ABSTRACT
Although the strategies used by bacteria to adapt to specific environmental conditions are widely reported, fewer studies have addressed how microbes with a cosmopolitan distribution can survive in diverse ecosystems. Exiguobacterium is a versatile genus whose members are commonly found in various habitats. To better understand the mechanisms underlying the universality of Exiguobacterium, we collected 105 strains from diverse environments and performed large-scale metabolic and adaptive ability tests. We found that most Exiguobacterium members have the capacity to survive under wide ranges of temperature, salinity, and pH. According to phylogenetic and average nucleotide identity analyses, we identified 27 putative species and classified two genetic groups: groups I and II. Comparative genomic analysis revealed that the Exiguobacterium members utilize a variety of complex polysaccharides and proteins to support survival in diverse environments and also employ a number of chaperonins and transporters for this purpose. We observed that the group I species can be found in more diverse terrestrial environments and have a larger genome size than the group II species. Our analyses revealed that the expansion of transporter families drove genomic expansion in group I strains, and we identified 25 transporter families, many of which are involved in the transport of important substrates and resistance to environmental stresses and are enriched in group I strains. This study provides important insights into both the overall general genetic basis for the cosmopolitan distribution of a bacterial genus and the evolutionary and adaptive strategies of Exiguobacterium.
IMPORTANCE The wide distribution characteristics of Exiguobacterium make it a valuable model for studying the adaptive strategies of bacteria that can survive in multiple habitats. In this study, we reveal that members of the Exiguobacterium genus have a cosmopolitan distribution and share an extensive adaptability that enables them to survive in various environments. The capacities shared by Exiguobacterium members, such as their diverse means of polysaccharide utilization and environmental-stress resistance, provide an important basis for their cosmopolitan distribution. Furthermore, the selective expansion of transporter families has been a main driving force for genomic evolution in Exiguobacterium. Our findings improve our understanding of the adaptive and evolutionary mechanisms of cosmopolitan bacteria and the vital genomic traits that can facilitate niche adaptation.
KEYWORDS: Exiguobacterium, cosmopolitan distribution, genomics, adaptation strategies, polysaccharide utilization, transporters
INTRODUCTION
Across the landscape, microbial communities are nonrandomly dispersed in terms of their composition and diversity (1). Physical and chemical factors in the environment significantly influence the distribution patterns of microbes (2). For example, marine and nonmarine habitats are separated by strong physiochemical differences in salinity, temperature, pH, dissolved oxygen, and water chemistry (3). As a result, most marine microbes belong to phylogenetic groups different from those of their freshwater and terrestrial relatives, and transitions between these two niches are rare (4, 5). It has frequently been reported that marine and freshwater/terrestrial bacterial members often utilize different strategies for niche adaptations (6). Comparative genomic analyses of ocean microbes have revealed that the genomes of many marine bacteria have been streamlined to reduce the metabolic costs of maintaining nonessential genetic material, which favors their adaptation to nutrient-poor ocean environments (7, 8). In contrast, free-living terrestrial bacteria usually have a normal genome size and exhibit frequent horizontal genetic-transfer events, which is believed to facilitate their capacity to use diverse nutrients and resist the stresses caused by complicated adverse environments (9, 10). The transition of a marine bacterium to a nonmarine bacterium or vice versa requires complex genomic evolution (11, 12). The gain and loss of genes involved in the transport, metabolism, and assimilation of different types of organic or inorganic nutrients are believed to play crucial roles during this process (11, 13). However, this knowledge is derived mainly from comparative genomic analyses of a few bacteria that are highly abundant in either marine or nonmarine microbiotas (14, 15). The strategies of evolution and adaptation for microbes that are widely distributed in both marine and nonmarine environments have not been well studied to date.
Bacteria of the genus Exiguobacterium are Gram-positive facultative anaerobes that have been frequently isolated from various habitats, including seawater, marine sediment, marine algae (16, 17), soil (18), freshwater (19), plant rhizosphere (20), and even extreme environments, such as a salt lake (21), glaciers, and hot springs (22). Previous studies also revealed that members of Exiguobacterium have transitioned between the terrestrial and marine niches (23, 24). Genomic analyses of these bacteria have provided some vital insights into their psychrophilic and thermophilic adaptations and resistances to multiple toxic compounds (25–28). However, we lack a comprehensive understanding of how these bacteria underwent evolutionary adaptation to marine and nonmarine habitats.
In this study, Exiguobacterium was used as a model to study the evolution and adaptive strategies of bacteria with a cosmopolitan distribution across diverse habitats. We first mined public databases for 16S rRNA gene sequences to reveal the diversity and distribution of members of this genus. We then isolated multiple strains from marine and terrestrial habitats and tested their adaptive and metabolic features. Furthermore, we sequenced the full genomes of 105 strains and performed large-scale phylogenomic and comparative genomic analyses using genomes of a total of 147 strains isolated from marine and nonmarine habitats worldwide. Special attention was paid to the genomic and metabolic characters that allow these organisms to respond to diverse environments.
RESULTS
Exiguobacterium members have a cosmopolitan distribution and share extensive abilities to adapt to survive in various environments.
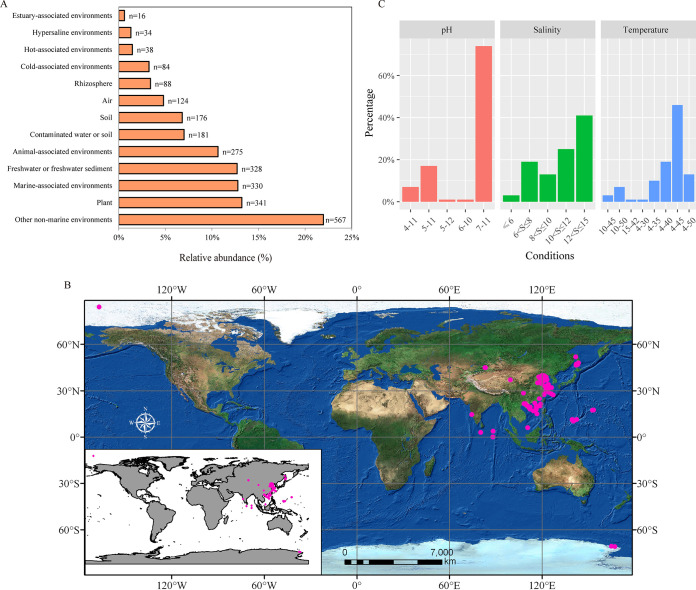
To explore the diversity and distribution of members belonging to the genus Exiguobacterium, we retrieved 16S rRNA gene sequences with >95% identities to those of reported type strains belonging to this genus from GenBank. A total of 2,582 Exiguobacterium 16S rRNA gene sequences with unambiguous information about isolation sources were collected (see Table S1 in the supplemental material). We found that members of this genus were frequently isolated from terrestrial environments (86.6%), including plants or rhizosphere (16.6%), animal skin or gut (10.7%), freshwater or freshwater sediments (12.7%), contaminated water or soil (7%), soil (6.8%), extreme environments (hot, cold, or hypersaline environments) (6%), air (4.8%), and other nonmarine environments (22%) (Fig. 1A; Table S1). The remaining members (13.4%) were isolated from marine-associated environments, including seawater, algae, and oceanic sediment. When we combined the location information, we found that Exiguobacterium can be found in ecosystems of all continents and oceans (Fig. S1A). This agrees with the current notion that Exiguobacterium is a cosmopolitan bacterial genus that includes many extremophiles capable of surviving in both marine and nonmarine environments worldwide (16).
FIG 1.
Cosmopolitan distribution of Exiguobacterium strains. (A) Relative abundances of various 16S rRNA gene sequences among 13 types of habitats. (B) Isolation sites (pink dots) of 97 Exiguobacterium strains examined in this study. The map was created with ArcGIS 10.6 software. (C) Temperature (°C), pH, and salinity (%) tolerance test results for 105 Exiguobacterium strains. The x axis represents the tolerance range of strains, and “S” represents salinity.
Geographic distribution of 16S rRNA gene sequences and pan-genome analysis. (A) Geographic distribution of 16S rRNA gene sequences with information about isolation sites (the pink dots represent the sampling locations, and the dots are colored by number of 16S rRNA gene sequences). The map was created using Tableau software (version 10.5) with map data from OpenStreetMap contributors. (B) Pan- and core genomes of the Exiguobacterium genus, i.e., the sizes of the pan- and core genomes in relation to numbers of genomes added into the gene pool. Box plots show the 25th and 75th percentiles, with medians shown as horizontal lines, and whiskers indicate the lowest and highest values within 1.5 times the interquartile range (IQR) from the first and third quartiles, respectively. The curve for the pan-genome is fitted by the power-law regression model, and the curve for the core genome is fitted by the exponential curve fit model. Download FIG S1, PDF file, 1.9 MB (1.9MB, pdf) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Information on the Exiguobacterium 16S rRNA gene sequences available in GenBank. Download Table S1, XLSX file, 0.10 MB (98.7KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To study the adaptation and evolution of Exiguobacterium, we performed large-scale phenotype tests and comparative genomics analysis. A total of 105 Exiguobacterium strains were collected here, including 86 isolates from marine niches (e.g., marine sediment, seawater, algae, marine cold springs, hydrothermal vents, seamounts, mangroves, marine fish, and coral), 11 strains from terrestrial environments (e.g., soil, salt lakes, coal mines, and pig farms), and eight type strains for which genomic data were previously unavailable (Fig. 1B; Table S2). We focused on strains isolated from marine habitats because most members of the Exiguobacterium genus with genomic data available in GenBank were from terrestrial environments.
Genome features and isolation sources of 147 Exiguobacterium strains. Download Table S2, XLSX file, 0.03 MB (27.8KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Previous studies showed that Exiguobacterium spp. can survive in a wide range of habitats, including cold, hot, hypersaline, and alkaline environments (17). However, it was unclear if such adaptability is shared by all members or if such capacities are strain/species specific. We thus performed large-scale phenotype tests and evaluated the growth potential of the 105 collected strains under different levels of pH, temperature, and salinity. We found that most Exiguobacterium members could survive and grow in a wide range of temperatures, salinities, and pH values (Fig. 1C; Table S3). The pH tests revealed that all strains were alkali resistant and able to survive in environments with pH values up to 11, and 24% of strains showed growth at pHs 5 and even 4. In the salinity test, most of the strains showed tolerance to salt, with 66% of strains showing growth in environments with NaCl concentrations above 10%. Our temperature tests showed that Exiguobacterium spp. can tolerate both high and low temperatures, with 91% of strains growing at 4°C and 61% surviving at high temperatures ranging from 40 to 50°C. We did not find any association between the growth abilities and source environments of analyzed strains. These results suggest that an extensive adaptability to survive in various environments is a general feature of this genus.
Temperature, pH, and salinity tolerance data for 105 Exiguobacterium strains. Download Table S3, XLSX file, 0.01 MB (12.5KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic analysis identified two genetic groups in the Exiguobacterium genus.
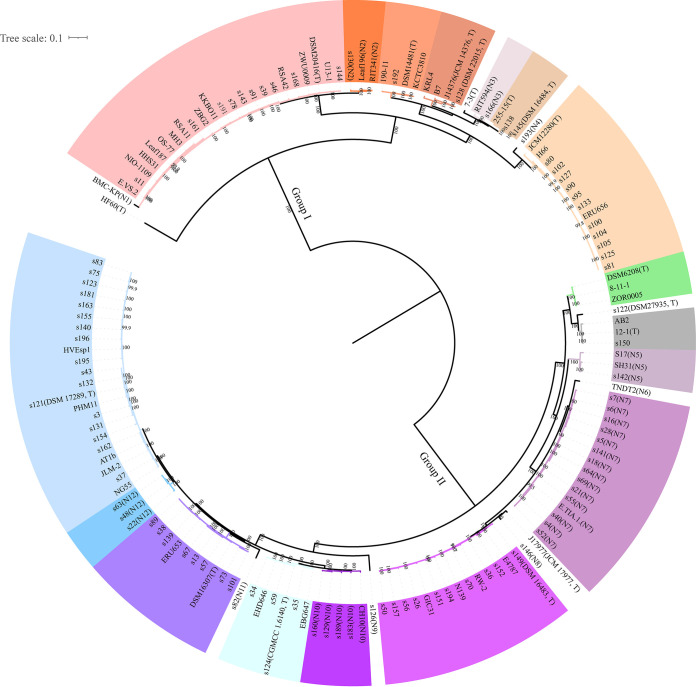
We conducted comparative genomic analyses to investigate the strategies that these microbes have used in their evolution and adaptation to diverse environments. All genomes of the 105 collected Exiguobacterium strains were sequenced and assembled. Forty-two Exiguobacterium genomes available in GenBank with more than 95% genome completeness and less than 5% contamination were also included (Table S2). All predicted protein-coding genes from the 147 genomes were clustered into 8,857 groups; the 1,316 groups that were shared across all of the studied genomes were classified as core gene families. A maximum-likelihood (ML) phylogenetic tree was constructed based on the concatenated single-copy core gene alignment (Fig. 2). Two genetic groups of Exiguobacterium spp. were classified and were well supported by bootstrapping analysis; this grouping was consistent with that from a previous analysis based on 16S rRNA gene sequences of type strains (16). Using an average nucleotide identity (ANI) of 95% as the threshold to define different species (29), we classified the analyzed genomes into a total of 27 species, including 12 putative new species (N1 to N12) (Fig. 2; Table S4). Of them, 11 and 16 species belonged to groups I and II, respectively (Table S4).
FIG 2.
Phylogenetic analysis of Exiguobacterium. The tree was built using IQ-TREE based on the concatenated amino acid sequence alignments of single-copy core genes. Bootstrap support values were calculated from 1,000 replicates. “T” represents the type strain for the following: NIO-1109 for E. enclense, HHS31 for E. indicum, DSM20416 for E. acetylicum, DSM14481 for E. undae, J14376 for E. soli, s128 for E. soli, 7-3 for E. sibiricum, 255-15 for E. sibiricum, s145 for E. artemiae, JCM12280 for E. oxidotolerans, DSM6208 for E. aurantiacum, s122 for “E. himgiriensis,” 12-1 for E. alkaliphilum, J17977 for E. aquaticum, s149 for E. mexicanum, s124 for E. aestuarii, DSM16307 for E. marinum, and s121 for E. profundum. N1 to N12 represent putative new species. Different colors represent different putative species, which were differentiated using the threshold ANI of 95%.
Average nucleotide identity (ANI) analysis of 147 Exiguobacterium strains. Download Table S4, XLSX file, 0.1 MB (117.3KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We next annotated the tree by adding the isolation environment for each strain and found that strains from the same species could be found in different environments (Table S5). For example, strains of E. acetylicum were found in seawater, ocean sediment, soil, rhizosphere, a glacier, and even animal gut. This suggested that a given species of Exiguobacterium may colonize different niches of terrestrial and marine environments. This finding contrasts with the reported behavior of some typical marine bacteria, different lineages of which were found to adapt to marine versus nonmarine habits (30).
Niche distribution of 147 Exiguobacterium strains based on phylogenetic analysis. Download Table S5, XLSX file, 0.01 MB (14.7KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Carbon and nitrogen source utilization for wide adaptation.
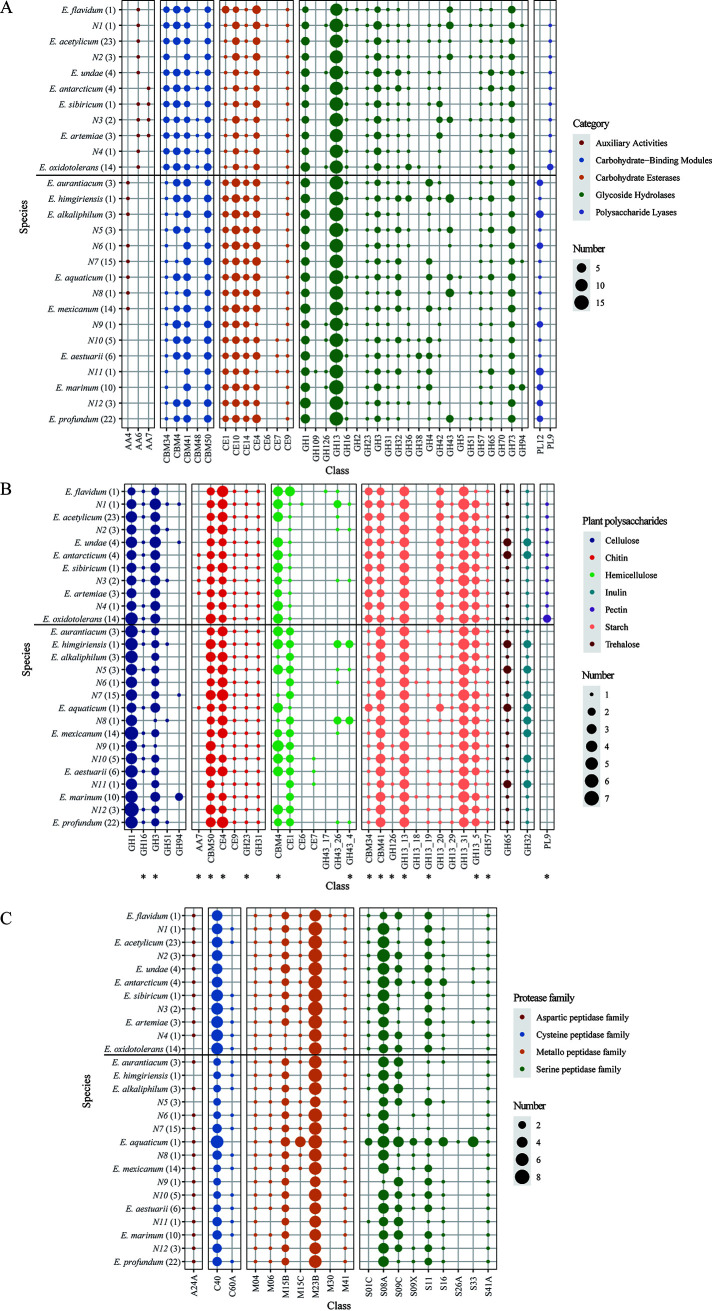
To explore a genetic basis for the ability of Exiguobacterium strains to survive in different niche types, we first focused on genes involved in nutrient metabolism. Carbohydrate-active enzymes (CAZymes) are involved in carbohydrate metabolism. A total of 7,976 genes belonging to five CAZyme superfamilies were identified from all of the studied genomes, with 61.5%, 18.7%, 15.9%, 2.7%, and 1.2% of these genes corresponding to glycoside hydrolase (GH), carbohydrate esterase (CE), the carbohydrate-binding module (CBM), polysaccharide lyase (PL), and auxiliary activities (AA), respectively (Fig. 3A; Table S6). Each of the studied genomic sequences encoded 43 to 68 of these enzymes.
FIG 3.
Carbon and nitrogen source utilization. (A to C) Numbers of carbohydrate-active enzymes (CAZymes) (A), plant polysaccharide degradation enzymes (B), and extracellular peptidases (C) encoded in Exiguobacterium genomes. Parentheses enclose the total number of genomes in each Exiguobacterium species. Asterisks indicate CAZymes with a potential secretion signal. The thick black thick line separates the species list into two parts; the species above the line belong to group I, while those below the line belong to group II.
Gene number of carbohydrate-active enzymes (CAZymes) and extracellular peptidases detected in each Exiguobacterium genome. Download Table S6, XLSX file, 0.1 MB (134.8KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Seven classes of enzyme for complex polysaccharide degradation were predicted from Exiguobacterium genomes (Fig. 3B; Table S6). The top three most numerous classes were those associated with the degradation of starch, cellulose, and chitin, and most of these CAZymes were found to have the potential to be secreted (Fig. 3B). Family GH13 represented the main amylolytic enzyme family; among the members of this family, GH13_31 (α-glucosidase) and GH13_13 (pullulanase) were the top two most numerous subfamilies (3.3 and 3 genes per genome, respectively). The most numerous family involved in cellulose degradation was GH1 (β-glucosidases, with more than 4 genes per genome). For chitin degradation, family CE4 (deacetylase) and CBM50 (chitin binding) showed significantly higher abundances.
Proteinaceous compounds are abundant forms of organic nitrogen that are found in aquatic and soil environments (31). Extracellular microbial peptidases are very important in both marine and terrestrial environments, as they play a crucial role in degrading organic nitrogen and thereby contribute to global nitrogen cycling (31). In this work, 3,912 putatively secreted peptidases were identified and assigned to 20 families; of them, 43.7%, 38.1%, 15%, and 3.2% belonged to the metallo, serine, cysteine, and aspartic peptidase families, respectively (Fig. 3C; Table S6). When normalized to the genome size, the average number of secreted peptidase-coding genes was nine genes per Mb, which is higher than the number (5.84) found in bacteria overall (31). Among these secreted peptidases, the metallo peptidase M23 and serine peptidase S08 represented the top two most abundant peptidases (Fig. 3C).
To validate the potential ability of Exiguobacterium spp. to degrade and metabolize complex carbohydrates and proteins, we tested the amylase and protease activities of the 105 studied strains on plates (Table S7). All of the strains effectively hydrolyzed starch, and approximately 70% could degrade proteins. Together, the results from our genomic analysis and activity testing provide strong evidence that most members of the Exiguobacterium genus can metabolize and utilize a wide range of polysaccharides and proteins from marine and nonmarine environments. This likely explains the genetic basis for the cosmopolitan distribution of these bacteria.
Validation of the abilities of Exiguobacterium spp. to degrade and metabolize complex carbohydrates and peptides. Download Table S7, XLSX file, 0.01 MB (13.9KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genetic basis of maintaining homeostasis in extreme environments.
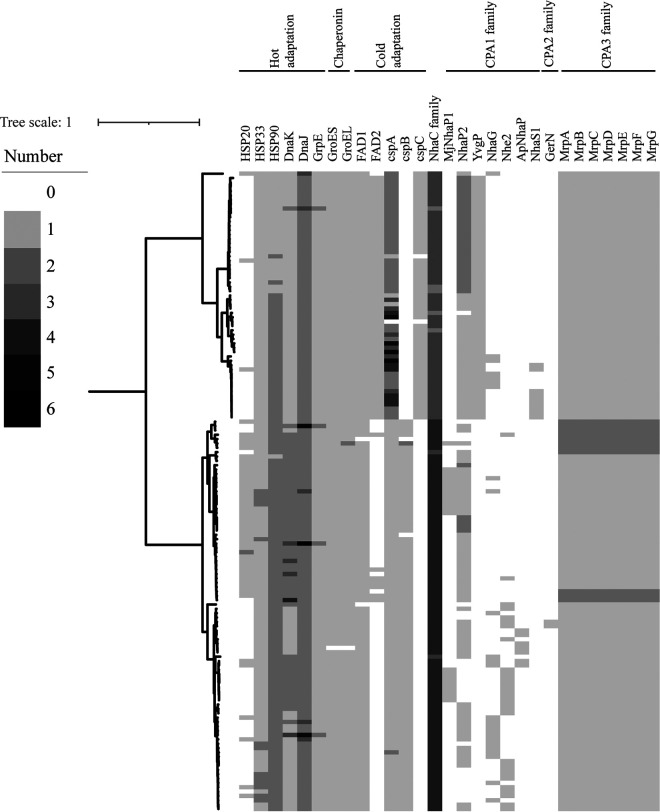
Bacteria use two main strategies to survive in cold environments; they use branched-chain fatty acids to maintain membrane fluidity and express cold shock proteins (Csps) that stabilize the bacterial cytosol at low temperatures (32, 33). Our genomic analysis suggested that all members of the Exiguobacterium genus could use both strategies to cope with low temperature. Two types of fatty acid desaturase (FAD) involved in unsaturated branched-chain fatty acid production were identified in Exiguobacterium genomes (Fig. 4; Table S8). All genomes except those of AB2 and s126 encoded at least one FAD1 protein, while genes for FAD2 were found mainly in strains belonging to group I. FADs can synthesize unsaturated fatty acids to maintain membrane fluidity at low temperatures, and potential cold-induced changes in the metabolic pathway of fatty acids were previously identified in E. antarcticum B7 (32–34). Three types of csp (cspA, cspB, and cspC) were predicted from all of the studied genomes (Fig. 4). Most genomes of group I members contained more than two cspA genes, while those from group II had only one, and cspB and cspC were specifically found in members of groups I and II, respectively. Csps are important for stabilizing the bacterial cytosol in cold environments and are induced upon a temperature downshift (35). It is worth noting that the psychrotrophic (able to grow in cold environments) strains were clustered mainly in group I; fad2 and cspA, enriched in group I, may contribute to the difference.
FIG 4.
Genes detected across Exiguobacterium genomes are vital for maintaining homeostasis in extreme environments. The heatmap represents the vital gene numbers and their distribution across the 147 genomes. The maximum-likelihood tree was constructed by IQ-TREE as described in Materials and Methods.
Genetic basis for adaptation of Exiguobacterium strains to diverse habitats. Download Table S8, XLSX file, 0.03 MB (27.4KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To survive in hot environments, Exiguobacterium spp. contain the heat shock gene cluster grpE-dnaJ-dnaK (Fig. 4; Table S8). DnaK plays a major role in maintaining protein homeostasis under thermal-stress conditions, and DnaJ and GrpE are cochaperones for DnaK (36). GroEL/GroES can reportedly cooperate with DnaK/DnaJ to prevent protein misfolding in bacteria (37). Thus, it is relevant that genes encoding GroEL and GroES were also discovered in all of the studied strains, with the exception of EHD646 (Fig. 4; Table S8). Both DnaK/DnaJ/GrpE and GroEL/GroES were reported to prevent aggregation and denaturation of proteins at high temperature in bacteria (38). Three types of heat shock proteins (HSPs; HSP20, HSP33, and HSP90) were also predicted. These HSPs can prevent irreversible protein denaturation and are important chaperones that help mediate appropriate responses to heat or oxidative stress (39–42). Together, the previous and present findings suggest that members of this genus utilize multiple strategies to cope with hot environments.
In bacteria, the Na+:H+ antiporters play crucial roles in maintaining intracellular pH homeostasis and the dynamic balance of cellular Na+. According to the Transporter Classification Database (TCDB), Na+:H+ antiporters contain mainly members of the large monovalent cation/proton antiporter (CPA) family, such as CPA1, CPA2, and CPA3, and the NhaC Na+:H+ antiporter family (43, 44). Seven types of CPA1 and one CPA2 were predicted from the genomes of Exiguobacterium spp. (Fig. 4; Table S8); the former were more frequently found in group I members, and the latter were more common in group II members. Compared to CPA1 and CPA2 antiporters, CPA3 antiporters are more structurally complex, as they have a multicomponent structure consisting of either seven or six members (45). The multicomponent Na+:H+ antiporter (Mrp) of CPA3 has been shown to provide Na+/H+ antiporter activity and function in the multiple compound resistance and pH homeostasis processes of Bacillus subtilis (45). In the present study, Mrp antiporters were identified in all Exiguobacterium genomes (Fig. 4; Table S8). In addition, an antiporter from the NhaC Na+:H+ antiporter (NhaC) family was identified in all Exiguobacterium genomes with copy numbers up to six (Fig. 4; Table S8). The presence of multiple types of Na+:H+ antiporter likely provides the basis for Exiguobacterium strains to maintain their osmotic and pH balances under various environments.
Together, the existence of diverse important proteins, including cold shock proteins, heat shock proteins, chaperonins, fatty acid desaturase, and diverse Na+:H+ antiporters, may explain the broad range of temperatures, pH, and salinity in which Exiguobacterium strains can survive to colonize diverse habitats.
Transporter expansion has driven the genomic expansion of Exiguobacterium spp.
We found that strains from marine environments were more frequently assigned to group II, while species from group I had more diverse terrestrial-niche distributions. Our analysis of 16S rRNA gene sequences from GenBank showed that of the 346 members from marine environments, 54 and 292 belonged to groups I and II, respectively (Table S1). Of the 86 marine strains isolated in this study, 24 and 62 belonged to groups I and II, respectively. Of the 11 species classified to group I, 10 contained strains that were isolated from both marine and terrestrial (e.g., soil, plant rhizosphere, fresh water, etc.) environments (Table S5). In group II, in contrast, most of the species were isolated solely from marine environments (Table S5).
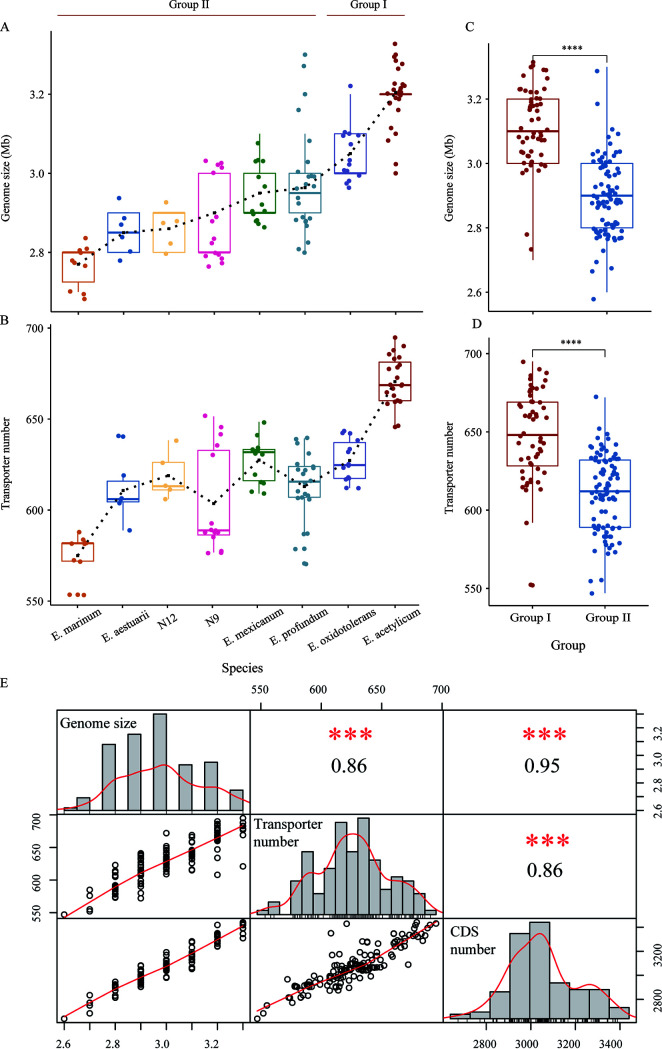
To understand the genetic background underlying the ecological differences of the two groups, we performed comparative genomic analysis. We found that the genome size and transporter number tended to increase for species of group I relative to those for group II (Fig. 5A and B and Table S9). The average genome size and transporter number of group I (3.12 Mb and 648, respectively) were significantly larger than those of group II (2.90 Mb and 610, respectively) (P < 0.0001 by the Wilcoxon test) (Fig. 5C and D). A pan-genome analysis suggested that there is a high level of genomic plasticity in this genus (Fig. S1B). The presence of an open pan-genome in Exiguobacterium spp. suggests that these species can change their genomes to facilitate their adaption to different habitats.
FIG 5.
Comparison and Spearman’s correlation analysis of genome sizes and transporter numbers. (A) Trends in the genome sizes of Exiguobacterium species (species with at least 5 strains were selected to show the trends). (B) Trends in the transporter numbers of Exiguobacterium species (species with at least 5 strains were selected to show the trends). (C) Comparison of genome sizes between group I and group II. (Black ****, significantly different at a P of <0.0001, as assessed by Wilcoxon tests). (D) Comparison of transporter numbers between group I and group II. (E) Spearman’s correlation analysis of genome sizes, CDS numbers, and transporter numbers. The genome size, CDS number, and transporter number are shown in the central diagonal, and the scatterplots are depicted with a fitted red line. On the corresponding side for each pairing is the Spearman rank correlation coefficient, rs. (Red ***, P < 0.01).
Transporter analysis of 147 Exiguobacterium genomes. Download Table S9, XLSX file, 0.2 MB (193.8KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
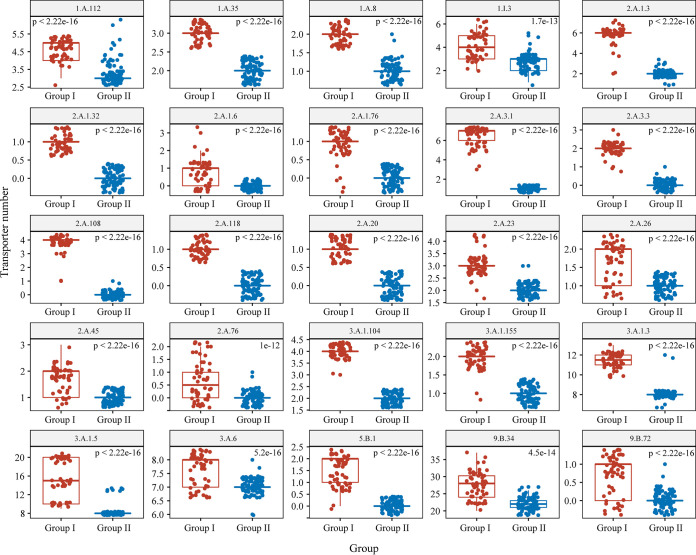
Spearman’s coefficient obtained for the correlation of transporter number with genome size was 0.86 (Fig. 5E). A significant correlation (P < 0.01) was found between genome size and the total number of transporter genes, suggesting that the expansion of transporters may have contributed to the difference in genomic contents between group I and group II strains. Among the 247 identified transporter families, 25 were predicted to have high degrees of correlation between their numbers for both genome size and protein coding sequence (CDS) (Spearman’s correlation coefficient > 0.6, P < 0.01) (Table 1). These 25 transporter gene families were significantly enriched in the genomes of group I members compared to group II members (Fig. 6; Table S9). Among these families, seven are associated with the transport of diverse amino acids, such as cationic, polar, branched-chain, and basic amino acids (Table 1), which are important components for nitrogen metabolism, protein synthesis, cell growth, and energy production or conversion (46). In addition, 2 of the 25 families are involved in the transport of Mg2+, namely, the cyclin M Mg2+ exporter family and the CorA metal ion transporter family. Mg2+ homeostasis is important in bacteria and has been reported to play a critical role in their thermotolerance (47, 48). The inorganic phosphate transporter family was also found to be expanded in group I relative to group II (Table 1). Phosphorus (P) is commonly associated with oxygen in the form of inorganic phosphates (PO4− and Pi) in the environment, and it is a crucial element involved in many energy production and metabolic pathways in bacteria (49).
TABLE 1.
Correlation analysis of transporter family with both genome size and CDS numbera
| Genome content | TC no. | Family | Correlation coefficient | P value |
|---|---|---|---|---|
| Genome size | 2.A.1 | Major facilitator superfamily (MFS) | 0.73 | 0 |
| CDS no. | 2.A.1 | 0.66 | 0 | |
| Genome size | 2.A.1.3 | Drug:H+ antiporter-2 (14-spanner) (DHA2) family | 0.73 | 0 |
| CDS no. | 2.A.1.3 | 0.66 | 0 | |
| Genome size | 2.A.1.76 | Uncharacterized major facilitator 24 family | 0.69 | 0 |
| CDS no. | 2.A.1.76 | 0.61 | 4.44E−16 | |
| Genome size | 2.A.1.32 | Putative aromatic compound/drug exporter (ACDE) family | 0.68 | 0 |
| CDS no. | 2.A.1.32 | 0.60 | 8.88E−16 | |
| Genome size | 2.A.1.6 | Metabolite:H+ symporter (MHS) family | 0.66 | 0 |
| CDS no. | 2.A.1.6 | 0.67 | 0 | |
| Genome size | 2.A.3 | Amino acid-polyamine-organocation (APC) superfamily | 0.67 | 0 |
| CDS no. | 2.A.3 | 0.61 | 2.22E−16 | |
| Genome size | 2.A.3.1 | Amino acid transporter (AAT) family | 0.71 | 0 |
| CDS no. | 2.A.3.1 | 0.66 | 0 | |
| Genome size | 2.A.3.3 | Cationic amino acid transporter (CAT) family | 0.68 | 0 |
| CDS no. | 2.A.3.3 | 0.60 | 8.88E−16 | |
| Genome size | 3.A.1 | ATP-binding cassette (ABC) superfamily | 0.75 | 0 |
| CDS no. | 3.A.1 | 0.69 | 0 | |
| Genome size | 3.A.1.5 | Peptide/opine/nickel uptake transporter (PepT) family | 0.72 | 0 |
| CDS no. | 3.A.1.5 | 0.68 | 0 | |
| Genome size | 3.A.1.3 | Polar amino acid uptake transporter (PAAT) family | 0.73 | 0 |
| CDS no. | 3.A.1.3 | 0.66 | 0 | |
| Genome size | 3.A.1.155 | Phage infection protein (PIP) family | 0.72 | 0 |
| CDS no. | 3.A.1.155 | 0.63 | 0 | |
| Genome size | 3.A.1.104 | Teichoic acid exporter (TAE) family | 0.68 | 0 |
| CDS no. | 3.A.1.104 | 0.60 | 1.33E−15 | |
| Genome size | 1.A.112 | Cyclin M Mg2+ exporter (CNNM) family | 0.72 | 0 |
| CDS no. | 1.A.112 | 0.70 | 0 | |
| Genome size | 1.A.8 | Major intrinsic protein (MIP) family | 0.70 | 0 |
| CDS no. | 1.A.8 | 0.61 | 2.22E−16 | |
| Genome size | 1.A.35 | CorA metal ion transporter (MIT) family | 0.68 | 0 |
| CDS no. | 1.A.35 | 0.60 | 8.88E−16 | |
| Genome size | 1.I.3 | Bacterial (Planctomycetes) nuclear pore-like complex (B-NPC) family | 0.63 | 0 |
| CDS no. | 1.I.3 | 0.62 | 2.22E−16 | |
| Genome size | 2.A.23 | Dicarboxylate/amino acid:cation (Na+ or H+) symporter (DAACS) family | 0.71 | 0 |
| CDS no. | 2.A.23 | 0.65 | 0 | |
| Genome size | 2.A.26 | Branched-chain amino acid:cation symporter (LIVCS) family | 0.61 | 4.44E−16 |
| CDS no. | 2.A.26 | 0.61 | 2.22E−16 | |
| Genome size | 2.A.45 | Arsenite-antimonite (ArsB) efflux family | 0.60 | 1.11E−15 |
| CDS no. | 2.A.45 | 0.62 | 0 | |
| Genome size | 2.A.118 | Basic amino acid antiporter (ArcD) family | 0.68 | 0 |
| CDS no. | 2.A.118 | 0.60 | 8.88E−16 | |
| Genome size | 2.A.20 | Inorganic phosphate transporter (PiT) family | 0.68 | 0 |
| CDS no. | 2.A.20 | 0.60 | 8.88E−16 | |
| Genome size | 2.A.108 | Iron/lead transporter (ILT) family | 0.69 | 0 |
| CDS no. | 2.A.108 | 0.61 | 4.44E−16 | |
| Genome size | 2.A.76 | Resistance to homoserine/threonine (RhtB) family | 0.61 | 8.88E−16 |
| CDS no. | 2.A.76 | 0.61 | 4.44E−16 | |
| Genome size | 3.A.6 | Type III (virulence-related) secretory pathway (IIISP) family | 0.62 | 0 |
| CDS no. | 3.A.6 | 0.62 | 0 | |
| Genome size | 5.B.1 | gp91phox phagocyte NADPH oxidase-associated cytochrome b558 (Phox) family | 0.73 | 0 |
| CDS no. | 5.B.1 | 0.67 | 0 | |
| Genome size | 9.B.34 | Kinase/phosphatase/cyclic GMP synthase/cyclic di-GMP hydrolase (KPSH) family | 0.75 | 0 |
| CDS no. | 9.B.34 | 0.76 | 0 | |
| Genome size | 9.B.72 | 4 TMS GlpM (GlpM) family | 0.63 | 0 |
| CDS no. | 9.B.72 | 0.63 | 0 |
TC, transporter classification. Boldface indicates superfamily.
FIG 6.
Comparison of 25 transporter family members between groups I and II. All pairwise comparisons were significantly different (Wilcoxon test). Each dot represents one strain.
The 25 selected transporter families also included three families involved in transporting heavy metal ions (the arsenite-antimonite efflux family, iron/lead transporter family, and peptide/opine/nickel uptake transporter family) and two major facilitator superfamilies (MFSs) associated with drug efflux (Table 1). These transporters have been shown to counteract the effects of toxic heavy metals and drugs (28, 50–52). Two of the 25 families are involved in the formation of the bacterial cell wall and biofilm, namely, the teichoic acid exporter family and the 4 TMS GlpM family. Teichoic acid is a major cell wall component of Gram-positive bacteria that plays crucial roles in bacterial resistance to antimicrobial agents and survival under disadvantageous conditions (53, 54). The 4 TMS GlpM family is required for normal production of alginate (55). Alginates are important polymeric substances that contribute to forming and developing the biofilm matrixes of numerous bacteria and enhancing colonization and thereby persistence under environmental stresses (56).
In all living organisms, transporters are vital to the uptake of nutrients, secretion of metabolites, maintenance of ion concentration gradients across membranes, and efflux of drugs and toxins (57). The enhanced ability of group I strains for transporting important substrates enables such strains to better meet demands for cellular metabolism and functions and thus appears to provide an important basis for their ability to survive in diverse terrestrial environments. Our results further suggest that the selective expansion of transporter families has been a major evolutionary strategy for members of the genus Exiguobacterium.
DISCUSSION
The Exiguobacterium genus comprises Gram-positive, non-spore-forming, and facultative anaerobic species. Members belonging to this genus have been isolated from a large array of environments, such as soils (18), freshwater (19), plant rhizospheres (20), salt lake sediment (21), a hot spring (22), glaciers, and permafrost (58). 16S rRNA marker gene surveys have uncovered Exiguobacterium spp. surviving in even more diverse habitats, including indoor air (59), skin (60), fish gut (61), and a cold desert (62). These results suggest that members of the genus Exiguobacterium are ubiquitous. In this study, we first explored the distribution of Exiguobacterium members through 16S rRNA gene analysis and correlated that with information on the collection location. We found that members of the Exiguobacterium genus are distributed in both marine and terrestrial environments worldwide but that GenBank contains more from terrestrial environments than from marine environments. We next set out to isolate additional Exiguobacterium members from marine ecosystems. We obtained 86 isolates from diverse marine niches, including marine sediment, seawater, algae, cold springs, hydrothermal vents, seamounts, mangroves, marine fish, and coral. This suggests that Exiguobacterium spp. are widespread in a variety of marine environments and, together with the existing data, shows that members of the Exiguobacterium genus have a cosmopolitan distribution.
Our comprehensive experimental results indicate that members of this genus have a wide range of metabolic and stress resistance capabilities, which may contribute to their wide distribution. The ability to degrade and absorb carbon and nitrogen nutrients is crucial for the ability of a microorganism to survive in diverse habitats (63). Genes encoding multiple types of proteins capable of directing carbohydrate hydrolysis were identified in each Exiguobacterium genome, suggesting that these bacteria have versatile abilities to metabolize carbohydrates. Among the predicted enzymes, those involved in degrading starch, cellulose, and chitin were enriched in almost all studied members of the Exiguobacterium genus. Starch is an important storage polysaccharide for plants of both terrestrial and marine environments (64, 65), Cellulose, which is the most prevalent polysaccharide in nature, makes up the cell walls of plants and algae (66, 67). Chitin, which is the second-most-common polysaccharide in nature (after cellulose), is also widely distributed in terrestrial and marine ecosystems; it functions as a major structural component of crustacean shells, arthropod exoskeletons, and diatom cell walls (68–70). The presence of abundant genes for chitin degradation may enable members of the Exiguobacterium genus to occupy different functional niches with regard to chitin breakdown. The potential to utilize the most plentiful polysaccharides from both marine and terrestrial environments likely supports the ability of Exiguobacterium members to survive and reproduce under an extensive range of environmental conditions. It is also notable that many secreted peptidases are predicted in Exiguobacterium genomes. As proteinaceous compounds are important nitrogen nutrients for microorganisms (71), the presence of numerous potentially secreted peptidases in Exiguobacterium genomes may allow them to exploit different niches for nitrogen source uptake in different environments. The metallo peptidase M23 and serine peptidase S08 were the most abundant peptidases. M23 peptidase reportedly degrades bacterial extracellular peptidoglycans and thereby contributes to the acquisition of nutrition or defense against competitors (72, 73). Serine peptidases are often used as marker enzymes for proteolytic activity in soil and play important roles in the ability of a microorganism to utilize nitrogen sources in the environment (74). The capacity of Exiguobacterium spp. to metabolize starches and proteins was validated on plates, further supporting our proposal that such genes form at least part of the genetic basis for the cosmopolitan distribution of these bacteria.
In addition to being collected from the common terrestrial and marine ecosystems, many Exiguobacterium isolates have been collected from extreme environments, such as glaciers, hot springs, and salt lakes (17). Thus, members of the Exiguobacterium genus can be classified as extremophiles. Consistently with this, our analysis showed that the genomes of Exiguobacterium spp. contain genes related to various classes of stress resistance systems and that members often have more than one strategy to fight against certain extreme environments. Here, we predict that Exiguobacterium spp. may use two strategies to survive in cold environments: the maintenance of membrane fluidity and the stabilization of their cytosol. To cope with hot environments, they appear to use two types of chaperone systems to prevent aggregation and denaturation of proteins at high temperature. Moreover, three types of antiporter families are utilized to maintain intracellular pH homeostasis and the dynamic balance of cellular Na+. In general, a bacterium is adapted to cope with only one particular extreme environment. For instance, members of the genus Psychrobacter have vital CSPs and thus are more frequently found in cold environments (75). The presence of multiple stress-responsive genes discussed herein might enable strains of Exiguobacterium spp. to adapt to a very wide range of niches, even extreme environments.
Although the members of the genus Exiguobacterium have a cosmopolitan distribution overall, we herein uncovered ecological differences related to the two groups defined for this genus based on our genomic analysis. More numerous transporters and a more diverse nonmarine environmental distribution were found for group I, which may suggest that the expansion of transporters contributes to these ecological differences. Most of the expanded transporter families were associated with important physiological metabolism and environmental-stress resistance processes. Compared with the marine environment, the terrestrial environment is more diverse and has a greater variety of complex microenvironments due to the influence of climates or seasons, leading to greater diversity in the experienced stress conditions and nutrient substrates (3, 76). In order to survive in these more diverse environments, bacteria must develop specific systems for survival, such as nutrient-sensing and transport systems (77). Efficient transport of substances related to metabolism, cellular function, and/or environmental-stress resistance is crucial for bacterial survival in a variety of environments (78). Therefore, the expanded families that contribute to environmental-stress resistance and the transport of organic or inorganic substrates may play crucial roles in the ability of Exiguobacterium species group I members to survive in more diverse terrestrial environments.
Conclusions.
The question of how microbes with a cosmopolitan distribution adapt to diverse habitats is an important topic in microbial ecology. Their wide distribution makes the genus Exiguobacterium a valuable system for studying the evolution and adaptive strategies that bacteria use to survive in multiple habitats. We herein reveal that Exiguobacterium spp. share extensive characteristics that enable them to adapt to various environments and exhibit a cosmopolitan distribution. Our genomic analysis revealed that Exiguobacterium members not only can utilize a variety of complex polysaccharides and proteins that are ubiquitous in both terrestrial and marine environments but also have a number of chaperonins and transporters that are expected to help them to survive in diverse extreme environments. We also revealed that the expansion of transporter families has driven genomic evolution in Exiguobacterium. The selective expansion of genes involved in transporting organic or inorganic substrates and environmental-stress resistance appears to have served as an evolutionary and adaptive strategy that has supported the extensive distribution of Exiguobacterium spp.
MATERIALS AND METHODS
Analysis of 16S rRNA gene sequences of Exiguobacterium spp.
The 16S rRNA gene sequences of Exiguobacterium spp. available in GenBank were retrieved. Sequences that had low quality (e.g., sequences that were not ribosomal or that contained N in the base sequence) or lacked isolation source tags were discarded, and 16S rRNA gene sequences with >95% identities to those of reported type strains were retained. Information on the isolation source of these sequences was collected, and the habitats were classified into 13 types: air, animal-associated environments, cold environments, estuary-associated environments, contaminated water or soil, freshwater or freshwater sediment, hot environments, hypersaline environments, marine environments, plants, rhizospheres, soil, and other inland environments.
Bacterial isolation, culture, and identification.
In this study, we collected a total of 105 strains, including 97 isolates that our group collected from terrestrial and marine environments worldwide and eight type strains obtained from the DSMZ, the Japan Collection of Microorganisms (JCM), and the China General Microbiological Culture Collection Center (CGMCC) (see Table S2 in the supplemental material). To obtain each target strain, initial samples from marine and terrestrial environments were macerated and mixed with sterile saline solution (0.8%) using a standard dilution plating method on marine agar 2216 (MA; Difco) and LB agar at 20°C. Each plate-grown colony was picked and subcultured three times to achieve a pure culture. 16S rRNA sequence analysis was performed to identify those belonging to the Exiguobacterium genus. All of the 105 strains were confirmed to grow in sea salt-free medium and were routinely cultivated on LB agar and in liquid LB for subsequent genomic sequencing.
Adaptive ability tests for pH, temperature, and salinity.
To assess the range of adaptation to pH, temperature, and salinity, all 105 strains were monitored for growth under the following conditions: for temperature-based assessments, at 4, 10, 25, 30, 40, 45, and 55°C on LB agar and in liquid LB at 150 rpm; for pH-based assessments, at 25°C in liquid LB medium in 1-unit steps from pH 4.0 to pH 11.0 using citrate-Na2HPO4 buffer (pH 4.0 to 7.0), Tris buffer (pH 7.5 to 9.0), and NaHCO3-Na2CO3 buffer (pH 9.5 to 11.0); and for salinity assessments, at 0 to 20% (in increments of 1%, wt/vol) NaCl after 2 to 3 days of cultivation at 25°C in medium containing 0.1% peptone, 0.1% yeast extract, 0.03% KCl, 0.25% MgSO4·7H2O, 0.05% CaCl2. Growth was evaluated by measuring the optical density at 600 nm (OD600) after 2 to 3 days of incubation at 150 rpm.
Degradation ability tests for starch and protein.
Enzyme activity screenings of Exiguobacterium members were performed as described by Margesin et al. (79). Protease and amylase activities were tested on LB agar supplemented with skim milk (2%, wt/vol) and starch (0.4%, wt/vol). After 3 days of incubation at 25°C, the plates were scored. A reaction was determined to be positive when transparent zones were readily visible around the colonies or detected after coloration was preformed using Lugol’s iodine solution.
Genome sequencing, assembly, and annotation.
The 105 Exiguobacterium strains were incubated at 25°C in LB liquid medium at 150 rpm for 2 days. Genomic DNA was extracted using a bacterial genomic DNA minikit (TaKaRa Bio) by following the manufacturer’s protocol. A paired-end library with an insert size of 350 bp was constructed for each genome and sequenced with an Illumina NovaSeq 6000 sequencer to generate 150 pair-end reads. The raw reads of each genome were processed to remove low-quality bases and adaptors by Trimmomatic v0.36 (80) and assembled with SPAdes v3.14.1 (81). The genomes of Exiguobacterium strains deposited in GenBank were collected and filtered based on the criterion that genomes were at least 95% complete and had <5% contamination based on CheckM v1.1.2 analysis (82). Gene predictions and annotations of all genomes were generated using Prokka v1.14.6 (83). The pairwise ANI values among all the studied genomes were computed using the FastANI v1.31 software (84).
Phylogenetic tree construction.
Analysis of orthologous clusters was performed using FastOrtho (http://enews.patricbrc.org/fastortho/), which is a faster reimplementation of OrthoMCL (85). In brief, an all-against-all BLAST search was performed with E values of <1 × 10−10. Ortholog groups were created with the MCL algorithm using an inflation value of 2, and single-copy gene families were obtained using custom-made Python scripts. The protein sequences of each family were aligned using MUSCLE v3.8.1551 (86) and trimmed with trimAl v1.4.rev22 (87). All trimmed alignments were concatenated into a new alignment by a local Python script. A phylogenetic tree based on this alignment was constructed using IQ-TREE v2.0.3 with 1,000 bootstrap replicates, employing the JTT+F+I+G4 model (88). iTOL was used to visualize the phylogenetic tree (89).
Identification of carbohydrate-active enzymes and proteases.
To identify genes encoding carbohydrate-active enzymes and proteases, all of the annotated genes were searched against the CAZy database (http://www.cazy.org) (90) and peptidase database (MEROPS) (91) using Diamond BLASTP (E value, 1E–5; ID 30; more sensitive). These carbohydrate-active enzymes were also predicted by HMMER using hidden Markov models (HMM) from dbCAN (92). Genes belonging to different carbohydrate-active enzymes or protease families were classified by in-house Python scripts according to the predictions. The potential secreted carbohydrate-active enzymes and peptidases were confirmed based on identification of extracellular transport signals, which was performed using SignalP (93).
Identification of vital genes for environmental-stress resistance.
Each protein predicted by Prokka was annotated using BLASTP and Hmmscan against the Clusters of Orthologous Groups (COG) database and PFAM database, respectively, with E values of <1 × 10−5. Transporters were predicted by using Diamond BLASTP (E value, 1E–10; sensitive) to search the Exiguobacterium protein sequences against all Transporter Classification Database (TCDB) sequences (94).
Data analyses.
Statistical analyses were performed using the Wilcoxon test. Correlation analyses for genome size, CDS number, and transporter number were performed using chart.Correlation from the PerformanceAnalytics package in R (https://cran.r-project.org/web/packages/PerformanceAnalytics/index.html).
Data availability.
The genomes supporting the reported results have been deposited in GenBank under BioProject accession no. PRJNA644789 (Table S2).
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grants 31670002, 31970003, and 31770003) and the Science & Technology Basic Resources Investigation Program of China (grant 2017FY100804).
We declare that we have no conflict of interest.
Contributor Information
Zhaolu Zhu, Email: zhaoluzhu@gmail.com.
Jinshui Zheng, Email: jszheng@mail.hzau.edu.cn.
Angela D. Kent, University of Illinois at Urbana-Champaign
REFERENCES
- 1.Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Øvreås L, Reysenbach A-L, Smith VH, Staley JT. 2006. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 2.Bock C, Jensen M, Forster D, Marks S, Nuy J, Psenner R, Beisser D, Boenigk J. 2020. Factors shaping community patterns of protists and bacteria on a European scale. Environ Microbiol 22:2243–2260. doi: 10.1111/1462-2920.14992. [DOI] [PubMed] [Google Scholar]
- 3.Valli S, Suvathi SS, Aysha OS, Nirmala P, Vinoth KP, Reena A. 2012. Antimicrobial potential of Actinomycetes species isolated from marine environment. Asian Pac J Trop Biomed 2:469–473. doi: 10.1016/S2221-1691(12)60078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logares R, Bråte J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, Rengefors K. 2009. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol 17:414–422. doi: 10.1016/j.tim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Logares R, Shalchian-Tabrizi K, Boltovskoy A, Rengefors K. 2007. Extensive dinoflagellate phylogenies indicate infrequent marine-freshwater transitions. Mol Phylogenet Evol 45:887–903. doi: 10.1016/j.ympev.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Simon M, Scheuner C, Meier-Kolthoff JP, Brinkhoff T, Wagner-Döbler I, Ulbrich M, Klenk H-P, Schomburg D, Petersen J, Göker M. 2017. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J 11:1483–1499. doi: 10.1038/ismej.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Getz EW, Tithi SS, Zhang LQ, Aylward FO. 2018. Parallel evolution of genome streamlining and cellular bioenergetics across the marine radiation of a bacterial phylum. mBio 9:e01089-18. doi: 10.1128/mBio.01089-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ, Rappé MS. 2012. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio 3:e00252-12. doi: 10.1128/mBio.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elser JJ, Bracken MES, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, Ngai JT, Seabloom EW, Shurin JB, Smith JE. 2007. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett 10:1135–1142. doi: 10.1111/j.1461-0248.2007.01113.x. [DOI] [PubMed] [Google Scholar]
- 10.Eichorst SA, Trojan D, Roux S, Herbold C, Rattei T, Woebken D. 2018. Genomic insights into the Acidobacteria reveal strategies for their success in terrestrial environments. Environ Microbiol 20:1041–1063. doi: 10.1111/1462-2920.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Yoshizawa S, Sun Y, Huang YJ, Chu X, González JM, Pinhassi J, Luo HW. 2019. Repeated evolutionary transitions of flavobacteria from marine to non-marine habitats. Environ Microbiol 21:648–666. doi: 10.1111/1462-2920.14509. [DOI] [PubMed] [Google Scholar]
- 12.Cabello-Yeves PJ, Rodriguez-Valera F. 2019. Marine-freshwater prokaryotic transitions require extensive changes in the predicted proteome. Microbiome 7:117. doi: 10.1186/s40168-019-0731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka M, Kumakura D, Mino S, Doi H, Ogura Y, Hayashi T, Yumoto I, Cai M, Zhou Y-G, Gomez-Gil B, Araki T, Sawabe T. 2020. Genomic characterization of closely related species in the Rumoiensis clade infers ecogenomic signatures to non-marine environments. Environ Microbiol 22:3205–3217. doi: 10.1111/1462-2920.15062. [DOI] [PubMed] [Google Scholar]
- 14.Giovannoni SJ. 2017. SAR11 bacteria: the most abundant plankton in the oceans. Annu Rev Mar Sci 9:231–255. doi: 10.1146/annurev-marine-010814-015934. [DOI] [PubMed] [Google Scholar]
- 15.Santoro AE, Richter RA, Dupont CL. 2019. Planktonic marine archaea. Annu Rev Mar Sci 11:131–158. doi: 10.1146/annurev-marine-121916-063141. [DOI] [PubMed] [Google Scholar]
- 16.Vishnivetskaya TA, Kathariou S, Tiedje JM. 2009. The Exiguobacterium genus: biodiversity and biogeography. Extremophiles 13:541–555. doi: 10.1007/s00792-009-0243-5. [DOI] [PubMed] [Google Scholar]
- 17.Kasana RC, Pandey CB. 2018. Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Crit Rev Biotechnol 38:141–156. doi: 10.1080/07388551.2017.1312273. [DOI] [PubMed] [Google Scholar]
- 18.Singh NK, Raichand R, Kaur I, Kaur C, Pareek S, Mayilraj S. 2013. Exiguobacterium himgiriensis sp. nov. a novel member of the genus Exiguobacterium, isolated from the Indian Himalayas. Antonie Van Leeuwenhoek 103:789–796. doi: 10.1007/s10482-012-9861-5. [DOI] [PubMed] [Google Scholar]
- 19.Raichand R, Pareek S, Singh NK, Mayilraj S. 2012. Exiguobacterium aquaticum sp. nov., a member of the genus Exiguobacterium. Int J Syst Evol Microbiol 62:2150–2155. doi: 10.1099/ijs.0.035790-0. [DOI] [PubMed] [Google Scholar]
- 20.Khandare RV, Rane NR, Waghmode TR, Govindwar SP. 2012. Bacterial assisted phytoremediation for enhanced degradation of highly sulfonated diazo reactive dye. Environ Sci Pollut Res Int 19:1709–1718. doi: 10.1007/s11356-011-0679-x. [DOI] [PubMed] [Google Scholar]
- 21.Jiang X, Xue Y, Wang L, Yu B, Ma Y. 2013. Genome sequence of a novel polymer-grade l-lactate-producing alkaliphile, Exiguobacterium sp. strain 8-11-1. Genome Announc 1:e00616-13. doi: 10.1128/genomeA.00616-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vishnivetskaya TA, Lucas S, Copeland A, Lapidus A, Glavina del Rio T, Dalin E, Tice H, Bruce DC, Goodwin LA, Pitluck S, Saunders E, Brettin T, Detter C, Han C, Larimer F, Land ML, Hauser LJ, Kyrpides NC, Ovchinnikova G, Kathariou S, Ramaley RF, Rodrigues DF, Hendrix C, Richardson P, Tiedje JM. 2011. Complete genome sequence of the thermophilic bacterium Exiguobacterium sp. AT1b. J Bacteriol 193:2880–2881. doi: 10.1128/JB.00303-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno Letelier A, Olmedo Alvarez G, Eguiarte LE, Souza V. 2012. Divergence and phylogeny of firmicutes from the Cuatro Ciénegas Basin, Mexico: a window to an ancient ocean. Astrobiology 12:674–684. doi: 10.1089/ast.2011.0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rebollar EA, Avitia M, Eguiarte LE, González González A, Mora L, Bonilla Rosso G, Souza V. 2012. Water-sediment niche differentiation in ancient marine lineages of Exiguobacterium endemic to the Cuatro Cienegas Basin. Environ Microbiol 14:2323–2333. doi: 10.1111/j.1462-2920.2012.02784.x. [DOI] [PubMed] [Google Scholar]
- 25.Gutiérrez-Preciado A, Vargas-Chávez C, Reyes-Prieto M, Ordoñez OF, Santos-García D, Rosas-Pérez T, Valdivia-Anistro J, Rebollar EA, Saralegui A, Moya A, Merino E, Farías ME, Latorre A, Souza V. 2017. The genomic sequence of Exiguobacterium chiriqhucha str. N139 reveals a species that thrives in cold waters and extreme environmental conditions. PeerJ 5:e3162. doi: 10.7717/peerj.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dias LM, Folador ARC, Oliveira AM, Ramos RTJ, Silva A, Baraúna RA. 2018. Genomic architecture of the two cold-adapted genera Exiguobacterium and Psychrobacter: evidence of functional reduction in the Exiguobacterium antarcticum B7 Genome. Genome Biol Evol 10:731–741. doi: 10.1093/gbe/evy029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castro-Severyn J, Pardo-Esté C, Mendez KN, Morales N, Marquez SL, Molina F, Remonsellez F, Castro-Nallar E, Saavedra CP. 2020. Genomic variation and arsenic tolerance emerged as niche specific adaptations by different Exiguobacterium strains isolated from the extreme Salar de Huasco environment in Chilean-Altiplano. Front Microbiol 11:1632. doi: 10.3389/fmicb.2020.01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castro Severyn J, Remonsellez F, Valenzuela SL, Salinas C, Fortt J, Aguilar P, Pardo Esté C, Dorador C, Quatrini R, Molina F, Aguayo D, Castro Nallar E, Saavedra CP. 2017. Comparative genomics analysis of a new Exiguobacterium strain from Salar de Huasco reveals a repertoire of stress-related genes and arsenic resistance. Front Microbiol 8:456. doi: 10.3389/fmicb.2017.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim M, Oh H-S, Park S-C, Chun J. 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 30.Logares R, Bråte J, Heinrich F, Shalchian-Tabrizi K, Bertilsson S. 2010. Infrequent transitions between saline and fresh waters in one of the most abundant microbial lineages (SAR11). Mol Biol Evol 27:347–357. doi: 10.1093/molbev/msp239. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen TTH, Myrold DD, Mueller RS. 2019. Distributions of extracellular peptidases across prokaryotic genomes reflect phylogeny and habitat. Front Microbiol 10:413–413. doi: 10.3389/fmicb.2019.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barria C, Malecki M, Arraiano C. 2013. Bacterial adaptation to cold. Microbiology (Reading) 159:2437–2443. doi: 10.1099/mic.0.052209-0. [DOI] [PubMed] [Google Scholar]
- 33.De Maayer P, Anderson D, Cary C, Cowan DA. 2014. Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep 15:508–517. doi: 10.1002/embr.201338170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawasaki R, Baraúna RA, Silva A, Carepo MSP, Oliveira R, Marques R, Ramos RTJ, Schneider MPC. 2016. Reconstruction of the fatty acid biosynthetic pathway of Exiguobacterium antarcticum B7 based on genomic and bibliomic data. BioMed Res Int 2016:7863706. doi: 10.1155/2016/7863706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ermolenko DN, Makhatadze GI. 2002. Bacterial cold-shock proteins. Cell Mol Life Sci 59:1902–1913. doi: 10.1007/pl00012513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feder ME, Hofmann GE. 1999. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 37.Gragerov A, Nudler E, Komissarova N, Gaitanaris GA, Gottesman ME, Nikiforov V. 1992. Cooperation of GroEL/GroES and DnaK/DnaJ heat shock proteins in preventing protein misfolding in Escherichia coli. Proc Natl Acad Sci U S A 89:10341–10344. doi: 10.1073/pnas.89.21.10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arsène F, Tomoyasu T, Bukau B. 2000. The heat shock response of Escherichia coli. Int J Food Microbiol 55:3–9. doi: 10.1016/s0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- 39.Narberhaus F. 2002. α-Crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev 66:64–93. doi: 10.1128/MMBR.66.1.64-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wholey WY, Jakob U. 2012. Hsp33 confers bleach resistance by protecting elongation factor Tu against oxidative degradation in Vibrio cholerae. Mol Microbiol 83:981–991. doi: 10.1111/j.1365-2958.2012.07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Honoré FA, Méjean V, Genest O. 2017. Hsp90 is essential under heat stress in the bacterium Shewanella oneidensis. Cell Rep 19:680–687. doi: 10.1016/j.celrep.2017.03.082. [DOI] [PubMed] [Google Scholar]
- 42.Segal N, Shapira M. 2015. HSP33 in eukaryotes—an evolutionary tale of a chaperone adapted to photosynthetic organisms. Plant J 82:850–860. doi: 10.1111/tpj.12855. [DOI] [PubMed] [Google Scholar]
- 43.Krulwich TA, Hicks DB, Ito M. 2009. Cation/proton antiporter complements of bacteria: why so large and diverse? Mol Microbiol 74:257–260. doi: 10.1111/j.1365-2958.2009.06842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee C, Kang HJ, von Ballmoos C, Newstead S, Uzdavinys P, Dotson DL, Iwata S, Beckstein O, Cameron AD, Drew D. 2013. A two-domain elevator mechanism for sodium/proton antiport. Nature 501:573–577. doi: 10.1038/nature12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kajiyama Y, Otagiri M, Sekiguchi J, Kosono S, Kudo T. 2007. Complex formation by the mrpABCDEFG gene products, which constitute a principal Na+/H+ antiporter in Bacillus subtilis. J Bacteriol 189:7511–7514. doi: 10.1128/JB.00968-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tesseraud S, Everaert N, Boussaid-Om Ezzine S, Collin A, Métayer-Coustard S, Berri C. 2011. Manipulating tissue metabolism by amino acids. World Poult Sci J 67:243–252. doi: 10.1017/S0043933911000274. [DOI] [Google Scholar]
- 47.Prasad D, Verma N, Bakshi M, Narayan OP, Singh AK, Dua M, Johri AK. 2019. Functional characterization of a magnesium transporter of root endophytic fungus Piriformospora indica. Front Microbiol 9:3231. doi: 10.3389/fmicb.2018.03231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Huang S, Wang J, Jan G, Jeantet R, Chen XD. 2017. Mg2+ improves the thermotolerance of probiotic Lactobacillus rhamnosus GG, Lactobacillus casei Zhang and Lactobacillus plantarum P-8. Lett Appl Microbiol 64:283–288. doi: 10.1111/lam.12716. [DOI] [PubMed] [Google Scholar]
- 49.Hudek L, Premachandra D, Webster WAJ, Bräu L. 2016. Role of phosphate transport system component PstB1 in phosphate internalization by Nostoc punctiforme. Appl Environ Microbiol 82:6344–6356. doi: 10.1128/AEM.01336-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grosse C, Scherer J, Koch D, Otto M, Taudte N, Grass G. 2006. A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol Microbiol 62:120–131. doi: 10.1111/j.1365-2958.2006.05326.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhao FJ. 2016. A novel pathway of arsenate detoxification. Mol Microbiol 100:928–930. doi: 10.1111/mmi.13395. [DOI] [PubMed] [Google Scholar]
- 52.Cavazza C. 2011. Extracytoplasmic nickel-binding proteins. In Encyclopedia of Inorganic and Bioinorganic Chemistry. Wiley Online Library. doi: 10.1002/9781119951438.eibc2438. [DOI] [Google Scholar]
- 53.Chen L, Hou WT, Fan T, Liu BH, Pan T, Li YH, Jiang YL, Wen W, Chen ZP, Sun LF, Zhou CZ, Chen YX. 2020. Cryo-electron microscopy structure and transport mechanism of a wall teichoic acid abc transporter. mBio 11:e02749-19. doi: 10.1128/mBio.02749-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swoboda JG, Campbell J, Meredith TC, Walker S. 2010. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 11:35–45. doi: 10.1002/cbic.200900557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schweizer HP, Po C, Bacic MK. 1995. Identification of Pseudomonas aeruginosa glpM, whose gene product is required for efficient alginate biosynthesis from various carbon sources. J Bacteriol 177:4801–4804. doi: 10.1128/jb.177.16.4801-4804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moradali MF, Rehm BHA. 2019. The role of alginate in bacterial biofilm formation, p 517–537. In Cohen E, Merzendorfer H (ed), Extracellular sugar-based biopolymers matrices. Springer International Publishing, Cham, Switzerland. doi: 10.1007/978-3-030-12919-4_13. [DOI] [Google Scholar]
- 57.Busch W, Saier MH. 2002. The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol 37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 58.Rodrigues DF, Goris J, Vishnivetskaya T, Gilichinsky D, Thomashow MF, Tiedje JM. 2006. Characterization of Exiguobacterium isolates from the Siberian permafrost. Description of Exiguobacterium sibiricum sp. nov. Extremophiles 10:285–294. doi: 10.1007/s00792-005-0497-5. [DOI] [PubMed] [Google Scholar]
- 59.Yuan L, Xu J, Millar BC, Dooley JSG, Rooney PJ, Alexander HD, Moore JE. 2007. Molecular identification of environmental bacteria in indoor air in the domestic home: description of a new species of Exiguobacterium. Int J Environ Health Res 17:75–82. doi: 10.1080/09603120601124199. [DOI] [PubMed] [Google Scholar]
- 60.Brockmann M, Aupperle-Lellbach H, Gentil M, Heusinger A, Müller E, Marschang RE, Pees M. 2020. Challenges in microbiological identification of aerobic bacteria isolated from the skin of reptiles. PLoS One 15:e0240085. doi: 10.1371/journal.pone.0240085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng X, Yang R, Hu J, Lin S, Gu Z, Ma Z. 2019. The gut microbiota community and antioxidant enzymes activity of barramundi reared at seawater and freshwater. Fish Shellfish Immunol 89:127–131. doi: 10.1016/j.fsi.2019.03.054. [DOI] [PubMed] [Google Scholar]
- 62.Yadav AN, Sachan SG, Verma P, Saxena AK. 2015. Prospecting cold deserts of north western Himalayas for microbial diversity and plant growth promoting attributes. J Biosci Bioeng 119:683–693. doi: 10.1016/j.jbiosc.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 63.Orsi WD, Richards TA, Francis WR. 2018. Predicted microbial secretomes and their target substrates in marine sediment. Nat Microbiol 3:32–37. doi: 10.1038/s41564-017-0047-9. [DOI] [PubMed] [Google Scholar]
- 64.Collinson SR, Thielemans W. 2010. The catalytic oxidation of biomass to new materials focusing on starch, cellulose and lignin. Coord Chem Rev 254:1854–1870. doi: 10.1016/j.ccr.2010.04.007. [DOI] [Google Scholar]
- 65.Prabhu M, Chemodanov A, Gottlieb R, Kazir M, Nahor O, Gozin M, Israel A, Livney YD, Golberg A. 2019. Starch from the sea: the green macroalga Ulva ohnoi as a potential source for sustainable starch production in the marine biorefinery. Algal Res 37:215–227. doi: 10.1016/j.algal.2018.11.007. [DOI] [Google Scholar]
- 66.Domozych DS, Ciancia M, Fangel JU, Mikkelsen MD, Ulvskov P, Willats WGT. 2012. The cell walls of green algae: a journey through evolution and diversity. Front Plant Sci 3:82–82. doi: 10.3389/fpls.2012.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Broxterman SE, Schols HA. 2018. Interactions between pectin and cellulose in primary plant cell walls. Carbohydr Polym 192:263–272. doi: 10.1016/j.carbpol.2018.03.070. [DOI] [PubMed] [Google Scholar]
- 68.Rinaudo M. 2006. Chitin and chitosan: properties and applications. Prog Polym Sci 31:603–632. doi: 10.1016/j.progpolymsci.2006.06.001. [DOI] [Google Scholar]
- 69.Nguyen STC, Freund HL, Kasanjian J, Berlemont R. 2018. Function, distribution, and annotation of characterized cellulases, xylanases, and chitinases from CAZy. Appl Microbiol Biotechnol 102:1629–1637. doi: 10.1007/s00253-018-8778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elieh-Ali-Komi D, Hamblin MR. 2016. Chitin and chitosan: production and application of versatile biomedical nanomaterials. Int J Adv Res (Indore) 4:411–427. [PMC free article] [PubMed] [Google Scholar]
- 71.Wongkiew S, Hu Z, Chandran K, Lee JW, Khanal SK. 2017. Nitrogen transformations in aquaponic systems: a review. Aquac Eng 76:9–19. doi: 10.1016/j.aquaeng.2017.01.004. [DOI] [Google Scholar]
- 72.Stohl EA, Chan YA, Hackett KT, Kohler PL, Dillard JP, Seifert HS. 2012. Neisseria gonorrhoeae virulence factor NG1686 is a bifunctional M23B family metallopeptidase that influences resistance to hydrogen peroxide and colony morphology. J Biol Chem 287:11222–11233. doi: 10.1074/jbc.M111.338830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baba T, Schneewind O. 1996. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J 15:4789–4797. doi: 10.1002/j.1460-2075.1996.tb00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fuka MM, Engel M, Gattinger A, Bausenwein U, Sommer M, Munch JC, Schloter M. 2008. Factors influencing variability of proteolytic genes and activities in arable soils. Soil Biol Biochem 40:1646–1653. doi: 10.1016/j.soilbio.2008.01.028. [DOI] [Google Scholar]
- 75.Zhang S, Song W, Yu M, Lin X. 2017. Comparative genomics analysis of five Psychrobacter strains isolated from world-wide habitats reveal high intra-genus variations. Extremophiles 21:581–589. doi: 10.1007/s00792-017-0927-1. [DOI] [PubMed] [Google Scholar]
- 76.DeMarco J, Mack MC, Bret-Harte MS. 2011. The effects of snow, soil microenvironment, and soil organic matter quality on N availability in three Alaskan Arctic plant communities. Ecosystems 14:804–817. doi: 10.1007/s10021-011-9447-5. [DOI] [Google Scholar]
- 77.Samyn DR, Persson BL. 2016. Inorganic phosphate and sulfate transport in S. cerevisiae, p 253–269. In Ramos J, Sychrová H, Kschischo M (ed), Yeast membrane transport. Springer International Publishing, Cham, Switzerland. doi: 10.1007/978-3-319-25304-6_10. [DOI] [Google Scholar]
- 78.Lorca GL, Barabote RD, Zlotopolski V, Tran C, Winnen B, Hvorup RN, Stonestrom AJ, Nguyen E, Huang L-W, Kim DS, Saier MH. 2007. Transport capabilities of eleven gram-positive bacteria: comparative genomic analyses. Biochim Biophys Acta 1768:1342–1366. doi: 10.1016/j.bbamem.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Margesin R, Gander S, Zacke G, Gounot AM, Schinner F. 2003. Hydrocarbon degradation and enzyme activities of cold-adapted bacteria and yeasts. Extremophiles 7:451–458. doi: 10.1007/s00792-003-0347-2. [DOI] [PubMed] [Google Scholar]
- 80.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 84.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rawlings ND, Waller M, Barrett AJ, Bateman A. 2014. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 94.Saier MH, Jr, Reddy VS, Tamang DG, Västermark Å. 2014. The transporter classification database. Nucleic Acids Res 42:D251–D258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Geographic distribution of 16S rRNA gene sequences and pan-genome analysis. (A) Geographic distribution of 16S rRNA gene sequences with information about isolation sites (the pink dots represent the sampling locations, and the dots are colored by number of 16S rRNA gene sequences). The map was created using Tableau software (version 10.5) with map data from OpenStreetMap contributors. (B) Pan- and core genomes of the Exiguobacterium genus, i.e., the sizes of the pan- and core genomes in relation to numbers of genomes added into the gene pool. Box plots show the 25th and 75th percentiles, with medians shown as horizontal lines, and whiskers indicate the lowest and highest values within 1.5 times the interquartile range (IQR) from the first and third quartiles, respectively. The curve for the pan-genome is fitted by the power-law regression model, and the curve for the core genome is fitted by the exponential curve fit model. Download FIG S1, PDF file, 1.9 MB (1.9MB, pdf) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Information on the Exiguobacterium 16S rRNA gene sequences available in GenBank. Download Table S1, XLSX file, 0.10 MB (98.7KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome features and isolation sources of 147 Exiguobacterium strains. Download Table S2, XLSX file, 0.03 MB (27.8KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Temperature, pH, and salinity tolerance data for 105 Exiguobacterium strains. Download Table S3, XLSX file, 0.01 MB (12.5KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Average nucleotide identity (ANI) analysis of 147 Exiguobacterium strains. Download Table S4, XLSX file, 0.1 MB (117.3KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Niche distribution of 147 Exiguobacterium strains based on phylogenetic analysis. Download Table S5, XLSX file, 0.01 MB (14.7KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene number of carbohydrate-active enzymes (CAZymes) and extracellular peptidases detected in each Exiguobacterium genome. Download Table S6, XLSX file, 0.1 MB (134.8KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Validation of the abilities of Exiguobacterium spp. to degrade and metabolize complex carbohydrates and peptides. Download Table S7, XLSX file, 0.01 MB (13.9KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genetic basis for adaptation of Exiguobacterium strains to diverse habitats. Download Table S8, XLSX file, 0.03 MB (27.4KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transporter analysis of 147 Exiguobacterium genomes. Download Table S9, XLSX file, 0.2 MB (193.8KB, xlsx) .
Copyright © 2021 Zhang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The genomes supporting the reported results have been deposited in GenBank under BioProject accession no. PRJNA644789 (Table S2).