Abstract
Enhancer elements potentiate the rearrangement of antigen receptor loci via changes in the accessibility of gene segment clusters to V(D)J recombinase. Here, we show that enhancer activity per se is insufficient to target T-cell receptor β miniloci for DβJβ recombination. Instead, a promoter situated 5′ to Dβ1 (PDβ) was required for efficient rearrangement of chromosomal substrates. A critical function for promoters in regulating gene segment accessibility was further supported by the ability of heterologous promoters to direct rearrangement of enhancer-containing substrates. Importantly, activation of a synthetic tetracycline-inducible promoter (Ptet) positioned upstream from the Dβ gene segment was sufficient to target recombination of miniloci lacking a distal enhancer element. The latter result suggests that DNA loops, generated by interactions between flanking promoter and enhancer elements, are not required for efficient recognition of chromosomal gene segments by V(D)J recombinase. Unexpectedly, the Ptet substrate exhibited normal levels of rearrangement despite its retention of a hypermethylated DNA status within the DβJβ cluster. Together, our findings support a model in which promoter activation, rather than intrinsic properties of enhancers, is the primary determinant for regulating recombinational accessibility within antigen receptor loci.
Precursor lymphocytes diversify immunoglobulin (Ig) and T-cell receptor (TCR) variable-region genes via a program of DNA recombination involving large arrays of variable (V), diversity (D), and joining (J) gene segments. All rearrangement events are mediated by a common V(D)J recombinase activity that targets conserved recognition sequences flanking each gene segment (32, 39). Despite these shared features, the rearrangement of antigen receptor loci proceeds in a tissue-, stage-, and allele-specific manner (39). For example, thymocytes specifically target TCR Dβ and Jβ gene segments for recombination upon commitment to the T-cell lineage. In turn, DβJβ joins rearrange with one of 30 upstream Vβ elements to complete assembly of a variable-region coding exon. The resultant expression of TCRβ protein signals for a cessation of TCRβ recombination and the initiation of TCRα gene assembly (42). Likewise, precursor B cells execute an ordered program of rearrangements at the Ig heavy-chain (IgH) and light-chain loci (8, 22). These observations indicate that precursor lymphocytes must direct and then redirect V(D)J recombinase activity to specific regions within antigen receptor loci at distinct stages of their development.
Recent studies have shown that the tissue- and stage-specific aspects of V(D)J rearrangement are governed by changes in the accessibility of gene segment clusters to recombinase proteins RAG-1 and RAG-2 (25, 41). An important role for enhancer elements in regulating the recombinational accessibility of linked gene segments has been deduced from numerous experimental approaches (reviewed in reference 39). For example, targeted deletion of Ig or TCR enhancers severely impairs recombination of gene segments specifically at the mutated alleles (1, 3, 6, 35, 40, 47). In addition, transgenic TCRβ miniloci undergo rearrangement in precursor lymphocytes only upon inclusion of Ig or TCR enhancers (4, 11, 12, 29). In studies using a recombinase-inducible cell line, the latter results have been extended to show that any active enhancer, including those derived from a viral genome, directs efficient DβJβ recombination within chromosomal miniloci (30). Despite these findings, the precise function of enhancer elements in regulating the rearrangement of associated gene segments remains unclear (39).
Prevailing models for enhancer-mediated control of V(D)J recombination invoke at least one of three effects exerted by these regulatory elements on neighboring gene segments. First, Ig and TCR enhancers activate transcription of germ line gene segments at developmental time points that coincide with their rearrangement (21, 31, 48). Second, the IgH enhancer (Eμ) has been shown to promote regional access to DNA-binding proteins, presumably via directed alterations in local chromatin configurations (17). Third, transcriptional enhancers protect chromosomal gene segments from active methylation and target hypermethylated sequences for demethylation (9, 19, 23). Each of these enhancer-dependent effects has been correlated with active recombination of linked gene segments (7, 24, 30, 34); however, their independent contributions to rearrangement efficiencies have not been established (39). Thus, it remains possible that enhancers directly regulate V(D)J recombination through their intrinsic abilities to potentiate the accessibility of neighboring chromatin. In this case, transcription and demethylation of gene segments would be simply by-products of enhancer function. Alternatively, enhancer-dependent activation of germ line promoters may be the critical parameter for directing efficient assembly of antigen receptor loci.
To dissect the role of transcriptional control elements in targeting V(D)J recombination, we have used a recombinase-inducible cell system to assess the rearrangement efficiency of chromosomal TCRβ miniloci. Here, we show that enhancer activity is insufficient to target these substrates for DβJβ recombination. Instead, a recently identified promoter situated 5′ to the Dβ1 gene segment (PDβ [37]) is absolutely required for enhancer-dependent rearrangement of chromosomal gene segments. Importantly, substitution of PDβ with a synthetic promoter completely restores recombination of substrates lacking a distal enhancer element, despite their retention of a hypermethylated DNA status. Together, these findings suggest that activation of germ line promoter elements is the primary mechanism by which enhancers initiate assembly of variable-region gene segments.
MATERIALS AND METHODS
Generation of stable TDR19 transfectants.
The recombinase-inducible cell line TDR19 was generated by cotransfection of linearized pTET-R1, pTET-R2, pTET-tTAk (36), and pSV-HIS vectors (15) into the recombinase-null B-cell line M12. Transfected cells were selected in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 0.01% penicillin-streptomycin, 50 μM β-mercaptoethanol, tetracycline (0.5 μg/ml), and histidinol (3 mM). Southern blot analyses demonstrated that the TDR19 clone contained more than 10 copies each of the pTET-R1 and pTET-R2 vectors (38).
To prepare stable transfectants of TCRβ recombination substrates, each minilocus (15 μg) was linearized with PvuI and electroporated (300 mV and 960 μF) together with linearized LTR-NEO expression vector (1.5 μg) into 107 TDR19 cells. Transfected cells were maintained in the presence of tetracycline and histidinol and were positively selected with G418 (1.5 mg/ml) after 48 h. The copy number and integrity of chromosomal miniloci in each clonal transfectant were determined by Southern blotting procedures as described previously (30). In most cases, analyses of germ line transcription and rearrangement were restricted to clones that harbored one to five copies of the test substrates.
Construction of TCRβ recombination substrates.
To dissect the components of recombinational accessibility, we generated a TCRβ parental vector (D−/E−) that contained unique cloning sites at locations 5′ (NotI) and 3′ (XhoI) to the Jβ1 and Jβ2 gene segments. For this purpose, the 4.8-kb HindIII fragment spanning murine Vβ14 and the 600-bp BglII/BamHI fragment containing murine Jβ1 and Jβ2 segments were sequentially inserted into the HindIII and BamHI sites, respectively, of pGEM 11Z creating the D−/E−/Cμ− construct. The Sμ/Cμ region of the D−/E− construct was prepared from a 7.1-kb XbaI/EcoRI genomic fragment that was modified to destroy the internal XhoI site and linker ligated to replace the 5′ XbaI site with a unique XhoI site. The modified Sμ/Cμ fragment was cloned into the corresponding XhoI/EcoRI sites in D−/E−/Cμ− polylinker sequences to yield the D−/E− vector. Finally, a blunt-ended 475-bp AluI/AluI fragment spanning iEκ was cloned into the blunted XhoI site of D−/E− to produce the D−/E+ construct. To generate the φ/iEκ minilocus, the AccI/BglII fragment spanning Dβ1 (430 bp), which lacks PDβ, was ligated to NotI linkers and inserted into the unique NotI site present in D−/E+. Likewise, other promoter-Dβ1 combinations were inserted as either blunt-ended or NotI-linkered fragments into the unique NotI site located between the Vβ14 and Jβ gene segments.
To prepare the promoter-Dβ1 combinations, each promoter element was positioned 5′ of a 430-bp AccI/BglII fragment spanning Dβ1 that was subcloned into the SmaI site of pBluescript (Stratagene, La Jolla, Calif.). The individual promoters 5′Dβ (2.3-kb HindIII/AccI fragment of the murine TCRβ situated immediately 5′ of Dβ1), PDβ (377-bp KpnI/AccI fragment from the p377/3′ vector [37]), PVκ (325-bp HindIII fragment from the pIM.Kp.LUC vector [13]), PGK (phosphoglycerate kinase; 540-bp EcoRI/XhoI fragment from the PGKPuro vector [44]), and teto (tetracycline operon; 300-bp XhoI/KpnI fragment from the pTET-R2 vector [36]) were isolated. The iEκ AluI/AluI fragment was generated by PCR amplification using the primers 5′Eκ (5′-ATG CGG ATC CGC TTT TGT GTT TGA CC-3′) and 3′Eκ (5′-ATG CGA ATT CAA CCT ACT GTA TGG AC-3′).
PCR analyses of coding and signal joins.
For coding join analyses, genomic DNA was harvested from transfectants that were propagated in the presence or absence of tetracycline for 5 days. To minimize amplification of unrearranged TCRβ miniloci, each DNA sample was digested with XbaI and ApaI, which both cleave at sites situated between the Dβ1 and Jβ1 gene segments. Extrachromosomal DNAs (30 μl) were prepared for signal join assays from 5 × 106 cells that were cultured in the presence or absence of tetracycline for 48 h (28).
Amplification of coding joins was performed in 50-μl reaction mixtures containing XbaI/ApaI-digested DNAs (500 ng for coding joins), 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 1 μg of bovine serum albumin per ml, 200 μM deoxynucleoside triphosphates, and 50 ng of each amplification primer (Table 1). Reaction mixtures were incubated at 72°C (3 min) prior to the addition of Taq polymerase (1.1 U) and amplified (94°C for 1 min, 60°C for 1 min, and 72°C for 1.5 min) for either 32 (DβJβ coding), 27 (VλJλ coding joins), or 25 (Cλ controls) cycles. The conditions for signal join amplifications were identical to those for DβJβ coding joins but used 5 × 105 cell equivalents (3 μl) of extrachromosomal DNA. All PCR products were separated on 1.2 or 2% agarose gels and transferred to ZetaProbe membranes (Bio-Rad) for probe hybridization (Table 1).
TABLE 1.
Oligonucleotide primers used for PCR amplification reactions and probes used for blot hybridizations
| Assay type | Primer or probe | Sequence or description |
|---|---|---|
| Coding join | ||
| DβJβ | ||
| Primer | 5′-DβTATA (primer A in Fig. 1) | 5′-GAC CTA TGG GAG GGT CCT-3′ |
| Primer | 3′-PFJβ2 (primer B in Fig. 1) | 5′-AAA GCC TGG TCC CTG AGC CGA-3′ |
| Probe | 3′DβTATA | 5′-GAA GAT CTC CCC ACA ATG TTA CAG C-3′ |
| VλJλ | ||
| Primer | 5′-Vλ1 | 5′-ACT GGT CTA ATA GGT GGT ACC AA-3′ |
| Primer | 3′-JλN | 5′-ACT TAC CTA GGA CAG TCA-3′ |
| Probe | Jλ1 | 5′-TGG GTG TTC GGT GGA GGA ACC-3′ |
| Signal join | ||
| DβJβ | ||
| Primer | 5′-600-2b (primer C in Fig. 6) | 5′-GGC TAC CTC ACT TTG ATG-3′ |
| Primer | 3′-600-2aa (primer E in Fig. 6) | 5′-TCT GGA TCT AAA CAC ATC TAG G-3′ |
| Probe | 3′Dβ2 | 5′-GCT AGT ATC TAG AGG ACC ATA GG-3′ |
| VλJλ | ||
| Primer | 5′-Jλ2/3-2 | 5′-TAC CAC CCA CTK CWW S-3′ |
| Primer | 3′-Vλ1-2 | 5′-TAT GTT GTG CCA AGT TGG-3′ |
| Probe | VλP | 5′-GTG TAG ATG GGG AAG TAG-3′ |
| Cλ control | ||
| Primer | 5′Cλ | 5′-CAG AAT TCA CCT TCC YCT GAR GAG-3′ |
| Primer | 3′Cλ | 5′-GAG TCG ACA RAC TCT TCT CCA C-3′ |
| Probe | 350-bp genomic amplification product of the 5′Cλ and 3′Cλ primers | |
| RT-PCR | ||
| DβJβCμ | ||
| Primer | 5′-600-2b (primer C in Fig. 1) | See above |
| Primer | 3′-Cμ exon 1 (primer D in Fig. 1) | 5′-TGA AGG AAA TGG TGC TGG G-3′ |
| Probe | Jβ1 | 5′-AGG AAC CAG ACT CAC AGT TG-3′ |
| β-Actin | ||
| Primer | 5′-Act | 5′-AGA GCT ATG AGC TGC CTG ACG GCC-3′ |
| Primer | 3′-Act | 5′-AGT AAT CTC CTT CTG CAT CCT GTC-3′ |
| Probe | 450-bp cDNA amplification product of the 5′-Act and 3′-Act primers |
Reverse transcription-PCR (RT-PCR) assay for germ line DβJβ transcription.
Total cellular mRNA was harvested by the LiCl method from clonal transfectants that were propagated for 72 h subsequent to tetracycline withdrawal. Reaction mixtures (20 μl) consisting of mRNA (3 μg), deoxynucleoside triphosphates (250 μM), random hexanucleotides (5 pmol), dithiothreitol (8.75 mM), and RNAsin (20 U; Promega) were preincubated at 65°C for 10 min to denature the RNAs and cooled rapidly to 42°C. Reverse transcription was initiated by immediate addition of MuLV reverse transcriptase (100 U; Perkin-Elmer) to each sample. The reaction mixtures were incubated at 42°C for 60 min, heat inactivated (75°C for 15 min), and stored at −20°C. To examine germ line expression, the resultant cDNAs (3 μl) were amplified with oligonucleotide primers specific for either DβJβCμ (27 cycles) or β-actin (25 cycles) transcripts, using the conditions described for DβJβ coding join assays. Amplification products were separated on a 1% agarose gel, blotted to ZetaProbe membranes, and hybridized to the appropriate radiolabeled probes (Table 1).
Primers and probes.
The sequences of oligonucleotide primers used for PCR amplification reactions and probes used for blot hybridizations are shown in Table 1.
DNA methylation analysis.
The methylation status of chromosomal Ptet (see below) substrates was evaluated in transfectants maintained in the absence of tetracycline (in the presence of tTAk [see below]) for 5 days. Genomic DNAs (10 μg) were digested with appropriate combinations of restriction enzymes HindIII, HpaII, and MspI, separated on a 1% agarose gel, and transferred to ZetaProbe membranes. Southern blots were probed with a radiolabeled XbaI/BamHI fragment spanning the Jβ1 and Jβ2 gene segments as specified by the manufacturer (Bio-Rad).
RESULTS
Enhancer-dependent regulation of DβJβ rearrangement in recombinase-inducible cells.
To expedite analyses of the molecular mechanisms that govern V(D)J recombination, we developed a B-cell system (TDR19) in which recombinase activity is expressed in an inducible manner. This strategy circumvents rearrangement of transfected substrates prior to their stable integration and allows us to specifically monitor the recombination potential of chromosomal gene segments. Prior studies have shown that coexpression of the recombination-activating genes RAG-1 and RAG-2 is sufficient to generate V(D)J recombinase activity in most mammalian cell lines (28, 32). Therefore, we prepared the TDR19 cell system by stable transfection of a recombinase-null B cell (M12) with RAG-1/2 expression vectors that were placed under the transcriptional control of a tetracycline-inducible promoter (Ptet [36]). In addition, the RAG cDNAs were cotransfected with an autoregulated expression vector encoding the chimeric Ptet-activating protein (tTAk [14, 36]), which binds to Ptet upon removal of tetracycline from the culture medium. The resultant TDR19 clone expressed undetectable levels of RAG transcripts in the presence of tetracycline. In contrast, 24 h after tetracycline withdrawal, levels of RAG gene expression in TDR19 were similar to those observed in primary thymocytes (38).
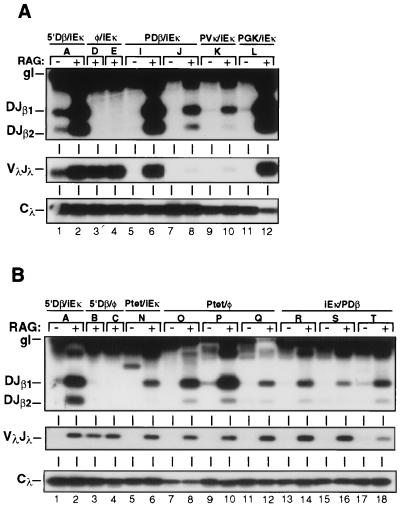
To validate their utility for studies of recombinational control, TDR19 cells were stably transfected with TCRβ miniloci containing a single Vβ14 element linked to a portion of the Dβ1/Jβ gene cluster (Fig. 1). Prior studies in transgenic mice have demonstrated that these miniloci faithfully recapitulate the enhancer dependence of TCRβ gene assembly. Specifically, enhancer-containing miniloci were targeted for DβJβ rearrangement in both B- and T-lineage cells, whereas enhancerless transgenes were recombinationally inert in developing lymphocytes (4, 12, 29). Based on these findings, we compared levels of substrate rearrangement in stable TDR19 transfectants harboring either an enhancerless TCRβ minilocus (5′Dβ/φ) or a version containing the Ig kappa intronic enhancer (5′Dβ/iEκ) (Fig. 1). Multiple independent clones for each substrate were cultured in the presence (without RAG) or absence (with RAG) of tetracycline for 5 days and analyzed for DβJβ rearrangement in a semiquantitative PCR assay (Fig. 1, primers A and B). Subsequent to RAG gene induction, DβJβ rearrangement was readily detected in all transfectants containing the iEκ minilocus (Fig. 2, top panel, lanes 1 to 4). In contrast, enhancerless miniloci were refractory to recombinase activity, regardless of substrate integration site or copy number (Fig. 2, top panel, lanes 5 and 6; Table 2). Control PCR assays specific for VλJλ rearrangement (2) at the constitutively accessible Igλ locus confirmed that similar levels of recombinase activity were induced in each of the TDR19 transfectants (Fig. 2, middle panel). As such, TDR19 cells reproduce the enhancer-dependent regulation of DβJβ rearrangement observed in animal models and provide a physiologically relevant system to test the effects of substrate alterations on recombinational accessibility.
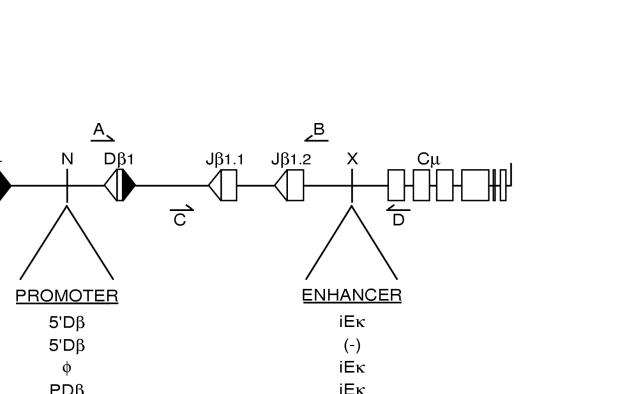
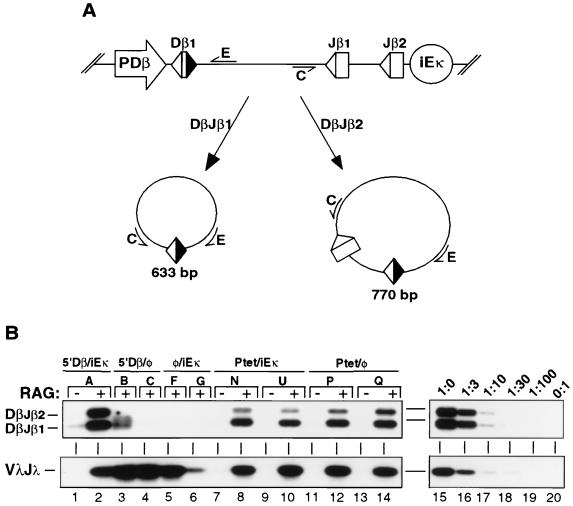
FIG. 1.
Schematic depiction of modified TCRβ miniloci. Unique restriction sites used for cloning promoter (NotI [N]) and enhancer (XhoI [X]) sequences are indicated. Arrows represent the relative positions of primers used for PCR amplification of DβJβ coding joins (A and B) or DβJβCμ cDNAs (C and D). Transcriptional regulatory elements: 5′Dβ, 2.3-kb fragment of endogenous Dβ1 sequences; PDβ, minimal 377-bp Dβ1 germ line promoter (37); PVκ, the murine Vκ21C promoter (13); PGK, the rat PGK promoter (44); teto, a heptamer of the bacterial tetracycline operon (14); iEκ, the murine Igκ intronic enhancer (13).
FIG. 2.
Enhancer-dependent recombination of TCRβ miniloci requires 5′ Dβ1 sequences. Levels of DβJβ rearrangements in chromosomal TCRβ miniloci were analyzed by a semiquantitative PCR assay using primers A and B (Fig. 1). Letters above the lanes identify independent TDR19 clones harboring the substrates 5′Dβ/iEκ (lanes 1 to 4), 5′Dβ/φ (lanes 5 and 6), and φ/iEκ (lanes 7 to 11). Transfectants were incubated in the presence (without RAG) or absence (with RAG) of tetracycline for 5 days. The relative positions of amplification products corresponding to germ line miniloci (gl), as well as DβJβ1 (DJβ1) and DβJβ2 (DJβ2) rearrangements, are shown at the left. Control assays for recombinase activity (VλJλ rearrangement) and total DNA content (Cλ) in each sample are shown in the middle and bottom panels, respectively. The linearity of the DβJβ coding join assay was confirmed by serial dilutions (lanes 12 to 17) of the 5′Dβ/iEκ sample shown in lane 2 with DNA harvested from the 5′Dβ/φ sample shown in lane 6.
TABLE 2.
Summary of germ line expression and Dβ-Jβ rearrangement in TCRβ minilocus transfectantsa
| TCRβ minilocus
|
No. positive/no. tested
|
|||
|---|---|---|---|---|
| Promoter | Enhancer | DβJβ expression | DβJβ recombination | DβJβ recombination in single-copy clones |
| 5′Dβb | iEκ | 6/6 | 6/6 | 3/3 |
| 5′Dβb | (−) | 0/7 | 0/7 | 0/3 |
| (−) | iEκ | 0/7 | 0/7 | 0/3 |
| PDβ | iEκ | 6/6 | 6/6 | 2/2 |
| PVκ | iEκ | 6/6 | 6/6 | 2/2 |
| PGK | iEκ | 5/7c | 5/7c | 1/1 |
| iEκ/PDβ | (−) | 6/8c | 6/8c | NAc |
| Ptet | iEκ | 7/8c | 7/8c | NA |
| Ptet | (−) | 7/7 | 7/7 | 2/2 |
Independent transfectants containing distinct copy numbers of the TCRβ miniloci were examined for DβJβCμ germ line transcription and DβJβ recombination as described in the legends to Fig. 2 and 4, respectively.
Represents 2.3-kb HindIII/AccI fragment upstream from Dβ1, including the 377-bp PDβ.
All transcriptionally active transfectants were positive for DβJβ recombination.
NA, not applicable.
Distal enhancer activity is insufficient to target rearrangement of TCRβ miniloci.
In recent studies, we have shown that a promoter located directly 5′ to the Dβ1 gene segment (PDβ) regulates germ line transcription of Dβ1/Jβ gene segments in an enhancer-dependent manner (37). As an initial attempt to dissect the individual roles of promoter and enhancer elements in targeting recombination, we deleted 2 kb of 5′ Dβ1 sequences from a TCRβ minilocus that contained iEκ. The resulting construct (φ/iEκ [Fig. 1]), which lacks PDβ, was stably transfected into TDR19 cells and assayed for DβJβ rearrangement subsequent to induction of recombinase activity. Deletion of the 5′Dβ1 sequences severely impaired recombination of the TCRβ minilocus in all clones examined (Fig. 2, lanes 7 to 11; Table 2). In addition to PDβ, the deleted 5′Dβ sequences span two regions that display DNase hypersensitivity in developing thymocytes (5). To test whether these regions were required for rearrangement of TCRβ miniloci, we inserted the 377-bp minimal PDβ element (37) at its native position in φ/iEκ to generate the PDβ/iEκ substrate. Importantly, restoration of PDβ was sufficient to support normal levels of DβJβ joining following recombinase induction (Fig. 3A, lanes 5 to 8). Together, these data clearly demonstrate that enhancer activity per se is insufficient to direct recombination of linked gene segments. Instead, positive regulation of TCRβ accessibility requires enhancer-dependent activation of the Dβ1 germ line promoter.
FIG. 3.
Promoter activation mediates DβJβ rearrangement in chromosomal TCRβ substrates. (A) Levels of DβJβ rearrangements within TCRβ/iEκ substrates containing the minimal PDβ (lanes 5 to 8), the Vκ21C (lanes 9 and 10), or the PGK (lanes 11 and 12) promoter element. Control PCRs with TDR19 transfectants containing the 5′Dβ/iEκ (lanes 1 and 2) or the promoterless (lanes 3 and 4) substrates are included for comparison. (B) Rearrangement levels in TDR19 transfectants harboring the Ptet/iEκ (lanes 5 and 6), Ptet/φ (lanes 7 to 12), or iEκ/PDβ (lanes 13 to 18) minilocus. Other notation is as for Fig. 2.
The assembly of TCRβ gene segments can be mediated by a diverse set of transcriptional enhancers, including those derived from viral genomes (1, 4, 30). Our finding that PDβ also was required for DβJβ rearrangement (Fig. 2) led us to test whether this promoter was unique in its ability to regulate recombination. For this purpose, we replaced PDβ with either tissue-specific or generally active promoter elements and measured the recombination potential of each substrate in the TDR19 cell system. In enhancer-containing miniloci, a B-lymphocyte-specific promoter derived from the Vκ21C gene segment (PVκ) as well as a promoter that drives expression of the housekeeping gene encoding PGK directed normal levels of DβJβ recombination (Fig. 3A, lanes 9 to 12). Thus, heterologous promoters can functionally replace PDβ to support enhancer-dependent rearrangement of chromosomal miniloci.
Promoter activation targets DβJβ rearrangement.
The results presented in Fig. 2 and 3 favor a model in which promoter activation is the critical parameter for mediating efficient DβJβ rearrangement. Alternatively, physical interactions between promoter- and enhancer-bound proteins might act to loop out intervening gene segments from the chromosome and thereby facilitate their recognition by recombinase. Indeed, prior studies have shown that promoter-enhancer interactions can significantly alter the structure of intervening chromatin (10). Further support for this loop model is provided by the regulatory architecture of all antigen receptor loci, in which germ line gene segments are flanked by 5′ promoter and 3′ enhancer elements.
To test the loop model of recombinational accessibility, we repositioned iEκ in the TCRβ substrate to a location upstream from the PDβ element (Fig. 1). This configuration eliminates the potential for loops spanning the DβJβ cluster but should retain transcriptional activation of the target gene segments. As shown in Fig. 3B, the iEκ/PDβ substrate was efficiently rearranged in TDR19 upon RAG gene induction (lanes 13 to 18). As an independent test of the requirement for DNA loops, we generated a TCRβ minilocus that contained both the teto and iEκ regulatory elements (Ptet/iEκ [Fig. 1]). Because teto is activated solely by binding to the exogenous factor tTAk (14), the enhancer and promoter within Ptet/iEκ should function independently. Consistent with results obtained for the iEκ/PDβ substrate, induction of tTAk expression in Ptet/iEκ transfectants produced normal levels of DβJβ joins (Fig. 3B, lanes 5 and 6).
Our results with the iEκ/PDβ and Ptet/iEκ substrates suggested that efficient V(D)J recombination does not require participating gene segments to be flanked by enhancer-promoter pairs. However, all iEκ/PDβ transfectants harbored multiple copies of the recombination substrate. As such, we could not exclude the potential for interactions between promoter and enhancer elements situated within separate copies of tandemly integrated miniloci. Moreover, it remained possible that the tTAk activator could interact with factors bound to iEκ, resulting in DNA loops. To directly address the requirement for distal enhancers in minilocus recombination, we generated the Ptet/φ substrate, which lacks iEκ (Fig. 1). Removal of the downstream enhancer from Ptet substrate had no significant effect on induced levels of DβJβ rearrangement in either single- or multiple-copy transfectants (Fig. 3B, lanes 7 to 12; Table 2). From these data, we conclude that interactions between flanking promoter and enhancer elements are dispensible for targeting recombination of chromosomal miniloci.
Germ line DβJβ transcription correlates precisely with substrate recombination potential.
The activation of enhancer elements within antigen receptor loci has been linked temporally with the onset of germ line transcription and V(D)J recombination at participating gene segments (4, 33). To assess whether DβJβ joining in modified TCRβ miniloci correlated with their germ line transcription, we analyzed TDR19 transfectants in an RT-PCR assay that specifically detected hybrid DβJβCμ transcripts derived from unrearranged substrates (Fig. 1, primers C and D). In multiple independent transfectants, removal of either iEκ or 5′Dβ sequences spanning the PDβ element abolished not only substrate rearrangement but germ line DβJβ transcription as well (Fig. 4, lanes 1 to 7). In contrast, DβJβCμ transcripts were readily detected in recombinationally active substrates containing iEκ linked to either the minimal PDβ element or heterologous promoters (lanes 8 to 16). These findings are fully consistent with prior studies, which have shown that PDβ is functional in B cells when linked to active enhancer elements (30, 37). Importantly, germ line transcription and rearrangement were restored in miniloci lacking a distal enhancer by positioning teto upstream from the consensus TATA element within the Dβ1 recognition sequence (lanes 17 to 20). These functional correlations held true even for rare clones in which promoter/iEκ substrates lacked both DβJβ rearrangement and germ line transcripts (Table 2). As shown previously, these transfectants likely harbor substrate integrations into regions of heterochromatin (18). Thus, the results presented in Fig. 4 provide a direct correlation between the transcriptional activity of DβJβ gene segments and their rearrangement potential.
FIG. 4.
Germ line expression of TCRβ miniloci correlates with DβJβ recombination. Total cellular RNAs were harvested from individual transfectants maintained in the presence (without tTAk) or absence (with tTAk) of tetracycline (TET) for 3 days. The resultant RNAs were subjected to RT-PCR amplification with primers C and D (Fig. 1), and the reaction products were analyzed by Southern blotting using an oligonucleotide probe derived from Jβ1 coding sequences (top panel). The relative positions of amplification products corresponding to germ line transcripts that were processed at either Jβ1 (DβJβ1Cμ) or Jβ2 (DβJβ2Cμ) 3′ splice sites are shown at the left. Total cDNA levels were controlled in each sample by using a PCR assay specific for β-actin transcripts (bottom panel). The linearity of each assay was confirmed with serial dilutions of cDNA (lanes 21 to 26) derived from the 5′Dβ/iEκ transfectant shown in lane 1.
Transcription and rearrangement of TCRβ miniloci are independent of methylation status.
In addition to transcription and recombination, enhancers direct the active demethylation of linked antigen receptor gene segments (7, 23). Several groups have proposed that demethylation leads to alterations in regional chromatin structure that promote accessibility to nuclear proteins, including RNA polymerase and V(D)J recombinase (26, 27). However, the lack of tractable experimental model systems has hampered previous efforts to establish causal relationships between enhancer activity, demethylation, transcription, and recombination of gene segments.
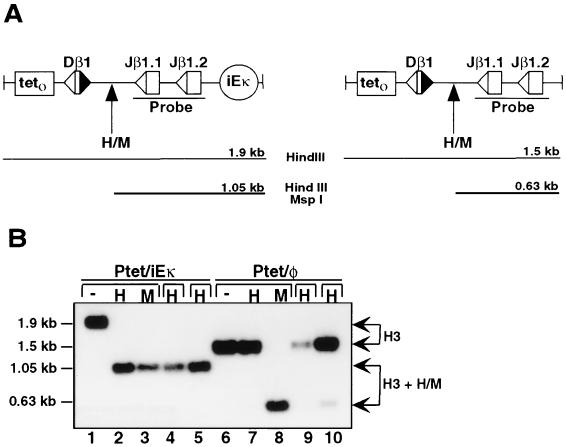
Our analyses clearly demonstrated that Ptet provides access to RNA polymerase and V(D)J recombinase in the absence of a distal enhancer (Fig. 3B and 4). However, these findings did not address the possibility that Ptet possesses demethylating activities associated with antigen receptor enhancer elements (20). To explore this possibility, we subjected Ptet/φ and Ptet/iEκ transfectants to Southern blot analyses using the methylation-sensitive restriction enzyme HpaII and a probe spanning the Jβ gene segment cluster. These analyses were designed to measure the relative degree of substrate DNA methylation at a CpG site that is equidistant from the Dβ1 and Jβ1 gene segments (Fig. 5A). As shown in Fig. 5B, HpaII completely digested this CpG site in all Ptet/iEκ transfectants, indicating a hypomethylated status (lanes 2, 4, and 5). The HpaII site was also hypomethylated in Ptet/iEκ substrates prior to induction of tTAk expression (data not shown), a finding consistent with the dominant role of Ig enhancer elements in protecting linked sequences from DNA methylation (7). In sharp contrast, the vast majority of HpaII sites were hypermethylated in Ptet miniloci that lacked iEκ (lanes 7, 9, and 10). As a control, the CpG sites were efficiently digested in both substrates with the methylation-insensitive isoschizomer MspI (lanes 3 and 8). Similar results were obtained with blotting strategies that probed the methylation status of sequences located 3′ to the Jβ gene segments (data not shown).
FIG. 5.
Ptet activation targets DβJβ rearrangement independent of substrate demethylation. (A) Schematic depiction of the DβJβ regions within the Ptet/iEκ and Ptet/φ substrates. The relative positions of HpaII/MspI (H/M) restriction sites within the parental HindIII fragments are highlighted. The sizes of predicted restriction fragments are shown below each diagram. (B) Methylation status of independent transfectants harboring Ptet/iEκ (lanes 1 to 5) or Ptet/φ (lanes 6 to 10) substrates. Genomic DNA from each clone was digested with HindIII alone (−; lanes 1 and 6) or in combination with either HpaII (H; lanes 2, 4, 5, 7, 9, and 10) or MspI (M; lanes 3 and 8). Digested DNAs were analyzed by Southern blotting procedures using a radiolabeled probe spanning the Jβ1/Jβ2 gene segments (Fig. 6A). The relative positions (arrows) and sizes (left) of restriction fragments resulting from digestion at the HindIII sites (H3) or from further digestion by HpaII and MspI (H3 + H/M) are indicated.
In mammalian cells, stable methylation patterns of CpG sequences are established subsequent to DNA replication (26). However, the failure of Ptet/φ substrates to undergo demethylation could not be attributed to insufficient rounds of DNA replication, since genomic DNAs were derived from transfectants that had completed at least four rounds of cell division following promoter activation. As such, the data presented in Fig. 5 indicate that Ptet lacks at least one function associated with enhancer activity—the ability to control demethylation of neighboring chromosomal sequences. Coupled with our previous results, we conclude that efficient DβJβ recombination can be dissociated from the regional methylation status of chromosomal miniloci.
Germ line promoter activation is required for generation of DβJβ signal joins.
Emerging studies suggest that in addition to controlling the initial access of DβJβ gene segments to recombinase, the TCRβ enhancer (Eβ) may affect the efficiency of coding join formation (16). A potential mechanistic explanation for these findings invokes transcriptional regulatory elements in the recruitment of DNA repair complexes to chromosomal breaks generated by recombinase cleavage. In contrast to coding join formation, DβJβ signal ends were resolved with similar efficiencies in mice harboring wild-type or Eβ −/− loci (16). As such, we reasoned that the levels of signal joins generated from each TCRβ substrate would provide a more direct readout for gene segment accessibility. Moreover, since the DβJβ intervening sequences are identical in all modified TCRβ miniloci, the kinetics of signal join formation should be similar regardless of their substrate origin.
To test whether transcriptional regulatory elements were required for the generation of DβJβ signal joins, we isolated extrachromosomal DNA from a panel of TDR19 transfectants 48 h after tetracycline withdrawal. The DNA from each transfectant was analyzed by a semiquantitative PCR assay that specifically detects the circular deletion products containing either DβJβ1 or DβJβ2 signal joins (Fig. 6A, primers C and E). Using this assay, we found that removal of either iEκ or PDβ severely impaired the generation of signal joins from TCRβ miniloci (Fig. 6B, lanes 2 to 6). Importantly, signal junctions were efficiently formed in TDR19 transfectants harboring either the Ptet/iEκ or Ptet/φ substrate (lanes 8 to 15). As an independent control for recombinase activity, we observed similar levels of VλJλ signal joins in all of the induced clones (Fig. 6B, bottom panel). The parallel between levels of coding and signal joins in modified substrates strongly suggests that transcriptional promoters mediate initial access of chromosomal miniloci to the recombinase complex.
FIG. 6.
Promoterless and enhancerless substrates do not generate DβJβ signal joins. (A) Diagram of the PCR assay used for signal join detection. The locations of amplification primers (C and E) as well as the predicted sizes of PCR products from DβJβ1 and DβJβ2 rearrangements are indicated. The relative positions of flanking promoter and enhancer elements are shown in the top diagram. (B) Levels of signal joins in TDR19 transfectants harboring the indicated miniloci 48 h after tetracycline withdrawal. The relative positions of amplification products corresponding to DβJβ1 and DβJβ2 signal joins are shown at the left. Control assays for recombinase activity (VλJλ signal joins) are presented in the bottom panel. The linearity of each assay was confirmed by serial dilutions (lanes 15 to 20) of the 5′Dβ/iEκ sample shown in lane 2.
DISCUSSION
Promoter activation targets V(D)J recombination.
The tissue, stage, and allele specificity of antigen receptor gene assembly is achieved through programmed alterations in the efficiency of V(D)J recombination at individual gene segment clusters (39). The results presented in this report provide novel insights into the critical role played by transcriptional control elements in targeting recombinase to chromosomal gene segments. Specifically, we show that positive regulation of DβJβ rearrangement within TCRβ miniloci can be dissociated from intrinsic properties of enhancer elements (Fig. 2). Instead, efficient recombination of these gene segments requires the presence of a germ line promoter located directly upstream from the DβJβ cluster (Fig. 3A). In light of these findings, we propose that the observed inhibition of endogenous TCRβ rearrangement by targeted deletion of Eβ (1, 3) may indirectly result from the strict enhancer dependence of PDβ activity (37). Indeed, deletion of sequences spanning PDβ1 specifically impairs Dβ1 rearrangement at the endogenous TCRβ locus without altering levels of Dβ2Jβ recombination (46). Similarly, removal of a regulatory region located 5′ to the Jα cluster that includes a functional germ line promoter has been shown to preferentially impair rearrangement of proximal Jα gene segments (45). Taken together, these studies suggest that the primary function of Ig and TCR enhancers for targeting V(D)J recombination is to provide cell type and stage specificity to the activity of individual germ line promoters.
The dual requirement for PDβ and enhancers suggested that direct interactions between these regulatory elements may be essential for targeting DβJβ rearrangement, perhaps via the formation of DNA loops. This requirement may underlie the unique functional architecture of antigen receptor loci, in which germ line promoter and enhancer elements are segregated to positions flanking gene segment clusters. A requirement for DNA loops would also be consistent with previous observations that selectable marker genes positioned adjacent to enhancer elements exert an inhibitory effect on the rearrangement of antigen receptor loci (1, 6, 43). In these loci, the flanking transcriptional unit may perturb enhancer-promoter interactions, squelching both germ line transcription and loop formation.
To test the requirement for DNA loops in mediating DβJβ rearrangement, we generated a panel of substrates that either repositioned or removed the enhancer element from its distal position. Unexpectedly, no differences were observed in the recombination potential of miniloci that contained promoter and enhancer elements at flanking positions (PDβ/iEκ) versus those in which both elements were colocalized upstream from the DβJβ cluster (iEκ/PDβ). Although interactions between regulatory elements in neighboring substrates could not be formally discounted in multicopy iEκ/PDβ transfectants, promoter activation by the adjacent enhancer should be strongly favored relative to long-range (>23 kb) activation by iEκ. In this regard, we have previously shown that the simian virus 40 enhancer directed PDβ activity when positioned within the TCRβ minilocus but failed to activate transcription or rearrangement of 5′Dβ/φ substrates when present in cointegrated drug resistance vectors (30). Consistent with the activity of iEκ/PDβ miniloci, rearrangement of single-copy substrates lacking a distal enhancer was restored by placement of teto 5′ to the Dβ1 gene segment (Fig. 3). Thus, formation of DNA loops between flanking promoter and enhancer elements is not an absolute requirement for targeting efficient rearrangement of chromosomal gene segments. However, these findings do not exclude a potential role for DNA looping in subsequent stages of TCRβ gene assembly, which may require the juxtaposition of more distant Vβ and Dβ gene segments in order to facilitate their recombination.
We observed a strict requirement for promoter activation to generate DβJβ coding and signal joins in TCRβ miniloci (Fig. 6), indicating that accessibility is the primary factor regulating recombination in these substrates. These data are fully consistent with recent findings that alterations in gene segment accessibility underlie the stage- and tissue-specific control of Igκ and TCRδ rearrangements (25, 41). What are the molecular features of promoter activation that potentiate access of chromosomal substrates to the recombinase complex? In this study, we clearly show that heterologous promoters, including PVκ, PGK, and Ptet, can replace PDβ to direct germ line transcription and rearrangement of chromosomal miniloci (Fig. 3 and 4). Because each of these promoters is regulated by a distinct set of transcription factors (37), their similar effects on DβJβ recombination cannot be readily explained by a recruitment of specific transactivating proteins that are essential for mediating accessibility. Thus, it is tempting to invoke transcriptional initiation or readthrough as critical components of the mechanisms that control substrate accessibility to V(D)J recombinase. This model is further supported by striking correlations that exist between the expression of sterile transcripts and the rearrangement of corresponding gene segments (Fig. 4 and references 21, 30, 34, and 48). Prior studies have demonstrated that factor binding to DNA motifs, including promoters, induces localized alterations in chromatin configuration (10). However, more general access to enzymatic complexes may require transcriptional readthrough in order to accentuate or propagate these chromatin alterations (10, 25). Thus, the presence of germ line promoters, rather than simple factor-binding sites, may be essential for conferring maximum recombinational accessibility to gene segments situated within complex antigen receptor loci. Although final confirmation of any regulatory model awaits the targeted modification of endogenous loci, TCRβ minilocus substrates provide a tractable experimental system to define the precise mechanisms by which promoters govern the initial access of gene segments to V(D)J recombinase.
Uncoupling V(D)J recombination from substrate demethylation.
Prior studies have established a correlation between the demethylation of antigen receptor loci and their recombination (7, 9, 19, 23). In turn, demethylation of gene segments, as well as their transcription and recombination, have been firmly linked to the activation of enhancer elements in cis. For example, Eμ and iEκ protect TCRβ transgenes from de novo methylation in murine lymphocytes (11) and promote demethylation of substrates in cell models (7, 23). Despite extensive correlations, it has been difficult to dissect the individual contributions of these distinct processes to recombinational accessibility. We now demonstrate that a synthetic promoter (Ptet) can efficiently direct germ line transcription and DβJβ rearrangement but not regional demethylation of TCRβ miniloci (Fig. 5B). These results indicate that Ptet lacks a hallmark feature associated with enhancers that drive Ig and TCR gene expression—the ability to direct demethylation of neighboring sequences in a chromosomal context. More importantly, these data indicate that hypomethylation of TCRβ miniloci is not essential for conferring recombinational accessibility to its composite gene segments. In this regard, prior studies have shown that demethylated IgH gene segments may be refractory to recombinase activity in the absence of germ line transcription (6). Overall, these findings strongly suggest that demethylation is a functional consequence of enhancer activation within a given locus, rather than a prerequisite for antigen receptor gene assembly.
Regulatory studies in recombinase-inducible cell models.
The physiological relevance of the TDR19-TCRβ model system is supported by its ability to recapitulate enhancer- and promoter-dependent recombination of the endogenous TCRβ locus (1, 3, 46). Although targeted deletion of endogenous regulatory elements can be used to judge their relative contributions to locus rearrangement, the TDR19-TCRβ system provides several distinct advantages for dissecting the molecular determinants of recombinational accessibility. For example, the framework TCRβ minilocus, which lacks PDβ and iEκ, is devoid of elements that direct either germ line transcription or rearrangement, but the substrate can be manipulated to include a broad panel of regulatory sequences. In contrast, endogenous loci harbor numerous promoter and enhancer elements that may partially overlap in their regulatory functions (1, 16, 35, 46). Furthermore, the TDR19 system permits transcriptional analysis of gene segments at the precise time point that rearrangement occurs (i.e., upon induction of recombinase activity). Analogous studies in murine models are complicated by fluctuations in promoter-enhancer activities that accompany the developmental progression of lymphocyte populations (29). Thus, our ability to directly compare transcription and rearrangement of substrates in TDR19 will be extremely valuable for future studies designed to dissect the molecular mechanisms by which promoter activation targets V(D)J recombination.
ACKNOWLEDGMENTS
We thank Rey Gomez and Guo Ming Zhang for technical assistance and David Schatz (Yale University) for the pTET-R1 and pTET-R2 constructs. We also thank D. Ballard, W. Khan, J. Hawiger, L. Van Kaer, S. Sessoms, and H. Bendall for valuable comments.
This work was supported by NIH grants AI36944, AI01412 (E.M.O.), and GM19597 (M.L.S.). E.M.O. is a Joe C. Davis Scholar.
REFERENCES
- 1.Bories J-C, Demengeot J, Davidson L, Alt F W. Gene-targeted deletion and replacement mutations of the T-cell receptor β-chain enhancer: the role of enhancer elements in controlling V(D)J recombination accessibility. Proc Natl Acad Sci USA. 1996;93:7871–7876. doi: 10.1073/pnas.93.15.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boudinot P, Drapier A M, Cazenave P A, Sanchez P. Conserved distribution of lambda subtypes from rearranged gene segments to immunoglobulin synthesis in the mouse B cell repertoire. Eur J Immunol. 1994;24:2013–2017. doi: 10.1002/eji.1830240912. [DOI] [PubMed] [Google Scholar]
- 3.Bouvier G, Watrin F, Naspetti M, Verthuy C, Nazuet P, Ferrier P. Deletion of the mouse T-cell receptor β gene enhancer blocks αβ T-cell development. Proc Natl Acad Sci USA. 1996;93:7877–7881. doi: 10.1073/pnas.93.15.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capone M, Watrin F, Fernex C, Horvat B, Krippl B, Wu L, Scollay R, Ferrier P. TCRβ and TCRα gene enhancers confer tissue- and stage-specificity on V(D)J recombination events. EMBO J. 1993;12:4335–4346. doi: 10.1002/j.1460-2075.1993.tb06118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chattopadhay S, Whitehurst C E, Schwenk F, Chen J. Biochemical and functional analyses of chromatin changes at the TCRβ gene locus during CD4−CD8− to CD4+CD8+ thymocyte differentiation. J Immunol. 1998;160:1256–1267. [PubMed] [Google Scholar]
- 6.Chen J, Young F, Bottaro A, Stewart V, Smith R K, Alt F W. Mutations of the intronic IgH enhancer and its flanking sequences differentially affect accessibility of the JH locus. EMBO J. 1993;12:4635–4645. doi: 10.1002/j.1460-2075.1993.tb06152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demengeot J, Oltz E M, Alt F W. Promotion of V(D)J recombinational accessibility by the intronic Eκ element: role of the κB motif. Int Immunol. 1995;7:1995–2003. doi: 10.1093/intimm/7.12.1995. [DOI] [PubMed] [Google Scholar]
- 8.Ehlich A, Schaal S, Gu H, Kitamura D, Muller W, Rajewsky K. Immunoglobulin heavy and light chain genes rearrange independently at early stages of B cell development. Cell. 1993;72:695–704. doi: 10.1016/0092-8674(93)90398-a. [DOI] [PubMed] [Google Scholar]
- 9.Engler P, Weng A, Storb U. Influence of CpG methylation and target spacing on V(D)J recombination in a transgenic substrate. Mol Cell Biol. 1993;13:571–577. doi: 10.1128/mcb.13.1.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V. Chromatin structure and gene expression. Proc Natl Acad Sci USA. 1996;93:9384–9388. doi: 10.1073/pnas.93.18.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernex C, Capone M, Ferrier P. The V(D)J recombinational and transcriptional activities of the immunoglobulin heavy-chain intronic enhancer can be mediated through distinct protein-binding sites in a transgenic substrate. Mol Cell Biol. 1995;15:3217–3226. doi: 10.1128/mcb.15.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrier P, Krippl B, Blackwell T K, Furley A J W, Suh H, Winoto A, Cook W D, Hood L, Constantini F, Alt F W. Separate elements control DJ and VDJ rearrangements in a transgenic recombination substrate. EMBO J. 1990;9:117–125. doi: 10.1002/j.1460-2075.1990.tb08087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fulton R, Van Ness B. Kappa immunoglobulin promoters and enhancers display developmentally controlled interactions. Nucleic Acids Res. 1993;21:4941–4947. doi: 10.1093/nar/21.21.4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartman S C, Mulligan R C. Two dominant selectable markers for gene transfer studies in mammalian cells. Proc Natl Acad Sci USA. 1988;85:8047–8051. doi: 10.1073/pnas.85.21.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hempel W M, Stanhope-Baker P, Mathieu N, Huang F, Schlissel M S, Ferrier P. Enhancer control of V(D)J recombination at the TCRβ locus: differential effects on DNA cleavage and joining. Genes Dev. 1998;12:2305–2317. doi: 10.1101/gad.12.15.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenuwein T, Forrester W C, Qiu R-G, Grosscheld R. The immunoglobulin μ enhancer core establishes local factor access in nuclear chromatin independent of transcriptional stimulation. Genes Dev. 1993;7:2016–2032. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- 18.Jenuwein T, Forrester W C, Fernandez-Herrero L A, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 19.Kelley D E, Pollock B A, Atchison M L, Perry R P. The coupling between enhancer activity and hypomethylation of kappa immunoglobulin genes is developmentally regulated. Mol Cell Biol. 1988;8:930–939. doi: 10.1128/mcb.8.2.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-κB in B-cell-specific demethylation of the Igκ locus. Nat Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 21.Lennon G G, Perry R P. The temporal order of appearance of transcripts from unrearranged and rearranged Ig genes in murine fetal liver. J Immunol. 1990;144:1983–1987. [PubMed] [Google Scholar]
- 22.Li Y S, Hayakawa K, Hardy R R. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J Exp Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichtenstein M, Keini G, Cedar H, Bergman Y. B cell-specific demethylation: a novel role for the intronic κ chain enhancer sequence. Cell. 1994;76:913. doi: 10.1016/0092-8674(94)90365-4. [DOI] [PubMed] [Google Scholar]
- 24.Mather E L, Perry R P. Methylation status and DNaseI sensitivity of immunoglobulin genes: changes associated with rearrangement. Proc Natl Acad Sci USA. 1983;80:4689–4693. doi: 10.1073/pnas.80.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMurry M T, Hernandez-Munain C, Lauzurica P, Krangel M S. Enhancer control of local accessibility to V(D)J recombinase. Mol Cell Biol. 1997;17:4553–4561. doi: 10.1128/mcb.17.8.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mostoslavsky R, Bergman Y. DNA methylation: regulation of gene expression and role in the immune system. Biochim Biophys Acta. 1997;1333:F29–F50. doi: 10.1016/s0304-419x(97)00010-3. [DOI] [PubMed] [Google Scholar]
- 27.Mostoslavsky R, Singh N, Kirillov A, Pelanda R, Cedar H, Chess A, Bergman Y. κ chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 1998;12:1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 29.Okada A, Mendelsohn M, Alt F. Differential activation of transcription versus recombination of transgenic T cell receptor β variable region gene segments in B and T lineage cells. J Exp Med. 1994;180:261–272. doi: 10.1084/jem.180.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oltz E M, Alt F W, Lin W-C, Chen J, Taccioli G, Desiderio S, Rathbun G. A V(D)J recombinase-inducible B-cell line: role of transcriptional enhancer elements in directing V(D)J recombination. Mol Cell Biol. 1993;13:6223–6230. doi: 10.1128/mcb.13.10.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Royer H D, Ramarli D, Acuto O, Campen T J, Reinherz E L. Genes encoding the T-cell receptor α and β subunits are transcribed in an ordered manner during intrathymic ontogeny. Proc Natl Acad Sci USA. 1985;82:5510–5514. doi: 10.1073/pnas.82.16.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schatz D G, Oettinger M A, Schlissel M A. V(D)J recombination: molecular biology and regulation. Annu Rev Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 33.Scherer D, Brockman J, Bendall H, Zhang G, Ballard D, Oltz E. Corepression of RelA and c-Rel inhibits immunoglobulin kappa gene transcription and rearrangement in pre-B lymphocytes. Immunity. 1996;5:563–574. doi: 10.1016/s1074-7613(00)80271-x. [DOI] [PubMed] [Google Scholar]
- 34.Schlissel M S, Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989;58:1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- 35.Serwe M, Sablitzky F. V(D)J recombination in B cells is impaired but not blocked by targeted deletion of the immunoglobulin heavy chain intron enhancer. EMBO J. 1993;12:2321–2327. doi: 10.1002/j.1460-2075.1993.tb05886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shockett P, Difilippantonia M, Hellman N, Schatz D G. A modified tetracycline-regulated system provides autoregulatory, inducible gene expression in cultured cells and transgenic mice. Proc Natl Acad Sci USA. 1992;92:6522–6526. doi: 10.1073/pnas.92.14.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikes M L, Gomez R J, Song J, Oltz E M. A developmental stage-specific promoter directs germline transcription of DβJβ gene segments in precursor T lymphocytes. J Immunol. 1998;161:1399–1405. [PubMed] [Google Scholar]
- 38.Sikes, M. L., and E. M. Oltz. An inducible cell model for studies of V(D)J recombinational control. J. Immunol. Methods, in press. [DOI] [PubMed]
- 39.Sleckman B P, Gorman J R, Alt F W. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 40.Sleckman B P, Bardon C G, Ferrini R, Davidson L, Alt F W. Function of the TCRα enhancer in αβ and γδ T cells. Immunity. 1997;7:505–515. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- 41.Stanhope-Baker P, Hudson K M, Shaffer A L, Constantinescu A, Schlissel M S. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- 42.Strominger J L. Developmental biology of T cell receptors. Science. 1989;244:943–950. doi: 10.1126/science.2658058. [DOI] [PubMed] [Google Scholar]
- 43.Takeda S, Zou Y-R, Bluethmann H, Kitamura D, Muller U, Rajewsky K. Deletion of the immunoglobulin κ chain intron enhancer abolishes κ chain gene rearrangement in cis but not λ chain gene rearrangement in trans. EMBO J. 1993;12:2329–2336. doi: 10.1002/j.1460-2075.1993.tb05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tucker K L, Beard C, Dausman J, Jackson-Grusby L, Laird P W, Lei H, Li E, Jaenisch R. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 1996;10:1008–1020. doi: 10.1101/gad.10.8.1008. [DOI] [PubMed] [Google Scholar]
- 45.Villey I, Caillol D, Selz F, Ferrier P, de Villartay J-P. Defect in rearrangement of the most 5′ TCR-J alpha following targeted deletion of T early alpha (TEA): implications for TCR alpha locus accessibility. Immunity. 1996;5:331–342. doi: 10.1016/s1074-7613(00)80259-9. [DOI] [PubMed] [Google Scholar]
- 46.Whitehurst, C., and J. Chen. Personal communication.
- 47.Xu Y, Davidson L, Alt F W, Baltimore D. Deletion of the Igκ light chain intronic enhancer/matrix attachment region impairs but does not abolish VκJκ rearrangement. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- 48.Yancopoulos G D, Alt F W. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. [PubMed] [Google Scholar]