ABSTRACT
Candidate Phyla Radiation (CPR) bacteria and nanoarchaea populate most ecosystems but are rarely detected in soil. We concentrated particles of less than 0.2 μm in size from grassland soil, enabling targeted metagenomic analysis of these organisms, which are almost totally unexplored in largely oxic environments such as soil. We recovered a diversity of CPR bacterial and some archaeal sequences but no sequences from other cellular organisms. The sampled sequences include Doudnabacteria (SM2F11) and Pacearchaeota, organisms rarely reported in soil, as well as Saccharibacteria, Parcubacteria, and Microgenomates. CPR and archaea of the phyla Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, and Nanohaloarchaeota (DPANN) were enriched 100- to 1,000-fold compared to that in bulk soil, in which we estimate each of these organisms comprises approximately 1 to 100 cells per gram of soil. Like most CPR and DPANN sequenced to date, we predict these microorganisms live symbiotic anaerobic lifestyles. However, Saccharibacteria, Parcubacteria, and Doudnabacteria genomes sampled here also harbor ubiquinol oxidase operons that may have been acquired from other bacteria, likely during adaptation to aerobic soil environments. We conclude that CPR bacteria and DPANN archaea are part of the rare soil biosphere and harbor unique metabolic platforms that potentially evolved to live symbiotically under relatively oxic conditions.
IMPORTANCE Here, we investigated overlooked microbes in soil, Candidate Phyla Radiation (CPR) bacteria and Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, and Nanohaloarchaeota (DPANN) archaea, by size fractionating small particles from soil, an approach typically used for the recovery of viral metagenomes. Concentration of these small cells (<0.2 μm) allowed us to identify these organisms as part of the rare soil biosphere and to sample genomes that were absent from non-size-fractionated metagenomes. We found that some of these predicted symbionts, which have been largely studied in anaerobic systems, have acquired aerobic capacity via lateral transfer that may enable adaptation to oxic soil environments. We estimate that there are approximately 1 to 100 cells of each of these lineages per gram of soil, highlighting that the approach provides a window into the rare soil biosphere and its associated genetic potential.
KEYWORDS: CPR, DPANN, archaea, bacteria, environmental microbiology, metagenomics, soil, soil microbiology, symbiosis
OBSERVATION
Interactions among soil microorganisms impact biogeochemical cycling and overall ecosystem function. A recent metagenomic analysis of soil microbial communities revealed that many steps of key reaction pathways central to transformations in soil are partitioned among coexisting organisms (1). Other interactions are mediated by molecules such as vitamins and antimicrobial compounds (2–4). Furthermore, there is the potential for a variety of symbiotic interactions, including those that involve obligate reliance on coexisting organisms, for even the most basic requirements (5, 6). Candidate Phyla Radiation (CPR) bacteria and DPANN archaea (an acronym of the names of the first included phyla: Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, and Nanohaloarchaeota) are detected across ecosystems and are often predicted to be obligate anaerobic (epi)symbionts that depend on other organisms for basic cellular building blocks (5, 7, 8). However, CPR bacteria and DPANN archaea have rarely been studied in relatively oxic environments or identified in soil (1, 9, 10). Genome-resolved metagenomic analyses circumvent the limitations of isolation-based methods that fail for organisms unable to grow alone and for bacteria that evade detection by primers used in 16S rRNA gene surveys (11); yet, there are few reports of CPR metagenome-assembled genomes (MAGs) from soil (12–15) almost certainly because of the rarity of these bacteria.
Prior studies of groundwater have taken advantage of the observation that CPR bacteria have ultrasmall cells that pass through 0.2-μm filters and enable genome recovery for these organisms (11). Studies of other systems reveal that size fractionation of particles prior to sequencing impacts the composition and function of metagenomes (16). However, to our knowledge, studies of 0.2-μm filtrates from soil have focused on viromes and have not assessed their microbial contents (17–19). Here, we took advantage of the expected very small sizes of CPR bacteria and DPANN archaeal cells (20) to concentrate them from soil. Thus, we could test the hypothesis that these anaerobic organisms are understudied parts of the rare soil biosphere, where they may have evolved pathways to persist in relatively oxic environments. We sequenced concentrated soil effluent that had passed through a 0.2-μm filter used to remove larger cells and recovered a diversity of bacterial and archaeal sequences.
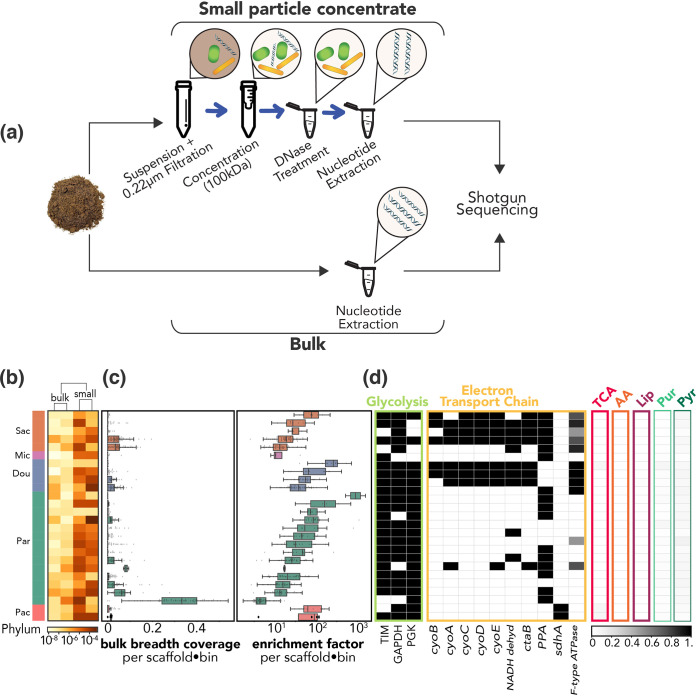
We sampled rhizosphere-associated soil from the top 10 cm of an annual grassland from the Hopland Research and Extension Center in February 2018. For a subset of the soil samples, we added a potassium citrate-based buffer and collected the effluent, which was passed through a 0.2-μm filter, concentrated, and treated with DNase to remove extracellular DNA that could have derived from larger lysed cells (Fig. 1a; see also Text S1 in the supplemental material) (21, 22). To evaluate enrichment, bulk DNA was extracted from the same soil samples for whole-community shotgun DNA sequencing, generating what are here referred to as “bulk metagenomes.” Approximately 20 Gbp of sequence was obtained from each of six concentrates and two bulk samples. In addition to recovering viral sequences and mobile elements from these small-particle-concentrate metagenomes, we reconstructed sequences from CPR and nanoarchaeal genomes. From these data, we resolved 26 draft genomes that were >70% complete (estimated using a CPR-specific single copy gene set [11]), with <10% contamination derived from either CPR or DPANN. No CPR or DPANN genomes were recovered from the bulk metagenomes.
FIG 1.
Enrichment and metabolic profiles of CPR bacteria in soil concentrate metagenomes. (a) Method for concentration of small particles from soil for metagenomic sequencing (top) compared to sample preparation methods for bulk soil metagenomes (bottom). (b) Heat map showing relative abundance of 26 organisms by phylum across bulk metagenomes and concentrate metagenomes. Sac, Saccharibacteria; Mic, Microgenomates; Dou, Doudnabacteria; Par, Parcubacteria; Pac, Pacearchaeota. (c) Coverage-based metrics showing recovery and enrichment in all concentrates combined relative to that in bulk fractions, combined as boxplots. (Left) Breadth of coverage of scaffolds comprising each genome (bin) in the bulk fraction. (Right) Enrichment factor (i.e., relative abundance of a scaffold from the concentrate metagenome over a scaffold’s bulk metagenome relative abundance) for each genome. (d) Metabolic analysis of each genome, including (i) presence of each of three glycolysis genes that are highly conserved among CPR bacteria, (ii) genes involved in the electron transport chain (NADH dehydrogenase; ctaB, heme O synthase [EC 2.5.1.141]; PPA, inorganic pyrophosphatase [EC 3.6.1.1]; sdhA, succinate dehydrogenase/fumarate reductase, flavoprotein subunit [EC 1.3.5.1 1.3.5.4]), and (iii) percentage completeness (grayscale) of F-type ATPase, the TCA cycle (tricarboxylic acid cycle), and pathways for amino acid biosynthesis (AA), lipid biosynthesis (Lip), purine biosynthesis (Pur), and pyrimidine biosynthesis (Pyr).
Detailed methods for generating small-particle concentrates from soil and subsequent DNA extraction in addition to metagenomic and phylogenetic methods used. Download Text S1, DOCX file, 0.04 MB (40.8KB, docx) .
Copyright © 2021 Nicolas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
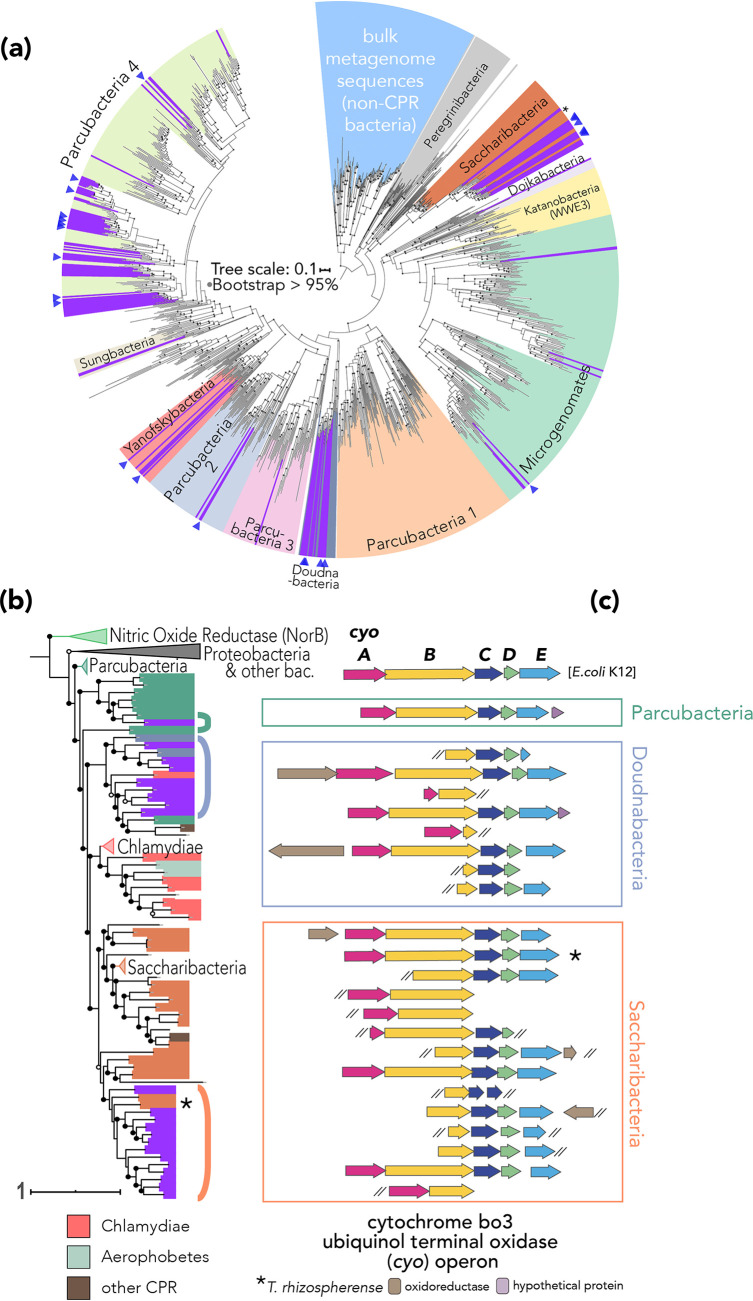
Sequences from cells of <0.2 μm in size were almost exclusively from 15 lineages of CPR bacteria and one DPANN archaeal phylum (Fig. 2; see also Fig. S1). Importantly, CPR and DPANN sequences were completely absent in bulk metagenome samples and were only detectable at very low, if any, coverage via read mapping to assembled sequences from the small-size-fraction metagenomes (Fig. 1c). Furthermore, from the 74 bacterial 16S rRNA sequences recovered from the concentrate metagenomes, all of which were assigned to CPR lineages, we predict that more than half (42 16S rRNA gene sequences) would not have been detected using standard amplicon sequencing primers. Notable was the phylum-level diversity of CPR lineages in the concentrate metagenomes. Previously, a genome of TM7 (Saccharibacteria) was reported from the same soil but sampled at less than 1× coverage from bulk soil (6), and Microgenomates and Parcubacteria have been genomically sampled at low abundance (13, 23). To our knowledge, this is one of very few reports of Pacearchaeota and Doudnabacteria (14, 15) in soil and the first report of a novel clade of Saccharibacteria.
FIG 2.
Soil CPR phylogeny and cytochrome operon synteny. Sequences assembled from the small concentrate metagenomes are shown in purple. (a) RpS3 tree of CPR bacteria rooted using RpS3 sequences that were assembled from bulk metagenomes, in light blue. Blue triangles denote draft genome recovered. Nodes with bootstrap values greater than or equal to 0.95 are marked as filled black circles. (b) Phylogenetic relationships of cytochrome bo3 ubiquinol terminal oxidase subunit I across bacterial phyla. Circles overlaid on nodes correspond to support values (unfilled, >0.50; filled, >0.70). *, the placement of T. rhizospherense CyoB (b) and its operon in (c) (6). Brackets next to tree tips correspond to phyla by color (green, Parcubacteria; blue, Doudnabacteria; orange, Saccharibacteria) and to sequence order in the synteny diagram (c). Tree rooted using a heme-copper oxidase superfamily member, the nitric oxide reductase (NorB). (c) Synteny diagram of cytochrome ubiquinol oxidase operon genes (cyoA, cyoB, cyoC, cyoD, and cyoE) with operon from E. coli K-12 as a reference. Scale bars correspond to the average number of substitutions per site across alignment.
Soil archaea phylogeny. Maximum likelihood tree of RpS3 sequences of archaea. Purple-colored sequences were assembled from the small-concentrate metagenomes reported. Tree was rooted using RpS3 sequences of Diapherotrites phylum sequences. Blue triangles represent RpS3 sequences for which draft genomes were recovered. Nodes with bootstrap values, >85%, are marked as filled circles. Tree scale bar corresponds to the average number of substitutions per site across alignment. Download FIG S1, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2021 Nicolas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparing sequence coverage from concentrates to that from the bulk metagenomes, we calculate that filtration enriched the relative abundance of genomes by 100 to 1,000× (Fig. 1c). We approximate, given the relative abundances of the most- and least-abundant CPR genomes in each of the bulk and concentrate metagenomes, that CPR cells may comprise on the order of 1 to 100 cells per gram of soil. Given estimates of 109 microbial cells per gram of soil, this would equate to, at maximum, ∼10−5% of microbial cells in a gram of soil (Text S1).
Given the unique challenges of the soil environment for microbes, we next assessed whether these soil CPR and DPANN organisms exhibited similar traits to those of their counterparts in other environments. Recent studies show that CPR bacteria generally appear to have the capacity for glycolysis and fermentation (24) but often lack complete pathways to synthesize nucleotides de novo and have many gaps in metabolism that suggest an obligate symbiotic lifestyle (8). We find that most of the genomes from this sampling effort encode the three central glycolysis enzymes reportedly found in nearly all CPR bacteria: triose phosphate isomerase (TIM), glyceraldehyde 3-phosphate (GAPDH), and phosphoglycerate kinase (PGK) (24). The genomes also contain few if any tricarboxylic acid (TCA) cycle genes and lack the vast majority of genes of the electron transport chain and for synthesis of lipids and nucleotides (Fig. 1d), suggesting they live anaerobic lifestyles and depend on resources from other organisms (8). However, we identified an operon encoding a multisubunit cytochrome bo3 ubiquinol terminal oxidase in three Doudnabacteria genomes, eight Saccharibacteria genomes, and one Parcubacteria sequence as well as in unbinned CPR phylum sequences from the concentrate metagenomes. We then performed a synteny analysis (Fig. 2c) to compare these loci to a related one from the first Saccharibacteria genome described from soil, Candidatus Teamsevenus rhizospherense (6). The comparison shows a gene order for the cyo operon identical to that in the highly studied Escherichia coli K-12 operon (25) and in the T. rhizospherense genome (6), although some CPR loci were incomplete due to assembly fragmentation. Several CPR loci also included an open reading frame (ORF) annotated as an oxidoreductase or a conserved hypothetical protein. While the genomes recovered do not contain quinone biosynthesis genes, the combination of this ubiquinol oxidase and the associated oxidoreductase (Fig. 2c), which often co-occur in genomes encoding an NADH dehydrogenase and, to varied completeness, F-type ATPase (Fig. 1d), suggests the possibility of some aerobic respiratory capacity. Perhaps, some form of aerobic respiration may be common in soil-associated Saccharibacteria specifically and perhaps in soil CPR more broadly. We thus hypothesize that this operon may confer an adaptive advantage for CPR bacteria to live in aerophilic environments such as surface soil.
Next, we generated a maximum-likelihood tree of subunit 1 (CyoB) of the cyo operon to test whether the operon exhibited a pattern of vertical inheritance in our CPR genomes (Text S1; Fig. 2b). This analysis suggests that this gene cluster has been laterally transferred from other bacteria, such as Proteobacteria or Chlamydiae, into these CPR bacteria at least once, with perhaps different origins for gene clusters in Parcubacteria and Doudnabacteria from those in Saccharibacteria. Furthermore, based on CyoB phylogeny, the sequences from T. rhizospherense appear more closely related to Saccharibacteria sequences from this study than to the RpS3 phylogeny, which may further underscore local adaptation to soil.
Here, we conducted a targeted study of CPR bacteria and nanoarchaea in a soil ecosystem to expand our understanding of rare soil-dwelling microbes. Using typical sequencing allocations for soil metagenomics, we were only able to recover genomes for these understudied community members through size fractionating of buffered soil. Our results indicate that CPR bacteria and DPANN archaea are relatively rare in soil, as they can be difficult to recover with typical metagenomic sequencing allotments.
While the precise ecological roles of these organisms remain unclear, their predicted requirement for interaction with nearby community members to satisfy their metabolic needs and their previously reported close physical association with other cells (5, 7) suggest that they may play still undescribed roles in soil microbial interaction networks.
The ability to selectively filter soil solutions to recover CPR and DPANN genomes suggests that either these organisms attach to larger microbial cells and the association can be physically disrupted or they are, at times, not attached to other cells. The approach enabled us to sample genetic inventories of rare soil-adapted microbes and uncover numerous genes and pathways, some of which likely evolved to handle symbiotic lifestyles under relatively oxic conditions. Specifically, we expanded the known diversity of genes and pathways in soil-adapted CPR bacteria and found that these inventories could explain the presence of these organisms, widely understood to be anaerobic, in soil. More generally, our approach provides a route to expand the known diversity of genes and pathways in the soil biosphere.
Data availability.
Curated genomes described in this study are available from ggKbase (https://ggkbase.berkeley.edu/soilcpr; please note that it is necessary to register for an account by provision of an email address before download) and are available under NCBI BioProject accession number PRJNA744897. NCBI accession numbers for metagenome-assembled genomes are provided in Table S2B.
Estimated count of CPR or nanoarchaea cells in soil. (Left) Approximation of the number of CPR or DPANN cells of a given lineage in 1 g of soil using bulk metagenome parameters. (Right) Same estimate using parameters derived from the small-particle-concentrate metagenome. Download Table S1, TIF file, 0.6 MB (597.7KB, tif) .
Copyright © 2021 Nicolas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sample metadata and genome relative abundances. (a) For 6 individual small-particle-concentrate metagenomes and 2 bulk metagenomes, sequencing metadata and statistics from their corresponding coassemblies. (b) Phylum-level classification, NCBI accession completeness, and relative abundance information for each metagenome-assembled genome resolved in this study. Download Table S2, XLSX file, 0.02 MB (19.7KB, xlsx) .
Copyright © 2021 Nicolas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research Genomic Science program under awards DE-SC0020163 and DE-SC0016247 to M.K.F. and the NIH grant DP2AI117984 to M.E.T. Work conducted at Lawrence Livermore National Laboratory was supported by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research Genomic Science program under award SCW1678, LLNL Lab Directed Research and Development award 18-ERD-041, and under the auspices of the U.S. DOE under contract DE-AC52-07NA27344. We acknowledge funding support to J.F.B. from the Chan Zuckerberg Biohub and the Innovative Genomics Institute at UC Berkeley.
We thank Cindy J. Castelle for support inferring archaea phylogenetic relationships and Katerina Estera-Molina for her expertise and management of the Firestone Lab field work site at the Hopland Research and Extension Center (HREC). We acknowledge that HREC sits on traditional, unceded land of the Pomo Indians.
J.F.B. is a founder of Metagenomi. The other authors declare no competing interests.
Contributor Information
Jillian F. Banfield, Email: jbanfield@berkeley.edu.
Haiyan Chu, Institute of Soil Science, Chinese Academy of Sciences.
Kiel Hards, University of Otago.
REFERENCES
- 1.Diamond S, Andeer PF, Li Z, Crits-Christoph A, Burstein D, Anantharaman K, Lane KR, Thomas BC, Pan C, Northen TR, Banfield JF. 2019. Mediterranean grassland soil C–N compound turnover is dependent on rainfall and depth, and is mediated by genomically divergent microorganisms. Nat Microbiol 4:1356–1367. doi: 10.1038/s41564-019-0449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang X, Zerfaß C, Feng S, Eichmann R, Asally M, Schäfer P, Soyer OS. 2018. Impact of spatial organization on a novel auxotrophic interaction among soil microbes. ISME J 12:1443–1456. doi: 10.1038/s41396-018-0095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abrudan MI, Smakman F, Grimbergen AJ, Westhoff S, Miller EL, van Wezel GP, Rozen DE. 2015. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci USA 112:11054–11059. doi: 10.1073/pnas.1504076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abreu NA, Taga ME. 2016. Decoding molecular interactions in microbial communities. FEMS Microbiol Rev 40:648–663. doi: 10.1093/femsre/fuw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu S-Y, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, Nelson KE, Lux R, Shi W. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci USA 112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starr EP, Shi S, Blazewicz SJ, Probst AJ, Herman DJ, Firestone MK, Banfield JF. 2018. Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially-processed plant-derived carbon. Microbiome 6:122. doi: 10.1186/s40168-018-0499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He C, Keren R, Whittaker ML, Farag IF, Doudna JA, Cate JHD, Banfield JF. 2021. Genome-resolved metagenomics reveals site-specific diversity of episymbiotic CPR bacteria and DPANN archaea in groundwater ecosystems. Nat Microbiol 6:354–365. doi: 10.1038/s41564-020-00840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castelle CJ, Brown CT, Anantharaman K, Probst AJ, Huang RH, Banfield JF. 2018. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat Rev Microbiol 16:629–645. doi: 10.1038/s41579-018-0076-2. [DOI] [PubMed] [Google Scholar]
- 9.Portillo MC, Leff JW, Lauber CL, Fierer N. 2013. Cell size distributions of soil bacterial and archaeal taxa. Appl Environ Microbiol 79:7610–7617. doi: 10.1128/AEM.02710-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK, Fierer N. 2018. A global atlas of the dominant bacteria found in soil. Science 359:320–325. doi: 10.1126/science.aap9516. [DOI] [PubMed] [Google Scholar]
- 11.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 12.Alteio LV, Schulz F, Seshadri R, Varghese N, Rodriguez-Reillo W, Ryan E, Goudeau D, Eichorst SA, Malmstrom RR, Bowers RM, Katz LA, Blanchard JL, Woyke T. 2020. Complementary metagenomic approaches improve reconstruction of microbial diversity in a forest soil. mSystems 5:e00768-19. doi: 10.1128/mSystems.00768-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroeger ME, Delmont TO, Eren AM, Meyer KM, Guo J, Khan K, Rodrigues JLM, Bohannan BJM, Tringe SG, Borges CD, Tiedje JM, Tsai SM, Nüsslein K. 2018. New biological insights into how deforestation in amazonia affects soil microbial communities using metagenomics and metagenome-assembled genomes. Front Microbiol 9:1635. doi: 10.3389/fmicb.2018.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavy A, McGrath DG, Matheus Carnevali PB, Wan J, Dong W, Tokunaga TK, Thomas BC, Williams KH, Hubbard SS, Banfield JF. 2019. Microbial communities across a hillslope‐riparian transect shaped by proximity to the stream, groundwater table, and weathered bedrock. Ecol Evol 9:6869–6900. doi: 10.1002/ece3.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matheus Carnevali PB, Lavy A, Thomas AD, Crits-Christoph A, Diamond S, Méheust R, Olm MR, Sharrar A, Lei S, Dong W, Falco N, Bouskill N, Newcomer ME, Nico P, Wainwright H, Dwivedi D, Williams KH, Hubbard S, Banfield JF. 2021. Meanders as a scaling motif for understanding of floodplain soil microbiome and biogeochemical potential at the watershed scale. Microbiome 9:121. doi: 10.1186/s40168-020-00957-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson SJ, Allen LZ, Lorenzi HA, Fadrosh DW, Brami D, Thiagarajan M, McCrow JP, Tovchigrechko A, Yooseph S, Venter JC. 2012. Metagenomic exploration of viruses throughout the Indian Ocean. PLoS One 7:e42047. doi: 10.1371/journal.pone.0042047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segobola J, Adriaenssens E, Tsekoa T, Rashamuse K, Cowan D. 2018. Exploring viral diversity in a unique South African soil habitat. Sci Rep 8:111. doi: 10.1038/s41598-017-18461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han L-L, Yu D-T, Zhang L-M, Shen J-P, He J-Z. 2017. Genetic and functional diversity of ubiquitous DNA viruses in selected Chinese agricultural soils. Sci Rep 7:45142. doi: 10.1038/srep45142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bi L, Yu D, Du S, Zhang L, Zhang L, Wu C, Xiong C, Han L, He J. 2021. Diversity and potential biogeochemical impacts of viruses in bulk and rhizosphere soils. Environ Microbiol 23:588–599. doi: 10.1111/1462-2920.15010. [DOI] [PubMed] [Google Scholar]
- 20.Luef B, Frischkorn KR, Wrighton KC, Holman H-YN, Birarda G, Thomas BC, Singh A, Williams KH, Siegerist CE, Tringe SG, Downing KH, Comolli LR, Banfield JF. 2015. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun 6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 21.Trubl G, Solonenko N, Chittick L, Solonenko SA, Rich VI, Sullivan MB. 2016. Optimization of viral resuspension methods for carbon-rich soils along a permafrost thaw gradient. PeerJ 4:e1999. doi: 10.7717/peerj.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williamson KE, Corzo KA, Drissi CL, Buckingham JM, Thompson CP, Helton RR. 2013. Estimates of viral abundance in soils are strongly influenced by extraction and enumeration methods. Biol Fertil Soils 49:857–869. doi: 10.1007/s00374-013-0780-z. [DOI] [Google Scholar]
- 23.Sharrar AM, Crits-Christoph A, Méheust R, Diamond S, Starr EP, Banfield JF. 2020. Bacterial secondary metabolite biosynthetic potential in soil varies with phylum, depth, and vegetation type. mBio 11:e00416-20. doi: 10.1128/mBio.00416-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaffe AL, Castelle CJ, Matheus Carnevali PB, Gribaldo S, Banfield JF. 2020. The rise of diversity in metabolic platforms across the Candidate Phyla Radiation. BMC Biol 18:69. doi: 10.1186/s12915-020-00804-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abramson J, Riistama S, Larsson G, Jasaitis A, Svensson-Ek M, Laakkonen L, Puustinen A, Iwata S, Wikström M. 2000. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat Struct Biol 7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed methods for generating small-particle concentrates from soil and subsequent DNA extraction in addition to metagenomic and phylogenetic methods used. Download Text S1, DOCX file, 0.04 MB (40.8KB, docx) .
Copyright © 2021 Nicolas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Soil archaea phylogeny. Maximum likelihood tree of RpS3 sequences of archaea. Purple-colored sequences were assembled from the small-concentrate metagenomes reported. Tree was rooted using RpS3 sequences of Diapherotrites phylum sequences. Blue triangles represent RpS3 sequences for which draft genomes were recovered. Nodes with bootstrap values, >85%, are marked as filled circles. Tree scale bar corresponds to the average number of substitutions per site across alignment. Download FIG S1, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2021 Nicolas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Estimated count of CPR or nanoarchaea cells in soil. (Left) Approximation of the number of CPR or DPANN cells of a given lineage in 1 g of soil using bulk metagenome parameters. (Right) Same estimate using parameters derived from the small-particle-concentrate metagenome. Download Table S1, TIF file, 0.6 MB (597.7KB, tif) .
Copyright © 2021 Nicolas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sample metadata and genome relative abundances. (a) For 6 individual small-particle-concentrate metagenomes and 2 bulk metagenomes, sequencing metadata and statistics from their corresponding coassemblies. (b) Phylum-level classification, NCBI accession completeness, and relative abundance information for each metagenome-assembled genome resolved in this study. Download Table S2, XLSX file, 0.02 MB (19.7KB, xlsx) .
Copyright © 2021 Nicolas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Curated genomes described in this study are available from ggKbase (https://ggkbase.berkeley.edu/soilcpr; please note that it is necessary to register for an account by provision of an email address before download) and are available under NCBI BioProject accession number PRJNA744897. NCBI accession numbers for metagenome-assembled genomes are provided in Table S2B.
Estimated count of CPR or nanoarchaea cells in soil. (Left) Approximation of the number of CPR or DPANN cells of a given lineage in 1 g of soil using bulk metagenome parameters. (Right) Same estimate using parameters derived from the small-particle-concentrate metagenome. Download Table S1, TIF file, 0.6 MB (597.7KB, tif) .
Copyright © 2021 Nicolas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sample metadata and genome relative abundances. (a) For 6 individual small-particle-concentrate metagenomes and 2 bulk metagenomes, sequencing metadata and statistics from their corresponding coassemblies. (b) Phylum-level classification, NCBI accession completeness, and relative abundance information for each metagenome-assembled genome resolved in this study. Download Table S2, XLSX file, 0.02 MB (19.7KB, xlsx) .
Copyright © 2021 Nicolas et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.