ABSTRACT
Methane and ammonia have to be removed from wastewater treatment effluent in order to discharge it to receiving water bodies. A potential solution for this is a combination of simultaneous ammonia and methane oxidation by anaerobic ammonia oxidation (anammox) bacteria and nitrite/nitrate-dependent anaerobic methane oxidation (N-damo) microorganisms. When applied, these microorganisms will be exposed to oxygen, but little is known about the effect of a low concentration of oxygen on a culture containing these microorganisms. In this study, a stable coculture containing anammox and N-damo microorganisms in a laboratory scale bioreactor was established under oxygen limitation. Membrane inlet mass spectrometry (MIMS) was used to directly measure the in situ simultaneous activity of N-damo, anammox, and aerobic ammonia-oxidizing microorganisms. In addition, batch tests revealed that the bioreactor also harbored aerobic methanotrophs and anaerobic methanogens. Together with fluorescence in situ hybridization (FISH) analysis and metagenomics, these results indicate that the combination of N-damo and anammox activity under the continuous supply of limiting oxygen concentrations is feasible and can be implemented for the removal of methane and ammonia from anaerobic digester effluents.
IMPORTANCE Nitrogen in wastewater leads to eutrophication of the receiving water bodies, and methane is a potent greenhouse gas; it is therefore important that these are removed from wastewater. A potential solution for the simultaneous removal of nitrogenous compounds and methane is the application of a combination of nitrite/nitrate-dependent methane oxidation (N-damo) and anaerobic ammonia oxidation (annamox). In order to do so, it is important to investigate the effect of oxygen on these two anaerobic processes. In this study, we investigate the effect of a continuous oxygen supply on the activity of an anaerobic methane- and ammonia-oxidizing coculture. The findings presented in this study are important for the potential application of these two microbial processes in wastewater treatment.
KEYWORDS: anaerobic methane oxidation, anammox, nitrification, methane oxidation, oxygen exposure, wastewater treatment, ammonia oxidation
INTRODUCTION
Aerobic removal of organic carbon and nitrogen from wastewater requires large amounts of oxygen, energy, and space, which make it an energy intensive and costly process (1). Furthermore, organic carbon is oxidized to the climate-active waste product CO2 or converted to biomass, contributing to global warming and solid waste generation, respectively. Conversely, in anaerobic wastewater treatment, most of the organic carbon is converted into valuable biogas (methane), resulting in less sludge production. Consequently, for anaerobic wastewater systems, operational costs are considerably lower and the produced biogas can be used for electricity generation (1). However, one drawback of using anaerobic wastewater treatment is the remaining dissolved methane after methane is harvested from the gas phase (2). This dissolved methane escapes from these anaerobic treatment systems to the atmosphere and has a significant environmental impact because methane is a greenhouse gas with radiative forcing that is 28 times higher than that of CO2 on a 100-year scale (3).
Recently, several anaerobic microbial processes, which oxidize methane or ammonia while reducing nitrogen oxides, were discovered. In the anaerobic ammonia oxidation (anammox) process, bacteria use nitrite as their terminal electron acceptor to convert ammonia into mainly N2 and some nitrate (equation 1) (4). In parallel with this, two distinct groups of microorganisms, collectively termed nitrite/nitrate-dependent anaerobic methane oxidation (N-damo) microorganisms, can oxidize methane coupled to nitrite or nitrate reduction. Nitrite-dependent methane oxidation is performed by bacteria (“Candidatus Methylomirabilis spp.”), which reduce nitrite to N2 via NO without producing N2O (equation 2) (5, 6). On the other hand, nitrate-dependent methane oxidation is carried out by archaea (“Candidatus Methanoperedens spp.”) by coupling the oxidation of methane to the reduction of nitrate to nitrite (equation 3) (7). A combination of these microbial methane and ammonia oxidation processes can be applied for the simultaneous removal of ammonia and methane from wastewater streams (8, 9).
| (1) |
| (2) |
| (3) |
Under laboratory conditions, anammox and N-damo microorganisms can be grown together in stable cocultures, which are able to convert ammonia and methane simultaneously (10–13). In these laboratory-scale systems, the electron acceptor in the form of nitrate and/or nitrite is fed externally to the cultures. However, these compounds cannot be added to full-scale wastewater treatment plants due to economic and environmental concerns. Furthermore, N-oxide concentrations present in wastewater cannot support anaerobic ammonia and methane oxidation in full-scale installations. Therefore, nitrate and/or nitrite has to be microbially produced in situ for a successful application of these processes (8). It was previously shown that nitrite can be produced by aerobic ammonia-oxidizing bacteria (AOB), which form stable cocultures with anammox bacteria in both laboratory-scale bioreactors and full-scale wastewater treatment plants (14, 15). In such bioreactors, limiting amounts of oxygen are added for the conversion of part of the ammonia to nitrite (partial nitritation), which is then subsequently used by anammox bacteria. The application of these partial-nitritation anammox (PNA) bioreactors at full scale has been accepted as a more cost-effective and environmentally friendly method to remove ammonia from wastewater (1, 16). It has recently been put forward that, with the addition of methane-oxidizing microorganisms, such bioreactors can simultaneously remove ammonia and methane (8, 11, 12). Furthermore, several modeling studies have explored the feasibility of such bioreactors in laboratory and full-scale applications (17–21).
The key to the successful application of a combined N-damo/anammox system lies in the controlled addition of oxygen and the resulting conversion of ammonia to nitrite, which would then support both anaerobic methane and ammonia oxidation. However, this oxygen addition should be strictly controlled, since too much oxygen would induce the growth of aerobic nitrite-oxidizing (NOB) and methane-oxidizing (MOB) microorganisms (11, 22). Furthermore, high concentrations of oxygen inhibit N-damo and anammox microorganisms (11, 23). Altogether, the efficient and successful application of these systems depends on the controlled addition of oxygen and the constant and sufficient supply of nitrite/nitrate for anammox and N-damo bacteria. The supply of oxygen, ammonium, and methane to a culture that contains aerobic and anaerobic ammonia-, nitrite-, and methane-oxidizing microorganisms will result in a competition for all supplied and produced substrates, thereby creating an interesting interplay between several clades of microorganisms. In order to apply the simultaneous removal of ammonia and methane under oxygen-limited conditions, it is imperative to understand the microbial interactions in these bioreactors.
Here, we describe a laboratory-scale sequencing batch reactor (SBR) containing an anammox/N-damo coculture. Limiting amounts of oxygen were introduced to this culture, and the microbial interactions between the different clades of ammonia- and methane-oxidizing microorganisms were investigated using stable 15N-isotopes, ex situ batch assays, and membrane inlet mass spectrometry (MIMS). The community composition was studied by fluorescence in situ hybridization (FISH) and 16S rRNA gene analysis. Taken together, results of this study show that aerobic ammonia oxidizers and methanotrophs can be simultaneously active with anammox and N-damo microorganisms in the presence of limited amounts of oxygen.
RESULTS
Establishing an oxygen-limited anammox/N-damo coculture.
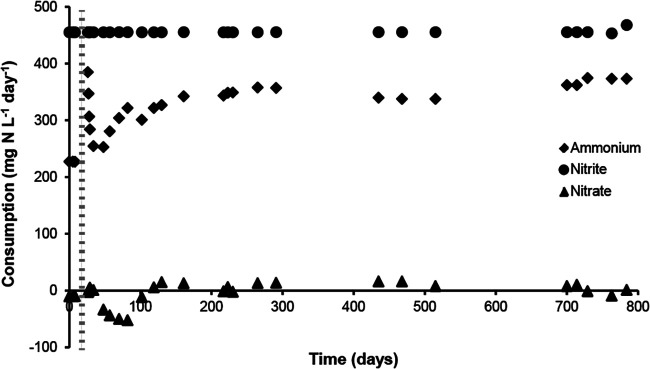
A 3-liter laboratory-scale SBR containing both nitrate- and nitrite-dependent anaerobic methane-oxidizing microorganisms (N-damo) and anammox bacteria was used to establish an oxygen-limited coculture. The initial SBR bioreactor simultaneously removed ammonium, nitrite, and nitrate; most nitrite was removed by anammox bacteria (a detailed description of the culture can be found in Stultiens et al. [13]). In order to stimulate the growth of aerobic ammonia-oxidizing bacteria for in situ nitrite production, oxygen was added via the gas phase. The effect on the overall performance and microbial composition of the culture was studied by combining activity assays with FISH and metagenomics. Over time, the oxygen concentration was gradually increased to a final concentration of 5.4% in the influent gas. The dissolved oxygen concentration remained below the detection limit of the oxygen probe (<3.1 μM), indicating that virtually all supplied oxygen was consumed by the culture. The ammonium concentration in the medium was increased parallel to the increasing oxygen supply in order to support the growth of aerobic ammonia oxidizers in the culture, and ammonia and nitrite were immediately consumed by the culture (Fig. 1). Initially, nitrate accumulation was observed after increasing the ammonium concentration in the medium (see Fig. S1 in the supplemental material). Most likely, the increased production of nitrate was due to nitrite oxidation carried out by anammox bacteria, which received more ammonium and consequently produced more N2 and nitrate. After 82 days, nitrate reduction increased, resulting in consumption of the produced nitrate (Fig. 1). The nitrate supplied externally via the medium was also consumed, which corresponded to a rate of 27 ± 2 mg N-NO3− · liter−1 · day−1. Ammonium was consumed at an average rate of 325 ± 44 mg N-NH4+ · liter−1 · day−1. The concentration of nitrite was always below the detection limit of our assay (2 μM); all supplied nitrite was thus consumed by the culture, which corresponds to a conversion of 456 ± 2 mg N-NO2− · liter−1 · day−1.
FIG 1.
Consumption of ammonium (diamonds), nitrite (circles), and nitrate (triangles) by the reactor. Directly after the addition of oxygen and the increase of the ammonium concentration in the medium, nitrate was produced and is plotted here as a negative consumption. The start of the addition of oxygen is indicated with the dotted line.
Microbial composition of the oxygen-limited coculture.
Before the addition of oxygen, the culture was dominated by N-damo bacteria (Fig. 2). The addition of oxygen resulted in the presence of two types of aggregates/flocs. The flocs settled within 5 to 10 min after the stirring stopped, whereas the granules immediately began settling during the settling phase of the bioreactor. The granules mainly consisted of methane-oxidizing Methanoperedens-like archaea and anammox bacteria. After the addition of oxygen, the Methanoperedens-like archaea increased in abundance, whereas Methylomirabilis-like bacteria slowly disappeared from the culture. The presence of N-damo bacteria and archaea and anammox bacteria was confirmed by analysis of the amplicon sequencing of 16S rRNA gene sequences at two different time points (2 and 9 months after the start of the supply of oxygen, respectively) (Table 1). The relative number of Methylomirabilis-like bacterial reads in the sequencing data decreased, confirming the FISH analysis, while Methanoperedens-like archaea increased in number. Aerobic ammonia oxidizers were not visible with FISH, but their presence was confirmed by the presence of Nitrosomonas 16S rRNA sequences in the metagenome at both time points (Table 1).
FIG 2.

Representative fluorescence in situ hybridization (FISH) pictures of the enrichment culture under anaerobic (A and B) and oxygen-limited (C to H) conditions. Bars, 20 μm. (A and B) Samples taken from an N-damo/anammox coculture before oxygen addition. In red, anammox bacteria are visible (Cy3, AMX820), and archaea are visualized in blue (Cy5, ARCH915). Methylomirabilis spp. constitute the majority of the biomass and are visible in green (FLUOS, DAMO1027). (C) Sample taken 292 days after oxygen addition. The percentage of Methanoperedens-like archaea (red, DARCH641) and anammox bacteria (blue, AMX820) increased, while the relative abundance of Methylomirabilis spp. (green, DAMO1027) decreased. (D) Sample taken 732 days after oxygen addition. The percentage of Methylomirabilis spp. (green, DAMO1027b) decreased further. Methanoperedens-like archaea (red, DARCH641), and anammox bacteria (blue, AMX820) became the dominant microorganisms in the culture. (E) Sample of granules sampled 800 days after the start of oxygen-limited culturing conditions. Anammox bacteria (green, AMX820) and archaea (blue, ARCH915) are the dominant microorganisms in the granules. Methylomirabilis spp. (red, DAMO1027) could not be detected. (F) Sample of flocculent biomass taken 800 days after the start of oxygen-limited culturing conditions. Archaea (blue, ARCH915) are nearly absent in the flocculent, while anammox biomass (green, AMX820) is less dense than in the granules. Like in the granules, no Methylomirabilis spp. (red, DAMO1027) was detected in the flocs. (G) Sample of flocculent biomass taken 800 days after the start of oxygen-limited culturing conditions. Next to anammox bacteria, visible in yellow/orange (Cy3, PLA46; Fluos, AMX820), other bacteria present in the flocculent biomass are visible in blue (EUBmix). (H) Negative control.
TABLE 1.
16S rRNA sequences obtained from nitrogen- and methane-cycling microorganismsa
| Group | No. of 16S rRNA reads at: |
Group or genus | Normalized for total reads at: |
||
|---|---|---|---|---|---|
| T1 | T2 | T1 | T2 | ||
| Archaea | 296 | 7,588 | Methanoperedens spp. | 688 | 2,920 |
| Methanogens | 22 | 125 | |||
| Other Archaea | 12 | 66 | |||
| Nitrifiers | 11 | 37 | Nitrosomonadacea | 27 | 15 |
| Methanotrophs | 47 | Methylobacterium | 2 | 0 | |
| Methylomonas | 0 | 17 | |||
| Methylocystis | 105 | 107 | |||
| Methylosinus | 2 | 9 | |||
| Methylovirgua | 5 | 0 | |||
| Anammox | 2,948 | 4,831 | Brocadiaceae | 7,169 | 1,979 |
| Scalinduaceae | 22 | 2 | |||
| N-damo bacteria | 3,963 | 1,716 | Methylomirabilis spp. | 9,666 | 704 |
aSamples were taken 2 (T1) and 9 (T2) months after the start of the supply of oxygen. For T1, 13.757 reads were analyzed and for T2, 33.555 reads.
In situ activity measurements in the reactor.
In order to determine the performance of different microbial groups within the N-damo/anammox coculture when supplied with oxygen, one of the nitrogen substrates was added to the medium as a 15N-labeled compound (ammonium, nitrite, or nitrate), and production of isotopically labeled gasses (dinitrogen gas and nitrous oxide) was monitored in the liquid phase of the reactor using membrane inlet mass spectrometry (MIMS). When 15N-labeled ammonium was added, single-labeled 14N15N (29N2) dinitrogen gas was produced, indicating the activity of anammox bacteria. In addition, double-labeled 15N15N (30N2) dinitrogen gas was detected, indicating combined activity of aerobic ammonia-oxidizing and anammox bacteria that resulted in the production of 30N2 (Fig. 3A). When 15N-labeled nitrite was fed to the reactor, both 29N2 and 30N2 were detected, indicating that anammox and other N2-producing microorganisms were active (Fig. 3B). Addition of 15N-labeled nitrate (Fig. 3C) led to the production of 29N2, whereas almost no 30N2 production was observed, suggesting that nitrate-dependent N2 production (complete denitrification processes) was negligible in the coculture. In addition, N2O production was not detected in any of these activity assays (Fig. S1).
FIG 3.
Labeled dinitrogen gas production by the culture measured using membrane inlet mass spectrometry (MIMS) when the culture was fed medium containing 15N-labeled ammonium (A), 15N-labeled nitrite (B), or 15N-labeled nitrate (C). The reactor was running normally, and sequencing batch reactor (SBR) cycles are indicated with gray bars. Time points at which medium containing 15N-labeled substrates was connected and disconnected are indicated by the dotted lines and symbols (+ and −). Production of both 29N2 (black) and 30N2 (gray) was measured and plotted as ratio of total nitrogen gas (28+29+30N2).
Batch activity assays.
To determine the potential for aerobic ammonia and methane oxidation by the coculture, batch assays were performed. In incubations supplemented with oxygen (6%) and ammonium, nitrite was produced at a rate of 50 ± 0.001 nmol NO2− ml · culture−1 · h−1 (Fig. 4A). Aerobic methane oxidizers were immediately active in batch incubations at a maximum rate of 2.0 ± 0.4 μmol CH4 · ml culture−1 · h−1 (Fig. 4B). Interestingly, batch incubations also showed production of some methane (Fig. 5), which could be inhibited by the addition of 25 mM 2‐bromoethanesulfonate (BES; a known inhibitor of methanogenesis [24]).
FIG 4.

Potential ammonium and methane oxidation by the culture. (A) In the presence of ammonium and oxygen, nitrite (circles) was produced by the culture. (B) In the presence of oxygen, methane (closed symbols) was rapidly consumed by the culture; methane was not consumed when oxygen was not present (open symbols). After 2.5 h, additional methane was added (indicated by the dotted line).
FIG 5.

Production of methane in batch incubations with and without 25 mM 2‐bromoethanesulfonate (BES). BES was added after 3 h of incubation (indicated by the dotted line).
DISCUSSION
Several studies have explored the feasibility of a combination of partial nitritation, anammox, and N-damo (25–27), but little is known about the effect of oxygen on the stability of such a coculture. This knowledge is important; when a combination of N-damo and anammox processes is applied, oxygen is required for the internal production of nitrite from ammonium, which is needed for both anammox and N-damo activity. In order to study the effect of oxygen on these anaerobic microbial processes and thus the feasibility of the application of anammox and N-damo, limiting amounts of oxygen were supplied to a coculture containing anammox and N-damo microorganisms. Virtually all supplied oxygen was consumed by the culture; ammonium, nitrite, and nitrate were removed by the culture with average rates of 325 ± 44 mg N-NH4+ · liter−1 · day−1, 456 ± 2 mg N-NO2− · liter−1 · day−1, and 27 ± 2 mg N-NO3− · liter−1 · day−1, respectively. After the increase of the ammonium concentration in the medium and the consequent increase of anammox activity, an accumulation of nitrate was observed. However, this additional nitrate was eventually consumed by the culture as well, most probably by N-damo archaea, which also increased in number. N-damo bacteria decreased in abundance, potentially because of the competition for nitrite with anammox bacteria; it has been shown before that Methylomirabilis-like bacteria have a lower affinity for nitrite than that of anammox bacteria (17, 28).
The active addition of oxygen to the culture made it possible for aerobic microorganisms to become more active. Using MIMS, production of 30N2 upon the addition of 15N-labeled ammonium could be detected, which can only occur if ammonia-oxidizing microorganisms are actively producing 15N-labeled nitrite that can subsequently be used by anammox bacteria. These microorganisms are indeed present in the culture, since nitrite production was observed in aerobic batch incubations with ammonium. However, the observed nitrite production was very low, and no significant decrease in the concentration of ammonium could be measured in these batch incubations. The supplied conditions in the batch incubation and in the reactor were very different, possibly explaining the low activity. In the reactor, the activity of the ammonia-oxidizing microorganisms must be higher in order to explain the observed production of 30N2. The potential methane oxidation activity was measured in batch assays; however, it is difficult to conclude how much methane and oxygen are consumed by aerobic methanotrophs in the bioreactor. Surprisingly, batch incubations of the coculture also showed production of some methane. Anammox bacteria have been observed before in methanogenic sludge, but bacteria in these reactor systems were kept under strict anoxic conditions (29, 30). Apparently, the granules provide an environment without oxygen in which the methanogens can thrive.
It is known that aerobic nitrifiers can produce N2O under oxygen-limited conditions. In our incubations, we did not observed production of N2O. This is in contrast to a study by Liu et al. (31), in which some production of N2O was observed in a coculture containing anaerobic and aerobic ammonia- and methane-oxidizing microorganisms. The reactor setups in their study and in ours were different, Liu et al. separated the addition of methane and oxygen by using a membrane reactor in which methane was added via the membrane and oxygen via the gas supply. No 30N2 production was detected upon the addition of labeled nitrate, implying that there is only little or even no activity of denitrifying microorganisms, which has been described before in cocultures containing N-damo and anammox microorganisms (32). In contrast, 30N2 production was observed when nitrite was added as the labeled substrate. This might be due to the activity of denitrifying microorganisms; however, these microorganisms would most likely also produce 30N2 when 15N-labeled nitrate would be supplied. 30N2 from labeled nitrite can also be produced from N-damo bacteria. However, FISH analysis has showed that the relative abundance of these microorganisms is very low. Another explanation would be that part of the 15N-labeled nitrite is converted into nitrate by anammox bacteria. This labeled nitrate can subsequently by used by N-damo archaea, which can perform dissimilatory nitrate reduction to ammonium (DNRA), resulting in the production of labeled ammonium (7, 33).
In order to apply a combination of N-damo and anammox processes in wastewater treatment, nitrite has to be produced microbially in the treatment plant, a process which requires oxygen. In this study, we show that combined activity of N-damo and anammox organisms is feasible under the continuous supply of oxygen without the production of N2O. This study thus shows that a combination of these microbial processes can possibly be applied to simultaneously remove nitrogen and methane from wastewater.
MATERIALS AND METHODS
Ethics statement.
This article does not contain any studies with human participants or animals performed by any of the authors.
Oxygen-limited anaerobic methane- and ammonia-oxidizing coculture.
An anaerobic methane- and ammonia-oxidizing culture was cultivated in a 3-liter sequencing batch reactor, as described by Stultiens et al. (13). During the filling period, the culture was continuously flushed at 10 ml · min−1 with a mixture of CH4/CO2 (95%/5%) and compressed air. The final oxygen concentration in the supplied gas was 5.4%, which resulted in a measured dissolved oxygen concentration that was below the detection limit of the dissolved O2 sensor (≤3.1 μM). The mineral medium fed to the culture was supplemented with 20 to 35 mM NH4Cl, 1.5 mM NaNO3, and 40 mM NaNO2, and 1.3 liters were fed to the culture per day. Roughly 1 month after the start of oxygen addition, the ammonium concentration in the medium was increased from 20 mM to 40 mM to stimulate the growth of aerobic ammonia oxidizers. Liquid samples were taken regularly and were centrifuged for 5 min at maximal speed. The supernatant was stored at −20°C until further analysis.
Fluorescence in situ hybridization (FISH).
To fix cells for FISH, biomass was harvested by centrifugation for 5 min at 20,000 × g. The obtained pellet was washed with phosphate-buffered saline (PBS; 10 mM Na2HPO4/NaH2PO4 [pH 7.5] and 130 mM NaCl). Subsequently, the biomass was fixed in paraformaldehyde (4%) and hybridized with fluorescently labeled probes as described by Ettwig et al. (6). Probes were Cy3, Cy5, or Fluos labeled and are specified in Table 1. Eub 338, Eub 338 II, and Eub 338 III were mixed in equimolar solution (EUBmix) and used as such. For Bet42a, Gam42a, and Ntspa712, unlabeled competitor probes were used to prevent unspecific hybridization. After hybridization, slides were air-dried and embedded in Vectashield that included 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Inc., Burlingame, CA). Subsequently, slides were examined and images obtained by utilization of an Axioplan 2 epifluorescence microscope equipped with a digital camera, in combination with the AxioVision software package (Zeiss, Germany).
Metagenome analysis.
Biomass samples (1.5 ml) were taken from the oxygen-limited anaerobic methane- and ammonia-oxidizing culture (1.6 liters) and centrifuged for 5 min at 20,000 × g. Subsequently, genomic DNA was isolated from the pellet using the PowerSoil kit (MO Bio, Carlsbad, CA), the ammonium acetate method (34), and the cetyltrimethylammonium bromide (CTAB) extraction method (35). Libraries were prepared using the Nextera XT kit (Illumina, San Diego, CA) according to the manufacturer’s instructions. Genomic DNA (1 ng) was used for enzymatic fragmentation, which was followed by incorporation of the indexed adapters and amplification. The prepared libraries were purified using the AMPure XP beads (Beckman Coulter, Indianapolis, IN) and checked for size distribution and quality using a high-sensitivity DNA kit in combination with the Agilent 2100 Bioanalyzer. Quantity of the DNA was determined using the Qubit double-stranded DNA (dsDNA) high-sensitivity (HS) assay kit (Thermo Fisher Scientific, Inc., Waltham, MA). Subsequently, libraries were pooled, denatured, and sequenced using the Illumina MiSeq sequence machine (San Diego, CA). Paired-end sequencing of 2 × 300 bp was performed using the MiSeq reagent kit v3 (San Diego, CA) according to the manufacturer’s protocol. To study biodiversity, 16S rRNA gene sequences were extracted and analyzed using CLC Workbench with the Silva Database version 132 as a reference.
Batch assay determining methanogenic activity.
The headspace of 120-ml serum bottles was exchanged with argon, and 20 ml biomass from the oxygen-limited anaerobic methane- and ammonia-oxidizing culture (1.6 liters) was added to each bottle. Subsequently, the headspace was flushed with argon for another 10 min. CO2 was added to the anaerobic headspace to a final headspace concentration of 4%. In parallel with incubations without additions, incubations were carried out with either 10 mM or 25 mM BES, added after 3 h of incubation. All incubations were performed in duplicate. Bottles were incubated in a shaking incubator at 30°C and 150 rpm. Gas samples were taken every hour and liquid samples every 2 h. At each time point, presence of nitrite and/or nitrate was also checked using test strips (Merck, Germany).
Batch assay determining aerobic ammonia- and methane-oxidizing activity.
The headspace of 60-ml serum bottles containing 10 ml biomass from the oxygen-limited anaerobic methane- and ammonia-oxidizing culture (1.6 liters) was exchanged with argon/CO2 (90%/10%). Subsequently, O2 was added to the headspace to a final headspace concentration of 6%. Incubations for the determination of ammonia-oxidizing activity were supplemented with 2 to 3 mM NH4+, and incubations for testing aerobic methanotrophy with 2% CH4. In addition, incubations without oxygen were prepared as negative controls. All treatments were done in triplicate. Bottles were incubated in a shaking incubator at 30°C and 150 rpm. Gas and liquid samples were taken every one to 2 h. For liquid samples, 0.5 ml liquid was extracted from the incubations with a syringe and transferred to an Eppendorf cup. Samples were centrifuged for 1 min at 20,000 × g, and the supernatant was transferred to a clean cup and stored at −20°C until further analysis. At each time point, the presence of nitrite was also checked using test strips (Merck, Germany). Afterwards, protein samples (2 × 1 ml) were taken per bottle and spun down for 5 min at 20,000 × g, and the pellet was stored at −20°C until further analysis.
Real-time gas measurements using membrane inlet spectrometry.
An HPR-40 membrane inlet mass spectrometer (Hiden Analytical, UK) was used for real-time gas measurements by inserting the membrane inlet mass spectrometry (MIMS) probe directly in the bioreactor. Reactor configuration remained normal to avoid any artifacts in the activity due to changes in substrate supply (both via the gas and medium supply). Labeled substrates (15N-labeled ammonium, nitrite, or nitrate) were added by replacing the unlabeled substrate by an isotopically labeled substrate in the standard medium. Medium was than supplied to the reactor under the standard configuration, after which the production of gasses was measured.
Analytical methods.
Liquid samples taken during activity tests were centrifuged for 1 min at 20,000 × g, and the supernatant was stored at −20°C until further analysis. Ammonium was determined colorimetrically using a modified orthophataldialdehyde assay (36), and nitrite by the sulfanilamide reaction. Nitrate was measured by reducing it to nitrite at 60°C (30 min) using a saturated solution of VCl3 in HCl. The produced nitrite was measured using the sulfanilamide reaction. To measure the protein content of the biomass, 1.5-ml biomass samples were taken in triplicate and centrifuged for 5 min at maximum speed. Supernatant was removed, and the pellet was subsequently homogenized in 0.5 M NaOH, boiled at 90°C for 30 min, and neutralized with 0.5 M HCl. Next, protein content was determined by the bicinchoninic acid assay (Pierce, USA). Methane was measured using gas chromatography (HP 5890 gas chromatograph with a flame ionization detector and Porapak Q column). N2, O2, CO2, and N2O production was analyzed using gas chromatography (Agilent 6890 and Porapak Q column, 80°C) in combination with mass spectrometry (Agilent 5975, quadruple inert MS).
Data availability.
The obtained 16S rRNA reads have been submitted to the Sequence Read Archive (SRA) under BioProject accession number PRJEB37137.
ACKNOWLEDGMENTS
We thank Katinka van de Pas-Schoonen and Guylaine Nuijten for their help with bioreactor maintenance. We thank Jeroen Frank and Geert Creemers for help with the metagenomics.
K.S. and M.A.H.J.V.K. were funded by the Technology Foundation STW (grant 13146). M.A.H.J.V.K. is further supported by an NWO Veni Grant (016.veni.192.062). H.J.M.O.D.C., M.S.M.J., and B.K. are supported by the European Research Council (ERC 669371, ERC 339880, and ERC 640422). M.S.M.J. is also supported by the Netherlands Organization for Scientific Research (NWO SIAM 024002002).
We declare that we have no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Maartje A. H. J. van Kessel, Email: maartje.vankessel@science.ru.nl.
Rebecca E. Parales, University of California, Davis
REFERENCES
- 1.Kartal B, Kuenen JG, van Loosdrecht MCM. 2010. Sewage treatment with anammox. Science 328:702–703. 10.1126/science.1185941. [DOI] [PubMed] [Google Scholar]
- 2.Daelman MRJ, van Voorthuizen EM, van Dongen UGJM, Volcke EIP, van Loosdrecht MCM. 2012. Methane emission during municipal wastewater treatment. Water Res 46:3657–3670. 10.1016/j.watres.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 3.Myhre G, Shindell D, Pongratz J, et al. 2013. Anthropogenic and natural radiative forcing, p 659.–. InStocker TF, et al. (ed), Climate change 2013: the physical science basis. Contribution of Working Group I to the fifth assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 4.Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MSM. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446–449. 10.1038/22749. [DOI] [PubMed] [Google Scholar]
- 5.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen TA, Luesken FA, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–549. 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 6.Ettwig KF, Shima S, van de Pas-Schoonen KT, Kahnt J, Medema MH, Op den Camp HJM, Jetten MSM, Strous M. 2008. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ Microbiol 10:3164–3173. 10.1111/j.1462-2920.2008.01724.x. [DOI] [PubMed] [Google Scholar]
- 7.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 8.van Kessel MAHJ, Stultiens K, Slegers MFW, Guerrero Cruz S, Jetten MSM, Kartal B, Op den Camp HJM. 2018. Current perspectives on the application of N-damo and anammox in wastewater treatment. Curr Opin Biotechnol 50:222–227. 10.1016/j.copbio.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 9.Kuypers MMM, Marchant HK, Kartal B. 2018. The microbial nitrogen cycling network. Nat Rev Microbiol 16:263–276. 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 10.Luesken FA, Wu ML, Op den Camp HJM, Keltjens JT, Stunnenberg H, Francoijs KJ, Strous M, Jetten MSM. 2012. Effect of oxygen on the anaerobic methanotroph ‘Candidatus Methylomirabilis oxyfera’: kinetic and transcriptional analysis. Environ Microbiol 14:1024–1034. 10.1111/j.1462-2920.2011.02682.x. [DOI] [PubMed] [Google Scholar]
- 11.Ding ZW, Lu YZ, Fu L, Ding J, Zeng RJ. 2017. Simultaneous enrichment of denitrifying anaerobic methane-oxidizing microorganisms and anammox bacteria in a hollow-fiber membrane biofilm reactor. Appl Microbiol Biotechnol 101:437–446. 10.1007/s00253-016-7908-7. [DOI] [PubMed] [Google Scholar]
- 12.Lu YZ, Na L, Ding ZW, Fu L, Bai YN, Sheng GP, Zeng RJ. 2017. Tracking the activity of the Anammox-DAMO process using excitation emission matrix (EEM) fluorescence spectroscopy. Water Res 122:624–632. 10.1016/j.watres.2017.06.036. [DOI] [PubMed] [Google Scholar]
- 13.Stultiens K, Guerrero Cruz S, van Kessel MAH, Jetten MSM, Kartal B, Op den Camp HJM. 2019. Interactions between anaerobic ammonium- and methane-oxidizing microorganisms in a laboratory-scale sequencing batch reactor. Appl Microbiol Biotechnol 103:6783–6795. 10.1007/s00253-019-09976-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sliekers AO, Derwort N, Campos GJ, Strous M, Kuenen JG, Jetten MSM. 2002. Completely autotrophic nitrogen removal over nitrite in one single reactor. Water Res 36:2475–2482. 10.1016/S0043-1354(01)00476-6. [DOI] [PubMed] [Google Scholar]
- 15.van der Star WRL, Abma WR, Blommers D, Mulder J-W, Tokutomi T, Strous M, Picioreanu C, van Loosdrecht MCM. 2007. Startup of reactors for anoxic ammonium oxidation: experiences from the first full-scale anammox reactor in Rottterdam. Water Res 41:4149–4163. 10.1016/j.watres.2007.03.044. [DOI] [PubMed] [Google Scholar]
- 16.Lackner S, Gilbert EM, Vlaeminck SE, Joss A, Horn H, van Loosdrecht MCM. 2014. Full-scale partial nitritation/anammox experiences—an application survey. Water Res 55:292–303. 10.1016/j.watres.2014.02.032. [DOI] [PubMed] [Google Scholar]
- 17.Winkler MKH, Ettwig KF, Vannecke TPW, Stultiens K, Bogdan A, Kartal B, Volcke EIP. 2015. Modelling simultaneous anaerobic methane and ammonium removal in a granular sludge reactor. Water Res 73:323–331. 10.1016/j.watres.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 18.Bhattacharjee AS, Motlagh AM, Jetten MSM, Goel R. 2016. Methane dependent denitrification- from ecosystem to laboratory-scale enrichment for engineering applications. Water Res 99:244–252. 10.1016/j.watres.2016.04.070. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Guo J, Xie GJ, Liu Y, Yuan Z, Ni BJ. 2015. A new approach to simultaneous ammonium and dissolved methane removal from anaerobic digestion liquor: a model-based investigation of feasibility. Water Res 85:295–303. 10.1016/j.watres.2015.08.046. [DOI] [PubMed] [Google Scholar]
- 20.Castro-Barros CM, Ho L, Winkler MKH, Volcke EIP. 2018. Integration of methane removal in aerobic anammox-based granular sludge reactors. Environ Tech 39:1615–1625. 10.1080/09593330.2017.1334709. [DOI] [PubMed] [Google Scholar]
- 21.Cogert K, Ziels R, Winkler MKH. 2019. Reducing costs and environmental impact on wastewater treatment with denitrifying methanotrophs, anammox and mainstream anaerobic treatment. Environ Sci Technol 53:12935–12944. 10.1021/acs.est.9b04764. [DOI] [PubMed] [Google Scholar]
- 22.Allegue T, Arias A, Fernandez-Gonzalez N, Omil F, Garrido JM. 2018. Enrichment of nitrite-dependent methane oxidizing bacteria in a membrane bioreactor. Chem Eng J 347:721–730. 10.1016/j.cej.2018.04.134. [DOI] [Google Scholar]
- 23.Guerrero Cruz S, Cremers G, van Alen TA, Op den Camp HJM, Jetten MSM, Rasigraf O, Vaksmaa A. 2018. Respons of the anaerobic methanotroph “Candidatus Methanoperedens nitroreducens” to oxygen stress. Appl Environ Microbiol 84:e01832-18. 10.1128/AEM.01832-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Wang J, Wang A, Chen J. 2011. Chemical inhibitors of methanogenesis and putative applications. Appl Microbiol Biotechnol 89:1333–1340. 10.1007/s00253-010-3066-5. [DOI] [PubMed] [Google Scholar]
- 25.Liu T, Hu S, Yuan Z, Guo J. 2019. High-level nitrogen removal by simultaneous partial nitritation, anammox and nitrite/nitrate-dependent anaerobic methane oxidation. Water Res 166:115057. 10.1016/j.watres.2019.115057. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y, Hu S, Lou J, Lu P, Keller J, Yuan Z. 2013. Nitrogen removal from wastewater by coupling anammox and methane-dependent denitrification in a membrane biofilm reactor. Environ Sci Technol 47:11577–11583. 10.1021/es402775z. [DOI] [PubMed] [Google Scholar]
- 27.Xie GJ, Cai C, Hu S, Yuan Z. 2017. Complete nitrogen removal from synthetic anaerobic sludge digestion liquor through integrating anammox and denitrifying anaerobic methane oxidation in a membrane biofilm reactor. Environ Sci Technol 51:819–827. 10.1021/acs.est.6b04500. [DOI] [PubMed] [Google Scholar]
- 28.Guerrero-Cruz S, Stultiens K, van Kessel MAHJ, Versantvoort W, Jetten MSM, Op den Camp HJM, Kartal B. 2019. Key physiology of a nitrite-dependent methane-oxidizing enrichment culture. Appl Environ Microbiol 85:e00124-19. 10.1128/AEM.00124-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang CJ, Zheng P, Zhang L, Chen JW, Mahmood Q, Chen XG, Hu BL, Wang CH, Yu Y. 2010. Enrichment features of anammox consortia from methanogenic granules loaded with high organic and methanol contents. Chemosphere 79:613–619. 10.1016/j.chemosphere.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez-Martinez A, Morillo JA, Garcia-Ruiz MJ, Gonzalez-Lopez J, Osorio F, Martinez-Toledo MV, van Loosdrecht MCM. 2015. Archaeal populations in full-scale autotrophic nitrogen removal bioreactors operated with different technologies: CANON, DEMON and partial nitritation/anammox. Chem Eng J 277:194–201. 10.1016/j.cej.2015.04.137. [DOI] [Google Scholar]
- 31.Liu P, Liu T, Ni BJ, Guo J, Yuan Z, Hu S. 2019. Growth kinetics of Candidatus “Methanoperedens nitroreducens” enriched in a laboratory reactor. Sci Tot Environ 659:442–450. 10.1016/j.scitotenv.2018.12.351. [DOI] [PubMed] [Google Scholar]
- 32.Nie W-B, Xie G-J, Ding J, Peng L, Lu Y, Tan X, Yue H, Liu B-F, Xing D-F, Meng J, Han H-J, Ren N-Q. 2020. Operation strategies of n-DAMO and anammox process based on microbial interactions for high rate nitrogen removal from landfill leachate. Environ Int 139:105596. 10.1016/j.envint.2020.105596. [DOI] [PubMed] [Google Scholar]
- 33.Nie W-B, Ding J, Xie G-J, Yang L, Peng L, Tan X, Liu B-F, Xing D-F, Yuan Z, Ren N-Q. 2020. Anaerobic oxidation of methane coupled with dissimilatory nitrate reduction to ammonium fuels anaerobic ammonium oxidation. Environ Sci Technol 55:1197–1208. 10.1021/acs.est.0c02664. [DOI] [PubMed] [Google Scholar]
- 34.Kowalchuk GA, de Bruijn FJ, Head IM, Akkermans AD, van Elsas JD. 2004. Molecular microbial ecology manual (MMEM), 2nd ed, vol 1. Kluwer Academic Publishing, London, United Kingdom.
- 35.Zhou J, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322. 10.1128/AEM.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor S, Ninjoor V, Dowd DM, Tappel AL. 1974. Cathepsin B2 measurement by sensitive fluorometric ammonia analysis. Anal Biochem 60:153–162. 10.1016/0003-2697(74)90140-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 and S2. Download AEM.00043-21-s0001.pdf, PDF file, 0.4 MB (423.7KB, pdf)
Data Availability Statement
The obtained 16S rRNA reads have been submitted to the Sequence Read Archive (SRA) under BioProject accession number PRJEB37137.