hematopoietic cells by signaling through the IL-3/GM-CSF receptors. EMBO J. 1995;14:266–275. doi: 10.1002/j.1460-2075.1995.tb07000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinoshita T, Shirouzu M, Kamiya A, Hashimoto K, Yokoyama S, Miyajima A. Raf/MAPK and rapamycin-sensitive pathways mediate the anti-apoptotic function of p21Ras in IL-3-dependent hematopoietic cells. Oncogene. 1997;15:619–627. doi: 10.1038/sj.onc.1201234. [DOI] [PubMed] [Google Scholar]
- 25.Kroemer G. The proto-oncogene Bcl-2 and its role in regulating apoptosis. Nat Med. 1997;3:614–620. doi: 10.1038/nm0697-614. [DOI] [PubMed] [Google Scholar]
- 26.Kuo C J, Chung J, Fiorentino D F, Flanagan W M, Blenis J, Crabtree G R. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature. 1992;358:70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- 27.Leverrier Y, Thomas J, Perkins G R, Mangeney M, Collins M K L, Marvel J. In bone marrow derived Baf-3 cells, inhibition of apoptosis by IL-3 is mediated by two independent pathways. Oncogene. 1997;14:425–430. doi: 10.1038/sj.onc.1200845. [DOI] [PubMed] [Google Scholar]
- 28.Metzstein M M, Hengartner M O, Tsung N, Ellis R E, Horvitz H R. Transcriptional regulator of programmed cell death encoded by Caenorhabditis elegans gene ces-2. Nature. 1996;382:545–547. doi: 10.1038/382545a0. [DOI] [PubMed] [Google Scholar]
- 29.Nicholson D W, Thornberry N A. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 30.Ohta T, Kinoshita T, Naito M, Nozaki T, Masutani M, Tsuruo T, Miyajima A. Requirement of the caspase-3/CPP32 protease cascade for apoptotic death following cytokine deprivation in hematopoietic cells. J Biol Chem. 1997;272:23111–23116. doi: 10.1074/jbc.272.37.23111. [DOI] [PubMed] [Google Scholar]
- 31.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier L L, Gorman D M, Nolan G P, Miyajima A, Kitamura T. Application of retrovirus-mediated expression cloning. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 32.Packham G, White E L, Eischen C M, Yang H, Parganas E, Ihle J N, Grillot D A, Zambetti G P, Nunez G, Cleveland J L. Selective regulation of Bcl-XL by a Jak kinase-dependent pathway is bypassed in murine hematopoietic malignancies. Genes Dev. 1998;12:2475–2487. doi: 10.1101/gad.12.16.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato N, Sakamaki K, Terada N, Arai K, Miyajima A. Signal transduction by the high-affinity GM-CSF receptor: two distinct cytoplasmic regions of the common β subunit responsible for different signaling. EMBO J. 1993;12:4181–4189. doi: 10.1002/j.1460-2075.1993.tb06102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirouzu M, Koide H, Fujita Y-J, Oshio H, Toyama Y, Yamasaki K, Fuhrman S A, Villafranca E, Kaziro Y, Yokoyama S. Mutations that abolish the ability of Ha-Ras to associate with Raf-1. Oncogene. 1994;9:2153–2157. [PubMed] [Google Scholar]
- 35.Songyang Z, Baltimore D, Cantley L C, Kaplan D R, Franke T F. Interleukin 3-dependent survival by the Akt protein kinase. Proc Natl Acad Sci USA. 1997;94:11345–11350. doi: 10.1073/pnas.94.21.11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terada N, Lucas J J, Szepesi A, Franklin R A, Takase K, Gelfand E W. Rapamycin inhibits the phosphorylation of p70 S6 kinase in IL-2 and mitogen-activated human T cells. Biochem Biophys Res Commun. 1992;186:1315–1321. doi: 10.1016/s0006-291x(05)81549-9. [DOI] [PubMed] [Google Scholar]
- 37.Yoshihara T, Inaba T, Shapiro L H, Kato J, Look A T. E2A-HLF-mediated cell transformation requires both the trans-activation domains of E2A and the leucine zipper dimerization domain of HLF. Mol Cell Biol. 1995;15:3247–3255. doi: 10.1128/mcb.15.6.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Serine phosphorylation of death agonist Bad in response to survival factor results in binding to 14-3-3 not Bcl-xL. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 39.Zhang W, Zhang J, Kornuc M, Kwan K, Frank R, Nimer S D. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol Cell Biol. 1995;15:6055–6063. doi: 10.1128/mcb.15.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]