ABSTRACT
Nε-lysine acetylation is an important, dynamic regulatory posttranslational modification (PTM) that is common in bacteria. Protein acetylomes have been characterized for more than 30 different species, and it is known that acetylation plays important regulatory roles in many essential biological processes. The levels of acetylation are enzymatically controlled by the opposing actions of lysine acetyltransferases and deacetylases. In bacteria, a second mechanism of acetylation exists and occurs via an enzyme-independent manner using the secondary metabolite acetyl-phosphate. Nonenzymatic acetylation accounts for global low levels of acetylation. Recently, studies concerning the role of protein acetylation in bacterial virulence have begun. Acetylated virulence factors have been identified and further characterized. The roles of the enzymes that acetylate and deacetylate proteins in the establishment of infection and biofilm formation have also been investigated. In this review, we discuss the acetylomes of human bacterial pathogens. We highlight examples of known acetylated virulence proteins and examine how they affect survival in the host. Finally, we discuss how acetylation might influence host-pathogen interactions and look at the contribution of acetylation to antimicrobial resistance.
KEYWORDS: posttranslational modification, acetylation, acetylome, virulence, pathogens, biofilm, antibiotic resistance, bacteria
INTRODUCTION
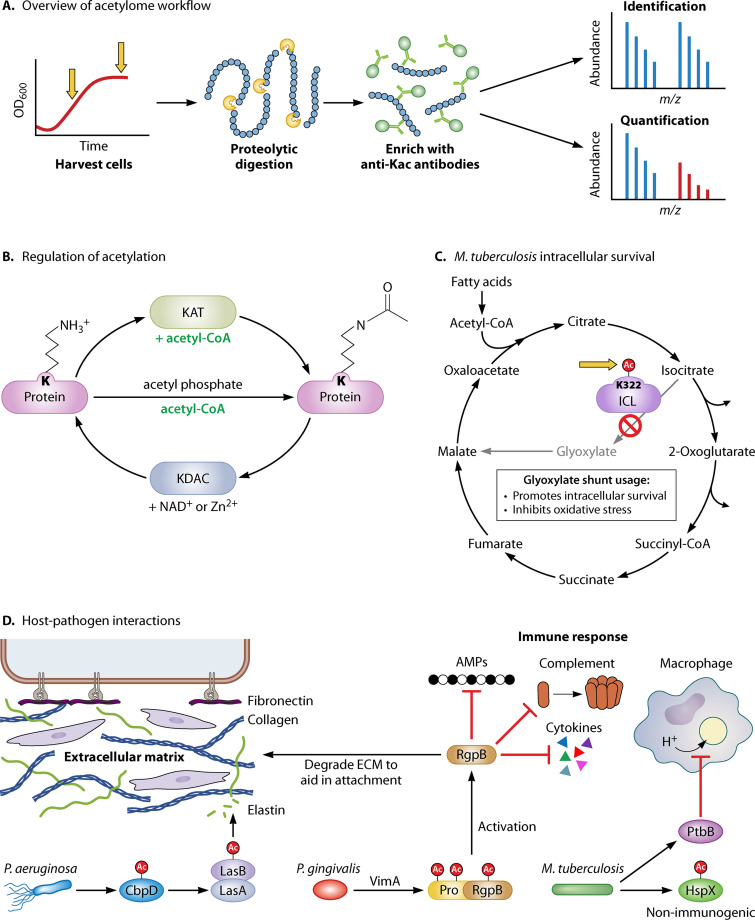
Nε-lysine acetylation is widely accepted as an important regulatory posttranslational modification (PTM) in bacteria. In the past decade, there has been an explosion of interest centered on protein acetylation in bacteria. Following the initial characterization of the Escherichia coli acetylome (1, 2), the acetylomes of more than 30 different species have been analyzed (reviewed in references 3–5). The typical workflow for acetylome analysis occurs in three main steps (Fig. 1A): (i) proteolytic digestion of proteins that were harvested following growth in a specific defined medium, (ii) enrichment of acetylated peptides through use of anti-acetyllysine antibodies, and (iii) identification or quantification of acetylation sites using mass spectrometry (MS)-based proteomics. The success of this workflow is largely due to the continuous improvements of MS-based technologies and the quality of anti-acetyllysine antibodies, enabling the identification of low-abundance acetylation sites and a growing number of identified acetylated proteins (3, 4).
FIG 1.
(A) Typical acetylome workflow. Cells are harvested at different growth phases or under different conditions, and proteins are digested, typically with trypsin. Often, acetylated peptides are enriched using anti-acetyllysine antibodies conjugated to agarose beads. The acetylated peptides are identified and quantified by MS. OD600, optical density at 600 nm. (B) Summary of the regulation of acetylation in bacteria. Proteins are acetylated either enzymatically by lysine acetyltransferases (KATs) or nonenzymatically via the high-energy intermediate acetyl-CoA or acetyl-phosphate. Deacetylation occurs by the action of NAD+-dependent sirtuins or Zn2+-dependent lysine deacetylases (KDACs). (C) For intracellular pathogens such as M. tuberculosis, intracellular metabolism is altered by acetylating key enzymes to control usage of the glyoxylate shunt. The glyoxylate shunt avoids the CO2-producing steps of the TCA cycle and replenishes intermediates. Acetylation of isocitrate lyase (ICL) at K322 inhibits its enzymatic activity and likely blocks usage of the shunt. Thus, deacetylation of ICL may be a critical step for intracellular survival. (D) Many secreted virulence factors are acetylated, which suggests that acetylation mediates pathogen-host interactions. P. aeruginosa secretes the acetylated proteins CbpD and LasB, which lead to the degradation of the human extracellular matrix (ECM) component elastin and aid in tissue invasion. In P. gingivalis, acetylation of the inactive pro-RgpB is required for enzyme activation as a protease. The acetylated pro-RgpB is secreted, where it is activated and degrades ECM components and immune system components, including cytokines, antimicrobial peptides (AMPs), and complement proteins. In M. tuberculosis, heat shock protein X is secreted and, when acetylated, is nonimmunogenic. The activity of the secreted protein tyrosine phosphatase PtpB is controlled by acetylation. PtpB promotes intracellular survival by inhibiting acidification inside the phagolysosome.
In bacteria, lysines are acetylated by two different mechanisms, enzymatic and nonenzymatic (Fig. 1B). Bacterial lysine acetyltransferases (KATs) are members of the Gcn5 N-acetyltransferase (GNAT) family that catalyze the addition of an acetyl group from acetyl-CoA (Ac-CoA) to a target lysine residue. This action is opposed by lysine deacetylases (KDACs), mostly members of the NAD+-dependent sirtuin family (5). Nonenzymatic acetylation by the high-energy intermediate acetyl-phosphate (AcP) and, possibly, Ac-CoA occurs at a low level when the local environment surrounding target lysine residues is favorable. Nonenzymatic acetylation is dependent upon intracellular AcP levels, which are controlled by the coordinated action of the enzymes phosphotransacetylase (Pta) and acetate kinase (AckA). Pta catalyzes the reversible reaction to convert Ac-CoA to AcP, and AckA catalyzes the reversible reaction to convert AcP to acetate (reviewed in reference 6). In Escherichia coli, it was demonstrated that global acetylation occurs via this nonenzymatic mechanism (7, 8). This finding is also true in Bacillus subtilis, Neisseria gonorrhoeae, and possibly all bacteria (9, 10). Nonenzymatic acetylation can be reversible, as it was shown in E. coli that the sirtuin CobB can deacetylate lysine residues irrespective of the mechanism of acetylation. However, while CobB is the major deacetylase in E. coli, only a small fraction of sites are subjected to this regulation and are reversible (11). Because two recent reviews have been published (5, 12), the mechanisms of acetylation will not be covered further here.
The next step for the bacterial acetylation field is to focus on understanding the physiological relevance of the thousands of identified acetylation sites to determine which of them represent important regulatory events. To study the effects of acetylation in vivo, lysine is commonly mutated to glutamine to mimic the acetylated state and to arginine to mimic the unacetylated state, which maintains the positive charge but cannot be acetylated. In addition, MS techniques are being developed to investigate the proportion of acetylation occupancy, or stoichiometry (4, 13–18), which is crucial to help prioritize further investigations. Currently, essential processes have been the subject of such investigations. For example, extensive work has been done assessing the role of acetylation in controlling cellular metabolism in response to environmental cues (reviewed in references 5 and 12). In addition, the functional relevance of lysine acetylation of selected individual proteins is being explored. The chemotaxis regulatory protein CheY, the regulator of capsule synthesis B (RcsB), and acetyl-CoA synthetase (Acs; reviewed in references 5 and 12) were some of the first proteins with identified functional roles for lysine acetylation (19–28). One emerging focus has been on investigating acetylated proteins involved in bacterial virulence, a topic that was recently reviewed in 2017 (29). The contribution of CheY and RcsB acetylation to bacterial virulence has been previously reviewed (12, 29) and will not be discussed here. In this review, we will first highlight new examples of how acetylated proteins contribute to bacterial survival in the host, including in the presence of antibiotics. We will then discuss how acetylation of bacterial proteins may influence interactions with host proteins. Finally, we will end with a discussion on how we can use this information to design novel treatment strategies to deal with troublesome infections.
ACETYLOME ANALYSIS IN PATHOGENIC BACTERIA
The characterization of the acetylomes of clinical pathogens has increased significantly in recent years, including in Staphylococcus aureus (30), Borrelia burgdorferi (31), Leptospira interrogans (32), and Acinetobacter baumannii (33). Some acetylome analyses revealed important acetylated virulence factors. The acetylated proteins involved in the virulence of A. baumannii were discussed in a prior review (29). Recently, it was found that the virulence of Streptococcus pneumoniae also might be regulated by acetylation (34). Seventeen proteins that are known virulence factors were acetylated, including enzymes involved in capsular polysaccharide biosynthesis. Production of capsule is a mechanism for immune evasion, and acetylation may be important to modulate this process (35). Vibrio cholerae contains 68 acetylated proteins that are known virulence factors (36), including important transcriptional regulators, such as AphB and LuxU. Structural examination of AphB predicted that acetylation site K103 lies in the dimerization interface and may influence protein-protein interactions (36). For the phosphorelay protein LuxU, acetylation occurred on K53, which is physically close to the phosphoacceptor site (H57), and acetylation was proposed to impact the phosphorylation state. However, none of these predictions have been experimentally confirmed; therefore, the physiologic significance of these observations remains unclear. The acetylomes of other pathogenic Vibrio species have also been characterized, including V. vulnificus (37), V. parahaemolyticus (38), and V. alginolyticus (39), but investigation of potential virulence factors was not extensively explored.
For the majority of these species, the acetylome characterizations were performed in defined chemical media under laboratory conditions. To fully understand how acetylation influences bacterial virulence, these studies on specific virulence factors must be performed in tissue culture or animal infection models. The challenge will be obtaining enough material to accurately quantify the acetylome, but the currently available mass spectrometers with the latest technological improvements should make this possible.
THE ROLE OF ACETYLATION ON INTRACELLULAR SURVIVAL
Virulence factors are proteins or other substances that are required for establishment of infection, acquisition of nutrients in the host environment, and immune evasion. For the remainder of this review, we will discuss examples of acetylated virulence factors with experimental validation (Table 1). Various large-scale acetylome analyses have been performed in Mycobacterium tuberculosis, and numerous acetylated proteins have been linked to virulence (40–42). Xie et al. found 20 acetylated proteins that were involved in virulence (40). One of these proteins is the metabolic enzyme isocitrate lyase (ICL1). During latent infection, M. tuberculosis alters carbon metabolism and relies on fatty acids from the host as their predominant carbon source (43). Some fatty acids are oxidized to produce Ac-CoA, which then enters the tricarboxylic acid (TCA) cycle by using the glyoxylate shunt, where ICL1 is involved (Fig. 1C). The glyoxylate shunt is primarily utilized to assimilate carbon when the source is 2C or 3C in length, such as acetate, which bypasses the CO2 generating steps of the TCA cycle and replenishes intermediates for biosynthetic processes (44). Thus, icl mutants cannot grow on fatty acids and display attenuated virulence in mouse infection models (45, 46). ICL1 is acetylated at K322, and mutation of this site to glutamine to mimic the acetylated state resulted in a decrease of enzymatic activity (40, 42). Therefore, deacetylation may be an important mechanism to regulate a shift toward usage of the glyxoylate shunt during host infection (Fig. 1C).
TABLE 1.
Acetylated virulence factorsa
| Species | Acetylated protein | Functional acetylation site | Function | Consequence of acetylation | Reference(s) |
|---|---|---|---|---|---|
| A. hydrophila | LuxS | K165 | Production of AI-2 | Inhibits enzymatic activity | 57 |
| B. pertussis | BvgA | Multiple | Response regulator, virulence expression | Unknown | 48 |
| E. coli | CheY | 92, 109 | Chemotaxis and motility regulator | Alters interactions, promotes CW rotation | 22, 25–27 |
| RcsB | 154, 180 | Regulates capsule synthesis and biofilm | Inhibits DNA binding | 19 – 21 | |
| M. tuberculosis | ICL1 | K322 | Central metabolism, glyoxylate shunt | Inhibits enzymatic activity | 40, 42 |
| PtpB | K224 | Protein tyrosine phosphatase | Inhibits enzymatic activity | 81 | |
| DosR | K182 | Response regulator induced by hypoxia | Inhibits DNA binding | 54 | |
| MtrA | K110 | Repressor of cell division | Inhibits DNA binding | 49, 50 | |
| HspX | Multiple | Heat shock protein X, immunogenic | Decreases immunogenicity | 41 | |
| P. gingivalis | RgpB | Multiple | Cysteine protease, required for host survival | Required for processing to mature, active enzyme | 76 |
| RprY | Unknown | Response regulator, controls expression of the T9SS | Inhibits DNA binding and reduces phosphorylation | 77 | |
| S. mutans | GtfB, GtfC, GtfD | Multiple | Glucan synthesis, biofilm formation | Decreases enzymatic activity | 61 |
| P. aeruginosa | LasB | Multiple | Elastase, degrades elastin | Unknown | 64 |
| CpbD | Multiple | Chitin binding protein, staphylolytic activity | Unknown | 64 | |
| F. novicida | ChiA and ChiB | Multiple | Chitinases | Inhibits activity, promotes biofilm | 71 |
| S. Typhimurium | PhoP | 201 | Response regulator, responds to low Mg2+ or acidic pH | Inhibits DNA binding | 91 |
| HilD | 297 | Transcription factor, regulates SPI-1 | Increases stability, reduces DNA binding | 92 | |
| AcrB | K1037 | Multidrug efflux pump | Regulates activity of pump | 82 |
Abbreviations: AI-2, autoinducer-2; CW, clockwise; T9SS, type IX secretion system; SPI-1, Salmonella pathogenicity island 1.
THE ROLE OF ACETYLATION ON TRANSCRIPTIONAL FACTORS THAT REGULATE VIRULENCE
One area that has been extensively studied is the role of acetylation in the modulation of transcription factor activity, typically by blocking DNA binding activity. Some examples that have been previously reviewed (13) are the acetylation of the transcription factors HilD and PhoP in Salmonella enterica serovar Typhimurium. In Bordetella pertussis, BvgA, a response regulator of the BvgAS two-component system (47), contains eight acetylation sites (48). It was determined that acetylation of BvgA does not alter protein levels or phosphorylation state; however, the effect on DNA binding was not assessed (48). Since at least two acetylation sites occur in the helix-turn-helix DNA binding domain, it is likely that acetylation inhibits the DNA binding activity of BvgA, similar to what was observed for other transcription factors. It would be interesting to generate glutamine substitutions to confirm this suggestion and determine the effect of such mutations in in vivo mouse models.
In M. tuberculosis H37Rv, MtrA of the MtrAB two-component system (49, 50) enhances its interaction with its cognate sensor kinase MtrB and reduces DNA binding activity. The phosphorylated form of MtrA is a repressor that sequesters oriC to block cell division and blocks expression of resuscitation promoting factor (Rpf), which hydrolyzes peptidoglycan during cell division and is required for reactivation (51–53). Therefore, active MtrA leads to latency. Acetylation of K111, which is located in the N-terminal receiver domain, was proposed to be a mechanism to turn off the MtrA repressor, possibly in response to nutrient availability, which would allow for resumption of bacterial growth and escape from latency. Interestingly, DosR, another response regulator involved in latency control, is regulated in the same fashion (54). Acetylation of DosR, which is induced under hypoxic conditions, is acetylated at K182, which inhibits DNA binding. DosR activates expression of 48 genes that are required for survival during latency (55). Because it has been demonstrated that the sirtuin Rv1151c deacetylates DosR and MtrA, drugs that specifically target Rv1151c may prevent the development of latent tuberculosis infections.
THE ROLE OF ACETYLATION DURING BIOFILM FORMATION
The majority of bacterial clinical infections are biofilm based, owing to the fact that biofilms offer bacteria protection from host immune responses and antibiotics (56). One protein involved in biofilm formation that was identified as acetylated in Aeromonas hydrophila is the S-ribosylhomocysteine lyase LuxS, which is involved in production of the quorum-sensing molecule autoinducer-2 (57, 58). LuxS is acetylated at K165, and, interestingly, this site is also succinylated (57). Acetylation of K165 inhibits enzymatic activity, while succinylation has the opposite effect, and the sirtuin CobB removes both modifications. In addition, a luxS deletion mutant exhibits enhanced biofilm production and increased virulence in mouse models (59). Taken together, acetylation of LuxS decreases enzymatic activity, which in turn may enhance biofilm formation and, therefore, virulence. To test this prediction, the biofilm properties and effects on virulence in a mouse model of luxSK165Q and cobB deletion strains should be determined. Identification of the LuxS acetylation mechanism may represent a new drug target to limit biofilm-based infections.
Streptococcus mutans produces plaque biofilms that contain glucans, which allow for adherence to the tooth surface (60). The three major glucosyltransferase enzymes, GtfB, GtfC, and GtfD, synthesize glucans and have decreased acetylation levels during biofilm growth compared to free-living, planktonic bacteria. Moreover, the enzymatic activity of these enzymes increases during biofilm growth, suggesting that acetylation is a mechanism to turn off glucan biosynthesis (61) when bacteria are not present in biofilms.
Increasing global acetylation often leads to large alterations in biofilm properties. For example, in M. tuberculosis H37Ra, an attenuated strain, deletion of the known sirtuin (MRA_1161, an ortholog of Rv1151c) results in defective biofilm formation (41), possibly due, in part, to inhibition of fatty acid metabolic enzymes via acetylation of key lysine residues in their active sites (62). In Neisseria gonorrhoeae, an acetate kinase mutant (ackA) displayed marked defects in maintaining biofilm structure over time (10). AckA is an enzyme involved in the reversible conversion of AcP to acetate, and in its absence, AcP accumulates (6, 12). As AcP is the main acetyl donor for nonenzymatic acetylation (12), this suggests that key biofilm regulatory proteins are increasingly acetylated, which leads to the observed defects. Further analysis is required to determine which important biofilm regulatory proteins are acetylated in N. gonorrhoeae.
THE ROLE OF ACETYLATION DURING INTERACTION WITH THE HOST
Proteins that are secreted often interact with the host to invade cells and destroy immune cells or other antimicrobial products (Fig. 1D). In Pseudomonas aeruginosa, several virulence factors were identified as acetylated or succinylated, including components of secretion systems and secreted factors (63). For example, two secreted virulence factors, CbpD and LasB, were found to be modified by multiple different PTMs in the extracellular environment (64). CbpD is a chitin binding protein with staphylolytic activity and is needed to process and activate LasA, which is involved in the degradation of the extracellular matrix protein elastin (65, 66). LasA enhances the elastase activity of LasB (67), which leads to tissue destruction in the host. Nine different lysine modifications, including acetylation, butyrylation, crotonylation, di- and tri-methylation, malonylation, methylation, propionylation, and succinylation, were identified in these proteins in the extracellular environment (64), but the effect of these PTMs on enzymatic activity or protein-protein interactions is not known. The mechanism of acylation is also unclear, as it may occur intracellularly or extracellularly by an acyltransferase or a nonenzymatic mechanism. This raises the interesting possibility that protein speciation, the presence of multiple PTMs on a single protein arising from one gene (68, 69), gives a broad functional diversity to a limited number of secreted proteins in the host environment, referred to as moonlighting (68, 70), information that may be essential for our understanding of bacterial virulence.
Francisella novicida contains 12 acetylated secreted proteins that are essential for proliferation and survival (71). Two chitinases, ChiA and ChiB, were among those secreted proteins. In vitro chemical acetylation with AcP resulted in a decrease in chitinase activity. These enzymes negatively regulate biofilm formation (72), so acetylation may inhibit chitinase activity to promote biofilm formation, aiding in host colonization. Acetylome profiling of Porphyromonas gingivalis revealed numerous interesting acetylated secreted proteins that play a role in bacterial virulence (73). A major group of virulence factors is the gingipains, which are secreted cysteine proteases that are required for invasion of tissues, inactivation of cytokines, and acquisition of essential nutrients (74). The gingipains consist of an arginine-specific protease (encoded by rgpA and rgpB) and a lysine-specific protease (75). The inactive, preprocessed proenzyme of RgpB (pro-RgpB) is acetylated by VimA and its paralog, PG1842. This acetylation is required for enzyme activation and, therefore, virulence (76). The transcription factor RprY, an orphan response regulator, was shown to be enzymatically regulated by the KAT Pat and sirtuin CobB, which inhibits its DNA binding activity and reduces phosphorylation (77). RprY regulates the expression of the type IX secretion system (T9SS), which exports many virulence factors. In support of this, RprY is required for virulence in a murine model (78). Thus, inhibition of RprY DNA-binding by acetylation is a mechanism to directly control secretion of virulence factors.
In M. tuberculosis, 45 secreted proteins were found to be acetylated, many of which stimulate host immune responses. Heat shock protein X (HspX) is secreted and has been shown to stimulate interferon gamma production in the host (79). Interestingly, blood samples from tuberculosis patients were collected and used to test the immunogenicity of acetylated and unacetylated fragments of HspX. The immune responses to the acetylated form of the protein were much weaker or absent (41). Therefore, acetylation of secreted proteins may be a way for bacteria to alter the host immune response and increase intracellular survival. Another secreted protein is the protein tyrosine phosphatase PtpB, which dephosphorylates various host proteins and is responsible for promoting intracellular survival by inhibiting acidification inside the phagolysosome (80). Acetylation of PtpB is controlled by the KAT (Pat) and deacetylase Rv1151c, and acetylation at site K224 decreases the rate of the phosphatase reaction (81). Again, Rv1151c may be an attractive drug target, as its inhibition would increase acetylation of PtpB and may have significant phenotypic consequences on downstream host targets, which may lead to less intracellular survival.
THE ROLE OF ACETYLATION IN ANTIBIOTIC RESISTANCE
The role of lysine acetylation in antibiotic resistance has not been well studied, but hints of its importance are emerging. For example, in Salmonella Typhimurium, 15 resistance-related proteins were acetylated, including a multidrug efflux transporter (AcrB) and various outer membrane proteins (OMPs), which may decrease outer membrane permeability (82). Following the development of fluoroquinolone resistance, the acetylation abundance increased on some OMPs and decreased on AcrB, suggesting that acetylation regulates the activity of these proteins and contributes to resistance development. In M. smegmatis, the histone-like protein HupB is acetylated at multiple sites. Mutation of K86 to arginine (unmodified mimic) resulted in the specific loss of the small-colony variant, isoniazid-tolerant subpopulation (83). As discussed before, in M. tuberculosis the metabolic enzyme isocitrate lyase (ICL) is acetylated. When exposed to isoniazid, rifampin, and streptomycin, the ICL enzymes are activated (84). Indeed, icl mutants are 100- to 1,000-fold more susceptible to these drugs, and this can be rescued by growth in the presence of an antioxidant, such as thiourea. This suggests that the ICLs participate in the defense against antibiotic-induced oxidative stress; therefore, deacetylation of ICL may be critical for drug tolerance.
CONCLUDING REMARKS
The physiological relevance of protein acetylation in bacteria is an important topic that is just beginning to be addressed, as there remain thousands of uncharacterized sites. So far, many important basic biological processes are regulated by acetylation, and the initial focus has largely been on transcription, translation, and metabolism (3–5, 12, 29). Recently, we have begun to investigate virulence, but there is still much to discover. Large-scale acetylome analysis can reveal hints of additional virulence proteins to investigate. For example, in E. coli the bifunctional acetaldehyde-CoA dehydrogenase and alcohol dehydrogenase AdhE was identified as acetylated at >10 sites in multiple studies (1, 7, 8, 13, 14, 17, 19, 85, 86). AdhE regulates host-cell binding and establishment of infection of enterohemorrhagic E. coli, and an adhE mutant displays attenuated virulence in rabbit models of infection (87). Therefore, AdhE acetylation may be an essential regulatory mechanism for survival in the host and is worthy of further investigation.
Our understanding of how acetylation regulates virulence factors could aid in the design of novel therapeutics. In theory, we could design drugs that specifically target the known enzymes of acetylation, either KATs, KDACs, or even acetate kinase, which could alter the levels of acetylation and limit virulence. For example, the M. tuberculosis sirtuin Rv1151c is a promising target. Inhibiting this enzyme may affect the establishment of M. tuberculosis latency by increasing the acetylation of MtrA and DosR (49, 54) while also decreasing the activity of the secreted phosphatase PtpB (81), mitigating intracellular survival. Recently, it was demonstrated that using a combination of the antibiotic fusidic acid followed by a KAT inhibitor (EIS 1a*) led to increased killing of M. smegmatis compared to antibiotic alone, a strategy that could be optimized for use with M. tuberculosis (88).
Interestingly, many secreted factors are modified by acetylation, but it is unclear if they are modified in the cytoplasm before secretion or in the extracellular environment. Some Nα-acetyltransferases (NATs), which acetylate the amino group of the N terminus of proteins, are known to be secreted. For example, in M. tuberculosis, members of the ESAT6 family of NATs are critical virulence factors and are present in the extracellular environment (89, 90). Currently, there are no examples of extracellular bacterial KATs or KDACs. However, if it turns out that extracellular KATs and KDACs do exist, it is intriguing to think of the possibility of designing drugs that block interactions with these enzymes to possibly interfere with host-pathogen interactions. This could make an infection less severe or easier for the immune system to clear. Because emerging evidence indicates that acetylation is involved in the development of antibiotic resistance, inhibiting acetylation may become a weapon to control the emergence of drug-resistant bacteria. As we learn more about the contributions of acetylation toward bacterial pathogenesis, exciting new avenues for novel therapeutics will emerge.
ACKNOWLEDGMENTS
We thank D. Dubnau, E. Dubnau, T. Greco, and M. Neiditch for critical reading of the manuscript. We thank Patrick Lane, ScEYEnce Studios, for assistance with the illustrations in Fig. 1.
This work was supported by grant GM138303, awarded to V.J.C.
Contributor Information
Valerie J. Carabetta, Email: carabetta@rowan.edu.
Frank Schmidt, Weill Cornell Medicine-Qatar.
REFERENCES
- 1.Yu BJ, Kim JA, Moon JH, Ryu SE, Pan JG. 2008. The diversity of lysine-acetylated proteins in Escherichia coli. J Microbiol Biotechnol 18:1529–1536. [PubMed] [Google Scholar]
- 2.Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu C-F, Grishin NV, Zhao Y. 2009. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics 8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carabetta VJ, Cristea IM. 2017. The regulation, function, and detection of protein acetylation in bacteria. J Bacteriol 199:e00107-17. doi: 10.1128/JB.00107-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen DG, Baumgartner JT, Xie X, Jew KM, Basisty N, Schilling B, Kuhn ML, Wolfe AJ. 2019. Mechanisms, detection, and relevance of protein acetylation in prokaryotes. mBio 10:e02708-18. doi: 10.1128/mBio.02708-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.VanDrisse CM, Escalante-Semerena JC. 2019. Protein acetylation in bacteria. Annu Rev Microbiol 73:111–132. doi: 10.1146/annurev-micro-020518-115526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfe AJ. 2005. The acetate switch. Microbiol Mol Biol Rev 69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinert BT, Iesmantavicius V, Wagner SA, Schölz C, Gummesson B, Beli P, Nyström T, Choudhary C. 2013. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell 51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Kuhn ML, Zemaitaitis B, Hu LI, Sahu A, Sorensen D, Minasov G, Lima BP, Scholle M, Mrksich M, Anderson WF, Gibson BW, Schilling B, Wolfe AJ. 2014. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One 9:e94816. doi: 10.1371/journal.pone.0094816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosono S, Tamura M, Suzuki S, Kawamura Y, Yoshida A, Nishiyama M, Yoshida M. 2015. Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLoS One 10:e0131169. doi: 10.1371/journal.pone.0131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Post DMB, Schilling B, Reinders LM, D’Souza AK, Ketterer MR, Kiel SJ, Chande AT, Apicella MA, Gibson BW. 2017. Identification and characterization of AckA-dependent protein acetylation in Neisseria gonorrhoeae. PLoS One 12:e0179621. doi: 10.1371/journal.pone.0179621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.AbouElfetouh A, Kuhn ML, Hu LI, Scholle MD, Sorensen DJ, Sahu AK, Becher D, Antelmann H, Mrksich M, Anderson WF, Gibson BW, Schilling B, Wolfe AJ. 2015. The E. coli sirtuin CobB shows no preference for enzymatic and nonenzymatic lysine acetylation substrate sites. Microbiologyopen 4:66–83. doi: 10.1002/mbo3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen DG, Xie X, Basisty N, Byrnes J, McSweeney S, Schilling B, Wolfe AJ. 2019. Post-translational protein acetylation: an elegant mechanism for bacteria to dynamically regulate metabolic functions. Front Microbiol 10:1604. doi: 10.3389/fmicb.2019.01604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baeza J, Dowell JA, Smallegan MJ, Fan J, Amador-Noguez D, Khan Z, Denu JM. 2014. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem 289:21326–21338. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer JG, D’Souza AK, Sorensen DJ, Rardin MJ, Wolfe AJ, Gibson BW, Schilling B. 2016. Quantification of lysine acetylation and succinylation stoichiometry in proteins using mass spectrometric data-independent acquisitions (SWATH). J Am Soc Mass Spectrom 27:1758–1771. doi: 10.1007/s13361-016-1476-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayasu ES, Wu S, Sydor MA, Shukla AK, Weitz KK, Moore RJ, Hixson KK, Kim J-S, Petyuk VA, Monroe ME, Pasa-Tolic L, Qian W-J, Smith RD, Adkins JN, Ansong C. 2014. A method to determine lysine acetylation stoichiometries. Int J Proteomics 2014:730725. doi: 10.1155/2014/730725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyagi M. 2017. Site-specific quantification of lysine acetylation using isotopic labeling. Methods Enzymol 586:85–95. doi: 10.1016/bs.mie.2016.09.029. [DOI] [PubMed] [Google Scholar]
- 17.Weinert BT, Satpathy S, Hansen BK, Lyon D, Jensen LJ, Choudhary C. 2017. Accurate quantification of site-specific acetylation stoichiometry reveals the impact of sirtuin deacetylase CobB on the E. coli acetylome. Mol Cell Proteomics 16:759–769. doi: 10.1074/mcp.M117.067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei L, Meyer JG, Schilling B. 2018. Quantification of site-specific protein lysine acetylation and succinylation stoichiometry using data-independent acquisition mass spectrometry. J Vis Exp 134:e57209. doi: 10.3791/57209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castano-Cerezo S, et al. 2014. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol Syst Biol 10:762. doi: 10.15252/msb.20145227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu LI, Chi BK, Kuhn ML, Filippova EV, Walker-Peddakotla AJ, Bäsell K, Becher D, Anderson WF, Antelmann H, Wolfe AJ. 2013. Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J Bacteriol 195:4174–4186. doi: 10.1128/JB.00383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thao S, Chen CS, Zhu H, Escalante-Semerena JC. 2010. N-epsilon-lysine acetylation of a bacterial transcription factor inhibits its DNA-binding activity. PLoS One 5:e15123. doi: 10.1371/journal.pone.0015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barak R, Eisenbach M. 2001. Acetylation of the response regulator, CheY, is involved in bacterial chemotaxis. Mol Microbiol 40:731–743. doi: 10.1046/j.1365-2958.2001.02425.x. [DOI] [PubMed] [Google Scholar]
- 23.Barak R, Welch M, Yanovsky A, Oosawa K, Eisenbach M. 1992. Acetyladenylate or its derivative acetylates the chemotaxis protein CheY in vitro and increases its activity at the flagellar switch. Biochemistry 31:10099–10107. doi: 10.1021/bi00156a033. [DOI] [PubMed] [Google Scholar]
- 24.Barak R, Yan J, Shainskaya A, Eisenbach M. 2006. The chemotaxis response regulator CheY can catalyze its own acetylation. J Mol Biol 359:251–265. doi: 10.1016/j.jmb.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 25.Fraiberg M, Afanzar O, Cassidy CK, Gabashvili A, Schulten K, Levin Y, Eisenbach M. 2015. CheY's acetylation sites responsible for generating clockwise flagellar rotation in Escherichia coli. Mol Microbiol 95:231–244. doi: 10.1111/mmi.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R, Chen P, Gu J, Deng JY. 2013. Acetylation reduces the ability of CheY to undergo autophosphorylation. FEMS Microbiol Lett 347:70–76. doi: 10.1111/1574-6968.12224. [DOI] [PubMed] [Google Scholar]
- 27.Liarzi O, Barak R, Bronner V, Dines M, Sagi Y, Shainskaya A, Eisenbach M. 2010. Acetylation represses the binding of CheY to its target proteins. Mol Microbiol 76:932–943. doi: 10.1111/j.1365-2958.2010.07148.x. [DOI] [PubMed] [Google Scholar]
- 28.Ramakrishnan R, Schuster M, Bourret RB. 1998. Acetylation at Lys-92 enhances signaling by the chemotaxis response regulator protein CheY. Proc Natl Acad Sci USA 95:4918–4923. doi: 10.1073/pnas.95.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren J, Sang Y, Lu J, Yao YF. 2017. Protein acetylation and its role in bacterial virulence. Trends Microbiol 25:768–779. doi: 10.1016/j.tim.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Wu Z, Wan X, Liu P, Zhang J, Ye Y, Zhao Y, Tan M. 2014. Comprehensive profiling of lysine acetylome in Staphylococcus aureus. Sci China Chem 57:732–738. doi: 10.1007/s11426-014-5100-4. [DOI] [Google Scholar]
- 31.Bontemps-Gallo S, Gaviard C, Richards CL, Kentache T, Raffel SJ, Lawrence KA, Schindler JC, Lovelace J, Dulebohn DP, Cluss RG, Hardouin J, Gherardini FC. 2018. Global profiling of lysine acetylation in Borrelia burgdorferi B31 reveals its role in central metabolism. Front Microbiol 9:2036. doi: 10.3389/fmicb.2018.02036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao X-J, Dai J, Xu H, Nie S, Chang X, Hu B-Y, Sheng Q-H, Wang L-S, Ning Z-B, Li Y-X, Guo X-K, Zhao G-P, Zeng R. 2010. High-coverage proteome analysis reveals the first insight of protein modification systems in the pathogenic spirochete Leptospira interrogans. Cell Res 20:197–210. doi: 10.1038/cr.2009.127. [DOI] [PubMed] [Google Scholar]
- 33.Kentache T, Jouenne T, De E, Hardouin J. 2016. Proteomic characterization of N-alpha- and N-epsilon-acetylation in Acinetobacter baumannii. J Proteomics 144:148–158. doi: 10.1016/j.jprot.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y-T, Pan Y, Lai F, Yin X-F, Ge R, He Q-Y, Sun X. 2018. Comprehensive analysis of the lysine acetylome and its potential regulatory roles in the virulence of Streptococcus pneumoniae. J Proteomics 176:46–55. doi: 10.1016/j.jprot.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 35.AlonsoDeVelasco E, Verheul AF, Verhoef J, Snippe H. 1995. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol Rev 59:591–603. doi: 10.1128/mr.59.4.591-603.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jers C, Ravikumar V, Lezyk M, Sultan A, Sjöling Å, Wai SN, Mijakovic I. 2017. The global acetylome of the human pathogen Vibrio cholerae V52 reveals lysine acetylation of major transcriptional regulators. Front Cell Infect Microbiol 7:537. doi: 10.3389/fcimb.2017.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pang R, Li Y, Liao K, Guo P, Li Y, Yang X, Zhang S, Lei T, Wang J, Chen M, Wu S, Xue L, Wu Q. 2020. Genome- and proteome-wide analysis of lysine acetylation in Vibrio vulnificus Vv180806 reveals its regulatory roles in virulence and antibiotic resistance. Front Microbiol 11:591287. doi: 10.3389/fmicb.2020.591287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan J, Ye Z, Cheng Z, Peng X, Wen L, Zhao F. 2014. Systematic analysis of the lysine acetylome in Vibrio parahemolyticus. J Proteome Res 13:3294–3302. doi: 10.1021/pr500133t. [DOI] [PubMed] [Google Scholar]
- 39.Pang H, Li W, Zhang W, Zhou S, Hoare R, Monaghan SJ, Jian J, Lin X. 2020. Acetylome profiling of Vibrio alginolyticus reveals its role in bacterial virulence. J Proteomics 211:103543. doi: 10.1016/j.jprot.2019.103543. [DOI] [PubMed] [Google Scholar]
- 40.Xie L, Wang X, Zeng J, Zhou M, Duan X, Li Q, Zhang Z, Luo H, Pang L, Li W, Liao G, Yu X, Li Y, Huang H, Xie J. 2015. Proteome-wide lysine acetylation profiling of the human pathogen Mycobacterium tuberculosis. Int J Biochem Cell Biol 59:193–202. doi: 10.1016/j.biocel.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Liu F, Yang M, Wang X, Yang S, Gu J, Zhou J, Zhang X-E, Deng J, Ge F. 2014. Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis. Mol Cell Proteomics 13:3352–3366. doi: 10.1074/mcp.M114.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bi J, Wang Y, Yu H, Qian X, Wang H, Liu J, Zhang X. 2017. Modulation of central carbon metabolism by acetylation of isocitrate lyase in Mycobacterium tuberculosis. Sci Rep 7:44826. doi: 10.1038/srep44826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell DG, VanderVen BC, Lee W, Abramovitch RB, Kim M-j, Homolka S, Niemann S, Rohde KH. 2010. Mycobacterium tuberculosis wears what it eats. Cell Host Microbe 8:68–76. doi: 10.1016/j.chom.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhee KY, de Carvalho LPS, Bryk R, Ehrt S, Marrero J, Park SW, Schnappinger D, Venugopal A, Nathan C. 2011. Central carbon metabolism in Mycobacterium tuberculosis: an unexpected frontier. Trends Microbiol 19:307–314. doi: 10.1016/j.tim.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKinney JD, Höner zu Bentrup K, Muñoz-Elías EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Russell DG. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 46.Muñoz-Elías EJ, McKinney JD. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med 11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moon K, Bonocora RP, Kim DD, Chen Q, Wade JT, Stibitz S, Hinton DM. 2017. The BvgAS regulon of Bordetella pertussis. mBio 8:e01526-17. doi: 10.1128/mBio.01526-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novak J, Fabrik I, Jurnecka D, Holubova J, Stanek O, Sebo P. 2020. Bordetella pertussis acetylome is shaped by lysine deacetylase Bkd1. J Proteome Res 19:3680–3696. doi: 10.1021/acs.jproteome.0c00178. [DOI] [PubMed] [Google Scholar]
- 49.Singh KK, Bhardwaj N, Sankhe GD, Udaykumar N, Singh R, Malhotra V, Saini DK. 2019. Acetylation of response regulator proteins, TcrX and MtrA in M. tuberculosis tunes their phosphotransfer ability and modulates two-component signaling crosstalk. J Mol Biol 431:777–793. doi: 10.1016/j.jmb.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 50.Singh KK, Athira PJ, Bhardwaj N, Singh DP, Watson U, Saini DK. 2020. Acetylation of response regulator protein MtrA in M. tuberculosis regulates its repressor activity. Front Microbiol 11:516315. doi: 10.3389/fmicb.2020.516315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma AK, Chatterjee A, Gupta S, Banerjee R, Mandal S, Mukhopadhyay J, Basu J, Kundu M. 2015. MtrA, an essential response regulator of the MtrAB two-component system, regulates the transcription of resuscitation-promoting factor B of Mycobacterium tuberculosis. Microbiology 161:1271–1281. doi: 10.1099/mic.0.000087. [DOI] [PubMed] [Google Scholar]
- 52.Purushotham G, Sarva KB, Blaszczyk E, Rajagopalan M, Madiraju MV. 2015. Mycobacterium tuberculosis oriC sequestration by MtrA response regulator. Mol Microbiol 98:586–604. doi: 10.1111/mmi.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, Machowski EE, Tsenova L, Young M, Kaprelyants A, Kaplan G, Mizrahi V. 2008. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol 67:672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bi J, Gou Z, Zhou F, Chen Y, Gan J, Liu J, Wang H, Zhang X. 2018. Acetylation of lysine 182 inhibits the ability of Mycobacterium tuberculosis DosR to bind DNA and regulate gene expression during hypoxia. Emerg Microbes Infect 7:108. doi: 10.1038/s41426-018-0112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park H-D, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol 48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoiby N, et al. 2011. The clinical impact of bacterial biofilms. Int J Oral Sci 3:55–65. doi: 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun L, Yao Z, Guo Z, Zhang L, Wang Y, Mao R, Lin Y, Fu Y, Lin X. 2019. Comprehensive analysis of the lysine acetylome in Aeromonas hydrophila reveals cross-talk between lysine acetylation and succinylation in LuxS. Emerg Microbes Infect 8:1229–1239. doi: 10.1080/22221751.2019.1656549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yao Z, Guo Z, Wang Y, Li W, Fu Y, Lin Y, Lin W, Lin X. 2019. Integrated succinylome and metabolome profiling reveals crucial role of S-ribosylhomocysteine lyase in quorum sensing and metabolism of Aeromonas hydrophila. Mol Cell Proteomics 18:200–215. doi: 10.1074/mcp.RA118.001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozlova EV, Popov VL, Sha J, Foltz SM, Erova TE, Agar SL, Horneman AJ, Chopra AK. 2008. Mutation in the S-ribosylhomocysteinase (luxS) gene involved in quorum sensing affects biofilm formation and virulence in a clinical isolate of Aeromonas hydrophila. Microb Pathog 45:343–354. doi: 10.1016/j.micpath.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 60.Cross SE, Kreth J, Zhu L, Sullivan R, Shi W, Qi F, Gimzewski JK. 2007. Nanomechanical properties of glucans and associated cell-surface adhesion of Streptococcus mutans probed by atomic force microscopy under in situ conditions. Microbiology 153:3124–3132. doi: 10.1099/mic.0.2007/007625-0. [DOI] [PubMed] [Google Scholar]
- 61.Lei L, Zeng J, Wang L, Gong T, Zheng X, Qiu W, Zhang R, Yun L, Yang Y, Li Y. 2021. Quantitative acetylome analysis reveals involvement of glucosyltransferase acetylation in Streptococcus mutans biofilm formation. Environ Microbiol Rep 13:86–97. doi: 10.1111/1758-2229.12907. [DOI] [PubMed] [Google Scholar]
- 62.Nambi S, Gupta K, Bhattacharyya M, Ramakrishnan P, Ravikumar V, Siddiqui N, Thomas AT, Visweswariah SS. 2013. Cyclic AMP-dependent protein lysine acylation in Mycobacteria regulates fatty acid and propionate metabolism. J Biol Chem 288:14114–14124. doi: 10.1074/jbc.M113.463992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaviard C, Broutin I, Cosette P, Dé E, Jouenne T, Hardouin J. 2018. Lysine succinylation and acetylation in Pseudomonas aeruginosa. J Proteome Res 17:2449–2459. doi: 10.1021/acs.jproteome.8b00210. [DOI] [PubMed] [Google Scholar]
- 64.Gaviard C, Cosette P, Jouenne T, Hardouin J. 2019. LasB and CbpD virulence factors of Pseudomonas aeruginosa carry multiple post-translational modifications on their lysine residues. J Proteome Res 18:923–933. doi: 10.1021/acs.jproteome.8b00556. [DOI] [PubMed] [Google Scholar]
- 65.Folders J, Tommassen J, van Loon LC, Bitter W. 2000. Identification of a chitin-binding protein secreted by Pseudomonas aeruginosa. J Bacteriol 182:1257–1263. doi: 10.1128/JB.182.5.1257-1263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park S, Galloway DR. 1998. Pseudomonas aeruginosa LasD processes the inactive LasA precursor to the active protease form. Arch Biochem Biophys 357:8–12. doi: 10.1006/abbi.1998.0787. [DOI] [PubMed] [Google Scholar]
- 67.Peters JE, Galloway DR. 1990. Purification and characterization of an active fragment of the LasA protein from Pseudomonas aeruginosa: enhancement of elastase activity. J Bacteriol 172:2236–2240. doi: 10.1128/jb.172.5.2236-2240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jungblut PR, Thiede B, Schlüter H. 2016. Towards deciphering proteomes via the proteoform, protein speciation, moonlighting and protein code concepts. J Proteomics 134:1–4. doi: 10.1016/j.jprot.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 69.Jungblut PR, Holzhütter HG, Apweiler R, Schlüter H. 2008. The speciation of the proteome. Chem Cent J 2:16. doi: 10.1186/1752-153X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeffery CJ. 2016. Protein species and moonlighting proteins: very small changes in a protein's covalent structure can change its biochemical function. J Proteomics 134:19–24. doi: 10.1016/j.jprot.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 71.Marakasova E, Ii A, Nelson KT, van Hoek ML. 2020. Proteome wide profiling of N-epsilon-lysine acetylation reveals a novel mechanism of regulation of the chitinase activity in Francisella novicida. J Proteome Res 19:1409–1422. doi: 10.1021/acs.jproteome.9b00512. [DOI] [PubMed] [Google Scholar]
- 72.Chung MC, Dean S, Marakasova ES, Nwabueze AO, van Hoek ML. 2014. Chitinases are negative regulators of Francisella novicida biofilms. PLoS One 9:e93119. doi: 10.1371/journal.pone.0093119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Butler CA, Veith PD, Nieto MF, Dashper SG, Reynolds EC. 2015. Lysine acetylation is a common post-translational modification of key metabolic pathway enzymes of the anaerobe Porphyromonas gingivalis. J Proteomics 128:352–364. doi: 10.1016/j.jprot.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 74.Guo Y, Nguyen KA, Potempa J. 2010. Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon's knife to a meat chopper-like brutal degradation of proteins. Periodontol 2000 54:15–44. doi: 10.1111/j.1600-0757.2010.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lamont RJ, Jenkinson HF. 1998. Life below the gum line: pathogenic mechanisms Porphyromonas gingivalis. Microbiol Mol Biol Rev 62:1244–1263. doi: 10.1128/MMBR.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mishra A, et al. 2018. Role of acetyltransferase PG1842 in gingipain giogenesis in Porphyromonas gingivalis. J Bacteriology 200:e00385-18. doi: 10.1128/JB.00385-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Krishnan K, Duncan MJ. 2018. Post-translational regulation of a Porphyromonas gingivalis regulator. J Oral Microbiol 10:1487743. doi: 10.1080/20002297.2018.1487743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen D, Perpich JD, Stocke KS, Yakoumatos L, Fitzsimonds ZR, Liu C, Miller DP, Lamont RJ. 2020. Role of the RprY response regulator in P gingivalis community development and virulence. Mol Oral Microbiol 35:231–239. doi: 10.1111/omi.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Geluk A, Lin MY, van Meijgaarden KE, Leyten EMS, Franken KLMC, Ottenhoff THM, Klein MR. 2007. T-cell recognition of the HspX protein of Mycobacterium tuberculosis correlates with latent M. tuberculosis infection but not with M. bovis BCG vaccination. Infect Immun 75:2914–2921. doi: 10.1128/IAI.01990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong D, Bach H, Sun J, Hmama Z, Av-Gay Y. 2011. Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc Natl Acad Sci USA 108:19371–19376. doi: 10.1073/pnas.1109201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singhal A, Arora G, Virmani R, Kundu P, Khanna T, Sajid A, Misra R, Joshi J, Yadav V, Samanta S, Saini N, Pandey AK, Visweswariah SS, Hentschker C, Becher D, Gerth U, Singh Y. 2015. Systematic analysis of mycobacterial acylation reveals first example of acylation-mediated regulation of enzyme activity of a bacterial phosphatase. J Biol Chem 290:26218–26234. doi: 10.1074/jbc.M115.687269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li L, Wang W, Zhang R, Xu J, Wang R, Wang L, Zhao X, Li J. 2018. First acetyl-proteome profiling of Salmonella Typhimurium revealed involvement of lysine acetylation in drug resistance. Vet Microbiol 226:1–8. doi: 10.1016/j.vetmic.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 83.Ghosh S, Padmanabhan B, Anand C, Nagaraja V. 2016. Lysine acetylation of the Mycobacterium tuberculosis HU protein modulates its DNA binding and genome organization. Mol Microbiol 100:577–588. doi: 10.1111/mmi.13339. [DOI] [PubMed] [Google Scholar]
- 84.Nandakumar M, Nathan C, Rhee KY. 2014. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun 5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 85.Zhang K, Zheng S, Yang JS, Chen Y, Cheng Z. 2013. Comprehensive profiling of protein lysine acetylation in Escherichia coli. J Proteome Res 12:844–851. doi: 10.1021/pr300912q. [DOI] [PubMed] [Google Scholar]
- 86.Schilling B, Basisty N, Christensen DG, Sorensen D, Orr JS, Wolfe AJ, Rao CV. 2019. Global lysine acetylation in Escherichia coli results from growth conditions that favor acetate fermentation. J Bacteriol 201:e00768-18. doi: 10.1128/JB.00768-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beckham KSH, Connolly JPR, Ritchie JM, Wang D, Gawthorne JA, Tahoun A, Gally DL, Burgess K, Burchmore RJ, Smith BO, Beatson SA, Byron O, Wolfe AJ, Douce GR, Roe AJ. 2014. The metabolic enzyme AdhE controls the virulence of Escherichia coli O157:H7. Mol Microbiol 93:199–211. doi: 10.1111/mmi.12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scutigliani EM, Scholl ER, Grootemaat AE, Khanal S, Kochan JA, Krawczyk PM, Reits EA, Garzan A, Ngo HX, Green KD, Garneau-Tsodikova S, Ruijter JM, van Veen HA, van der Wel NN. 2018. Interfering with DNA decondensation as a strategy against Mycobacteria. Front Microbiol 9:2034. doi: 10.3389/fmicb.2018.02034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brodin P, de Jonge MI, Majlessi L, Leclerc C, Nilges M, Cole ST, Brosch R. 2005. Functional analysis of early secreted antigenic target-6, the dominant T-cell antigen of Mycobacterium tuberculosis, reveals key residues involved in secretion, complex formation, virulence, and immunogenicity. J Biol Chem 280:33953–33959. doi: 10.1074/jbc.M503515200. [DOI] [PubMed] [Google Scholar]
- 90.Lange S, Rosenkrands I, Stein R, Andersen P, Kaufmann SHE, Jungblut PR. 2014. Analysis of protein species differentiation among mycobacterial low-Mr-secreted proteins by narrow pH range Immobiline gel 2-DE-MALDI-MS. J Proteomics 97:235–244. doi: 10.1016/j.jprot.2013.06.036. [DOI] [PubMed] [Google Scholar]
- 91.Ren J, Sang Y, Tan Y, Tao J, Ni J, Liu S, Fan X, Zhao W, Lu J, Wu W, Yao Y-F. 2016. Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PLoS Pathog 12:e1005458. doi: 10.1371/journal.ppat.1005458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sang Y, Ren J, Qin R, Liu S, Cui Z, Cheng S, Liu X, Lu J, Tao J, Yao Y-F. 2017. Acetylation regulating protein stability and DNA-binding ability of HilD, thus modulating Salmonella Typhimurium virulence. J Infect Dis 216:1018–1026. doi: 10.1093/infdis/jix102. [DOI] [PubMed] [Google Scholar]