Abstract
Background
Increasing infiltration of CD8+ T cells within tumor tissue predicts a better prognosis and is essential for response to checkpoint blocking therapy. Furthermore, current clinical protocols use unfractioned T cell populations as the starting point for transduction of chimeric antigen receptors (CARs)-modified T cells, but the optimal T cell subtype of CAR-modified T cells remains unclear. Thus, accurately identifying a group of cytotoxic T lymphocytes with high antitumor efficacy is imperative. Inspired by the theory of yin and yang, we explored a subset of CD8+ T cell in cancer with the same phenotypic characteristics as highly activated inflammatory T cells in autoimmune diseases.
Methods
Combination of single-cell RNA sequencing, general transcriptome sequencing data and multiparametric cytometric techniques allowed us to map CXCR6 expression on specific cell type and tissue. We applied Cxcr6−/− mice, immune checkpoint therapies and bone marrow chimeras to identify the function of CXCR6+CD8+ T cells. Transgenic Cxcr6−/− OT-I mice were employed to explore the functional role of CXCR6 in antigen-specific antitumor response.
Results
We identified that CXCR6 was exclusively expressed on intratumoral CD8+ T cell. CXCR6+CD8+ T cells were more immunocompetent, and chimeras with specific deficiency on CD8+ T cells showed weaker antitumor activity. In addition, Cxcr6−/− mice could not respond to anti-PD-1 treatment effectively. High tumor expression of CXCR6 was not mainly caused by ligand-receptor chemotaxis of CXCL16/CXCR6 but induced by tumor tissue self. Induced CXCR6+CD8+ T cells possessed tumor antigen specificity and could enhance the effect of anti-PD-1 blockade to retard tumor progression.
Conclusions
This study may contribute to the rational design of combined immunotherapy. Alternatively, CXCR6 may be used as a biomarker for effective CD8+ T cell state before adoptive cell therapy, providing a basis for tumor immunotherapy.
Keywords: CD8-positive T-lymphocytes, gastrointestinal neoplasms, immunotherapy, adoptive, lymphocytes, tumor-infiltrating, receptors, immunologic
Background
CD8+ T cells are the main immune surveillance cells that can detect antigens derived from developing malignant cells.1 2 High infiltration of CD8+ T cells in cancer predicts a good prognosis, which is also a key factor in response to checkpoint blocking therapy.3–5 However, the transient changes in the rejuvenation of effector cells and the widespread occurrence of recurrences together indicate the defect of persistent immune memory after checkpoint blockade. At present, no immunotherapeutic strategy that can reliably establish a stable function memory T cell bank that can prevent recurrence has been designed. This indicates that there is a lack of deeper research in tumorigenesis and antitumor T cell response disorders.
Compared with other forms of cancer immunotherapy, adoptive cell therapy (ACT) relies on the active development of a sufficient number of antitumor T cells in the body, which have necessary functions to mediate cancer regression. ACT enables the host to manipulate before cell transfer to provide a favorable microenvironment, thereby better supporting antitumor immunity.6 The clinical success of chimeric antigen receptors (CARs) targeting CD19 has been approved by the US Food and Drug Administration and the European Medical Agency and has triggered extensive investment in CAR T cell research by academia and private companies.7 As a monotherapy, CAR T cells have limited efficacy, partly due to the redundant regulatory mechanisms inherent in all T cells and our incomprehensive understanding of which features for T cells are essential and has antitumor activity.8 Current clinical protocols use unfractioned T cell populations as the starting point for transduction of CAR-modified T cells, but few studies investigate whether the optimal T cell subtype should be chosen when generating CAR T cells. Therefore, many key questions have been raised: which cell states are related to ongoing tumor-specific T cell responses? How does current immunotherapy affect these different T cell states? How does the presence of a single T cell state predict the response to antitumor therapy?
C-X-C motif chemokine receptor 6 (CXCR6) is preferentially expressed on memory T cells and activated Th1 and Tc1 effector T cell subsets.9 10 Its unique ligand, CXCL16, is expressed on the surface of antigen presenting cells (B cells, macrophages, and dendritic cells).11 CXCR6/CXCL16 are overexpressed in many cancers. It has been reported that both CXCR6 and CXCL16 are overexpressed in breast cancer tissues and cell lines12 and pancreatic ductal adenocarcinoma.13 Mossanen et al14 propose that CXCR6 inhibits the occurrence of liver cancer by promoting natural killer T and CD4+ T cell dependent senescence control. Intranasal vaccination with a cancer vaccine is less effective in Cxcr6−/− mice than wild-type mice.15 Much about the function of CXCR6 in tumors still need to be further studied. For example, whether CXCR6 is related to the prognosis of cancer patients, how environmental factors regulate CXCR6 positive cells and more research is needed to confirm the role of CXCR6 in tumor development. Interestingly, Hou et al16 reveals that in multiple sclerosis (MS) and mouse experimental encephalomyelitis (EAE) models, CXCR6 identifies a subcluster of T cells with inflammatory cytokine secretion, a cytolytic system, and extremely rapid proliferation. Therefore, we wonder whether there still exist the same phenotypic immunocompetent T cells in tumors according to the phenotypic characteristics of highly activated inflammatory T cells in autoimmune diseases.
To address this question, this study comprehensively combined the data of single-cell RNA sequencing, spatial transcriptome sequencing, immune checkpoint therapy, bone marrow chimeric mouse models, and (ovalbumin) OVA-specific activation system to prove that CXCR6 is specifically and highly expressed in intratumoral CD8+ T cell. CXCR6+CD8+ T cells were more immunocompetent and more active in response to checkpoint blocking therapy than negative counterpart. This may contribute to the rational design of combined immunotherapy. In addition, high tumor expression of CXCR6 is not primarily caused by ligand-receptor chemotaxis of CXCL16/CXCR6 but induced by tumor tissue self. Induced CXCR6+CD8+ T cells were tumor antigen specific and can retard the progression of tumors. Thus, CXCR6 may be used as a biomarker for effective CD8+ T cell before ACT.
Methods
Mice
Female C57BL/6 wild-type mice and Rag1−/− mice were purchased from GemPharmatech. CXCR6gfp/gfp mice (Stock No. 005693), homozygous CXCR6-deficient mice, were purchased from the Jackson Laboratory. Cd8−/− mice were obtained from Professor Lilin Ye (Third Military Medical University). B6.SJL-Ptprca Pepcb /Boy (CD45.1) were obtained from Professor Xuetao Cao (Nankai University). CXCR6gfp/gfp OT-I mice were obtained by crossing OT-I TCR transgenic mice with CXCR6gfp/gfp mice. Mice were used in experiments at 6–8 weeks of age. All mice were housed in animal facility under specific pathogen-free conditions.
Generation of chimeras
For CD8Cxcr6−/− mice, total bone marrow cells from Cd8−/− mice and from Cxcr6−/− or wild-type mice were mixed and adoptively transferred intravenously at a 7:3 ratio into lethally irradiated (9 Gry) naive Cd8−/− mice. A total of 5×106 bone marrow cells were transferred per mouse. For Cxcr6+/+:Cxcr6−/− chimeras, bone marrow cells from WT (CD45.2) or Cxcr6gfp/gfp (CD45.2) and bone marrow cells from C57BL/6 (CD45.1) mice were mixed and adoptively transferred intravenously at a 1:2 ratio into lethally irradiated Rag1−/− mice. Recipient mice were fed antibiotics for 2 weeks and allowed to reconstitute for at least 8 weeks before challenged with tumor cells.
Tumor cell lines
MC38 colon cancer cell line was obtained from Cell Resource Center of the Institutes of Biomedical 387 Sciences at Fudan University (Shanghai, China). CT26.WT colon cancer and B16F10 melanoma cell lines were purchased from American Type Culture Collection. B16 OVA melanoma cell line was kindly provided by Shengdian Wang (Institute of Biophysics, Chinese Academy of Sciences). All cell lines were cultured in Dulbecco’s modified Eagle medium or RPMI-1640 medium supplemented with 10% fetal bovine serum, 2 mmol/L glutamax, 100 U/mL penicillin, and 100 mg/mL streptomycin.
Immunofluorescence staining
Paraffin-embedded slides of samples were deparaffinized, rehydrated, antigen retrieved with sodium citrate and blocked with 5% bovine albumin. Primary antibodies used to stain the sections included: rabbit antimouse CD8 (Abcam Cat# ab217344, RRID:AB_2890649), goat antimouse CXCR6 (Abcam Cat# ab125115, RRID:AB_10974584), mouse antihuman CD8 (Cell Signaling Technology Cat# 70306, RRID:AB_2799781), rabbit antihuman CXCR6 (Novus Cat# NLS1102, RRID:AB_10000951), goat antimouse CXCL16 (R&D, Cat# AF503-SP), goat antirabbit IgG (H+L) Alexa Fluor 647 (Thermo Fisher Scientific Cat# A-21244, RRID:AB_2535812), goat antirabbit IgG (H+L) Alexa Fluor 488 (Thermo Fisher Scientific Cat# A-11008, RRID:AB_143165), goat antimouse IgG (H+L) Alexa Fluor 594 (Thermo Fisher Scientific Cat# A-11005, RRID:AB_2534073) and donkey antigoat Alexa Fluor 647 (Thermo Fisher Scientific Cat# A32849TR, RRID:AB_2866498).
Flow cytometric analysis
Flow cytometry was performed on Attune NxT Flow Cytometer and analyzed using FlowJo software. Cell-sorting experiments were performed on BD Aria II. Prepared single cells were stained by fixable viability stain reagents (BD Biosciences Cat# 565388, RRID: AB_2869673). Staining was performed at 4°C in the presence of FACS buffer (PBS, 0.5% BSA, 2 mM EDTA, and 0.1% sodium azide) after Fc block (BD Biosciences Cat# 558636, RRID: AB_1645217). In the case of intracellular cytokine staining, cell stimulation cocktail (plus protein transport inhibitors) (500×) (eBioscience Cat# 00-4975-93) was added for 4 hours before staining with the fixation/permeabilization solution kit (BD Biosciences Cat# 554714, RRID: AB_2869008). Antibodies used for staining were: antimouse IFN-γ-PE (Thermo Fisher Scientific Cat# 12-7311-81, RRID:AB_466192), antimouse IFN-γ-BV421 (BD Biosciences Cat# 563376, RRID: AB_2738165), antimouse Perforin-APC (Thermo Fisher Scientific Cat# 17-9392-80, RRID:AB_469514), antimouse Granzyme B-AF647 (BioLegend Cat# 515405, RRID:AB_2294995), antimouse CXCR6-BV421 (BioLegend Cat# 151109, RRID:AB_2616760), antimouse CXCR6-PE (BioLegend Cat# 151103, RRID:AB_2566545), antimouse CD8a-BV605 (BD Biosciences Cat# 563152, RRID:AB_2738030), antimouse CD45-AF700 (Thermo Fisher Scientific Cat# 56-0451-82, RRID:AB_891454), antimouse CD3-AF488 (BioLegend Cat# 100210, RRID:AB_389301), antimouse TNF-α-BV711 (BioLegend Cat# 502939, RRID:AB_2562740), antimouse CD8a-PE-Cy7 (BD Biosciences Cat# 552877, RRID:AB_394506), and antimouse NK1.1-PE-Cy7 (Thermo Fisher Scientific Cat# 25-5941-81, RRID:AB_469664)
Induction of CXCR6 expression
For induction in vivo, naïve T cells from OT-I mice were transferred intravenously into mice challenged with B16OVA tumor cells in advance. OT-I+CXCR6+CD8+ T cells were detected on days 2, 4 and 7, respectively. For induction in vitro by tumor cells, MC38 tumor cells and naïve T cells were mixed at a ratio of 1:100 and plated in 24-well plates. Recombinant murine 10 ng/mL IL-2 (Peprotech Cat# 212–12) and 5 µg/mL CD3 (Thermo Fisher Scientific Cat# 16-0032-38, RRID: AB_2865578) and 5 µg/mL CD28 (Thermo Fisher Scientific Cat# 16-0281-81, RRID: AB_468920) functional antibodies were added. For induction in vitro by tumor tissues, shredded tumor tissues were in upper wells and naïve T cells in lower wells with the presence of IL-2 recombinant cytokine and CD3/CD28 functional antibodies. The application of transwell was to eliminate the interference of CXCR6+ T cells that already existed in tumor tissues. In the experiment of ACT, the chamber was withdrawn when large batches of CXCR6+ T cells were induced and cultured.
Western blot
Tumor tissues were lysed in radio immunoprecipitation assay buffer supplemented with protease and phosphatase inhibitor (MedChemExpress). Proteins were quantified by the Pierce BCA Protein Assay Kit (ThermoFisher Scientific, Cat# 23227). The proteins were then separated by SDS-polyacrylamide gel electrophoresis and electrophoretically transferred onto polyvinylidene difluoride membranes. The membranes were probed with antibodies overnight at 4°C, and then incubated with a horseradish peroxidase-coupled secondary antibody. Detection was performed using a LumiGLO chemiluminescent substrate system. Antibodies were: goat antimouse CXCL16 (R&D, Cat# AF503-SP) and rabbit anti-β-actin (Abmart, Cat# M20011).
Tumor model and in vivo treatments
For anti-PD-1 treatment, MC38 cells (1×106) or B16F10 cells (2×105) were injected subcutaneously into female C57BL/6 mice at 6–8 weeks old. On day 10, tumor-bearing mice with similar size were randomly divided into two groups (n=8–10). Mice were treated with 200 µg isotype (Bio X Cell Cat# BE0089, RRID: AB_1107769) or anti-PD-1 antibody (Bio X Cell Cat# BE0146, RRID: AB_10949053) intraperitoneally every 3 days. For combination therapy strategy, the administration dose was reduced to 100 µg. For CXCR6+ cell deletion experiment, rabbit antimouse CXCR6 monoclonal antibody (Edelweiss Immune, clone: 19A5) or isotype was administrated 1 day before MC38 cells (1×106) were injected subcutaneously. Mice were treated with 200 µg anti-CXCR6 or isotype intraperitoneally every 3 days. Tumor volume was analyzed by V=1/2 (a × b2).
Adoptive transfer of CD8+ T cells
For CXCR6+CD8+ T cells induced in vitro transfer, MC38 cells (1×106) were injected subcutaneously into female C57BL/6 mice at age of 6–8 weeks. On day 7, 5×106 induced CXCR6+CD8+ T cells or CXCR6-CD8+ T isolated by FACS were transferred intravenously into tumor load mice. For OT-I CTL transfer, splenocytes isolated from Cxcr6−/− OT-I mice or wild-type OT-I mice were stimulated with 2 nM OVA257–264 (SIINFEKL) for 3 days with the presence of 10 ng/mL IL-2. Cells were centrifuged and cultured in fresh medium containing 10 ng/mL IL-2 for two more days, then the cells in the culture were CTLs. 5×106 Cxcr6−/− OT-I CTLs or WT OT-I CTLs were injected intravenously into B16OVA bearing mice.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (GraphPad Software). Statistical significance was determined as indicated in the figure legends. Data are generally shown as mean±SEM unless otherwise stated. P<0.05 was considered significant: *p<0.05; **p<0.01; ***p<0.001. All t-test analyses are two-tailed unpaired t-tests. Statistical significance in multiple comparison determined by one-way analysis of variance with Tukey’s multiple comparison test.
Results
CXCR6 is specifically and highly expressed on CD8+ T cells in tumor tissues
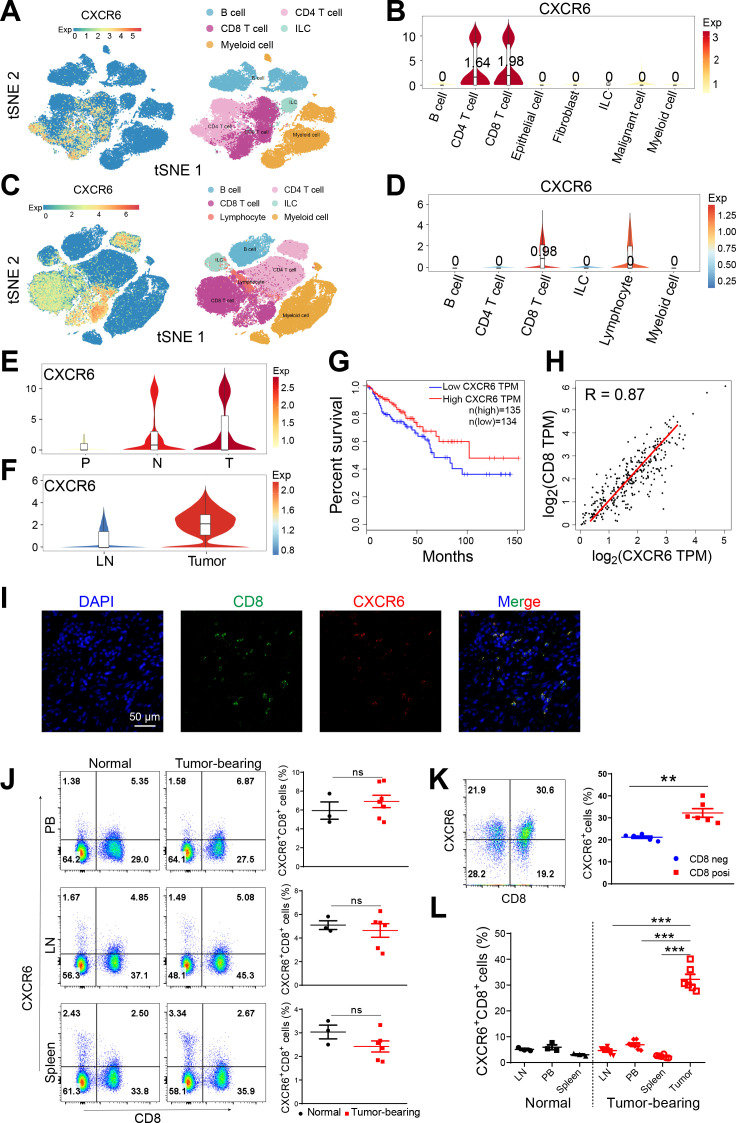
To understand the expression atlas of CXCR6 in tumor, we first analyzed the single cell RNA sequencing data of colon cancer from patients and mice reported by Zhang et al.17 We found that CXCR6 was mainly expressed on T cells, especially on CD8+ T cells (figure 1A–D). Compared with peripheral tissues like peripheral blood (P), lymph node (LN) and adjacent normal (N) tissues, CXCR6 was significantly enriched in tumors (T) (figure 1E, F). Spatial transcriptome sequencing of clinical samples we performed previously also showed that the expression of CXCR6 in tumors was higher than that in normal tissues (data not shown). To verify the single cell RNA sequencing results, we also analyzed transcriptome sequencing datasets in TCGA and identified an increased tumor expression of CXCR6 similarly (online supplemental figure S1A). In addition, high expression of CXCR6 in tumor showed a good prognosis and positively correlated with the expression of CD8 (figure 1G, H, online supplemental figure S1C, D).
Figure 1.
CXCR6 is highly expressed on CD8+ T cells in tumor tissues. (A) t-SNE plot showing immune cell clusters (right) and expression of CXCR6 (left) from human CRC. (B) Violin plot showing expression of CXCR6 on cell clusters from human CRC. (C) t-SNE plot showing immune cell clusters (right) and expression of CXCR6 (left) from MC38 mouse model. (D) Violin plot showing expression of CXCR6 on immune cell clusters from MC38 mouse model. (E) Violin plot showing expression of CXCR6 in PBMCs (P), tumor (T) and adjacent normal (N) tissues from patients. (F) Violin plot showing expression of CXCR6 in lymph node (LN) and tumor (T) tissues from MC38 mouse model. (G and H) Kaplan-Meier survival curves comparing the high and low expression of CXCR6 (G) and scatterplots of correlation between CXCR6 and CD8a expression (H) in TCGA database determined by gene expression profiling interactive analysis website (GEPIA, http://gepia.cancer-pku.cn/). (I) Immunofluorescence staining showing DAPI (blue)/CD8 (green)/CXCR6 (red) and their overlay in tumor tissues from human CRC samples. (J–L) Representative plots and frequencies of CXCR6+CD8+ T cells in peripheral blood (PB), lymph node (LN), spleen and tumor from normal or MC38 tumor-bearing mice (n=3–7). Error bars represent SEM. **P<0.01 and ***p<0.001. Single-cell RNA sequencing data were analyzed on website http://crcleukocytecancer-pkucn/. CXCR6, C-X-C motif chemokine receptor 6; NS, no significance.
jitc-2021-003100supp001.pdf (1.2MB, pdf)
We validated our findings using immunofluorescence staining in colon cancer slides, which exhibited high colocalization of CXCR6 and CD8 (figure 1I, online supplemental figure S1B). Next, we detected the expression profile of CXCR6 in mice from peripheral blood (PB), LN, spleen and tumor tissues in MC38 and CT26 mouse model (figure 1J, K, online supplemental figure S1E–G). We observed that CXCR6 almost exclusively presented in tumor (figure 1L). Paust et al18 proposed that CXCR6 expressing NK cell could mediate antigen-specific memory of haptens and viruses. Hence, we examined the expression of CXCR6 on NK cell. As shown in online supplemental figure S1H, intratumoral NK cells rarely expressed CXCR6. Taken together, we identified that CXCR6 was predominantly expressed on CD8+ T cell in tumor tissue.
CXCR6+CD8+T cells are more immunocompetent
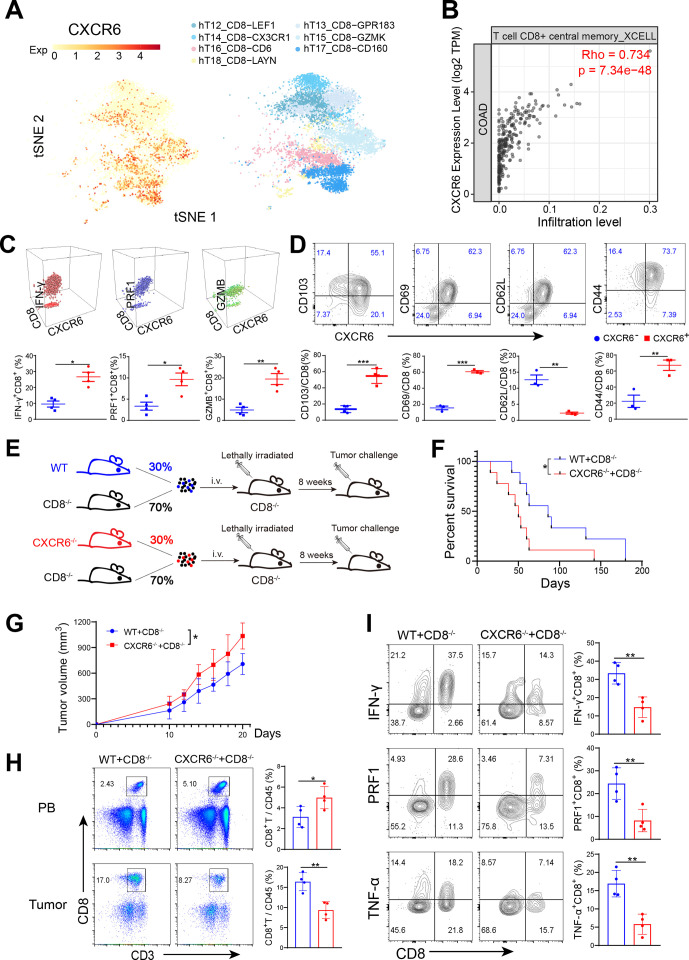
To characterize CXCR6+CD8+ T cell, we mapped CXCR6 expression on the CD8+ T cell subclusters using the single-cell sequencing data. We revealed that CXCR6 was mainly enriched in cytotoxic CD8+ T cell populations such as CD6+CD8+ T, GZMK+CD8+ T and CD160+CD8+ T cells (figure 2A). Furthermore, there was a strong correlation between the expression of CXCR6 and the infiltration level of central memory CD8+ T (TCM) (figure 2B). CXCR6+CD8+ T cells in tumor also expressed cytotoxic and activation markers such as IFN-γ, PRF1, GZMB, CD69 and CD44 but were negative for CD62L, a maker naïve T cell highly expressed (figure 2C, D, online supplemental figure S2A). These phenotypic characteristics indicated that CXCR6+CD8+ T cell subset possessed a higher degree of activation and killing ability.
Figure 2.
CXCR6-deficient CD8+ T cells show weaker antitumor activity. (A) t-SNE plot showing CD8+ T cell clusters (right) and expression of CXCR6 (left) from human CRC. (B) Scatterplots showing correlation between CXCR6 expression and CD8+ T cell infiltration level in COAD in TCGA database determined by tumor immune estimation resource (timer, http://cistrome.dfci.harvard.edu/TIMER/) website. (C and D) Phenotypic characterization of CXCR6+CD8+ T and CXCR6−CD8+ T cells from MC38 tumors. (E) Set-up of bone marrow chimeric mice. Total bone marrow obtained from Cxcr6−/− or wild-type mice were mixed with that obtained from Cd8−/− mice and then transferred to lethally irradiated Cd8−/− recipients. Eight weeks later, challenged chimeras with MC38 tumor cells. (F and G). survival (F) and tumor growth (G) of tumor load chimeric mice (n=9–10 mice per group). (H) Quantification of CD8+ T cells from tumors. (I) Frequencies of cytokine-producing CD8+ T cells. *P<0.05 and **p<0.01. CXCR6, C-X-C motif chemokine receptor 6.
To investigate whether CXCR6 expression on intratumoral CD8+ T cells was required for their antitumor efficacy, we constructed bone marrow chimeric mice in which CD8+ T cells were deficient for CXCR6 (named Cd8CXCR6−/− mice), while in most of the other immune cells, CXCR6 expression was intact (figure 2E, online supplemental figure S2B, C). After tumor cells inoculation, survivals were reduced, and tumor growth aggravated significantly in Cd8CXCR6−/− mice (figure 2F, G). Compared with mice reconstituted with wild-type and Cd8−/− mice BM, the percentage of intratumoral CD8+ T cell in recipients reconstituted with BM derived from Cxcr6−/− and Cd8−/− mice was only half. This suggested that CXCR6 was required for CD8+ T cells infiltration into tumor (figure 2H). Strikingly, when we evaluated the function of infiltrating CD8+ T cells, we found that CXCR6-deficient CD8+ T cells exhibited lower IFN-γ, PRF1 and TNF-α secretion than wild-type counterparts (figure 2I, online supplemental figure S2D). We applied anti-CXCR6 monoclonal antibody to delete CXCR6+CD8+ cells in vivo (online supplemental figure S2E) and found tumor progression aggravated (online supplemental figure S2F). These data demonstrated CXCR6 was crucial for CD8+ T cells to enrich in tumor tissues and exert antitumor function.
CXCR6 is essential for effective response to immune checkpoint therapy
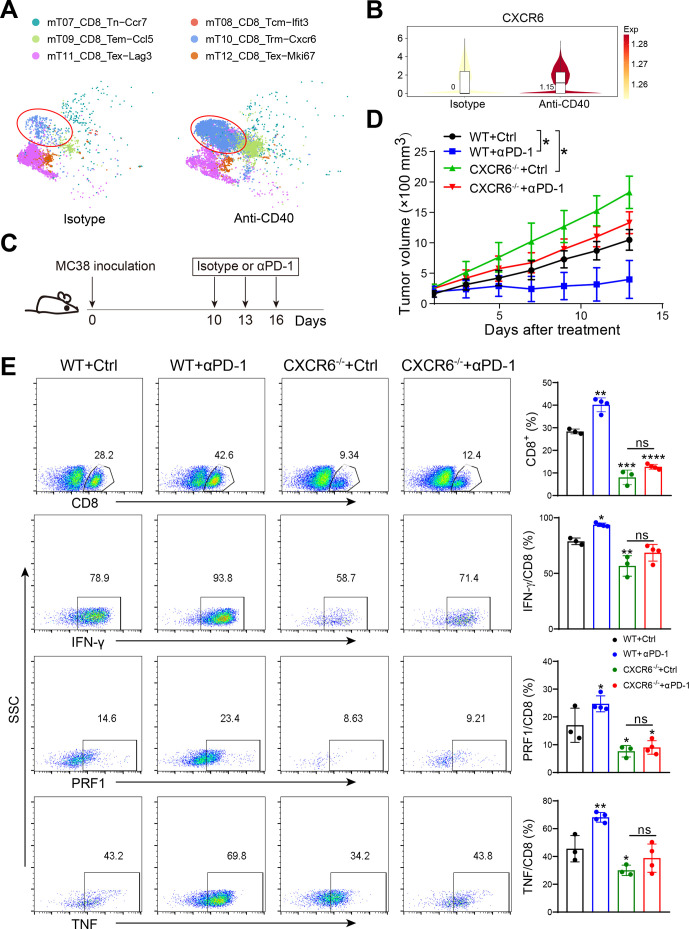
Given that after anti-CD40 treatment the CXCR6+CD8+ T cell subpopulation was significantly increased (figure 3A, B), we speculated that CXCR6+CD8+ T cells might be the main CD8+ T cell subset that responded to immune checkpoint therapy. To validate our hypothesis, we used an anti-PD-1-responsive transplantable tumor model: the MC38 tumor cell line19 20 (figure 3C). We observed that in vivo recall responses to anti-PD-1 therapy in Cxcr6−/− mice were markedly compromised (figure 3D). The percentages of intratumoral CD8+ T cells and cytokines production were rapidly decreased in Cxcr6−/− mice, even treated with PD-1 blockade therapy (figure 3E, online supplemental figure S3A). To determine whether the effects of CXCR6 expression could be extended to other cancers, we also examined the effect of anti-PD-1 treatment strategy in the melanoma model. A similar result was observed that anti-PD-1 treatment significantly increased CXCR6 expression on infiltrating CD8+ T cells (online supplemental figure S3B). Moreover, intracellular cytokine staining showed an increase in CD8+ T cells expressing GZMB and IFN-γ, especially in CXCR6+CD8+ T cells, which had a greater variation (online supplemental figure S3C). Taken together, CXCR6 played an essential role in mediating the efficacy of immune checkpoint therapy and CXCR6+CD8+ T cell was the main subset response to this treatment strategy.
Figure 3.
In vivo recall responses to anti-PD-1 in Cxcr6−/− mice are markedly compromised. (A) t-SNE plot showing distribution of T cell subsets before (left) or after (right) treatment with isotype control or anti-CD40 agonist. (B) Violin plot showing the expression of CXCR6 after treatment with isotype control or anti-CD40 agonist. (C) Schematic diagram of the anti-PD-1 treatment schedule. Mice were inoculated subcutaneously with MC38 tumor cells and were intraperitoneally treated with 200 µg of isotype control or anti-PD-1 antibodies on days 10/13/16 after tumor inoculation. (D) Tumor growth (n=6–9). (E) Proportions of intratumoral CD8+ T cells and cytokines produced was quantified from WT and Cxcr6−/− mice treated with isotype or anti-PD-1. Data are presented as the mean±SEM. *P<0.05, **p<0.01, and ***p<0.001, ****p<0.0001; single cell RNA sequencing data were analyzed on website http://crcleukocytecancer-pkucn/.
Intratumoral CXCR6+CD8+ T cells are critical to mediate cancer regression
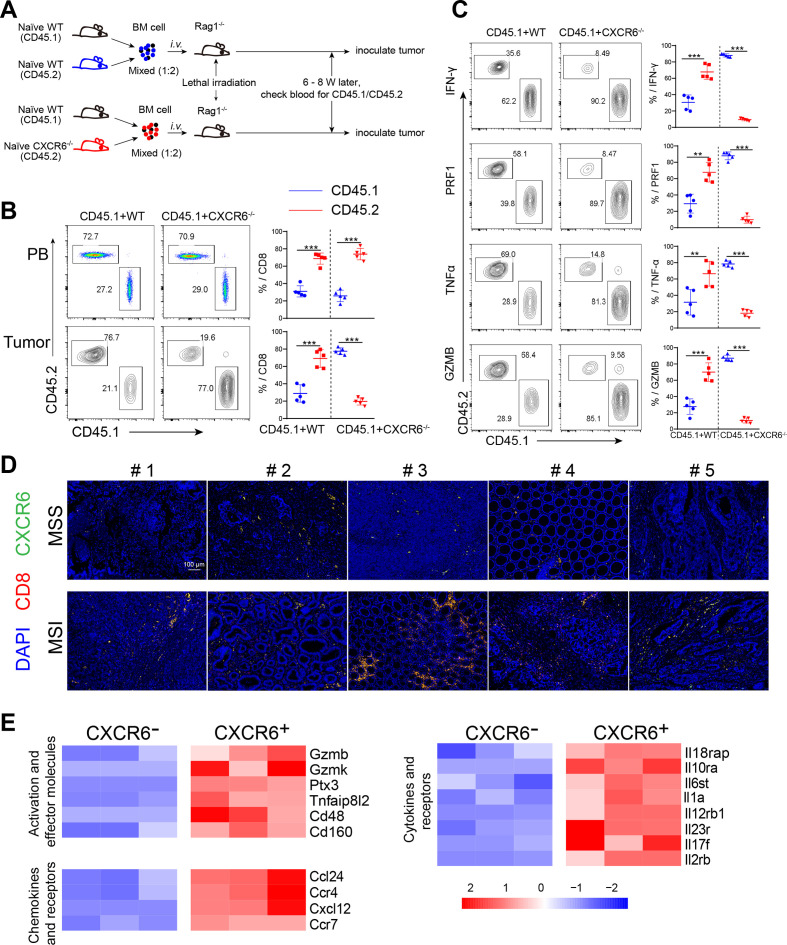
To further elucidate the role of the CXCR6 expression on CD8+ T cell, we transferred bone marrow from naïve CD45.1 mixed with WT or CXCR6−/− mice in a ratio of 1:2 to lethally irradiated Rag1−/− recipients (figure 4A, online supplemental figure S4A). Thus, in hosts that received bone marrow from both CD45.1 and CXCR6−/− mice, CD45.1 positive cells were intact, while CD45.2 positive cells were CXCR6 deficient. After inoculated with tumor cells, the percentages of CD8+ T cells from both chimeras were in line with the initial adoptive ratio in peripheral blood. In contrast, tumor tissues exhibited the opposite proportions or more, suggesting the key role of CXCR6 for intratumoral CD8+ T cell enrichment (figure 4B). Similar results were observed in melanoma model (online supplemental figure S4B). Moreover, the cytolytic capacity and cytokine production of the transferred CD45.2 positive (CXCR6 deficiency) cells was also compromised in recipients (figure 4C, online supplemental figure S4C). These results together indicated that CXCR6+CD8+ T cells represented tumor preference of CD8+ T cells and preserves their effector functions during cancer progression.
Figure 4.
Intratumoral CXCR6+CD8+ T cells are pivotal for antitumor efficacy. (A) Set-up of bone marrow chimeric mice. Total bone marrow obtained from naïve CD45.1+ mice were mixed in 1:2 ratio with that from CD45.2+ WT or Cxcr6−/− mice and then were injected into lethally irradiated Rag1−/− recipient mice. Eight weeks later, MC38 cells were injected subcutaneously into chimeras. (B) Quantification of CD45.1 and CD45.2 positive cells from peripheral blood (PB) and tumor gated on CD8+ cells. (C) Quantification of CD45.1 and CD45.2 positive cells from tumors gated on effectors-producing CD8+ T cells. (D) Immunofluorescence staining of DAPI (blue)/CD8 (green)/CXCR6 (red) and their overlay in MSS and MSI tumor tissues. (E) Heatmap showing the relative expression of genes in intratumoral CXCR6+CD8+ T and CXCR6−CD8+ T cells. data are presented as the mean±SEM. *P<0.05, **p<0.01, and ***p<0.001. MSI, microsatellite instability; MSS, microsatellite stability.
According to mismatch repair, colon cancer can be classified into microsatellite instability (MSI) and microsatellite stability (MSS) subtypes.21 Patients with MSI tended to have a better prognosis and higher infiltration of activated CD8+ CTL.22–24 Therefore, we applied immunofluorescence staining to detect the colocalization of CXCR6 and CD8 in colon cancer from patients with MSI and MSS, respectively. Consistent with CD8+ T cell infiltration, both the frequency of CXCR6 and colocalization with CD8 were significantly higher in MSI group than MSS group (figure 4D, online supplemental figure S4D). This suggested that the abundance of CD8+ T cells in MSI colon cancer closely related to the CXCR6 expression. To determine the differential gene expression between CXCR6+CD8+ T and CXCR6−CD8+ T cells, we performed transcriptome sequencing on both subsets of CD8+ T cells isolated from tumor by FACS (figure 4E, online supplemental figure S4E, F). Transcriptional profiling revealed that CXCR6+CD8+ T cells expressed higher levels of several effector molecules (Gzmk and Gzmb), activation molecules (Ptx3 and Cd48), chemokines and chemokine receptors (Ccl24, Ccr4, and Cxcl12), classical inflammatory factors or inflammatory factor receptors (Il17f, Il1a, Il23r, and Il6st), compared with CXCR6-CD8+ T cells (figure 4E). Taken together, transcriptional profiling indicated a more terminally differentiated state and innate immune responses of CXCR6+CD8+ T cells.
Expression of CXCR6 on CD8+ T cells can be induced by tumor
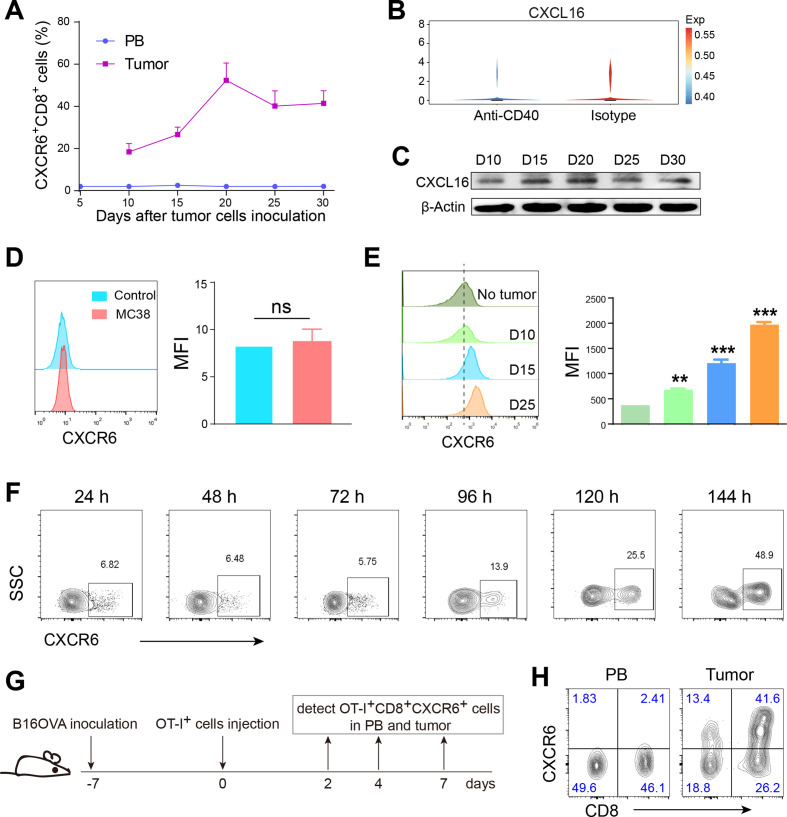
We hypothesized that it might be chemotaxis by its unique ligand, CXCL16, that was response for enrichment of CXCR6 in tumor. Thus, we checked the expression of CXCL16 in TCGA database and found high expression of CXCL16 predicted a good prognosis (online supplemental figure S5A). Especially interesting, when we tested the timing course of proportions of CXCR6+CD8+ T cells in PB and tumor in mice, we observed that CXCR6+CD8+ T cells were rarely detected in PB and almost maintained no changes, while intratumoral CXCR6+CD8+ T cells increased dramatically with cancer progression (figure 5A, online supplemental figure S5B, C). Furthermore, after treatment with anti-CD40, the expression of CXCL16 remained unchanged, even slightly lower than control group, while CXCR6 increased significantly (figures 3A, B and 5B). To time-dependently evaluate intratumoral CXCL16 expression, we examined its expression from day 10 to 30 after tumor cells inoculation. We found that either the mRNA expression level (online supplemental figure S5D, E) or protein level (figure 5C, online supplemental figure S5F) rarely changed with tumor progression. So we inferred that high tumor expression of CXCR6 was dependent on ligand-receptor chemotaxis and induction, the latter being even more primary.
Figure 5.
High tumor expression of CXCR6 was induced by tumor tissue rather than tumor cells. (A) Timing course of proportions of CXCR6+CD8+ T cells in peripheral blood (PB) and tumor tissues from tumor burden mice. (B) Violin plot showing the expression of CXCL16 after treatment with isotype control or anti-CD40 agonist. (C) Western blot showing intratumoral CXCL16 expression. (D and E) Expression of CXCR6 on CD8+ T cells was detected after coculture with MC38 tumor cells (D) or conditional coculture with disrupted tumor tissues (E). Shredded tumor tissues were in upper wells and naïve T cells in lower wells with the presence of IL-2 recombinant cytokine and CD3/CD28 functional antibodies. (F) Timing course of expression of induced CXCR6 in vitro after conditional coculture with shredded tumor tissues. (G) Schematic diagram of inducing expression of CXCR6 in vivo. Mice were inoculated subcutaneously with B16OVA tumor cells. A week later, naïve OT-I T cells were injected into tumor load mice by tail vein and then detected OT-I+CXCR6+CD8+ T cells in PB and tumor tissue on day 2/4/7. (H) Representative density showing the proportion of CXCR6+CD8+ T cells in Pb and tumor tissue on day 4. Displayed flow graph were gated on OT-I positive cells. Data are presented as the mean±SEM. *P<0.05, **p<0.01, and ***p<0.001. Single-cell RNA sequencing data were analyzed on website http://crcleukocytecancer-pkucn/. CXCR6, C-X-C motif chemokine receptor 6; NS, no significant.
To explore what induced high expression of CXCR6 in tumor, we cocultured tumor cells with naïve T cells in vitro. Unexpectedly, the expression of CXCR6 on CD8+ T cells was not induced successfully (figure 5D). Next, we constructed conditional coculture of the entire disrupted tumor tissue and naïve T cells. To avoid confusion with infiltrating CXCR6+CD8+ T cells in the tumor tissue, we used a cell culture chamber in which appropriate amount of tumor tissue was placed on the upper layer, while naïve T cells on lower layer. Gratifyingly, CXCR6+CD8+ T cells were successfully induced and the later stage of tumor, the higher of CXCR6 expression (figure 5E, F).
To determine the dynamic changes of CXCR6 abduction in vivo, we transferred naïve T cells from normal OT-I mice to WT mice challenged with B16OVA tumor cells (figure 5G). We revealed that on day 4 after T cells injection, CXCR6+CD8+ T cells in tumor tissue accounted for 40% of OT-I T cells but was poorly maintained in PB (figure 5H). Taken together, the high expression of CXCR6 in tumor tissue was mainly induced by the tumor tissue itself, not just chemotaxis.
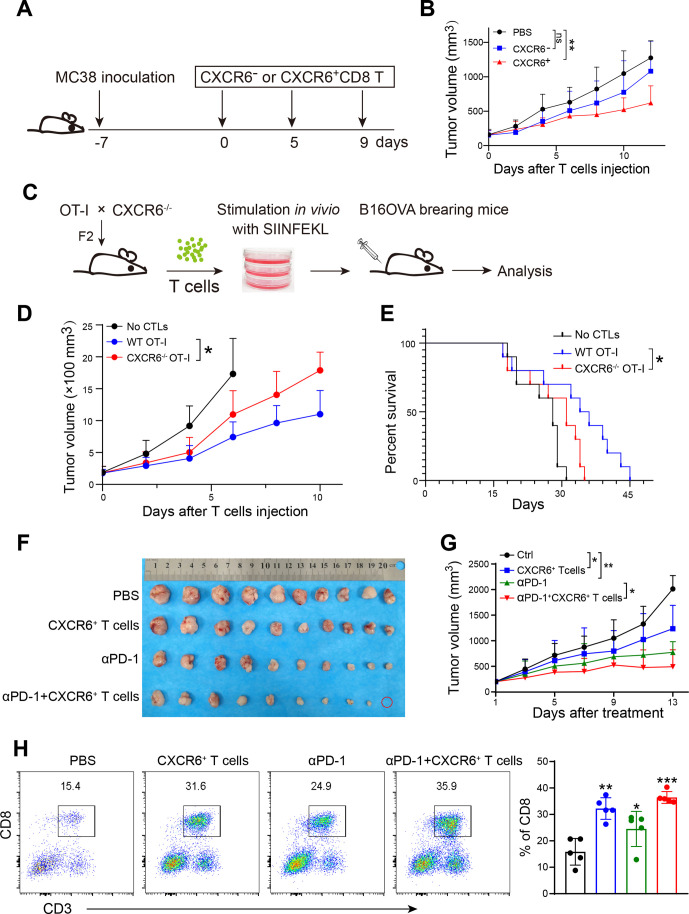
Induced CXCR6+CD8+ T cells retard tumor progression
We isolated CXCR6+CD8+ T and CXCR6-CD8+ T cells induced in vitro and transferred them to tumor-bearing mice, respectively (figure 6A). Recipient mice showed significantly alleviative tumor volume (figure 6B, online supplemental figure S6A). Nevertheless, body weight of recipient mice decreased, (online supplemental figure S6B) due to the excessive inflammatory response that damages the body.25
Figure 6.
Induced CXCR6+CD8+ T cells inhibit tumor growth in mouse model. (A) Schematic diagram showing treatment using CXCR6+CD8+ T cells induced in vitro. Mice were inoculated subcutaneously with MC38 tumor cells. A week later, tumor load mice were treated with CXCR6-CD8+ T or CXCR6+CD8+ T cells that were induced in vitro and isolated by FACS. (B). Tumor growth (n=9–11). (C). Schematic diagram showing treatment using OT-I +CXCR6+ CTL induced in vitro. OT-I and CXCR6−/− mice were crossed to obtain CXCR6−/− OT-I mice. SIINFEKL pulsed T cells were stimulated by IL-2 and then were injected into B16OVA tumor load mice by tail vein. (D) Tumor growth (n=10–11). (E) Survival curves of tumor burden mice. (F) Images of tumors. (G) Tumor growth (n=10). (H) Representative plots showing the ratio of CD8+ T cells in each group. *P<0.05 and **p<0.01.
We crossed CXCR6gfp/gfp mice with OT-I TCR transgenic mice to generate CXCR6 deletion OT-I mice (named CXCR6−/− OT-I mice) (figure 6C, online supplemental figure S6C). To confirm the intrinsic role of CXCR6 in CD8+ T cell function further, we treated B16OVA melanoma with adoptive T cell transfer therapy, which was stimulated in advance by peptide OVA257-264 (figure 6C). Compared with wild-type, recipient mice transferred with CXCR6−/− OT-I CTLs showed weaker antitumor activity, evidenced by larger tumor volume and a shorter survival time (figure 6D, E). These data indicated that CXCR6 was required for antitumor efficacy of intratumoral CD8+ T cells. When combined CXCR6+CD8+ T cells and anti-PD-1 antibody treatment strategies, we found that CXCR6+ T cells could enhance the effect of anti-PD-1 treatment (figure 6F, G). Even one mouse was completely cured and the tumor disappeared (figure 6F, red circle). The proportion of intratumoral CD8+ T cells also rose in combination group although slightly (figure 6H).
Discussion
We demonstrated that CXCR6+CD8+ T cell subset was exclusively enriched in tumor tissue and was more immunocompetent. High infiltration of CD8+ T cells in tumor tissue predicts a good prognosis.4 24 Thus, increasing the abundance of CD8+ T cells as well as activating them helps alleviate tumor progression and even eliminate tumors. Given that T cells in autoimmune diseases are often highly activated and inflammatory, while exhausted or repressed in tumor, we question that whether the same phenotypic T cells are still immunocompetent in tumors according to the phenotypic characteristics of highly activated inflammatory T cells in autoimmune diseases. In MS and mouse EAE models, CXCR6 identifies a group of pathogenic cells with excessive immune function,16 so this study focused on the expression of CXCR6 in the tumor microenvironment and the relationship between CXCR6 expression and CD8+ T cell function. We started with single-cell sequencing data of clinical clone cancer samples17 and combined transcriptome sequencing in the TCGA database and experimental results to validate CXCR6 was almost exclusively expressed in tumor tissues and highly expressed on CD8+ T cells (figure 1). Phenotypic characteristics showed that CXCR6+CD8+ T cells were more immunocompetent than the negative counterpart. Chimeras with CXCR6-specific deficiency on CD8+ T cell told us that CXCR6 was essential for CD8+ T cells to accumulate in tumor and exert antitumor function (figure 2E–I). These results indicate that CXCR6 still identifies highly activated and cytolytic T cell subset, consistent with its role in autoimmune diseases.16
Immune checkpoint therapy targeting the CTLA-4 or PD-1/PD-L1 pathways can induce inflammatory toxicities such as checkpoint inhibitor-induced colitis.26 27 Single-cell sequencing data reported recently that a striking accumulation of CD8+ T cells with highly cytotoxic and proliferative states is observed in checkpoint inhibitor-induced colitis, which is highly expressed CXCR6.28 We revealed that CXCR6+CD8+ T cells were more responsive to anti-PD-1 blockade and CXCR6 deficient CD8+ T cells almost lost their antitumor function, which could not be rescued even treated with anti-PD-1 therapy. In addition, the frequency of CXCR6 and colocalization with CD8 were significantly higher in MSI patients than MSS group (figure 4D). These results show again that CXCR6+CD8+ T are the main cell subset to response to immune checkpoint therapy.
We wondered why CXCR6+CD8+ T only accumulated in tumors, and how CXCR6 was highly expressed. As a chemokine receptor, CXCR6 made us think of its unqiue ligand CXCL16 unavoidably.11 Many studies report that CXCR6/CXCL16 signaling axis regulates localization of CXCR6+ T cells.29–31 Interestingly, timing course of proportions of CXCR6+CD8+ T cells from PB and tumor tissues showed that CXCR6+CD8+ T cells almost maintained no changes, while intratumoral CXCR6+CD8+ T cells increased dramatically with cancer progression. Furthermore, the expression of CXCL16 remained unchanged, even slightly lower than control group, while CXCR6 increased significantly after treatment with anti-CD40. Thus, high tumor expression of CXCR6 was not only dependent on ligand-receptor chemotaxis and might be mainly induced. Unexpectedly, the expression of CXCR6 on naïve CD8+ T cells was induced successfully by tumor tissue rather than tumor cells (figure 5C–G). When we induced CXCR6 expression in vitro, we applied transwell to eliminate the interference of CXCR6+ T cells that already existed in tumor tissues. So we inferred that certain soluble factor induced its increase, and this factor might originate from tumor cells or immune cells, even T cells themselves. Pallett et al32 report that sequential IL-15 or antigen exposure followed by TGFβ induced CXCR6 and CXCR3 expression, robust and cell autonomous IL-2 and IFN-γ production, to equip liver CD8TRM to survive while exerting local noncytolytic immunosurveillance in hepatotropic infection. Our transcriptome sequencing data showed that IL-2Rβ was upregulated in CXCR6+CD8+ T cells compared with CXCR6-CD8+ T cells (figure 4E, online supplemental figure S4F). Furthermore, IL-15/IL-2Rβ can induce the activation of JAK kinases, as well as the phosphorylation and activation of transcription activators STAT3, STAT5, and STAT6.33 CXCR6 is involved in STAT3, PI3K/AKT, ERK/MAPK pathway.34 Meanwhile, PI3K/AKT pathway is also the downstream of JAK kinases. BAIAP3, a C2 domain-containing Munc13 protein, was also upregulated in CXCR6+CD8+ T cells (online supplemental figure S4F). BAIAP3 can promote cell proliferation and the recycling of secretory vesicle transmembrane proteins. Munc13-4 is essential for the Ca2+-dependent priming of secretory granules in immune cells.35 Thus, IL-15/IL-2Rβ may be possible factor to induce CXCR6 expression. BAIAP3 may be related to enrichment for cytokine production and high cell proliferation of CXCR6+CD8+ T cells. Nevertheless, what factors in the tumor tissue cause the expression of CXCR6 need further investigation.
So the next question comes, what can CXCR6 on CD8+ T cell induced by tumor do? The induced CXCR6+CD8+ T cells are tumor antigen specific, can they be used for cell therapy to eliminate tumors? We found that transferred CXCR6+CD8+ T cells for treatment could alleviative tumor burden and prolong survival significantly. Moreover, combination with anti-PD-1 showed superior efficiency (figure 6). Srivastava et al demonstrate that Cxcr6−/− CAR-T cells show poorer tumor infiltration compared with their WT counterparts.36 Similarly, we also revealed that recipient mice transferred with CXCR6−/− OT-I CTLs showed weaker antitumor activity than WT OT-I CTLs. CXCR6 deficiency also impairs the efficacy of cancer vaccine and the recruitment of CD8+ resident memory T cell in head and neck and lung tumors.15 Thus, CXCR6 is essential for antitumor efficacy of intratumoral CD8+ T cells.
The expression of CXCR6 varies greatly in different patients with colon cancer (online supplemental figure S4G, H). High expression of CXCR6 indicates good prognosis. Both the frequency of CXCR6 and CD8 are significantly higher in MSI group than MSS group. Thus, we can first induce high CXCR6 expression in colon cancer patients with low level, and then apply immune checkpoint therapy. Furthermore, we can also design a combination of CXCR6 positive cell adoptive and immune checkpoint therapy to slow down tumor progression and even cure eventually.
In summary, we demonstrated that CXCR6 is specifically and highly expressed on CD8+ T cell in tumor. This high expression is not mainly caused by ligand-receptor chemotaxis of CXCL16/CXCR6 but induced by tumor tissue self. In addition, induced CXCR6+CD8+ T cells have tumor antigen specificity and can retard the progression of tumors. This study may contribute to the rational design of combined immunotherapy. Alternatively, CXCR6 may be used as a biomarker for effective CD8+ T cell before cell adoptive therapy, providing a basis for clinical transfusion therapy.
Acknowledgments
We would like to thank Professor Lilin Ye (Third Military Medical University) for Cd8-/- mice support. We would also like to thank Professor Xuetao Cao (Nankai University) for B6.SJL-Ptprca Pepcb /Boy (CD45.1) mice support.
Footnotes
BW and YW contributed equally.
Contributors: YS, HC and QX designed the study. BW and YW performed the experiments and analyzed the data. XS and GD helped collected samples. WH helped transferred intravenously. XW, YG, ZT and ZF gave methodological support and conceptual advice. BW and YS wrote the manuscript with the help of all other authors. All authors reviewed and approved the manuscript.
Funding: This work was supported by National Natural Science Foundation of China (Nos. 91853109, 81872877, 81730100, 81673436) and Mountain-Climbing Talents Project of Nanjing University.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplemental information. Data used for this analysis are available upon reasonable request to the corresponding author.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
Our study was approved by the Research and Ethical Committee of Nanjing University (IACUC-2005018).
References
- 1.Thompson ED, Enriquez HL, Fu Y-X, et al. Tumor masses support naive T cell infiltration, activation, and differentiation into effectors. J Exp Med 2010;207:1791–804. 10.1084/jem.20092454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity 2004;21:137–48. 10.1016/j.immuni.2004.07.017 [DOI] [PubMed] [Google Scholar]
- 3.Gubin MM, Zhang X, Schuster H, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014;515:577–81. 10.1038/nature13988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naito Y, Saito K, Shiiba K, et al. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res 1998;58:3491–4. [PubMed] [Google Scholar]
- 5.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960–4. 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 2015;348:62–8. 10.1126/science.aaa4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kochenderfer JN, Wilson WH, Janik JE, et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood 2010;116:4099–102. 10.1182/blood-2010-04-281931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinrichs CS, Rosenberg SA. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol Rev 2014;257:56–71. 10.1111/imr.12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim CH, Kunkel EJ, Boisvert J, et al. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J Clin Invest 2001;107:595–601. 10.1172/JCI11902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unutmaz D, Xiang W, Sunshine MJ, et al. The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern is conserved between human and mouse. J Immunol 2000;165:3284–92. 10.4049/jimmunol.165.6.3284 [DOI] [PubMed] [Google Scholar]
- 11.Matloubian M, David A, Engel S, et al. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol 2000;1:298–304. 10.1038/79738 [DOI] [PubMed] [Google Scholar]
- 12.Matsumura S, Wang B, Kawashima N, et al. Radiation-induced CXCL16 release by breast cancer cells attracts effector T cells. J Immunol 2008;181:3099–107. 10.4049/jimmunol.181.5.3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaida MM, Günther F, Wagner C, et al. Expression of the CXCR6 on polymorphonuclear neutrophils in pancreatic carcinoma and in acute, localized bacterial infections. Clin Exp Immunol 2008;154:216–23. 10.1111/j.1365-2249.2008.03745.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mossanen JC, Kohlhepp M, Wehr A, et al. CXCR6 Inhibits Hepatocarcinogenesis by Promoting Natural Killer T- and CD4+ T-Cell-Dependent Control of Senescence. Gastroenterology 2019;156:1877–89. 10.1053/j.gastro.2019.01.247 [DOI] [PubMed] [Google Scholar]
- 15.Karaki S, Blanc C, Tran T, et al. CXCR6 deficiency impairs cancer vaccine efficacy and CD8+ resident memory T-cell recruitment in head and neck and lung tumors. J Immunother Cancer 2021;9:e001948. 10.1136/jitc-2020-001948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou L, Rao DA, Yuki K, et al. SerpinB1 controls encephalitogenic T helper cells in neuroinflammation. Proc Natl Acad Sci U S A 2019;116:20635–43. 10.1073/pnas.1905762116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Li Z, Skrzypczynska KM, et al. Single-Cell analyses inform mechanisms of Myeloid-Targeted therapies in colon cancer. Cell 2020;181:442–59. 10.1016/j.cell.2020.03.048 [DOI] [PubMed] [Google Scholar]
- 18.Paust S, Gill HS, Wang B-Z, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol 2010;11:1127–35. 10.1038/ni.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo S-R, Turnis ME, Goldberg MV, et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res 2012;72:917–27. 10.1158/0008-5472.CAN-11-1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow MT, Ozga AJ, Servis RL, et al. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity 2019;50:1498–512. 10.1016/j.immuni.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganesh K, Stadler ZK, Cercek A, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol 2019;16:361–75. 10.1038/s41575-019-0126-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science 1993;260:816–9. 10.1126/science.8484122 [DOI] [PubMed] [Google Scholar]
- 23.Bao X, Zhang H, Wu W, et al. Analysis of the molecular nature associated with microsatellite status in colon cancer identifies clinical implications for immunotherapy. J Immunother Cancer 2020;8:e001437. 10.1136/jitc-2020-001437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips SM, Banerjea A, Feakins R, et al. Tumour-infiltrating lymphocytes in colorectal cancer with microsatellite instability are activated and cytotoxic. Br J Surg 2004;91:469–75. 10.1002/bjs.4472 [DOI] [PubMed] [Google Scholar]
- 25.Fajgenbaum DC, June CH. Cytokine storm. N Engl J Med 2020;383:2255–73. 10.1056/NEJMra2026131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 2015;373:23–34. 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postow MA, Sidlow R, Hellmann MD. Immune-Related adverse events associated with immune checkpoint blockade. N Engl J Med 2018;378:158–68. 10.1056/NEJMra1703481 [DOI] [PubMed] [Google Scholar]
- 28.Luoma AM, Suo S, Williams HL, et al. Molecular pathways of colon inflammation induced by cancer immunotherapy. Cell 2020;182:655–71. e22. 10.1016/j.cell.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wein AN, McMaster SR, Takamura S, et al. CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J Exp Med 2019;216:2748–62. 10.1084/jem.20181308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato T, Thorlacius H, Johnston B, et al. Role for CXCR6 in recruitment of activated CD8+ lymphocytes to inflamed liver. J Immunol 2005;174:277–83. 10.4049/jimmunol.174.1.277 [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Lu Y, Wang J, et al. CXCR6 induces prostate cancer progression by the Akt/mammalian target of rapamycin signaling pathway. Cancer Res 2008;68:10367–77. 10.1158/0008-5472.CAN-08-2780 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Pallett LJ, Davies J, Colbeck EJ, et al. IL-2high tissue-resident T cells in the human liver: Sentinels for hepatotropic infection. J Exp Med 2017;214:1567–80. 10.1084/jem.20162115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnston JA, Kawamura M, Kirken RA, et al. Phosphorylation and activation of the Jak-3 Janus kinase in response to interleukin-2. Nature 1994;370:151–3. 10.1038/370151a0 [DOI] [PubMed] [Google Scholar]
- 34.Korbecki J, Bajdak-Rusinek K, Kupnicka P, et al. The role of CXCL16 in the pathogenesis of cancer and other diseases. Int J Mol Sci 2021;22:3490. 10.3390/ijms22073490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Jiang S, Mitok KA, et al. BAIAP3, a C2 domain-containing Munc13 protein, controls the fate of dense-core vesicles in neuroendocrine cells. J Cell Biol 2017;216:2151–66. 10.1083/jcb.201702099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivastava S, Furlan SN, Jaeger-Ruckstuhl CA, et al. Immunogenic chemotherapy enhances recruitment of CAR-T cells to lung tumors and improves antitumor efficacy when combined with checkpoint blockade. Cancer Cell 2021;39:193–208. 10.1016/j.ccell.2020.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003100supp001.pdf (1.2MB, pdf)
Data Availability Statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as supplemental information. Data used for this analysis are available upon reasonable request to the corresponding author.