ABSTRACT
Microbial specialized metabolites are key mediators in host-microbiome interactions. Most of the chemical space produced by the microbiome currently remains unexplored and uncharacterized. This situation calls for new and improved methods to exploit the growing publicly available genomic and metabolomic data sets and connect the outcomes to structural and functional knowledge inferred from transcriptomics and proteomics experiments. Here, we first describe currently available approaches that support the comprehensive mining of metabolomics and genomics data. Next, we provide our vision on how to move forward toward the automated linking of omics data of specialized metabolites to their structures, biosynthesis pathways, producers, and functions.
KEYWORDS: computational biology, computational metabolomics, data mining, genomics, integrative omics, mass spectrometry, microbiome, natural products, specialized metabolites
COMMENTARY
Microbially produced and metabolized small molecules are everywhere: in the soil, plants, microbes, and our body. They constitute many functions ranging from simply providing nutrition to more specialistic tasks such as conveying messages or selectively killing organisms. These microbial specialized metabolites have been instrumental for humankind in medical applications such as antibiotics. The emerging threat of antimicrobial resistance is challenging our current medical advances. This has sparked a renewed interest in mining and elucidating the microbiome chemical diversity to find bioactive molecules.
The four main omics technologies are increasingly used to study microbial chemistry present in natural extracts. Advanced genome mining provides us with an organism’s biosynthetic potential, while transcriptomics and proteomics allow insight into pathway activity through the regulation of transcript and protein levels. Finally, untargeted tandem mass spectrometric (MS/MS) metabolomics records mass spectral data for many microbial natural products. Today, the comprehensive study of the microbial specialized metabolome is mainly hampered by our ability to structurally and functionally annotate omics features.
Technical, analytical, and software advances in the four omics technologies have been impressive over the last 2 decades, yet their integrated analysis remains very challenging. Thus, it is still difficult to rapidly assess the novelty of a metabolite, find the organism that produces it, and learn its function within an ecosystem (1). The Integrated Omics for Metabolomics and Genomics Annotation (iOMEGA) project (see https://github.com/iomega and https://www.esciencecenter.nl/projects/integrated-omics-analysis-for-small-molecule-mediated-host-microbiome-interactions/) led by our group enabled us to explore the current obstacles and opportunities to first improve these omics pillars separately and then build connections to link producers to molecular products (1).
In this perspective, we highlight our contributions to the emerging field of computational metabolomics, how these developments are foundational to performing integrated omics analyses, and how they will accelerate natural product discovery through improved structural and functional annotation of omics profiles.
Metabolome mining tools have been developed that mostly use the collection of MS/MS spectra (or election impact spectra for volatiles or derivatized metabolites [2]) as a representative of natural extracts. Alongside, repositories have emerged to archive the annotated spectra or spectral patterns that these mining tools recognize (3–5). In addition, multiple tools have appeared that mine genomes for biosynthetic gene clusters (BGCs) (6, 7), and precomputed mining results for all publicly available genomes are now also available for large-scale analyses. Experimentally characterized BGCs linked to structural information can be stored in a dedicated repository (8).
In currently existing omics annotation workflows (Fig. 1), matching to repositories is the most reliable step to add structural information to metabolomics profiles enabling biochemical interpretation. Moreover, structure databases with well-curated (meta)data (i.e., first isolation paper, validated biosynthetic gene cluster, and complete and computer-readable structural information, etc.) are also key to enable the accurate annotation of omics profiles with microbial metabolites (3, 8, 9). While increasing numbers of reference spectra and validated BGCs are deposited in public repositories, the resulting rates of matching to omics profiles remain low, and the elucidation of full structures thus remains very challenging. This has sparked the recent development of other approaches based on substructure-based, chemical compound class-based, and network-based techniques, which are all highlighted below.
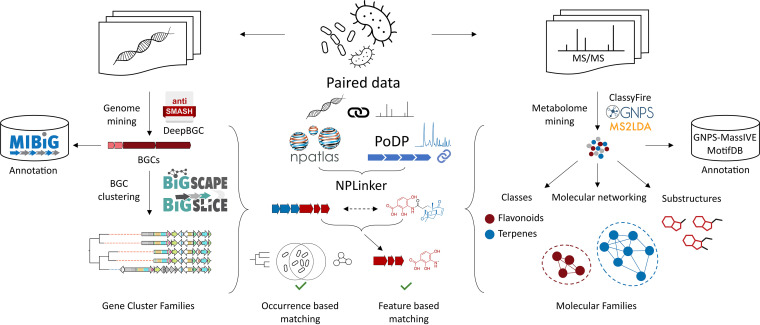
FIG 1.
Current state-of-the-art ecosystem of genomics (left) and metabolomics (right) natural product research, brought together by paired omics approaches (middle). Genomes are mined for biosynthetic gene clusters (BGCs) through tools such as antiSMASH and DeepBGC, and BGCs with structurally characterized products are stored in databases like MIBiG. BGCs are clustered into families with BiG-SCAPE and BiG-SLiCE. To infer compound classes, molecular families, and substructures, metabolomes (represented by collections of MS/MS spectra) are mined with tools such as ClassyFire, GNPS, MS2LDA, and MolNetEnhancer. Structural annotations relevant for microbiome research are stored in databases such as NP Atlas and MotifDB, and reference spectra are available in repositories such as GNPS-MassIVE. Paired data stored in platforms such as the Paired Omics Data Platform (PoDP) combine the two sides, which facilitates multi-omics approaches such as NPLinker that links gene cluster families (GCFs) to molecular families (MFs) through sample occurrence (also known as strain correlation) and feature-based matching.
Substructure-based metabolomics workflows use the idea that the basic building blocks that are shared by different naturally occurring structures will yield similar spectral signals. It is now possible to mine for substructure patterns in metabolomics profiles and store annotated patterns in a repository for reuse in future experiments (4, 5). For example, annotated substructures of Salinispora and Streptomyces bacteria are now available to accelerate substructure analysis of bacterial extracts from related strains.
Chemical compound class annotations can also provide useful information about metabolites that can be used to obtain a high-level overview of the type of chemistry present in natural extracts. For example, specific compound classes such as macrolides or lanthipeptides are likely to be microbially derived. In both genomics and metabolomics workflows, tools have emerged to assign chemical compound class information to BGCs or mass spectra (6, 10).
Network-based analysis is beneficial as it facilitates the large-scale analysis of BGC and MS/MS spectrum ensembles by grouping them into families (3, 11, 12) and allows the propagation of spectral annotations within molecular families. Various approaches to capture structural information at the structural, chemical class, and substructure levels have emerged, and for metabolomics data, MolNetEnhancer (10) was the first tool to integrate and visualize all that information in one place.
Multi-omics approaches facilitate structural and functional annotations by combining complementary information about microbial chemistry. Paired data sets are needed to perform integrative omics mining analysis (1). Recently, the Paired Omics Data Platform (PoDP) was developed, which already holds >4,800 links between (meta)genomes and metabolomics data sets (13). This will allow the detection of new links between BGCs, MS/MS spectra, and compounds, for example, through platforms such as NPLinker that facilitate the computation of various strain correlation-based and feature-based linking scores (1, 14) (Fig. 1).
Looking into the future, based on early successes in omics analysis (4, 7, 15), we envision that machine learning (ML) algorithms will become increasingly important. For example, in metabolomics analysis, mass spectral similarity metrics play a pivotal role across many tasks, including library matching and analogue searching. Our group applied ML to this task for the first time, resulting in the unsupervised Spec2Vec algorithm (16), which showed increased performance in library matching and analogue searching through the learning of relationships between mass features in many MS/MS spectra. Furthermore, we recently proposed the supervised MS2DeepScore algorithm (17), which was trained to learn molecular structural similarities based on MS/MS spectral pairs, resulting in an even better overall performance.
We expect that the learned unsupervised and trained supervised mass spectral embeddings to compute these novel similarity metrics will serve as the input for novel scores to facilitate integrated omics analysis in the recently established NPLinker platform (14). Furthermore, where existing annotation pipelines often struggle for sizable specialized metabolites, analyses based on these mass spectral embeddings are fast, scalable, and thus compatible with an integrated analysis framework for natural products (Fig. 2). Here, it is noteworthy that ML also allowed the development of the natural product-compatible structural classification scheme NPClassifier, which considers structural, functional, and biosynthetic relationships as historically defined by natural product researchers (18).
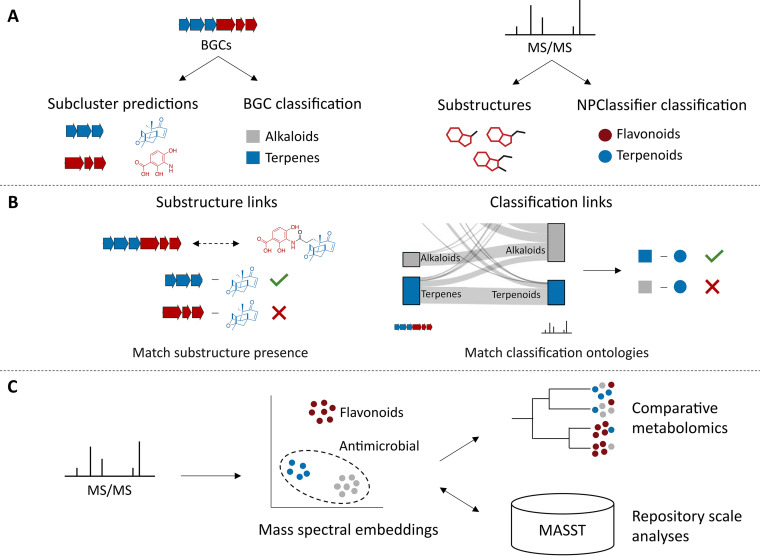
FIG 2.
Current and envisioned advances in multi-omics natural product discovery research. (A) Improved detection of subclusters and relevant natural product-related chemical compound classes in BGCs and MS/MS spectra will become possible based on machine learning-based computational tools. (B) We envision combining the existing BGC-metabolite matching approaches with substructure and chemical class predictions in platforms such as NPLinker. NPClassifier is a novel ML-based class predictor that considers both structural features and historical relationships between metabolites as defined by natural product researchers. (C) Mass spectral embeddings learned by Spec2Vec and trained with MS2DeepScore will enable fast and improved spectral similarity scoring. The bases for these mass spectral embeddings are the relationships between mass fragments and neutral losses based on their presence/absence in a large set of mass spectra. We expect that these embeddings will allow the rapid annotation of classes, substructures, or other labels such as pathways or functions based on clustering techniques. Finally, the developed workflows can also form the basis for improved comparative and repository-wide metabolomics approaches that highlight shared and novel chemistry produced by microbiomes.
In integrative omics for natural product discovery, one of the central aims is the linking of BGCs with the MS/MS spectra of the products that they encode, to facilitate the structural elucidation of the metabolite product(s), establish the producer(s), and infer the function of the specialized metabolites through annotated genes neighboring the BGC. We hypothesize that metabolite annotations can be used to improve the linking of BGC and metabolome information (Fig. 2). By comparing chemical compound classes with BGC classes, it would be possible to rerank BGC-MS/MS links based on the likelihood of occurrence, thereby removing implausible links such as a peptidic compound being produced by a terpene BGC. Similarly, we think that links could be reranked based on shared substructure content inferred from metabolomics and genomics data. Substructures can be annotated by metabolome mining tools from MS/MS spectra and predicted from BGCs by identifying subclusters, which can currently be done through either a targeted or a statistical approach (19). We anticipate that ML approaches for subcluster detection will further facilitate this.
To understand the function of specialized metabolites, comparative analyses between multiple relevant conditions or phenotypes and the linking of functional information inferred from transcriptomics or proteomics experiments will be key. To support such analyses, metabolome mining workflows were linked to statistical approaches through the coupling of metabolite feature recognition tools (20), even in a chemically informed manner (21). When grouped in metabolic pathways or metabolite sets, comparative analyses at the pathway activity level linked to BGC abundance profiles from (meta)transcriptomics can yield further information about which functional pathways or metabolite groups specialized metabolites are part of. To facilitate such analyses in the future, recording expression data through transcriptomics or proteomics in paired data repositories like the PoDP will be essential.
With vastly growing public databases, repository-scale analyses become increasingly relevant to assess the novelty of discovered metabolites by comparing experimental omics profiles not only to validated data (i.e., BGCs and MS/MS spectra assigned to metabolite products) but also to data from all publicly available omics profiles (22, 23). We envision that ML-based (and in particular mass spectral embedding-based) approaches will accelerate current approaches even further (24). It is important to realize that for reliable omics annotations and comparative analyses, consistent and curated metadata are key, for example, in the form of a controlled vocabulary for metabolomics metadata (25) and BGC metadata (8).
We expect that in the near future, the above-described toolset will become more accurate and user-friendly. Microbiome and natural product researchers will then be able to rapidly prioritize novel chemistry in omics profiles. Through accurate genome-metabolome linking, the genetic machinery and mass spectral data will be easily connected. This will boost the structural elucidation of novel metabolite products and enable the recognition of their producers in complex communities such as those originating from soil or our gut. This in turn will allow researchers, i.e., through functional omics profiling and BGC-neighboring gene annotations, to select potential novel antibiotics in their samples, e.g., based on resistance-associated annotations. We anticipate that such applications will help to combat the currently looming antimicrobial resistance pandemic.
To conclude, advances in computational metabolomics and genome mining have enabled natural product-targeted multi-omics analyses, and tools are starting to be in place to exploit recorded paired data sets and annotate omics profiles with structural and functional information to accelerate natural product discovery.
The views expressed in this article do not necessarily reflect the views of the journal or of ASM.
This article is part of a special series sponsored by Floré.
REFERENCES
- 1.van der Hooft JJJ, Mohimani H, Bauermeister A, Dorrestein PC, Duncan KR, Medema MH. 2020. Linking genomics and metabolomics to chart specialized metabolic diversity. Chem Soc Rev 49:3297–3314. doi: 10.1039/d0cs00162g. [DOI] [PubMed] [Google Scholar]
- 2.Aksenov AA, Laponogov I, Zhang Z, Doran SLF, Belluomo I, Veselkov D, Bittremieux W, Nothias LF, Nothias-Esposito M, Maloney KN, Misra BB, Melnik AV, Smirnov A, Du X, Jones KL, II, Dorrestein K, Panitchpakdi M, Ernst M, van der Hooft JJJ, Gonzalez M, Carazzone C, Amézquita A, Callewaert C, Morton JT, Quinn RA, Bouslimani A, Orio AA, Petras D, Smania AM, Couvillion SP, Burnet MC, Nicora CD, Zink E, Metz TO, Artaev V, Humston-Fulmer E, Gregor R, Meijler MM, Mizrahi I, Eyal S, Anderson B, Dutton R, Lugan R, Boulch PL, Guitton Y, Prevost S, Poirier A, Dervilly G, Le Bizec B, Fait A, et al. 2021. Auto-deconvolution and molecular networking of gas chromatography-mass spectrometry data. Nat Biotechnol 39:169–173. doi: 10.1038/s41587-020-0700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu W-T, Crüsemann M, Boudreau PD, Esquenazi E, Sandoval-Calderón M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu C-C, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel J, Murphy BT, Gerwick L, Liaw C-C, Yang Y-L, Humpf H-U, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, et al. 2016. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol 34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Hooft JJJ, Wandy J, Barrett MP, Burgess KEV, Rogers S. 2016. Topic modeling for untargeted substructure exploration in metabolomics. Proc Natl Acad Sci USA 113:13738–13743. doi: 10.1073/pnas.1608041113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogers S, Ong CW, Wandy J, Ernst M, Ridder L, van der Hooft JJJ. 2019. Deciphering complex metabolite mixtures by unsupervised and supervised substructure discovery and semi-automated annotation from MS/MS spectra. Faraday Discuss 218:284–302. doi: 10.1039/c8fd00235e. [DOI] [PubMed] [Google Scholar]
- 6.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannigan GD, Prihoda D, Palicka A, Soukup J, Klempir O, Rampula L, Durcak J, Wurst M, Kotowski J, Chang D, Wang R, Piizzi G, Temesi G, Hazuda DJ, Woelk CH, Bitton DA. 2019. A deep learning genome-mining strategy for biosynthetic gene cluster prediction. Nucleic Acids Res 47:e110. doi: 10.1093/nar/gkz654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kautsar SA, Blin K, Shaw S, Navarro-Muñoz JC, Terlouw BR, van der Hooft JJJ, van Santen JA, Tracanna V, Suarez Duran HG, Pascal Andreu V, Selem-Mojica N, Alanjary M, Robinson SL, Lund G, Epstein SC, Sisto AC, Charkoudian LK, Collemare J, Linington RG, Weber T, Medema MH. 2020. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res 48:D454–D458. doi: 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Santen JA, Jacob G, Singh AL, Aniebok V, Balunas MJ, Bunsko D, Neto FC, Castaño-Espriu L, Chang C, Clark TN, Cleary Little JL, Delgadillo DA, Dorrestein PC, Duncan KR, Egan JM, Galey MM, Haeckl FPJ, Hua A, Hughes AH, Iskakova D, Khadilkar A, Lee J-H, Lee S, LeGrow N, Liu DY, Macho JM, McCaughey CS, Medema MH, Neupane RP, O’Donnell TJ, Paula JS, Sanchez LM, Shaikh AF, Soldatou S, Terlouw BR, Tran TA, Valentine M, van der Hooft JJJ, Vo DA, Wang M, Wilson D, Zink KE, Linington RG. 2019. The Natural Products Atlas: an open access knowledge base for microbial natural products discovery. ACS Cent Sci 5:1824–1833. doi: 10.1021/acscentsci.9b00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst M, Kang KB, Caraballo-Rodríguez AM, Nothias L-F, Wandy J, Chen C, Wang M, Rogers S, Medema MH, Dorrestein PC, van der Hooft JJJ. 2019. MolNetEnhancer: enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 9:144. doi: 10.3390/metabo9070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navarro-Muñoz JC, Selem-Mojica N, Mullowney MW, Kautsar SA, Tryon JH, Parkinson EI, De Los Santos ELC, Yeong M, Cruz-Morales P, Abubucker S, Roeters A, Lokhorst W, Fernandez-Guerra A, Cappelini LTD, Goering AW, Thomson RJ, Metcalf WW, Kelleher NL, Barona-Gomez F, Medema MH. 2020. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol 16:60–68. doi: 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kautsar SA, van der Hooft JJJ, de Ridder D, Medema MH. 2021. BiG-SLiCE: a highly scalable tool maps the diversity of 1.2 million biosynthetic gene clusters. Gigascience 10:giaa154. doi: 10.1093/gigascience/giaa154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schorn MA, Verhoeven S, Ridder L, Huber F, Acharya DD, Aksenov AA, Aleti G, Moghaddam JA, Aron AT, Aziz S, Bauermeister A, Bauman KD, Baunach M, Beemelmanns C, Beman JM, Berlanga-Clavero MV, Blacutt AA, Bode HB, Boullie A, Brejnrod A, Bugni TS, Calteau A, Cao L, Carrión VJ, Castelo-Branco R, Chanana S, Chase AB, Chevrette MG, Costa-Lotufo LV, Crawford JM, Currie CR, Cuypers B, Dang T, de Rond T, Demko AM, Dittmann E, Du C, Drozd C, Dujardin J-C, Dutton RJ, Edlund A, Fewer DP, Garg N, Gauglitz JM, Gentry EC, Gerwick L, Glukhov E, Gross H, Gugger M, Guillén Matus DG, et al. 2021. A community resource for paired genomic and metabolomic data mining. Nat Chem Biol 17:363–368. doi: 10.1038/s41589-020-00724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eldjárn GH, Ramsay A, van der Hooft JJJ, Duncan KR, Soldatou S, Rousu J, Daly R, Wandy J, Rogers S. 2021. Ranking microbial metabolomic and genomic links in the NPLinker framework using complementary scoring functions. PLoS Comput Biol 17:e1008920. doi: 10.1371/journal.pcbi.1008920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dührkop K, Fleischauer M, Ludwig M, Aksenov AA, Melnik AV, Meusel M, Dorrestein PC, Rousu J, Böcker S. 2019. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat Methods 16:299–302. doi: 10.1038/s41592-019-0344-8. [DOI] [PubMed] [Google Scholar]
- 16.Huber F, Ridder L, Verhoeven S, Spaaks JH, Diblen F, Rogers S, van der Hooft JJJ. 2021. Spec2Vec: improved mass spectral similarity scoring through learning of structural relationships. PLoS Comput Biol 17:e1008724. doi: 10.1371/journal.pcbi.1008724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber F, van der Burg S, van der Hooft JJJ, Ridder L. 2021. MS2DeepScore—a novel deep learning similarity measure for mass fragmentation spectrum comparisons. bioRxiv 10.1101/2021.04.18.440324. [DOI] [PMC free article] [PubMed]
- 18.Kim H, Wang M, Leber C, Nothias L-F, Reher R, Kang KB, van der Hooft JJJ, Dorrestein P, Gerwick W, Cottrell G. 2020. NPClassifier: a deep neural network-based structural classification tool for natural products. ChemRxiv 12885494. [DOI] [PMC free article] [PubMed]
- 19.Del Carratore F, Zych K, Cummings M, Takano E, Medema MH, Breitling R. 2019. Computational identification of co-evolving multi-gene modules in microbial biosynthetic gene clusters. Commun Biol 2:83–10. doi: 10.1038/s42003-019-0333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nothias L-F, Petras D, Schmid R, Dührkop K, Rainer J, Sarvepalli A, Protsyuk I, Ernst M, Tsugawa H, Fleischauer M, Aicheler F, Aksenov AA, Alka O, Allard P-M, Barsch A, Cachet X, Caraballo-Rodriguez AM, Da Silva RR, Dang T, Garg N, Gauglitz JM, Gurevich A, Isaac G, Jarmusch AK, Kameník Z, Kang KB, Kessler N, Koester I, Korf A, Le Gouellec A, Ludwig M, Christian MH, McCall L-I, McSayles J, Meyer SW, Mohimani H, Morsy M, Moyne O, Neumann S, Neuweger H, Nguyen NH, Nothias-Esposito M, Paolini J, Phelan VV, Pluskal T, Quinn RA, Rogers S, Shrestha B, Tripathi A, van der Hooft JJJ, et al. 2020. Feature-based molecular networking in the GNPS analysis environment. Nat Methods 17:905–908. doi: 10.1038/s41592-020-0933-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripathi A, Vázquez-Baeza Y, Gauglitz JM, Wang M, Dührkop K, Nothias-Esposito M, Acharya DD, Ernst M, van der Hooft JJJ, Zhu Q, McDonald D, Brejnrod AD, Gonzalez A, Handelsman J, Fleischauer M, Ludwig M, Böcker S, Nothias L-F, Knight R, Dorrestein PC. 2021. Chemically informed analyses of metabolomics mass spectrometry data with Qemistree. Nat Chem Biol 17:146–151. doi: 10.1038/s41589-020-00677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M, Jarmusch AK, Vargas F, Aksenov AA, Gauglitz JM, Weldon K, Petras D, da Silva R, Quinn R, Melnik AV, van der Hooft JJJ, Caraballo-Rodríguez AM, Nothias LF, Aceves CM, Panitchpakdi M, Brown E, Di Ottavio F, Sikora N, Elijah EO, Labarta-Bajo L, Gentry EC, Shalapour S, Kyle KE, Puckett SP, Watrous JD, Carpenter CS, Bouslimani A, Ernst M, Swafford AD, Zúñiga EI, Balunas MJ, Klassen JL, Loomba R, Knight R, Bandeira N, Dorrestein PC. 2020. Mass spectrometry searches using MASST. Nat Biotechnol 38:23–26. doi: 10.1038/s41587-019-0375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kautsar SA, Blin K, Shaw S, Weber T, Medema MH. 2021. BiG-FAM: the biosynthetic gene cluster families database. Nucleic Acids Res 49:D490–D497. doi: 10.1093/nar/gkaa812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, De Vijlder T, Bittremieux W, Laukens K, Heyndrickx W. 6 May 2021. Current and future deep learning algorithms for MS/MS-based small molecule structure elucidation. Rapid Commun Mass Spectrom 10.1002/rcm.9120. [DOI] [PubMed]
- 25.Jarmusch AK, Wang M, Aceves CM, Advani RS, Aguirre S, Aksenov AA, Aleti G, Aron AT, Bauermeister A, Bolleddu S, Bouslimani A, Rodriguez AMC, Chaar R, Coras R, Elijah EO, Ernst M, Gauglitz JM, Gentry EC, Husband M, Jarmusch SA, Jones KL, Kamenik Z, Le Gouellec A, Lu A, McCall L-I, McPhail KL, Meehan MJ, Melnik AV, Menezes RC, Giraldo YAM, Nguyen NH, Nothias LF, Nothias-Esposito M, Panitchpakdi M, Petras D, Quinn RA, Sikora N, van der Hooft JJJ, Vargas F, Vrbanac A, Weldon KC, Knight R, Bandeira N, Dorrestein PC. 2020. ReDU: a framework to find and reanalyze public mass spectrometry data. Nat Methods 17:901–904. doi: 10.1038/s41592-020-0916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]