Abstract
The function of conserved regions of the metazoan U5 snRNA was investigated by reconstituting U5 small nuclear ribonucleoprotein particles (snRNPs) from purified snRNP proteins and HeLa or Xenopus U5 snRNA mutants and testing their ability to restore splicing to U5-depleted nuclear extracts. Substitution of conserved nucleotides comprising internal loop 2 or deletion of internal loop 1 had no significant effect on the ability of reconstituted U5 snRNPs to complement splicing. However, deletion of internal loop 2 abolished U5 activity in splicing and spliceosome formation. Surprisingly, substitution of the invariant loop 1 nucleotides with a GAGA tetraloop had no effect on U5 activity. Furthermore, U5 snRNPs reconstituted from an RNA formed by annealing the 5′ and 3′ halves of the U5 snRNA, which lacked all loop 1 nucleotides, complemented both steps of splicing. Thus, in contrast to yeast, loop 1 of the human U5 snRNA is dispensable for both steps of splicing in HeLa nuclear extracts. This suggests that its function can be compensated for in vitro by other spliceosomal components: for example, by proteins associated with the U5 snRNP. Consistent with this idea, immunoprecipitation studies indicated that several functionally important U5 proteins associate stably with U5 snRNPs containing a GAGA loop 1 substitution.
Nuclear pre-mRNA splicing proceeds via a two-step mechanism. In the first step, the pre-mRNA is hydrolyzed at the 5′ splice site and the 5′ end of the intron interacts concomitantly with an adenosine at the so-called branch point. The splicing intermediates thus generated include exon 1 and a lariat structure comprised of the intron and exon 2. In the second step, hydrolysis at the 3′ splice site and the concomitant ligation of exons 1 and 2 give rise to the mRNA and the excised intron in the form of a lariat. Both reactions are catalyzed by the spliceosome, a large ribonucleoprotein complex formed by the ordered interaction of numerous splicing factors and the four small nuclear ribonucleoprotein particles (snRNPs), U1, U2, U5, and U4/U6, with conserved regions of the pre-mRNA (reviewed in references 19, 27, and 34). Spliceosome assembly is initiated by the interaction of the U1 and U2 snRNPs with the 5′ splice site and branch site, respectively, thereby generating the so-called prespliceosome, or complex A. Mature spliceosomes (i.e., complexes B and C) are ultimately formed by the subsequent interaction of the U4/U6 and U5 snRNPs, in the form of a preassembled U4/U6.U5 tri-snRNP complex (reviewed in references 19 and 34).
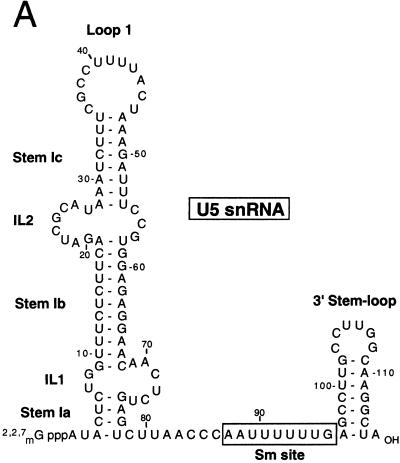
The assembly of a catalytically active spliceosome requires the formation of a network of RNA-RNA interactions which favorably position the chemically reactive groups of the pre-mRNA for catalysis (for reviews, see references 26 and 38). The U5 snRNP has been proposed to play a central role in recognizing and aligning the 5′ and 3′ splice sites for catalysis, and its function appears to be mediated, at least in part, by base pairing interactions between the U5 small nuclear RNA (snRNA) and the pre-mRNA. In particular, at least 3 of the 9 nucleotides (nt) present in its absolutely conserved loop 1 sequence (see Fig. 1A) were shown by several methods, including cross-linking and yeast genetic studies, to interact with exon nucleotides at the 5′ and/or 3′ splice site (9, 28, 29, 30, 37, 45). The interaction of loop 1 with exon 1 is observed both prior and subsequent to the first step of splicing, whereas its interaction with exon 2 is detectable only after step 1 (30, 37). Loop 1 was thus originally proposed to play an essential role in both catalytic steps of splicing in both higher and lower eukaryotes. Recent in vitro studies with yeast have demonstrated that the first, but not the second step of splicing can occur in its absence (31). More detailed mutational analyses in vitro have also revealed that only large loop 1 deletions or insertions, as opposed to minor ones, affect the efficiency of the second step of splicing in yeast (32). Loop 1 of the U5 snRNP is currently proposed to bind and favorably position excised exon 1 for its nucleophilic attack at the 3′ splice site during the second step of splicing (31). However, since the interaction of loop 1 nucleotides with either exon is limited to 2 to 3 bp and these are often non-Watson-Crick in nature, other components of the U5 snRNP, in particular U5-specific proteins (see below), have been proposed to help stabilize U5 snRNP interactions at both the 5′ and 3′ splice site (41).
FIG. 1.
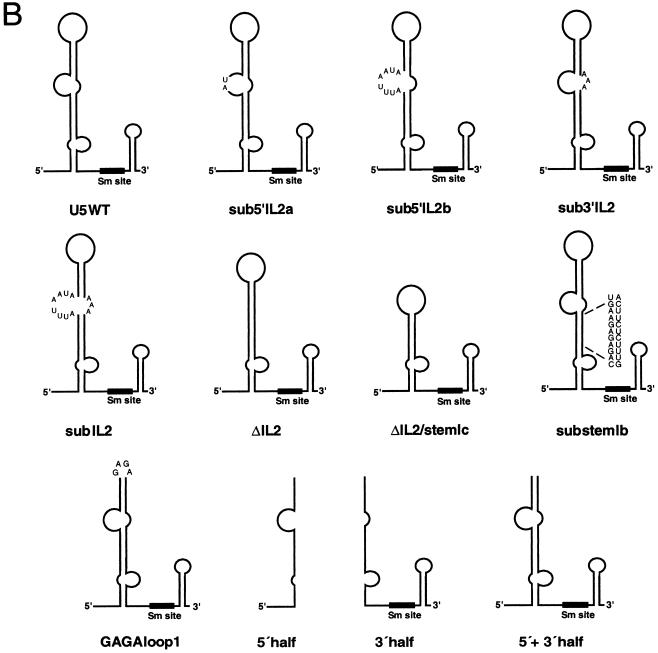
Secondary structure models of wild-type and mutant human U5 snRNAs. (A) Sequence and secondary structure model of the human U5a snRNA as originally proposed by Krol et al. (20). The conserved, single-stranded region of the Sm site is boxed. (B) The putative secondary structure of the human U5 snRNA mutants is shown schematically. All nucleotide substitutions are shown in detail.
In addition to a single U5 snRNA molecule, mammalian U5 snRNPs possess eight so-called Sm or core proteins (B, B′, D1, D2, D3, E, F, and G), common to all spliceosomal snRNP species, and nine U5-specific proteins (reviewed in reference 44). Three of these U5-specific proteins, with molecular masses of 116, 200, and 220 kDa, have been shown to be evolutionarily conserved and to carry out essential functions during splicing (2, 12, 17, 23, 24). The human 220-kDa protein and its yeast homolog, Prp8p, have been shown by site-specific cross-linking experiments to interact with the 5′ and 3′ splice sites as well as the branch site and polypyrimidine tract (8, 25, 35, 41, 42, 45). The interaction between Prp8p and the 5′ and 3′ splice sites was observed even in the absence of U5 loop 1 (11). This protein has thus been proposed to partially mediate the interaction of the U5 snRNP with both splice sites and thereby help position reactive groups of the pre-mRNA for catalysis (11, 41). The HeLa U5-specific 200-kDa protein and its yeast homolog, Snu246p, have been identified as members of the DEXH box family of putative RNA helicases (23). Consistent with the idea that it catalyzes RNA conformational changes during splicing, this U5 snRNP protein has recently been shown to possess RNA duplex unwinding activity in vitro (21, 33). Finally, the HeLa 116-kDa protein and its yeast homolog, Snu114p, were shown to possess all of the sequence motifs characteristic of GTP binding proteins, and, in the case of the human protein, to bind GTP (12). This putative GTPase has thus been proposed to act as a molecular switch, modulating RNA conformational changes within the spliceosome (12). Interestingly, these three proteins, together with the U5 40-kDa protein, interact in the absence of U5 RNA to form a stable heteromeric complex, suggesting that they associate concomitantly with U5 snRNPs during assembly (1).
Comparison of the U5 snRNAs across evolution has revealed only limited regions of sequence conservation, which include loop 1, internal loop 2 (IL2), and the Sm protein binding site (13, 14, 20). Despite this limited conservation, a general U5 snRNA secondary structure model can be generated (Fig. 1A). The Sm site, which is also present in the U1, U2, and U4 snRNAs, consists of a single-stranded uridylic acid-rich region typically flanked by two hairpin loops and serves as the primary binding site of the Sm proteins (7). Whereas the interaction of the Sm proteins with the U5 snRNA has been investigated in detail, relatively little is known about the sites of interaction of the U5-specific proteins (18). Based on chemical and nuclease accessibility studies, IL2 and its adjacent stems have been proposed to serve as binding sites for one or more U5-specific protein (4, 6). Indeed, studies performed in vivo with human U5 snRNA mutants suggest that IL2, stems Ib and Ic, and loop 1 are either directly or indirectly involved in the interaction of the 220-kDa protein with the U5 snRNA (16). More recent site-specific cross-linking experiments with yeast have also demonstrated that Prp8p (U5 220-kDa protein) interacts with multiple sites within the 5′ stem-loop of U5, including IL2 and loop 1 (11). These studies also revealed an interaction between IL2 and the yeast homolog of the U5 116-kDa protein (Snu114p).
Detailed analyses of the contribution of the various U5 snRNA structural domains to U5 snRNP function during splicing have been limited to the yeast Saccharomyces cerevisiae. A minimal U5 snRNA capable of complementing the lethal phenotype of a yeast U5 gene disruption was shown to require the presence of loop 1, IL2 plus an adjacent stem, and the Sm protein binding site (13). In vitro studies with yeast have investigated in detail the role of loop 1 sequences in splicing and the functional effects of deletions in other regions of the U5 snRNA (11, 31). Mutational analyses of the metazoan U5 snRNA have, on the other hand, focused on the involvement of its structural domains in the assembly of U5 snRNPs, U4/U6.U5 tri-snRNPs and the spliceosome (16, 18). The effect of U5 snRNA mutations on pre-mRNA splicing has been limited to in vivo studies employing cotransfection assays which investigated the effect of loop 1 point mutations on splice site selection (9). Here, we have investigated the function of conserved regions in the major stem-loop of the metazoan U5 snRNA in both splicing complex formation and splicing. To this end, we have reconstituted in vitro U5 snRNPs from human or Xenopus U5 snRNA mutants and tested their ability to restore splicing to U5-depleted nuclear extracts. The data presented here demonstrate that two of the most highly conserved regions of the U5 snRNA (i.e., loop 1 and IL2) are surprisingly amenable to mutation. U5 snRNPs unexpectedly retained their ability to efficiently complement both steps of splicing even after complete deletion of loop 1. These results thus indicate that, in metazoans, the function of U5 loop 1 during the second step of splicing in vitro can be compensated for by other factors in its absence.
MATERIALS AND METHODS
Construction of U5 snRNA mutants.
Human and Xenopus U5 snRNA deletion and substitution mutants were constructed as previously described by Jarmolowski and Mattaj (18). ΔIL2 and sub-stem Ib were kindly provided by Albrecht Bindereif and constructed as described by Hinz et al. (16). The 5′ (nt 1 to 35) and 3′ (nt 47 to 116) halves of U5 were transcribed from PCR products containing a T7 and SP6 promoter, respectively. Oligonucleotides used for PCR of these two U5 snRNA gene fragments were as follows: 5′ half forward primer, 5′ GCGCTAATACGACTCACTATAGGATACTCTGGTTTCTC 3′; 5′ half reverse primer, 5′ GGAGATTTATGCGAT 3′; 3′ half forward primer, 5′ GCGCATTTAGGTGACACTATAGGAGATTTCCGTGGAGAGG 3′; and 3′ half reverse primer, 5′ TAGCCTTGCCAAGGCAAGG 3′. The 5′ and 3′ halves were annealed in buffer containing 20 mM HEPES–KOH (pH 7.9), 100 mM KCl, and 10 mM MgCl2 by incubation at 70°C for 15 min and being allowed to slowly cool to room temperature.
Preparation of snRNAs, pre-mRNA, and native snRNP proteins.
Native, RNA-free snRNP proteins (TPs) were isolated from a mixture of m3G immunoaffinity-purified U1, U2, U5, and U4/U6 snRNPs by dissociation in the presence of EDTA and the anion-exchange resin DE53 (39). HeLa U5 snRNA was isolated from purified snRNPs as described previously (39). In vitro-transcribed human and Xenopus U5 snRNAs, as well as MINX pre-mRNA, were prepared as previously described (36).
U5 snRNP depletion and splicing complementation.
Nuclear extracts were prepared from HeLa cells (Computer Cell Culture Center, Mons, Belgium) as described by Dignam et al. (10). U5-depleted nuclear extract was prepared by affinity selection with a 2′-O-alkyl, biotinylated RNA oligonucleotide complementary to nt 36 to 47 of the human U5 snRNA (22, 36). Mock-depleted extract was handled in an identical manner, except that oligonucleotide was omitted. Complementation with in vitro-reconstituted particles was accomplished by combining 2.6 pmol (100 ng) of authentic or in vitro-transcribed U5 snRNA, or the annealed mixture containing 100 ng each of the 5′ and 3′ halves of U5 RNA and 3.3 pmol (650 ng) of purified native snRNP proteins (TPs). RNA and TPs were incubated for 60 min at 0°C in the presence of splicing reaction mixtures lacking pre-mRNA, and splicing was initiated by the addition of the pre-mRNA. In vitro splicing and the analysis of splicing intermediates and products were performed as described previously (36). No differences in complementation efficiency were observed when reconstitution was carried out either directly in splicing extract or by additionally preincubating the U5 snRNA and TPs in the absence of extract. Splicing complex formation was analyzed by native gel electrophoresis as described by Behrens et al. (5).
Immunoprecipitation of reconstituted U5 snRNPs.
32P-labelled U5 snRNA was prepared by in vitro transcription as described above and incubated under standard reconstitution conditions. Immunoprecipitations were performed with rabbit sera directed against the U5 116-kDa protein (12), essentially as previously described (15). Briefly, protein A-Sepharose (PAS)-bound antibody was incubated for 2 h at 4°C with 12.5 μl of a splicing reaction mixture containing 105 cpm (10 ng) of 32P-labelled U5 snRNA in 200 μl of IPP150 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.05% [vol/vol] Nonidet P-40) and subsequently washed four times with IPP buffer containing 300 mM NaCl. Immunoprecipitated RNA was extracted with phenol-chloroform, precipitated with ethanol, fractionated on a 10% polyacrylamide–7 M urea gel, and visualized by autoradiography.
RESULTS
Loop 1 of the U5 snRNA is dispensable for both steps of splicing in vitro.
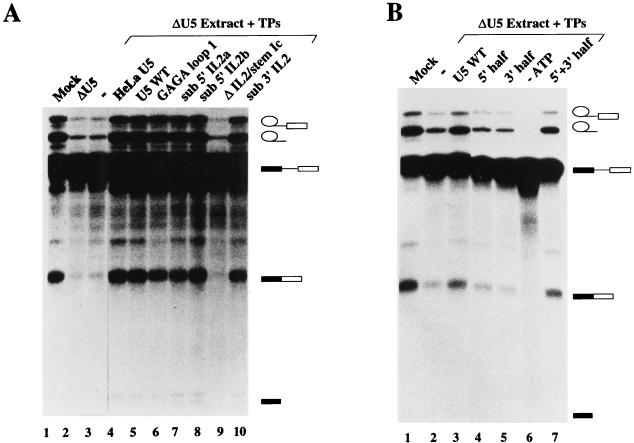
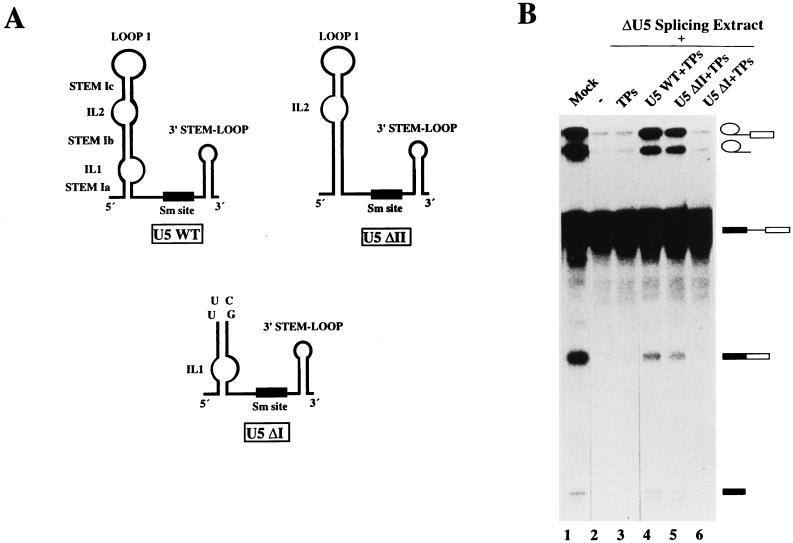
We previously reported the establishment of an in vitro reconstitution-splicing complementation system for HeLa U5 snRNPs (36). In this system, HeLa nuclear extracts are specifically depleted of U5 snRNPs by affinity selection with a biotinylated 2′-O-alkyl RNA oligonucleotide complementary to loop 1 of the U5 snRNA. U5 snRNPs are reconstituted by incubating purified U5 snRNA and native snRNP proteins (TPs) in the presence of splicing extract. TPs, which are essentially free of any snRNA, consist predominantly of the snRNP Sm proteins B, B′, D1, D2, D3, E, F, and G (36, 39), and the reconstitution of functional U5 snRNPs was previously shown to require their addition to the reconstitution mixture (36). The splicing activity of reconstituted U5 snRNPs is assayed directly in the reconstitution mixture by the addition of pre-mRNA. As shown in Fig. 2A, the splicing efficiency of an adenovirus major late II pre-mRNA (MINX) was significantly reduced in U5-depleted extract when compared to the mock-depleted extract (Fig. 2A, compare lanes 1 and 2). Consistent with previous results, splicing could be complemented by the addition of either authentic or in vitro-transcribed HeLa U5 snRNA plus native snRNP Sm proteins (TPs) (lanes 4 and 5). In contrast, the addition of RNA (not shown) or TPs alone (lane 3) had little or no effect on the splicing activity of U5-depleted extract.
FIG. 2.
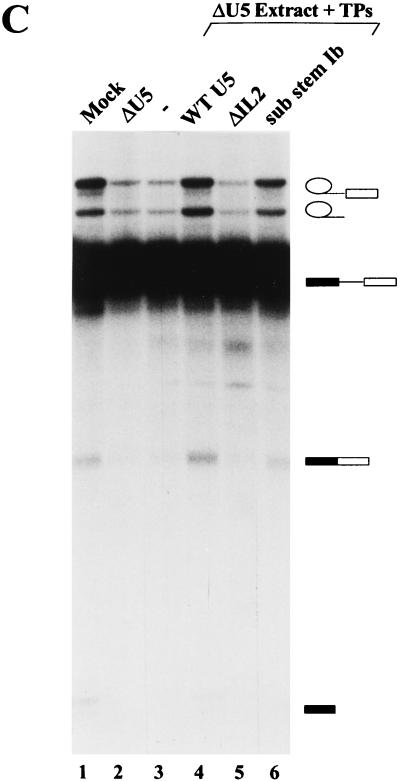
Conserved loop 1 of U5 snRNA, but not internal loop 2, is dispensable for both steps of splicing. Complementation of U5-depleted extracts with U5 snRNPs reconstituted from various human U5 snRNA mutants. U5 snRNP reconstitutions were performed in the presence of extract, and splicing was performed for 90 min with MINX pre-mRNA as described in Materials and Methods. (A) In vitro splicing reactions were performed with mock-depleted extract (lane 1), U5-depleted extract (lane 2), U5-depleted extract plus 3.3 pmol of native snRNP proteins (TPs) (lane 3), or U5-depleted extract plus TPs and 2.6 pmol of the U5 snRNA species indicated above each lane (lanes 4 to 10). HeLa U5 snRNA was isolated from purified U5 snRNP particles, whereas wild-type (WT) and mutant U5 snRNAs were transcribed in vitro. (B) In vitro splicing reactions were performed with mock-depleted extract (lane 1), U5-depleted extract plus 3.3 pmol of native snRNP proteins (TPs) (lane 2), or U5-depleted extract plus TPs and 2.6 pmol of the U5 snRNA species indicated above each lane (lanes 3 to 5 and 7). In lane 7, the 5′ and 3′ halves of U5 were annealed as described in Materials and Methods prior to reconstitution. In lane 6, splicing was performed in the absence of energy. (C) In vitro splicing reactions were performed with mock-depleted extract (lane 1), U5-depleted extract (lane 2), U5-depleted extract plus 3.3 pmol of TPs (lane 3) or U5-depleted extract plus TPs and 2.6 pmol of the U5 snRNA species indicated above each lane (lanes 4 to 6). Splicing intermediates and products as well as unspliced pre-mRNA (indicated schematically on the right) were fractionated on a 13% polyacrylamide–7 M urea gel and visualized by autoradiography.
The ability to complement splicing with in vitro-transcribed U5 snRNA allowed us to investigate the effect of U5 snRNA mutations on the activity of in vitro-reconstituted U5 snRNPs. To this end, we constructed a number of human U5 snRNA mutants with alterations primarily in either of two conserved regions, namely loop 1 or IL2. These mutants are depicted schematically in Fig. 1B, and a more precise description of deleted and/or substituted nucleotides is presented in Table 1. As a first step, we constructed a U5 snRNA mutant in which the invariant loop 1 sequence GCCUUUUAC was substituted with a GAGA tetraloop (designated GAGA loop 1). Loop 1 was replaced by a tetraloop rather than completely deleted in order to preserve the folding of stem Ic. The structure of this RNA was verified by nuclease susceptibility assays (data not shown). The activity of U5 snRNPs reconstituted with this mutant was then assayed in our in vitro splicing complementation system. Surprisingly, the GAGA tetraloop mutant restored both steps of splicing to U5-depleted extract to a level similar to that obtained with wild-type U5 snRNA (Fig. 2A, lanes 5 and 6). (Note that the slight reduction in spliced mRNA compared to that in the wild-type is due to experimental variability.) In addition, no differences in the migration behavior of the mRNA or excised exon 1 were observed when comparing the wild type to the GAGA tetraloop mutant, suggesting that substitution of loop 1 also had no effect on the accuracy of splicing. Complementation of both catalytic steps after substitution of loop 1 with a GAGA tetraloop was also observed with a second adenovirus pre-mRNA containing a different 5′ splice site, as well as with a β-globin pre-mRNA, demonstrating that the dispensability of conserved loop 1 nucleotides is not restricted to the MINX pre-mRNA substrate (data not shown).
TABLE 1.
Human U5 snRNA mutants examined in this study
| Designation | Mutation |
|---|---|
| Sub 5′ IL2a | nt 23 and 24 substituted with AU |
| Sub 5′ IL2b | nt 20–27 substituted with AUUUAAUA |
| Sub 3′ IL2 | nt 55–57 substituted with AAA |
| Sub IL2 | nt 20–27 substituted with AUUUAAUA and 55–57 substituted with AAA |
| ΔIL2 | nt 20–27 and 55–57 deleted |
| ΔIL2/stem 1c | nt 20–35 and 47–57 deleted |
| Sub-stem 1b | nt 9–19 substituted with CAGAGAGAAGU and 58–68 substituted with ACUUCUCUUUG |
| GAGA loop 1 | nt 36–46 substituted with GAGA |
| 5′ half | nt 36–116 deleted and 34 and 35 substituted with CC |
| 3′ half | nt 1–47 deleted and 47 and 48 substituted with GG |
| 5′+3′ half | 5′ and 3′ halves annealed |
Since the GAGA tetraloop could conceivably still interact with 5′ and/or 3′ splice site nucleotides, we next tested whether loop 1 was altogether dispensable for splicing. We thus transcribed separately the 5′ and 3′ halves of the human U5 snRNA, deleting all loop 1 sequences, and then annealed them. The 5′ stem-loop structure of a U5 snRNA formed in this manner is predicted to end with stem Ic (Fig. 1B). As shown in Fig. 2B, the addition of the 5′ or 3′ half of the U5 snRNA alone to the reconstitution-splicing complementation mixture had no effect on splicing efficiency (Fig. 2B, compare lane 2 with lanes 4 and 5). In contrast, particles reconstituted after annealing both halves of the U5 snRNA complemented both steps of splicing nearly as efficiently as wild-type U5 (Fig. 2B, compare lanes 3 and 7). These results conclusively demonstrate that U5 loop 1 is not essential for efficient pre-mRNA splicing in HeLa nuclear extracts.
IL2 is required for the formation of functional U5 snRNPs.
We next tested whether mutation of IL2, a second conserved region of the major stem-loop of U5 snRNA, or stem Ib, affected the splicing activity of reconstituted U5 particles. Compared to the wild type, substitution of nucleotides in either the 5′ half (sub 5′ IL2a or IL2b), 3′ half (sub 3′ IL2), or both bulged halves of IL2 (sub IL2) had no significant effect on the complementation efficiency of in vitro-reconstituted U5 snRNPs (Fig. 2A, lanes 7 and 8 and 10 [data not shown]). Thus, the precise sequence of IL2 does not appear to be relevant to U5 snRNP function. Similarly, substitution of stem Ib with a stem in which essentially the 5′ and 3′ halves of stem Ib were swapped (sub-stem Ib), resulted in only a slight reduction in the splicing activity of U5 snRNPs (Fig. 2C, lane 6). However, deletion of IL2 and stem Ic (ΔIL2/stemIc) abolished the ability of U5 snRNPs to complement both steps of splicing (Fig. 2A, lane 9). To distinguish whether this loss of activity was due either to deletion of IL2 or to stem Ic (which, in contrast to ΔIL2, would shorten the overall length of the major U5 5′ stem-loop), reconstitutions were performed with a U5 snRNA lacking solely IL2 (ΔIL2). Interestingly, the latter U5 snRNPs were unable to restore splicing activity to U5-depleted extracts (Fig. 2C, compare lane 5 with lanes 2 and 3). Because all of these U5 snRNA mutants exhibit similar stabilities during in vitro reconstitution and splicing, the observed losses in activity cannot be attributed to an increase in the turnover of the ΔIL2 or ΔIL2/stem Ic mutants. These results indicate that structural elements other than loop 1, namely IL2, are absolutely required for U5 snRNP function.
IL1 is dispensable for splicing in vitro.
To determine whether other regions of the major U5 stem-loop are essential for splicing activity, we extended our investigation to include a Xenopus U5 deletion mutant which lacked IL1 (designated ΔII). As a negative control, the activity of a mutant lacking both IL2 and stem Ic and possessing a UUCG tetraloop substitution of loop 1 (designated ΔI) was also assayed. These mutants are shown schematically in Fig. 3A. Since only minor changes in primary sequence are observed between human and Xenopus U5 snRNAs, the latter were expected to assemble into functional hybrid U5 snRNPs in our reconstitution system. Indeed, the majority of splicing could be restored to U5-depleted extracts by the addition of U5 snRNPs reconstituted from wild-type Xenopus U5 snRNA and HeLa snRNP proteins (Fig. 3B, compare lanes 1 through 4). Compared to the wild type, deletion of IL1 had no significant effect on the level of splicing complementation (Fig. 3B, lane 5), demonstrating that it is dispensable for U5 snRNP function. In contrast, consistent with the results described above, deletion of IL2 and stem Ic, as well as substitution of loop 1 with a UUCG tetraloop, abolished the in vitro splicing activity of reconstituted U5 snRNPs (Fig. 3B, lane 6).
FIG. 3.
Splicing active U5 snRNPs are formed in the absence of IL1. (A) Secondary structure models of wild-type and mutant Xenopus U5 snRNAs. The ΔII mutant was generated by deleting nt 7 and 8 and 70 to 75, which comprise the bulged halves of ILI. In the ΔI mutant, nt 19 to 59, which encompass IL2, stem Ic, and loop 1, were deleted and replaced by the tetraloop UUCU. (B) In vitro splicing reactions were performed with mock-depleted extract (lane 1), U5-depleted extract (lane 2), or U5-depleted extract plus the following: TPs alone (lane 3), wild-type Xenopus U5 snRNA plus TPs (lane 4), or the mutant Xenopus U5 snRNAs, ΔII and ΔI, plus TPs (lanes 5 and 6, respectively). In vitro reconstitution and in vitro splicing assays were performed as described in the legend to Fig. 2.
The splicing block observed upon deletion of IL2 occurs prior to or during splicing complex B formation.
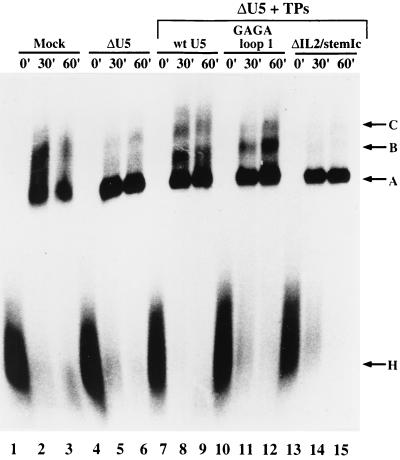
To determine whether U5 mutants lacking IL2 support the assembly of U4/U6.U5 snRNPs active in spliceosome assembly, splicing complex formation was analyzed by subjecting the in vitro splicing reaction mixtures to native gel electrophoresis. For comparison, splicing reactions performed with U5 snRNPs reconstituted from wild-type or U5 snRNA, whose loop 1 sequence had been substituted by a GAGA tetraloop, were also analyzed. Consistent with the known function of U5 during spliceosome assembly, the formation of splicing complexes B and C, but not A (which contains only the U1 and U2 snRNPs), was significantly reduced in U5-depleted extract (Fig. 4, compare lanes 1 to 3 with 4 to 6). The assembly of complexes B and C could, however, be restored by the addition of U5 snRNPs reconstituted from in vitro-transcribed wild-type or GAGA loop 1 U5 snRNA (lanes 7 to 12). In contrast, U5 snRNPs reconstituted from the ΔIL2/stem Ic mutant were unable to support complex B and C formation (lanes 13 to 15). Similar results were obtained with the ΔIL2 mutant (data not shown), suggesting that the deletion of IL2 inhibits either the assembly of the U4/U6.U5 tri-snRNP complex or its association with the prespliceosome (i.e., complex A).
FIG. 4.
Effect of U5 snRNA mutation on splicing complex formation. In vitro reconstitution and in vitro splicing assays were carried out with mock-depleted extract (lanes 1 to 3), U5-depleted extract (lanes 4 to 6), or U5-depleted extract plus TPs and the following U5 snRNAs: wild type (lanes 7 to 9), GAGA loop 1 (lanes 10 to 12), and ΔIL2/stem Ic (lanes 13 to 15). Splicing reactions were stopped by the addition of heparin after 0, 30, or 60 min, as indicated above each lane, and splicing complexes were fractionated by native gel electrophoresis as described in Materials and Methods and visualized by autoradiography. The positions of the H, A, B, and C complexes are indicated on the right. Note that the formation of B and C complexes is generally less efficient in the mock or depleted extracts than in an extract not subjected to the depletion procedure (not shown).
The U5 116-kDa protein associates with U5 snRNPs lacking loop 1, but not IL2.
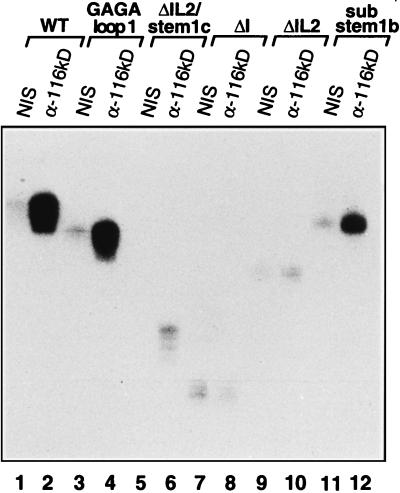
To determine whether alterations in the U5 snRNA affected the protein composition of the U5 snRNP, immunoprecipitation studies were performed, subsequent to reconstitution with radiolabeled U5 snRNA, with antibodies reacting specifically with the 116-kDa U5-specific protein. Recent studies have demonstrated that the 116-kDa protein forms a very tight complex with the U5 220-kDa protein (1). This dimer also interacts with two other U5-specific proteins, namely of 200 and 40 kDa (1). The presence of the 116-kDa protein in a particular U5 snRNP is thus a good indication for the presence of the U5 220-kDa protein, as well as these other U5 proteins. Wild-type, GAGA loop 1, ΔI, and ΔIL2/stem Ic U5 snRNAs were quantitatively precipitated by the anti-Sm monoclonal antibody Y12, demonstrating that each supports the association of the core or common snRNP proteins (data not shown). Only minimal background precipitation of each of these RNAs, as well as ΔIL2 and substem Ib, was observed when immunoprecipitations were performed with nonimmune serum (Fig. 5, lanes 1, 3, 5, 7, 9, and 11). In contrast, a significant amount (compared to nonimmune serum) of wild-type, GAGA loop 1, or sub-stem Ib U5 snRNA was precipitated by antibodies directed against the U5 116-kDa protein (Fig. 5, lanes 2, 4, and 12); however, in keeping with its slightly reduced splicing activity, precipitation of substem Ib was, by comparison, somewhat less efficient. Consistent with the fact that they were inactive in splicing, U5 mutants lacking IL2, either alone or in combination with other deletions, were not appreciably precipitated (Fig. 5, lanes 6, 8, and 10). Thus IL2, but not the conserved nucleotides of loop 1, is required for the stable association of the U5 116-kDa protein with the U5 snRNP. Because the U5 116-kDa protein is tightly associated with the U5 220-kDa protein, U5 snRNPs reconstituted from the GAGA loop 1 U5 snRNA most likely also contain the 220-kDa protein, which has been shown, like loop 1, to interact with both splice sites of the pre-mRNA. These results are therefore consistent with the idea that the U5 220-kDa protein and/or other U5 snRNP proteins might functionally substitute for loop 1 in its absence.
FIG. 5.
The U5 116-kDa protein stably interacts with U5 snRNPs harboring a GAGA loop 1 substitution, but not with those lacking IL2. The association of the U5 116-kDa protein was determined by immunoprecipitation with anti-U5 116-kDa protein antibodies (α-116 kD). Reconstitutions were performed with 32P-labelled wild-type (WT) (lanes 1 and 2), GAGA loop 1 (lanes 3 and 4), ΔIL2/stem Ic (lanes 5 and 6), ΔI (lanes 7 and 8), ΔIL2 (lanes 9 and 10), or sub-stem Ib U5 (lanes 11 and 12) snRNA in the presence of native snRNP proteins (TPs) and nuclear extract as described in Materials and Methods. Immunoprecipitations were performed with nonimmune serum (NIS) (lanes 1, 3, 5, 7, 9, and 11) or anti-116-kDa antiserum (lanes 2, 4, 6, 8, 10, and 12). Coimmunoprecipitated U5 snRNA was fractionated on a 10% polyacrylamide–7 M urea gel and visualized by autoradiography.
DISCUSSION
We have employed an in vitro reconstitution-splicing complementation system to investigate the effect of U5 snRNA mutations on both the structure and function of the metazoan U5 snRNP. Surprisingly, substitution or deletion of the invariant U5 loop 1 sequence had no effect on the ability of reconstituted U5 snRNPs to complement either step of splicing in HeLa nuclear extracts (Fig. 2). The ability of U5 snRNPs lacking loop 1 to efficiently support the first step of splicing in HeLa nuclear extracts is consistent with results recently obtained in the yeast S. cerevisiae. In this instance, mutant U5 snRNAs containing substituted or deleted loop 1 nucleotides were also shown to complement the first step of splicing in U5-inactivated yeast extracts (31). Thus, although loop 1 nucleotides have been shown to base pair with exon nucleotides at the 5′ splice site prior to step 1, contrary to previous models, this U5 snRNA–pre-mRNA base pairing interaction appears to be generally dispensable for the first step of splicing in both higher and lower eukaryotes.
However, in contrast to in vitro studies with yeast (31, 32) the presence of loop 1 was also not absolutely required for the second step of splicing in HeLa nuclear extracts (Fig. 2A and B). These results indicate that, in metazoans, loop 1 is not an essential component of the active sites responsible for either step of splicing in vitro. The basis for this fundamental difference between higher and lower eukaryotes is not clear. The fact that the yeast splicing machinery is generally considered to be less flexible than that of higher eukaryotes might explain the apparent difference in their requirement for U5 loop 1 during the second step of splicing. Based on our results, the function of loop 1 appears to be redundant in higher eukaryotes. That is although loop 1 may normally participate in the second step of splicing, other spliceosomal factors (e.g., the U5 220-kDa protein [see below]) apparently can compensate for it when it is absent. Alternatively, under normal circumstances base pairing interactions involving loop 1 nucleotides could simply play a secondary role in tethering exon 1 and aligning both splice sites for the second catalytic step of splicing. Nonetheless, the fact that U5 loop 1 nucleotides are absolutely, evolutionarily conserved, including their posttranscriptional modification (40), suggests that they contribute in some way to either the efficiency or accuracy of the splicing reaction. This function is, however, not readily apparent in our in vitro splicing system. Previous in vivo studies with HeLa cells suggested that loop 1 may contribute to 5′ splice site selection (9). The substrate used here had a single 5′ splice site, and based on the unchanged migration behavior of excised exon 1 in the presence of the GAGA tetraloop mutant (Fig. 2A) and the fact that a single nucleotide change in the length of exon 1 should be detectable under our gel electrophoresis conditions, we can exclude the possibility that the absence of loop 1 leads to aberrant 5′ splice site cleavage. Similarly, substitution or deletion of loop 1 of the yeast U5 snRNA also had no effect on the accuracy of 5′ splice site selection in yeast splicing extracts (31). However, it is conceivable that the fidelity of 3′ splice site cleavage may be altered in the absence of the invariant U5 loop 1 sequence, because small changes in the length of the mRNA or excised lariat would not be detectable under our experimental conditions. Loop 1 could also be absolutely required for some aspect of U5 snRNP or U4/U6.U5 tri-snRNP morphogenesis, such as the recycling of the tri-snRNP complex, which is thought to be dispensable in HeLa splicing extracts.
Our in vitro splicing complementation studies indicate that the functions previously attributed to loop 1 of the U5 snRNA, namely tethering of exon 1 subsequent to step 1 of splicing, as well as aligning the chemically reactive groups for the second step of splicing, can be compensated for by other spliceosomal components when loop 1 is absent. One likely candidate for this substitute is the U5 220-kDa protein. This highly conserved U5 snRNP protein (designated Prp8 in S. cerevisiae) has been shown to be in close proximity to both splice sites, as well as to the branch site and polypyrimidine tract (8, 25, 35, 41, 42, 45). Cross-linking studies further demonstrated that its interaction with the pre-mRNA substrate persists throughout the splicing reaction (41, 42, 45). The U5 220-kDa protein has also been implicated in 3′ splice site selection (42, 43). Based on these findings, the U5 220-kDa protein was proposed to assist the limited base pairing interactions between U5 loop 1 and the 5′ and 3′ splice sites (41). Consistent with the idea that U5 220-kDa can functionally compensate for the loss of loop 1, Prp8p has been shown to interact with the 5′ and 3′ splice sites even in the absence of U5 loop 1 (11).
The results of our immunoprecipitation studies are also consistent with the idea that the U5 220-kD protein could functionally replace loop 1. In particular, the GAGA tetraloop mutant was shown to stably associate with the U5 116-kDa protein (Fig. 5), which in turn has recently been shown to form a tight protein complex with the U5 220-kDa protein (1). These results suggest that U5 snRNPs reconstituted from the GAGA loop 1 U5 snRNA also contain the U5 220-kDa protein. Indeed, immunoprecipitation studies with anti-U5 220-kDa protein antibodies suggest that this protein probably does interact with U5 snRNPs containing a GAGA loop 1 substitution (data not shown), but due to the inefficiency of immunoprecipitation, as well as high levels of background precipitation, we have not been able to demonstrate conclusively that the U5 220-kDa protein is stably associated. Consistent with our observations, recent in vivo studies employing the transient transfection of mutant U5 genes into mammalian cells detected only a 60% reduction in U5 220-kDa protein binding upon replacement of U5 loop 1 with a UUCG tetraloop (16). Furthermore, the association of Prp8p with the yeast U5 snRNA in splicing extracts was observed even in the absence of loop 1 (11). These results support the idea that the presence of loop 1 is not necessarily a prerequisite for U5 220-kDa protein association with mammalian U5 snRNPs. Of course we cannot presently rule out whether U5 proteins besides or in addition to the U5 220-kDa protein, or even non-U5 snRNP spliceosomal components (including other RNAs), could also functionally substitute for loop 1.
In addition to loop 1, other regions of the metazoan U5 snRNA were shown to be dispensable for splicing in vitro. For example, consistent with previous in vivo studies of yeast (13), deletion of IL1 had little effect on the splicing activity of U5 snRNPs (Fig. 3). On the other hand, in vitro splicing in yeast was severely inhibited by deletion or substitution of the 5′ half of IL1 (11). However, this apparent difference could be attributed to differences in the IL1 mutants analyzed. Furthermore, despite its evolutionary conservation, substitutions in the sequence of either bulged half of IL2 also had no effect on U5 function, indicating it has no sequence-specific role (Fig. 2A). Similarly, substitution of stem Ib nucleotides had only a moderate effect on splicing (Fig. 2C). However, deletion of IL2 abolished U5 snRNP activity in splicing, demonstrating that this structural element is required for the formation of functional U5 snRNPs (Fig. 2C). Because the IL2 deletion, in contrast to the ΔIL2/stem Ic deletion, has little effect on the overall length of the major 5′ stem-loop of the U5 snRNA, its negative phenotype is probably not simply due to the shortening of this stem-loop structure. IL2 could play an important role in determining the tertiary structure of the U5 snRNA; indeed, it has been proposed to act as a hinge which would, for example, allow folding between stems Ic and Ib (3). Consistent with our result, in yeast, IL2 has been shown in vivo to provide an essential function for the yeast U5 snRNP and to be required for efficient splicing in vitro (11, 13).
The inability of U5 snRNA mutants lacking IL2 to support splicing suggests that these deleted nucleotides play either a direct or indirect role in the splicing process. U5 snRNA mutations could directly inhibit U5 snRNP function by altering or inhibiting the association of a U5-specific protein that is involved in either catalytic step of splicing. Alternatively, splicing could be indirectly affected if the protein in question were required for the proper assembly of the U5 snRNP or its subsequent interaction with U4/U6 to form the U4/U6.U5 tri-snRNP complex. Significantly, U5 snRNPs reconstituted from ΔIL2 or ΔIL2/stem Ic U5 snRNA did not allow the formation of splicing complex B (Fig. 4 and data not shown), consistent with the idea that the assembly of the U5 snRNP and/or U4/U6.U5 tri-snRNP complex was in some way compromised in the absence of IL2. Indeed, U5 snRNPs lacking IL2 did not support the stable association of the U5 116-kDa protein, as evidenced by immunoprecipitation studies (Fig. 5). Based on the recent demonstration that the U5 220-, 116-, 200-, and 40-kDa proteins form a highly stable heteromeric complex (1), these results suggest that ΔIL2 U5 snRNPs may also lack several proteins in addition to the U5 116-kDa protein. Indeed, deletion of IL2 was shown to abolish the interaction of the U5 220-kDa protein with U5 snRNPs in vivo (16). These results are also consistent with previous nuclease and chemical protection studies which suggested that one or more U5 proteins interact with IL2 (4, 6). Whether the U5 116-kDa protein directly interacts with IL2 is presently not clear. Because protein-protein interactions appear to predominate in the U5 snRNP, immunoprecipitation studies of this kind are rather limited in their potential for drawing conclusions about RNA-protein interactions. More detailed information regarding intermolecular interactions within the U5 snRNP is clearly needed to clarify this issue.
ACKNOWLEDGMENTS
We thank Michael Krause for preparing biotinylated 2′-O-alkyl oligonucleotides, and Peter Kempkes and Winfried Lorenz for expert technical assistance. We are grateful to Joan Steitz for kindly providing Y12 antibodies and Albrecht Bindereif for providing the ΔIL2 and sub-stem Ib U5 snRNA mutants.
This work was supported by the Deutsche Forschungsgemeinschaft SFB 397, the French Centre National de la Recherche Scientifique, and an EC Human Capital and Mobility Network grant (ERBCHRXCT 930191).
REFERENCES
- 1.Achsel T, Ahrens K, Brahms H, Teigelkamp S, Lührmann R. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of the ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol Cell Biol. 1998;18:6756–6766. doi: 10.1128/mcb.18.11.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson G J, Bach M, Lührmann R, Beggs J D. Conservation between yeast and man of a protein associated with U5 small nuclear ribonucleoprotein. Nature. 1989;342:819–821. doi: 10.1038/342819a0. [DOI] [PubMed] [Google Scholar]
- 3.Ast G, Weiner A M. Antisense oligonucleotide binding to U5 snRNP induces a conformational change that exposes the conserved loop of U5 snRNA. Nucleic Acids Res. 1997;25:3508–3513. doi: 10.1093/nar/25.17.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bach M, Lührmann R. Protein-RNA interactions in 20S U5 snRNPs. Biochim Biophys Acta. 1991;1088:139–143. doi: 10.1016/0167-4781(91)90164-h. [DOI] [PubMed] [Google Scholar]
- 5.Behrens S-E, Galisson F, Legrain P, Lührmann R. Evidence that the 60-kDa protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosomes. Proc Natl Acad Sci USA. 1993;90:8229–8233. doi: 10.1073/pnas.90.17.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black D L, Pinto A L. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol Cell Biol. 1989;9:3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branlant C, Krol A, Ebel J P, Lazar E, Haendler B, Jacob M. U2 RNA shares a structural domain with U1, U4 and U5 RNAs. EMBO J. 1982;1:1259–1265. doi: 10.1002/j.1460-2075.1982.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiara M D, Palandjian L, Kramer R F, Reed R. Evidence that U5 snRNP recognizes the 3′ splice site for the catalytic step II in mammals. EMBO J. 1997;16:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes J J, Sontheimer E J, Seiwert S D, Steitz J A. Mutations in the conserved loop of human U5 snRNA generate use of novel cryptic 5′ splice sites in vivo. EMBO J. 1993;12:5181–5189. doi: 10.1002/j.1460-2075.1993.tb06213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dix I, Russell C S, O’Keefe R T, Newman A J, Beggs J D. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA. 1998;4:1239–1250. doi: 10.1017/s1355838298981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabrizio P, Laggerbauer B, Lauber J, Lane W S, Lührmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank D N, Roiha H, Guthrie C. Architecture of the U5 small nuclear RNA. Mol Cell Biol. 1994;14:2180–2190. doi: 10.1128/mcb.14.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guthrie C, Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- 15.Hackl W, Fischer U, Lührmann R. A 69-kD protein that associates reversibly with the Sm core domain of several spliceosomal snRNP species. J Cell Biol. 1994;124:261–272. doi: 10.1083/jcb.124.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinz M, Moore M J, Bindereif A. Domain analysis of human U5 RNA. Cap trimethylation, protein binding and spliceosome assembly. J Biol Chem. 1996;271:19001–19007. doi: 10.1074/jbc.271.31.19001. [DOI] [PubMed] [Google Scholar]
- 17.Jackson S P, Lossky M, Beggs J D. Cloning of the RNA8 gene of Saccharomyces cerevisiae, detection of the RNA8 protein, and demonstration that it is essential for nuclear pre-mRNA splicing. Mol Cell Biol. 1988;8:1067–1075. doi: 10.1128/mcb.8.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarmolowski A, Mattaj I W. The determinants for Sm protein binding to Xenopus U1 and U5 snRNAs are complex and non-identical. EMBO J. 1993;12:223–232. doi: 10.1002/j.1460-2075.1993.tb05648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krämer A. The biochemistry of pre-mRNA splicing. In: Lamond A L, editor. Pre-mRNA processing. R. G. Austin, Tex: Landes Co.; 1995. pp. 35–64. [Google Scholar]
- 20.Krol A, Gallinaro H, Lazar E, Jacob M, Branlant C. The nuclear 5S RNAs from chicken, rat and man. U5 RNAs are encoded by multiple genes. Nucleic Acids Res. 1981;9:769–787. doi: 10.1093/nar/9.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laggerbauer B, Achsel T, Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci USA. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamond A I, Sproat B S. Isolation and characterization of ribonucleoprotein complexes. In: Rickwood D, Hames B D, editors. RNA processing. A practical approach. New York, N.Y: Oxford University Press; 1994. pp. 103–140. [Google Scholar]
- 23.Lauber J, Fabrizio P, Teigelkamp S, Lane W S, Hartmann E, Lührmann R. The HeLa 200 kDa U5 snRNP-specific protein and its homologue in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases. EMBO J. 1996;15:4001–4015. [PMC free article] [PubMed] [Google Scholar]
- 24.Lossky M, Anderson G J, Jackson S P, Beggs J D. Identification of a yeast snRNP protein and detection of snRNP-snRNP interactions. Cell. 1987;51:1019–1026. doi: 10.1016/0092-8674(87)90588-5. [DOI] [PubMed] [Google Scholar]
- 25.MacMillan A M, Query C C, Allerson C R, Chen S, Verdine G L, Sharp P A. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev. 1994;8:3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- 26.Madhani H D, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 27.Moore M J, Query C C, Sharp P A. Splicing of precursors to messenger RNAs by the spliceosome. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 28.Newman A, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- 29.Newman A J, Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:1–20. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 30.Newman A J, Teigelkamp S, Beggs J. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- 31.O’Keefe R T, Norman C, Newman A J. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- 32.O’Keefe R T, Newman A J. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J. 1998;17:565–574. doi: 10.1093/emboj/17.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raghunathan P, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and DEIH-box splicing factor Brr2. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 34.Reed R, Palandjian L. Spliceosome assembly. In: Krainer A, editor. Eukaryotic mRNA processing. New York, N.Y: Oxford University Press; 1997. pp. 103–129. [Google Scholar]
- 35.Reyes J L, Kois P, Konforti B B, Konarska M M. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA. 1996;2:213–225. [PMC free article] [PubMed] [Google Scholar]
- 36.Ségault V, Will C L, Sproat B S, Lührmann R. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 1995;14:4010–4021. doi: 10.1002/j.1460-2075.1995.tb00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sontheimer E J, Steitz J A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 38.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 39.Sumpter V, Kahrs A, Fischer U, Kornstädt U, Lührmann R. In vitro reconstitution of U1 and U2 snRNPs from isolated proteins and snRNA. Mol Biol Rep. 1992;16:229–240. doi: 10.1007/BF00419662. [DOI] [PubMed] [Google Scholar]
- 40.Szkukalek A, Myslinski E, Mougin A, Lührmann R, Branlant C. Phylogenetic conservation of modified nucleotides in the terminal loop 1 of the spliceosomal U5 snRNA. Biochimie. 1995;77:16–21. doi: 10.1016/0300-9084(96)88099-0. [DOI] [PubMed] [Google Scholar]
- 41.Teigelkamp S, Newman A J, Beggs J D. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umen J G, Guthrie C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev. 1995;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- 43.Umen J G, Guthrie C. Mutagenesis of the yeast gene, PRP8, reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Will C L, Lührmann R. snRNP structure and function. In: Krainer A R, editor. Eukaryotic mRNA processing. New York, N.Y: Oxford University Press; 1997. pp. 130–173. [Google Scholar]
- 45.Wyatt J R, Sontheimer E J, Steitz J A. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]