Abstract
The clinical significance of severe acute respiratory syndrome coronavirus 2 (SARS CoV-2) RNA in stool remains uncertain. We found that extrapulmonary dissemination of infection to the gastrointestinal tract, assessed by the presence of SARS-CoV-2 RNA in stool, is associated with decreased coronavirus disease 2019 (COVID-19) survival. Measurement of SARS-CoV-2 RNA in stool may have utility for clinical risk assessment.
Keywords: SARS CoV-2, mortality, fecal, RNA, organoids, GI epithelium, phylogenetics
Coronavirus disease 2019 (COVID-19) is a global pandemic causing high morbidity and mortality. The infection predominantly results in respiratory manifestations [1, 2], but extrapulmonary complications are increasingly being recognized as a critical part of the disease [3–5]. The presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in stool has been reported [6, 7]; however, no studies have described associations between stool SARS-CoV-2 viral load and patient outcomes. Additionally, characterization of the ability of SARS-CoV-2 to infect gastrointestinal (GI) tissue and remain infectious in stool is incomplete.
METHODS
We enrolled 64 hospitalized patients with COVID-19 confirmed by nasopharyngeal (NP) swab. Clinical information was obtained in collaboration with the Massachusetts General Hospital COVID Registry. Stool samples were collected from all hospitalized patients who consented to participate in the study at the first available time point, regardless of time since admission. Continuous data were compared by Mann-Whitney test; Fisher exact test was performed for categorical data. Survival was summarized using a Kaplan-Meier curve with significance and hazard ratio assessed by log-rank test.
Stool samples underwent quantitative reverse transcription polymerase chain reaction (qRT-PCR) to detect SARS-CoV-2 N1, N2, E, and RdRp RNA with a limit of detection of 50 copies/mL and cycle threshold value cutoff of 39. A standard curve of RNA generated from SARS-CoV-2 of known titer was used to quantify viral load. Viral particles from a subset of stool samples were isolated by filtering stool and applying 200 µL of filtrate to Vero E6 cells. Cell lysates were then assessed for SARS-CoV-2 subgenomic E RNA by qRT-PCR [8]. GI tissue specimens were obtained from a subset of patients (autopsy samples and GI surgical resections) for detection of SARS-CoV-2 by immunofluorescence microscopy, in situ hybridization, and qRT-PCR. SARS-CoV-2 whole genome sequencing was performed using the Artic protocol, a 20-amplicon nested PCR amplification strategy followed by maximum-likelihood phylogenetic reconstruction [3]. The organoids were isolated and cultured from colon tissues collected by surgical resection from COVID-19 patients who presented with ischemic bowel. Tissue specimens underwent manual dissection of the colon epithelium and were processed as described in the Supplementary Materials [9].
RESULTS
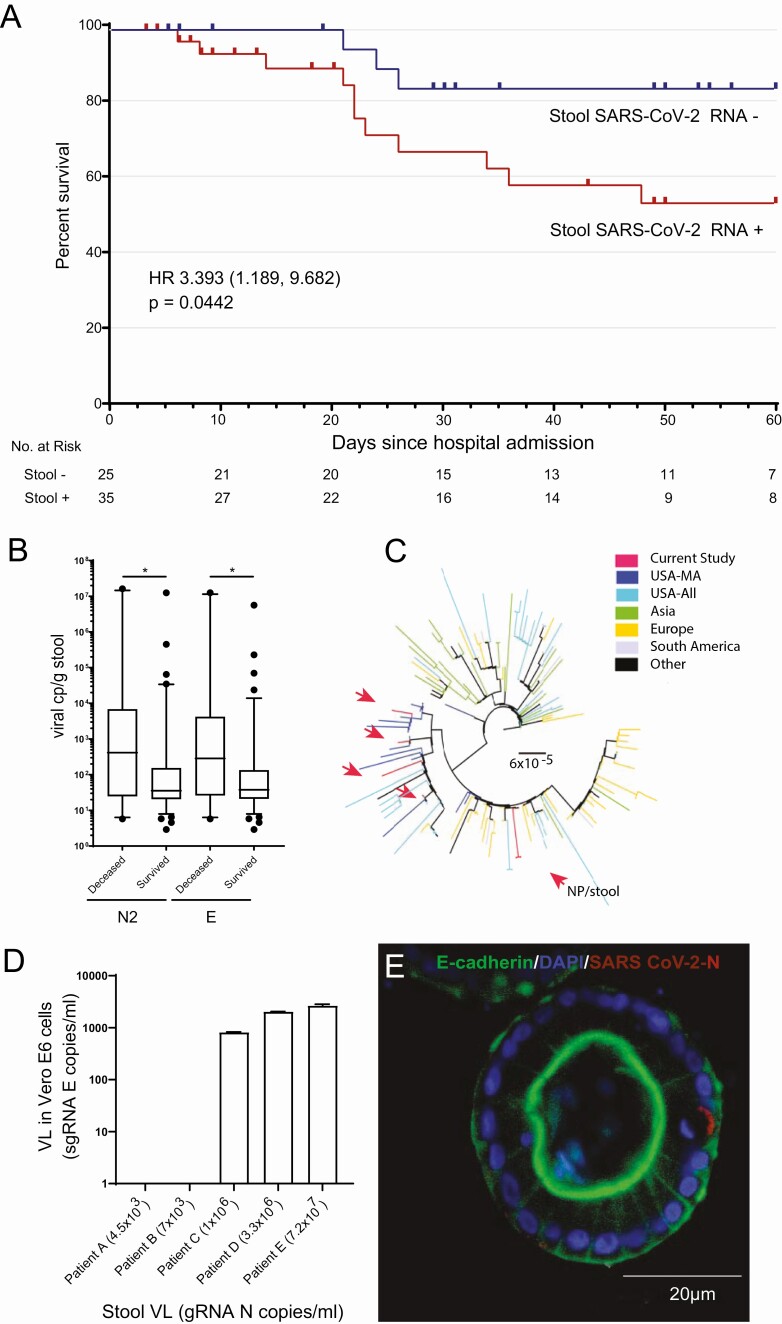
Fifty-eight percent (35/60) of hospitalized patients with COVID-19 demonstrated detectable SARS-CoV-2 RNA in stool samples by qRT-PCR. Clinical and demographic characteristics, including GI symptoms, on admission did not differ between patients with and without detectable SARS-CoV-2 RNA in feces (Supplementary Table 1). Since the timing of stool sample collection might impact associations with disease outcomes, we examined potential correlations between time of sample collection and viral load, but failed to see any significant correlations for stool, NP swabs, or plasma (Supplementary Figure 1). However, patients with detectable stool SARS-CoV-2 RNA at any time during their hospitalization had a significantly increased risk of mortality (hazard ratio, 3.4 [95% confidence interval: 1.189–9.862]; P = .0442; Figure 1A). Stool samples from patients who died also had significantly higher SARS-CoV-2 RNA levels compared to those who survived to discharge (N2 RNA: median of 415 copies/g for those who died and 35 copies/g for survivors, P = .0308; E RNA: median of 286 copies/g for those who died with COVID-19 and 38 copies/g for survivors, P = .0432) (Figure 1B). Analysis of stool and NP swabs demonstrated that patients maintained viral shedding from their stool for a median of 16 days from the first positive NP swab test and 20 days after symptom onset. Of note, 8.6% of patients continued to have detectable SARS-CoV-2 RNA in stool despite negative prior or concurrent testing by NP swab. To assess potential relationships between respiratory and GI levels of SARS-CoV-2, matched stool and NP specimens were collected from 25 patients and no correlation was observed (Supplementary Figure 2A). There was also no correlation observed between viral load in plasma and stool from 8 matched patient specimens and overall plasma viral load was very low (Supplementary Figure 2B). To assess relatedness of viruses found in the respiratory tract and stool, SARS-CoV-2 whole genome sequencing was performed from 5 stool samples, which showed close phylogenetic relationship to viral strains observed from NP swabs of U.S. patients (Figure 1C). Paired stool and NP viral sequences from a single patient were also highly related to each other.
Figure 1.
Association of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the gastrointestinal (GI) tract with coronavirus disease 2019 (COVID-19) survival and tissue inflammation. A, Kaplan-Meier survival curve shows a significant increase in the mortality of COVID-19 patients with detectable stool SARS-CoV-2 RNA compared to those without detectable RNA. B, SARS-CoV-2 viral load was higher in those who died from COVID-19 than those who survived as measured by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) of N2 and E gene RNA levels. *P <0.05. C, Phylogenetic tree showing stool-derived SARS-CoV-2 isolates (red arrows), along with representative sequences from Massachusetts (USA-MA), the United States (USA-All), Asia, Europe, South America, and Africa/Canada (Other). The scale represents 0.00006 nucleotide substitutions per site. D, SARS-CoV-2 isolated from COVID-19 patient stool replicated in Vero E6 cells. Subgenomic E RNA copies/mL was measured in Vero E6 cell lysates by qRT-PCR following inoculation with stool filtrates from COVID-19 patients with varying viral load. E, Immunofluorescence image of a representative colon organoid derived from surgically resected colon of a COVID-19 patient stained for DAPI (blue), E cadherin (green), and SARS-CoV-2 N (red). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; gRNA, genomic RNA; HR, hazard ratio; NP, nasopharyngeal; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; sgRNA, subgenomic RNA; VL, viral load.
Although SARS-CoV-2 RNA was readily detectable in stool, we further assessed the infectious potential of fecal virions. Viral particles were isolated from stool specimens of 5 patients with stool viral loads ranging from 4.5 × 103–7.2 × 107 nucleocapsid (N) gene copies/mL of stool filtrate. Vero E6 epithelial cell monolayers were inoculated with stool virions and productive SARS-CoV-2 infection was detected from stool samples with >106 N gene copies/mL (Figure 1D). Among our cohort, 6.25% (7/112) of hospitalized patients had stool viral load >106 N gene copies/mL, indicating that only a small subset of patients likely had measurable infectious virus in stool despite detectable SARS-CoV-2 RNA.
To further assess SARS-CoV-2 infection of intestinal tissue, colon specimens resected from patients with COVID-19 were stained for SARS-CoV-2 N protein (Supplementary Figure 2C and 2D). A median of 15% of colonic epithelial cells were observed to be infected in specimens from patients with COVID-19 (interquartile range, 21.5%). SARS-CoV-2 spike (S) RNA was also identified in colon epithelial cells by in situ hybridization (Supplementary Figure 2E), along with S antisense staining, indicating active viral replication in colon epithelium (Supplementary Figure 2F). Moreover, we generated organoid cultures from a collection of surgically resected colon from COVID-19 patients (n = 4) who underwent surgery due to ischemic bowel (stool viral load and clinical data reported in Supplementary Figure 2G and Supplementary Table 2). N protein staining of these organoids after 2 weeks of culture in which organoids were passaged confirmed the presence of patient-derived SARS-CoV-2 virus (Figure 1E and Supplementary Figure 2G), indicating that endogenously derived SARS-CoV-2–infected colon epithelial cells can be maintained within colon organoids ex vivo.
DISCUSSION
Although studies have indicated that COVID-19 patients with critical illness are more likely to have GI symptoms [10, 11], a recent study suggested that COVID-19 patients with GI symptoms have significantly reduced disease severity and mortality [12]. However, no study has identified an association between stool viral RNA and outcomes. In this study, we demonstrated that hospitalized COVID-19 patients with detectable SARS-CoV-2 RNA in stool had significantly higher mortality, despite indistinguishable laboratory value, GI symptoms, comorbidities, and demographic factors on admission. Patients who died from COVID-19 also had significantly higher stool SARS-CoV-2 RNA viral levels than those who survived. Furthermore, stool SARS-CoV-2 viral load did not correlate with RNA levels from plasma or NP swabs. Together, these observations suggest that assessment of SARS-CoV-2 RNA in stool may have utility as an independent noninvasive method to assess COVID-19 mortality risk.
Recovery of full-length genomic SARS-CoV-2 RNA sequence from stool indicated that stool RNA was not entirely due to degraded virus ingested from the respiratory tract. Additionally, we also observed the same viral strain in both GI and respiratory tissues, suggesting that the same viral strain can infect both sites. Importantly, our findings also indicate that infectious SARS CoV-2 can be recovered from stool, but only in those patients with high stool viral loads (>106 N gene copies/mL), which are found only in a minority of patients. Evidence of in vivo infection of GI tissues was observed using multiple approaches. The presence of SARS-CoV-2 antisense S RNA in the epithelium was specifically indicative of active viral replication in colon mucosa, which has not been previously reported. Detection of SARS-CoV-2–infected epithelial cells from resected colon of COVID-19 patients in ex vivo colon organoids cultured for 2 weeks was also consistent with in vivo infection. Overall, these findings suggest that dissemination of SARS-CoV-2 to the GI tract can occur and result in active infection of intestinal epithelial cells.
Overall, our study indicates that GI tissues are infected by SARS-CoV-2 and detection of extrapulmonary viral dissemination to the GI tract, reflected by the presence of SARS-CoV-2 RNA in stool, is associated with decreased COVID-19 survival. Although we identified infectious virus in stool, this only occurred in a small percentage of COVID-19 patients, suggesting that fecal-oral routes are unlikely to significantly contribute to transmission. This work informs our understanding of COVID-19 pathogenesis in the GI tract and identifies potential ways to better diagnose and risk-assess those with COVID-19.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. U. D. A., G. E., and M. F. performed experiments, analyzed, and interpreted data. M. C. C., A. N. H., A. G., L. R., D. L., and L. E. A. provided data and reagents. M. F., A. E. S., I. G., L. M. F., and T. J. D. helped in acquisition of data. V. A. T., G. C. L., S. A. R., D. E., G. V., J. Z. L., R. H., and J. R. S. helped with technical or material support. U. D. A. and G. E. designed the study and wrote the manuscript. D. S. K. and O. Y. supervised the study and critically revised the manuscript.
Financial support. This work was supported by the National Institutes of Health (NIH) - T32 CA009216 to G.E. and the Ragon Insitute of MGH, MIT and Harvard.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Huang C, Wang Y, Li X, et al. . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020; 323:1239–42. [DOI] [PubMed] [Google Scholar]
- 3. Choi B, Choudhary MC, Regan J, et al. . Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020; 383:2291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gupta A, Madhavan MV, Sehgal K, et al. . Extrapulmonary manifestations of COVID-19. Nat Med 2020; 26:1017–32. [DOI] [PubMed] [Google Scholar]
- 5. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020; 158:1831–3.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parasa S, Desai M, Thoguluva Chandrasekar V, et al. . Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019: a systematic review and meta-analysis. JAMA Netw Open 2020; 3:e2011335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheung KS, Hung IFN, Chan PPY, et al. . Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology 2020; 159:81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wölfel R, Corman VM, Guggemos W, et al. . Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 9. Ooft SN, Weeber F, Dijkstra KK, et al. . Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med 2019; 11:eaay2574. [DOI] [PubMed] [Google Scholar]
- 10. El Moheb M, Naar L, Christensen MA, et al. . Gastrointestinal complications in critically ill patients with and without COVID-19. JAMA 2020; 324:1899–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaafarani HMA, El Moheb M, Hwabejire JO, et al. . Gastrointestinal complications in critically ill patients with COVID-19. Ann Surg 2020; 272:e61–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Livanos EA, Jha D, Cossarini F, et al. . Intestinal host response to SARS CoV-2 infections and COVID-19 outcomes in patients with gastrointestinal symptoms. Gastroenterology 2021; 160:2435–50.e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.