Abstract
The Saccharomyces cerevisiae gene PHO5 is an excellent system with which to study regulated changes in chromatin structure. The PHO5 promoter is packaged into four positioned nucleosomes under repressing conditions; upon induction, the structure of these nucleosomes is altered such that the promoter DNA becomes accessible to nucleases. We report here the development and characterization of an in vitro system in which partially purified PHO5 minichromosomes undergo promoter chromatin remodeling. Several hallmarks of the PHO5 chromatin transition in vivo were reproduced in this system. Chromatin remodeling of PHO5 minichromosomes required the transcription factors Pho4 and Pho2, was localized to the promoter region of PHO5, and was independent of the chromatin-remodeling complex Swi-Snf. In vitro chromatin remodeling also required the addition of fractionated nuclear extract and hydrolyzable ATP. This in vitro system should serve as a useful tool for identifying the components required for this reaction and for elucidating the mechanism by which the PHO5 promoter chromatin structure is changed.
The packaging of eukaryotic DNA into nucleosomes presents a barrier to cellular processes that require specific contacts with DNA. During transcription, sequence-specific DNA binding proteins and the basal transcription apparatus must recognize and bind to appropriate promoter elements. Biochemical and genetic analyses demonstrate that the packaging of DNA into nucleosomes inhibits its stable association with transcription factors (2, 29, 37, 57). A number of cellular activities capable of facilitating factor binding to chromatin have been identified (3). These activities are thought to function by directly modifying chromatin structure.
The Saccharomyces cerevisiae gene PHO5 is a well-characterized system with which to study regulated gene expression. PHO5 encodes a secreted acid phosphatase whose transcription is regulated in response to environmental phosphate levels (for a review, see reference 32). When phosphate is plentiful, PHO5 expression is repressed; when phosphate is limiting, PHO5 expression is induced. Activation of PHO5 transcription requires two transcription factors: Pho4, a basic helix-loop-helix protein, and Pho2, a homeodomain protein (58). In vitro, Pho2 enhances the binding of Pho4 to two regulatory sequences in the PHO5 promoter, UASp1 and UASp2 (4, 5).
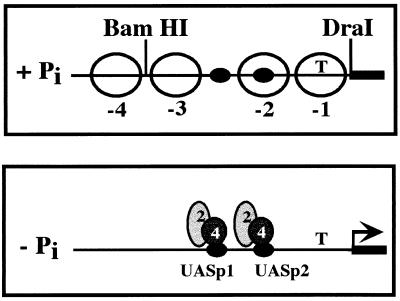
When transcription of PHO5 is activated, its promoter undergoes a dramatic change in chromatin structure (for a review, see reference 51). When yeast cells are grown in high-phosphate medium, two pairs of positioned nucleosomes flank a DNase I-hypersensitive site, which contains UASp1 (Fig. 1, +Pi). UASp2 and the TATA box are packaged into nucleosomes −2 and −1, respectively. In vivo footprinting experiments indicate that Pho4 does not bind the PHO5 promoter under repressing conditions (57). When environmental phosphate is limiting, the positioned nucleosomes no longer protect the PHO5 promoter, and Pho4 binds to UASp1 and UASp2 (Fig. 1, −Pi). In vivo footprinting of Pho2 at the PHO5 promoter has not been performed, but in vitro experiments indicate that Pho2 binds to this region in coordination with Pho4 (4, 5). The process by which the four positioned nucleosomes become undetectable and the PHO5 promoter is rendered sensitive to nucleases is termed the chromatin transition.
FIG. 1.
Chromatin structure of the PHO5 promoter from yeast grown in high- and low-phosphate media. Open circles, positioned nucleosomes; dark ovals, identified upstream activating sequences (UASs); T, location of the TATA box.
The mechanism by which PHO5 chromatin structure is changed during induction is unknown. However, a number of in vivo studies have provided some clues. The PHO5 chromatin transition is independent of transcription and DNA replication, as the loss of nucleosome positioning is unaffected by deletion of the PHO5 TATA box (18) and occurs when cell division is prevented (44). The Pho4 transcriptional activation domain is required for the PHO5 chromatin transition (52) but is not required for binding to naked DNA (19). If the activation domain is dispensable for binding to chromatin as well as to naked DNA, it may be required for interaction with a chromatin-remodeling activity or may be capable of changing chromatin structure itself.
It remains to be determined if factors besides Pho4 and Pho2 are required for PHO5 chromatin rearrangement. Several activities known to modify chromatin structure have been identified in yeast, and a few have been tested for a role in PHO5 induction. Loss-of-function mutations in several components of the ATP-dependent chromatin-remodeling complex Swi-Snf do not affect induction of acid phosphatase activity (10, 20, 45) or changes in PHO5 chromatin structure (20). PHO5 mRNA levels in high- and low-phosphate media are unaffected by mutations in the histone acetylase gene GCN5 (40), although gcn5 mutants have an unusual PHO5 promoter chromatin structure under partially inducing conditions (21). The PHO5 chromatin transition may involve the RNA polymerase holoenzyme, as artificial recruitment of the holoenzyme to PHO5 results in a promoter that is constitutively nuclease sensitive (20).
The reconstitution of chromatin rearrangement in vitro has allowed the isolation of several chromatin-remodeling activities from Drosophila embryo (22, 54, 56) and HeLa cell (35) extracts. It has been difficult, however, to obtain genetic evidence that these activities are involved in the transcriptional regulation of specific genes in vivo. In contrast, studies employing S. cerevisiae have a singular advantage in that this organism is easily manipulated in both biochemical and genetic experiments. This allows any result obtained in vitro to be rapidly tested for relevance in vivo. We describe here the reconstitution of the S. cerevisiae PHO5 chromatin transition in vitro. We propose to use this biochemical system to identify the components required for the chromatin rearrangement at the PHO5 promoter and to elucidate the mechanism by which it occurs.
MATERIALS AND METHODS
Plasmid construction.
pTA-PHO5 was constructed in two steps. First, a BamHI-to-SpeI fragment from pACD5 (62), containing PHO5 sequence from nucleotide −542 to 1466, was inserted into pBluescript II KS to create pBSPHO5. Next, a 1.5-kb EcoRI fragment consisting of the TRP1/ARS1 locus was released from pTA-R (41) and inserted into the EcoRI site of pBSPHO5 such that PHO5 and TRP1 are transcribed in opposite directions, creating plasmid pTA-PHO5. To obtain pTA-p1p2, the hexanucleotide Pho4 binding sites at UASp1 (CACGTT) and UASp2 (CACGTG) in pTA-PHO5 were replaced precisely with a SpeI site (ACTAGT) and a BamHI site (GGATCC), respectively. In pTA-PHO5-ATGΔ, the sequence AATGTT containing the translational start site was replaced with the sequence AGATCT, creating a BglII restriction site. Bacterial replication and selection sequences were removed from all minichromosome constructs by digestion with NotI, and the minichromosome circles were self-ligated before introduction into yeast. pRSPHO4 was constructed by inserting a BamHI-to-HindIII fragment of pACD4 (62) containing the PHO4 promoter and open reading frame into pRS426.
Strains.
S. cerevisiae YS18 (47) was used in all experiments. Northern analysis was performed with strains EY0244 (wild type), EY0168 (pho3Δ pho5Δ), and EY0168 harboring pTA-PHO5. For indirect end labeling, EY0255 (pho2Δ pho4Δ pho80Δ) harboring pTA-PHO5 was used. For analysis of chromatin remodeling of episomal PHO5 in vivo, we used strains EY0246 (pho3Δ pho5Δ pho4Δ pho80Δ) and EY0243 (pho3Δ pho5Δ pho80Δ), harboring either pTA-PHO5-ATGΔ or both pTA-PHO5-ATGΔ and pRSPHO4. For minichromosome purification and nuclear extract preparation, either EY0255 or EY0579 (pho2Δ pho4Δ pho80Δ snf6Δ) was used. To make EY0579, SNF6 was disrupted in a diploid by two-step gene replacement with pEY110 (16). After sporulation, snf6Δ haploid strains were identified by Southern blotting.
Northern blotting.
Cell cultures were grown in medium lacking inorganic phosphate for 6 h as described previously (25), and total RNA was prepared as described previously (12). RNA was quantitated, and 20 μg of each sample was loaded on 6.7% formaldehyde–1.5% agarose gels and run in 1× E buffer (20 mM MOPS [morpholinepropanesulfonic acid] [pH 7.0], 5 mM Na acetate, 0.5 mM EDTA). The RNA was blotted to nylon and probed as described for Southern blotting below.
Preparation of PHO5 minichromosomes.
PHO5 minichromosomes were prepared by the first steps of the procedure described by Simpson and colleagues (13, 42), with some modification. EY0255 or EY0579 cells harboring pTA-PHO5 or pTA-p1p2 were grown in 9 liters of synthetic medium to an A600 of 1.0. Cells were pelleted, washed with water, and incubated at 30°C for 30 min in a freshly prepared solution of 0.7 M β-mercaptoethanol–20 mM EDTA. The cells were washed once with 1 M sorbitol and resuspended in 500 ml of lyticase buffer (1.2 M sorbitol, 50 mM Tris-Cl [pH 8.0], 5 mM β-mercaptoethanol). Spheroplasting was performed with 1 ml of crude recombinant lyticase (46) per g (wet weight) of cells for 30 min at 30°C. All subsequent manipulations were at 0 to 4°C. A swinging-bucket rotor (Sorvall HB-6) was used in all centrifugation steps unless otherwise noted. The spheroplast pellet was washed two times with 1 M sorbitol and then thoroughly resuspended in 240 ml of Ficoll buffer (18% Ficoll, 20 mM MOPS-NaOH [pH 6.8], 1 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride [PMSF]) and Dounce homogenized by hand (Wheaton 40-ml Dounce homogenizer), 10 times with the loose pestle and 5 times with the tight pestle. The lysate was layered over an equal volume of glycerol-Ficoll buffer (20% glycerol, 7% Ficoll, 20 mM MOPS-NaOH [pH 6.8], 1 mM MgCl2, 1 mM PMSF) and spun at 11.5 krpm for 30 min. The pellet was resuspended in 20 ml of Ficoll buffer and centrifuged at 4.5 krpm for 15 min. The supernatant was transferred to a fresh chilled tube, and nuclei were collected by centrifugation at 11.5 krpm for 25 min. Pelleted nuclei were flash frozen at −80°C, thawed on ice, and incubated for 1 to 2 h in 9 ml of elution buffer {200 mM NaCl, 5 mM MgCl2, 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)]-NaOH [pH 7.3], 0.5 mM EGTA, 5 mM β-mercaptoethanol, 1 mM PMSF}. Nuclei were pelleted by centrifugation at 11.5 krpm for 10 min, and the eluate was split between two 35-ml 0.4 to 1 M sucrose gradients made in elution buffer supplemented to a final NaCl concentration of 250 mM. The gradients were spun at 45 krpm for 80 min in a VTi50 rotor, braked to 10 krpm, and then allowed to coast to a stop. DNA was purified from 50 μl of each 1-ml fraction and assayed on ethidium bromide-stained agarose gels. Minichromosome-containing fractions were pooled, concentrated 10-fold on Centri-Prep concentrators that had been preblocked with insulin, and stored at −80°C in aliquots.
The average yield from this procedure was approximately 40%. The greatest loss occurred at the nuclear elution step, where 50 to 80% of the PHO5 minichromosomes were recovered in the eluate. The final PHO5 minichromosome fraction contained approximately 1 μg of minichromosomal DNA per ml in a final volume of approximately 0.75 ml. This fraction contained a significant amount of cellular RNA but was free of genomic DNA.
Southern blotting.
Samples were loaded on 1.2% agarose gels and run in 0.5× Tris-borate-EDTA at 4 V per cm for 4 h. Gels were prepared as described previously (43) and blotted to nylon membranes (Amersham) overnight in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Prehybridization and hybridization with random-prime-labeled probes were performed in Rapid Hyb Buffer (Amersham). Typically, restriction fragments of 100 to 300 bp embedded in low-melting-point agarose were random prime labeled overnight at room temperature.
Micrococcal nuclease digestion and indirect end labeling of chromosomal or episomal PHO5 in vivo.
Cells collected from 500 ml of cell culture grown to an A600 of 1.0 were spheroplasted as in the minichromosome preparation, washed three times with 1 M sorbitol, and resuspended in 2 ml of digestion buffer A (1 M sorbitol, 50 mM NaCl, 10 mM Tris-Cl [pH 7.5], 5 mM MgCl2, 1 mM CaCl2, 1 mM β-mercaptoethanol). Aliquots of 200 μl were placed in tubes containing 0.1 to 100 U of micrococcal nuclease (Worthington). Two hundred microliters of buffer B (buffer A plus 0.15% Nonidet P-40) was added to each tube, and the reaction mixtures were incubated at 37°C for 5 min. Reactions were stopped with 1/10 volume of 5% sodium dodecyl sulfate (SDS)–250 mM EDTA. DNA was purified by digestion with 200 μg of proteinase K per ml at 37°C for 2 h, followed by phenol-chloroform extraction, RNase treatment, and ethanol precipitation. For indirect end labeling of minichromosomes, 1/10 of each sample was digested with NgoMI or XmnI; genomic PHO5 DNA was digested with StuI. The Southern blot probes for analysis of episomal PHO5 and TRP1 nucleosome positioning were derived from an NgoMI-to-BglII fragment of the TRP1 gene and an XmnI-to-ScaI fragment of PHO5, respectively. As pTA-PHO5 is maintained in high copy, hybridization of this probe to the chromosomal TRP1 or PHO5 locus did not interfere with analyses of minichromosomal chromatin structure. For analysis of chromosomal PHO5, the probe was derived from a StuI-to-ApaI fragment of the PHO5 upstream region.
Chromatin remodeling in vivo.
EY0246 and EY0243, harboring either pTA-PHO5-ATGΔ or both pTA-PHO5-ATGΔ and pRSPHO4, were cultured, spheroplasted, treated with micrococcal nuclease, and Southern blotted as described above. Probes for analysis of nucleosome −2 and nucleosome +1 correspond to ClaI-to-BstEII (probe A in Fig. 2) and DraI-to-SalI (probe B in Fig. 2) fragments of pBSPHO5, respectively. Southern blots were analyzed by phosphor screen autoradiography, and quantitative area analysis was performed with ImageQuant software.
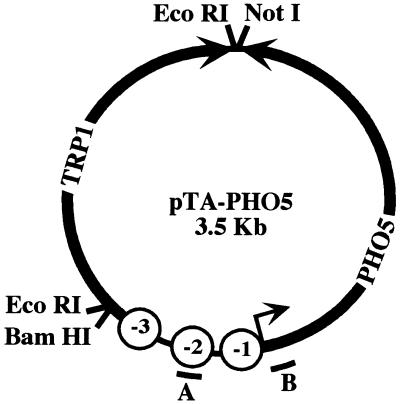
FIG. 2.
Map of pTA-PHO5. Bacterial selection and replication sequences were removed, and a 3.5-kb circle containing PHO5 and the TRP1/ARS1 locus were religated to form pTA-PHO5. Black arrows, directions of transcription of PHO5 and TRP1; open circles, locations of three positioned nucleosomes on the PHO5 promoter; bars, sequences from which probes A and B were derived.
Preparation of recombinant Pho4.
Escherichia coli BL21 harboring a T7-PHO4 expression vector (24) was grown in 3 liters of L broth supplemented with 50 μg of carbenicillin per ml to an A600 of 0.4. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 0.4 mM, and the culture was grown for 2 h at 37°C. Cells were harvested, washed with B(0.1) (10% glycerol, 20 mM PIPES-NaOH [pH 7.3], 1 mM dithiothreitol, 1 mM PMSF, 1 μg of pepstatin A per ml, 0.1 M NaCl), and resuspended in 40 ml of B(0.1). Cells were lysed on ice by sonication and centrifuged at 16 krpm for 20 min in a Sorvall SS-34 rotor at 4°C. The lysate was treated with 10 U of DNase I, clarified through a 0.22-μm-pore-size filter, and loaded onto a 10-ml SP-Sepharose High Performance (Pharmacia) column. Pho4 was eluted with a linear gradient from 100 to 1,000 mM NaCl. Fractions containing Pho4 were pooled, adjusted to the conductivity of B(0.1) by dilution with B(0) [equivalent to B(0.1) except containing 0 M NaCl], loaded onto a BioScale S5 column (Bio-Rad), and eluted as before. This procedure yielded approximately 15 mg of Pho4, which appeared as a single band in SDS-polyacrylamide gel electrophoresis with Coomassie blue staining.
Preparation of S(0.3) extract.
Nuclear extract was prepared from EY0255 or EY0579 as described previously (33), with the following modifications. Recombinant lyticase was used for spheroplasting, and lysis was performed with a hand-held glass Dounce homogenizer, as described above. The final protein pellet was resuspended in S(0.1) (20 mM HEPES-KOH [pH 7.9], 10% glycerol, 1 mM EDTA, 0.1 M K acetate, 1 μg of pepstatin A per ml, 1 mM PMSF). For fractionation, 5 mg of nuclear extract was applied to a 1-ml SP-Sepharose Fast Flow (Pharmacia) column, washed with S(0.1), and step eluted with S(0.3). [S(0.3) is equivalent to S(0.1) except that it contains 0.3 M K acetate.] Fractions containing protein were pooled and concentrated 10-fold with Centricon concentrators.
In vitro chromatin remodeling.
Ten microliters of minichromosomes (approximately 10 ng of DNA in 250 mM NaCl–5 mM MgCl2–10 mM PIPES-NaOH [pH 7.3]–0.5 mM EGTA) was incubated in 50-μl reaction mixtures containing 12 mM HEPES-NaOH (pH 7.5), 6 mM MgCl2, 1 mM dithiothreitol, 5% glycerol, and 0.5 mM CaCl2; 2 μg of poly(dC)-poly(dG) per ml and 0.1 mg of bovine serum albumin per ml were also added as nonspecific competitors. After addition of Pho4 (90 nM), Pho2 (approximately 20 nM), and 1 μg of S(0.3), the reaction mixtures were incubated at room temperature for 15 min. An ATP regeneration mix (final concentrations of 0.2 μg of creatine kinase per ml in 10 mM glycine [pH 8], 30 mM creatine phosphate, and 0.5 mM ATP or adenylyl imidi-diphosphate [AMP-PMP] was added, and reaction mixtures were incubated at 30°C for 30 min. The reaction mixtures were split and digested with either 0.1 or 0.05 U of micrococcal nuclease for 5 min at 37°C. Digestion was stopped with 1/10 volume of 5% SDS–250 mM EDTA. DNA was purified by overnight treatment with 200 μg of proteinase K per ml at 37°C, two phenol extractions, chloroform extraction, and ethanol precipitation. Samples were analyzed by Southern blotting as described above and probed as described for in vivo chromatin remodeling.
RESULTS
Episomal PHO5 is transcribed in response to phosphate starvation.
Minichromosomes, or circular plasmids packaged into chromatin, have been purified from S. cerevisiae for the analyses of transcription (48), retroviral integration (41), centromere function (27), and chromatin structure (7, 53). We modified a 1.5-kb TRP1/ARS1 circle by inserting the PHO5 promoter and open reading frame to form pTA-PHO5 (Fig. 2). The PHO5 promoter fragment that we inserted includes sequence that is packaged into nucleosomes −1 through −3. This sequence is sufficient for phosphate-regulated transcription of PHO5 from a plasmid and for appropriate positioning of the three promoter nucleosomes (6, 18).
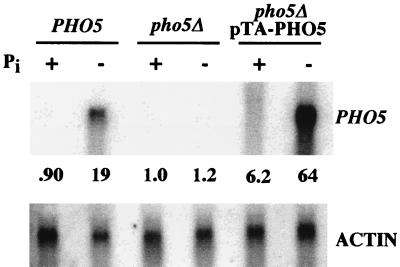
When yeast cells are grown in medium lacking inorganic phosphate, transcription of PHO5 is induced (36). We therefore tested if phosphate starvation induces transcription of episomal PHO5. A wild-type strain, a pho5Δ strain, and a pho5Δ strain carrying pTA-PHO5 were grown in medium lacking inorganic phosphate for 6 h. Total RNA was isolated from these cultures, and Northern analysis was performed (Fig. 3). PHO5 transcript levels were quantified and normalized to actin transcript levels. Transcription of chromosomal PHO5 increased approximately 20-fold upon starvation for phosphate, whereas transcription from episomal PHO5 was induced approximately 10-fold. Therefore, chromosomal PHO5 and episomal PHO5 were regulated by environmental phosphate levels to approximately the same degree.
FIG. 3.
Northern analysis of chromosomal and episomal PHO5 expression in response to phosphate starvation. A PHO5+ strain, a pho5Δ strain, and a pho5Δ strain harboring pTA-PHO5 were grown for 6 h in medium either containing (+) or lacking (−) inorganic phosphate (Pi). PHO5 transcript levels were quantified and normalized to the actin signal.
pTA-PHO5 was maintained at approximately 20 copies per cell (data not shown). If starvation for phosphate causes induction of every copy of pTA-PHO5 to the same extent as the chromosomal copy, a strain harboring pTA-PHO5 should express 20 times as much transcript as a wild-type strain. However, induced levels of PHO5 transcript from pTA-PHO5 were 3.4-fold higher than those measured for the chromosomal copy of PHO5. A three- to fourfold difference was reproducibly observed, as early as 3 h and as late as 12 h after transfer to medium lacking phosphate. These data suggest that some factor necessary for PHO5 transcription is limiting under these conditions, and expression of all 20 copies of pTA-PHO5 in each cell is prevented. The limiting factor could be Pho4, Pho2, a putative chromatin-remodeling activity, or components of the general transcription machinery.
Episomal PHO5 has correctly positioned nucleosomes under repressing conditions.
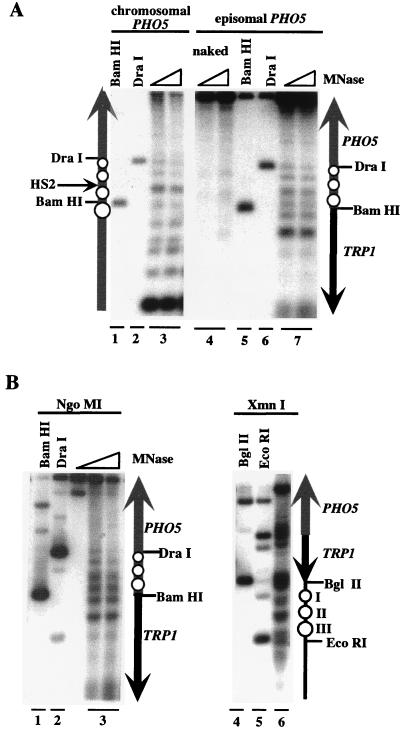
Micrococcal nuclease digestion followed by indirect end labeling is used as an assay for positioned nucleosomes in vivo (34, 61). We used this technique to compare the chromatin structure of chromosomal PHO5 with that of episomal PHO5 (Fig. 4). Spheroplasts with an intact chromosomal PHO5 locus harboring pTA-PHO5 were treated with micrococcal nuclease, and indirect end labeling was performed. Analysis of chromosomal PHO5 revealed four positioned nucleosomes (Fig. 4A, lane 3), which correspond to those mapped previously (1). Three similarly positioned nucleosomes were detected on the PHO5 promoter on pTA-PHO5 in vivo (compare lanes 3 and 7). The sequence upstream of nucleosome −3 on pTA-PHO5 (starting at the BamHI site) is the start of the TRP1 gene, which had a noticeably different pattern than the corresponding region of the chromosomal PHO5 gene.
FIG. 4.
Nucleosomes on the PHO5 promoter have similar positions on the chromosome and on pTA-PHO5 and are not changed upon PHO5 minichromosome preparation. (A) Spheroplasts were treated with micrococcal nuclease (MNase), and the DNA was purified. For size standards, untreated DNA was digested with BamHI (lanes 1 and 5) or DraI (lanes 2 and 6). For analysis of chromosomal PHO5, samples were digested with StuI, Southern blotted, and hybridized to a probe derived from a StuI-to-ApaI fragment of the PHO5 upstream region. For analysis of episomal PHO5, samples were digested with NgoMI, and the probe used was derived from an NgoMI-to-BglII fragment from TRP1. (B) Partially purified PHO5 minichromosomes were treated with MNase, and the DNA was purified. Samples digested with NgoMI were probed with an NgoMI-to-BglII fragment from TRP1; samples digested with XmnI were probed with an XmnI-to-ScaI fragment from PHO5. The schematics show inferred locations of nucleosomes (open circles) on PHO5 and pTA-PHO5. The grey and black arrows represent PHO5 and TRP1 sequences, respectively. The location of the hypersensitive site HS2 on chromosomal PHO5 is indicated with an arrow.
There is a detectable difference between the chromatin structures of chromosomal and episomal copies of PHO5. The nuclease-hypersensitive site (HS2), visible on the chromosomal copy of PHO5, is not apparent on pTA-PHO5 (Fig. 4A). This may be explained by the observation that nucleosome −3 appeared to be slightly shifted in position towards nucleosome −2. However, nucleosomes −1 and −2 (incorporating the TATA box and UASp2) appeared to be correctly positioned, thereby reproducing the appropriate repressed state. Micrococcal nuclease digestion followed by indirect end labeling thus indicates that the PHO5 promoter on pTA-PHO5 is incorporated into positioned nucleosomes with positioning that is very similar to that observed on chromosomal PHO5.
pTA-PHO5 is remodeled in vivo.
To test if the promoter chromatin of episomal PHO5 could be remodeled in vivo, we analyzed changes in chromatin structure by digestion with micrococcal nuclease followed by Southern blotting. This assay has been previously employed for analysis of the PHO5 chromatin transition (1, 44, 50). To analyze changes in chromatin structure at the PHO5 promoter, we used a probe derived from the PHO5 sequence packaged into nucleosome −2 (Fig. 2, probe A). A pattern of nucleosomal bands implies that PHO5 promoter sequence complementary to probe A is packaged into nucleosome −2 and is thereby protected from digestion. The disappearance of these bands implies that nucleosome −2 no longer protects the underlying DNA.
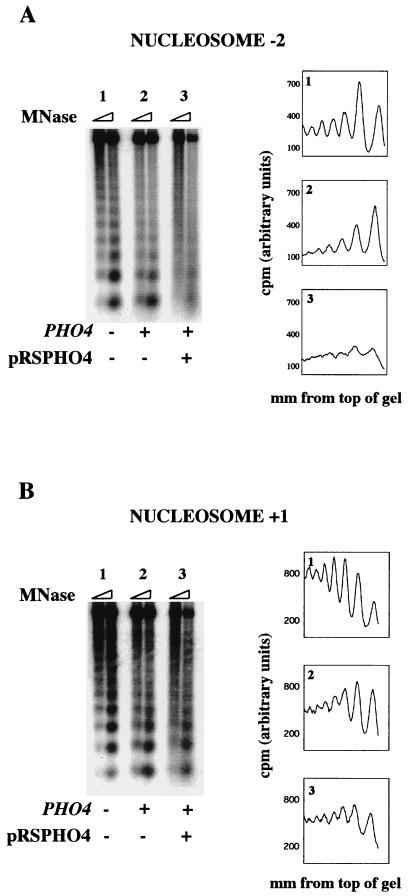
For this experiment, we compared the chromatin structures of the PHO5 promoter on pTA-PHO5-ATGΔ in a pho4Δ strain, a PHO4+ strain, and a PHO4+ strain carrying a high-copy-number plasmid expressing Pho4 (pRSPHO4). pTA-PHO5-ATGΔ is a derivative of pTA-PHO5 in which the PHO5 ATG was replaced with a restriction site. This derivative was used to prevent production of Pho5, a secreted acid phosphatase, as high-level PHO5 expression inhibits cell growth by disrupting the normal function of the secretory pathway (28). All strains lacked chromosomal PHO5, and all were also pho80Δ, which causes constitutive expression of PHO5 (36).
In a strain lacking Pho4, the sequence underlying nucleosome −2 produced a pattern of bands, indicating that it is protected from micrococcal nuclease digestion (Fig. 5A, sample 1). In a PHO4+ strain, minimal remodeling of the episomal PHO5 promoter was observed (sample 2). This is consistent with the results of Northern analysis (Fig. 3) and supports the hypothesis that a factor required for PHO5 expression is limiting in the cell, allowing only a subset of the pTA-PHO5 templates to be transcribed. To test if a limiting factor was Pho4, we assayed in vivo remodeling of pTA-PHO5-ATGΔ in a PHO4+ strain carrying the high-copy-number plasmid pRSPHO4. As shown in Fig. 5A, sample 3, remodeling of nucleosome −2 was observed under these conditions. This suggests that the concentration of Pho4 in the nucleus under inducing conditions is insufficient to support chromatin remodeling and activation of transcription of the majority of the copies of episomal PHO5. It should be noted that in the strain carrying pRSPHO4, the pTA-PHO5-ATGΔ copy number drops to approximately five. Thus, the drop in template number may also allow remodeling of a higher proportion of the templates in each cell.
FIG. 5.
In vivo chromatin remodeling of episomal PHO5. Spheroplasts from the indicated strains were treated with micrococcal nuclease (MNase), and the DNA was purified and Southern blotted. (A) The blot was probed with probe A (Fig. 2). Data from each sample were quantified, and the distance from the top of the gel was graphed against the signal density. (B) The blot shown in panel A was stripped and reprobed with probe B (Fig. 2).
To test if this remodeling was localized to the promoter region of episomal PHO5, the blot was stripped and reprobed with a probe derived from nucleosome +1 (Fig. 2, probe B). This sequence is mostly nucleosomal, even under conditions that allowed remodeling of nucleosome −2 (compare Fig. 5A and B). The small amount of in vivo remodeling that is apparent at nucleosome +1 was also observed at another nucleosome in the PHO5 open reading frame, as well as at a nucleosome in the TRP1 gene (data not shown). Thus, episomal PHO5 chromatin is remodeled in vivo, when high enough levels of Pho4 are present, and this remodeling is predominantly restricted to the promoter region. For the purposes of this report, an increase in micrococcal nuclease sensitivity at nucleosome −2 defines chromatin remodeling in our in vitro system (see Discussion).
Preparation of PHO5 minichromosomes with intact chromatin structure.
With evidence that the PHO5 promoter on pTA-PHO5 has correctly positioned nucleosomes, that its transcription is regulated by environmental phosphate levels, and that it can be remodeled in vivo, we developed a procedure to prepare PHO5 minichromosomes for in vitro study. Our protocol was based on the first steps of a purification procedure developed by Simpson and colleagues (13, 42). Our goals were to remove genomic DNA and cellular debris in a manner gentle enough to leave minichromosomal chromatin intact.
PHO5 minichromosomes were prepared from a pho2Δ pho4Δ strain to prevent contamination of the chromatin template with the Pho4 and Pho2 transcription factors. Cells harboring pTA-PHO5 were spheroplasted with lyticase (46), and lysis was performed with a hand-held Dounce homogenizer. Nuclei were purified away from cell debris and other organelles by spinning through a glycerol cushion, and unlysed spheroplasts and whole cells were removed by differential centrifugation. Minichromosomes were eluted from the purified nuclei, presumably by diffusing through fissures in the nuclear envelope created by flash freezing. The resulting eluate was further purified on a linear sucrose gradient, and fractions containing minichromosomes were pooled and concentrated.
We tested if the chromatin structure of PHO5 minichromosomes changed during their preparation by digesting PHO5 minichromosomes with micrococcal nuclease in vitro and then analyzing nucleosome positioning by indirect end labeling. As shown in Fig. 4B, lane 3, the digestion pattern observed with PHO5 minichromosomes was unchanged from that observed on pTA-PHO5 in vivo.
Three positioned nucleosomes (named I, II, and III) are positioned on the TRP1/ARS1 circle, both in vivo (63) and after purification (53). We therefore tested if positioned nucleosomes are present at these positions on purified PHO5 minichromosomes. As shown in Fig. 4B, lane 6, three appropriately positioned nucleosomes were detected on the TRP1/ARS1 sequence. These data indicate that partially purified PHO5 minichromosomes contain correctly positioned nucleosomes, both over the PHO5 promoter and on the TRP1/ARS1 sequence, and are therefore appropriate chromatin templates for biochemical analysis of PHO5 chromatin remodeling.
In vitro remodeling of PHO5 minichromosomes requires Pho4 and Pho2, hydrolyzable ATP, and a fraction of nuclear extract.
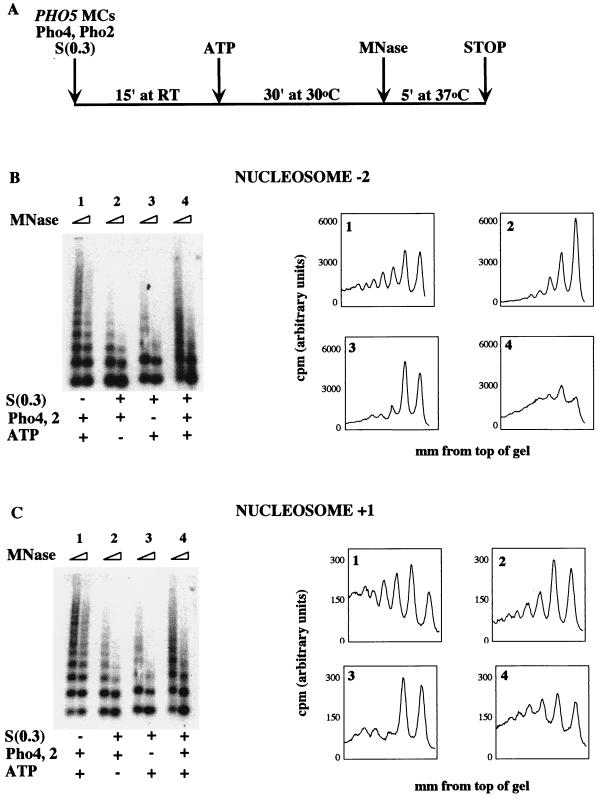
The scheme of our in vitro chromatin-remodeling experiments is outlined in Fig. 6A. PHO5 minichromosomes were mixed with transcription factors and nuclear extract in a reaction mixture. A source of energy was then added, and the remodeling reaction was allowed to proceed. Reaction mixtures were split, digested with micrococcal nuclease, transferred to nylon, and probed with sequence corresponding to nucleosome −2 as for analysis of in vivo remodeling.
FIG. 6.
In vitro chromatin remodeling of PHO5 minichromosomes requires Pho2, Pho4, S(0.3), and ATP and is localized predominantly to the PHO5 promoter. (A) Schematic of the in vitro remodeling reaction. MC, minichromosome; MNase, micrococcal nuclease. (B) Reaction mixtures were assembled and incubated as shown in panel A and then split and digested with micrococcal nuclease. Samples were purified, electrophoresed, and transferred to nylon. Southern blotting was performed with probe A, and data were graphed as in Fig. 5. (C) The blot shown in panel B was stripped and reprobed with probe B.
The chromatin transition and transcriptional activation of PHO5 in vivo require the transcription factors Pho4 and Pho2 (17). We therefore tested if these transcription factors are sufficient to support in vitro chromatin remodeling of PHO5 minichromosomes. As shown in Fig. 6B, sample 1, recombinant Pho4 and Pho2 were not sufficient for remodeling of nucleosome −2 in vitro.
The inability of Pho2 and Pho4 to remodel chromatin in vitro suggested that remodeling of PHO5 promoter chromatin requires an additional activity. We therefore tested if a fraction of S. cerevisiae nuclear extract, termed S(0.3), could provide a chromatin-remodeling activity. Addition of S(0.3) had no effect in our assay (Fig. 6B, sample 2). By analogy with previously identified chromatin-modifying complexes, such an activity might require either ATP or acetyl coenzyme A. When we tested this possibility by incubating Pho4 and Pho2, S(0.3), and an ATP regeneration system with PHO5 minichromosomes, chromatin remodeling was observed (sample 4). Under these conditions, chromatin remodeling was also observed when a probe derived from nucleosome −3 or a probe derived from all three positioned nucleosomes (−1 through −3) was used (data not shown). These data indicate that the PHO5 promoter nucleosomes are remodeled in our in vitro system.
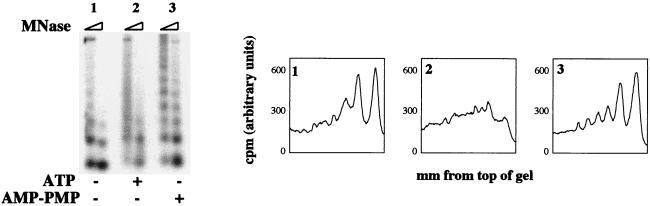
To test if ATP hydrolysis is required for S(0.3)- and Pho4-dependent remodeling of PHO5 minichromosome promoter chromatin, a nonhydrolyzable ATP analog was included in the regeneration system in place of ATP (Fig. 7). Whereas remodeling is observed when ATP is added (sample 2), there is no change in the PHO5 promoter chromatin structure when ATP is omitted (sample 1) or when an equivalent amount of AMP-PMP is substituted (sample 3). Samples containing acetyl coenzyme A as an energy source showed no remodeling under these conditions (data not shown).
FIG. 7.
Hydrolyzable ATP is required for in vitro chromatin remodeling of PHO5 minichromosomes. Either buffer (sample 1), ATP (sample 2), or AMP-PMP (sample 3) was included in chromatin-remodeling reactions with PHO5 minichromosomes, Pho4 and Pho2, and S(0.3). Samples were analyzed as described for Fig. 6. MNase, micrococcal nuclease.
Thus, the transcription factors Pho2 and Pho4, S(0.3), and ATP hydrolysis were all necessary for in vitro chromatin remodeling of nucleosome −2. No remodeling was observed if any of these three components were withheld. These data imply that Pho2, Pho4, and an ATP-dependent activity can remodel PHO5 promoter chromatin.
S(0.3)-, ATP-, and Pho4-dependent chromatin remodeling is restricted to the PHO5 promoter region.
In vivo, the loss of positioned nucleosomes in response to phosphate starvation is restricted to the four positioned nucleosomes on the PHO5 promoter (1). We tested if this was true of the remodeling observed in vitro by stripping and reprobing the Southern blot shown in Fig. 6B with a sequence underlying nucleosome +1. When analyzed with this probe, all samples produced a largely nucleosomal pattern (Fig. 6C). Thus, the dramatic change in chromatin structure observed in the presence of Pho4 and Pho2, S(0.3), and ATP at the PHO5 promoter does not extend significantly into the PHO5 open reading frame.
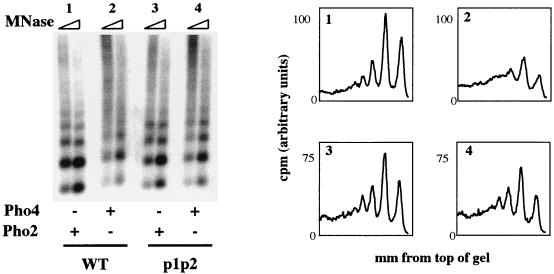
Pho4 can partially remodel PHO5 chromatin in the absence of Pho2.
Overexpression of Pho4 can partially suppress the PHO5 expression defect of a pho2Δ strain (17). We therefore tested if Pho4 was capable of supporting chromatin remodeling of PHO5 minichromosomes in vitro without Pho2. As indicated in Fig. 8, Pho4 is capable of supporting partial remodeling of PHO5 minichromosomes in the presence of S(0.3) and ATP without Pho2 (sample 2). However, when Pho2 is included in the remodeling reaction mixture and Pho4 is left out, no remodeling is observed (sample 1). These results demonstrate that the transcription factor Pho4 is required for S(0.3)- and ATP-dependent chromatin remodeling of PHO5 minichromosomes.
FIG. 8.
In vitro chromatin remodeling of PHO5 minichromosomes in the absence of Pho2 requires Pho4, UASp1, and UASp2. Wild-type (WT) PHO5 minichromosomes were tested for chromatin remodeling in the presence of either Pho2 (sample 1) or Pho4 (sample 2). Minichromosomes lacking the Pho4 binding sites at UASp1 and UASp2 (p1p2) were purified and tested in parallel (samples 3 and 4). S(0.3) and ATP were included in all reaction mixtures. MNase, micrococcal nuclease.
Pho4-dependent chromatin remodeling without Pho2 is incomplete, and a nucleosomal pattern is visible (Fig. 8, sample 1). When Pho2 is included in the reaction with Pho4, S(0.3), and ATP, remodeling is more complete (Fig. 6, sample 4). Analyses of the DNA binding and in vivo transcriptional activation properties of a version of Pho4 unable to interact with Pho2 suggest that Pho2 may affect the function of Pho4 in two ways: by modulating its ability to bind DNA and by enhancing its ability to activate transcription (4). By analogy, Pho2 may facilitate chromatin remodeling in our in vitro system by enhancing the binding of Pho4 to PHO5 minichromosomes or by enhancing the ability of Pho4 to support chromatin remodeling in a manner independent of DNA binding. When higher concentrations of Pho4 were used, remodeling was complete in the absence of Pho2 (data not shown); however, the relevance of remodeling at these concentrations is not clear. Importantly, at the Pho4 concentrations used in this study, Pho2 is required for complete remodeling of PHO5 minichromosomes in vitro, consistent with its role in vivo.
Pho4-dependent chromatin remodeling requires UASp1 and UASp2.
Deletion of a 26-bp region encompassing the Pho4 binding site in UASp1 has no effect on the assembly of repressive PHO5 chromatin structure, but it prevents Pho4 binding (57) and the chromatin transition (18) in vivo. These observations indicate that binding of Pho4 to the hypersensitive region is not required for nucleosome positioning yet is required for changes in chromatin structure upon induction. We wished to test if the Pho4-dependent remodeling observed in the absence of Pho2 requires specific binding by Pho4 to UASp1 and UASp2. A version of pTA-PHO5, pTA-p1p2, in which the two Pho4 binding sites in UASp1 and UASp2 were replaced precisely with restriction sites, was constructed. Wild-type and p1p2 minichromosomes were prepared in parallel and tested for remodeling in the in vitro system. Whereas the PHO5 promoter of wild-type minichromosomes was partially remodeled in the presence of Pho4 (Fig. 8, sample 2), no Pho4-dependent remodeling of p1p2 minichromosomes was observed (Fig. 8, sample 4). This result demonstrates that one or both of the Pho4 binding sites in UASp1 and UASp2 are required for Pho4-dependent chromatin remodeling of PHO5 minichromosomes in vitro.
Unexpectedly, p1p2 minichromosomes were remodeled when Pho2 was included in the remodeling reaction with Pho4, S(0.3), and ATP (data not shown). Yeast strains harboring pTA-p1p2 express low levels of acid phosphatase activity upon phosphate starvation (data not shown), suggesting that the deletion of UASp1 and UASp2 is not sufficient to prevent transcription of episomal PHO5. It is possible that binding of Pho2 to its site in UASp1 (which is intact in the p1p2 minichromosomes) can stabilize Pho4 DNA binding. In contrast, the 26-bp deletion analyzed in previous studies partially destroys this Pho2 binding site, which may account for the completely unremodeled state of this mutant promoter in vivo.
PHO5 chromatin remodeling in vitro does not require the Swi-Snf complex.
We reasoned that the factor(s) supplied by the S(0.3) extract might have ATPase activity, since ATP is required for in vitro remodeling. The yeast remodeling factor Swi-Snf contains an ATPase, which is required for its function (30). It was therefore possible that the putative ATP-dependent activity in S(0.3) was the Swi-Snf complex. As Swi-Snf is not required for PHO5 chromatin remodeling in vivo (10, 20, 45), it was of interest to determine if PHO5 minichromosome remodeling required this complex in vitro.
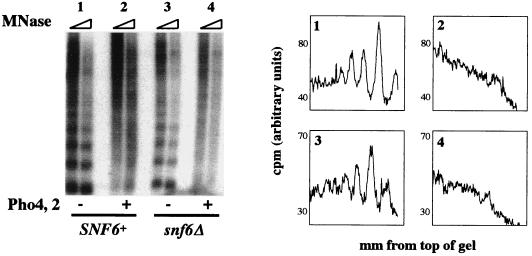
The Snf6 protein is a component of the Swi-Snf complex (9, 38). We tested the requirement for Swi-Snf in our in vitro system by preparing in parallel S(0.3) extract and PHO5 minichromosomes from SNF6+ and snf6Δ strains. In vitro remodeling with components derived from a snf6Δ strain occurred to the same extent as with those derived from SNF6+ cells (Fig. 9, compare samples 2 and 4), demonstrating that SNF6 is not required for remodeling in vitro. We can therefore infer that the 2-MDa Swi-Snf complex, which requires Snf6 for its structural integrity (38), is not required for PHO5 chromatin remodeling in vitro.
FIG. 9.
Swi-Snf is not required for in vitro chromatin remodeling of PHO5 minichromosomes. S(0.3) extract and minichromosomes were purified from a SNF6+ (samples 1 and 2) or snf6Δ (samples 3 and 4) strain and assayed for chromatin remodeling in the presence and absence of Pho2 and Pho4. An ATP regeneration mix was added to all samples. MNase, micrococcal nuclease.
DISCUSSION
We have developed an in vitro system in which PHO5 minichromosomes undergo promoter chromatin remodeling. To obtain chromatin templates with positioned nucleosomes, an important feature of the repressed PHO5 promoter in vivo, we purified minichromosomes carrying the PHO5 gene from yeast. Chromatin remodeling in our system required the presence of the transcriptional activators Pho2 and Pho4, a fraction of S. cerevisiae nuclear extract, and hydrolyzable ATP.
In vitro chromatin remodeling of PHO5 minichromosomes was localized to the PHO5 promoter and did not require the Swi-Snf complex. Specific binding by Pho4 to UASp1 and UASp2 was required for Pho4-dependent chromatin remodeling. As these are also characteristics of the PHO5 chromatin transition in vivo, we believe that this system will allow the identification of physiologically relevant chromatin-remodeling activities.
Choice of chromatin template.
The chromatin structures of the PHO5 promoter under repressing and inducing conditions have been characterized (51). In high-phosphate medium, four nucleosomes are positioned on the PHO5 promoter such that UASp1 is in a hypersensitive region between nucleosomes −2 and −3, UASp2 is packaged into nucleosome −2, and the TATA box is packaged into nucleosome −1. These four nucleosomes lose their positioning upon PHO5 induction and no longer protect the promoter from nuclease digestion (Fig. 1).
Many studies suggest that the stability and placement of positioned nucleosomes on the PHO5 promoter are important for regulation of PHO5 expression. In vivo depletion of histone H4 causes the disappearance of positioned nucleosomes from the PHO5 promoter and weak expression under repressing conditions (26). UASp1 and UASp2 are not required for this effect. These data suggest that the presence of nucleosome −1, which packages promoter sequence including the TATA box, is required for appropriate repression of basal PHO5 expression. Another experiment demonstrated that a Pho4 mutant lacking the activation domain binds UASp2 when it is in the hypersensitive site but not when it is packaged into nucleosome −2 (52). This implies that nucleosome −2 presents a barrier to Pho4 binding to UASp2 under repressing conditions. Both of these experiments suggest that the packaging of UASp2 and the TATA box into nucleosomes is an important characteristic of the repressed PHO5 promoter.
For the reasons described above, we sought templates that have appropriately positioned nucleosomes for our in vitro study of PHO5 remodeling. Reconstitution of PHO5 promoter chromatin with purified histones and Drosophila embryo extracts did not produce templates with positioned nucleosomes (data not shown). We therefore chose to purify chromatin templates from yeast cells. By using this strategy, we ensured that PHO5 promoter chromatin would be assembled with native histones, appropriately acetylated or otherwise modified. Additionally, any nonhistone proteins required for nucleosome positioning would be present during chromatin assembly.
Definition of chromatin remodeling.
To describe and characterize our in vitro system, we have defined chromatin remodeling of PHO5 minichromosomes as an increase in micrococcal nuclease sensitivity at the DNA sequence packaged into nucleosome −2. Samples probed with sequence from nucleosome −2 (Fig. 6B) showed the same loss of nucleosomal bands as when they were probed with sequence from nucleosome −3 or sequence from all three nucleosomes (data not shown). In contrast, samples probed with sequence from nucleosome +1 (Fig. 6C) or +5 (data not shown) were mostly nucleosomal. Remodeling as defined here thus extends from nucleosome −1 to nucleosome −3, but it does not extend into the PHO5 open reading frame.
We used two other assays to analyze the change in chromatin structure on PHO5 minichromosomes after in vitro remodeling: restriction enzyme accessibility and micrococcal nuclease digestion followed by indirect end labeling. The ClaI restriction site, located near UASp2 in the PHO5 promoter, became more accessible in the presence of ATP and S(0.3) but was unaffected by the presence of Pho2 or Pho4. Loss of nucleosome positioning, as detected by the indirect end-labeling technique, was also observed in the presence of ATP and S(0.3) but did not require Pho2 or Pho4. Thus, the changes in chromatin structure that are detected by these assays occur in the absence of sequence-specific DNA binding proteins and may therefore be ascribed to nonspecific remodeling activities provided by the S(0.3). Thus, restriction enzyme accessibility and indirect end labeling were not useful assays for the identification of a PHO5-specific chromatin-remodeling activity.
The nature of the alteration in histone-DNA contacts that produces an increase in micrococcal nuclease sensitivity is not known. One interpretation is that the nucleosomes are removed from the promoter upon induction, and as a result the entire promoter becomes accessible to nuclease digestion. Proposed mechanisms for histone removal include nucleosome sliding, transfer of nucleosomes to an acceptor molecule, and disassembly of the histone octamer (49). Alternatively, the PHO5 promoter may still be packaged into chromatin under inducing conditions but in such a way that the DNA is no longer protected from interaction with nucleases.
Role of Pho4 in PHO5 chromatin remodeling.
The activation domain of Pho4 is required for the PHO5 chromatin transition in vivo. A version of Pho4 that lacks the activation domain is capable of binding to UASp1, but not to UASp2, under inducing conditions in vivo (52). When this Pho4 mutant is expressed in place of full-length Pho4, the PHO5 promoter remains packaged into positioned nucleosomes, and there is no expression of PHO5 under inducing conditions.
One explanation for these observations is that Pho4 directly remodels PHO5 chromatin, and its activation domain is required for this activity. To date, however, no transcription factor has been shown to be sufficient for chromatin rearrangement in vitro. Furthermore, Pho4 and Pho2 cannot by themselves remodel PHO5 minichromosomes in our in vitro remodeling system (Fig. 6B, sample 1). It is therefore likely that an activity in addition to these transcription factors is required to change the PHO5 promoter chromatin structure in vivo.
If Pho4 itself does not remodel chromatin, it may recruit a remodeling factor to the PHO5 promoter through its activation domain. Artificial recruitment of the RNA polymerase holoenzyme to the PHO5 promoter causes constitutive PHO5 expression and prevents the assembly of positioned nucleosomes (20), a tantalizing result given recent evidence that the holoenzyme may associate with Swi-Snf (59) (but see reference 11). Although the PHO5 chromatin transition does not require Swi-Snf (20), another ATP-dependent chromatin-remodeling complex may be associated with the holoenzyme, or chromatin remodeling may be an function intrinsic to the holoenzyme itself. It is also possible that the activation domain of Pho4 interacts directly with a chromatin-remodeling activity. Substantial evidence supports a model whereby yeast transcriptional regulators recruit histone-acetylating (14, 55) and -deacetylating (23) complexes to the promoters they regulate. Similarly, an ATP-dependent chromatin-remodeling complex may be recruited to the PHO5 promoter through interaction with the activation domain of Pho4.
In the models described above, Pho4 first binds DNA and then mediates chromatin remodeling in a second step. In contrast, it is possible that chromatin remodeling must occur before Pho4 can bind to its recognition sites in the PHO5 promoter. According to this model, a nonspecific chromatin-remodeling activity acts constitutively, in an ATP-dependent manner, to make chromatin more accessible. Subsequently, binding of Pho4 and Pho2 to the PHO5 promoter prevents nucleosome positioning and stabilizes a nuclease-sensitive state. A requirement for the Pho4 activation domain may be explained if it is necessary for stable association with PHO5 promoter chromatin.
Identity of the remodeling activity in the S(0.3) extract.
The activity contained in the S(0.3) extract may be a member of the rapidly growing family of ATP-dependent chromatin-remodeling machines, each of which contains a member of the Snf2 family of helicase-like ATPases (15). These complexes are capable of altering the DNase I digestion pattern of a mononucleosome, facilitating factor binding to sites within nucleosomes, and potentiating activation of transcription from chromatin templates (8). Members of this family thus appear to have in common the ability to modify histone-DNA contacts in an ATP-dependent manner.
Two remodeling complexes in this family have been defined in yeast: Swi-Snf and RSC (remodels the structure of chromatin). Components of Swi-Snf are required for transcription of a small number of regulated genes, and none identified to date are essential (39, 60). It has been established that Swi-Snf is not required for the PHO5 chromatin transition in vivo (10, 20, 45), and we show here that it is not required for chromatin remodeling in vitro. RSC was identified on the basis of its homology to Swi-Snf and contains several essential subunits (11, 31). It is not known if RSC is required for PHO5 expression.
Thus, the activity contained in the S(0.3) extract could be RSC or a subcomplex of either Swi-Snf or RSC. In addition to those that are contained in Swi-Snf and RSC, there are several other members of the Snf2 family in yeast, and these may be components of novel chromatin-remodeling machines that function in a similar manner. The S(0.3) extract might contain one of these putative activities or a completely novel type of ATP-dependent chromatin-remodeling activity, unrelated to Swi-Snf or RSC. Identification of this activity through conventional fractionation and reconstitution experiments is under way.
Reconstitution of the PHO5 chromatin transition.
We describe here the reconstitution of PHO5 chromatin remodeling in vitro. The PHO5 chromatin transition was studied in S. cerevisiae, a model organism that is amenable to both genetic and biochemical experiments. This allowed us to compare the in vivo PHO5 chromatin transition with our in vitro remodeling reaction, and it provides a way to confirm the physiological relevance of future in vitro results.
We used reagents that resembled their in vivo counterparts to assemble our in vitro chromatin-remodeling system. The template contained relevant sequences for PHO5 expression and had appropriately positioned nucleosomes, the transcription factors used were those required in vivo for PHO5 expression, and the source of remodeling activity was an S. cerevisiae extract. We used an in vitro assay to detect changes in minichromosomal chromatin structure that occur in vivo at the PHO5 locus upon induction.
In vitro chromatin remodeling of PHO5 minichromosomes recapitulates many hallmarks of the PHO5 chromatin transition in vivo. In addition, we show that the transcription factors Pho2 and Pho4 are not sufficient to change the PHO5 chromatin structure and that an additional ATP-dependent activity is required. This extends our knowledge of the PHO5 chromatin transition and illustrates the power of our in vitro approach. We anticipate that our system will prove to be a useful tool to provide insight into the mechanism of the PHO5 chromatin transition. We hope to use our in vitro system to identify the components required for PHO5 chromatin remodeling and to characterize the alteration of histone-DNA contacts that occurs during the PHO5 chromatin transition.
ACKNOWLEDGMENTS
We thank Mary Maxon for Pho2, Wolfram Horz for yeast strain YS18, Yasuji Oshima for pACD5 and pACD4, and Marian Carlson for pEY110. We also acknowledge Sandy Johnson, Keith Yamamoto, Joachim Li, Mary Maxon, and members of the O’Shea lab for insightful comments on the manuscript and Beverly Emerson for a particularly useful suggestion.
This work was supported by NIH grant GM51377 to E.K.O. and by an NSF Predoctoral Fellowship to E.S.H. E.K.O. is a Packard Foundation Fellow and an NSF Presidential Faculty Fellow.
REFERENCES
- 1.Almer A, Rudolph H, Hinnen A, Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer T K, Lefebre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong J A, Emerson B M. Transcription of chromatin: these are complex times. Curr Opin Genet Dev. 1998;8:165–172. doi: 10.1016/s0959-437x(98)80137-8. [DOI] [PubMed] [Google Scholar]
- 4.Barbaric S, Munsterkotter M, Goding C, Horz W. Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol Cell Biol. 1998;18:2629–2639. doi: 10.1128/mcb.18.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbaric S, Munsterkotter M, Svaren J, Horz W. The homeodomain protein Pho2 and the basic-helix-loop-helix protein Pho4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Res. 1996;24:4479–4486. doi: 10.1093/nar/24.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergman L W, Kramer R A. Modulation of chromatin structure associated with derepression of the acid phosphatase gene of Saccharomyces cerevisiae. J Biol Chem. 1983;258:7223–7227. [PubMed] [Google Scholar]
- 7.Bergman L W, Stranathan M C, Preis L A. Structure of the transcriptionally repressed phosphate-repressible acid phosphatase gene (PHO5) of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:38–46. doi: 10.1128/mcb.6.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 9.Cairns B R, Kim Y J, Sayre M H, Laurent B C. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns B R, Levinson R S, Yamamoto K R, Kornberg R D. Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 1996;10:2131–2144. doi: 10.1101/gad.10.17.2131. [DOI] [PubMed] [Google Scholar]
- 11.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 12.Cox J S, Shamu C E, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident protein requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 13.Dean A, Pederson D S, Simpson R T. Isolation of yeast plasmid chromatin. Methods Enzymol. 1989;170:26–41. doi: 10.1016/0076-6879(89)70041-0. [DOI] [PubMed] [Google Scholar]
- 14.Drysdale C M, Jackson B M, McVeigh R, Klebanow E R, Bai Y, Kokubo T, Swanson M, Nakatani Y, Weil P A, Hinnebusch A G. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol Cell Biol. 1998;18:1711–1724. doi: 10.1128/mcb.18.3.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisen J A, Sweder K S, Hanawalt P C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estruch F, Carlson M. SNF6 encodes a nuclear protein that is required for expression of many genes in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2544–2553. doi: 10.1128/mcb.10.6.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fascher K-D, Schmitz J, Horz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990;9:2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fascher K-D, Schmitz J, Horz W. Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J Mol Biol. 1993;231:658–667. doi: 10.1006/jmbi.1993.1317. [DOI] [PubMed] [Google Scholar]
- 19.Fisher F, Jayaraman P-S, Goding C R. c-Myc and the yeast transcription factor Pho4 share a common CACGTG-binding motif. Oncogene. 1991;6:1099–1104. [PubMed] [Google Scholar]
- 20.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Horz W. RNA polymerase holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 21.Gregory P D, Schmid A, Zavari M, Lui L, Berger S L, Horz W. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol Cell. 1998;1:495–505. doi: 10.1016/s1097-2765(00)80050-7. [DOI] [PubMed] [Google Scholar]
- 22.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 23.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 24.Kaffman A, Herskowitz I, Tjian R, O’Shea E K. Phosphorylation of the transcription factor Pho4 by a cyclin-CDK complex, Pho80-Pho85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 25.Kaffman A, Rank N M, O’Shea E K. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 1998;12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim U-J, Kayne P, Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 1988;7:2221–2228. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kingsbury J, Koshland D. Centromere function on minichromosomes isolated from budding yeast. Mol Biol Cell. 1993;4:859–870. doi: 10.1091/mbc.4.8.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleene R, Janes M, Meyhack B, Pulfer K, Hinnen A. High-level expression of endogenous acid phosphatase inhibits growth and vectorial secretion in Saccharomyces cerevisiae. J Cell Biochem. 1995;57:238–250. doi: 10.1002/jcb.240570207. [DOI] [PubMed] [Google Scholar]
- 29.Kornberg R D, Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–887. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- 30.Laurent B C, Treich I, Carlson M. The yeast SNF2/SWI2-protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 31.Laurent B C, Yang X, Carlson M. An essential Saccharomyces cerevisiae gene homologous to SNF2 encodes a helicase-related protein in a new family. Mol Cell Biol. 1992;12:1893–1902. doi: 10.1128/mcb.12.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenburg M E, O’Shea E K. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- 33.Lue N F, Flanagen P M, Kelleher R J, Edwards A M, Kornberg R D. RNA polymerase II transcription in vitro. Methods Enzymol. 1991;194:545–550. doi: 10.1016/0076-6879(91)94041-a. [DOI] [PubMed] [Google Scholar]
- 34.Nedospasov S A, Shakhov A N, Georgiev G P. Analysis of nucleosome positioning by indirect end-labeling and molecular cloning. Methods Enzymol. 1989;170:408–420. doi: 10.1016/0076-6879(89)70059-8. [DOI] [PubMed] [Google Scholar]
- 35.Orphanides G, LeRoy G, Chang C H, Luse D S, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1988;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 36.Oshima Y. Regulatory circuits for gene expression: the metabolism of galactose and phosphate. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. pp. 159–180. [Google Scholar]
- 37.Paranjape S M, Kamakaka R T, Kadonaga J T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 38.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodeling machine. Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 40.Pollard K J, Peterson C L. Role of ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pryciak P M, Sil A, Varmus H E. Retroviral integration into minichromosomes in vitro. EMBO J. 1992;11:291–303. doi: 10.1002/j.1460-2075.1992.tb05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roth S Y, Simpson R T. Yeast minichromosomes. Methods Cell Biol. 1991;35:289–314. [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Schmid A, Fascher K-D, Horz W. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell. 1992;71:853–864. doi: 10.1016/0092-8674(92)90560-y. [DOI] [PubMed] [Google Scholar]
- 45.Schneider K. Ph.D. thesis. University of California, San Francisco; 1995. [Google Scholar]
- 46.Scott J H, Scheckman R. Lyticase: endoglucanase and protease activities that act together in yeast cell lysis. J Bacteriol. 1980;142:414–423. doi: 10.1128/jb.142.2.414-423.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Senstag C, Hinnen A. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual amino acid composition. Nucleic Acids Res. 1987;15:233–246. doi: 10.1093/nar/15.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silva J, Zinker S, Gariglio P. Isolation and partial characterization of 2-microns yeast plasmid as a transcriptionally active minichromosome. FEBS Lett. 1987;214:71–74. doi: 10.1016/0014-5793(87)80015-7. [DOI] [PubMed] [Google Scholar]
- 49.Steger D J, Workman J L. Remodeling chromatin structure for transcription: what happens to the histones? Bioessays. 1996;18:875–884. doi: 10.1002/bies.950181106. [DOI] [PubMed] [Google Scholar]
- 50.Straka C, Horz W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 1991;10:361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svaren J, Horz W. Transcription factors vs. nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 52.Svaren J, Schmitz J, Horz W. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thoma F, Bergman L W, Simpson R T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984;177:715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- 54.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 55.Utley F T, Ikeda K, Grant P A, Cote J, Steger D, Eberharter A, John S, Workman J. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 56.Varga-Weiss P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodeling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 57.Venter U, Svaren J, Schmitz J, Schmid A, Horz W. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 1994;13:4848–4855. doi: 10.1002/j.1460-2075.1994.tb06811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogel K, Horz W, Hinnen A. The two positively acting regulatory proteins Pho2 and Pho4 physically interact with PHO5 upstream activation regions. Mol Cell Biol. 1989;9:2050–2057. doi: 10.1128/mcb.9.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 60.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 61.Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 62.Yoshida K, Ogawa N, Oshima Y. Function of the PHO regulatory genes for repressible acid phosphatase synthesis in Saccharomyces cerevisiae. Mol Gen Genet. 1989;217:40–46. doi: 10.1007/BF00330940. [DOI] [PubMed] [Google Scholar]
- 63.Zakian V A, Scott J F. Construction, replication, and chromatin structure of TRP1 R1 circle, a multicopy synthetic plasmid derived from Saccharomyces cerevisiae chromosomal DNA. Mol Cell Biol. 1982;2:221–232. doi: 10.1128/mcb.2.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]