Abstract
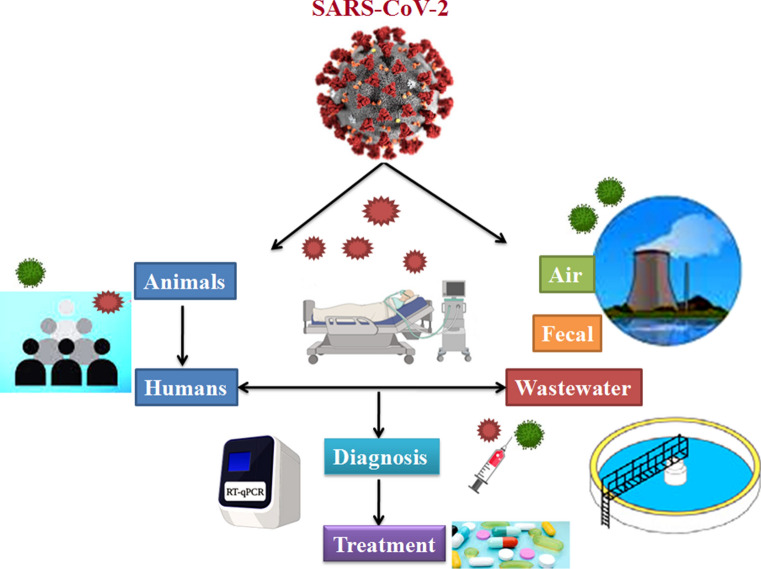
The novel coronavirus disease (COVID-19), caused by severe acute respiratory coronavirus-2 (SARS-CoV-2), was first identified in China and subsequently spread globally, resulting in a severe pandemic, and officially declared a significant health emergency by World Health Organization (WHO). Genetic analysis of coronavirus isolated from bats, snakes, and Malay pangolins suggested that they could be intermediate hosts for SARS-CoV-2. The transfer of virus from person to person has been confirmed widely, while the actual source of origin is still unknown. COVID-19 is a highly contagious and infectious disease, and the worldwide transmission of coronavirus has intense effects on the lives of human beings. The spread of the virus is observed mainly through close contact with the infected person due to coughing, sniffing or indirectly through the contaminated surfaces. If people touch contaminated surfaces through their hands, mouth, nose, or eye, it enters the body and causes disease. Also, the virus may transmit through air droplets, water, food, fecal–oral transmission, etc. The infection of virus in human beings could be detected by direct symptoms, or different diagnostic tools are available to determine the viral load. Various safety measures are used to contain the virus, including disinfectants, antiviral drugs, vaccines, wearing masks, social distancing, etc. In the present review, we have focused on transmission of COVID-19 through air and wastewater as environmental transmission modes. We have also discussed the origin of the virus, its mode of action, host immune response, vulnerability, varying symptoms and diagnosis, prevention and control. Further, we have discussed the various treatment options to cope with this viral outbreak.
Graphical abstract
Keyword: COVID-19, Transmission, Environment, Diagnosis, Mitigation
Introduction
Coronavirus disease (COVID-19) is Asia’s third coronavirus related outbreak in the last 2 decades, after severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) (Morens et al. 2020). In the beginning, 2019 novel coronavirus or 2019-nCoV was the newest of these coronaviruses. It originated when several patients reported unexplained pneumonia symptoms in Chinese health care facilities in Wuhan, China (Zhu et al. 2020). The COVID-19 outbreak was declared a global public health emergency on January 30, 2020. The International Committee on Taxonomy of Viruses (ICTV) has assigned the name to 2019-nCoV as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the disease as COVID-19 (WHO 2020; Lai et al. 2020). On March 2020, the coronavirus outbreak was declared a global pandemic as COVID-19 had become a major global health concern. As of May 10, 2021 COVID-19 had influenced over 157,973,438 confirmed cases in 223 nations and regions worldwide and around 3,288,455 deaths worldwide (https://www.worldometers.info/coronavirus/).
Coronaviruses generally affect the respiratory tracts of human beings, animals, and birds. Early studies suggested the spread of SARS-CoV-2 through animal to humans and humans to humans through direct contact or respiratory droplets (Li et al. 2020). Ciencewicki and Jaspers (2007) reported that air pollutants, including particulate matter (PM-25 and PM-10), carbon monoxide (CO), ozone (O3), nitrogen dioxide (NO2), and sulphur dioxide (SO2), could affect airways, increases the sensitivity to a viral respiratory illness, and also severity of the disease. Constant presence of air contaminants hinders the revival and tends to be more severe and fatal (Conticini et al. 2020; Wu et al. 2020a). Studies showed coronavirus shedding in the human stool (Gao et al. 2020; Jiehao et al. 2020; Zhang et al. 2020a) and also identified in the industrial wastewater from the United States, Netherlands, and Australia (Ahmed et al. 2020; Medema et al. 2020; Lodder and de Husman 2020). The pathogens transmission in wastewater study by Gormley et al. (2020) indicated that airborne spread of SARS-CoV-2 could be enabled through the wastewater treatment system. The contamination could also spread through hospital wastewater with positive SARS-CoV-2 virus particles (China Citywater 2020).
Different treatments for the SARS-CoV-2 are used for patients depending upon the symptoms, including oxygen therapy for critically ill patients. Supplementary treatments, including drugs and therapeutics, are used in severely ill patients (Zumla et al. 2016; WHO 2020). The study on the transmission of COVID-19 by environmental sources is not sufficient. Therefore in this study, we review the recent and updated transmission information of the SARS-CoV-2 through the environmental source, its prevention, monitoring, and potential therapeutic options for COVID-19.
Origin of coronaviruses
Coronaviruses were first described in the 1960s and belonged to the family Coronaviridae. Coronaviruses were categorised into four genera based on phylogenetic relationships and genomic structure analysis; Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Woo et al. 2012). α and β Coronaviruses are recognized to infect the mammals; primarily causing respiratory and intestinal infection while γ- and δ- coronaviruses infect birds. SARS-CoV-2 belongs to Betacoronavirus generation, and it has 84% nucleic acid similarity to SARS-like coronavirus and 78% and 50% nucleic acid similarity to SARS-CoV and MERS-CoV, respectively (Lu et al. 2020; Chan et al. 2020a). Including SARS-CoV-2, 7 coronavirus strains are believed to infect humans, such as HCoV-229E (human coronavirus 229), HCoV-OC43 (human coronavirus OC43), HCoV-NL63 (human coronavirus NL63), HCoVHKU1 (human coronavirus HKU1), SARS-CoV (severe acute coronavirus syndrome), and MERS-CoV (Middle East coronavirus syndrome) (Chan et al. 2020b; Cui et al. 2019; Ng and Hiscox 2020).
The first SARS-CoV-2 case was found in connection with China’s seafood market, although its source is not yet disclosed. SARS-CoV-2 is a zoonotic virus. Individuals from Wuhan seafood marketplace were initially infected and developed viral pneumonia, thereby suggesting that the virus is transmitted directly from animals to human beings. Subsequently, persons contaminated with SARS-CoV-2 without any associations or business contact with the seafood market confirmed that the virus could be passed from humans to humans. Rhinolophus affins bat coronavirus (BatCoV RaTG13) showed 96% full-genome similarity with SARS-CoV-2 signifying that the bat could be the origin of SARS-CoV-2 (Zhou et al. 2020a). While SARS-CoV-2 is close to SARS-CoV and MERS-CoV, Zhou et al. (2020b) hypothesized that the latest virus might have been transmitted directly to humans from an unspecified transitional host.
Ji et al. (2020) analysed the genetic sequence of SARS-CoV-2 similarity with all other known coronaviruses, suggesting that SARS-CoV-2 has the most analogous codon usage with the snake. According to reports, snakes were sold in China's domestic seafood market, increasing the risk of SARS-CoV-2 spreading from animals, such as bats to snakes, and then to humans during coronavirus outbreaks. However, how this infection may transform into cold and warm-blooded hosts is still a mystery (Ji et al. 2020). The study published by researchers showed that SARS-CoV-2 isolated from dead Malay pangolins (Manis jabanica) had SARS-CoV-2-like genomes, indicating that pangolins might be probable SARS-CoV-2 intermediate hosts (Lam et al. 2020). As a consequence, research is required to confirm whether SARS-CoV-2 spread from bats or other intermediate hosts.
Patho-physiology of SARS-CoV-2
SARS-CoV-2 encodes a non-structural polyprotein (ORF1a/b), which produces 15–16 proteins, four structural proteins, and five accessory proteins (Ramaiah and Arumugaswami 2020; Chan et al. 2020a; Wu et al. 2020b). Structural proteins of SARS-CoV-2 are essential for their assembly, consisting of the spike (S), i.e., surface glycoprotein, membrane protein (M), an envelope protein (E), and nucleocapsid protein (N). The surface glycoprotein is essential to its attachment, and entry into the host cell proteases can cleave into an N-terminal subunit (S1) and C-terminal (S2) membrane-bound region (Yuan et al. 2017). The spike protein contains a three-dimensional structure in the receptor-binding domain (RBD) region, loosely attached among viruses with van der waals forces, and may lead to infecting multiple hosts (Perlman and Netland 2009; Raj et al. 2013; Xu et al. 2020a). According to a study, SARS-CoV-2 spike links to angiotensin-converting enzyme 2 (ACE2), a membrane exopeptidase in the receptor, present in the respiratory tract of human beings (Jia et al. 2005) with higher affinity than SARS-CoV spike and regulates both spread from human to human and cross-species (Wan et al. 2020a; Wrapp et al. 2020; Letko and Munster 2020). The patho-physiology and virulence mechanisms for coronaviruses and SARS-CoV-2 are linked to non-structural polyproteins and the function of structural proteins. For instance, research has emphasised that non-structural polyproteins inhibit innate immune responses (Cascella et al. 2020). In order to promote viral assemblage and release, the envelope plays a crucial role in viral pathogensis. Upon infection, the infection median incubation time is about 4–5 days before the appearance of symptoms (Guan et al. 2020; Pung et al. 2020; Lauser et al. 2020), and 97.5% of symptoms occur within 11–12 days (Lauser et al. 2020). Also, at the hospitalization phase, patients face complications in breathing, diarrhea, dizziness/headache, nausea, joint pain, and coughing up of blood (Guan et al. 2020; Huang et al. 2020; Chen et al. 2020; Liu et al. 2020a). SARS-CoV-2 viral load reaches its peak within 5–6 days of symptom onset, significantly higher than SARS-CoV, where the viral load peaks about 10 days after the onset of symptoms (Pan et al. 2020; Kim et al. 2020; Zou et al. 2020). On average, extreme COVID-19 cases progress to ARDS (acute respiratory distress syndrome) in about 8–9 days after the onset of symptoms (Huang et al. 2020; Wang et al. 2020a).
Transmission of SARS-CoV-2
In general, the infection extends from person to person, usually in close contact or by respiratory droplets formed by the contaminated person while coughing or sniffing; due to this, distance of ~ 6 feet, i.e., more than two meters from an infected person, is recommended (Li et al. 2020). Other than coughing and wheezing, infections can spread from one person to the next through contacted areas (fomites) touched by contaminated individuals. The route of the virus transmission from fomites is relevant because the virus can live for several days on certain surfaces (van Doremalen et al. 2020; Phan et al. 2020; Riou and Althaus 2020). If people touch these fomites through their mouth, nose, or eyes, they will develop COVID-19 disease. Also, research suggested that SARS-CoV-2 is spread mainly by interaction with airborne drops rather than by air. A person with a gentle cough who does not experience infection might be infected by it.
Researchers suggested that the patients of COVID-19 have experienced gastrointestinal symptoms, such as vomiting, nausea, and diarrhea (Holshue et al. 2020; Zhang et al. 2020b). Huang et al. (2020) reported that the gastrointestinal symptoms of COVID-19 are linked with invaded ACE2-expressing enterocytes. The study was signifying that the digestive tract is a possible way of SARS-CoV-2 contamination as well as the respiratory system (Gu et al. 2020; Xiao et al. 2020). According to studies, SARS-CoV-2 can live for 4 days in samples of feces (Weber et al. 2016) and RNA of the virus was detected in sewage (Wu et al. 2020c; Wang et al. 2020b) thus, raising the possible route of fecal–oral transmission (Ahmed et al. 2020; Medema et al. 2020). Patients with COVID-19 will shed the virus in their feces for days after all the respiratory symptoms have vanished (Wu et al. 2020c). Vectors such as cockroaches and flies may serve as a mode of spread in environments and bring viruses from the feces in their bodies and intestinal tracts, thus infecting the surfaces (Dehghani and Kassiri 2020).
The importance of viral spread by air droplets (aerosols) was intensively discussed in the COVID-19 pandemic context (Lewis 2020; Morawska and Cao 2020). This one is the most widely accepted mode of viral diffusion, with larger respiratory droplets and close interaction with fomites. Although studies are currently underway on the airborne transmission of COVID-19, however, some hospital-based studies have conceded the presence of SARS-COV-2 in air (Ong et al. 2020; Chia et al. 2020; Ding et al. 2020; Jiang et al. 2020; Liu et al. 2020b; Santarpia et al. 2020). Using the PCR (polymerase chain reaction) test tool, several studies have confirmed SARS-CoV-2 genome (RNA) in air samples (Chia et al. 2020; Jiang et al. 2020; Liu et al. 2020b). The aerosols created by cough and particulates (dust) suspended in the air containing viruses (1–4 μm and > 4 μm and 1.8–3.4 viral RNA copies per L of air, Chia et al. 2020) remain suspended for a long time and are thus transported over long distances (usually greater than 6 feet) (Coleman et al. 2018; Knibbs et al. 2012; Nishiura et al. 2020). The chances of infection by airborne transmission are exceptionally high in enclosed spaces (buildings, public transport) and/or in poorly ventilated areas or where there is prolonged exposure to respiratory droplets produced during shouting, loud singing, and exercise (He et al. 2020; Leclerc et al. 2020; Park et al. 2020). Air conditioning systems used in enclosed spaces recirculate indoor air and help the virus stay suspended in the air for a more extended time (Goodman 2020; Morawska et al. 2020; Aggarwal 2020).
Impact of air and water pollution on the spread of COVID-19
Effect of air pollution
Together with the specific environmental situation and high air pollutant content, an atmosphere could endorse a longer persistence of the virus in the air (Frontera et al. 2020) and promote indirect diffusion of SARS-CoV-2. Martelletti and Martelletti (2020) indicated that SARS-CoV-2 might be an appropriate carrier for particulate air pollutants. People who live in areas with elevated air pollution are more likely to experience respiratory problems (Domingo et al. 2020) and are ideal for viral contamination (Xie et al. 2019). Conticini et al. (2020) reported that those living in the air contaminated regions were more to die of COVID-19 due to their possible health condition caused by air pollutants. It could be suggested that the elevated level of air contaminants is considered to be a supplementary part of the high lethality reported in the extremely polluted environments. The positive association of air pollutant concentrations with daily COVID-19 confirmed cases in China and Italy was reported by Zhu et al. (2020) and Martelletti and Martelletti (2020), respectively.
Frontera et al. (2020) suggested a “double hit hypothesis” showing overexpression of the alveolar ACE-2 receptor due to chronic PM 2.5 exposures, which in turn raises the viral load in patients exposed to contaminants that obstruct host defense and deplete ACE-2 receptors. Higher atmospheric NO2 will cause the second strike, resulting in an acute form of SARS-CoV-2 in ACE-2 depleting lungs. The association amid air contamination and SARS-CoV-2 infection has immediate and long-term consequences and directly influence the elevated rate of infection and lethality.
Effect of water pollution
Recent researchers have isolated live SARS-CoV-2 from feces and urine of infected people (Holshue et al. 2020; Collivignarelli et al. 2020; Bilal et al. 2020; Michael-Kordatou et al. 2020); so, it became apparent that the virus might enter the municipal wastewater or wastewater treatment plant (Holshue et al. 2020; RIVM 2020). The studies showed that wastewater might be an early warning and a sensitive surveillance system, as was previously demonstrated for poliovirus (Lodder et al. 2012).
Ahmed et al. (2020) track infection frequency of SARS-CoV-2 by quantifying wastewater within the population through wastewater-based epidemiology (WBE). About 22% of the samples collected from wastewater were screened positive for SARS-CoV-2 by the N_Sarbeco assay. Hart and Halden (2020) performed computational modeling using WBE to examine the economy, possibilities, feasibility, and difficulties of local and global enumeration of coronavirus infections. Randazzo et al. (2020) reported positive SARS-CoV-2 RNA in wastewater samples of influent and secondary treated effluents from wastewater treatment plants of Murcia, Spain. Casanova et al. (2009) stated that coronavirus remains infectious for days to weeks in pure water, room temperature, or settled sewage. During treatment process, wastewater contaminated with infected stools may generate more transmission of virus-laden aerosols. Therefore, the role of the aerosol from contaminated sewage in the COVID-19 spread requires to be investigated. The potential dissemination of SARS-CoV-2 by wastewater has yet to be thoroughly demonstrated (CDC and WHO 2020) and appears dubious due to less viral constancy and high sensitivity to disinfectants under environmental conditions (Lodder and de Husman 2020; Chin et al. 2020).
During the pandemic, genome sequencing led to the understanding of the distribution and degree of viral genetic diversity. Usually, nasopharyngeal or oral swabs of patients were collected for sequencing of viral genomes to track the spread of viruses. RT-qPCR (real time quantitative reverse-transcriptase-polymerase chain reaction) analysis of sewage wastewater has recently been used to assess the SARS-CoV-2 in many regions worldwide. In a research by Christoph et al. (2020), authors sequenced RNA from sewage samples gathered from the bay area of San Francisco to produce complete SARS-CoV-2 genomes. They characterised SARS-CoV-2 alleles in samples of wastewater using a single nucleotide variant (SNV) pipeline called in a metagenomic context and identified viral genotypes that were also present within clinical genomes in California (CA). The variants of virus genotypes in wastewater originating from patients were more akin to local CA than to other areas of the United States or anywhere in the world. Patients' genomes obtained from outside of California have identified viruses in wastewater, suggesting that wastewater sequencing may provide evidence for a recent viral lineage prior to local clinical sequencing. This result suggests that in a disease outbreak, epidemiological surveillance can help to track different viral strains by sequencing of wastewater samples.
In another study by Nemudryi et al. (2020), they use a nearly complete sequence of wastewater SARS-CoV-2 genomes (98.5%) and phylogenetic analysis to deduce viral ancestry. The samples were obtained from the urban wastewater treatment facility (Bozeman, Montana, USA). For RNA extraction, non-treated samples of wastewater were collected over 17 days, and extracted RNA was used for RT-qPCR. In particular, 11 variants of single nucleotides (SNVs) are contained in the assembled genome that differentiates the SARS-CoV-2 sequence of Bozeman wastewater from the reference sequence of Wuhan-Hu-1/2019. This genome was compared to 14,970 SARS-CoV-2 genomes from 74 countries. The resulting alignment was used to establish a phylogenetic tree reveals that in Bozeman’s wastewater, genomes from California, Victoria, and Australia are most closely related to the SARS-CoV-2 genome. The study indicated that the sequencing of wastewater from the temporal genome might help to differentiate viral strains in a specific community that circulates over time.
Host immune response against COVID-19
Several studies reported essential changes in COVID-19 patients that occur in both innate and adaptive immune systems (Huang et al. 2020; Wan et al. 2020b; Wang et al. 2020a). The molecular patterns of pathogen recognition in subsequent cytolytic immune responses are usually through the natural killer cells and type I interferons (INF). Adaptive immunity aids viral removal by inducing cytotoxic T cells that destroy virus-infected cells and antibody-producing B cells that target virus-specific antigens. Patients, mainly those with severe pneumonia, observed significantly lower lymphocyte counts and higher plasma concentrations with inflammatory cytokines such as IL-6 and tumor necrosis factor (TNF) (Huang et al. 2020; Wan et al. 2020b; Wang et al. 2020a). A significant proportion of COVID-19 patients (around 80%) are asymptomatic or have intermediate manifestations; however, approximately 15% experience extreme pneumonia, and approximately 5% develop to extraordinary respiratory distress syndrome (ARDS), septic shock, or likely organ failure (Huang et al. 2020; Xu et al. 2020b). In contrast to those with mild disease symptoms, seriously ill patients reported a marked decrease in natural killers (NK) cells, B cells, CD8 + cells, and CD4 + cells (Wan et al. 2020a; Huang et al. 2020; Xu et al. 2020b; Shi et al. 2020), as well as a decrease in basophils, eosinophils and monocytes (Shi et al. 2020; Qin et al. 2020; Zhang et al. 2020d). Patients with the infection secreted more pro-inflammatory cytokines and chemokines into their blood, including IL-6, IFN, monocyte chemoattractant protein-1 (MCP1), and interferon gamma-induced protein (IP-10) (Huang et al. 2020).
Vulnerability to COVID-19
Karnam et al. (2012) revealed that sex host factors and CD200-CD200R glycoprotein collectively establish the viral disease. Another study found that women are at increased risk of autoimmune diseases, especially during their reproductive years, but are more immune to viral infections than men. This is probably mediated by a number of variables, including sex hormones (Ghazeeri et al. 2011). Elgendy and Pepine (2020) described sex-based immunological differences driven by sex hormone and X chromosome, where blocking estrogen receptors increased mortality among female mice infected with SARS-CoV, indicating the involvement of estrogen receptors signaling may present some defensive outcome in female mice. In the European country, most COVID-19 deaths were observed in Italy (Onder et al. 2020) were male, while almost double number of men than women have passed away in Spain (http://globalhealth5050.org/covid19/ Accessed 19 April, 2020). Data from China revealed 41.9% of the admitted cases were female, with the majority of deaths were among men (Guan et al. 2020). In the US (Faure et al. 2020), South Korea (Shim et al. 2020), Germany, and the UK (http://globalhealth5050.org/covid10/ Accessed 19 April, 2020), similar mortality rates have been registered. In contrast to women, this trend may be part of genetic and immunology and different gender-specific lifestyles, such as drinking, smoking, being more prevalent amongst men (Shim et al. 2020). Furthermore, a recent study undertaken in Spain discovered that women took a more responsible approach to the COVID-19 pandemic than men (De La Vega et al. 2020) while taking preventive measures such as remaining at home, using face masking, and regular hand washing. Moreover, it has also been documented that the prevalence of clinical cases seems to be linked to age factor (> 70 years), co-morbidity such as hypertension, chronic obstructive pulmonary disease (COPD), obesity, and diabetes, rather than gender (Zhou et al. 2020b; Wu et al. 2020d; Han et al. 2020).
Symptoms and diagnosis
The general medical manifestations include sputum production (33.4%), cough (67.8%), fever (88.7%), fatigue (38.1%), sore throat (13.9%), shortness of breath (18.6%), and headache (13.6%) (Guan et al. 2020). In addition, some patients have gastrointestinal signs with vomiting (5.0%) and diarrhea (3.8%). In laboratory examination results, the majority of patients had normal or reduced lymphocytopenia numbers and white blood cell counts (Guan et al. 2020; Kui et al. 2020). A significantly higher level of blood urea, neutrophil count, creatinine, and d-dimer is displayed by critically ill patients, whereas the lymphocyte count steadily decreases. Additionally, rise in TNF-α (tumor necrosis factor-α), and IL-6, IL-10 (interleukin-inflammatory factors) demonstrates the immune status of patients. The data from the patients of intensive care units (ICU) showed having higher plasma levels of IL-2, 7, and 10, GCSF (granulocyte colony-stimulating factor), IP-10 (10 kD interferon gamma-induced protein), MCP-1 (monocyte chemoattractant protein-1), MIP-1α (macrophage inflammatory protein 1-α), and TNF-α (Huang et al. 2020).
Different methods of PCRs are used for SARS-CoV-2 detection in routine diagnostics worldwide. Although the RT-PCR takes around 24 h of the average time for the outcome, and also it is an expensive method. Besides, it cannot be carried out in any laboratory because of a lack of molecular virology facilities and technical expertise (Mohanty et al. 2020a). Serological antibody tests have been added but have not been deemed suitable for diagnosis since these antibodies appear only after 2 weeks of symptom initiation and are unable to diagnose acute infections. As a result, these antibody-based experiments were shown only for surveillance purposes (Mohanty et al. 2020b). Because of the above, a reliable rapid antigen test (RAT) was required. The virus's basic marker is supposedly a viral antigen and precedes the presence of antibodies within the infected individuals. Therefore, viral antigen detection may have a rapid screening effect and help achieve early diagnosis of this deadly disease. The computerized tomography (CT) imaging on the chest showed bilateral patchy shadowing (51.8%) and ground-glass opacity (56.4%) (Guan et al. 2020). Chest computed tomography is a valuable procedure for recognizing SARS-CoV-2 in the suspected patient (Huang et al. 2020). However, CT alone might restrict SARS-CoV-2 infection with a negative predictive value (Chung et al. 2020), as at early stages, some patients might possibly have normal radiology.
The CRISPR-based SHERLOCK (Specific High-Sensitivity Enzymatic Reporter UnLOCKing) method for SARS-CoV-2 diagnosis has been described by Zhang et al. (2020c). Using RNA fragments of synthetic SARS-CoV-2, the authors observed that this method can constantly identify target sequences of SARS-CoV-2 in a range between 20 and 200 aM (10–100 copies/microlitre of input). Without requiring extensive instrumentation, this test can be read out using a dipstick within 1 h. The SHERLOCK approach is more reliable compared to RT-qPCR, and the time of detection is also reduced. Mei et al. (2020) integrated chest CT findings along with artificial intelligence (AI) algorithms to rapidly detect COVID-19 patients with clinical symptoms, laboratory testing, and exposure history. In this investigation, they demonstrated the possible role of an extremely accurate AI algorithm for the rapid detection of COVID-19 patients, which combined clinical information and CT imaging and showed comparable precision with senior chest radiology, thus indicating that it may be a practical screening mechanism to quickly detect infectious COVID-19 and other diseases without physical tests or radiologist input.
COVID-19 control and prevention
Community health and disease prevention initiatives are desperately required to limit the virus’s global dissemination and the harm caused by COVID-19 (Song et al. 2020). SARS-CoV-2 might be vulnerable to proven disinfectants, such as 70% ethanol, quaternary ammonium compounds, bleach (sodium hypochlorite), phenolic compounds, and 0.5% hydrogen peroxide. Several vaccinations have been shown to be safe and effective and countries have approved various COVID-19 vaccines for the prevention of SARS-CoV-2 (Sharma et al. 2021). Some of the effective preventive methods have been summarised in Table 1. The World Health Organization and the United Nations Children's Fund suggest normal breast feeding within an hour of birth for babies, using adequate infection prevention precautions (WHO 2020; UNICEF 2020). Also, the child should stay ~ 6 feet apart while the mother is not breastfeeding. To reduce the contamination of viruses spread by the aerosol created by air conditioners, the American Society of Heating, Refrigerating and Air-Conditioning Engineers (ASHRAE 2020) proposes seven techniques, including dilution (high ventilation speeds, i.e., air changes per hour), airflow patterns (how the air flows), pressurisation (negative pressure concerning the atmosphere in insulation rooms), temperature, relative humidity, filtration, and ultraviolet germicidal irradiation as techniques to reduce exposure to infectious aerosols. Enhancing ventilation with outside air is also a way to reduce virus transmission. Indoor air recirculation in closed spaces for longer durations (such as in offices, trains, ships, etc.) can spread the infection. Therefore, adequate ventilation is critical in reducing indoor infection risk.
Table 1.
List of preventive measures against COVID 19
Source: Adopted and modified from World Health Organization (2020)
| Health care | |
| Protective device |
Wear a self-made “gastroscope mask” during gastroscopy Medical mask N95 respirator |
| Infection prevention |
Propofol anesthesia maintenance, common extubation Propofol anesthesia was maintained, and the extubation strategy of “circulatory induced respiration” was adopted after the operation Sevoflurane anesthesia was maintained, and the extubation strategy of “circulatory induced respiration” was adopted after the operation |
| Mouthwash |
Povidone-Iodine Hydrogen peroxide Neem extract (Azadirachta indicia) |
| Education | Training for wearing the mask |
| Non-healthcare | Wash your hands frequently and thoroughly with soap and water and dry them thoroughly |
| Cover the mouth and nose while coughing and sneezing with a flexed elbow or tissue | |
| Avoid contact with eyes, nose, and mouth | |
| Hold within physical ranges of others | |
| Clean and disinfect frequently touched surfaces every day |
Effective treatment strategies for COVID-19
Various treatment methods can be used against COVID depending on the seriousness of the patient and the local epidemiology (Chan et al. 2020b; Chinese Association of Rehabilitation Medicine 2020) (Table 2). To treat infected individuals and reduce disease burden during the current COVID-19 pandemic, successful production of antivirals and vaccines is critical. Bed rest and daily care, sufficient intake of energy, safeguard of a constant internal environment such as electrolytes, water, and other factors, monitoring of vital signs (respiratory rate, blood pressure, heart rate, pulses and oxygen saturation level, etc.) (Li et al. 2020) are important. The antiviral treatments used include lopinavir/ritonavir, interferon-alpha (IFN α), ribavirin, chloroquine, arbidol, and remedesivir (Ng and Hiscox 2020; Wang and Fish 2019; Wang et al. 2020c). However, only remdesivir was shown to have a significant effect against the virus (Agostini et al. 2018). Remdesivir, in combination with interferon or chloroquine, significantly inhibited SARS-CoV-2 replication, and patients were considered clinically restored (Sheahan et al. 2020; Holshue et al. 2020; Wang et al. 2020c).
Table 2.
List of potential experimental treatments of COVID 19
Source: Adopted and modified from World Health Organization (2020)
| Treatment type | Treatment name |
|---|---|
| Drugs | |
| Antiviral, non-specific | Interferons, interferon lambda alpha 1a, immunoglobulin, interleukin-2 |
| Antiviral, broad spectrum |
Favipiravir, ribavirin, triazavirin, umifenovir xiyanping, nitazoxanide, galidesivir |
| Antiviral, antiretrovirals |
Danoprevir, darunavir, darunavir + cobicistat lopinavir + ritonavir, remdesivir, baricitinib in combination with remdesivir Danoprevir + ritonavir |
| Antiviral combination (when combining antivirals from different groups) |
Interferon alpha + lopinavir Umifenovir + interferon alpha Lopinavir + ritonavir + ribavirin + interferon beta, ASC09F + oseltamivir ritonavir + oseltamivir, lopinavir + ritonavir + xiyanping lopinavir + ritonavir + interferon beta1 |
| Antimalaria |
Chloroquine, hydroxychloroquine, dihydroartemisinin, mefloquine GNS561 (chloroquine analog) |
| Antimalaria + antibiotics | Hydroxychloroquine + azithromycin |
| Antibiotics + antimalaria + monoclonal antibodies | Azithromycin + hydroxychloroquine + tocilizumab |
| Antibiotics |
Carrimycin, ceftriaxone, levofloxacin OR moxifloxacin, piperacillin + tazobactam Ceftaroline fosamil, amoxicillin + potassium clavulanate, standard course macrolide teicoplanin, azithromycin |
| Antimalaria + antiparasitics | Hydroxychloroquine + ivermectin |
| Non-specific anti-inflammatory |
Methylprednisolone, ciclesonide dexamethasone, budesonide + formoterol hydrocortisone, other corticosteroids prednisone |
| Immunosuppressant |
Fingolimod, leflunomide thalidomide |
| Immunosuppressant + antiviral | Thalidomide + umifenovir |
| Monoclonal antibodies |
Adalimumab, bevacizumab, camrelizumab eculizumab, mepolizumab, PD-1 mAb sarilumab, tocilizumab, adamumab + tozumab Ixekizumab, nivolumab, IFX-1, leronlimab (PRO 140), clazakizumab, bamlanivimab (LY-CoV555 and LY3819253), casirivimab + imdevimab |
| Antiviral + monoclonal antibodies | Favipiravir + tocilizumab |
| Antiviral + antimalaria |
Darunavir + cobicistat + hydroxychloroquine favipiravir + chloroquine phosphate |
| Immunomodulator | CD24, anakinra |
| ACE inhibitor |
Losartan, recombinant human angiotensin-converting enzyme 2 (rhACE2), telmisartan valsartan, renin-angiotensin-system-blockade captopril |
| Vasodilator | Angiotensin 1–7 |
| Cardioprotective | Aspirin + clopidogrel + rivaroxaban + atorvastatin + omeprazole |
| Anticoagulant | Enoxaparin sodium, rivaroxaban, tinzaparin |
| Vaccines |
COVID-19 vaccines (whole virus, protein subunit, viral vector, and nucleic acid-RNA and DNA), BCG vaccine, recombinant novel coronavirus (2019-nCOV) vaccine (adenovirus vector) bacTRL-spike vaccine, ChAdOx1 nCoV-19 vaccine, CIGB 2020 vaccine |
| Non-drug | |
| Advanced therapy medicinal products (ATMP) |
Aerosol inhalation of vMIP: viral macrophage inflammatory protein, ankylosaurus; M1 macrophages target, anti-2019-nCoV inactivated convalescent plasma, anti-SARS-CoV-2 inactivated convalescent plasma, biological preparation of human placenta, convalescent plasma treatment, cord blood mesenchymal stem cells Human menstrual blood-derived stem cells, immunoglobulin from cured patients, inactivated mycobacterium vaccine, infusion of convalescent plasma, mRNA-1273, NK cells, plasma treatment, recombinant cytokine gene-derived protein injection, regulating intestinal flora therapeutic antibody from recovered novel coronavirus pneumonia patients, umbilical cord blood mononuclear cells, umbilical cord mesenchymal stem cells (hucMSCs) recombinant super-compound interferon (rSIFN-co), convalescent anti-SARS-CoV-2 plasma mycobacterium, bone marrow-derived mesenchymal stem cell |
| Rehabilitation |
Shadowboxing, pulmonary rehabilitation lung rehabilitation training, exercise rehabilitation under remote monitoring patient education |
| Gas inhalation |
Nitric oxide gas, oxygen concentrator hydrogen–oxygen nebulize, hydrogen inhalation, continuous positive airway pressure hyperbaric oxygen |
| Respiratory support |
high-flow therapy by nasal cannulae (HFNC) bag- valve mask oxygenation (SMO), High flow nasal oxygen, postural positioning |
| Life support | Renal replacement therapy, artificial liver therapy, ozonated autohemotherapy, ECMO implantation, cytosorb-therapy, HA330 |
Cellular therapy includes natural killer (NK) cells and mesenchymal stem cells (MSCs). Human NK cells lyse virus-infected cells coated with antibodies using the antibody-dependent cellular cytotoxicity process (ADCC) (Hammer et al. 2018). For the treatment of SARS-CoV-2 infection, researchers have declared the extending use of CYNK-001, an off-the shelf, allogenic, umbilical cord blood-derived natural killer cell therapy (Sorrento and Celularity 2020). Placenta and umbilical cord blood are rich in mesenchymal stem cells. Several studies have reported that infiltration of immune cells to pulmonary tissues and pro-inflammatory cytokine secretion is suppressed with MSCs, thus can improve chronic/severe lung injury and ARDS (Ortiz et al. 2007; Gupta et al. 2007; Moodley et al. 2009; Matthay et al. 2010).
Convalescent plasma therapy and monoclonal antibodies are used in immunotherapy. Convalescent plasma therapy has been widely used in infectious diseases such as poliomyelitis, influenza A (H5N1), and Ebola (Zhou et al. 2007; van Griensven et al. 2016; Rinaldo 2005). Patients with viral infections can be treated effectively by antibodies including IgG, IgA, IgM, IgE, and IgD observed in the convalescent plasma of clinically improved patients. Doctors in Shanghai collected the blood plasma from medically recovered COVID-19 patients and injected it into the infected patients, showing promising results with rapid recovery (Derebail and Falk 2020). A study showed that the receptor-binding domain (RBD) of SARS-CoV-2 (KD of 6.3 nM) could effectively combine with the CR3022 antibody (Tian et al. 2020). However, the binding site of ACE-2 in the SARS-CoV-2 RBD does not overlap with the epitope of CR3022 (Tian et al. 2020). Thus, CR3022 could be a promising therapy for the treatment of COVID-19 pneumonia alone or in conjunction with other neutralising monoclonal antibodies. Some traditional Chinese medicines were also tested in mice and humans for infection caused by SARS-CoV-2 (Sheahan et al. 2020) showed positive results.
Conclusion
This study is to set one's eyes on the current research status of COVID-19 disease and its spread through environment. We have focused on the transmission of the virus by air droplets or aerosol, fecal matter, and wastewater. The diagnosis of SARS-CoV-2 using different types of testing methods has also been discussed. Various effective treatments, preventions, and controls, including therapeutics, antibodies against COVID-19, have been reported. The overall conclusion is that the spread of coronavirus is till increasing rapidly through different ways and poses threat to the human. Thus, combating this disease is a long-term task that will necessitate the cooperation and coordination of scientists, authorities, international organisations and general public.
Acknowledgements
The authors acknowledge the Department of Civil Engineering, Indian Institute of Technology Guwahati, and Director, Defence Institute of Bio-Energy Research-DRDO, to provide facilities for the research and literature review.
Declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aggarwal KK (2020) COVID-19 and air-conditioners: fresh and filtered. Columns. https://www.indialegallive.com/column-news/covid-19-ventilation-airbornetransmission-respiratory-droplets/
- Agostini ML, Andres EL, Sims A, Graham RL, Sheahan TP, Lu X, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. Mbio. 2018;9(2):e00221–e318. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Angel N, Edson J, Bibby K, Bivin A, O'Brien JW, Choi PM, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138764.(InPress). [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASHRAE . COVID-19 (Coronavirus) preparedness resources. American Society of Heating, Ventilating, and Air-Conditioning Engineers; 2020. [Google Scholar]
- Bilal M, Nazir MS, Rasheed T, Parra-Saldivar R, Iqbal HMN. Watermatrices as potential source of SARS-CoV-2 transmission—an overview from environmental perspective. Case Stud Chem Environ Eng. 2020 doi: 10.1016/j.cscee.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L, Rutala WA, Weber DJ, Sobsey MD. Survival of surrogate coronaviruses in water. Water Res. 2009;43(7):1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M, Rajnik M, Aleem A, Dulebohn SC, Di Napoli R (2021) Features, evaluation and treatment coronavirus (COVID-19). In: Stat Pearls [Internet]. StatPearls Publishing, Treasure Island, FL. PMID: 32150360 [PubMed]
- CDC and WHO water transmission and COVID-19: questions and answers. https://www.cdc.gov/coronavirus/2019-ncov/php/water.html. Accessed 21 Apr 2020
- Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KW, Wong VT, Tang SCW. COVID-19: an update on the epidemiological, clinical, preventive and therapeutic evidence and guidelines of integrative Chinese-Western medicine for the management of 2019 novel coronavirus disease. Am J Chin Med. 2020;13:1–26. doi: 10.1142/S0192415X20500378. [DOI] [PubMed] [Google Scholar]
- Chen G, et al. Clinical and immunologic features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020 doi: 10.1172/jci137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia PY, Coleman KK et al (2020) Detection of air and surface contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in hospital rooms of infected patients. medRxiv 2020
- Chin AWH, Chu JTS, Perera MRA, et al. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- China Citywater (2020) Measurement to reveal the transmission of SARS-CoV-2 from wards to the drainage system. http://www.chinacitywater.org/zwdt/swyw/102892.shtml
- Chinese Association of Rehabilitation Medicine; Respiratory Rehabilitation Committee of Chinese Association of Rehabilitation Medicine; Cardiopulmonary Rehabilitation Group of Chinese Society of Physical Medicine and Rehabilitation. [Recommendations for respiratory rehabilitation of coronavirus disease 2019 in adult] (2020). Zhonghua Jie He He Hu Xi Za Zhi 43(4):308–314. Chinese. 10.3760/cma.j.cn112147-20200228-00206 [DOI] [PubMed]
- Christoph et al (2020) Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. medRxiv 2020. [DOI] [PMC free article] [PubMed]
- Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciencewicki J, Jaspers I. Air pollution and respiratory viral infection. Inhal Toxicol. 2007;19:1135–1146. doi: 10.1080/08958370701665434. [DOI] [PubMed] [Google Scholar]
- Coleman KK, Nguyen TT, Yadana S, et al. Bioaerosol sampling for respiratory viruses in Singapore’s mass rapid transit network. Sci Rep. 2018;8:1–7. doi: 10.1038/s41598-018-35896-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collivignarelli MC, et al. SARS-CoV-2 in sewer systems and connected facilities. Process Saf Environ Prot. 2020 doi: 10.1016/j.psep.2020.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticini E, Frediani B, Caro D. Can atmospheric pollution be considered a co-factor in extremely high level of SARS-CoV-2 lethality in Northern Italy? Environ Pollut. 2020;261:114465. doi: 10.1016/j.envpol.2020.114465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Vega R, Barquín RR, Boros S, Szabo A (2020) Could attitudes toward COVID-19 in Spain rendermen more vulnerable than women? Preprint. 10.31234/osf.io/dyxqn [DOI] [PubMed]
- Dehghani R, Kassiri H. A brief review on the possible role of houseflies and cockroaches in the mechanical transmission of coronavirus disease 2019 (COVID-19) Arch Clin Infect Dis. 2020;15:e102863. doi: 10.5812/archcid.102863(COVID-19). [DOI] [Google Scholar]
- Derebail VK, Falk RJ. ANCA-associated vasculitis—refining therapy with plasma exchange and glucocorticoids. N Engl J Med. 2020;382(7):671–673. doi: 10.1056/NEJMe1917490. [DOI] [PubMed] [Google Scholar]
- Ding Z, Qian H, Xu B, Huang Y, Miao T et al (2020) Toilets dominate environmental detection of SARS-CoV-2 virus in a hospital. medRxiv 2020
- Domingo JL, Marquès M, Nadal M, Schuhmacher M. Health risks for the population living near petrochemical industrial complexes. 1. Cancer risks: a review of the scientific literature. Environ Res. 2020;186:109495. doi: 10.1016/j.envres.2020.109495. [DOI] [PubMed] [Google Scholar]
- Elgendy IY, Pepine CJ. Why are women better protected from COVID19: clues for men? Sex and COVID 19. Int J Cardiol. 2020 doi: 10.1016/j.ijcard.2020.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure E, Kipnis E, Bortolotti P, Salik J. Clinical characteristics of COVID-19 in New York City. N Engl J Med. 2020;29(1):2016–2017. [Google Scholar]
- Frontera A, Marti C, Vlachos K, Sgubin G. Regional air pollution persistence links to COVID-19 infection zoning. J Infect. 2020;81(2):318–356. doi: 10.1016/j.jinf.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao QY, Chen YX, Fang JY. 2019 Novel coronavirus infection and gastrointestinal tract. J Dig Dis. 2020 doi: 10.1111/1751-2980.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazeeri G, Abdullah L, Abbas O. Immunological differences in women compared with men: overview and contributing factors. Am J Reprod Immunol. 2011;66(3):163. doi: 10.1111/j.1600-0897.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Goodman B. Air conditioning may be spreading COVID. Webmed Health News; 2020. [Google Scholar]
- Gormley M, Aspray TJ, Kelly DA. COVID-19: mitigating transmission via wastewater plumbing systems. Lancet Glob Health. 2020;8(5):e643. doi: 10.1016/S2214-109X(20)30112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Han B, Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020;159:1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W-J, Ni Z-Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Eng J Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Su X, et al. Intrapulmonary delivery of bone marrow-derived mesenchymal stem cells improves survival and attenuates endotoxin-induced acute lung injury in mice. J Immunol. 2007;179:1855–1863. doi: 10.4049/jimmunol.179.3.1855. [DOI] [PubMed] [Google Scholar]
- Hammer Q, Ruckert T, Romagnani C. Natural killer cell specificity for viral infections. Nat Immunol. 2018;19:800–808. doi: 10.1038/s41590-018-0163-6. [DOI] [PubMed] [Google Scholar]
- Han W, Quan B, et al. The course of clinical diagnosis and treatment of a case infected with coronavirus disease 2019. J Med Virol. 2020;92:461–463. doi: 10.1002/jmv.25711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart OE, Halden RU. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci Total Environ. 2020;730:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Holshue ML, DeBolt C, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, et al. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92(4):433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HP, Look DC, et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79(23):14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wang H et al (2020) Clinical data on hospital environmental hygiene monitoring and medical staff protection during the coronavirus disease 2019 outbreak. medRxiv 2020
- Jiehao C, Jing X, et al. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnam G, Rygiel TP, et al. CD200 receptor controls sex-specific TLR7 responses to viral infection. PLoS Pathog. 2012;8(5):1–8. doi: 10.1371/journal.ppat.1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, et al. Viral load kinetics of SARS-CoV-2 infection in first two patients in Korea. J Korean Med Sci. 2020;35:e86. doi: 10.3346/jkms.2020.35.e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibbs LD, Morawska L, Bell SC. The risk of airborne influenza transmission in passenger cars. Epidemiol Infect. 2012;140:474–478. doi: 10.1017/S0950268811000835. [DOI] [PubMed] [Google Scholar]
- Kui L, Fang YY, et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J. 2020 doi: 10.1097/CM9.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT-Y, Shum MH-H, et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lauser SA, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020 doi: 10.7326/m20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc QJ, Fuller NM, Knight LE, et al. What settings have been linked to SARS-CoV-2 transmission clusters? Wellcome Open Res. 2020;5:83. doi: 10.12688/wellcomeopenres.15889.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M, Munster V (2020) Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV. bioRxiv. 915660 [DOI] [PMC free article] [PubMed]
- Lewis D. Is the coronavirus airborne? Experts can't agree. Nature News; 2020. [DOI] [PubMed] [Google Scholar]
- Li Q, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ning Z et al (2020b) Aerodynamic characteristics and RNA concentration of SARS-CoV-2 aerosol in Wuhan Hospitals during COVID-19 outbreak. bioRxiv 2020
- Lodder W, de Husman AMR. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol Hepatol. 2020 doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder WJ, Buisman AM, et al. Feasibility of quantitative environmental surveillance in poliovirus eradication strategies. Appl Environ Microbiol. 2012;78:3800–3805. doi: 10.1128/AEM.07972-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelletti L, Martelletti P. Air pollution and the novel Covid-19 disease: a putative disease risk factor. SN Compr Clin Med. 2020;15:1–5. doi: 10.1007/s42399-020-00274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay MA, Goolaerts A, Howard JP, Lee JW. Mesenchymal stem cells for acute lung injury: pre-clinical evidence. Crit Care. 2010;38:569–573. doi: 10.1097/CCM.0b013e3181f1ff1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G, Heijnen L, Elsinga G, Italiaander R. Presence of SARS-Coronavirus-2 in Sewage. Medrxiv. 2020 doi: 10.1101/2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- Mei X, et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat Med. 2020 doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael-Kordatou I, Karaolia P, Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J Environ. 2020;8:1043062. doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty A, et al. Role of rapid antigen test in the diagnosis of COVID-19 in India. JAMMR. 2020;32(18):77–80. doi: 10.9734/jammr/2020/v32i1830657. [DOI] [Google Scholar]
- Mohanty A, et al. Laboratory diagnosis of COVID-19 infection: current issues and challenges: an Indian perspective. JAMMR. 2020;32(14):10–17. doi: 10.9734/jammr/2020/v32i1430559. [DOI] [Google Scholar]
- Moodley Y, Atienza D, et al. Human umbilical cord mesenchymal stem cells reduce fibrosis of bleomycin-induced lung injury. Am J Pathol. 2009;175:303–313. doi: 10.2353/ajpath.2009.080629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L, Tang JW, Bahnfleth W, et al. How can airborne transmission of COVID-19 indoors be minimised? Environ Int. 2020;2020:142105832. doi: 10.1016/j.envint.2020.105832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L, Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ Int. 2020;139:105730. doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens DM, Breman JG, Calisher CH, Doherty PC, Hahn BH, et al. The origin of COVID-19 and why it matters. Am J Trop Med Hyg. 2020 doi: 10.4269/ajtmh.20-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A, Nemudraia A, Surya K, Wiegand T, Buyukyoruk M, Wilkinson R et al (2020) Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. medRxiv. 2020 [DOI] [PMC free article] [PubMed]
- Ng LFP, Hiscox JA. Coronaviruses in animals and humans. BMJ. 2020;368:m634. doi: 10.1136/bmj.m634. [DOI] [PubMed] [Google Scholar]
- Nishiura H, Oshitani H, Kobayashi T, Saito T, Sunagawa T, Matsui T et al (2020) Closed environments facilitate secondary transmission of coronavirus disease 2019 (COVID-19). medRxiv
- Onder G, Rezza G, Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020;2019:2019–2020. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- Ong SWX, Tan YK, et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323:1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz LA, Dutreil M, et al. Interleukin 1 receptor antagonist mediates the anti inflammatory and antifibrotic effect of mesenchymal stem cells during lung injury. Proc Natl Acad Sci USA. 2007;104:11002–11007. doi: 10.1073/pnas.0704421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020 doi: 10.1016/s1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Kim YM, Yi S, et al. Coronavirus disease outbreak in call center, South Korea. Emerg Infect Dis. 2020;26(8):1666–1670. doi: 10.3201/eid2608.201274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan LT, et al. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382(9):872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pung R, et al. Investigation of three clusters of COVID-19 in Singapore: implications for surveillance and response measures. Lancet. 2020;395:1039–1046. doi: 10.1016/S0140-6736(20)30528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis Publ Infect Dis Soc Am. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj VS, Mou H, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiah A, Arumugaswami V. Insights into cross-species evolution of novel human coronavirus 2019-nCoV and defining immune determinants for vaccine development. bioRxiv. 2020 doi: 10.1101/2020.01.29.925867. [DOI] [Google Scholar]
- Randazzo W, Truchado P, et al. SARS-CoV-2 RNA titers in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020 doi: 10.1101/2020.04.22.20075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo CR., Jr Passive immunization against poliomyelitis: the Hammon gamma globulin field trials, 1951–1953. Am J Public Health. 2005;95:790–799. doi: 10.2105/AJPH.2004.040790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou J, Althaus CL (2020) Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill 25(4):2000058. 10.2807/1560-7917.ES.2020.25.4.2000058. Erratum in: Euro Surveill. 2020 Feb;25(7) [DOI] [PMC free article] [PubMed]
- RIVM. Novel coronavirus found in wastewater. https://www.rivm.nl/en/news/novelcoronavirus-found-in-wastewater. Accessed 28 Mar 2020
- Santarpia JL, Rivera DN et al (2020) Transmission potential of SARS-CoV-2 in viral shedding observed at the University of Nebraska Medical Center. medRxiv 2020
- Sharma K, Koirala A, et al. Vaccines for COVID-19: where do we stand in 2021? Paediatr Respir Rev. 2021 doi: 10.1016/j.prrv.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan TP, Sims AC, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11:1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, et al. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Medrxiv. 2020 doi: 10.1101/2020.03.12.20034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim E, Tariq A, Choi W, Lee Y, Chowell G. Transmission potential and severity of COVID-19 in South Korea. Int J Infect Dis. 2020;93:339–344. doi: 10.1016/j.ijid.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Shi N, Shan F, Zhang Z, et al. Emerging coronavirus 2019-nCoV pneumonia. Radiology. 2020 doi: 10.1148/radiol.2020200274. [DOI] [Google Scholar]
- Sorrento and Celularity (2020) Sorrento and Celularity to initiate emergency allogeneic natural killer (NK) cell therapy development for coronavirus infection. https://seekingalpha.com/pr/17762358-sorrento-and-celularity-to-initiate-emergency-allogeneic-naturalkiller-nk-cell-therapy. Accessed 1 Apr 2020
- Tian X, Li C, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNICEF (2020) Coronavirus disease (COVID-19): What parents should know. How to protect yourself and UNICEF. Hand washing. UNICEFPacific/2018/Holmberg. Accessed 31 Mar 2020
- van Doremalen N, Bushmaker T, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Griensven J, Edwards T, et al. Evaluation of convalescent plasma for Ebola virus disease in Guinea. N Engl J Med. 2016;374:33–42. doi: 10.1056/NEJMoa1511812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol. 2020 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S, Yi Q, Fan S, et al. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP) medRxiv. 2020 doi: 10.1101/2020.02.10.20021832. [DOI] [Google Scholar]
- Wang BX, Fish EN, editors. Global virus outbreaks: Interferons as 1st responders. Seminars in immunology. Elsevier; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Cao R, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:1–3. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hu B, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber DJ, Rutala WA, et al. Emerging infectious diseases: Focus on infection control issues for novel coronaviruses (Severe Acute Respiratory Syndrome-CoV and Middle East Respiratory Syndrome-CoV), hemorrhagic fever viruses (Lassa and Ebola), and highly pathogenic avian influenza viruses, A(H5N1) and A(H7N9) Am J Infect Control. 2016;44(5):E91–E100. doi: 10.1016/j.ajic.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Lam CS, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2020) WHO R&D blueprint COVID 19 experimental treatments. https://www.who.int/teams/blueprint/covid-19. Accessed 30 Jan 2020
- Wrapp D, Wang N, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Chen X, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Guo C, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Nethery RC, Benjamin M, Sabath BM, Danielle Braun D, Dominici F (2020c) Exposure to air pollution and COVID-19 mortality in the United States: a Nationwide cross-sectional study. medRxiv. 10.1101/2020.04.05.20054502
- Wu A, Peng Y, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Teng J, Fan Y, Xie R, Shen A. The short-term effects of air pollutants on hospitalizations for respiratory disease in Hefei, China. Int J Biometeorol. 2019;63:315–326. doi: 10.1007/s00484-018-01665-y. [DOI] [PubMed] [Google Scholar]
- Xu X, Chen P, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Cao D, et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, et al. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. Front Mol Biosci. 2020 doi: 10.1101/2020.03.12.20035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Abudayyeh OO, Gootenberg JS (2020b) A protocol for detection of COVID-19 using CRISPR diagnostics. https://broad.io/sherlockprotocol
- Zhang JC, Bin WS, Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J Med Virol. 2020 doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Gong Y, Meng F, Bi Y, Yang P, Wang F. Virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. Infectious diseases (except HIV/AIDS) medRxiv. 2020 doi: 10.1101/2020.03.28.20043059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Zhong N, Guan Y. Treatment with convalescent plasma for influenza A (H5N1) infection. N Engl J Med. 2007;357:1450–1451. doi: 10.1056/NEJMc070359. [DOI] [PubMed] [Google Scholar]
- Zhou P, Yang XL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Yu T, et al. Clinical course and risk factors for mortality of adult in patients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Xie J, Huang F, Cao L. Association between short-term exposure to air pollution and COVID-19 infection: evidence from China. Sci Total Environ. 2020;727:138704. doi: 10.1016/j.scitotenv.2020.138704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A, et al. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug DiScov. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]