Abstract
Background
The urgent need for massively scaled clinical testing for SARS-CoV-2, along with global shortages of critical reagents and supplies, has necessitated development of streamlined laboratory testing protocols. Conventional nucleic acid testing for SARS-CoV-2 involves collection of a clinical specimen with a nasopharyngeal swab in transport medium, nucleic acid extraction, and quantitative reverse-transcription PCR (RT–qPCR). As testing has scaled across the world, the global supply chain has buckled, rendering testing reagents and materials scarce. To address shortages, we developed SwabExpress, an end-to-end protocol developed to employ mass produced anterior nares swabs and bypass the requirement for transport media and nucleic acid extraction.
Methods
We evaluated anterior nares swabs, transported dry and eluted in low-TE buffer as a direct-to-RT–qPCR alternative to extraction-dependent viral transport media. We validated our protocol of using heat treatment for viral inactivation and added a proteinase K digestion step to reduce amplification interference. We tested this protocol across archived and prospectively collected swab specimens to fine-tune test performance.
Results
After optimization, SwabExpress has a low limit of detection at 2–4 molecules/µL, 100% sensitivity, and 99.4% specificity when compared side by side with a traditional RT–qPCR protocol employing extraction. On real-world specimens, SwabExpress outperforms an automated extraction system while simultaneously reducing cost and hands-on time.
Conclusion
SwabExpress is a simplified workflow that facilitates scaled testing for COVID-19 without sacrificing test performance. It may serve as a template for the simplification of PCR-based clinical laboratory tests, particularly in times of critical shortages during pandemics.
Introduction
Since the first reported cases in the winter of 2019, the spread of the novel beta-coronavirus SARS-CoV-2 has grown into a global pandemic. The virus spreads easily from person to person and is often carried by asymptomatic individuals (1). These viral properties, in conjunction with a lack of an effective centralized response or societal adherence to public health recommendations, has led to a continued persistence of the pandemic throughout the USA (2). It is widely recognized that increased testing capacity can ameliorate the outbreak (3, 4), but the prohibitive cost of testing materials and reagents as well as global supply chain problems continue to thwart efforts to reach the required scale.
Since the beginning of the pandemic, the gold standard for SARS-CoV-2 detection has been RNA extraction followed by reverse-transcription quantitative polymerase chain reaction (RT–qPCR). Specimens are traditionally collected as nasopharyngeal (NP) specimens (1) by healthcare professionals and transported in viral media [e.g., Universal Transport Media (UTM)]. Worldwide reliance on this template protocol has led to global shortages in swabs, viral media, and laboratory reagents. These shortages continue to plague testing laboratories and impede efforts to scale. Previous literature (5, 6) and the work of United Health/Quantigen (7) have established that swabs collected without transport media are acceptable for nucleic acid detection-based diagnostics, eliminating the reliance on UTM. Extraction-free protocols have also been developed to remove the need for RNA extraction reagents and streamline testing protocols. Saliva specimens have been shown to be particularly amenable to extraction-free testing protocols. For example, SalivaDirect™—a protocol for performing SARS-CoV-2 RT–qPCR on saliva specimens without extraction (8)—had a sensitivity of 89% compared to traditionally processed anterior nares (AN) or oropharyngeal (OP) swabs, demonstrating the viability of extraction-free protocols. Unlike saliva, extraction-free methods for nasal swabs have been less sensitive than conventional protocols—likely due to PCR inhibition from transport media or saline (8–14).
Here we describe the development of an UTM and extraction-free protocol for anterior nasal dry swabs that is compatible with RT–qPCR and does not sacrifice test performance. This protocol, which we have coined “SwabExpress,” has a low limit of detection, high sensitivity, high specificity, and superior test performance when compared to conventional extraction-based RT–qPCR protocols. We further identify and ameliorate 2 distinct failure modes for extraction-free RT–qPCR-based testing. Widespread adoption of this approach and others like it could result in a dramatic increase in testing capacity, decrease consumables used during testing, and ultimately help curb the spread of SARS-CoV-2.
Methods
Collection of Nasal Swabs
For preliminary studies, individuals who tested positive for SARS-CoV-2 through clinical testing were identified and recruited into a study of home-based, self-collected home swabs (15). After providing consent, enrolled participants were supplied a Swab-and-Send kit (16) containing 2 swabs (Copan FloqSwab 56380CS01) delivered to their home via 2-hour delivery and were provided instructions to self-collect 2 mid-turbinate swabs. Participants placed one swab in a tube with UTM (Becton Dickinson PN 220220) and the other in an empty, dry 15-mL conical tube for transport. For all other studies, AN (US Cotton #3, distributed by Steripack) swabs were collected by the Seattle Flu Study, Husky Coronavirus Testing Program (HCT) (17) or the Seattle Coronavirus Assessment Network (SCAN) (18 ). Anterior nares swabs were transported in a sterile, empty conical tube directly to the laboratory by HCT technicians. SCAN swabs were packaged by the participant according to kit instructions and sent to the Brotman Baty Institute/Northwest Genomics Center, using standard International Air Transport Association shipping procedures by courier at ambient temperature. These IRB-supervised studies were public health surveillance programs and enrolled both symptomatic and asymptomatic participants. Informed consent was obtained from adult participants and parents/permanent legal guardians of participant children. Archived and fresh convenience specimens from these studies were chosen at random for use in the current study.
Usability Study
To recruit a sufficient number of children for the prospective usability study, participants were recruited that met broad eligibility criteria: (a) no COVID-19 symptoms, (b) no prior self-swab experience, and (c) no prior medical or laboratory training. We obtained informed consent from adult participants and parents/permanent legal guardians of participant children.
Swab Rehydration and Elution
All work was performed within a class II biosafety cabinet with appropriate precautions. For preliminary studies each mid-turbinate dry swab was placed into a 1.5-mL microfuge tube, then cut using a sterile razor blade. Next, 200 µL of low-TE [10 mM Tris-HCl pH 7.5 (T2319-1L, Sigma), 0.1 mM EDTA (15575020, Invitrogen)] was added to each tube and vortexed for 30 seconds. To test various buffers, 45 µL of this solution was removed and added to either 5 µL of low-TE or 5 µL of 10% Triton-X (X100-500ML, Sigma Aldrich). These 2 specimens constitute the undiluted eluate from the dry swabs.
For all other studies, AN swabs were rehydrated in 1 mL of low-TE prepared in UltraPure Water (Life Technologies PN 10977023). Specimens were vortexed for 30 seconds or shaken for 1 minute and allowed to incubate at room temperature for at least 10 minutes before transfer to matrix tubes (Thermo Fisher).
RNA Extraction of Specimens
Here, 200 µL of eluate was extracted on the Magna Pure 96 using a DNA and Viral NA Small Volume Kit (Roche, 06543588001) with the universal small volume protocol and eluted into 50 µL of proprietary elution buffer. Or 200 µL of eluted AN specimens were extracted on the KingFisher Flex using the MagMAX Viral Pathogen II Nucleic Acid Isolation Kit with MagMAX™ Viral/Pathogen Ultra Enzyme Mix (Thermo Fisher A48383 and A42366) and eluted in 50 µL (although roughly 35 µL is eluted).
SwabExpress Specimen Preparation
Here, 50 µL of 94 specimens were transferred to a LoBind 96 well plate (Eppendorf 30129512) using a manual 96-well pipetting system (Rainin Liquidator) with low retention tips (Rainin 17014402) with or without 5 µL of Proteinase K (Thermo Fisher A42363, proprietary concentration). The plate was sealed with foil (Eppendorf 0030127854 and 5392000013). Specimens with Proteinase K were incubated at 37 ˚C for 15 minutes in a convection oven (Across International 0853924003042) and then transferred to a second oven for heat inactivation at 95 ˚C for 15 minutes. Specimens without Proteinase K were heat inactivated at 95 ˚C for 30 minutes.
RT–qPCR
Each RT–qPCR reaction was performed at a final volume of 10 µL and containing 1× TaqPath RT–qPCR MasterMix (PN A15300, Life Technologies), 0.125× RNAse P TaqMan VIC assay (A30064, Life Technologies) or 1× RNAse P HEX assay (IDT), 1× SARS-CoV-2 ORF1b FAM assay (PN 4332079, Life Technologies assay no. APGZJKF) or 1× Spike (S) gene (PN 4332079, Life Technologies assay no. APXGVC4) and nuclease-free water (1907076, Thermo Fisher). Then 5 µL of specimen was added to each well. Primer sequences were designed against Wuhan-Hu-1 sequence (MN908947.3) and are proprietary to Thermo Fisher. Plates were sealed using optically clear microseal B (Biorad). Each assay was performed in technical duplicate for a total of 4 RT–qPCR wells per sample. RT–qPCR was then performed on the Applied Biosystems QuantStudio 6 Pro (25 ˚C for 2 minutes, 50 ˚C for 15 minutes, 98 ˚C for 3 minutes, followed by 40 cycles of 98 ˚C for 3 seconds and 60 ˚C for 30 seconds). Reported cycle threshold (Ct ) values were obtained from the onboard analysis using predetermined thresholds. Positive controls contained purified nucleic acid with sequence that was amplified by the ORF1b and Spike-gene assays.
The RT–qPCR reaction for the CDC COVID-19 diagnostic test was performed at a final volume of 20 µL. Reactions contained 1× TaqPath RT–qPCR MasterMix, nCOV-N1 FAM, or nCOV-N2 FAM primer and probe mix (10006713, IDT) and nuclease-free water (1907076, Thermo Fisher), and 5 µL of specimen was added to each well. RT–qPCR was then performed on the QuantStudio 6 Pro as above. Reported Ct values were obtained from the onboard analysis using the autodetermined thresholds. Data were analyzed using Excel and R v.3.5.
Preparation of Inactivated Viral Controls
Contrived SARS-CoV-2 positive swabs were generated by collecting clinical matrix from a confirmed healthy volunteer and loaded with 2 µL of diluted heat-inactivated virion [VR-1986HK (1.6 × 106 virion/µL), ATCC].
Viral Inactivation Studies
Viral inactivation studies were performed at the Seattle Children’s Research Institute biosafety level 3 facility. 25 μL of viral stock (isolate USA-WA1/2020 obtained from ATCC BEI Resources) with a titer of 5.8 × 106 pfu/mL was incubated in 200 μL of TE or TE + 0.25% Triton for 10 minutes at room temperature, or in TE at 65 ˚C for 10 minutes. Untreated and treated SARS-CoV-2 was then added neat and at 10-fold dilutions through 10−7 to confluent cultures of Vero E6 cells (CRL-1586, ATCC), and 48 hours later cytopathic effects were scored after staining with crystal violet. RNA was isolated from Vero cells using a TRIzol Plus RNA Purification Kit (ThermoFisher) and the amount of SARS-CoV-2 was quantified by RT–qPCR.
Retrospective Comparison Studies
Remnant participant specimens were stored either at 4 ˚C or −80 ˚C and prepared for RT–qPCR by extraction or heat treatment or SwabExpress digestion as described above. Technicians performing testing and clinical directors interpreting results were both blinded to previous test results.
Prospective Comparison Studies
Freshly acquired specimens from the SCAN and HCT studies were prepared by extraction or heat treatment or SwabExpress and tested by RT–qPCR in parallel. For prospective analyses, both technicians and clinical directors performed testing and interpretation blinded to results from the comparator method.
Results
Usability and Reliability of Anterior Nares (AN) Swabs for at-Home Specimen Collection
We first explored the use of anterior nares (AN) swabs for specimen collection. For mass testing purposes, a swab that is widely available, inexpensive, easy to manufacture, and simple for self-collection is critical. The US Cotton #3 swabs fit these specifications; a polyester AN swab that resembles consumer-brand Q-tips (19). For the purposes of scaled observed or at-home self-specimen collection or specimen collection for a child, swabbing the anterior nares anatomical site would be more comfortable, accessible, and easier to describe to test users leading to fewer mistakes and better specimen collection (16, 20).
Therefore, we conducted a usability study to determine both the accuracy and ease of AN swabs in a Swab-and-Send program where at-home specimen collection kits were delivered to participant residences, the participants swabbed themselves or a child while being virtually monitored by clinical study coordinators and then packaged the specimen for return to the molecular testing laboratory (16, 21). After using the specimen collection kit, study participants completed a survey reporting their level of confidence, the kit’s ease of use, and the level of discomfort experienced during swabbing. Participants were recruited from the greater Seattle area and spanned a range of ages, races, household income, and educational attainment (Fig. S1 in the online Data Supplement; Supplemental Table S1, A–D).
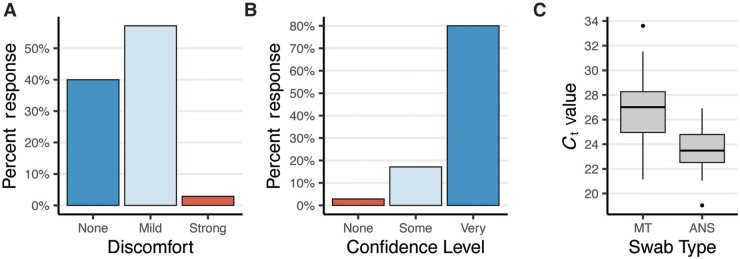
The results of the usability study were very encouraging. Most participants reported only mild discomfort during specimen collection with 40% of participants reporting no discomfort at all (Fig. 1, A). Most study participants also found the instructions clear and felt confident that they had correctly collected their specimen (Fig. 1, B). This was confirmed by low observed rates of error during specimen collection using the AN swabs and during packaging for return (online Supplemental Table S2, A and B). Molecular testing performed on these self-collected specimens confirmed this; RT–qPCR detected the human marker RNase P mRNA in 100% of swabs with an average Ct value of 23.5 (SD 1.7). The amount of RNase P mRNA recovered from the AN swabs was higher than for unsupervised collection of mid-turbinate swabs, which had an average Ct value of 26.9 (SD 2.5) (Fig. 1, C). Together, these data indicate that the use of widely available polyester swabs in the anterior nares is a viable and preferable alternative for at-home specimen collection.
Fig. 1.
Polyester anterior nares swabs are both comfortable and easy to use. (A, B), Study participants’ (n = 35) self-reported (A) discomfort and (B) confidence during self-administration of an anterior nares swab at home. (C), Boxplot depicting the RT–qPCR Ct values for RNaseP from self-administered anterior nares swabs (ANS) and mid-turbinate (MT) swabs.
Handling Dry Swabs in the Clinical Laboratory
Standard viral media such as UTM (e.g., COPAN Diagnostics) have been in short supply over the course of the pandemic. These salt-rich media inhibit direct RT–qPCR, making RNA extraction a necessity and thus create an additional bottleneck in the testing process. Furthermore, automated extraction systems are expensive and their reagents and consumables are also subject to global shortages. Therefore, we focused on eliminating UTM and extraction from our testing platform. To bypass UTM, we adopted a dry-swab transport and rehydration method validated by Quantigen that has been explored by other clinical testing laboratories (13, 22). Next, to eliminate RNA extraction and enable direct RT–qPCR, we tested rehydration solutions for their ability to elute contrived SARS-CoV-2 specimens, compatibility with direct RT–qPCR, and simplicity. We determined that elution in low-TE (10 mM Tris pH 7.5, 0.1 mM EDTA) without other detergents was best suited for direct RT–qPCR (online Supplemental Fig. S2). Unlike UTM and other saline solutions, the low ionic strength of low-TE does not inhibit PCR amplification. Moreover, low-TE can be quickly prepared using reagents commonly found in laboratories.
Bypassing nucleic acids extraction poses another problem; instead of the virus being inactivated by the denaturing agents during nucleic acid extraction, the specimen eluted from the swab remains potentially infectious for SARS-CoV-2 or other pathogens and poses a risk to laboratory staff. Accordingly, specimens from both conventional UTM and rehydrated dry swabs are processed inside a class II biosafety (BSL-2) cabinet, in accordance with federal regulatory guidance. However, it is practical and beneficial for downstream steps (such as preparing RT–qPCR reactions) to take place on a BSL-2 designated bench. Therefore, we compared several inactivation methods to determine which would be easiest without inhibiting PCR or causing a loss of sensitivity. Viral inactivation of coronaviruses can be achieved through the use of either detergent or heat (23). Our previous results demonstrate the negative impact of detergents on RT–qPCR (online Supplemental Fig. S2); therefore, we opted to deploy heat inactivation (online Supplemental Fig. S3). We used a protocol to heat inactivate at higher temperatures (95 ˚C) for 30 minutes to increase the safety margins. We also determined that this high-heat protocol had the added benefit of stabilizing the sample over time, a result concordant with another SARS-CoV-2 testing protocol in saliva (24).
Performance of Extraction-Free RT–qPCR
Having developed an extraction-free RT–qPCR protocol (EF-RT–qPCR), we set out to determine its performance on both contrived and clinical specimens. To assess analytical sensitivity, we first determined this assay’s limit of detection (LoD), the minimum number of SARS-CoV-2 RNA molecules that could be detected in greater than 95% of RT–qPCR reactions. To generate these contrived specimens, we inoculated AN swabs with clinical matrix collected from a healthy volunteer with dilutions of heat-inactivated SARS-CoV-2. These experiments determined the EF-RT–qPCR analytical sensitivity to be 2 molecules/µL of eluate for the Orf1b assay and 4 molecules/µL of eluate for the S-gene (Spike-gene) assay (online Supplemental Table S3). This LoD is comparable to the LoD of many other RT–qPCR-based tests that have been issued Emergency Use Authorization from the FDA (25).
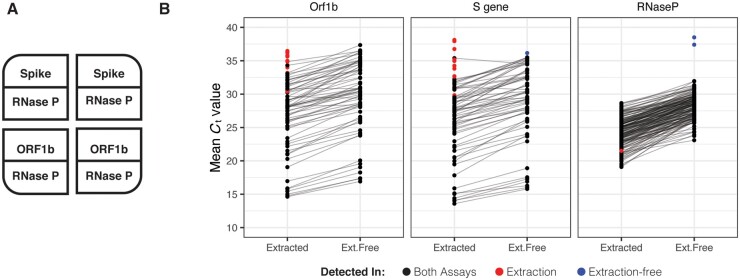
Next, we tested the performance of EF-RT–qPCR compared to our clinically validated RT–qPCR laboratory-developed test on archived AN specimens. In this assay, each sample is tested in 4 independent RT–qPCR reactions, comprising 2 SARS-CoV-2 assays (Orf1b and Spike) in duplicate, and is multiplexed with a RNase P assay in every well (Fig. 2, A and online Supplemental Fig. S4). Following RT–qPCR, a clinical result is determined by the number of replicates displaying SARS-CoV-2 amplification: positive (3 or 4 of 4 wells), low-positive/inconclusive (2 of 4 wells) and negative (0 or 1 of 4 wells). Head-to-head comparison between EF-RT–qPCR and a reference standard extraction-based RT–qPCR assay on matched specimens established that EF-RT–qPCR was 100% specific (56/56 negative specimens) and 91.0% sensitive (61/67–56 positive and 5 low-positive) (Fig. 2, B). Comparison of the mean delta Ct (ΔCt) values between the 2 assays showed that eliminating extraction did decrease analytical sensitivity. We observed an average increase of 1.96, 2.45, and 4.00 cycles for Orf1b, Spike, and RNase P assays, respectively. Indeed, the 6 specimens not detected by EF-RT–qPCR had an average Ct with the extraction-based RT–qPCR assay of 34.13 for Orf1b and 35.29 for Spike.
Fig. 2.
Extraction-free RT–qPCR set-up and test performance. (A), Assay layout of the EF-RT–qPCR test. One sample is assayed in 4 wells on a 384-well plate. Each sample is tested for 2 probes, in duplicate. RNase P is assayed in each well. (B), Mean Ct values for 67 specimens processed by EF-RT–qPCR and extraction-based RT–qPCR. Reactions with no amplification by one preparation protocol are demarcated with red or blue points as indicated.
Owing to an unstable supply chain, while validating the EF-RT–qPCR protocol, our clinical laboratory was forced to switch from the Roche Magna Pure 96 to the Thermo Fisher KingFisher Flex automated nucleic acids extraction platform. The relative sensitivity, specificity and ΔCt values between 619 prospective specimens run in parallel on both the KingFisher Flex (extraction) and EF-RT–qPCR were comparable to results of the retrospective study on stored specimens. EF-RT–qPCR detected SARS-CoV-2 in 100% of specimens that were positive by the extraction method with a 99.4% specificity (Supplemental Tables S5–S7).
Addition of Proteinase K Reduces Amplification Interference
After deploying EF-RT–qPCR as our clinical testing platform, we repeatedly observed 2 undesirable outcomes that were not observed in our validation studies. First, for 0.9% of specimens (n = 383/43 539), amplification of the human RNase P internal control was undetected in 2 or more of the 4 reactions (Fig. 2, A; online Supplemental Table S7). These specimens were classified as “failures” and each test was repeated before releasing the result. Second, for 0.5% of specimens (229/43 539), we sporadically observed the presence of strong amplification (Ct < 30) in a single well for one of the SARS-CoV-2 targets in specimens where the 3 other wells were undetected (online Supplemental Table S8). However, on repeat RT–qPCR, both with and without extraction, all 4 wells of the SARS-CoV-2 reactions for these specimens were undetected.
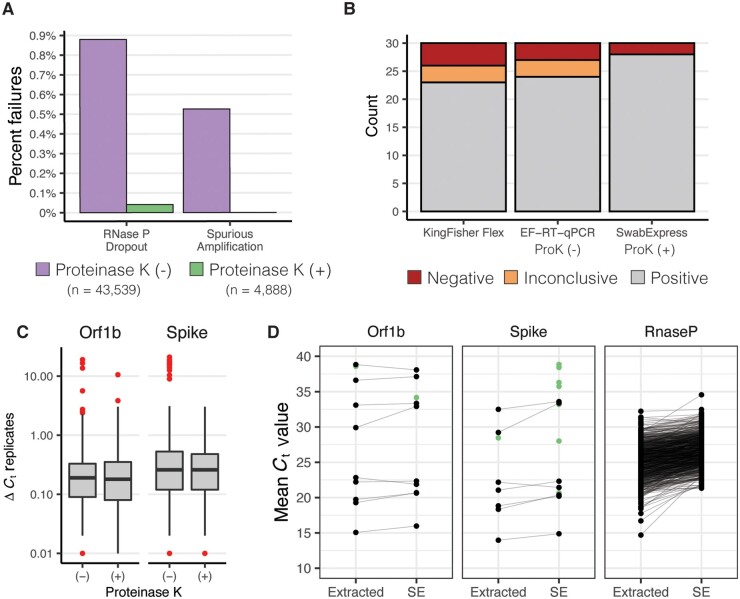
We noted that some of the specimens that produced these problematic outcomes had excess mucous or other nasal secretions. Therefore, we hypothesized that the addition of proteinase K (ProK) digestion could ameliorate both RNase P failures and the spurious SARS-CoV-2 amplification by digesting mucins and other potentially interfering proteins in the nasal specimens (26). We compared RT–qPCR results for 1222 clinical specimens prepared by the 30-minute 95 ˚C heat treatment with those digested with ProK for 15 minutes before heat treatment at 95 ˚C for 15 minutes. We observed approximately 10-fold fewer RT–qPCR reactions with failed RNase P amplification—27 of 4888 without ProK vs 2 of 4888 with ProK—reducing the failure rate to 0.04% (Fig. 3, A, online Supplemental Table S9), and improved RNase P detection (ΔCt −0.88) (online Supplemental Fig. S5). Furthermore, the addition of a ProK digestion step eliminated spurious amplification of SARS-CoV-2 targets.
Fig. 3.
Addition of Proteinase K improves test performance. (A), Observed percentage of test failures with and without the addition of proteinase K. (B), Archived samples with Ct > 28 reprocessed with either KingFisher Flex Extraction (left), Extraction-Free RT–PCR (middle), or SwabExpress (Extraction-Free RT–PCR + ProK) (right). Colors signify the number of samples and their classifications. (C), Box and whisker plots depicting the average delta Ct between replicate wells for SARS-Cov2 positive specimens. Red points indicate outliers. ΔCt values were more consistent on addition of proteinase K. (D), Mean Ct values of matched specimens run through the automated KingFisher extraction system (left) or using SwabExpress (SE). Specimens that were detected in only 1 of the 2 protocols are displayed as green points.
In the 4888 specimens processed both with and without ProK, ProK-treated specimens had decreased Ct values (mean decrease of 1.22 for Orf1B, and 0.97 for Spike). This increased sensitivity was also reflected in the ability to accurately classify archived SARS-CoV-2 positive specimens with Ct values >28 (Fig. 3, B). Repeatability and reproducibility were also improved with the addition of ProK (Fig. 3, C). On addition of ProK, on SARS-CoV-2 positive samples, our protocol had a higher concordance (93.3%) versus without ProK (90%) or specimens extracted on the KingFisher Flex (86.6%) (online Supplemental Table S10). After this optimization we named our final protocol “SwabExpress”—consisting of a dry AN swab, followed by ProK digestion and direct RT–PCR. Finally, we prospectively compared performance on 1169 specimens run in parallel on the SwabExpress and KingFisher Flex (extraction) platforms. Positive and negative clinical concordance was excellent; there was 100% concordance for positives results, 99.91% concordance across negatives with a small ΔCt value of 0.37 for the Orf1b target and 1.46 for the S target between the 2 assays (Fig. 3, D, online Supplemental Table S11).
SwabExpress is Compatible with Other SARS-CoV-2 RT–qPCR Assays
Our laboratory-developed test uses custom Orf1b and Spike-gene assays for detecting SARS-CoV-2. To establish that the SwabExpress protocol was compatible with the widely used CDC N1 and N2 assays, we performed RT–qPCR on 75 positive specimens and 92 negative specimens with the N1 and N2 assays performed in parallel on the SwabExpress platform and extraction-based RT–qPCR platform. The results were 100% concordant between our custom assays and the CDC assays. Ct values for positive samples were delayed when prepared by SwabExpress protocol compared to the Roche Magna Pure 96. However, this difference did not change the clinical interpretation of these samples (Supplemental Fig. S6). For the N1 assay, extracted specimens had an average Ct of 19.22 ± 3.67 versus 21.79 ± 4.33 with SwabExpress (ΔCt of 2.57). For the N2 assay, extracted specimens had an average Ct of 18.31 ± 3.73 versus Cts of 19.80 ± 3.72 for SwabExpress (ΔCt of 1.49) (Supplemental Table S12).
SwabExpress is Time and Cost Effective
A dry-swab, extraction-free RT–qPCR protocol comprises the minimal components of a diagnostic test. Although the addition of a proteinase K digestion adds $0.14 to the reagent cost for each sample, this cost is warranted. The addition of proteinase K reduces the repeat rate, reduces the chances of a false positive result from interfering substances during PCR amplification, improves the performance of the test, and, in our hands, outperformed a suboptimal yet widely used automated extraction system (Thermo KingFisher Flex™).
On adoption, SwabExpress approximately doubled laboratory capacity. First, hands-on technician time, previously spent preparing and running extraction systems, went toward accessioning and processing additional samples. Second, the SwabExpress protocol increases scale by using a convection oven that can process up to 6 96-well plates simultaneously. This throughput greatly exceeds the single 96-well plate processed by commercial automated extraction systems. Further scaling of the SwabExpress protocol can be accomplished through the purchase of additional or larger ovens, although RT–qPCR instruments used during amplification and readout still pose a substantial bottleneck in the testing protocol.
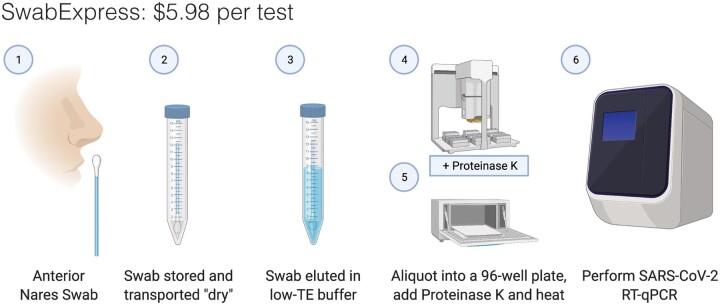
Along with the substantial cost of purchasing automated extractors, the consumables required for their operation cost between $4 and $5 per sample. By eliminating extraction and transport medium, SwabExpress reduces the associated costs by more than 90% (∼$0.20 per sample). In all, SwabExpress offers a time and cost-saving alternative to nucleic acids extraction using readily available reagents, which reduces dependence on a heavily burdened supply chain (Fig. 4, Supplemental Table S13).
Fig. 4.
SwabExpress workflow. (1) Anterior nares swabs are collected and (2) transported dry to the lab. On receipt, (3) each swab is then hydrated with low-TE buffer, aliquoted into a 96-well plate, and (4) proteinase K is added to every well. (5) The eluted specimens are digested and heat-inactivated in a laboratory oven before (6) they are loaded as the template in a RT–qPCR reaction. The cost listed includes reagents and consumables.
Discussion
Here we present SwabExpress, an end-to-end diagnostic platform optimized for faster and simpler low-cost detection of SARS-CoV-2 from nasal swabs without the use of nucleic acid extraction (Fig. 4). This protocol was so named for its ease, rapid turnaround, and simplicity—dry swabs, without extraction, enhanced with proK digestion. By eliminating transport media and extraction from the workflow, we have decreased cost per sample and reduced supply chain pressure for the laboratory. Because of the reduced cost and the ability to process many more specimens in parallel, our laboratory’s capacity markedly increased with its adoption. Importantly, we gained efficiency without sacrificing accuracy; our results suggest that the simplified SwabExpress protocol (direct elution from dry swab into low-TE + proteinase K → RT–qPCR) is as sensitive as the conventional PCR protocol (swab → UTM → RNA extraction → RT–qPCR). SwabExpress has supported scaled testing in our laboratory with over 91 000 tests performed to date and allowed us to support large testing endeavors such as thHusky Coronavirus Testing Program for the University of Washington (17).
There are some caveats to consider. Even with the addition of proteinase K, specimens with excess mucous fail to amplify RNase P. Since adding this proteinase digestion step, 18/12 991 specimens have had 2 or more RNase P reactions fail (0.1%) and our laboratory reflexes these few specimens to an extraction protocol. However, 0.1% compares favorably when compared to a protocol with extraction where the failure rate due to failed RNase P is 1% (215/22 546). In addition, the unknown presence of inhibitors precludes comparison of Ct values between specimens; therefore, studies directly comparing Ct values from different specimens may not yield accurate results.
We have observed a marked loss of viral RNA after freeze–thaw cycles for specimens stored in low-TE compared to specimens stored in commercial UTM. We detect a ΔCt of about 2.5 for specimens after −80 °C storage in low-TE, whereas the ΔCt for specimens retested after storage at −80 °C in UTM has historically been negligible. This affects the ability to use these specimens for downstream applications such as genomic sequencing.
Several improvements can be incorporated into the SwabExpress platform. First, the ability to detect multiple pathogens from one assay can be explored. It is likely that SwabExpress will be compatible with other enveloped viruses such as influenza and respiratory syncytial virus. Multiple targets can be detected in many qPCR systems and the reemergence of these viruses as COVID-19 prevention control measures are relaxed will be of interest to monitor. Second, the most labor-intensive part of the SwabExpress protocol is sample accessioning. Receiving individual 10-mL tubes and transferring the eluate to 96-well format takes approximately 2.5 minutes per sample. Receiving nasal swabs in 96-well compatible, laboratory-ready transport tubes would streamline the process considerably (27). Third, incubation times for proteinase K digestion, heat inactivation, and RT–qPCR could be further optimized to save additional time during the testing process.
Massive scaling and deployment of SARS-CoV-2 testing is essential to curtailing the COVID-19 pandemic, and will likely be necessary well into the future. The protocol evaluated here, including thousands of real-world, self-collected nasal swabs, would markedly simplify the workflow for RT–qPCR, the most widely deployed testing paradigm, by eliminating the need for viral transport media and RNA extraction, both of which are currently experiencing significant supply chain challenges. Looking forward, we envision that nasal swabs—self-collected into laboratory-ready barcoded tubes and transported dry—could potentially serve as a common input to a range of SARS-CoV-2 nucleic acid tests for public health surveillance applications. This includes gold-standard tests such as RT–qPCR, but also potentially new modalities such as SwabSeq (9). The operationalization of the mass distribution and return of such laboratory-ready collection devices is a significant effort that should begin now.
Ethics Approval
Sequencing and analysis of specimens from the Seattle Flu Study, the Hospitalized and Ambulatory Adults with Respiratory Viral Infections (HAARVI) study and the SCAN study were approved by the Institutional Review Board at the University of Washington (protocols STUDY00006181, STUDY00000959, STUDY00007628, STUDY00010432, STUDY00011148). Informed consent was obtained for all participant specimens.
Supplemental Material
Supplemental material is available at Clinical Chemistry online.
Supplementary Material
Contributor Information
Seattle Flu Study Investigators:
Helen Y Chu, Michael Boeckh, Janet A Englund, Michael Famulare, Christina M Lockwood, Barry R Lutz, Deborah A Nickerson, Mark J Rieder, Lea M Starita, Matthew Thompson, Cécile Viboud, Jay Shendure, and Trevor Bedford
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership
S. Kosuri is an employee of Octant Inc., who have developed SwabSeq, a method that will likely benefit from dry swab and extraction-free protocols becoming standard. E.Q. Konnick, Association for Molecular Pathology, College of American Pathologists, Washington State Society of Pathologists.
Consultant or Advisory Role
H.Y. Chu reported consulting with Ellume, Pfizer, The Bill and Melinda Gates Foundation, Glaxo Smith Kline, and Merck. J. Shendure is a consultant with Guardant Health, Maze Therapeutics, Camp4 Therapeutics, Nanostring, Phase Genomics, Adaptive Biotechnologies, and Stratos Genomics.
Stock Ownership
S. Kosuri, Octant Inc.
Honoraria
L.M. Starita, AMP.
Research Funding
The Seattle Flu Study and SCAN are administered by the Brotman Baty Institute for Precision Medicine and funded by Gates Ventures, the private office of Bill Gates. The funder was not involved in the design of the study and does not have any ownership over the management and conduct of the study, the data, or the rights to publish. Funding for the HAARVI study comes from DARPA HR001117S0019, the Bill and Melinda Gates Foundation, Emergent Ventures grant to H.Y. Chu. L.M. Starita, and J. Shendure are funded by 1RM1HG010461-01 from the NHGRI and J. Shendure is an Investigator of the Howard Hughes Medical Institute. J.S. Debley is funded by NIH/NIAID K24AI150991-01S1, which supported the viral inactivation studies. Additional funding for this project came from the University of Washington in support of the Husky Coronavirus Testing Program with funds from the United States Senate and House of Representative Bill 748, Coronavirus Aid, Relief, and Economic Security Act (CARES Act). H.Y. Chu has received research funding from Gates Ventures, Sanofi Pasteur, and support and reagents from Ellume and Cepheid outside of the submitted work. J. Shendure has a research collaboration with Illumina. S. Kosuri, Testing for America supporting separate work on SwabSeq development, Ginkgo Bioworks supporting separate work on CLIA laboratory formation.
Expert Testimony
S. Kosuri, Broad Institute.
Patents
S. Kosuri, Ginkgo Bioworks—Licensing for SwabSeq multiplexed assay development, Harvard U—Royalties from patents relating to genome editing, UCLA—Royalties from patents relating to multiplexed assays.
Role of Sponsor
The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Seattle Flu Study Principal Investigators
Helen Y. Chu1,7, Michael Boeckh1,2,7, Janet A. Englund3,7, Michael Famulare4, Christina M. Lockwood1,7, Barry R. Lutz5,7, Deborah A. Nickerson6,7, Mark J. Rieder7, Lea M. Starita6,7, Matthew Thompson9, Cécile Viboud10, Jay Shendure6,7,8, and Trevor Bedford2,6,7.
Affiliations
1 Department of Medicine, University of Washington, Seattle, WA
2 Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Center, Seattle, WA
3 Seattle Children’s Research Institute, Seattle, WA
4 Institute for Disease Modeling, Seattle, WA
5 Department of Bioengineering, University of Washington, Seattle, WA
6 Department of Genome Sciences, University of Washington, Seattle, WA
7 Brotman Baty Institute for Precision Medicine, Seattle, WA
8 Howard Hughes Medical Institute, Seattle, WA
9 Department of Family Medicine, University of Washington, Seattle, WA
10 National Institutes of Health, Bethesda, MD.
Acknowledgments
We would like to thank the Seattle Flu Study, HAARVI, SCAN, and Husky Coronavirus Testing program study participants for their invaluable contributions to this research. We thank Sydney Floth, Jefferson Nguyen, Ashley Gate, and Gift Nwanne for help with the usability study, Catherine Moore of Public Health Wales and Dan Wattendorf, Emily Turner, and Karen Heichman of the Bill and Melinda Gates Foundation for helpful advice.
References
- 1. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A.. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2021;2:e13–22–e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brauner JM, Mindermann S, Sharma M, Johnston D, Salvatier J, Gavenčiak T, et al. Inferring the effectiveness of government interventions against COVID-19. Science 2021;371:eabd9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Larremore DB, Wilder B, Lester E, Shehata S, Burke JM, Hay JA, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 surveillance. Preprint at medRxiv 10.1101/2020.06.22.20136309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellewell, J., Russell, T.W., The SAFER Investigators and Field Study Team. et al. Estimating the effectiveness of routine asymptomatic PCR testing at different frequencies for the detection of SARS-CoV-2 infections. BMC Med 19, 106 (2021). 10.1186/s12916-021-01982-x.et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moore C, Corden S, Sinha J, Jones R.. Dry cotton or flocked respiratory swabs as a simple collection technique for the molecular detection of respiratory viruses using real-time NASBA. J Virol Methods 2008;153:84–9. [DOI] [PubMed] [Google Scholar]
- 6. Emerson J, Cochrane E, McNamara S, Kuypers J, Gibson RL, Campbell AP.. Home self-collection of nasal swabs for diagnosis of acute respiratory virus infections in children with cystic fibrosis. J Pediatric Infect Dis Soc 2013;2:345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.U.S. Department of Health and Human Services Food and Drug Administration. Policy for coronavirus disease-2019 tests during the public health emergency. US Food and Drug Administration. 2020. https://www.fda.gov/media/135659/download (Accessed June 2020).
- 8.Vogels CBF, Watkins AE, Harden CA, Brackney DE, Shafer J, Wang J, Caraballo C, Kalinich CC, Ott IM, Fauver JR, Kudo E, Lu P, Venkataraman A, Tokuyama M, Moore AJ, Muenker MC, Casanovas-Massana A, Fournier J, Bermejo S, Campbell M, Datta R, Nelson A; Yale IMPACT Research Team, Dela Cruz CS, Ko AI, Iwasaki A, Krumholz HM, Matheus JD, Hui P, Liu C, Farhadian SF, Sikka R, Wyllie AL, Grubaugh ND., SalivaDirect: A simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med (N Y). 2021; 2:263-280.e6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smyrlaki I, Ekman M, Lentini A, Rufino de Sousa N, Papanicoloau N, Vondracek M, Aarum J, Safari H, Muradrasoli S, Rothfuchs AG, Albert J, Högberg B, Reinius B.. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat Commun 2020;11:4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lübke N, Senff T, Scherger S, Hauka S, Andrée M, Adams O, et al. Extraction-free SARS-CoV-2 detection by rapid RT-qPCR universal for all primary respiratory materials. J Clin Virol 2020;130:104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bruce EA, Huang M-L, Perchetti GA, Tighe S, Laaguiby P, Hoffman JJ, et al. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. PLoS Biol 2020;18:e3000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Merindol N, Pépin G, Marchand C, Rheault M, Peterson C, Poirier A, et al. SARS-CoV-2 detection by direct rRT-PCR without RNA extraction. J Clin Virol 2020;128:104423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Padgett LR, Kennington LA, Ahls CL, Samarasinghe DK, Tu Y-P, Wallander ML, et al. Polyester nasal swabs collected in a dry tube are a robust and inexpensive, minimal self-collection kit for SARS-CoV-2 testing. Available from: 10.1101/2020.10.09.20210302. [DOI] [PMC free article] [PubMed]
- 14. Ñique AM, Coronado-Marquina F, Mendez Rico JA, García Mendoza MP, Rojas-Serrano N, Simas PVM, et al. A faster and less costly alternative for RNA extraction of SARS-CoV-2 using proteinase k treatment followed by thermal shock. PLoS ONE 2021;16:e0248885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCulloch DJ, Kim AE, Wilcox NC, Logue JK, Greninger AL, Englund JA, et al. Comparison of unsupervised home self-collected midnasal swabs with clinician-collected nasopharyngeal swabs for detection of SARS-CoV-2 infection. JAMA Netw Open 2020;3:e2016382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim AE, Brandstetter E, Wilcox N, Heimonen J, Graham C, Han PD, et al. Evaluating specimen quality and results from a community-wide, home-based respiratory surveillance study. J Clin Microbiol 2021;59:e02934-20. 10.1128/JCM.02934-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weil AA, Sohlberg SL, O’Hanlon JA, Casto AM, Emanuels AW, Lo NK, et al. SARS-CoV-2 epidemiology on a public university campus in Washington State. Preprint at medRxiv http://medrxiv.org/lookup/doi/10.1101/2021.03.15.21253227 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greater Seattle Coronavirus Assessment Network (SCAN). 2021. https://scanpublichealth.org/ (Accessed June 2021).
- 19.U.S. Department of Health and Human Services Food and Drug Administration. FDA, Gates Foundation, UnitedHealth Group, Quantigen, and U.S. Cotton Collaborate to address testing supply needs. US Food and Drug Administration. 2020. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-gates-foundation-unitedhealth-group-quantigen-and-us-cotton (Accessed June 2020).
- 20. Truong M, Pfau B, McDermot E, Han PD, Brandstetter E, Richardson M, et al. Comparable specimen collection from both ends of at-home mid-turbinate swabs. J Clin Microbiol 2021;59. 10.1128/JCM.03073-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chu HY, Englund JA, Starita LM, Famulare M, Brandstetter E, Nickerson DA, et al. Early detection of Covid-19 through a citywide pandemic surveillance platform. N Engl J Med 2020;383:185–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parikh BA, Wallace MA, McCune BT, Burnham C-AD, Anderson NW.. The effects of “dry swab” incubation on SARS-CoV-2 molecular testing. J Appl Lab Med 2021; as doi: 10.1093/jalm/jfab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Darnell MER, Taylor DR.. Evaluation of inactivation methods for severe acute respiratory syndrome coronavirus in noncellular blood products. Transfusion 2006;46:1770–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ranoa DRE, Holland RL, Alnaji FG, Green KJ, Wang L, Brooke CB, et al. Saliva-based molecular testing for SARS-CoV-2 that bypasses RNA extraction. Cold Spring Harbor Lab 2020. https://www.biorxiv.org/content/10.1101/2020.06.18.159434v1 [Google Scholar]
- 25.Resilience Health. https://www.resiliencehealth.com/tests.html (2021) (Acessed October 2020).
- 26. Mallmann L, Schallenberger K, Demolliner M, Eisen AKA, Hermann BS, Heldt FH, et al. Pre-treatment of the clinical sample with Proteinase K allows detection of SARS-CoV-2 in the absence of RNA extraction. bioRxiv 2020.05.07.083139. 10.1101/2020.05.07.083139. [DOI]
- 27.Rhinostics. Rhinostics product information. https://rhinostics.com (2021) (Accessed June 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.