Abstract
Biological oscillations often cycle at different harmonics of the 24-h circadian rhythms, a phenomenon we coined “Musica Universalis” in 2017. Like the circadian rhythm, the 12-h oscillation is also evolutionarily conserved, robust, and has recently gained new traction in the field of chronobiology. Originally thought to be regulated by the circadian clock and/or environmental cues, recent new evidences support the notion that the majority of 12-h rhythms are regulated by a distinct and cell-autonomous pacemaker that includes the unfolded protein response (UPR) transcription factor spliced form of XBP1 (XBP1s). 12-h cycle of XBP1s level in turn transcriptionally generates robust 12-h rhythms of gene expression enriched in the central dogma information flow (CEDIF) pathway. Given the regulatory and functional separation of the 12-h and circadian clocks, in this review, we will focus our attention on the mammalian 12-h pacemaker, and discuss our current understanding of its prevalence, evolutionary origin, regulation, and functional roles in both physiological and pathological processes.
Supplementary Information
The online version of this article (10.1007/s00018-020-03730-5) contains supplementary material, which is available to authorized users.
Keyword: 12-h clock, Ultradian rhythm, XBP1s, Unfolded protein response, Circadian rhythm, ER stress
The history of the discovery of biological 12-h rhythms in mammals
Biological rhythms are a central and indispensable component of life on earth and have provided evolutionary advantages for almost all life forms ranging from the earliest bacteria to the most complicated mammals including humans [1, 2]. Rhythms and clocks of all types confer an organismal advantage by allowing plants, animals, and unicellular organisms to anticipate changes in their environment and adapt accordingly. This advantage has led to the evolution of diverse oscillators regulating all aspects of biology, including the cell cycle oscillator controlling cell-division, infradian (period longer than 24-h) hormone cycles regulating the timing of organismal development, and circadian (period close to 24-h) and ultradian (period shorter than 24-h) clocks regulating cycles of organismal activity, physiology, metabolism, and cellular activity [1].
Clocks regulating daily cycles include the well-known circadian clock, in which the molecular transcriptional-translational feedback loop (TTFL) present in most known cell types regulates the 24-h rhythms of cellular gene expression, metabolic processes, and organismal behavior [1]. In addition to 24-h circadian rhythms, biological rhythms have also been shown to have ‘harmonics’—rhythms with frequencies that are positive integer multiplier of 1.15*10e−5 Hz (24 h period) [2]. Harmonics oscillation in mammals were first comprehensively characterized in mouse liver using high-resolution temporal microarray in 2009 by Hughes and colleagues [3]. Analysis of these data with both Fisher’s G and COSOPT methods yielded a few hundred genes cycling with a 12-h period, and a smaller number of genes cycling with an 8-h period [3]. Notably, one of the 12-h cycles, Hspa1b, also showed 12-h rhythms with the same phase of peak expression in tissues outside of the liver, implying a wider prevalence of mammalian 12-h rhythms [3]. This study further identified a few key features of 12-h harmonics that distinguish from the circadian rhythm, which would be validated by later studies. These include the coincidence of the majority of 12-h gene expression peaks with the two ‘rush hours’: dawn and dusk; while 24-h cycling genes have peak expression across more varied time points [3]. Furthermore, it was noted that 12-h genes often have differential peak heights at their first and second peaks [3]. While this served as a strong argument against the existence of a separate 12-h pacemaker back then, as demonstrated in details later, an alternative explanation is that this differential height results from the superimposition of a 12-h rhythm (with two equal peaks) with additional oscillations, such as a circadian rhythm: the superimposition of the peak of the circadian rhythm with one peak of the 12-h oscillation leads to the larger peak and the superimposition of the nadir of the circadian rhythm with the other peak of the 12-h oscillation results in a smaller peak [4]. Thus, by fine-tuning the phases of the 12-h and circadian rhythms, it appears that the cells can alter peak gene expressions at dawn and dusk to accommodate differing needs in gene expression at these two rush hours.
In the same study, the authors did not observe apparent cell-autonomous 12-h or 8-h rhythms of gene expression in human U2OS cells synchronized with dexamethasone or mouse NIH3T3 cells synchronized with forskolin using the Fisher’s G or COSOPT methods [3] and therefore concluded that extracellular cues, probably related to feeding, were necessary to regulate these ultradian rhythms [3]. However, it was later found that by using a more powerful analytical method: the eigenvalue/pencil [4, 5], harmonic oscillations at both gene expression and metabolites level cycling at both 12-h and 8-h periods were uncovered from mouse embryonic fibroblasts (MEFs), U2OS cells, and mouse liver cell line MMH-D3 in response to serum shock and dexamethasone [4, 6]. The eigenvalue/pencil method assumes that most oscillations can be approximated by a linear combination of exponentials (standard sine waves with a decay factor) plus noise and therefore can be applied to any situations where such assumptions are valid. As a result, the biggest difference between the eigenvalue/pencil method and other cycle-identification methods (such as JTK_CYCLE [7], Fisher’s G [3], COSOPT [3], ARSER [8] and RAIN [9]) lies in the former’s ability to identify all superimposed oscillation in an unbiased manner simultaneously, while the latter ones require the users to define a narrow period range and thus can only reveal one single dominant oscillation from any given time series dataset [4, 10]. However, both theories and real-life experiences taught us that pure oscillation with a single frequency rarely exists in nature. Rather, physical rhythms are often presented as superpositions of basic waves such as the harmonic resonances found in music, light, and planetary motions. In a sense, the eigenvalue/pencil analysis thus enables the biologists to tackle time series data on a par with what physicists and engineers do: performing spectrum analysis to deconvolute ‘noisy’ data and uncover multiple hidden frequencies (for a more detailed understanding of the strengths and limitations of the eigenvalue/pencil method, its comparison with other cycle-identification methods, and its wider applications, please refer to [5]). The implications of such an analysis are multifaceted. Philosophically, it places biology as part of “Musica Universalis” [2]; empirically, it reveals that “non-sinusoidal” periodic waveforms actually reflect a confluence of multiple period inputs, and even irregular waveforms might have biological significance in their own right, a fact also highlighted by a recent study examining ultradian rhythms in Neurospora crassa [11], although the latter point warrants further experimental validation.
Mammalian 12-h rhythms are widespread in both central and peripheral tissues
Using the eigenvalue/pencil method, Zhu et al. re-analyzed the high-resolution hepatic microarray data published by Hughes et al. [3] and uncovered prevalent ultradian gene expression oscillations cycling at the periods of 12-h, 8-h, and even 4.8-h (5th harmonic) in mouse liver [4]. In particular, a total of 3,652 (~ 20% of total hepatic mRNA) hepatic 12-h mRNAs were identified, of which 760 are dominant ones (whose 12-h amplitudes are the greatest among all identified oscillations) [4]. This is a significant increase of the repertoire of 12-h transcriptome compared to the original study, as most of these genes are often superimposed with other oscillations with larger amplitude and thus evaded detection by the Fisher’s G and COSOPT methods [4]. The wider prevalence of hepatic 12-h rhythms of gene expression was further validated in a high resolution hepatic RNA-Seq study [6]. In this paper, using both eigenvalue/pencil and RAIN methods, Pan et al. uncovered ~ 3,650 high confidence 12-h hepatic mRNA (~ 27% of all hepatic transcriptome) in mouse liver under constant darkness conditions [6]. While the prevalence of hepatic 12-h transcriptome matches that of hepatic circadian rhythm [12], their amplitudes, on average, are smaller compared to that of the circadian rhythm, often in the range of 1 to fourfold change [6]. Consistent with the hepatic microarray study [3, 4], the acrophases (the time period in a cycle during which the cycle crests or peaks) of the hepatic 12-h rhythms identified by RNA-Seq are also heavily biased toward CT0 and CT12 [CT stands for circadian time. By convention, the onset of activity of diurnal organisms defines circadian time zero (CT0), while the onset of activity of nocturnal organisms defines circadian time twelve (CT12). Therefore, for a nocturnal animal such as mice, they are active from CT12 to CT24], corresponding to the dawn and dusk in a diurnal cycle [6].
In a most recent perspective, Zhu applied the eigenvalue/pencil method to analyze a previously published atlas of twelve mouse tissues profiled at 2 h resolution over 48 h under constant darkness condition [13] and discovered that brown and white adipose tissues, as well as skeletal muscle, had the highest number of 12-h cyclers [14]. By contrast, there were very few genes exhibiting 12-h rhythms of expression in the hypothalamus, brainstem, and cerebellum in this analysis [14]. The adrenal gland, aorta, lung, liver, heart, and kidney had intermediate level of 12-h cyclers [14]. Three hundred and eighty 12-h-cycling genes were present in at least eight tissues, with Zbtb16 being the top commonly shared gene found in ten tissues [14]. Acrophases at ~ CT0 and ~ CT12 were predominantly observed in all tissues, except for brown adipose tissue and skeletal muscle, which had most peaks observed at ~ CT6 and ~ CT18 instead [14]. It was hypothesized that the reason for the shift of rush hour peaks in brown adipose and skeletal muscle is the high level of fat catabolism and mitochondrial activity in these tissues [14]. Importantly, using the harmonic regression method to identify dominant oscillations, El-Athman and colleagues reported similar findings in an independent study [15]. In a separate study, prevalent 12-h rhythms of gene expression were also uncovered in mouse cornea [16].
El-Athman et al. further analyzed diurnal primate transcriptome data in 64 olive baboon tissues [17] using harmonic regression and found prevalent 12-h transcriptome in peripheral tissues including mesenteric white adipose tissue, bone marrow, mesenteric lymph nodes, smooth muscle, kidney, prostate, esophagus, skin, and pancreas [15]. More intriguingly, a few brain regions exhibit widespread 12-h rhythms of gene expression, with the hippocampus and cerebellum topping the list [17]. By contrast, the suprachiasmatic nucleus (SCN), which harbors the central circadian clock pacemaker [18], is largely deprived of 12-h transcriptome [15]. While the significance of these findings remains unknown at this point, it is tempting to speculate that 12-h rhythms of gene expression enriched in the primate hippocampus may be functionally related to the two neural activity peaks associated with daytime memory acquisition and nighttime memory consolidation [19, 20].
Besides transcriptome, prevailing 12-h hepatic oscillations are also identified at the proteome and metabolome level (occupying 20% ~ 30% of all hepatic proteome and metabolome) [2]. Further, coordinated 12-h rhythms of transcriptome, proteome, and metabolome are overrepresented in several metabolic processes, including nucleotide, amino, and nucleotide sugar, polyamine, glycerophospholipid, and sphingolipid metabolism [2, 4, 10, 14]. Systemically, 12-h rhythm of respiratory exchange ratio (RER) was also identified in mice fed ad libitum [4]. Taken together, the highly coupled 12-h rhythms of hepatic gene expression and metabolism ensure precisely timed orchestration of metabolic flux [6].
The vehicle-cargo hypothesis describes the distinct functions of 12-h versus the circadian rhythms
Gene Ontology (GO) analysis of mouse hepatic 12-h transcriptome revealed top-enriched genes involved in the entire central dogma information flow (CEDIF) process, ranging from transcription initiation (such as Tbp, Gtf2h3 and Cdk7), mRNA processing and export (such as Cstf1, Sf3a1 and Nxf1), ribosome biogenesis (such as Rpl13, Rpl15 and Rps28), translation initiation (such as Eif2ak3, Eif3a, and Eif3g) to protein folding, processing and sorting in the endoplasmic reticulum (ER) and Golgi (such as Xbp1, Sec23b, and Gosr2) and include both anabolic and catabolic processes [6]. Intriguingly, detailed phase analysis further unveiled an approximately 1 h acrophase delay from genes involved in mRNA processing (peaking at CT1 and CT13) to those participating in protein sorting in the ER/Golgi (peaking at CT2 and CT14), consistent with the one way traffic of genetic information flow [6]. In addition, 12-h-cycling genes enriched in numerous metabolic pathways were uncovered in mouse liver [10], including purine and pyrimidine de novo synthesis, hexosamine, and UDP-GlcNAc biosynthesis, glycerophospholipids and sphingolipids synthesis. The 12-h metabolic enzyme expression closely matches those of 12-h hepatic metabolome [2, 4, 6] and couples with the 12-h rhythm of the CEDIF process [2, 4]. For instance, 12-h rhythm of purine and pyrimidine synthesis precedes 12-h rhythms of transcription and mRNA processing [14]; 12-h rhythm of nucleotide and amino sugar metabolism coincides with 12-h rhythm of protein N-linked glycosylation occurring in the ER and Golgi [14]; 12-h rhythm of glycerophospholipid and sphingolipid metabolism is further expected to contribute to 12-h rhythm of ER/Golgi membrane homeostasis (likely by regulating membrane lipid composition, permeability, and fluidity), the integrity of which is central to protein sorting and vesicle trafficking in the ER and Golgi [21]. Last but not the least, the enriched CEDIF and metabolic gene signatures are commonly found in the 12-h transcriptomes of all tissues in mice [14].
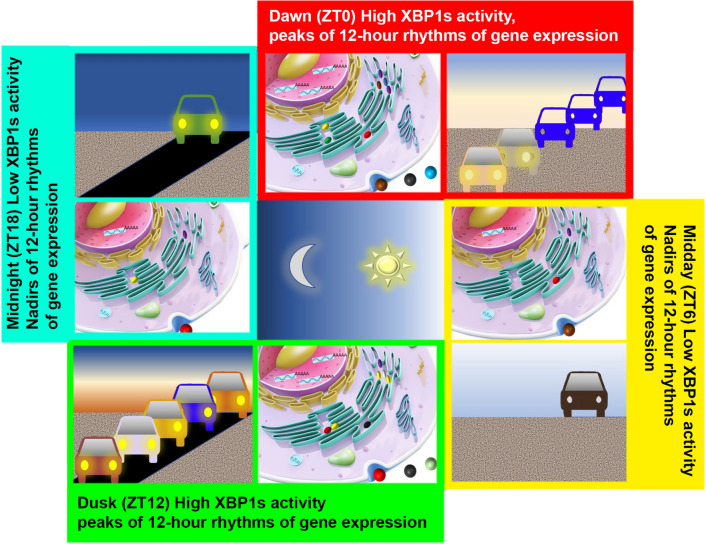
In light of these findings, a vehicle-cargo hypothesis was proposed by us to elucidate the distinct functions of 12-h versus the circadian rhythms [6] (Fig. 1). It was argued that the 12-h rhythm accommodates increased demands for gene expression/processing at the two biological ‘rush hours’ (dawn and dusk) by elevating the global traffic capacity of the central dogma information flow. This connects and tunes rates of mRNA and protein processing to the 12-h cycle of metabolic stress (thus acting as the vehicle). The circadian clock, on the other hand, dictates the particular genes/gene products processed at each rush hour (thus acting as the cargo). An everyday metaphor would be the fluctuating daily traffic on the highway: the 12-h biological clock increases capacity at dawn and dusk rush hours, just as the highway expand its capacity during rush hours at times of peak traffic (also one in the morning and one in the evening) through opening HOV lanes; whereas the circadian clock determines the specific genes transcribed and translated at each rush hour, just as the ‘cargos’ traversing highways differ between morning and evening.
Fig. 1.
The Vehicle Cargo Hypothesis. Likened to increasing traffic at the two “rush” hours at each dawn and dusk on the highway, 12-h rhythms of CEDIF gene expression peaking at dawn (ZT0) and dusk (ZT12) imply the existence of a 12-h oscillation of the trafficking capacity of the genetic information flow that also peaks at ZT0 and ZT12 in mammalian cells. The CEDIF encompasses the progressive molecular processing steps from transcription initiation, mRNA processing, ribosome biogenesis, translation, all the way to protein folding/processing/sorting in the ER and Golgi. At this point, it remains to be determined whether it is the total number of molecules that undergo processing or the metabolic rate of processing (or both) that exhibits a 12-h oscillation. The 12-h rhythms of CEDIF gene expression is in turn transcriptionally regulated by the XBP1s expression/activity, which also peaks at ZT0 and ZT12
Findings from two independent studies further supported the vehicle-cargo hypothesis [15, 22]. Genov et al. characterized the genomic and proteomic features of the clusters of genes oscillating with harmonics of circadian periodicity in mouse liver and also noted that genes with 12-h oscillations were enriched for RNA splicing, protein translation, and ER protein processing pathways [22]. A second study by El-Athman et al. comprehensively profiled rhythmic alternative splicing events in 12 mouse tissues and 64 baboon tissues and identified wide-spread rhythmic alternative spliced transcripts cycling at either 24-h or 12-h period [15]. Intriguingly, for 24-h alternative splicing events in the baboon, pairs of differentially rhythmic splice isoforms of the same gene often peaked at opposing times of day (phase shift greater than 10 h) [15]. We reason that these two peaks of splicing events are mediated by the 12-h rhythm of CEDIF (the vehicle), although the precise splicing sites (the cargo) may differ at the two peaks.
The existence of a cell-autonomous mammalian 12-h pacemaker regulating 12-h rhythms of gene expression, independent from the circadian clock
Several theories have arisen over the past decade on the possible molecular mechanisms governing 12-h rhythms of gene expression. Early studies favor the hypothesis that the mammalian 12-h rhythms are not cell-autonomous and instead are established by the circadian clock and/or fasting-feeding cues [3, 23, 24]. As mentioned in the first section, due to the lack of detection of cell-autonomous 12-h rhythms of gene expression, Hughes et al. [3] proposed that feeding and/or other extracellular signals are responsible for the generation of 12-h rhythms. This hypothesis was partially supported by a 2010 study by Cretenet et al. [24]. In this paper, the authors observed 12-h rhythms of XBP1s expression as well as 12-h rhythms of IRE1α phosphorylation (IRE1α is an ER membrane-anchored endoribonuclease that when phosphorylated can alternatively splice Xbp1 mRNA into Xbp1s [25]) in mouse liver under normal physiological conditions. However, perturbed 12-h rhythm of XBP1s and 12-h rhythms of output genes observed in Cry1/Cry2 double knockout mice (CRY1 and CRY2 are key proteins involved in the negative limb of the circadian clock control [18]) as well as during altered feeding regimen in wild-type mice led the authors to conclude that the hepatic 12-h rhythms are generated by the combined effects of the circadian clock and fasting/feeding cycles [24]. Nevertheless, as pointed out later, since the 12-h rhythms are sensitive to the disruption of normal metabolic homeostasis, it is highly likely that the observed changes of 12-h rhythms in Cry1/Cry2 knockout mice are a consequence of the metabolic deficits observed in these mice, rather than due to the absence of a functional circadian clock per se. A third study also by Hughes et al. observed conversion of 12-h to an apparent 24-h hepatic gene expression in a mouse model of brain-specific rescue of Clock function [23] (CLOCK is a transcription factor involved in the positive limb of the circadian clock control [18]). While this observation did initially substantiate the notion that the hepatic 12-h rhythms are likely regulated by the central circadian clock in the SCN, in light of the new findings supporting the existence of a dedicated 12-h pacemaker (as seen below), we believe this phenomenon can be accounted for by at least two alternative explanations. First, the brain-specific recue of Clock may exert a larger than normal circadian gene amplitude in the liver, thus masking the original 12-h of gene expression. Secondly, if the neuroendocrine signaling coming from the brain of the brain Clock rescue mice indeed can perturb the cell-autonomous 12-h rhythm in the liver, then it suggests the presence of cross-talk between the 24-h circadian and 12-h ultradian pacemakers mediated by humoral factors or neural circuits in a non-cell autonomous manner.
In 2013, Westermark and Herzel raised an alternative hypothesis. By performing mathematical modeling, they reported that two circadian transcription activators or repressors appearing in anti-phase are theoretically capable of establishing 12-h rhythms of gene expression in a cell-autonomous manner [26], although no experimental evidences were provided to corroborate their computational predictions.
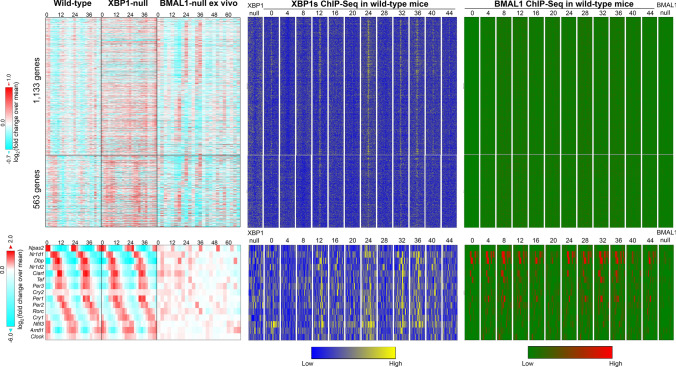
In sharp contrast to these earlier theories, our group proposed the existence of a dedicated cell-autonomous 12-h pacemaker that is responsible for the establishment and maintenance of the mammalian 12-h rhythms, which is independent of the 24-h circadian clock [2, 4, 6, 14]. The initial clue came from the observed mathematical orthogonality of different superimposed oscillations uncovered from the same gene [4, 5]. Namely, forcing the eigenvalue/pencil method to uncover additional oscillations of smaller period has negligible effects on the amplitude/phase/period of the oscillations of larger period that are previously uncovered [4, 5]. Subsequent post-hoc analysis of hepatic RNA-Seq data in wild-type and whole-body BMAL1 knock-out mice (BMAL1 is absolutely essential for establishing mammalian circadian rhythms, as BMAL1 knock-out mice showed a complete loss of circadian rhythms in locomotive activity under constant darkness condition [27]) fed ad libitum (both conventional and adult BMAL1 deletion mice [28]) confirmed the independence of 12-h rhythms of CEDIF and metabolism gene expressions from the circadian clock [4]. Specifically, for 12-h genes lacking superimposed circadian rhythms (such as Eif2ak3 and Gfpt1), their 12-h rhythms of gene expression are almost identical between wild-type and BMAL1 knock-out mice under either a 12-h light/12-h dark schedule or a constant darkness condition [4, 28]. For 12-h genes with superimposed circadian rhythms (such as Gck), more perceptible 12-h rhythms were observed in BMAL1 knock-out mice [4, 28]. The independent relationship between 12-h and 24-h rhythms also was further verified in ClockΔ19 mutant mice (which have a point mutation that causes a deficiency in the 19th exon of the Clock gene) [4, 29], and bolstered by a most recent high-resolution RNA-Seq data profiled from ex vivo liver slices of BMAL1 knock-out mice [30]. While the circadian clock gene expression is profoundly disrupted in BMAL1-null ex vivo tissue, as expected [30]; Xbp1 12-h expression is unchanged, and 12-h rhythmicity is maintained in the absence of BMAL1 in the vast majority of 12-h cycles (although these rhythms seem to dampen out by two days post synchronization) [30] (Figs. 2 and S1). Since the liver slices are cultured and synchronized by dexamethasone ex vivo, these data also indicate that 12-h rhythms of gene expression are cell-autonomous.
Fig. 2.
The XBP1s-dependent 12-h pacemaker is separate from the BMAL1-dependent circadian clock in the mouse. Heatmap illustrating 12-h hepatic transcriptome (upper) and core circadian clock TFs circadian expression (lower) in wild-type, XBP1 liver-specific knockout [6], and BMAL1-knockout ex vivo liver slices [30]. Chromatin binding of XBP1s [6] and BMAL1 [12] on the proximal promoter regions of these genes are shown on the right
In addition to ex vivo liver slices, cell-autonomous 12-h rhythms of gene expression are also prevalently found in murine hepatocyte MMH-D3 cells synchronized by serum shock [6, 31]. More importantly, Eif2ak3 (a canonical 12-h pacemaker output gene) promoter-driven-destabilized green fluorescent protein (dGFP) intensity revealed robust 12-h rhythmicity in unsynchronized single mouse embryonic fibroblasts (MEF) in a BMAL1-independent fashion [4, 6]. Besides dexamethasone and serum shock, which can also synchronize the circadian clock, 12-h rhythms can be uniquely synchronized by ER stressors. A lower dose of tunicamycin shock (25 ng/mL for 2 h) induces 12-h oscillations of key UPR and metabolic gene expression in MEFs [4, 6]. Metabolic stressors, specifically the excess or deprivation of glucose, also synchronizes 12-h rhythms in vitro [4]. Notably, 12-h rhythms of gene expression (including Xbp1s itself) induced by tunicamycin or glucose deprivation are not impaired by Bmal1 knockdown conditions [4, 6]. These evidences collectively support the existence of a circadian clock-independent, cell-autonomous mammalian 12-h pacemaker regulating the majority of 12-h rhythms of gene expression, although the other two hypotheses cannot be ruled out for certain 12-h genes.
XBP1s is a major transcriptional regulator of the mammalian 12-h pacemaker
Emerging data indicate that mammalian 12-h cycles are regulated by XBP1s. XBP1s is particularly intriguing, as it represents a molecular link between ER stress sensed in the ER and the transcriptional UPR response occurring in the nucleus [32, 33]. During ER stress, IRE1α oligomerizes in the ER membrane, activates its ribonuclease domain through auto (self) phosphorylation, and cleaves a 26 bp unconventional intron from the unspliced form of Xbp1 (Xbp1us) mRNA, leading to a frameshift in Xbp1 coding sequence and the translation of XBP1s in the cytosol [34, 35]. XBP1s then translocate into the nucleus and activates the transcription of UPR genes, including many that were found to have 12-h rhythms [6, 14]. XBP1s also transcriptionally regulates its own expression (Xbp1us), therefore creates a self-generating and self-regulating gene activation feedback loop [4, 6, 14, 36, 37].
Rigorous investigation of Xbp1 function revealed a central role in orchestrating the 12-h pacemaker. Liver-specific deletion of XBP1 in mice drastically impaired 12-h rhythms of gene expression (abolished 54.5%, and dampened the amplitude of another 31.6% of 12-h transcriptome) compared to control mice, while the core transcriptional-translational feedback loop of the circadian clock and the vast majority of circadian clock-controlled output genes remained intact (Figs. 2 and S1) [6]. Chromatin immunoprecipitation sequencing (ChIP-Seq) revealed a robust 12-h rhythm of hepatic XBP1s recruitment (peaking at CT0 and CT12) to the proximal promoter regions of 12-h genes that harbor consensus XBP1s binding sites CCACGTCA [6] (Figs. 2 and S1). Overall, a positive correlation among the amplitude of 12-h mRNA oscillation and the XBP1s binding motif stringency score was observed [6]. On the other hand, core circadian clock transcription factors (TF) bindings are largely absent from the promoters of dominant 12-h genes [6] (Figs. 2 and S1). For dual 12-h-clock and circadian clock-regulated genes (such as Spcs2 and Txndc5), XBP1s and core circadian clock TFs chromatin binding sites are usually spatially separated [6] (Fig. S1). In the case of Spcs2, XBP1s binding localizes at the promoter, while core circadian TFs bind at the transcription termination sites. As a result, the loss of XBP1s abolishes the superimposed 12-h rhythm of Spcs2, with the remaining 24-h rhythm still present. Similarly, loss of BMAL1 abolishes the superimposed 24-h rhythm, but the 12-h rhythm remains [6, 12, 30] (Figure S1). A similar scenario is found for Txndc5 gene, although in this case, both XBP1s and core circadian clock TFs bind at the proximal promoter region with their respective binding sites spatially separate [6, 12, 30] (Figure S1). XBP1s further transcriptionally regulates the 12-h rhythms of gene expression in a cell-autonomous manner. Knocking down Xbp1 in MEFs impaired the cell-autonomous 12-h rhythms of gene expression synchronized by serum shock or tunicamycin, without affecting the circadian gene expression of core circadian clock TFs [4, 6].
It is worth noting that of those over 3,000 hepatic 12-h transcriptomes, direct targeting of XBP1s to promoters was only observed for 550 genes [6], which are significantly enriched in protein homeostasis/ER stress pathways, and to a lesser extent, mRNA metabolism [6]. This result was largely consistent with previously published XBP1s ChIP-Seq in other tissues [38, 39]. XBP1s-dependent 12-h transcriptome lacking XBP1s proximal promoter binding, on the other hand, is strongly enriched in mRNA metabolism, and motif analysis of the promoter regions of these genes revealed novel binding motifs including E26 transformation-specific (ETS) transcription factors (which will be discussed in detail later). These data, thus, imply the presence of a hierarchy of 12-h pacemaker transcriptional control: while XBP1s can directly transcriptionally regulate the 12-h oscillation of many protein homeostasis genes, it also controls the 12-h rhythmic gene expression of several transcriptional factors (like GABPA), which can, in turn, modulate a much larger repertoire of 12-h transcriptome with more diverse functions [6].
One question that remains elusive, nonetheless, is the identification of TFs mediating the negative feedback loop that is required for sustaining XBP1s-mediated cell-autonomous oscillations of the 12-h pacemaker. The top candidate is an unspliced form of XBP1 (XBP1us), which has been shown previously to antagonize XBP1s-mediated UPR [40]. Providing support for this hypothesis is the observed anti-phase relation of their 12-h mRNA expression: hepatic Xbp1us mRNA peak at CT4 (and CT16), compared to the CT0 (and CT12) peaking of Xbp1s [14], although future studies are needed to conclusively demonstrate the potential implication of Xbp1us in the negative limb of 12-h pacemaker.
ETS and NFY transcription factor family members are putative novel regulators of 12-h rhythm
To search for potential novel transcriptional regulators of mammalian 12-h rhythms, several studies have resorted to computational/bioinformatic approaches. Specifically, transcription factors with 12-h rhythms of gene expression observed in multiple tissues as well as their DNA binding motifs enriched at the gene regulatory regions of 12-h transcriptome are likely candidates. Using this approach, several TFs belonging to E26 transformation-specific (ETS) [6, 14, 26, 41] and Nuclear transcription factor Y (NFY) [14] family members were identified, in addition to the Basic Leucine Zipper Domain (bZIP) family of TFs to which XBP1s belongs [14]. For the bZIP family, in addition to XBP1s, ATF6 (both α and β isoforms), CREB3, and CREB3L2 bind to similar DNA motifs, and their expression also exhibit 12-h rhythms of gene expression in multiple mouse tissues [6, 14]. As a side note, being one of three branches of canonical UPR, although ATF4 protein expression also exhibits a 12-h rhythm in mouse liver [4], its DNA binding motif is not as strongly enriched at gene promoters of 12-h transcriptome as other bZIP (including ATF6, XBP1s, CREB3), ETS and NFY TFs [6]. ETS is a large family of TFs that bind to consensus sequence CCGGAAG, known as ETS-binding site (EBS) [42], which also often occur in cis in the proximity of XBP1s cistrome in mouse liver [6]. Its family member Gabpa, Elf1, Erg, Etv1, Etv5, and Ets2 all exhibit dominant 12-h rhythms of mRNA expression in mouse liver (although other members like Elf2, Erf, and Spib have both superimposed circadian rhythm and 12-h rhythms of gene expression, with circadian rhythms being the dominant ones) (Table S1), with Gabpa and Elf1 further being direct target genes of XBP1s (which are defined as 12-h cycling genes with 12-h XBP1s chromatin recruitment to promoters and impaired 12-h oscillation in the absence of XBP1) [14]. 12-h rhythms of hepatic GABPA expression was further confirmed at the nuclear protein level as well as in an in vitro assay where hepatic nuclear GABPA/GABPB2 binding to an artificial DNA fragment harboring GABP consensus sequence also follows a 12-h rhythm [6, 43]. NFY family includes three members: NFYA, NFYB, and NFYC and binds with high affinity to CCAAT motif in the promoter region of target genes, which are also found to be highly enriched at XBP1s binding sites [14, 39]. In mouse liver, Nfyb is also a direct target gene of XBP1s and exhibit a 12-h rhythm of expression [14]. Nyfb further exhibits 12-h rhythms of gene expression in additional tissues in mice, including skeletal muscle, brown adipose tissue, and adrenal gland [14].
The putative implication of Gabpa in the regulation of mammalian 12-h pacemaker was further tested using Manf (a 12-h output gene) promoter-driven luciferase-expressing MEFs. Real-time luminescence measurement identified a lengthened period to 14 h in Gabpa knocking down cells, although the amplitude remains similar [6]. While this is the first study to investigate the potential role of ETS TF in the regulation of 12-h rhythm, whether its role in regulating the ultradian period can be generalized remains to be seen.
Despite the lack of concrete experimental evidence supporting additional bZIP TFs like ATF6, CREB3, CREB3L2 and NFY family in the regulation of 12-h ultradian rhythms, a theoretical tripartite regulatory network comprising of ETS, bZIP and NFY family of TFs with intricate interconnected positive feedforward and negative feedback loops was recently proposed to be the foundation of the mammalian 12-h pacemaker [14], although the detailed molecular circuits have yet to be elucidated.
The mammalian 12-h pacemaker is evolutionarily conserved and may be circatidal in origin
The prevalent mammalian 12 h rhythm of gene expression and metabolism is reminiscent of the ~ 12-h circatidal rhythms of coastal and estuarine animals that modulate their behavior in tune to the ~ 12.4 h ebb and flow of the tides, which is in turn orchestrated by the gravitational pull of the moon [44, 45]. The circatidal clock, therefore, has a separate genesis from the circadian clock, which is adapted to the 24-h cycles of light and darkness produced by the earth’s rotation on its axis [2]. Given that life first evolved in the ocean, it follows that the tides influence the timing of the behavior, metabolism, and gene expression of simple organisms [45].
Small, evolutionarily ancient, ocean-dwelling creatures have dominant 12-h rhythms of activity and gene expression [45]. For example, peak locomotor activity in crustaceans including C. maenas and E. pulchra coincides with high tide, and their 12-h rhythmicity in behaviors can be maintained in the lab under constant conditions [44, 46–49]. In addition, the 12-h behavior rhythms of these circatidal marine animals can also be entrained by individual components related to tides, such as salt concentration, osmolarity, and hydrostatic pressure [44, 50]. Intriguingly, it has been shown that in E. pulchra, the 12-h circatidal clock is dissociated from the circadian timekeeping system [48]. Sea creatures eventually evolved to survive on land, and organisms that live at the juncture of sea and land in tidal zones also have strong 12-h pacemakers [44, 51–53]. The mangrove cricket is an exemplar of these. Mangrove crickets have 12-h rhythms that persist in constant darkness, and are maintained even following removal of the optic lobe. Furthermore, these 12-h rhythms survive even when the expression of components of the circadian TTFL such as Clock or per are lowered through gene knock-down [51–53].
Comparing 12-h gene signature of circatidal animals and mice revealed stunning similarities. In E. pulchra, robust 12 h circatidal mRNA rhythms for 10 mitochondrial DNA (mtDNA)-encoded protein-coding genes (Mt-Nd1 ~ 6, Mt-Cox1 ~ 3 and Cytb) that encode mitochondrial components of complexes I (NADH dehydrogenase) and IV (cytochrome c oxidase) were previously described [49]. Intriguingly, in mouse liver, post hoc analysis of two Gro-Seq [54] and Nascent-Seq [55] datasets that measure nascent RNA transcription rate revealed conserved 12-h rhythms of global mtDNA-encoded gene transcription [4]. These 12-h rhythms of mtDNA-encoded gene expression were further confirmed at mature mRNA and protein levels in mouse liver [4, 56, 57]. While BMAL1 is dispensable for establishing 12-h rhythms of these genes [4], their relationship with XBP1s-mediated CEDIF is unclear at this moment. In another recent endeavor, Pan et al. compared the 12-h transcriptome of the mouse with those of two marine animals with circatidal clock: aposymbiotic sea anemone Aiptasia diaphaha [58] and one limpet Cellana rota [59], and found strong overlap among their 12-h transcriptomes that are enriched in CEDIF pathways [6]. Intriguingly, 12-h rhythms of Xbp1, Gabpa and Gabpb2 expression were also identified in these two marine species [2, 6]. Further, 12-h rhythms of gene expression involved in ER stress and protein homeostasis was recently identified in the circatidal mangrove cricket Apteronemobius asahinai [60]. Collectively, these data strongly suggest that the mammalian 12-h pacemaker evolved from the circatidal clock of coastal and estuarine animals [2, 4, 6]. This hypothesis is further supported by recent reports of an earlier evolutionary origin for genes cycling with a 12-h period than circadian genes [41], and higher conservation of chromosomal location of 12-h genes compared to circadian genes [22].
Beyond the prevalence of behavioral and physiological 12-h rhythms beginning with sea creatures and extending to mice, there are 12-h rhythms of gene expression conserved in many other organisms. Indeed, 12-h rhythms of gene expression involved in nuclear DNA-encoded CEDIF and mtDNA-encoded mitochondria metabolism were identified in temperature (but not light)-entrained as well as free-running C. elegans [4, 61]. Transcription factor Xbp1, Gabpa, and Nyfb orthologs in C. elegans also exhibited robust 12-h rhythms [4, 61]. In addition to the nematode, 12 h oscillations of ER homeostasis and metabolism gene expression also were found in light-entrained and free-running zebrafish [2, 62] as well as in different tissues of light-entrained olive baboons, as previously mentioned [15, 63]. Lending further evidence to the tidal origin of mammalian 12-h pacemaker is the recent discovery that XBP1 activity can respond to hyperosmolarity in mammals [64].
Maybe the most important question is whether a 12-h pacemaker exists in humans. Due to the technical difficulty and ethical constraint of obtaining time series tissue samples from humans, to the best of our knowledge, 12-h rhythms of high throughput “omics” data have not been reported in humans so far. Nonetheless, using Fourier transform and/or wavelet analysis, superimposed 12-h rhythms of human body temperature [65–70], heart rate variability and blood pressure [71–76], cognitive performance [70, 77–83], cerebrospinal fluid sodium level [84], circulating serum, urine metal and hormones levels [85–87] and sleep patterns [79, 80, 88–95] were all reported, therefore implying the very likely existence of a similar 12-h pacemaker in us too.
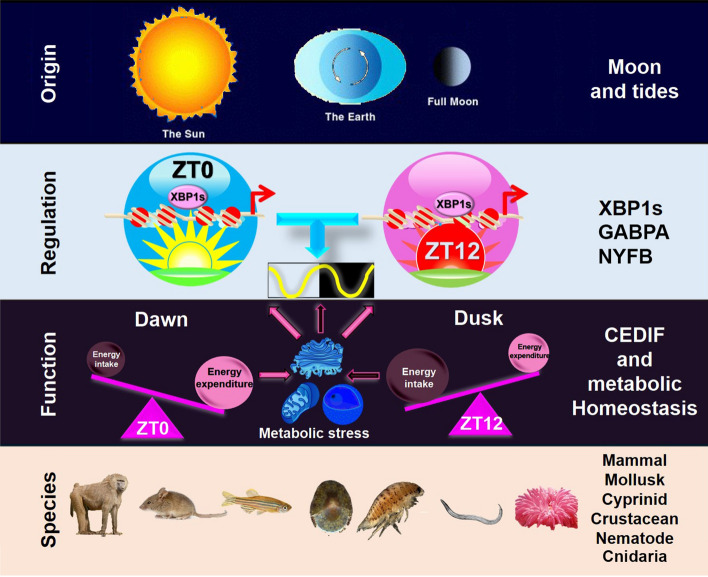
The conservation of XBP1s-dependent 12-h pacemaker in mammals shows that mammals retained their 12-h pacemaker after evolving to live on land. How the 12-h pacemaker provides an evolutionary advantage for mammals that no longer respond to tidal cues is an important question. We reason that the mammalian 12-h pacemaker is coopted to adapt to the 12-h cycle of metabolic stress that peak at transition periods: from sleep/fast to activity/feeding in the subjective dawn and the opposite in the subjective dusk. At the subjective dawn, the prolonged absence of energy intake combined with still significant energy expenditure from the subjective night leads to a peak of energy ‘shortage’, while at the subjective dusk, sufficient energy intake during the subjective day combined with gradually decreasing energy expenditure gives rise to a peak of energy ‘excess’. Both energy ‘shortage’ and energy ‘excess’ can impose metabolic stress and activate XBP1s to elicit UPR [14, 96, 97] (Fig. 3). 12-h pacemaker-mediated 12-h rhythms of CEDIF and related metabolic process, therefore, may provide increased metabolic capacity at a 12-h interval in mammals to anticipate and respond to the two transitions between rest and activity states under physiological conditions. It is further reasonable to deduce that multiple pathological conditions can deviate this system from its normal 12-h cycle, and if uncorrected, will contribute to increased disease susceptibility [98], which we will briefly discuss next.
Fig. 3.
A diagram summarizing the origin, regulation, function, and evolutionary conservation of mammalian 12-h. Please see the main text for a detailed description of each section
12-h pacemaker and the canonical ER stress/UPR
Although XBP1s is the major transcriptional regulator for both the mammalian 12-h pacemaker and the canonical ER stress/UPR, at least two features distinguish between these two processes. First, while XBP1s-dependent 12-h oscillating genes include most of the canonical genes regulated in response to ER stress, the repertoire of genes controlled by the former is much larger and more diverse, including those involved in mRNA metabolism, immune response, and numerous metabolic pathways that were not commonly associated with the canonical UPR [6]. As previously mentioned, these non-UPR-related pathways are very likely indirectly regulated by XBP1s via additional TFs. Secondly, even for genes commonly regulated by the two processes, the amplitude of gene expression in a 12-h cycle is much smaller than what was observed during canonical ER stress. For example, while the pathological level of ER stress can induce near-complete Xbp1 splicing [99], in mouse liver, the percentage of Xbp1 spliced (the ratio of Xbp1s to total Xbp1) oscillates between 3 to 12% in a given diurnal cycle [6]. Consequently, the amplitude of most 12-h transcriptome falls below fourfold change [6]. As a matter of fact, the pathological level of both acute and chronic ER stress has deleterious effects on the 12-h pacemaker in mouse. Acute induction of severe ER stress by tunicamycin injection alters the 12-h rhythmic gene expression in the mouse liver [24]. Chronic ER stress induced by nutritional challenges with a high fat diet also significantly disrupts the 12-h oscillation of hepatic transcriptome [2, 56]. Similarly, only low dose of tunicamycin treatment (25 ng/ml) can synchronize the 12-h rhythms of gene expression in vitro, while a higher dose severely impairs the oscillation [4]. In sum, whereas one of the functions of the 12-h pacemaker may be considered as regulating the previously proposed “endogenous ER stress/UPR cycle” [98], the full spectrum of its functions are expected to extend beyond the simple regulation of ER stress-related biological pathways.
12-h pacemaker and nonalcoholic fatty liver diseases (NAFLD)
IRE1α/XBP1s signaling cascade is critical for metabolic health on both the cellular and organismal levels. Particularly, numerous studies have shown that activating ER stress-sensing or ER quality control pathways, including Xbp1s and IRE1α, are protective against hepatic steatosis [25, 100–102]. One of such mechanisms is through regulating the membrane fluidity by controlling lipid composition. For example, Lysophosphatidylcholine Acyltransferase 3 (Lpcat3) gene expression, which promotes the incorporation of polyunsaturated fatty acid into phosphatidylcholines (PC) in the ER membrane, is induced during high-fat diet-mediated ER stress [4, 103]. The elevated level of polyunsaturated PC in turn increases the ER membrane fluidity and ameliorates the original ER stress [103]. Interestingly, robust 12-h rhythms of Lpcat3 gene expression and levels of various 2-Lyso-PC species (LPCAT3 conjugates 2-Lyso-PC and unsaturated Acyl-CoA to form PC) was found in mouse liver [2, 4]. More importantly, liver-specific ablation of XBP1 dampened the hepatic 12-h oscillation of Lpcat3 gene expression, reduced the hepatic polyunsaturated PC level, leading to a drastic reduction of membrane fluidity and impaired lipid metabolism, and subsequently accelerated NAFLD development, manifested further with impaired glucose tolerance and hyperinsulinemia in mice [104].
Interestingly, downregulation of the average expression of 12-h genes is also associated strongly with progression to hepatic steatosis and nonalcoholic steatohepatitis (NASH) in humans, although whether the 12-h rhythms were also disrupted in human NAFLD subjects remains to be determined [4, 105]. Albeit still in the early stage of research, these data have already started to reveal a potential causal role of the mammalian 12-h pacemaker in maintaining metabolic homeostasis and protecting against metabolic syndromes.
12-h pacemaker and immune regulation
In addition to genes involved in CEDIF and metabolism, genes implicated in immune regulation were also shown to exhibit 12-h rhythms of gene expression both in mouse liver in vivo and in MMH-D3 cells in vitro [6]. 12-h genes such as Rela, Nfkb1, and Tnfaip3 are involved in NFκb and tumor necrosis factor (TNF) signaling [6]. While surprising at first glance, these findings are in fact in line with a recently published study reporting two daily peaks of bone marrow TNF level in mice peaking at the morning (CT0) and evening (CT12) [106]. Interestingly, the two peaks of TNF regulate distinct biological processes. TNF morning peak augments hematopoietic stem and progenitor cell differentiation and increases vascular permeability to replenish the blood, while the evening peak exerts opposite effects by driving the renewal of hematopoietic stem cells and diminishing the vascular permeability [106].
Summary and future directions
12-h rhythms have existed for eons but have only recently been discovered by bold scientific efforts. Circatidal rhythmic activity of simple sea creatures such as C. rota and other crustaceans have long been known. However, 12-h rhythms in mammals, particularly regarding their regulation and function, are a relatively recent discovery.
Emerging studies of the 12-h pacemaker highlight its ancient origins as adaptations to the tides. 12-h rhythms of genes involved in ER stress, UPR, and mitochondrial genes show conservation in species as divergent as crustaceans, mollusk, nematode, and mammals. The purpose of these rhythms was found to be regulating the capacity of CEDIF: the orchestration of increased capacity for RNA and protein production and processing at the rush hours of dawn and dusk. These rush hours allow for increased gene expression at transitions between rest and activity. A newly proposed vehicle-cargo hypothesis offers an explanation for how the 12-h pacemaker and the circadian clock complement each other: the 12-h pacemaker increases overall gene expression capacity at dawn and dusk, with the circadian clock determining which genes are expressed at each rush hour (Fig. 3).
Since the original discovery of 12-h rhythms of gene expression in mouse liver, many more 12-h rhythmic genes have been found in several regions of the brain and in many peripheral tissues. Furthermore, analysis of the regulatory regions of these genes has provided novel candidates for a molecular clockwork interlocking with XBP1s, which opens new avenues for further study (Fig. 3). The complete molecular underpinnings of the 12-h clock, as well as the roles of XBP1s outside the liver, also long for future investigations.
It is our sincere hope that this review will attract more scientists into the nascent field of mammalian 12-h rhythms, as many more outstanding questions remain to be answered. For instance, it remains undetermined whether there is a central 12-h pacemaker in the brain that serves as the SCN equivalent for the circadian clock (hippocampus is one candidate). Future directions further include the identification of mRNA and protein substrates subject to 12-h CEDIF regulation, potential endocrine/paracrine factors capable of synchronizing the 12-h rhythms in different organs/cells, the characterization of the functions of the mammalian 12-h pacemakers in different diseases, and the discovery of the modes of crosstalk among the 12-h rhythm, other ultradian rhythms, and the circadian rhythms, just to name a few.
Supplementary Information
Below is the link to the electronic supplementary material.
Figure S1. The XBP1s-dependent 12-hour pacemaker is separate from the BMAL1-dependent circadian clock in mouse. Representative gene expression of 12-hour and/or circadian genes in wild-type, XBP1 liver specific knockout [6], and BMAL1-knockout ex vivo liver slices [30]. Temporal cistrome of XBP1s as well as key core circadian clock TFs are further shown. (tif 11670 kb)
Table S1. Rhythms uncovered for ETS TFs in mouse liver. For everyone EST TF, whether a 12-hour or circadian rhythm exist is shown, as well as the dominant rhythm identified (xlsx 12 kb)
Acknowledgement
We want to thank Dr. Akhilesh Reddy for kindly sharing with us the raw RNA-seq data to analyze ultradian rhythm in BMAL1-less ex vivo liver slices. We also would like to thank Dr. Baby Anjum for assistance with initial literature search. We apologize for any potential omission of relevant works and citations due to space constraints. We thank the American Diabetes Association junior faculty development award 1-18-JDF-025 to B.Z. B.Z. was further supported by the National Institute Of General Medical Sciences of the National Institutes of Health under Award Number DP2GM140924. Research reported in this publication was further supported by the National Institute of Diabetes And Digestive And Kidney Diseases of the National Institutes of Health under award number P30DK120531 to Pittsburgh Liver Research Center, in which B.Z. is a member. H.B. is further supported by a T32 training grant T32AG021885 from National Institute of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rijo-Ferreira F, Takahashi JS. Genomics of circadian rhythms in health and disease. Genome Med. 2019;11(1):82. doi: 10.1186/s13073-019-0704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu B, Dacso CC, O'Malley BW. Unveiling "Musica Universalis" of the cell: a brief history of biological 12-hour rhythms. J Endocr Soc. 2018;2(7):727–752. doi: 10.1210/js.2018-00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu B, Zhang Q, Pan Y, Mace EM, York B, Antoulas AC, Dacso CC, O'Malley BW (2017) A cell-autonomous mammalian 12 h clock coordinates metabolic and stress rhythms. Cell Metab 25(6):1305–1319 e1309. 10.1016/j.cmet.2017.05.004 [DOI] [PMC free article] [PubMed]
- 5.Antoulas AC, Zhu B, Zhang Q, York B, O'Malley BW, Dacso CC. A novel mathematical method for disclosing oscillations in gene transcription: a comparative study. PLoS ONE. 2018;13(9):e0198503. doi: 10.1371/journal.pone.0198503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Y, Ballance H, Meng H, Gonzalez N, Kim S-M, Abdurehman L, York B, Chen X, Schnytzer Y, Levy O, Dacso CC, McClung CA, O’Malley BW, Liu S, Zhu B. 12-h clock regulation of genetic information flow by XBP1s. PLoS Biol. 2020;18(1):e3000580. doi: 10.1371/journal.pbio.3000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25(5):372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang R, Su Z. Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics. 2010;26(12):i168–174. doi: 10.1093/bioinformatics/btq189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thaben PF, Westermark PO. Detecting rhythms in time series with RAIN. J Biol Rhythms. 2014;29(6):391–400. doi: 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan Y, Ballance H, Schnytzer Y, Chen X, Levy O, Coarfa C, Zhu B (2019) 12h-clock control of central dogma information flow by XBP1s. bioRxiv:559039. 10.1101/559039
- 11.Ananthasubramaniam B, Diernfellner A, Brunner M, Herzel H (2018) Ultradian Rhythms in the Transcriptome of Neurospora crassa. iScience 9:475–486. doi:10.1016/j.isci.2018.11.012 [DOI] [PMC free article] [PubMed]
- 12.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu B. Decoding the function and regulation of the mammalian 12h-clock. J Mol Cell Biol. 2020 doi: 10.1093/jmcb/mjaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Athman R, Knezevic D, Fuhr L, Relogio A (2019) A computational analysis of alternative splicing across mammalian tissues reveals circadian and ultradian rhythms in splicing events. Int J Mol Sci. 10.3390/ijms20163977 [DOI] [PMC free article] [PubMed]
- 16.Jiao X, Wu M, Lu D, Gu J, Li Z. Transcriptional profiling of daily patterns of mRNA expression in the C57BL/6J mouse cornea. Curr Eye Res. 2019;44(10):1054–1066. doi: 10.1080/02713683.2019.1625408. [DOI] [PubMed] [Google Scholar]
- 17.Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, Ngotho M, Kariuki T, Dkhissi-Benyahya O, Cooper HM, Panda S. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. 2018;359(6381):1232. doi: 10.1126/science.aao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoon Kim Y, Lazar MA. Transcriptional control of circadian rhythms and metabolism: a matter of time and space. Endocr Rev. 2020 doi: 10.1210/endrev/bnaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koyanagi I, Akers KG, Vergara P, Srinivasan S, Sakurai T, Sakaguchi M. Memory consolidation during sleep and adult hippocampal neurogenesis. Neural Regen Res. 2019;14(1):20–23. doi: 10.4103/1673-5374.243695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadel L, Hupbach A, Gomez R, Newman-Smith K. Memory formation, consolidation and transformation. Neurosci Biobehav Rev. 2012;36(7):1640–1645. doi: 10.1016/j.neubiorev.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 21.McMaster CR. Lipid metabolism and vesicle trafficking: more than just greasing the transport machinery. Biochem Cell Biol. 2001;79(6):681–692. doi: 10.1139/o01-139. [DOI] [PubMed] [Google Scholar]
- 22.Genov N, Castellana S, Scholkmann F, Capocefalo D, Truglio M, Rosati J, Turco EM, Biagini T, Carbone A, Mazza T, Relogio A, Mazzoccoli G (2019) A multi-layered study on harmonic oscillations in mammalian genomics and proteomics. Int J Mol Sci. 10.3390/ijms20184585 [DOI] [PMC free article] [PubMed]
- 23.Hughes ME, Hong HK, Chong JL, Indacochea AA, Lee SS, Han M, Takahashi JS, Hogenesch JB. Brain-specific rescue of clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet. 2012;8(7):e1002835. doi: 10.1371/journal.pgen.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11(1):47–57. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, Chang L, Xu W, Miao H, Leonardi R, Chen YE, Jackowski S, Kaufman RJ. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J. 2011;30(7):1357–1375. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westermark PO, Herzel H. Mechanism for 12 hr rhythm generation by the circadian clock. Cell Rep. 2013;3(4):1228–1238. doi: 10.1016/j.celrep.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang G, Chen L, Grant GR, Paschos G, Song WL, Musiek ES, Lee V, McLoughlin SC, Grosser T, Cotsarelis G, FitzGerald GA (2016) Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med 8(324):324ra316. 10.1126/scitranslmed.aad3305 [DOI] [PMC free article] [PubMed]
- 29.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, Takahashi JS. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104(9):3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray S, Valekunja UK, Stangherlin A, Howell SA, Snijders AP, Damodaran G, Reddy AB. Circadian rhythms in the absence of the clock gene Bmal1. Science. 2020;367(6479):800–806. doi: 10.1126/science.aaw7365. [DOI] [PubMed] [Google Scholar]
- 31.Atwood A, DeConde R, Wang SS, Mockler TC, Sabir JS, Ideker T, Kay SA. Cell-autonomous circadian clock of hepatocytes drives rhythms in transcription and polyamine synthesis. Proc Natl Acad Sci USA. 2011;108(45):18560–18565. doi: 10.1073/pnas.1115753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 33.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 34.Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. J Biochem. 2009;146(6):743–750. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- 35.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90(6):1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 36.Majumder M, Huang C, Snider MD, Komar AA, Tanaka J, Kaufman RJ, Krokowski D, Hatzoglou M. A novel feedback loop regulates the response to endoplasmic reticulum stress via the cooperation of cytoplasmic splicing and mRNA translation. Mol Cell Biol. 2012;32(5):992–1003. doi: 10.1128/MCB.06665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 38.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27(1):53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 39.Pramanik J, Chen X, Kar G, Henriksson J, Gomes T, Park JE, Natarajan K, Meyer KB, Miao Z, McKenzie ANJ, Mahata B, Teichmann SA. Genome-wide analyses reveal the IRE1a-XBP1 pathway promotes T helper cell differentiation by resolving secretory stress and accelerating proliferation. Genome Med. 2018;10(1):76. doi: 10.1186/s13073-018-0589-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172(4):565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castellana S, Mazza T, Capocefalo D, Genov N, Biagini T, Fusilli C, Scholkmann F, Relogio A, Hogenesch JB, Mazzoccoli G. Systematic analysis of mouse genome reveals distinct evolutionary and functional properties among circadian and ultradian genes. Front Physiol. 2018;9:1178. doi: 10.3389/fphys.2018.01178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper CD, Newman JA, Gileadi O. Recent advances in the structural molecular biology of Ets transcription factors: interactions, interfaces and inhibition. Biochem Soc Trans. 2014;42(1):130–138. doi: 10.1042/BST20130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Song L, Liu M, Ge R, Zhou Q, Liu W, Li R, Qie J, Zhen B, Wang Y, He F, Qin J, Ding C. A proteomics landscape of circadian clock in mouse liver. Nat Commun. 2018;9(1):1553. doi: 10.1038/s41467-018-03898-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilcockson D, Zhang L. Circatidal clocks. Curr Biol. 2008;18(17):R753–R755. doi: 10.1016/j.cub.2008.06.041. [DOI] [PubMed] [Google Scholar]
- 45.Andreatta G, Tessmar-Raible K. The still dark side of the moon: molecular mechanisms of lunar-controlled rhythms and clocks. J Mol Biol. 2020;432(12):3525–3546. doi: 10.1016/j.jmb.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.NAYLOR E (1958) Tidal and Diurnal Rhythms of Locomotory Activity in Carcinus maenas (L.). J Exp Biol 35 (3):602–610
- 47.Naylor E. Crab clockwork: the case for interactive circatidal and circadian oscillators controlling rhythmic locomotor activity of Carcinus maenas. Chronobiol Int. 1996;13(3):153–161. doi: 10.3109/07420529609012649. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L, Hastings MH, Green EW, Tauber E, Sladek M, Webster SG, Kyriacou CP, Wilcockson DC. Dissociation of circadian and circatidal timekeeping in the marine crustacean Eurydice pulchra. Curr Biol. 2013;23(19):1863–1873. doi: 10.1016/j.cub.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Neill JS, Lee KD, Zhang L, Feeney K, Webster SG, Blades MJ, Kyriacou CP, Hastings MH, Wilcockson DC. Metabolic molecular markers of the tidal clock in the marine crustacean Eurydice pulchra. Curr Biol. 2015;25(8):R326–327. doi: 10.1016/j.cub.2015.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akiyama T (2004) Entrainment of the circatidal swimming activity rhythm in the cumacean Dimorphostylis asiatica (Crustacea) to 12.5-hour hydrostatic pressure cycles. Zool Sci 21 (1):29–38. 10.2108/0289-0003(2004)21[29:EOTCSA]2.0.CO;2 [DOI] [PubMed]
- 51.Takekata H, Matsuura Y, Goto SG, Satoh A, Numata H. RNAi of the circadian clock gene period disrupts the circadian rhythm but not the circatidal rhythm in the mangrove cricket. Biol Lett. 2012;8(4):488–491. doi: 10.1098/rsbl.2012.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takekata H, Numata H, Shiga S, Goto SG. Silencing the circadian clock gene Clock using RNAi reveals dissociation of the circatidal clock from the circadian clock in the mangrove cricket. J Insect Physiol. 2014;68:16–22. doi: 10.1016/j.jinsphys.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 53.Takekata H, Numata H, Shiga S. The circatidal rhythm persists without the optic lobe in the mangrove cricket Apteronemobius asahinai. J Biol Rhythms. 2014;29(1):28–37. doi: 10.1177/0748730413516309. [DOI] [PubMed] [Google Scholar]
- 54.Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, Roy A, Gerhart-Hines Z, Sun Z, Lazar MA. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159(5):1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155(7):1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10(1):e1004047. doi: 10.1371/journal.pgen.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sorek M, Schnytzer Y, Ben-Asher HW, Caspi VC, Chen CS, Miller DJ, Levy O. Setting the pace: host rhythmic behaviour and gene expression patterns in the facultatively symbiotic cnidarian Aiptasia are determined largely by Symbiodinium. Microbiome. 2018;6(1):83. doi: 10.1186/s40168-018-0465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schnytzer Y, Simon-Blecher N, Li J, Ben-Asher HW, Salmon-Divon M, Achituv Y, Hughes ME, Levy O. Tidal and diel orchestration of behaviour and gene expression in an intertidal mollusc. Sci Rep. 2018;8(1):4917. doi: 10.1038/s41598-018-23167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Satoh A, Terai Y. Circatidal gene expression in the mangrove cricket Apteronemobius asahinai. Sci Rep. 2019;9(1):3719. doi: 10.1038/s41598-019-40197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Linden AM, Beverly M, Kadener S, Rodriguez J, Wasserman S, Rosbash M, Sengupta P. Genome-wide analysis of light- and temperature-entrained circadian transcripts in Caenorhabditis elegans. PLoS Biol. 2010;8(10):e1000503. doi: 10.1371/journal.pbio.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Li G, Wang H, Du J, Yan J. Analysis of a gene regulatory cascade mediating circadian rhythm in zebrafish. PLoS Comput Biol. 2013;9(2):e1002940. doi: 10.1371/journal.pcbi.1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, Ngotho M, Kariuki T, Dkhissi-Benyahya O, Cooper HM, Panda S. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. 2018 doi: 10.1126/science.aao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Casali C, Malvicini R, Erjavec L, Parra L, Artuch A, Fernández Tome MC (2020) X-box binding protein 1 (XBP1): a key protein for renal osmotic adaptation. Its role in lipogenic program regulation. Biochim Biophys Acta Mol Cell Biol Lipids 1865(4):158616. 10.1016/j.bbalip.2020.158616 [DOI] [PubMed]
- 65.Colquhoun WP, Blake MJ, Edwards RS. Experimental studies of shift-work I: a comparison of 'rotating' and 'stabilized' 4-hour shift systems. Ergonomics. 1968;11(5):437–453. doi: 10.1080/00140136808930993. [DOI] [PubMed] [Google Scholar]
- 66.Colquhoun WP, Paine MW, Fort A. Circadian rhythm of body temperature during prolonged undersea voyages. Aviat Space Environ Med. 1978;49(5):671–678. [PubMed] [Google Scholar]
- 67.Colquhoun WP, Paine MW, Fort A. Changes in the temperature rhythm of submariners following a rapidly rotating watchkeeping system for a prolonged period. Int Arch Occup Environ Health. 1979;42(3–4):185–190. doi: 10.1007/BF00377772. [DOI] [PubMed] [Google Scholar]
- 68.Moore-Ede MC, Czeisler CA. Mathematical models of the circadian sleep-wake cycle. New York: Raven Press; 1984. [Google Scholar]
- 69.Kronauer RE, Jewett ME. The relationship between circadian and hemicircadian components of human endogenous temperature rhythms. J Sleep Res. 1992;1(2):88–92. doi: 10.1111/j.1365-2869.1992.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 70.Monk TH, Buysse DJ, Reynolds CF, 3rd, Kupfer DJ. Circadian determinants of the postlunch dip in performance. Chronobiol Int. 1996;13(2):123–133. doi: 10.3109/07420529609037076. [DOI] [PubMed] [Google Scholar]
- 71.Wan C, Wang Z, Cornelissen G, Halberg F. Age, gender and circadian or circasemidian blood pressure and heart rate variation of children. Chronobiologia. 1992;19(3–4):121–129. [PubMed] [Google Scholar]
- 72.Otsuka K, Murakami S, Kubo Y, Yamanaka T, Mitsutake G, Ohkawa S, Matsubayashi K, Yano S, Cornelissen G, Halberg F. Chronomics for chronoastrobiology with immediate spin-offs for life quality and longevity. Biomed Pharmacother. 2003;57(Suppl 1):1s–18s. doi: 10.1016/j.biopha.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 73.Lee JS, Lee MS, Lee JY, Cornelissen G, Otsuka K, Halberg F. Effects of diaphragmatic breathing on ambulatory blood pressure and heart rate. Biomed Pharmacother. 2003;57(Suppl 1):87s–91s. doi: 10.1016/j.biopha.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 74.Otsuka K, Oinuma S, Cornelissen G, Weydahl A, Ichimaru Y, Kobayashi M, Yano S, Holmeslet B, Hansen TL, Mitsutake G, Engebretson MJ, Schwartzkopff O, Halberg F. Alternating light-darkness-influenced human electrocardiographic magnetoreception in association with geomagnetic pulsations. Biomed Pharmacother. 2001;55(Suppl 1):63s–75s. doi: 10.1016/s0753-3322(01)90007-1. [DOI] [PubMed] [Google Scholar]
- 75.Otsuka K, Cornelissen G, Furukawa S, Kubo Y, Hayashi M, Shibata K, Mizuno K, Aiba T, Ohshima H, Mukai C. Long-term exposure to space's microgravity alters the time structure of heart rate variability of astronauts. Heliyon. 2016;2(12):e00211. doi: 10.1016/j.heliyon.2016.e00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Otsuka K, Cornelissen G, Halberg F. Circadian rhythmic fractal scaling of heart rate variability in health and coronary artery disease. Clin Cardiol. 1997;20(7):631–638. doi: 10.1002/clc.4960200710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bjerner B, Holm A, Swensson A. Diurnal variation in mental performance; a study of three-shift workers. Br J Ind Med. 1955;12(2):103–110. doi: 10.1136/oem.12.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitler MM, Carskadon MA, Czeisler CA, Dement WC, Dinges DF, Graeber RC. Catastrophes, sleep, and public policy: consensus report. Sleep. 1988;11(1):100–109. doi: 10.1093/sleep/11.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dinges DF, Broughton RJ. Sleep and alertness: chronobiological, behavioral, and medical aspects of napping. New York: Raven Press; 1989. [Google Scholar]
- 80.Broughton RJ. SCN controlled circadian arousal and the afternoon "nap zone". Sleep Res Online. 1998;1(4):166–178. [PubMed] [Google Scholar]
- 81.Reinberg A, Bicakova-Rocher A, Nouguier J, Gorceix A, Mechkouri M, Touitou Y, Ashkenazi I. Circadian rhythm period in reaction time to light signals: difference between right- and left-hand side. Brain Res Cogn Brain Res. 1997;6(2):135–140. doi: 10.1016/S0926-6410(97)00024-4. [DOI] [PubMed] [Google Scholar]
- 82.Shub Y, Lewy H, Ashkenazi IE. Circadian pattern of simulated flight performance of pilots is derived from ultradian components. Chronobiol Int. 2001;18(6):987–1003. doi: 10.1081/CBI-100107973. [DOI] [PubMed] [Google Scholar]
- 83.Iskra-Golec I. Ultradian and asymmetric rhythms of hemispheric processing speed. Chronobiol Int. 2006;23(6):1229–1239. doi: 10.1080/07420520601077922. [DOI] [PubMed] [Google Scholar]
- 84.Harrington MG, Salomon RM, Pogoda JM, Oborina E, Okey N, Johnson B, Schmidt D, Fonteh AN, Dalleska NF. Cerebrospinal fluid sodium rhythms. Cerebrospinal Fluid Res. 2010;7:3. doi: 10.1186/1743-8454-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ayala DE, Hermida RC, Garcia L, Iglesias T, Lodeiro C. Multiple component analysis of plasma growth hormone in children with standard and short stature. Chronobiol Int. 1990;7(3):217–220. doi: 10.3109/07420529009056977. [DOI] [PubMed] [Google Scholar]
- 86.Francis SJ, Walker RF, Riad-Fahmy D, Hughes D, Murphy JF, Gray OP. Assessment of adrenocortical activity in term newborn infants using salivary cortisol determinations. J Pediatr. 1987;111(1):129–133. doi: 10.1016/S0022-3476(87)80359-1. [DOI] [PubMed] [Google Scholar]
- 87.Kanabrocki EL, Sothern RB, Ryan MD, Kahn S, Augustine G, Johnson C, Foley S, Gathing A, Eastman G, Friedman N, Nemchausky BA, Kaplan E. Circadian characteristics of serum calcium, magnesium and eight trace elements and of their metallo-moieties in urine of healthy middle-aged men. Clin Ter. 2008;159(5):329–346. [PubMed] [Google Scholar]
- 88.Broughton RJ. Biorhythmic variations in consciousness and psychological functions. Can Psychol Rev. 1979;16(4):217–239. doi: 10.1037/h0081810. [DOI] [Google Scholar]
- 89.Broughton R, Mullington J. Circasemidian sleep propensity and the phase-amplitude maintenance model of human sleep/wake regulation. J Sleep Res. 1992;1(2):93–98. doi: 10.1111/j.1365-2869.1992.tb00017.x. [DOI] [PubMed] [Google Scholar]
- 90.Hayashi M, Morikawa T, Hori T. Circasemidian 12 h cycle of slow wave sleep under constant darkness. Clin Neurophysiol. 2002;113(9):1505–1516. doi: 10.1016/S1388-2457(02)00168-2. [DOI] [PubMed] [Google Scholar]
- 91.Billiard M, Carlander B, Besset A. Circadian rhythm in normal and pathological sleep. Pathol Biol (Paris) 1996;44(6):509–517. [PubMed] [Google Scholar]
- 92.Nobili L, Besset A, Ferrillo F, Rosadini G, Schiavi G, Billiard M. Dynamics of slow wave activity in narcoleptic patients under bed rest conditions. Electroencephalogr Clin Neurophysiol. 1995;95(6):414–425. doi: 10.1016/0013-4694(95)00138-7. [DOI] [PubMed] [Google Scholar]
- 93.Broughton R, Mullington J. Chronobiological aspects of narcolepsy. Sleep. 1994;17(8 Suppl):S35–44. doi: 10.1093/sleep/17.suppl_8.S35. [DOI] [PubMed] [Google Scholar]
- 94.Broughton R, Dunham W, Newman J, Lutley K, Duschesne P, Rivers M. Ambulatory 24 hour sleep-wake monitoring in narcolepsy-cataplexy compared to matched controls. Electroencephalogr Clin Neurophysiol. 1988;70(6):473–481. doi: 10.1016/0013-4694(88)90145-9. [DOI] [PubMed] [Google Scholar]
- 95.De Koninck GC, Hebert M, Carrier J, Lamarche C, Dufour S. Body temperature and the return of slow wave activity in extended sleep. Electroencephalogr Clin Neurophysiol. 1996;98(1):42–50. doi: 10.1016/0013-4694(95)00215-4. [DOI] [PubMed] [Google Scholar]
- 96.Deng Y, Wang ZV, Tao C, Gao N, Holland WL, Ferdous A, Repa JJ, Liang G, Ye J, Lehrman MA, Hill JA, Horton JD, Scherer PE. The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. J Clin Invest. 2013;123(1):455–468. doi: 10.1172/JCI62819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shao M, Shan B, Liu Y, Deng Y, Yan C, Wu Y, Mao T, Qiu Y, Zhou Y, Jiang S, Jia W, Li J, Li J, Rui L, Yang L, Liu Y. Hepatic IRE1alpha regulates fasting-induced metabolic adaptive programs through the XBP1s-PPARalpha axis signalling. Nat Commun. 2014;5:3528. doi: 10.1038/ncomms4528. [DOI] [PubMed] [Google Scholar]
- 98.Fu S, Watkins SM, Hotamisligil GS. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 2012;15(5):623–634. doi: 10.1016/j.cmet.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 99.Uemura A, Oku M, Mori K, Yoshida H. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J Cell Sci. 2009;122(Pt 16):2877–2886. doi: 10.1242/jcs.040584. [DOI] [PubMed] [Google Scholar]
- 100.Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21(17):2975–2986. doi: 10.1091/mbc.E09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15(6):829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Herrema H, Zhou Y, Zhang D, Lee J, Salazar Hernandez MA, Shulman GI, Ozcan U. XBP1s is an anti-lipogenic protein. J Biol Chem. 2016;291(33):17394–17404. doi: 10.1074/jbc.M116.728949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rong X, Albert CJ, Hong C, Duerr MA, Chamberlain BT, Tarling EJ, Ito A, Gao J, Wang B, Edwards PA, Jung ME, Ford DA, Tontonoz P. LXRs regulate ER stress and inflammation through dynamic modulation of membrane phospholipid composition. Cell Metab. 2013;18(5):685–697. doi: 10.1016/j.cmet.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meng H, Gonzales NM, Lonard DM, Putluri N, Zhu B, Dacso CC, York B, O’Malley BW. XBP1 links the 12-hour clock to NAFLD and regulation of membrane fluidity and lipid homeostasis. Nat Commun. 2020;11(1):6215. doi: 10.1038/s41467-020-20028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ahrens M, Ammerpohl O, von Schonfels W, Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M, Hinrichsen H, Erhart W, Egberts J, Sipos B, Schreiber S, Hasler R, Stickel F, Becker T, Krawczak M, Rocken C, Siebert R, Schafmayer C, Hampe J. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18(2):296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 106.Golan K, Kumari A, Kollet O, Khatib-Massalha E, Subramaniam MD, Ferreira ZS, Avemaria F, Rzeszotek S, Garcia-Garcia A, Xie S, Flores-Figueroa E, Gur-Cohen S, Itkin T, Ludin-Tal A, Massalha H, Bernshtein B, Ciechanowicz AK, Brandis A, Mehlman T, Bhattacharya S, Bertagna M, Cheng H, Petrovich-Kopitman E, Janus T, Kaushansky N, Cheng T, Sagi I, Ratajczak MZ, Mendez-Ferrer S, Dick JE, Markus RP, Lapidot T (2018) Daily onset of light and darkness differentially controls hematopoietic stem cell differentiation and maintenance. Cell Stem Cell 23(4):572–585 e577. 10.1016/j.stem.2018.08.002 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The XBP1s-dependent 12-hour pacemaker is separate from the BMAL1-dependent circadian clock in mouse. Representative gene expression of 12-hour and/or circadian genes in wild-type, XBP1 liver specific knockout [6], and BMAL1-knockout ex vivo liver slices [30]. Temporal cistrome of XBP1s as well as key core circadian clock TFs are further shown. (tif 11670 kb)
Table S1. Rhythms uncovered for ETS TFs in mouse liver. For everyone EST TF, whether a 12-hour or circadian rhythm exist is shown, as well as the dominant rhythm identified (xlsx 12 kb)