Abstract
Background
Recurrent respiratory papillomatosis (RRP) is a human papillomavirus (HPV) driven neoplastic disorder of the upper aerodigestive tract that causes significant morbidity and can lead to fatal airway obstruction. Prior clinical study demonstrated clinical benefit with the programmed death-ligand 1 (PD-L1) monoclonal antibody avelumab. Bintrafusp alfa is a bifunctional inhibitor of PD-L1 and transforming growth factor-beta (TGF-b) that has shown clinical activity in several cancer types.
Methods
We conducted a phase II clinical trial evaluating bintrafusp alfa in adults with RRP. Papilloma samples before and after treatment with bintrafusp alfa were assessed for correlates of response with multiplex immunofluorescence as well as immunological and genomic analyses. Post hoc analyses of papilloma samples before and after treatment with avelumab were assessed for comparison.
Results
Dual PD-L1/TGF-b inhibition failed to abrogate papilloma growth in most subjects and increased the frequency of clinically indicated interventions after treatment in four of eight subjects based on each subject’s own historical control. TGF-b neutralization consistently decreased pSMAD3 and p21 and increased Ki67 expression within the basal layers of papillomas, indicating that TGF-b restrained proliferation. These alterations were not observed in papillomas treated with PD-L1 blockade alone. Dual PD-L1/TGF-b inhibition did not enhance anti-HPV immunity within papillomas beyond that observed with PD-L1 blockade. Genomic alterations in TGF-b superfamily genes were infrequent in papillomas and normal mucosa but present in a significant fraction of head and neck carcinomas.
Conclusions
Intact TGF-b signaling restrains proliferation within papillomas, and the use of clinical agents that abrogate this pathway should be avoided in patients with RRP.
Trial registration numbers
NCT03707587 and NCT02859454.
Keywords: immunotherapy, tumor microenvironment, therapies, investigational, immunohistochemistry, head and neck neoplasms
Introduction
Transforming growth factor-beta (TGF-b) is a multifunctional cytokine that controls heterogeneous processes within distinct cell types. Within normal stratified squamous epithelia, TGF-b regulates cellular proliferation.1 In the context specific of genetic or epigenetic aberrations affecting canonical antiproliferative signaling pathways, TGF-b may promote tumor development or progression.2 3 TGF-b is potently immunosuppressive, at least through immune cell exclusion from the tumor microenvironment and direct suppression of T lymphocyte and natural killer cell cytotoxic function.4–7 Additionally, TGF-b drives epithelial-to-mesenchymal transition in malignant cells.8 Neutralization or blockade of TGF-b signaling can lead to tumor growth inhibition both through direct tumor cell effects as well as through tumor cell independent alteration of the tumor microenvironment and enhancement of antitumor immunity.9 10 Patients with relapsed high-risk human papillomavirus (HPV) associated carcinoma benefit from treatment with agents that block TGF-b, including bintrafusp alfa, a bifunctional fusion protein linking a PD-L1 monoclonal antibody (mAb) with a TGF-b trap to neutralize soluble TGF-b.11
Recurrent respiratory papillomatosis (RRP) is caused by chronic infection with low-risk HPV types 6 or 11 and manifests as papillomatous growths in the upper and sometimes lower respiratory tract that can disrupt voice and breathing. Patients with RRP benefit from PD-L1 blockade with the PD-L1 mAb avelumab, with treatment-associated reduction in disease burden and the need for fewer surgical interventions after completion of treatment.12 Given promising clinical data in patients with relapsed HPV-associated malignancy, we hypothesized that patients with RRP would further clinically benefit from dual PD-L1 and TGF-b blockade with bintrafusp alfa. Here, we report the clinical results of a phase II study of bintrafusp alfa in adult subjects with RRP. Correlative studies of papilloma tissues from patients treated with dual PD-L1/TGF-b inhibition were performed alongside papillomas from patients treated with PD-L1 blockade alone as a comparison.
Methods
Patients
Subjects aged 18 years or greater with a pathologically confirmed diagnosis of papilloma and with a clinical diagnosis of recurrent respiratory papillomatosis were eligible for both studies. Eligible subjects were required to have tracheal disease, or laryngeal disease with an anatomic Derkay score of 10 or greater, that required two or more clinically indicated interventions in the prior 12 months. Inclusion criteria included adequate organ function as measured by standard laboratory tests and an Eastern Cooperative Oncology Group performance status of 0 or 1. Exclusion criteria included evidence of infection with HIV, hepatis B or C, history of solid organ or bone marrow transplant, autoimmune disease or immunosuppression.
Study design and oversight
Both phase II studies, Avelumab for People with Recurrent Respiratory Papillomatosis (NCT 02859454) and M7824 for People with Recurrent Respiratory Papillomatosis (NCT03427411) were approved by the National Cancer Institute (NCI) Institutional Review Board, and informed consent was obtained from all subjects. Treatment consistent of intravenous avelumab (10 mg/kg) or intravenous bintrafusp alfa (1200 mg) administered every 2 weeks. On-treatment disease response was assessed by clinical endoscopic examination every 2 weeks for the duration of the study. Papilloma disease burden was quantified via calculation of an anatomic Derkay score,13 a validated objective disease measurement tool. At any point, if a subject developed clear disease progression, treatment was halted and surgical debridement of papilloma was performed. If subjects achieved a partial response after three doses, as defined by a reduction in anatomic Derkay score of 30% or greater, subjects received an additional three doses for a maximum of six doses. Adverse events were graded according to Common Terminology Criteria for Adverse Events V.4.03. Papilloma and normal mucosa biopsies for correlative studies were obtained before treatment and at the time of surgical debridement following treatment with either avelumab or bintrafusp alfa. Both avelumab and M7824 were provided by EMD Serono through a Cooperative Research and Drug Agreement with the NCI. Following completion of treatment with avelumab or bintrafusp alfa, all subjects underwent surgical debridement of papilloma with standard-of-care techniques and returned to their referring otolaryngologist to resume care. Instruction was given to each referring specialist to resume standard of care after the clinical trial, with decisions of when to proceed with a clinically indicate intervention after the trial to occur in a similar fashion as before the clinical trial. Documentation of clinically indicated interventions before and after each clinical trial was determined from medical records obtained from referring centers for each patient following consent.
Multiplex immunofluorescence
Staining
Five micron sections from formalin-fixed paraffin-embedded papilloma blocks were deparaffinized and rehydrated using the Leica Bond Rx automated system. Antigen retrieval was performed using Leica Bond citrate based or EDTA-based solutions (Leica AR9961, AR9640). The OPAL technique (Akoya Biosciences) was used for multiplex staining (online supplemental methods). Briefly, primary antibody was applied for 60 min, secondary HRP-conjugated antibody was applied for 10 min and tyramide signal amplification was used to amplify fluorescent signal for each protein target sequentially. Primary antibodies included pSMAD2S465/S467 (Bioss Antibodies #bs-3419R; 1:100 dilution), pSMAD3S423/S425 (Abcam #ab5293; 1:200), p21 (Cell Signaling Technology #2947; 1:50), Ki67 (Ventana #790–4286; 1:1) and cytokeratin (Santa Cruz #sc81714, 1:400). Secondary antibody staining was performed with ImmPRESS HRP anti-rabbit and anti-mouse IgG Polymer detection kits (Vector MP-7451; MP-7402) per manufacturer recommendations. Staining was performed with OPAL-480,–520, −570 to –620 and −690 conjugates. Individual subject pretreatment and post-treatment samples were always stained in the same batch with appropriate positive and negative controls. All individual staining and multiplex conditions were optimized on human tonsil sections.
jitc-2021-003113supp003.pdf (452.6KB, pdf)
Analysis
Images of stained slides were acquired on a Akoya Vectra Polaris using seven-color whole slide unmixing filters. Analysis of images was performed using QuPath digital pathology software.14 H-scores of separately annotated basal and suprabasal regions from entire FFPE sections were calculated using common fluorescence thresholds for each protein target for each subject’s paired pretreatment and post-treatment biopsies. Individual cell detection was based on nuclear staining with DAPI. Cytokeratin was used to distinguish epithelial from stromal or vascular core cells. H-score was calculated as the sum of % of 3+ cells plus % of 2+ cells plus % of cells 1+ (maximum score of 300). Nuclear staining was scored for Ki67, p21, pSMAD3, and pSMAD2.
TGF-b ELISA
TGF-b ELISA (R&D Systems, #DB100B) was performed on pretreatment and post-treatment peripheral blood serum per manufacturer recommendations.
Immunohistochemistry
Five micron sections from formalin-fixed paraffin-embedded papilloma blocks were deparaffinized and rehydrated using the Leica ST5020 Autostainer. Antigen retrieval was performed using a Leica Bond citrate-based solution (Leica AR9961). Single color automated staining was performed using anti-human CD4 (Abcam ab133616; 1:400 dilution) or CD8 (Abcam ab182729; 1:200) antibodies applied for 60 min. After washing, secondary antibody staining and colorimetric development was perfumed using the Leica BOND Polymer Refine Detection kit (#DS9800) per manufacturer recommendations. Coverslipping was performed with the Leica Automated Coverslipper (#CV5030). Individual subject pretreatment and post-treatment samples were always stained in the same batch with appropriate positive and negative controls. All conditions were optimized on human tonsil sections. Images of stained slides were acquired on a Vectra Polaris using brightfield illumination. Analysis of images was performed using QuPath. Percentage positivity of stained cells for entire FFPE sections was calculated using common detection thresholds for each subject’s paired pretreatment and post-treatment biopsies.
Correlative assays on papilloma infiltrating lymphocytes
T lymphocyte cultures were initiated from fresh pretreatment and post-treatment papilloma fragments using interleukin-2 containing media. Autologous B lymphocytes were isolated from the Peripheral blood mononuclear cells (PBMC) using negative magnetic selection (StemCell) and expanded in the presence of irradiated (6000 rad) NIH3T3-CD40L feeder cells (1:1 ratio) and rhIL-4 (200 U/mL). Expanded papilloma T lymphocytes were assayed for reactivity against autologous B lymphocytes (1:2 ratio) electroporated with in vitro transcribed full-length mRNA from HPV 6 or 11 E2, E6 or E7 (mMESSAGE T7 ULTRA Transcription Kit, Thermo). T lymphocyte reactivity against these products or PMA/Ionomycin (positive control) was measured via ELISpot assay (R&D Systems). Spot counts were measured on an Immunospot ELISpot reader (Cellular Technology).
Genomic analysis of normal mucosa, papillomas and The Cancer Genome Atlas (TCGA) samples
Sequencing
DNA was extracted from snap frozen papilloma, normal mucosa or cryopreserved PBMC using the QIAamp DNA Mini Kit (Qiagen #51304) per manufacturer recommendations. Fifty nanograms of genomic DNA was tagmented, enriched through two rounds of probe-based hybridization and made into whole exome libraries using the Nextera DNA Exome kit (Illumina). Equimolar library pools were sequenced on a NextSeq 500 or a HiSeq 4000 (Illumina) with either 75 or 150 base paired end read configuration.
TCGA data access
WES data from TCGA15 was downloaded following approved access to the head and neck cohort within the National Center for Biotechnology Information database of Genotypes and Phenotypes.
WES data processing
This study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Maryland, USA (http://biowulf.nih.gov). WES paired-end reads were subjected to adapter trimming using TrimGalore (https://github.com/FelixKrueger/TrimGalore). The resulting reads were aligned to the hg38 reference genome using bwa-mem2 (DOI 10.1109/IPDPS.2019.00041) with default parameters. Fixmate, sort and markdup of the SAMtools toolkit16 were used to convert SAM files to BAM format and include pairing information, sorted by genomic coordinates and mark duplicates using default parameters. Base quality scores were recalibrated using GATK,17 BaseRecalibrator and ApplyBQSR. Single nucleotide variants (SNVs) and INDELs were identified using an ensemble calling strategy.18 SNV calling was performed using four variant callers: LoFreq v2.1.5,19 MuSE v1.0rc,20 Mutect2 v4.1.9.0 (https://doi.org/10.1101/861054) and Strelka2 v2.9.1021 using default parameters unless stated otherwise. In each comparison, tumor and the corresponding control BAM files were provided as input. Before running lofreq somatic using –call-indels, input BAM files were subjected to lofreq indelqual --dindel. Strelka was run using --indelCandidates obtained from Manta v1.6.0.22 Furthermore, GATK Mutect2 calls were filtered using GATK FilterMutectCalls. Final SNV calls were retained that were detected by at least three of the four variant callers. INDELs were retained that were detected by at least two of the three variant callers LoFreq, Mutect2 and Strelka2. Common SNPs in dbsnp_146.hg38.vcf.gz of the GATK resource bundle were used as needed. Variant effect prediction was performed using VEP v101.23 The resulting VCF files were converted to MAF using vcf2maf (10.5281/zenodo.593251). Downstream analysis was performed using R Core Team 2020 (https://www.R-project.org/) and the R packages maftools24 and tidyverse (10.21105/joss.01686). Copy number analysis was performed using the R package SuperFreq25 using default parameters. For each analysis, a panel of normals consisting of up to 10 control samples was used.
Statistical analyses
The Wilcoxon matched-pairs signed-rank test was used to test for significant differences between pretreatment and post-treatment clinical and correlative outcomes. The Holm-Sidak method was used to compare multiple means over multiple timepoint to test for significant difference between Derkay scores. A p value of <0.05 was considered statistically significant.
Results
Dual PD-L1/TGF-b blockade failed to consistently limit the growth of papillomas
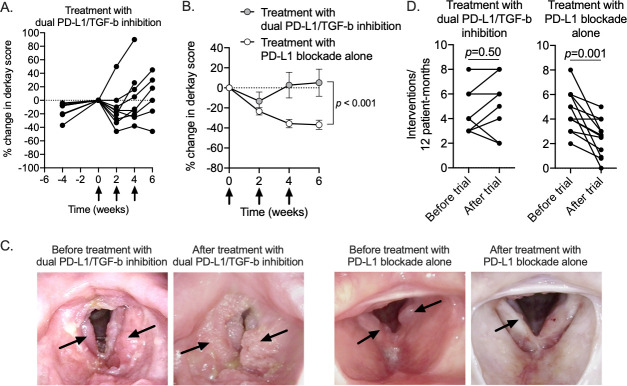
Nine subjects with RRP were treated with the dual PD-L1/TGF-b inhibitor bintrafusp alfa (online supplemental table 1). Based on frequent clinical endoscopic examination, subjects experienced initial disease regression after the first treatment, then papilloma regrowth (figure 1A). In five of nine subjects, the rate of papilloma growth from 2 to 4 or 6 weeks after treatment was greater than the rate of papilloma growth in the 4 weeks prior to starting treatment (online supplemental figure 1). A previously reported clinical trial treated a similar patient population with the PD-L1 mAb avelumab following the same treatment schedule.12 Comparing clinical endoscopy results from these two separate clinical trials revealed that treatment with dual PD-L1/TGF-b inhibition in the currently reported trial controlled the growth or papillomas to a lesser degree compared with treatment with PD-L1 blockade alone in the previously completed trial (figure 1B, C). Peripheral TGF-b levels were significantly reduced following treatment with PD-L1/TGF-b inhibition but not PD-L1 blockade alone (online supplemental figure 2). Based on a follow-up period of 1 year after completion of the trial, four of eight (50%) evaluable subjects treated with dual PD-L1/TGF-b inhibition required more frequent surgical intervention compared with the 1 year prior to the study (figure 1D). In comparison, 11 of 12 (92%) subjects treated with PD-L1 blockade alone experienced durable responses with increased intersurgery interval after completion of treatment in the previously completed trial. These data indicated that the addition of TGF-b neutralization to PD-L1 blockade abrogated the clinical benefit observed with PD-L1 blockade alone and was associated with reduced papilloma growth control in adult subjects with recurrent respiratory papillomatosis.
Figure 1.
Clinical responses following treatment with PD-L1/TGF-b inhibition or PD-L1 blockade alone in patients with recurrent respiratory papillomatosis. (A) Spider plot of change in laryngeal disease burden following treatment with dual PD-L1/TGF-b inhibition (n=9) as measured by anatomic Derkay score are shown. Negative time points on the x-axis indicate disease burden at the time of clinical trial screening. Treatment was initiated at time 0. Per cent change relative to disease burden at time 0. (B) Summary results demonstrating change in laryngeal disease burden following treatment with dual PD-L1/TGF-b inhibition (gray circles) or PD-L1 blockade alone (white circles) as previously reported. P value determined by comparing multiple means over multiple timepoints using the Holm-Sidak method. (C) Representative endoscopic still images from clinic laryngoscopy of patients treated with dual PD-L1/TGF-b inhibition or PD-L1 blockade alone. Black arrows indicate papillomatous disease. (D) Changes in the number of clinically indicated interventions per 12 patient months for patients treated with dual PD-L1/TGF-b inhibition or PD-L1 blockade alone are shown. P value determined by Wilcoxon matched-pairs signed-rank test. PD-L1, programmed death-ligand 1; TGF-b, transforming growth factor-beta.
jitc-2021-003113supp001.pdf (4.1MB, pdf)
Treatment with bintrafusp alfa was well tolerated. No subject experienced grade three or four adverse events. One subject experienced grade 2 diarrhea that quickly resolved spontaneously, and one subject experienced grade 2 dry eyes that spontaneously resolved after 6 months. Five of nine (56%) subjects treated with bintrafusp alfa developed grade 1 bleeding from the upper aerodigestive tract mucosa that resolved spontaneously within 3 months of completing treatment. Two of nine subjects (22%) developed a keratoacanthoma that resolved within 6 months of completing treatment.
TGF-b inhibition increased proliferation within basal papilloma cells
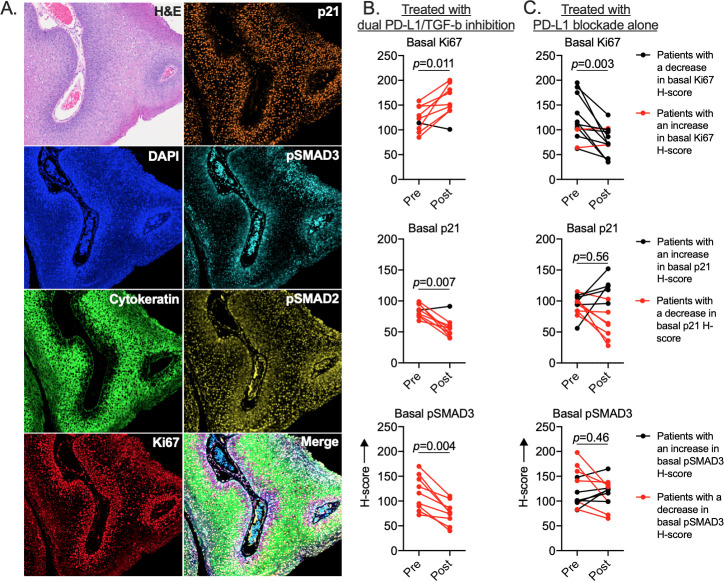
Given the discordant clinical outcomes observed with the addition of TGF-b neutralization to PD-L1 blockade in these two similar clinical studies, correlative analysis of pretreatment and post-treatment papillomas treated with dual PD-L1/TGF-b inhibition was performed. As a comparison, post hoc analysis of papillomas treated with PD-L1 blockade alone in a previous clinical trial was performed concurrently. To determine if canonical TGF-b signaling was intact within papilloma cells and possibly altered by TGF-b neutralization, changes in expression or phosphorylation of key signaling components was assessed in samples from subjects treated with dual PD-L1/TGF-b inhibition or PD-L1 blockade alone via multiplex immunofluorescence (figure 2A). Clear differences in histologic architecture, expression of TGF-b signaling components and proliferative state were evident between the basal and suprabasal layers within papillomas. Basal and suprabasal layers within each papilloma were separately annotated and digitally scored for immunofluorescence positivity and intensity (online supplemental figure 3). The basal layers of post-treatment papillomas from subjects treated with dual PD-L1/TGF-b inhibition consistently demonstrated increased expression of the proliferation marker Ki67, decreased p21 cell cycle checkpoint and decreased pSMAD3 compared with pretreatment (figure 2B). One subject with slightly decreased basal layer Ki67 also demonstrated slightly increased basal layer p21. Increased post-treatment basal Ki67 positively correlated with increased papilloma disease burden (Derkay score) in subjects treated with PD-L1 blockade with or without TGF-b neutralization (online supplemental figure 4). Basal Ki67 decreased in 10 of 12 (83%) subjects treated with PD-L1 blockade alone. No consistent changes in basal p21 or pSMAD3 were measured in these samples (figure 2C). Furthermore, no consistent changes in basal pSMAD2 or suprabasal Ki67, p21, pSMAD3 or pSMAD2 were observed in papillomas from subjects treated with or without TGF-b neutralization (online supplemental figure 5). These data indicated that TGF-b signaling is intact and may be restraining proliferation within basal layer papilloma cells and that TGF-b neutralization correlates with decreased cell cycle checkpoint expression and increased papilloma cell proliferation.
Figure 2.
Assessment of papilloma TGF-b signaling in patients treated with PD-L1/TGF-b inhibition or PD-L1 blockade alone. (A) Representative photomicrographs of papilloma tissue sections and TGF-b signaling pathway protein expression or phosphorylation measured by multiplex immunofluorescence are shown. Changes in quantification of TGF-b signaling pathway protein expression or phosphorylation within the basal layers of papillomas in patients treated with (B) dual PD-L1/TGF-b inhibition (n=9) or (C) PD-L1 blockade alone (n=12) are shown. H-scores were determined by quantifying annotated basal cell fluorescence across entire biopsy sections. P value determined by Wilcoxon matched-pairs signed-rank test. PD-L1, programmed death-ligand 1; TGF-b, transforming growth factor-beta.
jitc-2021-003113supp002.pdf (5.5MB, pdf)
Dual PD-L1/TGF-b inhibition did not enhance HPV-specific immunity beyond that observed with PD-L1 blockade alone
Immunosuppression mediated by TGF-b represents a mechanism of resistance to effective immune checkpoint blockade immunotherapy in patients with malignancy.4 5 To determine if TGF-b neutralization enhanced anti-HPV immunity within papillomas, samples from subjects treated with dual PD-L1/TGF-b inhibition or PD-L1 blockade alone were assessed for changes in T lymphocyte infiltration and specificity for HPV6 or 11. Determined by T lymphocyte IHC (online supplemental figure 6), papillomas from patients treated with dual PD-L1/TGF-b inhibition did not consistently increase CD8 or CD4 T lymphocyte infiltration into papillomas (figure 3A). No subjects harbored HPV specific T lymphocytes pretreatment, and treatment with bintrafusp alfa induced HPV immunity in one of eight (13%) evaluable subjects. In comparison, papillomas from subjects treated with PD-L1 blockade alone revealed significantly increased papilloma accumulation of CD8 T lymphocytes and a trend toward increased accumulation of CD4 T lymphocytes (figure 3B). Assessment of specificity of cultured papilloma infiltrating lymphocytes for HPV type specific E2, E6 or E7 revealed that no subjects harbored HPV specific T lymphocytes pretreatment, and treatment with avelumab induced HPV immunity in 3 of 11 (27%) evaluable subjects. These data indicated that dual PD-L1/TGF-b inhibition did not enhance HPV specific immunity beyond that observed with PD-L1 blockade alone within the papillomas of subjects with recurrent respiratory papillomatosis.
Figure 3.
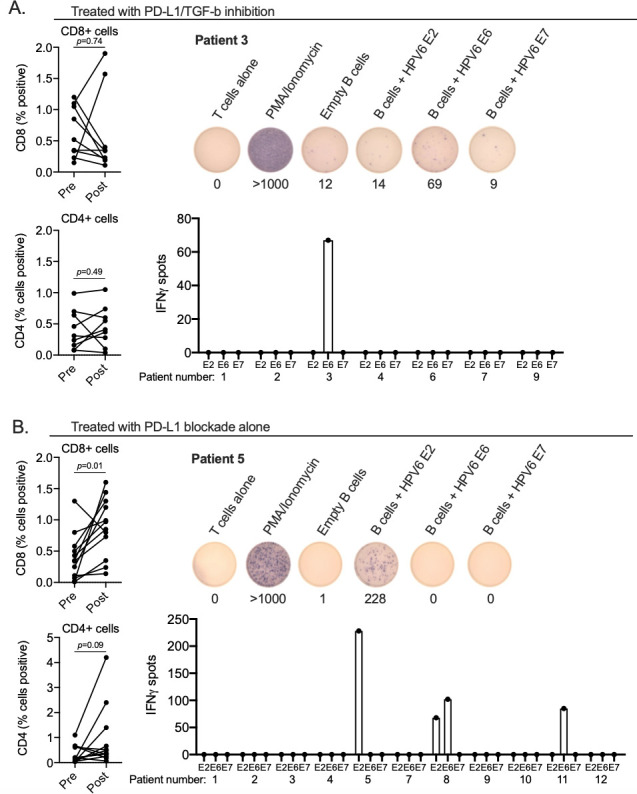
Quantification of papilloma infiltrating lymphocytes and HPV specificity. spider plots (left panels) of change in papilloma infiltration of CD8 or CD4 T-lymphocytes following treatment with (A) dual PD-L1/TGF-b inhibition (n=8 evaluable paired samples) or (B) PD-L1 blockade alone (n=12 evaluable paired samples) as measured by immunohistochemistry. Entire papilloma sections were scored for percentage of positive cells for each sample. P value determined by Wilcoxon matched-pairs signed-rank test. Right panels show quantification of the number of IFN spots following stimulation of papilloma infiltrating T-lymphocytes cultured from papillomas biopsied after treatment with dual PD-L1/TGF-b inhibition (A, n=7 evaluable cultures) or PD-L1 blockade alone (B, n=11 evaluable cultures) with autologous B-lymphocytes electroporated with full-length mRNA corresponding to HPV E2, E6 or E7 from HPV 6 or 11 as measured by ELISpot is shown. Also shown are representative ELISpot well photomicrographs from a patient treated with dual PD-L1/TGF-b inhibition (patient 3) or PD-L1 blockade alone (patient 5). HPV, human papillomavirus; PD-L1, programmed death-ligand 1; TGF-b, transforming growth factor-beta.
Genetic alterations of TGF-b signaling components were rare in papillomas but common in carcinomas
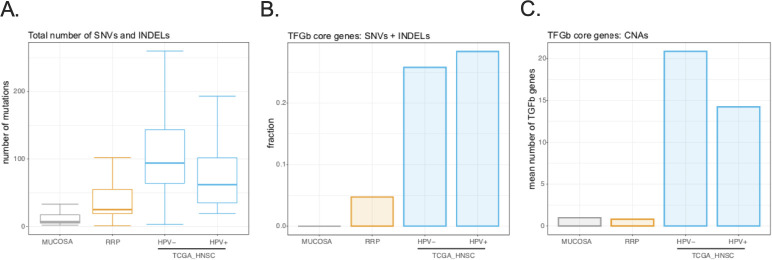
Dual PD-L1/TGF-b inhibition with bintrafusp alfa provides clinical benefit for patients with relapsed malignancy.26 Genomic alterations in TGF-b superfamily genes are frequent events in cancers,27 suggesting that genetic or epigenetic alterations in malignant cells may abrogate the antiproliferative effects of TGF-b signaling observed in non-malignant epithelial cells. We hypothesized that the rate of genomic alterations in key TGF-b superfamily genes27 would be less frequent in normal mucosa and papillomas from subjects with recurrent respiratory papillomatosis compared with head and neck squamous cell carcinoma (HNSCC). First, total somatic SNVs and small insertions and deletions (INDELs) were quantified in pretreatment papillomas (n=21) and laryngeal normal mucosa (n=8) using peripheral leukocytes as a germline reference and compared with HPV-positive HNSCC (n=74) and HPV-negative HNSCC (n=314) from TCGA.15 Both HNSCC cohorts harbored a greater number of mutations compared with mucosa or papilloma (figure 4A). Considering SNVs or INDELS, greater than 25% of all HNSCC samples harbored mutations in one or more TGF-b superfamily genes27 (online supplemental table 2) compared with infrequent mutations in these genes within normal mucosa or papilloma (figure 4B). Study of copy number alterations revealed a greater number of TGF-b superfamily genes either amplified or deleted in carcinomas compared with papillomas or mucosa (figure 4C). These data revealed that genetic alterations in TGF-b superfamily genes are infrequent events in normal mucosa or papilloma consistent with intact antiproliferative TGF-b signaling. Given the high frequency, genetic alterations in TGF-b superfamily genes may be advantageous for carcinoma cells, possibly through loss of TGF-b antiproliferative signaling.
Figure 4.
Assessment of genomic alterations in papillomas and head and neck carcinomas. (A) Boxplot shows the total number of single nucleotide variants (SNVs) and small insertions and deletions (INDELs) called from whole exome sequencing using an ensemble approach was determined for normal laryngeal mucosa (n=8), papillomas (n=21), HPV-negative HNSCC (n=74) and HPV-positive HNSCC (n=314). (B) Barplot shows the fraction of samples from each cohort harboring an SNV or INDEL in one or more of the 43 TGF-b superfamily genes was determined. (C) Barplot shows the mean number of TGF-b superfamily genes amplified or deleted within each cohort was determined. HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus; TGF-b, transforming growth factor-beta.
Discussion
Clinical and laboratory correlative data are presented from a clinical study treating adult subjects with recurrent respiratory papillomatosis with the bifunctional PD-L1 mAb/TGF-b trap compound bintrafusp alfa. Concurrent correlative analysis of papillomas from a previously reported clinical trial treating a similar population with the PD-L1 mAb avelumab was performed for comparison. This approach allowed assignment of differential clinical and correlative outcomes in this specific population of subjects to TGF-b neutralization. Subjects treated with dual PD-L1/TGF-b inhibition did not experience the degree of papilloma growth control observed with PD-L1 blockade alone, and half of the subjects treated with dual PD-L1/TGF-b inhibition need for more frequent interventions after completion of treatment compared with before treatment. Protein level analysis revealed that TGF-b neutralization inhibited basal papilloma cell specific SMAD3/p21 and increased cell proliferation as measured by Ki67, suggesting that antiproliferative TGF-b signaling is intact within papilloma cells and is reversed by TGF-b neutralization. Therapeutic agents that block TGF-b signaling should not be used clinically in patients with recurrent respiratory papillomatosis.
TGF-b plays multiple distinct roles within the complex microenvironment of epithelial neoplasms, including epithelial cell dependent and independent effects.9 Within normal skin and mucosa, TGF-b signaling results in inhibition of epithelial cell proliferation through defined mechanisms.1 During tumor progression and possibly tumorigenesis, genetic and epigenetic changes appear to lead to a switch in TGF-b signaling from antiproliferative to tumor promoting through mechanisms that appear to be cancer-type dependent.27–30 This appears to be particularly true for HPV-associated carcinomas, where genome-wide association studies have identified dysregulated TGFBR1 expression across cancers associated with high-risk HPV infection.31 Accordingly, patients with relapsed high-risk HPV-associated malignancies experience clinical benefit from treatment with bintrafusp alfa.26 32 Data presented here suggest that this is not the case in patients with neoplasms driven by infection with low-risk HPV, such as recurrent respiratory papillomatosis.
The rationale to use bintrafusp alfa in a clinical study for subjects with recurrent respiratory papillomatosis was also based on the presence of TGF-b within papillomas12 33 and the established role of TGF-b in mediating immunosuppression and resistance to immune checkpoint blockade efficacy in patients with cancer.2 4 5 11 32 Deal PD-L1/TGF-b inhibition failed to enhance HPV-specific immunity beyond that observed with PD-L1 blockade alone. It is possible that immune enhancement beyond PD-L1 blockade following TGF-b neutralization indeed occurred within papillomas but was overcome by papilloma-mediated effects resulting from increased cellular proliferation. Whether TGF-b mediates immunosuppression within the microenvironment of papillomas remains unclear. TGF-b is also known to play a role in the transition of normal fibroblasts to cancer associated fibroblasts that support the development of neoplasms.34 The role of fibroblasts exposed to TGF-b within papillomas is poorly studied, and whether neutralization of TGF-b alters the function of fibroblasts within papillomas requires further study.
Parallels can be drawn between the enhanced proliferation and growth of papillomas and the development of skin lesions such as keratoacanthomas following TGF-b neutralization. Roughly one-quarter of patients treated with agents that disrupt TGF-b signaling develop keratoacanthomas,26 35 and loss-of-function TGFBR1 mutations have been causally linked to a familial keratoacanthoma syndrome.36 The development of hyperproliferative lesions with TGF-b neutralization appears localized to skin as patients receiving such treatment do not develop mucosal lesions. This suggests that upper aerodigestive tract mucosa may harbor one or more mechanisms independent of TGF-b to limit uncontrolled proliferation and that infection with low-risk HPV may abrogate one or more of these mechanisms to sensitize papillomas to uncontrolled proliferation following TGF-b neutralization. Disruption of p53 or retinoblastoma function by E6 and E7 respectively are clear possibilities. Further study of these concepts may provide new insights into the biology of how chronic infection with low-risk HPV perturbs control of epithelial cell proliferation and differentiation within aerodigestive tract mucosa.
The lack of genomic alterations in key TGF-b signaling genes and intact TGF-b signaling within papilloma cells as measured by immunofluorescence provides a plausible explanation for why TGF-b neutralization accelerated papilloma growth. The high frequency of genomic alterations in these same TGF-b superfamily genes may help explain the clinical benefit observed with bintrafusp alfa treatment in patients with HPV-associated carcinomas.26 32 The overall outcome of TGF-b signaling disruption may reflect effects on both tumor cells and non-tumor cells of the immune microenvironment within complex heterogeneous tumors. In papillomas where epithelial compartment TGF-b signaling is intact and antiproliferative, the net result of TGF-b neutralization could be accelerated proliferation, as was observed in papillomas. In lesions such as carcinoma where TGF-b signaling is already disrupted within the malignant cell compartment due to the presence of genomic alterations, and antiproliferative function may be lost and inhibition of TGF-b may not lead to increased proliferation. In this context, effects on the immune compartment may dominate, resulting in clinical benefit. This leads to the intriguing hypothesis that the presence of one or more genomic alterations in TGF-b superfamily genes correlates with response to bintrafusp alfa in patients with relapsed HPV-associated carcinoma. This hypothesis requires further study.
In conclusion, genomically and functionally intact TGF-b signaling appears to retrain cellular proliferation within papillomas associated with low-risk HPV infection, and clinical agents that disrupt TGF-b signaling should be avoided in patients with recurrent respiratory papillomatosis. Whether or not a beneficial clinical response to bintrafusp alfa in patients with high-risk HPV-associated carcinomas is correlated with the presence of genomic alterations in TGF-b superfamily genes should be studied prospectively.
Acknowledgments
The authors would like to thank Drs Julius Strauss and Lalage Wakefield for their critical review of this manuscript. This work was supported by the Center for Cancer Research, National Cancer Institute, National Institute on Deafness and Other Communication Disorders and the Intramural Research Program of the National Institutes of Health. Bintrafusp alfa was provided by Merck KGaA, Darmstadt, Germany, as part of an alliance with GlaxoSmithKline. Merck KGaA, Darmstadt, Germany, and GlaxoSmithKline reviewed the manuscript for medical accuracy only before journal submission. The authors are fully responsible for the content of this manuscript, and the views and opinions described in the publication reflect solely those of the authors.
Footnotes
Twitter: @HsaterMD, @gulleyj1
Correction notice: This article has been corrected since it was first published. The author order has been amended and the spelling of 'bintrafusp alpha' has been corrected to 'bintrafusp alfa' throughout the article.
Contributors: YR, KB, SN, CSH and CA conceived and designed the studies. YR, JF, PEC, CS, KB, RND, JS, AS, HAS, JLG, CS, CSH and CA generated data, key reagents, and samples. YR, JF, PEC, CS, JS, HAS, JLG, CSH and CA analyzed and interpreted the data. YR, JF, PEC, CS, KB, RND, JS, AS, HAS, JLG, SC, CSH and CA wrote and revised the manuscript. All authors approved the final version of the manuscript.
Funding: Center for Cancer Research, National Cancer Institute, and the Intramural Research Program of the National Institute on Deafness and Other Communication Disorders, National Institutes of Health.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. All data relevant to the study are included in the article or uploaded as supplementary information. All experimental data associated with this work is included in the main report and supplements. Whole exome sequencing data from both clinical trials will be deposited in DBGaP according to the protocol data plans.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
Both phase II studies, Avelumab for People with Recurrent Respiratory Papillomatosis (NCT 02859454) and M7824 for People with Recurrent Respiratory Papillomatosis (NCT03427411) were approved by the National Cancer Institute Institutional Review Board, and informed consent was obtained from all subjects.
References
- 1.Zhang Y, Alexander PB, Wang X-F. TGF-β Family Signaling in the Control of Cell Proliferation and Survival. Cold Spring Harb Perspect Biol 2017;9:a022145. 10.1101/cshperspect.a022145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity 2019;50:924–40. 10.1016/j.immuni.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakefield LM, Stuelten C. Keeping order in the neighborhood: new roles for TGFbeta in maintaining epithelial homeostasis. Cancer Cell 2007;12:293–5. 10.1016/j.ccr.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 4.Mariathasan S, Turley SJ, Nickles D, et al. Tgfβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tauriello DVF, Palomo-Ponce S, Stork D, et al. Tgfβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554:538–43. 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- 6.Thomas DA, Massagué J. Tgf-Beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 2005;8:369–80. 10.1016/j.ccr.2005.10.012 [DOI] [PubMed] [Google Scholar]
- 7.Viel S, Marçais A, Guimaraes FS-F, et al. TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci Signal 2016;9:ra19. 10.1126/scisignal.aad1884 [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Tian X-J, Zhang H, et al. TGF-β-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci Signal 2014;7:ra91. 10.1126/scisignal.2005304 [DOI] [PubMed] [Google Scholar]
- 9.Canè S, Van Snick J, Uyttenhove C, et al. Tgfβ1 neutralization displays therapeutic efficacy through both an immunomodulatory and a non-immune tumor-intrinsic mechanism. J Immunother Cancer 2021;9:e001798. 10.1136/jitc-2020-001798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmgaard RB, Schaer DA, Li Y, et al. Targeting the TGFβ pathway with galunisertib, a TGFβRI small molecule inhibitor, promotes anti-tumor immunity leading to durable, complete responses, as monotherapy and in combination with checkpoint blockade. J Immunother Cancer 2018;6:47. 10.1186/s40425-018-0356-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan Y, Zhang D, Xu C, et al. Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci Transl Med 2018;10. 10.1126/scitranslmed.aan5488. [Epub ahead of print: 17 01 2018]. [DOI] [PubMed] [Google Scholar]
- 12.Allen CT, Lee S, Norberg SM, et al. Safety and clinical activity of PD-L1 blockade in patients with aggressive recurrent respiratory papillomatosis. J Immunother Cancer 2019;7:119. 10.1186/s40425-019-0603-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hester RP, Derkay CS, Burke BL, et al. Reliability of a staging assessment system for recurrent respiratory papillomatosis. Int J Pediatr Otorhinolaryngol 2003;67:505–9. 10.1016/S0165-5876(03)00007-7 [DOI] [PubMed] [Google Scholar]
- 14.Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. 10.1038/s41598-017-17204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Network . Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Handsaker B, Wysoker A. Genome project data processing S: the sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M, Luo W, Jones K, et al. SomaticCombiner: improving the performance of somatic variant calling based on evaluation tests and a consensus approach. Sci Rep 2020;10:12898. 10.1038/s41598-020-69772-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilm A, Aw PPK, Bertrand D, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 2012;40:11189–201. 10.1093/nar/gks918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Y, Xi L, Hughes DST, et al. Muse: accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome Biol 2016;17:178. 10.1186/s13059-016-1029-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Scheffler K, Halpern AL, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 2018;15:591–4. 10.1038/s41592-018-0051-x [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016;32:1220–2. 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- 23.McLaren W, Gil L, Hunt SE, et al. The Ensembl variant effect predictor. Genome Biol 2016;17:122. 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayakonda A, Lin D-C, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747–56. 10.1101/gr.239244.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flensburg C, Sargeant T, Oshlack A, et al. SuperFreq: integrated mutation detection and clonal tracking in cancer. PLoS Comput Biol 2020;16:e1007603. 10.1371/journal.pcbi.1007603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strauss J, Gatti-Mays ME, Cho BC, et al. Bintrafusp alfa, a bifunctional fusion protein targeting TGF-β and PD-L1, in patients with human papillomavirus-associated malignancies. J Immunother Cancer 2020;8:e001395. 10.1136/jitc-2020-001395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korkut A, Zaidi S, Kanchi RS, et al. A pan-cancer analysis reveals high-frequency genetic alterations in mediators of signaling by the TGF-β superfamily. Cell Syst 2018;7:e427:422–37. 10.1016/j.cels.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Yang HH, Tang B, et al. The Outcome of TGFβ Antagonism in Metastatic Breast Cancer Models In Vivo Reflects a Complex Balance between Tumor-Suppressive and Proprogression Activities of TGFβ. Clin Cancer Res 2020;26:643–56. 10.1158/1078-0432.CCR-19-2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeh H-W, Lee S-S, Chang C-Y, et al. A new switch for TGFβ in cancer. Cancer Res 2019;79:3797–805. 10.1158/0008-5472.CAN-18-2019 [DOI] [PubMed] [Google Scholar]
- 30.Cohen J, Chen Z, Lu S-L, et al. Attenuated transforming growth factor beta signaling promotes nuclear factor-kappaB activation in head and neck cancer. Cancer Res 2009;69:3415–24. 10.1158/0008-5472.CAN-08-3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levovitz C, Chen D, Ivansson E, et al. Tgfβ receptor 1: an immune susceptibility gene in HPV-associated cancer. Cancer Res 2014;74:6833–44. 10.1158/0008-5472.CAN-14-0602-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho BC, Daste A, Ravaud A, et al. Bintrafusp alfa, a bifunctional fusion protein targeting TGF-β and PD-L1, in advanced squamous cell carcinoma of the head and neck: results from a phase I cohort. J Immunother Cancer 2020;8:e000664. 10.1136/jitc-2020-000664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatam LJ, Devoti JA, Rosenthal DW, et al. Immune suppression in premalignant respiratory papillomas: enriched functional CD4+Foxp3+ regulatory T cells and PD-1/PD-L1/L2 expression. Clin Cancer Res 2012;18:1925–35. 10.1158/1078-0432.CCR-11-2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, Zhou L, Li D, et al. Cancer-Associated fibroblasts build and secure the tumor microenvironment. Front Cell Dev Biol 2019;7:60. 10.3389/fcell.2019.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacouture ME, Morris JC, Lawrence DP, et al. Cutaneous keratoacanthomas/squamous cell carcinomas associated with neutralization of transforming growth factor β by the monoclonal antibody fresolimumab (GC1008). Cancer Immunol Immunother 2015;64:437–46. 10.1007/s00262-015-1653-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goudie DR, D'Alessandro M, Merriman B, et al. Multiple self-healing squamous epithelioma is caused by a disease-specific spectrum of mutations in TGFBR1. Nat Genet 2011;43:365–9. 10.1038/ng.780 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003113supp003.pdf (452.6KB, pdf)
jitc-2021-003113supp001.pdf (4.1MB, pdf)
jitc-2021-003113supp002.pdf (5.5MB, pdf)
Data Availability Statement
Data are available in a public, open access repository. All data relevant to the study are included in the article or uploaded as supplementary information. All experimental data associated with this work is included in the main report and supplements. Whole exome sequencing data from both clinical trials will be deposited in DBGaP according to the protocol data plans.